-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
Understanding how genomes encode complex cellular and organismal behaviors has become the outstanding challenge of modern genetics. Unlike classical screening methods, analysis of genetic variation that occurs naturally in wild populations can enable rapid, genome-scale mapping of genotype to phenotype with a medium-throughput experimental design. Here we describe the results of the first genome-wide association study (GWAS) used to identify novel loci underlying trait variation in a microbial eukaryote, harnessing wild isolates of the filamentous fungus Neurospora crassa. We genotyped each of a population of wild Louisiana strains at 1 million genetic loci genome-wide, and we used these genotypes to map genetic determinants of microbial communication. In N. crassa, germinated asexual spores (germlings) sense the presence of other germlings, grow toward them in a coordinated fashion, and fuse. We evaluated germlings of each strain for their ability to chemically sense, chemotropically seek, and undergo cell fusion, and we subjected these trait measurements to GWAS. This analysis identified one gene, NCU04379 (cse-1, encoding a homolog of a neuronal calcium sensor), at which inheritance was strongly associated with the efficiency of germling communication. Deletion of cse-1 significantly impaired germling communication and fusion, and two genes encoding predicted interaction partners of CSE1 were also required for the communication trait. Additionally, mining our association results for signaling and secretion genes with a potential role in germling communication, we validated six more previously unknown molecular players, including a secreted protease and two other genes whose deletion conferred a novel phenotype of increased communication and multi-germling fusion. Our results establish protein secretion as a linchpin of germling communication in N. crassa and shed light on the regulation of communication molecules in this fungus. Our study demonstrates the power of population-genetic analyses for the rapid identification of genes contributing to complex traits in microbial species.
Published in the journal: Genome Wide Association Identifies Novel Loci Involved in Fungal Communication. PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003669
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003669Summary
Understanding how genomes encode complex cellular and organismal behaviors has become the outstanding challenge of modern genetics. Unlike classical screening methods, analysis of genetic variation that occurs naturally in wild populations can enable rapid, genome-scale mapping of genotype to phenotype with a medium-throughput experimental design. Here we describe the results of the first genome-wide association study (GWAS) used to identify novel loci underlying trait variation in a microbial eukaryote, harnessing wild isolates of the filamentous fungus Neurospora crassa. We genotyped each of a population of wild Louisiana strains at 1 million genetic loci genome-wide, and we used these genotypes to map genetic determinants of microbial communication. In N. crassa, germinated asexual spores (germlings) sense the presence of other germlings, grow toward them in a coordinated fashion, and fuse. We evaluated germlings of each strain for their ability to chemically sense, chemotropically seek, and undergo cell fusion, and we subjected these trait measurements to GWAS. This analysis identified one gene, NCU04379 (cse-1, encoding a homolog of a neuronal calcium sensor), at which inheritance was strongly associated with the efficiency of germling communication. Deletion of cse-1 significantly impaired germling communication and fusion, and two genes encoding predicted interaction partners of CSE1 were also required for the communication trait. Additionally, mining our association results for signaling and secretion genes with a potential role in germling communication, we validated six more previously unknown molecular players, including a secreted protease and two other genes whose deletion conferred a novel phenotype of increased communication and multi-germling fusion. Our results establish protein secretion as a linchpin of germling communication in N. crassa and shed light on the regulation of communication molecules in this fungus. Our study demonstrates the power of population-genetic analyses for the rapid identification of genes contributing to complex traits in microbial species.
Introduction
In most filamentous ascomycete species, hyphae form an interconnected network or syncytium of multi-nucleate cells known as a mycelium [1]. In nature, the formation of a mycelium often occurs via the germination of wind-dispersed asexual spores (conidia) [2]. Upon landing on a suitable substrate, conidia germinate to form germlings that are capable of fusion via specialized structures called conidial anastomosis tubes (CATs) to form the interconnected mycelial network common in this group of organisms [3], [4]. The formation of mycelial networks by germling fusion increases cytoplasmic flow and is important for the distribution of nutrients, signals and organelles within the colony [5], [6].
Similar to cell fusion in other organisms, the process of germling fusion in the filamentous ascomycete fungus Neurospora crassa requires cell recognition and attraction, adhesion, cell wall remodeling and membrane merger [7]. Genetically identical germlings of N. crassa exhibit remarkable chemotropism to each other, which enhances the formation of the inter-connected hyphal network [8], [9]. A number of mutants have been identified in N. crassa that fail to undergo germling and hyphal fusion, including nrc-1, mek-2 and mak-2, which are components of a conserved MAP kinase pathway [3], [10], [11], [12]. Other mutants of unknown biochemical function, such as soft (so), also show defects in chemosensing and cell fusion [13], [14]. The components of the MAP kinase pathway (NRC1, MEK2 and MAK2) and SO are recruited in a rapid and oscillatory manner to the plasma membranes of germling pairs undergoing chemotropic interactions [12], [14]. The oscillation of MAK2 and SO to CAT tips has been proposed to allow genetically identical cells to alternate between two different physiological states associated with signal delivery or response [14], [15], [16]. Given the complex physiology of cell communication and fusion, many other genes and proteins likely play a role in this process.
N. crassa is a heterothallic, obligate outbreeding species that has been a model for the study of population structure and genetic variability of fungi in the wild [17], [18], [19], [20], [21]. Recent advances in nucleic acid sequencing technologies have allowed for large-scale sampling of wild populations in this model microbe, and we recently harnessed this strategy in a population structure analysis of N. crassa by RNA-seq [21]. Data from such a sequencing survey provides a dense map of genetic variants across the genome and raises the possibility of genome-wide association studies in N. crassa. Association mapping is a powerful tool to identify candidate cases in which genetic variation at the DNA level underlies differences between wild individuals in a trait of interest. This approach is in common use in human genetics but has had little application to date in model organism systems, although recent work has established the power of association studies in mapping the genetic basis of trait variation across wild individuals in Drosophila [22], [23], [24], Arabidopsis [25], [26], [27], [28] and sunflower [29]. In fungi [30], [31] and in most other organisms beside humans, studies seeking to use natural variation as a screening tool to map genotype to phenotype have been largely limited to experimental cross designs, which survey polymorphisms in only a few wild individuals.
Here we describe the results of the first genome-wide association analysis used to identify novel loci underlying trait variation in a microbial eukaryote. We applied an association strategy using wild isolates of N. crassa to identify the genetic basis of the complex trait of germling communication. Developing a detailed, quantitative assay well-suited to the medium-throughput association-mapping paradigm, we surveyed germling communication across wild N. crassa strains and mapped differences in this trait to DNA sequence variants. We subsequently tested the function of genes mapped in our association study by assessing the germling communication phenotype of deletion strains, revealing mutants that showed both decreased and increased germling fusion frequency. We also tested the effect of some gene deletions on MAK2 and SO oscillation during chemotropic interactions. And we localized within hyphae the protein product of the gene that showed the most significant association with germling communication phenotype, a homolog of mammalian neuronal calcium sensor-1 (NCS-1).
Results
Efficiency of germling communication varies across individuals in a wild population of N. crassa
Our previous study of the relatedness of wild N. crassa isolates from the Western hemisphere by RNA-seq revealed a well-defined population of 20 individuals from Louisiana [21]. To establish a larger set of genotyped Louisiana strains suitable for use in association mapping, we transcriptionally profiled an additional 92 Louisiana strains (Table S1). Analysis of the regulatory variation across the Louisiana population detected in these data will be reported elsewhere; here we used the RNA-seq reads to identify 1.09 million single-nucleotide polymorphisms (SNPs) in coding regions of the seven N. crassa chromosomes (Dataset S1). Phylogenetic analysis of these SNPs (Figure S1) indicated a set of 100 strains with little population substructure, including the smaller sample of Louisiana isolates that we had previously characterized [21]. We identified 81,614 SNPs at which the minor allele was present in >25% of strains, and which were equally distributed throughout the euchromatic regions of all seven chromosomes of N. crassa (Figure S2 and Dataset S2). Across the 9,730 protein-coding genes of the N. crassa genome (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html), the average gene harbored ∼10 high-frequency SNPs.
To use our genotyped Louisiana strains to dissect the genetics of germling communication, we first developed a communication assay as follows. When genetically identical macroconidia of the N. crassa laboratory strain FGSC 2489 germinate near each other, ∼89% of the germlings within 15 µm of other germlings sense their neighbors, reorient their growth, and engage in cell fusion via CATs [3] (Figure 1A). The remaining germlings ignore each other, do not show chemotropism, do not form CATs and do not fuse (Figure 1B). We thus quantified communication by isolating macroconidia from each given wild strain, plating them on agarose minimal medium, and tabulating the percent of germling pairs exhibiting redirected CAT growth (communication) or fusion after 3–4 hours of incubation. Applying this procedure to 24 Louisiana strains showed that the germling communication trait varied among the wild isolates, from a high of 90% communication/cell fusion efficiency to a low of less than 40% communication (Table S2 and Figure 2).
