-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting
a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing
Machinery
Symptoms on virus-infected plants are often very specific to the given virus. The
molecular mechanisms involved in viral symptom induction have been extensively
studied, but are still poorly understood. Cucumber mosaic virus
(CMV) Y satellite RNA (Y-sat) is a non-coding subviral RNA and modifies the
typical symptom induced by CMV in specific hosts; Y-sat causes a bright yellow
mosaic on its natural host Nicotiana tabacum. The Y-sat-induced
yellow mosaic failed to develop in the infected Arabidopsis and
tomato plants suggesting a very specific interaction between Y-sat and its host.
In this study, we revealed that Y-sat produces specific short interfering RNAs
(siRNAs), which interfere with a host gene, thus inducing the specific symptom.
We found that the mRNA of tobacco magnesium protoporphyrin chelatase subunit I
(ChlI, the key gene involved in chlorophyll synthesis) had
a 22-nt sequence that was complementary to the Y-sat sequence, including four
G-U pairs, and that the Y-sat-derived siRNAs in the virus-infected plant
downregulate the mRNA of ChlI by targeting the complementary
sequence. ChlI mRNA was also downregulated in the transgenic
lines that express Y-sat inverted repeats. Strikingly, modifying the Y-sat
sequence in order to restore the 22-nt complementarity to
Arabidopsis and tomato ChlI mRNA resulted
in yellowing symptoms in Y-sat-infected Arabidopsis and tomato,
respectively. In 5′-RACE experiments, the ChlI transcript
was cleaved at the expected middle position of the 22-nt complementary sequence.
In GFP sensor experiments using agroinfiltration, we further demonstrated that
Y-sat specifically targeted the sensor mRNA containing the 22-nt complementary
sequence of ChlI. Our findings provide direct evidence that the
identified siRNAs derived from viral satellite RNA directly modulate the viral
disease symptom by RNA silencing-based regulation of a host gene.
Published in the journal: A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing Machinery. PLoS Pathog 7(5): e32767. doi:10.1371/journal.ppat.1002021
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002021Summary
Symptoms on virus-infected plants are often very specific to the given virus. The
molecular mechanisms involved in viral symptom induction have been extensively
studied, but are still poorly understood. Cucumber mosaic virus
(CMV) Y satellite RNA (Y-sat) is a non-coding subviral RNA and modifies the
typical symptom induced by CMV in specific hosts; Y-sat causes a bright yellow
mosaic on its natural host Nicotiana tabacum. The Y-sat-induced
yellow mosaic failed to develop in the infected Arabidopsis and
tomato plants suggesting a very specific interaction between Y-sat and its host.
In this study, we revealed that Y-sat produces specific short interfering RNAs
(siRNAs), which interfere with a host gene, thus inducing the specific symptom.
We found that the mRNA of tobacco magnesium protoporphyrin chelatase subunit I
(ChlI, the key gene involved in chlorophyll synthesis) had
a 22-nt sequence that was complementary to the Y-sat sequence, including four
G-U pairs, and that the Y-sat-derived siRNAs in the virus-infected plant
downregulate the mRNA of ChlI by targeting the complementary
sequence. ChlI mRNA was also downregulated in the transgenic
lines that express Y-sat inverted repeats. Strikingly, modifying the Y-sat
sequence in order to restore the 22-nt complementarity to
Arabidopsis and tomato ChlI mRNA resulted
in yellowing symptoms in Y-sat-infected Arabidopsis and tomato,
respectively. In 5′-RACE experiments, the ChlI transcript
was cleaved at the expected middle position of the 22-nt complementary sequence.
In GFP sensor experiments using agroinfiltration, we further demonstrated that
Y-sat specifically targeted the sensor mRNA containing the 22-nt complementary
sequence of ChlI. Our findings provide direct evidence that the
identified siRNAs derived from viral satellite RNA directly modulate the viral
disease symptom by RNA silencing-based regulation of a host gene.Introduction
Plants infected with viruses often display various symptoms, which can be very specific to given viruses. Despite past efforts, the molecular bases underlying virus-induced diseases symptoms are still poorly understood. Subviral non-coding RNA molecules such as satellite RNAs (satRNAs) or defective interfering (DI) RNAs are often associated with plant viruses and can modify the symptoms induced by helper viruses [1], [2], [3]. Because such subviral RNAs dramatically modify the symptoms induced by helper viruses, they are potential tools for gaining insights into the molecular mechanisms of symptom development.
SatRNAs of Cucumber mosaic virus (CMV) are dependent on helper viruses for their replication and encapsidation and often attenuate the disease symptoms induced by CMV. Specifically, Y-satellite RNA (Y-sat) modifies the symptoms and exacerbates the pathogenicity of CMV in specific hosts; Y-sat induces a bright yellowing of leaves of Nicotiana tabacum (the natural host) and other related species (i.e., N. benthamiana), which is yellower than a typical chlorosis, whereas it induces systemic necrosis on tomato [4], [5], [6], [7]. The sequence domains on Y-sat, which are responsible for the symptom induction, have been identified in our previous and several other reports [6], [7], [8], [9], [10]. We also suggested that a single, nuclear-encoded, incompletely dominant gene in tobacco controls the Y-sat-mediated yellowing in tobacco plants [11], but no such host genes have ever been shown to be involved in the symptom modification nor has the molecular mechanism been reported. An attractive model based on RNA silencing has been suggested [2], [12], but the solid experimental data are still needed.
RNA silencing is a conserved, sequence-specific gene regulation system, which has an essential role in development and maintenance of genome integrity. RNA silencing relies on short RNA (sRNA) molecules (21–24 nt), which are the key mediators of RNA silencing-related pathways in almost all eukaryotic organisms [13], [14], [15]. In plants, similar to other eukaryotic organisms, there are two main classes of sRNAs: microRNAs (miRNAs) and short interfering RNAs (siRNAs), but the latter class contains several different types [16], [17]. These sRNAs are produced from double-stranded RNA (dsRNA) or from folded structures by Dicer-like (DCL) proteins and guide Argonaute (AGO) proteins to target cognate RNA or DNA sequences [13], [18]. In higher plants, RNA silencing also operates as an adaptive inducible antiviral defense mechanism. As a counter-defense strategy, plant viruses have evolved viral suppressors of RNA silencing (VSRs) [19] that interfere with the RNA silencing pathway at different steps by binding to viral siRNA and/or dsRNAs or directly interacting with AGO1 [20], [21].
