-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Quorum Sensing Inhibition Selects for Virulence and Cooperation in
With the rising development of bacterial resistance the search for new medical treatments beyond conventional antimicrobials has become a key aim of public health research. Possible innovative strategies include the inhibition of bacterial virulence. However, consideration must be given to the evolutionary and environmental consequences of such new interventions. Virulence and cooperative social behaviour of the bacterium Pseudomonas aeruginosa rely on the quorum-sensing (QS) controlled production of extracellular products (public goods). Hence QS is an attractive target for anti-virulence interventions. During colonization, non-cooperating (and hence less virulent) P. aeruginosa QS-mutants, benefiting from public goods provided by wild type isolates, naturally increase in frequency providing a relative protection from invasive infection. We hypothesized that inhibition of QS-mediated gene expression removes this growth advantage and selection of less virulent QS-mutants, and maintains the predominance of more virulent QS-wild type bacteria. We addressed this possibility in a placebo-controlled trial investigating the anti-QS properties of azithromycin, a macrolide antibiotic devoid of bactericidal activity on P. aeruginosa, but interfering with QS, in intubated patients colonized by P. aeruginosa. In the absence of azithromycin, non-cooperating (and hence less virulent) lasR (QS)-mutants increased in frequency over time. Azithromycin significantly reduced QS-gene expression measured directly in tracheal aspirates. Concomitantly the advantage of lasR-mutants was lost and virulent wild-type isolates predominated during azithromycin treatment. We confirmed these results in vitro with fitness and invasion experiments. Azithromycin reduced growth rate of the wild-type, but not of the lasR-mutant. Furthermore, the lasR-mutant efficiently invaded wild-type populations in the absence, but not in the presence of azithromycin. These in vivo and in vitro results demonstrate that anti-virulence interventions based on QS-blockade diminish natural selection towards reduced virulence and therefore may increase the prevalence of more virulent genotypes in the Hospital environment. More generally, the impact of intervention on the evolution of virulence of pathogenic bacteria should be assessed.
Trial Registration: ClinicalTrials.gov NCT00610623
Published in the journal: Quorum Sensing Inhibition Selects for Virulence and Cooperation in. PLoS Pathog 6(5): e32767. doi:10.1371/journal.ppat.1000883
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000883Summary
With the rising development of bacterial resistance the search for new medical treatments beyond conventional antimicrobials has become a key aim of public health research. Possible innovative strategies include the inhibition of bacterial virulence. However, consideration must be given to the evolutionary and environmental consequences of such new interventions. Virulence and cooperative social behaviour of the bacterium Pseudomonas aeruginosa rely on the quorum-sensing (QS) controlled production of extracellular products (public goods). Hence QS is an attractive target for anti-virulence interventions. During colonization, non-cooperating (and hence less virulent) P. aeruginosa QS-mutants, benefiting from public goods provided by wild type isolates, naturally increase in frequency providing a relative protection from invasive infection. We hypothesized that inhibition of QS-mediated gene expression removes this growth advantage and selection of less virulent QS-mutants, and maintains the predominance of more virulent QS-wild type bacteria. We addressed this possibility in a placebo-controlled trial investigating the anti-QS properties of azithromycin, a macrolide antibiotic devoid of bactericidal activity on P. aeruginosa, but interfering with QS, in intubated patients colonized by P. aeruginosa. In the absence of azithromycin, non-cooperating (and hence less virulent) lasR (QS)-mutants increased in frequency over time. Azithromycin significantly reduced QS-gene expression measured directly in tracheal aspirates. Concomitantly the advantage of lasR-mutants was lost and virulent wild-type isolates predominated during azithromycin treatment. We confirmed these results in vitro with fitness and invasion experiments. Azithromycin reduced growth rate of the wild-type, but not of the lasR-mutant. Furthermore, the lasR-mutant efficiently invaded wild-type populations in the absence, but not in the presence of azithromycin. These in vivo and in vitro results demonstrate that anti-virulence interventions based on QS-blockade diminish natural selection towards reduced virulence and therefore may increase the prevalence of more virulent genotypes in the Hospital environment. More generally, the impact of intervention on the evolution of virulence of pathogenic bacteria should be assessed.
Trial Registration: ClinicalTrials.gov NCT00610623Introduction
Anti-virulence therapies have been recently suggested as alternative strategies to circumvent the growing problem of antibiotic resistance [1], [2]. In P. aeruginosa inhibition of Quorum-Sensing (QS) seems particularly attractive as QS regulates many virulence determinants of this pathogen [3]. Azithromycin is a widely used macrolide antibiotic without significant bactericidal activity on P. aeruginosa [4]. Recent studies suggest azithromycin might be of benefit against this bacterium because it interferes with the QS-circuit and thereby inhibits the expression of a wide range of extracellular virulence factors [5]. Inhibition of QS is likely to have important evolutionary consequences for P. aeruginosa. Both in vitro and in vivo studies suggest that mutants (QS-cheats) that don't respond to QS (specifically, mutants that are defective in one of the QS-receptors, LasR) can have a selective advantage in the presence of QS-wildtypes [6], [7]. This has been recently demonstrated during colonization of untreated colonized patients in whom QS-cheats accumulated over time [8]. The most likely explanation for this advantage is that the mutants exploit the wild type public goods, without paying the metabolic cost of their production [9]–[11]; although other direct costs of QS in clinical contexts can't be ruled out [12]–[14]. Regardless of the reasons why QS-mutants have a fitness advantage, this advantage is unlikely to be realised if QS is blocked in wild type bacteria. Azithromycin (or any QS-blocker) will therefore reduce, or remove, selection for less virulent QS-cheats and maintain the predominance of more virulent QS-wild type bacteria.
We tested this hypothesis by following the evolutionary dynamics of QS (lasR) mutants in intubated patients colonised by P. aeruginosa, during a placebo controlled clinical trial evaluating the prevention of pneumonia by azithromycin. Whereas the proportion of lasR mutants rapidly increased in the untreated control patients, the proportion did not change in the azithromycin-treated patients. This fitness advantage in the absence, but not the presence, of azithromycin was similarly observed in vitro. More generally, the impact of intervention on the evolution of virulence of pathogenic bacteria should be assessed [15].
Results/Discussion
We tested the hypothesis that azithromycin reduces selection for QS-cheats by following prospectively 92 intubated patients (colonization times of three to twenty days), colonized by P. aeruginosa and hospitalized in intensive care units of seventeen European Hospitals, participating in a placebo controlled azithromycin pneumonia prevention trial (see material and methods). Importantly, antibiotic treatments active against P. aeruginosa were forbidden during the trial. We collected a single P. aeruginosa isolate per patient per day from tracheal aspirates and estimated total density of P. aeruginosa bacteria in the aspirates through genomic copy numbers. Adequate microbiological sampling for bacterial population analysis was available for 61 patients (31 placebo and 30 azithromycin) of the initial 92 randomized patients in the intention-to-treat protocol (Figure 1).
Fig. 1. Patient enrollment and follow-up. 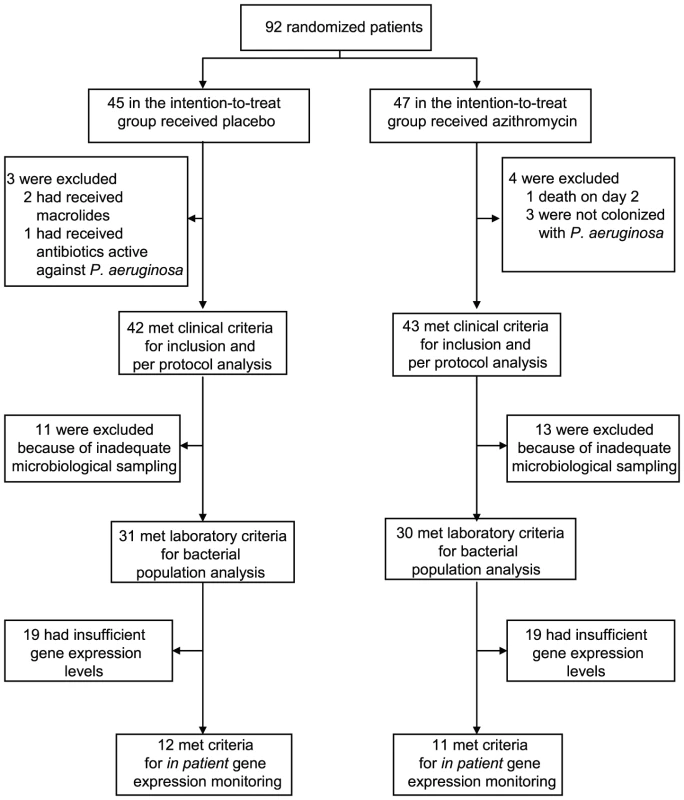
QS-inhibition in patient
We monitored QS-gene expression directly in tracheal aspirates to document the “in patient” QS-inhibition by azithromycin. Azithromycin significantly reduced the expression of both QS-circuit (lasI; Mann-Whitney test, P = 0.006) as well as QS-target (rhlA; P = 0.005) genes, whereas it did not affect expression of the QS-independent gene trpD (P>0.2) (Figure 2). It is of course possible that azithromycin inhibited the expression of some other genes unrelated to QS. However microarray data have shown that QS-regulated genes were among those whose expression was most severely affected by azithromycin [16].
Fig. 2. In patient QS-inhibition in azithromycin-treated patients. 
In patient QS-gene expression was determined as described. Tracheal aspirates from both day −1 and day x with bacterial RNA of adequate quality were available for twelve placebo and eleven azithromycin patients. Expression of QS-circuit gene lasI, QS-target gene rhlA and QS-independent gene trpD measured in tracheal aspirates is shown as the relative value (%) of the last accessible day (Dx) compared to day −1 (set as 100%). A horizontal line indicates the median expression levels. P values were calculated using Mann-Whitney tests. We determined the evolution of P. aeruginosa QS in patients by first measuring the production of elastase, which is under the control of the lasR QS-system [17] from the 650 collected isolates (data not shown). Variations in elastase activity correlated with mutations in lasR between independent wild type and mutant lasR alleles (Mann-Whitney: P<0.001), as determined by sequencing this gene in the first isolate obtained from each patient, and then in subsequent isolates presenting a different QS-phenotype (Figure 3). Mutations in lasR were therefore reducing the expression of elastase, and by inference, other lasR-regulated genes. Whereas the proportion of lasR mutants significantly increased through time in the 31 control group patients (Figure 3a; logistic regression: F1,10 = 65.36, P<0.001), there was a small, decline in the proportion of lasR mutants in the 30 azithromycin treated patients (Figure 3a; F1,10 = 32.58, P<0.001; test of whether slopes differ (treatment by time interaction) in full model: F1, 20 = 77.6; P<0.001). In agreement with this observation, isolates from placebo patients showed decreasing mean elastase levels in vitro (Figure 3b; F1,10 = 41.12, P<0.001), while isolates from the azithromycin treated group showed a concomitant increase through time (Figure 3b; F1,10 = 41.12, P<0.001; treatment by time interaction in full model: F1,20 = 26.46, P<0.001).
Fig. 3. Evolution of lasR mutants and elastase production in azithromycin-treated and untreated patients. 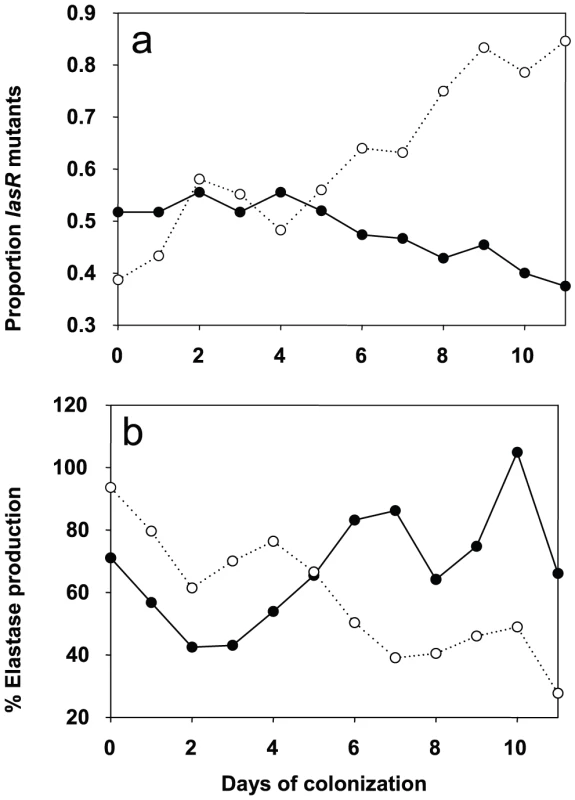
Change in the proportion of lasR mutants (a) and mean elastase production (b) through time. Solid lines and closed symbols indicate azithromycin-treated patients, and dashed lines and open symbols indicate placebo group. Note that data is presented to day 11 of colonization, despite some samples being collected up to 20 days, because of very small sample sizes (six isolates) by day 12 in the azithromycin-treated group. However, qualitatively identical results were obtained when the whole data set was analysed. The change in the proportion of lasR mutants and elastase through time was analysed using logistic regression, corrected for under-dispersion, and General linear Modelling, respectively in GenStat 10. These data are consistent with the hypothesis that azithromycin treatment removes any advantage of QS-mutants, because QS is blocked in the wildtype population. There are, however, a number of alternative explanations, particularly as bacterial densities, based on mean P. aeruginosa genomic copy numbers, were twice as large in the placebo compared to the azithromycin group (7×106 and 1.2×107; t = 1.96, P = 0.06). First, azithromycin-imposed reduction in densities could reflect reductions in growth rate, and this could simply have slowed down the rate at which lasR mutants change in frequency. We can however rule this out as a primary explanation for our data, because azithromycin did not only cause a quantitative change in the frequency of lasR mutants, but also a qualitative change: lasR mutants decreased in frequency during azithromycin treatment, whereas they increased in the placebo group (Figure 3). Second, it is possible that azithromycin may reduce selection for lasR mutants if reductions in QS-mediated public goods production results from reductions in bacterial density caused by azithromycin. Third, azithromycin may directly inhibit the growth of lasR mutants more than wildtype bacteria, explaining why there was a small reduction in the frequency of lasR mutants following azithromycin treatment. We address these possibilities below. Furthermore we cannot exclude that azithromycin influenced the structure of the resident bacterial flora of the patients which could in turn influence the P. aeruginosa population and its virulence properties [18].
QS-inhibition in vitro
To aid the interpretation of the clinical data, we carried out in vitro experiments with wild type P. aeruginosa (PAO1) and an isogenic lasR mutant in the presence and absence of azithromycin. Danesi et al. [19] measured azithromycin concentrations of 9 mg/kg in lung tissue of patients receiving a comparable dosing regimen as those in our study, hence we used similar concentrations (5 and 10 mg/l) for our in vitro experiments. We first measured growth rates in media where the primary nutrient source is protein (BSA), making lasR-controlled expression of proteases necessary for the production of useable amino acids [6].
Consistent with previous studies [6], growth rate of the lasR mutant in monoculture was reduced relative to the wildtype (by approximately 50%) in the absence of azithromycin, demonstrating an advantage of QS in this environment (Figure 4a; 2-sample t-test: P<0.05). The addition of azithromycin reduced densities of both genotypes (linear effect of azithromycin: F1,29 = 71.2, P<0.001), but this reduction was much greater for the wildtype than the lasR mutant (interaction between concentration and genotype: F1,29 = 6.92, P = 0.013), confirming a role of azithromycin in suppressing the production of QS-controlled exoproducts. Given that azithromycin inhibits wildtype growth more than that of the lasR mutant, we are unable to explain the slight drop in the frequency of lasR mutants in the patients.
