-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
In this work we seek to further the understanding of bone genetics by mapping bone traits and gene expression in the chicken. Bone in female birds is special due to egg production. In this study, we combine the genetic mapping of bone traits with bone gene expression to find candidate quantitative trait genes that explain the differences between wild and domestic chickens in terms of bone production. The concept of combining genetic mapping and gene expression mapping is not new, and has already been successful in isolating bone-related genes in mammals, however this is the first time it has been applied to an avian system with such unique bone modelling processes. We aim to reveal new molecular mechanisms of bone regulation, and many of the candidates we find are new, highlighting the potential this technique has to identify the potential differences between avian and mammalian bone biology.
Published in the journal: Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems. PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005250
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005250Summary
In this work we seek to further the understanding of bone genetics by mapping bone traits and gene expression in the chicken. Bone in female birds is special due to egg production. In this study, we combine the genetic mapping of bone traits with bone gene expression to find candidate quantitative trait genes that explain the differences between wild and domestic chickens in terms of bone production. The concept of combining genetic mapping and gene expression mapping is not new, and has already been successful in isolating bone-related genes in mammals, however this is the first time it has been applied to an avian system with such unique bone modelling processes. We aim to reveal new molecular mechanisms of bone regulation, and many of the candidates we find are new, highlighting the potential this technique has to identify the potential differences between avian and mammalian bone biology.
Introduction
The genetic basis of bone mineral density in the chicken is of both theoretical and practical importance. On the theoretical side, domestication is an example of intense directional selection causing wide-ranging differences in the phenotype of domestic animals compared to their wild progenitors. In the case of the domestic chicken, descended from the wild Red Junglefowl, bone metabolism is intrinsically coupled with the production of eggs. Hence, bone and fecundity phenotypes form a suite of correlated traits that have likely been tied with fitness during domestication. In addition, the comb, a sexual ornament in both male and female chickens [1,2], appears to be tied with bone mineral density. In terms of practical applications, bone mineral density is a predictor of osteoporosis, a debilitating condition in both chickens [3,4] and humans [5]. Describing quantitative trait genes for bone mineral density can give new clues about the biological processes underlying osteoporosis. Also, to the extent that the variants are still segregating in commercial chicken flocks, they may provide targets for selection.
Bone is made up of a mineral matrix, mostly hydroxyapatite, in an organic matrix dominated by collagen, lipids, proteoglycans and other bone structural proteins. Bone is continually being remodelled by osteoblasts, which produce bone components, and osteoclasts, which break them down. Bone metabolism in female birds is special in that they produce a medullary bone, which serves as a reservoir for calcium used in production of the eggshell [6,7]. Avian bone remodelling is quicker than mammalian and coordinated with the laying cycle [8,9]. Osteoporosis, loss of bone density, and bone fractures are a major health issue for layer chickens in production, likely exacerbated by the strains that high egg production and quick growth put on domestic chickens [10]. There is substantial genetic variation in bone traits in domestic layer chickens [11–13]. Studies involving dietary treatments suggest that the cause of osteoporosis in hens is not merely calcium deficiency [13,14], and that there is a genetic basis to bone strength independent of calcium metabolism.
With osteoporosis also being a serious health problem in humans, genetic mapping studies in humans and animal models have been performed to search for genes and processes involved in bone density. As is the case with heritable quantitative traits in general, the bone mineral density loci that have been identified so far leave most of the genetic variance unexplained (see review by [15]). Linkage mapping rarely has the resolution to isolate single molecular genes, but linkage mapping in humans has found variants affecting bone density in genes such as LRP5 [16–18] and BMP2 [19]. Genome-wide association studies, on the other hand, bring associated regions down to few or sometimes a single gene. In recent years, several such association studies have been published, implicating common genetic variants in human bone density variation. The hits include known bone-related genes such as osteoblast regulator osterix, osteoclast regulators RANKL and osteoprotegerin, and Wnt pathway genes such as β-catenin and WNT16 [20–31]. However, even with the high resolution of a GWAS, it is not necessarily the gene closest to the variant that is the causal gene. Authors have successfully employed eQTL mapping or genetical genomics and systems genetics approaches such as coexpression networks, to isolate quantitative trait genes for bone density in mice, including Alox5 [32], Alox15 [33], Bicc1 [34], Asxl2 [35] and Darc [36].
Our aim is to study the genetics of bone traits in the chicken using modern genomics methods. Due to the divergence of domestic chickens from Red Junglefowl during domestication, we can use a wild by domestic chicken cross as a study system for bone genetics. Quantitative trait locus (QTL) mapping as a top down mapping approach has been widely successful when it comes to finding chromosomal regions associated with various traits. However, going from the quantitative trait locus, usually a broad chromosomal region harbouring many genes, to causative genes or even variants, is difficult. The main obstacle to quantitative trait gene identification is the long linkage blocks in the pedigrees employed for mapping experiments. Making an advanced intercross line is one strategy to increase informative meioses and mapping resolution [37]. The chicken has a high recombination rate and a relatively compact genome compared other vertebrate models such as the mouse [38].
To aid quantitative trait gene identification, we turn to genetical genomics or expression QTL (eQTL) mapping to female femoral gene expression measured by microarrays. While gene regulation is complex, regulatory variation in molecular intermediates can be assumed to have simpler genetic architecture than the phenotypic trait, which is affected by the combined output of many such molecular pathways. In cases where the causative variant acts by means of gene expression, expression QTL mapping helps by reducing the number of candidate genes to the ones whose expression map to the QTL region. Beyond overlapping bone QTL and expression QTL, we consider the regression between bone trait and gene expression level of these positional candidate genes. In this way, we only retain candidates whose gene expression is associated with the bone trait.
Results
We find a total of 94 QTL for 21 traits (see Table 1 and S1 Table). When we combine loci for different variables they form 25 separate genomic regions associated with female bone traits, and 26 separate regions for male bone traits. We detect a considerable amount of digenic epistasis in the form of 40 epistatic pairs. As can be seen in Fig 1, the female and male QTL are largely separate. Additionally, medullary traits in females and trabecular traits in males are largely distinct from the cortical and total bone traits. We compare the results with previously published QTL from the F2 generation of this intercross. The advanced intercross (AIL) female and male QTL together form 41 genomic intervals, while the F2 QTL form 19 intervals. There are 11 regions of overlap between the sets on chromosomes 1, 2, 3, 6, 7, 13, and 18 (S6 Table). Variances explained by individual QTL and full QTL models including epistasis are given in S1 and S7 Tables.
