-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Transition of Sporozoites into Liver
Stage-Like Forms Is Regulated by the RNA Binding Protein Pumilio
Many eukaryotic developmental and cell fate decisions that are effected
post-transcriptionally involve RNA binding proteins as regulators of translation
of key mRNAs. In malaria parasites (Plasmodium spp.), the
development of round, non-motile and replicating exo-erythrocytic liver stage
forms from slender, motile and cell-cycle arrested sporozoites is believed to
depend on environmental changes experienced during the transmission of the
parasite from the mosquito vector to the vertebrate host. Here we identify a
Plasmodium member of the RNA binding protein family PUF as
a key regulator of this transformation. In the absence of Pumilio-2 (Puf2)
sporozoites initiate EEF development inside mosquito salivary glands
independently of the normal transmission-associated environmental cues.
Puf2- sporozoites exhibit genome-wide transcriptional
changes that result in loss of gliding motility, cell traversal ability and
reduction in infectivity, and, moreover, trigger metamorphosis typical of early
Plasmodium intra-hepatic development. These data
demonstrate that Puf2 is a key player in regulating sporozoite developmental
control, and imply that transformation of salivary gland-resident sporozoites
into liver stage-like parasites is regulated by a post-transcriptional
mechanism.
Published in the journal: Transition of Sporozoites into Liver Stage-Like Forms Is Regulated by the RNA Binding Protein Pumilio. PLoS Pathog 7(5): e32767. doi:10.1371/journal.ppat.1002046
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002046Summary
Many eukaryotic developmental and cell fate decisions that are effected
post-transcriptionally involve RNA binding proteins as regulators of translation
of key mRNAs. In malaria parasites (Plasmodium spp.), the
development of round, non-motile and replicating exo-erythrocytic liver stage
forms from slender, motile and cell-cycle arrested sporozoites is believed to
depend on environmental changes experienced during the transmission of the
parasite from the mosquito vector to the vertebrate host. Here we identify a
Plasmodium member of the RNA binding protein family PUF as
a key regulator of this transformation. In the absence of Pumilio-2 (Puf2)
sporozoites initiate EEF development inside mosquito salivary glands
independently of the normal transmission-associated environmental cues.
Puf2- sporozoites exhibit genome-wide transcriptional
changes that result in loss of gliding motility, cell traversal ability and
reduction in infectivity, and, moreover, trigger metamorphosis typical of early
Plasmodium intra-hepatic development. These data
demonstrate that Puf2 is a key player in regulating sporozoite developmental
control, and imply that transformation of salivary gland-resident sporozoites
into liver stage-like parasites is regulated by a post-transcriptional
mechanism.Introduction
Puf (Pumilio and fem-3 mRNA binding factor) proteins are an evolutionarily highly conserved family of proteins present from yeast to humans and plants characterized by a highly conserved C-terminal RNA-binding domain, composed of eight tandem Pumilio (PUM) repeats. Puf proteins typically decrease expression of targeted mRNAs by enhancing their decay or repressing their translation [reviewed in 1]. The conserved biochemical features and genetic function of Puf family members have emerged from studies of model organisms and although Puf proteins have been shown to play diverse functions, the one frequently shared throughout evolution relates to the maintenance of stemness [2], [3], [4] and control of differentiation [5], [6], [7].
The Plasmodium parasite alternates between mosquito vector and vertebrate host, with transmission relying on highly specialized parasite stages. Once inside the new host, developmental progression quickly gives rise to fundamentally different parasite forms adapted to their new environment [8]. For example, cell-cycle arrested gametocytes, transmitted from the mammalian host to the Anopheles vector during a mosquito blood meal, fertilize and generate the motile ookinete in the mosquito midgut. Similarly, a single slender, motile and cell-cycle arrested sporozoite, transmitted by a mosquito bite, while inside a liver cell will develop into a round, non-motile and replicating exo-erythrocytic form (EEF) and go on to generate thousands of merozoites [9], [10], [11]. Developmental progression of both gametocytes and sporozoites requires clear environmental cues; for gametocytes these include xanthurenic acid and a drop in temperature [12], while sporozoites need a rise in temperature and the presence of bicarbonate [13], [14], [15].
The sudden transition between hosts that have very different physiological environments requires a rapid molecular and cellular re-programming, which may only be realized by parasites that are in a state of molecular preparedness, while maintaining a quiescent state until transmission occurs. Indeed, successful development of the mosquito-infective ookinete relies on the availability of translationally repressed mRNAs previously transcribed in female gametocytes in the blood stage, which are only translated following fertilization [16], [17], as well as stored proteins [18]. Although suggested [19], [20] it is unknown whether equivalent post-transcriptional RNA-mediated events facilitate developmental progression during the parasite's exit from the mosquito and initiation of EEF development in the mammalian host liver. Still, Plasmodium sporozoites remain viable and transmission-competent for weeks in mosquito salivary glands [21].
The roles of Puf (Pumilio and fem-3 mRNA binding factor) proteins are diverse yet intimately involved in the post-transcriptional regulation of developmental and differentiation factors in organisms as diverse as yeast, Caenorhabdites elegans, Drosophila and humans [22], [23], [24], [25]. Two such proteins, Puf1 (PFE0935c) and Puf2 (PFD0825c), are known in the human malaria parasite P. falciparum [26], [27], [28], with orthologs in all Plasmodium species characterized, including the rodent-infectious species P. berghei. The Plasmodium Puf proteins have the typical highly conserved organization that includes the eight tandem copies of the PUM RNA binding domain (or Pumilio homology domain, PHD) at the carboxyterminus of the protein (Fig. S1) and P. falciparum Puf2 was shown to bind the Nanos Response Element RNA in vitro [26]. In P. falciparum evidence has been reported for a role for Puf2 in gametocyte development although puf2 is most highly transcribed in sporozoites [28], [29].
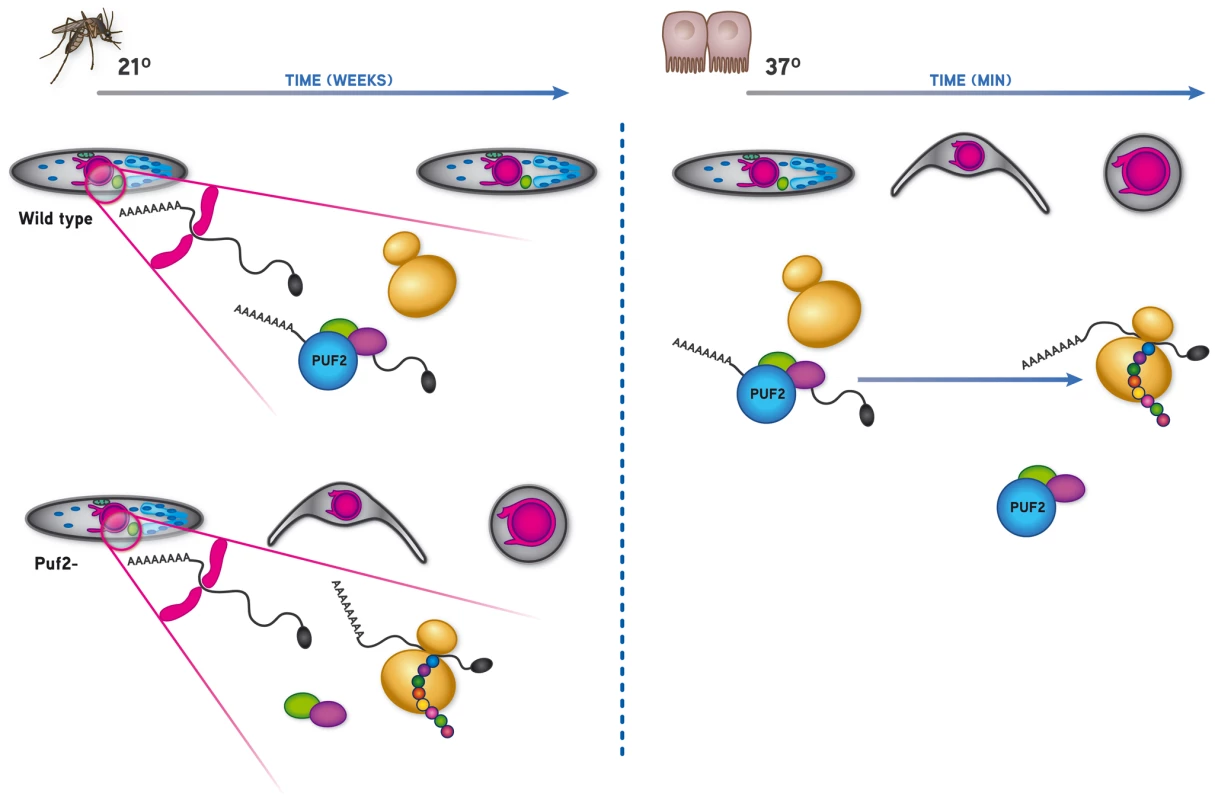
Here we provide strong evidence for an RNA-mediated regulatory event in the rodent malaria parasite P. berghei that relies on the RNA binding protein Pumilio 2 (PBANKA_071920) to maintain salivary gland sporozoites in a stand-by mode prior to transmission. The absence of the highly conserved protein Pumilio-2 is necessary and sufficient to enable the slow and progressive morphological transformation of P. berghei sporozoites into EEF-like forms while still inside the lumen of the mosquito salivary gland. This transformation is characteristic of EEFs both functionally and in respect to their gene expression repertoire and dissociates the transformation of sporozoites to EEF-like forms from its requirement for environmental cues.