Fig. 4. lasR mutant growth rates and invasion of wild type populations in the presence and absence of azithromycin. 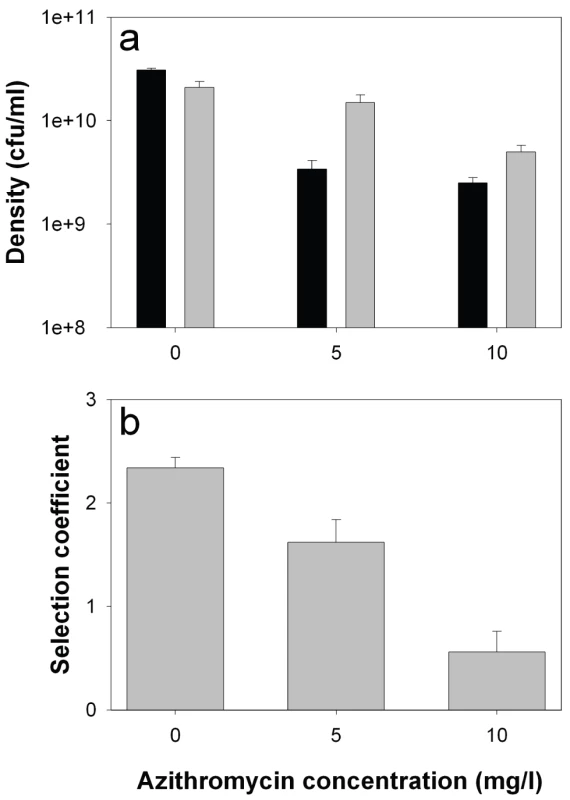
In vitro densities of wildtype (black) and lasR mutant (grey) after 72 hours growth in M9 salts BSA medium (a), and selection coefficients of lasR mutant relative to wildtype (b) as a function of azithromycin (AZM). Bars show means (± SEM) of six replicates. All differences (wildtype versus lasR) in the presence of azithromycin are statistically significant (p<0.05). We next measured the fitness of the lasR mutant invading wildtype populations (1∶100 ratio). Consistent with the in vivo results, we found that the fitness advantage of lasR mutants was decreased with increasing azithromycin concentration (Figure 4b; Linear effect of azithromycin concentration of fitness of lasR mutant: F1,16 = 48.41, P<0.001). Unlike in the clinical context, the lasR mutant still had a slight fitness advantage over the wildtype at the highest concentration of azithromycin used (10 mg/l), suggesting, unsurprisingly, that additional variables influence the fitness of lasR mutants in vivo. These results strongly suggest that the major advantage of lasR mutants in vitro (and presumably in vivo) is their ability to exploit wildtype public goods [8], and that azithromycin removes this advantage because less public good (elastase) is produced. However, the data do not distinguish between azithromycin directly inhibiting elastase production, or indirectly through density reductions, or both.
Conclusions
We have shown that azithromycin treatment can prevent selection for lasR mutants, and consequently increase the proportion of wild type P. aeruginosa in colonized patients. A number of not mutually exclusive factors may help to explain this pattern, but the data suggests that the primary reason is because azithromycin blocks QS. Blocking QS prevents lasR mutants from exploiting the public goods provided by the wildtype, and reduces any direct costs associated with QS, such as production of extracellular products that are not of benefit in this particular environment [8]. Both in vitro and in patient data obtained from the clinical trial suggest a key role of QS-dependent virulence for the development of infection (Köhler et al., submitted), and support the general consensus from studies using animal models showing that QS-expression (and public goods production in general) is associated with increased virulence [2], [6], [20]–[23]. Azithromycin is therefore likely to be of clinical benefit to a treated patient by inhibiting the QS-dependent virulence during the course of the treatment. However, when treatment is discontinued the patient is at risk of being colonized by highly virulent bacteria, with the potential of late onset infections. Moreover a wider use of such anti-virulence interventions may also increase the prevalence of highly virulent QS-wild type isolates within the hospital. More generally, any intervention that reduces bacterial densities is also likely to result in a reduced selective advantage of less virulent mutants that do not make public goods [24]. The study highlights the need to carefully consider both the short and longer term implications of anti-virulence therapy and other interventions (such as vaccines [15]) on pathogen virulence.
Materials and Methods
Ethic statement
We obtained approval for this study by the “Commission Centrale d'Ethique de la Recherche sur l'Etre Humain des Hôpitaux Universitaire de Genève”. Written informed consent from all patients or their legal representatives was obtained according to legal and ethical considerations.
Patients and clinical collection
This randomized, placebo-controlled, double blind study (ANB 006#2001, ClinicalTrials.gov ID#NCT00610623) was designed to assess the efficacy of azithromycin as a quorum-sensing inhibitor in preventing the occurrence of P. aeruginosa pneumonia in ventilated patients with documented colonization. Twenty-one European centers participated in this trial; eight in France, four in Spain, three in Belgium, three in Poland, two in Serbia and one in Switzerland. We screened mechanically-ventilated patients for respiratory tract colonization by P. aeruginosa every 48 hours. Neutropenic patients and patients treated with immunosuppressive drugs were not eligible. Patients with ongoing P. aeruginosa infection, having received macrolides or antibiotics active against the colonizing P. aeruginosa isolate during the last 14 days were excluded. Patients with proven colonization by P. aeruginosa were randomized (D-1) and received either placebo or 300 mg per day iv azithromycin in a double blind fashion for a maximum of 20 days (D1 to Dx). During the study, only the administration of antibiotics inactive against P. aeruginosa was allowed. Detailed information on study design is available in supporting information Protocol S1 and Checklist S1. Patient characteristics and clinical outcome of the study are published elsewhere (van Delden et al., submitted). Starting the first day of proven colonization (D-1), we collected tracheal aspirates (0.3 to 5 ml) and one P. aeruginoa isolate (collection period: 3–20 days) at 24 hours intervals. Samples were frozen at −80°C on site within 15 minutes, and sent on dry ice to the reference research laboratory at the University Hospital Geneva, where all analyses were performed in a blind fashion. The logit-transformed proportion of patients whose isolate was a lasR mutant was analyzed by logistic regression, with time (a covariate), treatment (placebo or azithromycin) and the interaction fitted in GenStat v10. Data were over dispersed, so a scaling factor to equalize the residual error and degrees of freedom was employed.
In patient gene expression analysis
From prospectively collected tracheal aspirates we extracted total genomic DNA and total RNA (for details see supporting information Text S1). We detected (>104 genomic copies/g aspirate) P. aeruginosa DNA in 98% of the aspirates, confirming the colonization by this organism. In the RNA extractions, we detected expression (>5×104 copies/g aspirate) of the rpsL housekeeping gene in 80% of the aspirates. This indicates that quality of both sample handling and RNA extractions were sufficient to detect bacterial gene expression in the majority of the tracheal aspirates. As a second control for the quality of the RNA extracts from clinical samples we plotted the amount of P. aeruginosa bacteria as determined by qRT-PCR from the genomic DNA extractions against the expression of the rpsL housekeeping gene. We observed a good correlation between these two variables (r = 0.69, P<0.001).
Determination of bacterial loads
The number of P. aeruginosa in aspirates was determined by qRT-PCR of genomic DNA preparations. Aliquots of genomic DNA preparations were diluted 10 fold into H2O and 3 µl of this dilution were added to the PCR reaction mix containing 1× Quantitect Sybr Green Master Mix and 600 nM primers in a total volume of 15 µl. PCR conditions were as described below for cDNA analysis. A standard curve was obtained by addition of 10-fold dilutions of a P. aeruginosa culture to an aspirate collected from a patient not colonized by this organism. Genomic DNA was then isolated as described above and quantified by qRT-PCR. Under these conditions, we detected 104 CFU/g aspirate. Standard curves yielded reproducible values during the 3-month analysis period. P. aeruginosa was found in the aspirates at levels varying from 4×104–1.8×108 CFU/g.
In vitro experiments
P. aeruginosa strain PAO1 was competed against a rare invading isogenic lasR knockout mutant [25] in 200 µl M9 minimal salts medium supplemented with 1% BSA [6] in 96-well plates, shaken at 200 rpm at 37°C in the presence or absence of azithromycin (5 and 10 mg/l) for 72 hours. Six wells per environment were inoculated with 107 cells of overnight cultures (grown in LB medium at 37°C), at a ratio of 1∶100 lasR mutant: wild type. Selection coefficients of the lasR mutants was calculated as the differences in malthusian parameters (ln(final density/starting density) as previously described [26], with cell counts determined by plating on LB agar and LB supplemented with 50 mg/l tetracycline. A selection coefficient of zero indicates that strains have equal fitness. Selection coefficients were regressed against azithromycin concentration. Densities (colony forming units) of pure cultures (6 replicates per treatment) under the same conditions were determined at the same time. Densities were log10-transformed, to meet assumption of general linear models, and concentration (a covariate), strain and the interaction fitted in GenStat.
Supporting Information
Zdroje
1. HentzerM
WuH
AndersenJB
RiedelK
RasmussenTB
2003 Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22 3803 3815
2. BjarnsholtT
GivskovM
2007 Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos Trans R Soc Lond B Biol Sci 362 1213 1222
3. Van DeldenC
IglewskiBH
1998 Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis 4 551 560
4. MolinariG
GuzmanCA
PesceA
SchitoGC
1993 Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J Antimicrob Chemother 31 681 688
5. TatedaK
ComteR
PechereJC
KöhlerT
YamaguchiK
2001 Azithromycin Inhibits Quorum Sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 45 1930 1933
6. DiggleSP
GriffinAS
CampbellGS
WestSA
2007 Cooperation and conflict in quorum-sensing bacterial populations. Nature 450 411 414
7. SandozKM
MitzimbergSM
SchusterM
2007 Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A 104 15876 15881
8. KöhlerT
BucklingA
Van DeldenC
2009 Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci U S A 106 6339 6344
9. BrownSP
HochbergME
GrenfellBT
2002 Does multiple infection select for raised virulence? Trends Microbiol 10 401 405
10. WestSA
BucklingA
2003 Cooperation, virulence and siderophore production in bacterial parasites. Proc Biol Sci 270 37 44
11. BucklingA
BrockhurstMA
2008 Kin selection and the evolution of virulence. Heredity 100 484 488
12. SmithEE
BuckleyDG
WuZ
SaenphimmachakC
HoffmanLR
2006 Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103 8487 8492
13. D'ArgenioDA
WuM
HoffmanLR
KulasekaraHD
DezielE
2007 Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64 512 533
14. HeurlierK
DenervaudV
HaenniM
GuyL
KrishnapillaiV
2005 Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J Bacteriol 187 4875 4883
15. GandonS
MackinnonMJ
NeeS
ReadAF
2001 Imperfect vaccines and the evolution of pathogen virulence. Nature 414 751 756
16. NalcaY
JanschL
BredenbruchF
GeffersR
BuerJ
2006 Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob Agents Chemother 50 1680 1688
17. PearsonJP
PesciEC
IglewskiBH
1997 Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179 5756 5767
18. DuanK
DammelC
SteinJ
RabinH
SuretteMG
2003 Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol 50 1477 1491
19. DanesiR
LupettiA
BarbaraC
GhelardiE
ChellaA
2003 Comparative distribution of azithromycin in lung tissue of patients given oral daily doses of 500 and 1000 mg. J Antimicrob Chemother 51 939 945
20. Le BerreR
NguyenS
NowakE
KipnisE
PierreM
2008 Quorum-sensing activity and related virulence factor expression in clinically pathogenic isolates of Pseudomonas aeruginosa. Clin Microbiol Infect 14 337 343
21. SmithRS
IglewskiBH
2003 P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol 6 56 60
22. RumbaughKP
DiggleSP
WattersCM
Ross-GillespieA
GriffinAS
2009 Quorum sensing and the social evolution of bacterial virulence. Curr Biol 19 341 345
23. HarrisonEF
BrowningL
VosM
BucklingA
2006 Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biology 4 21
24. BrownSP
JohnstoneRA
2001 Cooperation in the dark: signalling and collective action in quorum-sensing bacteria. Proc Biol Sci 268 961 965
25. KöhlerT
Kocjancic-CurtyL
BarjaF
Van DeldenC
PechèreJC
2000 Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182 5990 5996
26. LenskiRE
RoseMR
SimpsonSC
TadlerSC
1991 Long term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2000 generations. Am Nat 138 1315 1341
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing ActivityČlánek The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced MucositisČlánek Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External RegionsČlánek Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The HMW1C Protein Is a Glycosyltransferase That Transfers Hexose Residues to Asparagine Sites in the HMW1 Adhesin
- Analysis of Virion Structural Components Reveals Vestiges of the Ancestral Ichnovirus Genome
- Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing Activity
- Global Migration Dynamics Underlie Evolution and Persistence of Human Influenza A (H3N2)
- The Type III Effectors NleE and NleB from Enteropathogenic and OspZ from Block Nuclear Translocation of NF-κB p65
- VEGF Promotes Malaria-Associated Acute Lung Injury in Mice
- Identification of a Mutant PfCRT-Mediated Chloroquine Tolerance Phenotype in
- The Early Stage of Bacterial Genome-Reductive Evolution in the Host
- Host-Detrimental Role of Esx-1-Mediated Inflammasome Activation in Mycobacterial Infection
- Elevation of Intact and Proteolytic Fragments of Acute Phase Proteins Constitutes the Earliest Systemic Antiviral Response in HIV-1 Infection
- The Pleiotropic CymR Regulator of Plays an Important Role in Virulence and Stress Response
- Alternative Sigma Factor σ Modulates Prophage Integration and Excision in
- Effect of Neuraminidase Inhibitor–Resistant Mutations on Pathogenicity of Clade 2.2 A/Turkey/15/06 (H5N1) Influenza Virus in Ferrets
- Massive APOBEC3 Editing of Hepatitis B Viral DNA in Cirrhosis
- NK Cells and γδ T Cells Mediate Resistance to Polyomavirus–Induced Tumors
- Is Genetically Diverse in Animals and Appears to Have Crossed the Host Barrier to Humans on (At Least) Two Occasions
- Adenylate Cyclase Toxin Mobilizes Its β Integrin Receptor into Lipid Rafts to Accomplish Translocation across Target Cell Membrane in Two Steps
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- HIV-1 Transmitting Couples Have Similar Viral Load Set-Points in Rakai, Uganda
- Few and Far Between: How HIV May Be Evading Antibody Avidity
- Galectin-9/TIM-3 Interaction Regulates Virus-Specific Primary and Memory CD8 T Cell Response
- Perforin Expression Directly by HIV-Specific CD8 T-Cells Is a Correlate of HIV Elite Control
- The Set3/Hos2 Histone Deacetylase Complex Attenuates cAMP/PKA Signaling to Regulate Morphogenesis and Virulence of
- Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing
- Combining ChIP-chip and Expression Profiling to Model the MoCRZ1 Mediated Circuit for Ca/Calcineurin Signaling in the Rice Blast Fungus
- Internalin B Activates Junctional Endocytosis to Accelerate Intestinal Invasion
- A Complex Small RNA Repertoire Is Generated by a Plant/Fungal-Like Machinery and Effected by a Metazoan-Like Argonaute in the Single-Cell Human Parasite
- Opc Invasin Binds to the Sulphated Tyrosines of Activated Vitronectin to Attach to and Invade Human Brain Endothelial Cells
- Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa
- PdeH, a High-Affinity cAMP Phosphodiesterase, Is a Key Regulator of Asexual and Pathogenic Differentiation in
- Isolates with Antimony-Resistant but Not -Sensitive Phenotype Inhibit Sodium Antimony Gluconate-Induced Dendritic Cell Activation
- The Microbiota and Allergies/Asthma
- Environmental Factors Determining the Epidemiology and Population Genetic Structure of the Group in the Field
- Prolonged Antigen Presentation Is Required for Optimal CD8+ T Cell Responses against Malaria Liver Stage Parasites
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
- Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
- Effective, Broad Spectrum Control of Virulent Bacterial Infections Using Cationic DNA Liposome Complexes Combined with Bacterial Antigens
- High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men
- The -Specific Human Memory B Cell Compartment Expands Gradually with Repeated Malaria Infections
- EBV Promotes Human CD8 NKT Cell Development
- Persistent Growth of a Human Plasma-Derived Hepatitis C Virus Genotype 1b Isolate in Cell Culture
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