Subviral RNAs such as satRNA and DI RNA of tombusvirus have been also used to understand the roles of RNA silencing in viral replication and in symptom development. The DI RNA-induced RNA silencing response is known to control the level of helper virus, facilitating the long-term co-existence of the host and the viral pathogen [20], [22], [23], [24]. In addition, progress in understanding plant antiviral RNA silencing has revealed cross relationships between RNA silencing and viral pathogenicity. Recent studies suggest the possibility that virus-derived siRNA (vsiRNA) could mediate virus–host interactions through a shared sequence identity with the host mRNA, resulting in silencing of the host genes and subsequent viral symptom development. A few interactions between host mRNAs and vsiRNAs that resulted in the vsiRNA-guided cleavages of host mRNAs have been experimentally shown [25], [26], although their roles in the virus–host interaction have not been determined to date.
Magnesium (Mg)-chelatase is the key enzyme in chlorophyll biosynthesis, and three subunits (ChlI, ChlH and ChlD) of the tobacco magnesium protoporphyrin chelatase are required for the proper function of the enzyme [27]. Indeed, tobacco plants defective for ChlI have the yellow phenotype [28], suggesting that chlorophyll biosynthesis is impaired. The same yellow phenotype was observed when the ChlI gene of tobacco or cotton was targeted by virus-induced gene silencing (VIGS) [29], [30], [31]. Furthermore, an Arabidopsis mutant defective for ChlI also had pale-green to yellow leaves [32]. Importantly, the plants defective in the function of the Mg-chelatase enzyme had a very similar yellow phenotype to plants infected with CMV and Y-sat. Thus, these results raised the possibility that the ChlI is downregulated by Y-sat in the virus-infected plants.
In this study, we show that transgenic N. benthamiana plants develop a yellow phenotype when expressing the inverted-repeat sequence of Y-sat, similar to the symptoms of the Y-sat-infected plants. Moreover, we provided evidence that Y-sat targets the ChlI gene using the host RNA silencing machinery in such a way that Y-sat-derived siRNAs efficiently downregulate ChlI mRNA through RNA silencing-mediated cleavage. Our findings strongly suggest that this yellow phenotype is the result of a disorder in chlorophyll synthesis caused by the downregulation of the ChlI gene.
Results
Biosynthesis of chloroplast pigments is impaired in Nicotiana benthamiana plants that express the dsRNA of Y-sat
To identify host genes involved in the Y-sat-induced symptom modification, we created transgenic N. benthamiana plants that express the Y-sat sequence, expecting the yellow phenotype to be induced without CMV as a helper virus. We have used this strategy to avoid any effect of virus replication on host gene expression, because virus infection itself has been shown to regulate the expression of numerous genes [33]. We first created transgenic plants that expressed the Y-sat sequence either in the sense or antisense orientation, but these transgenic plants failed to have any phenotypic changes (data not shown). However, when the Y-sat inverted-repeat (IR) sequence-expressing cassette (Figure 1A) was introduced into N. benthamiana plants, we observed that the transgenic N. benthamiana lines (16c:YsatIR) had a yellow phenotype (Figure 1B), although the yellow phenotype was less pronounced in the 16c:YsatIR lines than in the Y-sat-replicating system. Of four transgenic lines that we obtained, two had phenotypes with distinct yellowing; line 1 had vein yellowing, and line 2 had a yellow mosaic. No yellow phenotype was observed on the N. benthamiana that expressed dsRNA of GUS (16c:GUSIR), demonstrating that the expression of dsRNA of an unrelated sequence in the same Y-sat IR transformation cassette does not cause a yellow symptom (Figure 1B and 1C). We also confirmed the lack of viral contamination in the 16c:YsatIR lines by RT-PCR using primers that are specific to CMV genomes (data not shown).
To identify putative plant genes responsible for the yellow phenotype, we carried out microarray analyses of RNA extracted from the 16c:YsatIR plants (Text S1). In 16c:YsatIR plants, 134 genes were significantly downregulated to levels that are at least 40% lower than in their wild-type counterparts (N. benthamiana 16c) (Table S1). Among them, 31 genes were actually involved in chlorophyll biosynthesis and chloroplast biogenesis (Table S1), further supporting the hypothesis that the yellow phenotype could be the result of downregulation of the host gene(s) involved in the biosynthesis pathway of chloroplast pigments. Indeed, proteome analyses showed that several chloroplast-related proteins, such as RuBisCo small subunit, RuBisCo activase and glyceraldehyde-3-phosphate dehydrogenase were significantly affected in 16c:YsatIR plants (Text S1, Figure S1). More interestingly, the mobility of the RuBisCo small subunits was shifted in a two-dimensional gel (Figure S1), indicating that the proteins had been modified. All together, these results suggest that the expression of chloroplast-related genes and subsequent synthesis of proteins were altered in the 16c:YsatIR plants.
ChlI mRNA is downregulated in 16c:YsatIR plants, in N. benthamiana plants infected with Y-sat, and in Y-sat dsRNA-transfected protoplasts
When we aligned the sequences of the 31 genes involved in chlorophyll biosynthesis and chloroplast biogenesis identified by microarray analysis with the Y-sat sequence, we found a high degree of sequence complementarity (22 nt in a row including four G-U pairs) between the yellow-inducing domain of Y-sat [7], [34] and the tobacco magnesium (Mg) protoporphyrin chelatase subunit I (ChlI) gene (accession AF014053). Because ChlI is a component of the primary enzyme that catalyzes the first step in chlorophyll synthesis via the tetrapyrrole biosynthesis pathway [32], this evidence encouraged us to clone and sequence the ChlI gene of N. benthamiana. We then found that both the ChlI genes from N. tabacum and N. benthamiana had the 22-nt sequence complementary to the Y-sat sequence (Figure 2A). Hereafter, we called the 22-nt complementary sequence for the ChlI gene and the Y-sat sequence as the yellow region (YR) and satellite yellow region (SYR), respectively (Figure 2A). We then examined the mRNA levels of the ChlI gene by Northern blot analysis and quantitative real-time RT-PCRs in 16c:YsatIR and Y-sat-infected N. benthamiana plants. The outputs of these analyses showed that the ChlI mRNA was markedly downregulated in both plants (Figure 2B and C) and confirmed the results of the microarray analysis. To confirm that the downregulation of the ChlI mRNA was due to the satRNA itself, we further conducted a quantitative real-time RT-PCR using RNAs from N. benthamiana protoplasts transfected with the dsRNA of Y-sat. As controls, we transfected protoplasts with dsRNA of three other CMV satRNAs; S19-sat, T73-sat [35] and CM-sat [36]. These satRNAs are different from Y-sat in the corresponding SYR sequences and do not induce any yellow phenotypes in tobacco plants [35]. As shown in Figure 2D, the ChlI mRNA level was lower in protoplasts treated with dsRNA of Y-sat than in those treated with dsRNA of the other satRNAs. In addition, the mRNA level of another chloroplast-related gene, CAB3, decreased in the Y-sat dsRNA-treated protoplasts (Figure 2D), confirming our findings from the microarray analysis. In the proteome analysis, many chloroplast-related proteins were affected in the transgenic 16c plants expressing Y-sat dsRNA; thus, it is conceivable that the down-regulation of the ChlI gene caused a decrease in other chloroplast-related genes expression in the Y-sat dsRNA-treated protoplasts.
