-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
The liverwort Marchantia polymorpha has a minimal but required repertoire of components for auxin-mediated transcriptional regulation, which is sufficient for achieving the simple yet multi-dimensional body plan.
Published in the journal: Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in. PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005084
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005084Summary
The liverwort Marchantia polymorpha has a minimal but required repertoire of components for auxin-mediated transcriptional regulation, which is sufficient for achieving the simple yet multi-dimensional body plan.
Introduction
In angiosperms, the plant hormone auxin regulates many aspects of growth and development such as axis formation during embryogenesis [1], initiation of leaf primordia at the shoot apical meristem [2], root development [3], and tropic responses to light or gravity [4,5]. A major auxin-signaling pathway is transcriptional regulation mediated by a co-receptor consisting of TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) and AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA). In addition, AUXIN BINDING PROTEIN 1 (ABP1), which is evolutionarily conserved from charophyte green algae to seed plants, is known as an extracellular auxin receptor involved in rapid re-orientation of microtubules [6–9]. Studies in Arabidopsis and other angiosperms have revealed that auxin perception by the TIR1/AFB-AUX/IAA co-receptor triggers transcriptional regulation mediated by the AUXIN RESPONSE FACTOR (ARF) transcription factors, which directly bind to cis-elements (auxin responsive elements, or AuxREs) of auxin responsive genes and positively or negatively regulates the expression [10]. In the absence of auxin, AUX/IAAs bind to ARFs through their C-terminal regions, called domains III/IV, of the respective proteins and the complex represses the expression of auxin responsive genes [11,12]. Auxin functions as “molecular glue” that stabilizes the interaction between the F-box protein TIR1/AFB and the transcriptional repressor AUX/IAA [13,14]. This interaction promotes ubiquitination of AUX/IAA by the ubiquitin ligase complex that contains TIR1/AFB and subsequent degradation of AUX/IAA by the 26S proteasome [15], which liberates the ARFs and allows them to play their roles in transcriptional regulation.
In Arabidopsis, an ensemble of 29 AUX/IAAs and 23 ARFs is believed to regulate various auxin responses [16,17]. However, the high level of genetic redundancy of these transcription factors and the complex body plan composed of various organs make it difficult to depict a comprehensive picture of auxin regulatory events consisting of interactions and feedback among multiple factors.
Auxin responses are also observed in basal land plant lineages, such as the bryophytes (liverworts, mosses, and hornworts), and green algal lineages related to land plants, the charophytes [18]. Whole genome sequencing approaches of the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii, a member of the basal lineage of vascular plants, have revealed that these two species have orthologues of basic components for auxin-mediated transcriptional regulation with relatively lower redundancy than observed in flowering plants [19,20]. Additionally, it was reported that P. patens possesses the auxin perception mechanism mediated by TIR1/AFB and AUX/IAA, which regulates the chloronema-caulonema transition and rhizoid formation [21]. On the other hand, there is limited knowledge about auxin-mediated regulatory systems in green algae. It is generally accepted that the ancestor of land plants was closely related to charophytes [22]. A recent study on the draft genome sequence of Klebsormidium flaccidum, a filamentous charophyte lacking differentiation of specialized cells, reported the absence of TIR1/AFB, AUX/IAA, and ARF genes in its genome [23], suggesting that auxin-mediated transcriptional regulation evolved subsequent to the divergence of Klebsormidium and the lineage leading to land plants.
Marchantia polymorpha is a liverwort species belonging to the earliest diverging clade of extant land plants [22] and has long history as an experimental organism. M. polymorpha is a complex thalloid liverwort and spends most of its life cycle as a haploid flat thallus which grows apically and has distinct dorsiventrality. On the dorsal side, air chambers are regularly arranged [24], and asexual reproductive organs, gemma cups and gemmae, are repeatedly formed [25]. On the ventral side of thallus, scales and rhizoids are produced [26,27]. M. polymorpha is dioecious and produces gametangiophores (archegoniophores that produce egg cells and antheridiophores that produce sperms) for sexual reproduction under certain environmental conditions [28,29]. Following fertilization, a diploid zygote develops into a multicellular sporophyte, or embryo, on archegoinophores, in which a set of cells undergo meiosis, resulting in a sporangium producing single-celled haploid spores [30].
The most common endogenous auxin, indole-3-acetic acid has been detected in M. polymorpha [31]. Application of exogenous auxin has revealed that auxin is involved in rhizoid initiation and elongation [32–34], thallus growth [34,35], regeneration from excised thalli [36], and apical dominance [37] in M. polymorpha and in dormancy of gemma in Lunularia, a related liverwort genus [38].
Recently, M. polymorpha has received attention for its critical evolutionary position [39]. Molecular genetic tools including transformation techniques [40,41], homologous recombination [42], and CRISPR/Cas9-mediated genome editing [43] have been developed. By applying molecular techniques, we demonstrated that the auxin response reporter which expresses β-glucuronidase under the soybean-derived GH3 promoter (proGH3:GUS) specifically responds to auxin in a dose-dependent manner [34], suggesting a possible conservation of regulatory machinery for auxin-mediated transcriptional activation in M. polymorpha.
In this study, we performed an in silico search for auxin signaling factors in M. polymorpha, and demonstrate that it has a minimal but complete auxin-mediated transcriptional system. We also describe critical roles of AUX/IAA-mediated auxin signaling in M. polymorpha development throughout its life cycle. Moreover, we investigated protein interactions and functional diversities of ARFs in M. polymorpha. From these results, we discuss how various auxin responses are regulated using the minimal set of auxin signaling factors in M. polymorpha.
Results
Identification of auxin signaling factors in M. polymorpha
To investigate whether M. polymorpha has basic components of auxin signaling, genes for known auxin signaling factors were surveyed by BLAST searches against M. polymorpha transcriptome and genome databases.
For AUX/IAA, only a single gene was found in the M. polymorpha genome (MpIAA), that contains four conserved domains (domains I to IV). In the N-terminus, the predicted MpIAA protein has a domain I motif (LxLxL), which is predicted to bind directly to the transcriptional co-repressor TOPLESS (TPL; Figs 1A and S1) [44]. In the C-terminus, MpIAA contains a domain II sequence, the AUX/IAA degron, and domains III and IV, which comprise a protein-protein interaction domain (Figs 1A and S1). In the domains III/IV of MpIAA, invariant lysine and acidic residues, which are shown to be important for AUX/IAA-ARF oligomerization in Arabidopsis, are conserved (S1 Fig) [45,46]. The amino acid sequence of MpIAA is considerably longer (825 amino acid residues) as compared to AUX/IAA proteins of vascular plants, and contains an additional glutamine-rich stretch between domains I and II. To understand if this structure is evolutionarily conserved, we obtained partial sequence of AUX/IAA homologues from two other Marchantiales species, Conocephalum conicum and C. japonicum, via degenerate RT-PCR. The Conocephalum sequences also contain long glutamine-rich regions in their N-termini, implying that this region has a conserved function among at least the Marchantiales (S1 Fig). In phylogenetic analyses, MpIAA resided in a clade with P. patens and S. moellendorffii distinct from the clade of Arabidopsis AUX/IAA sequences (Fig 1B).
Fig. 1. Auxin signaling factors in M. polymorpha. 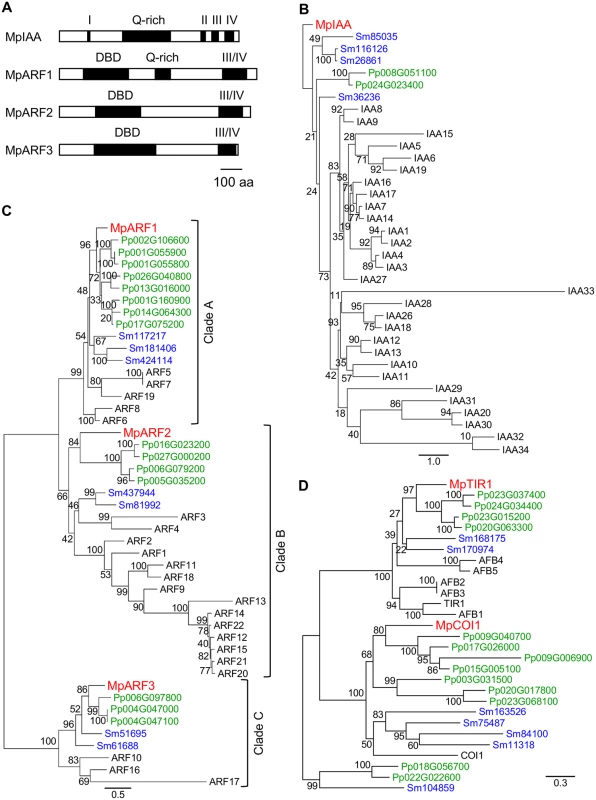
(A) Diagrams of domain structures of MpIAA and MpARFs. DBD: DNA-binding domain, Q-rich: glutamine-rich region, Roman numbers: domains I through IV. (B-D) Phylogenetic positions of MpIAA (B), MpARFs (C) and MpTIR1 (D) using the amino acid sequences from M. polymorpha (red), P. patens (green), S. moellendorffii (blue), and Arabidopsis (black). See S2 Table for sequence information. Numbers on the branches indicate bootstrap values. Bar in (A): 100 amino acid residues in length. Bars in (B-D): number of amino acid changes per branch length. The M. polymorpha genome encodes three ARFs (MpARF1, MpARF2, and MpARF3). The three MpARFs have a structure common to typical ARFs, containing a B3-like DNA binding domain (DBD) and a protein-protein interaction motif, called domains III and IV (Figs 1A and S2). Between the DBD and domain III, MpARF1 harbors a glutamine-rich region (Fig 1A), which is known as a feature of activator ARF members [10]. Finet et al. (2013) classified 224 ARF proteins across diverse land plants into three clades: clade A, clade B and clade C [47]. Phylogenetic analysis showed that MpARF1 belonged to the clade A, which represents activator ARF proteins such as ARF5/MONOPTEROS (MP) of Arabidopsis (Fig 1C). MpARF2 resided in clade B (Fig 1C). In Arabidopsis, some ARF members in this clade have been shown to function as transcriptional repressor [10,48]. MpARF3 has a relatively longer DBD, and its domains III and IV show lower similarity to ARFs in the clades A and B (Figs 1A, S2A, and S2B). MpARF3 mRNA contains the possible target sequence of microRNA160 (S2C Fig). These features are common with Arabidopsis ARF10 and ARF16, which possibly function as repressors [49]. Phylogenetic analysis supported the placement of MpARF3 in the clade C including Arabidopsis ARF10, ARF16, and ARF17 (Fig 1C). In summary, MpARF1, MpARF2, and MpARF3 were phylogenetically classified into the clades A, B, and C, respectively, suggesting that three functionally diverged types of ARFs existed in the common ancestor of extant land plants.
