-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Protective Efficacy of Centralized and Polyvalent Envelope Immunogens in an Attenuated Equine Lentivirus Vaccine
Our best effort for containment of the global HIV epidemic is the development of a broadly protective vaccine. Current research has focused on vaccines that can generate a protective immune response against numerous strains of the virus. For this reason, vaccines with centralized HIV genes as immunogens, which merge HIV genetic information and potentially protect against multiple viral strains in a single inoculation, are an increasing area of interest to the field. Existing published studies have not evaluated centralized immunogens in the context of attenuated vaccines, which to date, have demonstrated the highest level of vaccine protection in lentiviral studies. Furthermore, centralized immunogen studies have also not included protective efficacy findings accomplished through challenge with highly pathogenic virus strains. In this study we not only examine the immunogenicity of these immunogens in an animal model, but we also for the first time evaluate the ability of centralized immunogens to induce protection against virulent virus challenge.
Published in the journal: Protective Efficacy of Centralized and Polyvalent Envelope Immunogens in an Attenuated Equine Lentivirus Vaccine. PLoS Pathog 11(1): e32767. doi:10.1371/journal.ppat.1004610
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004610Summary
Our best effort for containment of the global HIV epidemic is the development of a broadly protective vaccine. Current research has focused on vaccines that can generate a protective immune response against numerous strains of the virus. For this reason, vaccines with centralized HIV genes as immunogens, which merge HIV genetic information and potentially protect against multiple viral strains in a single inoculation, are an increasing area of interest to the field. Existing published studies have not evaluated centralized immunogens in the context of attenuated vaccines, which to date, have demonstrated the highest level of vaccine protection in lentiviral studies. Furthermore, centralized immunogen studies have also not included protective efficacy findings accomplished through challenge with highly pathogenic virus strains. In this study we not only examine the immunogenicity of these immunogens in an animal model, but we also for the first time evaluate the ability of centralized immunogens to induce protection against virulent virus challenge.
Introduction
The scientific community has aggressively sought after the development of a universal HIV vaccine that can prevail over the extraordinary levels of antigenic diversity in the fight against HIV and AIDS. The considerable extent of genomic variation found between isolates and within clades, and to a larger extent within the circulating recombinant forms, make for an effectual blockade to vaccine protection. Different strategies of vaccine composition and delivery have been proposed that are actively and widely being examined. A majority of these vaccines target the Env protein, as lentiviral antigenic variation is most pronounced in the viral Env proteins that serve as initial primary targets for host immune responses [1]–[5].
Centralized Env immunogens are one of the more promising contemporary approaches to overcoming HIV antigenic diversity [1], [6]. Centralized sequences attempt to minimize the genetic distance between vaccine proteins and the circulating isolates that pose a threat to public health. The centralized genes are generated through the computational determination of consensus genes (the most common amino acid at each position), ancestral genes (modelling ancestral states through phylogenetics), or center of the tree sequences (phylogenetic determination of a central isolates) [1], [4], [7], [8]. Centralized genes have been investigated as effective vaccine approaches in the HIV field both as DNA and/or protein immunogens [6], [9]–[19]. To date, however, the efficacy of centralized immunogens has not been fully explored in the context of an attenuated lentiviral vaccine model that could provide both immunogenicity data as well as protective efficacy data via virulent challenge in an animal model.
Equine infectious anemia virus (EIAV), a macrophage-tropic lentivirus, causes a persistent infection and chronic disease in equids [20]. Infection, transmitted via blood-feeding horse flies, occurs in three stages: acute, chronic and inapparent. Acute and chronic stages are defined by episodes of clinical disease that are triggered by waves of viremia, and distinguished by fever, anemia, thrombocytopenia, edema, and various wasting signs. By one year post-infection animals typically progress to life-long inapparent carriers, but continue to harbor steady-state levels of viral replication in monocyte-rich tissue reservoirs [20]–[22]. Stress or immune suppression of inapparent carriers can induce increases in viral replication and potentially a recrudescence of disease [20], [23]. Among virulent lentiviruses, EIAV is unique in that, despite aggressive virus replication and associated rapid antigenic variation, greater than 90% of infected animals progress from chronic disease to an inapparent carrier stage, by a strict immunologic control over virus replication [20]. The EIAV system hence serves as a unique animal model for the natural immunologic control of lentiviral replication and disease. Further, EIAV inapparent carriers have proven to be resistant to subsequent virus exposure to diverse viral strains, indicating the development of a high level of prophylactic immunity. Thus, the EIAV system provides a valuable model for identifying critical immune correlates of protection and ascertaining the potential for developing effective prophylactic lentivirus vaccines [24].
While the disease processes for EIAV and HIV have distinguishing dynamics, key similarities between the two virus systems make EIAV an extremely valuable tool and model for AIDS vaccine development [24]–[26]. EIAV and HIV are transmitted parenterally and share a macrophage/monocyte tropism [26], [27]. EIAV quasispecies also possess high levels of antigenic heterogeneity and their Env proteins share architectural characteristics such as extensive glycosylation and immune decoys [24], [25], [28], [29]. These features, all of which are critical elements associated with initial virus exposure, coupled to a very similar immune maturation process of the EIAV-infected equine to HIV-infected humans, are fundamental factors relevant to vaccine efficacy [24], [30].
We previously reported serial studies evaluating the efficacy of an attenuated EIAV proviral vaccine containing a mutation in the viral S2 accessory gene (EIAVD9) [31]–[33]. The results of these studies indicate that horses inoculated with the EIAVD9 viral vaccine were 100% protected from disease by virulent, albeit homologous, EIAV challenge. Thus, the EIAV system mirrors other animal lentivirus vaccine models, which consistently identify attenuated vaccines as producing the highest level of vaccine protection. [31]–[38]. Our latest EIAVD9 data demonstrated the effects of challenge virus Env sequence variation on vaccine protection [39]–[41]. We identified a significant, inverse, linear correlation between vaccine efficacy and increasing divergence of the challenge virus Env surface protein, gp90, compared to the vaccine virus gp90 protein. The study demonstrated that the 100% protection of immunized horses from disease after challenge by virus with a homologous gp90 (EV0), dropped to approximately 60% protection when a challenge virus gp90 was 6% divergent (EV6), and nose-dived to less than 50% protection against challenge with a gp90 that was 13% (EV13) divergent from the vaccine strain. Most recently, we demonstrated that the attenuated vaccine strain progressively evolved during the seven-month pre-challenge period and that the observed protection from disease was significantly associated with divergence from the original vaccine strain, not the overall diversity of the vaccine Env quasispecies present on the day of challenge (DOC) [39].
Despite numerous studies on the immunogenicity of centralized Env proteins, use of these noteworthy immunogens in an attenuated vaccine model, accompanied by virulent virus challenge, has yet to be reported. In the current study, we sought to directly build upon our current model and the series of described EIAVD9 vaccine studies. Our attenuated vaccine model, coupled with the well-characterized genomic and phylogenetic ancestry of the Env gene of our EIAV strain, enabled a thorough, unparalleled evaluation of centralized sequence vaccine efficacy not as readily modelled in other lentiviral systems. The presented studies evaluated multiple derivatives of centralized Env immunogens, both consensus and ancestral, in our proviral attenuated vaccine backbone. The studies were designed to first, develop and test a consensus immunogen for functionality through examination of replication and pathogenic potential in proviral backbones; second, compare the protective efficacy of the consensus immunogen in an attenuated backbone to attenuated strains containing our ancestral Env [34] as well as a polyvalent Env attenuated strain mixture; and third, to develop and utilize a more stringent challenge model in the form of an engineered quasispecies.
Results
Development of consensus Env
Consensus gene development of the EIAV Env protein focused on the gp90 region of the gene as genomic evolution and antigenic variation in the transmembrane (gp45) protein has been shown to be minimal among characterized longitudinal EIAV isolates [42], [43]. To engineer a consensus Env, the gp90 genes of approximately 90 naturally occurring isolates from an experimental infection [42], [44] were aligned. Virus isolates included the inoculum strain Env as well as isolates from all three stages of disease (acute, chronic, and inapparent). Isolates therefore included an ancestral strain (EV0) and its descendant strains that evolved innately between day zero and 1200 days post-infection (DPI). Consensus sequences were derived primarily from codon-aligned nucleotide sequences and secondarily from amino acid alignments. The consensus sequence from the nucleotide alignment was translated, compared to the consensus sequence from the amino acid alignment for congruence and the resolution of ambiguities, and termed ConEnv. To evaluate the veracity of the derived ConEnv sequence and discriminate it against other potential consensus Env proteins, additional consensus sequences were designed. Consensus Envs representing the individual febrile episodes (six) and inapparent stage isolates were generated from the isolate amino acid sequences and thereafter a consensus from those engineered sequences was constructed as well. This “consensus from all consensus” method is similar to creating consensus Envs from each HIV clade and subsequently creating a consensus of those clades. The ConEnv consensus sequence was then examined by phylogenetic comparison with these control consensus sequences, the Env sequences involved in ConEnv construction, and sequences targeted as partner Envs for vaccine and challenge strains of this study (Fig. 1). The “consensus from all consensus” sequence was phylogenetically closely related to the ancestral Env (EV0) emerging on the main ancestral root amongst the acute disease Env genes. ConEnv shared the same ancestral root as the EV6 strain, manifesting ancestrally between EV0 and EV6. Genetic distance calculations demonstrated the ConEnv gp90 sequence was 4%, 6%, and 11% divergent from the EV0, EV6, and EV13 strains, respectively. The majority of the disparate residues occurred within the designated variable regions, specifically V3 through V7 (Fig. 2). Hence, the ConEnv sequence was a strong consensus sequence representative of the 90 isolates, capturing epitopes broadly characteristic of the family of EIAV isolates.
