-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus,
Fusarium graminearum is an important plant pathogen that causes head blight of major cereal crops. The fungus produces mycotoxins that are harmful to animal and human. In this study, a systematic analysis of 17 phenotypes of the mutants in 657 Fusarium graminearum genes encoding putative transcription factors (TFs) resulted in a database of over 11,000 phenotypes (phenome). This database provides comprehensive insights into how this cereal pathogen of global significance regulates traits important for growth, development, stress response, pathogenesis, and toxin production and how transcriptional regulations of these traits are interconnected. In-depth analysis of TFs involved in sexual development revealed that mutations causing defects in perithecia development frequently affect multiple other phenotypes, and the TFs associated with sexual development tend to be highly conserved in the fungal kingdom. Besides providing many new insights into understanding the function of F. graminearum TFs, this mutant library and phenome will be a valuable resource for characterizing the gene expression network in this fungus and serve as a reference for studying how different fungi have evolved to control various cellular processes at the transcriptional level.
Published in the journal: A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus,. PLoS Pathog 7(10): e32767. doi:10.1371/journal.ppat.1002310
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002310Summary
Fusarium graminearum is an important plant pathogen that causes head blight of major cereal crops. The fungus produces mycotoxins that are harmful to animal and human. In this study, a systematic analysis of 17 phenotypes of the mutants in 657 Fusarium graminearum genes encoding putative transcription factors (TFs) resulted in a database of over 11,000 phenotypes (phenome). This database provides comprehensive insights into how this cereal pathogen of global significance regulates traits important for growth, development, stress response, pathogenesis, and toxin production and how transcriptional regulations of these traits are interconnected. In-depth analysis of TFs involved in sexual development revealed that mutations causing defects in perithecia development frequently affect multiple other phenotypes, and the TFs associated with sexual development tend to be highly conserved in the fungal kingdom. Besides providing many new insights into understanding the function of F. graminearum TFs, this mutant library and phenome will be a valuable resource for characterizing the gene expression network in this fungus and serve as a reference for studying how different fungi have evolved to control various cellular processes at the transcriptional level.
Introduction
Transcription factors (TFs) orchestrate gene expression under the control of cellular signaling pathways and are key mediators of cellular function [1]. Understanding the regulatory mechanisms of each TF family and their function could provide valuable insight into gene expression changes underpinning cellular and developmental responses to environmental cues. Modification of TF activity through either gene disruption or overexpression can help determine the function and interconnectedness of individual TFs based on resulting cellular changes.
The filamentous fungus Fusarium graminearum (teleomorph: Gibberella zeae) is an important plant pathogen that causes head blight of major cereal crops, such as wheat, barley, and rice [2]. The fungus also produces mycotoxins in the infected cereals, posing grave threats to the health of animals and humans [3]. Complete genome sequencing of F. graminearum [4] has allowed for genome-wide gene functional studies, and transcriptome data from F. graminearum generated using the Affymetrix platform (http://www.plexdb.org/) can be used to predict gene function based on gene expression profiles [5], [6], [7], [8], [9], [10], [11], [12]. In addition, TFs from 64 fungal species, including F. graminearum, have been annotated in the Fungal Transcription Factor Database (FTFD, http://ftfd.snu.ac.kr/) [13]. The majority of F. graminearum TFs belong to the Zn(II)2Cys6 fungal binuclear cluster family; other types of TFs include C2H2 zinc finger, GATA, bHLH, B-ZIP, CBF, CCAAT-binding factor, homeobox, RING finger, PHD finger, and MIZ zinc finger [13]_ENREF_6. Although a few TFs that regulate pigmentation [14]_ENREF_6, mycotoxins biosynthesis [15], [16], [17]_ENREF_7_ENREF_7, sexual development, and virulence [18], [19], [20], [21], [22] have been characterized in F. graminearum, the function and regulation of most TFs remain to be determined.
In this study, we constructed F. graminearum deletion mutants in 657 genes encoding various types of TFs through homologous recombination to dissect their functions; resulting mutants were analyzed under 17 phenotypic categories to build a comprehensive phenotypic dataset (phenome). This phenome, which can be accessed at FgTFPD (Fusarium graminearum Transcription Factor Phenotype Database, http://ftfd.snu.ac.kr/FgTFPD), helped understand the mechanisms underpinning sexual and asexual development, toxin production, stress responses, and pathogenicity. The observed phenotypes were analyzed using a combination of data from published genome-wide fungal resources and bioinformatics analysis. Our mutant library will be a valuable source through distribution of mutants and easy access to our phenotypic and genetic data.
Results
Construction of genome-wide putative TF deletion mutants in F. graminearum
In order to construct F. graminearum TF mutants, we employed the FTFD [13] to choose the genes annotated as TFs in F. graminearum. The FTFD is a standardized pipeline for annotating fungal TFs using the InterPro database that is based on DNA-binding motifs. Among the 64 classified TF families present in the 61 fungal and three oomycete species, 693 putative F. graminearum TFs, belonging to 45 families, were annotated and selected for deletion. We also added 16 manually identified TFs based on sequence homology or conserved motifs (Table S1). We finally selected 709 putative TFs, consisting of 6.1% of all the F. graminearum genes [4]_ENREF_6. It has been reported that TFs cover 3–6% of the genes in Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana [23]. In the fungal kingdom the percentage of TFs is also variable, ranging from 2.3% in Pneumocystis carinii to 7.2% in Rhizopus oryzae [13]. Approximately 60% of the total TFs in F. graminearum belong to TFs bearing zinc finger DNA-binding domains, including Zn(II)2Cys6, C2H2, and CCCH (Figure 1A and Table S2). In contrast, approximately 20% of TFs in A. thaliana and C. elegans belong to this group_ENREF_10. The higher percentage of zinc finger TFs in fungi is mainly due to the existence of fungal-specific Zn(II)2Cys6 zinc finger TFs (e.g., 25% of total TFs in Saccharomyces cerevisiae) [23]. In addition to the zinc finger-related TFs, the other major TF families in the F. graminearum genome include OB-fold nucleic acid binding, high mobility group, winged helix repressor DNA-binding, and B-ZIP domains containing proteins (Figure 1A).
Fig. 1. Classification and deletion strategy of putative transcription factors (TFs) in Fusarium graminearum. 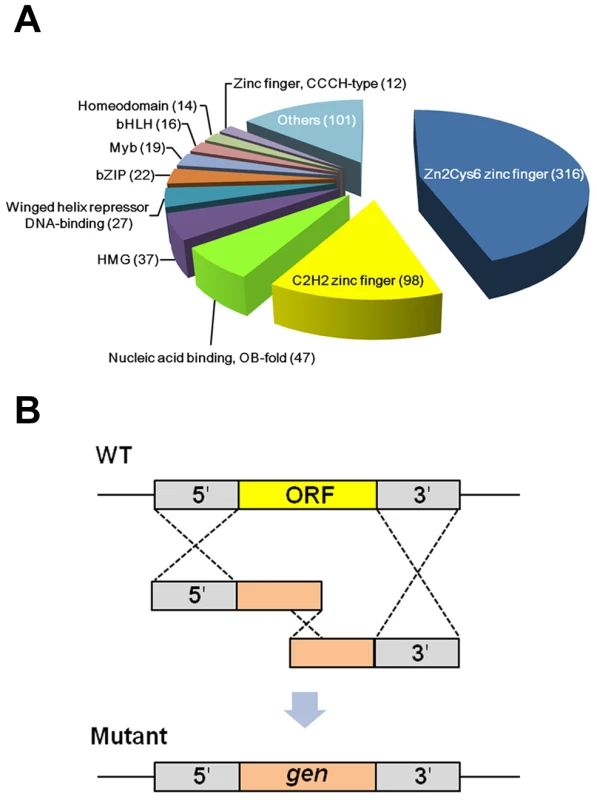
(A) Total TFs were classified based on nucleic acid binding domains. More than half of the total TFs are Zn6Cys6 zinc finger and C2H2 zinc finger proteins. (B) Each target TF gene was deleted using the split marker method based on triple homologous recombination. Using homologous recombination, we successfully disrupted 657 TF genes out of 709 (Figure 1B). Eight genes were excluded because of failure to amplify their 5′ or 3′ flanking region under various PCR conditions and primer sets. Repeated attempts to delete the remaining 44 genes were unsuccessful likely due to lethality. Disruption of 132 genes was confirmed by co-dominant PCR and Southern hybridization with a 5′ or 3′ flanking region probe, as previously reported [24]. The remaining mutants were confirmed by co-dominant PCR screening followed by Southern hybridization with a geneticin resistant gene cassette (gen) probe (Figure S1). The FgFSR1-deletion mutant was generated in our previous study [21]. Only co-dominant PCR screening was used to confirm disruption of two TF mutants (FGSG_06071 and FGSG_10716). We found an extra copy of the mutant allele integrated ectopically in 49 mutants (Figure S1 and Table S2). We screened more than 10,000 transformants using co-dominant PCR to identify desired mutants, resulting in 2,183 positive mutants. Among them, approximately 65% were single deletion mutants without ectopic integration and 23% were deletion mutants with multiple integrations. In a high-throughput gene deletion study in Neurospora crassa, more than 90% deletion efficiency was obtained using a strain with deletion of genes required for non-homologous end-joining DNA repair (mus-51 or mus-52) [25]. The split marker method based on triple homologous recombination to disrupt genes in F. graminearum was efficient enough that similar mutations of the parental strain were not required for mutagenesis.
Phenotypic analyses of resulting mutants
We analyzed 657 TF mutants for changes in 17 phenotypes (Table S2), which fall into six major phenotypic categories including mycelial growth (Figure S2 and Figure S3), sexual development (Figure S4), conidia production (Table S3), virulence (Figure S5), toxin production (Figure S6), and stress responses (Figure S7). A substantial fraction of the mutants (26%, 170/657) displayed clearly visible mutant phenotypes, with 73% (124/170) of these mutants exhibiting multiple mutant phenotypes and 27% (46/170) with single mutant phenotype (Figure 2A, Table S4, and Table S5). Analysis of TF locations in the F. graminearum genome revealed that the 170 TFs exhibiting phenotype changes were randomly distributed throughout the genome (Figure 2B). Among 103 TF deletion mutants in N. crassa [25], 43% (44/103) exhibited phenotype changes with 50% (22/44) of these exhibiting multiple defects in growth and asexual/sexual development. Deletion of members of the Zn(II)2Cys6 TF family in N. crassa, resulted in mutant phenotypes in 42% (30/72) of the mutants [25]. However, we observed mutant phenotypes in only 16% (46/296) of the F. graminearum mutants defective in Zn(II)2Cys6 TFs. Deletion of the genes encoding winged helix repressor DNA-binding TFs resulted in the highest percentage of mutants with phenotype change (48%, 12/25), whereas Nucleic acid-binding, OB-fold TF deletion mutants displayed the lowest percentage of phenotype change (13%, 5/40) (Figure 2C and Table S4). The percentages of phenotype change among the mutants of the C2H2 zinc finger, Myb, and B-ZIP TF family members were 41% (37/91), 41% (7/17), and 41% (9/22), respectively.
Fig. 2. Analysis of transcription factor (TF) mutant phenotypes. 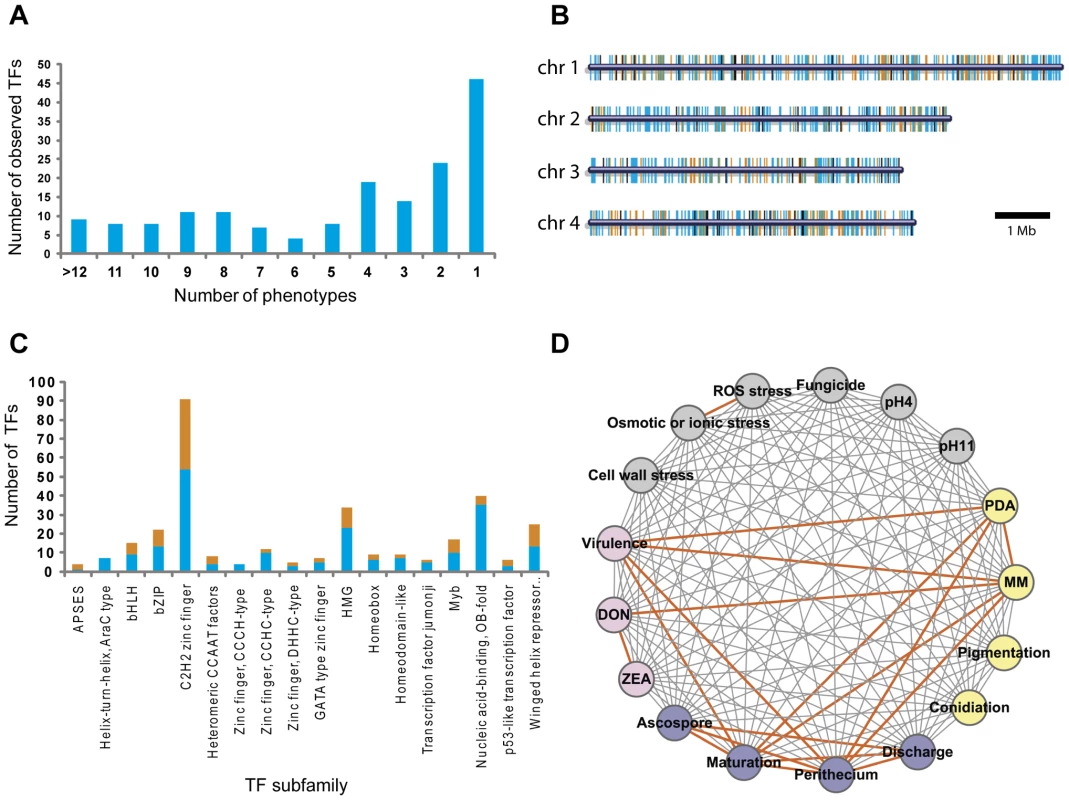
(A) Number of TFs showing multiple mutant phenotypes. TFs without mutant phenotype were omitted. (B) Chromosome distribution of 709 putative TFs. TFs that exhibit mutant phenotypes (orange bars) and those that do not (blue bars) are randomly distributed throughout the genome. Black bars represent the TF genes that could not be disrupted. (C) Number of mutants showing mutant phenotypes in each TF subfamily. Orange and blue bars represent the number of TFs whose deletion results in a mutant phenotype or no phenotype change, respectively. Data representing the Zn2Cys6 subfamily are not shown in this graph. (D) Phenotype network based on PCC (Table S6). PCC values were calculated from the results of tested phenotypes. Orange edges indicate positive correlations (PCC>0.60). To determine correlations between phenotypes, we calculated Pearson's correlation coefficient (PCC) from mutants that exhibit multiple mutant phenotypes (Table S6). The PCC between two phenotypes was calculated to determine which phenotypes are most highly correlated (Figure 2D). PCC>0.6 (p = 3.25×10−12) was chosen as the cut-off, and 18 phenotype pairs were selected and marked with orange edges between nodes (Figure 2D). This analysis revealed that sexual development, virulence, growth, and toxin production were highly correlated. The representative TFs are listed in Table 1. The phenotypes under sexual development, including perithecia formation and maturation, and ascospore production, were highly correlated with the phenotypes in other categories.
Tab. 1. List of key TF mutants showing no perithecia development and multiple defects in virulence, growth, and toxin production. 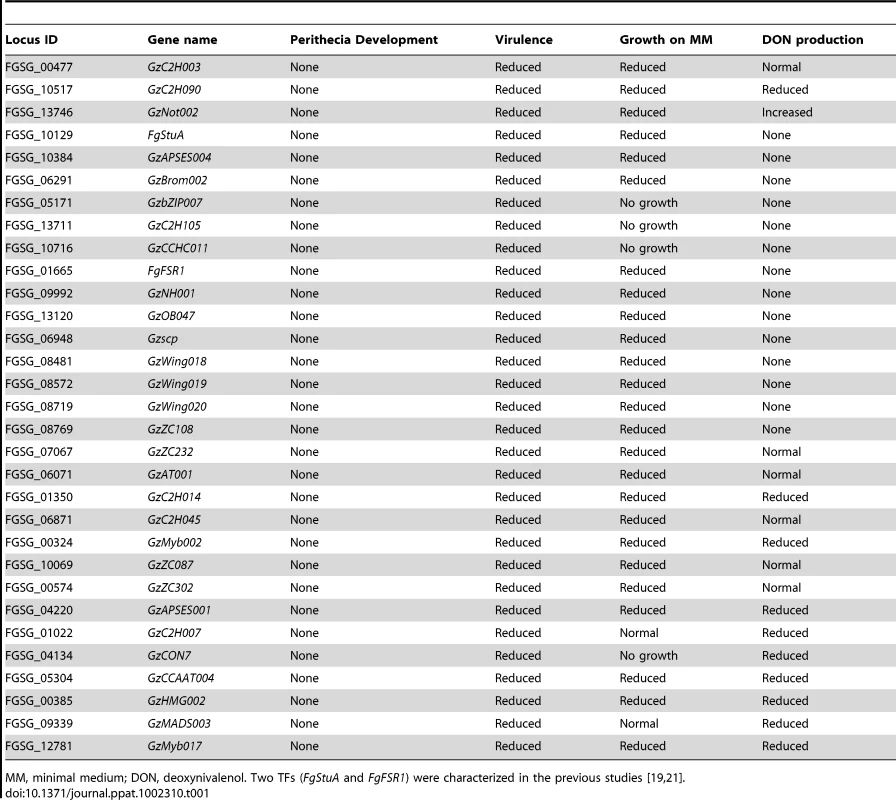
MM, minimal medium; DON, deoxynivalenol. Two TFs (FgStuA and FgFSR1) were characterized in the previous studies [19], [21]. We also performed in-depth studies of F. graminearum phenotypes under diverse environmental stresses. This analysis, including osmotic (or ionic), reactive oxygen species (ROS), fungicide, cell wall, and acidic (pH = 4) and basic (pH = 11) conditions, indicates there is no correlation between the stresses, except between osmotic and ROS stress (Figure 2D and Table S6).
We identified the F. graminearum orthologs of the 103 TF genes studied in N. crassa [25] to compare the resulting phenotypes between these species (Table S7A). Among 88 F. graminearum TFs identified, 26% of the mutants (23/88) exhibited mutant phenotypes with 43% (10/23) of these mutants showing multiple defects in growth and asexual/sexual development. However, only nine mutants showed the similar phenotype changes in growth, asexual, and/or sexual development in both species (Table S7B). This comparison of mutant phenotypes between F. graminearum and N. crassa suggests that the large majority of these TFs evolved to have unique, rather than conserved, functions in controlling growth and/or asexual and sexual developments.
Classification of TFs associated with sexual development
Among the 170 TF mutants exhibiting at least one mutant phenotype, 105 mutants showed considerable defect in sexual development. Based on the nature and severity of the phenotype change in perithecia development (number of perithecia and perithecia maturation) and ascospore production (existence and morphology) among these 105 mutants, we classified them into seven groups representing (Table 2 and Table S8). Four mutants produced more perithecia than the wild-type strain (Group 1). Group 2 contained 44 mutants that completely lack perithecia development and could not produce any initial structure for perithecium. The mutants showing decreased number of perithecia or delayed perithecia maturation, which eventually developed normally, were further divided into three groups based on ascospore formation: normal-shaped ascospore formation (23 TFs, Group 3), abnormal-shaped ascospore (9 TFs, Group 4), and no ascospore formation (19 TFs, Group 5). Among the Group 5 mutants, 12 TF mutants (GzCCAAT002, GzHMG010, MYT1, GzOB038, FgFlbA, GzWing015, GzRFX1, GzWing027, GzZC246, MAT1-1-3, MAT1-1-1, and MAT1-2-1) formed protoperithecia but failed to differentiate further and the other 7 TF mutants belonging to Group 5 still made initial structures of asci or rosettes (Figure S4). Five of the six mutants with normal perithecia development produced abnormally-shaped ascospores (Group 6), and one mutant produced neither asci nor ascospores (Group 7) (Table 2 and Figure 3). In total, 96 mutants had defects in perithecia development, five mutants exhibited defects in ascospore production with normal perithecia development, and four mutants showed enhanced sexual reproduction.
Fig. 3. Representative images of wild-type or transcription factor (TF) mutants. 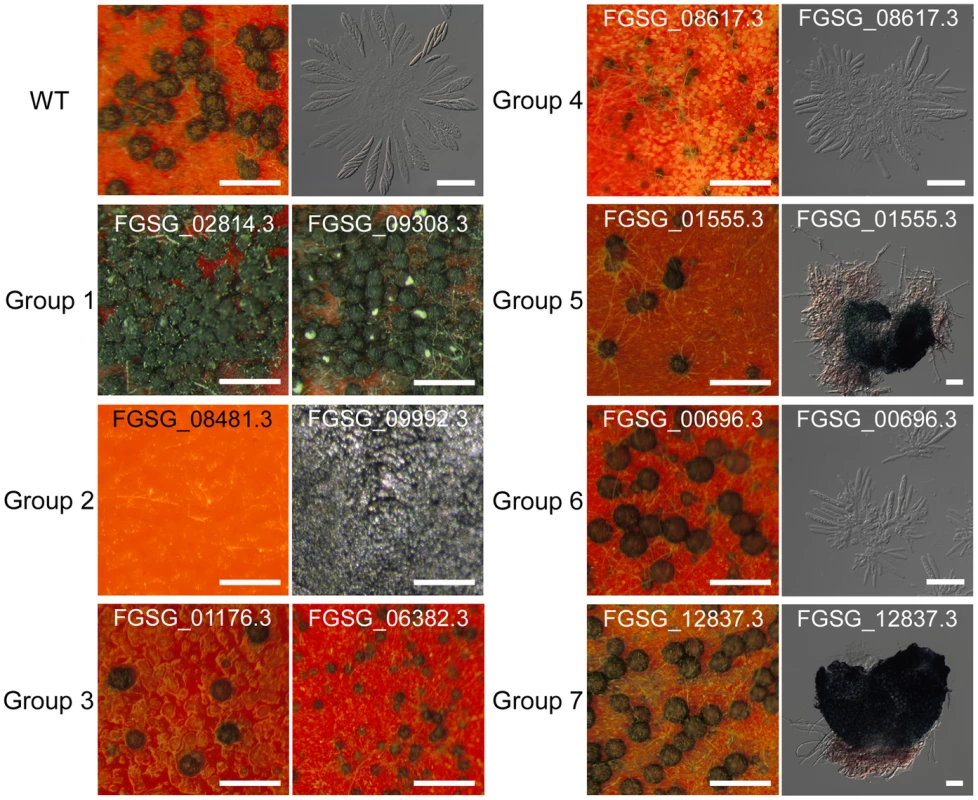
TF mutants are defects in sexual development 10 days after sexual induction using dissecting or differential interference contrast (DIC) microscopy. Scale bar = 0.5 mm (left panel in wild-type, for dissecting microscopic pictures), 40 µm (right panel in wild-type, for DIC images). Tab. 2. Classification of transcription factors (TFs) that are crucial for sexual development. 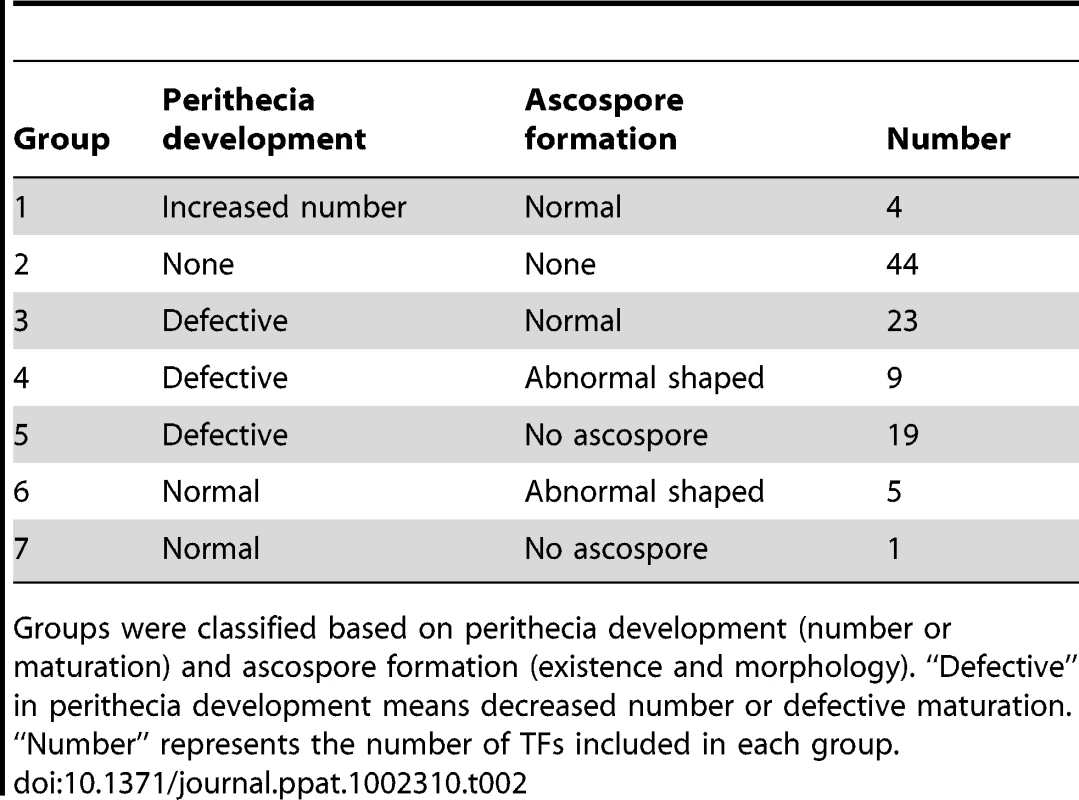
Groups were classified based on perithecia development (number or maturation) and ascospore formation (existence and morphology). “Defective” in perithecia development means decreased number or defective maturation. “Number” represents the number of TFs included in each group. Relationship between phenotype, TF conservation, and TF expression level
We utilized BLASTMatrix tool that is available on the Comparative Fungal Genomics Platform (CFGP, http://cfgp.riceblast.snu.ac.kr/) [26] to investigate whether TFs whose deletion resulted in a mutant phenotype are conserved in other fungal species. The distribution of each F. graminearum TF in other fungal species was surveyed using BLAST search scores. Total BLAST scores across the 164 species (159 fungi and five oomycetes) were obtained (Table S9). Higher scores for TFs imply that it is more distributed across fungal species. We divided the TF deletion mutants showing mutant phenotypes into subgroups based on the number of mutant phenotypes as well as phenotypic categories. The TFs whose mutants exhibited mutant phenotypes have higher BLAST scores than those TFs without mutant phenotypes (p = 1.01×10−11). In particular, TF mutants with more than six mutant phenotypes have significantly higher BLAST scores than those of TF mutants without a mutant phenotype (p<0.05, Fisher LSD test, Figure 4A and Table S10). Loss of TFs unique to F. graminearum showed mutant phenotypes in only 6% of the resulting mutants (2/31) compared to 26% of total tested F. graminearum TFs, and their expression levels were relatively low compared to the other 626 F. graminearum TFs. These unique TFs may function under specific conditions which we may not have tested in this study (Table S10).
Fig. 4. Relationship between multiple phenotypes, gene conservation in fungal kingdom, and transcription factor (TF) expression levels. 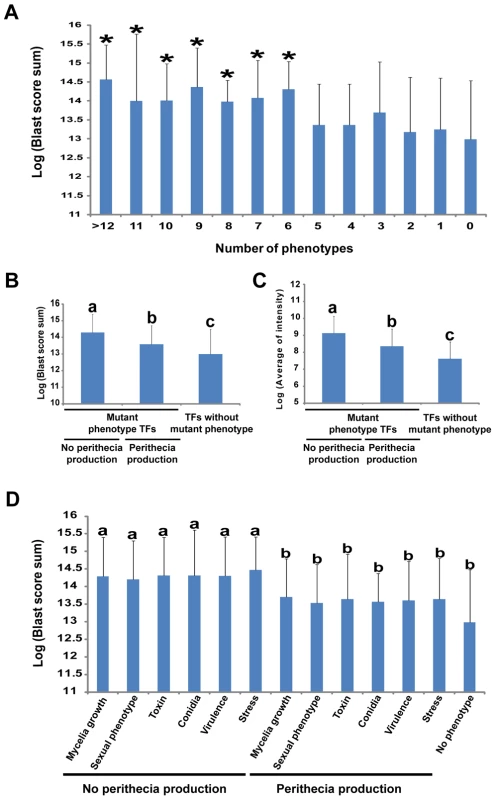
(A) Relationship between BLAST score sum and number of phenotype changes caused by a single TF deletion. Statistical analysis was performed using the Fisher LSD test. *p<0.05. (B) Correlation between TF conservation and various phenotypes with and without perithecia formation. (C) Comparison of expression level and mutant phenotype TFs with and without perithecia formation. Statistical analysis for (B) and (C) was performed using the Duncan multiple range test. Values with different superscript letters are significantly different (p<0.01). (D) Relationship between BLAST score sum and the no perithecia formation phenotype. Statistical analysis was performed using the Duncan multiple range test. Values with different superscript letters are significantly different (p<0.05). Phenotypic analysis revealed that perithecia development was identified as central among the tested phenotypes. Thus, we examined whether TFs that show perithecia development defects when deleted are widely distributed in other fungal species. Interestingly, TFs whose deletion results in no perithecia production are more widely distributed in other fungal species than those of TF whose loss results in no phenotype change (p<0.01, Duncan multiple range test, Figure 4B). Furthermore, TFs whose deletion results in no perithecia production showed significantly higher BLAST scores than those of TF whose loss results in normal perithecia production (p<0.05, Duncan's multiple range test, Figure 4D).
The relationship between function and gene expression of F. graminearum TFs was examined through comparison of the phenotype dataset with the normalized spot intensity of F. graminearum. We downloaded 121 slides from pLEXdb (http://www.plexdb.org/) and normalized spot intensity to measure the expression level of each gene. The normalized spot intensity of TFs was calculated for each slide and then an average intensity was determined as the level of gene expression (Table S11). The TF mutants with mutant phenotypes had higher levels of gene expression than those of TF whose deletion results in no mutant phenotype (p = 1.60×10−7). Mutant phenotype TFs showed higher levels of gene expression than those of TFs whose deletion results in no mutant phenotype, and mutant phenotype TFs with no perithecia production exhibited higher levels of gene expression than those of mutant phenotype TFs with normal perithecia production (p<0.01, Duncan's multiple range test, Figure 4C). However, no different expression patterns were observed among the TFs with mutant phenotypes in sexual development even in perithecia development-related microarrays (Figure S8) [5], [6]. Overall, phenotype and expression level of TFs as well as phenotype and distribution of TFs in other fungal species is highly correlated with F. graminearum TFs.
Interconnection of mutant phenotype TFs through phenotype, expression, and sequence
Dissecting transcriptional regulatory networks (TRNs) and determining which TFs participate in individual cellular processes and how they interact are main goals in the field of TF research. Analyses of mutants showing mutant phenotypes provide valuable information for addressing these questions. We attempted to identify a TRN in F. graminearum via construction of TF connections through patterns of gene expression and predicted protein-protein interaction. In order to analyze TF gene expression, we built F. graminearum gene transcript co-expression networks for the putative TFs based on PCC, calculated using the publicly available Affymetrix microarray data (Table S11). The use of PCC was successfully applied previously to validate other biological data [27]. We defined correlated and anticorrelated interactions through values of PCC (PCC>|0.75|) that is corresponding to approximately 0.6% portion of all PCC pairs (Table S12). We identified 426 TF pairs that have PCC>|0.75| (Table S12 and Figure 5A). Next, we predicted protein-protein interaction (PPI) among TF pairs using interologs in S. cerevisiae from the PPI database. Using this approach, 243 PPI pairs were predicted for F. graminearum TFs (Table S13 and Figure 5B). These co-expression and predicted PPI data were incorporated into our phenome dataset containing 170 TFs whose deletion results in at least one mutant phenotype (Figure 5C). Deletion of 35 out of 426 co-expressed TF pairs and 25 out of 243 predicted PPI pairs resulted in a mutant phenotype (Figure 5D). These interconnected 48 TFs have higher BLAST scores (p = 2.23×10−4) than the remaining 122 TFs whose deletion results in a mutant phenotype. Thus, these interconnected TFs, which are members of various subfamilies, may coregulate diverse functions.
Fig. 5. Transcription factors (TFs) with phenotype changes are interconnected either by co-expression or predicted protein-protein interaction (PPI). 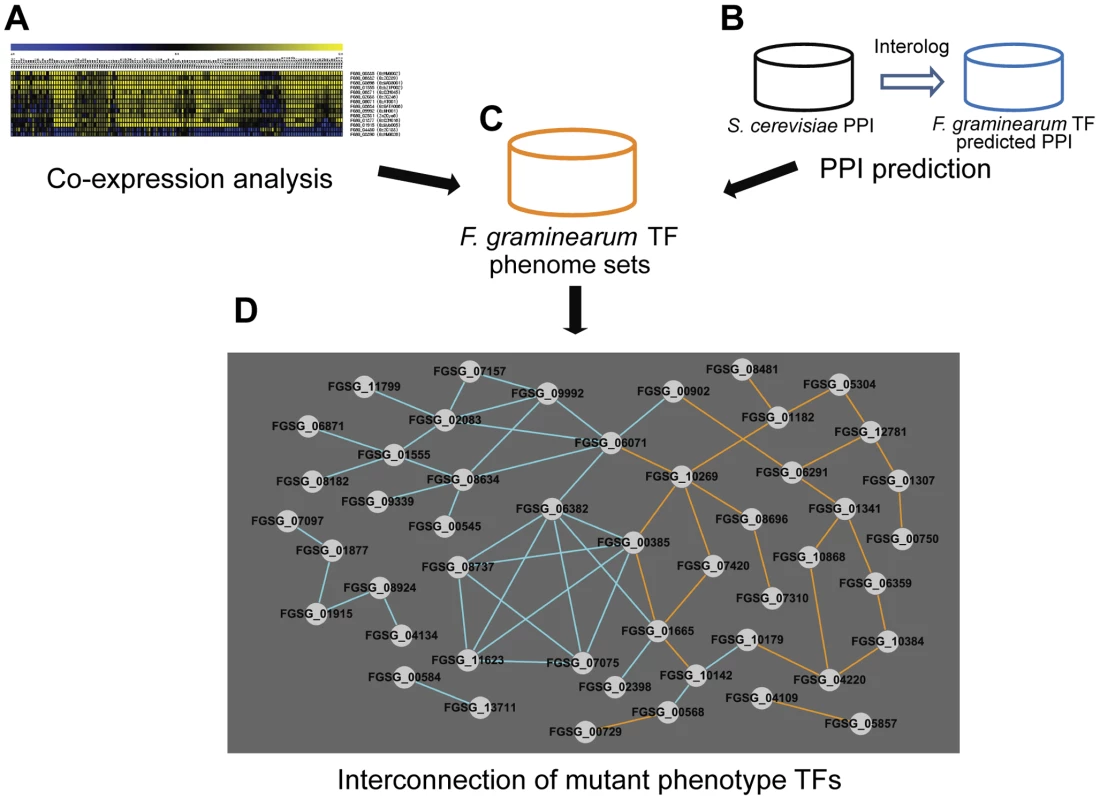
(A) Co-expression analysis (PCC>0.75 cut-off) of Fusarium graminearum TFs using public microarray data. PCC values were calculated using the 121 F. graminearum Affymetrix array data (Table S11). (B) Predicted PPI of F. graminearum TFs using the interolog map of the S. cerevisiae PPI dataset. (C) Phenome of F. graminearum TFs was generated. (D) Interconnection of mutant phenotype TFs through integration of co-expression (orange lines) and predicted PPI (blue lines). The nodes represent TFs whose knock-out displays a mutant phenotype. Discussion
Taking advantage of genome sequences of several model organisms, genome-wide high-throughput genetic screens have been performed through the use of gene disruption methods [28], [29] or RNA interference [30], [31]. For fungi, the construction of genome-wide gene deletion sets for the budding yeast S. cerevisiae [28], [32] and the fission yeast Schizosaccharomyces pombe [33] serve as valuable resources. In phytopathogenic fungi, genome-wide functional analysis of rice blast fungus, Magnaporthe oryzae, using insertional mutagenesis mediated by Agrobacterium transformation [34] facilitated analysis of the genetic basis of fungal pathogenicity.
In this study, we generated a comprehensive phenome of F. graminearum 657 TFs in 17 traits and analyzed shared and unique phenotypes of individual TF mutants. In most case, deletion of the mutant phenotype TFs resulted in pleiotropic phenotype changes compared to the wild-type strain, whereas 46 mutants were defected in single phenotypic category (Table S5). Especially, specific genes functional for virulence (7 TFs) or conidiation (4 TFs) had rarely been found in G. zeae. Research on these 46 TFs will expand our understanding on fungal biology.
Integration of data from other genome-wide analyses, such as transcriptome analysis and predicted PPI, into the constructed phenome, as well as investigation of the distribution pattern of TFs across the fungal kingdom revealed that deletion of single TFs that are widely-distributed in fungal species exhibited a greater chance of causing phenotypic defects. This observation was also supported by phenotypic analysis of 31 TFs unique to F. graminearum (Table S10). The result is similar with a hypothesis that essential genes are more evolutionary conserved than nonessential genes in bacteria because negative selection acted on more stringent essential genes [35]. Conserved TFs among fungal species might have more important functions in survival, development, and adaption.
The interaction and coordination of multiple TFs provides the ability to regulate cellular processes through fine-tuned transcriptional control. A recent study reported that high correlation between two phenotypes is highly predictive by known indicators of biological data such as protein-interactions [36]. Among the 170 TFs whose deletion resulted in mutant phenotypes, the 26 TFs whose connection was predicted using PPI are widely distributed among fungi and have higher gene expression levels compared to the other 144 TFs. Thus, these 26 TFs could be essential for the control of development and virulence in F. graminearum, as well as other fungi, and will be good resources to study the F. graminearum transcriptional network.
Our phenotypic data are mostly consistent with previously reported TF studies except the phenotype of Tri6 deletion mutant [37]. The previous report demonstrated that DON-nonproducing mutants showed reduced virulence on wheat head [37]. However, phenotype network in this study showed weak correlation (p-value = 3.25×10−12 and PCC = 0.5162) between virulence and DON production (cut-off with p-value<10−12 and PCC>0.6, Table S6 and Figure 2D). This discrepancy may be due to differences in experimental conditions and wheat cultivars. Differences in virulence between wheat plants inoculated with the wild-type strain and a DON-nonproducing mutant were more dramatic in the field tests than in controlled environment studies [37], [38]. In our study, we used a growth chamber for virulence test where the condition highly favors for disease development. In addition, the cultivar Eunpamil used for our study has been known to be highly susceptible to Fusarium head blight. Further virulence test in different field condition is necessary to resolve this discrepancy.
We revealed that there is no correlation between the stresses except between osmotic and ROS stress. The correlation between osmotic and ROS stress may be due to associated changes in membrane physiology. Further analysis of diverse stress responses in the 657 mutants generated in this study could provide a novel control strategy for F. graminearum.
Comparison of phenotypes between F. graminearum and N. crassa TF orthologs suggested that these TFs evolved divergently in how they control growth, asexual, and sexual development, rather than keeping the same function. These divergences may have been caused by differences in the life cycles between the two fungi. N. crassa is an obligate saprophyte that obtains its nutrients from dead or decaying matter, and F. graminearum is a facultative saprophytic plant pathogen that additionally needs specialized functions in order to obtain nutrients from plants. In addition, although both fungi might have similar features during sexual reproduction, i.e. repeat-induced point mutation, F. graminearum completes sexual reproduction homothallically, while N. crassa is heterothallic and the homothallic nature of F. graminearum may require unique pathways to complete sexual reproduction. The availability of a genome-wide TF deletion mutant library for F. graminearum, may provide a valuable resource for investigating sexual development and a reference for studying how different fungi have evolved to control various cellular processes at the transcriptional level.
Materials and Methods
Manual identification of 16 putative TFs
These TFs were identified based on the presence of specific motif associated with TFs or sequence homology with known TFs as listed in Table S1.
Fungal strains and media
G. zeae strain GZ3639 [39] was used for generating deletion mutants. All strains were stored as conidial suspensions in 20% glycerol at −70°C and deposited to the Center for Fungal Genetics Resources (CFGR, http://genebank.riceblast.snu.ac.kr/).Media were prepared and used according to the Fusarium laboratory manual [2].
DNA extraction, Southern blot, and PCR
Genomic DNA was extracted as previously described [2]. Restriction endonuclease digestion and Southern blot were performed following standard techniques [40]. PCR primers used in this study (Table S14) were synthesized by Bionics (Seoul, Korea).
Targeted gene deletion
Mutant alleles for targeted gene deletion were generated using the double-joint (DJ) PCR method [41]. A geneticin resistance gene cassette (gen) and the 5′ and 3′ flanking region of each TF gene were amplified from pII99 and GZ3639, respectively, and fused by DJ PCR using the conditions previously described [42]. Finally, split marker recombination was performed using primer sets from Table S14 [43]. At least three mutants were isolated for each gene deletion via co-dominant PCR screening. To exclude wild-type strain and ectopic mutants, (TF name)-5F/(TF name)-with 5F primers set was used. (TF name)-5F and Gen-with 5F primer set was used for positive selection of deletion mutants. In order to confirm a single copy integration of the mutant allele, Southern hybridization was performed (Figure S1).
Asexual development, stress test, and virulence test
In most cases, three independent mutants for each gene deletion were used for phenotyping with two replications. Radial growth on potato dextrose agar (PDA), minimal media (MM), and carrot agar were measured at three and five days after inoculation with freshly grown culture plugs from MM. The percentage of the average radial growth of mutants compared to the wild-type strain was calculated. Pigmentation of the bottom of PDA plates representing aurofusarin accumulation after culturing for five days was compared to wild-type strain and scored intensity of red color (0–6) with naked eyes.
Conidia production was measured by counting the number of conidia after culturing culture plugs from MM in 5 ml of carboxymethylcellulose medium (CMC) [44] for five days at 25°C on a rotary shaker (150 rpm). The virulence of fungal strains was determined on the susceptible wheat cultivar Eunpamil as previously described [24].
Various stress conditions were established in which the wild-type strain showed about one half of the radial growth observed on CM medium without any stress inducing agent: oxidative stress (5 mM hydrogen peroxide and 0.1 mM menadione) [45], osmotic stress (1 M NaCl, 1 M KCl, 1.5 M sorbitol, and 6 mM FeSO4), pH stress (pH = 4 and pH = 11), cell wall stress (60 mg/L Congo Red and 5 mg/L sodium dodecyl sulfate), DNA synthesis inhibition (8.6 mg/L fungicide iprodione), inhibition of mitogen-activated protein kinase pathway (0.023 mg/L fungicide fludioxonil). Radial growth of mutants on CM amended with each stress-inducing agent was evaluated compared to growth on CM.
Sexual development and mycotoxin analysis
Mycelia grown on carrot agar for five days were rubbed with a glass spreader after applying 2.5% sterilized Tween-60 solution to induce sexual reproduction [2]. After this treatment, all cultures were incubated under UV light (365 nm; HKiv Import & Export Co., Ltd., Xiamen, China) at 25°C. Perithecium growth was scored. Maturation of perithecia was determined by measuring their size. The presence of ascospores, their morphology, and ascospore discharge were also checked. In order to determine the effect of TF deletion on mycotoxin production, fungal strains were grown on 50 g of rice substrate for three weeks at 25°C in the dark. Rice cultures were harvested, and mycotoxins were extracted as previously described [46]. A portion of each extract was analyzed using TLC to quantify toxin production.
Construction of the co-expression network
We calculated Pearson correlation coefficient (PCC) scores to measure the correlation between expression of individual TF genes based on two sets of publicly available Affymetrix and NimbleGen microarray data. The raw Affymetrix data was downloaded from PLEXdb (http://www.plexdb.org/). We analyzed the raw Affymetrix data using MAS 5.0 R-package [47]. The trimmed mean target intensity of each array was arbitrarily set to 500, and the data were then log2 transformed. The mapping of Affymetrix probe sets onto F. graminearum genes was performed using the Fusarium graminearum Genome Database (FGDB).
TF conservation analysis
We utilized the BLASTMatrix tool that is available on the Comparative Fungal Genomics Platform (CFGP, http://cfgp.riceblast.snu.ac.kr/) for this analysis [26]. In brief, ungapped BLASTP with BLOSUM62 alignment scoring matrix was used without any filtering and BLAST scores calculated by “score = −log (expect)” equation were downloaded from the BLASTMatrix analysis results.
Prediction of F. graminearum TF protein-protein interactions (PPI)
Yeast interactome datasets were obtained from DIP (release 2010-6-14), BIOGRID (version 3.0.67), IntAct (release 2010-7-30), MINT (release 2010-07-27) and SGD (downloaded on 2010-8-2). The number of nonredundant PPIs is 88,720. Orthologs from yeast and F. graminearum were identified using Inparanoid version 4.1 [48], [49].
Microscopic observation
Microscopic observation was performed with a DE/Axio Imager A1 microscope (Carl Zeiss, Oberkochen, Germany).
Supporting Information
Zdroje
1. ShelestE 2008 Transcription factors in fungi. FEMS Microbiol Lett 286 145 151
2. LeslieJFSummerellBA 2006 The Fusarium laboratory manual Ames, IA Blackwell Pub
3. DesjardinsAEProctorRH 2007 Molecular biology of Fusarium mycotoxins. Int J Food Microbiol 119 47 50
4. CuomoCAGuldenerUXuJ-RTrailFTurgeonBG 2007 The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317 1400 1402
5. HallenHEHuebnerMShiuS-HGüldenerUTrailF 2007 Gene expression shifts during perithecium development in Gibberella zeae (anamorph Fusarium graminearum), with particular emphasis on ion transport proteins. Fungal Genet Biol 44 1146 1156
6. HallenHETrailF 2008 The L-type calcium ion channel Cch1 affects ascospore discharge and mycelial growth in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot Cell 7 415 424
7. LysøeESeongK-YKistlerHC 2011 The transcriptome of Fusarium graminearum during the infection of wheat. Mol Plant Microbe Interact 24 995 1000
8. GuentherJCHallen-AdamsHEBückingHShachar-HillYTrailF 2009 Triacylglyceride metabolism by Fusarium graminearum during colonization and sexual development on wheat. Mol Plant Microbe Interact 22 1492 1503
9. StephensAEGardinerDMWhiteRGMunnALMannersJM 2008 Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat. Mol Plant Microbe Interact 21 1571 1581
10. GardinerDMKazanKMannersJM 2009 Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol Plant Microbe Interact 22 1588 1600
11. SeongK-YZhaoXXuJ-RGüldenerUKistlerHC 2008 Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet Biol 45 389 399
12. GüldenerUSeongK-YBodduJChoSTrailF 2006 Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta. Fungal Genet Biol 43 316 325
13. ParkJParkJJangSKimSKongS 2008 FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24 1024 1025
14. KimJ-EJinJKimHKimJ-CYunS-H 2006 GIP2, a putative transcription factor that regulates the aurofusarin biosynthetic gene cluster in Gibberella zeae. Appl Environ Microbiol 72 1645 1652
15. SeongK-YPasqualiMZhouXSongJHilburnK 2009 Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol Microbiol 72 354 367
16. KimY-TLeeY-RJinJHanK-HKimH 2005 Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol Microbiol 58 1102 1113
17. MerhejJRichard-ForgetFBarreauC 2011 The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet Biol 48 275 284
18. LeeJLeeTLeeY-WYunS-HTurgeonBG 2003 Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol Microbiol 50 145 152
19. LysøeEPasqualiMBreakspearAKistlerHC 2011 The transcription factor FgStuAp influences spore development, pathogenicity, and secondary metabolism in Fusarium graminearum. Mol Plant Microbe Interact 24 54 67
20. SeongK-YHouZTracyMKistlerHCXuJ-R 2005 Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology 95 744 750
21. ShimW-BSagaramUSChoiY-ESoJWilkinsonHH 2006 FSR1 is essential for virulence and female fertility in Fusarium verticillioides and F. graminearum. Mol Plant Microbe Interact 19 725 733
22. WangYLiuWHouZWangCZhouX 2011 A novel transcriptional factor important for pathogenesis and ascosporogenesis in Fusarium graminearum. Mol Plant Microbe Interact 24 118 128
23. RiechmannJLHeardJMartinGReuberLJiangC-Z 2000 Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290 2105 2110
24. SonHLeeJParkARLeeY-W 2011 ATP citrate lyase is required for normal sexual and asexual development in Gibberella zeae. Fungal Genet Biol 48 408 417
25. ColotHVParkGTurnerGERingelbergCCrewCM 2006 A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103 10352 10357
26. ParkJParkBJungKJangSYuK 2008 CFGP: a web-based, comparative fungal genomics platform. Nucleic Acids Res 36 D562 D571
27. SeoY-SChernMBartleyLEHanMJungK-H 2011 Towards establishment of a rice stress response interactome. PLoS Genet 7 e1002020
28. TongAHYEvangelistaMParsonsABXuHBaderGD 2001 Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 2364 2368
29. BevanMWalshS 2005 The Arabidopsis genome: A foundation for plant research. Genome Res 15 1632 1642
30. BernsKHijmansEMMullendersJBrummelkampTRVeldsA 2004 A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428 431 437
31. AshrafiKChangFYWattsJLFraserAGKamathRS 2003 Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421 268 272
32. WinzelerEAShoemakerDDAstromoffALiangHAndersonK 1999 Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901 906
33. KimD-UHaylesJKimDWoodVParkH-O 2010 Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotech 28 617 623
34. JeonJParkS-YChiM-HChoiJParkJ 2007 Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet 39 561 565
35. JordanIKRogozinIBWolfYIKooninEV 2002 Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res 12 962 968
36. NicholsRJSenSChooYJBeltraoPZietekM 2011 Phenotypic landscape of a bacterial cell. Cell 144 143 156
37. ProctorRHHohnTMMcCormickSP 1995 Reduced virulence of Gibberella zeae caused by disruption of a trichthecene toxin biosynthetic gene. Mol Plant Microbe Interact 8 593 601
38. DesjardinsAEProctorRHBaiGMcCormickSPShanerG 1996 Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol Plant Microbe Interact 9 775 781
39. BowdenRLLeslieJF 1999 Sexual recombination in Gibberella zeae. Phytopathology 89 182 188
40. SambrookJRussellDW 2001 Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press
41. YuJ-HHamariZHanK-HSeoJ-AReyes-DominguezY 2004 Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41 973 981
42. HongS-YSoJLeeJMinKSonH 2010 Functional analyses of two syntaxin-like SNARE genes, GzSYN1 and GzSYN2, in the ascomycete Gibberella zeae. Fungal Genet Biol 47 364 372
43. CatlettNLLeeBNYoderOCTurgeonBG 2003 Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newsl 50 9 11
44. CappelliniRAPetersonJL 1965 Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia 57 962 966
45. BaiZHarveyLMMcNeilB 2003 Oxidative stress in submerged cultures of fungi. Crit Rev Biotechnol 23 267 302
46. SeoJ-AKimJ-CLeeD-HLeeY-W 1996 Variation in 8-ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia 134 31 37
47. Affymetrix 2001 Affymetrix Microarray Suite User Guide. Version 5 Santa Clara, CA Affymetrix
48. O'BrienKPRemmMSonnhammerELL 2005 Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33 D476 D480
49. YuHLuscombeNMLuHXZhuXXiaY 2004 Annotation transfer between genomes: Protein–protein interologs and protein–DNA regulogs. Genome Res 14 1107 1118
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Quorum Sensing in Fungi: Q&AČlánek Blood Feeding and Insulin-like Peptide 3 Stimulate Proliferation of Hemocytes in the MosquitoČlánek The DEAD-box RNA Helicase DDX6 is Required for Efficient Encapsidation of a Retroviral GenomeČlánek A Wide Extent of Inter-Strain Diversity in Virulent and Vaccine Strains of AlphaherpesvirusesČlánek The Anti-Sigma Factor TcdC Modulates Hypervirulence in an Epidemic BI/NAP1/027 Clinical Isolate ofČlánek Critical Roles for LIGHT and Its Receptors in Generating T Cell-Mediated Immunity during InfectionČlánek Frequent and Recent Human Acquisition of Simian Foamy Viruses Through Apes' Bites in Central Africa
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Quorum Sensing in Fungi: Q&A
- Discovery of an Ebolavirus-Like Filovirus in Europe
- Toll-like Receptor 7 Controls the Anti-Retroviral Germinal Center Response
- Tubule-Guided Cell-to-Cell Movement of a Plant Virus Requires Class XI Myosin Motors
- Herpesvirus Telomerase RNA (vTR) with a Mutated Template Sequence Abrogates Herpesvirus-Induced Lymphomagenesis
- Mitochondrial Peroxiredoxin Plays a Crucial Peroxidase-Unrelated Role during Infection: Insight into Its Novel Chaperone Activity
- Sustained CD8+ T Cell Memory Inflation after Infection with a Single-Cycle Cytomegalovirus
- Novel Mouse Xenograft Models Reveal a Critical Role of CD4 T Cells in the Proliferation of EBV-Infected T and NK Cells
- Toll-8/Tollo Negatively Regulates Antimicrobial Response in the Respiratory Epithelium
- Exhausted Cytotoxic Control of Epstein-Barr Virus in Human Lupus
- Structural and Functional Analysis of Laninamivir and its Octanoate Prodrug Reveals Group Specific Mechanisms for Influenza NA Inhibition
- Infection Drives IL-17-Mediated Neutrophilic Allergic Airways Disease
- Blood Feeding and Insulin-like Peptide 3 Stimulate Proliferation of Hemocytes in the Mosquito
- HIV-1 Replication in the Central Nervous System Occurs in Two Distinct Cell Types
- Deep Molecular Characterization of HIV-1 Dynamics under Suppressive HAART
- Fitness Landscape of Antibiotic Tolerance in Biofilms
- The DEAD-box RNA Helicase DDX6 is Required for Efficient Encapsidation of a Retroviral Genome
- Preventing Sepsis through the Inhibition of Its Agglutination in Blood
- A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus,
- IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry
- Targeting Cattle-Borne Zoonoses and Cattle Pathogens Using a Novel Trypanosomatid-Based Delivery System
- A Wide Extent of Inter-Strain Diversity in Virulent and Vaccine Strains of Alphaherpesviruses
- Coordinated Destruction of Cellular Messages in Translation Complexes by the Gammaherpesvirus Host Shutoff Factor and the Mammalian Exonuclease Xrn1
- Signal Transduction through CsrRS Confers an Invasive Phenotype in Group A
- Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex
- Histone Deacetylase 8 Is Required for Centrosome Cohesion and Influenza A Virus Entry
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis
- Co-opts the FGF2 Signaling Pathway to Enhance Infection
- IRAK-2 Regulates IL-1-Mediated Pathogenic Th17 Cell Development in Helminthic Infection
- Trafficking of Hepatitis C Virus Core Protein during Virus Particle Assembly
- The Anti-interferon Activity of Conserved Viral dUTPase ORF54 is Essential for an Effective MHV-68 Infection
- A Viral Nuclear Noncoding RNA Binds Re-localized Poly(A) Binding Protein and Is Required for Late KSHV Gene Expression
- Suppression of Methylation-Mediated Transcriptional Gene Silencing by βC1-SAHH Protein Interaction during Geminivirus-Betasatellite Infection
- ISG15 Is Critical in the Control of Chikungunya Virus Infection Independent of UbE1L Mediated Conjugation
- Non-Hematopoietic Cells in Lymph Nodes Drive Memory CD8 T Cell Inflation during Murine Cytomegalovirus Infection
- RNA Polymerase II Stalling Promotes Nucleosome Occlusion and pTEFb Recruitment to Drive Immortalization by Epstein-Barr Virus
- Noninfectious Retrovirus Particles Drive the / Dependent Neutralizing Antibody Response
- Endophytic Life Strategies Decoded by Genome and Transcriptome Analyses of the Mutualistic Root Symbiont
- An Integrated Approach to Elucidate the Intra-Viral and Viral-Cellular Protein Interaction Networks of a Gamma-Herpesvirus
- as an Animal Model for the Study of Biofilm Infections
- Homeostatic Proliferation Fails to Efficiently Reactivate HIV-1 Latently Infected Central Memory CD4+ T Cells
- The Anti-Sigma Factor TcdC Modulates Hypervirulence in an Epidemic BI/NAP1/027 Clinical Isolate of
- Enhances Protective and Detrimental HLA Class I-Mediated Immunity in Chronic Viral Infection
- The Mouse IAPE Endogenous Retrovirus Can Infect Cells through Any of the Five GPI-Anchored EphrinA Proteins
- The Urgent Need for Robust Coral Disease Diagnostics
- HacA-Independent Functions of the ER Stress Sensor IreA Synergize with the Canonical UPR to Influence Virulence Traits in
- A Novel Core Genome-Encoded Superantigen Contributes to Lethality of Community-Associated MRSA Necrotizing Pneumonia
- Critical Roles for LIGHT and Its Receptors in Generating T Cell-Mediated Immunity during Infection
- The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors
- Frequent and Recent Human Acquisition of Simian Foamy Viruses Through Apes' Bites in Central Africa
- Mechanisms of Trafficking to the Brain
- Defining Emerging Roles for NF-κB in Antivirus Responses: Revisiting the Enhanceosome Paradigm
- The Role of Sialyl Glycan Recognition in Host Tissue Tropism of the Avian Parasite
- Evolutionarily Divergent, Unstable Filamentous Actin Is Essential for Gliding Motility in Apicomplexan Parasites
- The Herpes Simplex Virus-1 Transactivator Infected Cell Protein-4 Drives VEGF-A Dependent Neovascularization
- Distinct Single Amino Acid Replacements in the Control of Virulence Regulator Protein Differentially Impact Streptococcal Pathogenesis
- Soluble Rhesus Lymphocryptovirus gp350 Protects against Infection and Reduces Viral Loads in Animals that Become Infected with Virus after Challenge
- A Genetic Screen Reveals Arabidopsis Stomatal and/or Apoplastic Defenses against pv. DC3000
- Hepatitis C Virus Reveals a Novel Early Control in Acute Immune Response
- Fumarate Reductase Activity Maintains an Energized Membrane in Anaerobic
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis
- The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors
- Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex
- Evolutionarily Divergent, Unstable Filamentous Actin Is Essential for Gliding Motility in Apicomplexan Parasites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