BLAST searches revealed that M. polymorpha genome harbors two genes that, respectively, exhibit high similarity to TIR1/AFB and a jasmonic acid receptor, CORONATINE INSENSITIVE1 (COI1), of Arabidopsis. One encodes a protein with 54% identity to Arabidopsis TIR1 and is phylogenetically classified in the TIR1/AFB clade (named MpTIR1, Figs 1D and S3). The other sequence is 44% identical to COI1, and phylogenetic analysis supported that this sequence belonged to the COI1 clade (Fig 1D). These results suggest that M. polymorpha has only one TIR1/AFB auxin receptor.
Taken together, M. polymorpha has all required components for auxin-mediated transcriptional regulation with minimal genetic redundancy. We also performed BLAST and HMMER searches using another auxin receptor, ABP1, as query. ABP1 is broadly conserved in the green lineage including charophytes green algae [7]. However, to our surprise, no homologue of ABP1 was found in the M. polymorpha genome.
Repression of auxin responses by domain II-modified MpIAA
Studies in angiosperms, including Arabidopsis, have shown AUX/IAA as the key component in auxin perception and signaling. Dominant mutations in domain II of AUX/IAA inhibit its auxin-dependent degradation and results in auxin resistance [50]. Because M. polymorpha has only the single AUX/IAA, we focused on MpIAA to investigate the mechanism and function of auxin signaling in M. polymorpha. To examine if MpIAA is involved in auxin signaling in M. polymorpha, we generated transgenic plants which expressed MpIAA with or without substitutions of two conserved proline residues in the domain II degron sequence into serines under the control of the MpELONGATION FACTOR 1α constitutively active promoter (proMpEF1α:MpIAAmDII and proMpEF1α:MpIAA; Fig 2A) [51] and analyzed their responses to exogenous auxin. In the absence of auxin, proMpEF1α:MpIAA plants were indistinguishable from wild type (WT). In contrast, in the absence of exogneous auxin proMpEF1α:MpIAAmDII plants displayed morphological defects such as dwarfism and curling of the thallus. In the presence of high concentrations of exogenous naphthaleneacetic acid (NAA), a synthetic auxin, proMpEF1α:MpIAA plants exhibited growth arrest and produced many rhizoids as reported for WT previously [34,52–55], while proMpEF1α:MpIAAmDII plants were insensitive to NAA treatment (Fig 2B). Thus, we conclude that MpIAA conveys auxin signaling through domain II function in a similar manner as that demonstrated in angiosperms.
Fig. 2. Effects of mutations in domain II of MpIAA on auxin sensitivity. 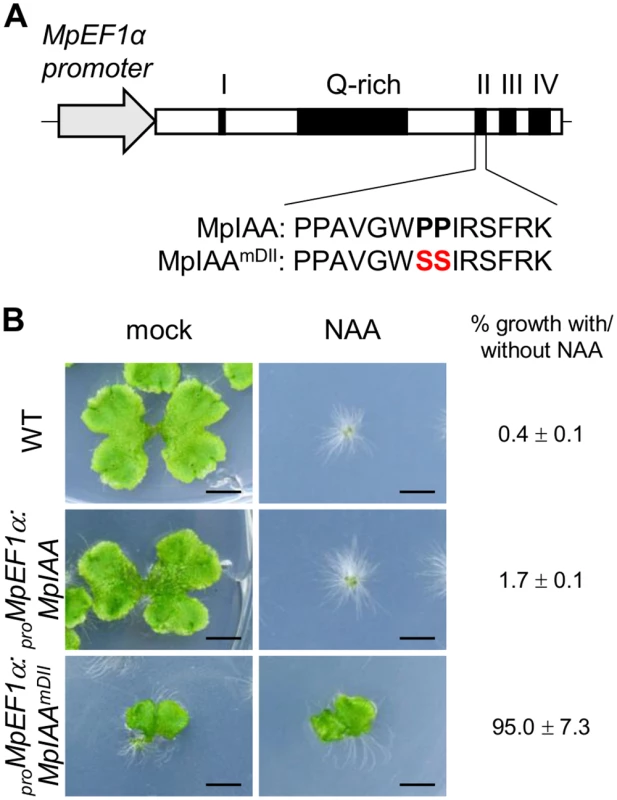
(A) Diagram of proMpEF1α:MpIAA and proMpEF1α:MpIAAmDII. Two conserved proline residues in domain II were substituted with serine. (B) Resistance to auxin by domain II-modified MpIAA expression. Photographs of WT, proMpEF1α:MpIAA and proMpEF1α:MpIAAmDII plants cultured with exogenous 3 μM NAA or under mock conditions for 2 weeks. Bars: 5 mm. The values indicate area ratios of 2-week-old thallus grown in the presence of NAA to that grown under mock conditions with SE (n = 12). We observed that the expression level of transgenes in the lines obtained was significantly lower in proMpEF1α:MpIAAmDII than that of proMpEF1α:MpIAA (S4 Fig), suggesting that high levels of expression of MpIAAmDII driven by the MpEF1α promoter might be deleterious. Therefore, we utilized an inducible system in which protein nuclear localization is modulated by a glucocorticoid receptor (GR) domain [56,57], to investigate in more detail the function of MpIAA and to determine developmental processes where auxin-mediated transcriptional regulation is involved in the life cycle of M. polymorpha. We generated transgenic plants expressing a chimeric protein of MpIAAmDII C-terminally fused with GR under the control of the MpIAA promoter (proMpIAA:MpIAAmDII-GR). In these transgenic plants auxin signaling should be repressed upon treatment with dexamethasone (DEX). To confirm that this experimental strategy works in plants, we first introduced proMpIAA:MpIAAmDII-GR into the M. polymorpha lines harboring the auxin response reporter proGH3:GUS [34]. Twelve hours of exogenous auxin treatment increased GUS activity in proGH3:GUS control plants. DEX treatment applied to proMpIAA:MpIAAmDII-GR/proGH3:GUS plants completely repressed auxin-dependent expression of the GUS reporter gene, whereas DEX treatment did not affect auxin-induced GUS activity in proGH3:GUS plants (Fig 3). These results suggest that the presumable accumulation of MpIAAmDII protein in the nucleus represses an auxin-dependent transcriptional response as has been shown in Arabidopsis [58]. Taken together, the mechanism of AUX/IAA-mediated auxin response is conserved in M. polymorpha.