Fig. 1. Phylogenetic analyses of the ConEnv, all consensus Envs, and the longitudinal isolates derived from experimental infection utilized in consensus gene development. 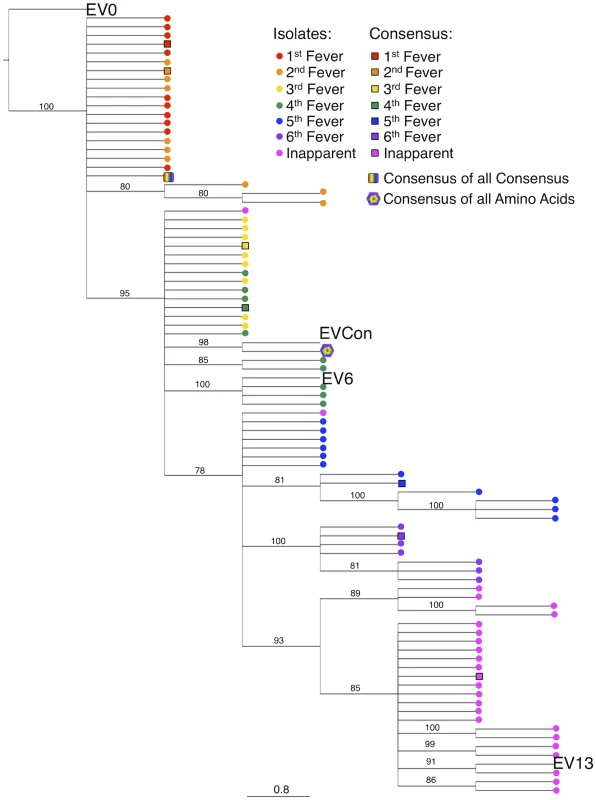
A phylogenetic tree of aligned plasma amino acid sequences (gp90 regions) was constructed by the neighbor joining method from Kimura corrected distances with the optimality criterion set to distance. The tree was rooted to the ancestral Env, EV0. Bootstrap values were determined over 1000 iterations and are indicated at the nodes of the branches. Branch lengths are proportional to the distance existing between the sequences. Fig. 2. Comparison of deduced amino acid gp90 variable region sequences from EIAV variant Envs (used in attenuated and challenge strain construction) and ConEnv. 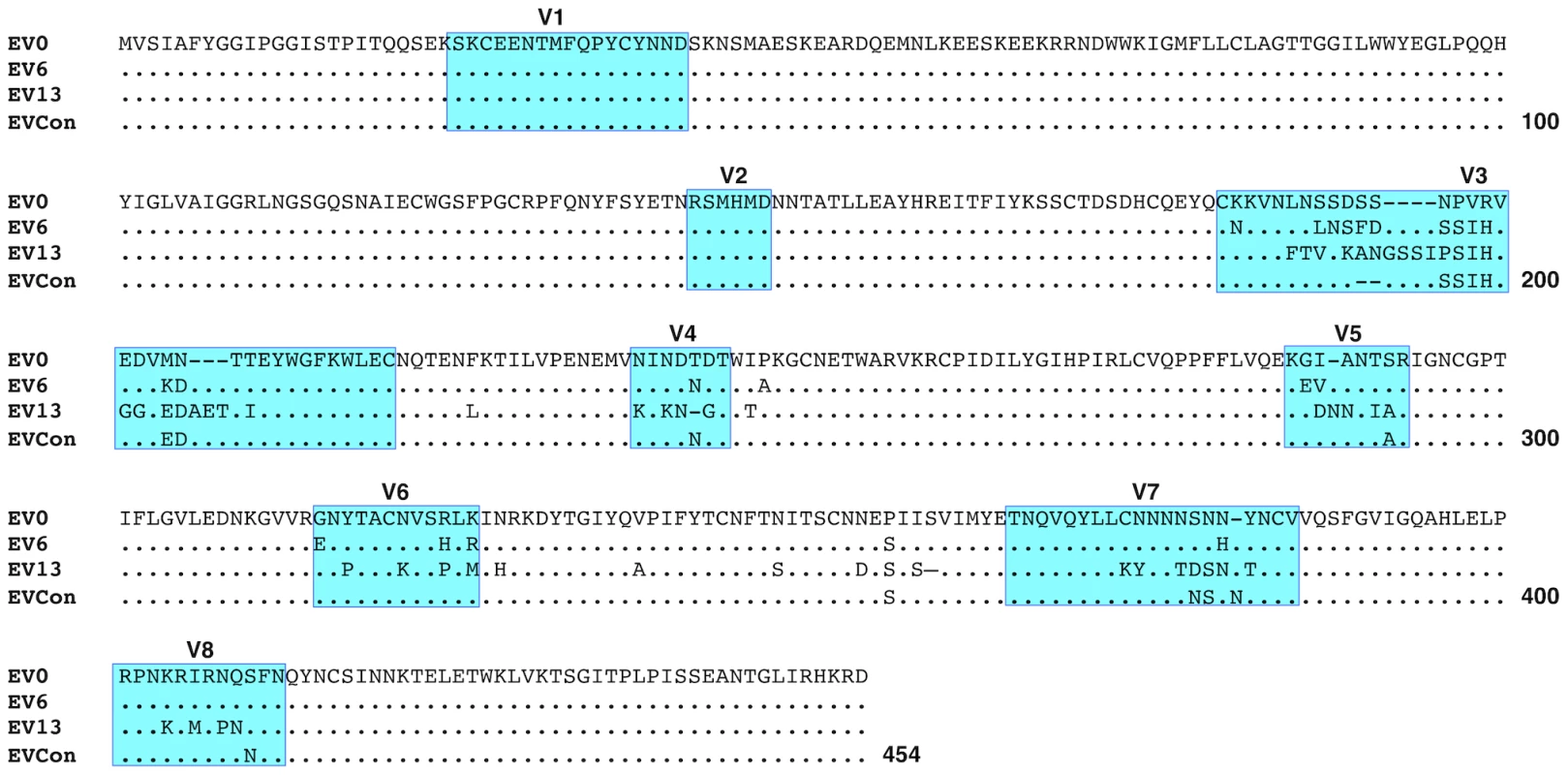
The region of env gene coding for the surface glycoprotein, gp90, of the Env variants, EV0, EV6, and EV13 (ref) was compared to the ConEnv sequence. Deduced amino acid sequences were aligned and compared to EV0 (ancestral Env that is analogous to the EIAVD9 Env). Only the amino acid residues different from EV0 are reported on the alignment. Dots (·) indicate residues identical to the EV0 sequence; dashes (-) indicate amino acid deletion; Previously described variable regions V1 through V8 are boxed and shaded in blue. Functional replication assessment and characterization of the consensus Env
To fully assess the competency of the ConEnv protein to function indistinguishably from a naturally occurring Env protein, ConEnv was evaluated in the context of both attenuated and pathogenic EIAV proviruses. Commercially synthesized ConEnv was cloned into the attenuated EIAVD9 backbone [31], [32] and the pathogenic EIAVUK3 backbone [45], with the resultant proviral strains termed ConD9, and EVCon, respectively. Attenuated and pathogenic proviral clones were sequenced to verify the ConEnv gene, and then transfected into equine dermal (ED) cells for production of infectious virus stocks. Virus stocks were titered and characterized for in vitro replication kinetics [46]–[48]. Both proviral strains, EVCon (pathogenic) and ConD9 (attenuated) demonstrated typical in vitro kinetics, emulating their parental and variant strain counterparts, and peaked in virus production at approximately ten DPI.
In vivo analysis of the proviral pathogenic and attenuated ConEnv strains, by experimental infections of equids, confirmed characteristic EIAV clinical and virological profiles of both pathogenic and attenuated infections (Fig. 3). Typical attenuated and avirulent replication properties were observed for the ConD9 strain. Low level, viral replication kinetics (averaging between 102–103 copies RNA/ml) which failed to progress to clinical disease over a 100 day observation period were observed. Conversely, pathogenesis and virulence, characterized by standard viremic replication kinetics (averaging between103–104 copies RNA/ml and peaking at 106 copies RNA/ml), including the induction of acute, and progression to chronic disease were observed with the EVCon strain [20], [43], [49]. Hence, for the first time, a synthetic consensus lentiviral Env was demonstrated to not only be fully functional in the context of replication competence in vitro and in vivo, but also capable of inducing traditional and virulent lentiviral disease.
Fig. 3. Clinical and virological profiles of ponies inoculated with the consensus Env in attenuated (ConD9) and pathogenic (EVCon) EIAV viral backbones. 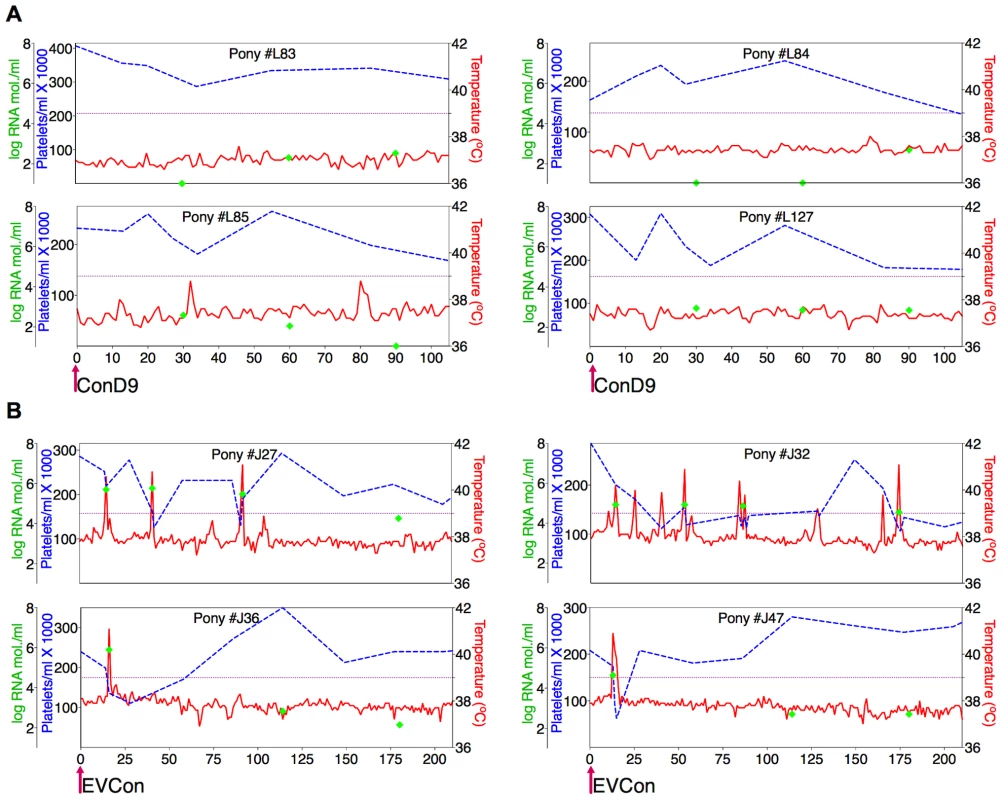
EIAV-naïve ponies (8 total) were inoculated intramuscularly with 1×103 TCID50 (pink arrow) of EIAV strains containing the consensus Env in either an (A) attenuated backbone, ConD9, or in (B) our pathogenic backbone, EVCon. Rectal temperature (red line, right Y axis) and platelet counts (blue dashed line, first left Y axis) were followed daily (X-axis). Quantification of the virus load (green diamond, second left Y axis) was performed on viral RNA extracted from plasma at periodic time points. Febrile episodes were defined by a combination of two-three features, including: rectal temperature above 39°C in conjunction with thrombocytopenia (platelet decrease of ≥70,000/µl of whole blood), EIAV viral load ≥105 as well as other clinical signs of EIA. The final pre-vaccine trial evaluation was an assessment of ConEnv's immunogenicity. Assays of antibody responses elicited by EVCon, EV0, EV6, and EV13 experimental infections, with variant and consensus viruses, indicated primarily distinct neutralization phenotypes for the individual variant Envs; each variant virus was neutralized by immune serum from homologous virus infections, but not from heterologous virus infections, except for marginal neutralization of the EVCon strain by the EV0 heterologous serum (Fig. 4). However, sera produced by the EVCon virus infections were capable of not only neutralizing its homologous strain, but also neutralized the EV0 and EV6 heterologous strains. Thus, these data demonstrate that the ConEnv was similar to EV0, EV6, and EV13 in replication and virulence properties [34], yet distinct in immune properties as a result of the defined Env sequence variation. Much like the HIV consensus Env recombinant proteins that have been reported [6], [15]–[18], this consensus Env, in the context of a fully functional virus, demonstrated immunogenicity induction of neutralizing antibodies with broader recognition of epitopes than that of the naturally occurring isolates from which the ConEnv was derived.
Fig. 4. Neutralization profiles of proviral variant strains with serum from animals experimentally infected with EV0, EV6, EV13, or EVCon. 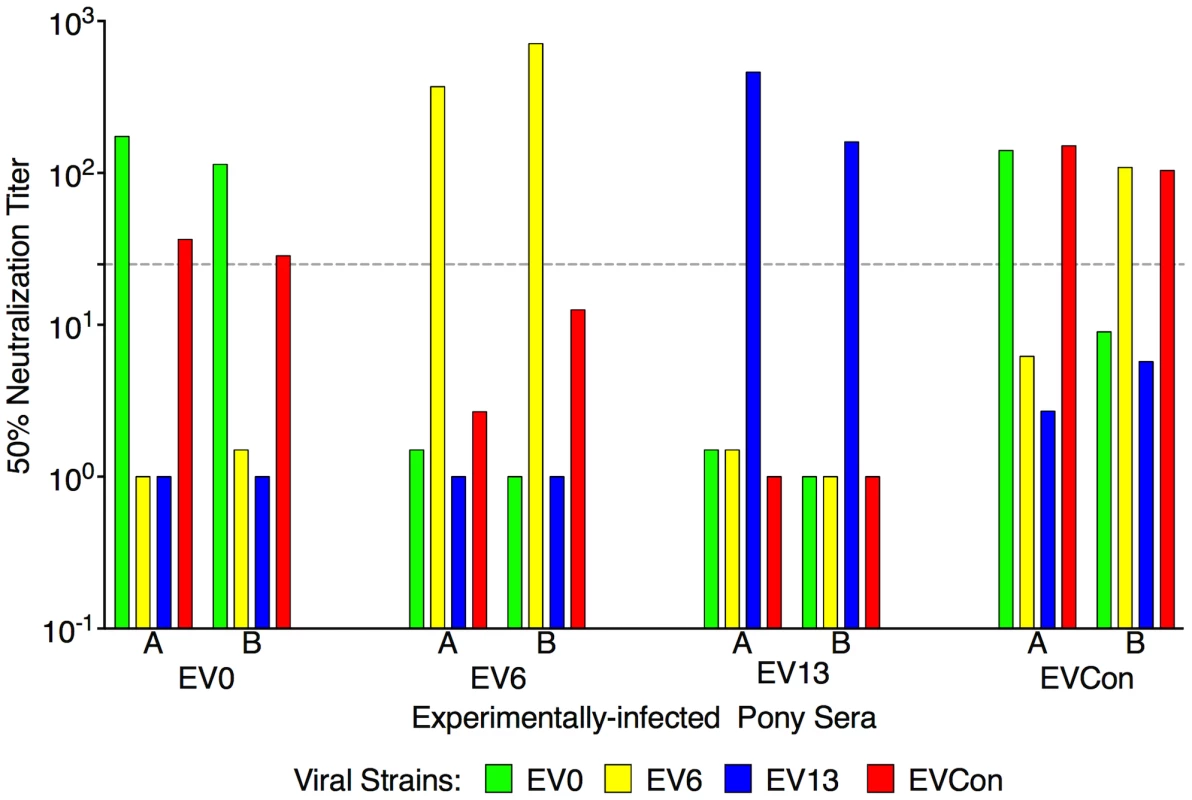
Variant proviral strains were tested for in vivo pathogenecity through experimental infections of ponies. Serum from the infections was utilized here to test for immunogenicity and characterize the variant Envs. At six-months post-infection serum from two representative animals (labeled A,B) was tested in our standard neutralization assay. The mean reciprocal dilutions of serum which neutralized 50% of input EV0, EV6, EV13, or EVCon as measured in an infectious center assay are presented. The dashed line denotes the cut off (≥25) value for valid 50% neutralization titers. Experimental vaccination and challenge
To directly evaluate the consensus Env as well as the general premise of centralized immunogens, we compared the consensus Env attenuated strain (ConD9) with an ancestral Env attenuated vaccine strain (EIAVD9 or D9). Proficiency of the centralized immunogens was further scrutinized by inclusion of a third attenuated vaccine regimen. The third arm of the study, a polyvalent attenuated strain mixture, was chosen as the most rigorous match to the centralized immunogens. The polyvalent attenuated quasispecies was constructed utilizing the D9 backbone. A trivalent attenuated mixture was assembled with the D9 as one of three strains. The EV6 and EV13 Envs were cloned into the D9 backbone to create 6D9 and 13D9, respectively, the final two strains of the polyvalent mix. The polyvalent attenuated mixture, a 1∶1∶1 mix of D9, 6D9, and 13D9, otherwise termed TriD9, was tested in vivo for TCID50 dosage verification in a group of eight ponies.
Twenty-four ponies of mixed age and gender were divided randomly into groups of eight animals and inoculated intravenously with two, 3×103 TCID50 doses of the ancestral D9 vaccine, the consensus ConD9 or polyvalent TriD9 vaccines, at four-week intervals. The inoculated ponies were monitored daily for clinical signs of EIA, and blood samples were taken at regular intervals for standard measurements of disease, virus replication, and host immune responses, as described previously [31]–[33], [50]. Figs. 5–7 display the clinical profiles of vaccinated animals. One of the eight animals in the polyvalent TriD9 group developed clinical complications pre-challenge that compromised continued use of the animal in the study and was thus removed from the trial. All twenty-three vaccinates (3 trial groups) exhibited no clinical signs of EIA disease from the attenuated vaccine strains during the seven month observation period, a time frame that allows complete maturation of vaccine immunity prior to virus challenge [22], [30], [51] (Figs. 5A–7A).
Fig. 5. Clinical and virological profiles of D9 vaccine trial group. 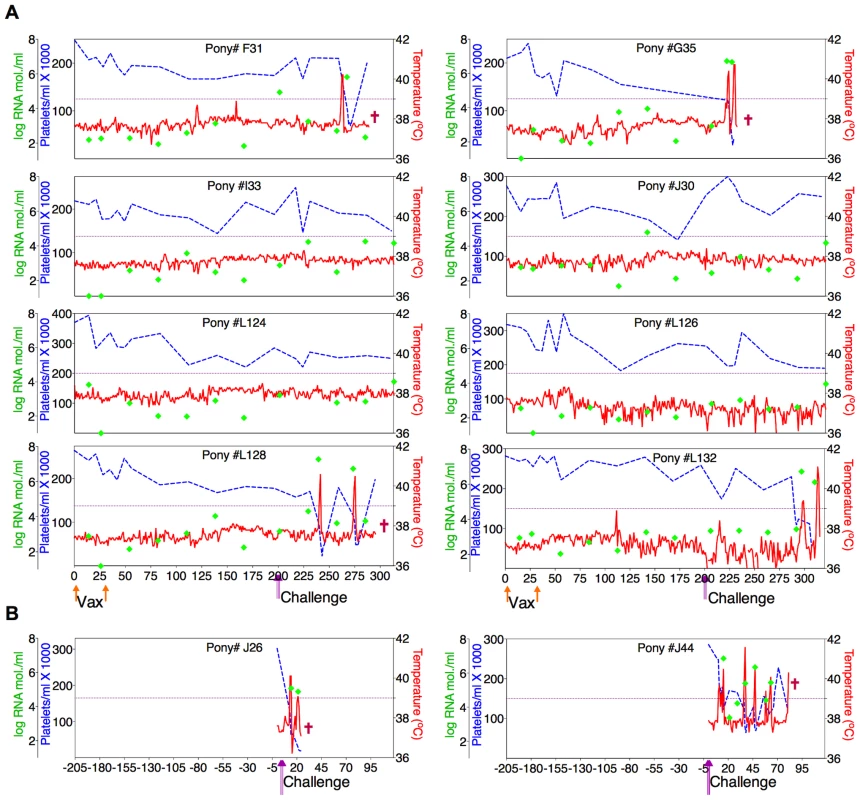
(A) Eight EIAV-naïve ponies were vaccinated with 3×103 TCID50 D9 I.M. (orange arrow, Vax). Rectal temperature (red line, right Y axis) and platelet counts (blue dashed line, first left Y axis) were followed daily for up to 320 days (X-axis) after the first vaccine dose. Quantification of the virus load (green diamond, second left Y axis) was performed on viral RNA extracted from plasma at periodic time points prior to and after virulent virus challenge seven months post-first vaccination with 3×103 TCID50 EVMX (1∶1∶1 - EV0, EV6, or EV13) I.M (pink arrows, Challenge). (B) Two EIAV-naïve ponies were also challenged with 3×103 TCID50 EVMX (1∶1∶1 - EV0, EV6, or EV13) I.M (pink arrows, Challenge). Febrile episodes were defined by a achieving a combination of two-three features such as: rectal temperature above 39°C in conjunction with thrombocytopenia (platelet decrease of ≥70,000/µl of whole blood), EIAV viral load ≥105 as well as other clinical signs of EIA. Burgundy cross symbol, animal euthanized due to severe disease or as endpoint of study. Fig. 6. Clinical and virological profiles of ConD9 vaccine trial group. 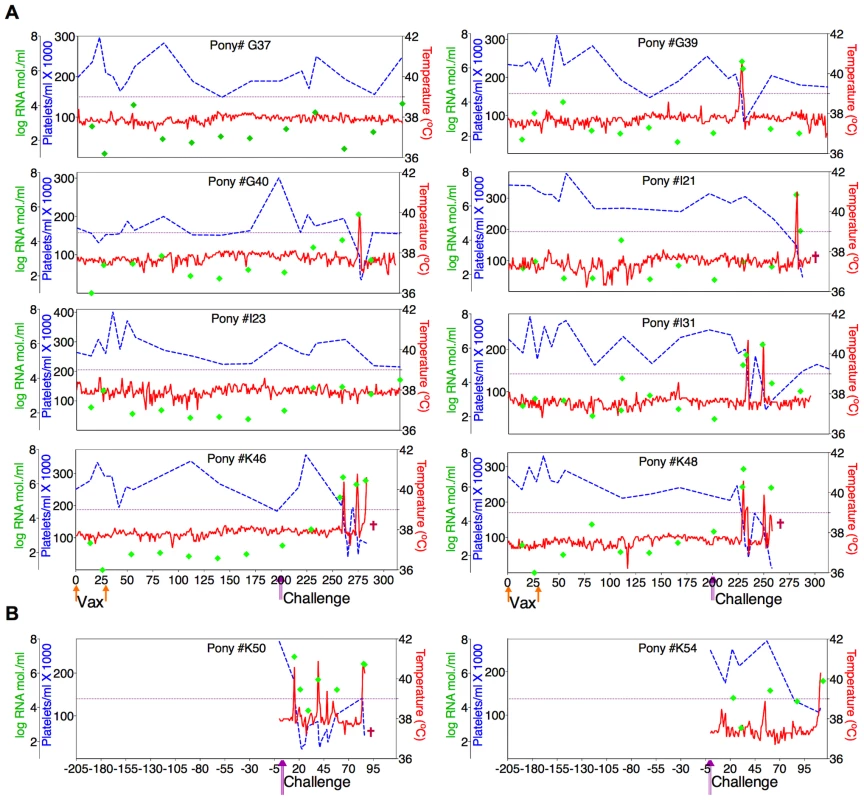
(A) Eight EIAV-naïve ponies were vaccinated with 3×103 TCID50 ConD9 I.M. (orange arrow, Vax). Rectal temperature (red line, right Y axis) and platelet counts (blue dashed line, first left Y axis) were followed daily for up to 320 days (X-axis) after the first vaccine dose. Quantification of the virus load (green diamond, second left Y axis) was performed on viral RNA extracted from plasma at periodic time points prior to and after virulent virus challenge seven months post-first vaccination with 3×103 TCID50 EVMX (1∶1∶1 - EV0, EV6, or EV13) I.M (pink arrows, Challenge). (B) Two EIAV-naïve ponies were also challenged with 3×103 TCID50 EVMX (1∶1∶1 - EV0, EV6, or EV13) I.M (pink arrows, Challenge). Febrile episodes were defined by a achieving a combination of two-three features such as: rectal temperature above 39°C in conjunction with thrombocytopenia (platelet decrease of ≥70,000/µl of whole blood), EIAV viral load ≥105 as well as other clinical signs of EIA. Burgundy cross symbol, animal euthanized due to severe disease or as endpoint of study. Fig. 7. Clinical and virological profiles of TriD9 vaccine trial group. 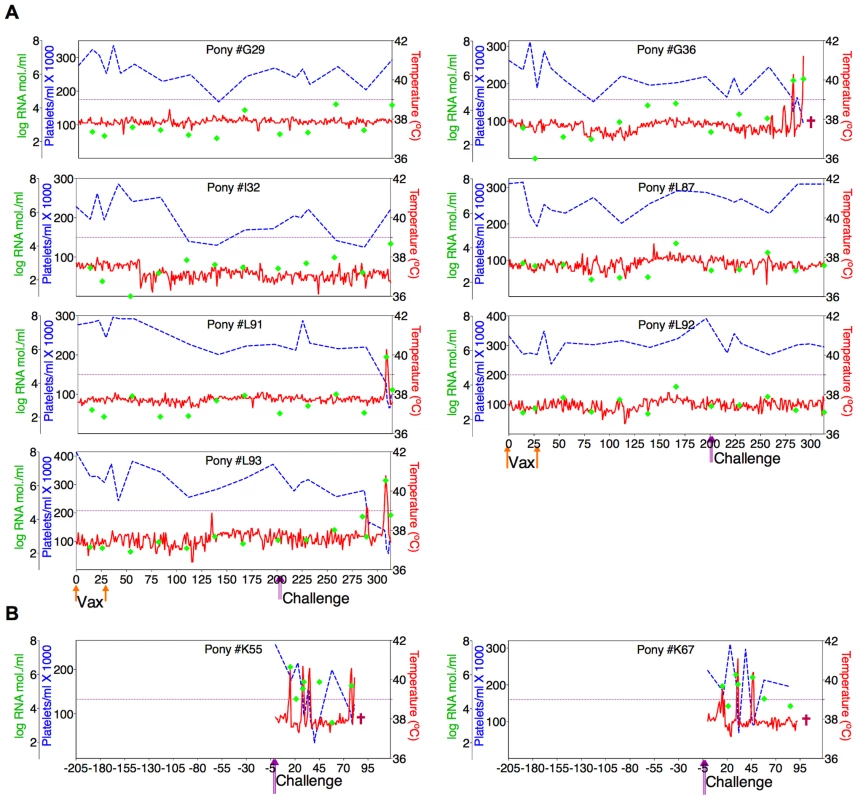
(A) Seven of eight EIAV-naïve ponies vaccinated with 3×103 TCID50 TriD9 I.M. (orange arrow, Vax) are presented. One of the eight original vaccinates experienced clinical complications and was removed from the trial. Rectal temperature (red line, right Y axis) and platelet counts (blue dashed line, first left Y axis) were followed daily for up to 320 days (X-axis) after the first vaccine dose. Quantification of the virus load (green diamond, second left Y axis) was performed on viral RNA extracted from plasma at periodic time points prior to and after virulent virus challenge seven months post-first vaccination with 3×103 TCID50 EVMX (1∶1∶1 - EV0, EV6, or EV13) I.M (pink arrows, Challenge). (B) Two EIAV-naïve ponies were also challenged with 3×103 TCID50 EVMX (1∶1∶1 - EV0, EV6, or EV13) I.M (pink arrows, Challenge). Febrile episodes were defined by a achieving a combination of two-three features such as: rectal temperature above 39°C in conjunction with thrombocytopenia (platelet decrease of ≥70,000/µl of whole blood), EIAV viral load ≥105 as well as other clinical signs of EIA. Burgundy cross symbol, animal euthanized due to severe disease or as endpoint of study. An engineered quasispecies challenge model, EVMX, was developed as a rigorous assessment of immune protection. Based on previous studies [28], [34], equivalent (1∶1∶1) TCID50 dosages of the virulent EV0, EV6, and EV13 strains combined to create the virulent, well-defined, swarm. Six months following the second vaccination, the immunized ponies were challenged intravenously every other day with three, 3×103 TCID50, EVMX inoculations. A control group consisting of six EIAV-naïve ponies was also challenged with EVMX (Figs. 5B–7B). Analyses of vaccinate day of challenge (DOC) viral loads demonstrated all three attenuated viral regimens replicated to similar levels, averaging, over the seven month pre-challenge period, between 2×103 and 3×103 copies RNA/ml plasma (Figs. 5–7). Despite these similarities in viral vaccine replication, however, trial groups displayed markedly different levels of EVMX-induced disease. Four ancestral D9, six consensus ConD9, and three polyvalent TriD9 animals displayed clinical signs of EIA disease during the observation period post challenge (Figs. 5A–7A). Chronic disease was observed in the majority of vaccinates that experienced initial acute disease. All six control animals of each variant virus challenge group developed clinical EIA disease, indicating 100% virulence of the quasispecies challenge under the current experimental infection conditions. DOC vaccine immune responses in all groups were also indicative of mature immune responses (Table 1, Table 2, S1 Fig.). The polyvalent TriD9 group demonstrated the highest Env-specific serum antibody titer and avidity, although it was only significantly different from the ancestral D9 regimen (P = 0.017, P<0.0001, respectively), and not from the consensus ConD9 regimen (S1 Fig.). While neutralizing antibodies were detectable in all three groups, they could not be associated with protection. Similarly, DOC cellular immunity levels were similar but not correlated with protection levels (Table 2). For example, measured in vivo cytokine responses were the lowest in the polyvalent TriD9 vaccinates that showed the highest level of protection.
Tab. 1. Characterization of vaccine Env-specific antibody prior to challenge.* 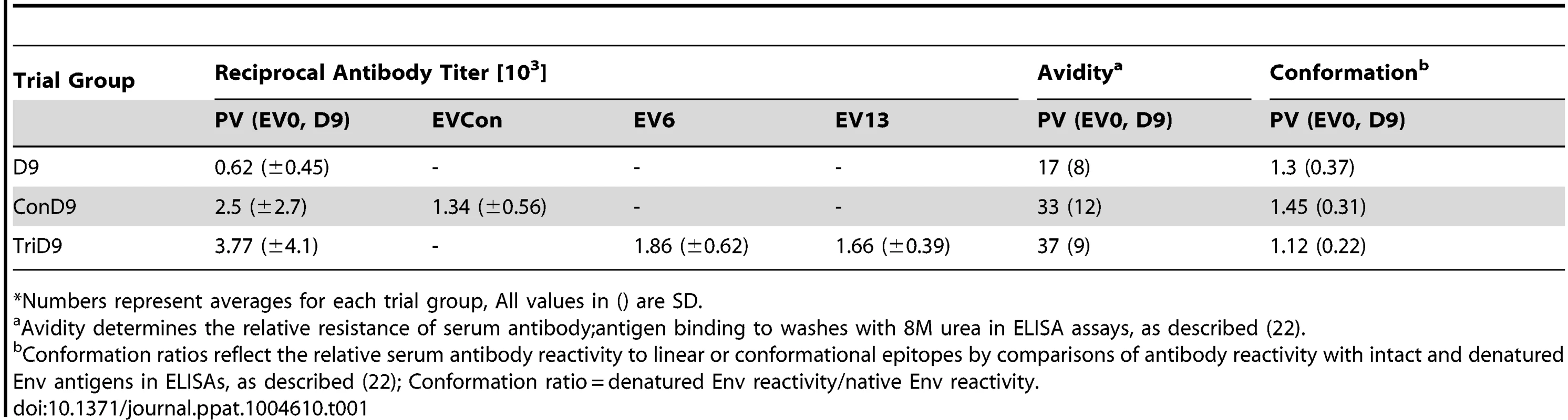
*Numbers represent averages for each trial group, All values in () are SD. Tab. 2. Characterization of vaccine immune responses prior to challenge.* 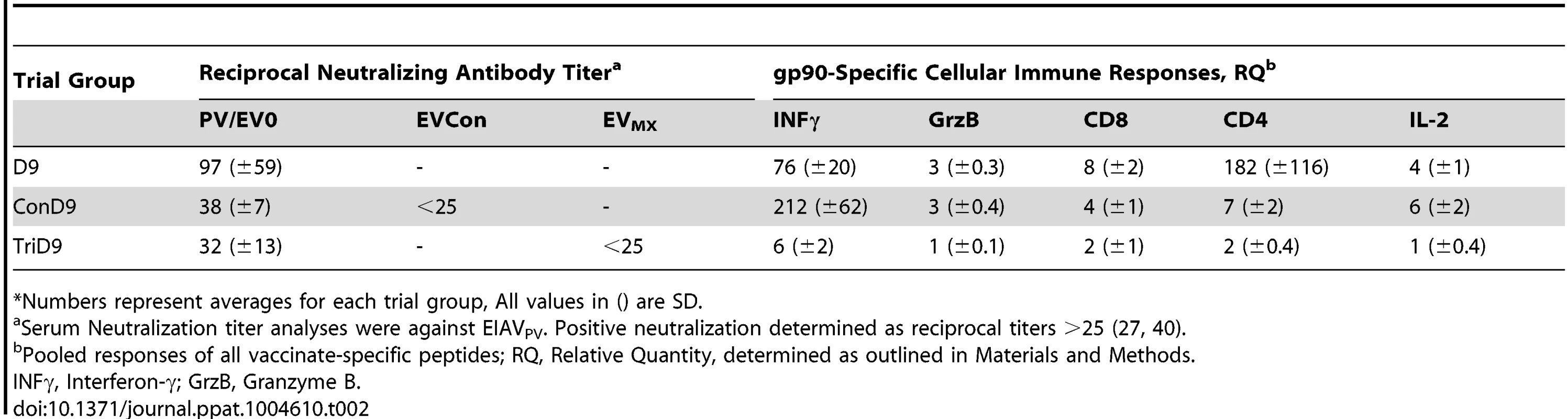
*Numbers represent averages for each trial group, All values in () are SD. The percentage of animals within each trial group protected from clinical EIA was plotted as a function of days post-challenge for survival analysis (Fig. 8). Both centralized Env vaccine groups had subjects that succumbed to EVMX disease within a typical time frame of 2–3 weeks post-challenge. The polyvalent, TriD9 group, however, demonstrated a delay in the onset of disease with the first animal not breaking until 81 days post-challenge (Fig. 8). Overall, protection curves of all three vaccine groups were significantly different from one another (ANOVA, P<0.0001). Polyvalent TriD9 vaccinates demonstrated the highest levels of protection that was significantly different from the unvaccinated controls and the consensus ConD9 curves (P<0.0001, P = 0.0002). While the consensus ConD9 group had the lowest level of protection, it was significant as compared to unvaccinated subjects (P = 0.001). Analysis of the trend revealed a significant relationship between the complexity of immunogen and protective efficacy (P = 0.02). Ultimately, the consensus ConD9 strain, while pre-trial appearing to be more broadly immunogenic, demonstrated the lowest level of protection. The polyvalent TriD9 regimen demonstrated the highest level of protection against a quasispecies challenge. The current study not only highlighted important information towards HIV vaccine development and highlighted the importance of rigorous challenge strain engineering.
Fig. 8. Protection from disease is associated with complexity of immunogen. 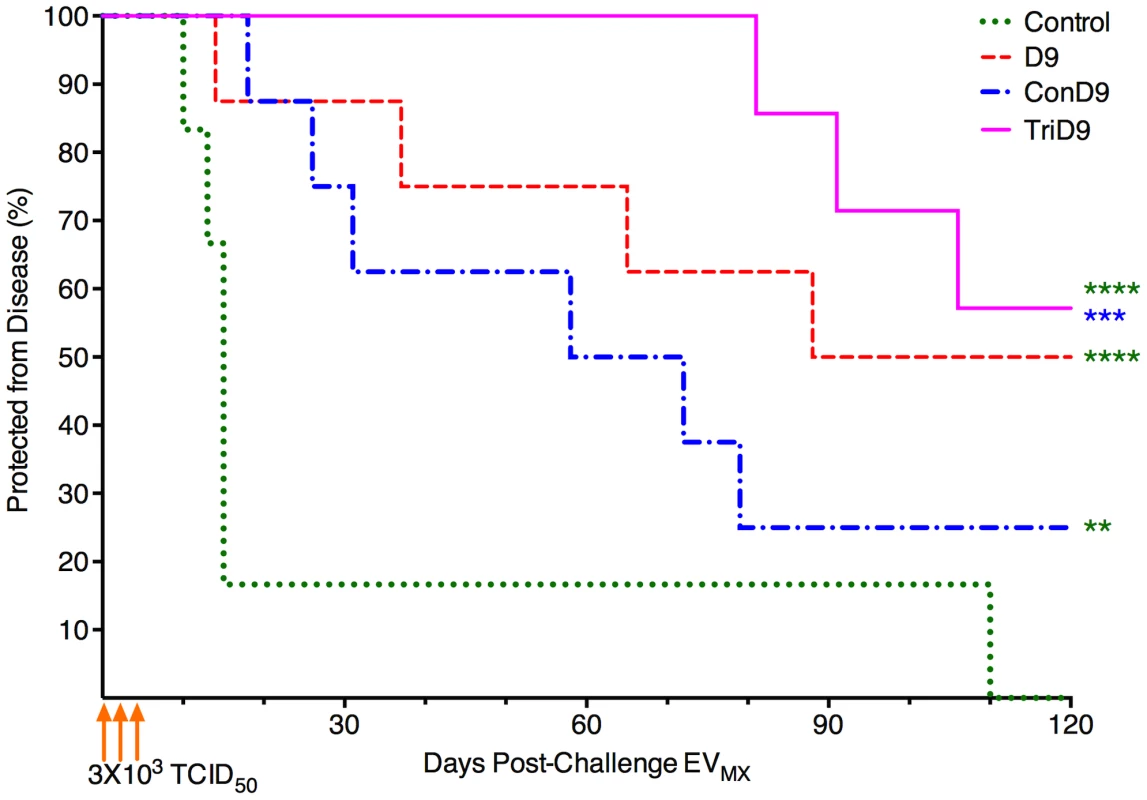
The percentage of animals within each trial group protected from clinical EIA was plotted as a function of days post-challenge with the engineered quasispecies EVMX. Overall resulting protection curves are significantly different from one another (ANOVA, P<0.0001). TriD9 demonstrated the highest level of protection (****), followed by D9 (****). The consensus Env, or ConD9 attenuated vaccine, conferred the lowest level of protection; however, it was significant as compared to unvaccinated subjects (**). The TriD9 vaccinated animals were also significantly more protected than the ConD9 animals (***). Logrank test for trend via survival curve analysis reveal a significant trend for the complexity of the immunogen and protective efficacy (P = 0.02.) Multiple comparisons performed via Bonferroni's multiple comparison test: ****, P<0.0001; ***, P = 0.0002, **, P = 0.001. Discussion
The collective success of attenuated vaccine regimens makes it the ideal modality to rigorously test novel vaccine concepts. Previous work by our group and others have demonstrated attenuated vaccines to be an extremely useful tool for vaccine development, regardless of the potential for marketable advancement of the attenuated platform due to the safety concerns associated with HIV vaccines. Consequently, we resolved to examine the efficacy of centralized Env immunogens in our well-established EIAV attenuated vaccine model. Results presented here reveal, for the first time, a consensus Env in a fully replication-competent attenuated virus backbone that can confer protective efficacy against virulent virus challenge; however, it does not induce the highest level of protection as compared to ancestral or polyvalent vaccines.
