-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Urgent Need for Robust Coral Disease Diagnostics
Coral disease has emerged over recent decades as a significant threat to coral reef ecosystems, with declines in coral cover and diversity of Caribbean reefs providing an example of the potential impacts of disease at regional scales. If similar trends are to be mitigated or avoided on reefs worldwide, a deeper understanding of the factors underlying the origin and spread of coral diseases and the steps that can be taken to prevent, control, or reduce their impacts is required. In recent years, an increased focus on coral microbiology and the application of classic culture techniques and emerging molecular technologies has revealed several coral pathogens that could serve as targets for novel coral disease diagnostic tools. The ability to detect and quantify microbial agents identified as indicators of coral disease will aid in the elucidation of disease causation and facilitate coral disease detection and diagnosis, pathogen monitoring in individuals and ecosystems, and identification of pathogen sources, vectors, and reservoirs. This information will advance the field of coral disease research and contribute knowledge necessary for effective coral reef management. This paper establishes the need for sensitive and specific molecular-based coral pathogen detection, outlines the emerging technologies that could serve as the basis of a new generation of coral disease diagnostic assays, and addresses the unique challenges inherent to the application of these techniques to environmentally derived coral samples.
Published in the journal: The Urgent Need for Robust Coral Disease Diagnostics. PLoS Pathog 7(10): e32767. doi:10.1371/journal.ppat.1002183
Category: Review
doi: https://doi.org/10.1371/journal.ppat.1002183Summary
Coral disease has emerged over recent decades as a significant threat to coral reef ecosystems, with declines in coral cover and diversity of Caribbean reefs providing an example of the potential impacts of disease at regional scales. If similar trends are to be mitigated or avoided on reefs worldwide, a deeper understanding of the factors underlying the origin and spread of coral diseases and the steps that can be taken to prevent, control, or reduce their impacts is required. In recent years, an increased focus on coral microbiology and the application of classic culture techniques and emerging molecular technologies has revealed several coral pathogens that could serve as targets for novel coral disease diagnostic tools. The ability to detect and quantify microbial agents identified as indicators of coral disease will aid in the elucidation of disease causation and facilitate coral disease detection and diagnosis, pathogen monitoring in individuals and ecosystems, and identification of pathogen sources, vectors, and reservoirs. This information will advance the field of coral disease research and contribute knowledge necessary for effective coral reef management. This paper establishes the need for sensitive and specific molecular-based coral pathogen detection, outlines the emerging technologies that could serve as the basis of a new generation of coral disease diagnostic assays, and addresses the unique challenges inherent to the application of these techniques to environmentally derived coral samples.
The Need for Improved Coral Disease Diagnostic Tools
The world's coral reefs are in decline, with hard coral cover on Caribbean reefs decreasing by an average of 80% in the last 30 years [1] and Indo-Pacific reefs suffering an estimated coral cover loss of 50% over the same period [2]. The causes of these declines are diverse and complex, including water pollution, habitat destruction, overfishing, invasive species, and global climate change [3]–[5]. In recent years, coral diseases have also emerged as a significant threat to the world's coral reef ecosystems [6], [7]. Since the first coral disease was described in 1973, evidence from field studies documenting the population and community-level impacts of disease on coral reef ecosystems worldwide has been accumulating (reviewed in [8]) [9]–[14] and it is now clear that coral diseases have the potential to cause widespread mortality and significantly alter reef community structure (e.g., [9], [15]–[17]).
Despite the serious threat that coral diseases pose to the health of reef ecosystems globally, little is known about many of these diseases, including their etiologies, transmission dynamics, and the steps that can be taken to prevent, control, or reduce their impacts. This work has been frustrated by the inability to determine etiological agents for many diseases (see Box 1), insufficient diagnostic tools, and limited application of established biomedical diagnostic methods [18]. Current diagnostics focus on documenting disease signs in situ, describing macroscopic characteristics such as species affected, extent and pattern of tissue loss [19], presence and appearance of microbial mats [15], abnormal coloration [20], or skeletal anomalies [21]. Corals display few macroscopic signs indicative of stress and consequently an array of maladies, including environmental stress, predation, and infectious disease, are often manifested as a paling or sloughing of the coral tissue. For example, more than six “white” diseases, which are characterized by a spreading zone of tissue loss, exposing white coral skeleton directly adjacent to asymptomatic coral tissue, have been described in the Caribbean alone (Figure 1) [16]. Because of their nearly identical appearance, several of these diseases (e.g., white plague I and white plague II) are differentiated almost exclusively by the rate of lesion progression over the infected colony [16]. Such difficulties have resulted in cases of misidentified diseases, repeated name changes for the same disease [22], and even classification of predation scars as disease [10], [23]. Currently, it is uncertain how many distinct coral diseases exist worldwide; in two articles published in the same year, one report identified 18 diseases [16], whereas another put the number at 29 [8]. This confusion underlines the need for more robust coral disease diagnostic methods.
Fig. 1. Examples of white diseases affecting Scleractinian corals. 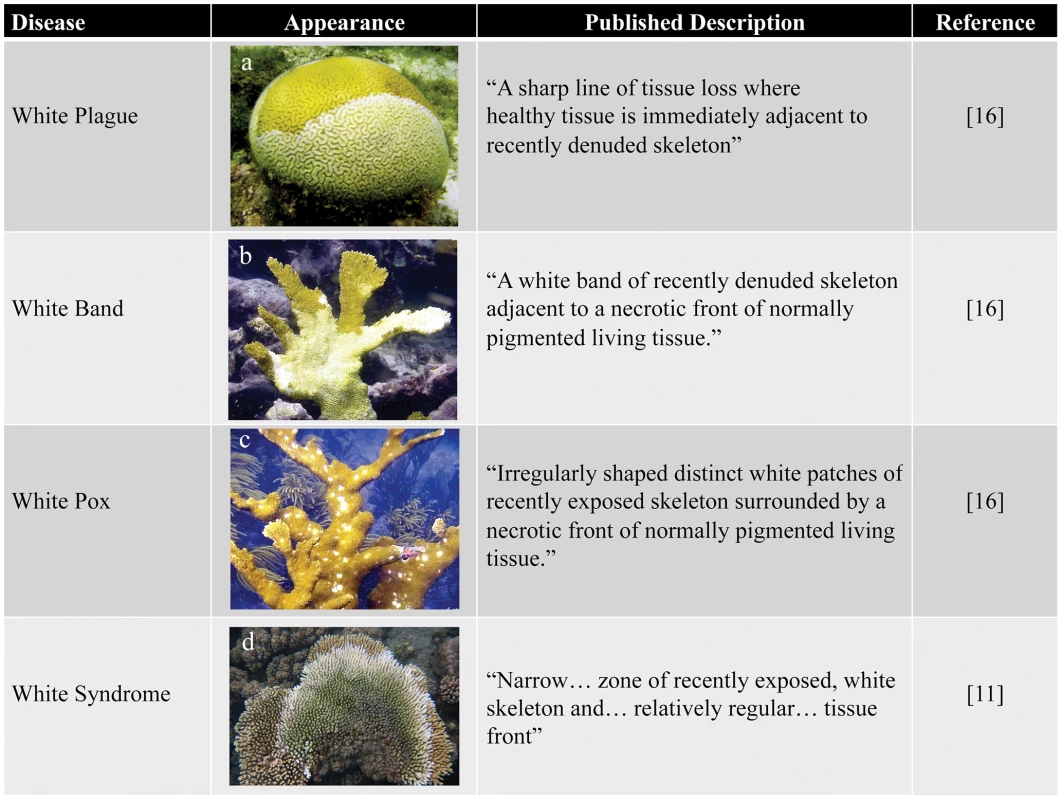
(a) White plague in Diploria labyrinthiformis; (b) white band in Acropora palmate; (c) white pox in A. palmate; and (d) white syndrome in A. millepora. Photos courtesy of Ernesto Weil. Box 1. The Need for Surrogate Models to Study Coral Disease
The development of effective coral disease diagnostics requires efficient methods for identifying microbial drivers of disease and an understanding of the pathogenesis of identified disease agents. In the field of human health, investigations using surrogate hosts, including the common rat (Rattus norvegicus), the house mouse (Mus musculus), and the common fruit fly (Drosophila melanogaster), have been integral to unraveling the intricate interactions between host, pathogen, and environment that lead to disease. A few coral species, including A. millepora in the Indo-Pacific and A. palmata in the Caribbean, have emerged as “lab rats” for the study of coral genetics [88], [89], physiology [90], [91], and health [92], [93]. However, our ability to study coral disease pathogenesis in the laboratory has been limited by: the complexity of the coral holobiont, which comprises animal, dinoflagellate, and microbial partners; a poorly understood coral immune system (see Box 2); and difficulties associated with sourcing and rearing these sensitive and often protected species [94]. For example, since A. palamata, which was once the dominant coral species throughout much of the Caribbean, was added to the IUCN Red List of critically endangered species [95], acquiring specimens for experimentation has become much more difficult both logistically and morally. By focusing disease investigations on surrogate models for the coral host, researchers may be able to overcome these limitations while still gaining valuable insights into the complex interactions between host, environment, and pathogen that lead to disease in corals.