Fig. 3. DEX-inducible system for repressing auxin responses. 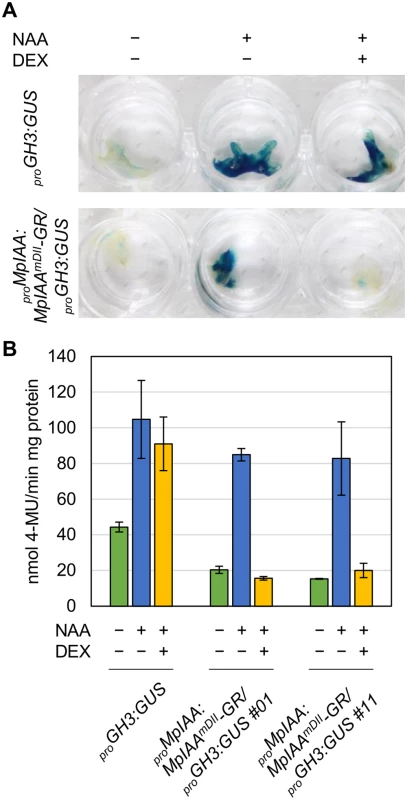
(A, B) GUS staining (A) and quantitative fluorometric assays (B) of proGH3:GUS and proMpIAA:MpIAAmDII-GR/proGH3:GUS transgenic plants. Each plant was treated with 10 μM NAA and/or 10 μM DEX for 12 h. Error bars: SE (n = 3). Regulation of cell expansion by MpIAA-mediated auxin signaling
We next examined the significance of MpIAA-mediated auxin signaling in the regulation of cellular morphology in M. polymorpha. We first analyzed the morphological and cellular responses of M. polymorpha thalli to exogenously supplied auxin. In the WT thallus, NAA treatment caused epinasty of thalli (Fig 4A and 4B), protrusion of air chambers (Fig 4D and 4E), and elongation of gemma cups (Fig 4G and 4H). Quantification of cell parameters revealed directional expansion of dorsal epidermal cells (S5 Fig), which therefore may be responsible for the above phenotypes. In the absence of DEX, proMpIAA:MpIAAmDII-GR plants responded to NAA as did WT (Fig 4J, 4K, 4M, 4N, 4P, and 4Q). Treatment of proMpIAA:MpIAAmDII-GR plants with DEX alleviated the NAA-induced phenotypes (Figs 4L, 4O, 4R, and S5), whereas the same treatment to WT thallus did not (Figs 4C, 4F, 4I, and S5). These results suggest that cell expansion in dorsal epidermal tissue is promoted by auxin through MpIAA-mediated transcriptional regulation.
Fig. 4. Effects of exogenous auxin on the morphology and cell shape. 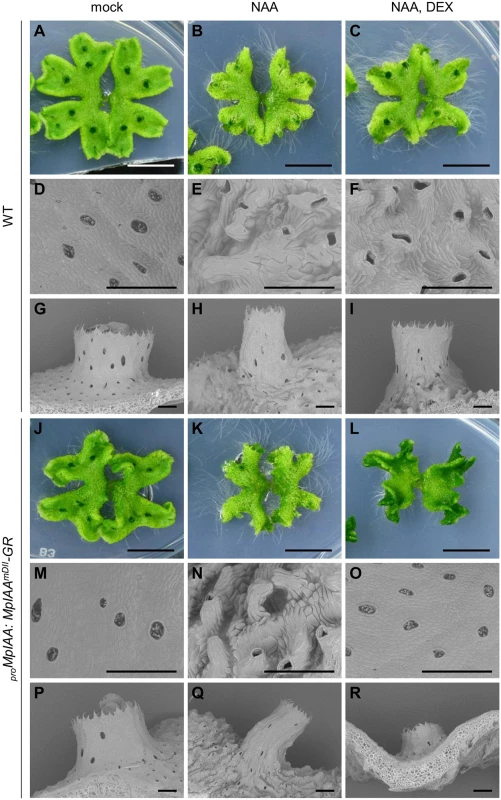
WT (A-I) and proMpIAA:MpIAAmDII-GR plants (J-R) were grown for 12 days in the absence of both NAA and DEX, then grown under mock condition (A, D, G, J, M, P), with 10 μM NAA (B, E, H, K, N, Q), or with 10 μM NAA and 10 μM DEX (C, F, I, L, O, R). At day 7 post treatment, photographs (A-C, J-L; bars: 10 mm) and scanning electron micrographs (D-I, M-R; bars: 500 μm) were taken. Expression specificity of MpIAA in the life cycle
The results described thus far suggest the central role of the sole AUX/IAA, MpIAA, in the auxin-mediated transcriptional regulation in M. polymorpha. To investigate spatiotemporal expression pattern of MpIAA, we generated transgenic plants expressing GUS reporter gene under the regulation of MpIAA promoter (proMpIAA:GUS). High levels of GUS activity was observed throughout proMpIAA:GUS thalli including the gemma cups (Fig 5A). Cross-sections of the proMpIAA:GUS thallus revealed that GUS staining was observed in all layers of thallus tissue, including dorsal air chambers, gemma cups and developing gemmae, internal parenchymatous tissue, and ventral scales and rhizoids (Fig 5B and 5C).
Fig. 5. Expression pattern of MpIAA throughout the life cycle. 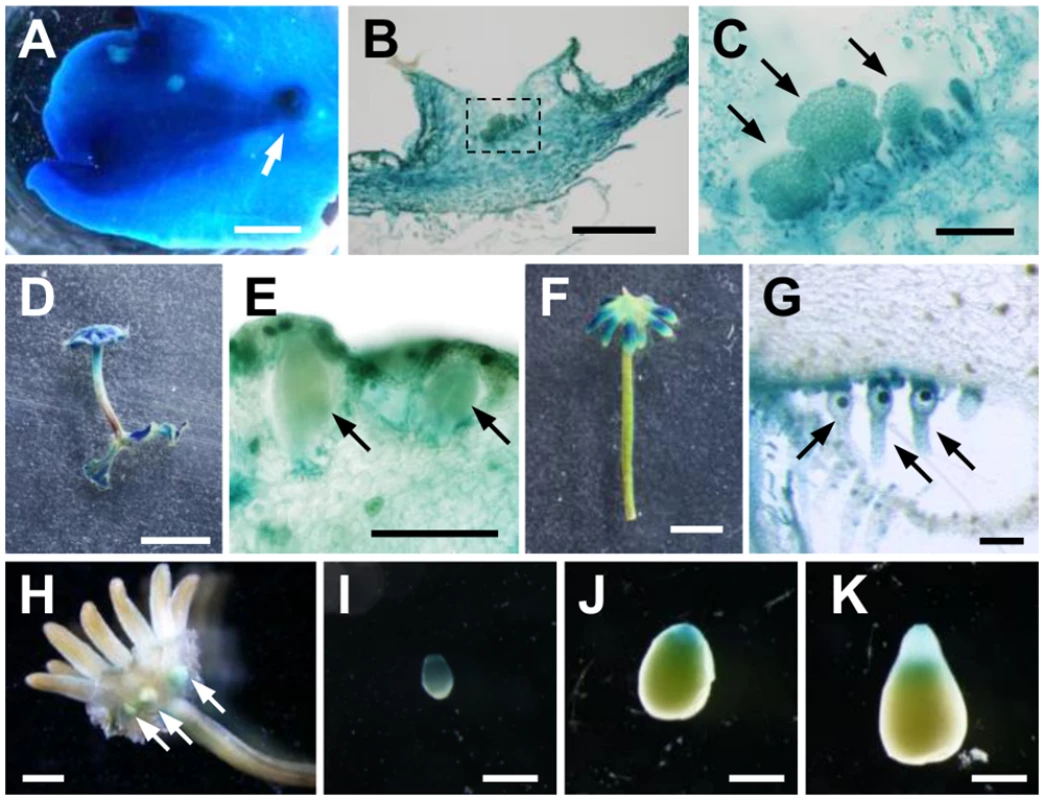
GUS staining of proMpIAA:GUS plants. (A) 3-week-old thallus. Arrow: gemma cup. (B) Longitudinal section of gemma cup. (C) Magnified view of the region shown by dotted line in (B). Arrows: developing gemmae. (D, E) Overview (D) and longitudinal section (E) of antheridiophore. The arrows represent antheridia. (F, G) Overview (F) and longitudinal section (G) of archegoniophore. Arrows: archegonia. (H-K) Sporophytes generated by crossing WT female and proMpIAA:GUS male. (H) Overview of fertilized archegoniophore containing developing sporophytes (arrows). (I-K) Isolated developing sporophytes. The apices of the sporophytes are directed downward. Scale bars: 2 mm (A, H), 0.5 mm (B, I-K), 0.1 mm (C, E, G), 5 mm (D, F). We also observed expression during the reproductive phase. Antheridiophores showed strong GUS staining (Fig 5D), including GUS signal in antheridia and surrounding tissues (Fig 5E). Compared with antheridiophores, archegoniophores showed relatively weak GUS staining, which was observed in the tips of digitate rays (Fig 5F). Cross-sectional analysis revealed that intensive GUS activity was observed in archegonia including the egg cells (Fig 5G).
We also observed expression in the diploid sporophyte generation following fertilization of the haploid gametes. Strong GUS staining was observed in young sporophytes (Fig 5H and 5I), and during sporophyte development, a gradient of GUS activity was evident along apical-basal axis. GUS staining of apical sporogenous tissue was relatively weaker, and diminished as sporophyte matured, whereas that of the basal region consisting of the foot and seta remained (Fig 5I–5K). These GUS staining patterns in the sporophyte were observed in reciprocal crosses between proMpIAA:GUS and WT.
These results suggest that MpIAA is widely expressed in both gametophyte and sporophyte generations with some tissue specificity, and that MpIAA-mediated auxin signaling would function in various tissues and organs in both generations of the life cycle.