A concept not articulated in most reports on centralized Env sequences are the specifics involved in the determination of the consensus sequence. Creation of an ideal consensus sequence requires careful consideration of various parameters. Our original attempts to construct a consensus gp90 gene focused on a more basic approach than what we utilized for the current study. Initial alignments were performed utilizing the three sequences that comprise the EVMX quasispecies challenge strain, EV0, EV6, and EV13 [34]. Phylogenetic analysis of this consensus gp90 demonstrated that the consensus sequence was located high on the ancestral root and would not have represented a good consensus sequence as it was highly related to the ancestral, or EV0 gp90. Likewise, the “consensus of all consensus” method utilized in both the HIV and influenza fields [6], [9], [15], [18], [19], [52]–[56] resulted in a gp90 sequence that phylogenetically was more related to early disease isolates on the ancestral root (Fig. 1) than to the consensus gene generated from all 90 isolate sequences (nucleotide and amino acid). In vitro analysis of the virus constructed from the second-generation consensus gp90, assembled from the 90-isolate alignment and cloned into a proviral backbone, was found to infect equine cells, but was not fully replication competent. Therefore, if we were to analyze this Env in a single-round infection assay it would falsely appear to be fully functional. Hence, the 90-isolate sequence alignment was re-evaluated and a higher level of hand-editing performed, especially in the highly variable V3 region, prior to consensus generation. This thorough computational analysis and consideration of the consensus Env limited the potential sampling bias that obfuscates computational engineering of protein immunogens [1], [57]. Likewise, our use of an actual ancestoral Env also reduced the potential sampling bias that is problematic to computationally constructed most recent common ancestors [1], [57]. Additionally, in the absence of replication analysis and study in an attenuated model, the incapability of the consensus Env to functional naturally would not have been observed and a gp90 less representative of a fully functional Env would have been examined. Ultimately, this is the first successful construction of a consensus envelope lentivirus construct with full replication and virulence properties.
The highest protective efficacy against disease was observed in the polyvalent TriD9 vaccinates. Survival analysis revealed the polyvalent TriD9 disease curve was significantly different from the naïve controls and importantly, the consensus ConD9 curve. Considering the EVMX challenge quasispecies virus strain composition, the polyvalent TriD9 regimen displaying the highest levels of protective efficacy might be anticipated; however, the ancestral D9 vaccinates also protected to a higher degree than the consensus ConD9 vaccinates. Pre-challenge analysis of clinical and virological factors would not have predicted these results. All three attenuated vaccine regimens displayed similar levels of pre-challenge viral strain replication. Pre-trial evaluation of the capability to induce neutralizing antibodies (Fig. 4) indicated that the ConEnv produced a broader immune response than the ancestral gp90 (EV0/D9) and also the EV6 and EV13 gp90 proteins. Immune analysis of DOC responses did not reveal a direct correlation of protection. Neutralizing antibodies and cellular immune responses were not associated with protective efficacy. Although a significantly higher level of Env-specific antibodies were found in the polyvalent TriD9 vaccinates, the consensus ConD9 vaccinates, similarly, had a higher antibody response as compared to the ancestral D9. In the case of the consensus ConD9 vaccinates, immune response data is suggestive of potential issues related to the artificial nature of the consensus Env and its ability to induce broadly protective responses. Protection in all three vaccine groups could be due to anamnestic responses that could be related to conserved regions or conformational epitopes that allow for protective response in lieu of sequence identity. Although total antibody binding does not reveal directly correlative data (S1 Fig.), mapping of the reactive epitopes will be key to determining if a region of the gp90 potentially conferred more protective responses. These studies are currently underway. A notable observation of the polyvalent TriD9 vaccinates was the delayed onset of disease (Fig. 8). The quasispecies challenge virus eventually broke through causing its first case of disease at the late time point of 81 days post-challenge. The nature of this break through is an interesting study of evolution: did the EVMX Env evolve at a faster rate than the polyvalent TriD9 and escape the protective immune responses or did recombination allow the escape. Current studies are being performed to enable future characterization of break-through febrile isolates.
As part of the current report we generated an engineered quasispecies challenge virus mix containing different degrees of Env variation. The majority of current lentiviral vaccine studies employ a single strain challenge model. Heterologous challenge models, while more rigorous than homologous challenge models, are commonly single viral strains. Data presented here indicate that more comprehensive challenge models, that include variable Env proteins, should be developed for the study of lentiviral vaccines to better test the vaccine modalities being investigated. The polyvalent TriD9 and EVMX vaccine and challenge strains were at their basic phenotype, a homologous pairing. However, the complexity of a pathogenic, diverse, challenge strain resulted in a notable difference and reduction in the protective efficacy as compared to previous studies where the vaccine and challenge strains were matched in their Env sequences. Improved challenge model development for animal lentiviral studies that comprehensively test protective efficacy are critical tools required for broadly protective vaccine development.
The studies presented here demonstrate definitively that polyvalent attenuated vaccine regimens have significantly higher levels of protection as compared to centralized immunogens. Although it is not possible with absolute confidence to extrapolate the results of vaccine studies in any single animal lentivirus system to other animal lentiviruses or to HIV, the data presented here certainly highlight the priority of ascertaining centralized immunogens on HIV vaccine efficacy in the context of higher animal models that include challenge studies which can inform on the true protective nature of the proposed immunogens.
Materials and Methods
Consensus Env and strains
Consensus gp90 Env protein sequence was determined by alignment of nucleotide (codon alignment) and amino acid sequences from naturally arisen EIAV isolates originated from an experimental infection (pony #567) [34], [42]–[44] in the Geneious Pro package of software (Biomatters, Ltd.). Alignments were hand edited where necessary, especially in the highly variable loop region, and ambiguities resolved through partner aligning of nucleotides and amino acids. Phylogenetic characterization of the consensus Env was were constructed by the neighbor-joining method of Jukes Cantor corrected distances with the optimality criterion set to distance as measured in PAUP [58] and implemented in the Geneious Pro 5.0.4 (Biomatters Ltd., NZ) package of software. Statistical significance of branchings and clustering was assessed by bootstrap resampling of 1000 pseudoreplicates on the complete data set; trees were rooted to the original infectious ancestral Env, EV0. The trees were edited for publication using FigTree Version 1.1.2. Resultant gp90 sequence was synthesized (GeneArt, Regensberg, Germany). The consensus gp90 was cloned into pathogenic and attenuated EIAV backbones using methods and restriction sites previously described [34], [47], [48]. Viral stocks were prepared as previously described [34], [48]. Viral stock titers were determined utilizing our infectious center assay (cell-based ELISA) in fetal equine kidney cells, described previously [46].
Experimental infections with ConD9 and EVCon
All equine procedures were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health at the Gluck Equine Research Center of the University of Kentucky according to protocols approved by the University of Kentucky IACUC (#01058A2006). The animals were monitored daily and maintained as described previously [31], [32], [34], [43], [49]. Eight outbred, mixed-breed ponies were separated into two groups of four and experimentally inoculated intravenously with 103 TCID50 of either chimeric strain ConD9 or EVCon. Rectal temperatures and clinical status were recorded daily. Platelet numbers were determined using the IDEXX VetAutoread Hematology Analyzer (IDEXX Laboratories Inc., Westbrook ME). Clinical EIA (fever) episodes were determined on the basis of rectal temperature and platelet count (rectal temperature >39°C; platelet number <100,000/µl of whole blood) in combination with the viremic presence of infectious plasma virus (≥105) [20], [26], [43], [49], [59]. Samples of whole blood, serum, and plasma were collected weekly as well as daily during fever episodes. Plasma samples were stored at −80°C until used to determine plasma viral RNA level.
Challenge strains
The challenge strain EVMX quasispecies was produced by combining equivalent infectious titers (TCID50) of the variant challenge strains EV0, EV6, and EV13 [34]. Viral stocks were prepared as previously described [34], [48]. Viral stock titers were determined utilizing our infectious center assay (cell-based ELISA) in fetal equine kidney cells, described previously [46].
Experimental subjects, vaccination, and challenge
Equine procedures were conducted at the Gluck Equine Research Center of the University of Kentucky according to protocols approved by the University of Kentucky IACUC. Thirty-six mixed age and gender outbred ponies, serognegative for EIAV, were utilized. Daily rectal temperatures and clinical status were recorded. CBC analysis of whole blood was performed using an IDEXX QBC Vet Autoreader. Hematocrit and platelet numbers were monitored weekly. The EIAVD9 stock was produced and vaccinations performed as described [31], [34]. Twenty-three vaccinated and six naïve ponies were challenged with 3×103 TCID50 of EVMX. The ponies were monitored daily for clinical symptoms of EIA, and blood was drawn at regular intervals (weekly, daily if febrile) for assays of platelets, viral replication, and virus-specific immune responses. During the course of these experiments ponies that demonstrated severe disease-associated symptoms resulting in distress as outlined by the University of Kentucky IACUC were euthanized.
Quantitative and Qualitative RT-PCR analysis of plasma virus RNA
Plasma samples from all subjects were analyzed for the levels of viral RNA per milliliter of plasma using a previously described quantitative real-time multiplex RT-PCR assay based on gag-specific amplification primers [60]. The standard RNA curve was linear in the range of 101 molecules as a lower limit and 108 molecules as an upper limit.
Quantitative assays of cellular immune responses
Our in vivo method for assessing immune reactivity to specific peptides, described previously [61], was used to explore cellular immune responses. Forty-four 20-mer peptides, overlapping by 10 residues, spanning the EIAVD9 gp90 were generated (GenScript USA Inc., Piscataway, NJ). An additional 15 peptides, specific for the variable regions ConD9 gp90, were included in the pool for analysis of those vaccinates. Vaccinates were screened for gp90 specific cellular immune responses one week prior to day of challenge. A 2 mm skin biopsy was collected and homogenized and RNA extracted. IFNγ gene expression was determined by real-time PCR, as previously described [61]. Amplification efficiencies were determined using Linreg [62] and samples with amplification efficiencies above 99% were included for further analyses. Beta-glucuronidase (β-GUS) was used as housekeeping gene and the ΔΔCT method [63] was used to determine relative gene expression with saline injection site for each vaccinate used as the calibrator. Relative quantity (RQ) was calculated as 2−ΔΔCT.
Quantitative and qualitative serological analyses
Serum IgG antibody reactivity to EIAV envelope glycoproteins was assayed quantitatively (end point titer) and qualitatively (avidity index, conformation ratio) using our standard concanavalin A (ConA) ELISA procedures as described previously [51]. Virus neutralizing activity to the historical reference strain EIAVPV, and vaccine-specific virus stocks EVCon and EVMX, mediated by immune sera, was assessed in an indirect cell-ELISA based infectious center assay using a constant amount of infectious EIAV and sequential 2-fold dilutions of serum [46], [51].
Statistical analyses
Significance of protection from disease was performed by survival curve analysis as implemented in GraphPad Prism version 6.0d (San Diego, CA). Significance of survival curves were determined utilizing One-way ANOVA with Bonferroni's multiple comparison's test as well as survival analysis of Kaplan Meier plots with Logrank test for trend.
Ethics statement
All equine procedures were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health at the Gluck Equine Research Center of the University of Kentucky according to protocols approved by the University of Kentucky IACUC (#01058A2006).
Supporting Information
Zdroje
1. GaschenB, TaylorJ, YusimK, FoleyB, GaoF, et al. (2002) Diversity considerations in HIV-1 vaccine selection. Science 296 : 2354–2360.
2. AnastassopoulouCG, KostrikisLG (2006) Global genetic variation of HIV-1 infection. Curr HIV Res 4 : 365–373.
3. BranderC, FrahmN, WalkerBD (2006) The challenges of host and viral diversity in HIV vaccine design. Curr Opin Immunol 18 : 430–437.
4. KorberB, GaschenB, YusimK, ThakallapallyR, KesmirC, et al. (2001) Evolutionary and immunological implications of contemporary HIV-1 variation. BrMedBull 58 : 19–42.
5. MullinsJI, JensenMA (2006) Evolutionary dynamics of HIV-1 and the control of AIDS. Curr Top Microbiol Immunol 299 : 171–192.
6. LiaoHX, TsaoCY, AlamSM, MuldoonM, VandergriftN, et al. (2013) Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol 87 : 4185–4201.
7. MullinsJI, NickleDC, HeathL, RodrigoAG, LearnGH (2004) Immunogen sequence: the fourth tier of AIDS vaccine design. Expert Rev Vaccines 3: S151–159.
8. NickleDC, JensenMA, GottliebGS, ShrinerD, LearnGH, et al. (2003) Consensus and ancestral state HIV vaccines. Science 299 : 1515–1518 author reply 1515-1518.
9. GaoF, WeaverEA, LuZ, LiY, LiaoHX, et al. (2005) Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol 79 : 1154–1163.
10. Doria-RoseNA, LearnGH, RodrigoAG, NickleDC, LiF, et al. (2005) Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol 79 : 11214–11224.
11. KotheDL, DeckerJM, LiY, WengZ, Bibollet-RucheF, et al. (2007) Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology 360 : 218–234.
12. KotheDL, LiY, DeckerJM, Bibollet-RucheF, ZammitKP, et al. (2006) Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology 352 : 438–449.
13. SantraS, MuldoonM, WatsonS, BuzbyA, BalachandranH, et al. (2012) Breadth of cellular and humoral immune responses elicited in rhesus monkeys by multi-valent mosaic and consensus immunogens. Virology 428 : 121–127.
14. SantraS, KorberBT, MuldoonM, BarouchDH, NabelGJ, et al. (2008) A centralized gene-based HIV-1 vaccine elicits broad cross-clade cellular immune responses in rhesus monkeys. Proc Natl Acad Sci U S A 105 : 10489–10494.
15. LiaoHX, SutherlandLL, XiaSM, BrockME, ScearceRM, et al. (2006) A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353 : 268–282.
16. GaoF, LiaoHX, HahnBH, LetvinNL, KorberBT, et al. (2007) Centralized HIV-1 envelope immunogens and neutralizing antibodies. Curr HIV Res 5 : 572–577.
17. WeaverEA, LuZ, CamachoZT, MoukdarF, LiaoHX, et al. (2006) Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group m consensus env immunogen. J Virol 80 : 6745–6756.
18. EugeneHS, Pierce-PaulBR, CragioJK, RossTM (2013) Rhesus macaques vaccinated with consensus envelopes elicit partially protective immune responses against SHIV SF162p4 challenge. Virol J 10 : 102.
19. McburneySP, RossTM (2009) Human immunodeficiency virus-like particles with consensus envelopes elicited broader cell-mediated peripheral and mucosal immune responses than polyvalent and monovalent Env vaccines. Vaccine 27 : 4337–4349.
20. Montelaro RC, Ball JM, Rushlow K (1993) Equine retroviruses. In: Levy JA, editor. The Retroviridae. New York, N.Y.: Plenum Press. pp. 257–360.
21. HarroldSM, CookSJ, CookRF, RushlowKE, IsselCJ, et al. (2000) Tissue sites of persistent infection and active replication of equine infectious anemia virus during acute disease and asymptomatic infection in experimentally infected equids. J Virol 74 : 3112–3121.
22. HammondSA, LiF, MckeonBMSr, CookSJ, IsselCJ, et al. (2000) Immune responses and viral replication in long-term inapparent carrier ponies inoculated with equine infectious anemia virus. J Virol 74 : 5968–5981.
23. KonoY, HirasawaK, FukunagaY, TaniguchiT (1976) Recrudesence of equine infectious anemia by treatment with immunosuppressive drugs. Nat Inst Anim Hlth Quart 16 : 8–15.
24. CraigoJK, MontelaroRC (2013) Lessons in AIDS vaccine development learned from studies of equine infectious, anemia virus infection and immunity. Viruses 5 : 2963–2976.
25. CraigoJK, MontelaroRC (2011) Equine Infectious Anemia Virus Infection and Immunity: Lessons for Aids Vaccine Development. Future Virol 6 : 139–142.
26. Craigo JK, Montelaro RC (2008) Equine Infectious Anemia Virus (Retroviridae). Encyclopedia of Virology. Third ed. Oxford: Elsevier. pp. 167–174.
27. Craigo JK, Montelaro RC (2010) Lentivirus Tropism and Disease. In: Desport M, editor. Lentiviruses and Macrophages: Molecular and Cellular Interactions: Horizon Academic Press. pp. 1–23.
28. CraigoJK, EzzelarabC, CookSJ, ChongL, HorohovD, et al. (2013) Envelope determinants of equine lentiviral vaccine protection. PLoS ONE 8: e66093.
29. CraigoJK, MontelaroRC (2010) EIAV envelope diversity: shaping viral persistence and encumbering vaccine efficacy. Curr HIV Res 8 : 81–86.
30. MontelaroRC, ColeKS, HammondSA (1998) Maturation of immune responses to lentivirus infection: implications for AIDS vaccine development. AIDS ResHumRetroviruses 14 Suppl 3: S255–S259.
31. CraigoJK, DurkinS, SturgeonTJ, TagmyerT, CookSJ, et al. (2007) Immune suppression of challenged vaccinates as a rigorous assessment of sterile protection by lentiviral vaccines. Vaccine 25 : 834–845.
32. CraigoJK, LiF, SteckbeckJD, DurkinS, HoweL, et al. (2005) Discerning an effective balance between equine infectious anemia virus attenuation and vaccine efficacy. J Virol 79 : 2666–2677.
33. LiF, CraigoJK, HoweL, SteckbeckJD, CookS, et al. (2003) A live attenuated equine infectious anemia virus proviral vaccine with a modified S2 gene provides protection from detectable infection by intravenous virulent virus challenge of experimentally inoculated horses. J Virol 77 : 7244–7253.
34. CraigoJK, ZhangB, BarnesS, TagmyerTL, CookSJ, et al. (2007) Envelope variation as a primary determinant of lentiviral vaccine efficacy. Proc Natl Acad Sci U S A 104 : 15105–15110.
35. MontelaroRC, WigzellH (1995) AIDS 1995. Vaccines and immunology: overview. AIDS 9 Suppl A: S111–S112.
36. MillsJ, DesrosiersRC, RudE, AlmondN (2000) Live Attenuated HIV Vaccines: A Proposal for Further Research and Development. AIDS ResHumRetroviruses 16 : 1453–1461.