Tab. 1. Table of high-confidence candidate eQTL with location of the gene, LOD score, and p-value for association between gene expression and bone trait. 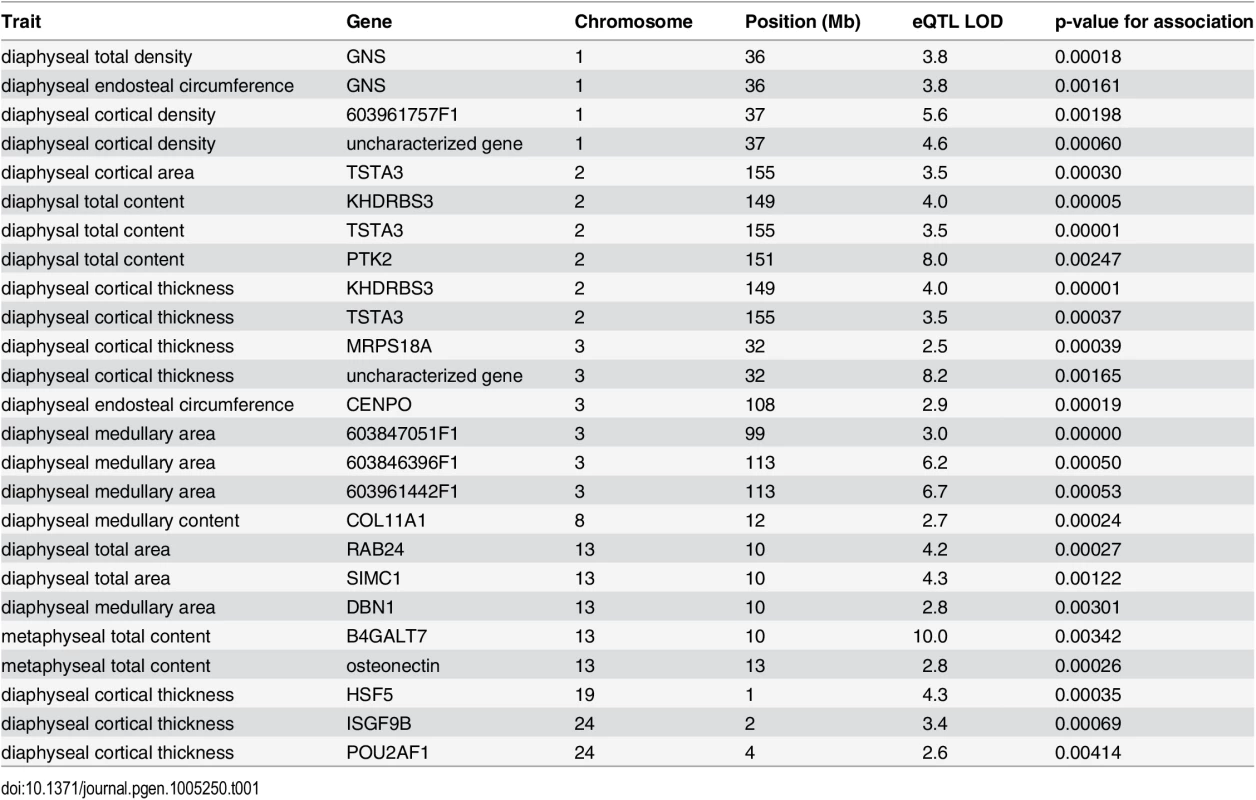
Fig. 1. Physical locations of QTL support intervals. 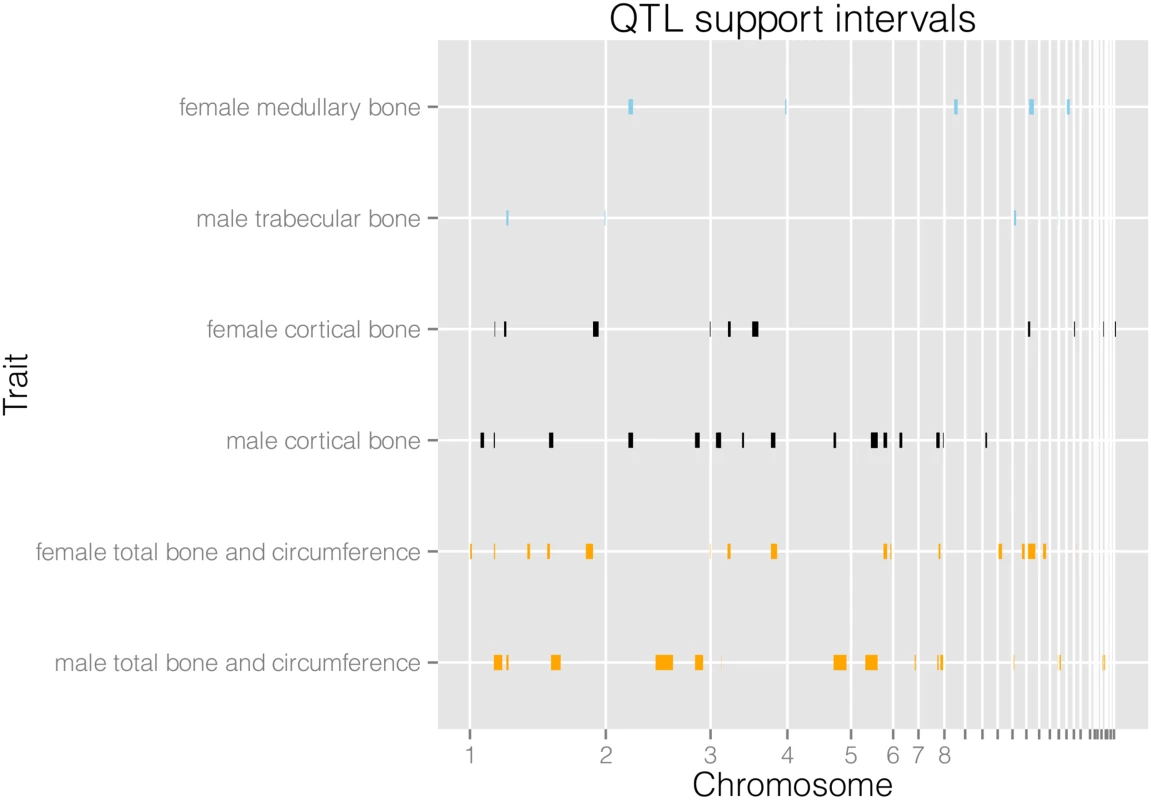
We display medullary, cortical and total area and circumference related traits for males and females separately. Their architectures are largely separate, with a few regions of overlap. We detect 6318 local cis-eQTL (see S3 Table) and 1470 distal trans-eQTL that affect female femoral gene expression. The eQTL study reveals a copious amount of gene-regulatory variation, but only a small subset of eQTL will be relevant for the phenotypic QTL that we detected. Therefore we find those local eQTL confidence intervals that overlap the confidence intervals of the female bone QTL and test for an association between gene expression level and trait value. In light of the difference between male and female genetic architecture, we do not consider the male QTL (gene expression only being measured in female bone samples). A total of 71 genes are candidates for bone QTL in the sense that they have a local eQTL that overlaps a bone QTL and that their expression is associated with the trait. In this way, we find 15 candidates for medullary traits, 20 for cortical traits, and 45 for total content, circumference and area traits. While it is possible that a QTL effect is actually made up of several linked variants, we have higher confidence in a candidate if there are few alternative candidates with significant associations in the same QTL region. The complete list of associations is shown in S2 Table, and here we outline the high confidence candidates (Figs 2 and 3, S1 and S2 Figs).
