-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
article has not abstract
Published in the journal: Infections in Humans and Animals: Pathophysiology, Detection, and Treatment. PLoS Pathog 11(1): e32767. doi:10.1371/journal.ppat.1004523
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004523Summary
article has not abstract
Introduction
The fungal genus Malassezia comprises lipid-dependent and lipophilic yeast species that are part of the normal skin microbiota [1]. The 14 species are classified in class Malasseziomycetes in the Ustilaginomycotina of Basidiomycota [2]. Malassezia species can be involved in skin disorders, such as pityriasis versicolor, seborrheic dermatitis, atopic eczema, and folliculitis, and occur at higher population densities on scalps with dandruff than on scalps without dandruff [3], [4]. Occasionally, invasive infections by Malassezia pachydermatis and lipid-dependent Malassezia spp. occur in neonates, most often in those who are receiving intravenous lipid supplementation, or in immunocompromised patients receiving parenteral nutrition via a catheter. Malassezia spp. have not yet been cultured from the environment, but metagenomics identified Malassezia phylotypes from terrestrial and marine habitats [5]. For instance, Malassezia ribosomal DNA (rDNA) has been reported from soil nematodes [6], sponges [7], and rocks [8]. Undeniably, much remains to be discovered about the spectrum of habitats exploited by Malassezia that would advance our knowledge on the ecological relationships between the Malassezia skin biotic community, their hosts, and the environment. The aim of this article is to review and discuss the literature available on the pathogenesis, detection, typing, and treatment of Malassezia infections in humans and animals.
Pathophysiology on Human Skin
The pathophysiology of Malassezia-caused or Malassezia-exacerbated skin conditions is largely unknown, owing to the complex interactions of this commensal with the skin, an organ that has been on the edge of extreme selection pressure during evolution. In healthy skin, Malassezia yeasts exploit essential nutrients for their growth without inflicting disease (Fig. 1). When this process is perturbed, Malassezia yeasts adapt by modifying the expression of enzymes involved in the acquisition of energy, such as lipases and phospholipases [9], [10], and at the same time synthesize an array of bioactive indoles that act through the aryl-hydrocarbon receptor (AhR), which is expressed on almost all cell types found in the epidermis [11].
Fig. 1. Model showing the putative interactions of Malassezia yeasts with the skin. 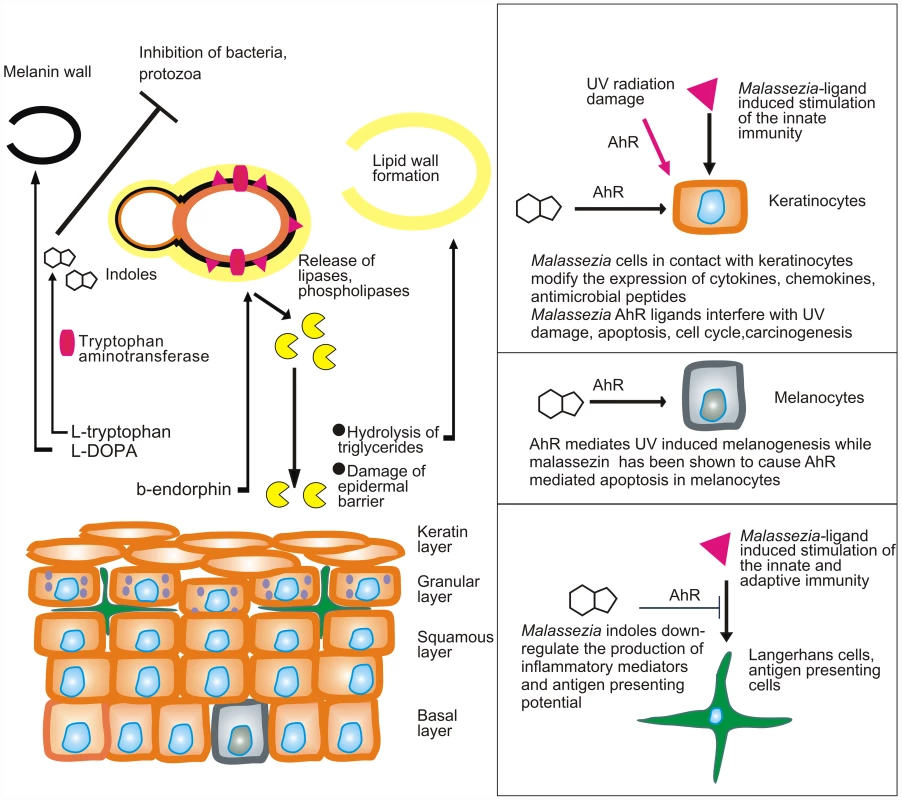
Malassezia yeasts take up nutrients as well as sebum lipids that are used to form the outer layer of the yeast or amino acids that are needed for the formation of melanin or the synthesis of AhR indolic ligands. In parallel they modify the expression of lipases and phospholipases under the action of β-endorphin. Cellular components (enzymes, proteins, glyceroglycolipids, and mannosyl fatty acids) are recognized by the innate and adaptive immune system and alter its function. AhR ligands potentially down-regulate immune stimulation, modify the function of epidermal cells, interfere with AhR-induced ultraviolet (UV) damage and melanogenesis, and probably inhibit antagonist microbes. A major challenge would be to comprehend the multifaceted interactions of Malassezia yeasts with the human skin during health and disease. These include (a) commensalism (healthy skin), as there is no strong evidence for a mutualistic or beneficial relationship of the Malassezia microbiome and the skin; (b) subtle alterations in the function of skin melanocytes, resulting in hypo - or hyperpigmented plaques with characteristic clinical absence of inflammation and mild alterations in the epidermal barrier function (pityriasis versicolor) (Fig. 1); (c) inflammation without generation of antibody-mediated immunity (seborrheic dermatitis and dandruff); (d) induction of specific immunity (atopic dermatitis); and (e) invasion and inflammation of the hair follicle (Malassezia folliculitis). Interestingly, the high lipase activity of M. globosa from folliculitis specimens during the summer months may be promoted by sweat components [12], such as sodium chloride and