37. UhlEW, Heaton-JonesTG, PuR, YamamotoJK (2002) FIV vaccine development and its importance to veterinary and human medicine: a review. FIV vaccine 2002 update and review. VetImmunolImmunopathol 90 : 113–132.
38. KoffWC, JohnsonPR, WatkinsDI, BurtonDR, LifsonJD, et al. (2006) HIV vaccine design: insights from live attenuated SIV vaccines. Nat Immunol 7 : 19–23.
39. CraigoJK, BarnesS, CookSJ, IsselCJ, MontelaroRC (2010) Divergence, not diversity of an attenuated equine lentivirus vaccine strain correlates with protection from disease. Vaccine 28 : 8095–8104.
40. TagmyerTL, CraigoJK, CookSJ, EvenDL, IsselCJ, et al. (2008) Envelope determinants of equine infectious anemia virus vaccine protection and the effects of sequence variation on immune recognition. J Virol 82 : 4052–4063.
41. TagmyerTL, CraigoJK, CookSJ, IsselCJ, MontelaroRC (2007) Envelope-specific T-helper and cytotoxic T-lymphocyte responses associated with protective immunity to equine infectious anemia virus. J Gen Virol 88 : 1324–1336.
42. LerouxC, CraigoJK, IsselCJ, MontelaroRC (2001) Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J Virol 75 : 4570–4583.
43. LerouxC, IsselC, MontelaroRC (1997) Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol 71 : 9627–9639.
44. CraigoJK, SturgeonTJ, CookSJ, IsselCJ, LerouxC, et al. (2006) Apparent elimination of EIAV ancestral species in a long-term inapparent carrier. Virology 344 : 340–353.
45. CookRF, CookSJ, BergerSL, LerouxC, GhabrialNN, et al. (2003) Enhancement of equine infectious anemia virus virulence by identification and removal of suboptimal nucleotides. Virology 313 : 588–603.
46. CraigoJK, EzzelarabC, MontelaroRC (2012) Development of a high throughput, semi-automated, infectious center cell-based ELISA for equine infectious anemia virus. J Virol Methods 185 : 221–227.
47. HoweL, LerouxC, IsselCJ, MontelaroRC (2002) Equine Infectious Anemia Virus Envelope Evolution In Vivo during Persistent Infection Progressively Increases Resistance to In Vitro Serum Antibody Neutralization as a Dominant Phenotype. The Journal of Virology 76 : 10588.
48. LiF, PufferBA, MontelaroRC (1998) The S2 gene of equine infectious anemia virus is dispensable for viral replication in vitro. J Virol 72 : 8344–8348.
49. LichtensteinDL, IsselCJ, MontelaroRC (1996) Genomic quasispecies associated with the initiation of infection and disease in ponies experimentally infected with equine infectious anemia virus. J Virol 70 : 3346–3354.
50. IsselCJ, HorohovDW, LeaDF, AdamsWVJr, HagiusSD, et al. (1992) Efficacy of inactivated whole-virus and subunit vaccines in preventing infection and disease caused by equine infectious anemia virus. J Virol 66 : 3398–3408.
51. HammondSA, CookSJ, LichtensteinDL, IsselCJ, MontelaroRC (1997) Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol 71 : 3840–3852.
52. WeaverEA, CamachoZT, GaoF (2010) Similar T-cell immune responses induced by group M consensus env immunogens with wild-type or minimum consensus variable regions. AIDS Res Hum Retroviruses 26 : 577–584.
53. GilesBM, BisselSJ, DealmeidaDR, WileyCA, RossTM (2012) Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol 19 : 128–139.
54. GilesBM, CrevarCJ, CarterDM, BisselSJ, Schultz-CherryS, et al. (2012) A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis 205 : 1562–1570.
55. GilesBM, RossTM (2012) Computationally optimized antigens to overcome influenza viral diversity. Expert Rev Vaccines 11 : 267–269.
56. GilesBM, RossTM (2011) A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29 : 3043–3054.
57. KotheDL, DeckerJM, LiY, WengZ, Bibollet-RucheF, et al. (2007) Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology 360 : 218–234.
58. Swofford DL (2001) PAUP: Phylogenetic Analysis Using Parsimony and Other Methods. Sunderlan, MA: Sinauer Associates.
59. LerouxC, CadoreJL, MontelaroRC (2004) Equine Infectious Anemia Virus (EIAV): what has HIV's country cousin got to tell us? Vet Res 35 : 485–512.
60. CookRF, CookSJ, LiFL, MontelaroRC, IsselCJ (2002) Development of a multiplex real-time reverse transcriptase-polymerase chain reaction for equine infectious anemia virus (EIAV). J Virol Methods 105 : 171–179.
61. LiuC, CookFR, CookSJ, CraigoJK, EvenDL, et al. (2012) The determination of in vivo envelope-specific cell-mediated immune responses in equine infectious anemia virus-infected ponies. Vet Immunol Immunopathol 148 : 302–310.
62. RamakersC, RuijterJM, DeprezRH, MoormanAF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 : 62–66.
63. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25 : 402–408.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and AdultsČlánek The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and HostČlánek Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for EntryČlánek Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 1- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Importance of Pathogen Load
- Implication of Gut Microbiota in Nonalcoholic Fatty Liver Disease
- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- Helminth-Induced Immune Regulation: Implications for Immune Responses to Tuberculosis
- The M3 Muscarinic Receptor Is Required for Optimal Adaptive Immunity to Helminth and Bacterial Infection
- An Iron-Mimicking, Trojan Horse-Entering Fungi—Has the Time Come for Molecular Imaging of Fungal Infections?
- Modulates the Unfolded Protein Response in during Infection
- Differential Reliance on Autophagy for Protection from HSV Encephalitis between Newborns and Adults
- Identification of HNRNPK as Regulator of Hepatitis C Virus Particle Production
- Parasite Biomass-Related Inflammation, Endothelial Activation, Microvascular Dysfunction and Disease Severity in Vivax Malaria
- : Trypanosomatids Adapted to Plant Environments
- Early Virus-Host Interactions Dictate the Course of a Persistent Infection
- TLR3 Signaling in Macrophages Is Indispensable for the Protective Immunity of Invariant Natural Killer T Cells against Enterovirus 71 Infection
- The Epstein-Barr Virus Encoded BART miRNAs Potentiate Tumor Growth
- Macrophage-Derived Human Resistin Is Induced in Multiple Helminth Infections and Promotes Inflammatory Monocytes and Increased Parasite Burden
- Dissemination of a Highly Virulent Pathogen: Tracking The Early Events That Define Infection
- Variability in Tuberculosis Granuloma T Cell Responses Exists, but a Balance of Pro- and Anti-inflammatory Cytokines Is Associated with Sterilization
- The Shear Stress of Host Cell Invasion: Exploring the Role of Biomolecular Complexes
- The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and Host
- Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for Entry
- Secreted Herpes Simplex Virus-2 Glycoprotein G Modifies NGF-TrkA Signaling to Attract Free Nerve Endings to the Site of Infection
- Preferential Use of Central Metabolism Reveals a Nutritional Basis for Polymicrobial Infection
- A New Family of Secreted Toxins in Pathogenic Neisseria Species
- A Human Type 5 Adenovirus-Based Therapeutic Vaccine Re-programs Immune Response and Reverses Chronic Cardiomyopathy
- Regulation of Oncogene Expression in T-DNA-Transformed Host Plant Cells
- GITR Intrinsically Sustains Early Type 1 and Late Follicular Helper CD4 T Cell Accumulation to Control a Chronic Viral Infection
- Cell Cycle-Independent Phospho-Regulation of Fkh2 during Hyphal Growth Regulates Pathogenesis
- Virus-Induced NETs – Critical Component of Host Defense or Pathogenic Mediator?
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Protective Efficacy of Centralized and Polyvalent Envelope Immunogens in an Attenuated Equine Lentivirus Vaccine
- Transmitted Virus Fitness and Host T Cell Responses Collectively Define Divergent Infection Outcomes in Two HIV-1 Recipients
- Systemic Expression of Kaposi Sarcoma Herpesvirus (KSHV) Vflip in Endothelial Cells Leads to a Profound Proinflammatory Phenotype and Myeloid Lineage Remodeling
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
- DNA Is an Antimicrobial Component of Neutrophil Extracellular Traps
- Uropathogenic Superinfection Enhances the Severity of Mouse Bladder Infection
- Well-Ordered Trimeric HIV-1 Subtype B and C Soluble Spike Mimetics Generated by Negative Selection Display Native-like Properties
- The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots
- Reprogramming of from Virulent to Persistent Mode Revealed by Complex RNA-seq Analysis
- Compartment-Specific and Sequential Role of MyD88 and CARD9 in Chemokine Induction and Innate Defense during Respiratory Fungal Infection
- Bacterial Flagella: Twist and Stick, or Dodge across the Kingdoms
- Elucidation of the RamA Regulon in Reveals a Role in LPS Regulation
- IL-1α Signaling Is Critical for Leukocyte Recruitment after Pulmonary Challenge
- Chronic Filarial Infection Provides Protection against Bacterial Sepsis by Functionally Reprogramming Macrophages
- Specificity and Dynamics of Effector and Memory CD8 T Cell Responses in Human Tick-Borne Encephalitis Virus Infection
- Promiscuous RNA Binding Ensures Effective Encapsidation of APOBEC3 Proteins by HIV-1
- Viral Activation of MK2-hsp27-p115RhoGEF-RhoA Signaling Axis Causes Cytoskeletal Rearrangements, P-body Disruption and ARE-mRNA Stabilization
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Infections in Humans and Animals: Pathophysiology, Detection, and Treatment
- : Trypanosomatids Adapted to Plant Environments
- Environmental Drivers of the Spatiotemporal Dynamics of Respiratory Syncytial Virus in the United States
- Dengue Virus RNA Structure Specialization Facilitates Host Adaptation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