CMV vector-based gene silencing of the ChlI induces downregulation of the ChlI mRNA and the yellow symptom
We next examined whether silencing of the ChlI gene using VIGS can induce similar yellow symptoms in the absence of Y-sat. The 150-bp of ChlI (817 to 966) was inserted into the two CMV vectors, CMV-A1 and CMV-H1; CMV-A1 lacks the C-terminal one-third of the intact 2b protein [37], while CMV-H1 vector lacks the entire 2b protein [38] (Figure 3A). In the VIGS experiments, we used a pseudorecombinant virus that contains RNA components derived from RNA1 and RNA3 of CMV strain L to avoid the severe mosaic symptoms induced by CMV-Y. N. benthamiana plants infected with either of the viral vectors had systemic yellow symptoms similar to those induced by the replicating Y-sat in the presence of the helper virus (Figure 3B). Although CMV-H1:ChlI150 induced the yellowing more slowly than CMV-A1:ChlI150 in the early stage of infection, the results of quantitative real-time RT-PCR confirmed that the ChlI mRNA was downregulated in both CMV-A1:ChlI150 - and CMV-H1:ChlI150-infected N. benthamiana plants compared to control plants infected with one of the empty vectors (Figure 3C). Using enzyme-linked immunosorbent assay (ELISA), we confirmed that both pseudorecombinant viruses carrying the inserted ChlI sequence replicated and accumulated to a similar level in the systemic leaves at 14 days post-inoculation (dpi) (Figure 3D).
Sequence complementarity between Y-sat and the ChlI gene is essential for the induction of the yellow phenotype
The ChlI genes of pepper, tomato and Arabidopsis thaliana were obtained from the gene database, and the 22-nt complementary sequences of the ChlI genes and Y-sat were aligned (Figure 4A). Pepper has the same YR sequence in the ChlI gene as those of tobacco and N. benthamiana. Conversely, several mismatches were found in the case of the tomato ChlI and Arabidopsis ChlI (ChlI1 and ChlI2) genes (Figure 4A). We next examined whether the Y-sat can induce yellow symptoms on pepper, tomato and Arabidopsis plants. As expected, infected pepper plants developed bright yellow symptoms (Figure 4B, right plant), whereas tomato plants did not (Figure 4C, right plant). By site-directed mutagenesis of the SYR, we generated three Y-sat derivatives having the 22-nt continuous sequence complementary to the corresponding YRs of tomato ChlI gene, Arabidopsis ChlI1 and ChlI2 genes (Y-sat-Tom, Y-sat-Ara1 and Y-sat-Ara2, respectively) (Figure 4A). When tomato plants were inoculated with the Y-sat mut-Tom and the helper virus, yellow symptoms appeared at 10 dpi (Figure 4C, left plant). However, some of the introduced mutations in individual plants had reverted to the original nucleotides at 21 dpi. Notably, the Y-sat mut-Tom did not induce yellow symptoms in N. benthamiana (Figure 4D, left plant). Similarly, when Arabidopsis plants were infected with CMV-Y and Y-sat mut-Ara1, yellow symptoms appeared (Figure 4E, right plant). On the other hand, Y-sat mut-Ara2 did not induce yellowing (data not shown). The last observation is consistent with the previous studies by Huang and Li [32], who reported that ChlI2 of Arabidopsis has lower functionality than ChlI1 due to a reduced level of expression. In addition, like Y-sat mut-Tom, Y-sat mut-Ara1 did not induce yellowing in N. benthamiana (Figure 4F). Quantitative real-time RT-PCRs confirmed that the mRNA levels of the ChlI gene in the Y-sat mutants-infected N. benthamiana plants were not downregulated, unlike in the Y-sat-infected plant (Figure 4G). There were little differences in satRNA or viral accumulation between Y-sat-infected - and Y-sat mut-Ara1-infected leaves of N. benthamiana (Figure 4H and 4I), confirming that the Y-sat mutant was replicated to a level similar to that of the original Y-sat in the systemic leaves of N. benthamiana. These results, all together, strongly suggest that a specific interaction between Y-sat and the ChlI host gene is involved in development of the yellow symptom.