Fig. 1. Chemotropic interactions and cell fusion between genetically identical conidial germlings. 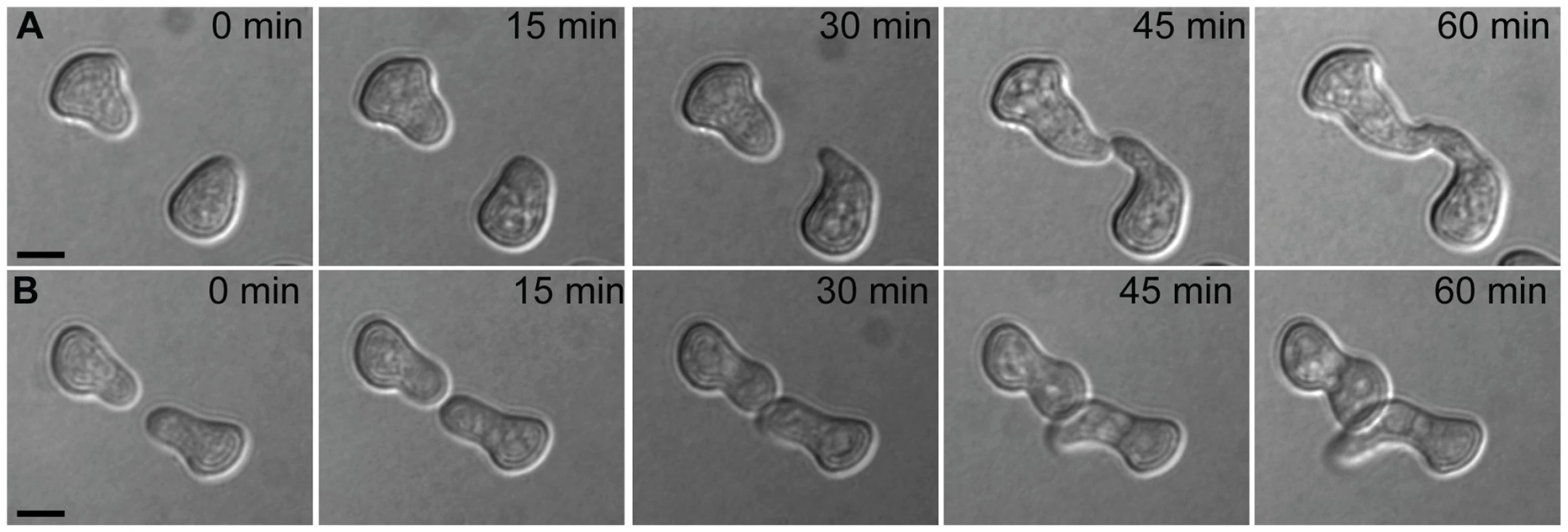
Each row shows one pair of germlings of the N. crassa laboratory strain FGSC 2489 during a fusion time-course. (A) Chemotropic interactions resulting in cell fusion. (B) Incomplete communication and fusion failure. Scale bar = 5 µm. Fig. 2. Germling communication frequency varies among wild N. crassa isolates from a single population in Louisiana. 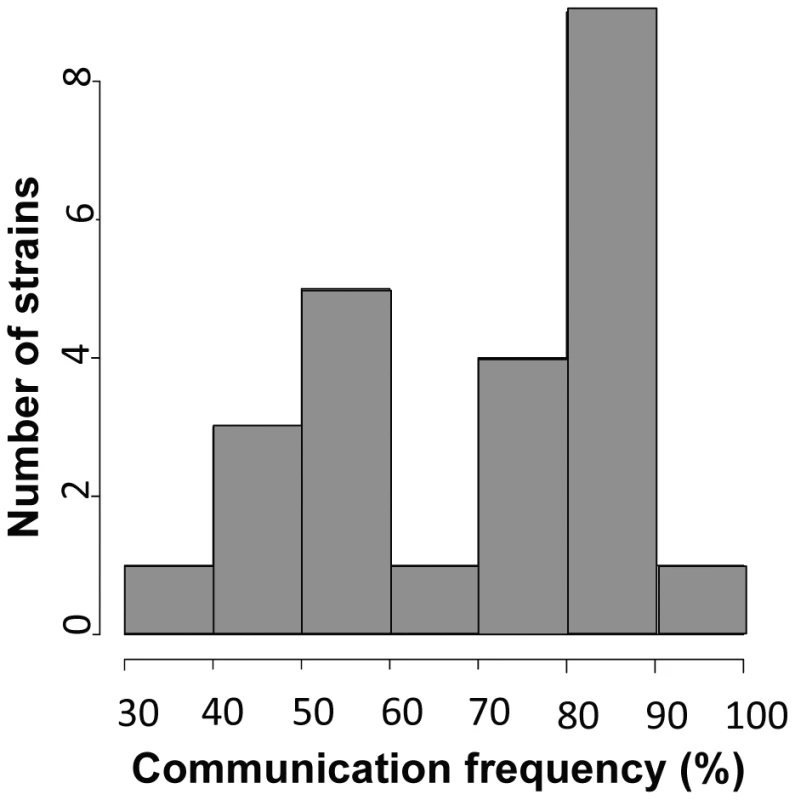
The x-axis shows the frequency of communication observed between genetically identical germling pairs for a given strain, and the y-axis reports the number of strains, among 24 Louisiana isolates, that exhibited the communication frequency denoted on the x axis. A Golgi-localized homolog of neuronal calcium sensor-1 is required for germling communication
To map loci underlying the variation in communication efficiency/cell fusion across our wild population, we first scored patterns of germling interactions as a qualitative, binary trait, such that the phenotype of a given individual was classified as either avidly or poorly communicating. We then used our set of genotypes at high-frequency SNPs to test each locus in turn for co-inheritance with the communication trait across the strains of the population, using a permutation strategy, described in Methods, to correct for multiple testing. This mapping calculation yielded 3 SNPs showing significant association with germling communication at a threshold at which we expected ∼0.01 SNP by chance (Figure 3 and Dataset S3). All three SNPs lay in the 3′ UTR of the gene NCU04379 with linkage disequilibrium decaying sharply around this peak (Figure 4); we detected no differential expression of NCU04379 between strains with avid germling communication and those whose germlings communicated poorly (data not shown).
Fig. 3. Genome-wide association of germling communication frequency on the seven chromosomes of N. crassa. 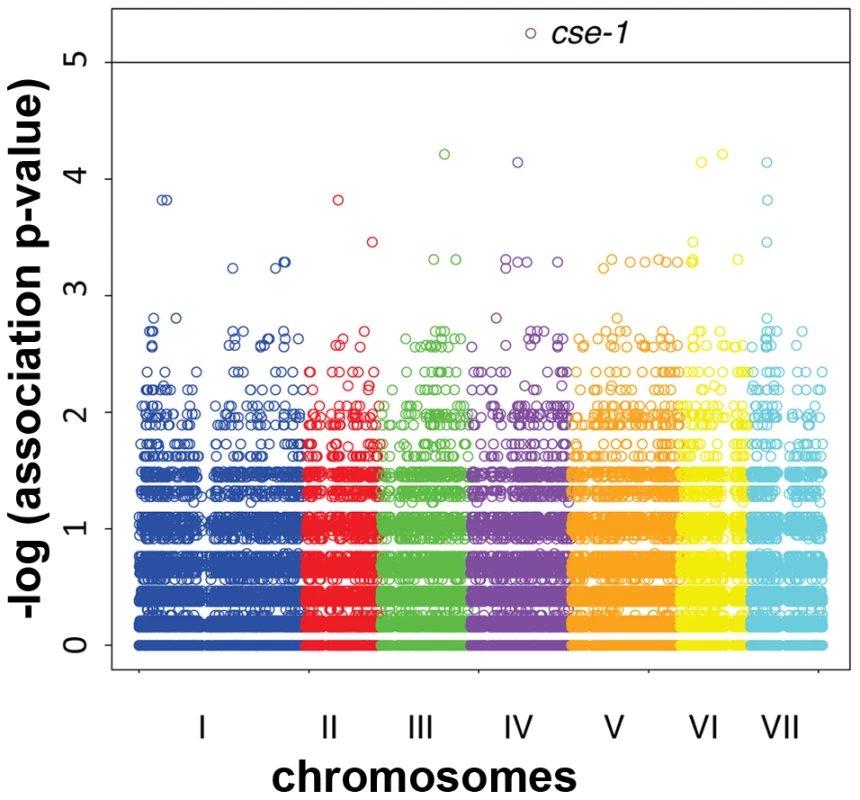
Each data point represents one single-nucleotide polymorphism (SNP) at which inheritance was tested for association with frequency of germling communication across 24 wild strains. The x-axis reports SNP position on each chromosome as labeled with Roman numerals, and the y-axis reports the negative logarithm of the nominal association p-value by Fisher's exact test. The horizontal line indicates the significance threshold at which we would expect 0.1 SNP to score by chance. Association signals from three SNPs in cse-1, whose chromosomal positions are not distinguishable on this scale (see Figure 4), are visible as a single point as indicated; in each case the association p-value is 5.6×10−6, where 0.011 SNP is expected by chance. Fig. 4. Variants in the 3′ UTR of cse-1 (NCU04379) are associated with germling communication frequency. 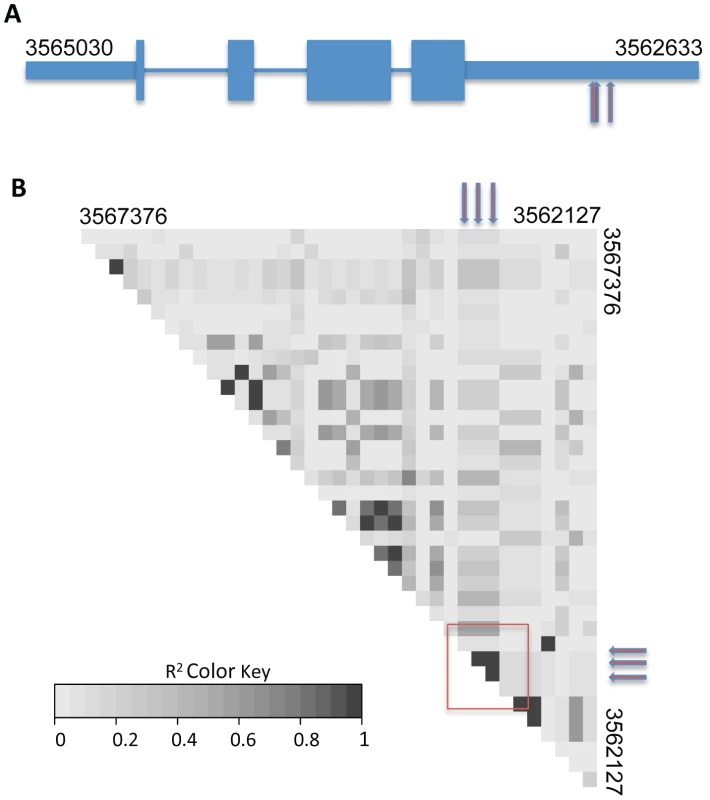
The remarkable precision of GWAS in the wild, Louisiana, N. crassa population is shown by identification of associated SNPs in just one region of one gene. (A) Cartoon of exon structure of CSE1. Narrow rectangles represent untranslated regions, wide rectangles represent exons and lines indicate introns. Red arrows represent positions of single-nucleotide polymorphisms at which inheritance associates strongly with germling communication in wild strains (SNP positions 3562999, 3562998, and 3562957 on chromosome IV). (B) Each grey box reports linkage disequilibrium (the degree to which one allele at one SNP preferentially appears in the population with one allele at a second SNP) as measured by r2 between one pair of high-frequency SNPs in a region of chromosome IV spanning cse-1. Red arrows represent the associating SNPs from (A), and the region of the heatmap reporting linkage disequilibrium between them is denoted with a red box. NCU04379 encodes CSE1, a homolog of the vertebrate neuronal calcium sensor-1 (NCS-1) and of Frq1p in Saccharomyces cerevisiae [32]. Deletion of cse-1 in N. crassa results in a mutant that is viable, but sensitive to calcium stress and ultraviolet light, and which shows slightly impaired growth [33]. Similar to NCS-1 and Frq1p, CSE1 harbors a consensus signal for N-terminal myristoylation and four EF-hand domains (PF00036) predicted to be involved in calcium binding [32], [34], [35]. We hypothesized that CSE1 played a role in germling communication and that mutations in this gene would impact cell fusion behavior. Germling CAT fusion experiments validated this prediction, revealing a striking 3.6-fold reduction in the frequency of communication and cell fusion between Δcse-1 germlings relative to communication between germlings of the wild-type, isogenic strain from which the Δcse-1 strain was derived (Figure 5A). The defect was rescued by integration of a wild-type copy of cse-1 at the his-3 locus in the Δcse-1 strain, confirming the specificity of the phenotype to the cse-1 lesion (Figure S3). To evaluate the ability of Δcse-1 germlings to respond to communication with wild-type isolates, we assayed Δcse-1 germlings positioned alongside those of the isogenic fusion-competent strain, and observed a defect similar to that of Δcse-1 germlings interacting with one another (Figure 5A). Thus, CSE1 is essential for chemotropic interactions, including the sensing of and response to the presence of a fusion-competent partner.
