-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Interdisciplinarity and Infectious Diseases: An Ebola Case Study
article has not abstract
Published in the journal: Interdisciplinarity and Infectious Diseases: An Ebola Case Study. PLoS Pathog 11(8): e32767. doi:10.1371/journal.ppat.1004992
Category: Opinion
doi: https://doi.org/10.1371/journal.ppat.1004992Summary
article has not abstract
High-profile epidemics such as Ebola, avian influenza, and severe acute respiratory syndrome (SARS) repeatedly thrust infectious diseases into the limelight. Because the emergence of diseases involves so many factors, the need for interdisciplinary approaches to studying emerging infections, particularly those originating from animals (i.e., zoonoses), is frequently discussed [1–4]. However, effective integration across disciplines is challenging in practice. Ecological ideas, for example, are rarely considered in biomedical research, while insights from biomedicine are often neglected in ecological studies of infectious diseases. One practical reason for this is that researchers in these fields focus on vastly different scales of biological organization (Fig 1), which are difficult to bridge both intellectually and methodologically. Nevertheless, integration across biological scales is increasingly needed for solving the complex problems zoonotic diseases pose to human and animal well-being. Motivated by current events, we use Ebola virus as a case study to highlight fundamental questions about zoonoses that can be addressed by integrating insights and approaches across scales.
Fig. 1. In the two leftmost panels, we depict the hierarchy of biological organization, from molecules and genes to ecosystems. 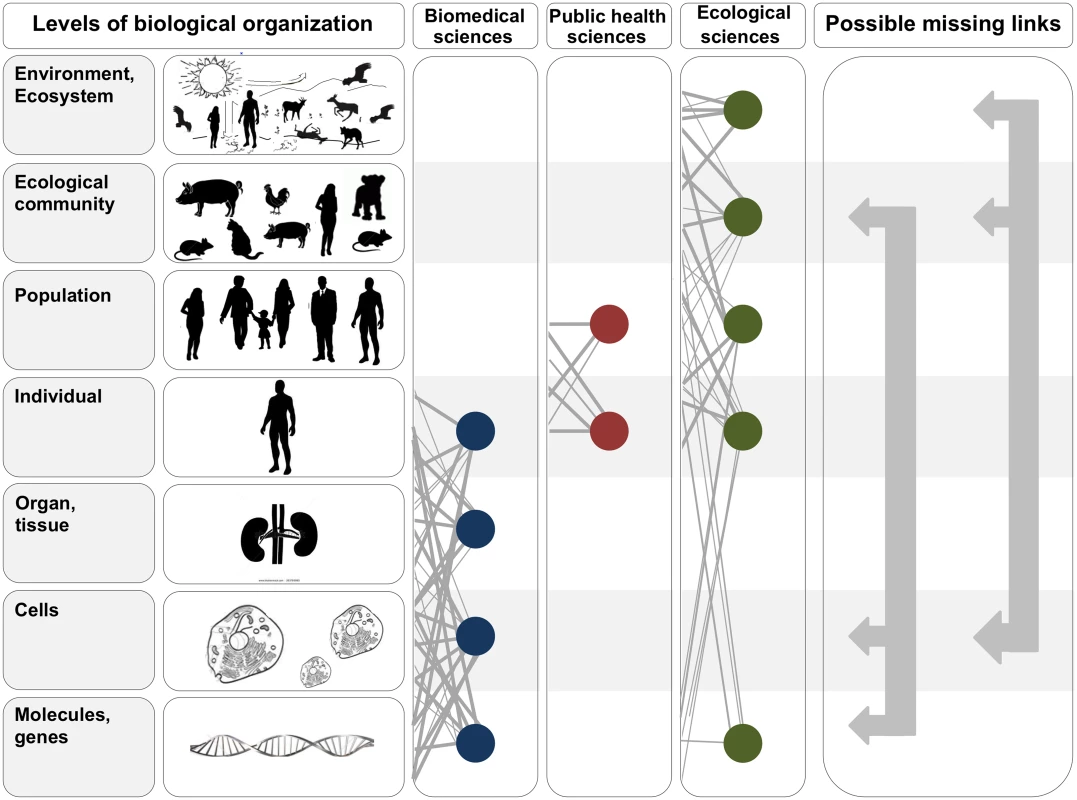
Each level of the hierarchy reflects an increase in organizational complexity, with each level being primarily composed of the previous level’s basic units. Middle panels illustrate how the study of interactions between infectious disease agents and their hosts differs across the biomedical, public health, and ecological sciences. Specifically, biomedical sciences typically focus on lower- and medium-scale levels of biological organization (e.g., molecules, genes, and organs). In contrast, public health and ecological sciences typically focus on medium- and higher-scale levels of organization (individual, population, community, ecosystem, and environment). The filled circles and solid lines connecting the circles illustrate key cross scale biological interactions studied within each field. The right panel shows example knowledge gaps that can emerge from the “typical” segregation of research activities across the three fields. To better integrate our understanding of the causes and consequences of zoonotic infectious diseases, researchers must begin focusing on these types of missing links. Ebola Severity: A Cell-to-Ecological Community Perspective