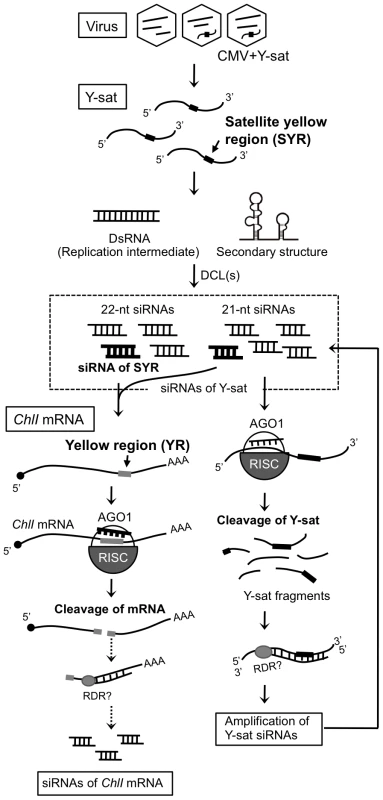
Massive amounts of small RNAs from SYR sequence accumulate in Y-sat-infected plants
Because Y-sat and the host ChlI gene seemed to have a specific interaction through their sequence complementarity, we then examined the possible involvement of RNA silencing in the Y-sat-mediated yellow phenotype. First, we tested whether the Y-sat-derived siRNAs can be hybridized and detected by ChlI mRNA probe. As shown in Figure S2, sense siRNAs from Y-sat in both Y-sat-infected and 16c:YsatIR plants were clearly detected in Northern blots using the ChlI sense RNA probe. On the other hand, we failed to detect antisense siRNAs from Y-sat by Northern blots using the ChlI antisense RNA probe. This result seems reasonable because the YR and SYR sequences do not share complementarity in the antisense orientation (Figure S2). In addition, we also detected siRNAs derived from Y-sat mut-Ara1 using the Arabidopsis ChlI1 sense RNA in Northern blots. As shown in Figure S3, 351-bp Arabidopsis ChlI1 sense RNA probe, which contains the 22-nt sequence complementary to Y-sat mut-Ara1, detected the siRNAs of Y-sat mut-Ara1 in the Arabidopsis leaves infected with CMV and Y-sat mut-Ara1. Assuming that the yellow symptoms are the result of post-transcriptional RNA silencing of host genes directed by Y-sat specific sequences, we further analyzed Y-sat-derived siRNAs profile to find whether Y-sat siRNAs targeting the ChlI mRNA accumulate in the Y-sat-infected plants. We thus conducted small RNA deep sequencing to map the small RNAs on the Y-sat sequence. As the result, Y-sat-derived siRNAs covered almost the entire Y-sat sequence, and the majority of Y-sat siRNAs accumulated in the sense orientation in the Y-sat-infected plants. In addition, 21-nt and 22-nt siRNAs were abundant among the Y-sat small RNAs populations (Figure 5A). Y-sat-derived siRNAs in both sense and antisense orientation were non-uniformly distributed along the sequence with a few small RNA-generating hot spots (Figure 5A). Abundant siRNAs were accumulated from the regions around positions 100, 180, 211 and 280 on the Y-sat. Northern hybridization confirmed that the most abundant siRNAs were generated from the region at positions 1–200 as opposed to 201–369 (Figure S4). Furthermore, we found abundant siRNAs homologous to the SYR (Figure 5B). The accumulation of siRNAs corresponding to SYR in 16c:YsatIR and Ysat-infected plans was confirmed by Northern hybridization using LNA probes specific to SYR of Y-sat (Figure 5C). In deep-sequencing analysis, we also identified the ChlI siRNAs in the Y-sat-infected tissues although the amounts were not very high (Figure S5). The profile of the ChlI siRNAs revealed a very unique feature; all siRNAs derived from ChlI were generated only from the 3′ region downstream of the cleavage site as described below.
Fig. 5. Small RNAs generated from SYR in Y-sat sequence. 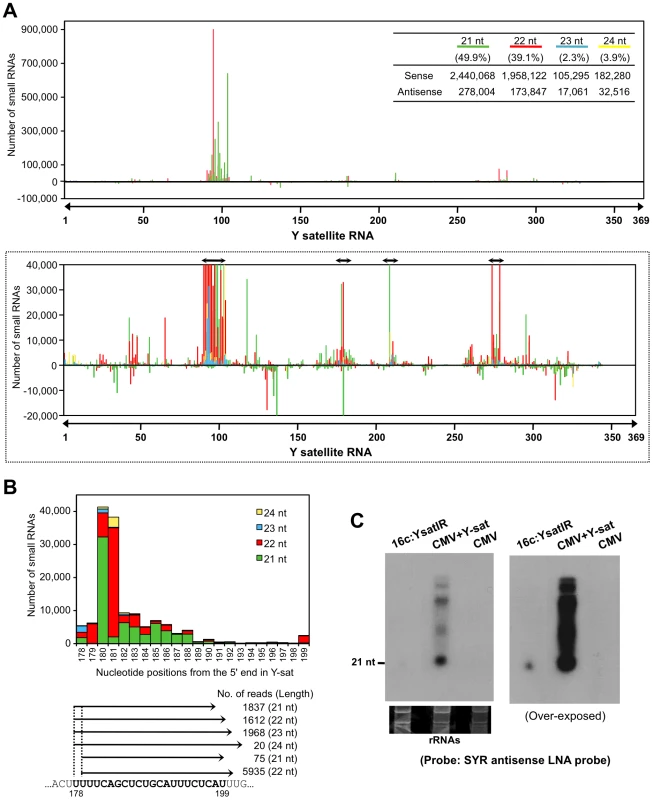
Small RNAs from Y-sat SYR cleave ChlI mRNA post-transcriptionally
To clarify whether the ChlI mRNA is cleaved in the Y-sat-infected plant, we analyzed the 5′ ends of the cleaved mRNA products with a 5′-RACE assay. Sequencing of the 5′-RACE products revealed two distinct cleavage sites in the YR of the ChlI mRNA. Almost all identified cleavage sites were mapped at the middle position in YR (between 890 and 891), which agrees with the expected cleavage site(s) driven by the 21-nt and 22-nt siRNAs (Figure 6A). To verify that Y-sat can direct sequence-specific cleavage, we created a GFP sensor construct in which the 3′ non-coding region contained the 22-nt YR sequence (Figure 6B). The construct was delivered by agroinfiltration into Y-sat-infected N. benthamiana leaves that had bright yellow symptoms (Figure 6B). GFP accumulation was monitored using UV light after agroinfiltration. As shown in Figure 6B, GFP fluorescence was reduced in the Y-sat-infected tissues, and this observation was supported by the results of quantitative real-time RT-PCR of the GFP mRNA (Figure 6C). The accumulation of GFP protein was also reduced in the Y-sat-infected tissues (Figure 6D). These results clearly demonstrated that the 22-nt YR sequence in the sensor mRNA was sufficient for the sequence-specific downregulation of GFP-YR mRNA in Y-sat-infected tissues.
Discussion
Plant RNA silencing has often been implicated as a molecular mechanism for symptom induction caused by viruses or viral subviral agents. Viral suppressors of RNA silencing (VSRs) are able to compromise the endogenous RNA silencing pathways [19], [20], and these virus-encoded silencing suppressors have also been identified as pathogenicity determinants. Indeed, virus-induced developmental abnormalities are often explained by the interference of virus-encoded VSRs with host miRNAs involved in the developmental processes [39], [40]. However, no explanation for specific symptoms caused by VSRs has ever been confirmed nor has any report explained the molecular basis for a specific viral symptom including yellowing and necrosis. In recent studies, host mRNAs were identified as potential targets of siRNAs and miRNAs in virus-infected tissues, and several have been proved to be downregulated [25], [26]. For example, Moissiard and Voinnet [25] demonstrated that the RCC1 gene in Arabidopsis infected with Cauliflower mosaic virus (CaMV) was downregulated by virus-derived siRNAs, but contrary to expectations, the decrease in gene expression did not affect either viral accumulation or symptoms. It is, in fact, quite difficult to clarify the relationship between such small RNAs and viral pathogenicity although the idea that host gene silencing against a particular gene might contribute to the specific expression of symptoms is very attractive.