Fig. 5. Nine novel genetic determinants of germling communication and fusion frequency. 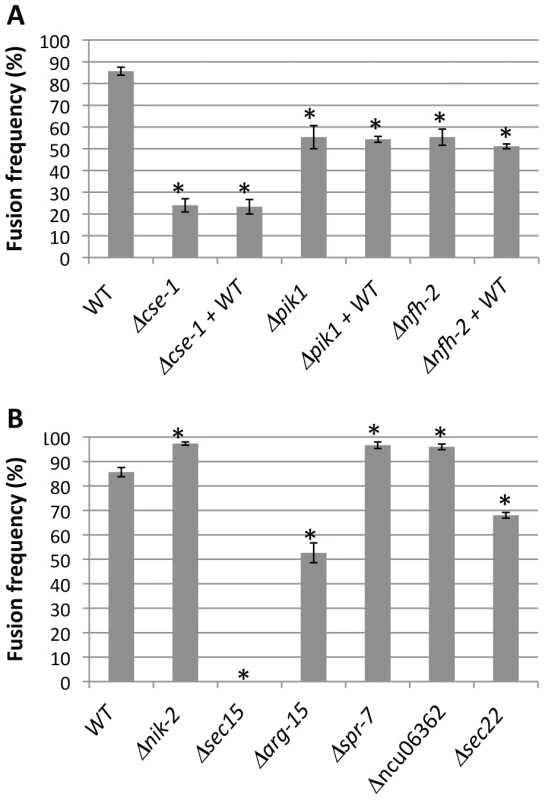
Each bar labeled with a single strain name reports the fusion frequency between germlings of wild-type FGSC 2489 (WT), or in a FGSC 2489 derivative harboring a deletion (Δ) of the indicated gene. Each bar labeled with a gene name+WT reports the fusion frequency observed when germlings of wild-type FGSC 2489 were mixed with germlings of the indicated deletion. (A) Germling communication by strains bearing deletions in the GWAS hit cse-1 (see Figures 3 and 4) and its predicted interaction partners, pik1 and nfh-2. (B) Germling communication in strains bearing deletions of genes ascertained using a permissive GWAS threshold (see Table 1). Asterisks indicate strains with communication significantly different from that of wild-type (Student's t-test, p<0.05). Bars indicate standard errors. We next sought to learn if CSE1 acts before or after a required, chemotropic interaction event in germling fusion, the observable oscillations of MAK2 and SO to the tips of communicating CATs [14]. To address this question, we obtained a wild-type strain expressing either MAK2-GFP or SO-GFP, and we visualized the subcellular localization of the latter proteins during interactions between wild-type germlings and those of the Δcse-1 mutant background. In the few cases in which a Δcse-1 germling showed chemotropic interactions toward a wild-type germling, we observed normal recruitment and oscillation of both MAK2 and SO to wild-type germling tips (every ∼4 minutes) (Figure 6). In the ∼75% of cases in which a Δcse-1 germling and a wild-type germling showed no evidence of chemotropic interactions, MAK2 and SO did not localize or oscillate to CAT tips, but remained in the cytoplasm. We conclude that CSE1 acts upstream of the signaling that underlies chemotropic interactions, because in the rare instances where Δcse-1 germlings commit to chemotropic interactions and cell fusion, they successfully drove MAK2 and SO oscillation.
Fig. 6. MAK2-GFP (A to C) and SO-GFP (D to F) oscillation in a wild type germling paired with Δcse-1 or Δnfh-2 germlings. 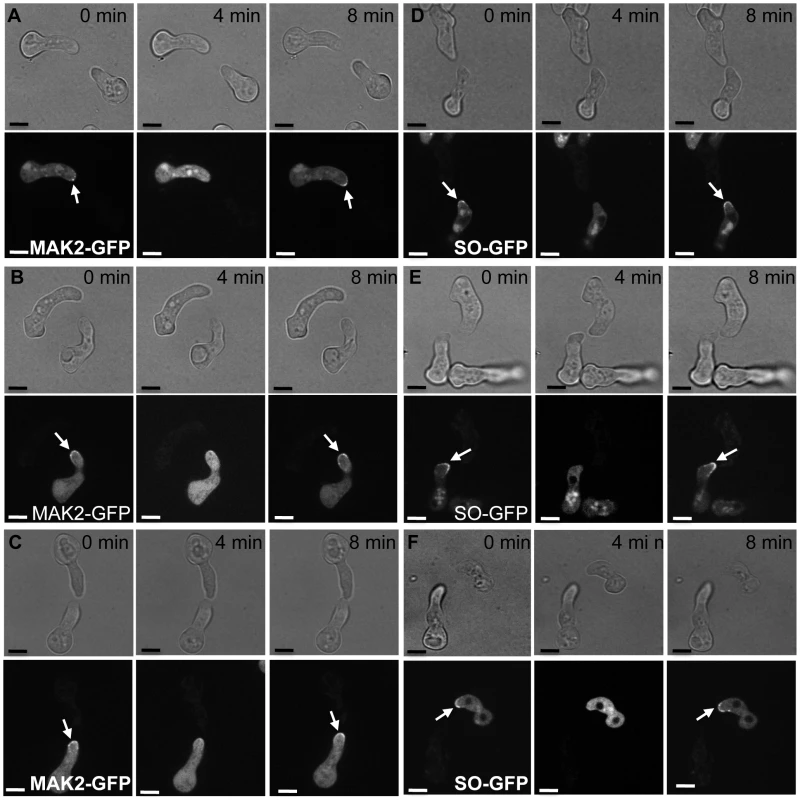
Each panel shows frames from time-lapse visualization of one representative pair of germlings, in which one cell is a wild-type FGSC 2489 derivative expressing a communication protein tagged with GFP and the other cell is unmarked, of the FGSC 2489 background or a FGSC 2489 derivative harboring a single-gene deletion (Δ). Arrows show localization of MAK2 to CATs in panels A–C and SO localization to CATs in panels D–F. (A) Oscillation of MAK2-GFP in a wild-type germling (his-3::Pccg1 mak-2-gfp; Δmak-2) when paired with unmarked wild type germling. All germlings participating in chemotropic interactions showed oscillation of MAK2. (B) Oscillation of MAK2-GFP in a wild-type germling (his-3::Pccg1 mak-2-gfp; Δmak-2) when paired with an unmarked Δcse-1 mutant, in one of the rare instances in which chemotropic interactions were observed. (C) Oscillation of MAK2-GFP in a wild-type germling (his-3::Pccg1 mak-2-gfp; Δmak-2) when paired with a nfh-2 germling in one of the few cases where chemotropic interactions were observed. (D) Oscillation of SO-GFP in a wild-type germling (his-3::Pccg1 SO-gfp; ΔSO) when paired with unmarked wild-type germling. Oscillation of SO was observed in all wild type-germlings participating in chemotropic interactions. (E) Oscillation of SO-GFP in a wild-type germling (his-3::Pccg1 SO-gfp; ΔSO) when paired with an unmarked Δcse-1 germling, in a case in which chemotropic interactions were observed. (F) Oscillation of SO-GFP in a wild-type germling (his-3::Pccg1 SO-gfp; ΔSO) when paired with unmarked Δnfh-2 germling, in one of the few cases in which chemotropic interactions were observed. Scale bar = 5 µm. The mammalian homolog of CSE1, NCS-1, functions during regulated exocytosis in response to calcium signaling [36], [37], and the yeast homolog Frq1p localizes to the Golgi membrane [38]. We reasoned that these attributes would likely be conserved in N. crassa. We first focused on the role of calcium; the Δcse-1 mutant shows growth sensitivity to excess calcium, as well as to calcium depletion [33]. We therefore hypothesized that calcium could be required for chemotropic interactions between N. crassa germlings, and to test this, we assayed fusion of wild-type germlings on growth medium depleted of Ca2+. The results (Figure 7B) bore out our prediction, with no detectable chemotropic interactions or CAT fusion in the absence of Ca2+. We next investigated the localization of CSE1 in N. crassa. For this purpose, we used a Δcse-1 strain in which the cse-1 allele with a C-terminal GFP tag had been integrated at the his-3 locus. The introduction of the GFP-tagged cse-1 allele restored wild-type growth and germling communication phenotype to the Δcse-1 strain (Figure S3). We compared the localization of CSE1-GFP to that of the late Golgi marker VSP52 tagged with RFP [39], [40]. The results, shown in Figure 8, revealed colocalization of the CSE1 and VPS52, with CSE1-GFP also present in the cytoplasm.
