-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
article has not abstract
Published in the journal: Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease. PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003768
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003768Summary
article has not abstract
This year, 2013, is the fiftieth anniversary of the discovery of alpha one antitrypsin deficiency (AATD), a disease caused by mutation in SERPINA1, which predisposes to early onset lung disease, specifically chronic obstructive pulmonary disease (COPD). In this issue of PLOS Genetics, the timely study of Thun et al. [1] investigated alpha one antitrypsin (AAT) levels with a genome-wide association study (GWAS) approach in a cohort recruited primarily for studies of asthma. They demonstrate that, although a genome-wide association is detected for a common variant in SERPINA1, the association disappears after adjusting the results for the presence of the known rare variants that cause AATD (PiZ and PiS). The observation of this “synthetic association” illustrates the potential for detection of rare genetic variants, with minor allele frequencies (MAF) of less than 1%, to revolutionize the understanding of pathogenesis.
Importantly, the approach used by Thun et al.—the sequencing of SERPINA1, fine mapping, and a conditional approach to statistical analysis in the regression model—does not depend on knowing the rare variants on which models need to be conditioned, and was able to reliably identify known variants with MAF 1%–5%. It is therefore a nice demonstration of the potential for sequencing to reveal rare variants and ascertain their true contribution to traits. Thun et al. have also compensated for known environmental exposure that interacts with genes, such as cigarette smoke exposure (“ever smoked”), enhancing this analysis by use of a proxy measure for smoke intensity (hsCRP). The robustness of the results was demonstrated using a second large cohort. The clinical relevance of this work was demonstrated by looking at lung function as the outcome, although this revealed a more complex scenario. The results leave some unanswered questions, such as the observation that variants influencing expression which lie outside SERPINA1 exons are not accounted for, and that recent expression quantitative trait loci (eQTL) data for SERPINA1 did not report their associated SNPs.
Since the advent of GWAS it has become increasingly obvious that this research design cannot detect the majority of the heritability of most studied traits. Several ideas have been proposed to explain this “missing heritability,” including gene–environment interaction, reduced power of GWAS to detect functional variants due to low levels of linkage disequilibrium with causative SNPs, and undetected rare variants. Thun et al. [1] provide an excellent realization of this last hypothesis. Most GWAS have picked up common variants that confer only a small measured increase in risk (increased odds ratios [ORs]), but have missed by design rare variants that confer larger ORs of disease. AATD and GWAS of COPD and lung function are perfect examples of this as they failed to detect both the PiZ and PiS variants, despite the well-established role of these mutations in lung disease. Only through a huge meta-analysis of over 20,000 individuals and smoking interaction modeling could the influence of PiZ [2] on lung function be detected. Thun et al. could use their analyses to begin to address the longstanding debate regarding the role of AATD in lung disease at the population level. Specifically, does carrying a single abnormal SERPINA1 allele increase risk of lung disease at the population level [3]? A demonstration of an association, even at low AAT levels not considered truly deficient, would mean that AAT pathways are relevant to a far larger proportion of the population than previously thought. For this reason, the work of Thun et al. is not only an interesting example of synthetic association, but has potential clinical importance since it suggests that even small variation in AAT level could affect lung function [1].
Since rare variant analysis is a topic of great interest to genetic epidemiologists at present, it is worth reprising how understanding AATD has moved clinical medicine forward. It was first described in 1963 by Laurell and Eriksson, who reported an absence of the α1 band on protein electrophoresis of serum taken from a patient at a local respiratory hospital [4]; this missing band was due to very low circulating levels of AAT. The observation that people with this deficiency developed early-onset emphysema and COPD suggested a role for pathways involving AAT in pathogenesis(summarized in Figure 1). The main function of AAT is as an anti-protease, which protects tissues against neutrophil elastase (NE) [5]; the protease–antiprotease hypothesis of emphysema resulted directly from this knowledge and has driven much of the research into COPD. Clinically significant AATD-related lung disease occurs in approximately 1 in 2,500 Caucasian individuals who usually carry the PiZ or PiS variants of SERPINA1; importantly, both SNPs confer an increased risk of lung disease in carriers who smoke and in homozygous individuals who do not smoke [6]. However, COPD is very common, affecting up to 20% of smokers and 11% of non-smokers worldwide, and is projected to be the third most common cause of death by 2030 [7]. Consequently, changing the direction of research in COPD had potential to greatly affect population health. At the time of detection of AATD, COPD as a term was not in common use; rather, patients were described as having emphysema or chronic bronchitis, both diagnosed by clinical features and chest radiography. It is now a common term and is diagnosed by a reduction in FEV1/FVC ratio on spirometry—a simple test of lung function—and may be further characterized by more complex lung function tests and computerized tomography (CT) scanning of the lungs.
Fig. 1. Simplified pathogenesis of lung disease in AATD. 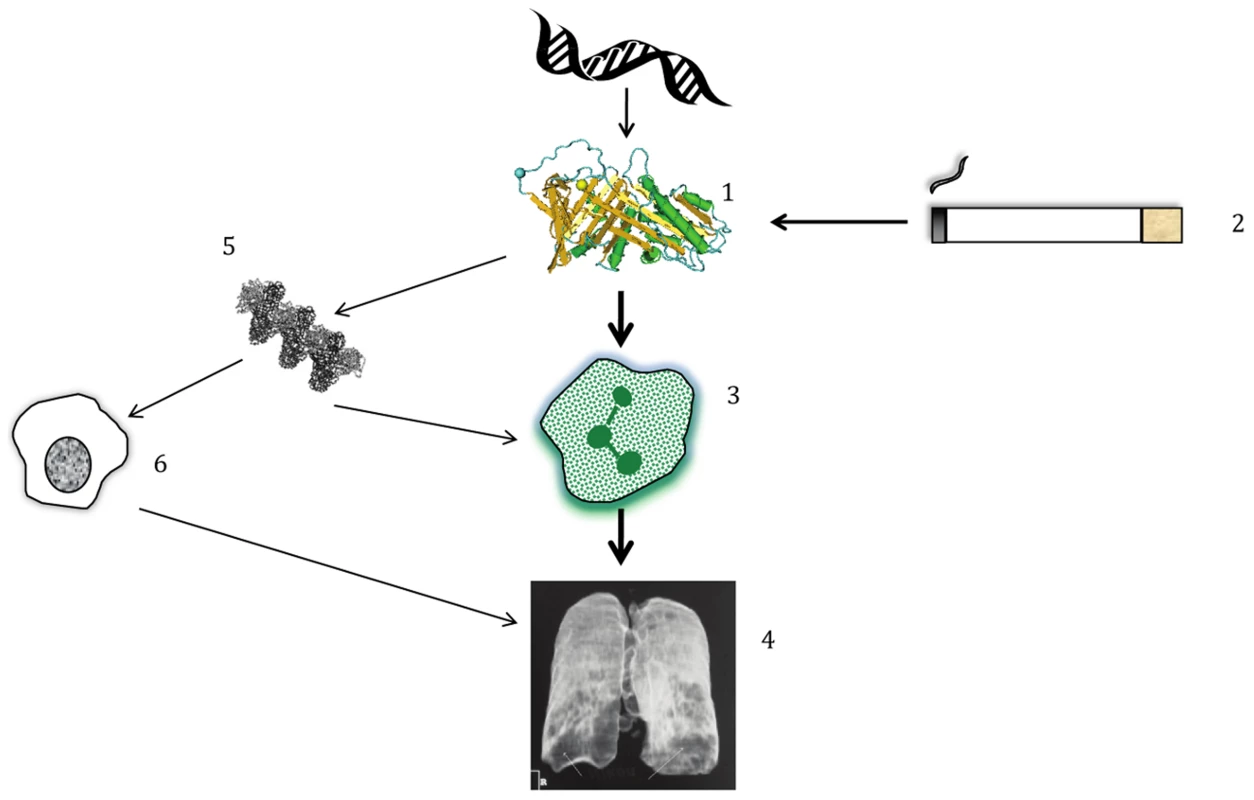
(1) Polymorphisms in DNA lead to structural changes in AAT which interact with (2) environmental exposure to cigarette smoke, amongst other influences. The combination of gene+environment leads to (3) neutrophilic inflammation in the lung; the main driver of disease is an inability to protect from the harmful effects of NE released by neutrophils. (4) Proteolytic destruction of lung tissue leads to the typical clinical pattern of emphysema, usually worst at the lung bases, as shown on this reconstructed image. (5 & 6) In addition, AAT polymers present in the lung, whose formation occurs due to the PiZ and to a lesser extent the PiS variant, play a smaller role in augmenting inflammation by attracting other inflammatory cells such as macrophages. Genetic research into the etiology of lung dysfunction in the general population [2] as well as the risk of COPD [8] has succeeded in identifying many variants that are significantly associated with these outcomes. Candidate gene approaches discovered other relevant proteases [9], and our increased understanding of the inflammatory cascade leading to neutrophil activation and NE release has guided the implementation of therapies targeting inflammation, the most common of which is inhaled corticosteroids (ICS). However, anti-inflammatory therapies have not been a magic bullet for COPD sufferers; indeed, the main trial seeking reduction in mortality showed only a trend in this direction (p = 0.052) when ICS were combined with long-acting beta agonists (LABA) [10]. However, lung function, flare-ups of the disease (known as exacerbations), and quality of life have been shown in many studies to improve with ICS/LABA combinations [7]. Relevant to the current paper on SERPINA1 variants as a predictor of AAT, specific treatment for AATD has been developed in the form of AAT augmentation therapy. A meta-analysis of 1,509 patients in observational studies and randomized controlled trials (RCTs) concluded that it was beneficial, because decline in lung function was slower upon treatment [11]. More sophisticated measures of lung disease using quantitative analysis of CT scans have shown that augmentation is beneficial in the RCTs alone [12]. Whether or not the recognition that rare variants explain common variant associations at SERPINA1 with AATD changes clinical practice remains to be seen, but the story should be a source of inspiration for genetic researchers to continue pursuing rare variants and their potential use in disease prediction and targeted therapy.
Zdroje
1. ThunGA, ImbodenM, FerrarottiI, KumarA, ObeidatM, et al. (2013) Causal and synthetic associations of variants in the serpina gene cluster with alpha1-antitrypsin serum levels. PLoS Genet 9 (8) e1003585 doi:10.1371/journal.pgen.1003585
2. ObeidatM, WainLV, ShrineN, KalshekerN, ArtigasMS, et al. (2011) A comprehensive evaluation of potential lung function associated genes in the SpiroMeta general population sample. PLoS One 6: e19382.
3. SorheimIC, BakkeP, GulsvikA, PillaiSG, JohannessenA, et al. (2010) alpha(1)-Antitrypsin protease inhibitor MZ heterozygosity is associated with airflow obstruction in two large cohorts. Chest 138 : 1125–1132.
4. LaurellCB, ErikssonS (1963) The electrophoretic alpha 1 globulin pattern of serum in alpha 1 antitrypsin deficiency. Scand J Clin Lab Invest 15 : 132–140.
5. CarrellRW, JeppssonJO, LaurellCB, BrennanSO, OwenMC, et al. (1982) Structure and variation of human alpha 1-antitrypsin. Nature 298 : 329–334.
6. LuisettiM, SeersholmN (2004) Alpha1-antitrypsin deficiency. 1: epidemiology of alpha1-antitrypsin deficiency. Thorax 59 : 164–169.
7. Global Initiative for Obstructive Lung Disease.Available: http://www.goldcopd.org. Accessed June 2013.
8. PillaiSG, GeD, ZhuG, KongX, ShiannaKV, et al. (2009) A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 5: e1000421.
9. HunninghakeGM, ChoMH, TesfaigziY, Soto-QuirosME, AvilaL, et al. (2009) MMP12, lung function, and COPD in high-risk populations. N Engl J Med 361 : 2599–2608.
10. CalverleyPM, AndersonJA, CelliB, FergusonGT, JenkinsC, et al. (2007) Salmeterol and fluticasone propionate and survival in chronic obstructive pulmonary disease. N Engl J Med 356 : 775–789.
11. ChapmanKR, StockleyRA, DawkinsC, WilkesMM, NavickisRJ (2009) Augmentation therapy for alpha1 antitrypsin deficiency: a meta-analysis. COPD 6 : 177–184.
12. StockleyRA, ParrDG, PiitulainenE, StolkJ, StoelBC, et al. (2010) Therapeutic efficacy of alpha-1 antitrypsin augmentation therapy on the loss of lung tissue: an integrated analysis of 2 randomised clinical trials using computed tomography densitometry. Respir Res 11 : 136.
Štítky
Genetika Reprodukčná medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