In the present study, we have shown that siRNAs derived from Y-sat induced bright yellow mosaics on tobacco by specifically targeting mRNA of the host ChlI gene, resulting in the inhibition of chlorophyll biosynthesis. Here we provide several lines of evidence that Y-sat-induced bright yellow mosaics are the outcome of specific interference between the pathogen-derived siRNAs and a host gene. First, the 22-nt long region of Y-sat (SYR) produces specific siRNAs that were complementary, including four G-U pairs, to the 22-nt long region of tobacco ChlI mRNA (YR). Second, the ChlI mRNA could detect Y-sat-derived siRNAs in Northern blots. Third, 5′-RACE experiments revealed that the ChlI mRNA was cleaved exactly in the expected middle of the YR. Fourth, the levels of the ChlI transcript significantly decreased in both Y-sat-infected plants and the transgenic plants expressing Y-sat dsRNA. Fifth, the Y-sat mutants that had the modified SYR to either Arabidopsis ChlI1 mRNA or tomato ChlI mRNA were able to induce yellow symptoms in these host plants. In contrast, these modified Y-sat lost the ability to induce yellow symptoms on tobacco. Sixth, the GFP sensor construct carrying the YR sequence was specifically targeted in Y-sat-infected plants. Considering all these results, we propose a model that explains that the Y-sat-mediated yellow symptom results from the cleavage of host ChlI mRNA by RNA silencing machinery (Figure 7).
In deep-sequencing analysis, we found abundant Y-sat-derived siRNAs in the Y-sat-infected N. benthamiana. Furthermore, we noticed that the ChlI-derived siRNAs also accumulated in the Y-sat-infected tissues although the amounts were not very high. The profile of the ChlI siRNAs was very unique because all siRNAs derived from ChlI were generated only from the 3′ region downstream of the cleavage site (Figure S5). Importantly, spread of RNA silencing beyond the targeting site in endogenous plant genes has not been shown [30], [41], [42], except for trans-acting siRNAs [43]. Whether secondary siRNAs can be generated from the ChlI mRNA after vsiRNA-directed cleavage, and whether such secondary siRNAs are involved in the downregulation of the ChlI gene still need careful studies.
Here we propose that Y-sat caused the yellow symptoms on tobacco by directing post-transcriptional RNA silencing against the ChlI mRNA. However, yellow symptoms appeared much brighter in Y-sat-infected plants than in 16c:YsatIR plants (Figure 1B). With regard to the observation, the amount of Y-sat-derived siRNAs in 16c:YsatIR plants was lower than in Y-sat-infected plants (Figure 5C), probably leading to different yellow phenotype between 16c:YsatIR plants and Y-sat-infected plants. Indeed, the level of the ChlI transcript analyzed by the Northern blot was higher in the 16c:YsatIR plants than in the Y-sat-infected plants (Figure 2B). Alternatively, as suggested by Du et al. [44], Y-sat siRNAs from secondary structures (T-shaped hairpins) may predominate over the Y-sat siRNAs generated from perfect dsRNA forms. Thus it is likely that RNA silencing against ChlI and subsequent yellow phenotype can vary depending on the qualities and amounts of siRNAs derived from satRNA.
In conclusion, we discovered the molecular basis of the symptom modifications induced by Y-sat: the involvement of RNA silencing mechanism in the pathogenicity of Y-sat. But the molecular mechanism underlying the synergistic and/or antagonistic interaction between satRNAs, helper viruses and host plants still remain to be explored. In addition, the origin(s) of satRNAs, their evolutionary strategy and biological significance have long been intriguing topics. Since the original isolation of Y-sat in Japan more than 30 years ago [4], no other satRNAs that induce yellow mosaics on tobacco have been isolated in the world, suggesting that Y-sat is a rare satRNA that specifically induces yellow mosaics on tobacco. We have observed that Y-sat cannot compete with other similar size satRNAs [35], and thus Y-sat may survive through a different strategy from other satRNAs; the Y-sat-induced yellowing of leaves, which could preferentially attract aphids (the vectors of CMV and its satRNAs), may have favored the transfer of CMV that harbors Y-sat during the its evolutionary history.
Materials and Methods
Plant materials
Nicotiana benthamiana, Capsicum annuum, Solanum lycopersicum and Arabidopsis thaliana were used as host plants for the analysis. Nicotiana benthamiana line 16c having a single copy of the GFP transgene [45] was obtained from Dr. D. Baulcombe (Sainsbury Laboratory, UK) and was also used for the analysis. All plants were grown in a plant growth room with a 16-h light/8-h dark at 24°C and 50% relative humidity.
Transgenic N. benthamiana lines expressing the inverted repeat (IR) of Y-sat were generated by transforming N. benthamiana 16c with the binary vector pIG121-Hm carrying the IR of Y-sat under the CaMV 35S promoter. In the sense and antisense orientations, the 317-bp (53 to 369) Y-sat sequence (GenBank accession D00542) was inserted in the pJM007 vector [46], then the inverted repeat (IR)-expressing cassette was transferred to a Ti-plasmid vector, pIG121-Hm. The Ti plasmid vector containing the IR (1004 nt) of the GUS sequence (GUS-IR) was previously constructed [47].
Virus materials and inoculation
CMV strain Y (CMV-Y) was used as a helper virus for satellite RNA. To induce gene silencing to the ChlI gene, we used two CMV-based vectors, CMV-A1 and CMV-H1. CMV-A1 and CMV-H1 are derived from RNA2 of CMV-Y, and CMV-A1 lacks the C-terminal one-third of the intact 2b protein as a consequence of introducing a multiple cloning site [37], while CMV-H1 vector lacks the entire 2b protein [38]. The 150-bp of the ChlI gene (817 to 966) was inserted into the CMV vectors to create CMV-A1:ChlI150 and CMV-H1:ChlI150, respectively. To avoid severe mosaic symptom induction by CMV-Y, we used a pseudorecombinant virus that contains RNA components derived from RNA1 and RNA3 of CMV strain L together with RNA2 of the vector. Each plasmid containing a full-length cDNA clone of RNA1 to RNA3 was transcribed in vitro after linearization with a restriction enzyme [37]. Infectious viruses were then created by mixing transcripts of RNAs 1 to 3. For virus propagation, leaves of 4-week-old plants of N. benthamiana were dusted with carborundum and rub-inoculated with the RNA transcripts. For inoculation of tomato plants, leaves of young plants were rub-inoculated with the sap from virus-infected tissues of N. benthamiana. Successful systemic infection with the virus containing the full insert sequence was confirmed by RT-PCRs. Viral accumulation was examined by conventional ELISA [48] using the antibodies raised against the CMV CP.
