-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
Cryptococcus neoformans is a human fungal pathogen with a defined sexual cycle. Nutrient-limiting conditions and pheromones induce a dimorphic transition from unicellular yeast to multicellular hyphae and the production of infectious spores. Sexual reproduction involves cells of either opposite (bisexual) or one (unisexual) mating type. Bisexual and unisexual reproduction are governed by shared components of the conserved pheromone-sensing Cpk1 MAPK signal transduction cascade and by Mat2, the major transcriptional regulator of the pathway. However, the downstream targets of the pathway are largely unknown, and homology-based approaches have failed to yield downstream transcriptional regulators or other targets. In this study, we applied insertional mutagenesis via Agrobacterium tumefaciens transkingdom DNA delivery to identify mutants with unisexual reproduction defects. In addition to elements known to be involved in sexual development (Crg1, Ste7, Mat2, and Znf2), three key regulators of sexual development were identified by our screen: Znf3, Spo11, and Ubc5. Spo11 and Ubc5 promote sporulation during both bisexual and unisexual reproduction. Genetic and phenotypic analyses provide further evidence implicating both genes in the regulation of meiosis. Phenotypic analysis of sexual development showed that Znf3 is required for hyphal development during unisexual reproduction and also plays a central role during bisexual reproduction. Znf3 promotes cell fusion and pheromone production through a pathway parallel to and independent of the pheromone signaling cascade. Surprisingly, Znf3 participates in transposon silencing during unisexual reproduction and may serve as a link between RNAi silencing and sexual development. Our studies illustrate the power of unbiased genetic screens to reveal both novel and conserved circuits that operate sexual reproduction.
Published in the journal: Genetic Circuits that Govern Bisexual and Unisexual Reproduction in. PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003688
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003688Summary
Cryptococcus neoformans is a human fungal pathogen with a defined sexual cycle. Nutrient-limiting conditions and pheromones induce a dimorphic transition from unicellular yeast to multicellular hyphae and the production of infectious spores. Sexual reproduction involves cells of either opposite (bisexual) or one (unisexual) mating type. Bisexual and unisexual reproduction are governed by shared components of the conserved pheromone-sensing Cpk1 MAPK signal transduction cascade and by Mat2, the major transcriptional regulator of the pathway. However, the downstream targets of the pathway are largely unknown, and homology-based approaches have failed to yield downstream transcriptional regulators or other targets. In this study, we applied insertional mutagenesis via Agrobacterium tumefaciens transkingdom DNA delivery to identify mutants with unisexual reproduction defects. In addition to elements known to be involved in sexual development (Crg1, Ste7, Mat2, and Znf2), three key regulators of sexual development were identified by our screen: Znf3, Spo11, and Ubc5. Spo11 and Ubc5 promote sporulation during both bisexual and unisexual reproduction. Genetic and phenotypic analyses provide further evidence implicating both genes in the regulation of meiosis. Phenotypic analysis of sexual development showed that Znf3 is required for hyphal development during unisexual reproduction and also plays a central role during bisexual reproduction. Znf3 promotes cell fusion and pheromone production through a pathway parallel to and independent of the pheromone signaling cascade. Surprisingly, Znf3 participates in transposon silencing during unisexual reproduction and may serve as a link between RNAi silencing and sexual development. Our studies illustrate the power of unbiased genetic screens to reveal both novel and conserved circuits that operate sexual reproduction.
Introduction
Sexual reproduction in eukaryotes facilities genetic diversity and eliminates deleterious mutations leading to better fit progeny. In fungi, sex often involves two cells of opposite mating type (heterothallism) that secrete pheromones in order to induce cell fusion and subsequently nuclear fusion and meiosis generate recombinant progeny. However, in other fungi, solo incubation of an individual isolate can result in sexual reproduction, and this selfing process is referred to as homothallism. Homothallism can involve 1) mating type switching, 2) the presence of both mating type alleles (fused or unlinked), or 3) unisexual reproduction of just one mating type [1]. Paradigmatic examples of fungi with both modes of sexual reproduction are Saccharomyces cerevisiae and Cryptococcus neoformans.
S. cerevisiae has served as a model for the exploration and elucidation of molecular mechanisms of the dimorphic switch and therefore serves as a framework for morphogenesis in other dimorphic fungi [2]. Pheromones produced by the opposite-mating type partner activate the mitogen-activated protein kinase (MAPK) signaling cascade (which is also called the pheromone response pathway) to induce mating, pseudohyphae, and invasive growth [3], [4]. Additional environmental cues are known to regulate pseudohyphae formation. Nutrients activate cellular receptors and the cAMP-dependent pathway to govern the expression of genes evoking the dimorphic transition [5]. Core components of these pathways are highly conserved throughout the fungal kingdom; however, the downstream targets are often species-specific.
Cryptococcus neoformans is a human fungal pathogen that grows as a yeast in the environment and inside the host. It causes severe central nervous system infections in HIV-infected patients and less frequently in immunocompetent individuals. Cryptococcal meningitis is an AIDS-defining illness in 30% of HIV infections worldwide and is uniformly fatal if untreated [6].
C. neoformans yeast cells undergo a dimorphic switch into hyphae during sexual development. The organism has a defined life cycle with α (common) and a (rare) mating type cells. Under nutrient-limiting conditions or in response to inositol, cells of the opposite mating type secrete pheromones leading to cell-cell fusion [7], [8], [9], [10]. The two cells fuse to form a stable heterokaryon, which undergoes the dimorphic switch to filamentous growth. The two haploid nuclei remain separate in the growing hyphae and migrate from one hyphal cell to the adjacent one through clamp cells that fuse to connect the neighboring hyphal cells. At the apex of the aerial filaments, specialized, swollen structures known as basidia form where nuclear fusion and meiosis occur [8]. Multiple rounds of mitosis and budding produce four chains of spores, which are dispersed by air and germinate to re-enter the life cycle [11]. The dimorphic switch during bisexual reproduction occurs in both the A and D serotypes of C. neoformans, as well as in the sibling species C. gattii [8], [12], [13], [14]. Meiosis and sporulation are integral to the C. neoformans life cycle and play important roles in virulence. Cryptococcal infections are acquired by the inhalation of infectious particles, spores or desiccated yeast cells, from the environment. Spores are of an ideal size to penetrate and colonize the alveoli of the lung and are pathogenic [15], [16].
The predominance of the α mating type in all varieties of Cryptococcus and the presence of highly clonal populations led to the hypothesis that this pathogen might have a predominantly asexual life cycle. However, previous studies have shown that the dimorphic switch can occur during unisexual reproduction involving cells of only one mating type [17], [18]. Unisexual reproduction has been directly observed under laboratory conditions mainly in the serotype D lineage. In response to nutrient limitation, α cells of a single haploid isolate grown in solo culture can form a haploid or a diploid monokaryotic hyphae. Like bisexual reproduction (which can produce either a dikaryotic mycelium with paired haploid nuclei or a diploid monokaryotic mycelium [19]), during unisexual reproduction the haploid nuclei can either diploidize early or later in the basidium just prior to meiosis (Figure S1). Diploidization can result from either cell-cell and then nuclear fusion, or via endoreplication. The resulting hyphae grow to form basidia where meiosis and repeated rounds of mitosis and budding occur to produce haploid meiotic progeny as long chains of spores [18]. That the meiotic recombinase Dmc1 is required for spore production and germination shows definitively that unisexual reproduction is a meiotic process [18]. Candida albicans also has been found to undergo unisexual reproduction in the absence the of Bar1 protease, or in ménage à trois matings [20], similar to α-α cell fusion stimulated by a cells in Cryptococcus [18]. Unisex may serve to introduce genetic diversity and confer adaptive benefits.
The MAPK pheromone response signaling cascade governs dimorphism during bisexual and unisexual reproduction in C. neoformans. This signaling circuit is structurally and functionally conserved among both closely (i.e. Ustilago maydis) and distantly related fungi (i.e. S. cerevisiae). The pathway is activated by pheromones and orchestrates signal transduction through sequential phosphorylations evoked by the Ste20α/a (p21-activated kinase, PAK), Ste11α/a (MAPK kinase kinase), Ste7 (MAPK kinase), and Cpk1 (MAPK) kinases [21], [22]. Deletion of the corresponding genes severely impairs hyphal development during sexual development [21], [23]. In S. cerevisiae, the main transcriptional target of the pathway is Ste12, and a conserved STE12α/a homolog is present in the mating-type locus of C. neoformans [24], [25]. Recent studies have shown that the MAPK pathway target is Mat2, an HMG protein essential for bisexual and unisexual reproduction [26], that binds Pheromone Response Elements (PRE) in the promoters of the pheromone genes and 11 other pheromone induced genes [27]. A conserved cAMP pathway and a MAPK signaling cascade govern mating, morphogenesis, and pathogenicity in the closely related basidiomycete Ustilago maydis. However, the downstream transcription factor is a distinct HMG protein, Prf1, which regulates virulence and is pheromone-induced [28]. Thus the molecular pathway governing sexual development has been rewired since the divergence of both U. maydis and C. neoformans from their last common ancestor shared with S. cerevisiae.
While a-α bisexual reproduction and α-α unisexual reproduction are related sexual cycles, there are distinguishing characteristics. Notably, the cell identity homeodomain Sxi1α/Sxi2a protein complex is specifically required for bisexual mating but dispensable for unisexual reproduction [18]. Other distinct elements that may specifically regulate the two sexual cycles, and which remained to be identified involve hyphal elongation, basidia formation, meiosis, and sporulation.
The aim of this study was to identify and characterize unique regulators of unisexual reproduction. We employed a high-throughput genetic screen using an Agrobacterium-transkingdom DNA delivery approach to isolate random insertional mutants [26], [29], [30], [31] in a genome sequenced self-fertile haploid strain and isolates with filamentation defects were identified by microscopy. We disrupted genes encoding three regulators of unisexual development: Spo11, Ubc5, and Znf3. Spo11 is the homolog of S. cerevisiae Spo11, which induces DNA double-strand breaks (DSBs) that promote meiotic recombination [32]. UBC5 encodes a ubiquitin-conjugating enzyme that degrades short-lived or abnormal proteins in S. cerevisiae and also has a role in sporulation [33]. Deletion of either the SPO11 or the UBC5 gene significantly reduced spore number and germination providing additional definitive evidence, beyond the established role of Dmc1, that Cryptococcus unisexual reproduction is a meiotic process. Znf3 is a novel zinc finger protein. Phenotypic analysis of Znf3 insertion and deletion mutants showed that this component regulates hyphal development during both bisexual and unisexual reproduction. znf3 mutations impaired cell-cell fusion and blocked hyphal development, indicating a role in the early steps of sexual reproduction. Based on epistasis analysis, Znf3 is not a transcriptional target of the pheromone signaling cascade, but rather contributes to modulate Mat2 expression and is required for pheromone expression. Surprisingly, deletion of ZNF3 increased expression of transposons and transposon-related genes during unisexual reproduction. These results support the conclusion that Znf3 regulates sexual development through a different circuit parallel to the pheromone response pathway and governs pheromone gene expression and silences transposons, either directly or indirectly. Taken together these results proffer new insights into the control of hyphal development during bisexual and unisexual reproduction.
Results
Mutants altered for self-filamentous unisexual reproduction
To identify genes encoding novel components governing unisexual reproduction we utilized the ability of A. tumefaciens to transfer and mutagenize its target by randomly introducing a known DNA sequence into the genome. A. tumefaciens-mediated transkingdom DNA delivery has been successfully applied in C. neoformans to identify and characterize novel virulence factors and components of other sensing pathways [26], [30], [31], [34]. Similar Agrobacterium-mutagenesis studies by Lin et al. investigated the transition from yeast to hyphae with a limited number of mutants and identified Mat2, the major transcriptional regulator of the pheromone response signaling cascade [26] whereas Fu et al. identified the scaffold protein Ste50 [35].
Here, we sought to dissect each of the sequential steps of unisexual reproduction. A large library of insertion mutants was screened to identify and characterize genes required for hyphal initiation, hyphal elongation, basidia formation, and spore production. We utilized the haploid serotype D strain XL280α that, when solo cultured on appropriate media (V8, MS, FA, SLAD), undergoes robust hyphal development via unisexual reproduction [36]. The nuclear content of the vegetative yeast cells budded from the hyphae (blastospores) reflect the ploidy of the hyphae. XL280 generates a mixture of diploid and haploid hyphae during unisexual reproduction based on FACS (Fluorescent-Activated Cell Sorting) analysis in two independent experiments (Figure S1). Diploid blastospores [5/19 (26%) and 9/24 (37.5%) hyphae examined] are produced by budding from a diploid monokaryotic hyphae that is generated by early nuclear diploidization. In the haploid monokaryotic hyphae [14/19 (74%) and 15/24 (62.5%) hyphae examined] late diploidization occurs in the basidium to generate a diploid nucleus ([37]. In both cases the homozygous diploid nucleus undergoes meiosis and sporulation to produce the basidiospores that form four chains of spores on the surface of the basidium. Ploidy was congruent in all cases with hyphal behavior: diploids were hyperfilamentous compared to haploids.
XL280α is a laboratory strain derived from a cross of two sibling strains, JEC20a (congenic with JEC21α) and B3501α, which share ∼50% genome identity. Both of the parental strains have been sequenced [38]. Whole genome sequence analysis of XL280α reveals that it shares ∼81% genome identity with the congenic strains JEC20a and JEC21α (Ni, Feretzaki, Li, Floyd-Averette, Mieczkowski, Dietrich, and Heitman, submitted to PLOS Biology).
A. tumefaciens mediated insertional mutagenesis generated approximately 6,100 mutants yielding ∼1× genomic coverage. The mutants were transferred onto unisex-inducing V8 medium and incubated at room temperature in the dark for 14 days. The mutant library was then screened microscopically for enhanced or diminished hyphal development during unisexual reproduction. The library was also incubated on V8 medium for 14 days under constant light conditions. Light inhibits sexual development in Cryptococcus and this process is genetically regulated by the two light sensing genes BWC1 and BWC2 [30], [39]. To identify candidate genes for light sensing, we also screened the library for mutants that filamented equally well in the dark and the light.
We identified 225 mutants that exhibited a stable filamentation difference resulting in no hyphal growth (16), short hyphae (131), abnormal hyphae (21), more abundant hyphae (46), sporulation defects (2), increased chlamydospore production (6), or altered light response (3) (Figure 1). Mutants with sporulation defects and no hyphal growth or a hyperfilamentous phenotype were subjected to PCR to identify insertions in known candidate genes. Our screen was successful in isolating mutations in the previously identified genes CRG1, STE7, MAT2 (two independent mutants), and ZNF2, whose products are key components of the pheromone signaling cascade and in BWC2, which encodes a major regulator of light sensing [13], [21], [26], [30]. Isolates with increased ploidy (2N diploid or 3N triploid) are known to exhibit more robust filamentation during unisexual reproduction compared to congenic haploid strains [18]. All of the hyperfilamentous mutants were subjected to FACS analysis to determine their ploidy state, and four hyperfilamentous mutants were found to be diploid (2N). Using a microdissection microscope, 20 basidiospores were isolated from each diploid isolate; hyphal development of all of these progeny was similar to the wild type XL280α parental strain. FACS analysis of 15 progeny from each diploid showed that they were all haploid, indicating that their hyperfilamentous phenotype was due to an increase in ploidy (2N) (and not to a mutation enhancing diploidization). These findings further confirm that 1) α/α diploid isolates undergo unisexual reproduction to produce haploid progeny and 2) diploid spore isolates can be recovered from the sexual cycle (see also [19]).
Fig. 1. Phenotypic analysis of insertion mutants altered in unisexual reproduction. 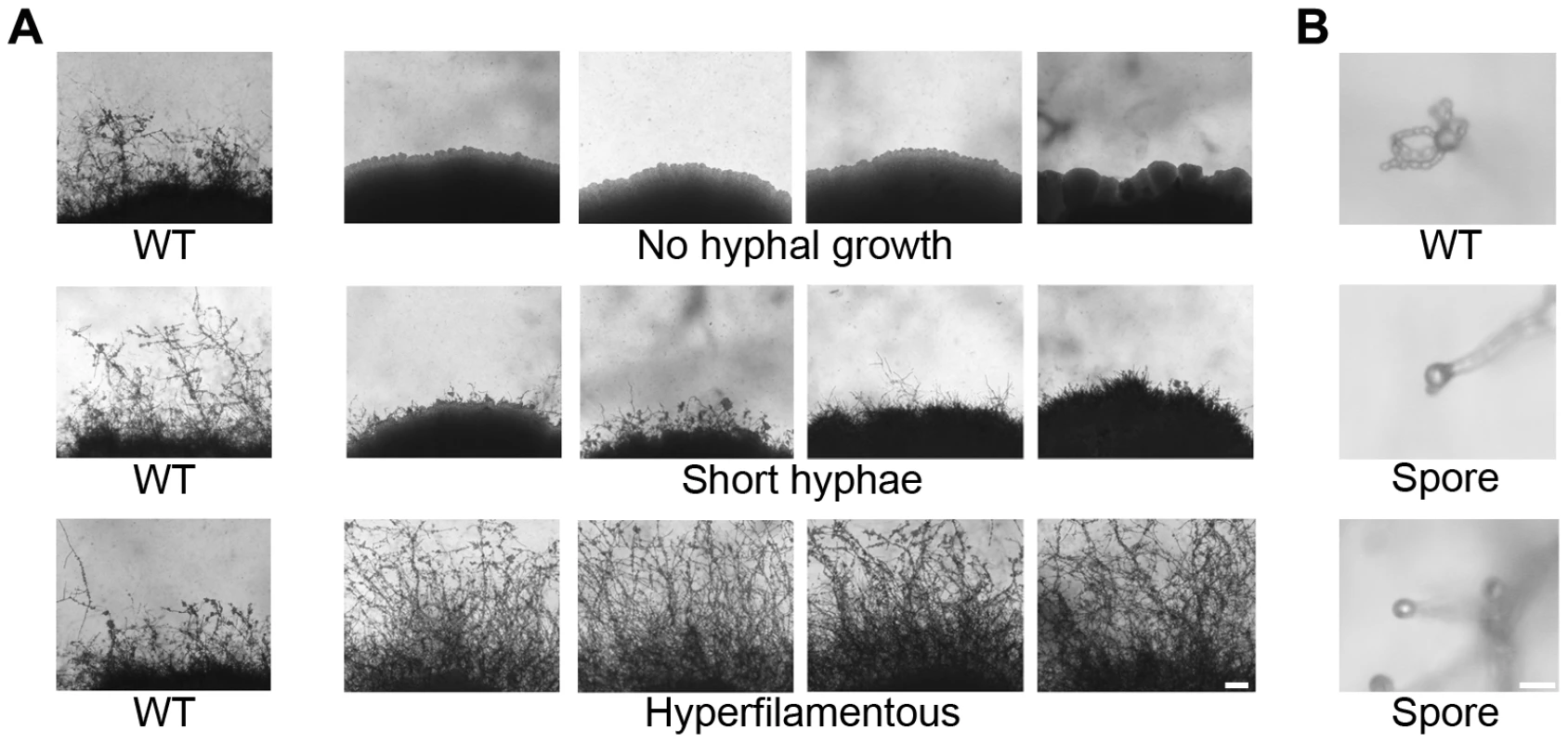
Nourseothricin (NAT)-resistant Agrobacterium transconjugant mutants of XL280α were grown on V8 solid agar for 14 days in the dark at room temperature to examine hyphal development during sexual reproduction by light microscopy. (A) The wild type strain XL280α undergoes robust hyphal growth during unisexual reproduction. The most extreme mutant phenotypes include no hyphal development, short hyphae, or hyperfilamentous strains. The scale bar represents 100 µm. (B) The progeny of wild type strain XL280α are organized in four long chains of spores that emerge from the surface of the basidium. Sporulation was severely impaired in several insertion mutants. The scale bar represents 10 µm. To identify insertions in unknown genes, the mutants were subjected to linkage analysis, inverse PCR, and sequence analysis. Linkage analysis of the mutants identified 12 in which the insertion was not linked with the mutant phenotype and these were excluded from further characterization. Of the remaining mutants 31 yielded an inverse PCR product. The sequences obtained were used in BLAST searches against the C. neoformans JEC21 genome database to identify the disrupted genes. Table 1 summarizes the insertion sites for 31 mutants. One of the newly identified genes from mutants that were afilamentous, CNK01880, encodes a protein with three zinc finger domains (Text S1, Figure S2). Zinc finger proteins are often involved in binding to nucleic acids (DNA and RNA), and the C2H2 motif is often a feature of transcription factors. The presence of three C2H2 zinc finger domains prompted us to name this protein Znf3 to distinguish it from two other genes encoding zinc finger transcription factors (Znf1 and Znf2) [26], [40]. Two sporulation defective mutants had insertions in two highly conserved genes. The first gene, CNH01330, encodes a topoisomerase homolog and the second, CND02770, encodes a ubiquitin-conjugating enzyme (see Text S1, Figure S2). Because these genes are the orthologs of the S. cerevisiae meiosis-specific endonuclease Spo11 and the ubiquitin-conjugating enzyme Ubc5, we named the Cryptococcus genes SPO11 and UBC5, respectively.
Tab. 1. Genes governing unisexual reproduction. 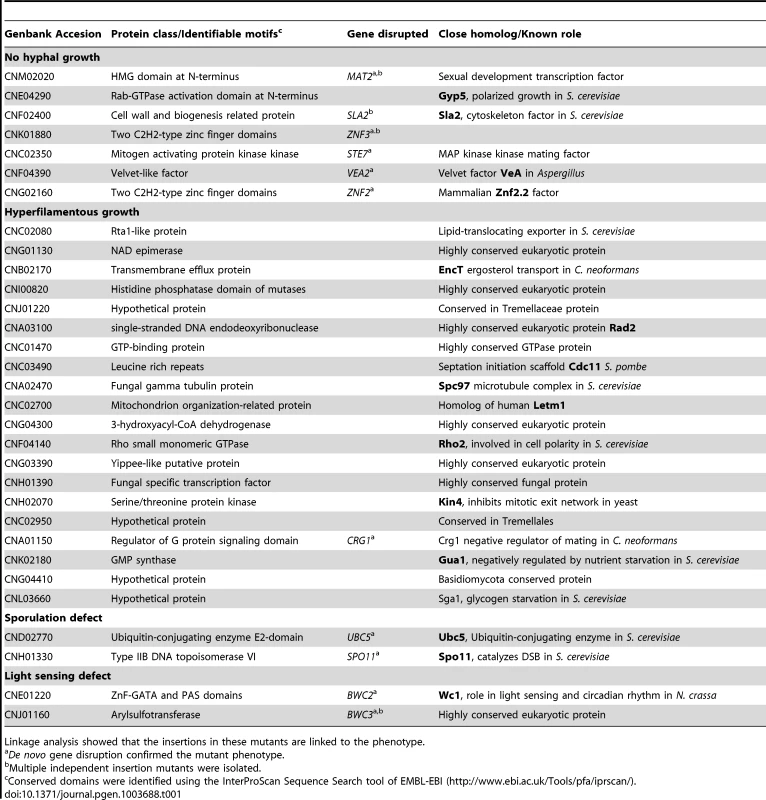
Linkage analysis showed that the insertions in these mutants are linked to the phenotype. Znf3 is essential for unisexual reproduction
The genetic screen revealed five different mutants with a similar filamentation defect, each with an insertion into a different site in the ZNF3 gene. One insertion was in the coding region of the gene, and four other insertions were in different positions of the promoter region. Insertion in promoters or terminators of a gene is a common feature of T-DNA insertions, which exhibit a bias towards promoters and 5′ and 3′ UTRs [41]. To elucidate the role of Znf3 in unisexual reproduction, we deleted the complete gene, from start to stop codon, in XL280α via biolistic transformation and homologous recombination. Two independent znf3Δ deletion mutants were generated, and the deletions were verified by Southern blot and PCR analyses. We assessed the ability of the insertion and deletion mutants to undergo unisexual reproduction by incubating the strains in the dark for two weeks on different filamentation-inducing media and examining them by light microscopy. The wild type XL280α strain formed hyphae along the entire periphery of the colony, while two znf3 insertion mutants were defective in hyphal production (Figure 2A). The znf3Δ mutation blocked hyphal development in both independent mutants (Figure 2A). Prolonged incubation on filamentation inducing media (≥3 weeks) did not result in the production of any hyphae, indicating that Znf3 is critical for the dimorphic transition during unisexual reproduction.
Fig. 2. Deletion of ZNF3 impairs hyphal development during bisexual and unisexual reproduction. 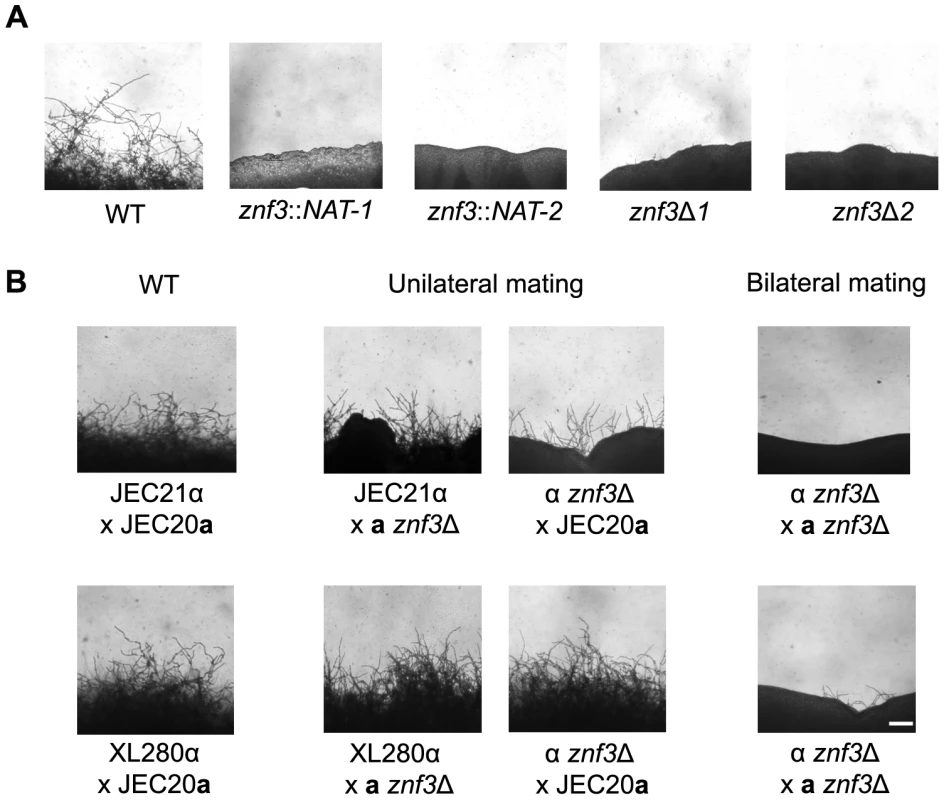
(A) XL280α, two mutants with independent insertions in ZNF3 (II-12, II-147), and two independent znf3Δ deletion mutants (MF01, MF38) were incubated on V8 medium in the dark at room temperature for 10 days and hyphal formation during unisexual reproduction was assessed by microscopy. (B) Bisexual wild type crosses (JEC21α×JEC20a and XL280α×JEC20a), unilateral mutant crosses (α znf3Δ×WTa and WTα×a znf3Δ), and bilateral mutant crosses (α znf3Δ×a znf3Δ) were conducted on V8 medium in the dark at room temperature for 7 days and photographed. The scale bar represents 100 µm. Znf3 plays a role in hyphal development during bisexual reproduction
Because the pheromone signaling cascade components are known to regulate both bisexual and unisexual reproduction [21], we hypothesized that Znf3 might also play a role in the dimorphic transition during bisexual reproduction. As mentioned earlier, the a-α sexual cycle is induced by the fusion of opposite-mating type cells to form a dikaryon, which initiates the development of dikaryotic hyphae and leads to the formation of basidia, where nuclear fusion and meiosis occur to produce chains of spores. To determine the role of Znf3 in bisexual reproduction, we deleted the gene in both strains of the serotype D congenic mating pair JEC21α and JEC20a via biolistic transformation. Because two different mating type partners are involved we used unilateral (one mating partner is mutant) and bilateral (both mating partners are mutant) mating assays to examine the role of Znf3. In the wild type JEC21α×JEC20a cross, filamentation was robust, with basidia and spores forming after 7 days of incubation on V8 medium. However, unilateral crosses of znf3Δ (α znf3Δ×WT or WT×a znf3Δ) produced significantly fewer hyphae, while in bilateral crosses (α znf3Δ×a znf3Δ), hyphal development was severely impaired (Figure 2B). Unilateral mutant crosses were able to complete the sexual cycle and form spores following prolonged incubation on V8 (∼2 weeks), while bilateral mutant crosses produced only sporadic hyphae. In contrast, the znf3Δ mutant phenotype was less severe in the hyperfilamentous XL280α background. Unilateral matings exhibited hyphae with basidia and spores at approximately wild type levels (Figure 2B). However, hyphal development was impaired in bilateral matings with sporadic short hyphae that eventually grew long enough to produce basidia and spores (∼3 weeks). Based on these findings, we conclude that Znf3 plays an important role during a-α mating.
Defects in hyphal development could be due to defects in fusion of α and a mating partners. Because the znf3Δ mutation blocked the dimorphic transition during bisexual reproduction, Znf3 could be involved in the initial step of mating. Cell-cell fusion assays were employed to determine the role of ZNF3 in cell-cell fusion. Wild type α and a strains and α znf3Δ and a znf3Δ mutant strains genetically marked with different drug resistance markers were mated on V8 medium for 15 or 24 hours in the dark. The cells were collected and plated in serial dilutions on media that select for doubly drug-resistant fusion products (to measure cell-cell fusion) or allow all cells to grow (to determine total CFU). In znf3Δ×WT unilateral matings, the relative fusion efficiency was ∼50% at 15 hours of co-incubation and ∼70% at 24 hours compared to wild type. Interestingly, in znf3Δ×znf3Δ bilateral matings, the fusion efficiency was reduced to ∼15% after 15 hours of co-incubation and ∼50% after 24 hours of co-incubation. In contrast, disruption of the CPK1 MAPK and MAT2 transcription factor genes essentially abolished cell-cell fusion in both unilateral and bilateral mating assays, indicating an essential role in the first steps of sexual reproduction [21], [26]. These observations support the hypothesis that Znf3 plays a modest role in cell-cell fusion events early in mating. However, with prolonged incubation times, znf3Δ mutant cells do undergo efficient cell-cell fusion. A possible explanation could be that Znf3 acts in parallel with the pheromone signaling cascade and that it is required to enhance the intracellular signal for the dimorphic transition during bisexual reproduction.
Znf3 regulates pheromone production
The dimorphic transition during bisexual and unisexual reproduction is in part induced by the production of peptide pheromones. The Cpk1 MAPK signaling cascade mediates sensing and pheromone production during sexual development [21], [26]. To determine whether Znf3 plays a role in the pheromone-sensing pathway, we generated expression profiles for the ZNF3, MAT2, SXI1α, and MFα1 genes in wild type, ste7Δ, mat2Δ, znf2Δ, sxi1αΔ, and znf3Δ mutant backgrounds. Gene expression was monitored during bisexual and unisexual reproduction by RT-PCR. We determined that expression of these genes in wild type bisexual reproduction reached the highest levels after 24 hours of incubation on filamentation-inducing V8 medium (Figure S3). MFα1 expression, which increased ∼2,500-fold, served as a positive control. During unisexual reproduction, expression of the genes in XL280α reached the highest level after 48 hours of incubation on V8. These time points were chosen to determine the expression of the genes in the different mutant backgrounds.
Both bilateral bisexual and unisexual reproduction were performed on V8 media and the cells were harvested at the designated times. RNA was isolated, and RT-PCR was employed to determine the transcript levels in each mutant. ZNF3 expression remained the same and similar to wild type, even when components of the MAPK signaling cascade were absent during crosses (Figure 3A). This indicates that Znf3 is not a transcriptional target of the pheromone-signaling cascade and that expression does not depend on the MAPK pathway. However, during bisexual reproduction, MAT2 expression was decreased in the znf3Δ mutant in an expression pattern similar to that of ste7Δ mutants previously observed (Figure 3B) [26]. The ste7Δ phenotype is in accord with the hypothesis that Mat2 is the direct target of the MAPK signaling cascade; however, Znf3 may moderately co-regulate the expression levels of Mat2 during hyphal development, and in fact MAT2 expression was also decreased in the znf3Δ mutant during unisexual reproduction. A similar phenotype was observed for Sxi1α. Previous studies have shown that Mat2 governs SXI1α through the Cpk1 MAPK cascade in bisexual reproduction [26]. SXI1α expression was abolished in ste7Δ and mat2Δ mutant backgrounds. Deletion of ZNF3 significantly decreased the transcript levels of SXI1α compared to wild type; however, the level of inhibition was less severe than in the ste7Δ and mat2Δ mutant backgrounds (Figure 3C). In unisexual reproduction, the znf3Δ mutant decreased SXI1α expression similar to ste7Δ and mat2Δ mutants (Figure 3C). Interestingly, the inhibition profile of SXI1α during bisexual reproduction was similar to unisexual reproduction, even though SXI1α does not participate in hyphal development during unisex.
Fig. 3. Znf3 regulates the expression of Mat2 and promotes pheromone production. 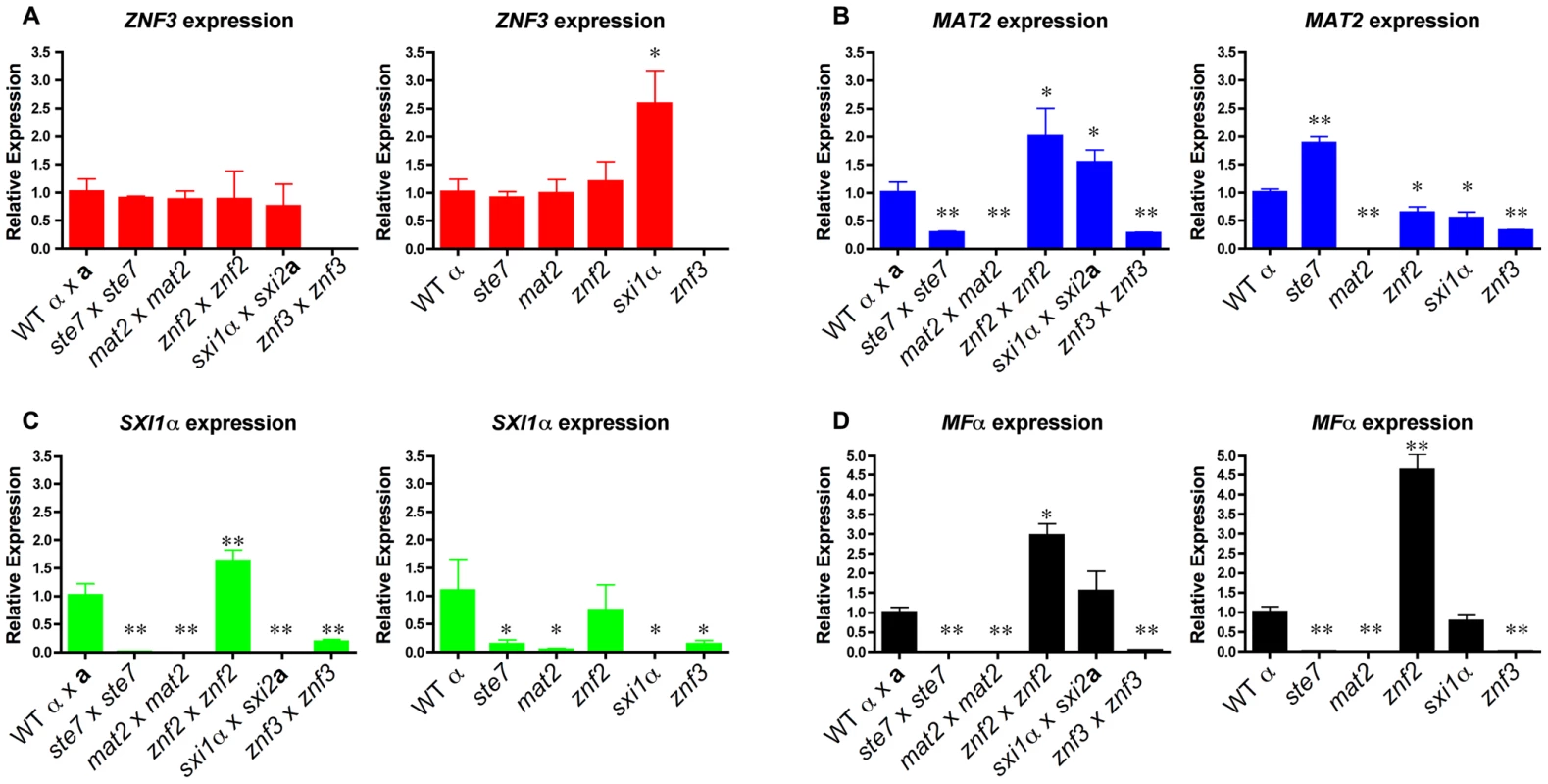
Cells were incubated for 24 hours on V8 medium for bisexual and 48 hours for unisexual reproduction. Yeast and hyphal cells were harvested and RNA was isolated. The expression of the (A) ZNF3, (B) MAT2, (C) SXI1α, and (D) MFα1 genes was measured by RT-PCR in wild type, ste7Δ, mat2Δ, znf2Δ, sxi1αΔ, and znf3Δ mutants (* indicates P<0.05 and ** indicates P<0.005 compared to the WT). The error bars represent the standard deviations from the mean for the three biological replicates. In further experiments, we determined the expression levels of other components of the MAPK signaling cascade. During bisexual reproduction the transcriptional profile of CPK1 was similar to MAT2 with modestly reduced levels in znf3Δ bilateral crossing cultures (Figure S4A). We also assessed the expression levels of two negative regulators of the pheromone pathway, CRG1 and CRG2, in the absence of components of the MAPK signaling cascade during unisexual reproduction (Figure S4B). CRG2 expression levels were slightly reduced in the deletion mutants. CRG1 transcription significantly increased in the znf2Δ mutant, which may contribute to the inhibition of hyphal development. However, expression of these negative regulators was not significantly altered in the znf3Δ mutant.
Pheromones are key activators of sexual development, as well as important targets. During vegetative growth the pheromones are expressed at basal levels and in a nutrient-limited environment the pathway is activated by a massive increase in pheromone production enforced by a positive feedback loop. Deletion of components of the Cpk1 MAPK cascade blocks expression of the pheromones and inhibits responses [21], [26]. By measuring the transcript levels of MFα1 in bilateral bisexual and unisexual reproduction in ste7Δ and mat2Δ mutant backgrounds, we found that the inducible expression level was completely blocked compared to the wild type (Figure 3D). In contrast, Znf2 functions further downstream and regulates hyphal formation without participating in the initiation of the pheromone signal [26]. MFα1 expression in the znf2Δ mutants was significantly increased in both sexual cycles, as previously observed (Figure 3D) [26]. On the other hand, the sxi1αΔ mutation modestly increased pheromone expression during bisexual reproduction as previously observed [42]. However, MFα1 production was significantly reduced in the znf3Δ mutant. During both bisexual and unisexual reproduction of znf3Δ mutants the expression of the pheromone gene was severely impaired (Figure 3D). This indicates that Znf3 is important for pheromone production, and the hyphal defect of znf3Δ mutants may result in part from impaired MFα1 expression. Nevertheless, the expression of the pheromone was not completely blocked and the expression profile pattern was similar to MAT2 and SXI1α expression in the znf3Δ mutant. Taking these results together, we can conclude that Znf3 is not regulated by the pheromone-signaling cascade; however, it may act in a parallel pathway that regulates the expression of the pheromone or Mat2 and consequently its targets, including Sxi1α.
Znf3 governs sexual reproduction through an independent pathway
Znf3 is required for hyphal growth during bisexual and unisexual reproduction, and we have shown that it plays an important role in pheromone expression. Although it is not a transcriptional target of the MAPK pathway, it may act downstream and contribute to or enhance the pheromone-induced signal. To investigate the possible role of Znf3 in the pheromone signaling cascade, we engineered an overexpression allele with ZNF3 under the control of the constitutively active GPD1 gene promoter. Wild type, a mat2Δ mutant, and a znf3Δ mutant were transformed with this overexpression allele using biolistic transformation. Based on RT-PCR results, expression of ZNF3 was increased ∼5-fold in the otherwise wild type PGPD1-ZNF3 background. Hyphal development was assessed on V8 medium by light microscopy. Hyphal growth in XL280α PGPD1-ZNF3 was similar but slightly more robust compared to wild type (Figure 4). Because a wild type ZNF3 complementation construct was not available, we transformed the znf3Δ mutant with the PGPD1-ZNF3 allele and found that this overexpression allele restored unisexual reproduction (Figure 4). Hyphal growth was slightly more condensed in the complementation strain due to the ∼5 fold increased ZNF3 expression, and it was moderately enhanced compared to the WT, possibly due to the expression from an exogenous locus.
Fig. 4. Znf3 is not a transcriptional target of the MAPK signaling cascade or Mat2. 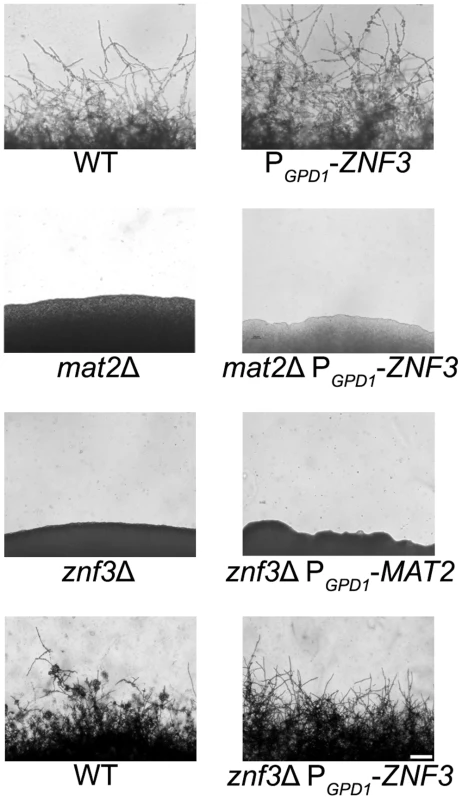
Wild type XL280α, mat2Δ, and znf3Δ mutants were transformed with the PGPD1-ZNF3 transgene, and a znf3Δ mutant was transformed with the PGPD1-MAT2 transgene. The cells were incubated on V8 medium for 14 days in the dark. The ZNF3 overexpression allele moderately increased hyphal development in the wild type and restored unisexual reproduction in the znf3Δ mutant; however, it failed to suppress the sterile phenotype of mat2Δ mutants. Similarly, the MAT2 overexpression allele failed to restore hyphal development of the znf3Δ mutant. The scale bar represents 100 µm. Mat2 is a major component of the MAPK pathway, and overexpression of a downstream target should bypass its requirement for mating and stimulate hyphal production. However, overexpression of ZNF3 failed to restore hyphal development in the afilamentous mat2Δ mutant, which indicates an independent function for Znf3 in unisexual reproduction (Figure 4). These observations provide further evidence that Znf3 does not act downstream of the pheromone signaling cascade.
Expression profiling showed that Znf3 moderately regulates the expression of MAT2 in C. neoformans, indicating a mode of control on the major regulator of sexual reproduction. Transcriptional control of mating and morphogenesis in the closely related species U. maydis involves additional transcription factors upstream of Prf1, the HMG target of the conserved MAPK pathway [28], [43]. Constitutive expression of PRF1 complements the severe defect caused by deletion of these factors. To determine if Znf3 functions upstream of Mat2 we used a MAT2 allele under the control of the constitutively active GPD1 promoter [26]. The overexpression MAT2 allele was introduced into znf3Δ mutants via biolistic transformation and hyphal growth was assessed by light microscopy. Overexpression of MAT2 in the znf3Δ background failed to induce the dimorphic switch from yeast to hyphae development (Figure 4). These results provide evidence that Znf3 and Mat2 may act in independent, parallel pathways. Transcription of MAT2 is tightly regulated by the presence of pheromone, and during mating a 12-fold increase in expression was observed (Figure S3). Decreased expression of MAT2 is possibly an indirect effect of the low pheromone levels observed in the znf3Δ mutant. The pathway that governs Znf3 and stimulates its control over pheromone expression warrants additional analysis.
Znf3 regulates the expression of retrotransposons during sexual development
A lack of any obvious link between Znf3 and the pheromone-signaling cascade prompted us to analyze the genome-wide transcriptional profile of the znf3Δ mutant. We performed a comparative transcriptome analysis between XL280α and the znf3Δ and mat2Δ mutants during bisexual and unisexual reproduction. In the first level of analysis, we considered genes that were regulated similarly in the znf3Δ and mat2Δ mutants. As expected, the majority of the genes that were downregulated in the mutants were involved in the pheromone signaling cascade during unisexual reproduction. The expression of the pheromone and pheromone receptor genes was significantly decreased in both znf3Δ and mat2Δ mutants compared to the WT (Table S1). However, the whole genome transcriptional profile of the znf3Δ mutant was different (Figure S5). A similar phenotype was observed during bisexual reproduction (Table S1). This result validates our previous findings and supports the hypothesis that Znf3 acts in an independent, parallel pathway.
To determine the role of Znf3 during unisexual reproduction and obtain further mechanistic insights, we extended our genome-wide expression analysis to genes that are upregulated specifically in the znf3Δ mutant. More than 500 independent tags yielded more than a two-fold increase in the znf3Δ mutant during unisexual reproduction (Table S2). We aligned the highly enriched tags to the C. neoformans JEC21α genome database and found that the majority encoded putative transposases, hypothetical endonucleases, and RNA-dependent DNA polymerases (Table 2). Surprisingly, we also found a significant increase in the expression of the T1 and T3 DNA transposons (Table 2). Previous studies have shown that transposable elements are highly expressed during bisexual and unisexual reproduction; however, the RNAi pathway inhibits transposon activity via Sex-Induced Silencing (SIS) and protects the genome against transposition and mutations [44], [45], [46]. Deletion of RDP1, a component of the RNAi pathway (the RNA-dependent RNA polymerase), induces the expression of retrotransposons during sexual development [44], [46]. The previously identified expression profile of the rdp1Δ mutant exhibits a similar increase in transposable elements and RNA-dependent DNA polymerases as the znf3Δ mutant (Table 2). This type of regulation was not observed in the mat2Δ mutant, indicating a unique role for Znf3 in transposon silencing. Taking these results together, we conclude that Znf3 is required for transposon silencing and pheromone expression during sexual development, and these interesting findings will provide the foundation for further analysis in future investigations.
Tab. 2. Transposon-related genes are upregulated during <i>znf3</i>Δ mutant unisexual reproduction. 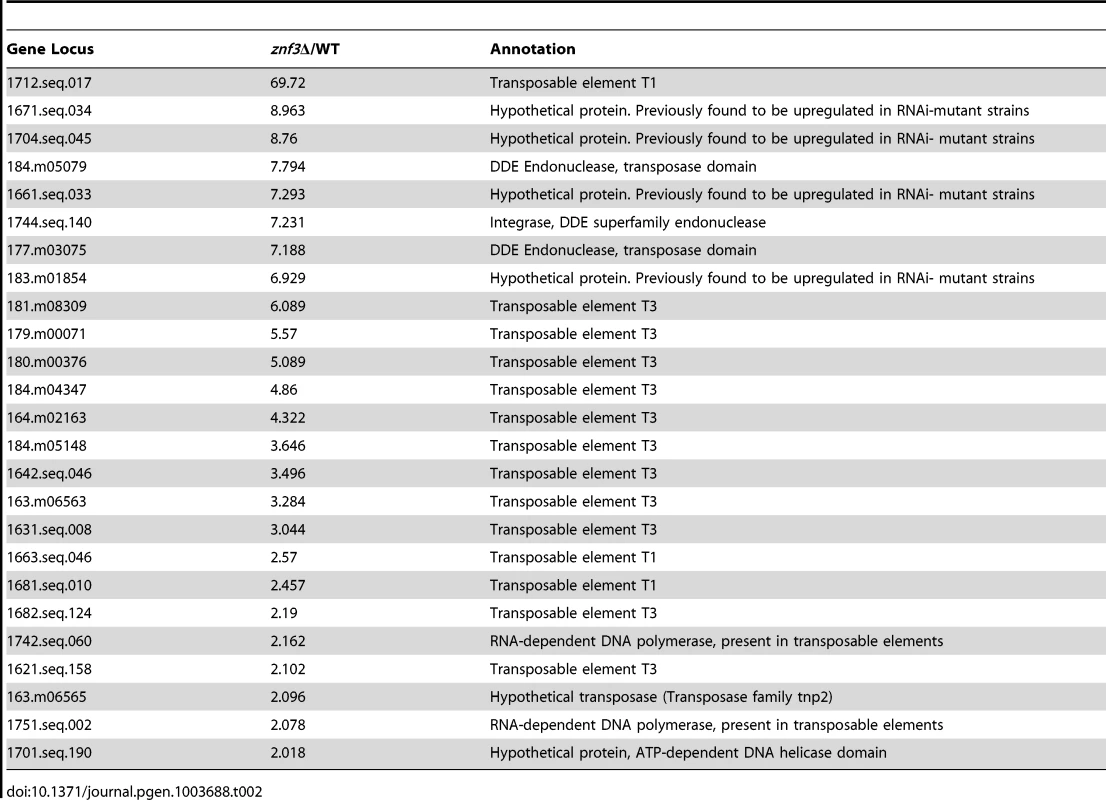
Spo11 and Ubc5 are required for sporulation
Pheromone production and sensing initiate sexual development by inducing cell fusion and hyphal development. At the termini of the hyphae, a bulb-like structure known as the basidium forms where meiosis and sporulation occur. Previous studies in model organisms have shown that the endonuclease Spo11 and members of the ubiquitin complex govern meiosis and sporulation. In S. cerevisiae homozygous spo11/spo11 mutants exhibit defects in meiosis and sporulation, while Ubc5 has been implicated in sporulation [33], [47], [48]. To establish the roles of Spo11 and Ubc5 in sporulation or meiosis in C. neoformans, both genes were deleted in XL280α via biolistic transformation and homologous recombination and two independent mutants were isolated for each. Unisexually reproducing cultures were incubated on filamentation-inducing media in the dark at room temperature for 10 days or longer. Hyphal development of the spo11Δ mutant was similar to wild type XL280α with hyphae covering the entire periphery of the colony (Figure 5A). Filamentation of the ubc5Δ mutants was also similar to spo11Δ and XL280α, albeit slower and slightly reduced. Deletion of either the SPO11 or UBC5 gene blocked sporulation during unisexual reproduction. In wild type XL280α, the majority of the basidia formed at the termini of the hyphae and were decorated with four long chains of basidiospores (Figure 5A). In the spo11Δ and ubc5Δ mutants, the basidia were either “bald” and completely lacked spores or had at most one or two spores and occasionally one very short chain of spores (Figure 5B). Complementation of spo11Δ and ubc5Δ mutants with SPO11 and UBC5 genes under the control of their native promoter and terminator, respectively, restored wild type sporulation.
Fig. 5. Spo11 and Ubc5 are required to complete sporulation during unisexual reproduction. 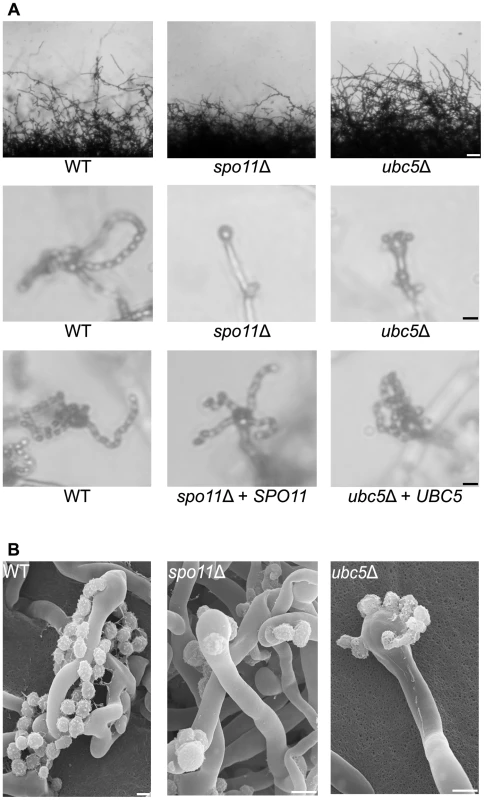
Wild type XL280α and spo11Δ and ubc5Δ mutants were incubated on V8 medium in the dark at room temperature for 14 days. (A) Hyphal development was evaluated by light microscopy. The edges of the colony produced abundant hyphae in WT, spo11Δ, and ubc5Δ mutants (upper row). In XL280α, basidia were decorated with four spore chains, whereas in spo11Δ and ubc5Δ mutants, spore production was severely impaired and naked basidia or at most one or two short spore chains were observed. The scale bars represent 100 µm for the upper row and 10 µm for the middle rows. (B) Scanning electron microscopic analysis of sporulation defects during unisexual reproduction. The scale bars represent 5 µm. Bisexual reproduction results in significantly longer basidiospore chains in a shorter incubation time compared to unisexual reproduction. We deleted the SPO11 and UBC5 genes in the congenic mating partners JEC21α and JEC20a to assess their role in unilateral and bilateral crosses. In wild type matings (JEC21α×JEC20a and XL280α×JEC20a), the basidia formed four long spore chains on filamentation-inducing media after 7 days incubation in the dark at room temperature (Figure 6A). In unilateral crosses spo11Δ and ubc5Δ mutants were able to form four spore chains, but they were significantly shorter and in some cases, the basidia had only two or three short chains or even as few as only two or three spores. In bilateral spo11Δ×spo11Δ or ubc5Δ×ubc5Δ crosses, both mutants exhibited a severe sporulation defect. Most of the basidia were bald or were decorated with only one or two spores on the hyphae of the entire periphery (Figure 6B). Upon prolonged incubation (∼3 weeks), hyphal development increased with longer and more dense hyphae; however, the basidia failed to yield more than at most two or three spores (Figure 6 and Figure S7). This indicates that Spo11 and Ubc5 are critical for sporulation. A similar phenotype has been observed in the absence of the meiotic-specific recombinase Dmc1 in C. neoformans [18]. Deletion of DMC1 results in basidia with a severe sporulation defect during bisexual and unisexual reproduction. In S. cerevisiae, Dmc1 is responsible for the repair of DNA double-strand breaks and homologous pairing during meiotic recombination [49]. Considering these results together, and based on the fact that SPO11 is highly conserved, we hypothesize that its requirement for sporulation is attributable to roles in meiotic recombination occurring during both bisexual and unisexual reproduction.
Fig. 6. Spo11 and Ubc5 are necessary for sporulation during bisexual reproduction. 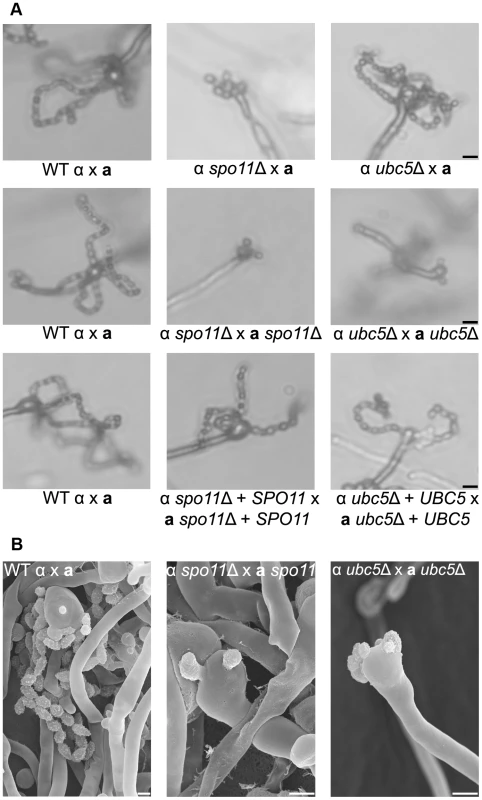
Sporulation was assessed in wild type (XL280α×JEC20a), unilateral (α spo11Δ×JEC20a, α ubc5Δ×JEC20a), and bilateral (α spo11Δ×a spo11Δ, α ubc5Δ×a ubc5Δ) mating assays on V8 medium at room temperature in the dark for 7 days. (A) Spore production was evaluated by light microscopy. In WT crosses basidia produce abundant spores organized in four chains. In unilateral mutant matings spore production was similar to WT although modestly diminished. Bilateral mutants exhibited a severe defect in sporulation with none, one, or two spores. The scale bar represents 10 µm. (B) Scanning electron microscopic analysis of sporulation defects during bisexual reproduction. The scale bar represents 5 µm. Spo11 is required for meiosis
An essential step prior to meiosis and sporulation is karyogamy. Karyogamy is characterized by two important steps: nuclear congression and nuclear fusion [50]. A series of genes are known to govern these steps in S. cerevisiae. In C. neoformans, how karyogamy is controlled remains largely unknown. A recent study identified a karyogamy gene, KAR7, which regulates nuclear fusion during bisexual and unisexual development [37]. Deletion of KAR7 blocks nuclear fusion and severely impairs sporulation, similar to spo11Δ and ubc5Δ. To determine possible roles of Spo11 and Ubc5 in karyogamy, bisexual and unisexual reproduction patches of wild type and mutant partners were incubated for 10 days on V8 medium in the dark. The sexual reproduction cultures were fixed in 4% paraformaldehyde and nuclear positioning was visualized with Hoechst staining by fluorescence microscopy. Nuclear fusion in spo11Δ and ubc5Δ was normal in both hyphae and basidia and similar to wild type (Figure S6), suggesting that the spo11Δ and ubc5Δ sporulation phenotype is unrelated to nuclear fusion and that it is caused by defects in a step subsequent to karyogamy.
During meiosis, recombination plays a crucial role in promoting pairing of homologous chromosomes in yeast and mammals. Meiotic recombination is initiated by the formation of DNA double strand breaks (DSBs) induced by Spo11. Spo11 is highly conserved in eukaryotic species and mutations can lead to sterility or offspring with an abnormal chromosome number. Fertility in mice is abolished in the absence of Spo11 [51]; in S. cerevisiae, the survival rate of spo11Δ meiotic progeny is extremely low with high levels of aneuploidy [52], [53]. To determine the survival rates in C. neoformans, unisexual reproduction cultures of wild type XL280α and spo11Δ and ubc5Δ mutants were incubated on filamentation-inducing V8 medium in the dark at room temperature for 10 days. Using a microscope with a microdissection needle, we dissected spores from four different mating unisexual development assays for each strain. The results are summarized in Table 3. For wild type XL280α, 30 spores were dissected from each assay. The germination rate of dissected spores ranged from 40% to 70%, as previously observed. Due to sporulation defects, spo11Δ and ubc5Δ generated only one or two spores per basidium, and thus the number of dissected spores was lower than wild type. However, the germination frequency of dissected spores for both spo11Δ and ubc5Δ was significantly lower than the wild type and ranged from 0% to 9%. Extremely low viability of the unisexual progeny is usually attributed to chromosomal abnormalities, which is indicative of abnormal meiosis. Thus the low germination frequency of spo11Δ and ubc5Δ could be due to high levels of aneuploidy in the spores.
Tab. 3. Viability of wild type, <i>spo11</i>Δ, and <i>ubc5</i>Δ unisexual reproduction progeny. 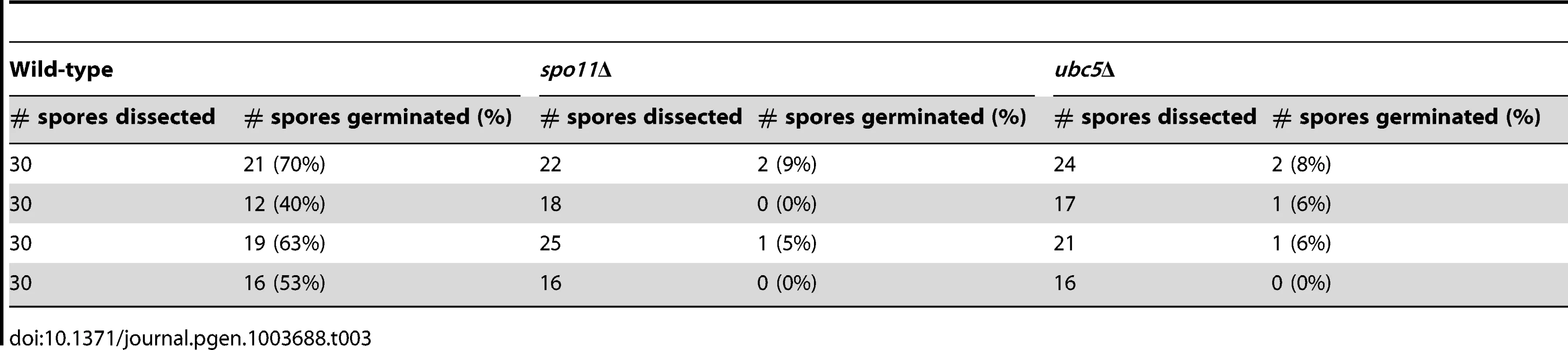
Based on sequence similarity, high homology, and the low germination rate of spo11Δ progeny, we reasoned that Spo11 may be required to catalyze DNA DSBs during meiosis in C. neoformans. Ionizing radiation has been found to suppress the meiotic defects of spo11Δ mutants and partially restore the wild type phenotype in S. cerevisiae and C. cinerea [54], [55]. To determine the role of Spo11 in DSBs during unisexual reproduction, cultures of XL280α, ubc5Δ, and spo11Δ were spotted on V8 medium to induce unisexual reproduction, incubated in the dark for 7 days, irradiated using X-rays at different doses (0, 1, 3, 5, 10, and 20 krad), and incubated for two additional days. Following irradiation, the spo11Δ mutant exhibited higher sporulation efficiency and a higher germination rate than the non-irradiated spo11Δ culture (Table 4 and Figure S8). A similar result was observed during spo11Δ×spo11Δ bisexual reproduction. Unlike the non-irradiated cultures of the spo11Δ bilateral cross (α spo11Δ×spo11Δ a), which produced very few spores, irradiated spo11Δ basidia had significantly more spores with considerably enhanced viability (Table 4 and Figure S8). Sporulation during both sexual cycles of WT and ubc5Δ was not significantly affected by irradiation, although viability was slightly lower, possibly due to higher rates of mutation (Table 4). XL280α, JEC21α, ubc5Δ, and spo11Δ mutants did not exhibit a growth defect in response to X-irradiation, and vegetative survival was similar between the WT and the mutants.
Tab. 4. X-irradiation-induced DSBs partially rescue the spore survival defect of spo11Δ mutants during bisexual and unisexual reproduction. 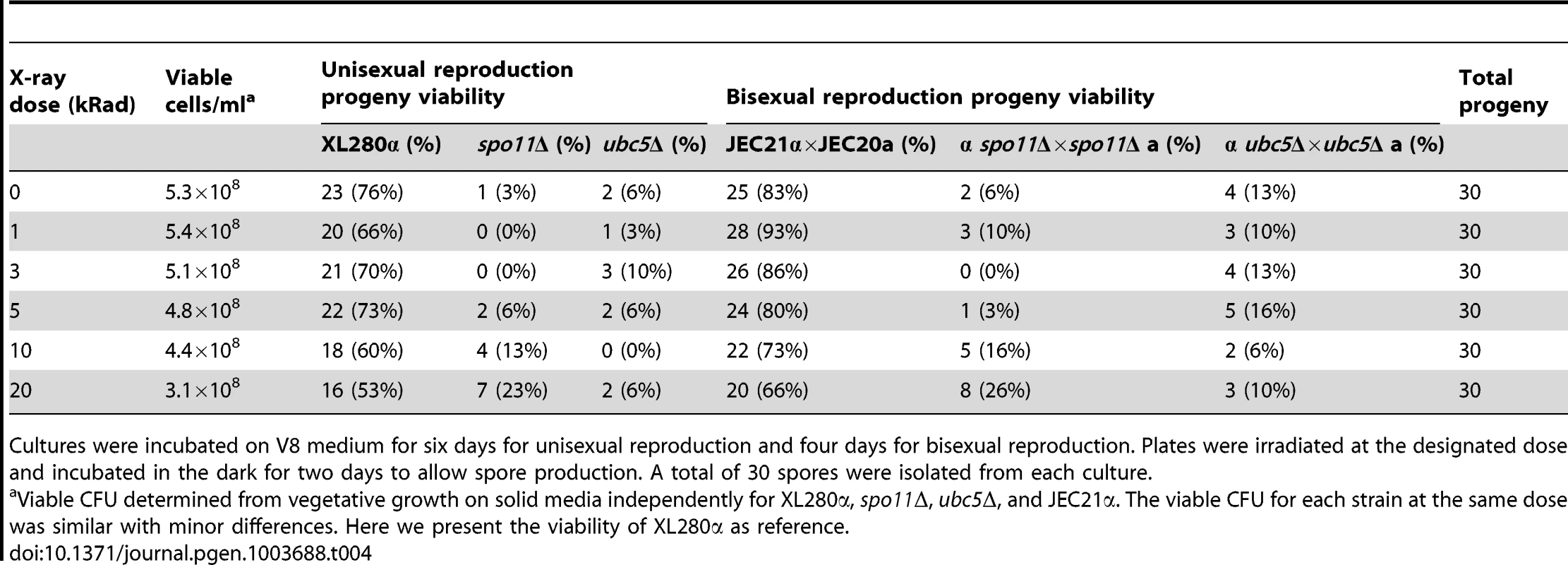
Cultures were incubated on V8 medium for six days for unisexual reproduction and four days for bisexual reproduction. Plates were irradiated at the designated dose and incubated in the dark for two days to allow spore production. A total of 30 spores were isolated from each culture. Taking these results together, we can conclude that 1) Spo11 is required for meiosis, likely by inducing DNA DSBs that promote recombination homologous pairing, and 2) Ubc5 plays an important role in sporulation that warrants further investigation. That Spo11 operates during unisexual reproduction provides further definitive evidence that this is a meiotic process.
Discussion
We employed a classical genetic approach to identify genes that orchestrate unisexual reproduction in C. neoformans. The screen identified a number of genes that may be directly or indirectly implicated in the dimorphic switch during mating. Importantly, we successfully isolated five previously identified regulators, CRG1, STE7, MAT2, ZNF2, and BWC2, validating this approach. Our findings provide evidence to implicate ZNF3, SPO11, and UBC5 in the control of hyphal development, meiosis, or sporulation during sexual differentiation. The severe defect in filamentation of the znf3Δ mutant provides evidence that Znf3 is a regulator of hyphal development during unisexual reproduction. The znf3Δ mutant phenotype is similar to that of ste7Δ and mat2Δ mutants and of other components of the pheromone pathway. In addition, RT-PCR analysis showed that the expression profiles of these mutants and znf3Δ followed a similar pattern during bisexual and unisexual reproduction. Hyphal development during bisexual reproduction in the znf3Δ mutant is also affected; however, the defect is less pronounced, and filamentation is mainly delayed rather than abolished. These defects are less severe than the defects observed in ste7Δ, mat2Δ, or znf2Δ α-a bilateral mutant sexual reproduction assays.
Regulation of pheromone expression is a key function of the components of the MAPK signaling cascade. Pheromone expression is dramatically attenuated in ste7Δ and mat2Δ mutants, as shown here and in previous studies [26]. A striking finding was the inhibition of MAT2 and pheromone expression in the znf3Δ mutant. Interestingly, we found that MAT2 transcription increases during mating (Figure S3), and znf3Δ and ste7Δ mutations have a similar impact on the transcriptional control of MAT2 and MFα1 during bisexual reproduction (Figure 3B), indicating that they may be in the same or related pathways. Epistasis analysis suggests Znf3 does not function upstream or downstream of Mat2. The lower expression of MAT2 could be attributable to the absence of pheromone in the ste7Δ and znf3Δ mutants. Pheromones activate the pathway that leads to expression of targets, including MAT2, whose transcription is regulated by itself, an unknown factor, or both. Based on these findings, we hypothesize that Znf3 imposes a mode of regulation on Mat2 via direct or indirect control of the pheromone through an independent signaling cascade that remains to be defined (Figure 7).
Fig. 7. Znf3 regulates pheromone expression activating the pheromone signaling cascade and promotes transposon silencing. 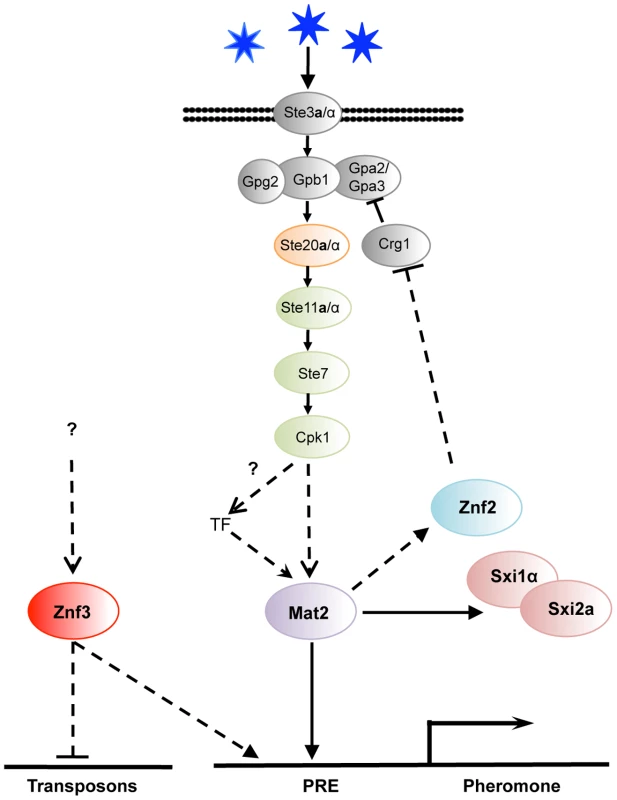
Mat2 is the transcription factor target of the pheromone signaling cascade that binds to the pheromone response element (PRE) in the promoter of the pheromone gene. Znf3 is required for induced levels of the pheromone and transposon silencing. Znf3 may bind additional cis-regulatory element to inhibit transposon mobilization during sexual development or function by binding to RNA targets. We found that ZNF3 transcription levels are similar during vegetative and hyphal growth, and ZNF3 may require activation through phosphorylation by the MAPK signaling cascade or via a different pathway. A recent study by Kruzel et al. showed that Mat2 is the pheromone response transcription factor, with a consensus binding motif present as the pheromone response element (PRE) in key mating genes [27]. The defined PRE sequence lies upstream of multiple genes (i.e. MFα1, MFα2, MFα3, STE3α, STE12α, SXI1α, and RAM1) and is recognized specifically by Mat2 [27]. However, extensive study of the MAPK signaling cascade in S. cerevisiae revealed that the promoter regions of several PRE-containing genes bear additional cis-regulatory elements that are recognized by different transcription factors or other elements, conferring additional levels of control [56], [57], [58]. Different environmental cues may govern these additional transcription factors, likely through pheromone-independent signaling cascades. It is possible that similar networks may contribute to PRE activation in C. neoformans. Znf3 may regulate the pheromone genes through another cis-regulatory element induced by a different environmental cue or an independent pathway that confers developmental specificity (Figure 7).
The identification and characterization of the targets of this novel factor will be essential to elucidate its molecular functions during mating. Surprisingly, deletion of ZNF3 increased the expression of transposons during unisexual reproduction. Combined with the fact that a similar transcriptional profile is observed in RNAi mutant strains, this leads to the conclusion that Znf3 may play a role in SIS and could be required to defend the genome against transposon mobilization. It is of interest that this protein appears to play a dualistic role during sex. It acts as an activator of pheromone expression and, simultaneously, as an inhibitor of transposon transcription (Figure 7). A possible explanation is that Znf3 plays a pivotal role in transposon silencing and is a molecular link between SIS and sexual development. Future studies will focus on the mechanism of transposon silencing by Znf3 and explore the links with sexual development.
Sexual reproduction has several stages that are each precisely regulated. Meiosis and sporulation are the ultimate steps. Our screen revealed two genes implicated in meiosis and/or sporulation. The first, UBC5, has a homolog in S. cerevisiae that is involved in ubiquitin-targeted degradation [33]. The second, SPO11, is considered a hallmark meiotic gene [59]. We found that deletion of either gene in C. neoformans results in barren basidia or basidia with only one or two spores with a very low germination rate. Deletion of DMC1, a previously identified meiosis-specific factor, also results in a severe sporulation defect, similar to the spo11Δ mutant [18]. We found that X-irradiation-induced DSBs partially restored the WT phenotype and enhanced the germination rate of the spo11Δ progeny. Along with the observation that Spo11 shares homology with the S. cerevisiae ortholog, we suggest that Cryptococcus Spo11 is required for initiating meiosis by catalyzing DSBs and inducing meiotic homologous recombination during bisexual and unisexual development.
Deletion of UBC5 shares a similar phenotype with the spo11Δ mutant, namely a severe sporulation defect and largely inviable meiotic progeny. This result is consistent with presumptive roles in meiosis or sporulation. In addition, Cryptococcus Ubc5 exhibits significant homology and identity throughout the fungal kingdom, possibly reflecting a conserved role. The S. cerevisiae Ubc5 homolog has been implicated in sporulation; however, the precise role(s) remains unknown. Ubiquitin-conjugating enzymes are associated with structurally similar but distinct recognition subunits that direct these enzymes to proteins that are targeted for degradation. Ubc5 is associated with the proteasome in S. cerevisiae, along with multiple E2 enzymes, and there are likely to be multiple physiological substrates, only some of which are involved in sporulation. Based on the highly conserved ubiquitin-conjugating enzyme E2 catalytic domain in Cryptococcus, we hypothesize that the role of Ubc5 involves degradation of multiple as yet unknown targets, and depletion of some of these targets is required for meiotic entry or sporulation.
Hallmarks of sexually reproductive species are the presence of recombinant progeny in the population and a group of highly conserved meiotic genes termed the “meiotic toolkit” in their genomes [59]. These genes represent the best markers for the capacity for a complete sexual cycle in a fungal species. Meiosis toolkit genes are present in C. neoformans and both our study and others have shown that two of these, SPO11 and DMC1, are required for meiosis and sporulation during bisexual and unisexual reproduction [18]. However the majority of meiotic gene candidates remain uncharacterized. S. cerevisiae represents an extensively studied meiotic system. Hop1 secures the synapses between homologous chromosome by binding DNA at sites where DSB will be induced by the Spo11 endonuclease [32], [60]. Dmc1 is required for DNA DSBs repair and catalyzes strand invasion and the formation of Holiday junctions that are stabilized by the Mnd1/Hop2 complex [61], [62]. Finally, Msh4 forms a dimer with Msh5 and facilitates crossing over, while Rec8 mediates cohesion between sister chromatids [63], [64]. Although the sequence of meiotic events is highly conserved in sexual species, the system exhibits exceptional plasticity in the fungal kingdom. For example, no true meiosis has been observed in C. albicans, which has a parasexual cycle, where random, concerted, chromosome loss occurs following mating, despite the presence of many meiosis-specific genes [65]. On the other hand, sporulation in C. lusitaniae is relatively efficient and involves Spo11-induced meiotic recombination, although it is missing multiple meiotic genes [66]. Moreover, in U. maydis, a basidiomycete, sexual development is efficient; however, it lacks key regulators of meiosis (Dmc1, Hop1, Hop2, and Mnd1) and has incorporated novel factors like homologs of H. sapiens BRCA2 and DSS1 [67]. In C. neoformans, we have found that Spo11 is required for meiosis, evidently for the induction of DSBs; however, taking into account the mechanistic and evolutionary plasticity of meiotic events in fungal species, the survey of the genome for the repertoire of meiotic-specific factors alone may well miss elements that require genetic approaches such as those applied here to unveil. Future research should focus on the genetic and evolutionary analysis of these and other unidentified genes to explore a possible conserved mechanism or a diverse rewiring network that controls meiosis and sporulation during C. neoformans unisexual reproduction.
Materials and Methods
Strains, media, and growth conditions
C. neoformans strains and plasmids used in this study are summarized in Table S3. Cryptococcus strains were maintained in −80°C glycerol stocks and grown at 30°C on rich YPD media (Yeast extract Peptone Dextrose). Strains harboring dominant selectable markers were grown on YPD containing nourseothricin (NAT) or G418 (NEO). Mating assays were performed on 5% V8 juice agar medium (pH = 7) in the dark at room temperature for the designated incubation period. To visualize spores and spore chains, mating assays were conducted on either V8 solid agar or on MS (Murashige and Skoog) medium minus sucrose (Sigma-Aldrich).
Insertional mutagenesis and phenotypic analysis of the mutants
Agrobacterium tumefaciens-mediated transformation was performed as previously described [29]. A. tumefaciens strain EHA105 transformed with the pPZP-NAT1cc plasmid was grown on Luria-Bertani (LB) medium containing kanamycin for 48 h at 25°C with shaking. The cells were washed briefly with sterile ddH2O and diluted in induction medium containing 100 µM acetosyringone to obtain an optical density of 0.15 at 660 nm. The cells were then incubated for 6 h while shaking at room temperature. The C. neoformans serotype D strain XL280α was grown overnight in YPD, washed with induction medium, and resuspended to a final density of 107 cells/ml. Equal aliquots of C. neoformans and A. tumefaciens (200 µl) were mixed and plated on induction medium agar containing acetosyringone. The cells were co-cultured for three days, scraped from the plates, and transferred to selective medium (YPD+NAT+100 µg/ml cefotaxime). The colonies were transferred into 96-well plates and grown in YPD+NAT+100 µg/ml cefotaxime as shaking cultures. A total of 6,100 transformants were spotted on V8 solid medium, incubated for 14 days in the dark at room temperature, and examined for hyphal development using light microscopy. Mutants with a stable filamentation defect were selected and clustered into groups based on similar phenotypes. Linkage between NAT resistance and the phenotype was assessed by crossing the insertional mutants with the JEC20a mating partner. Individual spores were isolated using a microdissection microscope, and the progeny were screened for the mutant phenotypes and NAT resistance. To identify the insertion sites, genomic DNA was digested with multiple restriction enzymes, purified, self-ligated, and subjected to inverse PCR using the pre-selected primers AI076/AI077. The PCR products were sequenced and the flanking regions were used in BLAST searches against the Cryptococcus genome to identify the mutated genetic loci [68].
Genomic DNA preparation
Cells were grown overnight in 50 ml YPD at 30°C in shaking cultures. The cells were harvested, washed three times with ddH2O, and frozen at −80°C overnight. The next day, they were lyophilized overnight and either stored at −20°C or genomic DNA was isolated using the CTAB protocol as previously described [69].
Gene disruption, complementation, and overexpression
Genes of interest were disrupted using an overlap PCR approach [14], [70]. The 5′ and 3′ flanking sequences of ZNF3, SPO11, and UBC5 were amplified from either XL280α or JEC21α genomic DNA. The dominant selectable markers NAT and NEO were amplified from plasmids pAI3 and pJAF1 respectively, with the universal primers M13F and M13R. The flanking sequences and the selectable markers were used to generate full-length deletion cassettes in an overlap PCR reaction. The construct was purified, precipitated onto gold microcarrier beads (0.6 mm, Bio-Rad), and introduced into strains XL280α or JEC20a via biolistic transformation as previously described [71]. Genes were replaced via homologous recombination and confirmed with PCR and Southern hybridization. Primers used to generate mutations are listed in Table S4. For complementation, the wild type SPO11 and UBC5 genes, along with their endogenous promoters and terminators, were amplified from XL280α genomic DNA with primer pairs JOHE37665/JOHE37666 and JOEH37667/JOHE37668, respectively, and cloned into the pJAF12 and pJAF13 vectors [14]. The plasmids were confirmed by sequenced, and spo11Δ and ubc5Δ mutants were biolistically transformed. The transformants were screened for spore production during mating by light microscopy. The wild type ZNF3 gene is exceptionally large (∼7,000 bp including promoter and terminator) and the construction of a complementation allele was challenging. To show that the hyphal defect was attributable to disruption of the gene, we screened and found a consistent afilamentous phenotype in several insertional mutants with independent insertions in ZNF3 and also in two independently derived znf3Δ mutants. To generate the ZNF3 overexpression plasmid pMF3, the wild type ZNF3 allele was amplified from XL280α genomic DNA using primer pair JOHE21470/JOHE21471. The gene was cloned into the pXL1 vector under the control of the constitutively expressed GPD1 promoter [72]. Cryptococcus strains were transformed with the circular plasmid via biolistic transformation. Independent transformants were selected for RNA extraction and screened to determine the expression levels of ZNF3 by RT-PCR.
RNA extraction and RT-PCR
RNA from cells undergoing bisexual reproduction was isolated by mixing equal amounts of a and α cells in an eppendorf tube. Then, 5 ml of cells were spotted on V8 medium and incubated for 24 hours at room temperature in the dark. RNA was isolated from unisexual reproduction cultures by spotting 5 ml of α cells on V8 for 48 hours at room temperature in the dark. Mating cultures were harvested, washed with ddH2O, and lyophilized overnight. RNA was extracted using TRIzol Reagent following the manufacturer's instructions (Invitrogen). Then, 5 µg of total RNA were subjected to DNAse treatment using Turbo DNAse (Ambion), and single-stranded cDNA was synthesized by AffinityScript RT-RNAse (Stratagene). Quantitative Real-Time PCR (RT-PCR) was performed on an Applied Biosystems 7500 Real-Time PCR System using Brilliant SYBR Green qRT-PCR master mix (Stratagene). For each target, a “no template control” was included and melting curves were analyzed to exclude primer artifacts and a single PCR product. All assays were conducted in triplicate. Data was normalized relative to the reference gene GPD1, and expression was determined using the 2−ΔΔCT approach. The primers used for RT-PCR are summarized in Table S4. The Student's t-test was employed to determine if the relative expression of the mutants was significantly different compared with the wild type (significance p<0.05).
Mating assays and microscopy
Each a and α mating partner was grown on YPD solid agar overnight at 30°C. The cells of each mating type were either mixed for bisexual reproduction or spotted individually for unisexual reproduction on V8 solid medium (pH = 7) for 7 or 14 days at room temperature in the dark. Hyphal structures and spores were visualized with an Eclipse E400 microscope (Nikon) equipped with a DXM1200F digital camera (Nikon) and interfaced with ACT-1 software (Nikon). Individual spores were isolated using a microdissection microscope equipped with a 25-µm microneedle (Cora Styles Needles ‘N Blocks, Dissection Needle Kit) as previously described [73].
To visualize nuclear position and fusion in basidia and the hyphae, mating assays were fixed in 4% paraformaldehyde for 20 minutes and then washed three times with sterile PBS. The cells were stained with 1 mg/ml Hoechst dye, incubated for 20 min, and washed with sterile PBS. The mating structures were mounted and captured by brightfield, differential interference (DIC), and fluorescence microscopy (DAPI channel) using a Zeiss Axioskop 2 PLUS equipped with an AxioCam MRm camera (Carl Zeiss Inc., Thornwood, NY).
For scanning electron microscopy, the mating samples were processed at the North Carolina State University Center for Electron Microscopy, Raleigh, NC, USA. Briefly, 1 mm3 blocks of matings performed on MS medium were excised from the agar and fixed with 0.1 M Na cacodylate buffer (pH = 6.8) containing 3% glutaraldehyde for several weeks at 4°C. The block was then rinsed with cold 0.1 M Na cacodylate buffer (pH = 6.8) three times and post-fixed in 2% osmium tetroxide in cold 0.1 M Na cacodylate buffer (pH = 6.8) for 2.5 hours at 4°C. Before viewing, the samples were critical-point dried with liquid CO2 and sputter coated with 50 Å of gold/palladium using a Hummer 6.2 sputter coater (Anatech). The samples were viewed at 15 kV with a JSM 5900LV scanning electron microscope (JEOL) and captured with a Digital Scan Generator (JEOL) image acquisition system. All of the microscopic images were processed with PhotoShop (Adobe).
Cell-cell fusion assay
To determine the cell-cell fusion frequency of wild type and mutants cells during bisexual reproduction, wild type strains XL561 (XL280α NAT) and XL491 (JEC20a NEO) and znf3Δ mutants MF38 (XL280α znf3Δ::NAT) and MF40 (JEC20a znf3Δ::NEO) were grown overnight in YPD liquid medium at 30°C. The cells were washed twice with ddH2O and diluted to a final density of 107 cells/ml. Both unilateral and bilateral mutant matings were assessed using XL926α (XL280α mat2Δ::NAT) and XL961a (JEC20a mat2Δ::NEO) mutants as negative controls. Equal amounts of cells were mixed (500 µl) and spotted on V8 medium (pH = 7) and incubated for 15 and 24 hours in the dark at room temperature. The cells were then removed, washed with ddH2O, and plated in serial dilution on YPD+NEO+NAT media to select for cell-cell fusion products. The cells were incubated for five days at room temperature. Cell-cell fusion efficiency was measured by counting the average number of double drug resistance CFU/total CFU in the wild type and mutant crosses.
Microarray and data analysis
To obtain RNA for microarray analysis, XL280α and the congenic znf3Δ mutant were grown in liquid cultures, washed in water, and spotted on V8 medium for 24 hrs. The unisexual reproduction cultures were harvested, washed in water, and lyophilized overnight. Three independent cultures were isolated for each strain and prepared for RNA isolation as biological replicates. The RNA was isolated using the RiboPure™-Yeast Kit (Ambion) according to the manufacturer's instructions (Life Technologies #AM1926). cDNA was synthesized using AffinityScript reverse transcriptase (Stratagene). cDNA was Cy3/Cy5-labeled and hybridized to a C. neoformans 70-mer microarray slide developed by the Cryptococcus Community Microarray Consortium (Washington University, St. Louis, MO). The arrays were washed and scanned with a GenePix 4000B scanner (Axon Instruments at Duke University DNA Microarray Core Facility) [74]. Three independent DNA microarrays were performed with three independent biological replicates. For statistical analysis, data was analyzed using GeneSpring software (Agilent) by employing Lowess normalization, reliable gene filtering, ANOVA analysis (significance p<0.05), and Microsoft Excel Software.