lactic acid, thus laying a framework for examining potential metabolome, structure and function relationships between M. globosa lipases and the human skin. In seborrheic dermatitis and dandruff, there is a difference in the quality of sebum lipids between healthy and diseased skin [13], while the expression and function of Malassezia lipases in addition to barrier function defects and individual susceptibility take part in the exacerbation of these conditions [14], [15]. Recently, culture and biopsy evidence supported an association of M. restricta and M. globosa with rare nipple hyperkeratotic lesions [16] in young women, who responded to a combination therapy of oral itraconazole and topical ketoconazole. This denotes that the metabolome of strains involved in rare presentations of skin diseases should be thoroughly investigated, clearly in conjunction with key host and environmental factors.
In that respect, at least two Malassezia yeast metabolic pathways, i.e., phospholipase production [17], [18] and indole pigment synthesis, have been associated with strains isolated from human and animal diseased skin. Malassezia produces potent indolic AhR ligands, such as indirubin and indolo [3,2-b] carbazole (ICZ) [19], which potentially modify the function of almost all cells found in the epidermis and express this receptor (Fig. 1). In view of the AhR participation in (a) carcinogenesis, (b) immune regulation, and (c) the mediation of ultraviolet radiation damage, a hypothesis on the potential contribution of Malassezia yeasts in skin carcinogenesis has been formulated [20].
Risk Factors for Malassezia Fungemia and Disseminated Disease
Patients under total parenteral nutrition (TPN) and immunocompromised patients with increased length of stay (LOS) in intensive care units are at risk for Malassezia infections. Risk for Malassezia infections is also high in very-low-birth-weight infants (<1500 g) and highest in premature infants [21]. The mechanism of transmission to the infant is vertical or horizontal [22]. After host exposure, the degree of prematurity, the corresponding skin condition, endotracheal intubation, central vascular access, diseases such as necrotizing enterocolitis or focal bowel perforation, and abdominal surgery contribute to colonization. Colonization is further enhanced by the pathogen's virulence factors, including adherence properties that favour colonization and proliferation followed by biofilm formation in central vascular catheters [23], [24]. These, in conjunction with iatrogenic factors, comprising invasive treatments and use of broad-spectrum antibiotics, parenteral nutrition, and administration of postnatal steroids and gastric acid inhibitors, contribute to the infection processes [23]. Compromised or immature host immunity, delayed diagnosis followed by persistent Malassezia fungemia and subsequent delayed vascular catheter removal, tissue or valve injury, insufficient antifungal dosing, or coinfection may lead to dissemination and occasionally result in poor prognosis.
Risk Factors for Otitis and Dermatitis in Animals
M. pachydermatis, normally present on the skin and in the ear canal of dogs and cats, frequently causes dermatitis and otitis in mammals. However, the pathogenic role of Malassezia in the occurrence of lesions seems to be related to the host immune system as well as to yeast virulence factors [25], [26]. Particular conditions, such as atopic or seborrheic dermatitis, parasitic infestation, diabetes mellitus in dogs, feline immunodeficiency virus, and feline leukaemia virus infections, and long-term antibiotic use associated with glucocorticoid treatment may predispose to Malassezia overgrowth, usually leading to development of lesions [27]. Additionally, lesions might appear as a consequence of hypersensitivity reaction to yeast allergens or might be prevented by active stimulation of the reticuloendothelial system, as previously shown in dogs infected with Leishmania spp. [25].
The zymogen proenzyme of the yeast cell wall may activate the complement system, instigating damage to keratinocyte integrity, and lead to epidermal spongiosis, inflammation, and pruritus. Additionally, the yeast produces esterase, lipase, phosphatase acid, lipoxygenase, protease, and phospholipase enzymes that are recognized as virulence factors [17], [28].
