-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen
Essentially all Gram-negative pathogens are reliant on specific transport machineries termed binding protein-dependent transporters (BPDTs) to transport solutes such as amino acids, sugars and metal ions across their membranes. In this study we investigated AfuABC, a predicted iron-transporting BPDT found in many bacterial pathogens. We show by structural and functional approaches that AfuABC is not an iron transporter. Instead, AfuABC is a trio of proteins that bind and transport sugar-phosphates such as glucose-6-phosphate (G6P). In doing so, we present the first structural solution of a G6P-specific transport protein and add to the few known unique machineries for sugar-phosphate uptake by bacteria. Furthermore, we show that AfuABC is required by the intestinal pathogen C. rodentium to effectively transmit between mice and re-establish infection, leading us to propose that the transport of sugar-phosphates is an important part of general bacterial pathogenesis.
Published in the journal: Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen. PLoS Pathog 11(8): e32767. doi:10.1371/journal.ppat.1005107
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005107Summary
Essentially all Gram-negative pathogens are reliant on specific transport machineries termed binding protein-dependent transporters (BPDTs) to transport solutes such as amino acids, sugars and metal ions across their membranes. In this study we investigated AfuABC, a predicted iron-transporting BPDT found in many bacterial pathogens. We show by structural and functional approaches that AfuABC is not an iron transporter. Instead, AfuABC is a trio of proteins that bind and transport sugar-phosphates such as glucose-6-phosphate (G6P). In doing so, we present the first structural solution of a G6P-specific transport protein and add to the few known unique machineries for sugar-phosphate uptake by bacteria. Furthermore, we show that AfuABC is required by the intestinal pathogen C. rodentium to effectively transmit between mice and re-establish infection, leading us to propose that the transport of sugar-phosphates is an important part of general bacterial pathogenesis.
Introduction
Competition between host cells and pathogenic bacteria for diverse substrates ranging from metals to vitamins can often influence the outcome of infection. As a result, pathogens have developed a variety of nutrient uptake mechanisms to increase their competitiveness and hence their ability to colonize and successfully infect their hosts [1]. One such example of these nutrient uptake mechanisms are the binding-protein dependent transporters (BPDTs). BPDTs are ubiquitous in bacteria and rely on the presence of a soluble ligand binding protein that directs a specific substrate to an integral membrane permease/ATPase complex. The ensuing tripartite complex then couples the hydrolysis of cytosolic ATP to the influx of the ligand into the cytosol for downstream uses [2]. BPDTs target diverse ligands such as metal ions, amino acids and sugars and are recognized as both virulence determinants and therapeutic targets in many bacterial infections [2].
In this study, we investigate AfuABC (Actinobacillus ferric uptake ABC), a BPDT originally identified as an iron-specific transporter in Actinobacillus pleuropneumoniae, a Gram-negative upper respiratory tract pathogen [3]. The locus afuABC encodes 3 polypeptides corresponding to a conventional BPDT–AfuA (a periplasmic binding protein or PBP), AfuB (an inner membrane permease), and AfuC (a cytosolic ATPase). Although AfuABC is annotated as an Fe3+-binding transporter, its ligand has been the subject of controversy [4–6]. Initially of interest given the central role of iron piracy to the success of bacterial pathogens, AfuABC has not been investigated on a functional level and the role of currently annotated AfuABC homologues in virulence of other bacterial species has not been characterized. Since several studies have identified afuABC as transcriptionally upregulated during bacterial infection [7–9], we investigated AfuABC on a structural and functional basis to definitively identify its ligand and further illuminate its role in pathogenesis. We found that AfuABC is not an iron transporter as previously described and instead that this BPDT is specific for phosphorylated sugars. AfuABC contributed to virulence and was key to transmission success in a mouse model of C. rodentium infection, highlighting its role in nutrient uptake during bacterial pathogenesis.
Results
AfuA is a sugar-phosphate specific periplasmic binding protein
To structurally characterize the ligand of AfuA, a 1.6Å crystal structure of ligand-bound A. pleuropneumoniae AfuA purified from E. coli was solved by use of sulfur single wavelength anomalous diffraction. AfuA is a class II/cluster D periplasmic binding protein with two globular α/β domains linked by a dual β-stranded hinge [10] (S1 Table and Fig 1A). In the binding cleft, we unexpectedly observed positive electron density and identified glucose-6-phosphate (β-G6P) as the bound ligand with a small (~5%) portion of density at carbon 2 corresponding to mannose-6-phosphate (β-M6P) (Fig 1B). In the binding cleft, His205, Asp206 and Glu229 interact multivalently with the sugar ring while Ser37 and Thr150 form hydrogen bonds with the phosphate moiety of G6P (Fig 1B). Isothermal titration calorimetry (ITC) validated the binding affinity of A. pleuropneumoniae AfuA for β-G6P at 24 nM with favourable values of enthalpy and entropy change (S2 Table and Fig 1C and 1D). In order to probe the ligand-binding site of AfuA, site-directed mutants were generated for putative binding residues. The mutations T150A and S37A lowered but did not eliminate binding to G6P, whereas mutations to the sugar-binding residues (H205A, D206A and E229A) completely abrogated the interaction (Table 1). This indicates that the hydrogen-bonding capacity of the sugar-coordinating residues is the critical determinant in mediating binding of potential sugar phosphates.
Fig. 1. AfuA is a sugar-phosphate specific periplasmic binding protein. 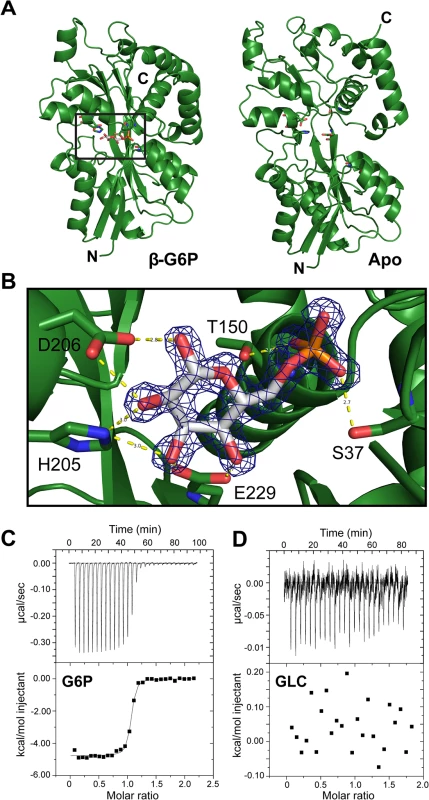
(A) Ribbon diagrams of A.pleuropneumoniae AfuA complexed with either no ligand (right) or β-G6P (left). (B) Cleft region of AfuA depicting 2F0-Fc map contoured at 2σ around β-G6P. Residues used for mutagenic studies are indicated with text labels. Predicted hydrogen bonds are drawn as yellow dashed lines with distances indicated in angstroms (Å). (C) Representative ITC curve for a positive AfuA-ligand interaction. The curve shown is from a titration of A.pleuropneumoniae AfuA with a 10:1 ratio of G6P. (D) Representative ITC curve illustrating lack of an AfuA-ligand interaction. The curve shown is from a titration of A.pleuropneumoniae AfuA with a 10:1 ratio of glucose. Tab. 1. Binding constants between A.pleuropneumoniae AfuA and G6P. 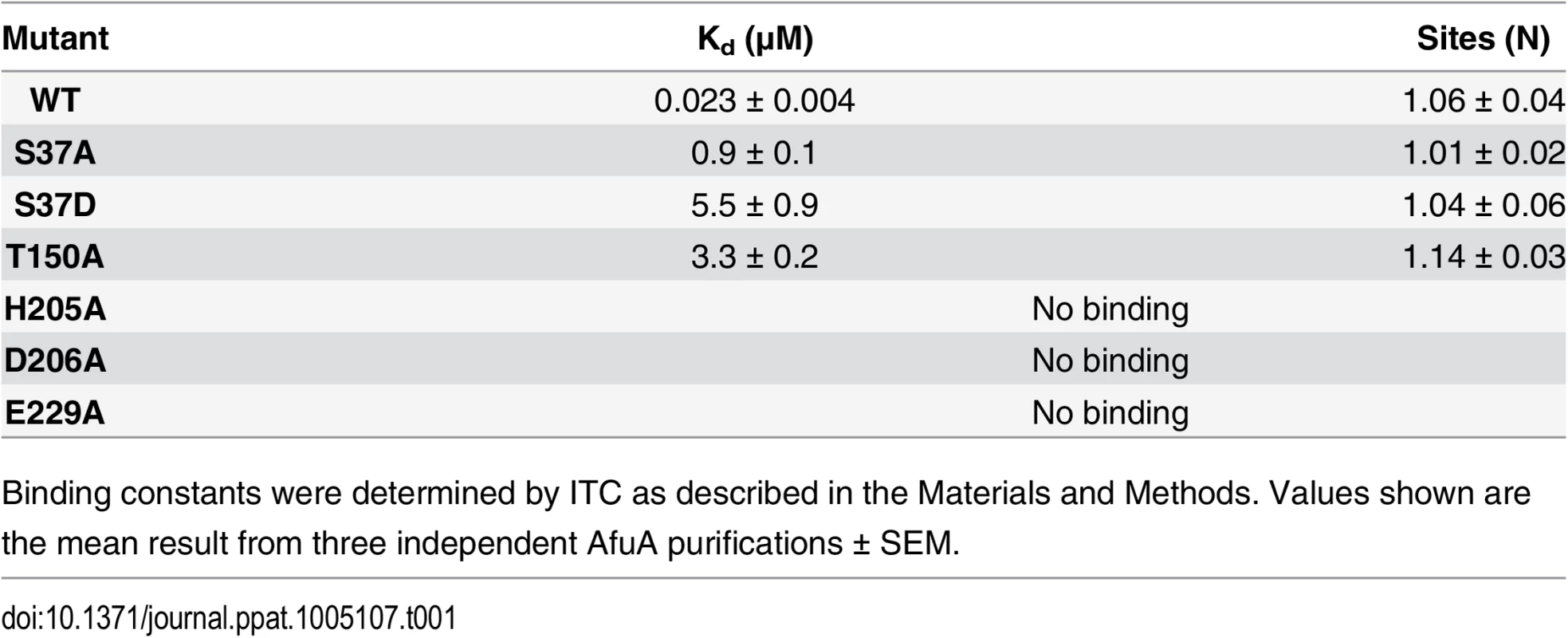
Binding constants were determined by ITC as described in the Materials and Methods. Values shown are the mean result from three independent AfuA purifications ± SEM. To gain insight into the mechanism of ligand capture by this PBP, a structure of apo (open)-AfuA was solved to 1.6Å. The globular domains of apo-AfuA are held at a 29.7° angle relative to the closed form (Fig 1A). The binding pocket of AfuA contains both electropositive and electronegative regions, corresponding to the regions of the pocket that bound the phosphate or sugar moiety of G6P respectively (Fig 2A, 2B and 2C). A subsequent ligand screen by ITC with structurally similar molecules revealed that three other sugar-phosphates including fructose-6-phosphate (F6P), sedoheptulose-7-phosphate (S7P) and M6P had affinities of 8, 57 and 960 nM respectively for AfuA (Table 2 and S1 Fig). F6P binding affinities of the single site AfuA mutants displayed the same trend as for G6P, suggesting a similar binding mechanism between the two ligands (S3 Table). Structures of AfuA bound to F6P (2.0Å) and S7P (1.3Å) were also solved to confirm ligand coordination. Similar to the G6P structure, both F6P and S7P were found in the β-anomer conformation (Fig 2D and 2E). Between the three holo-AfuA structures, strand and amino acid placement and conformation were identical (RMSD < 0.4Å between Cα chains). Interestingly, S7P was captured by AfuA in its furan form and not pyran as typically depicted. S7P and its derivatives can exist as furan forms in vivo [11], indicating that this ligand structure is biologically relevant. AfuA did not bind other sugar-phosphates such as glucose-1-phosphate and ribose-5-phosphate, demonstrating a high level of selectivity for ligands (Table 2 and S1 Fig). No interaction between AfuA and glucose, fructose or inorganic phosphate could be detected (Table 2 and S1 Fig). These data collectively show that AfuA is a PBP that is highly specific for four sugar-phosphates involved in central metabolism.