For the hierarchical clustering analysis the microarray expression data was carried out using the Limma Bioconductor package of the R statistical programming language [75], [76]. Briefly, normal exponential background subtraction was applied to the data followed by Lowess normalization. Significance of differential expression for the comparisons of the wild type versus mutant strains was then carried out using an empirical Bayes' moderated t-test. The False Discovery Rate method was applied to correct for multiple hypothesis testing [77]. Hierarchical clustering (complete-linkage, correlation distance) was accomplished using the R statistical programming language.
Fluorescence Activated Cell Sorting (FACS) analysis
Strains were processed for flow cytometry using XL280α as a haploid control and XL143 as a diploid control, as previously described [19]. Cells were grown overnight in YPD liquid media at 30°C, harvested, and washed with PBS buffer. They were fixed with 70% ethanol and incubated at 4°C overnight. The strains were washed with 1 ml of NS buffer [(10 mM Tris-HCl (pH = 7.2), 250 mM sucrose, 1 mM EDTA (pH = 8.0), 1 mM MgCl2, 0.1 mM CaCl2, and 0.1 mM ZnCl2], treated with 1 mg/ml RNase, and stained with 10 mg/ml propidium iodide in a final volume of 200 µl at 4°C overnight. Then, 50 µl of stained cells were diluted in 2 ml of 50 mM Tris-HCl (pH = 8.0) and sonicated for 1 min. Flow cytometry was performed on 10,000 cells and analyzed on the FL1 channel with a Becton-Dickinson FACScan at the Duke Cancer Institute Flow Cytometry Shared Resource.
X-irradiation procedures
Strains were grown overnight in liquid cultures, washed with water, and spotted on V8 medium pH = 7. It is crucial to impose irradiation treatment soon after karyogamy is completed and after the initiation of premeiotic DNA synthesis; however, the timing of these two processes are unknown in Cryptococcus. Therefore, we X-irradiated cultured crosses undergoing sexual reproduction at different time points (3, 4, 5, 6, 7, and 8 days incubation on V8 media) at a dose rate of 420 rad/min in an X-RAD 320 X-ray irradiator set at 320 kilovolts and 10 milliamps. Unirradiated cultures were plated in the same fashion and transferred to the X-ray machine to avoid inadvertent effects of these manipulations. Vegetative asexual cultures were plated in serial dilutions on solid YPD media and irradiated together with the sexual cultures to determine the vegetative survival rate and the presence of growth defects in response to radiation. After X-irradiation, crossing cultures undergoing sexual reproduction were incubated at room temperature in the dark for two additional days and assessed for sporulation efficiency with light microscopy. Germination rate was determined by isolating individual spores micro-manipulation from each culture and incubating these on YPD media to determine the number that formed a colony.
Supporting Information
Zdroje
1. Lin X, Heitman J (2007) Mechanisms of homothallism in fungi–transitions between heterothallism and homothallism. In: J. Heitman, J.W. Kronstad, J.W. Taylor, and L.A. Casselton, editor. Sex in Fungi: molecular determination and evolutionary implications. Washington, DC: ASM Press. pp. pp. 35–58.
2. MadhaniHD, FinkGR (1998) The control of filamentous differentiation and virulence in fungi. Trends Cell Biol 8 : 348–353.
3. LiuHP, StylesCA, FinkGR (1993) Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262 : 1741–1744.
4. RobertsRL, FinkGR (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes & Dev 8 : 2974–2985.
5. RuppS, SummersE, LoHJ, MadhaniH, FinkG (1999) MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO Journal 18 : 1257–1269.
6. Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A (2011) Cryptococcus: from human pathogen to model yeast: ASM Press.
7. Kwon-ChungKJ (1975) A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67 : 1197–1200.
8. Kwon-ChungKJ (1976) Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68 : 821–833.
9. HeitmanJ, AllenB, AlspaughJA, Kwon-ChungKJ (1999) On the origins of the congenic MATalpha and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet Biol 28 : 1–5.
10. HullCM, HeitmanJ (2002) Genetics of Cryptococcus neoformans. Annu Rev Genet 36 : 557–615.
11. IdnurmA, BahnYS, NielsenK, LinX, FraserJA, et al. (2005) Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol 3 : 753–764.
12. Kwon-ChungKJ (1976) A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68 : 943–946.
13. NielsenK, CoxGM, WangP, ToffalettiDL, PerfectJR, et al. (2003) Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 71 : 4831–4841.
14. FraserJA, SubaranRL, NicholsCB, HeitmanJ (2003) Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans variety gattii: implications for an outbreak on Vancouver Island. Eukaryot Cell 2 : 1036–1045.
15. VelagapudiR, HsuehYP, Geunes-BoyerS, WrightJR, HeitmanJ (2009) Spores as infectious propagules of Cryptococcus neoformans. Infect Immun 77 : 4345–4355.
16. GilesSS, DagenaisTR, BottsMR, KellerNP, HullCM (2009) Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect Immun 77 : 3491–3500.
17. WickesBL, MayorgaME, EdmanU, EdmanJC (1996) Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc Natl Acad Sci USA 93 : 7327–7331.
18. LinX, HullCM, HeitmanJ (2005) Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434 : 1017–1021.
19. SiaRA, LengelerKB, HeitmanJ (2000) Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol 29 : 153–163.
20. AlbyK, SchaeferD, BennettRJ (2009) Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460 : 890–893.
21. DavidsonRC, NicholsCB, CoxGM, PerfectJR, HeitmanJ (2003) A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol 49 : 469–485.
22. NicholsCB, FraserJA, HeitmanJ (2004) PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol Biol Cell 15 : 4476–4489.
23. WangP, NicholsCB, LengelerKB, CardenasME, CoxGM, et al. (2002) Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot Cell 1 : 257–272.
24. WickesBL, EdmanU, EdmanJC (1997) The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol 26 : 951–960.
25. ChangYC, WickesBL, MillerGF, PenoyerLA, Kwon-ChungKJ (2000) Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J Exp Med 191 : 871–882.
26. LinX, JacksonJC, FeretzakiM, XueC, HeitmanJ (2010) Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite - and same-sex mating in Cryptococcus neoformans. PLoS Genet 6: e1000953.
27. KruzelEK, GilesSS, HullCM (2012) Analysis of Cryptococcus neoformans sexual development reveals rewiring of the pheromone-response network by a change in transcription factor identity. Genetics 191 : 435–449.
28. KlostermanSJ, PerlinMH, Garcia-PedrajasM, CovertSF, GoldSE (2007) Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv Genet 57 : 1–47.
29. IdnurmA, ReedyJL, NussbaumJC, HeitmanJ (2004) Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell 3 : 420–429.
30. IdnurmA, HeitmanJ (2005) Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3: e95.
31. ChunCD, MadhaniHD (2010) Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS One 5: e12503.
32. KeeneyS, GirouxCN, KlecknerN (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 : 375–384.
33. SeufertW, JentschS (1990) Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 9 : 543–550.
34. WaltonFJ, IdnurmA, HeitmanJ (2005) Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol 57 : 1381–1396.
35. FuJ, MaresC, LizcanoA, LiuY, WickesBL (2011) Insertional mutagenesis combined with an inducible filamentation phenotype reveals a conserved STE50 homologue in Cryptococcus neoformans that is required for monokaryotic fruiting and sexual reproduction. Mol Microbiol 79 : 990–1007.
36. LinX, HuangJ, MitchellT, HeitmanJ (2006) Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα allele enhances filamentation. PLoS Genet 2: e187.
37. LeeSC, HeitmanJ (2012) Function of Cryptococcus neoformans KAR7 (SEC66) in karyogamy during unisexual and opposite-sex mating. Eukaryot Cell 11 : 783–794.
38. LoftusBJ, FungE, RoncagliaP, RowleyD, AmedeoP, et al. (2005) The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307 : 1321–1324.
39. LuYK, SunKH, ShenWC (2005) Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol Microbiol 56 : 480–491.
40. LengelerKB, FoxDS, FraserJA, AllenA, ForresterK, et al. (2002) Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot Cell 1 : 704–718.
41. AlonsoJM, StepanovaAN, LeisseTJ, KimCJ, ChenH, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 : 653–657.
42. HullCM, DavidsonRC, HeitmanJ (2002) Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev 16 : 3046–3060.
43. BrefortT, MullerP, KahmannR (2005) The high-mobility-group domain transcription factor Rop1 is a direct regulator of PRF1 in Ustilago maydis. Eukaryot Cell 4 : 379–391.
44. WangX, HsuehYP, LiW, FloydA, SkalskyR, et al. (2010) Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev 24 : 2566–2582.
45. WangX, DarwicheS, HeitmanJ (2013) Sex-induced silencing operates during opposite-sex and unisexual reproduction in Cryptococcus neoformans. Genetics 193 : 1163–1174.
46. JanbonG, MaengS, YangDH, KoYJ, JungKW, et al. (2010) Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet Biol 47 : 1070–1080.
47. JinR, DobryCJ, McCownPJ, KumarA (2008) Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol Biol Cell 19 : 284–296.
48. EnyenihiAH, SaundersWS (2003) Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163 : 47–54.
49. BishopDK, ParkD, XuL, KlecknerN (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69 : 439–456.
50. RoseMD (1996) Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu Rev Cell Dev Biol 12 : 663–695.
51. RomanienkoPJ, Camerini-OteroRD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6 : 975–987.
52. CaoL, AlaniE, KlecknerN (1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61 : 1089–1101.
53. KlapholzS, WaddellCS, EspositoRE (1985) The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110 : 187–216.
54. ThorneLW, ByersB (1993) Stage-specific effects of X-irradiation on yeast meiosis. Genetics 134 : 29–42.
55. CelerinM, MerinoST, StoneJE, MenzieAM, ZolanME (2000) Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J 19 : 2739–2750.
56. MadhaniHD, FinkGR (1997) Combinatorial control required for the specificity of yeast MAPK signaling. Science 275 : 1314–1317.
57. LalouxI, JacobsE, DuboisE (1994) Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res 22 : 999–1005.
58. Hwang-ShumJJ, HagenDC, JarvisEE, WestbyCA, SpragueGFJr (1991) Relative contributions of MCM1 and STE12 to transcriptional activation of a - and alpha-specific genes from Saccharomyces cerevisiae. Mol Gen Genet 227 : 197–204.
59. SchurkoAM, LogsdonJMJr (2008) Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. BioEssays 30 : 579–589.
60. WanL, de los SantosT, ZhangC, ShokatK, HollingsworthNM (2004) Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol Biol Cell 15 : 11–23.
61. ShinoharaA, GasiorS, OgawaT, KlecknerN, BishopDK (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2 : 615–629.
62. HenryJM, CamahortR, RiceDA, FlorensL, SwansonSK, et al. (2006) Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol Cell Biol 26 : 2913–2923.
63. NovakJE, Ross-MacdonaldPB, RoederGS (2001) The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158 : 1013–1025.
64. BuonomoSB, ClyneRK, FuchsJ, LoidlJ, UhlmannF, et al. (2000) Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103 : 387–398.
65. SherwoodRK, BennettRJ (2009) Fungal meiosis and parasexual reproduction–lessons from pathogenic yeast. Curr Opin Microbiol 12 : 599–607.
66. ReedyJL, FloydAM, HeitmanJ (2009) Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 19 : 891–899.
67. HollomanWK, SchirawskiJ, HollidayR (2008) The homologous recombination system of Ustilago maydis. Fungal Genet Biol 45 Suppl 1: S31–39.
68. McCullaghE, SeshanA, El-SamadH, MadhaniHD Coordinate control of gene expression noise and interchromosomal interactions in a MAP kinase pathway. Nat Cell Biol 12 : 954–962.
69. PitkinJW, PanaccioneDG, WaltonJD (1996) A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142 : 1557–1565.
70. DavidsonRC, BlankenshipJR, KrausPR, BerriosMd, HullCM, et al. (2002) A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148 : 2607–2615.
71. DavidsonRC, CruzMC, SiaRAL, AllenBM, AlspaughJA, et al. (2000) Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fung Genet Biol 29 : 38–48.
72. XueC, BahnYS, CoxGM, HeitmanJ (2006) G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol Biol Cell 17 : 667–679.
73. HsuehYP, IdnurmA, HeitmanJ (2006) Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet 2: e184.
74. KoYJ, YuYM, KimGB, LeeGW, MaengPJ, et al. (2009) Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 8 : 1197–1217.
75. Smyth GK (2005) Limma: limear models for microarray data. In: R. Gentleman VC, S. Dudoit, R. Irizarry, W. Huber editor. Bioinformatics and Computational Biology Solutions Using R and Bioconductor: Springer. pp. pp. 397–420.
76. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
77. BenjaminiY, HockbergY (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series 57 : 289–300.
Štítky
Genetika Reprodukčná medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