Results
puf2- P. berghei sporozoites undergo EEF-like metamorphosis inside mosquito salivary glands
Similar to P. falciparum, both P. berghei puf orthologs (puf1, PBANKA_123350 and puf2, PBANKA_071920) are not only transcribed in gametocytes but also in salivary gland sporozoites (SGS) (Fig. 1A, S3C). Antibodies raised against PbPuf2 confirmed the expression of the protein in SGS and localized the protein to a small number of discrete foci in the cytoplasm of the cell consistent with the localization of most Puf proteins (Fig. 1B). To address the roles of the two encoded proteins during parasite transmission we generated transgenic P. berghei that lack either puf1 or puf2, or both genes (Figs. S2–S4). All 3 gene deletion mutants (puf1-, puf2-, puf1-/2-) showed normal growth and multiplication of asexual blood stage parasites and in contrast to the reports for P. falciparum produced gametocytes comparable in number to wild type parasites; the transition of gametocytes into gametes and ookinete formation was also not affected (Table S1). Furthermore oocyst numbers per midgut and sporozoites reaching the salivary glands were not significantly altered when compared to wild type parasites (Fig. S5).
Together these data suggested that lack of either Puf1 or Puf2, or both proteins has no, or at most very minor effects on the majority of the different life cycle stages of P. berghei, including the number of sporozoites reaching mosquito salivary glands. However, microscopic examination of salivary gland 30 days after mosquito infection revealed aberrant morphology of puf2- parasites (Fig. 2; Videos S1 [wild type], S2 [puf1-], S3 [puf2-]). Sporozoites of both independent puf2- mutants at day 22 after mosquito infection and later, began to round up and progressively resembled early hepatic stages (Fig. 2A, B; Fig. S6); by day 24 after mosquito infection the majority of parasites in mosquito salivary glands were morphologically similar to early EEF's (76.49±2.43%) (Fig. 2C) with an average bulging area of 4.72±0.42 µm2 that increased to 6.02±1.14 µm2 on day 30 of infection. The bulging area of older puf2- parasites is comparable in size to 8–10 hours liver stage EEF's. On the other hand puf1 - and wild type salivary gland sporozoites (SGS) remained typically slender throughout the entire period (Fig. 2). puf1-/2 - parasites recapitulated the puf2- single KO phenotype (Fig. S6).
18-day puf2- sporozoites are defective in motility, cell traversal and infection
At day 18 puf2- sporozoites had reached the mosquito salivary glands in similar numbers as puf1-, puf1-/2 - and wild type parasites (Fig. S5) and did not present obvious morphological changes. Although superficially morphologically identical to wild type sporozoites, 18 day puf2- SGS displayed significantly reduced gliding motility and cell traversal ability when compared to puf1 - and wild type parasites (Fig. 3A, B; t-test p<0.05). Consequently, puf2- SGS were less infective in vitro to Huh7 hepatoma cells (Fig. 3C; t-test p<0.05) and parasites that had successfully invaded, showed delayed development (Fig. 3D; t-test p<0.05). When we compared parasite loads in mouse livers infected 44 hours earlier after intra-venous injection of identical numbers (n = 10,000) of day 18 puf1-, puf2- or wild type sporozoites we found a significant impairment of liver infection by puf2- when either compared to WT or puf1 - parasites (Fig. 3E; t-test p<0.05). Ten days after i.v. injection of SGS all mice infected with wild type and puf1 - SGS developed blood stage parasitemia, while only 32% of the mice infected with puf2- parasites did (Fig. 3F). During infection by mosquito bite, blood stage parasites became patent only in mice infected with wild type and puf1-, but never with puf2- parasites (Fig. 3G). Although mice infected with puf1 - SGS show a lower parasite liver load than mice infected with WT SGS (Fig. 3E), no differences were found in blood stage patency (Fig. 3F and G). Throughout, the behavior of the puf1-/2 - parasite was similar to the puf2- parasite, which suggests that all defects are attributable to the lack of Puf2, with no additional effects arising from the simultaneous deletion of both genes (Table S2).
Transcriptome changes precede visible morphological changes of puf2- sporozoites
The phenotypic analyses of day 18 puf2- SGS suggested that premature de-differentiation of sporozoites could already have been initiated prior to the visible manifestation of the morphological changes evident in older parasites; we reasoned the absence of Puf2 might affect the steady state transcriptome; intra-hepatic and axenic differentiation of SGS into EEF's is correlated with distinct transcriptome adaptations [13], [14], [30]. Therefore we compared in 18-day SGS by RT-PCR (data not shown) and RT-qPCR the expression profiles of genes known to be transcribed in sporozoites, or in EEF's but not in sporozoites (e.g. exp-2 - PBANKA_133430 and exp-1 -PBANKA_092670) in comparison to ama-1 (PBANKA_091500). Transcripts of the sporozoite genes gap45 (PBANKA_143760), myo-a (PBANKA_135570), spect2 (PBANKA_100630), celtos (PBANKA_143230) and spect1 (PBANKA_135560) – their protein products are important for gliding motility and cell traversal [8] – were less abundant in puf2-; uis-4 (PBANKA_050120) showed no marked difference (Fig. 4A). Conversely, the liver stage genes exp-1 and exp-2 (a constituent of the PTEX translocon of exported PEXEL-containing proteins [31]) were clearly more abundant in the absence of Puf2. uis-1/ik2 (PBANKA_020580), a kinase reported to regulate translational capacity of salivary gland sporozoites [32] was also down-regulated. Together these differences in mRNA indicated that the changes in morphology of the older puf2- salivary gland parasite are indeed preceded by changes in steady state mRNA levels in superficially normal, younger SGS. The down-regulation of myo-a, gap45, celtos, spect1 and spect2 could explain why day 18 puf2- sporozoites are deficient in gliding motility as well as infection (Fig. 3).
Fig. 4. Transcriptional changes in <i>puf2-</i>. 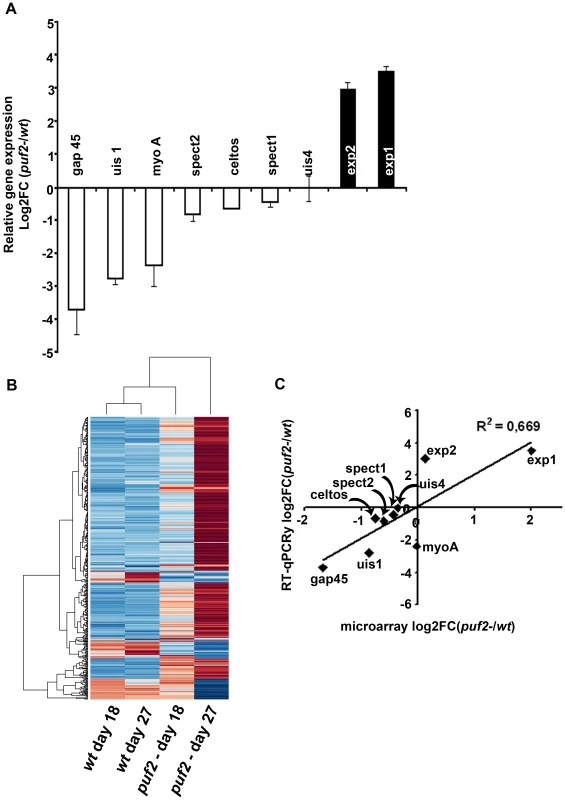
puf2- parasites in salivary glands show progressive transcriptome changes
The RT-PCR data prompted the analysis of the global transcriptome variations in puf2- sporozoites. We compared mRNA levels obtained from both day 18 and day 27 wild type and puf2- parasites by microarray hybridization using 3 biological replicates from each time-point. In total, our analyses showed that in the absence of Puf2, 267 genes were up-regulated (UR) at either days 18 or 27 or both, while 47 were down-regulated (DR) at least 1.5-fold (F-test, p<0.05; Fig. 4B; Table S3A); these genes included those that we initially identified in the RT-qPCR survey (Fig. 4C; Fig. S7).
