-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A PNPase Dependent CRISPR System in
The human bacterial pathogen Listeria monocytogenes is emerging as a model organism to study RNA-mediated regulation in pathogenic bacteria. A class of non-coding RNAs called CRISPRs (clustered regularly interspaced short palindromic repeats) has been described to confer bacterial resistance against invading bacteriophages and conjugative plasmids. CRISPR function relies on the activity of CRISPR associated (cas) genes that encode a large family of proteins with nuclease or helicase activities and DNA and RNA binding domains. Here, we characterized a CRISPR element (RliB) that is expressed and processed in the L. monocytogenes strain EGD-e, which is completely devoid of cas genes. Structural probing revealed that RliB has an unexpected secondary structure comprising basepair interactions between the repeats and the adjacent spacers in place of canonical hairpins formed by the palindromic repeats. Moreover, in contrast to other CRISPR-Cas systems identified in Listeria, RliB-CRISPR is ubiquitously present among Listeria genomes at the same genomic locus and is never associated with the cas genes. We showed that RliB-CRISPR is a substrate for the endogenously encoded polynucleotide phosphorylase (PNPase) enzyme. The spacers of the different Listeria RliB-CRISPRs share many sequences with temperate and virulent phages. Furthermore, we show that a cas-less RliB-CRISPR lowers the acquisition frequency of a plasmid carrying the matching protospacer, provided that trans encoded cas genes of a second CRISPR-Cas system are present in the genome. Importantly, we show that PNPase is required for RliB-CRISPR mediated DNA interference. Altogether, our data reveal a yet undescribed CRISPR system whose both processing and activity depend on PNPase, highlighting a new and unexpected function for PNPase in “CRISPRology”.
Published in the journal: A PNPase Dependent CRISPR System in. PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004065
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004065Summary
The human bacterial pathogen Listeria monocytogenes is emerging as a model organism to study RNA-mediated regulation in pathogenic bacteria. A class of non-coding RNAs called CRISPRs (clustered regularly interspaced short palindromic repeats) has been described to confer bacterial resistance against invading bacteriophages and conjugative plasmids. CRISPR function relies on the activity of CRISPR associated (cas) genes that encode a large family of proteins with nuclease or helicase activities and DNA and RNA binding domains. Here, we characterized a CRISPR element (RliB) that is expressed and processed in the L. monocytogenes strain EGD-e, which is completely devoid of cas genes. Structural probing revealed that RliB has an unexpected secondary structure comprising basepair interactions between the repeats and the adjacent spacers in place of canonical hairpins formed by the palindromic repeats. Moreover, in contrast to other CRISPR-Cas systems identified in Listeria, RliB-CRISPR is ubiquitously present among Listeria genomes at the same genomic locus and is never associated with the cas genes. We showed that RliB-CRISPR is a substrate for the endogenously encoded polynucleotide phosphorylase (PNPase) enzyme. The spacers of the different Listeria RliB-CRISPRs share many sequences with temperate and virulent phages. Furthermore, we show that a cas-less RliB-CRISPR lowers the acquisition frequency of a plasmid carrying the matching protospacer, provided that trans encoded cas genes of a second CRISPR-Cas system are present in the genome. Importantly, we show that PNPase is required for RliB-CRISPR mediated DNA interference. Altogether, our data reveal a yet undescribed CRISPR system whose both processing and activity depend on PNPase, highlighting a new and unexpected function for PNPase in “CRISPRology”.
Introduction
Listeria monocytogenes is a gram-positive foodborne pathogenic bacterium that has evolved two distinct lifestyles: a saprophytic one, primarily in decaying vegetation and a parasitic one in the tissues of mammals and birds, causing a disease known as listeriosis. Infection in humans starts by the ingestion of contaminated food products that deliver the bacteria in the intestinal lumen. In the course of the infection of susceptible individuals e.g. elderly and pregnant women, Listeria can cross three barriers of the organism: the intestinal, blood-brain and feto-placental barriers, causing meningitis, encephalitis and abortion. The main and best studied regulator that orchestrates the Listeria infectious process is PrfA (Positive regulatory factor A), a transcription factor that activates expression of the major known virulence genes [1]. In addition to protein determinants contributing to infection, Listeria possesses a virulence gene repertoire that expands to non-coding RNA (ncRNAs) molecules [2]–[4].
Bacterial ncRNAs are key regulatory molecules of metabolic, physiological and pathogenic processes and can be generally classified in four groups: a) the RNA regulatory elements located in the 5′ untranslated regions (5′UTRs) which regulate the expression of the corresponding mRNAs through the binding of various factors, like proteins (e.g. CsrA) and small metabolites (riboswitches) or by sensing environmental cues like temperature (thermosensors); b) the trans-acting small RNAs (sRNAs) regulating one or several target mRNAs located elsewhere on the chromosome; c) the sRNAs that sequester RNA-binding proteins; and d) the antisense transcripts (asRNAs), which overlap and are complementary to their target genes in the same genomic locus [5]. A novel class of non-coding RNAs, named CRISPRs (clustered regularly interspaced short palindromic repeats) has been shown to mediate bacterial adaptive immunity against invading bacteriophages and conjugative plasmids. A CRISPR is defined by the alternating array of identical 20–40 nucleotides (nt) long repeat sequences, interspaced by non-repetitive spacer sequences. In the proximity of the locus, are usually found gene clusters called CRISPR-associated (cas) genes. Cas genes form 23–45 different gene families (depending on the classification), encoding diverse proteins with nuclease, helicase, integrase, polymerase or nucleotide-binding activities, which are involved in the different steps of CRISPR generation, maintenance, processing and the interference mechanism. Analysis of the various sets of cas genes has revealed that CRISPR-Cas system generally cluster into three basic types (Type-I, Type-II and Type-III) which are further divided into at least ten subtypes (Types IA–F, Types IIA–C and Types IIIA–B) [6], [7]. The first clue about CRISPR function was brought about by the discovery that the different spacers were homologous to bacteriophage and plasmid sequences [8]–[10]. It was thus hypothesized that CRISPRs could play a role in immunity against invading genetic elements, which was later experimentally demonstrated in several elegant studies, e.g. in Streptococcus thermophilus [11], Escherichia coli [12] and Staphylococcus epidermidis [13]. The mechanism of action underlying the whole process is still not entirely understood, but can be roughly divided in three major stages: i) CRISPR adaptation that occurs when bacteria first encounter the foreign invader after transformation, conjugation or transduction. The CRISPR system recognizes the foreign element and incorporates parts of its DNA into what becomes a new spacer in the CRISPR locus; ii) CRISPR expression that generates a long poly-spacer precursor RNA, which is then cleaved by Cas proteins producing smaller, mature RNAs (crRNAs). Each crRNA generally contains part of the repeat and a single spacer that serves as a guide for the sequence specific recognition of the foreign invader; iii) CRISPR interference mediated by mature target-specific crRNAs that, with the help of cas gene products, inactivate the foreign, bacteriophage or plasmid nucleic acid [14], [15].
In Listeria, 14 plasmids [16] and 11 bacteriophages [17] have been sequenced so far. Bacteriophages infecting Listeria belong to the Siphoviridae and Myoviridae families in the Caudovirales order. They are either temperate integrating in the host genome by site-specific recombination or virulent actively replicating and forming virion particles that subsequently lyse the host cell. Comparative genomic analysis of Listeria bacteriophages revealed that their genomes are highly mosaic, characterized by interspecies homology as well as homology to bacteriophages infecting Bacillus, Enterococcus, Clostridium and Staphylococcus [17], [18]. Prophages are considered as the major source of diversity within the Listeria genus [18] and can constitute up to 7% of the Listeria coding genes [19], [20].
Recently, CRISPR-Cas systems have started to be analyzed in Listeria [18], [21]. We previously described in L. monocytogenes strain EGD-e, a small CRISPR RNA (RliB) exhibiting five identical repeats interspaced by non-related spacer sequences of similar size. Strikingly, no cas genes were found either in the proximity of RliB or elsewhere in the L. monocytogenes EGD-e genome [22]. Despite the absence of Cas proteins, RliB is expressed and significantly upregulated in bacteria isolated from the intestinal lumen of gnotobiotic mice, in bacteria grown in the human blood, or bacteria exposed to hypoxia. More importantly, we showed that RliB is involved in L. monocytogenes virulence [4].
Here, we characterized the cas-less RliB-CRISPR by first determining its secondary structure and analyzing its processing. Furthermore, we undertook a search for RliB protein ligands, to address the molecular machinery underlying RliB processing in the absence of Cas proteins. By using two different protein affinity purification approaches, we showed that RliB binds and is a substrate for polynucleotide phosphorylase (PNPase), a bi-functional enzyme harboring a 3′ to 5′ exoribonuclease and 3′ polymerase activities [23]. Furthermore, we performed a global analysis of CRISPR-Cas systems in all sequenced Listeria genomes, revealing a striking ubiquity of the RliB-CRISPRs in L. monocytogenes strains. Surprisingly, RliB-CRISPRs are never associated with cas gene clusters and we could demonstrate that even in Listeria strains harboring a complete set of cas genes, RliB-CRISPRs are processed by PNPase. Finally, we carried out a functional assay for RliB-CRISPR and demonstrated it requires presence of the cas genes of a second CRISPR system to lower the acquisition frequency of a plasmid carrying the matching protospacer. Moreover, we show that PNPase is required for this DNA interference activity. Together, our data highlight a novel type of CRISPR system that relies on the activity of PNPase, highlighting a new role for this enzyme in bacteria.
Results
RliB is a CRISPR RNA expressed and processed in the absence of Cas proteins
In L. monocytogenes EGD-e, RliB is located between the genes lmo0509 and lmo0510 that encode a protein similar to phosphoribosyl pyrophosphatase and a hypothetical protein, respectively. Its primary sequence resembles a typical CRISPR. It is composed of 5 identical 29 nt repeat sequences (GTTTTAGTTACTTATTGTGAAATGTAAAT) interspaced by four 35–37 nt long spacer sequences (S1, S2, S3 and S4 in the Figure 1A). The spacer 3 (S3) has identity with the Listeria temperate bacteriophage B054 sequence and spacer 4 (S4) identity to Listeria virulent bacteriophage P70 sequence [17], [24]. We analyzed the secondary structure of the full length RliB that is detectable in vivo, using RNase V1 (specific for helical regions), RNase T2 (specific for unpaired nucleotides with a preference for adenines) and dimethylsulfate (which methylates N1 of adenine and N3 of cytosine) (Figure S1). The secondary structure of RliB, that explains most of the probing data, involves six stem-loop structures among which five contain a GUUUU motif within the loops, followed by a hairpin terminator at the 3′ end (Figure 1B). In contrast to CRISPR systems where the repeat sequences form independent and stable palindromic structures [25], the RliB hairpin structures are mostly formed by base pairings between the repeat sequences and the adjacent spacer sequence. These data suggest that RliB structure largely depends on the nature of the incoming spacer DNA.