In search of alternative surrogates for the coral animal, researchers have explored cnidarians in the class Anthozoa, including the Symbiodinium-harboring, tropical anemone Aiptasia sp. [96], as well as more distantly related hydrozoans, such as freshwater Hydra species [97]. Research on Aiptasia, for example, has provided insights into the physiological responses of anthozoans and their algal symbionts to thermal stress and bleaching [98], [99], and Hydra species have been used to explore the development and maintenance of cnidarian-associated bacterial communities [97]. These readily available, easily reared, and phylogenetically closely related coral analogues could also provide insights into the role of coral-associated microbes as mutualistic, commensal, or pathogenic and potentially reveal the functional pathogenesis mechanisms of identified pathogens.
Once potential pathogens are identified and their virulence mechanisms determined in surrogate hosts, researchers must confirm these findings within the complex coral holobiont. Captive-bred coral juveniles and laboratory-maintained Symbiodinium cultures provide easily replicated and environmentally responsible alternatives to wild-harvested adult colonies for laboratory-based experimentation. Both brooding and mass spawning corals provide thousands of genotypically similar coral juveniles from just a single pair. Although many spawning species breed during short periods each year [100], limiting the availability of juveniles to researchers, some brooding corals release gametes much more frequently [101]. Furthermore, juveniles of some coral species can be maintained Symbiodinium free for weeks, allowing researchers to control the algal symbionts they uptake [102]. Many research laboratories currently possess pure and mixed cultures of coral-derived Symbiodinium, which could be used to seed juveniles or directly test the effect of putative pathogens on the Symbiodinium themselves. An experimental system comprised of Symbiodinium cultures, asymbiotic, and symbiotic juveniles (or other Symbiodinium-harboring cnidarians such as Aiptasia) would allow researchers to tease out the targets of specific pathogens within the coral holobiont. By focusing research on a few, well-chosen model systems, researchers will be better able to identify potential pathogens and study their virulence mechanisms in an efficient, environmentally friendly, and easily comparable manner.
In recent years, an increased interest in coral microbiology, in combination with the application of histology and biomedical approaches, has revealed several bacterial species linked to coral disease lesions [6], [24], [25]. Debate exists as to the primacy of a compromised coral host versus opportunistically proliferating bacteria in causing coral diseases [26], [27]. However, since disease is classically considered to be the outcome of interactions among a causative agent, susceptible host, and the environment (e.g., [28]), debating the status of an etiological agent as either a primary or secondary pathogen is diversionary and does not negate the need to understand its role in pathogenesis [18], [29]. While coral immunity plays a critical role in maintaining coral health and indicators of coral immune status can provide insight into the health state of the coral host (summarized in Box 2), this is not the focus of this review. Here we discuss the use of identified coral pathogens or disease indicators as targets for a new generation of sensitive and highly specific, molecular-based diagnostic assays that can begin to address many of the basic questions that plague the field of coral disease research.
Box 2. The Diagnostic Potential of Coral Immunity
Recent reviews have highlighted immunological indicators of coral stress and disease [16], [103], [104], thus here we briefly discuss coral immune response as a proxy for disease susceptibility and as an indicator of past or present exposure to pathogens or other stressors (e.g., high water temperatures, excessive UV exposure). Corals, like other invertebrates, are limited to innate immunity [105], which is defined as the ability of certain cells and cellular mechanisms to defend the host from infection by other organisms in a nonspecific manner [106]. Much work is currently focused on the use of coral host factors and immunological responses as indicators of coral stress and disease [107], [108]. For corals and other marine invertebrates, phagocytosis provides the first line of cellular defense [109], [110]. In response to invasion by a pathogen, corals increase production of motile phagocytic cells, also known as amoebocytes, that migrate from healthy coral tissues to the site of infection and either attack the invading pathogens via phagocytosis or contribute to healing and regeneration of the damaged tissue [104], [107]. Histology is a well-established technique for detection and quantification of amoebocytes in the coral host. The examination of specially stained histological slides has been used to detect amoebocyte accumulation in response to a range of insults including sedimentation [111], skeletal anomalies [112], and disease [107].
Exposure of corals to pathogens also induces production of antibiotic compounds, which may instill some resistance against invading microbes. Gorgonian corals have been shown to resist infection by the fungus A. sydowii, the causative agent of aspergillosis, through the production of antifungal agents that inhibit germination of A. sydowii spores [113], [114]. White syndrome and yellow band disease have also been shown to induce antimicrobial activity in scleractinian corals [115], [116]. Methods exist for the detection of antimicrobial residues in animals [117] and analogous assays could easily be adapted for corals.
Recent investigations have revealed the melanization cascade to be an integral component of coral immunity. The melanization cascade involves the production of prophenoloxidase (PPO), which is involved in wound healing, encapsulation, and disease resistance [107], [103]. PPO serves as the precursor molecule of phenoloxidase (PO), which is activated by proteases during active pathogen invasion and in turn induces the deposition of melanin, the endpoint of the cascade and a potent physiochemical barrier [107], [108]. Melanin has antimicrobial and cytotoxic attributes, and therefore its presence in stressed and diseased corals implies the activation of innate immune responses. Assays to detect PO and melanin in coral samples have been developed [108], [112], which could be included in future disease studies [103]. The ability to detect and quantify amoebocytes, antimicrobial compounds, melanin deposits, and the precursors of melanization, including PPO and PO, will provide proxies for immune response in corals. Although immune response is not a direct indicator of disease, these parameters could be used to assess coral health, disease susceptibility, and past or present exposure to pathogens or other stressors.
Benefits of Pathogen-Specific Detection Tools
In this section, we highlight the role that specific and sensitive pathogen detection will play in advancing our understanding of the etiology, spread, and ultimately management of coral diseases.
Detecting Shifts in Coral-Associated Microbial Communities
The coral holobiont comprises a complex association between the coral animal and its microbial partners, including symbiotic dinoflagellates (zooxanthellae) [30], bacteria [14], [31], archaea [32], [33]), viruses [34], endolithic algae [35], and fungi [36]. Numerous studies have examined these associations in both healthy and stressed corals and it has been suggested that shifts in microbial communities can act as indicators of coral stress [37]–[39]. For example, Pantos et al. [37] demonstrated bacterial community shifts throughout the entire coral colony, even when just a small part of the colony showed signs of disease, and Bourne et al. [38] reported shifts in coral-associated microbial communities well before the appearance of visual signs of thermal bleaching. Using metagenomic approaches, Vega-Thurber et al. [39] demonstrated functional gene shifts, including an increased abundance of virulence genes, in coral microbial partners during temperature, nutrient, and pH stress, although it should be noted that this study did not quantify expression of these genes. Additionally, Kimes et al. [40] observed significant differences in biogeochemical cycling-related genes between healthy and yellow-band infected Montastraea faveolata colonies. These community-level bacterial profiling approaches facilitate diagnosis at the earliest stages of infection when mitigation measures would be most effective [41]. Therefore, the development of rapid and sensitive assays to monitor coral-associated microbial communities as proxies for coral health should be a research focus.
Better Understanding of Disease Etiology
While some coral diseases are tightly linked with the presence of a specific pathogen, the causes of many other diseases and disease-like syndromes remain elusive [41]. Better tools with high specificity for target pathogens would enable investigations of the circumstances under which microbes that are normally found on corals become pathogenic and the conditions and mechanisms that trigger a switch from commensal or neutral to pathogenic. Moreover, there are cases where bacterial species, which were linked to specific diseases in early studies, no longer elicit the same response or are not associated with disease signs, potentially indicating development of disease resistance [6], [42]. For example, Vibrio shiloi, which was initially identified as the agent responsible for annual bleaching of the Mediterranean coral Oculina patagonica, no longer appears to cause bleaching in this coral species [27], [43]. Additionally, Aspergillus sydowii, which was shown to cause disease in gorgonians, has also been found on healthy coral colonies, leading Toledo-Hernandez et al. [44] to raise questions about its role in disease onset. The development of tools to detect and quantify putative pathogens in both controlled laboratory experiments and environmentally derived samples will help to establish the etiology of specific coral diseases and clarify the role of individual microbes in the onset of disease lesions. Once the link between a specific microbial entity and lesion onset is established, pathogen-specific assays can provide information on all aspects of the disease onset process.
