-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
In natural environments, bacteria often adhere to surfaces where they form complex multicellular communities. Surface adherence is determined by the biochemical composition of the cell envelope. We describe a novel regulatory mechanism by which the bacterium, Caulobacter crescentus, integrates cell cycle and nutritional signals to control development of an adhesive envelope structure known as the holdfast. Specifically, we have discovered a 68-residue protein inhibitor of holdfast development (HfiA) that directly targets a conserved glycolipid glycosyltransferase required for holdfast production (HfsJ). Multiple cell cycle regulators associate with the hfiA and hfsJ promoters and control their expression, temporally constraining holdfast development to the late stages of G1. HfiA further functions as part of a ‘nutritional override’ system that decouples holdfast development from the cell cycle in response to nutritional cues. This control mechanism can limit surface adhesion in nutritionally sub-optimal environments without affecting cell cycle progression. We conclude that post-translational regulation of cell envelope enzymes by small proteins like HfiA may provide a general means to modulate the surface properties of bacterial cells.
Published in the journal: A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion. PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004101
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004101Summary
In natural environments, bacteria often adhere to surfaces where they form complex multicellular communities. Surface adherence is determined by the biochemical composition of the cell envelope. We describe a novel regulatory mechanism by which the bacterium, Caulobacter crescentus, integrates cell cycle and nutritional signals to control development of an adhesive envelope structure known as the holdfast. Specifically, we have discovered a 68-residue protein inhibitor of holdfast development (HfiA) that directly targets a conserved glycolipid glycosyltransferase required for holdfast production (HfsJ). Multiple cell cycle regulators associate with the hfiA and hfsJ promoters and control their expression, temporally constraining holdfast development to the late stages of G1. HfiA further functions as part of a ‘nutritional override’ system that decouples holdfast development from the cell cycle in response to nutritional cues. This control mechanism can limit surface adhesion in nutritionally sub-optimal environments without affecting cell cycle progression. We conclude that post-translational regulation of cell envelope enzymes by small proteins like HfiA may provide a general means to modulate the surface properties of bacterial cells.
Introduction
The majority of bacteria in the biosphere exist within surface-attached communities [1]–[3] that facilitate metabolic cooperation, sharing of genetic information, and protect cells against stress (reviewed in [1]). Environmental signals including nutrient availability, pH, and ion concentrations influence surface community formation by modulating expression of adhesive cell envelope structures and extracellular polymers that determine surface attachment (reviewed in [4]). The Gram negative bacterium, Caulobacter crescentus, thrives in dilute freshwater ecosystems and has the ability to permanently attach [5], [6] to a chemically diverse range of surfaces [7]–[10] via a polysaccharide-rich, polar organelle known as the holdfast [9], [11]–[13]. As organic polymers and ions concentrate on material surfaces in aquatic environments [14], surface attachment likely provides C. crescentus a nutritional advantage. Given that holdfast surface attachment is permanent, C. crescentus should exhibit tight control over holdfast development to ensure that cells do not become perpetual residents of a poor environment. In this study, we have sought to elucidate the molecular regulatory determinants of holdfast development in C. crescentus.
Elaboration of the holdfast adhesin in C. crescentus is cell-cycle-regulated, though it is not requisite for cell-cycle progression [8], [15]–[17]. The cell cycle yields two cell types that are physiologically, morphologically and functionally distinct (Figure 1A). The flagellated and motile swarmer cell provides this species a means for dispersal; this cell type is arrested in G1 and incapable of replication. In order to initiate growth and replication, the swarmer relinquishes motility and differentiates into a stalked cell. The stalked cell, specialized for nutrient uptake, grows and divides asymmetrically to generate a new swarmer cell upon division [8], [18]. Development of the holdfast at the cell surface is temporally restricted to the late swarmer cell stage, where it emerges at the nascent stalked cell pole ([15], [17], Figure 1A). However, the timing of holdfast emergence within this developmental window can be hastened at the post-translational level by physical contact of the flagellum with surfaces [19]. Once constructed, the holdfast is a permanent feature of the cell surface that is not shed or reassimilated. Premature holdfast development at the nascent swarmer pole prior to cell division would hinder dispersal of newborn swarmer cells. Thus cell-cycle control of holdfast biogenesis helps to ensure appropriate cell dispersal.
Fig. 1. lovK-lovR–enhanced adhesion and transcriptional repression of CC_0817 (hfiA). 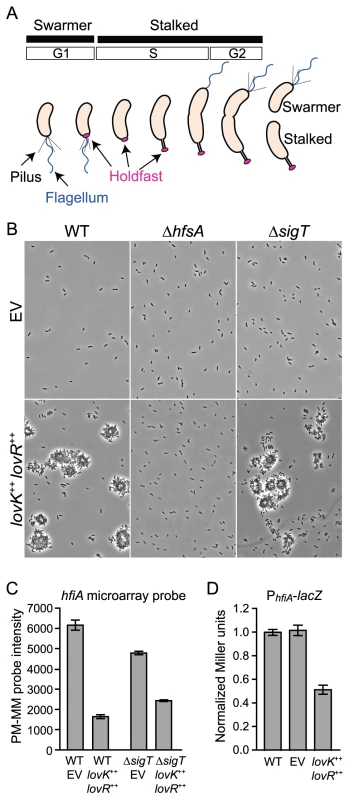
A. C. crescentus cell cycle yields two cell types: swarmer and stalked. Development of polar structures including pili (black), flagellum (blue) and the adhesive holdfast (pink) is cell-cycle regulated. B. Phase contrast micrographs of wild-type (WT), ΔhfsA or ΔsigT cells bearing either empty plasmids (EV) or lovK and lovR overexpression plasmids (lovK++, lovR++) grown in M2XV medium. C. Affymetrix microarray expression values (perfect match minus mismatch probes: PM-MM) indicating transcript level for hfiA in log phase cells grown in M2XV. Bars represent mean ± range of two independent cultures. hfiA transcript is significantly lower in lovK-lovR overexpression strains compared to isogenic empty vector controls (ANOVA followed by Tukey's post test; p<0.001). D. β-galactosidase activity from a PhfiA-lacZ transcriptional fusion (pRKlac290-PhfiA) in wild-type (WT), vector control (EV), and lovK–lovR overexpression strains grown in M2XV. Bars represent mean ± s.e.m. of 11 independent cultures assayed on 4 different days. PhfiA-lacZ activity in the lovK-lovR overexpression strain is significantly lower than WT and vector control strains (ANOVA followed by Tukey's post test; p<0.001). We have previously observed that a two-component regulatory system composed of the soluble sensor histidine kinase, LovK, and the single domain receiver, LovR, regulates the Caulobacter general stress response [20] and modulates cell adhesion [21]. We sought to understand the mechanism of adhesion control and have discovered a novel inhibitor of holdfast development, hfiA, that is regulated downstream of lovK-lovR. A forward genetic screen for HfiA-insensitive mutants identified suppressing mutations in a glycosyltransferase gene required for holdfast development, which we name hfsJ. We demonstrate a physical interaction between HfiA and HfsJ, and that suppressing mutations in HfsJ attenuate the HfsJ-HfiA interaction. These results support a model in which HfiA inhibits holdfast development via direct interaction with an enzyme required for holdfast biosynthesis.
Expression of hfiA is temporally regulated across the cell cycle, and is lowest during the period when the holdfast is elaborated at the cell surface. Multiple Caulobacter developmental regulators, CtrA, GcrA and StaR, physically occupy and control transcription from the hfiA promoter. The coordinate action of these regulators induces hfiA at the end of G1, thus restricting holdfast formation to the swarmer cell. However, not every cell makes a holdfast; the probability of holdfast emergence at the single cell level depends on the nutritional composition of the growth medium and is inversely correlated with hfiA expression. Our data thus support a model in which holdfast development is controlled by cell cycle and nutritional input signals that are integrated at the promoter of hfiA. As a negative regulator of an enzyme required for holdfast production, HfiA functions as a checkpoint protein that ensures holdfast development occurs within the appropriate cell cycle window and nutritional conditions.
Results
lovK-lovR-enhanced adhesion requires holdfast synthesis
We previously observed that coordinate overexpression of lovK and lovR increases cell-cell adhesion, and deletion of lovK or lovR reduces adhesion [21]. To understand the genetic basis of this adhesion phenotype, we first tested if the holdfast is required for lovK-lovR-enhanced adhesion. We overexpressed lovK and lovR in a strain lacking hfsA, a gene required for holdfast synthesis [13]. In a wild-type background, overexpression of lovK and lovR results in large cell aggregates that are readily visible in the culture (Figure 1B) and accumulate in a ring along the culture tube wall (Figure S1). Strains lacking hfsA do not exhibit enhanced cell-cell adhesion or tube ring formation upon lovK-lovR overexpression. Thus, holdfast is required for the lovK-lovR-enhanced adhesion phenotype.
We recently discovered that lovK and lovR are potent negative regulators of the general stress response (GSR) sigma factor σT and, concordantly, function as negative regulators of cell survival during osmotic and oxidative stress [20]. As σT controls expression of a number of genes involved in cell envelope function [20], [22], we hypothesized that lovK-lovR affects adhesion via σT-dependent modulation of the cell envelope. Our hypothesis predicts that cells lacking sigT should be hyper-adhesive. However, a ΔsigT null strain is not hyper-adhesive. Moreover, coordinate overexpression of lovK-lovR in ΔsigT background results in an equivalent cell adhesion phenotype as a strain expressing lovK-lovR in a wild-type background (Figures 1B & S1). These data demonstrate that lovK-lovR modulates adhesion independent of sigT, via a mechanism that requires holdfast development.