Pleiotropic roles of MpIAA-dependent auxin signaling throughout life cycle
In proMpIAA:MpIAAmDII-GR plants, auxin responses could be repressed in a DEX-dependent manner (see above). In order to investigate tissue - or stage-specific functions of MpIAA-mediated auxin signaling, we analyzed morphological phenotypes of proMpIAA:MpIAAmDII-GR plants caused by repression of auxin responses during various developmental stages. Without DEX treatment, proMpIAA:MpIAAmDII-GR plants developed normal thalli with gemma cups and regularly-arranged air pores on their dorsal sides (Fig 6A). Gemma cups formed serrated structures on their rims and produced many gemmae from their bases (Fig 6B and 6C). On the ventral side of thallus, ventral scales and numerous rhizoids were observed (Fig 6D).
Fig. 6. Morphological defects of proMpIAA:MpIAAmDII-GR plants. 
(A-D) proMpIAA:MpIAAmDII-GR plants grown for 14 days without DEX. (A) SEM of the dorsal side of the thallus. Air pores are visible as black dots. (B, C) SEM (B) and longitudinal section (C) of a gemma cup. (D) SEM of the ventral side of the thallus. Arrow: ventral scale. (E-H) proMpIAA:MpIAAmDII-GR gemmalings grown in the presence of 10 μM DEX for 14 days. SEM images are shown. Arrows: serrated structures reminiscent of gemma cup. Arrowheads: air pores. (I-L) proMpIAA:MpIAAmDII-GR plants grown in the absence of DEX for 7 days and then subsequently in the presence of 10 μM DEX for 7 days. (I, J) SEM images of the dorsal side of the thallus (I) and a gemma cup (J). (K) Longitudinal section of a gemma cup. (L) SEM image of the ventral side of the thallus. (M-P) Antheridiophores (M, N) and archegoniophores (O, P) of male and female plants, respectively, grown for 2 weeks under mock (M, O) or DEX-treated (N, P) conditions. (Q-T) Fertilized archegoniophores at 4 weeks (Q, R) or 2 weeks (S, T) after crossing without (Q, S) or with DEX treatment (R, T). DEX treatment was performed every one or two days, beginning the day after crossing. Arrows: developing sporophytes. Scale bars: 1 mm (A, D, E, I, L), 0.5 mm (B, F-H, J), 0.1 mm (C, K), 5 mm (M-R), 0.2 mm (S, T). Compared to the control condition, gemmalings grown in the presence of 10 μM DEX for 14 days showed severe growth inhibition (Fig 6A and 6E). The eight gemmalings observed exhibited some or all of the following morphological abnormalities: five produced a cellular mass lacking dorsiventrality (Fig 6F), four formed air pores ectopically, and five produced serrated structures, which were reminiscent of the gemma cup rim (Fig 6G). Adventitious gemma-like multicellular bodies were frequently (six of the eight) formed as clusters on the surface of gemmalings (Fig 6H). We could not find any ventral scales in the apical regions of gemmalings treated with DEX. These results suggest critical roles of MpIAA-mediated auxin signaling in gemmaling growth and differentiation, especially with respect to ventral structures.
We then applied DEX treatment to 7-day-old thallus precultured in the absence of DEX. At this stage, gemmalings had developed into mature thalli with organogenesis in a proper dorsiventral topology. DEX treatment for 7 subsequent days conferred hyponasty resulting in V-shaped thalli (Fig 6I). Although gemma cups were observed on dorsal side of the DEX-treated thalli, the cups were shallow and elongated along with the apical-basal axis, generating many serrated structures (Fig 6J). At the bottom of gemma cups, in spite of the normal development of gemma primordia, mature gemmae did not develop (Fig 6K). On the ventral side, the number of rhizoids was decreased, especially smooth rhizoids (Fig 6L). These results suggest involvement of endogenous auxin and MpIAA-mediated transcriptional regulation in the harmonized growth of dorsal and ventral thallus tissues. MpIAA-mediated auxin response also functions in the development of gemma cups, gemmae and rhizoids.
To investigate the role of MpIAA-mediated auxin signaling in gametangiophore growth, we started periodical DEX treatment to gametangiophores after they became visible (smaller than 5 mm in height). DEX treatment resulted in short stalks, while, in the control condition, male and female gametangiophores had vertically elongated stalks (Fig 6M–6P). Additionally, DEX treatment to proMpIAA:MpIAAmDII-GR plants compromised the tropic growth of gametangiophore stalks (Figs 6M–6P and S6). These results suggest involvement of MpIAA-mediated auxin signaling in both tropic and differential growth, which was reported as responses in gametangiophores to exogenous auxin application [59].
Finally, we investigated MpIAA-mediated auxin signaling in sporophyte development. Without DEX treatment, sporophytes developed on archegoniophores, producing yellow sporangia in approximately 4 weeks after crossing (Fig 6Q and 6S). Periodical DEX treatment that was initiated on the day following crossing conferred developmental arrest of the sporophyte (Fig 6T). We did not observe any mature sporangia 4 weeks after crossing (Fig 6R). These results suggest that proper MpIAA-mediated auxin signaling is critical for sporophyte development.
Protein-protein interaction between MpIAA and MpARFs
Phenotypic analysis of proMpIAA:MpIAAmDII-GR plants demonstrated that MpIAA-mediated auxin signaling regulates many aspects of growth and development of M. polymorpha. Therefore, our next question was how M. polymorpha generates various auxin responses using the simplified components for auxin-mediated transcriptional regulation. To tackle this question, we focused on the three ARF genes of M. polymorpha. We speculated that each MpARF might have a different specificity in protein-protein interactions and/or transcription activities. In Arabidopsis, interactions between AUX/IAA and ARF proteins have been examined by yeast two-hybrid (Y2H) assays and bimolecular fluorescence complementation (BiFC) [5,16]. To investigate if MpIAA and MpARFs have ability to form homo - or hetero-dimers, we first performed Y2H assays using C-terminal regions of MpIAA and MpARFs. Our Y2H assay showed that MpIAA could interact with all three MpARFs (Fig 7). Interactions between MpARFs were observed in all combinations except for MpARF3 homotypic interaction (Fig 7). We also examined the strengths of the observed Y2H interactions by quantitative measurements of 0β-galactosidase reporter activities (S1 Table). MpIAA showed high β-galactosidase activities with all MpARFs, but there were significant differences among the combinations. MpARF1 showed a notably higher activity in combination with MpARF1 than with the other MpARFs, while MpARF2 did with MpARF3, suggesting different affinities in the interactions among the three MpARFs.
Fig. 7. Protein-protein interactions between MpIAA and MpARFs in yeast. 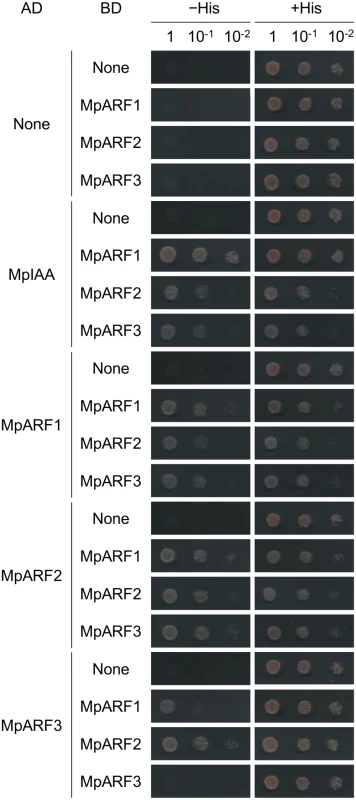
Yeast two-hybrid assays with the HIS3 reporter. Ten-fold serial dilutions of overnight cultures were spotted on either nonselective +His (-Trp/-Leu) or selective-His (-His/-Trp/-Leu) media and grown for 2 days at 22°C. AD: proteins fused to VP16 activation domain. BD: proteins fused to lexA DNA-binding domain. The interactions between MpIAA and MpARFs in planta were also examined by BiFC assay in Nicotiana benthamiana leaves. We analyzed all combinations of interactions between proteins fused to N-terminal and C-terminal halves of YFP under the condition where negative control experiments with empty vectors yielded no fluorescent signal. BiFC assays revealed that MpIAA interacted with MpIAA itself and all MpARFs. All combinations of MpARF-MpARF except for MpARF3-MpARF3 produced signal (Fig 8). These results confirmed protein interactions between MpIAA and MpARFs, as well as between MpARFs.