Fig. 2. Candidate quantitative trait genes (A) <i>DNB1</i> for female medullary and (B) <i>HSF5</i> for female cortical traits: LOD curves and confidence intervals of bone QTL and associated eQTL, and scatterplots of residual phenotypes against gene expression values. 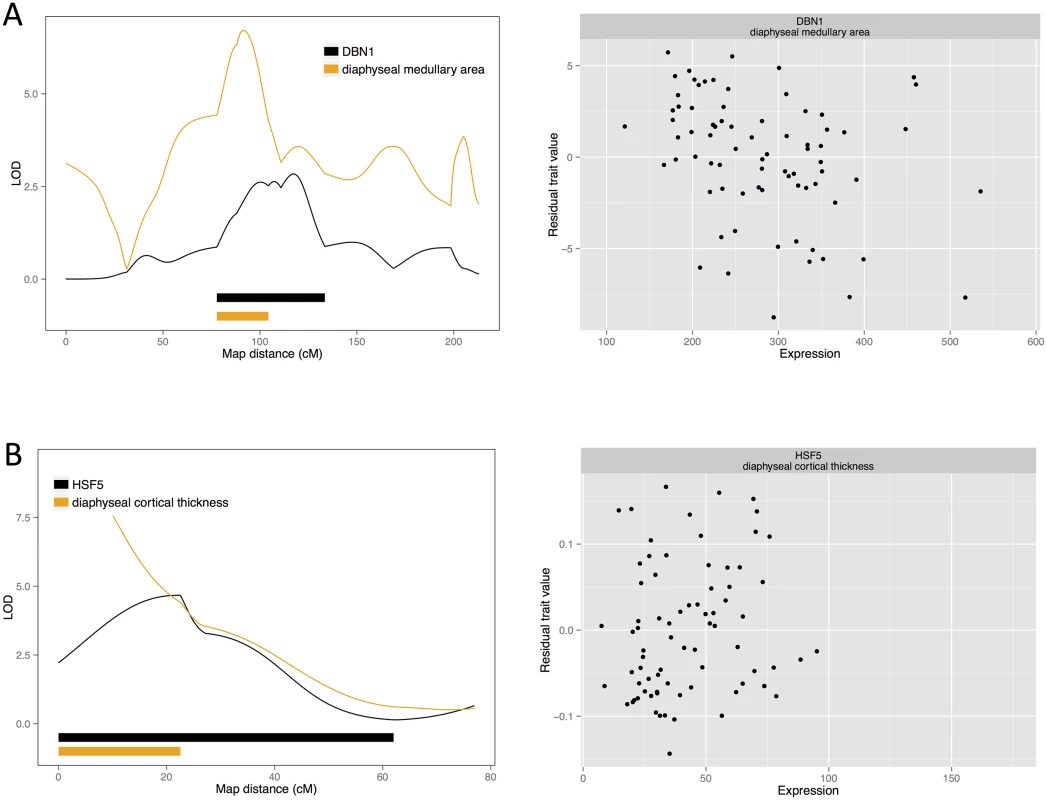
Fig. 3. Candidate quantitative trait genes osteonectin and <i>B4GALT7</i> for total bone content: LOD curves and confidence intervals of bone QTL and associated eQTL, and scatterplots of residual phenotypes against gene expression values. 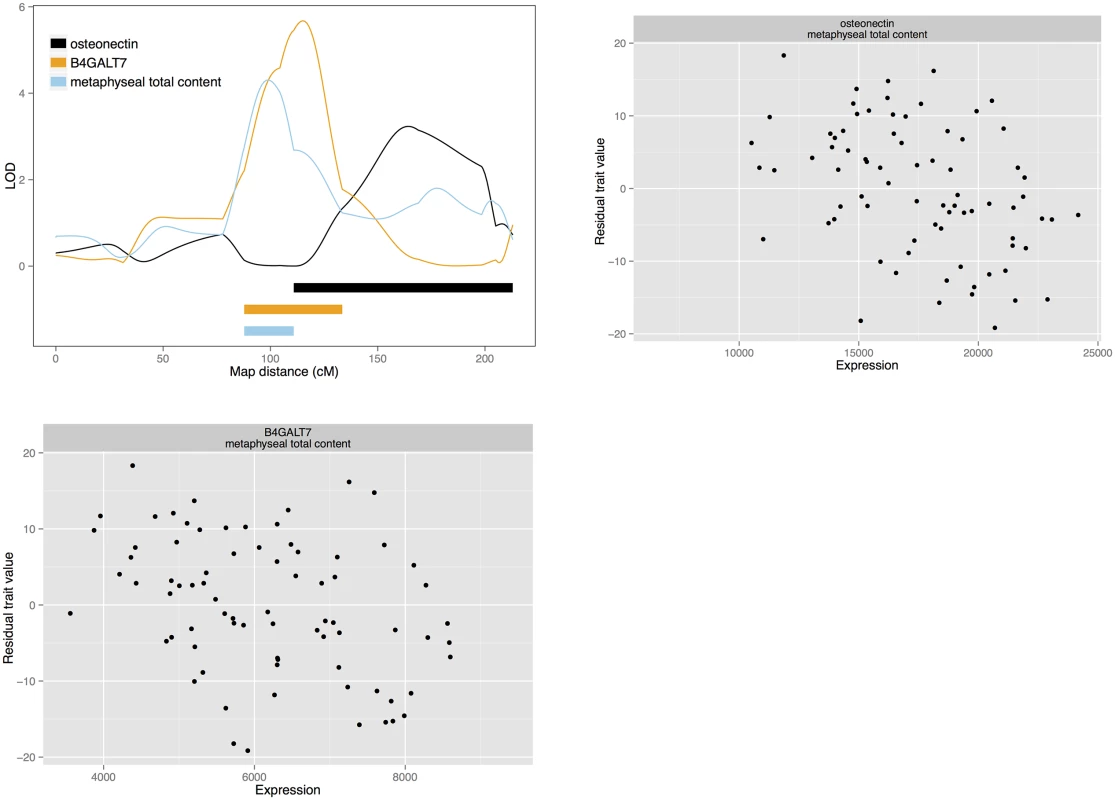
Candidate genes: Medullary traits
Beginning with the medullary traits, we find two candidate causal genes, each for a different QTL. These genes are drebrin (DBN1; NM_205499) for diaphyseal medullary area on chromosome 13, and collagen type XI alpha 1 (COL11A1; represented by the EST sequence 603372847F1) for diaphyseal medullary content on chromosome 8. Additionally, a QTL for diaphyseal medullary area on chromosome had three candidates: EST probesets 603847051F1, 603961442F1 and 603846396F1. These correspond to two unknown cDNA clones.
Candidate genes: Cortical traits
There are three cortical trait QTL with a single candidate each. Heat shock transcription factor family member 5 (HSF5; ENSGALG00000001031) is a candidate for diaphyseal cortical thickness on chromosome 19. Also, tissue specific transplantation antigen P35B (TST3A; ENSGALG00000016129) is a candidate for diaphyseal cortical area on chromosome 2. This QTL colocalises with another for diaphyseal cortical thickness, which has both TST3A and KH domain-containing, RNA-binding, signal transduction-associated protein 3 (KHDRBS3; ENSGALG00000016203) as candidates. Mitochondrial ribosomal protein S18A (MRPS18A; ENSGALG00000010296) and an uncharacterized gene from the Ensembl database (ENSGALG00000010303) are candidates for diaphyseal cortical thickness on chromosome 3. Immunoglobulin superfamily, member 9B (IGSF9B; ENSGALG00000001450) and POU class 2 associating factor 1 (POU2AF1; NM_204175) are also candidates for a diaphyseal cortical thickness QTL on chromosome 24. Finally, on chromosome 1 we find an uncharacterized Ensembl gene ENSGALG00000021181 and EST probeset 603234378F1 as candidates for diaphyseal cortical density. However, both these probesets correspond to the predicted gene LOC771935 (XM_001235144.2), which has since been removed from GenBank.
Candidate genes: Total bone and circumference
We also detect gene expression candidates for total bone and bone circumference. Glucosamine (N-acetyl)-6-sulfatase (GNS; NM_001199559) is the sole candidate for a diaphyseal total bone density and a diaphyseal endosteal circumference QTL on chromosome 1. Also, centromere protein O (CENPO), represented by EST 603256840F1, is a candidate for a diaphyseal endosteal circumference QTL on chromosome 3. On chromosome 13, secreted protein, acidic, cysteine-rich (osteonecin) represented by the EST probeset 603470410F1 and xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (B4GALT7; NM_001039911) are candidates for a metaphyseal total content QTL. This QTL colocalises with the metaphyseal medullary area QTL for which drebrin is a candidate. B4GALT7 and osteonectin are also among the candidates for a third, colocalising medullary content QTL. These genes may affect both total bone content and medullary content. In a similar fashion, TSTA3, KHDRBS3 are candidates for diaphyseal total bone content, as well as the cortical traits mentioned above. However, in the case of diaphyseal total content, the QTL also has a third candidate: protein tyrosine kinase 2 (PTK2), represented by EST probeset 603469577F1. We also detect Ras-related protein Rab-24 (RAB24) and SUMO-interacting motifs containing 1 (SIMC1) as candidates for diaphyseal total area on chromosome 13.
Translational comparisons
Finally, we compared our candidate quantitative trait genes with results from genome-wide association studies (GWAS) in mice and humans. 30 out of 76 candidate probesets could be mapped to a mouse ortholog. Three of them were associated with bone mineral density in the GWAS by Farber et al. [35]. These genes were somatostatin receptor 5 (SSTR5; NM_001024834), calcium channel, voltage-dependent, T type, alpha 1H subunit (CACNA1H; ENSGALG00000005215), mitochondrial ribosomal protein S18A (MRPS18A), and glycoprotein 1b, alpha polypeptide (GP1BA; ENSGALG00000021693). These genes are located in QTL that have other associated candidates, but these previous associations increase our confidence in them as potential causative genes. We also searched for our candidates among the top associated genes for bone mineral density in the dbGAP Association Results Browser, and found no overlap.
eQTL hotspots
Hotspots of trans-eQTL occurring in the same region could reflect regulatory variants with many downstream effects. We searched for such hotspots by comparing the number of overlapping trans-eQTL with the number of trans-eQTL observed with simulated uniformly distributed eQTL intervals. In this way we found 12 clusters of trans-eQTL, making up putative hotspots on chromosomes 1, 2, 3, 4, 5 and 25 (see S4 Table). Four of these hotspots overlap female bone QTL. In particular, the hotspot on chromosome 3 overlaps the cortical thickness QTL for which we detect candidate genes MRPS18A and ENSGALG00000010303. Further dissection of these loci would be necessary to know whether the clusters are genuine trans-regulatory hotspots involved in bone function.