Monitoring Pathogen Load
Emerging evidence suggests that the abundance of coral pathogens varies on reefs throughout the year and within coral hosts during the course of infection [45]–[47]. The ability to quantify pathogen load in coral and environmental samples will allow researchers and reef managers to gauge the health status of individual corals, assess the impact of environmental parameters (e.g., temperature, nutrient load, sedimentation rate) on pathogen load, and better predict large-scale disease outbreaks. Some efforts have been made to establish links between environmental parameters and coral disease prevalence. Using high-resolution satellite datasets and long-term coral disease surveys, Bruno et al. [2] established a link between coral disease outbreaks and warm temperature anomalies at sites with high coral cover. By monitoring bacterial communities in situ, Vezzulli et al. [47] also discovered a link between mass mortality events of the coral Paramuricea clavata and seawater temperatures, chlorophyll concentrations, and the presence of culturable Vibrio spp. in the surrounding seawater. Tools for monitoring pathogen density would provide a deeper understanding of how pathogen load and virulence respond to natural (e.g., seasonal, El Niño/La Niña) and anthropogenic (e.g., pesticide and nutrient influx, sedimentation) fluctuations, allowing researchers and managers to closely follow these dynamics and model pathogen response to environmental change.
Identifying Pathogen Sources, Vectors, and Reservoirs
It is currently unclear if the emergence of coral diseases on reefs is associated with the introduction of pathogenic organisms, or whether potentially pathogenic microbes are a normal component of reef ecosystems that increase in virulence because of altered environmental conditions and/or reduced host resistance. To better understand the dynamics of coral disease outbreaks and ensure that they are effectively managed, information regarding pathogen sources, vectors, and reservoirs is needed. Pathogen sources are the avenues through which a pathogen enters the environment, reservoirs are biotic or abiotic entities that harbor a pathogen, and vectors are living entities that do not cause or suffer from a disease, but transmit a pathogen from one host to another [28]. The identification of the marine fireworm as the winter reservoir and spring/summer vector of the coral pathogen V. shiloi nicely demonstrates the utility and importance of molecular-based pathogen detection techniques in the study of coral epidemiology [46].
Better Informed Management Decisions
To effectively manage coral disease outbreaks, a deeper understanding of the causes of observed diseases, how they are spread between colonies and populations, and how environmental parameters influence pathogen virulence and host susceptibility to infection is required [48]. Tools that increase our capacity to establish links between disease signs and the presence of specific microbial agents will improve coral disease classification and diagnosis. These capabilities will help reef managers to discern the threats that impact the occurrence, prevalence, and severity of diseases so their sources can be identified and possibly reduced through better management practices [48]. For example, habitat degradation, poor water quality, and warming seas are often speculated as causes of the recent rise in coral diseases [13], but few studies have directly linked specific factors with increases in coral disease. By understanding the relationship between various stressors and the occurrence of coral diseases, managers may be able to identify potential threats in a timely manner and develop strategies to lessen their impacts [48]. Several biological controls for coral diseases, including bacteriophage therapy and probiotic addition, have recently been proposed [41], [49]. Pathogen-specific diagnostics could be used to identify where and when these controls should be implemented and also assess their efficacy. In order to assist resource managers to combat disease epizootics, prevent future outbreaks, and reduce the time needed for recovery, the development of sensitive, specific, and robust coral disease diagnostics should be an essential research priority [7].
Pathogen Detection Methods
Effective diagnostic tools must be sensitive, reproducible, and specific in their detection of targeted microbial organisms. In the field of human pathogen detection, culture and colony counting, immunology, and nucleic acid-based methods are most commonly used [50]. Here, we provide a brief overview of these techniques and evaluate their potential for coral pathogen detection (summarized in Table 1].
Tab. 1. Summary of pathogen detection techniques and molecular diagnostics. 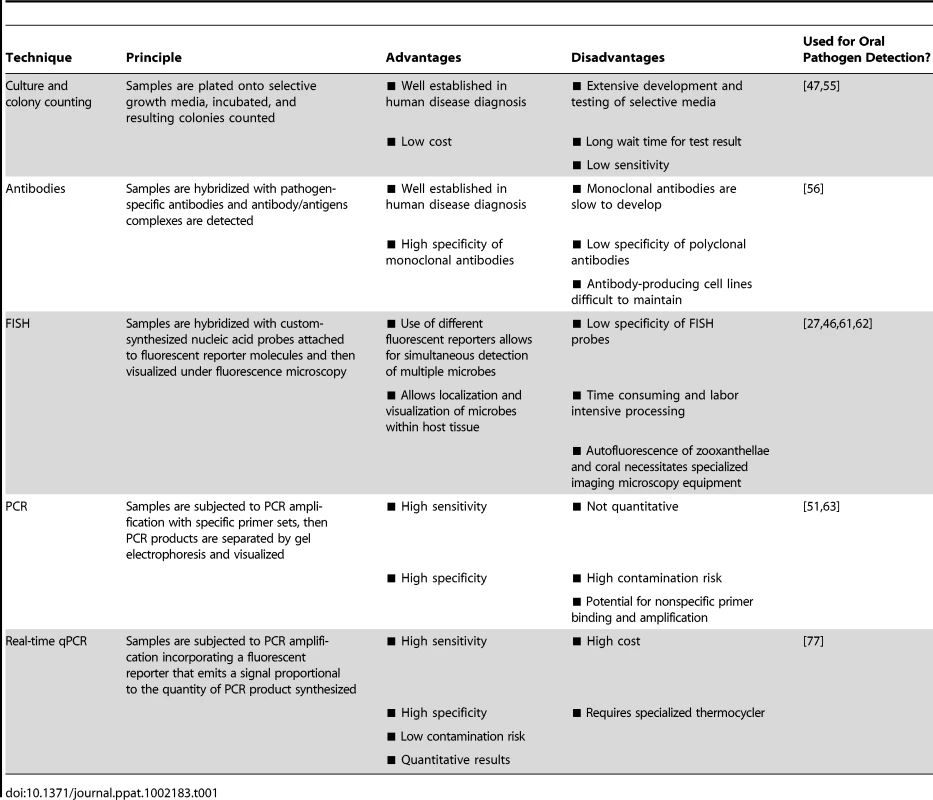
Culture-Based Detection
The culture and plating method is the oldest bacterial detection technique and remains a cornerstone of human pathogen detection. This method involves plating of samples onto selective growth media followed by an incubation period and then colony counting. Specialized growth media can contain inhibitors of nontarget species/strains, substrates that only the targeted microbe can degrade, and/or substances that confer a particular color to the growing colonies [50]. However, selective media take time to develop and test, and even when selective media are available for a pathogen of interest, culture and plating techniques are excessively time consuming and less sensitive than immunologic or genetic-based techniques [51], [52]. For corals, standardized diagnostics based on culture-dependent methods are limited, largely due to the lack of selective media capable of promoting growth of specific pathogens amongst the highly complex, diverse, and abundant microbial populations associated with compromised coral tissues. For example, selective media, such as thiosulfate citrate bile sucrose (TCBS), have been developed to discriminate Vibrio bacteria from other bacterial species. While known coral pathogens, including V. coralliilyticus and V. shiloi, can be grown on TCBS agar, they cannot be effectively discriminated from other Vibrio species [53] that coexist within the coral holobiont. In order to overcome this limitation, Ritchie et al. [54] developed a method to discriminate potential bacterial invaders from normal residents of the coral holobiont by including sterile coral mucus into the growth media. This innovative approach is based upon the assumption that symbiotic coral-associated bacteria will be resistant to the antibiotic properties of coral mucus while opportunistic pathogens will not. By comparing the bacterial strains growing on mucus-treated media plates to those growing on control media plates, it is theoretically possible to separate coral associated bacterial residents from potentially invasive visitors. While this technique is useful for identifying potential pathogens, the processes is extremely time consuming, requiring isolation of individual colonies, PCR amplification, and sequencing, and does not allow specific detection at the single pathogen level.
In a few cases, culture protocols have been developed to selectively grow specific coral-associated microbes. For example, Sutherland et al. [55] developed a technique to isolate Serratia marcescens, the presumed etiological agent of white pox in the Caribbean, involving two subsequent colorimetric culture steps followed by inoculation onto nonselective media. Interestingly, this method revealed human sewage to be a likely source of the pathogen on reefs in the Florida Keys. Where appropriate selective media exist, most probable number (MPN) methods can be used to estimate the concentration of bacteria [47]. MPN involves serially diluting samples into appropriate media, further dividing these dilutions into replicate aliquots, culturing, and assigning a binomial (growth versus no growth) score to the resulting cultures. This method can be used to estimate the concentration of certain bacterial groups in a given sample; however, the dilution and culturing steps can be time consuming and reproducibility is often an issue. Due to the high diversity of microbes present in coral samples, lack of appropriate media for many coral pathogens, and the low sensitivity and long processing time required, culture-based diagnostic methods are not the ideal platform for coral pathogen detection.