Fig. 8. Protein-protein interactions between MpIAA and MpARFs in planta. 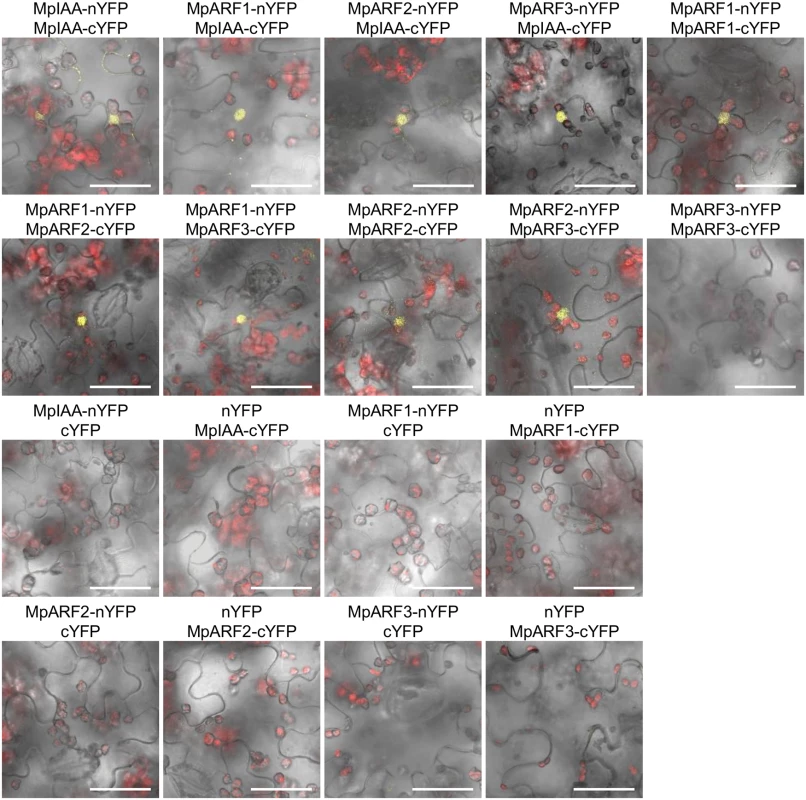
BiFC assays of MpIAA and MpARF using N. benthamiana leaves. Confocal images of YFP (yellow) and chloroplast auto-fluorescence (red) were merged with bright-field images. Vectors containing only nYFP or cYFP was used as negative controls. Bars: 50 μm. Transcription activity of MpARFs
To characterize the transcription activity of each MpARF, we performed transient transactivation assays using cultured tobacco BY-2 cells. The effector constructs carried full-length or the middle region sequences of MpARFs fused with the Gal4 DNA binding domain. The reporter vector expressed firefly luciferase (F-Luc) under the control of a promoter containing six repeats of the Gal4 binding site. As a transformation control, we prepared the plasmid carrying the Renilla luciferase (R-Luc) gene driven by the cauliflower mosaic virus 35S promoter (Fig 9A). These constructs were simultaneously introduced into BY-2 cells by particle bombardment. Transcriptional activity was evaluated by the relative activity of F-Luc to R-Luc. Both the middle region and full-length sequences of MpARF1 showed approximately two-fold higher activity than the effector expressing only Gal4 DBD. In the case of MpARF2, luciferase activity was lower than the control (Fig 9B). These results suggest that MpARF1 and MpARF2 can function as a transcriptional activator and repressor, respectively. We could characterize MpARF3 as neither an activator nor a repressor from this experiment, as the middle region and full-length sequences of MpARF3 showed just slightly lower and higher luciferase activities, respectively, than the control (Fig 9B). Taken together, our results suggest that M. polymorpha has three types of ARFs with different characteristics in their transcriptional activities.
Fig. 9. Tranactivation assay for MpARFs. 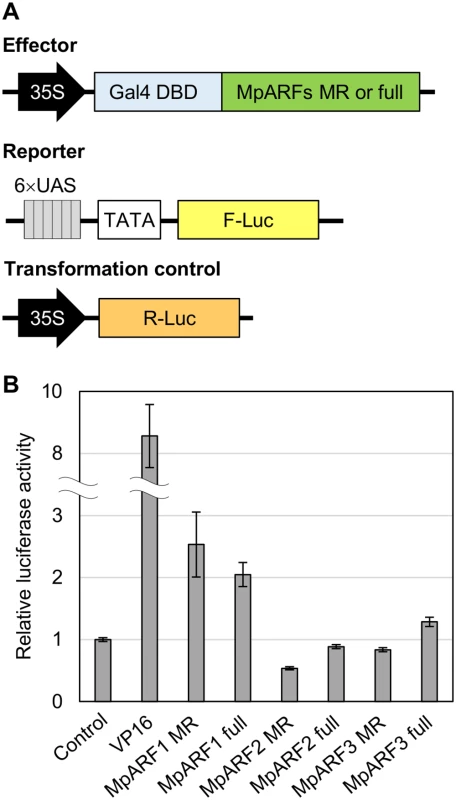
(A) Diagrams of the constructs for dual luciferase assay. (B) Relative luciferase activity elicited by effector plasmid. The vector expressing only Gal4 DBD was used as a control. A virus-derived activation domain, VP16, was used for a positive control. See Material and Methods for effector vectors. Error bars: SE (n = 3). Discussion
The origin and evolution of auxin-mediated transcriptional regulation in plants
Our results revealed that the liverwort M. polymorpha has a single TIR1/AFB, a single AUX/IAA, and three phylogenetically and functionally diverged ARF homologues. In Arabidopsis, it has been shown that AUX/IAA functions as a repressor through the interaction via domain I with the co-repressor TPL [44]. MpIAA has a conserved LxLxL motif in domain I (S1 Fig). The M. polymorpha genome encodes a homologue of TPL, and it is suggested that MpTPL is involved in auxin-mediated transcription (in an accompanying paper). In the present study, we showed that expression of domain II-modified MpIAA conferred an auxin-resistant phenotype and suppressed the transcriptional response to exogenously supplied auxin as monitored by proGH3:GUS (Figs 3 and 4). Additionally, it was reported that knock-down of MpIAA resulted in auxin hypersensitivity (in an accompanying paper). These results suggest that auxin-mediated degradation of MpIAA, presumably promoted by MpTIR1, is critical for transcriptional regulation. Our results also showed interaction between MpIAA and MpARFs through domains III/IV (Figs 7 and 8, S1 Table). Loss - and gain-of-function mutants of MpARF1 show auxin-resistance and hypersensitivity, respectively (in an accompanying paper) [43]. Taken together, these results suggested that M. polymorpha possesses an auxin-mediated transcriptional regulation system that involves AUX/IAA and ARFs. Previous genomic analyses revealed that the lycophyte S. moellendorffii and the moss P. patens have all the basic components for auxin-mediated transcription [19,20], and that the filamentous charophyte alga K. flaccidum has none of the components [23]. Expressed sequences showing high similarity to the DBD and domains III/IV of ARFs were found in two other charophyte species, Coleochaete orbicularis and Spirogyra pratensis, respectively, that in some analyses represent the sister lineage to extant land plants [60,61]. Although it is still controversial whether aquatic ancestors of land plants had acquired the auxin-mediated transcriptional regulation, these data suggest that the origin of auxin responses using the three types of ARFs dates back to at least the last common ancestor of extant land plants.
In comparison with AUX/IAAs of vascular plants, the predicted amino-acid sequence of MpIAA, is much longer, and contains a long glutamine-rich region between domains I and II, which is conserved at least in the Marchantiales (Figs 1A and S1). Glutamine-rich domains are known to activate transcription in eukaryotes [62]. Activator ARFs, including MpARF1, also contain a glutamine-rich domain (Fig 1A) [10]. Since ARFs and AUX/IAAs also exhibit similarity in their C-terminal interaction domains, these genes likely evolved from a common ancestral gene. It is possible that the glutamine-rich domain of Marchantiales AUX/IAAs might be a remnant of the ancestral gene and may retain an unknown function in this lineage.
We previously demonstrated that proGH3:GUS activity could reflect the sites of endogenous auxin responses [34]. Expression of MpIAA was observed in various tissues including those showing high proGH3:GUS activities, such as the base of gemma cups, stalks and lobes of archegoniophores, antheridia and developing sporophytes (Fig 5). The significance of MpIAA expression in these tissues was supported by observation of phenotypes of proMpIAA:MpIAAmDII-GR plants (Fig 6J, 6K, and 6M–6T). These results suggest that auxin responses monitored by proGH3:GUS can be accounted for by MpIAA function. Interestingly, developmental defects of DEX-treated proMpIAA:MpIAAmDII-GR plants were also observed where no proGH3:GUS activity was detected, such as gemmalings and rhizoids (Fig 6E–6I and 6L). This could be possibly due to a high threshold of proGH3:GUS expression in response to auxin, or limitation of this reporter only representing the transcriptional activity mediated by activator-type ARF (MpARF1). If the latter is the case, non-activator-type ARFs (MpARF2 and MpARF3) could have important developmental roles in M. polymorpha.
Evolutionarily conserved roles of auxin-mediated transcription in land plants
Auxin regulates many aspects of plant growth and development via modulating cell differentiation and expansion. In Arabidopsis, ARF10 belonging to the same clade as MpARF3 regulates cell totipotency in cultured cells [63]. In the moss P. patens, gain-of-function AUX/IAA mutants showed delayed caulonema differentiation from chloronema [21]. The present study showed that gemmaling produced undifferentiated cell mass by repression of MpIAA-mediated auxin signaling (Fig 6), suggesting that auxin-mediated transcription regulates cell differentiation in M. polymorpha. Consistently, transgenic plants expressing a bacterial auxin inactivating enzyme produced undifferentiated cell mass (in an accompanying paper). With respect to cell elongation, various mutants of AUX/IAA, ARF, and TIR1/AFB genes in Arabidopsis show defects in cell expansion and tropic responses [4,5,64–67]. Our study revealed that MpIAA-dependent auxin signaling regulated directional elongation of epidermal cells and tropic responses (Figs 4, 6M–6P, S5, and S6). These results suggest that regulation of cell differentiation and expansion by auxin-mediated transcription was already present in the common ancestor of land plants.