Discussion
We report here QTL mapping and genetical genomics of several aspects of chicken bone density under domestication. These QTL explain part of the difference in these traits between wild and domestic chickens. We map QTL affecting total femoral bone, cortical bone and medullary bone, which is particular to laying hens. We find 25 genomic regions in total for female traits and 26 for male traits. Also, we identify 76 candidate genes correlated with female bone traits. A handful of these are high confidence candidates for quantitative trait genes acting by changes in gene expression.
In this work, we have combined quantitative trait locus mapping and genetical genomics to illuminate the genetic underpinnings of changes in bone deposition under chicken domestication. To our knowledge this is the first eQTL study performed on bone tissue using the chicken as a model. Although our study is reasonably large, both for a phenotypic QTL scan and an eQTL scan, genetic mapping is always limited by power. Complex traits are often massively polygenic and gene-regulatory effects, particularly in trans, can be subtle. In addition, how large an effect a QTL needs to have to be relevant for domestication and how large mediating eQTL effects have to be, are open questions. We find a genetic architecture of bone with several moderately large QTL and digenic epistasis. The eQTL mapping reveals a rich landscape of expression QTL, both local cis - and distal trans-acting. Though most of these eQTL are likely to be inconsequential for bone phenotypes, this demonstrates wide ranging gene-regulatory evolution during chicken domestication.
Medullary bone candidates
Drebrin (DBN1) is the sole candidate for one of our medullary QTL. Drebrin binds actin filaments and is known for its role in the nervous system. However, it is also expressed in lymphocytes and involved in connecting chemokine receptor CXCR4 to the cytoskeleton [39]. This receptor, in turn, is involved in differentiation of osteoclasts from hematopoietic cells [40]. While drebrin has not been studied in connection with bone metabolism, we hypothesise that eQTL effects on drebrin expression could affect medullary bone through osteoclasts. We also detect collagen XI alpha 1 as a candidate for medullary content on chromosome 13 with multiple sources indicating effects on bone. Mutations in this gene cause the rare human chondrodysplasias fibrochondrogenesis [41], Marshall syndrome and Stickler syndrome [42,43], and may predispose to osteoarthritis [44]. Cell culture experiments suggest that it inhibits osteoblast differentiation [45].
Cortical bone candidates
We find HSF5 to be associated with cortical thickness. Heat shock factors (HSF) regulate heat shock protein (HSP) genes and activate them in response to cellular stressors. Furthermore, there is some evidence that heat shock responses may be involved in bone formation. Bone growth can be stimulated by heat, and heat treatment increases mineralisation and heat shock protein expression in cell culture [46]. Molecular studies of the RANKL promoter and HSF2 knock-down suggest that this osteoclast regulator is regulated by heat shock factors [47,48]. However, there is little known about HSF5 specifically. TSTA3, our candidate for a diaphyseal cortical area QTL, is involved in protein glycosylation by contributing to the synthesis of GDP-fucose, which is used by fucosyltransferases in glycosylation of cell adhesion proteins. Knockout studies of TSTA3 in mice suggest that fucosylation is needed to regulate myeloid cell differentiation by Notch signalling [49]. Notch also regulates both osteoblast and osteoclast differentiation [50,51]. There are two additional candidates for the colocalising cortical thickness QTL: KHDRBS3, which encodes an RNA binding protein, and PTK2, protein tyrosine kinase. PTK2 is involved in osteoblast differentiation [52,53]. Along with the unknown Ensembl gene, ENSGALG00000010303, MRPS18A is a candidate for diaphyseal cortical thickness, whilst MPRS18A encodes a mitochondrial ribosomal protein. These ribosomes translate proteins required for mitochondrial function. Hence mutations in mitochondrial ribosomal proteins can cause oxidative phosphorylation deficiencies (such as OMIM phenotypes 614582, 611719, 610498) [54]. Such defects can affect diverse organ systems, since mitochondrial function is crucial for cellular metabolism. To our knowledge, no such disorders with specific bone-related symptoms are known, although there is still the possibility of mitochondrial effects on bone. Active osteoclasts, for instance, are rich in mitochondria [55]. Quantitative changes in mitochondrial activity could influence the balance of bone remodelling. Finally, we find two candidates for a cortical thickness locus on chromosome 24: IGSF9B and POU2AF1. There is little known about IGSF9B, except that it is involved in cell adhesion at synapses [56]. POU2AF1, however, is known as a transcription factor in B cells of the immune system. It regulates B cell maturation, and is mutated in some forms of leukaemia [57,58]. While no direct connection between POU2AF1 expression and bone is known, the immune system affects bone regulation. For example, B cells produce cytokines with effects on bone, including osteoclast-stimulating RANKL [59–61].
Total bone and circumference candidates
GNS is our single candidate for a couple of colocalising QTL for diaphyseal total bone and endosteal circumference on chromosome 1. It encodes a glucosamine sulfatase that breaks down heparane sulphate. Heparan sulphates are polysaccharides that make up part of the extracellular matrix and are part of proteoglycan glycoproteins. As such, heparan sulphates have regulatory functions in development, including regulation of osteoblasts [62,63]. Mutations in the GNS gene cause mucopolysaccharidosis type IIID (Sanfilippo syndrome D; OMIM phenotype 252940) [64,65]. It is a lysosomal enzyme, and loss of function causes accumulation of heparan sulphate in the organelle with deleterious effects on several organ systems. Symptoms includes effects on bone, though these may be secondary effects [66]. Taken together, this leads to the speculation that the GNS eQTL may affect bone density through either a signaling effect of heparan sulphate or through a lysosome-dependent mechanism. CENPO, a candidate for an endosteal circumference QTL, encodes a centromere protein and is involved in chromosomal segregation during mitosis [67]. While proliferation of different bone cell populations is important for bone metabolism, its involvement in bone is unknown. Another locus for total bone content has two candidates: osteonectin and B4GALT7. Osteonectin is a glycoprotein produced by osteoblasts. It contributes to mineralisation by binding mineral crystals and linking to collagen, and it functions in bone remodelling [68,69]. B4GALT7 is another candidate involved in protein glycosylation, a glycosyltransferase that participates in synthesis of proteoglycans, and in particular heparane sulphate. Mutations in human cause Ehler—Danlos syndrome (OMIM phenotype 130070), affecting connective tissue and causing skeletal deformations [70–72]. Finally, a locus for diaphyseal total area has RAB24 and SMIC1 as candidates. RAB24 is one of the small GTPases of Ras-related proteins that are usually involved in intracellular trafficking. However, RAB24 seems to participate in cell division and chromosome segregation [73]. Potential functions in bone are unknown, but RAB24 was down-regulated in a gene expression study of stimulated mineralisation in the osteoblast-like mouse cell line MCT3-E1 [74]. The other candidate at the same QTL, SIMC1, regulates the protease calpain 3 in skeletal muscle [75].
Mouse and human candidates
The number of mouse GWAS candidates for bone density that were also found as candidates in our study was restricted to four shared associations. One of them is MRPS18A, which has been discussed above. The others are from QTL that contain several gene expression candidates. SSTR5 encodes one of the receptors for the hormone somatostatin. It regulates growth hormone secretion, and mutations in the gene are associated with acromegaly [76,77]. The site of action of growth hormone regulation should be the pituitary, but there is also evidence somatostatin and its receptors affect bone precursor cells [78,79]. CACNA1H encodes, Cav3.2, a voltage gated calcium channel subunit. It is expressed in osteoblasts and chondrocytes during bone development in mice [80,81]. GP1BA encodes a von Willebrand protein receptor expressed on the surface of platelets. Mutations cause bleeding diseases Bernard-Soulier syndrome (OMIM phenotype number 231200 and 153670) and platelet-type von Willebrand disease (OMIM phenotype number 177820). von Willebrand factor can interact with osteoprotegrin to regulate osteoclast differentiation [82], and a transgenic GP1BA von Willebrand disease model has increased bone mass and reduced osteoclast activity [83]. We found no overlap between our candidate quantitative trait genes and the top associations for human bone traits in the dbGAP Association Results Browser. However, the Framingham osteoporosis study found a region around KHDRBS3 as a potentially pleiotropic association with lumbar spine and femoral neck bone mineral density [84]. As mentioned previously in connection with the candidates, several of the genes highlighted here cause rare Mendelian diseases in humans, some of them with known skeletal phenotypes. Local cis-eQTL effects need not be as drastic as the loss-of-function mutations that cause Mendelian diseases, and could be restricted to the bone by tissue-specific regulatory elements.