LovK-LovR represses transcription of cc_0817 (hfiA), a novel inhibitor of holdfast development
We next sought to identify specific adhesion effector(s) regulated by LovK-LovR, independent of σT. We measured change in global transcript abundance upon lovK-lovR overexpression in ΔsigT and in wild-type genetic backgrounds. Only one transcript, cc_0817 (ccna_00860), exhibited differential steady-state levels in both experiments; showing a 2–3-fold reduction (Figure 1C). A lacZ transcriptional fusion to the cc_0817 promoter confirmed that overexpression of lovK-lovR represses cc_0817 transcription (Figure 1D). cc_0817 mRNA is among the top 10% of transcripts in the C. crescentus cell in terms of abundance, based on analysis of wild-type expression array data [20]. Thus modest fold changes in transcript abundance reflect large changes in absolute number of transcripts.
Our finding that lovK-lovR overexpression results in decreased transcription of cc_0817 and increased holdfast-dependent adhesion suggested that cc_0817 functions downstream of lovK-lovR as an inhibitor of holdfast development. To test this hypothesis, we assayed the effect of cc_0817 deletion and overexpression on holdfast development. Holdfast development was monitored by incubating cells with fluorescently-labeled wheat germ agglutinin (WGA-Alexa595), a lectin that binds N-acetylglucosamine and marks holdfast at the cell surface [9].
In minimal defined medium with xylose as the carbon source, about 3% of wild-type cells display a holdfast (Figure 2A&B). Overexpression of cc_0817 reduces the fraction of cells with a visible holdfast to near zero. lovK-lovR overexpression increases the fraction of cells with a holdfast ∼10-fold and cc_0817 overexpression in this background (lovK-lovR++ cc_0817++) attenuates this effect (Figure 2A&B). Conversely, deletion of cc_0817 results in elaboration of a holdfast on nearly every cell (Figure 2A&B). Expression of cc_0817 from the ectopic xylX locus in a Δcc_0817 null background restores wild-type holdfast levels. These data support a model in which cc_0817 functions to inhibit holdfast development. We have named this gene holdfast inhibitor A, hfiA.
Fig. 2. CC_0817 functions as a holdfast inhibitor (HfiA). 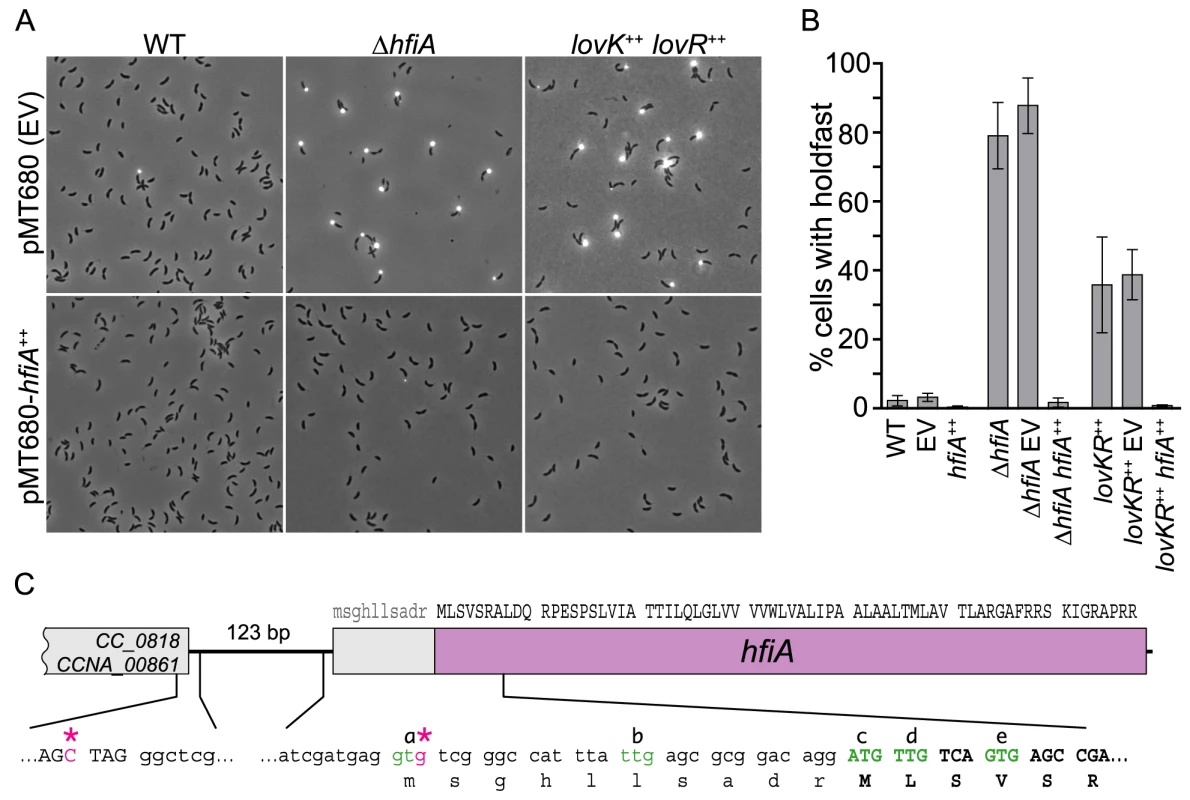
A. Representative micrographs of cells grown in M2XV medium and incubated with WGA-Alexa594 to visualize holdfast. Strains carry either empty vector (EV) control plasmid (top row) or a xylose inducible hfiA overexpression plasmid (bottom row). B. Quantification of cells displaying holdfast. Bars indicate mean ± s.d. of at least three independent samples of each genotype. At least 300 cells were counted in each sample. C. hfiA locus with DNA sequences of selected regions. Triplets indicate predicted coding sequence. Transcription start sites mapped using 5′ RACE are marked with pink asterisks (*). NTG codons that could function as translation start sites are green and annotated a-e. Reannotated coding sequence is uppercase. hfiA encodes a small novel protein
hfiA was annotated as a 78 aa hypothetical protein [23]; the central portion of the putative protein contains a hydrophobic stretch of 35 amino acids. A search of the Pfam and Conserved Domain Databases with the primary sequence of HfiA revealed no conserved domains. Given the small predicted size of hfiA and the lack of functional clues in the sequence, we sought to validate the prediction that hfiA is translated into a protein, and to define the length of the predicted open reading frame.
We identified two hfiA transcriptional starts by 5′ RACE. Seventy-five percent of the sequenced RACE products started at the third position of the predicted hfiA translational start codon and twenty-five percent started 126 bp upstream of the predicted translational start (Figure 2C). These data suggested the hfiA start codon was annotated incorrectly. The first 14 codons of the annotated hfiA coding sequence include 4 additional NTG codons (marked b, c, d, and e in Figure 2C) that could potentially function as translation start sites. To test if putative codons a or b function as translation start sites, we expressed from the hfiA promoter a translational fusion between the first 7 predicted hfiA codons and lacZ. We engineered a second translational fusion that also included codons c, d and e fused to lacZ. Only the second fusion yielded β-galactosidase activity above background (Figure S2), demonstrating that hfiA is translated and that translation initiates from codons c, d or e.
To identify the site of translation initiation we replaced the wild-type hfiA allele with mutant alleles in which codons c, d or e were mutated away from NTG, and thus could no longer function as translation start sites. We predicted that loss of the translation start would phenocopy the hyper-holdfast phenotype of the ΔhfiA null strain. However, no single codon mutant exhibited a null phenotype (Figure S2), suggesting hfiA translation can initiate at multiple sites. Furthermore, no double codon mutant exhibited a full hyper-holdfast phenotype. Only the strain bearing mutations in all three putative start codons (c, d and e) phenocopied the ΔhfiA null strain (Figure S2). Thus all three of these codons can likely function as sites of translation initiation, resulting in synthesis of 65–68 amino acid proteins (∼7 kDa). We have reannotated hfiA to reflect initiation at the ATG that was originally predicted to be codon 11.
A forward genetic screen identifies HfiA suppressor mutations
We hypothesized that the small protein, HfiA, functions to directly inhibit a protein required for holdfast development. To test this hypothesis, we designed an unbiased genetic screen to identify adhesive mutants that continue to produce holdfast when hfiA is overexpressed (Figure 3A). Several classes of adhesive mutants emerged from this screen: a) Mutants in which hfiA overexpression was disrupted by lesions in the xylose-inducible promoter, in the hfiA coding sequence (Table S1 & Figure S3), or in the xylose transport system; b) Mutants with increased surface adhesion but not increased holdfast including lesions in the gene encoding the S-layer protein or in genes involved in synthesis of lipopolysaccharide (LPS), which attaches the S-layer protein to the cell [24]–[26]; c) Mutants with an elevated number of holdfast-bearing cells in the presence of an intact hfiA overexpression system. We initially isolated two independent hfiA-suppressor strains, 256-39 and 261-15, with strongly enhanced surface adhesion (Figure 3C) and a high fraction of cells bearing a holdfast.
Fig. 3. HfiA suppressor screen identifies a novel holdfast synthesis gene, hfsJ. 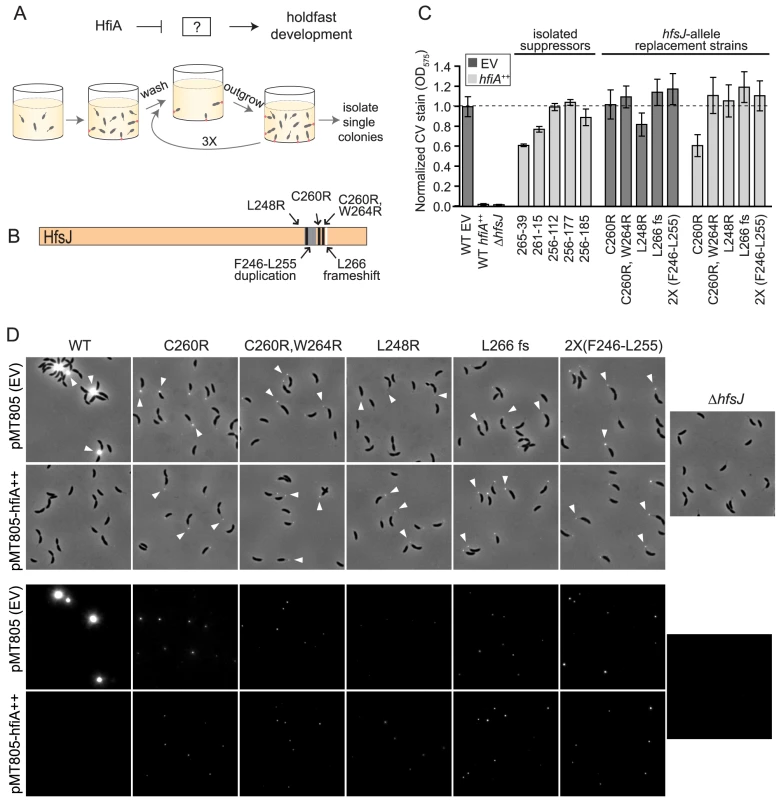
A. Schematic of the expected target of a forward genetic screen for HfiA suppressors, and a strategy to enrich for mutants that are insensitive to hfiA overexpression (see Materials and Methods for details). B. Summary of hfsJ mutations in the 5 strains that suppress the hfiA overexpression phenotype. Black lines, non-synonymous SNPs; grey, duplicated region; and white, out-of-frame deletion. C. Surface attachment after growth in polystyrene plates measured by crystal violet staining of attached cells for original suppressor strains, and strains bearing targeted mutations in hfsJ. All strains carry either the empty vector (pMT805; dark grey bars) or the hfiA overexpression plasmid (light grey bars). Data represents mean ± s.d.; n = 16. Data were collected over 4 days and normalized to the wild-type empty vector control on each day. D. Strains with directed mutations in the hfsJ locus elaborate a holdfast in the presence of hfiA overexpression. WGA-Alexa595 lectin staining of cells grown in PYE supplemented with 0.15% xylose to induce expression. Holdfast (white arrowheads) on wild-type and hfsJ mutants are shown for strains carrying the empty plasmid (EV) or hfiA overexpression plasmid. Top panel: phase contrast and fluorescence combined. Bottom panel: fluorescence signal alone. Intensity is scaled identically across all images in each panel. Whole genome sequencing of these suppressors revealed multiple mutations relative to the wild-type parent (Table S1). While each suppressor strain bore unique mutations, both 256-39 and 261-15 shared non-synonymous polymorphisms in gene cc_0095 (ccna_00094) that resulted in a C260R substitution and C260R,W264R substitutions, respectively.
We conducted additional enrichment screens and identified three other independent HfiA-supressors (256-112, 256-177 and 256-185) that exhibit near wild-type surface adhesion when hfiA is overexpressed (Figure 3C). Targeted sequencing of the cc_0095 locus in these strains revealed that each harbor mutations in the 3′ end of this gene which result in the following coding changes: L248R, a frame-shift after L266 and a duplication of F246-R254 respectively (Table S1, Figure 3B). The independent isolation of five unique lesions in the same region of cc_0095 strongly implicated this gene in hfiA-mediated control of holdfast development.
CC_0095 is annotated as a UDP-N-acetyl-D-mannosaminuronic acid transferase and is related to E. coli WecG (29% identity/45% similarity) and Bacillus subtilis TagA (27% identity/46% similarity) glycosyltransferases. This protein is strongly classified as a WecG/TagA–family glycosyltransferase in the Conserved Domain Database (E-value<e−84) [27], though the glyco-substrates are difficult to predict from primary sequence. This family of enzymes is widely distributed in Gram-negative and Gram-positive bacteria. Within the Caulobacterales, all sequenced species that encode the holdfast synthesis gene cluster, hfsEFGHCBAD, also encode proteins that are 50–80% identical to CC_0095.
cc_0095 (hfsJ) is required for holdfast development
WecG/TagA–family enzymes catalyze the transfer of an activated nucleotide sugar to a glycosylated membrane phospholipid, undecaprenyl pyrophosphate (Und-PP) [27]. Execution of this chemistry is a critical early step in the biosynthesis of extracellular sugar polymers including the holdfast material [12]. To test if cc_0095 functions in holdfast development, we generated a strain carrying an in-frame deletion of this gene. Strains lacking cc_0095 do not develop holdfast (Figure 3D) and are completely defective in surface adhesion (Figure 3C) consistent with the defective biofilm phenotype reported for a cc_0095 transposon insertion mutant [28]. Expression of cc_0095 from an ectopic locus restores holdfast synthesis and surface adhesion to the null mutant (Figure S4). Neither E. coli wecG nor B. subtilis tagA complements the Δcc_0095 adhesion or holdfast defects, although expression of E. coli WecG alters C. crescentus morphology resulting in cells that are longer, thinner, and no longer curved (Figure S4). Genes required for holdfast synthesis have been assigned the names hfsA through hfsI [12]. We have named gene cc_0095, hfsJ.
We note that hfsJ prediction is 10 codons shorter in the C. crescentus NA1000 (CCNA_00094) genome annotation compared to strain CB15 (CC_0095) annotation. To experimentally define the start codon, we generated strains in which each predicted start codon was mutated. Only mutation of the predicted start annotated in NA1000 phenocopied the null, supporting the annotation of the shorter open reading frame (Figure S4); this was used as the frame of reference for numbering the position of hfsJ mutations.
A fluorescent protein fusion, HfsJ-venus, expressed from the native hfsJ promoter complements the holdfast null ΔhfsJ phenotype (Figure S4). In contrast to holdfast export and anchoring proteins, which are localized to the stalked pole [11], [29], HfsJ-venus is distributed throughout the cell (Figure S4). Western blot analysis on this strain with antibodies to GFP/venus indicates that the HfsJ-venus fusion is not cleaved; no degradation products were detected (Figure S4). Thus the fluorescence signal observed throughout the cell reflects the distribution of HfsJ-venus. Moreover, HfsJ-venus was detected only in the pellet fraction but not the soluble fraction of the cell lysate (Figure S4), supporting a model in which this protein is membrane associated. We cannot rule out the possibility that the fluorescent tag either alters the localization of HfsJ, or stabilizes it so that a localization site becomes saturated. The chemistry that HfsJ is predicted to execute (modification of an inner membrane carrier glycolipid) occurs in the cytoplasm. Given the rapid two-dimensional diffusion of lipids in the inner membrane, such a lipid-modifying enzyme need not be spatially restricted to produce a product that is utilized by the holdfast synthesis and export machinery located at the nascent stalked cell pole.
Lesions near the C-terminus of HfsJ suppress holdfast inhibition by HfiA
To test if the hfsJ lesions identified in our genetic screen specifically suppress the holdfast inhibition function of hfiA, we constructed strains in which we replaced wild-type hfsJ with each of the mutant alleles. These hfsJ-mutant strains were transformed with either empty plasmid or the hfiA overexpression plasmid and assayed for surface adhesion and visible holdfast.
Each of the hfsJ-mutant strains exhibits surface adhesion in the presence of the empty control plasmid (Figure 3C, dark bars). Thus, the mutations in hfsJ do not compromise bulk adhesion. However, the holdfast in these strains do not stain as intensely as wild-type (Figure 3D) suggesting they may be smaller than wild type. While overexpression of hfiA nearly abolishes surface adhesion and holdfast development in wild-type cells (Figure 3C&D), the hfsJ-mutant strains are largely insensitive to the effect of hfiA overexpression on surface adhesion and holdfast formation (Figure 3C&D).
HfiA and HfsJ directly interact in vitro and in vivo
To test if HfiA and HfsJ physically interact, we assayed whether the proteins co-purify by serial affinity chromatography. We cloned hfiA and hfsJ into a tandem E. coli expression plasmid, with N-terminal maltose binding protein (MBP) and His6 affinity tags, respectively (Figure 4A). MBP-HfiA (51 kDa) and His6-HfsJ (33 kDa) co-eluted from amylose affinity resin (Figure 4B). We observed an additional band, the size of the MBP tag alone (42 kDa), suggesting that the MBP-HfiA fusion is partially unstable. Eluate from the amylose resin was then bound to Ni2+ resin. Only two proteins of sizes corresponding to His6-HfsJ and MBP-HfiA co-eluted from the Ni2+ resin (Figure 4B). We confirmed the identities of these proteins as His6-HfsJ and MBP-HfiA by mass spectrometry. As a control, we confirmed that MBP-HfiA does not bind to Ni2+ resin, nor does His6-HfsJ bind to amylose resin, nor is the interaction mediated by the MBP domain (Figure S5). Together these data provide strong support for a direct physical interaction between HfsJ and HfiA.
Fig. 4. HfiA and HfsJ directly interact in vitro and in vivo. 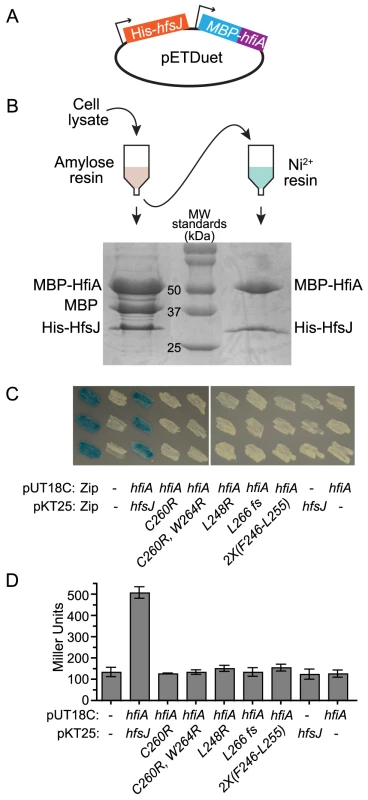
A. pETDuet plasmid used to co-express His6-hfsJ and MBP-hfiA from T7 promoters. B. Co-affinity purification scheme and Coomassie stained SDS-PAGE gel of proteins purified first with amylose resin followed by affinity purification with Ni2+ resin. C. Bacterial two-hybrid assay. E. coli cells bearing plasmids with fusions to either the T18c or T25 domains of adenylate cyclase were grown on medium containing X-gal. Interaction between the fusion proteins results in expression of lacZ and conversion of X-gal to yield blue colonies. T18c was fused to either a positive control (Zip) or hfiA. T25 was fused to a positive control (Zip), hfsJ or hfsJ mutant alleles. Dashes indicate empty vector controls. Three independent colonies of each combination are shown. D. β-galactosidase activity from strains expressing the fusions in C. Bars represent mean ± s.d. (n = 3). Only the strain with fusions to wild-type alleles of hfiA and hfsJ yielded β-galactosidase activity significantly (p<0.01) different from the control strain with split domains lacking fusions (one-way ANOVA followed by Dunnett's multiple comparison post-test). To provide additional support for a physical interaction between HfiA and HfsJ, we preformed a bacterial two-hybrid assay [30]. Co-expression of T25-hfsJ and T18-hfiA fusions results in blue colonies when grown on medium containing X-gal (Figure 4C), and significant β-galactosidase activity when grown in liquid (Figure 4D), demonstrating that HfsJ and HfiA interact and bring together the split T25/T18 adenylyl cyclase domains. Reconstitution of adenylyl cyclase activity required both fusions; neither fusion alone was sufficient. Importantly, none of the HfsJ mutant alleles interact with HfiA sufficiently to yield a positive result in this assay (Figure 4C&D).
These data support a model in which HfiA functions to inhibit holdfast development through direct interaction with HfsJ, a putative glycosyltransferase required for holdfast development.
hfiA expression is cell-cycle regulated and inversely correlated with holdfast development
Holdfast development is temporally regulated across the cell cycle [8], [15], [17], [19]: the holdfast is elaborated at the flagellated pole of the swarmer cell, before or during the swarmer-to-stalked cell transition (Figure 1A). Profiles of C. crescentus gene expression throughout the cell cycle reveal that transcription of the holdfast inhibitor hfiA is cell-cycle regulated, with a minimum at the period of holdfast development (Figure 5A), [31]–[33]). These results are consistent with a model in which cell-cycle regulation of hifA expression determines the developmental window for holdfast biogenesis.
Fig. 5. Transcription of hfiA is coordinately controlled by multiple regulatory proteins. 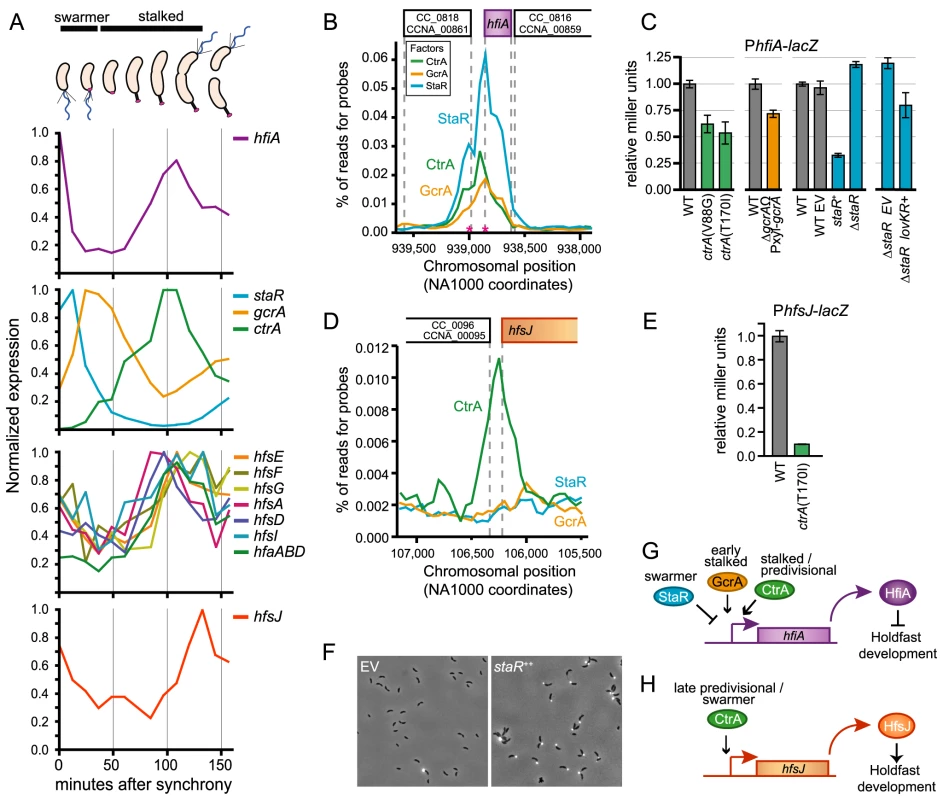
A. Cell cycle-dependent transcriptional regulation of genes involved in holdfast development. Data were extracted from global transcriptional profiling experiments [33] and normalized to maximal expression for each gene. Approximate developmental stage at each time point is shown at top. B. The core cell cycle regulators StaR, CtrA and GcrA occupy the hfiA promoter. ChIP-seq data of the hfiA locus for each regulator. Chromosomal position of DNA pulled down with each of the regulators is indicated below; the gene context is indicated above. Pink asterisks indicated mapped transcriptional start sites for hfiA. C. β-galactosidase activity from a PhfiA-lacZ transcriptional fusion (pRKlac290-PhfiA) assayed in strains bearing temperature sensitive alleles of ctrA outgrown in PYE at 37°C for 4 hours, in gcrA depletions strains grown in the absence of xylose (PYE+0.15% glucose) for 6 hours, and in strains in which staR was deleted or overexpressed from the Pxyl promoter (PYE+0.15% xylose). In each experiment, Miller units were normalized to WT grown in parallel conditions. Data represent mean ± s.d. of 6 independent cultures assayed on two different days. D. CtrA, but not GcrA or StaR, occupies the hfsJ promoter. ChIP-seq data for the hfsJ locus annotated as in B. E. CtrA activates transcription from the hfsJ promoter. β-galactosidase activity from a PhfsJ-lacZ transcriptional fusion in wild-type and ctrAts strains at restrictive temperature (37°C for 3 hours). Data represent mean ± s.d. of 4 independent cultures assayed on two different days. F. WGA-Alexa594 staining of holdfast in wild-type empty vector (EV) control strain or a staR++ overexpression strain grown in M2X. G. Model of the hfiA regulatory activities of CtrA, GcrA and StaR. H. Model of the hfsJ regulatory activities of CtrA. Holdfast development is connected to the cell cycle control network via hfiA
C. crescentus cell cycle progression is controlled via dynamic interplay between a number of developmental regulatory proteins (reviewed by [34]–[36]). Three known developmental regulators, CtrA, GcrA, and StaR directly control hfiA expression. These proteins are introduced briefly here: a) CtrA, an essential response regulator with a DNA-binding output domain [37], is a ‘master developmental regulator’ that directly or indirectly controls transcription of ∼25% of the C. crescentus cell cycle regulated genes [32], [37]. b) GcrA is critical developmental regulator required for efficient growth that forms a feedback control loop with CtrA [38], [39]. c) StaR is a non-essential developmental regulator that controls stalk biogenesis [40]. Transcription of these genes is temporally regulated across the cell cycle (Figure 5A; [31]–[33]).
StaR, CtrA and GcrA physically occupy the chromosomal region immediately upstream of hfiA (Figure 5B; Tables S2, S3, S4, S5, and [41]). To test whether these proteins affect hfiA expression, we assayed transcription from a PhfiA-lacZ transcriptional fusion in ctrA, gcrA, or staR mutant backgrounds. Cells lacking ctrA and gcrA are severely compromised and have developmental defects thus, we used temperature sensitive alleles [37], [42] or depletion strains [38] to evaluate the effects of protein loss on hfiA transcription. At restrictive temperatures, strains bearing ctrA ts alleles have 2-fold less PhfiA-lacZ activity than wild type; PhfiA-lacZ activity is reduced ∼25% upon GcrA depletion (Figure 5C). We conclude that CtrA and GcrA are transcriptional activators of hfiA (Figure 5G). Overexpression of staR from a xylose-inducible promoter reduced PhfiA-lacZ activity by ∼70%, deletion of staR enhanced PhfiA-lacZ activity by ∼20% (Figure 5C). These data provide evidence that StaR represses hfiA transcription. We note that our experiments with unsynchronized populations will mask the amplitude of temporally-restricted transcriptional change. Indeed, endogenous StaR is expected to affect only a subset of cells in the population at any given time. Conversely, overexpression of staR from an inducible promoter affects PhfiA in all of the cells at any given time.
Like many genes involved in holdfast biogenesis, transcription of hfsJ is also cell cycle regulated (Figure 5A; [31]–[33]). Sequences corresponding to the hfsJ locus are enriched by immunoprecipitation of CtrA, but not GcrA or StaR (Figure 5D). Transcription from the hfsJ promoter is diminished in a ctrA temperature sensitive (ctrAts) mutant (Figure 5E); we conclude that CtrA is a direct activator of hfsJ transcription (Figure 5H). We identified CtrA binding sites upstream of both hfiA and hfsJ (CTCttaaAGCTTTCtaaaCCT, 92 bp, p = 1.0e-04 and ATActtaGCGGGATttaaCCA, 66 bp, p = 6.3e-07 respectively).
We next asked whether the activity StaR on hfiA transcription affects holdfast development. Both ctrA and gcrA are essential, and mutant strains have pleiotropic defects that confound assessment of holdfast development. The function of StaR was initially investigated in a holdfast deficient genetic background; thus no holdfast phenotype was reported [40]. As StaR is a repressor of hfiA, we predicted that overexpression of StaR should result in an increase in holdfast development. Indeed, staR overexpression results in a dramatic enhancement of visible holdfast (Figure 5F).
LovR is a single domain response regulator lacking a DNA-binding output domain [21], thus the effects of LovK-LovR on hfiA transcription must be indirect. We investigated whether inhibition of hfiA transcription by lovK-lovR is dependent on CtrA, GcrA, or StaR. Deletion of staR had no effect on inhibition of hfiA transcription by lovK-lovR (Figure 5C). We further demonstrated that lovK-lovR does not affect the occupancy of StaR at the hfiA locus (Figure S6). GcrA and CtrA regulated genes are not differentially regulated in lovK-lovR transcriptional profiling experiments [20] suggesting that the activity of GcrA or CtrA is not perturbed by LovK-LovR. Similarly, transcription from known GcrA and CtrA regulated promoters is not affected when lovK and lovR are coordinately overexpressed (Figure S6). We conclude that LovK and LovR affect hfiA transcription via a mechanism that is independent of StaR, GcrA, and CtrA.
Holdfast development and hfiA transcription are regulated by the nutritional composition of the culture medium
The cell cycle expression profile of hfiA is coordinated with the timing of holdfast development. However, not every cell makes a holdfast. What determines whether a cell will elaborate a holdfast? Transcriptional profiling experiments suggest that culture environment affects hfiA transcription; in mid-log phase, cells grown in minimal medium (M2X) have 1.6 times more hfiA transcript than cells grown in complex medium (PYE) [43]. While this relative difference in hfiA transcript level is not large, hfiA is a highly expressed gene. Thus, the absolute difference in transcript levels is large. To test whether culture environment affects probability of holdfast development, we grew wild-type C. crescentus CB15 cells in either complex (PYE) or minimal defined medium (M2X) and quantified the fraction cells with visible holdfast. The culture environment has a dramatic impact on the probability that a cell displays a holdfast. In PYE medium, approximately 80% of cells develop a holdfast (Figure 6A) compared to 1–3% of cells in M2X.
Fig. 6. Nutrient environment affects hfiA transcription and the probability of holdfast development. 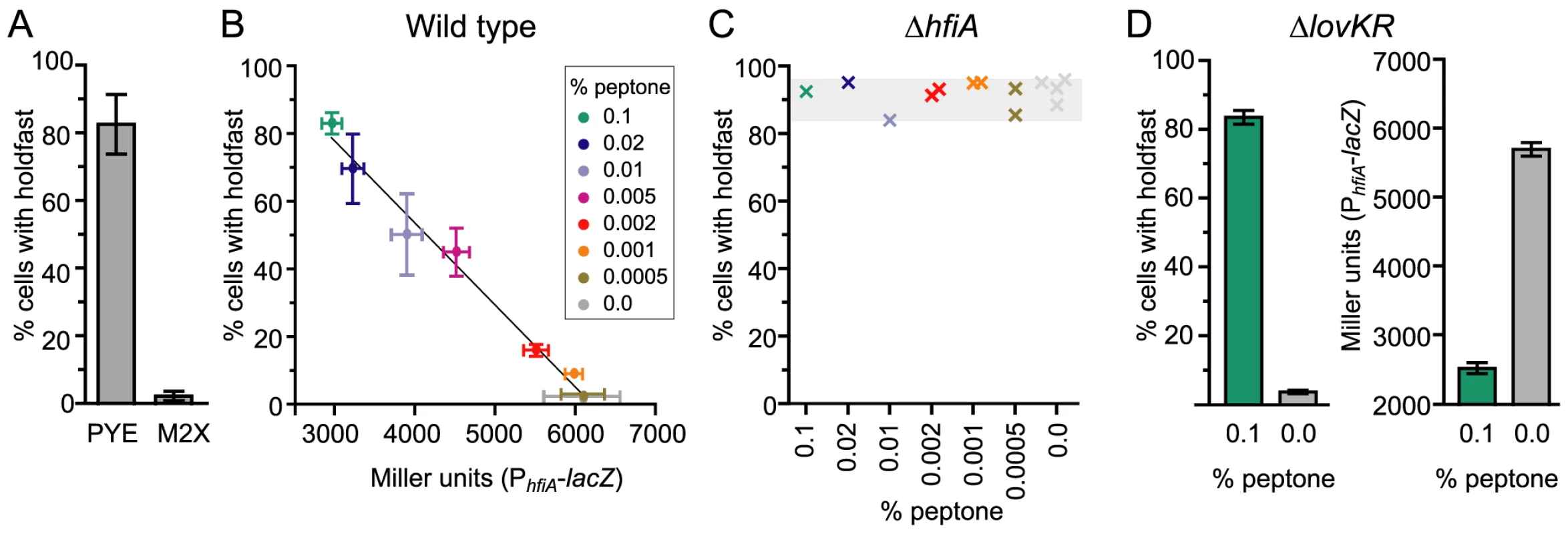
A. Quantification of WGA-Alexa595 stained holdfast on wild-type CB15 cells grown in complex medium (PYE) or defined minimal medium (M2X). Bars represent the mean ± s.d. of 12 (PYE) or 18 (M2X) independent samples. At least 300 cells per sample were counted. B. Holdfast development and hfiA transcription are inversely correlated and dependent on the growth medium. Cultures grown in M2X supplemented with increasing amounts of peptone were assayed at a final density of 0.05–0.15 OD660. Holdfast were assessed as above in 4–8 independent cultures of wild-type cells per condition (mean ± s.e.m). hfiA transcription was assessed in 6–12 independent cultures of wild-type carrying the pRKlac290-PhfiA reporter plasmid per condition (mean ± s.e.m.). C. HfiA is required for nutrient dependent regulation of holdfast development. Holdfast were counted, as above, on ΔhfiA cultures grown M2X supplemented with a range of peptone concentrations. Each x represents holdfast counts in an individual culture of ΔhfiA cells colored by growth medium. D. Nutrient-dependent regulation of PhfiA and holdfast probability does not require lovK and lovR. As above, holdfast were assessed in ΔlovKR cells in 6 independent cultures per condition (mean ± s.e.m.). β-galactosidase activity was measured in ΔlovKR cells bearing the pRKlac290-PhfiA reporter plasmid (n = 10 independent cultures per condition; mean ± s.e.m). We extended this analysis beyond these two standard growth media by analyzing both the probability of holdfast development and hfiA transcription from the PhfiA-lacZ reporter in a series of minimal media supplemented with increasing amounts of peptone, from 0.0005% to 0.1%. In this panel, we observed an approximately 2-fold change in activity from the hfiA promoter (Figure 6B). The probability of holdfast development in the population shows an inverse linear correlation with hfiA promoter activity (r2 = 0.99; Figure 6B). Cells cultured with little or no peptone exhibit the highest hfiA transcription and the lowest fraction of cells with holdfast. Increasing peptone concentration results in both decreased hfiA transcription and an increased fraction of cells with a holdfast. These data are consistent with a model in which modest relative changes in hfiA expression can have a large effect on holdfast development.
To test whether hfiA is required for regulated differences in holdfast between growth media, we evaluated holdfast development in the ΔhfiA null mutant. Regardless of the composition of the growth medium, ΔhfiA mutants elaborate a holdfast on nearly every cell (Figure 6C); only a small portion of swarmer cells can be found without holdfast. Transcription from the hfsJ promoter does not change between these growth conditions (Figure S7). We conclude that the capacity of a cell to elaborate a holdfast is controlled by the expression of the holdfast regulator, hfiA, and not by a change in expression of the holdfast synthesis gene hfsJ.
Finally, we tested if nutrient-dependent regulation of PhfiA and holdfast development requires the LovK-LovR sensory system. We measured the number of holdfast in strains lacking the lovKR locus. In minimal medium only small fraction of ΔlovKR cells display a holdfast. Upon supplementation with 0.1% peptone, the majority of ΔlovKR cells exhibit a holdfast (Figure 6D). Similarly, in a ΔlovKR background, the PhfiA-lacZ transcriptional reporter is reduced upon supplementation with 0.1% peptone (Figure 6D). Together these results indicate that LovK and LovR are not required for nutrient-dependent control of holdfast and suggest an additional, unknown regulator of hfiA.
Discussion
Holdfast adhesin development in C. crescentus is regulated by the developmental state of the cell and by the culture environment. This surface organelle emerges at the flagellated pole during the late swarmer cell stage ([8], [15]–[17], [19], Figure 1). We have discovered a novel small protein, HfiA, whose expression is developmentally and nutritionally regulated, and which functions as a potent inhibitor of holdfast. We demonstrate that the predicted glycosyltransferase, HfsJ, is a required component of the holdfast development machinery and that residues at the C-terminus of HfsJ mediate a direct interaction with HfiA. We propose a model in which HfiA functions as a cell cycle and nutritional checkpoint protein that prevents inappropriate holdfast development via post-translational inhibition of HfsJ (Figure 7).
Fig. 7. HfiA and HfsJ coordinately control holdfast development in response to cell cycle and environmental signals. 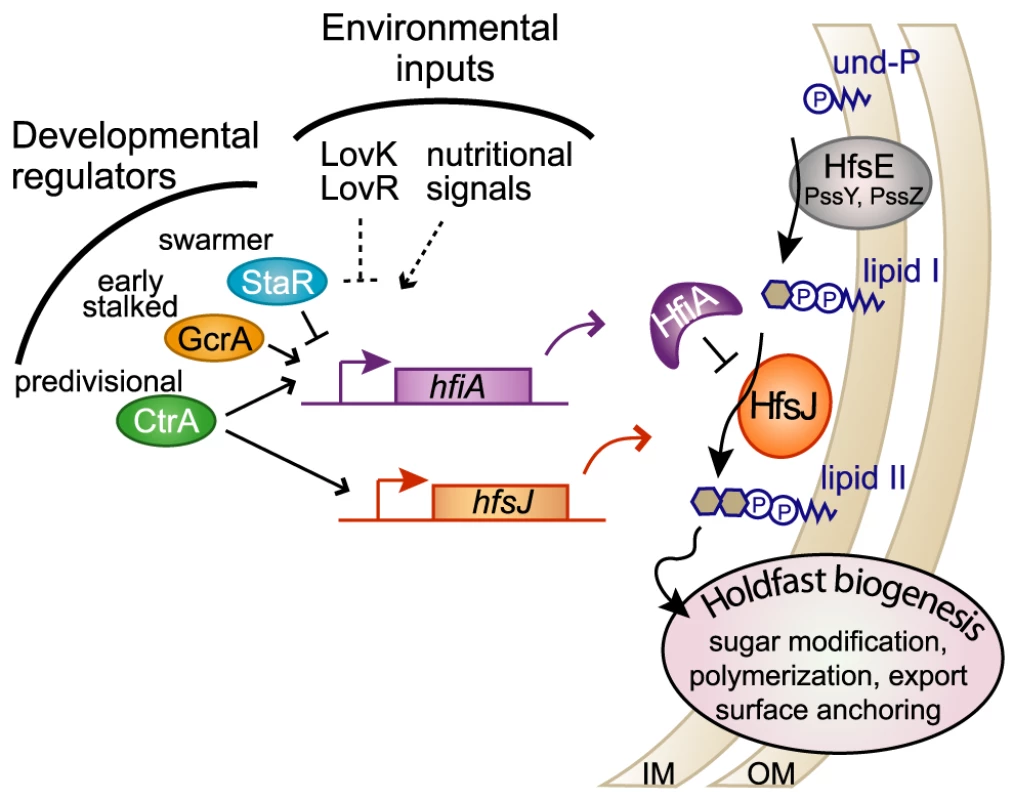
Proposed model in which HfiA directly inhibits HfsJ, a WecG/TagA-family glycosyltransferase required for holdfast production. Expression of hfsJ and hfiA are controlled by cell cycle and environmental input signals. Solid and dashed lines indicate direct and indirect regulation respectively. Notably, the dynamic range of hfiA transcriptional control (∼2-fold) is modest at the population level compared to the dynamic range of holdfast probability (∼2-log). One prediction from this observation is that the binding affinity and cellular concentrations of HfiA and HfsJ are tuned such that this regulatory system is responsive to small changes rather than robust to large changes. This predication is consistent with a highly responsive and sensitive regulatory system.
HfsJ has strong similarity to WecG/TagA–family glycosyltransferases (E-value<e−84) [27]. Enzymes in this family are known to catalyze the transfer of a nucleotide diphosphate (NDP)-activated sugar to monoglycosylated Und-PP (i.e. lipid I) [44], [45]. The product of this reaction is the phosphoglycolipid, Und-PP-disaccharide (i.e. lipid II). Varied forms of lipid II are precursors for extracellular polysaccharide structures in bacteria including lipopolysaccharide, wall teichoic acid, capsular polysaccharide, and holdfast. In C. crescentus, there is apparent redundancy in the holdfast synthesis enzymes predicted to catalyze formation of lipid I [12]. Our genetic data suggest that HfsJ is solely responsible for the production of holdfast lipid II. B. subtilis and Staphylococcus aureus TagA catalyze a specific transformation of lipid I to lipid II that commits the phosphoglycolipid for wall teichoic acid biosynthesis [46]. By analogy, we predict that HfsJ commits its phosphoglycolipid substrate to holdfast biosynthesis. Post-translational regulation of such a “gate-keeping” enzyme would enable specific control of lipid I commitment to holdfast development (Figure 7).
Temporally staggered cell-cycle transcription of hfiA and hfsJ correlates with the developmental timing of holdfast synthesis. Key developmental regulators physically interact with and regulate both of these genes, directly tying holdfast development to the core cell cycle control network. The master cell cycle regulator, CtrA, activates transcription of both hfiA and hfsJ. The methylation-responsive transcriptional regulator, GcrA [41], and the developmental regulator, StaR, provide additional layers of direct hfiA regulation. The activities of these regulators on hfiA, but not hfsJ can account for the temporal shift in hfsJ and hfiA expression. Consistent with the observation of hfs gene transcripts in late pre-divisional and swarmer cells ([31]–[33], Figure 5), swarmer cells are born preloaded with all the proteins required to synthesize a holdfast [17], [19]. staR is activated in swarmer cells ([31]–[33], [47], Figure 5) and as a functional repressor of hfiA, presumably drives the decrease in hfiA transcription prior to holdfast development. Decreased expression of the holdfast inhibitor is predicted to permit holdfast production as cells approach the swarmer-to-stalk cell transition. Upon this transition, cells accumulate GcrA [38], which can initiate activation of hfiA expression, but not hfsJ. As the cell cycle progresses, de novo synthesis and activation of CtrA [37] should reinforce expression of the inhibitor and also activate hfsJ in preparation for the next generation swarmer cell.
Notably, hfsJ is among the last holdfast synthesis genes to be transcriptionally activated; it does not reach maximum transcription until just prior or coincident with cell division (Figure 5). This delayed expression provides two intuitive mechanisms that should restrict premature holdfast synthesis. First, HfsJ is essential for holdfast biosynthesis, thus a preassembled machine will not be functional in the predivisional cell until hfsJ is expressed, just around the time of cell division. Second, peak hfiA expression precedes that of its target. A pool of accumulated inhibitor should block activity of nascent HfsJ in the late predivisional cell. Together these features ensure that the motile swarmer cells are not born with a holdfast and are able to fulfill a dispersal role.
What, then, relieves HfiA inhibition so that holdfast development can progress in the swarmer cell? One possibility is that HfiA is inherently unstable and rapidly degraded by cellular proteases. Indeed, proteins optimized for regulatory flexibility tend to have short half-lives [48]. If HfiA is unstable, high synthesis rates would be necessary to maintain an appreciable steady state concentration in the cell. In this scenario, the initial concentration of HfiA in the swarmer cell (where hfiA is not transcribed) could serve as a timer for the initiation of holdfast synthesis. Several efforts by our research group to quantify HfiA protein levels in the cell have been unsuccessful. These negative results provide indirect support for the hypothesis that HfiA is an unstable polypeptide, though we still seek direct experimental support for this hypothesis.
Alternatively, post-translational modification could affect HfiA stability or its binding affinity with HfsJ. Surface contact-dependent perturbation of the flagellum [19] could serve as a signal for HfiA modification or degradation. Another possibility is that cyclic-di-GMP (cdG) could serve as a second messenger that directly or indirectly inactivates HfiA or activates HfsJ. In many systems cdG serves as a developmental cue signaling the transition from motile to non-motile states [49]. Indeed the activity of the diguanylate cyclase, PleD, is cell cycle regulated and activated during the swarmer to stalk transition [50]; C. crescentus cells lacking pleD are delayed in holdfast development [17]. Thus, it is reasonable to predict that cdG may play a role in control of the HfiA-HfsJ adhesion checkpoint.
While the developmental circuitry of the cell directly controls hfiA expression, environmental signals provide an additional regulatory input that can override developmental control. A mixed population of cells grown in carbon replete minimal defined medium have 60% more hfiA transcript than cells grown in complex medium [43]; this correlates with the observed frequency of holdfast-bearing cells in a population (i.e. cells grown in minimal medium rarely elaborate a holdfast while the majority cells grown in complex medium possess holdfast) (Figure 6). Moreover, supplementation of minimal defined medium with peptone modulates both hfiA expression over a two-fold range and the probability of holdfast development over a 2-log range (Figure 6). A similar correlation is observed upon overexpression of the LovK-LovR two-component sensory system, which results in hfiA repression and increased probability of holdfast development (Figures 1 & 2). Notably, the nutrient-dependent control of hfiA transcription and holdfast development is independent of the LovK-LovR sensory system. The exact regulatory connection between hfiA transcription and either LovK-LovR signaling or the metabolic state of the cell remains unclear. Indeed, the data presented here speak to existence of at least one additional direct regulator of hfiA, as the repressive effect of LovK-LovR on hfiA transcription is necessarily indirect and also independent of CtrA, GcrA, and StaR.
Given the permanence of the cellular decision to adhere to a surface, it is not surprising that environmental and nutritional stimuli influence hfiA expression and holdfast adhesin development. Our study provides evidence that multiple developmental and environmental signals are integrated at the promoter of hfiA, which encodes a novel, small protein inhibitor of the required holdfast synthesis enzyme, HfsJ. C. crescentus employs a multi-level regulatory system that ensures proper timing of holdfast development, and safeguards against permanent cell adherence in a sub-optimal environment.
Materials and Methods
Strain construction and growth conditions
Standard cloning methods, strain construction techniques and growth conditions were employed and are detailed in the Text S1. Strains and primers used are in Table S6.
Microscopy
Cells were imaged with a DM5000 microscope (Leica) in phase contrast and fluorescence modes using a HCX PL APO 63×/1.4na Ph3 objective. Fluorescent samples were excited with an external mercury halide bulb in an EL6000 lamp (Leica). Standard filter sets were used to detect WGA-Alexa594 (Chroma set 41043) and the fluorescent protein, Venus (Chroma set 41028). Images were captured using an Orca-ER digital camera (Hamamatsu) controlled by Image-Pro (Media Cybernetics, Inc.).
Holdfast stain
To visualize holdfast, 100–500 µl of cells were incubated for 10–15 minutes with 50 µg/ml Wheat Germ Agglutinin, Alexa Fluor 594 Conjugate (Life Technologies, Molecular Probes), diluted with 1 ml water or media, collected by centrifugation for 3 minutes (14,000 g), and resuspended in 20–50 µl. For quantitative analyses, overnight cultures were diluted to an approximate OD660 of 0.00005 so that after 16–18 hours of growth the culture density was between 0.05 and 0.1 OD660. This approach minimized cell-cell adhesion and rosette formation and ensured that all cells were “born” into nutritionally replete conditions.
Transcriptional analysis
Global transcriptional profiling of ΔsigT xylX::pMT585 vanR::pMT528 (EV) and ΔsigT xylX::pMT585-lovR vanR::pMT528-lovK cultures were conducted as in [20]. β-galactosidase activity from promoter-lacZ fusions was measured colorimetrically [51] as in [20].
5′ RACE
Transcription start sites were identified by mapping 5′ ends of mRNA using the FirstChoice RLM-RACE kit (Life Technologies, Ambion) following the manufacturer's protocol. The RNA template was extracted from log phase cells grown in M2X medium using Trizol (Life Technologies, Invitrogen). 5′GTCGGTCGTGCGCATAGT and 5′GATCTTCGAGCGGCGAAA primers were used as hfiA specific primers.
ChIP-seq
Mid-log phase cells grown in PYE were cross-linked in 10 mM sodium phosphate (pH 7.6) and 1% formaldehyde at room temperature for 10 min and on ice for 30 min thereafter, washed three times in phosphate buffered saline (PBS) and lysed in a Ready-Lyse lysozyme solution (Epicentre, Madison, WI) according to the manufacturer's instructions. Lysates were sonicated (Sonifier Cell Disruptor B-30; Branson) on ice using 10 bursts of 20 sec at output level 4.5 to shear DNA fragments to an average length of 300–500 bp and cleared by centrifugation for 2 minutes (14,000 rpm, 4°C). Lysates were normalized by protein content, diluted to 1 mL using ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), 167 mM NaCl plus protease inhibitors (Roche, Switzerland) and pre-cleared with 80 µL of protein-A agarose (Roche, Switzerland) and 100 µg BSA. Ten percent of the supernatant was removed and used as total chromatin input DNA.
Polyclonal antibodies to StaR or CtrA were added to the remains of the supernatant (1∶1,000 dilution), incubated overnight at 4°C with 80 µL of protein-A agarose beads pre-saturated with BSA, washed once with low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 150 mM NaCl), high salt buffer (same as previous with 500 mM NaCl) and LiCl buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl (pH 8.1)) and twice with TE buffer (10 mM Tris-HCl (pH 8.1) and 1 mM EDTA). The protein•DNA complexes were eluted in 500 µL freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3), supplemented with NaCl to a final concentration of 300 mM and incubated overnight at 65°C to reverse the crosslinks. The samples were treated with 2 µg of Proteinase K for 2 h at 45°C in 40 mM EDTA and 40 mM Tris-HCl (pH 6.5). DNA was extracted using phenol∶chloroform∶isoamyl alcohol (25∶24∶1), ethanol-precipitated using 20 µg of glycogen as carrier and resuspended in 100 µL of water.
Illumina Genome Analyzer IIx or HiSeq 2000 runs of barcoded ChIP-Seq libraries yielded several million reads that were mapped to C. crescentus NA1000 (NC_011916) using the ELAND alignment algorithm (services provided by Fasteris SA, Switzerland). Analysis of sequences is described in Text S1.
HfiA suppressor screen
The goal of this screen was to identify mutants that are insensitive to HfiA and can develop holdfast even when hfiA is overexpressed. Our strategy to enrich the population with holdfast+ mutants is conceptually the opposite of that used by [7] to enrich a population with holdfast null mutants by removing holdfast bearing cells with cheese cloth. Here unattached cells are removed by aspiration and surface attached cells are allowed to populate the culture.
FC1935 and FC1936, strains that overexpress hfiA from either the xylose-inducible promoter on a mid-copy replicating plasmid or from the xylose promoter integrated at two independent sites on the chromosome, respectively, were used as the parental strains. Enrichment was carried out in complex medium, which promotes holdfast development in wild-type cells, supplemented with xylose to induce overexpression of hfiA. Explicit mutagenesis was unnecessary; spontaneous mutants arise in the course of the enrichment.
Starter cultures inoculated from freshly grown colonies into PYE supplemented with 0.15% xylose and appropriate antibiotics were diluted 1∶100 in 1 ml of fresh medium in each well of a sterile 24-well polystyrene plate with a lid. The lid was sealed with a strip of AeraSeal air-permeable sealing film (Excel Scientific) to prevent evaporation. Plates were incubated with gentle shaking (155 rpm) at 30°C overnight. The culture medium was removed by aspiration. Unattached cells were thoroughly washed away with a stream of sterile water expelled from a 20 cc syringe through a 22 G needle. The inoculum of attached cells was allowed to regrow to saturation in 1 ml of fresh medium under the same growth conditions. Washing and regrowth in fresh medium were repeated. After the first wash, the wells appear clear and regrowth requires 36–48 hours, but after 3–4 rounds of washing the wells are cloudy with attached cells and the outgrowth saturates in less than 18 hours. The culture was serially diluted and plated on solid medium to isolate single colonies.
Isolated colonies were subjected to several secondary screens. First, the Pxyl-hfiA region of the overexpression plasmids were amplified and re-sequenced to eliminate mutants in the overexpression system. Second, surface attachment to polystyrene was measured with crystal violet staining (see below) to confirm enhanced adhesion capacity of each isolate. Third, cells were grown in minimal medium with xylose as the sole carbon source (M2X) to ensure a functional xylose transport system. Fourth, WGA-Alexa594 binding was used to assess holdfast development.
Whole genome sequencing
Genomic DNA was isolated from individual suppressor strains using guanidium thiocyanate [52]. Bar-coded Next Generation Sequencing libraries were pooled and sequenced (50-bp single end reads) using the SOLiD 5500 xl sequencing platform (Applied Biosystems, Life Technologies) generating an average of 12 million reads per library. The Functional Genomics Facility at the University of Chicago provided sequencing services. Sequences were processed through an automated analysis pipeline by University of Chicago Center for Research Informatics. The analysis pipeline is described in more detail in Text S1.
Surface attachment
Cells were grown from 5 ul of an overnight starter culture in 1 ml of fresh growth medium in 24-well sterile polystryrene plates with lids. Lids were sealed with AeraSeal air-permeable sealing film (Excel Scientific) and plates were incubated with gentle shaking (155 rpm) at 30°C for 24 hours. Culture medium and planktonic cells were removed by aspiration and washed away with tap water. Surface attached cells were measured by crystal violet staining using a protocol similar to those outlined by [53], [54]. Briefly, wells were incubated with 1 ml 0.01% crystal violet for 5 minutes with gentle shaking then washed with tap water to remove unbound stain. Bound stain was extracted with 1.5 ml 95% ethanol while gently shaking for 5–10 minutes. Extracted stain was diluted 1∶6 and the optical density at 575 nm was measured spectroscopically.
Protein co-expression and co-purification
A 50 ml overnight culture (30°C, 220 rpm) was used to inoculate 500 ml of LB broth supplemented with appropriate antibiotics and 0.2% glucose to repress expression of endogenous maltose degradation genes. When the culture reached OD660∼0.8, 0.5 mM of IPTG was added to induce expression. After 2 hours (30°C, 220 rpm), cells were harvested by centrifugation for 20 min (12,000 g, 4°C) and the pellet was resuspended in 5 ml of Tris-NaCl buffer (10 mM Tris pH 7.4, 150 mM NaCl) supplemented with 10 mM imidazole, 5 µg/ml of DNase I and PMSF. Cells were disrupted by three passages in a French pressure cell, and cell debris was removed by centrifugation for 20 min (25,000 g, 4°C). The supernatant was then mixed with 500 µl of Amylose resin (New England Biolabs) pre-equilibrated with Tris-NaCl buffer, which allowed for binding of MBP domains. The resin was thoroughly washed with the Tris-NaCl buffer and bound proteins were eluted with 500 µl of Tris-NaCl buffer supplemented with 20 mM maltose. The eluted proteins were mixed with 100 µl of Ni2+-NTA Sepharose affinity resin (GE Life Sciences) pre-equilibrated with Tris-NaCl buffer to allow binding of His-tagged proteins. Two stringent washing steps were performed with Tris-NaCl buffer containing 10 mM or 75 mM imidazole followed by elution with 100 µl of 1 M imidazole Tris-NaCL buffer. To monitor proteins bound and eluted from each resin, samples were separated by electrophoresis on a 14% SDS-PAGE gel and stained with Coomassie. Polyacrylamide fragments containing purified proteins were excised and sent to the Pan Facility at Stanford University, Palo Alto, CA for Mass Mapping to confirm protein identity. In a similar way, the reverse experiment (purification first with Ni2+ Sepharose affinity resin and then with Amylose resin) was also performed.