In Arabidopsis, it is reported that early-phase auxin-induced hypocotyl elongation occurs independently of TIR1/AFB-mediated transcription [68]. ABP1 has been proposed to be another auxin receptor that rapidly activates cell expansion in transcription-independent manner in angiosperms [6,69–72]. ABP1 is also found in green algae, although its function in these taxa remains to be elucidated [7,23,60]. To our surprise, no homologue of ABP1 was found in the M. polymorpha genome, suggesting that M. polymorpha lost ABP1 during evolution. This brings up new open questions, such as when was ABP1 function in auxin-mediated cell elongation established and what is the ancestral role of ABP1 in the plant and land plant lineages?
One of the important roles of auxin in plant development is axis formation. The present study revealed that the expression pattern of MpIAA exhibited a gradient along the apical-basal axis in the sporophyte, similar to the auxin-response reporter proGH3:GUS (Fig 5I–5K). Repression of MpIAA-mediated auxin signaling caused arrest of sporophyte development (Fig 6Q–6T). In the moss P. patens, it has been reported that the expression pattern of proGH3:GUS changes dynamically along apical-basal axis during sporophyte development, and that defects in auxin transport causes abnormal morphology of sporophyte [73,74]. In Arabidopsis, IAA12/BODENLOS and ARF5/MP are involved in formation of apical-basal axis in embryogenesis [75,76]. These results suggest that land plants would have common auxin-mediated mechanism for apical-basal axis formation during embryogenesis after fertilization.
Past studies showed that in M. polymorpha excessive exogenous auxin treatment promoted of rhizoid formation both dorsal and ventral sides of gemmalings, and thus proposed involvement of auxin into dorsiventral patterning of thallus [32,34,35,55]. The present study revealed that repression of MpIAA-mediated auxin signaling inhibited development of rhizoids and ventral scales (Fig 6). These results at least suggest MpIAA-mediated auxin signaling promotes the development of ventral tissues. It would be intriguing to clarify whether auxin mediates it directly or through dorsiventral axis formation.
Diverse auxin responses by minimum components
In Arabidopsis, it is thought that the complex transcriptional regulation using 29 AUX/IAAs and 23 ARFs underlies robust auxin responses [16]. In addition, various combinations of TIR1 and AUX/IAA proteins form co-receptor complex with a wide range of auxin-binding affinities and show various auxin sensitivity of AUX/IAA degradation, which also contributes to complex auxin responses in angiosperms [77,78]. In this study, we demonstrated that the liverwort M. polymorpha regulates various developmental processes with a minimized auxin-mediated transcription system. Because M. polymorpha has only one AUX/IAA and one TIR1/AFB orthologue (Fig 1), it was expected that the variation in auxin responses could be attributed to the functional diversification of the three MpARFs. Our results showed that the three ARFs in M. polymorpha are phylogenetically diverged and have different transcriptional activities (Figs 1C and 9B). Furthermore, different binding affinities of MpIAA observed with the three MpARFs (S1 Table) suggest different auxin responsiveness, and MpARF-MpARF interactions (Figs 7 and 8) could add a higher level of regulation. This is also supported by the analyses of chimeric protein of TPL fused with domains III/IV of MpIAA and MpARFs (in an accompanying paper). Recently, crystal structure analyses revealed that AUX/IAA and ARF proteins multimerize through domain III/IV, and that the DBDs of ARFs dimerize and function as a “molecular caliper” when they bind to palindromically orientated AuxREs [45,46,79]. Although the physiological significance of interactions between MpARFs thorough domains III/IV and potentially between DBDs is unclear, it is possible that M. polymorpha can regulate diverse auxin responses via a combination of protein interactions of functionally diverged ARFs.
Still, would it be possible to explain such various auxin-response outputs only by the protein interaction variations? Recent work shows that functionally diverged ARF proteins from Arabidopsis have little differences in their DNA-binding specificity [79], raising the idea that the variation of ARFs may not contribute much to target specificities. Given the single-copy existence of the activator ARF in M. polymorpha, it might be more plausible that outputs are pre-determined depending on the respective cell types and that auxin just modulates switches via MpARF1. This idea is consistent with the recently proposed model, where auxin is viewed as a signal that provides “impetus” to processes [80]. It is expected that investigation on how three MpARFs regulate pleiotropic auxin responses will provide insights into the mechanisms of eliciting the variety of auxin responses observed in land plants.
Perspectives
The present study demonstrated that M. polymorpha has minimal but complete, relative to that known in flowering plants, auxin-mediated transcription system, regulating diverse morphological events including both cell expansion and differentiation. In addition to transcriptional regulation, various auxin responses can be generated by regulating auxin biosynthesis, metabolism, and transport. It is also necessary to investigate the regulation of these factors in M. polymorpha. We propose that M. polymorpha with a low genetic redundancy is a good model for investigating the evolution and mechanisms of morphogenesis controlled by auxin.
Materials and Methods
Plant material and growth condition
Male and female accessions of M. polymorpha, Takaragaike-1 (Tak-1) and Tak-2, respectively [28], were maintained asexually. F1 spores generated by crossing Tak-1 to Tak-2 or proGH3:GUS #21 [34], were used for transformation.
Gametangiophore formation was induced by far-red irradiation as described previously [28]. M. polymorpha was cultured on half strength Gamborg’s B5 medium [81] containing 1% agar under 50–60 μmol photons m-2 s-1 continuous white fluorescent light at 22°C unless otherwise defined.
Gene identification and phylogenetic analysis
A similarity search for M. polymorpha genes was performed using BLAST against transcriptome and genome databases from on-going project by US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/). The transcriptome data contained 3.0 × 106 reads by Roche 454 GS FLX and >1010 reads by Illumina Hi-Seq from >18 conditions/tissues in different growth stages. The genomic DNA was sequenced under the coverage of 26.7× and 54.0× by Roche 454 GS FLX and Illumina Hi-Seq, respectively. The protein sequences of MpIAA, MpARFs and MpTIR1 were aligned with sequences listed in S2 Table. Partial cDNA sequences of AUX/IAAs in C. conicum and C. japonicum were amplified by degenerate RT-PCR using the primer set, degenerate-IAA_L2 and degenerate-IAA_R1. Primers used in this study are listed in S3 Table. PCR fragments were subcloned into pBC-SK+ and sequenced. These sequences were aligned using the MUSCLE program [82] implemented in Geneious software version 6.1.6 (Biomatters; http://www.geneious.com/) with default parameters. For phylogenetic analysis we used the C-terminal region (domain II-stop) of AUX/IAA, the DNA-binding domain of ARFs, and full length sequences of TIR1/AFBs. Phylogenic trees were generated by PhyML program version 2.1.0 [83] implemented in the Geneious software using the LG model and four categories of rate substitution. Tree topology, branch length, and substitution rates were optimized, and the tree topology was searched using the nearest neighbor interchange method. Bootstrap values were computed from 1000 trials. HMMER search for ABP1 homologues was performed against a database of six-frame translation products derived from the M. polymorpha transcript database, using the hmmscan program in HMMER3.1 (http://hmmer.org) with the raw HMM file for ABP1 downloaded from the Pfam database (http://pfam.xfam.org; ID: PF02041).
Construction of plasmids for plant transformation
The coding sequence of MpIAA was amplified by RT-PCR using the primer set MpIAA_entry and MpIAA_stop, and cloned into pENTR/D-TOPO vector using the Gateway TOPO cloning kit (Life Technologies). Mutations in domain II were introduced by PCR using the primer set mDII_L3 and mDII_R3. Then the MpIAA and MpIAAmDII cassettes were transferred into pKIGWB2 using LR Clonase II (Life Technologies) according to the manufacture’s protocol, which generated proMpEF1α:MpIAA and proMpEF1α:MpIAAmDII constructs, respectively.
To generate a construct for proMpIAA:GUS, the genomic fragment covering from 5.2 kb upstream of putative start codon to the 52nd codon was amplified using the primer set MpIAA_usEntry and MpIAA_R9, and cloned into pENTR/D-TOPO vector. The resultant genomic fragment was transferred into pGWB3 [84] by LR Clonase II and translationally fused with GUS reporter gene.
To generate a construct for proMpIAA:MpIAAmDII-GR, the genomic fragment covering 5.2 kb upstream region and the coding sequence of MpIAA was amplified using the primer set MpIAA_usEntry and MpIAA_nonstop, and cloned into pENTR/D-TOPO vector. Mutations in domain II were introduced as described above. The glucocorticoid receptor hormone binding domain (GR) was amplified from pOpOn2.1 [85], and cloned into the AscI site of pENTR/D-TOPO vector. The resultant cassette containing genomic fragment of MpIAA fused with GR was transferred into pGWB1 [84] or pMpGWB201.