Our data indicate an overall increase in transcriptional activity in mutant parasites which is suggested by the larger number of UR versus DR genes (66 vs. 14 at day 18; 271 vs. 47 at day 27, on a pairwise basis, p<0.05). Still, the DR transcript data set contains genes that encode components of the inner membrane complex and enzymes of the TCA cycle (Fig. S8), as well as genes with a well-documented role in motility and invasion, reflecting the observed functional deficiencies (see Fig. 3) in the puf2- parasites. These genes include celtos, spect1, spect2, tlp1 (PBANKA_111600), trsp (PBANKA_020910), siap1 (PBANKA_100620), mtrap (PBANKA_051280), trep (PBANKA_130650), psop9/gama (PBANKA_070190), gap45, and p36p (PBANKA_100220; Table S3B); another 10 conserved, but uncharacterized Plasmodium proteins that contain a signal peptide, trans-membrane domain(s) or GPI anchor are also DR, maybe indicating a function during the hepatocyte invasion process.
On the other hand, UR genes fit in the categories of DNA metabolic processes, ribonucleoprotein complex, ribosome/translation and protein folding (Fig. S8). Of the 7 differentially expressed transcription factors found 6 are UR and include TFIIH (PBANKA_141340), the RNA polymerase II subunit (PBANKA_020330), 2 putative transcription factors (AP2's, PBANKA_083520 and PBANKA_010950), and 2 TFIIS Zinc-fingers (PBANKA_030420 and PBANKA_142110; Table S2C); concomitantly mRNA capping and splicing factors, and genes involved in ribosomal and transfer RNA processing (n = 16) are almost exclusively UR (Table S2D). Throughout, translation factors and ribosomal proteins (n = 52; Table S3E) are UR in puf2-, while ik2 (a negative regulator of translation through phosphorylation of eIF2α) is DR, consistent with the observed increase in protein translation in IK2 null mutant sporozoites [32]. In parallel, many chaperones (n = 17; Table S3F) and genes with protein transport-related functions (n = 29; Table S3G) are UR; these include for example plasmepsin V (PBANKA_133870) – the PEXEL-motif cleaving enzyme – and exp2 [31], [33]. 14 genes linked to the ubiquitin-proteasome system are UR at day 27 in puf2- parasites (Table S3H) which supports an involvement in the observed elimination of rhoptries and micronemes during metamorphosis [34]. Additional UR genes in the puf2- mutant include mitochondrial and fatty acid synthesis genes (Table S3I and J). Finally we observe an increase in replication factors, rad51 (PBANKA_093950), histone h2b (PBANKA_094180) and alba-3 (PBANKA_120440; n = 17, Table S3K).
In summary, the microarray analysis emphasizes genes involved in increased metabolic activity to be UR in EEF-like mutant parasites. The comparison between wild type SGS from days 18 and 27 post-mosquito infection on the other hand showed almost no transcriptome alterations. Only 6 and 16 genes, respectively, were UR or DR out of the total of approximately 5400 P. berghei genes; none of them however significantly (moderated t-test p>0.05; Table S3A). This clearly suggests that the wild type parasites' quiescent yet infective status with respect to transcription and mRNA abundance is maintained for at least 10 days while residing in the mosquito salivary gland. A comparison with the sporozoite and EEF proteomes of the related, rodent malaria species P. yoelii [35] showed that 89.7% (96/107) of UR P. berghei mRNAs in puf2- sporozoites (at day 18 and 27) are indeed detected only in P. yoelii liver stage parasites but not in SGS, corroborating the notion that puf2- sporozoites in fact resemble genuine, early liver stage parasites (Table S3A).
puf2- parasites in salivary glands exhibit ultrastructural features of early stage EEFs
Further evidence of the genuine nature of older puf2- parasites as early liver stage parasites arose from ultrastructural studies. The bulge formation in the center of parasite is a hallmark of the initial differentiation process during hepatic development. Importantly, it has been shown to coincide with a loss of the inner membrane complex (IMC) associated with motility and rounding-up [34]. When we analyzed wild type and puf2 - P. berghei by EM we found a clear sign of IMC disruption in day 27 puf2- parasites (Fig. 5) but not in day 18 puf2- SGS (Fig. S9). Neither wild type nor puf1 - showed any signs of IMC impairment (Fig. 5). The resulting protrusions mark the beginning of sporozoite differentiation into liver stage trophozoites and occur approximately 4 hours after liver cell infection, following degradation of IMC components [34] such as Alveolin 9 (PBANKA_124060) or MyoA. All these changes in puf2- SGS further confirm the profound developmental switch in response to the lack of Puf2.
Plasmodium sporozoite transformation into EEFs is protein translation dependent
Overall, our data show that Puf2 is a master regulator of Plasmodium developmental control during transmission from the mosquito vector to the mammalian host. The highly conserved nature of the Pumilio homology domain (PHD) of the Plasmodium proteins [27] (Fig. S1), its conserved function in many different organisms [5], [22], [36], [37], [38], the capacity of P. falciparum Puf2 to bind RNA in vitro [26] and its localization to few cytoplasmic speckles in P. berghei sporozoites (Fig. 1B) strongly suggested that sporozoite latency in Anopheles salivary glands relies on the control of protein translation through a post-transcriptional mechanism.
The notion that SGS to EEF transformation is dependent on de novo protein synthesis was supported by the ability of cycloheximide (a general protein synthesis inhibitor) to significantly reduce the metamorphosis of wild type SGS into EEF-like parasites (Fig. 6A) in an established in vitro transformation assay [15]. In this assay, sporozoites transform into EEF-like parasites within 1–2 h when placed at a temperature of 37°C (Fig. 6A). Importantly, comparison of transformation of WT and puf-2 sporozoites showed that puf-2 parasites produced almost twice as many early EEF's compared to WT after 4 h incubation at 37°C in a cycloheximide-insensitive manner (Fig. 6B). This result implies that, in puf2 - parasites, proteins required for SGS transformation into EEF-like parasites have already been produced by day 18 of mosquito infection. Indeed, Western blots performed with 18 day wild type and mutant SGS showed that transformation-associated changes in protein level had already occurred (Fig. 6C): proteins involved in sporozoite motility (MyoA) or IMC maintenance (Alveolin 9) are clearly less abundant in mutant SGS whilst proteins typical of EEF development (Exp1 and Exp2) are readily detected prior to any morphological changes.
Our data clearly show that Plasmodium sporozoite transformation into EEFs is protein synthesis dependent, and sporozoite quiescence relies on post-transcriptional control and the RNA binding protein Puf2.
Discussion
Following the mosquito blood meal, sporozoites that manage to invade a host hepatocyte quickly initiate differentiation into liver stage trophozoites; these early changes are characterized by a de-differentiation process that involves loss of the inner membrane complex, protrusions in the region of the parasites nucleus and loss of internal organelles [34]. A single parasite ultimately multiplies by a factor of a few 1000-fold within 48 hours in P. berghei to give rise to first generation merozoites. Plasmodium development in the liver is accompanied by clear host cell transcriptional changes and adaptations that reflect the parasite's needs [39]; these include generation and maintenance of the parasitophorous vacuole, the export of proteins such as circumsporozoite protein (CS) into the host cytoplasm [40] and possibly uptake of exogenous lipids [41], replication and differentiation to form merozoites.