Fig. 2. Electrostatic surface mapping of AfuA and additional ligand-bound structures. 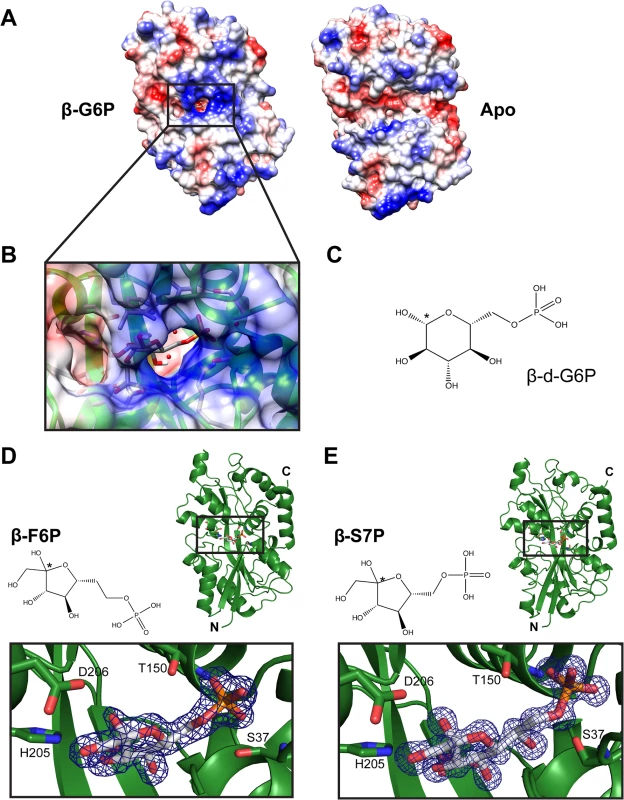
(A) Electrostatic surface potential of apo (right) and G6P-bound (left) AfuA. Potential values range from -10 (red) to +10 kJ/mol·e (blue). (B) Magnified view of G6P-AfuA binding cleft with electrostatic surface map overlay. Residues proximal to the G6P molecule are shown as sticks. Water molecules are shown as red spheres. (C) Chemical structure of β-G6P. * denotes the anomeric carbon. (D, E) Ribbon diagrams showing A.pleuropneumoniae AfuA in complex with (D) F6P or (E) S7P. Inset boxes below each ribbon structure show the 2F0-Fc map contoured at 2σ around each ligand. Tab. 2. Binding constants between WT A.pleuropneumoniae AfuA and screened molecules. 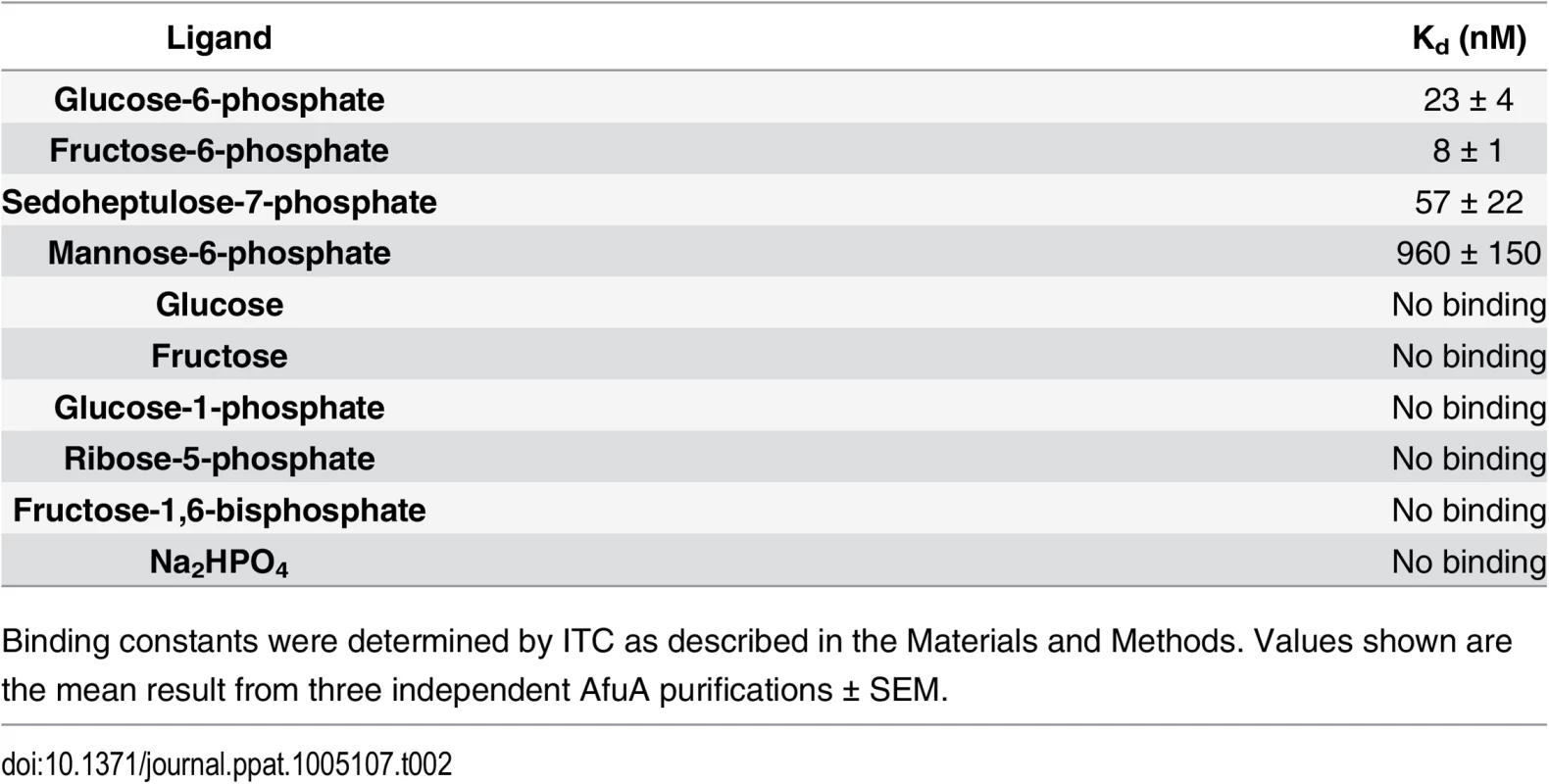
Binding constants were determined by ITC as described in the Materials and Methods. Values shown are the mean result from three independent AfuA purifications ± SEM. AfuABC is a bona fide sugar phosphate-specific BPDT
Since AfuA binds sugar-phosphates, we queried whether AfuABC could recapitulate sugar-phosphate transport in a bacterium that could not transport these substrates. The only other sugar-phosphate uptake system identified thus far in Gram-negative bacteria is the two component system UhpABCT. UhpT is an inner membrane antiporter that couples the influx of hexose phosphates such as G6P and F6P to the efflux of inorganic phosphate [12]. The expression of UhpT is regulated by the transcription factor UhpA, which is activated in response to periplasmic sugar-phosphates through the inner membrane sensor kinase complex UhpB/UhpC [13]. E. coli ΔuhpT were unable to replicate in medium where G6P or F6P were the only carbon source, confirming UhpT is the only hexose-phosphate specific transporter in BW25113 (Figs 3A, 3B and S2) [14]. When ΔuhpT cells were transformed with a vector carrying the A. pleuropneumoniae afuABC operon, a significant increase in growth rate on M9 + G6P or F6P agar media compared to empty vector-transformed cells was observed (S4 Table and Figs 3A and S2A). This growth complementation was reproduced with liquid media of the same supplementation (Fig 3B). These results demonstrate that AfuABC is sufficient to transport G6P and F6P as substrates for bacterial metabolism and growth. We did not observe complementation with afuA or afuBC alone, consistent with a model where AfuABC acts as a BPDT to transport substrates from the periplasm into the cytosol (Fig 3A).