Transformation of M. polymorpha
Transformation of M. polymorpha was performed as described previously [40]. Independent T1 lines were isolated, and single G1 lines from independent T1 lines were established by subcultivating single gemmae which arose asexually from single initial cells [25,86]. Plants grown from gemmae of G1 lines (termed the G2 generation) were used for experiments.
Histochemical assay for GUS activity
Histochemical assays for GUS activity were performed as described previously [34]. GUS stained gemma cups and gametangiophores were embedded in 6% agar block and sectioned into ~100-μm-thick slices with LinearSlicer PRO 7 (DOSAKA EM, Kyoto, Japan).
Quantitative measurement of GUS activity
NAA and DEX treatments were performed by submerging plants into half-strength Gamborg’s B5 liquid medium [81] containing 10 μM NAA and/or 10 μM DEX for 12 h. GUS activity was then measured by monitoring cleavage of the β-glucuronidase substrate 4-methylumbelliferyl β-D-glucuronide (MUG) as described previously [29] with some modifications. After adjusting the concentration of extracted protein to 10 μg/70 μl extraction buffer, 20 μl of methanol and 10 μl of 10 mM MUG were added. After incubation at 37°C for 30 min, 900 μl of 200 mM sodium carbonate was added to stop the reaction. Fluorescence (460 nm emission/360 nm excitation) of liberated 4-methylumbelliferone (MU) was measured on Powerscan4 (DS Pharma Biomedical, Osaka, Japan).
Phenotypic analysis of proMpIAA:MpIAAmDII-GR plants
In the vegetative phase, NAA and DEX treatments of proMpIAA:MpIAAmDII-GR plants were performed by growing on half-strength Gamborg’s B5 medium [81] containing 10 μM NAA and/or 10 μM DEX. In the reproductive phase, DEX treatment was performed by spraying 10 μM DEX solution every 1 or 2 days. For scanning electron microscopy, plant samples were frozen in liquid nitrogen and directly observed on a Miniscope TM3000 (HITACHI, Japan). Measurement of size and aspect ratio of epidermal cells, and curvature of gametangiophores were performed using ImageJ (http://imagej.nih.gov/ij/) from SEM and photographic images, respectively.
Protein interaction analyses
For Y2H analyses the C-terminal regions of MpIAA (aa 627–825), MpARF1 (aa 783–928), MpARF2 (aa 751–879) and MpARF3 (aa 730–821) were cloned into pBTM116SBEN and pVP16PS, which were modified from pBTM116 and pVP16PS [87], using EcoRI/BamHI and BamHI/NotI site, respectively. Resultant constructs were transformed into the L40 Saccharomyces cerevisiae reporter strain [87], and transformants were selected on SD medium lacking tryptophan and leucine. Protein interactions were checked by histidine requirement or ONPG assays in the conventional method [87].
For BiFC analyses, C-terminal regions of MpIAA and MpARFs described above were cloned into pENTR/D-TOPO. Each insert fragment was introduced into pB4CY2 and pB4NY2 by the LR reaction. pB4CY2 and pB4NY2 were obtained from S. Mano, National Institute of Basic Biology, Japan. Preparation and infiltration of Agrobacterium cultures were performed as described previously [88]. Fluorescence from YFP (observation, 520 to 560 nm; excitation, 515 nm) was observed 20 to 21 h after infiltration. Fluorescent signals, chloroplast autofluorescence and bright-field images were captured using a confocal laser scanning microscope, FluoView 1000 (Olympus).
Transient transactivation assay
cDNAs encoding full-length or middle region (MpARF1: aa 347–822, MpARF2: aa 388–764, MpARF3: aa 448–628) sequences of MpARF proteins were amplified with specific primers listed in S3 Table, and cloned into the vector to express them as a fusion protein with Gal4-DBD driven by the cauliflower mosaic virus 35S promoter [89] using the In-Fusion HD Cloning Kit (Clontech). A reporter plasmid containing six repeats of the Gal4 binding site and F-Luc, and the transformation control plasmid carrying R-Luc driven by the 35S promoter were described previously [89]. These constructs were introduced simultaneously into cultured tobacco BY-2 cells by bombardment using Biolistic PDS-1000/He Particle Delivery System (BIO-RAD). After 48 h incubation, F-luc and R-luc activities were assayed using the Dual-Luciferase Reporter Assay System (Promega) in accordance with the manufacture’s protocol. Luminescence was detected by Centro XS3 LB 960 Microplate Luminometer (Berthold). Luciferase activity was normalized by protein concentration.
Semi-quantitative RT-PCR
Total RNA was extracted from 2-week-old thalli using TRIZOL Reagent (Life Technologies). First-strand cDNA was synthesized from 0.5 μg of total RNA with ReverTra Ace reverse transcriptase (Toyobo) and oligo(dT) primer. PCR amplification of the transgene-specific sequence was performed using the primer set MpIAA_dN2 and attB2_R. PCR amplification of the cDNA encoding the EF1α was performed as described before [90], and served as a control. These reactions were performed using the C1000 Thermal Cycler (Bio-Rad).
Accession numbers
The genomic sequences from this article are available in DDBJ under the following accession numbers: AB981316 (MpIAA), AB981317 (MpARF1), AB981318 (MpARF2), AB981319 (MpARF3), AB981320 (MpTIR1), and AB981321 (MpCOI1).
Supporting Information
Zdroje
1. Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, et al. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 : 147–153. 14614497
2. Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 : 507–518. 10760240
3. Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, et al. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 : 591–602. 14651850
4. Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, et al. (2000) The NPH4 locus encodes the auxin response cactor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12 : 757–770. 10810148
5. Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, et al. (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 : 379–393. 14729917
6. Perrot-Rechenmann C (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol 2: a001446. doi: 10.1101/cshperspect.a001446 20452959
7. Tromas A, Paponov I, Perrot-Rechenmann C (2010) AUXIN BINDING PROTEIN 1: functional and evolutionary aspects. Trends Plant Sci 15 : 436–446. doi: 10.1016/j.tplants.2010.05.001 20605513
8. Xu T, Dai N, Chen J, Nagawa S, Cao M, et al. (2014) Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343 : 1025–1028. doi: 10.1126/science.1245125 24578577
9. Chen X, Grandont L, Li H, Hauschild R, Paque S, et al. (2014) Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature.
10. Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin response factors. Proc Natl Acad Sci U S A 96 : 5844–5849. 10318972
11. Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19 : 309–319. 10476078
12. Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci U S A 94 : 11786–11791. 9342315
13. Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 : 446–451. 15917798
14. Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, et al. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9 : 109–119. 15992545
15. Gray WG, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414 : 271–276. 11713520
16. Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7 : 508. doi: 10.1038/msb.2011.39 21734647
17. Piya S, Shrestha SK, Binder B, Stewart CN Jr., Hewezi T (2014) Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5 : 744. doi: 10.3389/fpls.2014.00744 25566309
18. Cooke TJ, Poli D, Sztein AE, Cohen JD (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49 : 319–338. 12036257
19. Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 : 64–69. 18079367
20. Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, et al. (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332 : 960–963. doi: 10.1126/science.1203810 21551031
21. Prigge MJ, Lavy M, Ashton NW, Estelle M (2010) Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 20 : 1907–1912. doi: 10.1016/j.cub.2010.08.050 20951049
22. Qiu YL, Li L, Wang B, Chen Z, Knoop V, et al. (2006) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci U S A 103 : 15511–15516. 17030812
23. Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, et al. (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5 : 3978. doi: 10.1038/ncomms4978 24865297
24. Barnes CR, Land WJG (1907) Bryological papers. I. The origin of air chambers. Bot Gaz 44 : 197–213.
25. Barnes CR, Land WJG (1908) Bryologifal papers. II. The origin of the cupule of Marchantia. Bot Gaz 46 : 401–409.
26. Schiffner V (1909) Studien über die rhizoïden der Marchantiales. Ann jard bot Buitenz 3 : 473–492.
27. McConaha M (1941) Ventral structures effecting capillarity in the Marchantiales. Am J Bot 28 : 301–306.
28. Chiyoda S, Ishizaki K, Kataoka H, Yamato KT, Kohchi T (2008) Direct transformation of the liverwort Marchantia polymorpha L. by particle bombardment using immature thalli developing from spores. Plant Cell Rep 27 : 1467–1473. doi: 10.1007/s00299-008-0570-5 18553085
29. Kubota A, Kita S, Ishizaki K, Nishihama R, Yamato KT, et al. (2014) Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat Commun 5 : 3668. doi: 10.1038/ncomms4668 24752248
30. Durand EJ (1908) The development of the sexual organs and sporogonium of Marchantia polymorpha. Bulletin of the Torrey Botanical Club 35 : 321–335.
31. Schneider MJ, Troxler RF, Voth PD (1967) Occurrence of indoleacetic acid in bryophytes. Bot Gaz 128 : 174–179.
32. Kaul KN, Mitra GC, Tripathi BK (1962) Responses of Marchantia in aseptic culture to well-known auxins and antiauxins. Ann Bot 26 : 447–466.