Chicken bone genetics
Based on the functional literature and the above associations, many of the genes found are plausible candidates for bone traits. However, we find a number of unexpected or unknown candidates. This is reasonable given the dearth of knowledge about the genetic machinery of complex traits in general. However, it could also be due to peculiarities of chicken bone metabolism as compared to mammals such as humans and mice. Previously unknown or poorly understood genes, coupled with the relative lack of human and mouse GWAS overlaps in our data, potentially indicate the novel aspect of chicken bone metabolism. In the female chicken femoral bone, calcium is continually removed from bone to supply the forming eggshell on a 24-hour basis throughout laying. This might require genetic mechanisms different from those in mammals. Understanding this mechanism will offer novel insights into bone metabolism.
We also find that several known genes involved in bone, including Matrix Gla protein, TRAF2, PDZRN3 and PDZRN4 display cis-eQTL effects. While they are not candidates for the phenotypic QTL detected in this study, this regulatory variation in bone genes suggest that there is more subtle variation in bone metabolism between wild and domestic chickens. Among the most highly expressed genes that also have an eQTL, we note BF1 and BF2 genes on chromosome 16, involved in immunity in the chicken, the MHC class II beta chain gene BLB1, the MHC B-G antigen, and cathepsin S involved in degrading protein antigens for MHC II presention. While evolution of the immune system under domestication is not the focus of this work, we note that this suggests changes to adaptive immunity.
Genetic variation affecting bone traits between wild and domestic genotypes is of interest to the evolutionary study of chicken domestication, and potentially provides insights into the molecular genetic regulation of bone mineral density. However, variation present within domestic breeds is most useful to breeders. Whether the QTL we detect segregate in contemporary domestic chickens can only be found by studies of these populations. We do at least find some replicability of the results of the F2 generation of the intercross used in this work [85,86]. When comparing the advanced intercross bone results to the previous F2 results, about half of the F2 QTL overlapped at least one of the F8 QTL. Possible reasons for differences between F2 and advanced intercross mapping include thee limited power to detect small-effect QTL and the larger linkage blocks causing the F2 study to detect the aggregate effect of several linked QTL rather than single QTL. The latter phenomenon should be more pronounced for polygenic traits with epistatic interactions, and bone traits display both a large number of loci and considerable epistasis.
Expression QTL mapping is an established genomics approach that has had several successes in quantitative trait gene identification in animals. The localisation of a QTL from a linkage study is always rough, due both to the inherently limited resolution of experimental crosses, and the statistical uncertainties of QTL mapping. Our candidate gene prioritisation builds on the overlap of 1.8 LOD drop intervals, corresponding to approximate 95% confidence intervals, between QTL and eQTL. In the next step, we test for an association between the gene expression level and the bone phenotype. We will suffer false negatives when variants act by other means than gene expression changes in adult bone tissue. Conversely, a candidate gene found might not be causative if the gene expression is correlated with the trait for some other reason, such as the gene being downstream of the causative gene in a regulatory pathway. For instance, if the bone QTL causes the proliferation of some population of bone cells, causing increases in mRNA of cell division related genes such as CENPO or RAB24 and mitochondrial protein genes such as MRPS18A. Some of the QTL intervals have many candidate genes that are highlighted by our approach (see S2 Table). While it is possible for a QTL region to harbour multiple causative genes, false positives are likely among these candidates. In the end, conclusive evidence for a quantitative trait gene can come through experimental manipulations in cell culture or transgenic animals. Our QTL and eQTL mapping results provide compelling candidates for such experiments.
Conclusions
In conclusion, we find several potential quantitative trait genes for bone traits in the chicken, using to our knowledge the first eQTL analysis of bone tissue in the chicken. They are supported by genetical genomics evidence in the form of QTL-eQTL overlap and trait-expression associations. While the genes are not particularly obvious candidates, some of them have connections to bone metabolism in the literature, and a handful have been associated with bone mineral density in mice or humans. Further investigation of the molecular mechanisms of these potential quantitative trait genes could reveal unexplored pathways of bone density regulation.
Materials and Methods
Ethics statement
The study was approved by the Regional Committee for Ethical Approval of Animal Experiments (Jordbruks verket DNR# 122–10). Birds were sacrificed by cervical dislocation and decapitation, as per the guidelines of the permit.
Wild x domestic advanced intercross
The intercross was established from one Red Junglefowl male originating from Thailand and three White Leghorn females of the L13 line. QTL mapping has previously been performed in the F2 generation (see [85,86]). The intercross was maintained with approximately a hundred individuals per generation at Linköping University and expanded for QTL mapping in generation eight. Five batches of eighth generation intercross chickens were hatched and kept at the Linköping University chicken facility. They originated from 107 pairings of 122 F7 individuals. The chickens lived in 3 x 3 meter pens with three levels, access to perches, and food and water ad libitum. Chickens were culled when they were 212 days old. The study was approved by the Regional Committee for Ethical Approval of Animal Experiments.
Phenotyping
Femurs were dissected out after sacrifice, the right stored frozen at -20°C for bone phenotyping while a piece cut from the left was frozen in liquid nitrogen and stored at -80°C for RNA isolation. We used peripheral quantitative computer tomography (Stratec XCT—Research SA, Stratec Medizintechnik, Germany) to measure a metaphyseal (6% of femur length) and a diaphysial (50% of femur length) cross section from each femur. We used the CORTMODE1 setting with a density threshold of >1000 mg/cm3 to measure cortical bone, PEELMODE2 with a threshold of 1000 mg/cm3 to measure endosteal area, density and bone content, PEELMODE2 with thresholds 1000 mg/cm3 and 150 mg/cm3 to measure medullary area, density and bone content, and PEELMODE2 with a threshold of 150 mg/cm3 to measure total area. In total, we gathered bone phenotypes from 227 male and 229 female chickens. Female chickens were also tested in egg-laying trials, as detailed in [87]. There were two fecundity trials of two weeks each. In the first, eggs were collected daily, and in the second, females were given two dummy eggs initially, and kept laid eggs. Since the second trial was closest to the time of sacrifice, we considered the number of eggs produced during this trial as a possible covariate for mapping of bone phenotypes. Females that produced no eggs at all were excluded. Bone and fecundity phenotypes from this subset of the intercross that was assayed for gene expression have previously been used to test for associations between phenotypes and gene expression of comb candidate genes; see [87,88]. Summary statistics for traits are given in S5 Table.
QTL mapping
The chickens were genotyped for 652 SNP markers spread across the sequenced part of the chicken genome. DNA was isolated from blood samples using a standard TRIS extraction. Genotyping was performed with the Illumina Golden Gate platform at Uppsala Seq and SNP platform. We performed QTL mapping with R/qtl [89] and Haley-Knott regression using forward-selection for multiple-QTL models and pairwise scans to search for epistasis. All analyses included sex, batch and body weight as covariates. For female traits we also considered egg laying in the second fecundity trial as a possible covariate. If there was a significant association between the bone phenotype in question and the total weight of eggs produced we included the egg covariate in QTL mapping. To adjust for cryptic relatedness structure, which can be a confounder in advanced intercrosses, we applied principal component analysis on the genotype matrix, and included principal components as covariates in QTL mapping. Empirical significance thresholds were established by means of permutation tests, shuffling the phenotypes while preserving the correlation structure of the genotypes. Genomic support intervals around the QTL were formed with the 1.8 LOD drop method [90]. See S1 Data for marker informativeness values, and S2 Data for phenotype and genotype data in R/qtl format.
Gene expression microarrays
At the time of dissection, a piece was cut from the middle of the left femur and stored in liquid nitrogen. Bone samples were disrupted with a hammer while frozen and then homogenised on a FastPrep 24 instrument using ceramic beads (Lysing matrix D, MP Biomedicals) and Tri reagent (Ambion). RNA was isolated using Tri reagent according to the manufacturer's protocol. RNA was further purified with the Fermentas GeneJet RNA purification kit (Thermo Scientific). After treatment with DNAse I, double-stranded cDNA was synthesised with a combination of Fermentas RevertAid Premium First-strand cDNA synthesis kit, DNA polymerase I, RNase H and T4 DNA polymerase (Thermo Scientific) according to protocols provided with the kit. The cDNA was labelled with the NimbleGen One Colour labelling kit (Roche) and hybridised to NimbleGen 12x135k custom gene expression microarrays. Scanning was performed with a NimbleGen microarray scanner (Roche). Subarrays were discarded due to low fluorescence intensity or visual uneven fluorescence resulting in 125 individual samples. Micoarrays were designed to cover all RefSeq and Ensembl genes as well as a database of ESTs. EST probesets were annotated by alignment to the chicken genome (version 2.1/galGal3) with BLAT [91] (see S3 Dataset). The name of each probeset used in tables contains the database accession (from RefSeq, Ensembl or the NCBI EST database) of the gene model or EST sequence used to design it. Selected gene expression data with bone and fecundity phenotypes from this subset of the intercross has previously been used for targeted genetical genomics of QTL regions for comb size; see [87,88]. Microarrays have been uploaded to ArrayExpress under accession number E-MTAB-3141.
eQTL mapping
Expression QTL mapping was performed with Haley-Knott regression as implemented in R/qtl [89]. Local, putative cis-acting, eQTL were mapped within an interval of 100 cM around the position of the probeset. The interval was expanded to the closest flanking markers spanning at least 50 cM in each direction. Global trans-acting eQTL were mapped using the entire genetic map. A probeset was assigned a cis-eQTL when the logarithm of odds passed the cis-threshold on any markers in the window around the location of the probeset, and a trans-eQTL if it passed the (higher) trans-threshold somewhere else in the genome. 1.8 LOD drop intervals, expanded to the closest markers, were used to form confidence intervals around eQTL. Thresholds were generated by permutation separately for cis and trans associations either using the whole-genome or the 100 cM regions around probeset locations. Each iteration, the individual identities were resampled, a 100 probesets were subsampled, and the maximum LOD score from an eQTL scan of this permuted dataset was saved. The process was repeated 10000 times, and the 95th percentile LOD score was used to generate the significance threshold.
Candidate quantitative trait genes
We used a two-phase method to search for quantitative trait genes for bone phenotypes: 1) we overlapped 1.8 LOD drop intervals from bone QTL and expression QTL; 2) tested for an association between probeset expression levels and bone traits with a regression model including body mass as covariate. Also, the same egg fecundity phenotype was included as covariate, in cases where the QTL in question was detected with a fecundity covariate. The regression between probeset expression level and bone trait included the body weight as a covariate. p-values from the t-test of the regression coefficient were adjusted by Bonferroni correction for the number of uncorrelated cis-eQTL (those where p-value for pairwise correlation test > 0.05) in the interval.
Expression QTL colocalisation
To investigate clustering of diastal trans-eQTL, we simulated uniformly placed eQTL on an interval the size of the sequenced chicken genome and counted the maximum coverage of simulated eQTL intervals in 1000 iterations. We regard any region with coverage above the 95th percentile of the simulated maximum coverage, which was 22, as an eQTL cluster.
Comparison with human and mouse GWAS results
We compared our candidate quantitative trait genes with previously published genome-wide association studies (GWAS) in human and mouse. We used the mouse GWAS data from [35] and the human top associations catalogued in the dbGAP Association Results Browser (http://www.ncbi.nlm.nih.gov/projects/gapplusprev/sgap_plus.htm). Using Ensembl version 70, we mapped Ensembl and RefSeq probesets to Ensembl gene identifiers and to orthologous human and mouse genes. In the mouse dataset we extracted the p-values for SNPs in 200 kb windows around the start of the Ensembl gene model. The threshold for genome-wide significance used in the original study was 4 * 10–6. For the human associations, we exported all associations for bone mineral density in humans from dbGAP Association Result Browser with a p-value less than 10–5. We mapped the pair of flanking genes listed in the Association Results Browser to chicken Ensembl gene identifiers and overlapped them with the list derived from our results.
Supporting Information
Zdroje
1. Pizzari T, Birkhead TR (2000) Female feral fowl eject sperm of subordinate males. Nature 405 : 787. 10866198
2. Pizzari T, Cornwallis CK, Lovlie H, Jakobsson S, Birkhead TR (2003) Sophisticated sperm allocation in male fowl. Nature 426 : 70–73. 14603319
3. Budgell K, Silversides F (2004) Bone breakage in three strains of end-of-lay hens. Canadian journal of animal science 84 : 745–747.
4. Webster A (2004) Welfare implications of avian osteoporosis. Poultry Science 83 : 184–192. 14979568
5. Ralston SH, de Crombrugghe B (2006) Genetic regulation of bone mass and susceptibility to osteoporosis. Genes & Development 20 : 2492–2506.
6. Bloom MA, Domm LV, Nalbandov AV, Bloom W (1958) Medullary bone of laying chickens. American Journal of Anatomy 102 : 411–453. 13617222
7. Mueller WJ, Schraer R, Scharer H (1964) Calcium metabolism and skeletal dynamics of laying pullets. The Journal of nutrition 84 : 20–26. 14210016
8. Van de Velde J, Vermeiden J, Touw J, Veldhuijzen J (1984) Changes in activity of chicken medullary bone cell populations in relation to the egg-laying cycle. Metabolic Bone Disease and Related Research 5 : 191–193. 6738357
9. Kerschnitzki M, Zander T, Zaslansky P, Fratzl P, Shahar R, et al. (2014) Rapid alterations of avian medullary bone material during the daily egg-laying cycle. Bone 69 : 109–117. doi: 10.1016/j.bone.2014.08.019 25204794
10. Whitehead C (2004) Skeletal disorders in laying hens: the probIem of osteoporosis and bone fractures. Welfare of the laying hen 27 : 259.
11. Bishop S, Fleming R, McCormack H, Flock D, Whitehead C (2000) Inheritance of bone characteristics affecting osteoporosis in laying hens. British Poultry Science 41 : 33–40. 10821520
12. Hocking P, Bain M, Channing C, Fleming R, Wilson S (2003) Genetic variation for egg production, egg quality and bone strength in selected and traditional breeds of laying fowl. British Poultry Science 44 : 365–373. 12964619
13. Fleming R, McCormack H, McTeir L, Whitehead C (2006) Relationships between genetic, environmental and nutritional factors influencing osteoporosis in laying hens. British Poultry Science 47 : 742–755. 17190683
14. Rennie J, Fleming R, McCormack H, McCorquodale C, Whitehead C (1997) Studies on effects of nutritional factors on bone structure and osteoporosis in laying hens. British Poultry Science 38 : 417–424. 9347152
15. Ferrari S (2008) Human genetics of osteoporosis. Best Practice & Research Clinical Endocrinology & Metabolism 22 : 723–735.
16. Little RD, Folz C, Manning SP, Swain PM, Zhao S-C, et al. (2002) A mutation in the LDL receptor—related protein 5 gene results in the autosomal dominant high—bone-mass trait. The American Journal of Human Genetics 70 : 11–19.
17. Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, et al. (2002) High bone density due to a mutation in LDL-receptor—related protein 5. New England Journal of Medicine 346 : 1513–1521. 12015390
18. Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, et al. (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107 : 513–523. 11719191
19. Styrkarsdottir U, Cazier J-B, Kong A, Rolfsson O, Larsen H, et al. (2003) Linkage of Osteoporosis to Chromosome 20p12 and Association to BMP2. PLoS Biol 1: e69. 14691541
20. Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, et al. (2008) Multiple genetic loci for bone mineral density and fractures. New England Journal of Medicine 358 : 2355–2365. doi: 10.1056/NEJMoa0801197 18445777
21. Rivadeneira F, Styrkársdottir U, Estrada K, Halldórsson BV, Hsu Y-H, et al. (2009) Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nature Genetics 41 : 1199–1206. doi: 10.1038/ng.446 19801982
22. Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y-H, Duncan EL, et al. (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nature Genetics 44 : 491–501. doi: 10.1038/ng.2249 22504420
23. Kung AW, Xiao S-M, Cherny S, Li GH, Gao Y, et al. (2010) Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. The American Journal of Human Genetics 86 : 229–239. doi: 10.1016/j.ajhg.2009.12.014 20096396
24. Timpson NJ, Tobias JH, Richards JB, Soranzo N, Duncan EL, et al. (2009) Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Human Molecular Genetics 18 : 1510–1517. doi: 10.1093/hmg/ddp052 19181680
25. Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, et al. (2011) Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genetics 7: e1001372. doi: 10.1371/journal.pgen.1001372 21533022
26. Koller DL, Ichikawa S, Lai D, Padgett LR, Doheny KF, et al. (2010) Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. Journal of Clinical Endocrinology & Metabolism 95 : 1802–1809.
27. Zheng H-F, Tobias JH, Duncan E, Evans DM, Eriksson J, et al. (2012) WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet 8: e1002745. doi: 10.1371/journal.pgen.1002745 22792071
28. Xiong D-H, Liu X-G, Guo Y-F, Tan L-J, Wang L, et al. (2009) Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. The American Journal of Human Genetics 84 : 388–398. doi: 10.1016/j.ajhg.2009.01.025 19249006
29. Richards J, Rivadeneira F, Inouye M, Pastinen T, Soranzo N, et al. (2008) Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. The Lancet 371 : 1505–1512. doi: 10.1016/S0140-6736(08)60599-1 18455228
30. Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, et al. (2009) A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nature Genetics 41 : 527–534. doi: 10.1038/ng.357 19396169
31. Paternoster L, Lorentzon M, Vandenput L, Karlsson MK, Ljunggren Ö, et al. (2010) Genome-wide association meta-analysis of cortical bone mineral density unravels allelic heterogeneity at the RANKL locus and potential pleiotropic effects on bone. PLoS Genetics 6: e1001217. doi: 10.1371/journal.pgen.1001217 21124946
32. Mehrabian M, Allayee H, Stockton J, Lum P, Drake T, et al. (2005) Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet 37 : 1224–1233. 16200066
33. Klein R, Allard J, Avnur Z, Nikolcheva T, Rotstein D, et al. (2004) Regulation of bone mass in mice by the lipoxygenase gene Alox15. Science 303 : 229–232. 14716014
34. Mesner LD, Ray B, Hsu Y-H, Manichaikul A, Lum E, et al. (2014) Bicc1 is a genetic determinant of osteoblastogenesis and bone mineral density. The Journal of clinical investigation 124 : 0–0.
35. Farber CR, Bennett BJ, Orozco L, Zou W, Lira A, et al. (2011) Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genetics 7: e1002038. doi: 10.1371/journal.pgen.1002038 21490954
36. Edderkaoui B, Baylink DJ, Beamer WG, Wergedal JE, Porte R, et al. (2007) Identification of mouse Duffy antigen receptor for chemokines (Darc) as a BMD QTL gene. Genome Research 17 : 577–585. 17416748
37. Darvasi A, Soller M (1995) Advanced Intercross Lines, an experimental population for fine genetic-mapping. Genetics 141 : 1199–1207. 8582624
38. Hillier LW, Miller W, Birney E, Warren W, Hardison RC, et al. (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432 : 695–716. 15592404
39. Pérez-Martínez M, Gordón-Alonso M, Cabrero JR, Barrero-Villar M, Rey M, et al. (2010) F-actin-binding protein drebrin regulates CXCR4 recruitment to the immune synapse. Journal of Cell Science 123 : 1160–1170. doi: 10.1242/jcs.064238 20215400
40. Hirbe AC, Rubin J, Uluçkan Ö, Morgan EA, Eagleton MC, et al. (2007) Disruption of CXCR4 enhances osteoclastogenesis and tumor growth in bone. Proceedings of the National Academy of Sciences 104 : 14062–14067. 17715292
41. Tompson SW, Bacino CA, Safina NP, Bober MB, Proud VK, et al. (2010) Fibrochondrogenesis results from mutations in the COL11A1 type XI collagen gene. The American Journal of Human Genetics 87 : 708–712. doi: 10.1016/j.ajhg.2010.10.009 21035103
42. Annunen S, Körkkö J, Czarny M, Warman ML, Brunner HG, et al. (1999) Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome, but other mutations cause overlapping Marshall/Stickler phenotypes. The American Journal of Human Genetics 65 : 974–983. 10486316
43. Griffith AJ, Sprunger LK, Sirko-Osadsa DA, Tiller GE, Meisler MH, et al. (1998) Marshall syndrome associated with a splicing defect at the COL11A1 locus. The American Journal of Human Genetics 62 : 816–823.
44. Jakkula E, Melkoniemi M, Kiviranta I, Lohiniva J, Räinä S, et al. (2005) The role of sequence variations within the genes encoding collagen II, IX and XI in non-syndromic, early-onset osteoarthritis. Osteoarthritis and Cartilage 13 : 497–507. 15922184
45. Kahler RA, Yingst SM, Hoeppner LH, Jensen ED, Krawczak D, et al. (2008) Collagen 11a1 is indirectly activated by lymphocyte enhancer-binding factor 1 (Lef1) and negatively regulates osteoblast maturation. Matrix biology 27 : 330–338. doi: 10.1016/j.matbio.2008.01.002 18280717
46. Shui C, Scutt A (2001) Mild Heat Shock Induces Proliferation, Alkaline Phosphatase Activity, and Mineralization in Human Bone Marrow Stromal Cells and Mg-63 Cells In Vitro. JBMR 16 : 731–741. 11316001
47. Roccisana JL, Kawanabe N, Kajiya H, Koide M, Roodman GD, et al. (2004) Functional role for heat shock factors in the transcriptional regulation of human RANK ligand gene expression in stromal/osteoblast cells. Journal of Biological Chemistry 279 : 10500–10507. 14699143
48. Kajiya H, Ito M, Ohshima H, Kenmotsu Si, Ries WL, et al. (2006) RANK ligand expression in heat shock factor-2 deficient mouse bone marrow stromal/preosteoblast cells. Journal of Cellular Biochemistry 97 : 1362–1369. 16365894
49. Zhou L, Li LW, Yan Q, Petryniak B, Man Y, et al. (2008) Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood 112 : 308–319. doi: 10.1182/blood-2007-11-115204 18359890
50. Engin F, Yao Z, Yang T, Zhou G, Bertin T, et al. (2008) Dimorphic effects of Notch signaling in bone homeostasis. Nature medicine 14 : 299–305. doi: 10.1038/nm1712 18297084
51. Bai S, Kopan R, Zou W, Hilton MJ, Ong C-t, et al. (2008) NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. Journal of Biological Chemistry 283 : 6509–6518. 18156632
52. Salasznyk RM, Klees RF, Williams WA, Boskey A, Plopper GE (2007) Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Experimental cell research 313 : 22–37. 17081517
53. Tamura Y, Takeuchi Y, Suzawa M, Fukumoto S, Kato M, et al. (2001) Focal Adhesion Kinase activity is required for Bone Morphogenetic Protein—Smad1 signaling and osteoblastic differentiation in murine MC3T3-E1 cells. JBMR 16 : 1772–1779. 11585340
54. O'Brien TW, O'Brien BJ, Norman RA (2005) Nuclear MRP genes and mitochondrial disease. Gene 354 : 147–151. 15908146
55. Dai XM, Zong XH, Akhter MP, Stanley ER (2004) Osteoclast deficiency results in disorganized matrix, reduced mineralization, and abnormal osteoblast behavior in developing bone. JBMR 19 : 1441–1451. 15312244
56. Woo J, Kwon S-K, Nam J, Choi S, Takahashi H, et al. (2013) The adhesion protein IgSF9b is coupled to neuroligin 2 via S-SCAM to promote inhibitory synapse development. The Journal of cell biology 201 : 929–944. doi: 10.1083/jcb.201209132 23751499
57. Luo Y, Roeder RG (1995) Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Molecular and Cellular Biology 15 : 4115–4124. 7623806
58. Auer RL, Starczynski J, McElwaine S, Bertoni F, Newland AC, et al. (2005) Identification of a potential role for POU2AF1 and BTG4 in the deletion of 11q23 in chronic lymphocytic leukemia. Genes, Chromosomes and Cancer 43 : 1–10. 15672409
59. Yeo L, Toellner K-M, Salmon M, Filer A, Buckley CD, et al. (2011) Cytokine mRNA profiling identifies B cells as a major source of RANKL in rheumatoid arthritis. Annals of the rheumatic diseases 70 : 2022–2028. doi: 10.1136/ard.2011.153312 21742639
60. Li Y, Toraldo G, Li A, Yang X, Zhang H, et al. (2007) B cells and T cells are critical for the preservation of bone homeostasis and attainment of peak bone mass in vivo. Blood 109 : 3839–3848. 17202317
61. Choi Y, Mi Woo K, Ko SH, Jung Lee Y, Park SJ, et al. (2001) Osteoclastogenesis is enhanced by activated B cells but suppressed by activated CD8+ T cells. European journal of immunology 31 : 2179–2188. 11449372
62. Turnbull J, Powell A, Guimond S (2001) Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends in cell biology 11 : 75–82. 11166215
63. Cool SM, Nurcombe V (2005) The osteoblast-heparan sulfate axis: control of the bone cell lineage. The international journal of biochemistry & cell biology 37 : 1739–1745.
64. Beesley C, Burke D, Jackson M, Vellodi A, Winchester B, et al. (2003) Sanfilippo syndrome type D: identification of the first mutation in the N-acetylglucosamine-6-sulphatase gene. Journal of medical genetics 40 : 192–194. 12624138
65. Mok A, Cao H, Hegele RA (2003) Genomic basis of mucopolysaccharidosis type IIID (MIM 252940) revealed by sequencing of GNS encoding N-acetylglucosamine-6-sulfatase. Genomics 81 : 1–5. 12573255
66. Rigante D, Caradonna P (2004) Secondary skeletal involvement in Sanfilippo syndrome. Qjm 97 : 205–209. 15028850
67. McClelland SE, Borusu S, Amaro AC, Winter JR, Belwal M, et al. (2007) The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. The EMBO journal 26 : 5033–5047. 18007590
68. Delany A, Amling M, Priemel M, Howe C, Baron R, et al. (2000) Osteopenia and decreased bone formation in osteonectin-deficient mice. Journal of Clinical Investigation 105 : 915. 10749571
69. Termine JD, Kleinman HK, Whitson SW, Conn KM, McGarvey ML, et al. (1981) Osteonectin, a bone-specific protein linking mineral to collagen. Cell 26 : 99–105. 7034958
70. Faiyaz-Ul-Haque M, Zaidi SHE, Al-Ali M, Al-Mureikhi MS, Kennedy S, et al. (2004) A novel missense mutation in the galactosyltransferase‐I (B4GALT7) gene in a family exhibiting facioskeletal anomalies and Ehlers—Danlos syndrome resembling the progeroid type. American Journal of Medical Genetics Part A 128 : 39–45.
71. Götte M, Spillmann D, Yip GW, Versteeg E, Echtermeyer FG, et al. (2008) Changes in heparan sulfate are associated with delayed wound repair, altered cell migration, adhesion and contractility in the galactosyltransferase I (ß4GalT-7) deficient form of Ehlers—Danlos syndrome. Human Molecular Genetics 17 : 996–1009. 18158310
72. Almeida R, Levery SB, Mandel U, Kresse H, Schwientek T, et al. (1999) Cloning and expression of a proteoglycan UDP-galactose: β-Xylose β1, 4-galactosyltransferase IA seventh member of the human β4-galactosyltransferase gene family. Journal of Biological Chemistry 274 : 26165–26171. 10473568
73. Militello RD, Munafó DB, Berón W, López LA, Monier S, et al. (2013) Rab24 is required for normal cell division. Traffic 14 : 502–518. doi: 10.1111/tra.12057 23387408
74. Kitching R, Qi S, Li V, Raouf A, Vary CP, et al. (2002) Coordinate gene expression patterns during osteoblast maturation and retinoic acid treatment of MC3T3-E1 cells. J Bone Miner Metab 20 : 269–280. 12203032
75. Ono Y, Iemura S-i, Novak SM, Doi N, Kitamura F, et al. (2013) PLEIAD/SIMC1/C5orf25, a novel autolysis regulator for a skeletal-muscle-specific calpain, CAPN3, scaffolds a CAPN3 substrate, CTBP1. Journal of molecular biology 425 : 2955–2972. doi: 10.1016/j.jmb.2013.05.009 23707407
76. Ciganoka D, Balcere I, Kapa I, Peculis R, Valtere A, et al. (2011) Identification of somatostatin receptor type 5 gene polymorphisms associated with acromegaly. European Journal of Endocrinology 165 : 517–525. doi: 10.1530/EJE-11-0416 21810856
77. Ren S-G, Taylor J, Dong J, Yu R, Culler MD, et al. (2003) Functional association of somatostatin receptor subtypes 2 and 5 in inhibiting human growth hormone secretion. The Journal of Clinical Endocrinology & Metabolism 88 : 4239–4245.
78. Bruns C, Dietl MM, Palacios JM, Pless J (1990) Identification and characterization of somatostatin receptors in neonatal rat long bones. Biochem J 265 : 39–44. 1967933
79. Weiss RE, Reddi A, Nimni M (1981) Somatostatin can locally inhibit proliferation and differentiation of cartilage and bone precursor cells. Calcified Tissue International 33 : 425–430. 6117356
80. Shao Y, Alicknavitch M, Farach-Carson MC (2005) Expression of voltage sensitive calcium channel (VSCC) L-type Cav1. 2 (α1C) and T‐type Cav3. 2 (α1H) subunits during mouse bone development. Developmental dynamics 234 : 54–62. 16059921
81. Fodor J, Matta C, Oláh T, Juhász T, Takács R, et al. (2013) Store-operated calcium entry and calcium influx via voltage-operated calcium channels regulate intracellular calcium oscillations in chondrogenic cells. Cell calcium 54 : 1–16. doi: 10.1016/j.ceca.2013.03.003 23664335
82. Baud'Huin M, Duplomb L, Téletchéa S, Charrier C, Maillasson M, et al. (2009) Factor VIII-von Willebrand factor complex inhibits osteoclastogenesis and controls cell survival. Journal of Biological Chemistry 284 : 31704–31713. doi: 10.1074/jbc.M109.030312 19758994
83. Suva LJ, Hartman E, Dilley JD, Russell S, Akel NS, et al. (2008) Platelet dysfunction and a high bone mass phenotype in a murine model of platelet-type von Willebrand disease. The American journal of pathology 172 : 430–439. doi: 10.2353/ajpath.2008.070417 18187573
84. Karasik D, Hsu Y-H, Zhou Y, Cupples LA, Kiel DP, et al. (2010) Genome-wide pleiotropy of osteoporosis‐related phenotypes: the Framingham study. JBMR 25 : 1555–1563. doi: 10.1002/jbmr.38 20200953
85. Wright D, Rubin CJ, Martinez Barrio A, Schütz K, Kerje S, et al. (2010) The genetic architecture of domestication in the chicken: effects of pleiotropy and linkage. Molecular Ecology 19 : 5140–5156. doi: 10.1111/j.1365-294X.2010.04882.x 21040053
86. Rubin CJ, Brändström H, Wright D, Kerje S, Gunnarsson U, et al. (2007) Quantitative trait loci for BMD and bone strength in an intercross between domestic and wildtype chickens. JBMR 22 : 375–384. 17181401
87. Johnsson M, Rubin CJ, Höglund A, Sahlqvist AS, Jonsson K, et al. (2014) The role of pleiotropy and linkage in genes affecting a sexual ornament and bone allocation in the chicken. Molecular Ecology 23 : 2275–2286. doi: 10.1111/mec.12723 24655072
88. Johnsson M, Gustafson I, Rubin C-J, Sahlqvist A-S, Jonsson KB, et al. (2012) A sexual ornament in chickens is affected by pleiotropic alleles at HAO1 and BMP2, selected during domestication. PLoS Genetics 8: e1002914. doi: 10.1371/journal.pgen.1002914 22956912
89. Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL maping in experimental crosses. Bioinformatics 19 : 889–890. 12724300
90. Manichaikul A, Dupuis J, Sen S, Broman KW (2006) Poor Performance of Bootstrap Confidence Intervals for the Location of a Quantitative Trait Locus. Genetics 174 : 481–489. 16783000
91. Kent W (2002) BLAT—the BLAST-like alignment tool. Genome Res 12 : 656–664. 11932250
Štítky
Genetika Reprodukčná medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