Fig. 3. afuABC rescues a nutrient-limited E.coli strain. 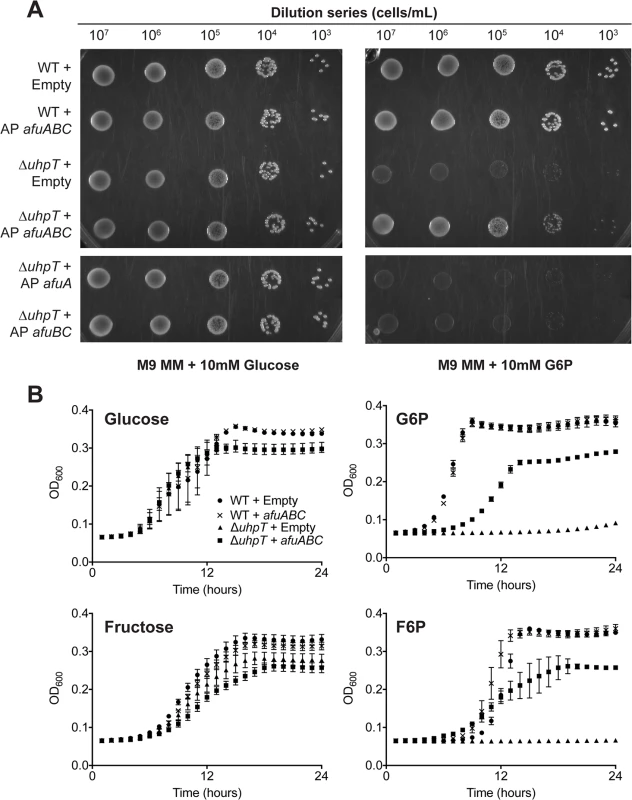
(A) Solid media complementation of ΔuhpT in M9 minimal media (M9 MM) supplemented with 10mM glucose or G6P. 5μL drops were plated of the indicated dilutions and grown at 37°C for 30 hours before imaging. Plates are representative of n = 3 transformations. (B): OD600 readings of E.coli growth over 24 hours at 37°C in M9 minimal medium supplemented with 10mM glucose, G6P, fructose or F6P. Readings were taken every 15 minutes–data shown is parsed to hourly readings for clarity. Curve legend is the same in all panels and is indicated in the first panel. Error bars represent SEM of cell growth from n = 3 transformations. Conservation of AfuABC in bacteria
Given the novel function of AfuABC as a sugar-phosphate transporter, we next sought to identify whether this machinery was conserved in other bacteria. The three crystal structures provided a means to identify the sugar-coordinating residues (H205, D206 and E229) as the critical determinants in mediating binding of sugar-phosphates. These residues were subsequently used to define a potential sugar-phosphate binding motif, which in turn was used to search for and identify putative homologues of AfuA and hence the AfuABC operon (S5 Table and Fig 4). The majority of hits were found in Gram-negative bacteria. In particular, AfuABC was identified in many human pathogens from the Pasteurellaceae (e.g. Haemophilus influenzae), the Vibrionaceae (e.g. Vibrio cholerae) and the Enterobacteriaceae (e.g. E. coli O157:H7). Surprisingly, a minority of putative afuABC loci were also found in Gram-positive bacteria such as Clostridium tetani and other members of the Firmicutes. The conservation of AfuABC over a broad spectrum of bacterial genera suggests sugar-phosphate transport is required across a wide range of colonization niches and conditions.
Fig. 4. Phylogenetic tree of AfuA. 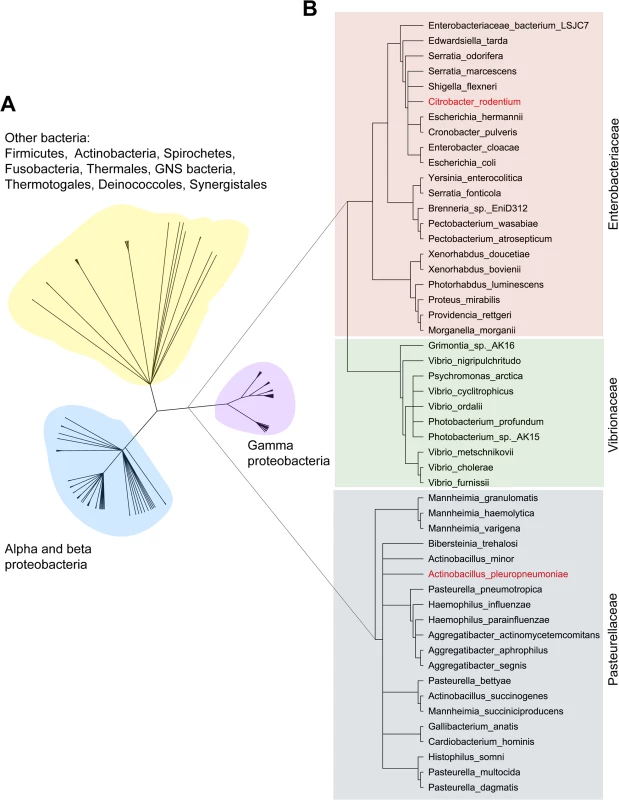
(A) The complete tree of all 268 putative AfuA homologues. Based on the tree, homologues were divided into: gammaproteobacteria (83 hits, purple), alpha and betaproteobacteria (121 hits, cyan) and other bacteria (64 hits, yellow). (B) The phylogenetic tree of AfuA in gammaproteobacteria. The 83 gammaproteobacteria AfuA homologues were reduced to 51 by removing sequences that shared >95% identity. These hits were divided into: 21 in the Enterobacteriaceae (red), 10 present in the Vibrionaceae (green), and 20 in the Pasteurellaceae (grey). The two AfuA homologues used in this study are highlighted in red. AfuABC contributes to the pathogenesis of C. rodentium
To examine whether AfuABC plays a role in bacterial pathogenesis, we utilized the intestinal murine pathogen C. rodentium, a commonly used model for EHEC O157:H7 as well as other attaching and effacing pathogens [15]. In EHEC, afuABC is located on the putative pathogenicity island OI-20. OI-20 also contains a recently identified fucose-sensing two-component regulatory system, suggesting this genomic island may play a role in carbohydrate sensing and/or acquisition [16]. OI-20 is conserved to >83% amino acid identity in C. rodentium ICC168 (Fig 5A), and ITC and growth assays confirmed that C. rodentium AfuABC functioned as a sugar-phosphate transporter [17,18] (S6 Table and S3 Fig). We generated an AfuA knockout strain of C. rodentium (ΔafuA) which did not display significant changes in planktonic growth, intestinal epithelial adherence/pedestal formation, type III effector secretion and bacterial tissue localization (Figs 5B–5E and S4).
Fig. 5. C. rodentium requires AfuA during host colonization. 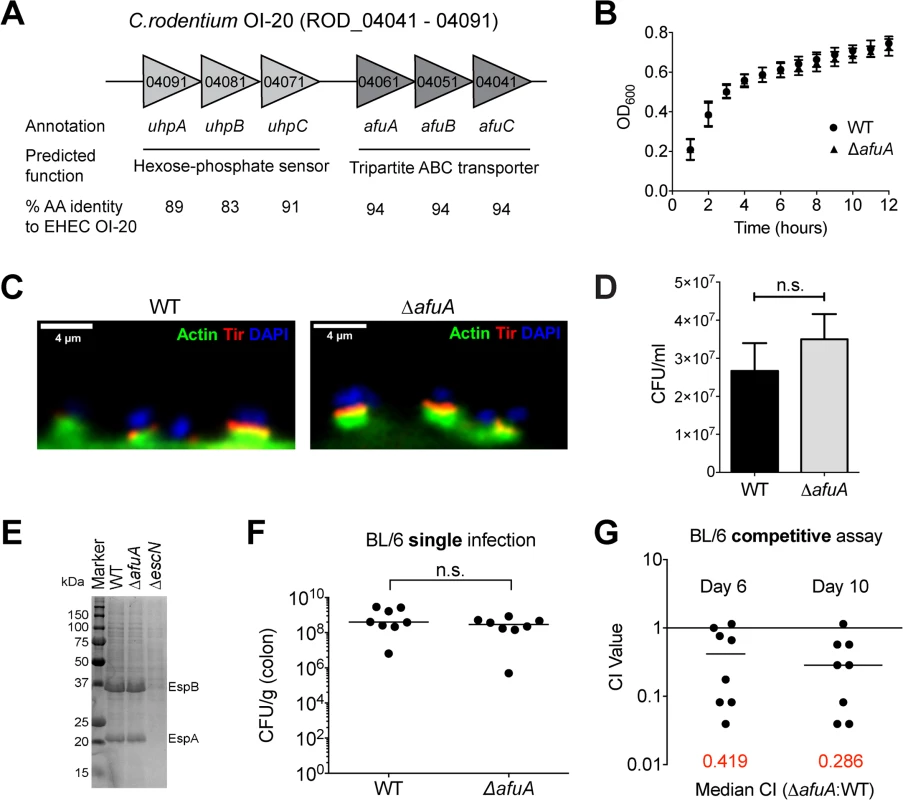
(A) Gene organization, annotation and predicted functions for genomic region corresponding to OI-20 in C. rodentium. Percent identity to EHEC O157:H7 was calculated with ClustalW2 using the entire predicted sequence of each protein. (B) Growth curves in LB broth for WT and ΔafuA C. rodentium. Cultures were tracked with OD600 readings for the indicated amount of time (n = 2). (C) Actin pedestal formation by C. rodentium WT or ΔafuA. HeLa cells were infected for 8hrs with C. rodentium then stained with phalloidin (green), anti-C. rodentium Tir (red) and DAPI to detect DNA (blue). (D) Adherence of C. rodentium WT and ΔafuA to HeLa cells after 8hrs infection. Counts shown represent adherent bacteria on a monolayer of cells (see Methods) and are mean values ± SEM (n = 3 infections). (E) SDS-PAGE of type-3 secretion effectors by C. rodentium WT, ΔafuA and ΔescN after growth in DMEM. ΔescN is a negative control strain that is secretion-deficient. (F) C57BL/6 mice were orally infected with C. rodentium WT or ΔafuA and bacterial colonic burdens determined at 10 dpi. Data were combined from two independent experiments. Each symbol represents one animal (n = 8 for each infection). The median is indicated. (G) Competitive index (CI) of simultaneous C. rodentium WT and ΔafuA infection in C57BL/6 mice in stool 6 dpi, and colonic tissue 10 dpi. Data were combined from two independent experiments (n = 8). A CI < 1 indicates the WT strain outcompeted the knockout (6 dpi median CI = 0.49, p = 0.0313; 10 dpi median CI = 0.38, p = 0.0156). Oral infection of C57BL/6 mice by C. rodentium WT or ΔafuA yielded no significant difference in bacterial colonic tissue burdens between the two groups at 10 days post infection (dpi) (Fig 5F). However, C. rodentium is a highly adapted mouse pathogen and strains lacking important virulence factors can still reach high colonic burdens in individual infections [19,20]. This prompted us to perform a competitive index (CI) assay to measure the comparative fitness between WT and ΔafuA in a simultaneous infection. ΔafuA was significantly impaired compared to WT in stool pellets at 6 dpi (median CI = 0.42, p = 0.031) as well as in colonic tissues at 10 dpi (CI = 0.29, p = 0.015) (Fig 5G) indicating that AfuABC is required for C. rodentium optimal colonization of the intestine. A competitive defect of this magnitude is comparable to those previously reported for C. rodentium strains lacking known secreted effectors [19–21].