Fig. 1. RliB in L. monocytogenes EGD-e. 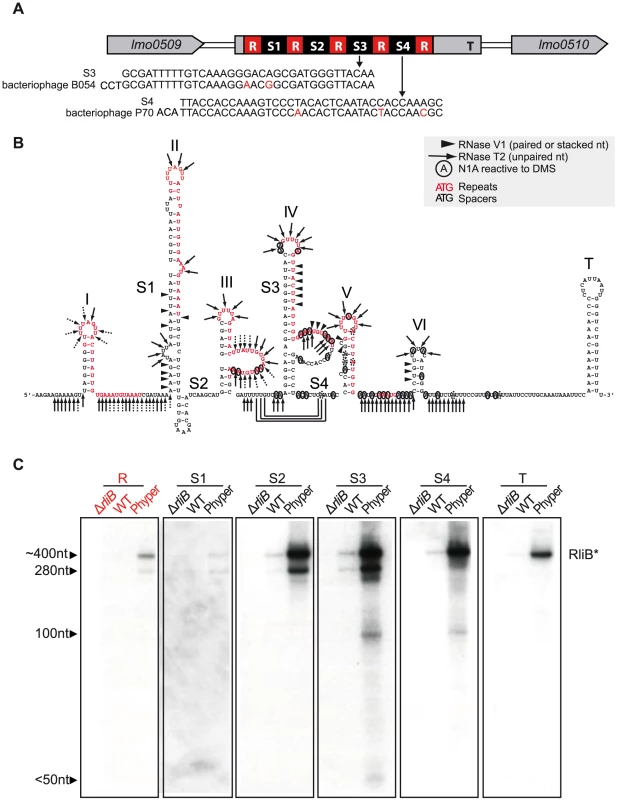
A) Schematic representation of the RliB locus. RliB is located between genes lmo0509 and lmo0510. It is composed of 5 identical repeat sequences (R), 4 spacer sequences (S1 to S4) and the terminator (T). The sequence alignments between the spacers and bacteriophage genomes are shown. B) RliB secondary structure. Repeat sequences are highlighted in red, spacers are numbered S1 to S4, the hairpin structures are numbered I to VI and a terminator hairpin is indicated T. C) RliB processing. Northern blot on total RNAs extracted from bacteria deleted for RliB (ΔrliB), wild type bacteria (WT) and bacteria overexpressing RliB (Phyper RliB). Expression of RliB was detected using P32-5′ labelled probes complementary to different parts of the RliB molecule: repeat (R), spacer 1 (S1), spacer 2 (S2), spacer 3 (S3), spacer 4 (S4), 5′end and the terminator (T). We had previously noticed that RliB in L. monocytogenes EGD-e is processed to a smaller fragment [22]. Here, we examined this processing by Northern blot analysis of total RNA isolated from the wild type (WT) L. monocytogenes EGD-e and bacteria expressing RliB from a constitutive promoter (Phyper-RliB). We used probes complementary to the repeat (R), to each unique spacer (S1, S2, S3 and S4) and to the 3′ end of the molecule including the terminator region (T) (Figure 1C). All the probes allowed detection of a 400 nt fragment, which corresponds to the full length RliB molecule. The probes for S1, S2, S3 and R regions detected an additional 280 nt long fragment. The probes for S3 and S4 regions showed a minor 100 nt long fragment and the probe for S3 region a fragment smaller than 50 nt.
Altogether, our data show that the RliB-CRISPR has a secondary structure largely determined by the interactions between each repeat and the adjacent spacer and, despite the absence of Cas proteins, it is processed and exists under two major forms: i) a 400 nt full length RliB molecule and ii) a shorter form of RliB molecule, approximately 280 nt long, comprising spacers S1, S2 and S3.
RliB interacts with the Polynucleotide Phosphorylase (PNPase) enzyme
Considering the complete absence of cas genes in L. monocytogenes EGD-e strain, we hypothesized that the RliB-CRISPR processing is governed by another bacterial ribonuclease. To identify which enzymes are involved in this process, we first searched for proteins that interact with RliB using the affinity purification method with a tagged RliB molecule. Given that addition of a tag may perturb the folding of the bait-RNA molecule and/or change its accessibility, resulting in a loss of interaction with its binding partners, we used two strategies using two different tags added either at the 5′ or at the 3′ end of the bait-RNA. The first affinity purification was performed with the 3′-biotinylated full length RliB (RliB-B) and a control RNA, the quorum sensing induced RNAIII from Staphylococcus aureus (Figure 2A). In the second approach, we used as a bait RliB tagged at the 5′ end with two hairpin structures constituting the “MS2 binding sequence” (RliB-MS2), i.e. the RNA binding sequence of bacteriophage MS2 coat protein (MS2) (Figure 2B). Structure probing using enzymes show that the MS2-tag did not change the structure of RliB (Figure S1). The RliB-B or RliB-MS2 RNAs were bound to streptavidin or MBP-MS2 coated beads, respectively and incubated with total Listeria cell extracts. After extensive washing of unspecific proteins, the bound fraction was eluted and loaded on SDS-polyacrylamide denaturing gel. To verify the integrity of the bait-RNA, we also analyzed the eluted tagged RNA using polyacrylamide-urea gel electrophoresis. For both experiments, we detected a single and major protein band of approximately 78 kDa, specific to RliB-bound elution fractions (RliB-B and RliB-MS2) (Figures 2A,B). The protein was identified by mass spectrometry to be the Listeria polynucleotide phosphorylase (PNPase) encoded by gene lmo1331 (pnpA), a bi-functional enzyme that acts as 3′-5′ exoribonuclease and a 3′-terminal polymerase [23], [26].
Fig. 2. RliB interacts with PNPase. 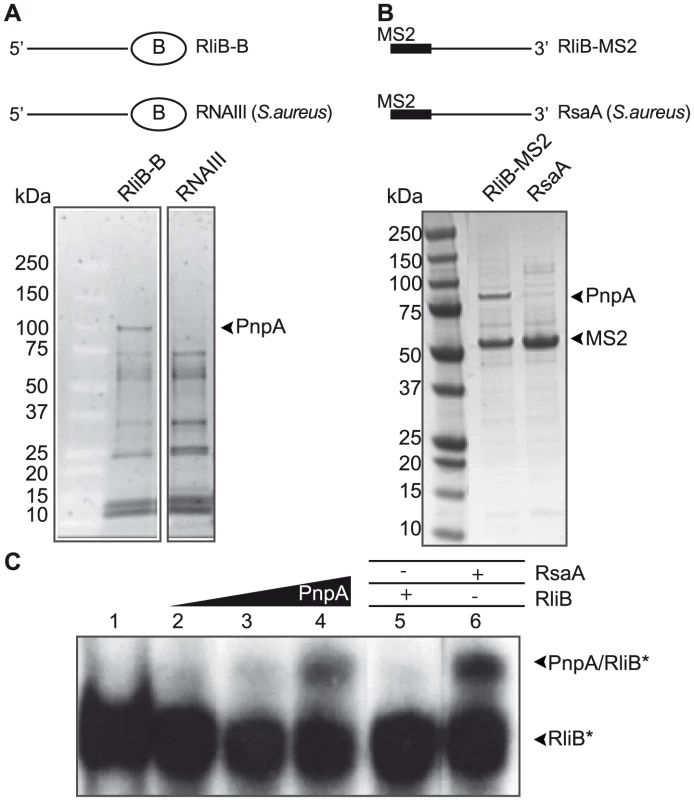
A) Protein affinity purification using 3′ biotinylated RliB. Two biotinylated bait RNAs were used: full length RliB (RliB-B) and a control RNA (S. aureus RNAIII). Coomassie stained SDS-PAGE gel of the two elution fractions is shown. B) Protein affinity purification using 5′ MS2 tagged RliB. Two MS2 tagged bait RNAs were used: full length RliB (RliB-MS2) and a control RNA (S.aureus RsaA). Coomassie stained SDS-PAGE gel of the fractions is shown. C) Interaction between purified PNPase and in vitro transcribed RliB. Gel retardation assay performed with P32-5′ labelled RliB (RliB) and increasing concentrations of the purified PNPase protein (lane 1, no PNPase added; lane 2, 100 nM PNPase; lane 3, 250 nM PNPase; lane 4, 400 nM PNPase). Competition experiments were performed in the presence of non-labelled RliB (lane 5, 400 nM RliB) and non-labelled RsaA (lane 6, 400 nM RsaA). In these experiments, the concentration of PNPase was 400 nM. We then analyzed whether the interaction between RliB and PNPase is direct or requires another binding partner. The L. monocytogenes PNPase protein carrying 6 histidines at its C-terminal end was purified and binding experiments were carried out using gel retardation assays with in vitro transcribed P32-labeled full length RliB and increasing amount of the purified PNPase. Formation of a complex between RliB and PNPase was observed with 400 nM PNPase, showing that the interaction is direct and does not require another binding partner. To demonstrate the specificity of PNPase binding, competition experiments were done with various non-labelled RNAs. The addition of non-labelled RliB outcompeted the interaction between PNPase and P32-labeled RliB in contrast to the addition of a non-labelled control RNA from S. aureus (RsaA) that did not affect the complex formation (Figure 2C). Altogether, our results show that PNPase specifically interacts with RliB.
RliB is a substrate for PNPase
PNPase is a bifunctional enzyme, which in vivo acts primarily as a 3′ to 5′ exoribonuclease of single stranded target RNAs [26]. We investigated if RliB is a substrate of PNPase. We first verified the activity of the purified PNPase protein and performed an in vitro assay where a 37 nt P32-end labeled substrate RNA (RNA37) was incubated alone or with 200 nM purified PNPase (Figure 3A). The presence of PNPase resulted in the degradation of the 5′ end P32-labeled RNA37 while no cleavage reaction was observed using a P32-pCp 3′ end labeled RNA37 (results not shown). These data demonstrate that the purified protein is active and able to degrade single-stranded RNA substrates. Three non-labeled competitor RNAs were then added to the reaction; i) a non-labeled RliB; ii) a non-labeled control RNA (RsaA); iii) and the non-labeled RNA37 substrate. As expected, the addition of non-labeled RNA37 substrate decreased the cleavage reaction. Strikingly, the addition of 100 nM non-labeled RliB resulted in the loss of PNPase mediated RNA37 degradation, whereas addition of RsaA did not alter the degradation of RNA37, indicating that RliB acts as a competitive inhibitor of PNPase.
Fig. 3. RliB is a substrate for PNPase. 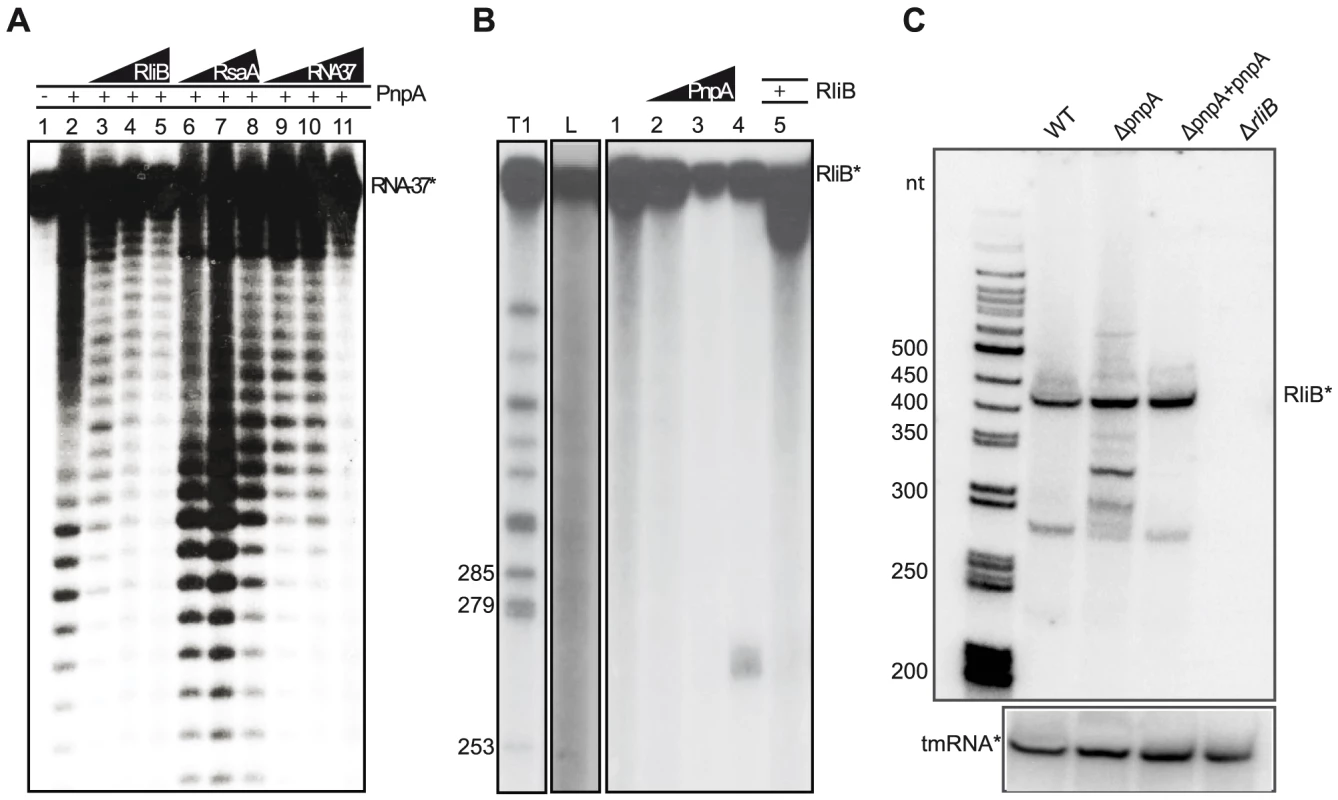
A) In vitro PNPase activity assay. A 37 nt short P32-labeled substrate RNA (RNA37*) was incubated alone (lane 1) or with 200 nM purified PNPase (lane 2). Three types of non-labeled competitor RNAs were then added: non-labeled RliB (lanes 3, 100 nM; lane 4, 200 nM; lane 5, 400 nM), non-labeled control RNA RsaA (lane 6, 100 nM; lane 7, 200 nM; lane 8, 400 nM) and non-labeled RNA37 substrate (lane 9, 100 nM; lane 10, 200 nM; lane 11, 400 nM). B) In vitro PNPase-mediated processing of RliB. On the left are indicated ladders T1 (RNase T1 hydrolysis) and L (alkaline hydrolysis). The full length P32-5′ labeled RliB (RliB*) was incubated with increasing concentrations of purified PNPase (lane 1, no PNPase; lane 2, 25 nM; lane 3, 100 nM; lane 4, 400 nM). Competition experiments were performed in the presence of 400 nM non-labelled RliB (lane 5). C) In vivo PNPase-mediated processing of RliB. Northern blot performed on the total RNAs extracted from the L. monocytogenes EGD-e wild type bacteria (WT), strain deleted for pnpA (ΔpnpA), complemented strain (ΔpnpA-pnpA) and strain deleted for RliB (ΔrliB). Expression of RliB was detected by the P32 labeled in vitro transcribed RNA probe complementary to the full length RliB. The membrane was reprobed with P32 labeled in vitro transcribed RNA probe complementary to the tmRNA. To investigate further the activity of PNPase on RliB, we incubated the full length 5′ end-labeled RliB with increasing concentrations of purified PNPase (Figure 3B). We observed on the gel the appearance of a band migrating around 270 nt, an RliB processing product generated by the PNPase-mediated degradation up to the stem-loop IV. This cleavage reaction was inhibited by the addition of the full length non-labeled RliB. Altogether, these data suggest that in vitro, PNPase is processing RliB until its exoribonuclease activity is stalled in the S4 repeat region.
To study the effect of PNPase on RliB in vivo, we constructed a pnpA deletion mutant (ΔpnpA) and compared by Northern blot the size of the RliB transcript in the ΔpnpA mutant and WT bacteria. In the absence of PNPase, two major bands migrating as 300 and 330 nt long RNAs, were observed. Upon complementation (ΔpnpA-pnpA), the RliB processing was restored, identical to that observed in the WT strain (Figure 3C). Our results thus strongly suggest that RliB is a substrate for PNPase in vivo.
RliB-CRISPR is ubiquitous among Listeria strains
CRISPR arrays are thought to evolve rapidly in prokaryotic genomes [14], [27], [28]. Therefore, we investigated the presence of RliB in other Listeria strains. For this, we searched for CRISPRs in 29 complete and 17 draft Listeria genomes (Table S1). As mentioned in the introduction, the highly diverse CRISPR-Cas systems are classified into three main types (I, II and III) each including several subtypes [6]. In Listeria, we found two types of CRISPR-Cas systems: i) CRISPR-Cas systems type-I (subtype I-B) with the cas operon composed of cas6-cas8a1-cas7-cas5-cas3-cas1, including also in some cases cas4, associated with the repeat sequence GTTTTAGTTACTTATTGTGAAATGTAAAT that is almost identical to the repeat of RliB-CRISPR; ii) CRISPR-Cas systems type-II (subtype II-A) associated with csn2-cas2-cas1-cas9 operon and the repeat sequence GTTTTGTTAGCATTCAAAATAACATAGCTCTAAAAC (Figure 4A).
Fig. 4. CRISPR-Cas systems in L.monocytogenes genomes. 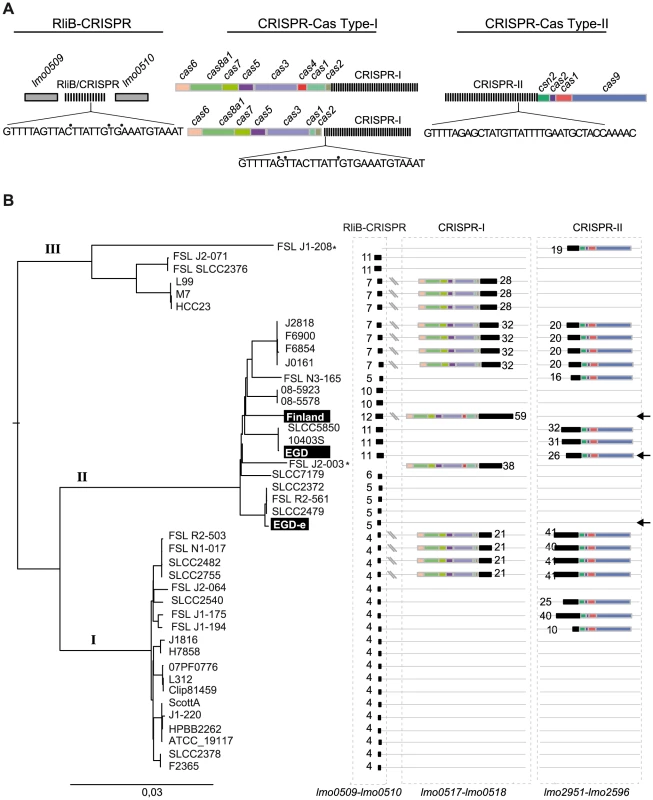
A) The three types of CRISPR-Cas systems found in L.monocytogenes. The sequences of the repeats are given. Points (•) indicate sites of repeat variability among Listeria strains. B) CRISPR-Cas systems in complete and draft L. monocytogenes genomes of the lineages I, II and III. For two strains indicated by an *, the sequencing results are too preliminary with a high number of contigs and RliB was not detected. However, these strains were included in the spacer analysis. CRISPR-I is present at the locus between lmo0517 and lmo0510 in 7 complete L. monocytogenes genomes, 10 draft L. monocytogenes genomes, in Listeria seeligeri and Listeria ivanovii (Figure 4B and Table S1) and it is always associated with a type-I cas operon located in close proximity. The CRISPR-II was detected between lmo2591 and lmo2596 in 6 complete L. monocytogenes genomes, 9 draft genomes and in Listeria innocua (Figure 4B, Table S1). The CRISPR-II is also found exclusively associated with type-II cas operon. The tight association of CRISPR-I and CRISPR-II with type-I and type-II cas genes, suggests that the function of those CRISPRs is dependent on the activity of the corresponding Cas protein machinery.
In contrast to CRISPR-I and CRISPR-II that are found in about 30% of the complete Listeria genomes, the RliB-CRISPR is present at the same genomic locus in all analyzed complete and draft L. monocytogenes genomes as well as in other Listeria species (Figure 4B, Table S1). This suggests a stronger selective pressure on this element relative to the cas-associated CRISPRs. In silico structure prediction performed on three representative RliB-CRISPRs carrying different number of repeats revealed a putative secondary structure that is highly similar to that experimentally determined in L. monocytogenes EGD-e strain (Figure S2). Cas operons have not been detected in the close proximity to the RliB-CRISPRs. Furthermore, 14 complete Listeria genomes completely lack cas genes. The number of repeats in RliB-CRISPRs range from 1 to 11 and does not correlate with the presence or absence of cas genes elsewhere in the genome (Table S3). Together, the conservation of RliB-CRISPRs among Listeria strains suggests that they may have a function despite the absence of Listeria Cas proteins.
Although RliB-CRISPR and CRISPR-I have different pattern of conservation the two systems share almost identical repeat sequences (Figure 4A). To investigate the correlation between the two systems, we compared their putative leader and upstream sequences. Multiple alignments of the DNA fragment preceding the identified RliB-CRISPR and CRISPR-I systems revealed a striking homology (Figure S3A). More interestingly, the putative leader sequences harbour a highly conserved sequence homologous to RpoD dependent promoter, that was previously reported upstream of RliB in the L. monocytogenes EGD-e strain [22] (Figure S3B). High homology of the repeats and the leader sequences of RliB-CRISPR and CRISPR-I systems suggest a possible close relationship between the two systems.
RliB-CRISPRs in Listeria strains carrying cas genes are also substrates for PNPase
To investigate if the PNPase-mediated processing of RliB-CRISPR is specific to L. monocytogenes EGD-e strain or is more general, we examined if PNPase is also involved in RliB-CRISPR processing in Listeria strains containing a complete set of cas genes. We constructed a pnpA deletion mutant in the L. monocytogenes Finland strain (ΔpnpA-Fin), which carries a complete CRISPR-Cas system type I and in the L. monocytogenes EGD strain (ΔpnpA-EGD), which has a complete CRISPR-Cas system type II. We also constructed the deletion mutants for the RliB-CRISPR in the same strains (ΔrliB-CRISPR-Fin and ΔrliB-CRISPR-EGD, respectively). The RliB-CRISPR processing was examined by northern blot in the corresponding strains (Figure 5).
Fig. 5. RliB-CRISPRs in EGD and Finland L. monocytogenes strains. 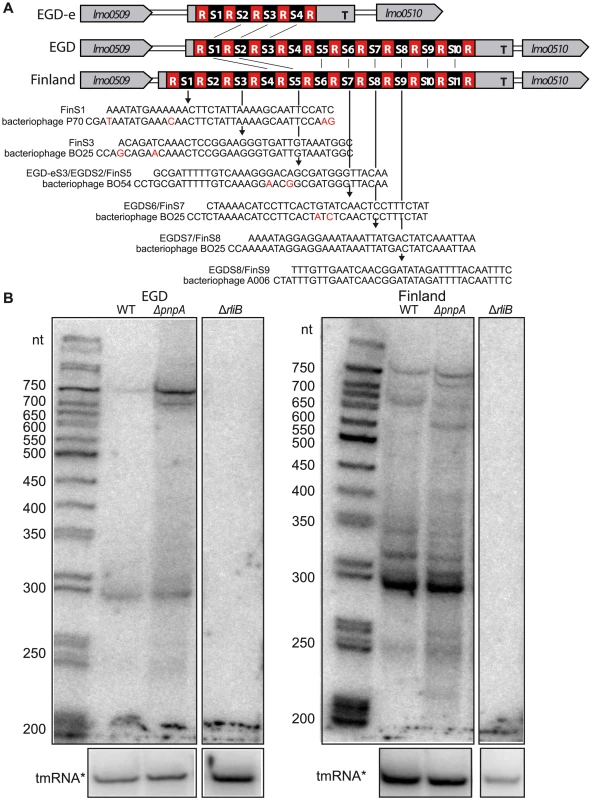
A) Schematic representation of the loci harbouring RliB-CRISPR in L.monocytogenes EGD-e, EGD and Finland strains. In all three strains, RliB-CRISPR is located between genes lmo0509 and lmo0510. Spacers that are identical between the strains are connected with black lines (S1EGD = S4Fin, S2EGD = S5Fin, S5EGD = S6Fin, S6EGD = S7Fin, S7EGD = S8Fin, S8EGD = S9Fin, S9EGD = S10Fin and S10EGD = S11Fin) and the sequence alignments between the spacers and bacteriophage genomes are shown below. B) Processing of the RliB-CRIPSRs. On the left is shown a northern blot performed on the total RNAs extracted from the wild type L. monocytogenes EGD strain (WT) and its isogenic mutants deleted for pnpA (ΔpnpA-EGD) or RliB-CRISPR (ΔRliB-EGD). On the right is shown a northern blot performed on the total RNAs extracted from the wild type L. moncytogenes Finland strain (WT) and its isogenic mutants deleted for pnpA (ΔpnpA-Fin) or RliB-CRISPR (ΔRliB-Fin). Expression of RliB-CRISPR was detected with the P32 labeled in vitro transcribed RNA probe complementary to the spacers S5 to S10 in EGD, and S6 to S11 in the Finland strain. The membranes were reprobed with P32 labeled in vitro transcribed RNA probe complementary to the tmRNA. The RliB-CRISPR in EGD strain (RliB-CRISPR-EGD) is composed of 11 identical repeats and 10 spacer sequences among which spacers S2, S6, S7 and S8 share similarity to Listeria temperate bacteriophages B054, B025 and A006 (Figure 5A, 6). In the WT EGD strain, RliB-CRISPR is expressed as a 750 nt long RNA that is processed into shorter fragments with the major processed form being 280 nt long. In the absence of PNPase, the total amount of full-length RliB-CRISPR-EGD increased and the transcript processing changed compared to the WT bacteria, i.e. we observed additional bands with a major one of 700 nt (Figure 5B).
Fig. 6. Spacer composition of CRISPR arrays in L. monocytogenes. 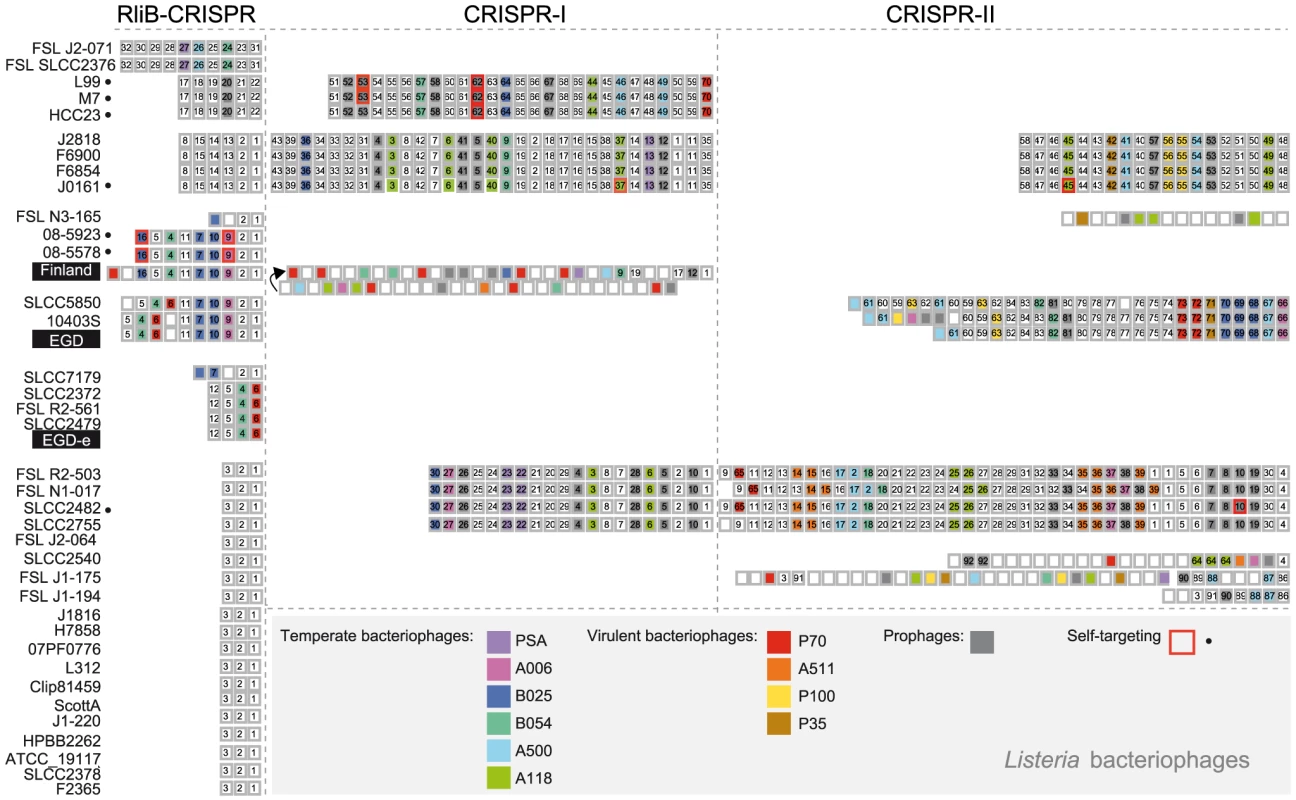
CRISPR systems present in complete and draft L. monocytogenes genomes are represented. For each L. monocytogenes strain spacers belonging to RliB-CRISPR, CRISPR-I and CRISPR-II are shown. Boxed numbers represent individual spacers conserved within an CRISPR array among the represented strains (for the RliB-CRISPR: 1-32, for the CRISPR-I: 1-70, for the CRISPR-II: 1- 92). Unique spacers are represented as not numbered boxes. Spacers matching bacteriophage sequences available in the Genbank are coloured according to the type of bacteriophage they target. The color code and corresponding bacteriophages are represented in the bottom part of the figure. Spacers matching prophages are highlighted as gray boxes. Self-targeting spacers, identified in complete Listeria genomes are highlighted with red box and the strain carrying the spacer is highlighted (•). The RliB-CRISPR in the Finland strain (RliB-CRISPR-Fin) is composed of 12 identical repeats and 11 spacer sequences among which 8 spacers are shared with RliB-CRISPR-EGD. Spacers S1, S3, S5, S7, S8 and S9 show high similarity to sequences in Listeria bacteriophages P70, B025, B054 and A006 (Figure 5A, 6). The full-length RliB-CRISPR-Fin is expressed in the WT bacteria, as a 780 nt long RNA that it is processed to several shorter fragments with the 280 nt being again the most abundant form. In the absence of PNPase, RliB-CRISPR-Fin processing changed as additional bands are observed compared to the WT bacteria (Figure 5B).
Together, our results suggest that PNPase contributes to the RliB-CRISPR processing in vivo, independently of the presence of either CRISPR-Cas system type I in the L. monocytogenes Finland strain, or the presence of CRISPR-Cas system type II in the L. monocytogenes EGD strain.
Spacers of the RliB - CRISPRs are directed against Listeria bacteriophages
We analyzed CRISPR spacers to compare the putative functions of cas-less RliB-CRISPR and cas-associated CRISPR-I and CRISPR-II. In total, we identified 978 spacers that correspond to 348 unique sequences (Table S2). These were used to search for the similarity with the sequences of all complete prokaryote, plasmid and virus genomes available in the Genbank as well as the sequences of integrated temperate bacteriophages (prophages) identified in complete Listeria genomes (Figure 6, Table S3).
We identified 142 (41%) spacers that share identity to bacteriophages known to infect Listeria species (Figure 6). They match sequences detected in 6 temperate (B054, B052, A118, A500, A006, PSA), 4 virulent phages (A115, P35, P70, P100) as well as 35 distinct prophages found in complete Listeria genomes (Figure S4). Overall, we found matching protospacers for 33% RliB-CRISPR spacers, 41% CRISPR-I spacers and 42% CRISPR-II spacers (Figure 6, Table S3). RliB-CRISPR and CRISPR-I systems share an identical protospacer adjacent motif (PAM) CCA at the 5′ of the protospacer, in contrast to CRISPR-II harboring NGG at the 3′ of the protospacer (Figure S5). Numerous spacers showed 100% identity with viral sequences (14% RliB-CRISPR spacers, 15% CRISPR-I spacers and 24% CRISPR-II spacers). None of the spacers matched bacterial (excluding prophages) or plasmid sequences. The high abundance of spacers perfectly matching bacteriophages in the RliB-CRISPRs and in the CRISPR-I and CRISPR-II, suggests that both cas-less and cas-associated CRISPR-Cas systems have a role in the immunity against bacteriophages.
To investigate the nature of the phage nucleic acid potentially targeted by the RliB-CRISPR, CRISPR-I and CRISPR-II systems, we first examined the orientation of the protospacers in respect to the corresponding spacers and then the function of the phage genes where the protospacers are located. Protospacers targeted by CRISPR-I and CRISPR-II systems originate both from sense and antisense DNA strand and are equally distributed along the phage genome, which suggests that these systems target phage DNA (Table S4, S5, Figure S6). Among 13 protospacers targeted by RliB-CRISPR, 9 protospacers are in the antisense orientation, 3 are positioned in intergenic regions and 2 are in the sense orientation. Moreover, among 11 protospacers for which the function of the targeted gene is known, 10 protospacers are located in the late phage genes encoding DNA packaging and structural proteins (Table S6, Figure S6). The occurrence of both sense and antisense oriented protospacers suggests RliB-CRISPR most probably targets DNA. However, higher occurrence of the antisense oriented protospacers and more interestingly, specificity for the function of the targeted bacteriophage gene suggests that RliB-CRISPR could potentially have a function in RNA interference.
Furthermore, we identified genomes containing a number of spacers matching their own prophages. For example, L. monocytogenes strain EGD has prophage B025 (our unpublished data) and carries one RliB-CRISPR spacer (S7) and three CRISPR-II spacers (S21, S22, S23) that match the same prophage with up to 97% identity (Figure S4A,B). We also identified two RliB-CRISPR spacers, three CRISPR-I spacers and two CRISPR-II spacers in 9 Listeria genomes that correspond to prophages in the same genome with 100% identity. In two cases, the strains (L. monocytogenes 08-5578 and 08-5923) lack the cas genes and in one case the repeat flanking the self-targeting spacer carries a point mutation (L. monocytogenes J0161), suggesting that in three instances the spacers are presumably inactive (Table S7, Figure 6). The remaining spacers are either in cas-associated CRISPRs or in the RliB-CRISPR. Furthermore, the PAMS corresponding to self-targeted protospacers do not significantly deviate from the consensus (Table S7). These results show that spacers matching the protospacer located in the same bacterial chromosome do not necessarily have strong negative fitness effects.
RliB-CRISPR DNA interference activity relies both on PNPase and trans encoded Cas proteins
To test if the cas-less RliB-CRISPR might provide Listeria with DNA interference activity, we designed an experiment using a conjugation system and two plasmids that differ in the presence or absence of protospacer: i) a protospacer plasmid (P) and ii) the control plasmid (C), as previously done by Almendros et al. [29] (Figure 7). The plasmid P carries a protospacer matching spacer 3 (S3) of the RliB-CRISPR in the L. monocytogenes EGD-e strain and spacer 5 (S5) of the RliB-CRISPR in the Finland strain. The plasmid C is identical to the plasmid P, but the protospacer sequence is shuffled in silico (C) and does not correspond to any known sequence in the NCBI database.
Fig. 7. CRISPR DNA interference activity assay. 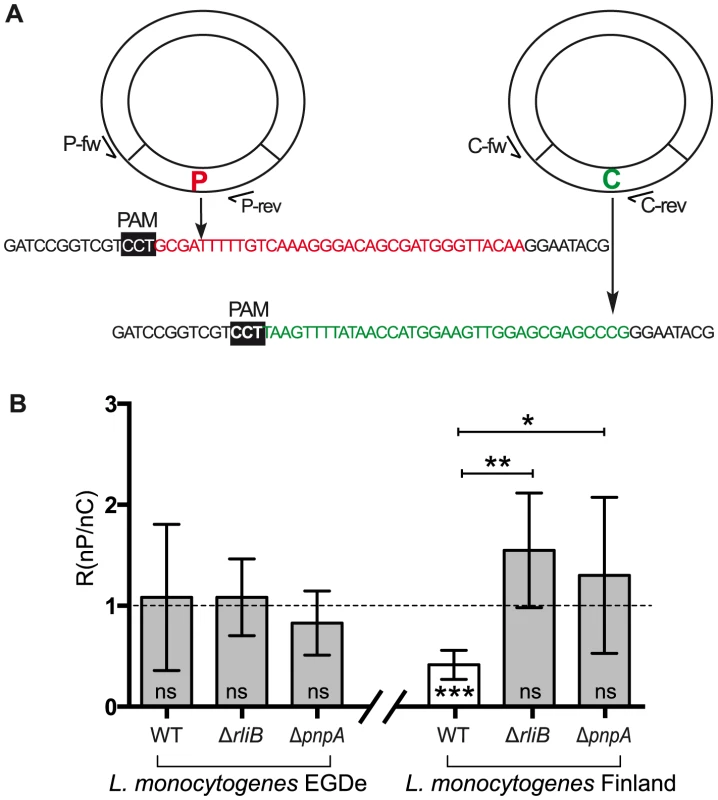
A) Design of the plasmids. Two types of plasmids were used: i) protospacer plasmid (P) carrying the protospacer (highlighted in red) matching the spacer 3 in L. monocytogenes EGD-e strain RliB-CRISPR and spacer 5 in L. monocytogenes Finland strain RliB-CRISPR; ii) control plasmid (C) carrying the DNA fragment corresponding to the shuffled protospacer (highlighted in green). Black box marks the position of the protospacer adjacent motif (PAM). Position of the specific oligonucleotides used for the Q-PCR screening is indicated (P-fw, P-rev, C-fw, C-rev) B) Ratio (R). The number of colonies carrying plasmid P and the number of colonies carrying the plasmid C in the different Listeria strains and genetic backgrounds is represented as the ratio (R = n (colony P)/n (colony C). Shown are the mean and the standard deviation of five experiments performed for each bacterial strain. Two statistical tests were used: i) a t-test measuring if the ratio in the given strain is significantly different from 1 (*** - p = 0,0008; ns- not significant); ii) a t-test comparing the ratios of the WT strain and the ΔrliB and ΔpnpA strains (**- p = 0,0025; *- p = 0,0334). Listeria is not naturally competent and the plasmid transformation efficiency in this bacterium is very low in comparison to other bacteria such as Bacillus or Streptococcus. Moreover, the Δpnp genetic background has a severe effect on bacterial growth, and plasmid transformation is even more difficult than in the WT strain. Therefore, the plasmids P and C were conjugated simultaneously via Escherichia coli S17 strains to L. monocytogenes EGD-e and Finland WT strains and their isogenic mutants deleted for RliB (ΔrliB) and PNPase (ΔpnpA). Quantitative PCR (Q-PCR) was used to determine the identity of the plasmids distributed among the transformants (see materials and methods). We then calculated for each individual strain the ratio (R) of the number of colonies carrying plasmid P and the number of colonies carrying plasmid C (R = nP/nC) (Figure 7B). The proportion of the transformants carrying the plasmid P for each experiment is an indication of the interference activity driven by the spacer, a lower proportion of transformants with the plasmid P (R<1) suggesting interference activity.
In the L. monocytogenes EGD-e in which no cas genes was identified, there is no significant difference in the R values between the strains carrying the RliB-CRISPR (WT) and the strains lacking either the RliB-CRISPR (ΔrliB-EGDe) or PNPase (ΔpnpA-EGDe), demonstrating that both plasmids are equally acquired and that the system is not able to provide any detectable DNA interference in the tested experimental conditions (Figure 7B). In contrast, in the L. monocytogenes Finland that bears an additional CRISPR-Cas Type-I system, the R ratio is significantly smaller than 1 in the WT strain whereas it reaches 1 in the strains lacking either the RliB-CRISPR (ΔRliB-Fin) or the PNPase (ΔpnpA-Fin). Thus, RliB-CRISPR can lower the plasmid P acquisition in the strain carrying the CRISPR-I system, suggesting that the RliB-CRISPR is able to use the trans encoded Cas proteins encoded by the CRISPR-I and confer to Listeria a DNA interference activity. Interestingly, PNPase is required for this process.
Discussion
Since the initial discovery that CRISPR-Cas systems function as an adaptable prokaryotic immune system, the CRISPR research has been flourishing and biochemical insights into the CRISPR-Cas systems have increased dramatically over the past few years. However, these systems are extremely diverse and the function and molecular mechanism of many of them are still unknown. In particular, little is known about CRISPR-Cas type I-B, I-C, and I-D systems [30]. Here, we studied RliB-CRISPR that has an unusual secondary structure comprised of basepair interactions between the repeat sequence and the adjacent spacer. We showed that RliB-CRISPR is expressed and processed even in the complete absence of Cas proteins, and demonstrated this event occurs under the guidance of PNPase. The RliB-CRISPR spacers match numerous temperate and virulent Listeria bacteriophages and in the presence of CRISPR-Cas system Type-I, RliB-CRISPR lowers the frequency of a plasmid carrying a matching protospacer. In addition, we show that this DNA interference is dependent on the presence of PNPase. Overall, our data demonstrate that PNPase is involved in Listeria RliB-CRISPR processing and its DNA interference activity.
Secondary structure of RliB
In silico analysis of the secondary structures of CRISPR repeats across bacterial and archaeal CRISPR-Cas systems suggested that some CRISPR repeats can form stable stem-loops due to the palindromic nature of their repeats, but that other lack any detectable conserved structure [25]. RliB repeats are only weakly palindromic and unlikely form a stable stem-loop structure. Here, we experimentally determined the secondary structure of RliB and surprisingly, discovered that RliB contained 6 hairpin structures formed mostly by base-pair interactions between the spacer sequences and the adjacent repeats, and with GUUU-rich apical loops (Figure 1B). The structure of RliB is thus dependent on the nature of the acquired spacer. In silico analysis of other representative RliB-CRISPRs showed their putative structures rely on the same principle, suggesting that base-pair interactions between the repeat and the spacer could be a common structural motif among RliB-CRISPRs (Figure S2). It is tempting to hypothesize that successful acquisition of a new spacer requires some degree of complementarity with the repeat. Spacer acquisition is the least understood step of CRISPR-Cas system function [31] and our data potentially highlight new aspects of the integration mechanism via homology with the repeat sequence.
RliB-CRISPR processing by PNPase
It is generally accepted that CRISPR arrays require Cas proteins for their processing and activity. A first example of CRISPR-Cas system that does not rely solely on the Cas proteins but requires also the activity of endogenously encoded enzymes has been recently reported in Streptococcus pyogenes. In this case, a CRISPR array type-II is processed by the widely conserved endoribonuclease III and a small trans-acting RNA tracrRNA [32]. In addition, a recent study of Zhang et al [33] revealed a CRISPR in Neisseria meningitidis, where crRNAs are transcribed from promoters that are present within each repeat and require RNase III and trans-encoded tracrRNA-mediated processing for their maturation. Surprisingly, the maturation processing is dispensable for the CRISPR interference [33].
Here, we characterized a CRISPR array that is processed in a bacterium completely devoid of cas genes. In contrast to other CRISPRs, RliB-CRISPR is present in all sequenced strains of L. monocytogenes, even in other members of the genus and never co-localizes with cas operons. We demonstrated that RliB binds to and is a substrate for endogenously encoded PNPase, both in cas-less Listeria strains and in those encoding a complete set of cas genes elsewhere in the genome. Generally, PNPase degrades single-stranded RNA in a processive manner along the substrate until it stops, stalled by a stable RNA structure [23]. For instance, a hairpin structure in a bacteriophage mRNA can block the processivity of PNPase to protect the RNA against degradation [34]. It remains to be understood at the molecular level how PNPase specifically recognizes the RliB-CRISPR and how the progression of the enzyme stops.
In the three analyzed L. monocytogenes strains, RliB-CRISPRs is expressed as a full length molecule that is processed to several fragments out of which a 280 nt fragment is the most abundant form. The consistency of processing that is independent of the number of the repeat/spacer units suggests that the molecular mechanisms guiding the processing in the tested CRISPRs is conserved. Interestingly, in the bacteria deleted for pnpA (Δpnp-EGDe, Δpnp -EGD, Δpnp -Fin), some processing still occurs, indicating there are other endogenously encoded ribonucleases contributing to this mechanism, particularly in the EGD-e strain that is devoid of cas genes (Figures 3C and 5B). Listeria encodes at least 17 different putative RNases identified by homology with closely related Bacillus subtilis [35]. Future work will have to determine which enzymes might function together with PNPase and also contribute to the CRISPR processing.
The DNA interference activity of the RliB-CRISPRs
We showed that cas-less RliB-CRISPRs are rich in spacers matching virulent and temperate bacteriophages. In addition, a large fraction of those spacers have 100% matches with phages, strongly suggesting a function for RliB-CRISPR even in the absence of cas (Figure 6, Table S3). Accordingly, we showed that cas-less RliB-CRISPR lowers the acquisition of a plasmid carrying the corresponding protospacer, provided a CRISPR-I system is present (Figure 7). The RliB-CRISPR and CRISPR-I share many similar features; almost identical repeat sequence (Figure 4), homologous putative leader sequences (Figure S3) and identical PAM motifs (Figure S5), indicating that these two systems are closely related and are possibly functionally linked. It is thus not surprising that RliB-CRISPR can share the Cas machinery with the CRISPR-I to acquire the DNA interference activity, however future analysis will be required to establish the exact mechanism by which this crosstalk occurs.
More interestingly, the DNA interference activity of the RliB-CRISPR is also dependent on the presence of PNPase (Figure 7), indicating that the processing by this enzyme is important for the activity of the RliB-CRISPR. PNPase is a highly complex enzyme with 3′ to 5′ exoribonuclase and RNA polymerase activities being the most studied up to date. However, it was recently shown that PNPase can degrade single stranded DNA (ssDNA) and also catalyze template independent polymerization of dNDPs into 3′ends of ssDNA, which established a molecular model for the role of PNPase in DNA repair [36], [37]. In Escherichia coli, PNPase affects the stability of several regulatory sRNAs [38], [39]. Here, we hypothesize that Listeria PNPase, potentially with other endogenously encoded enzymes, may contribute to the RliB-CRISPR maturation. Alternatively, PNPase may affect the RliB-CRISPR RNA stability and turnover, and hence, regulate the levels of its mature form. Finally, PNPase dependent processing of the RliB-CRISPR and the DNA interference might be uncoupled activities. Hence, this complex enzyme could use different enzymatic activities to contribute to different processes. It will be also important to determine if PNPase is involved in other CRISPR-Cas system activities, such as new spacer acquisition. Currently, our data do not provide evidence on which form of RliB molecule is active in the DNA interference. These and other mechanistic details, such as the role of PAMs are to be determined in the future.
Noticeably, the RliB-CRISPR mediated DNA interference is not 100% effective. This might be the consequence of our experimental design or this CRISPR-Cas system did not evolve to eradicate the bacteriophage from a population but rather to fine-tune its copy number in the bacterial cytoplasm.
The potential function of RliB-CRISPRs in the absence of Cas
Our functional assay showed that the RliB-CRISPR in the L.monocytogenes EGD-e strain that completely lacks cas genes, although processed by PNPase, is not able to provide DNA interference activity against a plasmid carrying a matching protospacer. This lack of activity is probably due to the absence of trans encoded CRISPR-I system required for RliB-CRISPR DNA interference activity, as shown in L. monocytogenes Finland strain. However, the conservation of the RliB-CRISPRs in Listeria is independent on the presence of CRISPR-I, strongly suggesting that it is a functional element with an important function even in the absence of Cas Type-I, as sequences lacking selection pressure for their maintenance are quickly lost in bacterial genomes [40]. Interestingly, RliB-CRISPRs in average possess a smaller number of repeats and the variability of their spacers is lower compared to the spacer content of the CRISPR-I and CRISPR-II. Have they evolved a new function? It is to be kept in mind the remarkable finding that all RliB-CRISPRs accumulate as a 280 nt fragment, which might be the functional form. In support for a functional role of RliB, our recent RNA-seq analysis has shown that RliB-CRISPR is not only conserved but also expressed in the more distant L. innocua species that also lacks CRISPR-I [41].
Although RliB-CRISPRs share many similarities with cas-associated CRISPR-I system, the identified RliB-CRISPR protospacers are more often in the antisense orientation with respect to the corresponding spacer and in addition they are mostly located in the late phage genes encoding DNA packaging and envelope proteins. It is tempting to speculate that “the” RliB-CRISPRs cas-independent activity might be RNA interference. In this scenario RliB-CRISPR would not destroy the bacteriophage DNA but would rather control the bacteriophage late gene expression i.e., it would prevent the formation of viral particles and lysis of the bacterial cell. RliB-CRISPR interference could be also based on transcription-dependent DNA targeting, as recently described in Sulfolobus islandicus REY15A [42]. Alternatively, RliB-CRISPR might have evolved a broader function relevant for Listeria physiology that is not related to the immunity. Such examples have been described in Pseudomonas aeruginosa, where a CRISPR appear to be involved in lysogeny dependent biofilm formation [43], in myxobacteria where CRISPR has been implicated in swarming motility [44] and more recently in Francisella novicida where a tracrRNA was shown to regulate an endogenous transcript encoding a lipoprotein important for the bacterial infection [45].
The interaction between bacteriophages and bacteria is mostly seen as a parasitic interaction where the virus exploits the host resources for its own benefit. However, there are some viruses that have a beneficial effect on their host [46]. In case of pathogenic bacteria, bacteriophages often carry virulence factors required for successful infection [47]. More recently, a study by Rabinovich et al. (2012) showed that during Listeria intracellular infection, a temperate prophage is excised, which reconstitutes a function of the gene where the bacteriophage was integrated, and promotes bacterial escape from macrophage phagosomes. Remarkably, the excision event does not lead to propagation and release of the progeny virions neither to the subsequent lysis of the bacterial cell. Hence, the virion production is actively aborted [48]. This example highlights an important crosstalk between the phage and the pathogenic bacteria during the infection of the mammalian cell, and more importantly, it emphasizes the conditional advantage for a bacterium to maintain a bacteriophage and control its virulence. Our previous studies have shown that RliB expression is upregulated in the bacteria grown in human blood and in the intestine of gnotobiotic mice and is important for Listeria virulence [4]. Whether RliB-CRISPR expression and prophage excision followed by aborted virion production are linked processes, remains to be examined. Our study thus paves the way for new regulatory studies on the interactions between bacteriophages and bacteria during saprophytic life or during infection.
Materials and Methods
Strains and plasmids
Strains used in this study are L. monocytogenes EGD-e (BUG1600) and its isogenic mutants ΔrliB(BUG2621) and ΔpnpA(CMA751), L. monocytogenes EGD (BUG600) and its isogenic mutant, ΔpnpA-EGD (BUG3415) and ΔrliB-EGD (BUG3243), L. monocytogenes Finland 1998 (BUG3297, CLIP2012/00396, FE49845/IHD42536) and its isogenic mutants ΔpnpA-Fin (BUG3465) and ΔrliB-Fin (BUG3466). Mutants were obtained by deletion of the corresponding ORF or non-coding RNA by PCR-ligation and amplicon cloning in the suicide vector pMAD as previously described [49]. Overexpression of RliB was obtained by cloning the rliB gene into the pAD vector carrying Phyper constitute promoter [50], resulting in the strain Phyper-RliB (BUG2987). PNPase complementation was obtained by cloning the PnpA ORF into the pPl2 vector [51] resulting in the strain ΔpnpA+pnpA (CMA752).
Bacterial growth
Bacteria were grown overnight in Brain heart infusion (BHI) medium (Difco) at 37°C with shaking at 200 rpm. Cultures were subsequently diluted 1/500 into 100 ml BHI and grown at 37°C until mid-exponential phase (OD600 = 1.0). When required, erythromycin and chloramphenicol were used at 5 µg/ml and 20 µg/ml, respectively as final concentration. For RNA extraction, bacteria were pelleted, by centrifugation at 10,000 X G for five minutes, flash frozen in liquid nitrogen and stored at −80°C.
RNA isolation
Bacterial pellets were resuspended in 400 µl solution A (½ volume Glucose 20%+½ volume Tris 25 mM pH 7.6+EDTA 10 mM) to which an additional 60 µl of 0.5M EDTA was added. Bacteria were lysed in FastPrep homogenizer (Bio101) and RNA was subsequently extracted using TRI reagent (Invitrogen) as described previously [4]. RNA integrity was verified using the Experion Automated Electrophoresis system (Biorad).
Northern blot analysis
10–20 µg of total RNA was mixed with two volumes of Formaldehyde Loading Buffer (Ambion) followed by denaturation at 65°C for 15 min. Samples were separated by electrophoresis on 5% TBE-Urea polyacrylamide gels (Criterion-Biorad) at 100 V for 2 hours in 1× TBE running buffer at RT, followed by an overnight transfer at 4°C/100 mA to Nytran membranes (Sigma). Membranes were UV-crosslinked and probed with RNA probes or DNA oligo probes. Briefly, RNA probes were synthesized and α32P-UTP labelled using the Maxiscript T7RNA polymerase kit (Ambion) with PCR generated templates according to the manufacturer's instructions. Oligonucleotide DNA probes were 5′ labelled with γ32P-ATP using the T4 Polynucleotide Kinase according to the manufacturer's protocol (New England Biolabs). Membranes were prehybridized for 60 min in Ultrahyb buffer (Ambion) and hybridizations were performed overnight at 64°C for RNA probes and at 37°C for oligonucleotide probes. Following hybridization, membranes were washed twice for 5 min with 2× SSC, 0.1% SDS at room temperature. When hybridized with RNA probes, membranes were additionally washed twice for 15 min in 0.1× SSC, 0.1% SDS at 60°C. The size marker was a 50-bp ladder (Invitrogen), which was 5′ end labelled with γ32P-ATP.
Affinity chromatography using MS2-RliB or biotinylated RliB
We first have optimized an affinity purification assay using 3′-biotinylated RliB and streptavidin sepharose modified as described in Jestin et al. [52]. As a negative control, we used the regulatory RNAIII from S. aureus. Total cell extract prepared from 500 ml of culture of L. monocytogenes ΔrliB mutant strain was first incubated with streptavidin sepharose beads to remove proteins unspecifically bound to the beads. The beads were first incubated with the 3′ biotinylated RNA and the pre-cleaned crude extract was passed through the column and washed with the binding buffer containing 50 mM Hepes-NaOH pH 7,5, 5 mM MgCl2, 1 mM DTT, and 150 mM KCl. The elution of the proteins was done with the same buffer containing 6 M urea, 2 M thiourea and 30 mM d-biotin. The fractions were then analyzed by 4–15% gradient SDS-PAGE, and the proteins were identified by mass spectrometry.
A second approach was used to purify proteins associated with RliB carrying at its 5′ end two hairpin motifs recognized by the coat protein of the MS2 bacteriophage. As a control we used the untagged RliB. Both RNAs were transcribed in vitro using homemade T7 RNA polymerase. The experimental conditions were as previously described [53]. The MS2 coat protein fused to Maltose binding protein was expressed in E. coli and purified on an amylose column followed by a monoQ column. The MS2-MBP coat protein was first immobilized on the amylose resin, and the tagged-RNA was loaded on the column, which was washed with 2 ml of the Binding Buffer. Subsequently, the pre-cleared bacterial lysate was loaded onto the column, followed by three washes with 2 ml Binding Buffer, and the proteins were eluted with the binding buffer containing 10 mM maltose. The fractions were loaded on a SDS-PAGE and the proteins were identified by mass spectrometry.
RNA structure probing
Enzymatic hydrolysis was performed with 1 pmol of RliB in 10 µl of a buffer containing 50 mM NaOH-Hepes pH 7.5, 10 mM MgCl2, 150 mM KCl, in the presence of 1 µg carrier tRNA at 20°C for 5 min: RNase T2 (0.01 units), RNase V1 (0.5 units). Chemical modifications were performed on 2 pmol of RliB at 20°C in 20 µl of the same buffer containing 2 µg of carrier tRNA. Methylation of C(N3) and A(N1) positions was done with 1 µl DMS (diluted 1/8 and 1/16 in ethanol) for 2 min at 20°C. Modification of U(N3) and G(N1) was performed with 2,5 µl and 5 µl of CMCT (40 mg/ml) for 20 min at 20°C. The cleavage or modification sites of unlabeled RNAs were detected by primer extension. Details for hybridization conditions, primer extension, and analysis of the data have been previously described [54].
PNPase cleavage assays
PNPase cleavage assays were done using a 5′ end-labelled RliB or RNA37. Reaction was performed in 10 µl of TMK buffer containing 20 mM Tris-HCl pH 7.5, 10 mM magnesium-acetate, 100 mM KCl, 1 mM DTT at 37°C for 15 min in the presence of PNPase 200 nM in the presence of 1 µg of carrier tRNA. Competition experiments were carried out in the presence of 200 nM, 400 nM of cold RliB and its derivatives (RliB-3′ domain or RliB-5′ domain). Reactions were stopped by phenol extraction followed by RNA precipitation. The assays were loaded on a denaturing 12% polyacrylamide-urea gel electrophoresis. The PNPase cleavage sites were assigned by running in parallel RNase T1 ladder and an alkaline ladder on a denatured end-labelled RNA [54].
Gel retardation assays
To perform gel retardation assays, 5′ end-labelled transcript (20000 cpm, <1 nM) was incubated in the presence of increasing concentrations of PNPase (100 to 800 nM) in TMK buffer containing 20 mM Tris-HCl pH 7.5, 10 mM magnesium-acetate, 100 mM KCl at 37°C for 15 min. At the end of the binding reaction 6X loading dye (30% glycerol, 0.25% bromophenol blue and 0.25% xylene cyanol) was added to the samples and they were analyzed on a 6% polyacrylamide gel under non-denaturing conditions.
CRISPR activity assay
Plasmid design
The DNA fragment of the protospacer plasmid (P) was reconstituted using two complementary, 5′-phosphorylated oligonucleotides with the flanking BamH-I restriction sites P-A (GATCCGGTCGTCCTGCGATTTTTGTCAAAGGGACAGCGATGGGTTACAAGGAATACG) and P-B-(GATCCGTATTCCTTGTAACCCATCGCTGTCCCTTTGACAAAAATCGCAGGACGACCG) whereas the DNA fragment of the control plasmid was reconstituted using the oligonucleotides C-A (GATCCGGTCGTCCTTAAGTTTTATAACCATGGAAGTTGGAGCGAGCCCGGGAATACG) and C-B (GATCCGTATTCCCGGGCTCGCTCCAACTTCCATGGTTATAAAACTTAAGGACGACCG). 45 µl (c = 100 µM) of each oligonucleotide was added to 10 µl NEB buffer 3. The mixture was heated to 95°C and then slowly cooled down to room temperature. The reconstituted DNA fragments were cloned to Ppl2 integrative vector [51] using the BamH-I restriction site. Obtained clones were isolated and sequenced in order to verify the sequence integrity and the orientation of the cloned fragment. Plasmids with only one orientation of the fragments P and C were used for all the performed experiments.
Conjugation
Plasmids P and C were heat shock transformed to a donnor E. coli S17 to create E.coli-P and E.coli-C strains. Overnight cultures of E. coli and L. monocytogenes strains were diluted 1/20 in 5 ml LB media supplemented with 35 µg/ml chloramphenicol and 5 ml BHI media, respectively. The cultures were grown until mid exponential phase (OD = 1.0). 1 ml of each E. coli culture was washed 3X with 1 ml BHI media to remove the remaining antibiotic. The equal amount of E. coli-P and E. coli-C cultures were mixed. Subsequently, 200 µl of E. coli-P/C mixture was added to 200 µl L. monocytogenes culture and placed on the milipore filter (0.45 mm) on top of the agar plate and incubated overnight at 37°C. The next day, the cultures were removed from the filter, resuspended in 1 ml of BHI and plated on BHI plates supplemented with 7 µg/ml chloramphenicol, 50 µg/ml nalidixic acid and 100 µg/ml colistin. The plates were incubated 72 h at 37°C.
Quantitative PCR (Q-PCR)
Colonies were picked on a new BHI plate supplemented with 7 µg/ml chloramphenicol and incubated overnight at 37°C. The next day, each colony was resuspended in 50 µl water and heated to 95°C during 20 min. After centrifugation, supernatant was used as the template for the Q-PCR. Oligonucleotides were used to specifically amplify the fragments in the P plasmid (P-fw CAGGGCAGGGTCGTTAAATAG and P-rev AGCGATGGGTTACAAGGAATAC) and C plasmid (C-fw CGACTCACTATAGGGCGAATTG and C-rev CGCTCCAACTTCCATGGTTAT). As the control, we used oligonucleotides corresponding to the chloramphenicol gene present in both plasmids (Cm-fw CACTCATCGCAGTACTGTTGTA and Cm-rev CAAACGGCATGATGAACCTG). The reaction was based on Sso Fast Eva Green Supermix (Biorad) using the CFX384 Real-Time System (Biorad). The Ct values corresponding to P or C specific amplification were compared to deduce the plasmid identity of each individual colony. To evaluate the E.coli-P/C conjugation mix and the levels of the plasmids P and C at the beginning of the experiment, the plasmids from the E.coli-P/C mix were extracted using the MiniPrep Kit (QIAGEN) and their relative quantity was compared using the Q-PCR with the aforementioned oligonucleotides. To calculate the ratio (R = n(P)/n(C)), at least 32 colonies of each strain were screened. Each experiment was repeated multiple times on independent conjugation reactions. Results were statistically analyzed using t-tests.
Bioinformatic analysis
We analyzed 45 Listeria genomes taken from GenBank, available at the time of analysis. These include 28 complete genomes and 17 draft genomes with less than 800 contigs (Table S1). We also added the genome of L. monocytogenes EGD strain (unpublished data). We used GenBank annotations, excluded genes with stops in phase and with lengths not multiple of three. We re-annotated the prophages in the genomes using a methodology described previously [55].
Cas gene identification
The Hidden Markov models (HMMs) for the Cas protein families described previously [6] were obtained from the TIGRFAM database, version 6.0 (http://www.tigr.org/TIGRFAMs/). To identify cas genes, all coding sequences within each complete and draft genomes were searched with the 57 Cas HMMs profiles using hmmpfam [56] (e-value <0.001) (Table S1). To identify cas pseudogenes, all Cas proteins previously detected were searched in all the genomic sequences using tblastn (e-value <0.001).
Identification of the core genome
A preliminary set of orthologs was defined by identifying unique pairwise reciprocal best hits, with at least 60% similarity in amino acid sequence and less than 20% of difference in protein length. The list was then refined using information on the distribution of similarity of these putative orthologs and data on gene order conservation (as in [57]). The analysis of orthology was made for every pair of L. monocytogenes and L. innocua genomes. The core genome was defined as the intersection of pairwise lists of positional orthologs and was used to build the phylogenetic tree (see below). We used L. innocua as out-group to root the tree of L. monocytogenes.
Phylogenetic analysis
The reference phylogenetic tree for the core genome of the 43 complete/draft genomes was reconstructed from the concatenated alignments of 513 proteins of the core genome obtained with muscle v3.6 [58] then back-translated to DNA, as is standard usage. We used Tree-puzzle 5.2 [59] to compute the distance matrix between all genomes using maximum likelihood under the HKY+G(8)+I model. The tree of the core genome was built from the distance matrix using BioNJ [60]. We made 100 bootstrap experiments on the concatenated sequences to assess the robustness of the topology. The topology of this tree is congruent with previous
CRISPR array identification
CRISPR arrays were identified using CRT (CRISPR Recognition Tool) with default parameter values [61], in all the Listeria complete/draft genomes publicly available at the time of analysis. In the complete genomes, loci bordered by the same core genes were identified as RliB-CRISPR (bounded by the core genes: lmo0509-lmo0510), CRISPR-I (lmo0517-lmo0518) and CRISPR-II (lmo2951-lmo2596) (Figure 4). For each array, the repeats were extracted and were aligned using Muscle [58]. Then we used Cons (http://bioweb.pasteur.fr/docs/EMBOSS/cons.html) to obtain consensus sequences from these multiple sequence alignments of the three arrays. In all cases, the consensus sequence corresponds to the most frequent sequence within a particular array. We used the sequence of the repeats as patterns to identify additional, smaller and/or degenerate repeat clusters in all available complete and draft genomes. This analysis was done with Fuzznuc (http://bioweb2.pasteur.fr/docs/EMBOSS/fuzznuc.html).
Protospacer identification
Blastn was used for similarity searches between CRISPR spacer sequences and all the complete prokaryote genomes (3703 replicons), complete plasmid genomes (2930), and virus genomes (1153) available in GenBank. Matches showing an E value lower than 10−5 and less than 10% difference in sequence length between query and hit were retained. Matches to sequences found within CRISPR loci were ignored.
Supporting Information
Zdroje
1. CossartP (2011) Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA 108 : 19484–19491.
2. MellinJR, CossartP (2012) The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNABiol 9 : 372–378.
3. SestoN, WurtzelO, ArchambaudC, SorekR, CossartP (2013) The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol 11 : 75–82.
4. Toledo-AranaA, DussurgetO, NikitasG, SestoN, Guet-RevilletH, et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 : 950–956.
5. StorzG, VogelJ, WassarmanKM (2011) Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol Cell 43 : 880–891.
6. MakarovaKS, HaftDH, BarrangouR, BrounsSJJ, CharpentierE, et al. (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9 : 467–477.
7. SangalV, FineranPC, HoskissonPA (2013) Novel configurations of Type I and II CRISPR/Cas systems in Corynebacterium diphtheriae. Microbiology Epub.
8. BolotinA, QuinquisB, SorokinA, EhrlichSD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading, Engl) 151 : 2551–2561.
9. MojicaFJ, Diez-VillasenorC, Garcia-MartinezJ, SoriaE (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60 : 174–182.
10. PourcelC, SalvignolG, VergnaudG (2005) CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology (Reading, Engl) 151 : 653–663.
11. BarrangouR, FremauxC, DeveauH, RichardsM, BoyavalP, et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 : 1709–1712.
12. BrounsSJJ, JoreMM, LundgrenM, WestraER, SlijkhuisRJH, et al. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321 : 960–964.
13. MarraffiniLA, SontheimerEJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322 : 1843–1845.
14. BarrangouR (2013) CRISPR-Cas systems and RNA-guided interference. Wiley interdisciplinary reviews RNA 4 : 267–278.
15. WiedenheftB, SternbergSH, DoudnaJA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482 : 331–338.
16. KuenneC, VogetS, PischimarovJ, OehmS, GoesmannA, et al. (2010) Comparative Analysis of Plasmids in the Genus Listeria. PLoS ONE 5: e12511.
17. DorschtJ, KlumppJ, BielmannR, SchmelcherM, BornY, et al. (2009) Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J Bacteriol 191 : 7206–7215.
18. KuenneC, BillionA, MraheilMA, StrittmatterA, DanielR, et al. (2013) Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14 : 47.
19. GlaserP, FrangeulL, BuchrieserC, RusniokC, AmendA, et al. (2001) Comparative genomics of Listeria species. Science 294 : 849–852.
20. NelsonKE, FoutsDE, MongodinEF, RavelJ, DeBoyRT, et al. (2004) Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32 : 2386–2395.
21. HainT, GhaiR, BillionA, KuenneCT, SteinwegC, et al. (2012) Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genomics 13 : 144.
22. MandinP, RepoilaF, VergassolaM, GeissmannT, CossartP (2007) Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35 : 962–974.
23. CondonC (2007) Maturation and degradation of RNA in bacteria. Curr Opin Microbiol 10 : 271–278.
24. SchmukiMM, ErneD, LoessnerMJ, KlumppJ (2012) Bacteriophage p70: unique morphology and unrelatedness to other listeria bacteriophages. J Virol 86 : 13099–13102.
25. KuninV, SorekR, HugenholtzP (2007) Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8: R61.
26. ArraianoCM, AndradeJM, DominguesS, GuinoteIB, MaleckiM, et al. (2010) The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34 : 883–923.
27. BhayaD, DavisonM, BarrangouR (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45 : 273–297.
28. BikardD, MarraffiniLA (2012) Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr Opin Immunol 24 : 15–20.
29. AlmendrosC, GuzmanNM, Diez-VillasenorC, Garcia-MartinezJ, MojicaFJ (2012) Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli. PloS ONE 7: e50797.
30. WestraER, SwartsDC, StaalsRH, JoreMM, BrounsSJ, et al. (2012) The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet 46 : 311–339.
31. FineranPC, CharpentierE (2012) Memory of viral infections by CRISPR-Cas adaptive immune systems Acquisition of new information. Virology 434 : 202–209.
32. DeltchevaE, ChylinskiK, SharmaCM, GonzalesK, ChaoY, et al. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 : 602–607.
33. ZhangY, HeidrichN, AmpattuBJ, GundersonCW, SeifertHS, et al. (2013) Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Mol Cell 50 : 488–503.
34. FarrrG, OussenkoI, BechhhoferD (1999) Protection against 3′-to-5′ RNA Decay in Bacillus subtilis. J Bacteriol 23 : 1–9.
35. Lehnik-HabrinkM, LewisRJ, MäderU, StülkeJ (2012) RNA degradation in Bacillus subtilis: an interplay of essential endo - and exoribonucleases. Mol Microbiol 84 : 1005–1017.
36. CardenasPP, CarrascoB, SanchezH, DeikusG, BechhoferDH, et al. (2009) Bacillus subtilis polynucleotide phosphorylase 3′-to-5′ DNase activity is involved in DNA repair. Nucleic Acids Res 37 : 4157–4169.
37. CardenasPP, CarzanigaT, ZangrossiS, BrianiF, Garcia-TiradoE, et al. (2011) Polynucleotide phosphorylase exonuclease and polymerase activities on single-stranded DNA ends are modulated by RecN, SsbA and RecA proteins. Nucleic Acids Res 39 : 9250–9261.
38. AndradeJM, PobreV, MatosAM, ArraianoCM (2012) The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18 : 844–855.
39. De LayN, GottesmanS (2011) Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17 : 1172–1189.
40. KuoC-H, OchmanH (2010) The extinction dynamics of bacterial pseudogenes. PLoS Genet 6: e1001050.
41. WurtzelO, SestoN, MellinJR, KarunkerI, EdelheitS, et al. (2012) Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 8 : 583.
42. DengL, GarrettRA, ShahSA, PengX, SheQ (2013) A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87 : 1088–1099.
43. ZegansME, WagnerJC, CadyKC, MurphyDM, HammondJH, et al. (2009) Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191 : 210–219.
44. ViswanathanP, MurphyK, JulienB, GarzaAG, KroosL (2007) Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol 189 : 3738–3750.
45. SampsonTR, SarojSD, LlewellynAC, TzengYL, WeissDS (2013) A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497 : 254–257.
46. RoossinckMJ (2011) The good viruses: viral mutualistic symbioses. Nat Rev Microbiol 9 : 99–108.
47. BoydEF, BrussowH (2002) Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol 10 : 521–529.
48. RabinovichL, SigalN, BorovokI, Nir-PazR, HerskovitsAA (2012) Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150 : 792–802.
49. ArnaudM, ChastanetA, DebarbouilleM (2004) New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. App Environ Microbiol 70 : 6887–6891.
50. BalestrinoD, HamonMA, DortetL, NahoriMA, Pizarro-CerdaJ, et al. (2010) Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. App Environ Microbiol 76 : 3625–3636.
51. LauerP, ChowMY, LoessnerMJ, PortnoyDA, CalendarR (2002) Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriology 184 : 4177–4186.
52. JestinJL, DèmeE, JacquierA (1997) Identification of structural elements critical for inter-domain interactions in a group II self-splicing intron. EMBO J 16 : 2945–2954.
53. SaidN, RiederR, HurwitzR, DeckertJ, UrlaubH, et al. (2009) In vivo expression and purification of aptamer-tagged small RNA regulators. Nucleic Acids Res 37: e133.
54. ChevalierC, GeissmannT, HelferA-C, RombyP (2009) Probing mRNA structure and sRNA-mRNA interactions in bacteria using enzymes and lead(II). Methods Mol Biol 540 : 215–232.
55. BobayL-M, RochaEPC, TouchonM (2013) The Adaptation of Temperate Bacteriophages to Their Host Genomes. Mol Biol Evol 30 : 737–757.
56. EddySR (2011) Accelerated Profile HMM Searches. PLoS Comput Biol 7: e1002195.
57. TouchonM, HoedeC, TenaillonO, BarbeV, BaeriswylS, et al. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344.
58. EdgarRC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 : 1792–1797.
59. SchmidtHA, StrimmerK, VingronM, von HaeselerA (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18 : 502–504.
60. GascuelO (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14 : 685–695.
61. BlandC, RamseyTL, SabreeF, LoweM, BrownK, et al. (2007) CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8 : 209.
Štítky
Genetika Reprodukčná medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