33. Maravolo NC, Voth PD (1966) Morphogenic effects of three growth substances on Marchantia gemmalings. Bot Gaz 127 : 79–86.
34. Ishizaki K, Nonomura M, Kato H, Yamato KT, Kohchi T (2012) Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. J Plant Res 125 : 643–651. doi: 10.1007/s10265-012-0477-7 22311005
35. Maravolo NC (1980) Control of development in hepatics. Bull Torrey Bot Club 107 : 308–324.
36. Binns AN, Maravolo NC (1972) Apical dominance, polarity, and adventitious growth in Marchantia polymorpha. Am J Bot 59 : 691–696.
37. Davidonis GH, Munroe MH (1972) Apical dominance in Marchantia: correlative inhibition of neighbor lobe growth. Bot Gaz 133 : 177–184.
38. LaRue CD, Narayanaswami S (1957) Auxin inhibition in the liverwort Lunularia. New Phytol 56 : 61–70.
39. Bowman JL, Floyd SK, Sakakibara K (2007) Green genes-comparative genomics of the green branch of life. Cell 129 : 229–234. 17448980
40. Ishizaki K, Chiyoda S, Yamato KT, Kohchi T (2008) Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol 49 : 1084–1091. doi: 10.1093/pcp/pcn085 18535011
41. Kubota A, Ishizaki K, Hosaka M, Kohchi T (2013) Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci Biotechnol Biochem 77 : 167–172. 23291762
42. Ishizaki K, Johzuka-Hisatomi Y, Ishida S, Iida S, Kohchi T (2013) Homologous recombination-mediated gene targeting in the liverwort Marchantia polymorpha L. Sci Rep 3 : 1532. doi: 10.1038/srep01532 23524944
43. Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, et al. (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55 : 475–481. doi: 10.1093/pcp/pcu014 24443494
44. Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319 : 1384–1386. doi: 10.1126/science.1151461 18258861
45. Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, et al. (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A 111 : 5427–5432. doi: 10.1073/pnas.1400074111 24706860
46. Nanao MH, Vinos-Poyo T, Brunoud G, Thevenon E, Mazzoleni M, et al. (2014) Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun 5 : 3617. doi: 10.1038/ncomms4617 24710426
47. Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F (2013) Evolution of the ARF gene family in land plants: old domains, new tricks. Mol Biol Evol 30 : 45–56. doi: 10.1093/molbev/mss220 22977118
48. Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15 : 533–543. 12566590
49. Liu X, Huang J, Wang Y, Khanna K, Xie Z, et al. (2010) The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J 62 : 416–428. doi: 10.1111/j.1365-313X.2010.04164.x 20136729
50. Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24 : 55–80. doi: 10.1146/annurev.cellbio.23.090506.123214 18631113
51. Althoff F, Kopischke S, Zobell O, Ide K, Ishizaki K, et al. (2013) Comparison of the MpEF1alpha and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic Res 23 : 235–244. doi: 10.1007/s11248-013-9746-z 24036909
52. Rousseau J (1950) Action de l'acide indol beta-acétique sur les propagules de marchantia polymorpha et Lunularia cruciata. Comptes rendus hebdomadaires des séances de l'Académie des sciences 230 : 675–676. 17597171
53. Rousseau J (1951) Action de l'acide alpha napthaléne acétique sur les corbeilles á propagules de Marchantia polymorpha L. et de Lunularia cruciata Adans. Comptes rendus hebdomadaires des séances de l'Académie des sciences 232 : 107–108. 17597171
54. Tarén N (1958) Factors regulating the initial development of gemmae in Marchantia polymorpha. The Bryologist 61 : 191–204.
55. Otto KR, Halbsguth W (1975) Die Förderung der Bildung von Primärrhizoiden an Brutkörpern von Marchantia polymorpha L. durch Licht und IES (Stimulation of primary rhizoid formation on gemmae of Marchantia polymorpha L. as caused by light and IAA). Z Pflanzenphysiol 80 : 197–205.
56. Lloyd AM, Schena M, Walbot V, Davis RW (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 266 : 436–439. 7939683
57. Schena M, Lloyd AM, Davis RW (1991) A steroid-inducible gene expression system for plant cells. Proc Natl Acad Sci U S A 88 : 10421–10425. 1961707
58. Park JY, Kim HJ, Kim J (2002) Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J 32 : 669–683. 12472684
59. Rousseau J (1953) Action des hetero-auxines sur les chapeaux du Marchantia polymorpha L. Bulletin de la Société Botanique de France 100 : 179–180.
60. Timme RE, Delwiche CF (2010) Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol 10 : 96. doi: 10.1186/1471-2229-10-96 20500869
61. Bowman JL (2013) Walkabout on the long branches of plant evolution. Curr Opin Plant Biol 16 : 70–77. doi: 10.1016/j.pbi.2012.10.001 23140608
62. Escher D, Bodmer-Glavas M, Barberis A, Schaffner W (2000) Conservation of glutamine-rich transactivation function between yeast and humans. Mol Cell Biol 20 : 2774–2782. 10733580
63. Qiao M, Zhao Z, Song Y, Liu Z, Cao L, et al. (2012) Proper regeneration from in vitro cultured Arabidopsis thaliana requires the microRNA-directed action of an auxin response factor. Plant J 71 : 14–22. doi: 10.1111/j.1365-313X.2012.04944.x 22335436
64. Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol.
65. Leyser HM, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10 : 403–413. 8811856
66. Perez-Perez JM, Candela H, Robles P, Lopez-Torrejon G, del Pozo JC, et al. (2010) A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol 51 : 1661–1673. doi: 10.1093/pcp/pcq123 20739302
67. Möller B, Schenck D, Lüthen H (2010) Exploring the link between auxin receptors, rapid cell elongation and organ tropisms. Plant Signal Behav 5 : 601–603. doi: 10.4161/psb.11501 21228634
68. Schenck D, Christian M, Jones A, Lüthen H (2010) Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol 152 : 1183–1185. doi: 10.1104/pp.109.149591 20071604
69. Rück A, Palme K, Venis MA, Napier RM, Felle HH (1993) Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J 4 : 41–46.
70. Steffens B, Feckler C, Palme K, Christian M, Böttger M, et al. (2001) The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J 27 : 591–599. 11576442
71. Chen JG, Shimomura S, Sitbon F, Sandberg G, Jones AM (2001) The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. Plant J 28 : 607–617. 11851907
72. Chen JG, Ullah H, Young JC, Sussman MR, Jones AM (2001) ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev 15 : 902–911. 11297513
73. Fujita T, Sakaguchi H, Hiwatashi Y, Wagstaff SJ, Ito M, et al. (2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evol Dev 10 : 176–186. doi: 10.1111/j.1525-142X.2008.00225.x 18315811
74. Bennett TA, Liu MM, Aoyama T, Bierfreund NM, Braun M, et al. (2014) Plasma Membrane-Targeted PIN Proteins Drive Shoot Development in a Moss. Curr Biol 24 : 2776–2785. doi: 10.1016/j.cub.2014.09.054 25448003
75. Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16 : 1610–1615. 12101120
76. Hamann T, Mayer U, Jürgens G (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126 : 1387–1395. 10068632
77. Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8 : 477–485. doi: 10.1038/nchembio.926 22466420
78. Shimizu-Mitao Y, Kakimoto T (2014) Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol 55 : 1450–1459. doi: 10.1093/pcp/pcu077 24880779
79. Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, et al. (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156 : 577–589. doi: 10.1016/j.cell.2013.12.027 24485461
80. Bennett T, Leyser O (2014) The Auxin Question: A Philosophical Overwiew. In: Zazimalova E, Petrasek J, Benkova E, editors. Auxin and Its Role in Plant Development. Wien: Springer. pp. 3–19.
81. Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50 : 151–158. 5650857
82. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 : 1792–1797. 15034147
83. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59 : 307–321. doi: 10.1093/sysbio/syq010 20525638
84. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, et al. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 : 34–41. 17697981
85. Moore I, Samalova M, Kurup S (2006) Transactivated and chemically inducible gene expression in plants. Plant J 45 : 651–683. 16441354
86. Mirbel M (1835) Researches anatomiques et physiologiques sur le Marchantia polymorpha. Mém Acad Roy Sc Inst France 13 : 337–436.
87. Vojtek AB, Hollenberg SM, Cooper JA (1993) Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74 : 205–214. 8334704
88. Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, et al. (2012) The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 24 : 3248–3263. 22904146
89. Shikata M, Takemura M, Yokota A, Kohchi T (2003) Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci Biotechnol Biochem 67 : 2495–2497. 14646219
90. Ishizaki K, Mizutani M, Shimamura M, Masuda A, Nishihama R, et al. (2013) Essential Role of the E3 Ubiquitin Ligase NOPPERABO1 in Schizogenous Intercellular Space Formation in the Liverwort Marchantia polymorpha. Plant Cell 25 : 4075–4084. doi: 10.1105/tpc.113.117051 24170128
Štítky
Genetika Reprodukčná medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