AfuABC substrates are present in the infected mouse gut
The primary role of sugar-phosphates as cytosolic metabolites suggests that they would not be present in the extracellular environment and hence available for an extracellular pathogen such as C. rodentium. However, recent evidence has emerged indicating that they are present in the intestine and their availability is dependent on the presence of the host microbiota [22,23]. Since previous studies have not defined the concentration of these molecules, we used targeted liquid chromatography/tandem mass spectrometry to quantify the level of AfuABC substrates accessible to C. rodentium in control and C. rodentium-infected mice colons. Both validated substrates of AfuABC (G6P and F6P) as well as the other AfuA ligands (M6P and S7P) were present in all locations sampled (luminal contents, stool pellets and mucus scrapings) at micromolar quantities (Table 3 and Fig 6). These amounts exceed the Kd of C. rodentium AfuA for G6P or F6P, indicating that these sugar-phosphates are present in the mouse intestine at levels sufficient to be utilized by the bacteria.
Fig. 6. AfuABC substrates are present in the intestine during C. rodentium infection. 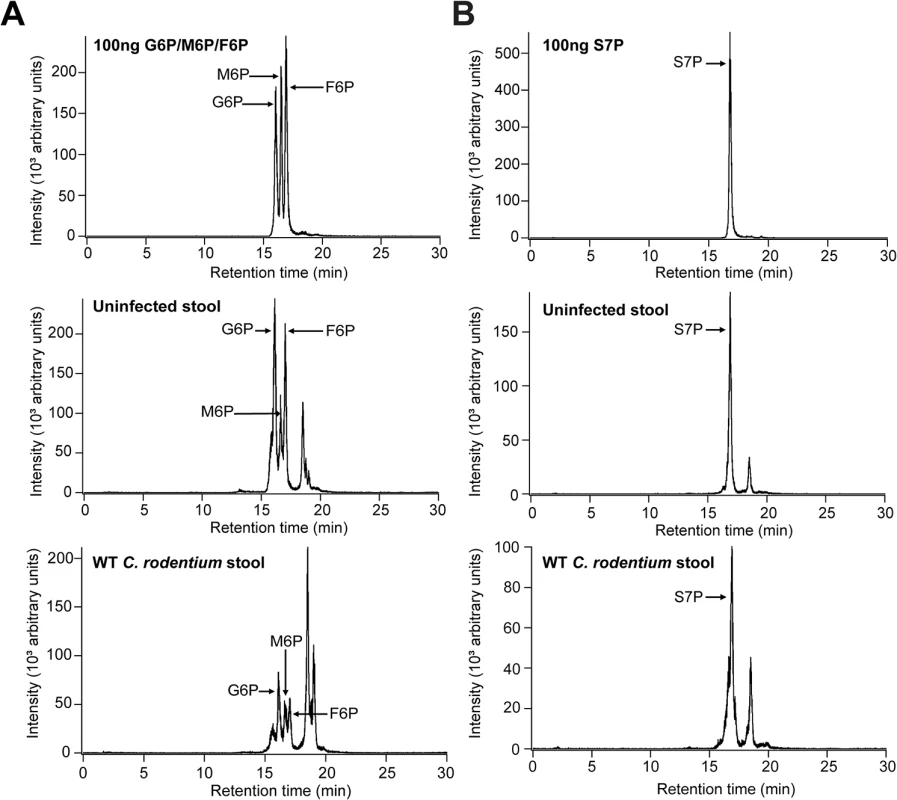
(A) Representative LC-MS/MS chromatograms with indicated peaks for 100ng G6P/F6P/M6P mix (top), uninfected stool (middle) and C. rodentium WT infected stool (bottom). (B) S7P peaks. G6P/F6P/M6P were identified with a 259→97 ion transition and S7P was identified with a 289→97 ion transition. Tab. 3. LC-MS/MS quantification of AfuA substrates across two sets of <i>C</i>. <i>rodentium</i>-infected mice. 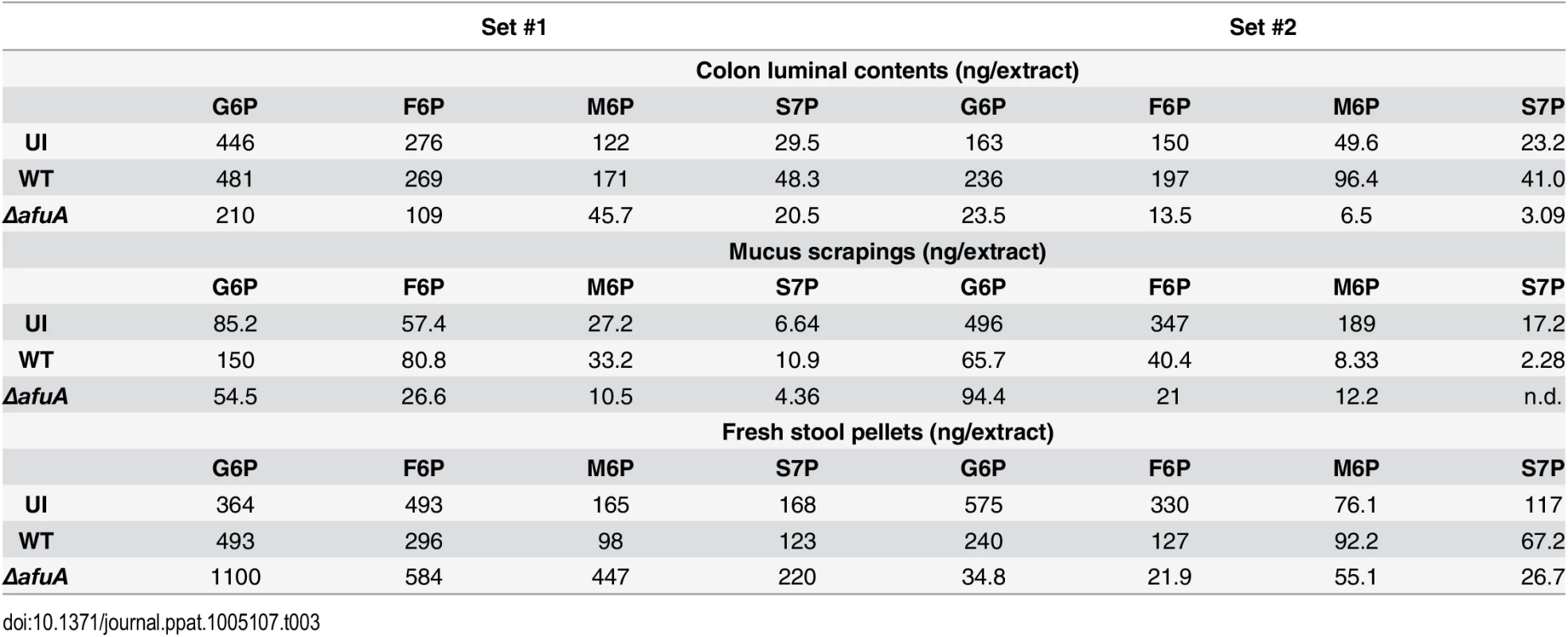
AfuABC plays a critical role in transmission success of C. rodentium
Typical assays with C. rodentium involve the oral delivery of large doses of bacteria to ensure reproducible infections. As such, they fail to mimic how C. rodentium and other enteric bacterial pathogens typically spread via the oral-fecal route in wild host populations. The high levels of sugar-phosphates present in shed stool pellets led us to examine a potential role for AfuA in the transmission of C. rodentium in a murine population. Index mice were orally infected with either WT or ΔafuA and after six days, co-housed with two naïve mice for 48 hours to allow transmission via coprophagy (Fig 7A). Seven out of eight co-housed mice exposed to WT-infected index mice were successfully infected, with heavy pathogen burdens recovered from both their colonic tissues and lumen (Fig 7B and 7C). In contrast, only half of the ΔafuA exposed mice showed any detectable C. rodentium within their luminal contents, and only two out of eight mice showed any C. rodentium adherent to their colonic tissues (Fig 7B and 7C). These results indicate that AfuABC does not directly affect the growth or infective capacity of C. rodentium but rather impacts its ability to compete and survive within the host intestinal environment and ultimately transmit to new hosts.