Bacterial two-hybrid assay
Based on the system developed by [30], plasmid bearing fusions to either the T18 or T25 domains of adenylate cyclase were co-transformed into the adenylate cyclase null strain, BTH101, by electroporation. The outgrowth was serially diluted and plated on LB-agar containing Amp100, Kan50, X-gal (40 µg/ml) and IPTG (0.5 mM). The color of single colonies from each transformation was evaluated after 48 hours growth at 30°C. Two colonies from each strain were repatched on identical medium for side-by-side comparisons.
Supporting Information
Zdroje
1. CostertonJW, LewandowskiZ, CaldwellDE, KorberDR, Lappin-ScottHM (1995) Microbial biofilms. Annu Rev Microbiol 49 : 711–745.
2. GeeseyGG, RichardsonWT, YeomansHG, IrvinRT, CostertonJW (1977) Microscopic examination of natural sessile bacterial populations from an alpine stream. Can J Microbiol 23 : 1733–1736.
3. Kjelleberg S, Givskov M (2007) The Biofilm Mode of Life. In: Kjelleberg S, Givskov M, editors. The Biofilm Mode of Life: Mechanisms and Adaptations. New York: Taylor & Francis.
4. PetrovaOE, SauerK (2012) Sticky situations: key components that control bacterial surface attachment. J Bacteriol 194 : 2413–2425.
5. LiG, SmithCS, BrunYV, TangJX (2005) The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J Bacteriol 187 : 257–265.
6. TsangPH, LiG, BrunYV, FreundLB, TangJX (2006) Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci U S A 103 : 5764–5768.
7. OngCJ, WongML, SmitJ (1990) Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J Bacteriol 172 : 1448–1456.
8. PoindexterJS (1964) Biological properties and classification of the Caulobacter group. Bacteriol Rev 28 : 231–295.
9. MerkerRI, SmitJ (1988) Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol 54 : 2078–2085.
10. BerneC, MaX, LicataNA, NevesBR, SetayeshgarS, et al. (2013) Physiochemical properties of Caulobacter crescentus holdfast: a localized bacterial adhesive. J Phys Chem B 117 : 10492–10503.
11. HardyGG, AllenRC, TohE, LongM, BrownPJ, et al. (2010) A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol 76 : 409–427.
12. TohE, KurtzHDJr, BrunYV (2008) Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol 190 : 7219–7231.
13. SmithCS, HinzA, BodenmillerD, LarsonDE, BrunYV (2003) Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol 185 : 1432–1442.
14. LoebGI, NeihofRA (1975) Marine conditioning films. Adv Chem 145 : 319–335.
15. BodenmillerD, TohE, BrunYV (2004) Development of surface adhesion in Caulobacter crescentus. J Bacteriol 186 : 1438–1447.
16. JanakiramanRS, BrunYV (1999) Cell cycle control of a holdfast attachment gene in Caulobacter crescentus. J Bacteriol 181 : 1118–1125.
17. LeviA, JenalU (2006) Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol 188 : 5315–5318.
18. ShapiroL, Agabian-KeshishianN, BendisI (1971) Bacterial differentiation. Science 173 : 884–892.
19. LiG, BrownPJ, TangJX, XuJ, QuardokusEM, et al. (2012) Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83 : 41–51.
20. ForemanR, FiebigA, CrossonS (2012) The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J Bacteriol 194 : 3038–3049.
21. PurcellEB, Siegal-GaskinsD, RawlingDC, FiebigA, CrossonS (2007) A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A 104 : 18241–18246.
22. Alvarez-MartinezCE, LourencoRF, BaldiniRL, LaubMT, GomesSL (2007) The ECF sigma factor sigma(T) is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol Microbiol 66 : 1240–1255.
23. MarksME, Castro-RojasCM, TeilingC, DuL, KapatralV, et al. (2010) The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol 192 : 3678–3688.
24. BingleWH, NomelliniJF, SmitJ (1997) Linker mutagenesis of the Caulobacter crescentus S-layer protein: toward a definition of an N-terminal anchoring region and a C-terminal secretion signal and the potential for heterologous protein secretion. J Bacteriol 179 : 601–611.
25. WalkerSG, KarunaratneDN, RavenscroftN, SmitJ (1994) Characterization of mutants of Caulobacter crescentus defective in surface attachment of the paracrystalline surface layer. J Bacteriol 176 : 6312–6323.
26. AwramP, SmitJ (2001) Identification of lipopolysaccharide O antigen synthesis genes required for attachment of the S-layer of Caulobacter crescentus. Microbiology 147 : 1451–1460.
27. Marchler-BauerA, ZhengC, ChitsazF, DerbyshireMK, GeerLY, et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41: D348–352.
28. Entcheva-DimitrovP, SpormannAM (2004) Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J Bacteriol 186 : 8254–8266.
29. JavensJ, WanZ, HardyGG, BrunYV (2013) Bypassing the need for subcellular localization of a polysaccharide export-anchor complex by overexpressing its protein subunits. Mol Microbiol 89 : 350–371.
30. KarimovaG, PidouxJ, UllmannA, LadantD (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95 : 5752–5756.
31. FangG, PassalacquaKD, HockingJ, LlopisPM, GersteinM, et al. (2013) Transcriptomic and phylogenetic analysis of a bacterial cell cycle reveals strong associations between gene co-expression and evolution. BMC Genomics 14 : 450.
32. LaubMT, McAdamsHH, FeldblyumT, FraserCM, ShapiroL (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290 : 2144–2148.
33. McGrathPT, LeeH, ZhangL, IniestaAA, HottesAK, et al. (2007) High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol 25 : 584–592.
34. CurtisPD, BrunYV (2010) Getting in the Loop: Regulation of Development in Caulobacter crescentus. Microbiology and Molecular Biology Reviews 74 : 13–41.
35. KirkpatrickCL, ViollierPH (2012) Decoding Caulobacter development. Fems Microbiology Reviews 36 : 193–205.
36. TsokosCG, LaubMT (2012) Polarity and cell fate asymmetry in Caulobacter crescentus. Current Opinion in Microbiology 15 : 744–750.
37. QuonKC, MarczynskiGT, ShapiroL (1996) Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84 : 83–93.
38. HoltzendorffJ, HungD, BrendeP, ReisenauerA, ViollierPH, et al. (2004) Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304 : 983–987.
39. MurraySM, PanisG, FumeauxC, ViollierPH, HowardM (2013) Computational and Genetic Reduction of a Cell Cycle to Its Simplest, Primordial Components. PLoS Biol 11: e1001749.
40. BiondiEG, SkerkerJM, ArifM, PrasolMS, PerchukBS, et al. (2006) A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Molecular Microbiology 59 : 386–401.
41. FioravantiA, FumeauxC, MohapatraSS, BompardC, BrilliM, et al. (2013) DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in Caulobacter crescentus and Other Alphaproteobacteria. PLoS Genet 9: e1003541.
42. JacobsC, DomianIJ, MaddockJR, ShapiroL (1999) Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97 : 111–120.
43. HottesAK, MeewanM, YangD, AranaN, RomeroP, et al. (2004) Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J Bacteriol 186 : 1448–1461.
44. BarrK, WardS, Meier-DieterU, MayerH, RickPD (1988) Characterization of an Escherichia coli rff mutant defective in transfer of N-acetylmannosaminuronic acid (ManNAcA) from UDP-ManNAcA to a lipid-linked intermediate involved in enterobacterial common antigen synthesis. J Bacteriol 170 : 228–233.
45. GinsbergC, ZhangYH, YuanY, WalkerS (2006) In vitro reconstitution of two essential steps in wall teichoic acid biosynthesis. ACS Chem Biol 1 : 25–28.
46. D'EliaMA, HendersonJA, BeveridgeTJ, HeinrichsDE, BrownED (2009) The N-acetylmannosamine transferase catalyzes the first committed step of teichoic acid assembly in Bacillus subtilis and Staphylococcus aureus. J Bacteriol 191 : 4030–4034.
47. ChenJC, HottesAK, McAdamsHH, McGrathPT, ViollierPH, et al. (2006) Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J 25 : 377–386.
48. BelleA, TanayA, BitinckaL, ShamirR, O'SheaEK (2006) Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A 103 : 13004–13009.
49. JenalU, MaloneJ (2006) Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40 : 385–407.
50. PaulR, WeiserS, AmiotNC, ChanC, SchirmerT, et al. (2004) Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18 : 715–727.
51. Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory.
52. PitcherDG, SaundersNA, OwenRJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Letters in Applied Microbiology 8 : 151–156.
53. MerrittJH, KadouriDE, O'TooleGA (2005) Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1 Unit 1B 1.
54. O'TooleGA, PrattLA, WatnickPI, NewmanDK, WeaverVB, et al. (1999) Genetic approaches to study of biofilms. Methods Enzymol 310 : 91–109.
Štítky
Genetika Reprodukčná medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