On the other hand, the transcription of Plasmodium genes essential for full intra-hepatic development is triggered by a temperature shift and contact with host cells [13], [14]. Transformation into early EEF's can to some degree be recapitulated in the absence of host cells; for this, wild type SGS require merely the presence of serum or bicarbonate and a temperature shift from the mosquito's body temperature (≈21°C) to the mammalian host's 37°C [13], [15]. However, our data clearly show that in the absence of the RNA binding protein Puf2, initiation of sporozoite to EEF metamorphosis takes place inside mosquito salivary glands without the need for environmental cues received during transmission from the mosquito vector to the mammalian host. Although the morphological changes observed at the light-microscope level could be interpreted as resulting from non-specific degenerative processes, our data on specific transcriptional changes, changes in protein synthesis and ultrastructural features indicate that the phenotype of puf2- SGS is the result of a specific differentiation process into EEFs. Indeed, the alterations in transcription, protein expression and ultrastructural features in puf2 - sporozoites match those occurring in early liver stage forms [30], [42] and are not easily reconciled with random degenerative processes. Importantly, the cellular and molecular events leading to the metamorphosis of puf2 - SGS into early hepatic stages occur prior to any apparent morphological changes, as shown by the loss of infectivity of puf2- SGS before any morphological changes are manifest.
In many organisms, Puf proteins inhibit translation of specifically recognized mRNAs (generally a small number), either by repressing their translation or enhancing decay [1]. Consistent with this conserved biological function, we show that Plasmodium Puf2 is localized to few cytoplasmic speckles and possesses a highly conserved Pumilio homology domain (PHD). Together with our transcriptional analyses, these data strongly suggest that Puf2 is regulating the translational efficiency of one or several unknown key factor(s); we hypothesize that once translationally activated such proteins quickly direct the developmental progression from SGS to early hepatic stages.
Recently, the sporozoite's latency status was reported to rely on mechanisms akin to mammalian and yeast stress granule formation with a phosphorylation dependent inhibition of protein translation [32]. Absence of eIF2α phosphorylation in a mutant lacking expression of the PBIK2 protein–the pbik2 gene was originally identified as upregulated in sporozoites 1 or uis1 [43]–was shown to result in an approximately 2-fold increase in translation as measured by 35S-Met/Cys incorporation in sporozoites at 25°C, and 3-fold at 37°C. However, close observation of ik2- parasites in Anopheles salivary glands revealed that only 11.6±8% parasites show signs of transformation by day 30 of infection, while more than 99% of puf2- parasites are already fully rounded up by that time (Fig. S10). Thus, despite a significant increase in protein translation in the absence of PBIK2, pbik2- sporozoites do not initiate the program of transformation as significantly as puf2- sporozoites. This difference in phenotype could be explained by a dominant role of Puf2 in binding to essential mRNAs repressing their translation into proteins that are needed for the transformation program to occur; in our proposed model (Fig. 7), we speculate the absence of Puf2 is consistent with the translation of these essential transcripts thereby triggering premature metamorphosis. This may alleviate the translational repression promoted by IK2 [32], perhaps involving protein phosphatase 2C (PBANKA_091340) [44] which is strongly up-regulated in puf2- parasites at day 18 of mosquito infection (Table S3L). Although we have very limited data on proteome changes in the puf2- parasites, our Western analyses indicate that changes (both up and down) in steady state protein levels do occur. It remains unclear whether Puf2 independent translation is mediated through eIF2α, although ik2 is already significantly decreased in puf2- parasites and protein phosphatase activity might be increased by day 18 of infection.
At present the nature of the mRNAs directly regulated by Puf2 are unknown; an exploratory MEME analysis of DR transcripts identified at all time points was inconclusive, most likely due to the fact that both the consensus Pumilio recognition motif (UGUAAA/UAU) and untranslated regions of P. berghei mRNAs are extremely AU-rich; out of the 374 transcripts detected to be significantly de-regulated in the puf2 gene deletion mutant, 106 have at least 1 NRE (Nanos Response Element) within 400 nucleotides of the stop codon. Statistically, there is no enrichment for NRE's in up or down-regulated genes (chi-square, df = 1, p = 0.2238; Table S3N) with the important caveat that actual 3′ UTRs have rarely been mapped in P. berghei; hence these bio-informatic results are very speculative and identified NRE's may not exist in mature mRNAs. Nonetheless, Puf2 clearly maintains SGS on a “stand-by”, quiescent mode until they have invaded mammalian host hepatocytes where they expand into first-generation, blood-infectious merozoites. Thus, Puf2 constitutes a key player in the developmental control during a critical time-point of the Plasmodium life cycle, the malaria parasites' transmission from the invertebrate to the vertebrate host. Altogether, and considering the highly conserved nature of PUFs, this shows that post-transcriptional events are central to the major developmental switches that are associated with host transition during the Plasmodium life cycle.
Materials and Methods
Laboratory animals
This study was carried out in strict accordance with the recommendations of both the Animal Experiment Committees governed by section 18 of the Experiments on Animals Act and registered by the Dutch Inspectorate for Health, Protection and Veterinary Public Health (Ministry of Health, Welfare and Sport), and the Portuguese official Veterinary Directorate, which complies with the Portuguese Law (Portaria 1005/92). The Dutch and Portuguese Experiments on Animal Act strictly comply with the European Guideline 86/609/EEC and follow the FELASA (Federation of European Laboratory Animal Science Associations) guidelines and recommendations concerning laboratory animal welfare. In The Netherlands, all animal experiments were approved by the Animal Experiments Committee of the LUMC (ADEC). In Portugal, all animal experiments were approved by the Portuguese official veterinary department for welfare licensing and the IMM Animal Ethics Committee. All experiments were carried out using Swiss-OF1 female mice (OF1-ico, Construct 242; age 6 weeks old, Charles River Laboratories International, Inc), C57Bl/6 and BALB/c mice (6–8 weeks of age; Harlan Laboratories, Inc. or Charles River Laboratories International, Inc). All efforts were made to ensure minimal suffering to the animals.
Generation of puf1 (gene model PBANKA_123350), puf2 (gene model PBANKA_071920) and puf1/puf2 P. berghei gene deletion mutants
puf1 and puf2 were targeted for disruption by standard double-crossover homologous recombination with linearized targeting plasmids. Transfection and drug selection of mutant parasites was performed using standard technology of genetic modification developed for P. berghei [45], [46]. Cloned parasite lines were obtained by limiting dilution. Plasmid integration into the genome was verified by Southern analysis of separated chromosomes and diagnostic PCR; the absence of transcript was confirmed by Northern analysis. For details of vectors, targeting regions, and primers used see Figs. S2 and S3, as well as Tables S4-7. Plasmids pL0001 and pL0035 can be obtained from http://www.mr4.org. Details for all Rodent Malaria genetically modified P. berghei lines used in this study can be found in the RMgm database (http://www.pberghei.eu).
For puf1 gene deletion, PCR-amplified 5′ and 3′ targeting regions were cloned into plasmid pL0001 yielding pAB60 (containing the pyrimethamine tgdhfr/ts selection marker), or plasmid pL0035 yielding pL1214 (containing the pyrimethamine/5-fluorocytosine hdhr/yfcu positive/negative selection marker). Mutant 351cl1 (pAB60; puf1-a; RMgm-513) was generated in the GFP-reference line cl15cy1 [45], mutant 900m2cl3 (pL1214; puf1-b; RMgm-514) was generated in the GFP+ reference line 507cl1 (RMgm-7 at http://www.pberghei.eu).
The selection cassette (hdhfr/yfcu) in 900m2cl3 was removed by negative selection [47]; four mice infected with parent population 900 were treated with 5-fluorocytosine (5-FC) at a parasitemia of 0.1–0.5% with a single, daily 20 mg/ml dose (0.5 ml) for a period of 4 days. Resistant parasites were collected at days 5–7 and analyzed by diagnostic Southern analysis to confirm removal of the drug-selectable marker hdhfr/yfcu by a recombination event between the two 3′-dhfr-ts sequences (Fig. S2). A PCR amplified fragment of the 3′-dhfr-ts region was used for Southern analysis (the primer sequences are provided in Table S5). Parasites from mouse 2 were cloned by limiting dilution, resulting in mutant 900m2cl3 (puf1-b).
For puf2 gene deletion, PCR-amplified 5′ and 3′ targeting regions were cloned into plasmid pL0001 yielding pAB70 (containing the pyrimethamine tgdhfr/ts selection marker), or plasmid pL0006 yielding pL1317 (containing the pyrimethamine hdhfr selection marker). Mutant 375cl1 (pAB70; puf2-a; RMgm-515) was generated in the GFP-reference line cl15cy1 [45], mutant 1267cl2 (pL1317; puf2-b; RMgm-516) was generated in the GFP+ reference line 507cl1 (RMgm-7 at http://www.pberghei.eu).