Immunology-Based Detection
The use of antibody technology is well established in human medical diagnostics and has been applied with some success to the detection of coral pathogens. Immunology-based pathogen monitoring involves the production of either polyclonal or monoclonal antibodies and the detection of antibody/antigen complexes that indicate the presence of the targeted pathogen within a sample. Specific anti-V. shiloi antibodies have provided insight into the dynamics of pathogen invasion and spread within the O. patagonica coral host, suggesting a temperature-dependent host defense against the pathogen [56]. However, immunology-based techniques can only be developed once specific pathogens have been identified and successfully cultured (see Box 1). Furthermore, polyclonal antibodies often have low specificity [57] and highly specific monoclonal antibodies are generally slow to develop and expensive to produce and maintain. While immunology-based coral pathogen detection is feasible once specific pathogens have been successfully isolated, the cost and effort required to develop and maintain antibody-producing cell lines may limit its utility in routine monitoring. However, if adapted into routine assays such as ELISA, common in many human health targeted kits such as pregnancy tests [58], [59], this approach has the potential to provide rapid coral pathogen detection.
Nucleic Acid-Based Detection
Nucleic acid-based techniques using molecular probes and/or PCR offer an appealing alternative to culture and immunology-based methods because of their potential for high specificity and sensitivity. Here we discuss the utility of fluorescent in situ hydridization (FISH) and PCR-based techniques in coral pathogen detection.
Fluorescent in situ hybridization
FISH allows identification, localization, and visualization of individual microbial cells within healthy and diseased tissue [60] by targeting these microbes with custom-synthesized nucleic acid probes attached to fluorescent reporter molecules. Ainsworth et al. [27], [61], [62] utilized FISH to assess the microbial composition of diseased corals in the Mediterranean [27], Red Sea [62], and on the Great Barrier Reef [61]. V. shiloi-specific FISH probes also revealed the marine fireworm Hermodice carunculata as the reservoir and transmission vector of this coral bleaching pathogen [46]. While these studies provide useful information on the spatial arrangement of microbes in healthy and diseased tissue, the low specificity of FISH probes can limit their utility in accurately detecting pathogenic microbes beyond the genus level. In addition, the method is time consuming, labor intensive, and requires specialized imaging microscopy equipment. Extensive processing of samples may also result in the loss of loosely attached microbes including the pathogen cells themselves. Therefore, although helping to elucidate disease etiology, the utility of FISH as a routine coral disease diagnostic is limited.
PCR-based methods
PCR-based methods allow high sensitivity and specificity by targeting and amplifying short nucleic acid (DNA or RNA) sequences within the genomes of coral-associated microbes [63]. These methods are far less time consuming than most culture or immunology-based approaches, yielding results in hours rather than days or even weeks with some culture-based techniques [64]. Several community-level PCR techniques, including denaturing gradient gel electrophoresis (DGGE) [38], terminal restriction fragment length polymorphism (T-RFLP) [65], [66], automated ribosomal intergenic spacer analysis (ARISA) [67], 16S rRNA clone libraries [38], and microarrays [40], [68] have provided insights into the microbial communities associated with healthy and stressed corals. Although this information can be used to detect shifts in community structure, these changes cannot be linked to specific pathogens. Even when specific pathogens have been identified, standard PCR-based methods do not provide accurate quantification of individual microbial species/strains.
Real-time quantitative PCR
The combination of high sensitivity and specificity, low contamination risk, ease of performance, and speed make real-time, quantitative PCR (qPCR) technology an appealing option for specific coral pathogen detection [69]. qPCR allows for accurate quantification of microbe densities by incorporating a fluorescent reporter in the PCR reaction that emits a signal proportional to the quantity of PCR product. This information can then be used to infer the amount of target gene and relative number of pathogen cells in a given sample [70]–[72]. qPCR assays have been designed for a number of bacterial [73], [74], fungal [75], and viral [76] pathogens. For example, a real-time PCR assay was developed to detect V. penaeicida in the prawn Litopenaeus stylirostris and aquaculture facilities in New Caledonia [64]. This single-day assay provided a research tool for understanding the dynamics of this pathogen within aquaculture facilities and served as a decision-making tool for prawn farmers. Analogous assays to detect and quantify coral pathogens in environmentally derived samples are beginning to emerge. For example, Pollock et al. [77] developed a qPCR assay to detect the identified coral pathogen V. coralliilyticus. This technique, which is capable of detecting the bacterium at concentrations as low as 1 CFU ml−1 in seawater and 103 CFU cm−2 on coral fragments, is currently being used to investigate the epidemiology of V. coralliilyticus, including information on its distribution and role in the initiation and spread of white syndrome lesions in the Indo-Pacific. This assay represents the first application of qPCR technology for the detection of an established coral pathogen.
qPCR technologies fall into two broad categories on the basis of their fluorescence chemistries: (1) intercalating dyes and (2) oligonucleotide-specific probes (Figure 2). Intercalating dye technologies, such as SYBR Green, fluoresce as they anneal to the double-stranded DNA (dsDNA) that is synthesized during PCR amplification. As the quantity of dsDNA increases during subsequent PCR cycles, the fluorescence signal increases proportionally (illustrated in Figure 2) [78]. Oligonucleotide probe technologies, including TaqMan and Molecular Beacon, add an additional layer of specificity to the qPCR assay by incorporating a sequence-specific probe that must anneal to a particular region within the PCR amplicon for fluorescence (illustrated in Figure 2). Intercalating dyes are more commonly used than probe technologies because they are less expensive and work with traditional PCR primer sets, negating the time and labor-intensive design of specific probes. However, since intercalating dyes fluoresce in the presence of any dsDNA, they are not specific and must be accompanied by melting curve analysis to differentiate PCR products on the basis of length and G-C content [69]. Oligonucleotide-specific probes are more specific, but also more expensive and require the design of custom synthesized probes. Probe technologies also allow inclusion of several distinct primer/probe sets labeled with different colored fluorescent reporters in a single reaction, facilitating the simultaneous detection of several pathogens.
Fig. 2. Summary of qPCR chemistries. 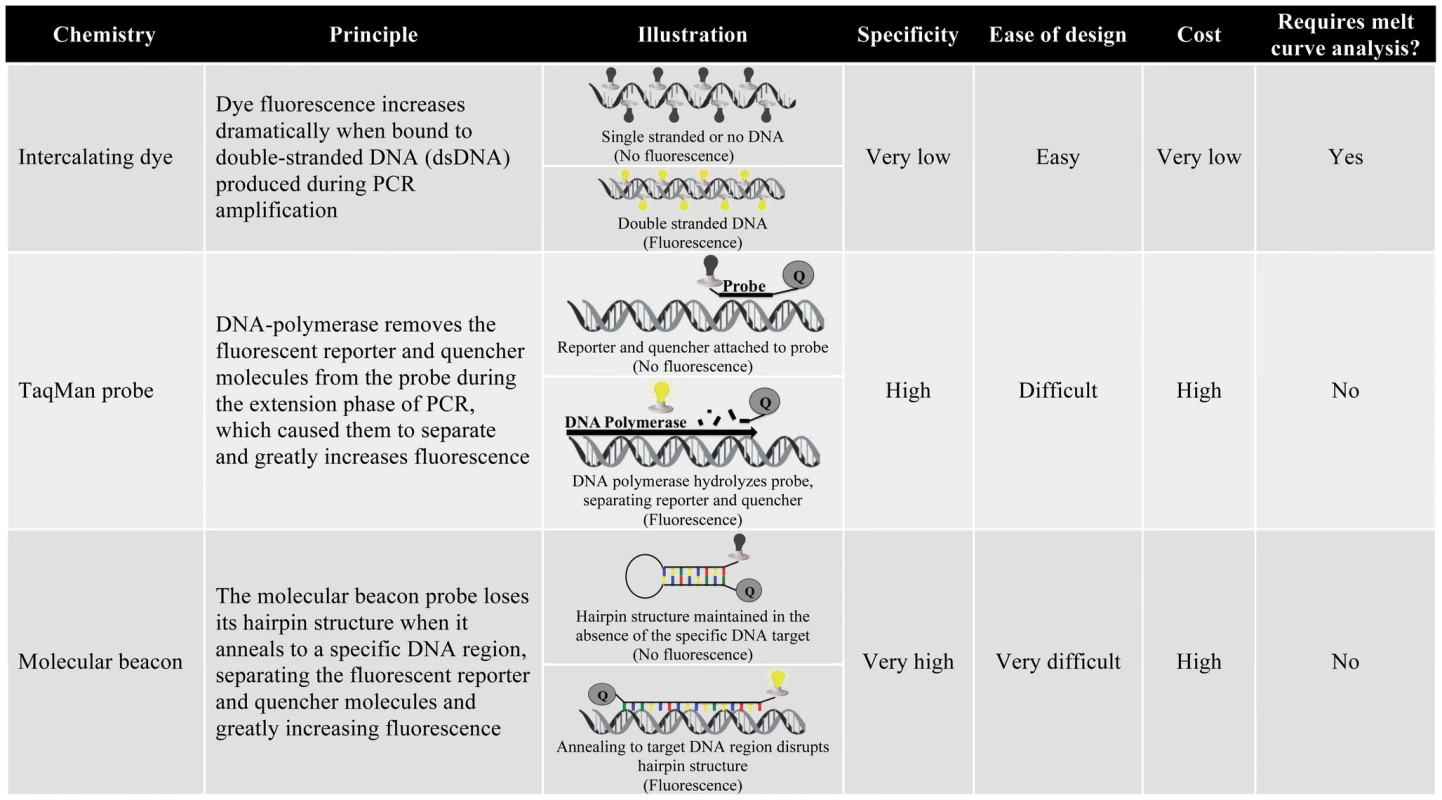
DNA Target Selection
For the development of molecular diagnostic assays, the choice of a nucleic acid target is just as important as the platform used to detect it. To allow for high specificity, nucleic acid targets must be both well-conserved within the genome of the target species/strain and distinct from nontarget sequences. Therefore, a great deal of care must be taken in genetic target selection, primer/probe design, and assay optimization. Ribosomal and mitochondrial DNA are the most common targets for nucleic acid-based microbe detection because of their genetic stability and high copy numbers within cells [79]. However, other genes, including housekeeping genes and virulence factors that are present as only a single or limited copy number in the genome, may also serve as useful targets.
Ribosomal genes
Several ribosomal RNA genes, including 16S, 18S, 23S, and internal transcribed spacer region (ITS) genes, have served as targets for nucleic acid-based detection. With the public availability of over 2 million 16S rRNA sequences (GenBank), spanning both the variable and more highly conserved regions of this ubiquitous bacterial gene, the 16S rRNA gene provides an obvious nucleic acid target. Since its introduction in the late 1980s, most FISH applications have targeted rRNA genes because of the large number of publically available sequences and its high copy number in bacterial cells [60]. However, low genetic divergence in closely related species/strains often hinders the utility of the 16S rRNA gene in differentiating beyond the genus level [80]. For example, the known coral pathogen V. coralliilyticus shares greater than 98% 16S sequence similarity with its closest phylogenetic neighbor, V. neptunius [80]. Although some variation (2%) exists between these closely related species, it is likely inadequate to design sufficiently specific primers and/or probes.
Genomic phylogenetic marker genes
The accelerated use of genetic sequencing as a means of differentiating closely related bacterial species and strains has led to the proliferation of sequence information from a large number of nonribosomal phylogenetic marker genes in a diverse sampling of microbial species. In some of the better-studied groups, such as the vibrios that contain four of the seven described coral pathogens, sequence data from several phylogenetic marker genes are available for all described species [80] and even multiple strains of the identified coral pathogen V. coralliilyticus [81]. This information is useful for selecting genes with the greatest discriminatory power based on phylogenetic reconstructions and also provides the raw sequence data to identify specific oligonucleotide sequences within these genes, which can be targeted by custom-designed molecular primers and probes.
Virulence factors
In the field of human medicine, an increasing number of molecular-based pathogen detection assays have targeted genes directly involved in virulence. For example, the thermostable direct hemolysin gene (tdh) has been used as a target for detection of the human pathogen V. parahaemolyticus and the gene is also inferred as a direct marker of its pathogenicity [82]. Similarly, the hemolysin gene (vvh) has been targeted for the specific detection of V. vulnificus in oysters [73]. Directly targeting strain-specific virulence factors provides a means of differentiating pathogenic and benign strains; by targeting a 2-kb fragment of the cytotoxin-coding gene (rtxA) unique to virulent strains of V. cholerae, Gubala [74] designed a qPCR-based assay capable of exclusively detecting potentially toxigenic strains.
Specific virulence factors have been described in two coral pathogens, the Zn-metalloprotease gene (vcpA) in V. coralliilyticus and the Toxin P gene in V. shiloi, both of which could serve as molecular targets [81], [83], [84]. As researchers develop a deeper understanding of the genetic basis of coral pathogen virulence, it is likely that more virulence targets will become available.
Emerging Diagnostic Techniques
Application of the technological advances outlined above will undoubtedly enhance our ability to study coral diseases; however, a variety of new and emerging technologies will further revolutionize the field in decades to come. High resolution microarrays offer one method for rapid assessment of shifts in coral-associated bacterial community structure. For example, Sunagawa et al. [68] utilized a 16S rRNA gene microarray (PhyloChip G2) to characterize the bacterial community structures of asymptomatic and diseased corals and investigate the etiology of the observed disease. If known bacterial groups or indicator organisms are indentified that are important to coral health, these shifts can be used to infer potential changes in coral health or, additionally, detect identified pathogens associated with disease. Vega Thurber et al. [39] assessed changes in overall bacterial community structure and abundance of functional genes in response to environmental stressors using a 454 pyrosequencing platform. Comparative genomic approaches such as these will continue to provide insights into the bacterial community-level changes that accompany coral stress and potentially facilitate coral disease outbreaks.
Transcriptomic approaches also have great potential for the identification of organisms actively involved in the infection process as well as virulence genes controlling disease progression. To date, the application of RNA-based expression studies on diseased coral samples is limited, except for certain band diseases, such as black band disease [85], where the microbial mat can first be separated from the coral. This limited application of transciptomic techniques is largely due to the inherent instability of mRNA, particularly in the presence of the extensive exogenous enzymes present within coral-derived samples.
Metabolomic techniques, which use NMR and mass spectroscopy to detect chemical fingerprints left behind by specific chemical processes, also show great promise for improving disease diagnosis and pathogen detection [86], [87]. While genomic, metagenomic, transcriptomic, and metabolomic approaches have the potential to generate extensive data, these techniques require expensive, specialized equipment and often the desired information is hidden within immense datasets that require specialized software and highly trained individuals to decipher. However, just as qPCR, which was only available to a handful of well-funded laboratories just a decade ago, is becoming increasingly affordable and accessible, the prohibitive cost of emerging technologies will certainly fall, increasing their availability to coral researchers.
Validation of Diagnostics for Coral Pathogen Detection
The validity of any diagnostic test is determined by its ability to distinguish host organisms that have the disease from those that do not. Validity is comprised of two key components: sensitivity and specificity. Sensitivity describes the test's ability to correctly identify those with the disease and is expressed as the proportion of affected animals that are correctly identified as disease positive by the test compared to the total number of diseased animals tested. Specificity is the ability of the test to correctly identify those that do not have the disease and is expressed as the proportion of animals that are correctly identified as disease negative to the total number of disease-free animals tested [28]. In order to calculate the specificity and sensitivity of a test, we must first know which animals are actually infected with the disease. Such knowledge is usually gained by comparing a test's results with the results of a so-called gold standard, which theoretically has both a sensitivity and specificity of 100% [28]. For example, the gold standard for Chlamydia diagnosis in humans is isolation of the causative agent, the bacteria C. trachomatis. It is important to realize that while gold standards are the best evidence available, they are not infallible and gold standards providing full certainty are rare, particularly in a young field like coral disease research. Generally, the challenge is to find a standard that is as close as possible to the theoretical gold standard, but until effective gold standards are established for coral pathogen detection, it may be useful to use several of the diagnostic techniques described previously to cross-validate test results.
Coral researchers are faced with a unique set of challenges when developing disease diagnostics for the detection of specific pathogenic microbes among the diverse and complex coral holobiont. One major challenge is reproducibly obtaining high purity microbial DNA (or RNA) from coral-derived samples. The complex nature of the coral holobiont, which contains genetic material from the coral host as well as its associated algae, bacteria, and viruses, in combination with the presence of high concentrations of PCR inhibitors (e.g., salts and DNAses) make successful DNA extraction and pathogen detection from coral tissue extremely difficult. Several extraction methods have been developed to overcome these limitations, but consistently obtaining high quality DNA from coral samples remains a persistent challenge to coral researchers. Furthermore, there is the potential for gene copy number variability even between closely related bacterial strains as well as horizontal gene transfer between distantly related species, which could confound accurate detection and quantification. Early pathogen detection assays will therefore require extensive testing to confirm their specificity and sensitivity.
Conclusions
Further development and application of diagnostic tools for coral pathogen detection is limited by a lack of knowledge of the organisms and genes involved in the onset and progression of most coral diseases. In particular, current knowledge of the causes of a large number of coral diseases is rudimentary, with only a few actual pathogens identified (reviewed in [7], [13]). Therefore, further research into coral disease ecology, in combination with robust biomedical approaches to describe diseases at gross and cellular levels is needed to develop an understanding of the pathogenesis of coral diseases and the interactions between agent, host, and the environment [18]. Only after pathogens are identified and their mechanisms of virulence determined can the development of diagnostics that target certain microbial groups or important genes proceed. Coral disease investigations, like other human, veterinary, or wildlife disease investigations, require an interdisciplinary approach, including the use of both traditional and developing technologies.
As coral diseases continue to threaten reefs worldwide, there is increasing urgency for tools to understand and control their spread. Several approaches, including phage therapy and probiotic addition, have been suggested to mitigate coral disease outbreaks [41]; however, the success of any of these strategies will depend upon rapid and reliable disease detection and diagnosis. With the extensive cost and potential environmental risk of certain control measures (e.g., phage therapy), it will be critical that diagnoses are made with an extremely high degree of certainty. Therefore, the development and testing of highly sensitive and specific coral disease diagnostics should be a major research priority. Accurate coral disease diagnosis will help to direct research and management strategies to address the true cause of disease on reefs and aid reef managers in their efforts to control the occurrence, prevalence, and severity of coral disease on reefs worldwide.
Zdroje
1. GardenerTCoteIMGillJAGrantAWatkinsonAR 2003 Long-term region-wide declines in Caribbean corals. Science 301 958 960
2. BrunoJFSeligER 2007 Regional decline of coral cover in the Indo-Pacific: timing, extent, and subregional comparisons. PLoS ONE 2 e711 doi:10.1371/journal.pone.0000711
3. WalkerDIOrmandRFG 1982 Coral death from sewage and phosphate pollution at Aqaba, Red Sea. Mar Pollut Bull 13 21 25
4. BryantDBurkeLMcManusJSpaldingM 1998 Reefs at risk: a map-based indicator of threats to the world's coral reefs. Washington (D.C.) World Resources Institute
5. BellwoodDRHughesTPFolkeCNystromM 2004 Confronting the coral reef crisis. Nature 429 827 833
6. RosenbergEKelloffCARohwerF 2007 Coral microbiology. Oceanography 20 146 154
7. BourneDGGarrenMWorkTMRosenbergESmithGW 2009 Microbial disease and the coral holobiont. Trends Microbiol 17 554 562
8. WeilE 2004 Coral reef diseases in the wider Caribbean. RosenbergELoyaY Coral health and disease Heidelberg Springer 35 68
9. AronsonRBPrechtWF 2001 White-band disease and the changing face of Caribbean coral reefs. Hydrobiologia 460 25 38
10. PattersonKLPorterJWRitchieKBPolsonSWMuellerE 2002 The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc Natl Acad Sci U S A 99 8725 8730
11. WillisBEPageCADinsdaleEA 2004 Coral disease on the Great Barrier Reef. Coral health and disease. Berlin Springer-Verlag Publishing 69 104
12. WeilESmithGGil-AgudeloDL 2006 Status and progress in coral reef disease research. Dis Aquat Organ 69 1 7
13. HarvellDJordán-DahlgrenEMerkelSRosenbergERaymundoL 2007 Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography 20 172 195
14. MouchkaMEHewsonIHarvellCD 2010 Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr Comp Biol 50 1 13
15. PorterJDustanPJaapWPattersonKKosmyninV 2001 Patterns of spread of coral disease in the Florida Keys. Hydrobiologia 460 1 24
16. SutherlandKPPorterJWTorresC 2004 Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar Ecol Prog Ser 266 273 302
17. MillerJMullerERogersCWaaraRAtkinsonA 2009 Coral disease following massive bleaching in 2005 causes 60% decline in coral cover on reefs in the US Virgin Islands. Coral Reefs 28 925 937
18. WorkTMRichardsonLLReynoldsTLWillisBL 2008 Biomedical and veterinary science can increase our understanding of coral disease. J Exp Mar Biol Ecol 362 63 70
19. AntoniusA 1977 Coral mortality in reefs: a problem for science and management. Proc 3rd Int Coral Reef Symp Miami 617 623
20. AntoniusA 1981 Coral reef pathology: a review Proc 4th Int Coral Reef Symp. Manila 3 6
21. LoyaYBullGPichonM 1984 Tumor formations in scleractinian corals. Helgol Mar Res 37 99 112
22. RichardsonLL 1998 Coral diseases: what is really known? Trends Ecol Evol 13 438 443
23. BrucknerAWBrucknerRJ 2002 Coral predation by Sparisima viride and lack of relationship with coral disease Bali 1245 1249
24. KushmaroARosenbergEFineMLoyaY 1997 Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser 147 159 165
25. SussmanMWillisBLVictorSBourneDG 2008 Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS ONE 3 e2393 doi:10.1371/journal.pone.0002393
26. LesserMPBythellJCGatesRDJohnstoneRWHoegh-GuldbergO 2007 Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Biol Ecol 346 36 44
27. AinsworthTDFineMRoffGHoegh-GuldbergO 2008 Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica. ISME J 2 67 73
28. WobeserGA 2006 Essentials of disease in wild animals. Ames (Iowa) Blackwell Pub 243
29. JubbKVFKennedyPCPalmerN 1993 Pathology of domestic animals. San Diego Academic Press Inc 653
30. MuscatineLMcCloskeyLRMarianRE 1981 Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnol Oceanogr 26 601 611
31. RowherFSeguritanVAzamFKnowltonN 2002 Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243 1 10
32. KelloggCA 2004 Tropical Archaea: diversity associated with the surface microlayer of corals. Mar Ecol Prog Ser 273 81 88
33. WegleyLYuYBreitbartMCasasVKlineDI 2004 Coral-associated Archaea. Mar Ecol Prog Ser 273 89 96
34. WilsonWHDaleALDavyJEDavySK 2005 An enemy within? Observations of virus-like particles in reef corals. Coral Reefs 24 145 148
35. ShasharNBanaszakATLesserMPAmramiD 1997 Coral endolithic algae: Life in a protected environment. Pac Sci 51 167 173
36. BentisCJKaufmanLGolubicS 2000 Endolithic fungi in reef-building corals (Order: Scleractinia) are common, cosmopolitan, and potentially pathogenic. Biol Bull 198 254 260
37. PantosOCooneyRPLe TissierMDBarerMRO'DonnellAG 2003 The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ Microbiol 5 370 382
38. BourneDIidaYUthickeSSmith-KeuneC 2008 Changes in coral-associated microbial communities during a bleaching event. ISME J 2 350 363
39. Vega-ThurberRWillner-HallDRodriguez-MuellerBDesnuesCEdwardsRA 2009 Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11 2148 2163
40. KimesNEVan NostrandJDWeilEZhouJMorrisPJ 2010 Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ Microbiol 12 541 556
41. TeplitskiMRitchieK 2009 How feasible is the biological control of coral diseases? Trends Ecol Evol 24 378 385
42. ReshefLKorenOLoyaYZilber-RosenbergIRosenbergE 2006 The coral probiotic hypothesis. Environ Microbiol 8 2068 2073
43. RosenbergEKushmaroAKramarsky-WinterEBaninEYossiL 2009 The role of microorganisms in coral bleaching. ISME J 3 139 146
44. Toledo-HernándezCZuluaga-MonteroABones-GonzálezARodríguezJASabatAM 2008 Fungi in healthy and diseased sea fans (Gorgonia ventalina): is Aspergillus sydowii always the pathogen? Coral Reefs 27 707 714
45. BaninEIsraelyTKushmaroALoyaYOrrE 2000 Penetration of the coral-bleaching bacterium Vibrio shiloi into Oculina patagonica. Appl Environ Microbiol 66 3031 3036
46. SussmanMLoyaYFineMRosenbergE 2003 The marine fireworm Hermodice carunculata is a winter reservoir and spring-summer vector for the coral-bleaching pathogen Vibrio shiloi. Environ Microbiol 5 250 255
47. VezzulliLPreviatiMPruzzoCMarcheseABourneDG 2010 Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ Microbiol 12 2007 2019
48. BrucknerAW 2002 Priorities for effective management of coral diseases. Commerce USDo National Oceanic and Atmospheric Administration 57
49. EfronyRAtadIRosenbergE 2009 Phage therapy of coral white plague disease: properties of phage BA3. Curr Microbiol 58 139 145
50. LazckaODel CampoFJMunozFX 2007 Pathogen detection: a perspective of traditional methods and biosensors. Biosens Bioelectron 22 1205 1217
51. RitchieKBPolsonSWSmithGW 2001 Microbial disease causation in marine invertebrates: problems, practices, and future prospects. Hydrobiologia 460 131 139
52. BrooksBWDevenishJLutze-WallaceCLMilnesDRobertsonRH 2004 Evaluation of a monoclonal antibody-based enzyme-linked immunosorbent assay for detection of Campylobacter fetus in bovine preputial washing and vaginal mucus samples. Vet Microbiol 103 77 84
53. Gomez-DiazE 2009 Linking questions to practices in the study of microbial pathogens: sampling bias and typing methods. Infect Genet Evol 9 1418 1423
54. RitchieKB 2006 Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322 1 14
55. SutherlandKPPorterJWTurnerJWThomasBJLooneyEE 2010 Human sewage identified as likely source of white pox disease of the threatened Caribbean elkhorn coral, Acropora palmata. Environ Microbiol 12 1122 1131
56. IsraelyTBaninERosenbergE 2001 Growth, differentiation and death of Vibrio shiloi in coral tissue as a function of seawater temperature. Aquat Microb Ecol 24 1 8
57. MichaudGASalciusMZhouFBanghamRBoninJ 2003 Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol 21 1509 1512
58. ArmstrongEGEhrlichPHBirkenSSchlattererJPSirisE 1984 Use of a highly sensitive and specific immunoradiometric assay for detection of human chorionic gonadotropin in urine of normal, nonpregnant, and pregnant individuals. J Clin Endocrinol Metab 59 867 874
59. BandiZLSchoenIDeLaraM 1987 Enzyme-linked immunosorbent urine pregnancy tests. Clinical specificity studies. Am J Clin Pathol 87 236 242
60. MoterAGobelUB 2000 Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods 41 85 112
61. AinsworthTDFineMBlackallLLHoegh-GuldbergO 2006 Fluorescence in situ hybridization and spectral imaging of coral-associated bacterial communities. Appl Environ Microbiol 72 3016 3020
62. AinsworthTDKramasky-WinterELoyaYHoegh-GuldbergOFineM 2007 Coral disease diagnostics: what's between a plague and a band? Appl Environ Microbiol 73 981 992
63. PolsonSHigginsJWoodleyC 2008 PCR-based assay for detection of four coral pathogens. Proc 11th Int Coral Reef Symp. Ft. Lauderdale 247 251
64. GoarantCMerienF 2006 Quantification of Vibrio penaeicida, the etiological agent of Syndrome 93 in New Caledonian shrimp, by real-time PCR using SYBR Green I chemistry. J Microbiol Methods 67 27 35
65. LunaGMBiavascoFDanovaroR 2007 Bacteria associated with the rapid tissue necrosis of stony corals. Environ Microbiol 9 1851 1857
66. SatoYWillisBLBourneDG 2009 Successional changes in bacterial communities during the development of black band disease on the reef coral, Montipora hispida. ISME J 4 203 214
67. DanielsCAZeifmanAHeymKRitchieKBWatsonCA 2011 Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar Ecol Prog Ser 426 29 40
68. SunagawaSDeSantisTZPicenoYMBrodieELDeSalvoMK 2009 Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J 3 512 521
69. EspyMJUhlJRSloanLMBuckwalterSPJonesMF 2006 Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev 19 165 256
70. HoughAJHarbisonSASavillMGMeltonLDFletcherG 2002 Rapid enumeration of Listeria monocytogenes in artificially contaminated cabbage using real-time polymerase chain reaction. J Food Prot 65 1329 1332
71. RuzsovicsAMolnaBUngerZTulassayZLaszloP 2002 Determination of Helicobacter pylori cagA, vacA genotypes with real-time PCR melting curve analysis. J Physiol 95 369 377
72. TondellaMLTalkingtonDFHollowayBPDowellSFCowleyK 2002 Development and evaluation of real-time PCR-based fluorescence assays for detection of Chlamydia pneumoniae. J Clin Microbiol 40 575 583
73. PanickerGBejAK 2005 Real-time PCR detection of Vibrio vulnificus in oysters: comparison of oligonucleotide primers and probes targeting vvhA. Appl Environ Microbiol 71 5702 5709
74. GubalaAJ 2006 Multiplex real-time PCR detection of Vibrio cholerae. J Microbiol Methods 65 278 293
75. HauglandRAVarmaMWymerLJVesperSJ 2004 Quantitative PCR analysis of selected Aspergillus, Penicillium and Paecilomyces species. Syst Appl Microbiol 27 198 210
76. ButlerSLHansenMSBushmanFD 2001 A quantitative assay for HIV DNA integration in vivo. Nat Med 7 631 634
77. PollockFJMorrisPJWillisBLBourneDG 2010 Detection and quantification of the coral pathogen Vibrio coralliilyticus by real-time PCR with TaqMan fluorescent probes. Appl Environ Microbiol 76 5282 5286
78. MorrisonTBWeisJJWittwerCT 1998 Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24
79. WoeseCR 1987 Bacterial evolution. Microbiol Rev 51 221 271
80. ThompsonFLGeversDThompsonCCDawyndtPNaserS 2005 Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 71 5107 5115
81. PollockFJWilsonBJohnsonWRMorrisPJWillisBL 2010 Phylogeny of the cosmopolitan coral pathogen Vibrio coralliilyticus. Environ Microbiol Rep 2 172 178
82. BlackstoneGMNordstromJLVickeryMCBowenMDMeyerRF 2003 Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J Microbiol Methods 53 149 155
83. BaninEKhareSKNaiderFRosenbergE 2001 Proline-rich peptide from the coral pathogen Vibrio shiloi that inhibits photosynthesis of Zooxanthellae. Appl Environ Microbiol 67 1536 1541
84. SussmanMMieogJCDoyleJVictorSWillisBL 2009 Vibrio zinc-metalloprotease causes photoinactivation of coral endosymbionts and coral tissue lesions. PLoS ONE 4 e4511 doi:10.1371/journal.pone.0004511
85. Frias-LopezJBonheyoGTFoukeBW 2004 Identification of differential gene expression in bacteria associated with coral black band disease by using RNA-arbitrarily primed PCR. Appl Environ Microbiol 70 3687 3694
86. GowdaGAZhangSGuHAsiagoVShanaiahN 2008 Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn 8 617 633
87. BoroujerdiAFBVizcainoMIMeyersAPollockECHuynhSL 2009 NMR-based microbial metabolomics and the temperature-dependent coral pathogen Vibrio coralliilyticus. Environ Sci Technol 43 7658 7664
88. BaumsIBMillerMWHellbergME 2005 Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol Ecol 14 1377 1390
89. Smith-KeuneCvan OppenM 2006 Genetic structure of a reef-building coral from thermally distinct environments on the Great Barrier Reef. Coral Reefs 25 493 502
90. BakRPM 1983 Neoplasia, regeneration and growth in the reef-building coral Acropora palmata. Mar Biol 77 221 227
91. HumphreyCWeberMLottCCooperTFabriciusK 2008 Effects of suspended sediments, dissolved inorganic nutrients and salinity on fertilisation and embryo development in the coral Acropora millepora (Ehrenberg, 1834). Coral Reefs 27 837 850
92. GladfelterWB 1982 White-band disease in Acropora palmata: implications for the structure and growth of shallow reefs. Bull Mar Sci 32 639 643
93. KvenneforsECELeggatWHoegh-GuldbergODegnanBMBarnesAC 2008 An ancient and variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev Comp Immunol 32 1582 1592
94. WeisVMDavySKHoegh-GuldbergORodriguez-LanettyMPringleJR 2008 Cell biology in model systems as the key to understanding corals. Trends Ecol Evol 23 369 376
95. AronsonRBrucknerAMooreJPrechtBWeilE 2008 Acropora palmata. IUCN 2010. IUCN Red List of Threatened Species. Version 2010.4
96. Belda-BaillieCABaillieBKMaruyamaT 2002 Specificity of a model cnidarian-dinoflagellate symbiosis. Biol Bull 202 74
97. FrauneSBoschTCG 2007 Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci U S A 104 13146
98. GatesRDBaghdasarianGMuscatineL 1992 Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol Bull 182 324
99. DunnSRBythellJCLe TissierMDABurnettWJThomasonJC 2002 Programmed cell death and cell necrosis activity during hyperthermic stress-induced bleaching of the symbiotic sea anemone Aiptasia sp. J Exp Mar Bio Ecol 272 29 53
100. BairdAHGuestJRWillisBL 2009 Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu Rev Ecol Evol Syst 40 551 571
101. HarrisonPLWallaceCC 1990 Ecosystems of the world: coral reefs. Coral reproduction Amsterdam Elsevier 133 208
102. AbregoDUlstrupKEWillisBLvan OppenMJH 2008 Species–specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proc R Soc Lond B Biol Sci 275 2273 2282
103. PalmerCVBythellJCWillisBL 2010 Levels of immunity parameters underpin bleaching and disease susceptibility of reef corals. FASEB J 24 1935 1946
104. MydlarzLDJonesLEHarvellCD 2006 Innate immunity, environmental drivers, and disease ecology of marine and freshwater invertebrates. Annu Rev Ecol Evol Syst 37 251 288
105. RochP 1999 Defense mechanisms and disease prevention in farmed marine invertebrates. Aquaculture 172 125 145
106. StedmanTL 2000 Stedman's medical dictionary. 27th edition Baltimore Lippincott Williams & Wilkins
107. MydlarzLDHolthouseSFPetersECHarvellCD 2008 Cellular responses in sea fan corals: granular amoebocytes react to pathogen and climate stressors. PLoS ONE 3 e1811 doi:10.1371/journal.pone.0001811
108. PalmerCVMydlarzLDWillisBL 2008 Evidence of an inflammatory-like response in non-normally pigmented tissues of two scleractinian corals. Proc Biol Sci 275 2687 2693
109. SindermannCJ 1990 Principal diseases of marine fish and shellfish.Volume 2. Diseases of marine shellfish. San Diego Academic Press
110. PetersEC 1997 Diseases of coral-reef organisms.Life and death of coral reefs. New York Chapman & Hall 114 139
111. Vargas-ÁngelBPetersECKramarsky-WinterEGilliamDSDodgeRE 2007 Cellular reactions to sedimentation and temperature stress in the Caribbean coral Montastraea cavernosa. J Invert Path 95 140 145
112. Domart-CoulonIJTraylor-KnowlesNPetersEElbertDDownsCA 2006 Comprehensive characterization of skeletal tissue growth anomalies of the finger coral Porites compressa. Coral Reefs 25 531 543
113. KimKHarvellCDKimPDSmithGWMerkelSM 2000 Fungal disease resistance of Caribbean sea fan corals (Gorgonia spp.). Mar Biol 136 259 267
114. DubeDKimKAlkerAPHarvellCD 2002 Size structure and geographic variation in chemical resistance of sea fan corals Gorgonia ventalina to a fungal pathogen. Mar Ecol Prog Ser 231 139 150
115. GochfeldDJAebyGS 2008 Antibacterial chemical defenses in Hawaiian corals provide possible protection from disease. Mar Ecol Prog Ser 362 119 128
116. MydlarzLDCouchCSWeilESmithGHarvellCD 2009 Immune defenses of healthy, bleached and diseased Montastraea faveolata during a natural bleaching event. Dis Aquat Org 87 67 78
117. PikkemaatMGDijkSOSchoutenJRapalliniMVan EgmondHJ 2008 A new microbial screening method for the detection of antimicrobial residues in slaughter animals: The Nouws antibiotic test (NAT-screening). Food Control 19 781 789
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Quorum Sensing in Fungi: Q&AČlánek Blood Feeding and Insulin-like Peptide 3 Stimulate Proliferation of Hemocytes in the MosquitoČlánek The DEAD-box RNA Helicase DDX6 is Required for Efficient Encapsidation of a Retroviral GenomeČlánek A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus,Článek A Wide Extent of Inter-Strain Diversity in Virulent and Vaccine Strains of AlphaherpesvirusesČlánek The Anti-Sigma Factor TcdC Modulates Hypervirulence in an Epidemic BI/NAP1/027 Clinical Isolate ofČlánek Critical Roles for LIGHT and Its Receptors in Generating T Cell-Mediated Immunity during InfectionČlánek Frequent and Recent Human Acquisition of Simian Foamy Viruses Through Apes' Bites in Central Africa
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Quorum Sensing in Fungi: Q&A
- Discovery of an Ebolavirus-Like Filovirus in Europe
- Toll-like Receptor 7 Controls the Anti-Retroviral Germinal Center Response
- Tubule-Guided Cell-to-Cell Movement of a Plant Virus Requires Class XI Myosin Motors
- Herpesvirus Telomerase RNA (vTR) with a Mutated Template Sequence Abrogates Herpesvirus-Induced Lymphomagenesis
- Mitochondrial Peroxiredoxin Plays a Crucial Peroxidase-Unrelated Role during Infection: Insight into Its Novel Chaperone Activity
- Sustained CD8+ T Cell Memory Inflation after Infection with a Single-Cycle Cytomegalovirus
- Novel Mouse Xenograft Models Reveal a Critical Role of CD4 T Cells in the Proliferation of EBV-Infected T and NK Cells
- Toll-8/Tollo Negatively Regulates Antimicrobial Response in the Respiratory Epithelium
- Exhausted Cytotoxic Control of Epstein-Barr Virus in Human Lupus
- Structural and Functional Analysis of Laninamivir and its Octanoate Prodrug Reveals Group Specific Mechanisms for Influenza NA Inhibition
- Infection Drives IL-17-Mediated Neutrophilic Allergic Airways Disease
- Blood Feeding and Insulin-like Peptide 3 Stimulate Proliferation of Hemocytes in the Mosquito
- HIV-1 Replication in the Central Nervous System Occurs in Two Distinct Cell Types
- Deep Molecular Characterization of HIV-1 Dynamics under Suppressive HAART
- Fitness Landscape of Antibiotic Tolerance in Biofilms
- The DEAD-box RNA Helicase DDX6 is Required for Efficient Encapsidation of a Retroviral Genome
- Preventing Sepsis through the Inhibition of Its Agglutination in Blood
- A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus,
- IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry
- Targeting Cattle-Borne Zoonoses and Cattle Pathogens Using a Novel Trypanosomatid-Based Delivery System
- A Wide Extent of Inter-Strain Diversity in Virulent and Vaccine Strains of Alphaherpesviruses
- Coordinated Destruction of Cellular Messages in Translation Complexes by the Gammaherpesvirus Host Shutoff Factor and the Mammalian Exonuclease Xrn1
- Signal Transduction through CsrRS Confers an Invasive Phenotype in Group A
- Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex
- Histone Deacetylase 8 Is Required for Centrosome Cohesion and Influenza A Virus Entry
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis
- Co-opts the FGF2 Signaling Pathway to Enhance Infection
- IRAK-2 Regulates IL-1-Mediated Pathogenic Th17 Cell Development in Helminthic Infection
- Trafficking of Hepatitis C Virus Core Protein during Virus Particle Assembly
- The Anti-interferon Activity of Conserved Viral dUTPase ORF54 is Essential for an Effective MHV-68 Infection
- A Viral Nuclear Noncoding RNA Binds Re-localized Poly(A) Binding Protein and Is Required for Late KSHV Gene Expression
- Suppression of Methylation-Mediated Transcriptional Gene Silencing by βC1-SAHH Protein Interaction during Geminivirus-Betasatellite Infection
- ISG15 Is Critical in the Control of Chikungunya Virus Infection Independent of UbE1L Mediated Conjugation
- Non-Hematopoietic Cells in Lymph Nodes Drive Memory CD8 T Cell Inflation during Murine Cytomegalovirus Infection
- RNA Polymerase II Stalling Promotes Nucleosome Occlusion and pTEFb Recruitment to Drive Immortalization by Epstein-Barr Virus
- Noninfectious Retrovirus Particles Drive the / Dependent Neutralizing Antibody Response
- Endophytic Life Strategies Decoded by Genome and Transcriptome Analyses of the Mutualistic Root Symbiont
- An Integrated Approach to Elucidate the Intra-Viral and Viral-Cellular Protein Interaction Networks of a Gamma-Herpesvirus
- as an Animal Model for the Study of Biofilm Infections
- Homeostatic Proliferation Fails to Efficiently Reactivate HIV-1 Latently Infected Central Memory CD4+ T Cells
- The Anti-Sigma Factor TcdC Modulates Hypervirulence in an Epidemic BI/NAP1/027 Clinical Isolate of
- Enhances Protective and Detrimental HLA Class I-Mediated Immunity in Chronic Viral Infection
- The Mouse IAPE Endogenous Retrovirus Can Infect Cells through Any of the Five GPI-Anchored EphrinA Proteins
- The Urgent Need for Robust Coral Disease Diagnostics
- HacA-Independent Functions of the ER Stress Sensor IreA Synergize with the Canonical UPR to Influence Virulence Traits in
- A Novel Core Genome-Encoded Superantigen Contributes to Lethality of Community-Associated MRSA Necrotizing Pneumonia
- Critical Roles for LIGHT and Its Receptors in Generating T Cell-Mediated Immunity during Infection
- The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors
- Frequent and Recent Human Acquisition of Simian Foamy Viruses Through Apes' Bites in Central Africa
- Mechanisms of Trafficking to the Brain
- Defining Emerging Roles for NF-κB in Antivirus Responses: Revisiting the Enhanceosome Paradigm
- The Role of Sialyl Glycan Recognition in Host Tissue Tropism of the Avian Parasite
- Evolutionarily Divergent, Unstable Filamentous Actin Is Essential for Gliding Motility in Apicomplexan Parasites
- The Herpes Simplex Virus-1 Transactivator Infected Cell Protein-4 Drives VEGF-A Dependent Neovascularization
- Distinct Single Amino Acid Replacements in the Control of Virulence Regulator Protein Differentially Impact Streptococcal Pathogenesis
- Soluble Rhesus Lymphocryptovirus gp350 Protects against Infection and Reduces Viral Loads in Animals that Become Infected with Virus after Challenge
- A Genetic Screen Reveals Arabidopsis Stomatal and/or Apoplastic Defenses against pv. DC3000
- Hepatitis C Virus Reveals a Novel Early Control in Acute Immune Response
- Fumarate Reductase Activity Maintains an Energized Membrane in Anaerobic
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis
- The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors
- Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex
- Evolutionarily Divergent, Unstable Filamentous Actin Is Essential for Gliding Motility in Apicomplexan Parasites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