RNA analyses
Total RNAs were extracted by either a conventional phenol/chloroform method [47] or a method using Trizol reagent (Invitrogen) following the manufacturer's instructions. The N. benthamiana ChlI clone including the entire ORF was amplified by RT-PCR using the primer pair designed from the tobacco ChlI sequence (5′-GCTCTAGAATGGCTTCACTACTAGGAAC-3′ for forward primer, 5′-GCCCAAGCTTAGGCGAAAACCTCATAAAATTTC-3′ for reverse primer). Quantitative real-time RT-PCR was performed essentially as described before [37]. Primers for quantitative real-time RT-PCR for the N. benthamiana ChlI gene were as follows: 5′-CTTATTGGTTCGGGTAATCCTG-3′ for forward primer and 5′-GCTGAGTCGATTTGGTTCTG-3′ for reverse primer. The N. benthamiana actin gene was amplified using 5′-GCGGGAAATTGTTAGGGATGT-3′ for forward primer and 5′-CCATCAGGCAGCTCGTAGCT-3′ for reverse primer and used for data normalization. Northern blot hybridization was performed essentially as previously described [49]. Specific probe for the ChlI gene was generated by PCR with the PCR DIG Probe Synthesis Kit (Roche Diagnostics) to amplify the 371 bp (634 to 1004) of 3′-terminal regions of the ChlI gene using the primer pair ChlI-634F (5′-GAGCCTGGTCTTCTTGCTAAAGC-3′) and ChlI-1004R (5′-GCTGAGTCGATTTGGTTCTG-3′). In the Northern blots of the small RNAs corresponding to the 22-nt complementary sequence region (satellite yellow region, SYR), the SYRs were detected by using 32P-labeled locked nucleic acid (LNA) oligonucleotide probes described previously [50].
The ChlI mRNA cleavage sites were analyzed by modified RNA-ligase mediated 5′-RACE [51]. Total RNA (10 µg) was purified using the MicroPoly(A) Purist Kit (Ambion), then the fractionated Poly(A)+ mRNA was ligated to the GeneRacer RNA Oligo adaptor using the GeneRacer Kit (Invitrogen). Ligated RNAs were reverse transcribed using the gene-specific reverse primer for the ChlI gene, ChlI-1004R (5′-GCTGAGTCGATTTGGTTCTG-3′). The 5′end of the cDNA was then amplified by PCR using the GeneRacer 5′ primer and the gene-specific reverse primer used for the reverse transcription for the first PCR. The GeneRacer 5′ nested primer was also used for the subsequent nested PCR. The amplified product from the nested PCR was excised from 1.2% agarose gel and cloned into pGEM-T Easy (Promega) for sequencing.
Protoplast experiments
Protoplasts were prepared from leaves of N. benthamiana as described before [52]. The dsRNA of four satRNAs (Y-sat, S19-sat and T73-sat [35] and CM-sat [36]) were used. DsRNA of satRNA was prepared by in vitro transcription using a PCR-amplified fragment containing the T7 promoter sequence as described previously [52]. The prepared protoplasts were transfected with the satRNA dsRNAs (2 µg) in a PEG–calcium solution as described [52] and then incubated for 20 h. Total RNA was extracted from the harvested protoplasts with Trizol reagent (Invitrogen), and the mRNA levels of the ChlI and CAB gene were measured by quantitative real-time RT-PCR (mean ± SE; n = 3). Primers for quantitative real-time RT-PCR for the CAB gene were 5′-CGGCCGATCCAGAAACTTT-3′ for forward primer and 5′-GCCCATCTGCAGTGAATAACC-3′ for reverse primer.
Deep-sequencing analysis
Total RNA was extracted from CMV and Y-sat-infected N. benthamiana plants. Small RNAs were isolated essentially as described [49] and submitted to Hokkaido System Science (Sapporo, Japan), where deep-sequencing analysis was performed on an Illumina Genome Analyzer using the standard protocol of the manufacturer. The 18–45-nt small RNA reads were extracted from raw reads and aligned with the Y-sat sequence using the program SOAP [53] to search for perfectly matched sequences.
GFP sensor experiments
The GFP-YR sensor gene was inserted between the BamHI and SacI sites in the pBE2113 vector. The Ti-plasmid construct was then introduced into Agrobacterium tumefaciens KYRT1 strain, which was supplied by Dr. G. B. Collins (University of Kentucky, USA). Agrobacterium infiltration was carried out essentially as described [49].
Western blot analysis
Total proteins were extracted from the sample tissues by grinding in Laemmli buffer, separated by SDS-PAGE, and /transferred onto a PVDF membrane (Immobilon, Millipore). Anti-GFP antibodies were purchased from Roche and used at a 1∶1000 dilution. For immunostaining, an alkaline phosphatase-conjugated goat anti-rabbit antibody was added to the blots at a 1∶3000 dilution followed by colorimetric development with BCIP and NBT.
Supporting Information
Zdroje
1. SimonAERoossinckMJHaveldaZ
2004
Plant virus satellite and defective interfering RNAs: new
paradigms for a new century.
Annu Rev Phytopathol
42
415
437
2. HuCCHsuYHLinNS
2009
Satellite RNAs and satellite viruses of plants.
Viruses
1
1325
1350
3. HuangYWHuCCLinNSHsuYH
2010
Mimicry of molecular pretenders: the terminal structures of
satellites associated with plant RNA viruses.
RNA Biol
7
162
171
4. TakanamiY
1981
A striking change in symptoms on Cucumber mosaic
virus-infected tobacco plants induced by a satellite
RNA.
Virology
109
120
126
5. XuPRoossinckMJ
2000
Cucumber mosaic virus D satellite RNA-induced
programmed cell death in tomato.
Plant Cell
12
1079
1092
6. MasutaCTakanamiY
1989
Determination of sequence and structural requirements for
pathogenicity of a Cucumber mosaic virus satellite RNA
(Y-satRNA).
Plant Cell
1
1165
1173
7. KuwataSMasutaCTakanamiY
1991
Reciprocal phenotype alterations between two satellite RNAs of
Cucumber mosaic virus.
J Gen Virol
72
2385
2389
8. DevicMJaegleMBaulcombeD
1989
Symptom production on tobacco and tomato is determined by two
distinct domains of the satellite RNA of Cucumber mosaic
virus (strain Y).