Fig. 7. C. rodentium requires AfuA for efficient host-to-host transmission. 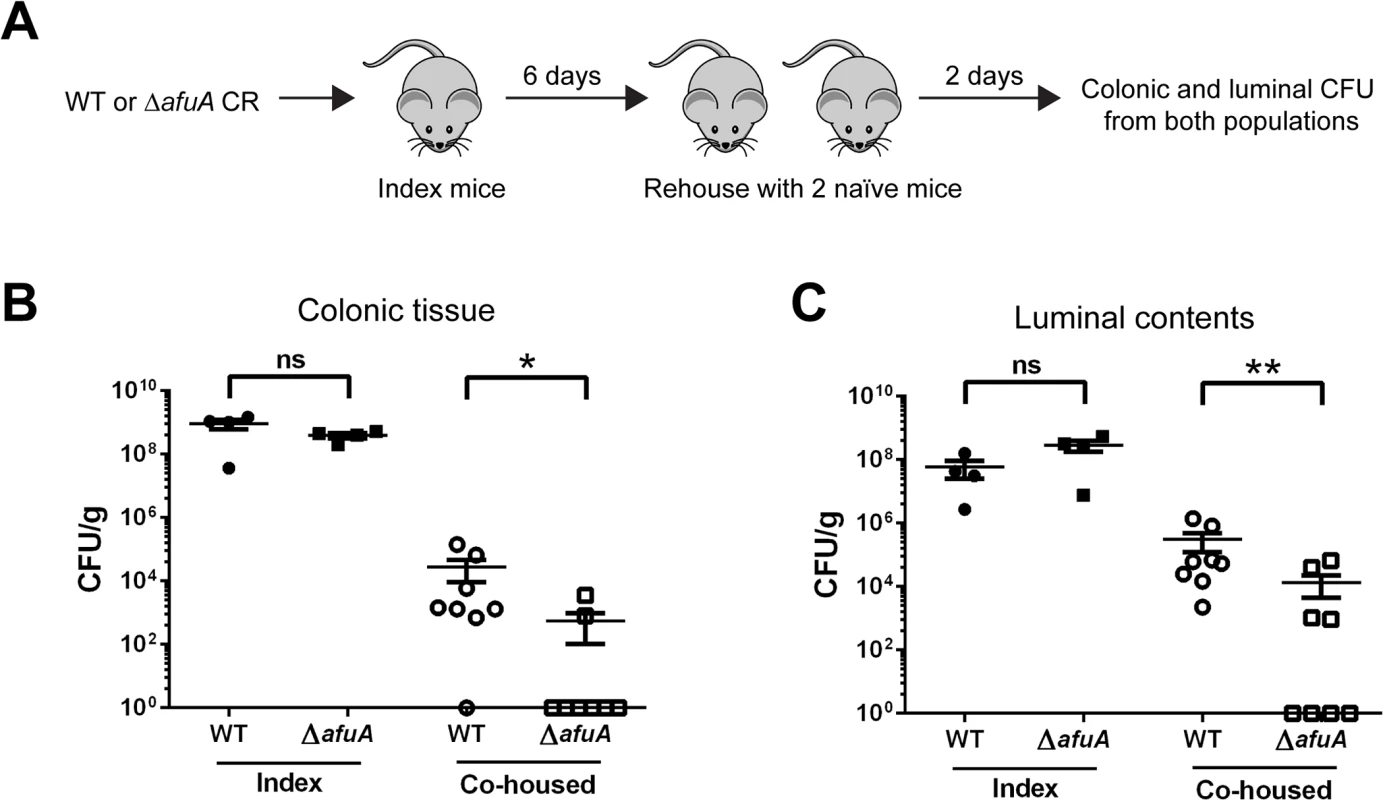
(A) Infection schematic for quantification of C. rodentium host-to-host transmission. C57BL/6 index mice were orally infected with C. rodentium WT or ΔafuA. At 6 dpi, index mice were added to a cage containing two naïve mice and 48h post-exposure, bacterial luminal and colonic burdens determined. (B, C) Ability of C. rodentium WT or ΔafuA to transmit between index and co-housed C57BL/6 mice. Bacterial colonic (B) and luminal (C) burdens were determined for both index and co-housed mice. Data were combined from two independent experiments. Each symbol represents one animal (n = 4 (index), 8 (co-housed) for each infection). The median is indicated.* = p<0.05. Discussion
We conclude that AfuABC has been misannotated and is a sugar-phosphate transporter that contributes to the virulence of an enteric pathogen. We have defined a potential sugar-phosphate binding motif in AfuA and used it to identify a broad conservation of AfuABC across bacteria, including both Gram-negative and-positive species. Although AfuA was not required for in vivo colonization by C. rodentium or functional type III secretion, loss of AfuA significantly impacted its ability to be transmitted between hosts, indicating that sugar-phosphate uptake is required for a functional C. rodentium infective life cycle.
Competition between the host and bacteria for iron is an established concept in bacterial pathogenesis [24]. Therefore, a locus such as afuABC that had been misannotated as a Fe3+ transporter might have been assumed to contribute to virulence without direct experimental evidence indicating such a function. Previous data implicating AfuABC in iron uptake was based on the afuABC-mediated rescue of ΔaroB (siderophore-deficient) E. coli on iron-limited media [3]. However, since aroB encodes a metabolic enzyme that has dual roles in both carbon metabolism (the shikimate pathway) and siderophore synthesis, it is not clear whether the afuABC complementation of ΔaroB observed was only attributable to a complementation of iron uptake in this strain. Since the sole function of UhpT is in hexose-phosphate uptake, the assay we used in this study is more specific and directly implicates AfuABC in transport of these substrates instead of iron. Based on the results presented here, AfuABC does contribute to bacterial pathogenesis but this is achieved through sugar-phosphate and not iron uptake. It remains an open question whether other predicted iron transporters may, akin to AfuABC, be misannotated.
The structures shown here are the first structural data for a non-enzymatic sugar-phosphate (i.e. G6P) binding protein. In AfuA, the essential residues His205 and Asp206 form multivalent hydrogen bonds to the sugar moiety of ligands and function as the primary determinants of ligand selectivity. On the opposing end of the pocket, redundant phosphate-coordinating residues such as Ser37 cooperate to hold the phosphate moiety of ligands. This bipartite binding cleft is able to discriminate between isomers of the same sugar-phosphate (G6P vs. G1P) and between differing orientations of side chains around the carbohydrate ring (F6P vs. R5P). Our observation that both furan (F6P and S7P) and pyran (G6P) ligands could bind AfuA suggests the position of hydroxyl determinants is more important than steric fitting of the overall ring structure into the binding pocket.
We propose a two-step mechanism for ligand capture by AfuA. Substrates such as G6P or F6P first dock to the protein via hydrogen bonding to residues in the electronegative region of the binding cleft. Next, the negatively charged phosphate group of the ligand serves as the target of a simultaneous rotation and closure of the two globular domains of AfuA, generating a stable AfuA-ligand complex. This cinching mechanism is driven by the electrostatic attraction of the Arg/Lys-rich regions of the binding pocket (which also includes residues such as Ser37 and Thr150). This mechanism is compatible with our mutagenesis data, as all mutations to sugar-coordinating residues completely eliminated binding, suggesting initial recognition of the sugar ring is the determining step in ligand capture. Other homologues of AfuA may display altered substrate specificity and thus might provide more insight into the mechanism of sugar recognition and coordination by PBPs.
We have confirmed AfuABC functions as a novel sugar-phosphate uptake mechanism. The only other known transporter in Gram-negative bacteria for sugar-phosphates is UhpABCT [4]. The primary distinction between these two systems is that AfuABC is an active transport system that relies on cytosolic hydrolysis of ATP, whereas UhpT-mediated transport is dependent on the maintenance of a phosphate/sugar-phosphate concentration gradient. Since AfuABC does not rely on a concentration gradient nor the concomitant phosphate efflux which is required by UhpT, uptake of sugar-phosphates through AfuABC may be preferable in a location where nutrients are constantly in flux (such as the intestinal lumen and mucosa). AfuABC-mediated transport could potentially allow a pathogen to simultaneously conserve phosphate stores and take advantage of nutrients for which other bacteria may lack an appropriate transport mechanism [4]. It is established that sugar-phosphate uptake is important for intracellular pathogen replication [25–27]. Our study provides evidence that the same is true for extracellular pathogens and provides reference values for these ligands in the mouse intestine. While the most likely downstream fate of transported sugar-phosphates through AfuABC is to support bacterial metabolism, recent studies have identified both phosphorylated and cognate sugars as molecules that can act as signaling intermediaries in the host-bacterial interaction [16,28]. The close genetic proximity of AfuABC to one of these systems in both C. rodentium and EHEC O157:H7, the FusKR system, implies that the substrates targeted by AfuABC could also be involved in other roles besides pure carbon and phosphate metabolism.
The identification of AfuABC homologues in bacteria that colonize a variety of niches, such as the gastrointestinal and respiratory tracts, suggests uptake of sugar-phosphates plays a role in their competitive establishment on these surfaces. Since the microbiota is known to modulate the nutritional environment of these areas, we propose that AfuABC could aid in pathogen competition with host-associated commensal microbes during colonization. Notably, the clear dependence of successful pathogen transmission on the presence of AfuABC supports this concept since our assays were performed at early time points, when C. rodentium would need to overcome colonization resistance by the resident commensal microbes in order to successfully infect the host [29]. The major contributing source of sugar-phosphates to the gut metabolome remains unclear. It is likely that a variety of host and microbial factors contribute to the pool of these substrates (e.g. from epithelial cell turnover and bacterial lysis). One potential source of sugar phosphates is the mucin family of glycosylated cell surface proteins. The contributions of mucin-derived substrates, in particular fucose and sialic acid, to bacterial infections have recently been recognized in several studies [16,30–32]. Intriguingly, when EHEC is grown in the presence of mucus, the OI-20 pathogenicity island is upregulated, indicating a potential connection between AfuABC and the presence of mucin [33]. Mice lacking the predominant intestinal mucin Muc2 mice are hypersusceptible to C. rodentium infection due to decreased mucin-dependent bacterial clearance and it will be interesting to determine if this and other mucins are involved in generation of free sugar-phosphates in the gut [34]. Further characterization of the source and flux of concentration of sugar-phosphates during dysbiosis will help to illuminate the potential role of the microbiota in generating substrates for AfuABC and the contribution of sugar-phosphate transport within mammalian hosts.
Materials and Methods
Bacterial strains and growth
All bacteria were grown at 37°C on Luria-Bertani (LB) agar plates or in LB broth with shaking. A list of strains and plasmids used in this study is included in S7 Table. Where appropriate, antibiotics were supplemented at the following concentrations: kanamycin – 50μg/mL, ampicillin – 50μg/mL, streptomycin—100μg/mL.
Cloning and expression of AfuA and AfuABC
AfuA was initially cloned into a pET26b expression vector from A.pleuropneumoniae genomic DNA. For the growth assays, complementation vectors with low (PSC101) copy numbers were used. afuA or afuABC were amplified from A. pleuropneumoniae genomic DNA and cloned into each backbone by restriction free or exponential megaprimer cloning, respectively [35,36]. Plasmid insert sequences were confirmed by forward and reverse capillary sequencing (The Centre for Applied Genomics, Toronto, Canada).
Purification of AfuA
E. coli transformed with an AfuA expression plasmid were grown overnight in LB-kanamycin and used to initiate liter-scale volumes of the same media. Population density was estimated by OD600 and IPTG was added to a concentration of 1mM when the OD600 reached 0.6. Cells were induced at 20°C for 16 hours, after which they were spun down at 4000rpm at 4°C. Pellets were resuspended in a 20% sucrose solution to initiate osmotic shock and spun down again at 4000rpm. Sucrose pellets were resuspended in a 5mM MgCl2 lysis solution with added protease inhibitors (Roche Applied Sciences, Canada) and allowed to incubate on ice for at least 20 minutes. The fluid was then centrifuged at 30,000rpm for 40 minutes at 4°C in an ultracentrifuge (Beckman Coulter, USA). The supernatant was passed through a 0.45μM syringe filter and batch bound for >1 hour to Ni-NTA resin (Pierce, USA) at 4°C with shaking. The solution was then passed through a gravity column and washed with wash buffer (20mM imidazole, 50mM Tris pH 8 and 300mM NaCl). A 6M solution of guanidine hydrochloride was used to denature bound protein after which a 3M Gd-HCl solution and finally pure wash buffer was applied to allow bound protein to refold. Protein was eluted with elution buffer (wash buffer with 400mM imidazole) and purity assessed with denaturing SDS-PAGE followed by Coomassie blue staining. Fractions with detectable AfuA were pooled and dialyzed with added thrombin (Sigma-Aldrich, USA) in wash buffer to simultaneously remove excess imidazole and cleave the N-terminal 6xHis tag from AfuA. The solution was spun down and concentrated in a 10kDa cutoff centrifugal filter (Millipore, USA). Samples were further purified on a Superdex S75 10/300 gel filtration column (GE Healthcare, USA) pre-equilibrated with a buffer consisting of 20mM Tris (pH 8) and 100mM NaCl. Fraction purity was assessed by SDS-PAGE with Coomassie Blue staining. Since we initially investigated AfuA in the context of iron capture, initial protein purifications in this study leading to the structure of G6P-AfuA were performed in the absence of guanidine. All other purifications used for ITC and crystallography were denaturing preparations.
Site-directed mutagenesis
To generate single-residue mutants of AfuA, site-directed mutagenesis was performed on the original pET26b-APAfuA vector. A short (17 cycle) PCR reaction was used to generate the desired mutation in the template plasmid. PCR products were digested with DpnI at 37°C for 4–6 hours to cleave undesired template plasmid and were then used to transform competent E. coli (NEB Turbo) for selection. Mutations were confirmed by forward and reverse capillary sequencing (The Centre for Applied Genomics, Toronto, Canada).
Isothermal titration calorimetry (ITC)
ITC was performed using a VP-ITC instrument (Microcal, GE Healthcare). Runs consisted of ligand loaded into the injection syringe titrated against purified protein loaded into the sample cell. All solutions were pre-dialyzed in buffer for at least 12 hours to mitigate effects of dilution. The buffer used was the same as the gel filtration buffer described above and power changes were measured relative to a reference cell filled with distilled water. Runs consisted of a 10 : 1 ligand:protein ratio, with efforts made to adhere to a 200μM:20μM concentration standard. Data used to calculate binding constants were referenced against runs performed with ligand alone to control for the heat of ligand solvation.
Crystallization, data collection and structure solution
For crystallization trials, AfuA was purified as described above and concentrated to at least 10mg/mL. A common crystallization condition consisting of 0.2M MgCl2, 0.1M Tris pH 6.5 and 25% PEG3350 was found for AfuA bound to either G6P, F6P or S7P. Apo-AfuA crystallized best in 0.2M MgCl2, 0.1M MES:NaOH pH 6.0 and 27.5% PEG 3350. Candidate crystals were looped in a cryoprotectant solution consisting of the mother liquor with 20% glycerol and flash frozen in liquid N2. The sulfur anomalous diffraction data for AfuA bound to G6P was collected at the Structural Genomics Consortium (SGC) (Toronto, Canada) using a chromium rotating anode X-ray source (λ= 2.29 Å) and R-AXIS IV++ detector (Rigaku). To improve the signal-to-noise ratio of the anomalous signal of sulfur, the crystal was scanned over 326° and a dataset was collected with an average I/σI of 59 with 11 fold redundancy to 2.3 Å resolution. Data was collected for AfuA-G6P, AfuA-F6P and apo-AfuA at beamline 08ID-1 (CMCF-ID) at the Canadian Light Source (CLS) (Saskatchewan, Canada). For AfuA-S7P, data was collected at NECAT beamline 24-ID-E at the Advanced Photon Source (APS) (Chicago, USA). Sulfur single wavelength anomalous dispersion (SAD) phasing was used to phase the initial G6P-bound structure of AfuA. This structure was then used as a molecular replacement (MR) search model for the solutions of AfuA-F6P and AfuA-S7P. For apo-AfuA, the AfuA-G6P structure was split into two domains (correlating to the two lobes of the protein), each of which was used as an independent search model for MR. All MR and automated refinement was performed with the PHENIX suite [37]. Manual refinement was undertaken with Coot [38]. Chimera and PyMol were used for molecular modeling, graphics (PyMol) and electrostatic surface mapping (Chimera) [39]. Atomic coordinate files were deposited in the Protein Data Bank under the accession numbers 4R72 (Apo), 4R73 (G6P), 4R74 (F6P) and 4R75 (S7P).
E. coli complementation assays
For growth assays, both the mutant (ΔuhpT) and the WT strain (BW25113) were obtained from the Keio collection of single gene knockouts [40]. Overnight cultures were grown in 5mL of M9 minimal media + 10mM glucose + antibiotics. Cells were washed twice in 1mL M9 salts (no carbon) and brought to a density of 109 cells/mL. For solid agar growth assays, a dilution series of 5μL spots from 107 to 103 cells/mL was used. Plates were incubated at 37°C for 30 hours and were imaged with a GelDoc XR system (BioRad). For liquid growth assays, cells were plated at dilutions ranging from 108 to 104 cells/mL in wells containing 100μL of the appropriate minimal media. Each well was duplicated to account for pipetting errors. Plates were incubated at 37°C with shaking in a Bioscreen C microplate reader (Growth Curves USA). Growth was measured by OD600 readings every 15 minutes for at least 24 hours. For data analysis, cell density of duplicate wells was averaged. All assays were performed in at least triplicate.
Bioinfomatics and phylogenetic tree construction
A. pleuropneumoniae AfuA was used as a template for a BlastP search. The results were filtered by removing homologs that did not possess the substrate specificity-determining residues. To reduce the tree size further, AfuA hits from multiple strains of the same organism were removed, keeping the top hit with highest sequence similarity. The selected hits used for further analysis came from 268 different organisms. The multiple sequence alignments and phylogenetic tree (neighbor-joining) construction was performed with Geneious R7 (Biomatters). MUSCLE [41] was used to generate the multiple sequence alignment. The tree was re-sampled 100 times using the bootstrap module and braches with less than 60% confidence were trimmed. The tree was confirmed by using the MrBayes [42] module.
C. rodentium growth curve
Strains from an overnight culture were diluted 1/100 into fresh LB, and 200 μl transferred into a sterile 96-well plate in triplicate (Costar). The optical density at 600 nm was measured every 15 min after 5s of orbital shaking using a Trilux Scintillation Counter (Wallec).
Generation of afuA mutant C.rodentium
DBS100 in-frame deletion mutants were generated using the suicide vector pRE118 via sacB-based allelic exchange [43]. A 1.5kb fragment upstream of afuA was amplified by primers (GTGGTACCTGCGCGAGCGCGTCAGCGCG and CCGCTAGCCGCCGCCAGCGCTACGGCAGAG) with flanking KpnI and NheI sites, while a 1.5kb fragment downstream of afuA was amplified by primers (CCGAGCTCTGCTGCCGTAGAGCAGGGCG and CCGCTAGCTACGGCTCAACGGAGGTGC) with flanking SacI and NheI sites (italicised text indicates restriction sites). Upstream and downstream fragments were digested alongside pRE118 with appropriate restriction enzyme and cloned into E. coli SY327 λpir to generate deletion vector pΔafuA. DBS100 was electroporated with pΔafuA and grown on kanamycin. Resulting colonies were inoculated on sucrose plates and a third set of primers (GTCGCTGGTTATTGAACGC and GGATGCAGAGCGAGTGTCTG) was used to confirm deletion of the gene for sucrose resistant, kanamycin sensitive colonies.
Mouse infections and competitive index (CI) assays
C57BL/6 mice (8–12 weeks old) were bred under specific pathogen-free conditions at the Child and Family Research Institute. C. rodentium WT (DBS100) and ΔafuA strains were grown shaking overnight at 37°C in 3 ml of LB. Cell density was measured by OD600 readings. Mice were orally gavaged with 100 μl of either WT, ΔafuA or a 1 : 1 mix of WT to ΔafuA (total 2.5 × 108 cfu). Stool was collected at day 6 pi, homogenized, serial diluted and plated on LB agar supplemented with 100 μg/ml of streptomycin. Animals were euthanized at 10 dpi and colonic tissues were removed, homogenized and plated. For CI, single colonies were picked and used as templates for colony PCR with deletion screening primers from ΔafuA generation. CI was calculated as the ratio of ΔafuA to WT colonies divided by the ratio of ΔafuA to WT from the input. All mouse experiments were performed in accordance with protocols approved by the University of British Columbia’s Animal Care Committee and in direct accordance with the Canadian Council on Animal Care’s guidelines.
Mouse transmission experiments
Assessment of the ability of C. rodentium to transmit between hosts was adapted from the protocol used by Wickham et al [44]. Index C57BL/6 mice were infected with WT or ΔafuA C. rodentium as outlined above. At 6 dpi, the index mouse plus two naïve mice (referred to as “co-housed”) were added to a new cage. Naïve mice remained co-housed with the index mouse for 48 h, at which time they were euthanized. Their colon tissues and luminal contents were then homogenized, serial diluted and plated on LB agar supplemented with 100 μg/ml of streptomycin for enumeration of C. rodentium burdens.
Profiling T3SS effectors in WT and ΔafuA C. rodentium
As previously described [45,46], C. rodentium WT, ΔafuA and ΔescN were streaked onto LB-streptomycin plates for single colony isolation. 5 ml LB-streptomycin was inoculated with a single colony of above mentioned strains and grown shaking O/N at 37°C. The strains were then subcultured at a 1 : 50 dilution into Dulbecco's modified Eagle's medium (DMEM). The bacteria were incubated statically at 37°C, 5% CO2 until the optical density of the cultures reached 0.7 (OD600 ~0.7). Bacteria were pelleted (13200 rpm, 4°C, 10 minutes) and proteins in the supernatant were precipitated using 10% trichloroacetic acid (TCA, Sigma) O/N at 4°C. Precipitated proteins were pelleted by centrifugation (13200 rpm, 4°C, 10 minutes) and resuspended in Laemmli buffer. Samples were resolved on 12% SDS-polyacrylamide gels and visualized by Coomassie R-250 Blue staining.
In vitro C. rodentium adherence and pedestal formation
HeLa cells were seeded at a concentration of 5x104 cells per well and incubated for 24 h prior to infection. To initialize bacterial infection, tissue cultures were pre-incubated in 1 ml of DMEM (Life Technologies) supplemented with 2% FBS for 30 min. Cells were then infected with an overnight culture of C. rodentium DBS100 at a MOI of 1 : 100 for 8 hours. Monolayers were washed three times with Dulbecco's PBS (Life Technologies) to remove nonadherent bacteria, and treated with 200μl of 0.1% Triton X-100 PBS for 5 min at room temperature. Samples were serially diluted and incubated on LB-streptomycin plates overnight at 37°C. Values are the mean CFU of three independent experiments repeated in triplicate, and statistical comparisons between groups were performed with two-tailed Student’s t tests. Sterile round coverslips were also seeded and infected as above. After infection, coverslips were washed three times with Dulbecco's PBS and fixed in 4% paraformaldehyde (Fisher Scientific) overnight. The fixed cells were washed three times in PBS and permeabilized by incubation in 0.1% Triton X-100 in PBS for 10 min. Cover slips were incubated in anti-Tir rat polyclonal IgG antibody diluted 1 : 2000 in 1% BSA in TBST for 1 hour. The coverslips were then washed three times in TBST and incubated in Alexa Fluor 568 goat anti-rat IgG (diluted 1 : 1000) and phalloidin-Alexa Fluor 488 conjugate (Pierce) in 1% BSA in TBST for 1 hour. After three washes in PBS, cells were mounted with ProLong Gold Mountant with DAPI (Life Technologies). Images were acquired on a Zeiss AxioImager Z1 with an AxioCam HRm camera operating through AxioVision software.
Fluorescence microscopy of C. rodentium-infected colonic tissue
Immunofluorescence staining of infected tissues was performed using previously described procedures [47]. In brief, colon tissues were fixed in 10% neutral buffered formalin (Fisher) for 16 hrs, rinsed in 70% ethanol and paraffin embedded. Sectioning was completed by the histology laboratory at the Child and Family Research Institute. Serial 5 μm sections were cut and deparaffinized by heating at 60°C, xylene treatment and rehydration through an ethanol gradient to water. Sections were treated with PBS containing 0.1% Triton X-100, followed by blocking buffer (PBS containing 0.05% Tween 20 and 1% normal donkey serum). Tissues were incubated with goat anti-cytokeratin 19 (1 : 300, Santa Cruz Biotechnology) and rabbit anti-C. rodentium Tir (1 : 5,000; gift from W. Deng) and subsequently probed with Alexa Fluor 568-conjugated donkey anti-goat IgG (1 : 1000; Life Technologies) and Alexa Fluor 488-conjugate donkey anti-rabbit IgG (1 : 1000; Life Technologies). Tissues were mounted and imaged in the same manner as the in vitro pedestal coverslips.
Quantification of sugar-phosphates in murine intestinal tissues
C57BL/6 mice were infected with C. rodentium WT or ΔafuA as described above for single-strain infections. At 6 dpi, shed stool pellets were collected and mice sacrificed. From each individual, colons were extracted, cut longitudinally, and luminal contents removed. Mucus was collected via gentle scraping along the apical colonic surface with a glass slide. Samples were immediately resuspended in 25mM HEPES pH 7.0 at a ratio of 1μL:1mg tissue, gently shaken to dissolve soluble extracellular species and stored on ice. Suspensions were centrifuged and equal volumes of supernatant from each sample were subjected to a conventional MeOH/CHCl3 extraction. Aqueous layers were kept and dried in a vacuum centrifuge and stored at -80°C for further analysis. Samples were analyzed with targeted tandem liquid chromatography/mass spectrometry (LC-MS/MS) at the Analytical Facility for Bioactive Molecules (Hospital for Sick Children, Canada) using an optimized protocol for LC separation of sugar-phosphates. Samples were separated on a Synergi Hydro-RP column (Phenomenex) and quantified with an API 4000 triple quadrupole mass spectrometer (Sciex). Quantification was performed against a standard curve subjected to the extraction process.
Statistical analysis
The mean values ± standard errors of the means (SEM) for at least two independent experiments is shown in all figures unless stated otherwise. p values were calculated using a Wilcoxon Rank Sum Test (CI) or a nonparametric Mann-Whitney t-test using GraphPad Prism software (v6.05). A p value less than 0.05 was considered statistically significant.
Supporting Information
Zdroje
1. Rohmer L, Hocquet D, Miller SI (2011) Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol 19 : 341–348. doi: 10.1016/j.tim.2011.04.003 21600774
2. Hollenstein K, Dawson RJP, Locher KP (2007) Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol 17 : 412–418. doi: 10.1016/j.sbi.2007.07.003 17723295
3. Chin N, Frey J, Chang CF, Chang YF (1996) Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol Lett 143 : 1–6. 8807793
4. Moisi M, Lichtenegger S, Tutz S, Seper A, Schild S, et al. (2013) Characterizing the hexose-6-phosphate transport system of Vibrio cholerae, a utilization system for carbon and phosphate sources. J Bacteriol 195 : 1800–1808. doi: 10.1128/JB.01952-12 23417487
5. Hsu Y-M, Chin N, Chang C-F, Chang Y-F (2003) Cloning and characterization of the Actinobacillus pleuropneumoniae fur gene and its role in regulation of ApxI and AfuABC expression. DNA Seq 14 : 169–181. doi: 10.1080/1042517031000089469 14509829
6. Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM (2006) Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol 188 : 6515–6523. doi: 10.1128/JB.00626-06 16952942
7. Klitgaard K, Friis C, Jensen TK, Angen Ø, Boye M (2012) Transcriptional portrait of Actinobacillus pleuropneumoniae during acute disease—potential strategies for survival and persistence in the host. PLoS One 7: e35549. doi: 10.1371/journal.pone.0035549 22530048
8. Whitby PW, Vanwagoner TM, Seale TW, Morton DJ, Stull TL (2006) Transcriptional profile of Haemophilus influenzae: effects of iron and heme. J Bacteriol 188 : 5640–5645. doi: 10.1128/JB.00417-06 16855256
9. Bergholz TM, Wick LM, Qi W, Riordan JT, Ouellette LM, et al. (2007) Global transcriptional response of Escherichia coli O157:H7 to growth transitions in glucose minimal medium. BMC Microbiol 7 : 97. doi: 10.1186/1471-2180-7-97 17967175
10. Berntsson RP-A, Smits SHJ, Schmitt L, Slotboom D-J, Poolman B (2010) A structural classification of substrate-binding proteins. FEBS Lett 584 : 2606–2617. doi: 10.1016/j.febslet.2010.04.043 20412802
11. Clasquin MF, Melamud E, Singer A, Gooding JR, Xu X, et al. (2011) Riboneogenesis in yeast. Cell 145 : 969–980. doi: 10.1016/j.cell.2011.05.022 21663798
12. Hall JA, Maloney PC (2005) Altered oxyanion selectivity in mutants of UhpT, the Pi-linked sugar phosphate carrier of Escherichia coli. J Biol Chem 280 : 3376–3381. doi: 10.1074/jbc.M409965200 15556940
13. Weston LA, Kadner RJ (1987) Identification of uhp polypeptides and evidence for their role in exogenous induction of the sugar phosphate transport system of Escherichia coli K-12. J Bacteriol 169 : 3546–3555. 3038843
14. Götz A, Goebel W (2010) Glucose and glucose 6-phosphate as carbon sources in extra - and intracellular growth of enteroinvasive Escherichia coli and Salmonella enterica. Microbiology 156 : 1176–1187. doi: 10.1099/mic.0.034744–0 20075042
15. Collins JW, Keeney KM, Crepin VF, Rathinam VAK, Fitzgerald KA, et al. (2014) Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 12 : 612–623. doi: 10.1038/nrmicro3315 25088150
16. Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, et al. (2012) Fucose sensing regulates bacterial intestinal colonization. Nature 492 : 113–117. doi: 10.1038/nature11623 23160491
17. Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, et al. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409 : 529–533. doi: 10.1038/35054089 11206551
18. Petty NK, Bulgin R, Crepin VF, Cerdeño-Tárraga AM, Schroeder GN, et al. (2010) The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J Bacteriol 192 : 525–538. doi: 10.1128/JB.01144-09 19897651
19. Ma C, Wickham ME, Guttman JA, Deng W, Walker J, et al. (2006) Citrobacter rodentium infection causes both mitochondrial dysfunction and intestinal epithelial barrier disruption in vivo: role of mitochondrial associated protein (Map). Cell Microbiol 8 : 1669–1686. doi: 10.1111/j.1462-5822.2006.00741.x 16759225
20. Wickham ME, Lupp C, Vázquez A, Mascarenhas M, Coburn B, et al. (2007) Citrobacter rodentium virulence in mice associates with bacterial load and the type III effector NleE. Microbes Infect 9 : 400–407. doi: 10.1016/j.micinf.2006.12.016 17317262
21. Kelly M, Hart E, Mundy R, Marchès O, Wiles S, et al. (2006) Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect Immun 74 : 2328–2337. doi: 10.1128/IAI.74.4.2328–2337.2006 16552063
22. Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, et al. (2014) Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5 : 3114. doi: 10.1038/ncomms4114 24445449
23. Antunes LCM, McDonald JAK, Schroeter K, Carlucci C, Ferreira RBR, et al. (2014) Antivirulence activity of the human gut metabolome. MBio 5: e01183–14. doi: 10.1128/mBio.01183-14 25073640
24. Ratledge C, Dover L (2000) Iron metabolism in pathogenic bacteria. Annu Rev Microbiol: 881–941. 11018148
25. Chico-Calero I, Suárez M, González-Zorn B, Scortti M, Slaghuis J, et al. (2002) Hpt, a bacterial homolog of the microsomal glucose - 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A 99 : 431–436. doi: 10.1073/pnas.012363899 11756655
26. Schwoppe C, Winkler HH, Neuhaus HE (2002) Properties of the Glucose-6-Phosphate Transporter from Chlamydia pneumoniae (HPTcp) and the Glucose-6-Phosphate Sensor from Escherichia coli (UhpC). J Bacteriol 184 : 2108–2115. doi: 10.1128/JB.184.8.2108–2115.2002 11914341
27. Engström P, Krishnan KS, Ngyuen BD, Chorell E, Normark J, et al. (2015) A 2-Pyridone-Amide Inhibitor Targets the Glucose Metabolism Pathway of Chlamydia trachomatis. MBio 6 : 1–9. doi: 10.1128/mBio.02304-14.Editor
28. Gaudet RG, Sintsova A, Buckwalter CM, Leung N, Cochrane A, et al. (2015) Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science 348 : 1251–1255. doi: 10.1126/science.aaa4921 26068852
29. Sham HP, Yu EYS, Gulen MF, Bhinder G, Stahl M, et al. (2013) SIGIRR, a Negative Regulator of TLR/IL-1R Signalling Promotes Microbiota Dependent Resistance to Colonization by Enteric Bacterial Pathogens. PLoS Pathog 9. doi: 10.1371/journal.ppat.1003539
30. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, et al. (2013) Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502 : 96–99. doi: 10.1038/nature12503 23995682
31. Siegel SJ, Roche AM, Weiser JN (2014) Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe 16 : 55–67. doi: 10.1016/j.chom.2014.06.005 25011108
32. Holmén Larsson JM, Thomsson KA, Rodríguez-Piñeiro AM, Karlsson H, Hansson GC (2013) Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol 305: G357–G363. doi: 10.1152/ajpgi.00048.2013 23832516
33. Bai J, McAteer SP, Paxton E, Mahajan A, Gally DL, et al. (2011) Screening of an E. coli O157:H7 Bacterial Artificial Chromosome Library by Comparative Genomic Hybridization to Identify Genomic Regions Contributing to Growth in Bovine Gastrointestinal Mucus and Epithelial Cell Colonization. Front Microbiol 2 : 168. doi: 10.3389/fmicb.2011.00168 21887152
34. Bergstrom KSB, Kissoon-Singh V, Gibson DL, Ma C, Montero M, et al. (2010) Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 6. doi: 10.1371/journal.ppat.1000902
35. Ulrich A, Andersen KR, Schwartz TU (2012) Exponential megapriming PCR (EMP) cloning—seamless DNA insertion into any target plasmid without sequence constraints. PLoS One 7: e53360. doi: 10.1371/journal.pone.0053360 23300917
36. van den Ent F, Löwe J (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67 : 67–74. doi: 10.1016/j.jbbm.2005.12.008 16480772
37. Adams PD, Afonine P V, Bunkóczi G, Chen VB, Echols N, et al. (2011) The Phenix software for automated determination of macromolecular structures. Methods 55 : 94–106. doi: 10.1016/j.ymeth.2011.07.005 21821126
38. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66 : 486–501. doi: 10.1107/S0907444910007493 20383002
39. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25 : 1605–1612. doi: 10.1002/jcc.20084 15264254
40. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2 : 2006.0008. doi: 10.1038/msb4100050
41. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 : 1792–1797. doi: 10.1093/nar/gkh340 15034147
42. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 : 1572–1574. doi: 10.1093/bioinformatics/btg180 12912839
43. Edwards RA, Keller LH, Schifferli DM (1998) Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207 : 149–157. doi: 10.1016/S0378-1119(97)00619-7 9511756
44. Wickham ME, Brown NF, Boyle EC, Coombes BK, Finlay BB (2007) Virulence Is Positively Selected by Transmission Success between Mammalian Hosts. Curr Biol 17 : 783–788. doi: 10.1016/j.cub.2007.03.067 17442572
45. Kenny B, Abe A, Stein M, Finlay BB (1997) Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun 65 : 2606–2612. 9199427
46. Newman J V, Zabel BA, Jha SS, Schauer DB (1999) Citrobacter rodentium espB is necessary for signal transduction and for infection of laboratory mice. Infect Immun 67 : 6019–6025. 10531262
47. Knodler LA, Crowley SM, Sham HP, Yang H, Wrande M, et al. (2014) Noncanonical Inflammasome Activation of Caspase-4/Caspase-11 Mediates Epithelial Defenses against Enteric Bacterial Pathogens. Cell Host Microbe 16 : 249–256. doi: 10.1016/j.chom.2014.07.002 25121752
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type GenesČlánek Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion IsolatesČlánek -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated GlycolipidsČlánek Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 8- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Long and Winding Road (Apologies to the Beatles)
- The Ebola Virus: From Basic Research to a Global Health Crisis
- Riding the R Train into the Cell
- The Two-Phase Emergence of Non Pandemic HIV-1 Group O in Cameroon
- Tumor Progression Locus 2 Promotes Induction of IFNλ, Interferon Stimulated Genes and Antigen-Specific CD8 T Cell Responses and Protects against Influenza Virus
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
- E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3
- The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type Genes
- Hyperinvasive Meningococci Induce Intra-nuclear Cleavage of the NF-κB Protein p65/RelA by Meningococcal IgA Protease
- Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen
- HTLV-1 Tax Stimulates Ubiquitin E3 Ligase, Ring Finger Protein 8, to Assemble Lysine 63-Linked Polyubiquitin Chains for TAK1 and IKK Activation
- Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion Isolates
- Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion
- Inhibiting the Recruitment of PLCγ1 to Kaposi’s Sarcoma Herpesvirus K15 Protein Reduces the Invasiveness and Angiogenesis of Infected Endothelial Cells
- Goblet Cell Derived RELM-β Recruits CD4 T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation
- HLA Class-II Associated HIV Polymorphisms Predict Escape from CD4+ T Cell Responses
- An siRNA Screen Identifies the U2 snRNP Spliceosome as a Host Restriction Factor for Recombinant Adeno-associated Viruses
- Extracellular Adenosine Protects against Lung Infection by Regulating Pulmonary Neutrophil Recruitment
- : Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with
- Which Way In? The RalF Arf-GEF Orchestrates Host Cell Invasion
- Intracellular Uropathogenic . Exploits Host Rab35 for Iron Acquisition and Survival within Urinary Bladder Cells
- A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane
- Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect
- -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
- The Activation of Effector Avr3b by Plant Cyclophilin is Required for the Nudix Hydrolase Activity of Avr3b
- A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in
- Double-Edge Sword of Sustained ROCK Activation in Prion Diseases through Neuritogenesis Defects and Prion Accumulation
- The Rsb Phosphoregulatory Network Controls Availability of the Primary Sigma Factor in and Influences the Kinetics of Growth and Development
- Inhibits Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
- Illuminating Targets of Bacterial Secretion
- Chemical Signals and Mechanosensing in Bacterial Responses to Their Environment
- Interdisciplinarity and Infectious Diseases: An Ebola Case Study
- Fungi That Infect Insects: Altering Host Behavior and Beyond
- Plasticity and Redundancy in Proteins Important for Invasion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
- A Novel Virus Causes Scale Drop Disease in
- STAT2 Knockout Syrian Hamsters Support Enhanced Replication and Pathogenicity of Human Adenovirus, Revealing an Important Role of Type I Interferon Response in Viral Control
- Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
- Twenty-Eight Years of Poliovirus Replication in an Immunodeficient Individual: Impact on the Global Polio Eradication Initiative
- AAV-Delivered Antibody Mediates Significant Protective Effects against SIVmac239 Challenge in the Absence of Neutralizing Activity
- Interferon-γ Promotes Inflammation and Development of T-Cell Lymphoma in HTLV-1 bZIP Factor Transgenic Mice
- Transgenic Rabbits Expressing Ovine PrP Are Susceptible to Scrapie
- Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of . Virulence Pathways
- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors
- Inhibition of mTORC1 Enhances the Translation of Chikungunya Proteins the Activation of the MnK/eIF4E Pathway
- Nanoformulations of Rilpivirine for Topical Pericoital and Systemic Coitus-Independent Administration Efficiently Prevent HIV Transmission
- Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection
- sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic
- Evolutionary and Functional Analysis of Old World Primate TRIM5 Reveals the Ancient Emergence of Primate Lentiviruses and Convergent Evolution Targeting a Conserved Capsid Interface
- Hepcidin and Host Defense against Infectious Diseases
- Type I IFN Induction via Poly-ICLC Protects Mice against Cryptococcosis
- Mucosal B Cells Are Associated with Delayed SIV Acquisition in Vaccinated Female but Not Male Rhesus Macaques Following SIV Rectal Challenge
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Illuminating Targets of Bacterial Secretion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