Fig. 7. Conidial germling fusion phenotypes. 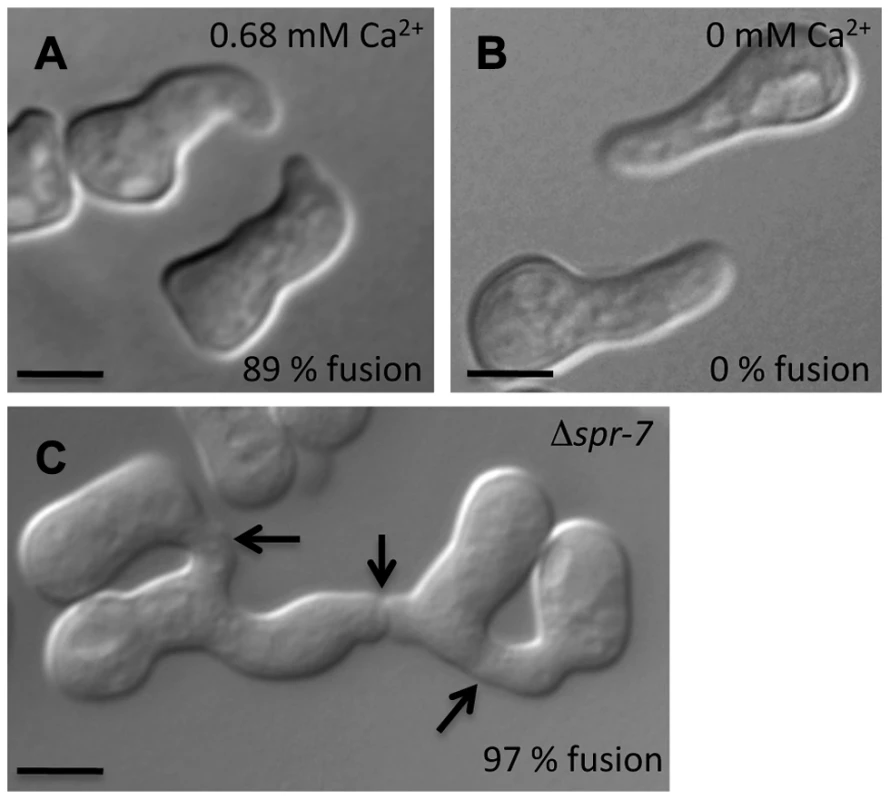
(A) Representative micrograph of wild-type (FGSC 2489) germlings undergoing chemotropic interactions in standard laboratory medium (0.68 mM Ca2+) [71]. (B) Representative micrograph of failure of chemotropic interactions and cell fusion in wild type (FGSC 2489) germlings in standard laboratory media lacking Ca2+. Fusion frequencies from large-scale replicate analyses are in black text at the lower right of each image. (C) Germlings bearing a deletion in spr-7 involved in multiple germling fusion events. Arrows indicate fusion points. Scale bar = 5 µm. Fig. 8. CSE1-GFP and RFP-VPS-52 colocalization. 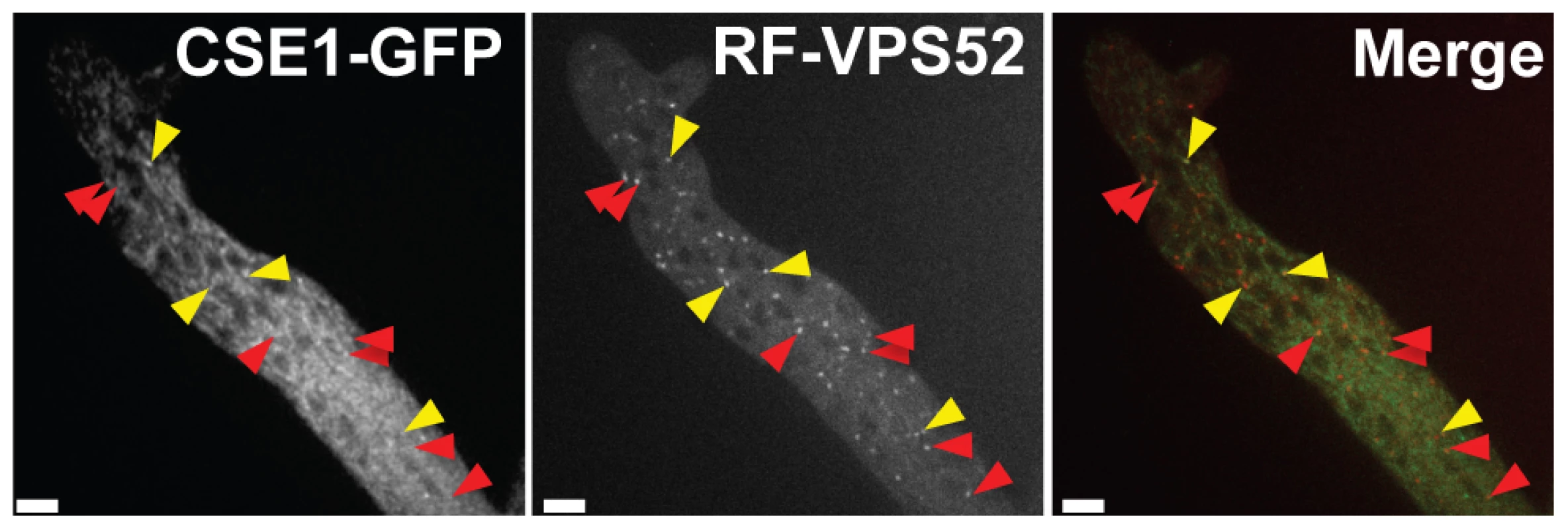
A heterokaryon between Δcse-1 (cse-1-gfp::his-3) A+rfp-vps-52::his-3 A showed co-localization of CSE1-GFP and the Golgi marker RFP-VPS-52 (yellow arrows). The red arrows indicate partial colocalization, likely due to cytoplasmic movement between frames. Scale bar = 5 µm. The Golgi secretion PI-4-kinase and a 14-3-3 regulator function in germling communication
Mammalian NCS-1 and S. cerevisiae Frq1p interact with phosphatidylinositol 4-kinase (Pik1p) [32], [37], a protein involved in secretion from the Golgi to the plasma membrane. As Frq1p is required for regulated exocytosis through Pik1p [38], we hypothesized that N. crassa homologs of components of this secretion pathway would play a role in germling communication. To test this hypothesis, we first assayed germlings carrying a deletion of the Pik1p homolog in N. crassa, NCU10397 (pik1), and observed a 1.5-fold reduction of germling communication (Figure 5A). A communication defect of similar magnitude was apparent when Δpik1 mutant germlings were assayed for interactions with wild-type fusion partners (Figure 5A). We next investigated 14-3-3 proteins, regulatory molecules that bind diverse signaling proteins [41] and in S. cerevisiae transport Pik1p from the nucleus to the cytoplasm [42]. Two members of this family have been identified in N. crassa, NCU03300 (nfh-1, encoding the DNA damage checkpoint component RAD24) and NCU02806 (nfh-2, encoding a 14-3-3 protein); we assayed germling communication in strains harboring deletions in each of these genes in turn. The results revealed no effect of the Δnfh-1 mutation (data not shown), but Δnfh-2 germlings communicated with one another at a frequency 1.5-fold less than that of isogenic wild-type germlings (Figure 5A), and Δnfh-2 conidia mixed with those of a wild-type strain exhibited a similar defect (Figure 5A). Echoing our findings from the Δcse-1 mutant, we observed normal oscillation of MAK2-GFP and SO-GFP to the CATs of wild-type germlings when they participated in chemotropic interactions with Δnfh-2 germlings, while wild-type germlings that did not communicate with those of the Δnfh-2 background showed uniquely cytoplasmic localization of MAK2-GFP and SO-GFP (Figure 6). Taken together, these data indicate that CSE1, PIK1, and NFH2 are each required for the calcium-dependent initiation of germling communication and chemotropic interactions, strongly suggesting their joint function in a Golgi secretion pathway involved in signaling to initiate germling fusion.