Further studies demonstrated that the expression of phospholipase in M. pachydermatis is modified by the endogenous opioid peptide β-endorphin [29], and this is mediated from mu-opiod receptors that are present on the cell wall of this species [30]. Thus, these receptors seem to impact the phenotype (commensal or pathogenic) of this species under the action of appropriate agonists (β-endorphin) or antagonists (naloxon) [30]. The pathogenic role of Malassezia yeasts seems to be related to changes in the chemical or immunological mechanisms of the skin, which may modify the composition of the Malassezia cell wall. The recently elucidated polysaccharide organization of the M. restricta cell wall showed that this is unique among the fungi with an average content of 5% chitin, 20% chitosan, 5% β-(1–3)-glucan, and 70% β-(1–6)-glucan that form a large alkali-insoluble complex [31].
Detection of Malassezia Infections in Humans and Animals
Isolation and enumeration of Malassezia cells from clinical specimens remains a challenge because of their lipid dependency. Since the clinical features, laboratory markers, and strategies for patient management do not differ between Candida and Malassezia fungemia, a more accurate etiological diagnosis is needed in high-risk patients by employing lipid-supplemented culture media in the current mycological routine [21], [32]. For septicaemia, contemporary paediatric aerobic lysis and centrifugation bottles supporting the growth of this yeast are recommended [32], followed by subculturing on lipid-supplemented media, such as modified Dixon or Leeming and Notman agars. Use of Sabouraud dextrose agar with the addition of a few drops of sterile olive oil does not support growth of all Malassezia spp. [33].
In contemporary clinical diagnostics, negative blood culture results obtained by the Candida QuickFISH BC platform (AdvanDx, Massachusetts, United States) for common yeasts and presence of the typically flask-like yeast cells in the gram-stained blood culture smear suggest possible infection by Malassezia spp., thus prompting employment of appropriate management and control strategies. Commercial molecular assays promise rapid and reliable detection of bloodstream and invasive Malassezia infections. Broad-spectrum real-time PCR platforms may prove useful tools for direct detection of Malassezia spp. in clinical specimens. The SeptiTest assay (Molzym, Bremen, Germany), engaging PCR followed by amplicon sequencing, can detect and discriminate M. furfur DNA in spiked clinical specimens with an analytical sensitivity corresponding to 2.5–3.5 M. furfur genomes per sample (A. Velegraki, unpublished data), as estimated based on the pulsed-field gel electrophoresis (PFGE)–generated M. furfur genome size range of 8.5–14 Mb [34]. Nonetheless, this assay needs validation for clinical use.
Malassezia species colonize a wide range of animals but may also cause disease to them [35]–[37]. Malassezia dermatitis is suspected in animals with inflammatory skin diseases characterized by erythematous or greasy lesions, especially in the intertriginous areas [36], [37]. As in humans, techniques for direct microscopy include the impression of cotton swab samples on glass slides and/or adhesive tape strips. Cultures are performed by inoculating specimens collected by cotton swabs or directly by contact plates containing lipid-supplemented media [26], [27]. Microscopy of swab specimens is useful for diagnosing animal and human dermatitis. The presence of ten or more yeast cells in five fields at 40× magnification from ear specimens of dogs indicate Malassezia otitis, whereas the presence of five cells from skin specimens suggests dermatitis [27]. Cultures are required only when direct microscopy is negative in animals with suspected infections. In this case, more than 70 colony-forming units per sample might be indicative of infection when the sample was collected by swabbing an area of 25 cm2 [27].
Identification and Genotyping
Identification of Malassezia isolates can to some extent be achieved by microbiological and physiological assays [32], [33], [38]. However, molecular diagnostic methods are preferred for both strain identification and typing. These may comprise PCR–restriction fragment length polymorphism (RFLP) analysis of the internal transcribed spacer 2 (ITS2) region of rDNA, sequence analysis of the ITS 1+2 regions (including the 5.8S rRNA gene) of rDNA, the 5′ end of the large-subunit (LSU or 26S) rDNA, and the β-tubulin gene, and terminal fragment length polymorphism analysis (tFLP) [4], [23], [26], [28], [39]–[41]. Recently, matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (MS) has been used to identify Malassezia isolates [42], [43]. Direct identification and quantification of Malassezia species from specimens obtained from skin by adhesive transparent dressings using multiplex real-time PCR [44] also provides reliable identification outcomes.
The unravelling of Malassezia biodiversity in 1996 [45] was followed by enthusiasm concerning the possible association of particular species with skin diseases; however, this was not confirmed by experimental data. Subsequent genotyping studies in conjunction with conventional identification methods displayed a certain degree of concordance regarding (a) geographic origin and specific Malassezia spp. genotypes, (b) the relationship between particular genotypes and certain skin conditions, and (c) the correlation of Malassezia spp. genotypes with host species. A number of worldwide epidemiological studies indicated that Malassezia species may have distinctive geographic associations. This is highlighted by higher frequencies of M. dermatis and M. japonica in East Asia, compared with their rarity in the rest of the world [35], [46], [47]. Recently, M. japonica was reported from lesional and non lesional psoriasis patients' skin in India, thus expanding the geographical distribution of this species to South Asia [48]. Multiple genotypes and subgenotypes occurring on skin showed distribution patterns related to the host geographical origin [46] and to host skin sites with a specific microenvironment [26], [28], [49]. Amplified fragment length polymorphism (AFLP) analysis confirmed geographical variability among M. furfur isolates from Southeast Europe compared with those from other European regions. Sequence analysis of the intergenic spacer (IGS1) distinguished specific M. globosa, M. restricta, and M. pachydermatis variants in seborrhoeic dermatitis, atopic eczema, and on healthy skin. Moreover, sequence analyses of the LSU rDNA showed distinct Malassezia spp. subtypes from different host species [23]. Sequence analysis of chitin synthase 2 (chs2) indicated that clinical isolates of M. pachydermatis from cats and dogs cluster in four distinct genotypes (i.e., A, B, C, and D) linked to skin lesions or otitis.