J Gen Virol
70
2765
2774
9. JaegleMDevicMLongstaffMBaulcombeD
1990
Cucumber mosaic virus satellite RNA (Y strain):
analysis of sequences which affect yellow mosaic symptoms on
tobacco.
J Gen Virol
71
1905
1912
10. SleatDEPalukaitisP
1992
A single nucleotide change within a plant virus satellite RNA
alters the host specificity of disease induction.
Plant J
2
43
49
11. MasutaCSuzukiMKuwataSTakanamiYKoiwaiA
1993
Yellow mosaic symptoms induced by Y satellite RNA of
Cucumber mosaic virus is regulated by a single
incompletely dominant gene in wild Nicotiana
species.
Phytopathology
83
411
413
12. WangMBBianXYWuLMLiuLXSmithNA
2004
On the role of RNA silencing in the pathogenicity and evolution
of viroids and viral satellites.
Proc Natl Acad Sci U S A
101
3275
3280
13. Ruiz-FerrerVVoinnetO
2009
Roles of plant small RNAs in biotic stress
responses.
Annu Rev Plant Biol
60
485
510
14. PhillipsJRDalmayTBartelsD
2007
The role of small RNAs in abiotic stress.
FEBS Lett
581
3592
3597
15. VoinnetO
2009
Origin, biogenesis, and activity of plant
microRNAs.
Cell
136
669
687
16. VaucheretHMalloryACBartelDP
2006
AGO1 homeostasis entails coexpression of MIR168 and AGO1 and
preferential stabilization of miR168 by AGO1.
Mol Cell
22
129
136
17. BrosnanCAVoinnetO
2009
The long and the short of noncoding RNAs.
Curr Opin Cell Biol
21
416
425
18. LlaveC
2004
MicroRNAs: more than a role in plant development?
Mol Plant Pathol
5
361
366
19. DingSWVoinnetO
2007
Antiviral immunity directed by small RNAs.
Cell
130
413
426
20. CsorbaTPantaleoVBurgyánJ
2009
RNA silencing: an antiviral mechanism.
Adv Virus Res
75
35
71
21. MlotshwaSPrussGJVanceV
2008
Small RNAs in viral infection and host defense.
Trends Plant Sci
13
375
382
22. HaveldaZHornyikCValocziABurgyanJ
2005
Defective interfering RNA hinders the activity of a
tombusvirus-encoded posttranscriptional gene silencing
suppressor.
J Virol
79
450
457
23. SzittyaGMolnarASilhavyDHornyikCBurgyánJ
2002
Short defective interfering RNAs of tombusviruses are not
targeted but trigger post-transcriptional gene silencing against their
helper virus.
Plant Cell
14
359
372
24. OmarovRTRezendeJAScholthofHB
2004
Host-specific generation and maintenance of Tomato bushy
stunt virus defective interfering RNAs.
Mol Plant-Microbe Interact
17
195
201
25. MoissiardGVoinnetO
2006
RNA silencing of host transcripts by cauliflower mosaic virus
requires coordinated action of the four Arabidopsis
Dicer-like proteins.
Proc Natl Acad Sci U S A
103
19593
19598
26. QiXBaoFSXieZ
2009
Small RNA deep sequencing reveals role for Arabidopsis
thaliana RNA-dependent RNA polymerases in viral siRNA
biogenesis.
PLoS One
4
e4971
27. MoulinMMcCormacACTerryMJSmithAG
2008
Tetrapyrrole profiling in Arabidopsis seedlings
reveals that retrograde plastid nuclear signaling is not due to
Mg-protoporphyrin IX accumulation.
Proc Natl Acad Sci U S A
105
15178
15183
28. FitzmauriceWPNguyenLVWernsmanEAThompsonWFConklingMA
1999
Transposon tagging of the sulfur gene of tobacco using engineered
maize Ac/Ds elements.
Genetics
153
1919
1928
29. KjemtrupSSampsonKSPeeleCGNguyenLVConklingMA
1998
Gene silencing from plant DNA carried by a
Geminivirus.
Plant J
14
91
100
30. PetersenBOAlbrechtsenM
2005
Evidence implying only unprimed RdRP activity during transitive
gene silencing in plants.
Plant Mol Biol
58
575
583
31. TuttleJRIdrisAMBrownJKHaiglerCHRobertsonD
2008
Geminivirus-mediated gene silencing from
Cotton leaf crumple virus is enhanced by low
temperature in cotton.
Plant Physiol
148
41
50
32. HuangYSLiHM
2009
Arabidopsis CHLI2 can substitute for
CHLI1.
Plant Physiol
150
636
645
33. HaveldaZVarallyayEValocziABurgyánJ
2008
Plant virus infection-induced persistent host gene downregulation
in systemically infected leaves.
Plant J
55
278
288
34. MasutaCKuwataSMatzuzakiTTakanamiYKoiwaiA
1992
A plant virus satellite RNA exhibits a significant sequence
complementarity to a chloroplast tRNA.
Nucleic Acids Res
20
2885
35. MasutaCHayashiYWangWQTakanamiY
1990
Comparison of four satellite RNA isolates of Cucumber
mosaic virus.
Ann Phytopath Soc Japan
56
207
212
36. KosakaYFukunishiT
1997
Multiple inoculation with three attenuated viruses for the
control of cucumber virus disease.
Plant Dis
81
733
738
37. OtagakiSAraiMTakahashiAGotoKHongJS
2006
Rapid induction of transcriptional and post-transcriptional gene
silencing using a novel Cucumber mosaic virus
vector.
Plant Biotechnol
23
259
265
38. MatsuoKHongJSTabayashiNItoAMasutaC
2007
Development of Cucumber mosaic virus as a vector
modifiable for different host species to produce therapeutic
proteins.
Planta
225
277
286
39. KasschauKDXieZAllenELlaveCChapmanEJ
2003
P1/HC-Pro, a viral suppressor of RNA silencing, interferes with
Arabidopsis development and miRNA
function.
Dev Cell
4
205
217
40. ChapmanEJProkhnevskyAIGopinathKDoljaVVCarringtonJC
2004
Viral RNA silencing suppressors inhibit the microRNA pathway at
an intermediate step.
Genes Dev
18
1179
1186
41. VaistijFEJonesLBaulcombeDC
2002
Spreading of RNA targeting and DNA methylation in RNA silencing
requires transcription of the target gene and a putative RNA-dependent RNA
polymerase.
Plant Cell
14
857
867
42. HimberCDunoyerPMoissiardGRitzenthalerCVoinnetO
2003
Transitivity-dependent and -independent cell-to-cell movement of
RNA silencing.
EMBO J
22
4523
4533
43. AllenEXieZGustafsonAMCarringtonJC
2005
MicroRNA-directed phasing during trans-acting siRNA biogenesis in
plants.
Cell
121
207
221
44. DuQSDuanCGZhangZHFangYYFangRX
2007
DCL4 targets Cucumber mosaic virus satellite RNA
at novel secondary structures.
J Virol
81
9142
9151
45. RuizMTVoinnetOBaulcombeDC
1998
Initiation and maintenance of virus-induced gene
silencing.
Plant Cell
10
937
946
46. SchattatMHKlosgenRBMarquesJP
2004
A novel vector for efficient gene silencing in
plants.
Plant Mol Biol Rep
22
145
153
47. SendaMMasutaCOhnishiSGotoKKasaiA
2004
Patterning of virus-infected Glycine max seed
coat is associated with suppression of endogenous silencing of chalcone
synthase genes.
Plant Cell
16
807
818
48. MasutaCTanakaHUeharaKKuwataSKoiwaiA
1995
Broad resistance to plant viruses in transgenic plants conferred
by antisense inhibition of a host gene essential in
S-adenosylmethionine-dependent transmethylation reactions.
Proc Natl Acad Sci U S A
92
6117
6121
49. GotoKKoboriTKosakaYNatsuakiTMasutaC
2007
Characterization of silencing suppressor 2b of Cucumber
mosaic virus based on examination of its small RNA-binding
abilities.
Plant Cell Physiol
48
1050
1060
50. ValocziAHornyikCVargaNBurgyánJKauppinenS
2004
Sensitive and specific detection of microRNAs by northern blot
analysis using LNA-modified oligonucleotide probes.
Nucleic Acids Res
32
e175
51. LlaveCXieZKasschauKDCarringtonJC
2002
Cleavage of Scarecrow-like mRNA targets directed by a class of
Arabidopsis miRNA.
Science
297
2053
2056
52. ShimuraHFukagawaTMeguroAYamadaHOh-HiraM
2008
A strategy for screening an inhibitor of viral silencing
suppressors, which attenuates symptom development of plant
viruses.
FEBS Lett
582
4047
4052
53. LiRLiYKristiansenKWangJ
2008
SOAP: short oligonucleotide alignment program.
Bioinformatics
24
713
714
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus SpeciesČlánek SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion ReleaseČlánek Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T CellsČlánek A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the CytosolČlánek Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- Infections Are Virulent and Inhibit the Human Malaria Parasite in
- A Gamma Interferon Independent Mechanism of CD4 T Cell Mediated Control of Infection
- MDA5 and TLR3 Initiate Pro-Inflammatory Signaling Pathways Leading to Rhinovirus-Induced Airways Inflammation and Hyperresponsiveness
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- An E2F1-Mediated DNA Damage Response Contributes to the Replication of Human Cytomegalovirus
- Quantitative Subcellular Proteome and Secretome Profiling of Influenza A Virus-Infected Human Primary Macrophages
- Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus Species
- Inhibition of Both HIV-1 Reverse Transcription and Gene Expression by a Cyclic Peptide that Binds the Tat-Transactivating Response Element (TAR) RNA
- A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing Machinery
- Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Investigating the Host Binding Signature on the PfEMP1 Protein Family
- Human Neutrophil Clearance of Bacterial Pathogens Triggers Anti-Microbial γδ T Cell Responses in Early Infection
- Septation of Infectious Hyphae Is Critical for Appressoria Formation and Virulence in the Smut Fungus
- A Family of Helminth Molecules that Modulate Innate Cell Responses via Molecular Mimicry of Host Antimicrobial Peptides
- Phospholipids Trigger Capsular Enlargement during Interactions with Amoebae and Macrophages
- CTL Escape Mediated by Proteasomal Destruction of an HIV-1 Cryptic Epitope
- Evolution of Th2 Immunity: A Rapid Repair Response to Tissue Destructive Pathogens
- Extensive Genome-Wide Variability of Human Cytomegalovirus in Congenitally Infected Infants
- The Antiviral Efficacy of HIV-Specific CD8 T-Cells to a Conserved Epitope Is Heavily Dependent on the Infecting HIV-1 Isolate
- Epstein-Barr Virus Infection of Polarized Epithelial Cells via the Basolateral Surface by Memory B Cell-Mediated Transfer Infection
- Reactive Oxygen Species Hydrogen Peroxide Mediates Kaposi's Sarcoma-Associated Herpesvirus Reactivation from Latency
- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- The Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication
- AAV Exploits Subcellular Stress Associated with Inflammation, Endoplasmic Reticulum Expansion, and Misfolded Proteins in Models of Cystic Fibrosis
- Suboptimal Activation of Antigen-Specific CD4 Effector Cells Enables Persistence of In Vivo
- SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion Release
- Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T Cells
- Transition of Sporozoites into Liver Stage-Like Forms Is Regulated by the RNA Binding Protein Pumilio
- A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the Cytosol
- Transcriptome Analysis of in Human Whole Blood and Mutagenesis Studies Identify Virulence Factors Involved in Blood Survival
- Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
- Structural Insights into Viral Determinants of Nematode Mediated Transmission
- Protective Efficacy of Serially Up-Ranked Subdominant CD8 T Cell Epitopes against Virus Challenges
- Viral CTL Escape Mutants Are Generated in Lymph Nodes and Subsequently Become Fixed in Plasma and Rectal Mucosa during Acute SIV Infection of Macaques
- Taking Some of the Mystery out of Host∶Virus Interactions
- Viral Small Interfering RNAs Target Host Genes to Mediate Disease Symptoms in Plants
- : An Emerging Cause of Sexually Transmitted Disease in Women
- Mitochondrial Ubiquitin Ligase MARCH5 Promotes TLR7 Signaling by Attenuating TANK Action
- The Hexamer Structure of the Rift Valley Fever Virus Nucleoprotein Suggests a Mechanism for its Assembly into Ribonucleoprotein Complexes
- Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages during Their Emergence in Birds in Egypt
- Stromal Down-Regulation of Macrophage CD4/CCR5 Expression and NF-κB Activation Mediates HIV-1 Non-Permissiveness in Intestinal Macrophages
- A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins
- Perturbs Iron Trafficking in the Epithelium to Grow on the Cell Surface
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- : An Emerging Cause of Sexually Transmitted Disease in Women
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy