-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
DNA Is an Antimicrobial Component of Neutrophil Extracellular Traps
Comprising the first line of the innate immune response, neutrophils combat infectious microorganisms through the release of toxic molecules, phagocytosis of invaders and the production of the recently characterized neutrophil extracellular traps (NETs). The antimicrobial activity of NETs has been attributed to proteins bound to the DNA backbone. Our results demonstrate that the DNA lattice of each NET is potently antibacterial and elicits upregulation of protective surface modifications by the opportunistic bacterial pathogen Pseudomonas aeruginosa. These modifications, previously shown to protect bacteria from antimicrobial peptides, confer greater bacterial tolerance to DNA and NET-mediated antibacterial activity. Treatments that quench the cation chelating capacity of DNA restore bacterial viability and suppress the expression of surface modifications even in the presence of intact NETs. These observations highlight the dual function of DNA as an antibacterial component of NETs, but also a signal perceived by bacteria to induce broad host resistance strategies. Therefore, the ability of P. aeruginosa to sense and defend against the antibacterial activity of neutrophil extracellular traps may contribute to long-term survival in chronic infection sites including the Cystic Fibrosis lung.
Published in the journal: DNA Is an Antimicrobial Component of Neutrophil Extracellular Traps. PLoS Pathog 11(1): e32767. doi:10.1371/journal.ppat.1004593
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004593Summary
Comprising the first line of the innate immune response, neutrophils combat infectious microorganisms through the release of toxic molecules, phagocytosis of invaders and the production of the recently characterized neutrophil extracellular traps (NETs). The antimicrobial activity of NETs has been attributed to proteins bound to the DNA backbone. Our results demonstrate that the DNA lattice of each NET is potently antibacterial and elicits upregulation of protective surface modifications by the opportunistic bacterial pathogen Pseudomonas aeruginosa. These modifications, previously shown to protect bacteria from antimicrobial peptides, confer greater bacterial tolerance to DNA and NET-mediated antibacterial activity. Treatments that quench the cation chelating capacity of DNA restore bacterial viability and suppress the expression of surface modifications even in the presence of intact NETs. These observations highlight the dual function of DNA as an antibacterial component of NETs, but also a signal perceived by bacteria to induce broad host resistance strategies. Therefore, the ability of P. aeruginosa to sense and defend against the antibacterial activity of neutrophil extracellular traps may contribute to long-term survival in chronic infection sites including the Cystic Fibrosis lung.
Introduction
Neutrophils are central mediators of the innate immune defense system and perform their role by killing invading microbes through phagocytosis, degranulation, and the release of neutrophil extracellular traps (NETs) [1, 2]. The scaffold of NETs is composed of genomic DNA, which is enmeshed with antimicrobial proteins normally found in the nucleus, granules, or cytoplasm of neutrophils [1, 2]. Although largely characterized ex vivo using purified human neutrophils and the chemical inducer phorbol-12-myristate 13-acetate (PMA), the process of NETosis resulting in the generation of NETs has been observed in vivo during infection where these structures function to trap bacteria, fungi, protozoa and viruses [1–5]. The mechanism by which NETs kill microbial invaders remains controversial [5–7]. Given the detection of known antimicrobial proteins that decorate the genomic lattice structure [1, 2, 8, 9], current models describing the antimicrobial function of NETs focus on the role of NET-bound proteins. However, most of these proteins are present in low abundance and evidence of their antimicrobial function while bound to the NET structure is limited to a few proteins [1, 8–11]. NET-bound calprotectin is a zinc-chelating protein with antimicrobial activity against Candida and Klebsiella that can be neutralized with excess zinc [3, 9, 10]. Histones, the most abundant NET-bound proteins [10], possess direct membrane-acting antibacterial activity [12, 13] and were shown to contribute to killing of Staphylococcus and Shigella [1]. Cathepsin G, a granular serine protease, is required for the clearance of Neisseria by NETs [8]. To demonstrate the antibacterial contribution of the latter two NET-bound proteins, antibodies raised to histones or cathepsin G were shown to limit the bactericidal capacity of NETs towards these pathogens [1, 8].
Immunocompromised individuals, including those with Cystic Fibrosis (CF), are particularly susceptible to P. aeruginosa infection, which is a major cause of morbidity and mortality in these patients. Chronic P. aeruginosa infection of the CF lung leads to an intense inflammatory immune response, resulting in the recruitment of large numbers of neutrophils to the site of infection [14, 15]. CF sputum is highly enriched in neutrophil-derived DNA, including that of NET origin, indicating that neutrophils deploy NETs in an effort to combat infection of the lung [16, 17]. However, persistence of P. aeruginosa in the midst of sustained neutrophil presence and NETosis suggests that the pathogen is capable of evading this host immune response [18].
An important virulence strategy adopted by successful microbial pathogens is the tolerance of NETs. In a number of cases described so far, microbial invaders accomplish this goal by either avoiding or disarming neutrophil extracellular traps. Modification of the bacterial capsule and surface-localized lipoteichoic acid reduces the trapping of Streptococcus pneumonia in NETs [19]. The secretion of extracellular nucleases by Staphylococcus aureus, Streptococcus pneumonia, group A Streptococcus and Vibrio cholerae highlight a shared virulence strategy that functions to degrade the DNA-backbone of NETs, enabling evasion or liberation of the bacteria from entrapment [2, 20–22].
Although microbial or exogenous DNase is proposed to dissolve NET structures to avoid capture, it has not been considered that the DNA backbone itself may be antimicrobial. We have previously demonstrated that extracellular DNA is an efficient chelator of divalent metal cations [23]. This cation chelation has pleiotropic effects on P. aeruginosa depending on the concentration of extracellular DNA. At subinhibitory levels, sequestration of cations by DNA leads to the expression of genes controlled by the two component systems PhoPQ and PmrAB that sense Mg2+ limitation [23–25]. However, at higher concentrations, extracellular DNA causes dramatic disruptions to the bacterial envelope integrity, which leads to lysis and rapid cell death [23]. It is predicted that the phosphodiester backbone is required for cation sequestration of metal cations on the bacterial surface and the membrane-destabilizing antimicrobial activity of extracellular DNA [23]. Therefore, we hypothesize that the DNA backbone of NETs contributes to their antibacterial activity. Here we show that neutralizing the membrane destabilizing activity of extracellular DNA by quenching the capacity of the phosphodiester backbone to chelate cations protected bacteria from NETs. P. aeruginosa is capable of detecting the DNA lattice of NETs, and in response, upregulates genes required to modify the bacterial outer membrane surface to tolerate the toxic effects of DNA-mediated NET killing.
Results
Pseudomonas aeruginosa induces and tolerates NETosis
P. aeruginosa has been shown to induce the formation of neutrophil extracellular traps in purified human neutrophils [18] and NETs have been observed in CF sputum [16, 17], where P. aeruginosa is a predominant pathogen. PMA-induced NETs contained known neutrophil proteins embedded in the DNA lattice, including myeloperoxidase (MPO) and histones [2] (Fig. 1A). We also show that Gfp-tagged P. aeruginosa is efficiently trapped and aggregated in NETs (Fig. 1A). However, in vivo NETosis during P. aeruginosa infection has not been reported. Therefore intravital confocal microscopy was used to determine whether P. aeruginosa elicited NETosis in the mouse skin infection model [26]. Infection with P. aeruginosa led to the production of large NET-like structures that stained with the DNA-binding dye Sytox green and entrapped ChFP-labeled P. aeruginosa (Fig. 1B and C). In addition to the presence of NETs, neutrophils remained chemotactic and phagocytosed bacteria, suggesting that multiple neutrophil clearance mechanisms are employed in vivo to combat P. aeruginosa (Fig. 1D and S1 Movie).
Fig. 1. P. aeruginosa PAO1 is trapped by human and mouse neutrophil extracellular traps. 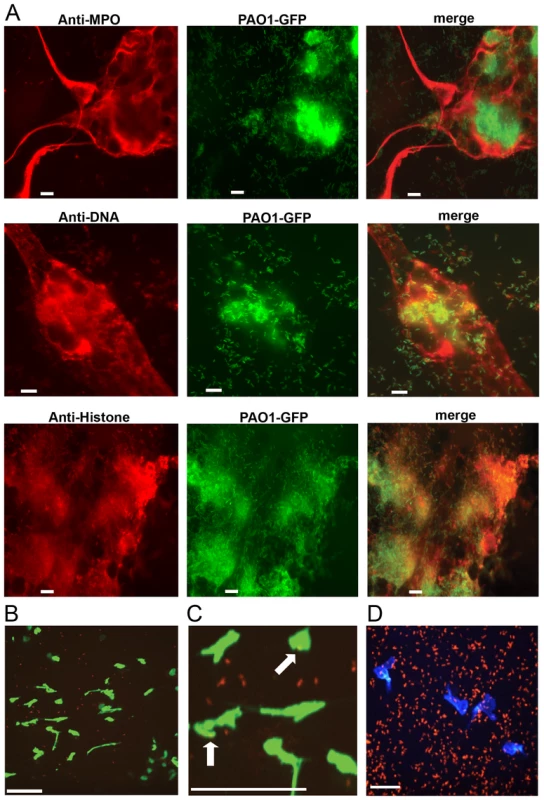
(A) PMA-induced NETs trapped P. aeruginosa Tn7::gfp and contained myeloperoxidase (MPO), DNA and histones when visualized by immunofluorescence with antibodies from autoimmune patient sera (see Methods). Representative images of the NET components (left), Gfp-tagged PAO1 (middle), and the merged (right) are presented. (B) NETosis in the skin of mice infected with ChFP-labeled P. aeruginosa as visualized by Sytox green stained extracellular DNA structures. Scale bar: 40 μm. (C) Arrows indicate ChFP-labeled P. aeruginosa trapped by Sytox green-stained NETs in vivo. (D) ChFP-labeled P. aeruginosa is phagocytosed by neutrophils during a skin infection model. Neutrophils (blue) are visualized with anti-mouse GR-1 antibody. Scale bar: 25 μm. Given that P. aeruginosa induces the production of NETs in vitro and in vivo (Fig. 1), we sought to compare the relative abilities of P. aeruginosa, S. aureus, E. coli and the chemical inducer PMA to elicit NETosis. In the presence of purified human neutrophils, PMA, E. coli and S. aureus induced significantly more NET formation, relative to P. aeruginosa within 1 hour of coincubation (Fig. 2A). However, at later time points (3h), P. aeruginosa elicited the formation of similar amounts of NET structures. Furthermore, quantification of the number of NETs and NET-area using the skin infection model confirmed that P. aeruginosa weakly induces NETosis relative to S. aureus in vivo (Fig. 2B and C).
Fig. 2. Quantification of NETosis in human neutrophils ex vivo and mouse neutrophils during an infection. 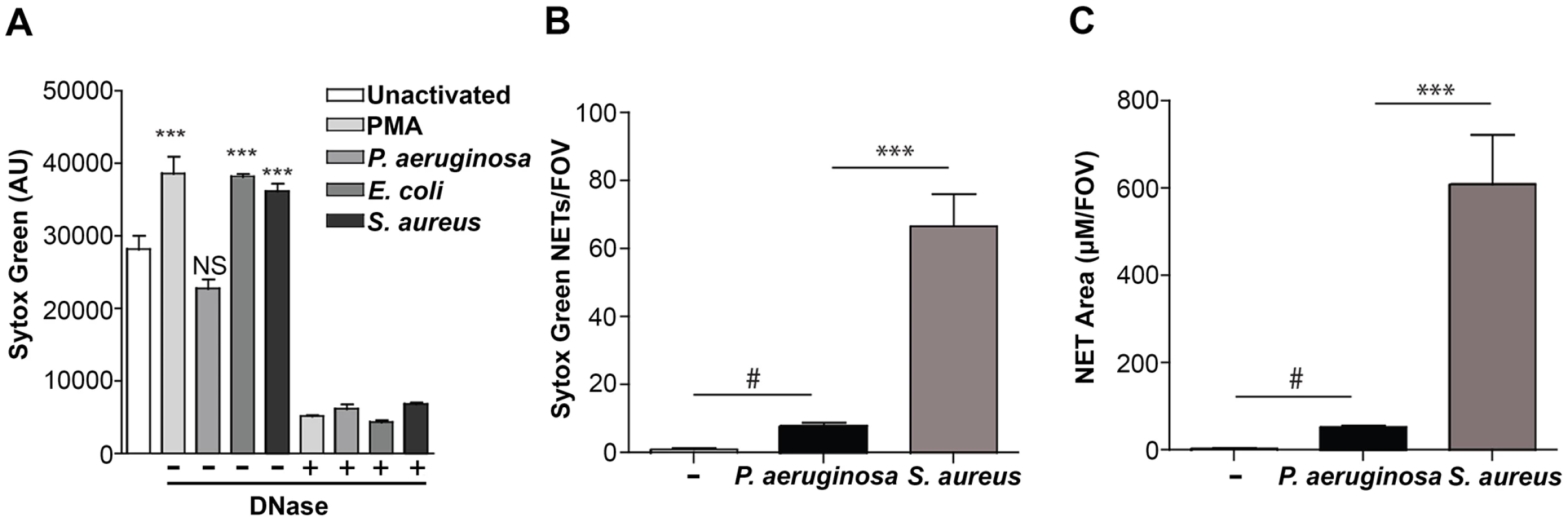
(A) Human neutrophils were stimulated with PMA, P. aeruginosa, E. coli and S. aureus and Sytox green fluorescence was measured as an indicator of DNA release by NETosis after 1 hr stimulation. Exogenous DNase was added as a control to confirm extracellular DNA presence in NETs, indicated by a plus sign (+). Asterisks denote a significant difference in extracellular DNA release between stimulated and unstimulated neutrophils (white bar) (**P<0.01, ***P<0.001). Each value shown is an average from 6 replicates with error bars representing the standard error. (B) The total number of NETs in uninfected mice, or infected mice with P. aeruginosa PAO1 or S. aureus and (C) the total NET area quantified. # denotes a significant difference in NET area and number in P. aeruginosa PAO1 infected mice compared to the uninfected control (#P<0.05). Asterisks denote a significant difference in NET area and number in mice infected with P. aeruginosa compared to S. aureus infected mice (***P<0.001). One possible explanation for the reduced NET formation in the presence of P. aeruginosa is the production of a microbial secreted DNase that degrades NETs more efficiently than other organisms. Therefore, we measured the DNase activity in overnight culture supernatants, as well as in supernatants isolated from coincubating bacteria with PMA-treated neutrophils. We demonstrate that there was little, if any, DNase activity produced by P. aeruginosa, S. aureus or E. coli under these conditions (S1 Fig.). Since cations are a requirement for DNase activity, we were able to restore DNase activity in S. aureus supernatants after the addition of excess cations (S1 Fig.). However, under the cation-free conditions used to quantitate antibacterial NET function, even in the presence of PMA-stimulated neutrophils, DNase activity was not detected and thus P. aeruginosa appears to limit NETosis through an uncharacterized mechanism.
In order to characterize the bactericidal capacity of NETs, neutrophils were treated with PMA to stimulate maximal NETosis and cytochalasin D to block phagocytosis, thus restricting bacterial killing to extracellular NET function [1, 8, 21, 22]. Importantly, the addition of cytochalasin D had no effect on NETosis induced in PMA-treated neutrophils (S2 Fig.). We used the conventional method of direct bacterial counts to enumerate the number of bacteria before and after challenge with PMA-induced NETs. Direct counts of NET-exposed bacteria revealed that P. aeruginosa was most tolerant to NET killing, whereas S. aureus and E. coli were significantly more sensitive (Fig. 3A). The addition of deoxyribonuclease (DNase) restored bacterial survival of the NET-sensitive organisms E. coli and S. aureus, confirming that killing was mediated by extracellular NET function (Fig. 3A). Further, the kinetics of bacterial killing by PMA-generated NETs was determined by measuring the loss of luminescence from chromosomally-tagged luminescent P. aeruginosa strain, PAO1::p16Slux [23], and plasmid-borne luminescent E. coli / pσ70-lux [27]. This approach confirmed that P. aeruginosa was more tolerant to NET killing than E. coli, where luminescence rapidly decreased upon neutrophil challenge (Fig. 3B).
Fig. 3. P. aeruginosa, E. coli and S. aureus differ in their ability to tolerate the bactericidal effects of NETs. 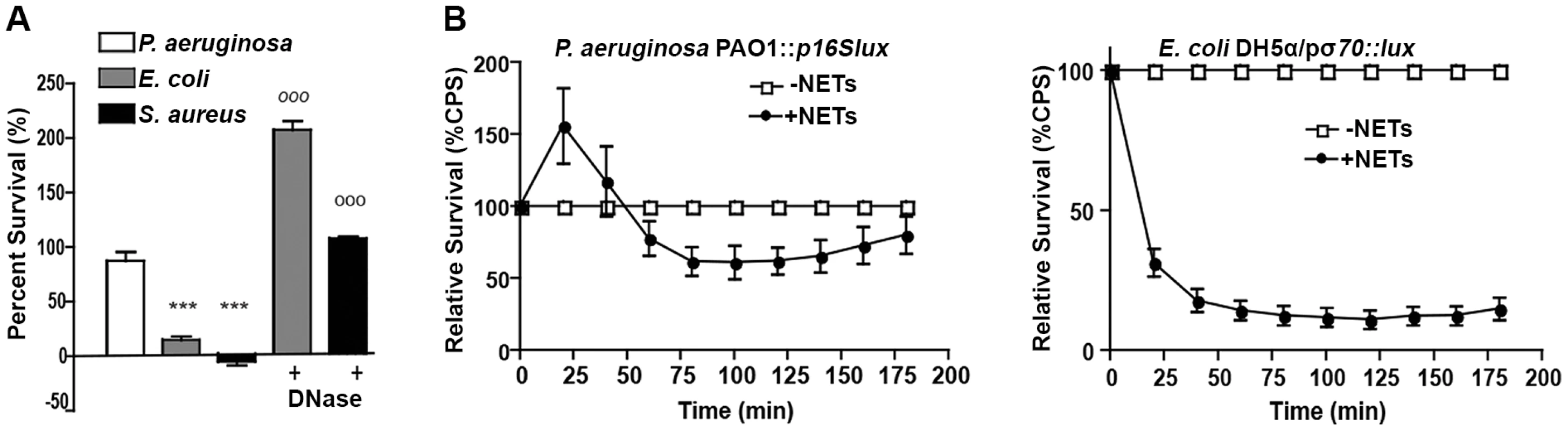
(A) Survival analysis of 1 × 107 CFU P. aeruginosa, S. aureus and E. coli upon exposure to NETs (MOI 10:1). Bacterial viability was determined by direct plate counts (CFU/ml) before and after 4 hour incubation with PMA-activated neutrophils and was normalized to bacterial counts in the absence of neutrophils. DNase I was added exogenously 0.5 hour prior to the end of the experiment to degrade NETs and ensure accurate counts of recoverable colonies. Results are representative of three independent replicates. ***P<0.001 versus P. aeruginosa PAO1. °°°P<0.001 versus non-DNase condition by one-way ANOVA with Bonferroni post tests. (B) Bacterial viability was determined by measuring luminescence from 1 × 107 CFU lux-tagged PAO1::p16Slux or E. coli DH5α/pσ70-lux in the absence or presence of PMA-induced NETs (MOI 10:1). Errors bars represent SEM from six replicates. All experiments were performed at least three times. Direct contact and the phosphate backbone are required for the antimicrobial, cation chelating activity of extracellular DNA
The addition of exogenous DNase I and the production of secreted DNases by bacteria are the most effective means to disable NET killing [1, 2, 20, 21]. It is thought that DNase treatment dissolves NET structures, thereby releasing and diluting the antimicrobial proteins bound to NETs. We have previously shown that extracellular DNA has a potent antibacterial activity, as purified salmon DNA (2% w/v) causes several log orders of bacterial killing within minutes and breaks the integrity of both the inner and outer membranes, leading to lysis [23]. DNA is a very efficient cation chelator and the antimicrobial activity of DNA can be blocked with addition of excess divalent metal cations [23]. It is predicted that the cation chelation is mediated by the phosphodiester backbone.
To confirm the mechanism by which extracellular DNA kills bacteria, we monitored the loss of P. aeruginosa viability in the presence of dilute extracellular DNA (0.125% w/v; Fig. 4A). Bacterial survival was restored if DNA was pretreated with DNase or excess 5mM Mg2+, which degrades DNA or saturates its cation chelating ability, respectively, thus neutralizing the antibacterial activity (Fig. 4A). Pretreatment of extracellular DNA with calf intestinal alkaline phosphatase (PTase), which cleaves 5’-phosphates, also blocked the observed antibacterial activity (Fig. 4A). The addition of decreasing amounts of DNase, PTase or Mg2+, resulted in marked, dose-dependent, decreases in bacterial survival when challenged with 0.15% DNA (Fig. 4B). The DNA-mediated damage to the outer membrane leads to the formation of ChFP-enriched outer membrane vesicle-like structures (OMVs; Fig. 4C) [23]. However, incubation of P. aeruginosa with extracellular DNA pretreated with DNase, PTase and Mg2+ greatly diminished the number of ChFP-labeled OMVs relative to control conditions (Fig. 4D), confirming that the bactericidal mechanism of extracellular DNA is through disruption of the bacterial membrane.
Fig. 4. Extracellular DNA exerts bactericidal activity through cation chelation-mediated disruption of the bacterial outer membrane. 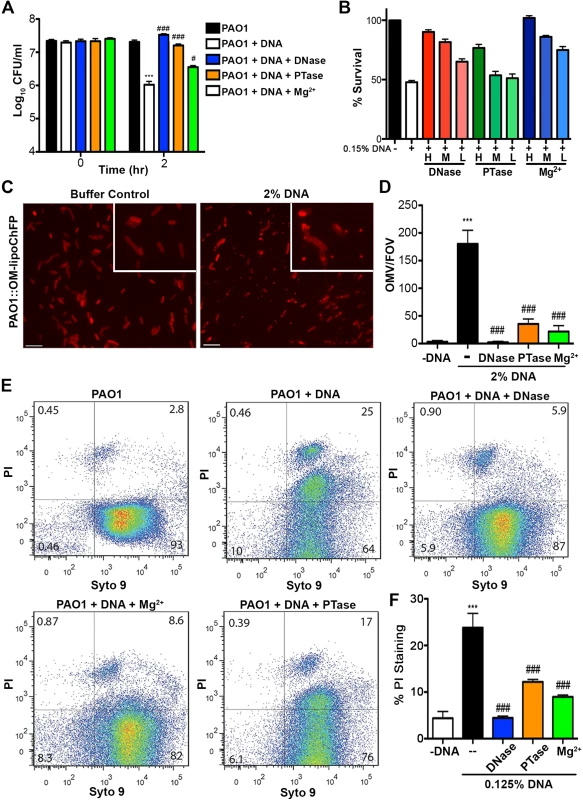
(A) Survival analysis of 1 × 107 CFU P. aeruginosa PAO1 coincubated with 0.125% (w/v) DNA or DNA pretreated with DNase I, PTase or 5 mM Mg2+. Bacterial counts were performed before (0) and after two hours treatment with DNA (2). Results are representative of three independent experiments. Error bars represent the standard deviation (SD) from eight replicates. (B) High, medium and low concentrations of DNase (430kU, 43kU, 4.3kU), PTase (50 U, 10 U, 1 U) or excess Mg2+ (5 mM, 500 μM, 5 μM) leads to increased levels of protection from killing with 0.15% DNA (w/v). (C) Visualization of the outer membrane integrity of P. aeruginosa PAO1::OM-lipoChFP expressing an outer membrane-localized mCherry fluorescent lipoprotein [38] immediately after 2% (w/v) DNA-exposure. Insets represent increased magnification of presented micrographs. Scale bar: 10 μM. (D) Quantification of ChFP-rich OMV generation in the field of view from 6 representative images generated from bacteria-DNA coincubation as described in (C) or DNA pretreated with DNase, PTase and Mg2+. Error bars represent SD from 6 fields of view. (E) Flow cytometry of DNA-exposed P. aeruginosa PAO1 using SYTO9-PI dual staining as a measure of membrane-compromised bacteria [28, 32]. 2.5 × 107 CFU P. aeruginosa PAO1 were exposed to 0.0125% DNA alone or pretreated as in (A) then immediately analyzed by the collection of positive events (N = 50 000) by BD LSRII. Numbers in corners represent the % of 50 000 events that fall into each quadrant gate. (F) Quantification of membrane-compromised, PI-stained P. aeruginosa PAO1 as measured by flow cytometry. Mean percent PI stained was derived from the average of three replicates (each with N = 50 000 for each plot) in each exposure condition as in (E). *** denotes a significant difference between the control and 0.125% DNA sample. ### and # denote a statistically significant difference, P<0.01 and P<0.05, respectively, between DNA alone sample and pretreated samples. Two-tailed student t-tests were performed to test for significant differences. Extracellular DNA-mediated damage to membrane integrity was confirmed by using flow cytometry (Fig. 4E). Treatment of P. aeruginosa with DNA resulted in a new population of cells that were dual positive for SYTO9 and PI, indicative of membrane damage and increased PI uptake, and possibly dead cells. The increased staining of DNA-exposed bacteria by membrane-impermeable propidium iodide (PI) was not observed in DNase and Mg2+ pretreatments and was reduced in PTase pretreated DNA samples (Fig. 4E and F). To ensure cation chelation was responsible for the observed bacterial membrane destabilization, we assessed whether the known cation chelator EDTA could cause membrane disruption. Like DNA, EDTA caused major outer membrane disruptions and the release of OMVs, as well as a dramatic increase in PI-staining of EDTA-treated cells monitored by flow cytometry (S3 Fig.) [28]. To determine whether the antibacterial capacity of DNA requires direct bacterial contact or can be mediated through passive cation sequestration, we exposed P. aeruginosa to high concentrations of DNA spatially separated by an ion-permeable barrier. Blocking direct interaction between P. aeruginosa and DNA resulted in bacterial survival after a prolonged exposure, compared to the rapid antibacterial activity of DNA in direct contact with the bacteria (S4 Fig.). Together, these results demonstrate that the antibacterial activity of extracellular DNA requires direct contact, and the phosphate backbone for cation chelation, leading to membrane disruption and bacterial cell death.
Bactericidal activity of NETs is blocked by neutralizing the cation chelation capacity of DNA
The bactericidal activity of neutrophil extracellular traps is attributed to direct contact and exposure of bacteria to the antimicrobial proteins embedded in the DNA scaffold of NETs [1, 2]. Given the antimicrobial activity of DNA, we propose that the DNA backbone of the NET itself is antibacterial. Therefore, if DNA contributes to bacterial killing, treatments that quench the cation chelation potential of the DNA backbone will block bactericidal activity of NETs. To address this possibility, PMA-activated neutrophils were treated with the addition of DNase, PTase or excess Mg2+ and bacterial viability was monitored. The DNA-targeted treatments completely protected P. aeruginosa and E. coli from killing by neutrophil extracellular traps (Fig. 5A). To confirm these results, we monitored the luminescence of P. aeruginosa PAO1::p16Slux co-incubated with PMA-activated neutrophils and observed that the antibacterial effects of NETs were neutralized by treatment with exogenous DNase, Mg2+ cations and PTase (Fig. 5B).
To determine whether the restored bacterial viability in the presence of NETs was due to preventing damage to the bacterial envelope, we performed flow cytometry to assess membrane integrity. Increased PI staining of NET-exposed bacteria is an indicator of membrane damage, which was completely blocked by addition of DNase (Fig. 5C). The addition of excess Mg2+ or treatment with exogenous PTase also limited membrane damage (Fig. 5C). Importantly, both Mg2+ and PTase treatments neutralized the antimicrobial activity of NETs (Fig. 5A and B) did not disrupt overall NET architecture, as MPO and histones were still present within the treated NET structures (Fig. 6). To assess the function of a NET-bound protein, we measured elastase activity in PMA-treated neutrophils and showed no difference in elastase activity when NETs were treated with exogenous PTase or Mg2+ (S5 Fig.). NET structures remain intact and contain functional proteins (elastase) after treatment with PTase or excess Mg2+, but are no longer antibacterial for E. coli and P. aeruginosa (Fig. 5). Together these results suggest an antibacterial mechanism wherein the DNA backbone of NETs target and destabilize the bacterial membrane and promotes cell death.
Fig. 5. Neutralizing the cation chelating activity of the DNA backbone of NETs protects bacteria. 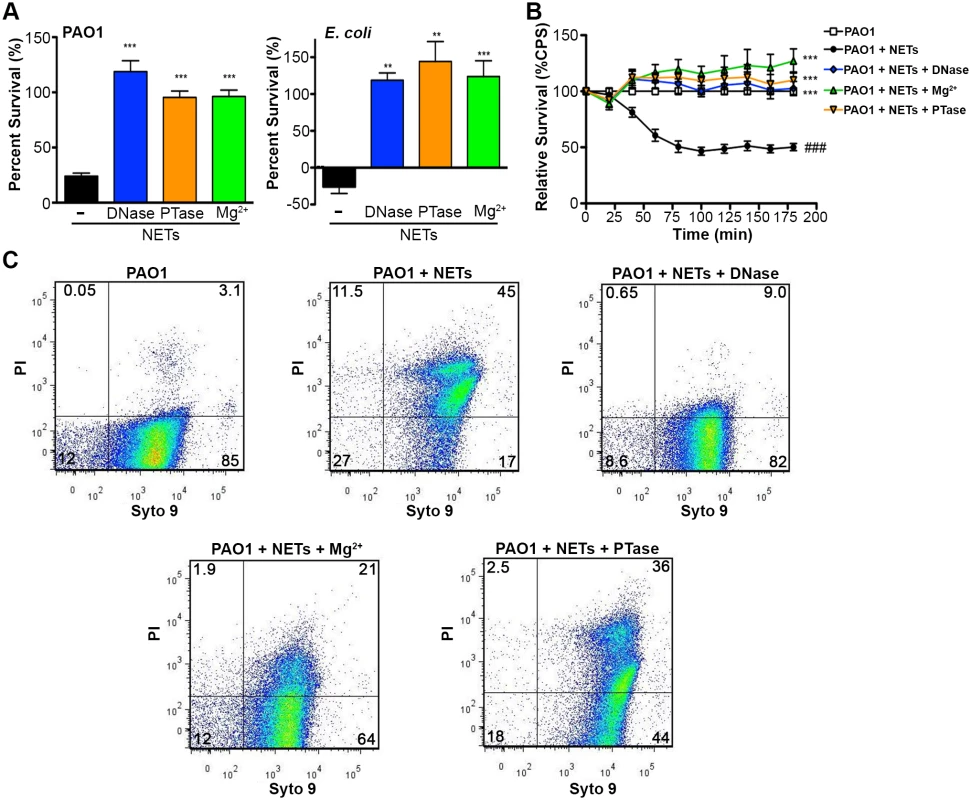
(A) Percent survival of P. aeruginosa PAO1 and E. coli DH5α as determined by direct plate counts (CFU/ml) before and after 4 hour incubation with PMA-activated neutrophils or combined treatment of NETS with DNase, PTase or Mg2+. Error bars are SEM from 6 replicates. ** or *** denotes a statistically significant difference (P<0.05 or P<0.01, respectively) between NET-alone versus NET and enzymatic or excess cation treatments, as determined by one-way ANOVA with Bonferroni post tests. (B) Luminescence-based viability as a real-time measure of P. aeruginosa PAO1::p16Slux survival in the presence of NETs alone, or combined treatment of NETS with DNase I, PTase or Mg2+. ### denotes a statistically significant difference of P<0.001 between NET-challenged PAO1 versus PAO1 alone (white). ***P<0.001 versus NET killed samples (black). (C) Flow cytometry of P. aeruginosa PAO1 (2 × 107 CFU) coincubated for four hours with PMA-stimulated neutrophils alone (1 × 106; MOI: 10) or with the addition of DNase I, PTase and 5 mM Mg2+. N = 50 000 for each plot. Numbers in each corner represent the % of 50 000 events that fall into each quadrant gate. Fig. 6. Structurally intact NETs still possess histones and MPO after treatment with excess Mg2+ and PTase. 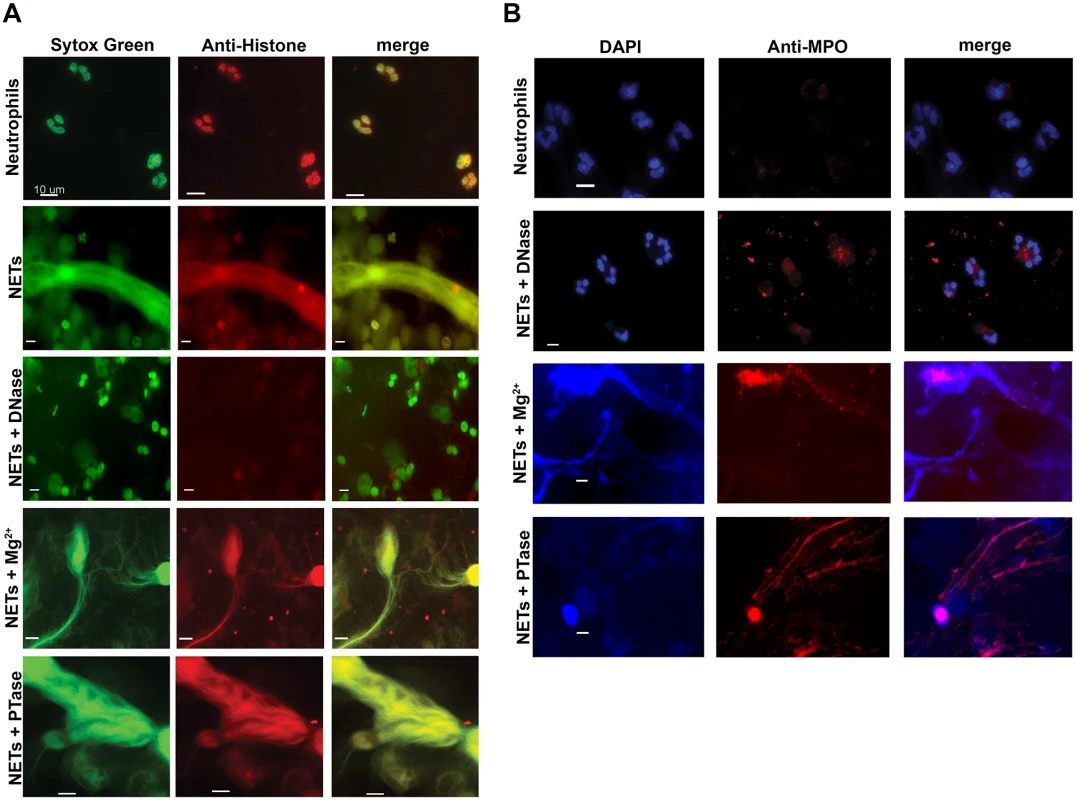
Neutrophil extracellular traps were visualized with (A) Sytox Green staining of DNA and anti-histone primary antibodies. (B) Neutrophil extracellular traps were visualized with DAPI staining of DNA and anti-MPO primary antibodies. NETs were not observed in unactivated neutrophils, but were present in PMA-induced NETs that were treated with either excess 5 mM Mg2+ or exogenous PTase. Alexa Fluor 647-conjugated secondary antibodies were used to visualize histone H1 and MPO. Representative immunofluorescence images were merged to show overlap of histone H1 and MPO with structurally intact NETs. Scale bars, 10 µm. Extracellular DNA elicits induction of surface modifications that protect P. aeruginosa from NETs
Subinhibitory concentrations of extracellular DNA sequester Mg2+ and trigger the expression of multiple surface modifications that are known to protect the bacterial outer membrane from antimicrobial peptide (AP) damage and killing [23–25]. The arn operon (PA3552-PA3559) is required for the covalent addition of aminoarabinose to the phosphates of lipid A and the spermidine synthesis genes (PA4773-PA4774; speDE homologs) are required for production of the polycation spermidine on the outer surface [23, 24]. Both modifications substitute for divalent metal cations, mask the negative charges of the outer surface and thus contribute to AP resistance [23, 24, 29–31]. Given that these modifications stabilize the bacterial envelope, we sought to determine whether these surface modification pathways provided a more general mechanism to resist bacterial membrane damage. We noted that P. aeruginosa strains with mutations in the arn or spermidine biosynthetic pathways were significantly less capable of surviving exposure to DNA (Fig. 7A).
Fig. 7. Pseudomonas aeruginosa responds to DNA in neutrophil extracellular traps and induces protective bacterial surface modifications. 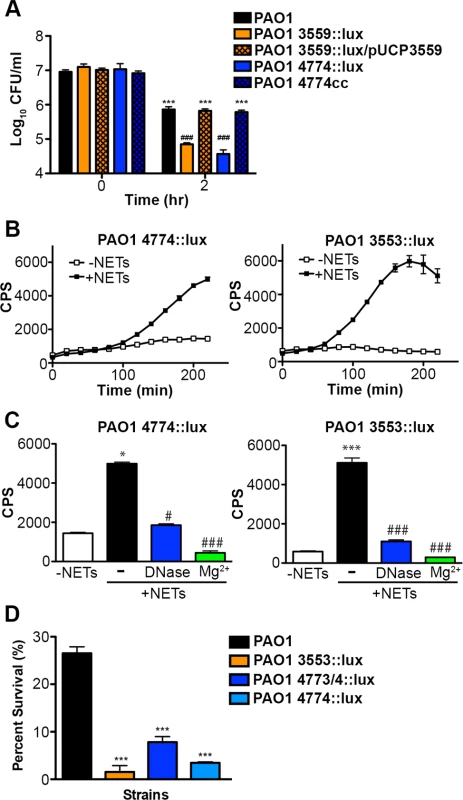
(A) Survival analysis of 1 × 107 CFU wild-type P. aeruginosa PAO1 or mutants with defects in the aminoarabinose-LPS modification (PA3553::lux) or in spermidine synthesis (PA4774::lux) after coincubation with 0.125% DNA. *** denotes a statistically significant difference between time 0 and 2 hours post coincubation with 0.125% DNA. ### denotes a statistically significant difference of P<0.001 between wild-type and mutant P. aeruginosa when exposed to 0.125% DNA. Two-tailed student t-tests were performed to test for significant differences. Error bars represent SD from eight replicates. (B) 2 × 107 CFU P. aeruginosa PAO1 spermidine synthesis (PA4774::lux) and aminoarabinose modification (PA3553::lux) transcriptional reporter strains were incubated with PMA-activated neutrophils (MOI 10:1) and gene expression (luminescence as quantified by CPS) was measured every 20 minutes in the absence (empty squares) and presence (solid circles) of NETs. Error bars represent SEM from six replicates. (C) The effect of DNase or 2 mM Mg2+ treatment on NET-mediated gene induction of 2 × 107 CFU PA4774::lux or PA3553::lux after four hours of coincubation (MOI 10:1). *P<0.05, ***P<0.001 versus bacteria alone (white bar), #P<0.05, ###P<0.001 versus NET exposure (black bar) as determined by one-way ANOVA with Bonferroni post tests. (D) Bacterial survival analysis of 2 × 107 CFU NET-exposed P. aeruginosa PAO1 wild-type, aminoarabinose modification mutant PA3553::lux, or the spermidine synthesis mutants PA47743/4::lux, PA4774::lux (MOI 10:1) Error bars represent the SEM from 6 replicates. *** denotes a statistically significant difference of P<0.001 versus wild-type survival as determined by one-way ANOVA with Bonferroni post tests. All assays were conducted at least three times and representative data is presented. Given the role of these pathways for tolerance of exogenous DNA, we then investigated whether the DNA component of NETs induced expression of the arn or spermidine synthesis genes in P. aeruginosa. Expression of both pathways was strongly induced 2–6 fold following co-incubation with NETs produced by PMA-activated neutrophils (Fig. 7B). To confirm that DNA was the component of NETs that led to induction of the bacterial gene expression response, the addition of excess Mg2+ cations and enzymatic treatment with DNase (Fig. 7C) and PTase (S6 Fig.) all blocked the induction of the arn and spermidine operons response to NETs. While these treatments specifically neutralize DNA, we also considered the possibility that NET-bound antimicrobial proteins including histones or LL-37 may elicit these protective responses. We have previously shown that sub-MIC concentrations of antimicrobial peptides induce both outer surface modifications [29]. Therefore, to assess the relative capacity of each NET component to act as a bacterial signal, we compared the ability of purified histones, the well-characterized APs polymyxin B and colistin, and DNA to induce the expression of the spermidine synthesis pathway. Although all NET components induced expression of the PA4773-PA4774 spermidine synthesis pathway (S7 Fig.), DNA was the most potent inducer of this bacterial response (S7 Fig.).
The upregulation of protective outer membrane modifications (aminoarabinose-modified LPS and surface spermidine production) by NETs, and by the individual NET components of DNA and histones, suggests that these modifications are required to defend the membrane against assault from multiple innate immune components that target the bacterial membrane. Both modifications result in stable substitutions for divalent metal cations in the outer membrane, and protect P. aeruginosa from antimicrobial peptides [23, 24, 29–31] and DNA killing (Fig. 7A). P. aeruginosa mutants in the arn and spermidine synthesis genes also exhibited increased susceptibility to the disruptive effects of NETs (Fig. 7D). We next compared the susceptibilities of P. aeruginosa, E. coli and S. aureus to histone and DNA killing. Surprisingly, P. aeruginosa was the most susceptible to DNA killing, while S. aureus was the most tolerant, the opposite pattern of NET susceptibility (Figs. 3A and 8A). The bactericidal capacity of purified histones was modest, where after 2 hours exposure P. aeruginosa was the more histone tolerant (Fig. 8B). Taken together, these results highlight the ability of P. aeruginosa to quickly respond and defend against the DNA and histone mediated-antibacterial effects of NETs by stabilizing the outer membrane.
Fig. 8. Bacterial species differ in their susceptibility to killing by DNA and histones. 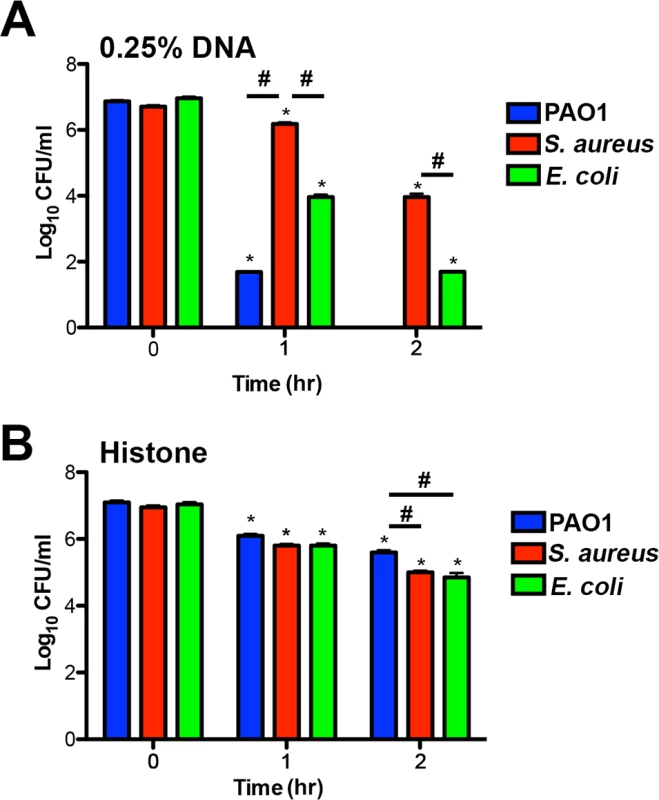
(A) P. aeruginosa PAO1, S. aureus and E. coli killing assay in the presence of 0.25% DNA (w/v). PAO1 was significantly more sensitive at both eDNA concentrations relative to the highly tolerant S. aureus. (B) P. aeruginosa PAO1, S. aureus and E. coli killing assay in presence of 1.25 µg/mL histones. Bacterial survival was quantified after 1 and 2 hours by colony count, and statistical significance assessed by 2-tailed student t-tests. * denotes statistical differences (P<0.05) between the indicated time point and the initial bacterial count, while # indicates significant differences (P<0.05) between bacterial species. The values shown are the mean plus standard deviation from 8 samples. Experiments were repeated three times and the data shown is from one representative experiment. Discussion
Given the observations that exogenous or secreted microbial DNases protect bacteria against NET killing and that extracellular DNA has rapid, membrane-damaging antibacterial activity, we sought to test the hypothesis that the DNA backbone of NETs contributes to their bactericidal function. Extracellular DNA possessed contact-mediated antibacterial activity that could be neutralized by enzymatic and cationic treatments that degrade or quench the capacity of DNA to chelate cations (Fig. 4 and S3 Fig.). NETs exposed to the same treatments that target the DNA scaffold were unable to cause bacterial membrane damage or to cause bacterial killing of P. aeruginosa and E. coli (Fig. 5). Therefore, we propose a novel bactericidal mechanism of NETs whereby the removal of surface-stabilizing cations by the DNA phosphodiester backbone results in bacterial lysis (Fig. 4B and C). To address the controversy surrounding the bacterial killing ability of NETs [5–7], we used multiple viability assays to measure NET killing and membrane damage, which included direct bacterial counts, luminescence viability assays and flow cytometry of PI-stained cells [32]. Taken together, these data support the general notion that NETs are directly antimicrobial.
Deciphering the specific antimicrobial mechanisms of NETs has been limited to a few candidate proteins [1, 8–11]. Most studies have focused on the NET-bound proteins as the antimicrobial components, given their important role during phagocytosis and degranulation. Although granular, cytoplasmic and nuclear proteins derived from neutrophils can be detected in NETs by immunofluorescence, the abundance of most NET-bound proteins is low (<1–6%), relative to histones, which comprise 65% of the total protein content [10]. Although classically characterized as chromatin structural proteins, NET-bound histones possess antimicrobial activity that can be neutralized through the addition of anti-histone antibodies [1, 12, 13]. However, other neutrophil proteins exhibit altered or reduced enzymatic activity when enmeshed in the NET backbone raising questions as to their antimicrobial capacity. For example, S. aureus tolerates myeloperoxidase in NETs unless supplemented with the addition of exogenous H2O2 [11]. Neutrophil elastase activity increases in DNase treated sputum from Cystic Fibrosis patients, suggesting that DNA may inhibit elastase activity, thus limiting the role of elastase as a potential NET-bound factor [17].
We observed that both the antibacterial activity of NETs and the ability to induce protective bacterial responses were blocked by treatments that target extracellular DNA, suggesting that the NET scaffold is not simply a passive structure (Figs. 5 and 7) [23–25, 31]. The most potent inducing triggers of the P. aeruginosa surface modifications are purified eDNA, followed by APs and purified histones (S7 Fig.). Since being widely induced by these components, it is not surprising that the aminoarabinose-modified LPS and spermidine synthesis pathways protect the outer membrane from DNA, NETs (Fig. 7) and antimicrobial proteins [23, 24, 29–31]. The low potency of the tested histones may be explained by the fact that our assays were performed with a mixture of full-length histones, which had modest antibacterial activity (Fig. 8). Recent evidence highlights that histones are proteolytically processed by proteases such as elastase during the process of nucleus decondensation, prior to NET release [33] It is therefore likely that potent bactericidal histone-derived peptides are present in NETs as an important antibacterial component of NETs.
P. aeruginosa appears to mount a multifunctional, outer membrane defense strategy to combat multiple antimicrobial components enmeshed in NETs. Consistent with this model, we noticed unexpected susceptibility patterns that also suggest that NET killing may be the result of the combinatorial effect of DNA and NET-bound proteins. The observation that S. aureus is susceptible to NET killing but tolerant to DNA (Figs. 3 and 8), suggests that NET killing of S. aureus is likely dependent on other anti-staphylococcal proteins enmeshed in the DNA lattice. The DNA susceptibility phenotype of P. aeruginosa may explain the potency of DNA as the strongest inducer of the protective outer surface modifications that contribute to the observed NET tolerance. It is intriguing to speculate that the NET-bound, antimicrobial proteins act in concert with the antibacterial activity of DNA to provide broad-spectrum protection against a wide range of microbial pathogens.
Modification of the bacterial cell surface and the production of secreted DNases are virulence strategies utilized by microbial pathogens to evade NET killing [19–22]. Here we report that the spermidine and the arn surface modification pathways are required to tolerate the antibacterial action of both DNA and NETs (Fig. 7). The covalent addition of aminoarabinose to the lipid A component of LPS masks the negative charges of core LPS phosphates, and the polycationic nature of spermidine (+3 charge) substitutes for surface divalent metal cations, and may also bind and neutralize DNA. In addition, spermidine possesses an antioxidant activity that protects bacterial membrane lipids from oxidative damage [24] and therefore may protect P. aeruginosa from NET-induced oxidative damage [11]. Combined, these results suggest that the spermidine and arn surface modifications possess multiple protective roles that may contribute to resisting a broad range of antimicrobial components present within NETs.
We propose that bacterial surface-bound, divalent metal cations are displaced by direct contact with extracellular DNA, and that DNA-induced surface modifications prevent outer membrane disruption and bacterial killing by NETs. Therefore, the antibacterial mechanism of cation chelation exerted by DNA is distinct from that of other previously characterized antimicrobial cation chelating proteins such as calprotectin. We have previously demonstrated that DNA chelates diverse metal cations (Mg2+, Ca2+, Zn2+, Mn2+) [23] while calprotectin chelates zinc and manganese [34]. Additionally, the antimicrobial function of calprotectin is contact-independent whereas the bactericidal function of DNA requires contact (S4 Fig.). Further, sequestration of zinc and manganese by calprotectin does not target the microbial membrane but rather sequesters cation cofactors required by bacterial enzymes such as superoxide dismutase, which protects bacteria from superoxide [34].
In summary, we have identified that the DNA backbone is a bona fide antibacterial component of neutrophil extracellular traps. The DNA scaffold structure also acts as a warning signal perceived by P. aeruginosa. Overall, these results support a model where the membrane-destabilizing activity of the DNA scaffold contributes to the bactericidal capacity of NETs, while the cation chelating activity acts as a signal perceived by P. aeruginosa that leads to upregulation of protective surface modifications. These results highlight a dynamic bacterial-host interaction between an opportunistic pathogen that causes chronic infections in the lungs of individuals with Cystic Fibrosis, an infection site known to be rich in neutrophil DNA and neutrophil extracellular traps [16, 17]. This ability to sense and defend against NETosis may help explain the long-term persistence of P. aeruginosa in CF lung infections.
Materials and Methods
Bacterial strains and growth conditions
All strains and plasmids used in this study are shown in S1 Table. Bacterial cultures were routinely grown at 37°C in LB or BM2 defined minimal media with 0.5 mM MgSO4, unless otherwise stated. S. aureus was grown overnight in BHI media. When necessary, the following antibiotics were used: 50 µg/mL tetracycline for P. aeruginosa mini-Tn 5-lux mutants, and 50 µg/mL kanamycin for E. coli DH5α/ pσ70-lux. Mid-log cultures were used for co-incubation experiments with neutrophils or extracellular DNA.
Human neutrophil isolation
Neutrophils were isolated from healthy donors as previously described [35]. Whole blood was collected and mixed 5 : 1 in acid citrate dextrose, followed by removal of red blood cells using dextran sedimentation and hypotonic lysis with KCl. After all red blood cells were lysed, the cell pellet was subjected to Ficol-Histopaque density centrifugation. The subsequent pellet was resuspended in 2 mL of HBSS (Hank’s balanced salt solution, no cations; Invitrogen 14175–095). The viable cell concentration was determined using a haemocytometer and Trypan blue staining.
NET imaging
Glass cover slips were HAS-coated and placed in 6-well tissue culture plates. Neutrophils were added at 2.0×106 cells/mL per well, adhered (30 min, 5% CO2, 37°C) and treated with cytochalasin D (10 µg/well) and PMA (25 nM) to activate NETosis [35]. Mid-log bacterial cultures were diluted in HBSS (no cations) (5.0x107 CFU/mL) for an MOI of 25 : 1, centrifuged to the neutrophils, and coincubated for 1–4 hours (5% CO2, 37°C). Cells on cover slips were fixed with 4% paraformaldehyde, washed with 250 µL of 10% FBS (Invitrogen) in PBS and stained with either DNA dyes and/or various primary and secondary antibodies (described below).
Immunofluorescence microscopy of NETs
For NET visualization with antibodies, the primary anti-human MPO antibody (DakoCytomation - A0398) was diluted into 10% FBS in PBS (1/500). 30 µL was added to adhered neutrophils, incubated (30 min, 37°C) and washed twice with sterile PBS. 40 µL of the secondary anti-rabbit Cy 5 antibody (Jackson ImmunoResearch 60354) (1/500 dilution) was added. After 15 min incubation in the dark, cover slips were washed twice with PBS, and prepared with mounting media. Anti-DNA and anti-histone antibodies where obtained from Dr. Marvin Fritzler. Either the anti-DNA (1 : 10) or anti-histone (1 : 500) antibodies [36, 37] were added as described above. The anti-human secondary antibodies with Alexa Flour 647 (Invitrogen A21445, 1/500) were added to cover slips and mounted as described above. Images of human NETs were acquired using the Leica DMI 4000B inverted microscope equipped with ORCA R2 digital camera and Metamorph software for image acquisition using the 63X or 100X objectives. The following excitation and emission filters were used for blue fluorescence (Ex 390/40; Em 455/50), red fluorescence (Ex 555/25; Em 605/52), far red fluorescence (Ex 645/30; Em 705/72) and green fluorescence (Ex 490/20; Em 525/36). Images were formatted and analyzed using the Imaris 7.0.0 imaging software. All images shown are representative of at least three experiments.
Mouse skin infection model
Mice were anaesthetized (10 mg/kg xylazine hydrochloride and 200 mg/kg ketamine hydrochloride) and body temperature was maintained using a rectal probe and heating pad. The mice were pretreated with intradermal MIP-2 (0.2µg/injection) diluted in sterile normal saline 30 minutes prior to imaging. The right jugular vein was cannulated to administer additional anesthetic and fluorescent dyes. The microcirculation of the dorsal skin was prepared for microscopy as previously described [26]. Briefly, after shaving the mouse’s back, a midline dorsal incision was made extending from the tail region up to the level of the occiput. The skin was separated from the underlying tissue, remaining attached laterally to ensure the blood supply remained intact. The area of skin was then extended over a viewing pedestal and secured along the edges using 5.0 sutures. The loose connective tissue lying on top of the dermal microvasculature was carefully removed by dissection under an operating microscope. The exposed dermal microvasculature was immersed in isotonic saline and covered with a coverslip held in place with vacuum grease. Alexa Fluor 649 conjugated anti-mouse GR-1 antibody (10µl per mouse i.v.; eBioscience) was used visualization of neutrophils. To visualize NETs in vivo the membrane impermeable dyes SYTOX-green or SYTOX-orange were administered (diluted 1 : 1000 with sterile saline, 100µl per mouse i.v.). MIP-2 injection (0.2µg/injection) was initiated 30 min prior to Pseudomonas aeruginosa or S. aureus administration. Following baseline visualization, all bacteria were directly administered into the field of view using a tuberculin needle (1×108 CFU/100µl of sterile saline, i.d.).
Intravital microscopy
Spinning disk confocal intravital microscopy was performed using an Olympus BX51WI (Olympus, Center Valley, PA) upright microscope equipped with a 20×/0.95 XLUM Plan Fl water immersion objective. The microscope was equipped with a confocal light path (WaveFx, Quorum, Guelph, ON) based on a modified Yokogawa CSU-10 head (Yokogawa Electric Corporation, Tokyo, Japan). Laser excitation at 488, 561 and 649nm (Cobalt, Stockholm, Sweden), was used in rapid succession and fluorescence in green, red and blue channels was visualized with the appropriate long pass filters (Semrock, Rochester, NY). Exposure time for all wavelengths was between 500 and 600ms. Sensitivity settings were maintained at the same level for all experiments. A 512×512 pixels back-thinned EMCCD camera (C9100–13, Hamamatsu, Bridgewater, NJ) was used for fluorescence detection. Volocity Acquisition software (Improvision Inc., Lexington, MA) was used to drive the confocal microscope. Images captured using the spinning disk were processed and analyzed in Volocity 6.0.1. NET area and NET number were quantified using the Volocity software.
Quantification of NET area and number
NET area was determined using Volocity imaging software. Briefly, in each field of view (FOV) the threshold of the corresponding fluorescent channel in which NET structures were stained was set to eliminate the background staining of the skin. The area and number of NET positive structures in the FOV was calculated and counted via the Volocity 6.0.1 software. Structures that showed no characteristic NET-like shape and resembled the staining of a nucleus of an obvious dead cell in the FOV were excluded from the quantification manually. NET image analysis was performed in at least two infected or uninfected animals and from 5 fields of view.
NET quantification from purified human neutrophils
NETs were quantitated by measuring the amount of extracellular DNA that stains with the cell impermeant dye Styox Green [35]. Cell culture media (CCM) consisted of 48.5 mL of RPMI 1640 (Invitrogen), 0.5 mL of 1.0 M HEPES and 1.0 mL of human serum albumin (HAS; Innovative Research). Neutrophils were diluted into CCM (2.0×105 cell/well) and added to an HSA-coated, 96-well black, clear-bottom plate (Thermo Scientific). As a positive control, PMA (25 nM/well) was added to activate the neutrophils [35]. For bacterial activation of NETs, mid-log bacterial cultures were diluted in CCM (2.0×106 CFU/well) for a multiplicity of infection (MOI) of 10 : 1 (bacteria to neutrophils). DNase (430 kU/well; VWR 31149) was added to degrade extracellular DNA in NETs. Bacteria were gently centrifuged onto the adhered neutrophils (800xg, 10 min). Sytox green (2.5 µM; Invitrogen) was added to each well and green fluorescence (Ex 490/8; Em 535/25) was measured with Perkin Elmer 1420 Multilabel Counter Victor3 between 1 and 4 hours. All values shown are the mean from at least three individual replicates and each experiment was performed at least 3 times. We noticed variation in the background level of NET staining between individual donors, but there was a reproducible and robust 2.5 to 10-fold increase in NETosis after PMA treatment of neutrophils from various donors (S2 Fig.).
Quantification of Bacterial Viability and Gene Expression Using Plate Counts and Luminescence NET killing was examined using direct plate counting methods where a reduction in cell number indicated bacterial killing. All NET killing experiments were performed in HBSS solution lacking divalent cations. Isolated human neutrophils were mixed with mid-log bacteria in HBSS (no cations) in black, clear-bottom 96-well plates with of 2.0 × 107 CFU bacteria and 2.0 × 106 neutrophils (MOI 10 : 1). After a 1–4 hr incubation, 50 µL of DNase I solution (430 kU/mL) was added to every well, mixed, and incubated for 30 min at 37°C, in order to release bacteria trapped in NETs for accurate plate counts. 15 µL of suspension was serially diluted (1/10) in 0.9% NaCl solution in a sterile 96-well plate and 5 µL from each well was stamped onto LB agar plates to obtain bacterial plate count data for time zero (T0) and after 4h (T4). CFU/ml values from T4 and T0 time points were used to calculate the percentage survival by subtracting the T4—T0 plate counts and dividing the ‘bacteria and neutrophil’ conditions by the ‘bacteria alone’ conditions and multiplying by 100. For lux viability and gene expression assays [23], bacteria were centrifuged onto the adhered neutrophils and placed in the Victor3 plate reader for luminescence (CPS) measurements every 20 minutes for 3–4 hours. All values shown are the mean from at least six individual replicates and each experiment was performed at least 3 times.
Flow cytometry analysis of SYTO9/propidium iodide stained bacteria
SYTO9 stains the DNA in all cells and propidium iodide (PI) stains the DNA in dead cells and cells with damaged membranes [28, 32]. The sample of bacteria and neutrophils (~200 µL) was placed in 5 mL polystyrene round-bottom sample tubes and stained with SYTO9 and propidium iodide at final concentrations of 0.02 mM and 0.2 mM, respectively. The tubes were centrifuged at 300x gravity and incubated (RT, 15 min). Bacterial cells were analyzed using the BD LSRII flow cytometer (BD Bioscience, San Jose, USA) equipped with a blue laser (488nm) and a green laser (532nm). Unstained, mid-log bacterial cells were used to gate the forward scatter (FSC) and size scatter (SSC) parameters. For green and red fluorescence profiles, SYTO9 was excited by blue (488nm) laser with emission filters 525/50BP and 505LP and PI was excited using the green (532 nm) laser with emission filters 610/20BP and 600LP. All detectors were set to the logarithmic amplification with the following voltages, 500, 240, 596, and 489 and threshold was set at 200 for both FSC and SSC. For each sample, 50 000 events were acquired using the BD FACSDiva software 6.1.3. The Hierarchical gating strategy was used to determine double positive population of bacterial cells (stained with both SYTO9 and PI) where gate P1 is the total population of FSC and SSC gated events, as determined from bacteria alone control and then applied to all other samples. P2 is the population of events stained by SYTO9 and P3 is the population stained with both SYTO9 and PI. Neutrophils and mid-log bacteria controls do not contribute any autofluorescence or PI-stained events when stained with either or both of the SYTO9/PI dyes. Values displayed in each density plot represent the percentage of 50 000 cells (N value) in each quadrant gate and each experiment was performed with at least 5 times.
DNase, PTase or Mg2+ treatment of NETs
During the coincubation experiments of bacteria and PMA-activated neutrophils, exogenous Mg2+ was added at a final concentration of 5 mM MgSO4. For the enzyme treatments, deoxyribonuclease (DNase I, VWR) was added at a final concentration of 430 kU/well and calf intestinal alkaline phosphatase (PTase, Invitrogen) at a final concentration of 16.6 U/well. Maximum enzyme amounts were added to bacterial-neutrophil mixtures that had no effect on bacterial viability and without the addition of enzyme buffers. The killing experiments were incubated for up to 4 hours in the 5% CO2 incubator at 37°C. All % survival values shown are the mean from at least three individual replicates and each experiment was performed at least 3 times.
Extracellular DNA and histone killing assays
P. aeruginosa was grown to mid-log in LB medium (OD600 = 0.2–0.4), washed and resuspended in 10 mM Tris buffer (pH 7.4; 1.0 × 107 CFU/well). Cells were incubated with fish sperm DNA (0.125%, w/v; USB) or with DNA that had been pretreated with exogenous DNase I (150 kU/well), PTase (50 U/well) or 5 mM MgSO4. DNA was pretreated for up to 3 hrs at 37°C in order to neutralize the antimicrobial activity. To determine if DNA killing required direct cell contact, 2% w/v fish sperm DNA (USB) was resuspended in 10 mM Tris pH 7.4 was placed in sealed dialysis membranes (MW cutoff 3500 Da) and allowed to dialyze into 10 mM Tris pH 7.4 for 4 hours, exchanging the buffer every hour. Cells from mid-log P. aeruginosa PAO1 cultures (1 × 107 CFU) were washed into 10 mM Tris pH 7.4 and coincubated directly with 1% or 0.125% dialyzed DNA (final concentration), with 1% eDNA maintained inside dialysis tubing, or 10 mM Tris pH 7.4 alone as a negative control. For histone killing experiments, 1 × 107 CFU mid-log growth phase P. aeruginosa PAO1, E. coli and S. aureus were washed into 10 mM Tris pH 7.4 and subsequently coincubated directly with 1.5 µg/mL calf thymus histones (Roche). Killing experiments were performed at RT in 96-well microplates and bacterial survival was assessed by colony counts (CFU/ml) every hour. All survival values shown are the mean from 4–8 individual replicates and each experiment was performed at least 3 times. Differences in bacterial survival were statistically analyzed by two-tailed student t-test.
Outer membrane damage visualization
PAO1::OM-lipoChFP was used as an indicator for outer membrane damage. This strain of PAO1 expresses a synthetic Cherry fluorescent lipoprotein (CSFPOmlA-ChFP) anchored to the outer membrane encoded on plasmid pCHAP6656 [38]. PAO1::OM-lipoChFP was exposed to a lethal concentration of 2% w/v (20 mg/ml) extracellular sperm DNA (USB) or 2 mM EDTA and red fluorescence of untreated and DNA killed cells were monitored as described above. Fluorescent outer membrane vesicles (OMVs) were counted in 6 fields of view by ImageJ quantification using a manually controlled threshold cutoff.
DNA, histone, and CAP-mediated induction of PA4774::lux
Overnight cultures were grown in LB medium, diluted 1/100 (approximately 1 × 107 CFU) into 100 µl of HBSS medium lacking cations (Life Technologies) in 96-well black plates with a transparent bottom (Thermo Scientific) and overlaid with 75 µl of mineral oil (Sigma Aldrich) to prevent evaporation. Microplate planktonic cultures were incubated at 37°C in a Wallac Victor3 luminescence plate reader (Perkin-Elmer) and optical density (growth, OD600) and luminescence (gene expression, CPS) readings were taken every 20 minutes in the presence of 0.2% salmon sperm DNA, 0.125 µg/mL polymyxin B and colistin, and 0.1 µg/mL calf thymus histones (Roche). Mean gene expression was derived from triplicate samples at 180 minutes after initial dilution and error bars represent the standard deviation from 4 individual replicates. Differences were statistically assessed by two-tailed student t-test.
Deoxyribonuclease assay
Overnight cultures of S. aureus and E. coli were grown in BHI medium and P. aeruginosa in BM2 medium, normalized to an OD600 = 1 and supernatants were collected by centrifugation at 8000 rpm for 3 minutes. 15 µL of supernatant was incubated with 5 µg of P. aeruginosa genomic DNA for 1 h at 37°C. Pseudomonas aeruginosa genomic DNA was purified using the Wizard Genomic DNA purification kit (Promega). DNA degradation was visualized on red safe (FroggaBio) stained 1% agarose gels. To test whether exposure to NETs induced DNase production, supernatants from S. aureus, E. coli and P. aeruginosa incubated in HBSS lacking cations with 106 PMA-stimulated human neutrophils (MOI 10 : 1, same method as described in the NET killing experiments section) were collected by centrifugation at 8000 rpm for 3 minutes. 100 µL of the supernatants were then coincubated at 37°C with 5 µg salmon sperm DNA stained with 2.5 µM Sytox green. 90 kU of DNase I was included as a positive control. Reactions were placed in 96-well black plates with a transparent bottom and Sytox green fluorescence quantified after 1 hour in a Wallac Victor3 luminescence plate reader.
Neutrophil elastase assay
To determine whether phosphatase treatment or the presence of excess Mg2+ altered NET-bound protein function, 2 × 105 human neutrophils were seeded in 96-well black plates with a transparent bottom and induced with 100 nM PMA. Immediately after PMA addition, 50 units of phosphatase (CIAP) and 5 mM MgSO4 were added to wells. The plate was then placed in cell culture conditions for 2 hours (37°C, 5% CO2). 300 µM elastase substrate I was added to all wells (Calbiochem), which were subsequently overlaid with 75 µL of mineral oil. The plate was then placed in a Wallac Victor3 luminescence plate reader at 37°C. Neutrophil elastase activity was monitored by measuring absorbance at OD410 nm every 20 minutes for 8 hours.
Statistical analysis
Statistical analysis was performed using GraphPad Prism v4.0 software. One-way ANOVA with Bonferroni posts tests and two-tailed students t-tests were used to calculate significant differences for plate counts, luminescence and flow cytometry analyses. Significant differences refer to P< 0.05 or less, or as otherwise denoted.
Ethics statement
Human neutrophils were isolated from human blood samples with ethical approval by the University of Calgary Research Ethics Committee (Ethics ID# 23187), where all subjects provided written informed consent. All animal protocols were approved by the animal care committee of the University of Calgary under the protocol number AC12–0222. All protocols used were in accordance with the guidelines drafted by the University of Calgary Animal Care Committee and the Canadian Council on the Use of Laboratory Animals.
Supporting Information
Zdroje
1. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, et al. (2004) Neutrophil extracellular traps kill bacteria. Science 303(5663): 1532–1535. doi: 10.1126/science.1092385 15001782
2. Brinkmann V, Zychlinsky A. (2012) Neutrophil extracellular traps: Is immunity the second function of chromatin? J Cell Biol 198(5): 773–783. doi: 10.1083/jcb.201203170 22945932
3. Urban CF, Reichard U, Brinkmann V, Zychlinsky A. (2006) Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol 8(4): 668–676. doi: 10.1111/j.1462-5822.2005.00659.x 16548892
4. Clark SR, Ma AC, Tavener SA, McDonald B, Goodarzi Z, et al. (2007) Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med 13(4): 463–469. doi: 10.1038/nm1565 17384648
5. Lu T, Kobayashi SD, Quinn MT, Deleo FR. (2012) A NET outcome. Front Immunol 3 : 365. doi: 10.3389/fimmu.2012.00365 23227026
6. Menegazzi R, Decleva E, Dri P. (2012) Killing by neutrophil extracellular traps: Fact or folklore? Blood 119(5): 1214–1216. doi: 10.1182/blood-2011-07-364604 22210873
7. Yipp BG, Kubes P. (2013) NETosis: How vital is it? Blood 122 (16): 2784–2794. doi: 10.1182/blood-2013-04-457671 24009232
8. Lappann M, Danhof S, Guenther F, Olivares-Florez S, Mordhorst IL, et al. (2013) In vitro resistance mechanisms of Neisseria meningitidis against neutrophil extracellular traps. Mol Microbiol 89 : 433. doi: 10.1111/mmi.12288 23750848
9. Achouiti A, Vogl T, Urban CF, Rohm M, Hommes TJ, et al. (2012) Myeloid-related protein-14 contributes to protective immunity in Gram-negative pneumonia derived sepsis. PLoS Pathog 8(10): e1002987. doi: 10.1371/journal.ppat.1002987 23133376
10. Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, et al. (2009) Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 5(10): e1000639. doi: 10.1371/journal.ppat.1000639 19876394
11. Parker H, Albrett AM, Kettle AJ, Winterbourn CC. (2012) Myeloperoxidase associated with neutrophil extracellular traps is active and mediates bacterial killing in the presence of hydrogen peroxide. J Leukoc Biol 91(3): 369–376. doi: 10.1189/jlb.0711387 22131345
12. Richards RC, O’Neil DB, Thibault P, Ewart KV. (2001) Histone H1: An antimicrobial protein of atlantic salmon (salmo salar). Biochem Biophys Res Commun 284(3): 549–555. doi: 10.1006/bbrc.2001.5020 11396934
13. Parseghian MH, Luhrs KA. (2006) Beyond the walls of the nucleus: The role of histones in cellular signaling and innate immunity. Biochem Cell Biol 84(4): 589–604. doi: 10.1139/o06-082 16936831
14. Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, et al. (1995) Early pulmonary inflammation in infants with cystic fibrosis. Am J Respir Crit Care Med 151(4): 1075–1082. doi: 10.1164/ajrccm.151.4.7697234 7697234
15. Konstan MW, Hilliard KA, Norvell TM, Berger M. (1994) Bronchoalveolar lavage findings in cystic fibrosis patients with stable, clinically mild lung disease suggest ongoing infection and inflammation. Am J Respir Crit Care Med 150(2): 448–454. doi: 10.1164/ajrccm.150.2.8049828 8049828
16. Manzenreiter R, Kienberger F, Marcos V, Schilcher K, Krautgartner WD, et al. (2012) Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J Cyst Fibros 11(2): 84–92. doi: 10.1016/j.jcf.2011.09.008 21996135
17. Papayannopoulos V, Staab D, Zychlinsky A. (2011) Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PLoS One 6(12): e28526. doi: 10.1371/journal.pone.0028526 22174830
18. Young RL, Malcolm KC, Kret JE, Caceres SM, Poch KR, et al. (2011) Neutrophil extracellular trap (NET)-mediated killing of Pseudomonas aeruginosa: Evidence of acquired resistance within the CF airway, independent of CFTR. PLoS One 6(9): e23637. doi: 10.1371/journal.pone.0023637 21909403
19. Wartha F, Beiter K, Albiger B, Fernebro J, Zychlinsky A, et al. (2007) Capsule and D-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell Microbiol 9(5): 1162–1171. doi: 10.1111/j.1462-5822.2006.00857.x 17217430
20. Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, et al. (2006) An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol 16(4): 401–407. doi: 10.1016/j.cub.2006.01.056 16488875
21. Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, et al. (2006) DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol 16(4): 396–400. doi: 10.1016/j.cub.2005.12.039 16488874
22. Seper A, Hosseinzade A, Gorkiewicz G, Lichtenegger S, Roier S, et al. (2013) Vibrio cholerae evades neutrophil extracellular traps by the activity of two extracellular nucleases. PLoS Pathog 9: e1003614. doi: 10.1371/journal.ppat.1003614 24039581
23. Mulcahy H, Charron-Mazenod L, Lewenza S. (2008) Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4(11): e1000213. doi: 10.1371/journal.ppat.1000213 19023416
24. Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. (2012) Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J Bacteriol 194(4): 813–826. doi: 10.1128/JB.05230-11 22155771
25. Johnson L, Horsman SR, Charron-Mazenod L, Turnbull AL, Mulcahy H, et al. (2013) Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol 13(1): 115. doi: 10.1186/1471-2180-13-115 23705831
26. Yipp BG, Petri B, Salina D, Jenne CN, Scott BN, et al. (2012) Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med 18(9): 1386–1393. doi: 10.1038/nm.2847 22922410
27. Davidson CJ, Narang A, Surette MG. (2010) Integration of transcriptional inputs at promoters of the arabinose catabolic pathway. BMC Syst Biol 4 : 75–0509–4–75. doi: 10.1186/1752-0509-4-75
28. Berney M, Hammes F, Bosshard F, Weilenmann HU, Egli T. (2007) Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl Environ Microbiol 73(10): 3283–3290. doi: 10.1128/AEM.02750-06 17384309
29. McPhee JB, Lewenza S, Hancock RE. (2003) Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50(1): 205–217. doi: 10.1046/j.1365-2958.2003.03673.x 14507375
30. Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, et al. (2005) Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: A tool for identifying differentially regulated genes. Genome Res 15(4): 583–589. doi: 10.1101/gr.3513905 15805499
31. Lewenza S. (2013) Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front Microbiol 4 : 21. doi: 10.3389/fmicb.2013.00021 23419933
32. Virta M, Lineri S, Kankaanpaa P, Karp M, Peltonen K, et al. (1998) Determination of complement-mediated killing of bacteria by viability staining and bioluminescence. Appl Environ Microbiol 64 : 512–519.
33. Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. (2010) Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol 191 : 677–691. doi: 10.1083/jcb.201006052 20974816
34. Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, et al. (2011) Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10(2): 158–164. doi: 10.1016/j.chom.2011.07.004 21843872
35. Pilsczek FH, Salina D, Poon KK, Fahey C, Yipp BG, et al. (2010) A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol 185(12): 7413–7425. doi: 10.4049/jimmunol.1000675 21098229
36. Fritzler M, Ryan P, Kinsella TD. (1982) Clinical features of systemic lupus erythematosus patients with antihistone antibodies. J Rheumatol 9(1): 46–51. 7086779
37. Tan EM, Fritzler MJ, McDougal JS, McDuffie FC, Nakamura RM, et al. (1982) Reference sera for antinuclear antibodies. I. antibodies to native DNA, sm, nuclear RNP, and SS-B/la. Arthritis Rheum 25(8): 1003–1005. doi: 10.1002/art.1780250814 6214260
38. Lewenza S, Mhlanga MM, Pugsley AP. (2008) Novel inner membrane retention signals in Pseudomonas aeruginosa lipoproteins. J Bacteriol 190(18):6119–6125. doi: 10.1128/JB.00603-08 18641140
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and AdultsČlánek The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and HostČlánek Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for EntryČlánek Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 1- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Importance of Pathogen Load
- Implication of Gut Microbiota in Nonalcoholic Fatty Liver Disease
- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- Helminth-Induced Immune Regulation: Implications for Immune Responses to Tuberculosis
- The M3 Muscarinic Receptor Is Required for Optimal Adaptive Immunity to Helminth and Bacterial Infection
- An Iron-Mimicking, Trojan Horse-Entering Fungi—Has the Time Come for Molecular Imaging of Fungal Infections?
- Modulates the Unfolded Protein Response in during Infection
- Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and Adults
- Identification of HNRNPK as Regulator of Hepatitis C Virus Particle Production
- Parasite Biomass-Related Inflammation, Endothelial Activation, Microvascular Dysfunction and Disease Severity in Vivax Malaria
- : Trypanosomatids Adapted to Plant Environments
- Early Virus-Host Interactions Dictate the Course of a Persistent Infection
- TLR3 Signaling in Macrophages Is Indispensable for the Protective Immunity of Invariant Natural Killer T Cells against Enterovirus 71 Infection
- The Epstein-Barr Virus Encoded BART miRNAs Potentiate Tumor Growth
- Macrophage-Derived Human Resistin Is Induced in Multiple Helminth Infections and Promotes Inflammatory Monocytes and Increased Parasite Burden
- Dissemination of a Highly Virulent Pathogen: Tracking The Early Events That Define Infection
- Variability in Tuberculosis Granuloma T Cell Responses Exists, but a Balance of Pro- and Anti-inflammatory Cytokines Is Associated with Sterilization
- The Shear Stress of Host Cell Invasion: Exploring the Role of Biomolecular Complexes
- The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and Host
- Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for Entry
- Secreted Herpes Simplex Virus-2 Glycoprotein G Modifies NGF-TrkA Signaling to Attract Free Nerve Endings to the Site of Infection
- Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
- A New Family of Secreted Toxins in Pathogenic Neisseria Species
- A Human Type 5 Adenovirus-Based Therapeutic Vaccine Re-programs Immune Response and Reverses Chronic Cardiomyopathy
- Regulation of Oncogene Expression in T-DNA-Transformed Host Plant Cells
- GITR Intrinsically Sustains Early Type 1 and Late Follicular Helper CD4 T Cell Accumulation to Control a Chronic Viral Infection
- Cell Cycle-Independent Phospho-Regulation of Fkh2 during Hyphal Growth Regulates Pathogenesis
- Virus-Induced NETs – Critical Component of Host Defense or Pathogenic Mediator?
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Protective Efficacy of Centralized and Polyvalent Envelope Immunogens in an Attenuated Equine Lentivirus Vaccine
- Transmitted Virus Fitness and Host T Cell Responses Collectively Define Divergent Infection Outcomes in Two HIV-1 Recipients
- Systemic Expression of Kaposi Sarcoma Herpesvirus (KSHV) Vflip in Endothelial Cells Leads to a Profound Proinflammatory Phenotype and Myeloid Lineage Remodeling
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
- DNA Is an Antimicrobial Component of Neutrophil Extracellular Traps
- Uropathogenic Superinfection Enhances the Severity of Mouse Bladder Infection
- Well-Ordered Trimeric HIV-1 Subtype B and C Soluble Spike Mimetics Generated by Negative Selection Display Native-like Properties
- The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots
- Reprogramming of from Virulent to Persistent Mode Revealed by Complex RNA-seq Analysis
- Compartment-Specific and Sequential Role of MyD88 and CARD9 in Chemokine Induction and Innate Defense during Respiratory Fungal Infection
- Bacterial Flagella: Twist and Stick, or Dodge across the Kingdoms
- Elucidation of the RamA Regulon in Reveals a Role in LPS Regulation
- IL-1α Signaling Is Critical for Leukocyte Recruitment after Pulmonary Challenge
- Chronic Filarial Infection Provides Protection against Bacterial Sepsis by Functionally Reprogramming Macrophages
- Specificity and Dynamics of Effector and Memory CD8 T Cell Responses in Human Tick-Borne Encephalitis Virus Infection
- Promiscuous RNA Binding Ensures Effective Encapsidation of APOBEC3 Proteins by HIV-1
- Viral Activation of MK2-hsp27-p115RhoGEF-RhoA Signaling Axis Causes Cytoskeletal Rearrangements, P-body Disruption and ARE-mRNA Stabilization
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- : Trypanosomatids Adapted to Plant Environments
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