Multilocus sequence analysis that included the D1/D2 domains of LSU rDNA, the chs2 gene, and the ITS1 region grouped M. pachydermatis strains from skin of healthy dogs and from skin lesions in three main genotypes (A, B, and C) with eight ITS1 subtypes [28], [40]. Genotype B included isolates from dogs of European origin and appears to be present on healthy dog skin, without producing phospholipase. The A and C genotypes and their subtypes seem to be predominantly associated with skin lesions and showed high phospholipase activity [28]. Similarly, IGS1 subtypes 3C and 3D displaying high phospholipase activity are more frequently isolated from skin lesions of dogs with atopic dermatitis [48].
A range of skin microenvironmental factors, such as the bacterial microbiota present, pH, salts, immune responses, biochemistry, and physiology, may play a role in adherence and growth of Malassezia species, favouring distinct genotypes depending on the geographical area and/or the skin sites [26], [40]. In addition, the biochemical composition of the skin selecting genetic populations of Malassezia yeasts can indirectly affect their drug susceptibility [50]. Indeed, M. pachydermatis genotype B, growing on skin enriched with lipids (i.e., healthy skin) showed lower fluconazole and higher ketoconazole, voriconazole, and posaconazole susceptibility than M. pachydermatis genotypes A and C [50]. Finally, the finding of different Malassezia genotypes or subtypes on distinct skin sites suggests that Malassezia yeasts may have a sexual or parasexual reproductive phase that might enhance its virulence, thus influencing the association between Malassezia spp. genetics and disease [28], [49], [51]. This possibility is strongly supported by the finding of mating type loci in both M. globosa and M. sympodialis [9], [10], [52].
Susceptibility Testing and Treatment
Malassezia systemic infections require prompt identification of the pathogenic agent, removal of the central venous catheter and discontinuation of lipid supplementation, and treatment with liposomal amphotericin B [23], [53]. On the contrary, topical antifungal agents are adequate for the management of localized skin disease, while extensive disease requires administration of systemic itraconazole or fluconazole [23], [54]. This is also suitable for Malassezia folliculitis with concomitant modification of predisposing factors such as occlusion or systemic immunosuppresion. For the characteristic inflammatory conditions, seborrheic and atopic dermatitis, the addition of local anti-inflammatory therapy (i.e., corticosteroids or calcineurin inhibitors) is a prerequisite for rapid and effective control of exacerbations. One should always bear in mind that Malassezia yeasts are integral components of the skin microbiota and therefore the therapeutic target should be controlling the Malassezia population with subsequent long-term antifungal treatment, rather than eradicating it. Likewise, the need for extended (>2 months) azole treatment is required for suppression of symptoms in the Malassezia-triggered head and neck variant of atopic dermatitis [55]. Although the in vitro susceptibility testing is not yet standardized for Malassezia spp., the Clinical and Laboratory Standards Institute (CLSI) broth microdilution protocol was adapted by modifying media, time of incubation, and inocula, showing that itraconazole, ketoconazole, and posaconazole are the most effective drugs [50], [56].
Malassezia infections in animals are frequently treated with topical and/or systemic azole antifungal drugs [36]–[38], [57]–[59] usually combined with antibiotics and glucocorticoids in dogs with otitis externa [37], [38]. The emergence of azole-resistant M. pachydermatis [57], [58], as well as the increasing number of Malassezia infections in both humans and animals, emphasizes the importance of susceptibility tests as a guide for proper antifungal treatment [56].
Alternative therapeutic protocols, i.e., desensitization to Malassezia by immunotherapy or administration of inhibitors of yeast adherence factors, have been proposed to avoid repeated administration of antifungals and the occurrence of drug resistance phenomena [60]. Recently, the daily administration (150 µl, 2 mg/ml for 8 days) of a killer decapeptide, engineered from the variable region of a single-chain recombinant anti-idiotypic antibody, was shown to be a safe and effective treatment for Malassezia otitis externa in dogs [60].
Conclusions
Over the last few decades, advances in research and technologies have greatly contributed to elucidating the role of Malassezia species in human and animal skin diseases and in human bloodstream infections. Molecular and alternative approaches have provided insights into the identification, taxonomy, and epidemiology of Malassezia species. In particular, PCR-RFLP, random amplified polymorphic DNA (RAPD), AFLP, PCR-single strand conformation polymorphism (SSCP) analysis, multilocus sequence typing (MLST, e.g., of ITS, IGS, chs2, and RNA polymerase 1 and 2), and MALDI-TOF MS resulted in the accurate identification and genotyping of Malassezia strains from humans or animals, thus resolving questions related to the geographical distribution of the infection agents and the characterization of strains causing outbreaks [61], [62]. Nevertheless, these studies showed that the diversity within a single Malassezia species can more likely be attributed to a high degree of evolution driven by ecology, host adaptation, and pathogenicity. In particular, the pathogenic role of Malassezia yeasts seems to be related to changes in the normal physical, chemical, or immunological processes in the skin, which may enhance or down-regulate the molecular production of yeast virulence factors or antigens [23], [39]. The chemical composition of host epidermis seems to play a pivotal role in influencing the pathogenic or commensal phenotype of Malassezia yeasts by selecting different genetic populations with specific physiological requirements, different cell wall compositions, and different antifungal susceptibility profiles. In addition, molecular and physiological studies suggest the possibility of sexual or parasexual reproduction that might have a role in the process of adaptation of different Malassezia genotypes on different hosts or skin sites. As a consequence, antifungal therapy in Malassezia infections requires careful appraisal of drugs chosen, especially in cases of unresponsiveness to the treatment or recurrent infections. So far, restoring the epidermal-barrier function and avoiding immunoglobulin E (IgE) sensitization seems to be useful for the prevention and treatment of skin diseases complicated by Malassezia [63], even if antifungal therapy remains the main effective treatment in the near future. Alternative future treatments seem to be the use of selected cell-penetrating peptides that are harmless for mammalian cells but have antifungal activity, as shown for Malassezia otitis in dogs [60].
Undoubtedly, proteomic and genomic studies are needed in order to better understand the relationship between particular species/genotypes of Malassezia and the host at molecular and biochemical levels. Detailed biochemical analysis of the cell wall of the various species, as recently performed for M. restricta [31], and studies on the genotypic variants and their interaction with the immune system seem important here. Such studies might be the base for designing methods for the prevention, treatment, and control of infections caused by these fungi.
Zdroje
1. FindleyK, OhJ, YangJ, ConlanS, DemingC, et al. (2013) Topographic diversity of fungal and bacterial communities in human skin. Nature 498 : 367–370.
2. WangQ-M, TheelenB, GroenewaldM, BaiF-Y, BoekhoutT (2014) Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia 33 : 41–47.
3. McGinleyKJ, LeydenJJ, MarplesRR, KligmanAM (1975) Quantitative microbiology of the scalp in non-dandruff, dandruff, and seborrheic dermatitis. J Invest Dermatol 64 : 401–405.
4. GemmerCM, De AngelisYM, TheelenB, BoekhoutT, DawsonTLJr, Jr (2002) Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J Clin Microbiol 40 : 3350–3357.
5. AmendA (2014) From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLOS Pathog 10: e1004277.
6. RenkerC, AlpheiJ, BuscotF (2003) Soil nematodes associated with the mammal pathogenic fungal genus Malassezia (Basidiomycota: Ustilaginomycetes) in Central European forests. Biol Fertil Soils 37 : 70–72.
7. GaoZ, LiB, ZhengC, WangG (2008) Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl Environ Microbiol 74 : 6091–6101.
8. BjellandT, EkmanS (2005) Fungal diversity in rock beneath a crustose lichen as revealed by molecular markers. Microb Ecol 49 : 598–603.
9. XuJ, SaundersCW, HuP, GrantRA, BoekhoutT, et al. (2007) Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci USA 104 : 18730–18735.
10. Sun S, Hagen F, Xu J, Dawson T, Heitman J, et al.. (2013) Ecogenomics of human and animal Basidiomycetous yeast pathogens. In: The Ecological Genomics of Fungi (Ed. F. Martin). Wiley-Blackwell, Oxford, UK, pp. 215–242.
11. GaitanisG, MagiatisP, StathopoulouK, BassukasID, AlexopoulosEC, et al. (2008) AhR ligands, malassezin, and indolo [3,2-b] carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J Invest Dermatol 128 : 1620–1625.
12. AkazaN, AkamatsuH, TakeokaS, SasakiY, MizutaniH, et al. (2012) Malassezia globosa tends to grow actively in summer conditions more than other cutaneous Malassezia species. J Dermatol 39 : 613–616.
13. PassiS, PicardoM, MorroneA, De LucaC, IppolitoF (1991) Skin surface lipids in HIV sero-positive and HIV sero-negative patients affected with seborrheic dermatitis. J Dermatol Sci 2 : 84–91.
14. DawsonTLJr (2007) Malassezia globosa and restricta: breakthrough understanding of the etiology and treatment of dandruff and seborrheic dermatitis through whole-genome analysis. J Investig Dermatol Symp Proc 12 : 15–19.
15. Patiño-UzcáteguiA, AmadoY, Cepero de GarcíaM, ChavesD, TabimaJ, et al. (2011) Virulence gene expression in Malassezia spp. from individuals with seborrheic dermatitis. J Invest Dermatol 131 : 2134–2136.
16. LiC, RanY, SugitaT, ZhangE, XieZ, et al. (2014) Malassezia associated hyperkeratosis of the nipple in young females: Report of three cases. Ind J Derm Ven Lepr 80 : 78–80.
17. CafarchiaC, OtrantoD (2004) Association between phospholipase production by Malassezia pachydermatis and skin lesions. J Clin Microbiol 42 : 4868–4869.
18. VlachosC, GaitanisG, AlexopoulosEC, PapadopoulouC, BassukasID (2013) Phospholipase activity after β-endorphin exposure discriminates Malassezia strains isolated from healthy and seborrhoeic dermatitis skin. J Eur Acad Dermatol Venereol 27 : 1575–1578.
19. MagiatisP, PappasP, GaitanisG, MexiaN, MelliouE, et al. (2013) Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J Invest Dermatol 133 : 2023–2030.
20. GaitanisG, VelegrakiA, MagiatisP, PappasP, BassukasID (2011) Could Malassezia yeasts be implicated in skin carcinogenesis through the production of aryl-hydrocarbon receptor ligands? Med Hypotheses 77 : 47–51.
21. IattaR, CafarchiaC, CunaT, MontagnaO, LaforgiaN, et al. (2014) Bloodstream infections by Malassezia and Candida species in critical care patients. Med Mycol 52 : 264–269.
22. NagataR, NaganoH, OgishimaD, NakamuraY, HirumaM, et al. (2012) Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr Int 54 : 350–355.
23. GaitanisG, MagiatisP, HantschkeM, BassukasID, VelegrakiA (2012) The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25 : 106–141.
24. KanekoT, MurotaniM, OhkusuK, SugitaT, MakimuraK (2012) Genetic and biological features of catheter-associated Malassezia furfur from hospitalized adults. Med Mycol 50 : 74–80.
25. CafarchiaC, GalloS, DanesiP, CapelliG, ParadiesP, et al. (2008) Assessing the relationship between Malassezia and leishmaniasis in dogs with or without skin lesions. Acta Trop 107 : 25–29.
26. MachadoML, CafarchiaC, OtrantoD, FerreiraRR, BianchiSP, et al. (2010) Genetic variability and phospholipase production of Malassezia pachydermatis isolated from dogs with diverse grades of skin lesions. Med Mycol 48 : 889–892.
27. CafarchiaC, GalloS, RomitoD, CapelliG, ChermetteR, et al. (2005) Frequency, body distribution, and population size of Malassezia species in healthy dogs and in dogs with localized cutaneous lesions. J Vet Diagn Invest 17 : 316–322.
28. CafarchiaC, GasserRB, LatrofaMS, ParisiA, CampbellBE, et al. (2008) Genetic variants of Malassezia pachydermatis from canine skin: body distribution and phospholipase activity. FEMS Yeast Res 8 : 451–459.
29. CafarchiaC, Dell'AquilaME, CapelliG, MinoiaP, OtrantoD (2007) Role of beta-endorphin on phospholipase production in Malassezia pachydermatis in dogs: new insights into the pathogenesis of this yeast. Med Mycol 45 : 11–15.
30. CafarchiaC, Dell'AquilaME, TraversaD, AlbrizioM, GuaricciAC, et al. (2010) Expression of the micro-opioid receptor on Malassezia pachydermatis and its effect in modulating phospholipase production. Med Mycol 48 : 73–78.
31. StalhbergerTh, SimenelC, ClavaudC, EijsinkVGH, JourdainR, et al. (2014) Chemical organization of the cell wall polysaccharide core of Malassezia restricta. J Biol Chem 289 : 12647–12656.
32. BaronEJ, MillerJM, WeinsteinMP, RichterSS, GilliganPH, et al. (2013) A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin Infect Dis. 57: e22–e121.
33. Guého-Kellermann E, Boekhout T, Begerow D. (2010) Biodiversity phylogeny and ultrastructure In: Malassezia and the Skin: Science and Clinical Practice. Berlin: Springer. Boekhout T, Guého E, Mayser P, Velegraki A (Editors), pp 17–63.
34. BoekhoutT, KampM, GuéhoE (1998) Molecular typing of Malassezia species with PFGE and RAPD. Med Mycol 36 : 365–372.
35. Sugita T, Boekhout T, Velegraki A, Guillot J, Hadina S, et al.. (2010). Epidemiology of Malassezia-related skin diseases. In: Malassezia and the Skin: Science and Clinical Practice. Berlin: Springer. Boekhout T, Gueho E, Mayser P, Velegraki A (Editors), pp 65–119.
36. Bond R, Guillot J, Cabañes FJ (2010) Malassezia yeasts in animal disease. In: Malassezia and the Skin: Science and Clinical Practice. Berlin: Springer. Boekhout T, Gueho E, Mayser P, Velegraki A (Editors), pp 271–299.
37. BondR (2010) Superficial veterinary mycoses. Clin Dermatol 28 : 226–236.
38. NegreA, BensignorE, GuillotJ (2009) Evidence-based veterinary dermatology: a systematic review of interventions for Malassezia dermatitis in dogs. Vet Dermatol 20 : 1–12.
39. CabañesFJ (2014) Malassezia yeasts: how many species infect humans and animals? PLOS Pathogens 10: e1003892.
40. CafarchiaC, GasserRB, FigueredoLA, LatrofaMS, OtrantoD (2011a) Advances in the identification of Malassezia. Mol Cell Probes 25 : 1–7.
41. GuptaAK, BoekhoutT, TheelenB, SummerbellR, BatraR (2004) Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J Clin Microbiol 42 : 4253–4260.
42. KoleckaA, KhayhanK, ArabatzisM, VelegrakiA, KostrzewaM, et al. (2014) Efficient identification of Malassezia yeasts by matrix-assisted laser desorptionionization-time off light mass spectrometry (MALDI-TOF MS). Br J Dermatol 170 : 332–341.
43. YamamotoM, UmedaY, YoA, YamauraM, MakimuraK (2014) Utilization of matrix-assisted laser desorption and ionization time-of-flight mass spectrometry for identification of infantile seborrheic dermatitis-causing Malassezia and incidence of culture-based cutaneous Malassezia microbiota of 1-month-old infants. J Dermatol. 41 : 117–123.
44. SugitaT, TajimaM, TsubukuH, TsuboiR, NishikawaA (2006) Quantitative analysis of cutaneous Malassezia in atopic dermatitis patients using real-time PCR. Microbiol Immunol 50 : 549–552.
45. GuéhoE, MidgleyG, GuillotJ (1996) The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek 69 : 337–355.
46. GaitanisG, VelegrakiA, AlexopoulosEC, Kapsanaki-GotsiE, ZisovaL, et al. (2009b) Malassezia furfur fingerprints as possible markers for human phylogeography. ISME J 3 : 498–502.
47. GiusianoG, Sosa MdeL, RojasF, VanacoreST, MangiaterraM (2010) Prevalence of Malassezia species in pityriasis versicolor lesions in northeast Argentina. Rev Iberoam Micol 27 : 71–74.
48. RudramurthySM, HonnavarP, ChakrabartiA, DograS, PankajS, et al. (2014) Association of Malassezia species with psoriatic lesions. Mycoses 57 : 483–488.
49. KobayashiT, KanoR, NagataM, HasegawaA, KamataH (2011) Genotyping of Malassezia pachydermatis isolates from canine healthy skin and atopic dermatitis by internal spacer 1 (IGS1) region analysis. Vet Dermatol 22 : 401–405.
50. CafarchiaC, FigueredoLA, IattaR, ColaoV, MontagnaMT, OtrantoD (2012) In vitro evaluation of Malassezia pachydermatis susceptibility to azole compounds using E-test and CLSI microdilution methods. Med Mycol 50 : 795–801.
51. MidreuilF, GuillotJ, GuéhoE, RenaudF, MalliéM, et al. (1999) Genetic diversity in the yeast species Malassezia pachydermatis analysed by multilocus enzyme electrophoresis. Int J Syst Bacteriol 49 : 1287–1294.
52. GiotiA, NystedtB, LiW, XuJ, AnderssonA, et al. (2013) Genomics insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. MBio 4: e00572–e00612.
53. ArendrupMC, BoekhoutT, AkovaM, MeisJF, CornelyOA, et al. (2014) ESCMID/ECMM Joint Clinical Guideline for the Diagnosis and Management of Rare Invasive Yeast Infections. Clin Microbiol Infect 20 : 76–98.
54. HaldM, ArendrupMC, SvejgaardEL, LindskovR, FogedEK, et al. (2014) Evidence-based Danish guidelines for the treatment of Malassezia-related skin diseases. Acta Derm Venereol E-pub ahead of print. doi:–10.2340/00015555–1825
55. KaffenbergerBH, MathisJ, ZirwasMJ (2014) A retrospective descriptive study of oral azole antifungal agents in patients with patch test-negative head and neck predominant atopic dermatitis. J Am Acad Dermatol 71 : 480–483.
56. VelegrakiA, AlexopoulosEC, KritikouS, GaitanisG (2004) Use of fatty acid RPMI 1640 media for testing susceptibilities of eight Malassezia species to the new triazole posaconazole and to six established antifungal agents by a modified NCCLS M27-A2 microdilution method and Etest. J Clin Microbiol 42 : 3589–3593.
57. NijimaM, KanoR, NagataM, HasegawaA, KamataH (2011) An azole resistant isolate of Malassezia pachydermatis. Vet Microbiol 149 : 288–290.
58. Al-SweihN, AhmadS, JosephL, KhanS, KhanZ (2014) Malassezia pachydermatis fungemia in a preterm neonate resistant to fluconazole and flucytosine. Med Mycol Case Rep. 5 : 9–11.
59. CafarchiaC, FigueredoLA, IattaR, MontagnaMT, OtrantoD (2012) In vitro antifungal susceptibility of Malassezia pachydermatis from dogs with and without skin lesions. Vet Microbiol 155 : 395–398.
60. CafarchiaC, ImmediatoD, Di PaolaG, MaglianiW, CiociolaT, et al. (2014) In vitro and in vivo activity of a killer peptide against Malassezia pachydermatis causing otitis in dogs. Med Mycol 52 : 350–355.
61. GaitanisG, BassukasID, VelegrakiA (2009) The range of molecular methods for typing Malassezia. Curr Opin Infect Dis 22 : 119–125.
62. CastelláG, CoutinhoSD, CabañesFJ (2014) Phylogenetic relationships of Malassezia species based on multilocus sequence analysis. Med Mycol 52 : 99–105.
63. SaundersCW, ScheyniusA, HeitmanJ (2012) Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS Pathog 8: e1002701.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and AdultsČlánek The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and HostČlánek Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for EntryČlánek Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 1- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Importance of Pathogen Load
- Implication of Gut Microbiota in Nonalcoholic Fatty Liver Disease
- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- Helminth-Induced Immune Regulation: Implications for Immune Responses to Tuberculosis
- The M3 Muscarinic Receptor Is Required for Optimal Adaptive Immunity to Helminth and Bacterial Infection
- An Iron-Mimicking, Trojan Horse-Entering Fungi—Has the Time Come for Molecular Imaging of Fungal Infections?
- Modulates the Unfolded Protein Response in during Infection
- Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and Adults
- Identification of HNRNPK as Regulator of Hepatitis C Virus Particle Production
- Parasite Biomass-Related Inflammation, Endothelial Activation, Microvascular Dysfunction and Disease Severity in Vivax Malaria
- : Trypanosomatids Adapted to Plant Environments
- Early Virus-Host Interactions Dictate the Course of a Persistent Infection
- TLR3 Signaling in Macrophages Is Indispensable for the Protective Immunity of Invariant Natural Killer T Cells against Enterovirus 71 Infection
- The Epstein-Barr Virus Encoded BART miRNAs Potentiate Tumor Growth
- Macrophage-Derived Human Resistin Is Induced in Multiple Helminth Infections and Promotes Inflammatory Monocytes and Increased Parasite Burden
- Dissemination of a Highly Virulent Pathogen: Tracking The Early Events That Define Infection
- Variability in Tuberculosis Granuloma T Cell Responses Exists, but a Balance of Pro- and Anti-inflammatory Cytokines Is Associated with Sterilization
- The Shear Stress of Host Cell Invasion: Exploring the Role of Biomolecular Complexes
- The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and Host
- Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for Entry
- Secreted Herpes Simplex Virus-2 Glycoprotein G Modifies NGF-TrkA Signaling to Attract Free Nerve Endings to the Site of Infection
- Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
- A New Family of Secreted Toxins in Pathogenic Neisseria Species
- A Human Type 5 Adenovirus-Based Therapeutic Vaccine Re-programs Immune Response and Reverses Chronic Cardiomyopathy
- Regulation of Oncogene Expression in T-DNA-Transformed Host Plant Cells
- GITR Intrinsically Sustains Early Type 1 and Late Follicular Helper CD4 T Cell Accumulation to Control a Chronic Viral Infection
- Cell Cycle-Independent Phospho-Regulation of Fkh2 during Hyphal Growth Regulates Pathogenesis
- Virus-Induced NETs – Critical Component of Host Defense or Pathogenic Mediator?
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Protective Efficacy of Centralized and Polyvalent Envelope Immunogens in an Attenuated Equine Lentivirus Vaccine
- Transmitted Virus Fitness and Host T Cell Responses Collectively Define Divergent Infection Outcomes in Two HIV-1 Recipients
- Systemic Expression of Kaposi Sarcoma Herpesvirus (KSHV) Vflip in Endothelial Cells Leads to a Profound Proinflammatory Phenotype and Myeloid Lineage Remodeling
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
- DNA Is an Antimicrobial Component of Neutrophil Extracellular Traps
- Uropathogenic Superinfection Enhances the Severity of Mouse Bladder Infection
- Well-Ordered Trimeric HIV-1 Subtype B and C Soluble Spike Mimetics Generated by Negative Selection Display Native-like Properties
- The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots
- Reprogramming of from Virulent to Persistent Mode Revealed by Complex RNA-seq Analysis
- Compartment-Specific and Sequential Role of MyD88 and CARD9 in Chemokine Induction and Innate Defense during Respiratory Fungal Infection
- Bacterial Flagella: Twist and Stick, or Dodge across the Kingdoms
- Elucidation of the RamA Regulon in Reveals a Role in LPS Regulation
- IL-1α Signaling Is Critical for Leukocyte Recruitment after Pulmonary Challenge
- Chronic Filarial Infection Provides Protection against Bacterial Sepsis by Functionally Reprogramming Macrophages
- Specificity and Dynamics of Effector and Memory CD8 T Cell Responses in Human Tick-Borne Encephalitis Virus Infection
- Promiscuous RNA Binding Ensures Effective Encapsidation of APOBEC3 Proteins by HIV-1
- Viral Activation of MK2-hsp27-p115RhoGEF-RhoA Signaling Axis Causes Cytoskeletal Rearrangements, P-body Disruption and ARE-mRNA Stabilization
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- : Trypanosomatids Adapted to Plant Environments
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