Six additional secretion and signaling genes are involved in germling communication
Given the robust genetic association between cse-1 genotype and germling communcation in wild strains (Figure 3), we reasoned that additional determinants of germling communication could be revealed by mining our genome-wide association data at lower significance levels. For this purpose, we re-examined our association results using a permissive threshold of p<0.015. Permutation testing estimated that 22% of loci reaching this level would be true positives (see methods); as such, independent gene-by-gene validation could uncover bona fide communication genes among this set, potentially both activators and repressors of the communication trait. We focused on genes annotated in secretion, kinase signalling pathways, or peptide hydrolysis in which SNPs showed association reaching our permissive significance cutoff. Of the 18 genes that fit this description and for which deletion strains were available and viable (Table 1), deletion of six genes had significant impact on communication frequencies as compared to a wild-type strain (Figure 5B). The most extreme phenotype, a complete failure of chemotropic interactions and CAT fusion, was observed in the deletion strain for the exocyst complex component sec15 (NCU00117) (Table 1; Figure 5B). The latter mutant also exhibited slower growth, reduced conidiation, and slower conidial germination. Deletion of two additional genes, the protein transporter sec22 (NCU06708) and the acetylornithine-glutamate transacetylase arg-15 (NCU05622) [43], also compromised fusion frequency (68%±2 and 53%±4, respectively) (Figure 5B). Remarkably, deletion of each of three genes heightened germling communication and fusion frequencies (Figure 5B): a GTPase activating protein (NCU06362; 96%±2), the nonidentical kinase-2 nik-2 (NCU01833; 97%±0.7), and the secreted subtilisin-like serine protease spr-7 (NCU07159; 97±1.3). The elevated fusion frequency in each of these strains contrasts with any known germling fusion mutant, all of which reduce or eliminate chemotropic interactions or cell fusion, and highlights the ability of association mapping to pinpoint negative regulators as well as genes with a positive role in cell communication. In each mutant with heightened fusion frequency, germlings were also often involved in fusion events with more than one germling (multiple fusion events) (26.33%±5.24 in ΔNCU06362, 21.33%±1.8 in Δnik-2, and 20.66%±4.07 in Δspr-7; Figure 7C). By contrast, multiple germling fusion events was a phenotype only observed at a low level in a wild-type strain (2%±2).
Tab. 1. GWAS of germling communication in wild N. crassa isolates and validation in laboratory deletion strains. 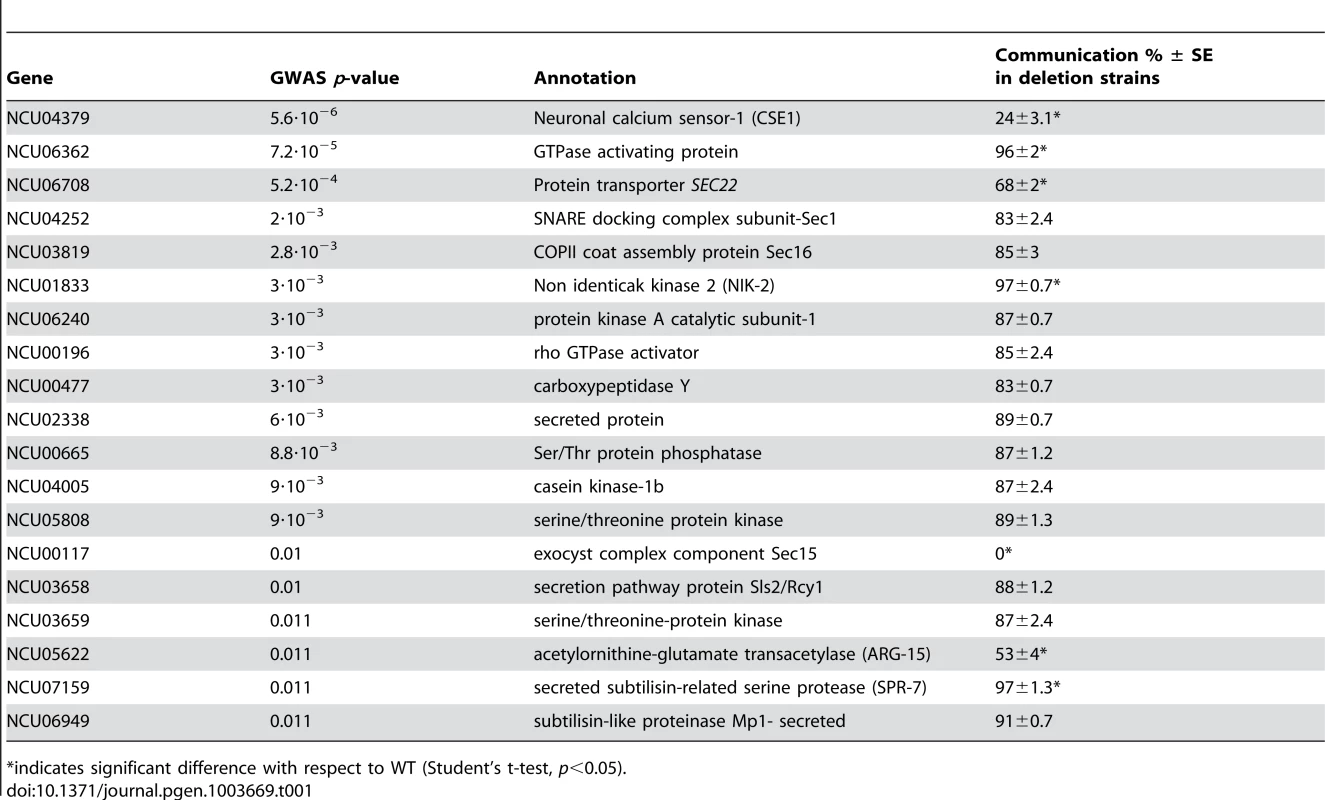
indicates significant difference with respect to WT (Student's t-test, p<0.05). To investigate further the novel gain-of-fusion phenotype, we focused on the putative secreted serine protease spr-7. We first confirmed that the introduction of an ectopic copy of spr-7 at the his-3 locus restored hyphal communication of the spr-7 deletion strain to wild-type levels, establishing the deletion as the sole cause of the increased communication phenotype (Figure S3). We next asked whether the presence of wild-type germlings would be sufficient to complement the Δspr-7 phenotype during communication. Assays of Δspr-7 germlings mixed with those of a wild-type strain confirmed this hypothesis, revealing a fully wild-type communication phenotype (fusion frequency 82%±3), a striking contrast to the failure to communicate with wild-type germlings we had noted in Δcse-1, Δpik1 and Δnfh-2 mutants (Figure 5 and see above). These results support a model in which secreted SPR-7 from wild-type germlings acts in a cell-non-autonomous fashion to restrict communication and CAT fusion between wild type germlings.
Discussion
In N. crassa, genetically identical germlings chemotropically sense partner cells and undergo mutual recognition-directed growth and cell fusion [14], [15], [16]. The molecular basis of this phenotype is only partly understood, and tools to identify candidate genes involved in fusion are at a premium in the field. In this work, we genotyped more than 100 wild N. crassa isolates, advanced our understanding of germling communication and fusion, and established this population as a powerful resource for high-resolution association mapping that can be used with any variable phenotype. Our study is the first to illustrate the utility of genome-wide association mapping to identify novel loci underlying trait variation in a microbe. We anticipate that this methodology will be a powerful and generally applicable tool in future genetic study of many eukaryotic microbes, owing to the small genome sizes and deeply-sampled populations of a number of species, particularly filamentous fungi.
The top hit from our association analysis was cse-1, which is homologous to a neuronal calcium sensor gene in animals that shows nervous-system-specific expression and neuron-specific phenotypes; neurons, like hyphae in filamentous fungi, are a highly polarized tissue. Neuronal calcium sensor-1 (Frequenin) is a myristolylated protein with four EF hands that functions as a calcium ion sensor for modulation of syntaptic activity and secretion [34], [44], [45], [46]. Our analysis revealed a near-complete loss of cellular communication during germling fusion in a N. crassa Δcse-1 mutant. In animals and in S. cerevisiae, NCS-1/Frq1p and Bmh1p-Bmh2p regulate phosphatidylinositol 4-kinase/Pik1p, with Bmh1p-Bmh2p mediating the nucleocytoplasmic shuttling of Pik1p [42]. NCS-1/Frq1p promotes association of Pik1p with the Golgi membrane, which is required for its role in regulated exocytosis [37], [38]. Our results established that in N. crassa, CSE1 localized to the Golgi and that deletion of pik1 or nfh-2 phenocopied a cse-1 deletion strain. These observations together support a model in which, in N. crassa, CSE1, PIK1 and NFH2 regulate exocytosis of an unidentified ligand and/or receptor, perhaps initiated via calcium signaling, which is important for establishing communication between cells and subsequent chemotropic interactions (Figure 9). Recently, an essential kinase (MSS-4) involved in the generation of phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P(2)) was found to localize to contact points between germlings during cell fusion [47], indicating that the generation of different phosphatidylinositol phosphate moieties may regulate different aspects of germling fusion.
Fig. 9. Schematic of a mechanistic model for the regulation of secretion by CSE1, PIK1 and NFH2 during germling chemotropic interactions and cell fusion. 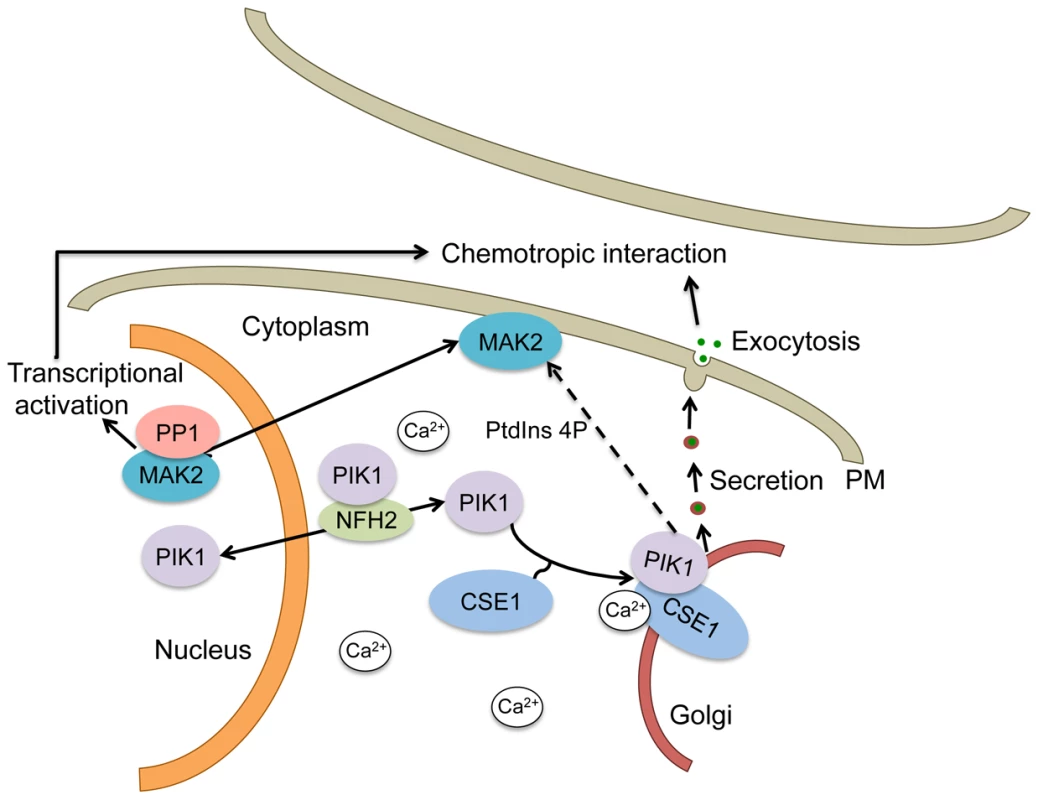
PIK1 interaction with CSE1 allows its recruitment to Golgi membranes, where it plays a role in PtdIns4p production and the regulated secretion of the signals needed for chemotropic interactions. In addition, PtdIns4p may play a role in MAK2 activation, which is required for chemotropic interactions and cell fusion. PIK1 is predicted to shuttle between the cytosol and the nucleus in a manner that is dependent on the BMH1 homolog NFH2. A role for phosphorylation is suggested by our finding that the defect in germling communication observed in the Δcse-1, Δpik1 and Δnfh-2 mutants correlates with an absence of oscillation of MAK2 and SO to CAT tips, because MAK2 kinase activity has been shown to be required for chemotropic interactions and MAK2 and SO oscillation [14]. In S. cerevisiae, Pik1p is required for full activation of the MAP kinases Fus3p and Hog1p and repression of Kss1p [48], and the Fus3p ortholog in N. crassa is MAK2 [10]. It is therefore tempting to speculate that the activation of PIK1 by CSE1 may play an important role in germling communication by affecting activation of MAK2, thus modulating MAK2 phosphorylation targets as well as downstream transcriptional targets required for germling fusion (Figure 9).
In addition to our mapping of cse-1 as a determinant of variation in germling communication across wild N. crassa, further mining of our association results led to the identification and validation of six other genes associated with CAT fusion. Of these, one gene, sec15, is a homolog of a component of the exocyst complex in S. cerevisiae, a multiprotein complex that localizes at the bud tip and is associated with exocytosis [49]. Our results indicated that sec15 is essential for CAT fusion in N. crassa. Likewise, our results revealed a defect in germling communication and fusion frequency in a strain bearing a deletion in a homolog of SEC22 in N. crassa, NCU06708; in S. cerevisiae, Sec22p assembles into a SNARE complex and plays a role in ER-Golgi protein trafficking [50]. Our demonstration that cse-1, pik1, nfh-2, sec15, and sec22 are all required for germling communication establishes the importance of protein secretion and trafficking for chemotropic interactions and cell fusion in N. crassa.
Our results also established that mutation of the acetylornithine-glutamate transacetylase arg-15 [43] confers a defect in germling communication. The homolog of arg-15 in S. cerevisiae, Dug2p, is involved in degradation of the antioxidant glutathione and other peptides containing a gamma-glu-X. dug2 mutants show deficient utilization of glutathione [51], which reacts non-enzymatically with reactive oxygen species and detoxifies oxidatively stressed cells [52]. A role for redox reactions in germling communication through arg-15 would dovetail with reports that mutants in components of the NADPH oxidase complex, which is involved in redox signaling, are defective in CAT fusion [9].
Our work has uncovered a new category of fusion mutants that exhibited germling fusion frequencies higher than those of wild-type, and which displayed multiple fusion events. Of the genes whose deletions gave rise to this striking phenotype, one encoded an uncharacterized predicted GTPase activating protein (GAP) (NCU06362). NCU06362 contains a TBC domain (PF00566) and is a paralog of GYP5 in S. cerevisiae; Gyp5p is involved in the recruitment to sites of polarized growth of the BAR domain protein Rvs167p, which has been implicated in exocytosis at the bud tip [53]. Rvs167p interacts with a second BAR domain protein, Rvs161p, and together this complex plays a role in receptor-mediated endocytosis [54]. Gyp5p also has in vitro GAP activity towards Ypt1p, which is involved in ER-to-Golgi trafficking, and towards Sec4p, which regulates exocytosis [55]. Thus, the increase in germling fusion frequencies observed in the ΔNCU06362 mutant could be due to alterations in secretion or in the reduction of endocytosis of a receptor involved in germling communication.
A second gene whose deletion enhanced hyphal communication, spr-7, encodes a secreted subtilisin-related serine protease, part of a family whose members carry out a wide range of peptidase activities [56]. The increase in fusion frequency and germlings involved in mutiple fusion events in the Δspr-7 mutant suggests that SPR-7 may be responsible for the degradation of a peptide required for extracellular communication (Figure 9). The nature of the extracellular ligand and receptor(s) that guide chemotropic interactions during cell fusion in N. crassa is currently unknown. In fungi, secreted peptides involved in extracellular communication have not been reported, apart from peptide pheromones involved in mating [57], [58] or small secreted proteins with antifungal properties [59], [60]. The genes we have uncovered here will serve as targets for future genetic and biochemical efforts to identify extracellular ligands and receptors involved in germling communication and cell fusion in N. crassa.
Our results also revealed an increase in germling communication in a nik-2 deletion strain. This gene encodes a histidine kinase, a member of a canonical two-component signal transduction pathway and part of an 11-member family in N. crassa. No phenotype for the Δnik-2 mutant has been previously reported [61]. However, other histidine kinases affect MAPK signal transduction pathways in fungi, including nik-1, a member of the osmoregulatory OS-2 pathway in N. crassa [62], and the histidine kinase Sln1p, which regulates the Hog1p MAPK pathway in S. cerevisiae [63]. We hypothesize that the increase in fusion frequencies in the absence of nik-2 may stem from a defect in the regulation of the MAK2 phosphorylation pathway, leading to a hyper-activated state during chemotropic interaction (Figure 9). Further research will be necessary to elucidate the specific role of nik-2 in this process.
By identifying multiple novel determinants of germling communication, our results underscore the power of association studies for the mapping of genes to phenotypes in wild populations. Importantly, our N. crassa population is particularly amenable to GWAS, with little discernable population structure and low linkage disequilibrium, allowing the detection of strong association to finely resolved loci. These attributes of N. crassa stand in contrast to S. cerevisiae, where GWA studies are hampered by a mosaic and heterogenous population structure [64]. Our relatively modest, medium-throughput phenotyping of a quantitative phenotype in wild individuals compares favorably with the high-throughput approach that would be required to survey the >9000 strains of the N. crassa deletion collection [65], not only by saving 98% of the labor, but in enabling analysis of all genes, including those that are essential. However, our molecular follow-up of GWAS hits was aided by the availability of a near-full genome deletion strain collection for N. crassa. When the central question, as in our work, is to infer novel function for poorly annotated genes, comparing a given gene's deletion strain and the isogenic wild-type strain is a straightforward and precise approach that obviates potential complications from epistasis in allele-swapping experiments. Our GWAS method also compares favorably to two-parent crossing schemes for the dissection of natural variation [66]: first, because linkage blocks in our outbreeding population often contain a single gene, whereas more than 50 can be contained in those resulting from just one cross [67], and second, because we sample phenotypes that vary among multiple individuals and not just those that differ between two parents. With the availability of our collection of 112 genotyped individuals to the fungal genetic community, future studies will require only phenotyping to map the molecular basis of trait variation using the strategy we have pioneered here. And as population-genomic resources are developed in many taxa, we anticipate that association mapping will be successfully applied in other species, within and outside the fungal kingdom.
Methods
Strains and growth conditions
All 112 strains used in this study were isolated from Louisiana, USA (Table S1) and are available from the Fungal Genetics Stock Center (FGSC) [68].
The deletion mutants used in these study were generated by the Neurospora Genome Project [65], [69] and are administered by the FGSC [70]. The rfp-vps-52 transformant was generously provided by Barry Bowman [40]. All strains were grown on Vogel's medium [71] and all crosses were performed on Westergaard's synthetic cross medium [72]. The his-3 A mutant (FGSC# 6103) and a his-3 a strain (FGSC #9716) were used as females in crosses with deletion mutants. Progeny bearing the deletion mutations and the his-3 mutation were isolated and used in complementation experiments.
RNA isolation and cDNA synthesis
Total RNA was isolated for each of the 112 strains listed in Table S1. Strains were grown for 16 hrs on cellophane on Bird medium [73]. Mycelia were harvested and immediately added to 1 mL of TRIzol reagent (Invitrogen Life Technologies) [74] and zirconia/silica beads (0.2 g, 0.5-mm diameter; Biospec Products). Cells were disrupted using a MiniBeadBeater instrument (Biospec Products) at maximum speed for 30 seconds twice in succession. Total RNA was extracted according to the manufacturer's protocol for TRIzol (Invitrogen) and quantified on a Bioanalyzer (Agilent).
For polyA RNA purification, 10 µg of total RNA was bound to dynal oligo(dT) magnetic beads (Invitrogen 610.02) two times, using the manufacturer's instructions. Purified polyA RNA was fragmented by metal-ion catalysis [75] using fragmentation reagents from Ambion (AM12450). For first strand cDNA synthesis 1 µg fragmented polyA RNA was incubated with 3 µg random hexamers (Invitrogen 48190-011), and incubated at 65°C for 5 minutes and then transferred to ice. 1st strand buffer (Invitrogen 18064-014) was added to 1× final concentration (4 µL). Dithiothreitol (DTT), dNTPs and RNAseOUT (Invitrogen 10777-019) were added to 100 mM, 10 mM, and 20 U/20 µL respectively, and the sample was incubated at 25°C for 2 minutes. 200 U of Superscript II (Invitrogen 18064-014) were added and the sample was incubated at 25°C for 10 minutes, 42°C for 50 minutes and 70°C for 15 minutes.
For second strand synthesis, 51 µL of H2O, 20 µL of 5× second strand buffer (Invitrogen 10812-014), and dNTPs (10 mM) were added to the first strand cDNA synthesis mix and incubated on ice for 5 minutes. RNaseH (2 U) (Invitrogen 18021-014), DNA pol I (50 U) (Invitrogen 18010-017) were then added and the mixture was incubated at 16°C for 2.5 hours.
Library construction and sequencing
End-repair was performed by adding 45 µL of H2O, T4 DNA ligase buffer with 10 mM ATP (NEB B0202S) (10 µL), dNTP mix (10 mM), T4 DNA polymerase (15 U) (NEB M0203L), Klenow DNA polymerase (5 U) (NEB M0210S), and T4 PNK (50 U) (NEB M0201L) to the sample and incubating at 20°C for 30 minutes. A single base was added each to cDNA fragment by adding Klenow buffer (NEB M0212L), dATP (1 mM), and Klenow 3′ to 5′ exo - (15 U) (NEB M0212L). The mixture was then incubated at 37°C in for 30 minutes.
Standard Illumina adapters (FC-102-1003) were ligated to the cDNA fragments using 2× DNA ligase buffer (Enzymatics L603-HC-L), 1 µL of adapters, and DNA ligase (5 U) (Enzymatics L603-HC-L). The sample was incubated at 25°C for 15 minutes. The sample was purified in a 2% low-melting point agarose gel, and a slice of gel containing 200-bp fragments was removed and the DNA purified. The polymerase chain reaction (PCR) was used to enrich the sequencing library. A 10-µL aliquot of purified cDNA library was amplified by PCR. PCR cycling conditions were: a denaturing step at 98°C for 30 seconds, 14 cycles of 98°C for 10 seconds, 65°C for 30 seconds, 68°C for 30 seconds, and a final extension at 68°C for 5 minutes. All libraries were sequenced using an Illumina Genome Analyzer-II using standard Illumina operating procedures. RNAseq data for all strains used in these analyses has been deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE45406; GSM1103708-GSM1103819).
SNP identification and phylogenetics
Mapping of RNA-seq reads to the genome sequence of N. crassa strain FGSC 2489 [76] and calling of single nucleotide polymorphisms (SNPs) was carried out with Maq [77]. All RNA-seq reads that mapped to multiple locations were eliminated from analysis, as were SNPs located in regions of low consensus read quality. These variants were further filtered to retain only those that were bi-allelic, yielding a complete data set of 1.09×106 SNPs (Dataset S1) which were used as input into phylogenetic inference with FastTree; because patterns of inheritance in one strain, JW168, were suggestive of misclassification (data not shown) we did not include this strain in the tree shown in Figure S1. For markers used as input into calculations of genetic association with the germling communication phenotype (see below), we filtered the complete SNP set to retain only sites at which the minor allele was present at >25% frequency (Dataset S2).
Conidial germling fusion frequency measurements
For germling communication assays, each strain was grown on Vogel's minimal media [71] in slant tubes for 4–6 days or until significant conidiation occurred. Conidial suspensions were prepared by collecting conidia with wood sticks and suspending in 600 µl of sterile distilled water. The conidial suspension was filtered by pouring over cheesecloth to remove hyphal fragments. Conidia were diluted to a concentration of 3×107 conidia/ml and 300 µl of this final mixture were spread either on an agar or agarose minimal-medium plates. The plates were incubated for 3–4 hours at 30°. At each of 2–3 timepoints for each strain, agar squares of 1 cm were excised and observed with a Zeiss Axioskop 2 using a 403 Plan-Neofluor oil immersion objective. For image acquisition DIC images were taken with a Hamamatsu Orca 03 camera (Hamamatsu, Japan) using the iVision Mac4.5 software and a Zeiss Axioimager microscope. Fusion events were counted for 50 germling pairs in each of 2–3 biological replicates.
Complementation analysis
Complementation experiments were done using the pMF272 plasmid system [78] to insert a wild type copy of the deleted gene into the intergenic region 3′ of the his-3 locus; transformants were subsequently analyzed for germling fusion frequencies. Wild type copies of genes were amplified using Taq polymerase from New England Biolabs (Ipswich, CA, USA). Primers were designed to amplify the coding regions and also contained an added restriction enzyme site. The amplified DNA fragments were TOPO (Invitrogen) cloned, cut with restriction enzymes and ligated into restriction enzyme-digested pMF272 plasmid. The ligated DNA was used to transform Escherichia coli (DH5a), and the plasmid isolated from individual transformants. The DNA sequence of each plasmid was determined; plasmids containing wild type copies of the genes were used for complementation experiments.
Confocal microscopy
Some mutants showing reduced fusion frequencies were further characterized by studying the ability of the mutant germlings to induce recruitment of MAK2-GFP or SO-GFP to the plasma membrane of opposing germlings as described by Fleißner et al [13]. Conidia from MAK2-GFP and SO-GFP strains were mixed with equal amounts of conidia from the respective deletion mutants and samples were prepared for microscopy as described above. Images were taken at two-minute intervals using a Leica SD6000 microscope with a 100×1.4 NA oil-immersion objective equipped with a Yokogawa CSU-X1 spinning disk head and a 488-nm laser controlled by Metamorph software (Molecular Devices, Sunnyvale, CA).
To visualize CSE1-GFP and RFP-VPS-52 localization, the strains were grown on Vogel's MM plates overnight and squares of 1 cm were excised and examined in the same confocal microscope explained above using the 488-nm laser for GFP and 563 nm laser for RFP. To study co-localization of both proteins, heterokaryons were made by mixing conidia from both strains in the center of a plate and incubating them overnight to allow cell fusion and cytoplasmic mixing from both strains. The samples were prepared and imaged as explained above.
Whole-genome association mapping
We used germling communication phenotype measurements in biological triplicate from 24 Louisiana strains in a genome-wide association analysis as follows. For each strain, we first calculated the average communication frequency across all replicates and timepoints to yield a final quantitative communication measurement. We then converted the latter value to a qualitative score: we calculated the grand mean and standard deviation of communication frequency across all strains, and we classified a given strain as low-communicating if its communication measurement was more than one standard deviation below the grand mean, and high-communicating otherwise. We then tested each marker in turn, from our set of SNPs with >25% minor allele frequency (see above), for co-inheritance with this qualitative communication score using Fisher's exact test [79]. To evaluate the experiment-wise false discovery rate at a given Fisher's p-value threshold pthresh, we shuffled the vector of phenotype category values among strains, repeated the association test, and tabulated the number of SNPs with Fisher's p-value<pthresh, in this null data set. Averaging over 1000 such permutations yielded an expectation of 0.011 SNPs called at pthresh = 5.6×10−6 and 652 SNPs at pthresh = 0.015, under a null model of no true association. Given the 3 and 837 SNPs, respectively, reaching these levels in the real data (Dataset S3), false discovery rates at these thresholds were 0.4% and 78%, respectively. Linkage disequilibrium in Figure 4 was calculated between all high-frequency SNPs in the region of cse-1 using the LDcorSV package in R.
Supporting Information
Zdroje
1. Read ND, Fleissner A, Roca MG, Glass NL (2010) Hyphal fusion. In: Cellular and Molecular Biology of Filamentous Fungi. Borkovich KA and Ebbole DJ, editors. Washington, D.C.: ASM Press. pp. 260–273.
2. SchmitJC, BrodyS (1976) Biochemical genetics of Neurospora crassa conidial germination. Bacteriol Revs 40 : 1–41.
3. RocaMG, ArltJ, JeffreeCE, ReadND (2005) Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot Cell 4 : 911–919.
4. RocaGM, ReadND, WhealsAE (2005) Conidial anastomosis tubes in filamentous fungi. FEMS Microbiol Letts 249 : 191–198.
5. SimoninA, Palma-GuerreroJ, FrickerM, GlassNL (2012) The physiological significance of network organization in fungi. Eukaryot Cell 11 : 1345–1352.
6. RoperM, SimoninA, HickeyPC, LeederA, GlassNL (2013) Nuclear dynamics in a fungal chimera. Proc Natl Acad Sci USA (in revision).
7. FleissnerA, SimoninAR, GlassNL (2008) Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol Biol 475 : 21–38.
8. WrightGD, ArltJ, PoonWC, ReadND (2007) Optical tweezer micromanipulation of filamentous fungi. Fungal Genet Biol 44 : 1–13.
9. ReadND, GoryachevAB, LichiusA (2012) The mechanistic basis of self-fusion between conidial anastomosis tubes during fungal colony initiation. Fungal Biol Rev 26 : 1–11.
10. PandeyA, RocaMG, ReadND, GlassNL (2004) Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot Cell 3 : 348–358.
11. MaerzS, ZivC, VogtN, HelmstaedtK, CohenN, et al. (2008) The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics 179 : 1313–1325.
12. DettmannA, IllgenJ, MarzS, SchurgT, FleissnerA, et al. (2012) The NDR kinase scaffold HYM1/MO25 is essential for MAK2 map kinase signaling in Neurospora crassa. PLoS Genet 8: e1002950.
13. FleissnerA, SarkarS, JacobsonDJ, RocaMG, ReadND, et al. (2005) The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot Cell 4 : 920–930.
14. FleissnerA, LeederAC, RocaMG, ReadND, GlassNL (2009) Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc Natl Acad Sci U S A 106 : 19387–19392.
15. GoryachevAB, LichiusA, WrightGD, ReadND (2012) Excitable behavior can explain the “ping-pong” mode of communication between cells using the same chemoattractant. BioEssays 34 : 259–266.
16. ReadND, LichiusA, ShojiJY, GoryachevAB (2009) Self-signalling and self-fusion in filamentous fungi. Curr Opin Microbiol 12 : 608–615.
17. PerkinsDD, TurnerBC, BarryEG (1976) Strains of Neurospora collected from nature. Evolution 30 : 281–313.
18. TurnerE, JacobsonDJ, TaylorJW (2010) Reinforced postmating reproductive isolation barriers in Neurospora, an Ascomycete microfungus. J Evol Biol 23 : 1642–1656.
19. DettmanJR, JacobsonDJ, TurnerE, PringleA, TaylorJW (2003) Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57 : 2721–2741.
20. DettmanJR, JacobsonDJ, TaylorJW (2003) A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57 : 2703–2720.
21. EllisonCE, HallC, KowbelD, WelchJ, BremRB, et al. (2011) Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci U S A 108 : 2831–2836.
22. MagwireMM, FabianDK, SchweyenH, CaoC, LongdonB, et al. (2012) Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet 8: e1003057.
23. WeberAL, KhanGF, MagwireMM, TaborCL, MackayTF, et al. (2012) Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster. PLoS One 7: e34745.
24. MackayTF, RichardsS, StoneEA, BarbadillaA, AyrolesJF, et al. (2012) The Drosophila melanogaster genetic reference panel. Nature 482 : 173–178.
25. BrachiB, FaureN, HortonM, FlahauwE, VazquezA, et al. (2010) Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet 6: e1000940.
26. ChanEK, RoweHC, CorwinJA, JosephB, KliebensteinDJ (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125.
27. AtwellS, HuangYS, VilhjalmssonBJ, WillemsG, HortonM, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465 : 627–631.
28. FiliaultDL, MaloofJN (2012) A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genet 8: e1002589.
29. MandelJR, NambeesanS, BowersJE, MarekLF, EbertD, et al. (2013) Association mapping and the genomic consequences of selection in sunflower. PLoS Genet 9: e1003378.
30. LitiG, LouisEJ (2012) Advances in quantitative trait analysis in yeast. PLoS Genet 8: e1002912.
31. Foulongne-OriolM (2012) Genetic linkage mapping in fungi: current state, applications, and future trends. Appl Microbiol Biotechnol 95 : 891–904.
32. HendricksKB, WangBQ, SchniedersEA, ThornerJ (1999) Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat Cell Biol 1 : 234–241.
33. DekaR, KumarR, TamuliR (2011) Neurospora crassa homologue of Neuronal Calcium Sensor-1 has a role in growth, calcium stress tolerance, and ultraviolet survival. Genetica 139 : 885–894.
34. BurgoyneRD (2007) Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci 8 : 182–193.
35. TamuliR, KumarR, DekaR (2011) Cellular roles of neuronal calcium sensor-1 and calcium/calmodulin-dependent kinases in fungi. J Basic Microbiol 51 : 120–128.
36. GromadaJ, BarkC, SmidtK, EfanovAM, JansonJ, et al. (2005) Neuronal calcium sensor-1 potentiates glucose-dependent exocytosis in pancreatic beta cells through activation of phosphatidylinositol 4-kinase beta. Proc Natl Acad Sci U S A 102 : 10303–10308.
37. Kapp-BarneaY, MelnikovS, SheflerI, JerominA, Sagi-EisenbergR (2003) Neuronal calcium sensor-1 and phosphatidylinositol 4-kinase beta regulate IgE receptor-triggered exocytosis in cultured mast cells. J Immunol 171 : 5320–5327.
38. StrahlT, HamaH, DeWaldDB, ThornerJ (2005) Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol 171 : 967–979.
39. ConibearE, StevensTH (2000) Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell 11 : 305–323.
40. BowmanBJ, DraskovicM, FreitagM, BowmanEJ (2009) Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot Cell 8 : 1845–1855.
41. BridgesD, MoorheadGB (2005) 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 2005: re10.
42. DemmelL, BeckM, KloseC, SchlaitzAL, GloorY, et al. (2008) Nucleocytoplasmic shuttling of the Golgi phosphatidylinositol 4-kinase Pik1 is regulated by 14-3-3 proteins and coordinates Golgi function with cell growth. Mol Biol Cell 19 : 1046–1061.
43. RadfordA (2004) Metabolic highways of Neurospora crassa revisited. Adv Genet 52 : 165–207.
44. BourneY, DannenbergJ, PollmannV, MarchotP, PongsO (2001) Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1). J Biol Chem 276 : 11949–11955.
45. De CastroE, NefS, FiumelliH, LenzSE, KawamuraS, et al. (1995) Regulation of rhodopsin phosphorylation by a family of neuronal calcium sensors. Biochem Biophys Res Commun 216 : 133–140.
46. DasonJS, Romero-PozueloJ, MarinL, IyengarBG, KloseMK, et al. (2009) Frequenin/NCS-1 and the Ca2+-channel alpha1-subunit co-regulate synaptic transmission and nerve-terminal growth. J Cell Sci 122 : 4109–4121.
47. MahsA, IschebeckT, HeiligY, StenzelI, HempelF, et al. (2012) The essential phosphoinositide kinase MSS-4 Is required for polar hyphal morphogenesis, localizing to sites of growth and cell fusion in Neurospora crassa. PloS One 7: e51454.
48. CappellSD, DohlmanHG (2011) Selective regulation of MAP kinase signaling by an endomembrane phosphatidylinositol 4-kinase. J Biol Chem 286 : 14852–14860.
49. TerBushDR, MauriceT, RothD, NovickP (1996) The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J 15 : 6483–6494.
50. NewmanAP, ShimJ, Ferro-NovickS (1990) BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol 10 : 3405–3414.
51. KaurH, GanguliD, BachhawatAK (2012) Glutathione degradation by the alternative pathway (DUG pathway) in Saccharomyces cerevisiae is initiated by (Dug2p-Dug3p)2 complex, a novel glutamine amidotransferase (GATase) enzyme acting on glutathione. J Biol Chem 287 : 8920–8931.
52. PocsiI, PradeRA, PenninckxMJ (2004) Glutathione, altruistic metabolite in fungi. Adv Microb Physiol 49 : 1–76.
53. PrigentM, Boy-MarcotteE, ChesneauL, GibsonK, Dupre-CrochetS, et al. (2011) The RabGAP proteins Gyp5p and Gyl1p recruit the BAR domain protein Rvs167p for polarized exocytosis. Traffic 12 : 1084–1097.
54. LombardiR, RiezmanH (2001) Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J Biol Chem 276 : 6016–6022.
55. De AntoniA, SchmitzovaJ, TrepteHH, GallwitzD, AlbertS (2002) Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J Biol Chem 277 : 41023–41031.
56. RawlingsND, BarrettAJ (1994) Families of serine peptidases. Methods Enzymol 244 : 19–61.
57. BardwellL (2005) A walk-through of the yeast mating pheromone response pathway. Peptides 26 : 339–350.
58. KimH, BorkovichKA (2006) Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot Cell 5 : 544–554.
59. TheisT, WeddeM, MeyerV, StahlU (2003) The antifungal protein from Aspergillus giganteus causes membrane permeabilization. Antimicrob Agents Chemother 47 : 588–593.
60. LeiterE, SzappanosH, OberparleiterC, KaisererL, CsernochL, et al. (2005) Antifungal protein PAF severely affects the integrity of the plasma membrane of Aspergillus nidulans and induces an apoptosis-like phenotype. Antimicrob Agents Chemother 49 : 2445–2453.
61. BorkovichKA, AlexLA, YardenO, FreitagM, TurnerGE, et al. (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68 : 1–108.
62. JonesCA, Greer-PhillipsSE, BorkovichKA (2007) The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol Biol Cell 18 : 2123–2136.
63. SaitoH, TatebayashiK (2004) Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem 136 : 267–272.
64. ConnellyCF, AkeyJM (2012) On the prospects of whole-genome association mapping in Saccharomyces cerevisiae. Genetics 191 : 1345–1353.
65. DunlapJC, BorkovichKA, HennMR, TurnerGE, SachsMS, et al. (2007) Enabling a community to dissect an organism: Overview of the Neurospora functional genomics project. Adv Genet 57 : 49–96.
66. ZhuC, GoreM, BucklerES, YuJ (2008) Status and prospects of association mapping in plants. Plant Genome 1 : 5–20.
67. TurnerE, JacobsonDJ, TaylorJW (2011) Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. PLoS Genet 7: e1002204.
68. McCluskeyK, WiestA, PlamannM (2010) The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci 35 : 119–126.
69. ColotHV, ParkG, TurnerGE, RingelbergC, CrewCM, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103 : 10352–10357.
70. McCluskeyK (2003) The fungal genetics stock center: From molds to molecules. Adv Appl Microbiol 52 : 245–262.
71. VogelHJ (1956) A convenient growth medium for Neurospora. Microbial Genetics Bulletin 13 : 42–46.
72. WestergaardM, MitchellHK (1947) Neurospora V. A synthetic medium favoring sexual reproduction. Amer J Bot 34 : 573–577.
73. MetzenbergRL (2004) Bird Medium: an alternative to Vogel Medium. Fungal Genet Newslett 51 : 19–20.
74. ChomczynskiP, SacchiN (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162 : 156–159.
75. BrowneKA (2002) Metal ion-catalyzed nucleic acid alkylation and fragmentation. J Am Chem Soc 124 : 7950–7962.
76. GalaganJE, CalvoSE, BorkovichKA, SelkerEU, ReadND, et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 : 859–868.
77. LiH, RuanJ, DurbinR (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18 : 1851–1858.
78. FreitagM, HickeyPC, RajuNB, SelkerEU, ReadND (2004) GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol 41 : 897–910.
79. FisherRA (1922) On the interpretation of χ2 from contingency tables, and the calculation of P. J Royal Stat Soc 85 : 87–94.
80. PriceMN, DehalPS, ArkinAP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 5: e9490.
81. LetunicI, BorkP (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucl Acids Res 39: W475–478.
Štítky
Genetika Reprodukčná medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