Zaire ebolavirus (EBOV), the virus responsible for the 2014 Ebola outbreak in West Africa, causes a deadly haemorrhagic disease in humans with case fatality rates ranging from 60%–88% [5]. Although well-known for its lethality, Ebola severity is variable at the individual level; some people die of infection, some survive, and some never develop symptoms [6–8]. Asymptomatic infection is poorly understood but may have important implications for how EBOV spreads. After a 1996 outbreak in Gabon, one study found that 45% of household contacts of symptomatic patients never developed disease symptoms despite becoming infected with the virus and mounting EBOV-specific immune responses [7]. Intriguingly, asymptomatic infection might also result from contact between humans and animals. As an example, a 2010 serological survey of over 4,000 people from 220 villages in Gabon found that 15% of people overall, and 19% of those in forested regions, had EBOV-specific immunoglobulin G (IgG) antibodies [9]. Detection of EBOV-specific T cell responses in a subset of IgG+ individuals corroborated that these individuals were exposed to EBOV. Based on the known epidemiology of Ebola in Gabon, the authors ruled out human-to-human transmission as a sufficient explanation for the high antibody prevalence. Instead, they hypothesized that human–animal contact, specifically human contact with noninfectious virus particles in the environment (e.g., by eating or handling fruit contaminated with the saliva of infected bats), may have triggered virus-specific immune responses. If the immune responses detected in Gabon are protective against subsequent EBOV infection, large-scale phenomena occurring at the level of the ecological community might interact with molecular and cellular-level processes to influence the severity of any given Ebola outbreak.
Using an epidemiological model, Bellan et al. [10] showed that accounting for asymptomatic infections that induce protective immunity reduced Ebola incidence projections for Liberia by 50%. Ultimately, the relative frequency of protective asymptomatic infections determines the size of this effect. Although the model was predicated on asymptomatic infection occurring during human-to-human transmission, asymptomatic cases that arise from environmental exposure, as hypothesized by Becquart et al. [9], could have similar dampening effects on epidemic spread. The frequency of such environmental exposure would depend on the animal community in a region. If certain bat species are the natural reservoirs of EBOV [11], their presence, relative abundance, and behaviour could all affect the frequency with which humans come into contact with them and thereby develop “environmentally-induced” immune protection. Of course, human contact with bats also triggers Ebola outbreaks [12], so understanding the context in which human–bat contact is protective (e.g., induces asymptomatic infection and immunity) rather than hazardous (e.g., causes symptomatic infection and epidemic spread) requires investigating phenomena occurring in both humans and bats, from the drivers and frequency of contact between humans, bats, and other relevant species to the characteristics of host cell–virus interactions upon contact.
Ecosystem Dynamics, Viral Evolution, and Human Epidemics
The Ebola outbreak in West Africa and punctuated outbreaks in Central Africa since the 1970s raise fundamental questions about what drives disease spillover to humans. Ebola outbreaks are not limited to human populations. Wildlife die-offs occur routinely before or during human epidemics, indicating that the virus circulates in a range of other mammal species, including great apes and forest antelopes [13–16]. Even though these species are not considered natural reservoirs, circulation of EBOV in these animals still has implications for human disease. First, human contact with these species can directly trigger disease outbreaks [17]. Second, these animals might affect spillover risk by influencing rates of virus evolution. Phylogenetic analysis of EBOV in great apes [18] suggests that genetic variation can accumulate rapidly during EBOV transmission in these populations. Importantly, virus evolution in animal hosts may facilitate the emergence of strains that spread more efficiently to humans or that cause more severe disease.
Although unknown for EBOV, the idea that virus circulation in wild species can drive changes that impact human–virus interactions has support for other RNA viruses such as SARS coronavirus and influenza A virus (see Table 1) [19,20]. Given evidence from these other viruses, understanding if and how animal hosts affect EBOV evolution is crucial. Doing this requires studies that connect large-scale environmental and ecosystem processes to small-scale genetic and molecular processes. For example, food web or habitat structure may determine the diets of target wildlife species, and host nutrition could affect rates of infection, virus replication, and shedding. Likewise, contact rates among species determine levels of cross species virus transmission, which may influence virus mutation or recombination rates. These examples, though speculative, highlight how cross scale chains of events might influence disease emergence in humans.
Tab. 1. Examples of zoonotic disease systems in which cross scale research has contributed to key insights about infection dynamics. 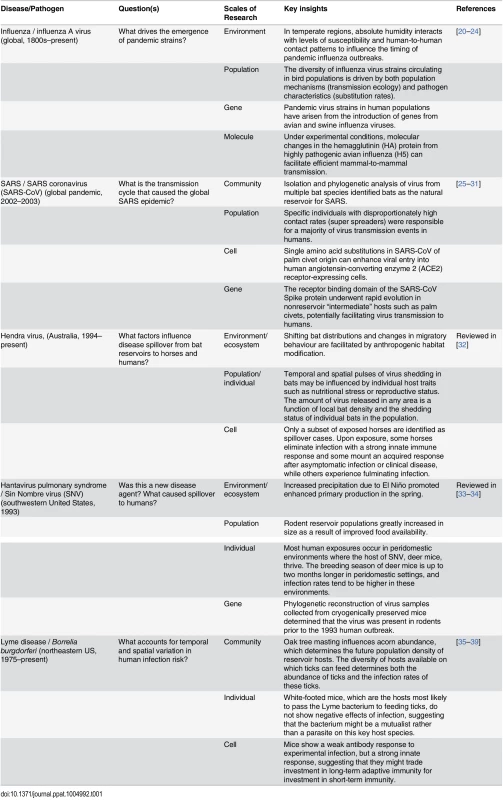
Towards a More Integrative Future
The Ebola outbreak in West Africa reminds us that zoonotic diseases continue to be a major threat. The benefits of cross scale research are evident for several high-profile zoonoses (Table 1). Nevertheless, this type of work is far from the norm, and successful integration of research approaches across vastly different biological scales remains challenging. A first step toward greater integration involves student training. Training programs in infectious disease typically focus on a single or narrow range of biological scales, but more crosscutting approaches are needed. Training grants focused on multiscale literacy in infectious disease research should be a priority for funding agencies, for example. Professional societies could also lead the way by sponsoring workshops, symposia, and other events on integration across disciplines. The involvement of professional societies has the added benefit of allowing infectious disease researchers to expand their perspectives beyond their years of formal training. Updating our collective mind-set in these and other ways will put us in a much better position to tackle the next zoonotic disease threat.
Zdroje
1. Morens DM, Folkers GK, Fauci AS (2004) The challenge of emerging and re-emerging infectious diseases. Nature 430 : 242–249. 15241422
2. Holmes EC (2013) What can we predict about viral evolution and emergence? Curr Opin Virol 3 : 180–184. doi: 10.1016/j.coviro.2012.12.003 23273851
3. Zinsstag J, Schelling E, Waltner-Toews D, Tanner M (2011) From "one medicine" to "one health" and systemic approaches to health and well-being. Prev Vet Med 101 : 148–156. doi: 10.1016/j.prevetmed.2010.07.003 20832879
4. Parkes MW, Bienen L, Breilh J, Hsu L-N, McDonald M, et al. (2005) All Hands on Deck: Transdisciplinary Approaches to Emerging Infectious Disease. EcoHealth 2 : 258–272.
5. Lefebvre A, Fiet C, Belpois-Duchamp C, Tiv M, Astruc K, et al. (2014) Case fatality rates of Ebola virus diseases: a meta-analysis of World Health Organization data. Med Mal Infect 44 : 412–416. doi: 10.1016/j.medmal.2014.08.005 25193630
6. Gonzalez JP, Nakoune E, Slenczka W, Vidal P, Morvan JM (2000) Ebola and Marburg virus antibody prevalence in selected populations of the Central African Republic. Microbes Infect 2 : 39–44. 10717539
7. Leroy EM, Baize S, Volchkov VE, Fisher-Hoch SP, Georges-Courbot MC, et al. (2000) Human asymptomatic Ebola infection and strong inflammatory response. Lancet 355 : 2210–2215. 10881895
8. Goeijenbier M, van Kampen JJ, Reusken CB, Koopmans MP, van Gorp EC (2014) Ebola virus disease: a review on epidemiology, symptoms, treatment and pathogenesis. Neth J Med 72 : 442–448. 25387613
9. Becquart P, Wauquier N, Mahlakoiv T, Nkoghe D, Padilla C, et al. (2010) High prevalence of both humoral and cellular immunity to Zaire ebolavirus among rural populations in Gabon. PLoS One 5: e9126. doi: 10.1371/journal.pone.0009126 20161740
10. Bellan SE, Pulliam JR, Dushoff J, Meyers LA (2014) Ebola control: effect of asymptomatic infection and acquired immunity. Lancet 384 : 1499–1500. doi: 10.1016/S0140-6736(14)61839-0 25390569
11. Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, et al. (2005) Fruit bats as reservoirs of Ebola virus. Nature 438 : 575–576. 16319873
12. Leroy EM, Epelboin A, Mondonge V, Pourrut X, Gonzalez JP, et al. (2009) Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic Dis 9 : 723–728. doi: 10.1089/vbz.2008.0167 19323614
13. Leroy EM, Rouquet P, Formenty P, Souquiere S, Kilbourne A, et al. (2004) Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science 303 : 387–390. 14726594
14. Rouquet P, Froment JM, Bermejo M, Kilbourn A, Karesh W, et al. (2005) Wild animal mortality monitoring and human Ebola outbreaks, Gabon and Republic of Congo, 2001–2003. Emerg Infect Dis 11 : 283–290. 15752448
15. Bermejo M, Rodriguez-Teijeiro JD, Illera G, Barroso A, Vila C, et al. (2006) Ebola outbreak killed 5000 gorillas. Science 314 : 1564. 17158318
16. Lahm SA, Kombila M, Swanepoel R, Barnes RF (2007) Morbidity and mortality of wild animals in relation to outbreaks of Ebola haemorrhagic fever in Gabon, 1994–2003. Trans R Soc Trop Med Hyg 101 : 64–78. 17010400
17. Pourrut X, Kumulungui B, Wittmann T, Moussavou G, Delicat A, et al. (2005) The natural history of Ebola virus in Africa. Microbes Infect 7 : 1005–1014. 16002313
18. Wittmann TJ, Biek R, Hassanin A, Rouquet P, Reed P, et al. (2007) Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc Natl Acad Sci USA 104 : 17123–17127. 17942693
19. Graham RL, Baric RS (2010) Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol 84 : 3134–3146. doi: 10.1128/JVI.01394-09 19906932
20. Worobey M, Han GZ, Rambaut A (2014) A synchronized global sweep of the internal genes of modern avian influenza virus. Nature 508 : 254–257. doi: 10.1038/nature13016 24531761
21. Shaman J, Goldstein E, Lipsitch M (2011) Absolute humidity and pandemic versus epidemic influenza. Am J Epidemiol 173 : 127–135. doi: 10.1093/aje/kwq347 21081646
22. Roche B, Drake JM, Brown J, Stallknecht DE, Bedford T, et al. (2014) Adaptive evolution and environmental durability jointly structure phylodynamic patterns in avian influenza viruses. PLoS Biol 12: e1001931. doi: 10.1371/journal.pbio.1001931 25116957
23. Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, et al. (2012) Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486 : 420–428. doi: 10.1038/nature10831 22722205
24. Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, et al. (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459 : 1122–1125. doi: 10.1038/nature08182 19516283
25. Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, et al. (2005) Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA 102 : 14040–14045. 16169905
26. Li W, Shi Z, Yu M, Ren W, Smith C, et al. (2005) Bats are natural reservoirs of SARS-like coronaviruses. Science 310 : 676–679. 16195424
27. Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM (2005) Superspreading and the effect of individual variation on disease emergence. Nature 438 : 355–359.5. 16292310
28. Qu XX, Hao P, Song XJ, Jiang SM, Liu YX, et al. (2005) Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J Biol Chem 280 : 29588–29595. 15980414
29. Kan B, Wang M, Jing H, Xu H, Jiang X, et al. (2005) Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol 79 : 11892–11900. 16140765
30. Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, et al. (2005) Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A 102 : 2430–2435. 15695582
31. Wang LF, Eaton BT (2007) Bats, civets and the emergence of SARS. Curr Top Microbiol Immunol 315 : 325–344. 17848070
32. Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, et al. (2015) Ecological dynamics of emerging bat virus spillover. Proc Biol Sci 282 : 20142124. doi: 10.1098/rspb.2014.2124 25392474
33. Yates TL, Mills JN, Parmenter CA, Ksiazek TG, Parmenter RR, et al. (2002) The ecology and evolutionary history of an emergent disease: hantavirus pulmonary syndrome. Bioscience 52 : 989–998.
34. Mills JN (2005) Regulation of rodent-borne viruses in the natural host: implications for human disease. Arch Virol Suppl: 45–57. 16355867
35. Ostfeld RS, Canham CD, Oggenfuss K, Winchcombe RJ, Keesing F (2006) Climate, deer, rodents, and acorns as determinants of variation in lyme-disease risk. PLoS Biol 4: e145. 16669698
36. LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F (2003) The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc Natl Acad Sci USA 100 : 567–571. 12525705
37. Schwanz LE, Voordouw MJ, Brisson D, Ostfeld RS (2011) Borrelia burgdorferi has minimal impact on the Lyme disease reservoir host Peromyscus leucopus. Vector Borne Zoonotic Dis 11 : 117–124. doi: 10.1089/vbz.2009.0215 20569016
38. Hersh MH, LaDeau SL, Previtali MA, Ostfeld RS (2014) When is a parasite not a parasite? Effects of larval tick burdens on white-footed mouse survival. Ecology 95 : 1360–1369. 25000767
39. Previtali MA., Ostfeld RS, Keesing F, Jolles AE, Hanselmann R, et al. (2012) Relationship between pace of life and immune responses in wild rodents. Oikos 121 : 1483–1492.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type GenesČlánek Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion IsolatesČlánek -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated GlycolipidsČlánek Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 8- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Long and Winding Road (Apologies to the Beatles)
- The Ebola Virus: From Basic Research to a Global Health Crisis
- Riding the R Train into the Cell
- The Two-Phase Emergence of Non Pandemic HIV-1 Group O in Cameroon
- Tumor Progression Locus 2 Promotes Induction of IFNλ, Interferon Stimulated Genes and Antigen-Specific CD8 T Cell Responses and Protects against Influenza Virus
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
- E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3
- The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type Genes
- Hyperinvasive Meningococci Induce Intra-nuclear Cleavage of the NF-κB Protein p65/RelA by Meningococcal IgA Protease
- Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen
- HTLV-1 Tax Stimulates Ubiquitin E3 Ligase, Ring Finger Protein 8, to Assemble Lysine 63-Linked Polyubiquitin Chains for TAK1 and IKK Activation
- Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion Isolates
- Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion
- Inhibiting the Recruitment of PLCγ1 to Kaposi’s Sarcoma Herpesvirus K15 Protein Reduces the Invasiveness and Angiogenesis of Infected Endothelial Cells
- Goblet Cell Derived RELM-β Recruits CD4 T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation
- HLA Class-II Associated HIV Polymorphisms Predict Escape from CD4+ T Cell Responses
- An siRNA Screen Identifies the U2 snRNP Spliceosome as a Host Restriction Factor for Recombinant Adeno-associated Viruses
- Extracellular Adenosine Protects against Lung Infection by Regulating Pulmonary Neutrophil Recruitment
- : Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with
- Which Way In? The RalF Arf-GEF Orchestrates Host Cell Invasion
- Intracellular Uropathogenic . Exploits Host Rab35 for Iron Acquisition and Survival within Urinary Bladder Cells
- A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane
- Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect
- -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
- The Activation of Effector Avr3b by Plant Cyclophilin is Required for the Nudix Hydrolase Activity of Avr3b
- A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in
- Double-Edge Sword of Sustained ROCK Activation in Prion Diseases through Neuritogenesis Defects and Prion Accumulation
- The Rsb Phosphoregulatory Network Controls Availability of the Primary Sigma Factor in and Influences the Kinetics of Growth and Development
- Inhibits Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
- Illuminating Targets of Bacterial Secretion
- Chemical Signals and Mechanosensing in Bacterial Responses to Their Environment
- Interdisciplinarity and Infectious Diseases: An Ebola Case Study
- Fungi That Infect Insects: Altering Host Behavior and Beyond
- Plasticity and Redundancy in Proteins Important for Invasion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
- A Novel Virus Causes Scale Drop Disease in
- STAT2 Knockout Syrian Hamsters Support Enhanced Replication and Pathogenicity of Human Adenovirus, Revealing an Important Role of Type I Interferon Response in Viral Control
- Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
- Twenty-Eight Years of Poliovirus Replication in an Immunodeficient Individual: Impact on the Global Polio Eradication Initiative
- AAV-Delivered Antibody Mediates Significant Protective Effects against SIVmac239 Challenge in the Absence of Neutralizing Activity
- Interferon-γ Promotes Inflammation and Development of T-Cell Lymphoma in HTLV-1 bZIP Factor Transgenic Mice
- Transgenic Rabbits Expressing Ovine PrP Are Susceptible to Scrapie
- Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of . Virulence Pathways
- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors
- Inhibition of mTORC1 Enhances the Translation of Chikungunya Proteins the Activation of the MnK/eIF4E Pathway
- Nanoformulations of Rilpivirine for Topical Pericoital and Systemic Coitus-Independent Administration Efficiently Prevent HIV Transmission
- Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection
- sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic
- Evolutionary and Functional Analysis of Old World Primate TRIM5 Reveals the Ancient Emergence of Primate Lentiviruses and Convergent Evolution Targeting a Conserved Capsid Interface
- Hepcidin and Host Defense against Infectious Diseases
- Type I IFN Induction via Poly-ICLC Protects Mice against Cryptococcosis
- Mucosal B Cells Are Associated with Delayed SIV Acquisition in Vaccinated Female but Not Male Rhesus Macaques Following SIV Rectal Challenge
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Illuminating Targets of Bacterial Secretion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