The following probes were used for Southern analysis: PCR-amplified fragments for the hdhfr and tgdhfr-ts genes (for primer sequences see Table S7) and a puf2 sequence consisting of the 0.4 kb EcoRI/HincII puf2 fragment; in puf2- this part is deleted.
In experiment 1081 we generated a mutant line in which both puf1 and puf2 were deleted. To generate mutant 1081cl1 (puf1-/2-; RMgm-591) the selectable marker cassette hdhfr-yfcu was first removed from mutant 900 (puf1-b) by negative selection essentially as described [47]. In brief, 4 mice infected with mutant 900 were treated with 5-fluorocytosine (5-FC) starting at a parasitemia of 0.1–0.5% with a daily single dose of 0.5 ml of a solution of 20 mg/ml day for a period of 4 days. Resistant parasites were collected between days 5–7 after start of the 5-FC treatment and the genotype analyzed by diagnostic Southern analysis to confirm removal of the drug-selectable marker hdhfr-yfcu by a recombination event between the two 3′ pbdhfr/ts sequences (Fig. S2). Parasites from one of the four mice (mouse 2) that had been treated with 5-FC were cloned by limiting dilution, resulting in mutant 900m2cl3 (puf1-b). Parasites of line 900m2cl3 were then transfected with vector pL1317 for disruption of puf2 (Fig. S3). Selection and cloning of transformed parasites resulted in mutant 1081cl1 (puf1-/2-) in which both pumilio genes are disrupted.
Gene expression analysis by Northern blot and RT-PCR
Total RNA was isolated from blood stage parasites from asynchronous and synchronized infections [48] and analyzed by Northern hybridization. Northern blots were hybridised with puf1 and puf2 PCR-amplified fragments (for primer sequences see Tables S5 and S7). As loading control, blots were hybridized with p28 (PBANKA_051490) or with primer L644R specific for the blood stage, large subunit ribosomal RNA [49].
For RT-PCR, total RNA was isolated from highly purified gametocytes and day 18 and 27 sporozoites and reverse transcribed with hexamers and oligo d(T) oligonucleotides; primers were 479 and 480 for puf1, and 477 and 478 for puf2, in both cases spanning an intron (Table S8).
Asexual growth rate, gametocytogenesis and gametogenesis
The in vivo multiplication rate of asexual blood stage parasites was determined during the cloning procedure and calculated as follows: the percentage of infected erythrocytes in Swiss OF1 mice injected with a single parasite is determined at day 8 to 11 by counting Giemsa stained blood films; the mean asexual multiplication rate per 24 h is then calculated assuming a total of 1.2×1010 erythrocytes per mouse (2 ml of blood). The percentage of infected erythrocytes in mice infected with wild type reference lines of the P. berghei ANKA strain typically ranges between 0.5–2% at day 8 after infection, resulting in a mean multiplication rate of 10 per 24 h [50], [51].
Gametocyte and gamete production were determined following standardized conditions [48]. Gametocyte production is defined as the Gametocyte Conversion Rate which is the percentage of ring forms that develop into mature gametocytes in synchronized infections in mice treated with phenylhydrazine. Male gamete formation is defined as the percentage of male gametocytes that form gametes after in vitro induction by exflagellation; exflagellating male gametocytes are counted in a Bürker cell counter 15 to 20 minutes after induction. Female gamete formation is defined as the percentage of female gametocytes that emerge from the red blood host cells after in vitro induction of gametogenesis; free female gametes were counted in Giemsa stained smears made 20 minutes after induction. The fertility of wild type and mutant gamete populations was analysed by standard in vitro fertilisation and ookinete maturation assays [52], [53] from highly pure gametocyte populations [54]; the fertilisation rate of gametes is defined as the percentage of female gametes that develop into mature ookinetes determined by counting female gametes and mature ookinetes in Giemsa stained blood smears 16–18 h after in vitro induction.
Human hepatoma cell line Huh7. Huh7 cells were cultured in RPMI medium supplemented with 10% fetal calf serum (FCS), 1% non-essential amino acids, 1% penicillin/streptomycin, 1% glutamine and 10 mM Hepes, pH 7 and maintained at 37°C with 5% CO2. All consumables were obtained from Gibco/Invitrogen.
Anopheles stephensi mosquito maintenance
A. stephensi were bred at the insectary of the Instituto de Medicina Molecular (IMM). All life cycle associated experiments (mosquito infection, in vitro Huh7 infection, in vivo mouse infection) presented in this paper were performed with GFP+ puf1 - clone 900m2cl3 and puf2 - clone 1267cl2 and confirmed with GFP - puf1- 351cl1 and puf2- 375cl1.
Anopheles stephensi mosquito infection and analysis of parasite development
1×106 infected red blood cells of P. berghei wild type (259cl2; RMgm-5; GFP+) [52] and mutant lines, puf1-and puf2- were intraperitoneally injected in BALB/C mice . Four to 5 days later, when at least one exflagellation event was observed per microscope field, mosquitoes were allowed to feed on anaesthetized mice for 0.5–1 h on two consecutive days. At day 10 post blood meal, 9 infected midguts were removed and the number of oocysts per midgut determined by fluorescence microscopy. Parasites per salivary gland (SG) were quantified in 3 independent transmission experiments in which 9 infected mosquitoes per experiment, from 19 to 22 days after mosquito infection for each genotype, were dissected; three groups of 3 SGs for each experiment for each genotype were smashed and the number of parasites per SG quantified in a Neubauer chamber.
Quantification and morphological analyses of mutant sporozoites
Three A. stephensi SGs infected with wild type, puf1 - or puf2 - parasites were removed on days 18, 22, 24, 27 and 30 after infection. SGs were smashed to release the parasites, and the proportion of sporozoites to EEFs-like quantified. On day 30 of mosquito infection, whole infected SGs were mounted in glass bottom culture dishes (MatTek Corporation); a Zeiss LSM 510 META confocal microscope (Zeiss, Oberkochen, Germany) was used to perform a Z-series scan followed by 3D reconstructions of the infected SGs using Imaris (Bitplane AG, Switzerland).
In vitro sporozoite hepatocyte infectivity
For all experiments, wild type, puf1- and puf2- salivary gland sporozoites were collected on day 18 after the mosquito blood meal.
Sporozoite gliding was evaluated with 3×104 sporozoites for 40 minutes in complete RPMI, at 37°C on glass cover slips covered with anti-circumsporozoite protein (CSP) monoclonal antibody [3D11; 53]. Sporozoites were subsequently fixed in 4% paraformaldehyde (PFA) for 10 minutes and stained with anti-CSP. The percentage of sporozoites associated with CSP trails was quantified by fluorescence microscopy.
Cell traversal assays were performed with 3×104 sporozoites added to 7×104 Huh7 cells (seeded on the previous day) in the presence of 1 mg/ml of cell-impermeable dextran tetramethylrhodamine (10 000 MW), lysine fixable (fluoro-ruby) (Molecular Probes/Invitrogen). After 2 hours, the percentage of dextran-positive cells was quantified by fluorescence-activated cell sorting (FACS)[55].
In order to quantify cell invasion, 3×104 sporozoites were added to Huh7 cells. Infection was stopped after 2 h by addition of PFA 4%; double staining with anti-CSP was performed according to [56] in order to distinguish extracellular from intracellular sporozoites. Intra-hepatic development was assessed by fixing infected Huh7 cells at 48 h p.i. with 4% PFA. Parasites were stained with anti-GFP antibody conjugated with FITC (Molecular Probes/Invitrogen). Pictures were taken on an Axiovert 200 M fluorescence microscope and EEF size measured using ImageJ 1.38 h software.
In vivo sporozoite infectivity
Male C57BL/6 mice (6–8 weeks) were intravenously (i.v.) injected with 1×104 18-day SG sporozoites (wild type, puf1- or puf2-). After 44 hours liver infection load was quantified by qRT-PCR analysis of P. berghei 18S rRNA normalized against hypoxanthine-guanine phosphoribosyltransferase (HPRT) (for primers see Table S4).
To assess mutant parasites capacity to pass through the liver and reach the blood, 1×104 sporozoites were injected i.v. into C57BL/6 mice.
To verify mutant sporozoites infectivity during natural infection, C57BL/6 mice were exposed to 4 infected mosquitoes for 30 minutes. All mice were bitten by at least by one infected mosquito. Parasitemia were checked by Giemsa-stained blood smear daily until 10 days post infection.
Electron microscopy
A. stephensi salivary glands infected with wild type, puf1 - or puf2 - parasites were removed and fixed in 2.5% gluteraldehyde in 0.1 M sodium cacodylate (pH = 7.3) for 48 hours at 4°C, followed by 3 10-minute washes in 0.1 M sodium cacodylate. All tissues were post fixed in 1% OsO4 in deionized water, washed and counterstained with uranil acetate for 30 minutes. After washing with de-ionized water for 10 minutes, dehydration with ethanol (70% and 96%, 1 minute each) was performed followed by 2 10-minute incubations in absolute ethanol and propylene oxide. Salivary glands were finally infiltrated with 1∶1 propylene oxyde and EPON resin for 30 minutes followed by overnight infiltration in 100% EPON's resin. The tissues were embedded in flat molds in 100 EPON for 48 hours at 70°C. Ultra-thin sections of 70 nm were cut with a diamond knife (Diatome 45°) in a ultra-microtome (Reichert Jung Ultracut-E), collected on copper grids (mesh 200 hexagonal) and stained with Reynolds lead citrate and 2% uranil acetate (5+5 minutes). The grids were observed on a Jeol JEM-100cxI transmission electron microscope.
Expression profiling Reverse Transcriptase (RT)-PCRs and RT-qPCR
Wild type, puf1- and puf2- sporozoites were extracted at days 18 and 27 post mosquito infection; total RNA was extracted with TRIzol, and 400 ng total RNA reverse transcribed in the presence of random hexamers and oligo d(T) oligonucleotides with Superscript II. 25 ng were used in a PCR using the following cycling parameters: 94°C 3 minutes, 35 cycles of 94°C 10 seconds and 1 minute at 60°C, with a final elongation step of 10 minutes. PCR amplicons were run on 2% agarose gels. Oligonucleotide primers are shown in Table S3. Negative controls were performed with RT-negative samples (data not shown). RT-qPCR analyses were performed on cDNA prepared from day 18 wild type and puf2- salivary gland sporozoites; oligonucleotide primers are shown in Table S8. qPCR was done with Power SYBR Green (Applied Biosystems) according to the manufacturer's instructions. Three independent biological replicate cDNA samples were tested for each parasite. ABI 7500 Fast Sequence Detection System. Cycling parameters for all genes were: 95°C for 15 minutes, followed by 50 cycles of 95°C|15 seconds, 55°C|15 seconds, 60°C|45 seconds, followed by melting curve analyses. Relative mRNA abundance for each transcript was determined by the 2−ΔΔCt method following ABI User Bulletin 2; expression data was normalised versus ama-1. Final values were log2 transformed to be comparable to subsequent microarray data.
Expression profiling by microarray hybridization
The RMSANGER Affymetrix custom tiling array was designed against the 8 x genome assemblies for P. berghei and P. chabaudi. Prior to analysis, all 6.3 million probes were remapped using the exonerate software (http://http://www.ebi.ac.uk/~guy/exonerate) against the latest P. berghei genome assembly available from the Wellcome Trust Sanger institute (ftp://ftp.sanger.ac.uk/pub/pathogens/P_berghei/February_2011); all non-exact matches and redundant probes were discarded. A custom CDF file was generated using a combination of Perl scripts to analyse gene expression profiles of all ≈5000 annotated genes. 18 and 27 days sporozoites were dissected from salivary glands of Anopheles stephensi mosquitoes infected with wild type (ANKA GFPcon 259cl2) or puf2- (1267cl2 ). RNA from 3 independent infections each was extracted with TRIzol according to the manufacturer's instructions. Double amplified cDNA was synthesized using the Ambion WT Expression kit starting with ≈400 ng of mRNA and labelled using the Affymetrix Genechip WT Terminal Labeling and Hybridisation Kit according to the manufacturers' protocols. 18 hours hybridisations, washing, and staining were done according to Affymetrix recommendations. Genechip arrays were scanned with an Affymetrix 7G scanner. Raw scanned images were acquired using Affymetrix software suite GCOS and raw CEL files transferred to R/Bioconductor for pre-processing. The 3×wild type 18 days pi, 3×wild type 27 days pi, 3×puf2- 18 days pi and 2×puf2- 27 days p.i. hybridised arrays were background subtracted, quantile normalised and median polished using RMA [57]. An overall F-test was used to select for 374 variant genes using an adjusted p-value <0.05 (after correction for false discovery rate using the Bonferroni-Hochberg adjustment). A linear modelling was used to extract differential expression (DE) for each pair wise comparison using the Limma package [58]. Gene Ontology enrichment was tested using GOstats [59], GO.db (M. Carlson, S. Falcon, H. Pages and N. Li. GO.db: A set of annotation maps describing the entire Gene Ontology. R package version 2.3.5.) and GohyperGall function as described in [60] using the GO terms annotated for P. falciparum orthologs (version 5/31/2010, downloaded from http://www.geneontology.org/GO.downloads.annotations.shtml). All microarray gene expression data are presented in Table S3. Microarray gene expression for selected genes was validated with RT-qPCR (Fig. S7). Microarray data have been submitted to ArrayExpress under the accession number E-TABM-1067.
Protein expression profiling by Western Blot
Wild type and puf2- sporozoites were extracted from mosquito salivary glands at day 18 post infection. An amount of protein corresponding to 300 000 sporozoites was loaded in each well of a 10% polyacrylamide gel and transferred to nitrocellulose membrane (Protran) by electroblotting. Protein expression levels were determined by incubating the membranes overnight at 4°C, with the following primary antibodies: anti-Exp1 (kindly provided by Volker Heussler), 1∶1000; anti-Exp2 (kindly provided by Paul Gilson and Brendan Crabb) 1∶1000; anti-Myo-A (kindly provided by Julian Rayner), 1∶300 and anti-Alveolin-9, 1∶300 and subsequent incubation with horseradish-peroxidase conjugated secondary antibody. Immunostained proteins were visualized with chemiluminescence detection (Thermo Scientific).
Immunofluorescence assay of Puf2
Red fluorescent protein (RFP)+ sporozoites from the wild type reference line 733cl1 (RMgm-86) were dissected at day 23 post infection and washed once in 1X PBS (9300 rcf, 7 minutes, 4°C). 6500 parasites in 10 µl were allowed to adhere to polylysine slides, fixed for 15 minutes with 4% PFA, and washed 3×5 minutes with 1X PBS. After a 10-minutes wash with fresh 0.1 M Glycine buffer, sporozoites were permeabilized with 0.1% Triton-X100 for 10 minutes followed by a 3x5 minutes wash with 1X PBS. Slides were blocked 20 minutes at RT in 1% Albumin and incubated O/N with polyclonal rabbit anti-Puf antiserum (dilution 1∶300) upside down. Sera 904 and 905 were raised in rabbits immunised against FKDNLYNLKELNSW and ENLDKLKEETYILR at Eurogentec. Slides were washed 3x 15minutes in 1X PBS and incubated with donkey anti-rabbit, Alexa 488-conjugated secondary antibody, 30 minutes, 37°C (1∶400) again upside down. Slides were washed 3x 15 minutes in PBS 1X, and then incubated 3 minutes with DAPI, RT. Prior to mounting, slides were washed for 5 minutes and analysed with a widefield Zeiss Axiovert 200M microscope, with 63x, 1.40 NA objective. To ascertain sera specificity, pre-adsorption experiments using 5 µg of peptides were used together with labelling using an unrelated rabbit polyclonal antibody (data not shown). Donkey anti-rabbit was used without a primary antibody to make sure no cross reaction was to be observed (data not shown).
Protein inhibitor experiment on salivary gland at 18 days post mosquito infection
Day 18 salivary gland sporozoites (SGS) were hand dissected from both wild type and puf2- parasite lines. 96-wells plate were seeded with 20,000 SGS in triplicate for both wild type and puf2- with or without 100 ug/ml (357.1 µM) of Cycloheximide (Sigma) in RPMI medium (without FBS supplement) and allowed to develop for 2 and 4 h at room temperature or 37°C. Three images were taken for each well using a widefield Zeiss Axiovert 200 M microscope, with 20x, 1.40 NA objective and parasites counted using ImageJ software to determine slender versus round.
List of accession numbers
Puf1/UIS9 (PFE0935c, PBANKA_123350), Puf2 (PFD0825c, PBANKA_071920), Exp-2 (PBANKA_133430), Exp-1 (PBANKA_092670), Ama-1 (PBANKA_091500), GAP45 (PBANKA_143760), Myo-A (PBANKA_135570), Spect2 (PBANKA_100630), CelTOS (PBANKA_143230), Spect1 (PBANKA_135560), UIS4 (PBANKA_050120), UIS1/IK2 (PBANKA_020580), TLP1 (PBANKA_111600), TRSP (PBANKA_020910), SIAP1 (PBANKA_100620), MTRAP (PBANKA_051280), TREP (PBANKA_130650), PSOP9/GAMA (PBANKA_070190), P36p (PBANKA_100220), TFIIH (PBANKA_141340), RNA polymerase II subunit (PBANKA_020330), AP2 (PBANKA_083520 and PBANKA_010950), TFIIS Zinc-fingers (PBANKA_030420 and PBANKA_142110), Plasmepsin V (PBANKA_133870), RAD51 (PBANKA_093950), Histone H2B (PBANKA_094180) and ALBA3 (PBANKA_120440), Alveolin 9 (PBANKA_124060), Protein phosphatase 2C, putative (PBANKA_091340).
Supporting Information
Zdroje
1. WickensMBernsteinDSKimbleJParkerR
2002
A PUF family portrait: 3′UTR regulation as a way of
life.
Trends Genet
18
150
157
2. CrittendenSLBernsteinDSBachorikJLThompsonBEGallegosM
2002
A conserved RNA-binding protein controls germline stem cells in
Caenorhabditis elegans.
Nature
417
660
663
3. LinHSpradlingAC
1997
A novel group of pumilio mutations affects the asymmetric
division of germline stem cells in the Drosophila ovary.
Development
124
2463
2476
4. MooreFLJaruzelskaJFoxMSUranoJFirpoMT
2003
Human Pumilio-2 is expressed in embryonic stem cells and germ
cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like
proteins.
Proc Natl Acad Sci U S A
100
538
543
5. MurataYWhartonRP
1995
Binding of pumilio to maternal hunchback mRNA is required for
posterior patterning in Drosophila embryos.
Cell
80
747
756
6. SzakmaryACoxDNWangZLinH
2005
Regulatory relationship among piwi, pumilio, and bag-of-marbles
in Drosophila germline stem cell self-renewal and
differentiation.
Curr Biol
15
171
178
7. WangZLinH
2004
Nanos maintains germline stem cell self-renewal by preventing
differentiation.
Science
303
2016
2019
8. VaughanAMO'NeillMTTarunASCamargoNPhuongTM
2009
Type II fatty acid synthesis is essential only for malaria
parasite late liver stage development.
Cell Microbiol
11
506
520
9. PrudencioMRodriguezAMotaMM
2006
The silent path to thousands of merozoites: the Plasmodium liver
stage.
Nat Rev Microbiol
4
849
856
10. MotaMMPradelGVanderbergJPHafallaJCFrevertU
2001
Migration of Plasmodium sporozoites through cells before
infection.
Science
291
141
144
11. MotaMMHafallaJCRodriguezA
2002
Migration through host cells activates Plasmodium sporozoites for
infection.
Nat Med
8
1318
1322
12. BillkerOLindoVPanicoMEtienneAEPaxtonT
1998
Identification of xanthurenic acid as the putative inducer of
malaria development in the mosquito.
Nature
392
289
292
13. KaiserKCamargoNKappeSH
2003
Transformation of sporozoites into early exoerythrocytic malaria
parasites does not require host cells.
J Exp Med
197
1045
1050
14. SiauASilvieOFranetichJFYalaouiSMarinachC
2008
Temperature shift and host cell contact up-regulate sporozoite
expression of Plasmodium falciparum genes involved in hepatocyte
infection.
PLoS Pathog
4
e1000121
15. HeggeSKudryashevMBarniolLFrischknechtF
2010
Key factors regulating Plasmodium berghei sporozoite survival and
transformation revealed by an automated visual assay.
FASEB J
24
5003
12
16. MairGRBraksJAGarverLSWiegantJCHallN
2006
Regulation of sexual development of Plasmodium by translational
repression.
Science
313
667
669
17. MairGRLasonderEGarverLSFranke-FayardBMCarretCK
2010
Universal features of post-transcriptional gene regulation are
critical for Plasmodium zygote development.
PLoS Pathog
6
e1000767
18. KhanSMFranke-FayardBMairGRLasonderEJanseCJ
2005
Proteome analysis of separated male and female gametocytes
reveals novel sex-specific Plasmodium biology.
Cell
121
675
687
19. SilvieOGoetzKMatuschewskiK
2008
A sporozoite asparagine-rich protein controls initiation of
Plasmodium liver stage development.
PLoS Pathog
4
e1000086
20. AlyASMikolajczakSARiveraHSCamargoNJacobs-LorenaV
2008
Targeted deletion of SAP1 abolishes the expression of infectivity
factors necessary for successful malaria parasite liver
infection.
Mol Microbiol
69
152
163
21. PorterRJLairdRLDusseauEM
1954
Studies on malarial sporozoites. II. Effect of age and dosage of
sporozoites on their infectiousness.
Exp Parasitol
3
267
274
22. ZhangBGallegosMPuotiADurkinEFieldsS
1997
A conserved RNA-binding protein that regulates sexual fates in
the C. elegans hermaphrodite germ line.
Nature
390
477
484
23. ZamorePDWilliamsonJRLehmannR
1997
The Pumilio protein binds RNA through a conserved domain that
defines a new class of RNA-binding proteins.
Rna
3
1421
1433
24. WangXZamorePDHallTM
2001
Crystal structure of a Pumilio homology domain.
Mol Cell
7
855
865
25. KennedyBKGottaMSinclairDAMillsKMcNabbDS
1997
Redistribution of silencing proteins from telomeres to the
nucleolus is associated with extension of life span in S.
cerevisiae.
Cell
89
381
391
26. FanQLiJKariukiMCuiL
2004
Characterization of PfPuf2, member of the Puf family RNA-binding
proteins from the malaria parasite Plasmodium falciparum.
DNA Cell Biol
23
753
760
27. CuiLFanQLiJ
2002
The malaria parasite Plasmodium falciparum encodes members of the
Puf RNA-binding protein family with conserved RNA binding
activity.
Nucleic Acids Res
30
4607
4617
28. MiaoJLiJFanQLiXLiX
2010
The Puf-family RNA-binding protein PfPuf2 regulates sexual
development and sex differentiation in the malaria parasite Plasmodium
falciparum.
J Cell Sci
123
1039
1049
29. Le RochKGZhouYBlairPLGraingerMMochJK
2003
Discovery of gene function by expression profiling of the malaria
parasite life cycle.
Science
301
1503
1508
30. WangQBrownSRoosDSNussenzweigVBhanotP
2004
Transcriptome of axenic liver stages of Plasmodium
yoelii.
Mol Biochem Parasitol
137
161
168
31. de Koning-WardTFGilsonPRBoddeyJARugMSmithBJ
2009
A newly discovered protein export machine in malaria
parasites.
Nature
459
945
949
32. ZhangMFennellCRanford-CartwrightLSakthivelRGueirardP
2010
The Plasmodium eukaryotic initiation factor-2alpha kinase IK2
controls the latency of sporozoites in the mosquito salivary
glands.
J Exp Med
207
1465
1474
33. BoddeyJAHodderANGuntherSGilsonPRPatsiourasH
2010
An aspartyl protease directs malaria effector proteins to the
host cell.
Nature
463
627
631
34. JayabalasinghamBBanoNCoppensI
2010
Metamorphosis of the malaria parasite in the liver is associated
with organelle clearance.
Cell Res
20
1043
59
35. TarunASPengXDumpitRFOgataYSilva-RiveraH
2008
A combined transcriptome and proteome survey of malaria parasite
liver stages.
Proc Natl Acad Sci U S A
105
305
310
36. KuoMWWangSHChangJCChangCHHuangLJ
2009
A novel puf-A gene predicted from evolutionary analysis is
involved in the development of eyes and primordial
germ-cells.
PLoS One
4
e4980
37. CrittendenSLEckmannCRWangLBernsteinDSWickensM
2003
Regulation of the mitosis/meiosis decision in the Caenorhabditis
elegans germline.
Philos Trans R Soc Lond B Biol Sci
358
1359
1362
38. BarkerDDWangCMooreJDickinsonLKLehmannR
1992
Pumilio is essential for function but not for distribution of the
Drosophila abdominal determinant Nanos.
Genes Dev
6
2312
2326
39. AlbuquerqueSSCarretCGrossoARTarunASPengX
2009
Host cell transcriptional profiling during malaria liver stage
infection reveals a coordinated and sequential set of biological
events.
BMC Genomics
10
270
40. SinghAPBuscagliaCAWangQLevayANussenzweigDR
2007
Plasmodium circumsporozoite protein promotes the development of
the liver stages of the parasite.
Cell
131
492
504
41. RodriguesCDHannusMPrudencioMMartinCGoncalvesLA
2008
Host scavenger receptor SR-BI plays a dual role in the
establishment of malaria parasite liver infection.
Cell Host Microbe
4
271
282
42. JayabalasinghamBBanoNCoppensI
2010
Metamorphosis of the malaria parasite in the liver is associated
with organelle clearance.
Cell Res
20
1043
1059
43. MatuschewskiKRossJBrownSMKaiserKNussenzweigV
2002
Infectivity-associated changes in the transcriptional repertoire
of the malaria parasite sporozoite stage.
J Biol Chem
277
41948
41953
44. MamounCBGoldbergDE
2001
Plasmodium protein phosphatase 2C dephosphorylates translation
elongation factor 1beta and inhibits its PKC-mediated nucleotide exchange
activity in vitro.
Mol Microbiol
39
973
981
45. JanseCJFranke-FayardBMairGRRamesarJThielC
2006
High efficiency transfection of Plasmodium berghei facilitates
novel selection procedures.
Mol Biochem Parasitol
145
60
70
46. JanseCJRamesarJWatersAP
2006
High-efficiency transfection and drug selection of genetically
transformed blood stages of the rodent malaria parasite Plasmodium
berghei.
Nat Protoc
1
346
356
47. BraksJAMairGRFranke-FayardBJanseCJWatersAP
2008
A conserved U-rich RNA region implicated in regulation of
translation in Plasmodium female gametocytes.
Nucleic Acids Res
36
1176
1186
48. JanseCJWatersAP
1995
Plasmodium berghei: the application of cultivation and
purification techniques to molecular studies of malaria
parasites.
Parasitol Today
11
138
143
49. van SpaendonkRMRamesarJvan WigcherenAElingWBeetsmaAL
2001
Functional equivalence of structurally distinct ribosomes in the
malaria parasite, Plasmodium berghei.
J Biol Chem
276
22638
22647
50. JanseCJHaghparastASperancaMARamesarJKroezeH
2003
Malaria parasites lacking eef1a have a normal S/M phase yet grow
more slowly due to a longer G1 phase.
Mol Microbiol
50
1539
1551
51. SpaccapeloRJanseCJCaterbiSFranke-FayardBBonillaJA
2010
Plasmepsin 4-deficient Plasmodium berghei are virulence
attenuated and induce protective immunity against experimental
malaria.
Am J Pathol
176
205
217
52. Franke-FayardBTruemanHRamesarJMendozaJvan der KeurM
2004
A Plasmodium berghei reference line that constitutively expresses
GFP at a high level throughout the complete life cycle.
Mol Biochem Parasitol
137
23
33
53. YoshidaNNussenzweigRSPotocnjakPNussenzweigVAikawaM
1980
Hybridoma produces protective antibodies directed against the
sporozoite stage of malaria parasite.
Science
207
71
73
54. BeetsmaALvan de WielTJSauerweinRWElingWM
1998
Plasmodium berghei ANKA: purification of large numbers of
infectious gametocytes.
Exp Parasitol
88
69
72
55. PrudencioMRodriguesCDAtaideRMotaMM
2008
Dissecting in vitro host cell infection by Plasmodium sporozoites
using flow cytometry.
Cell Microbiol
10
218
224
56. LabaiedMCamargoNKappeSH
2007
Depletion of the Plasmodium berghei thrombospondin-related
sporozoite protein reveals a role in host cell entry by
sporozoites.
Mol Biochem Parasitol
153
158
166
57. IrizarryRAHobbsBCollinFBeazer-BarclayYDAntonellisKJ
2003
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data.
Biostatistics
4
249
264
58. SmythGK
2005
Limma: linear models for microarray data.
Gentleman VCR DudoitS IrizarryR HuberW
Bioinformatics and Computational Biology Solutions using R and
Bioconductor
New York
Springer
397
420
59. FalconSGentlemanR
2007
Using GOstats to test gene lists for GO term
association.
Bioinformatics
23
257
258
60. HoranKJangCBailey-SerresJMittlerRSheltonC
2008
Annotating genes of known and unknown function by large-scale
coexpression analysis.
Plant Physiol
147
41
57
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus SpeciesČlánek SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion ReleaseČlánek Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T CellsČlánek A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the CytosolČlánek Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- Infections Are Virulent and Inhibit the Human Malaria Parasite in
- A Gamma Interferon Independent Mechanism of CD4 T Cell Mediated Control of Infection
- MDA5 and TLR3 Initiate Pro-Inflammatory Signaling Pathways Leading to Rhinovirus-Induced Airways Inflammation and Hyperresponsiveness
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- An E2F1-Mediated DNA Damage Response Contributes to the Replication of Human Cytomegalovirus
- Quantitative Subcellular Proteome and Secretome Profiling of Influenza A Virus-Infected Human Primary Macrophages
- Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus Species
- Inhibition of Both HIV-1 Reverse Transcription and Gene Expression by a Cyclic Peptide that Binds the Tat-Transactivating Response Element (TAR) RNA
- A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing Machinery
- Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Investigating the Host Binding Signature on the PfEMP1 Protein Family
- Human Neutrophil Clearance of Bacterial Pathogens Triggers Anti-Microbial γδ T Cell Responses in Early Infection
- Septation of Infectious Hyphae Is Critical for Appressoria Formation and Virulence in the Smut Fungus
- A Family of Helminth Molecules that Modulate Innate Cell Responses via Molecular Mimicry of Host Antimicrobial Peptides
- Phospholipids Trigger Capsular Enlargement during Interactions with Amoebae and Macrophages
- CTL Escape Mediated by Proteasomal Destruction of an HIV-1 Cryptic Epitope
- Evolution of Th2 Immunity: A Rapid Repair Response to Tissue Destructive Pathogens
- Extensive Genome-Wide Variability of Human Cytomegalovirus in Congenitally Infected Infants
- The Antiviral Efficacy of HIV-Specific CD8 T-Cells to a Conserved Epitope Is Heavily Dependent on the Infecting HIV-1 Isolate
- Epstein-Barr Virus Infection of Polarized Epithelial Cells via the Basolateral Surface by Memory B Cell-Mediated Transfer Infection
- Reactive Oxygen Species Hydrogen Peroxide Mediates Kaposi's Sarcoma-Associated Herpesvirus Reactivation from Latency
- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- The Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication
- AAV Exploits Subcellular Stress Associated with Inflammation, Endoplasmic Reticulum Expansion, and Misfolded Proteins in Models of Cystic Fibrosis
- Suboptimal Activation of Antigen-Specific CD4 Effector Cells Enables Persistence of In Vivo
- SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion Release
- Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T Cells
- Transition of Sporozoites into Liver Stage-Like Forms Is Regulated by the RNA Binding Protein Pumilio
- A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the Cytosol
- Transcriptome Analysis of in Human Whole Blood and Mutagenesis Studies Identify Virulence Factors Involved in Blood Survival
- Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
- Structural Insights into Viral Determinants of Nematode Mediated Transmission
- Protective Efficacy of Serially Up-Ranked Subdominant CD8 T Cell Epitopes against Virus Challenges
- Viral CTL Escape Mutants Are Generated in Lymph Nodes and Subsequently Become Fixed in Plasma and Rectal Mucosa during Acute SIV Infection of Macaques
- Taking Some of the Mystery out of Host∶Virus Interactions
- Viral Small Interfering RNAs Target Host Genes to Mediate Disease Symptoms in Plants
- : An Emerging Cause of Sexually Transmitted Disease in Women
- Mitochondrial Ubiquitin Ligase MARCH5 Promotes TLR7 Signaling by Attenuating TANK Action
- The Hexamer Structure of the Rift Valley Fever Virus Nucleoprotein Suggests a Mechanism for its Assembly into Ribonucleoprotein Complexes
- Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages during Their Emergence in Birds in Egypt
- Stromal Down-Regulation of Macrophage CD4/CCR5 Expression and NF-κB Activation Mediates HIV-1 Non-Permissiveness in Intestinal Macrophages
- A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins
- Perturbs Iron Trafficking in the Epithelium to Grow on the Cell Surface
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- : An Emerging Cause of Sexually Transmitted Disease in Women
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy