-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
Altered DNA methylation patterns in CD4+ T-cells indicate the importance of epigenetic mechanisms in inflammatory diseases. However, the identification of these alterations is complicated by the heterogeneity of most inflammatory diseases. Seasonal allergic rhinitis (SAR) is an optimal disease model for the study of DNA methylation because of its well-defined phenotype and etiology. We generated genome-wide DNA methylation (Npatients = 8, Ncontrols = 8) and gene expression (Npatients = 9, Ncontrols = 10) profiles of CD4+ T-cells from SAR patients and healthy controls using Illumina's HumanMethylation450 and HT-12 microarrays, respectively. DNA methylation profiles clearly and robustly distinguished SAR patients from controls, during and outside the pollen season. In agreement with previously published studies, gene expression profiles of the same samples failed to separate patients and controls. Separation by methylation (Npatients = 12, Ncontrols = 12), but not by gene expression (Npatients = 21, Ncontrols = 21) was also observed in an in vitro model system in which purified PBMCs from patients and healthy controls were challenged with allergen. We observed changes in the proportions of memory T-cell populations between patients (Npatients = 35) and controls (Ncontrols = 12), which could explain the observed difference in DNA methylation. Our data highlight the potential of epigenomics in the stratification of immune disease and represents the first successful molecular classification of SAR using CD4+ T cells.
Published in the journal: DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure. PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004059
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004059Summary
Altered DNA methylation patterns in CD4+ T-cells indicate the importance of epigenetic mechanisms in inflammatory diseases. However, the identification of these alterations is complicated by the heterogeneity of most inflammatory diseases. Seasonal allergic rhinitis (SAR) is an optimal disease model for the study of DNA methylation because of its well-defined phenotype and etiology. We generated genome-wide DNA methylation (Npatients = 8, Ncontrols = 8) and gene expression (Npatients = 9, Ncontrols = 10) profiles of CD4+ T-cells from SAR patients and healthy controls using Illumina's HumanMethylation450 and HT-12 microarrays, respectively. DNA methylation profiles clearly and robustly distinguished SAR patients from controls, during and outside the pollen season. In agreement with previously published studies, gene expression profiles of the same samples failed to separate patients and controls. Separation by methylation (Npatients = 12, Ncontrols = 12), but not by gene expression (Npatients = 21, Ncontrols = 21) was also observed in an in vitro model system in which purified PBMCs from patients and healthy controls were challenged with allergen. We observed changes in the proportions of memory T-cell populations between patients (Npatients = 35) and controls (Ncontrols = 12), which could explain the observed difference in DNA methylation. Our data highlight the potential of epigenomics in the stratification of immune disease and represents the first successful molecular classification of SAR using CD4+ T cells.
Introduction
The modest effects of genetic variants in inflammatory diseases indicate the importance of epigenetic mechanisms like DNA methylation to disease pathology. However, studies of inflammatory diseases have shown conflicting results. In monozygotic twins discordant for multiple sclerosis (MS), no significant differences in DNA methylation profile were found [1]. A more recent study of monozygotic twins discordant for psoriasis identified widespread differences between siblings [2]. Other studies of autoimmune diseases have reported varying findings [3]. Discordant monozygotic twin studies benefit from a constant genetic background on which to identify disease-associated epigenetic changes. However, intrinsically, such studies tend to involve small samples sizes, and may thus lack the power to detect small, rare, disease-associated changes in DNA methylation. The variation between studies may be due to disease heterogeneity, variations in disease course and the confounding effects of treatment, and as the disease causing agent is unknown, it is difficult to experimentally model disease pathogenesis.
By contrast, seasonal allergic rhinitis (SAR) occurs at defined time points each year and the disease causing agent, pollen, is known. These unique features of SAR permit analysis of CD4+ T-cells from SAR patients during and after the pollen season in vivo, and by allergen-challenge in vitro [4]. An epigenetic component in SAR is supported by its increasing prevalence in the developing world, failure of genome-wide association studies to identify a consistent genetic component to the disease, and frequent discordance for SAR between monozygotic twins [5], [6]. DNA methylation changes at numerous loci are required for appropriate differentiation of naïve CD4+ T-cells into CD4+ T effector cell subtypes [7].
We generated genome-wide expression (Npatients = 9, Ncontrols = 10) and DNA methylation (Npatients = 8, Ncontrols = 8) profiles for CD4+ T-cells from untreated SAR patients and healthy controls, both during and outside the pollen season. Consistent with previous studies, we found that CD4+ T-cell gene expression profiles were poor classifiers of SAR. However, we observed clear and robust separation of patients and controls by DNA methylation signature, both during and outside the pollen season. Separation by methylation (Npatients = 12, Ncontrols = 12), but not by gene expression (Npatients = 21, Ncontrols = 21) was also observed in an in vitro model system in which purified PBMCs were challenged with allergen in culture. Moreover, we found that these methylation profiles were significantly associated with disease severity in patients during season, and may be due to differing proportions of central memory CD4+ T-cells (Npatients = 35, Ncontrols = 12). This, to our knowledge, is the first successful molecular classification of SAR using CD4+ T-cells, and highlights the potential of genome-wide epigenetic technologies in the stratification of immune disease.
Results and Discussion
The DNA methylation profile of CD4+ T-cells separates allergic patients from healthy controls following allergen-challenge in vitro
Seasonal allergic rhinitis (SAR) is a powerful disease model because, (1) SAR's clear clinical manifestations make it easy to assess disease severity, (2) the affected cell type, CD4+ T-cells, can be obtained from patients when they are symptom-free (outside the pollen season) and compared to the same cell type in the same individual when symptomatic (during the pollen season). We aimed to test the ability of CD4+ T-cell DNA methylation to separate SAR patients from healthy controls. The cohorts used in the study are outlined in Supplemental Figure S1. In a previous study, we obtained PBMCs from adult SAR patients (N = 21; age = 25.4 years±7.8 SD) and healthy controls (N = 21; age = 25.7 years±10.1 SD) outside the pollen season, and challenged these cells with either grass pollen extract or diluent (PBS) (Figure 1A). Seven days post-challenge, total CD4+ T-cells were isolated by magnetic-activated cell sorting (MACS, positive-selection), and the mRNA expression profile determined by gene expression microarray (GEO: GSE50223). Consistent with several previous studies, we were unable to separate patients and controls after challenge with allergen by CD4+ T-cell mRNA expression profile [6], [8], [9] (Figure 1B).
Fig. 1. The DNA methylation profile of allergen-challenged CD4+ T-cells separates SAR patients from healthy controls. 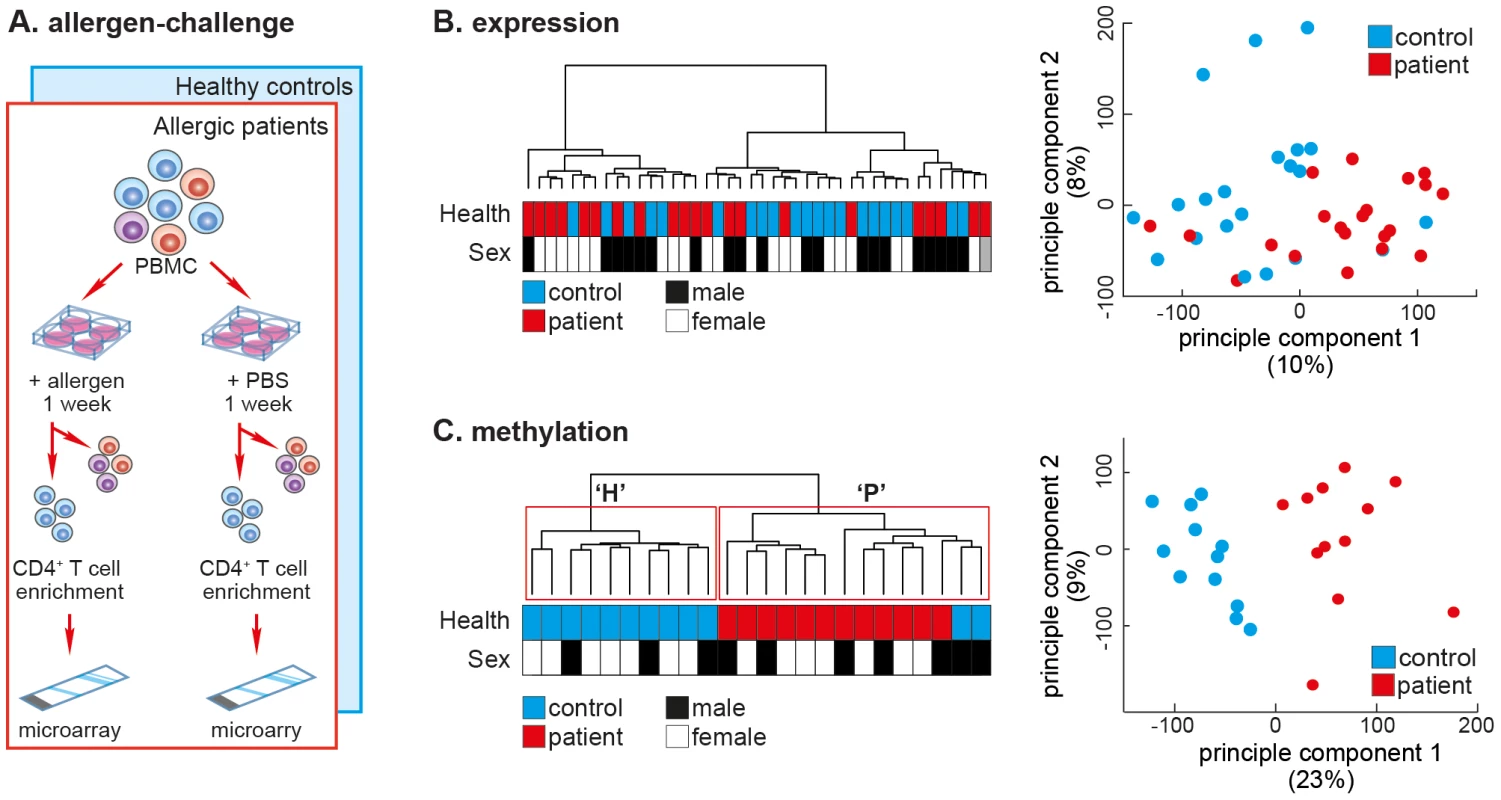
(A) Allergen-challenge assay. Peripheral blood mononuclear cells were isolated from healthy individuals and SAR patients and challenged in vitro with either allergen (pollen) or diluent (PBS). One-week post-challenge total CD4+ T-cells were isolated by MACS negative cell sorting. Genomic DNA and total RNA were isolated from the purified cells and cDNA or bisulfite-converted DNA was applied to gene expression or DNA methylation arrays, respectively. (B) Unsupervised hierarchical clustering of gene expression data of CD4+ T-cells isolated after allergen-challenge of PBMCs from SAR patients (N = 21) and healthy control subjects (N = 21) collected outside the pollen season (left panel).. Sample annotation is illustrated by colored boxes below the dendrogram. Principle components analysis of the same gene expression data fails to cluster data by disease status (right panel). (C) Unsupervised hierarchical clustering of quantitative genome-wide DNA methylation data of CD4+ T-cell DNA isolated after allergen-challenge of PBMCs from SAR patients (N = 12) and healthy control subjects (N = 12) collected outside the pollen season (left panel). Repeated bootstrap resampling of the data to calculate P-values for each cluster revealed that the two main clusters (H & P) were significantly supported by the data (P<0.05). Principle components analysis of the same DNA methylation data also revealed clear separation by disease state along the main principle component (right panel). Here, PBMCs from a new cohort of SAR patients (N = 12; age = 28.3 years±12.1 SD) and healthy controls (N = 12; age = 27.3 years±10.7 SD) were allergen-challenged, after which purified CD4+ T-cell DNA was analysed using Illumina HumanMethylation27 DNA methylation microarrays. Unsupervised hierarchical clustering of samples by genome-wide DNA methylation profiles resulted in two groups, ‘H’ (healthy controls) and ‘P’ (patients), clearly separating samples by disease-state (Figure 1C, left panel). Consensus clustering, whereby the data are repeatedly re-sampled and re-clustered, found that clusters ‘H’ and ‘P’ were reproducibly and stably identified (P<0.05). We confirmed this separation in the data using principal component analysis (PCA), which also revealed a clear separation between allergen-challenged CD4+ T-cells from patients versus controls (Figure 1C, right panel). A leave-one out (LOO) cross-validation found that methylation data accurately classified all samples as patient or healthy control (χ2;P<0.0001). DNA methylation and gene expression signatures did not separate patients and healthy controls after diluent challenge (Figure S2).
The DNA methylation profile of in vivo CD4+ T-cells separates allergic patients from healthy controls both during and outside the pollen season
Though striking, the results obtained from in vitro allergen-challenge of PBMCs may be confounded by cell culture effects. To verify the observations made after in vitro allergen-challenge, the mRNA expression and DNA methylation profiles of in vivo CD4+ T-cells were determined in a new cohort of SAR patients and healthy controls (GEO: GSE50387). In this experiment we used Illumina HumanMethylation450k methylation arrays, which quantitatively assess 450,000 CpG sites across the genome, with over half the probes targeting CpGs outside gene promoters and a quarter of all probes targeting CpGs in non-genic regions. CD4+ T-cells were purified from fresh blood collected from SAR patients and healthy controls by negative magnetic cell sorting both during and outside the pollen season in the same calendar year. Symptom scores for patients were recorded at the time of collection (Table S1). DNA and total RNA were harvested simultaneously from each sample. Subsequently, cDNA and bisulfite converted DNA was applied to Illumina HT12 expression microarrays (Npatients = 9, Ncontrols = 10; both during and outside the pollen season) and HumanMethylation450k microarrays microarrays (Npatients = 8, Ncontrols = 8; both during and outside the pollen season), respectively.
Unsupervised hierarchical clustering of samples by mRNA expression profile did not result in separation of samples by disease state either during or outside the pollen season (Figure 2A, left panel). No effect of array batch or sex was evident in the observed clusters (Figure 2A, left panel). These findings are in line with those reported here (Figure 1B) and in numerous previous studies of CD4+ T-cells in allergy and other autoimmune diseases [6], [8], [9]. PCA of the gene expression data also failed to distinguish between SAR patients and healthy controls (Figure 2A, right panel). Differentially expressed genes were identified as those showing differential expression between patients and controls (Mann-Whitney U-test; P<0.05, unadjusted) both during and outside the pollen season (357 genes identified). The relative changes observed across all genes were small (mean absolute fold change = 0.39, SEM ± 0.01). Gene Ontology (GO) term analysis failed to identify any significantly enriched gene annotation clusters after correction for multiple correction. However, four of the top 10 annotations related to lymphocyte activation (Figure S3A). All four clusters consisted of mixtures of the same seven genes, namely; HSPD1, TGFBR2, CD134 (TNFRSF4, OX40), CD45 (PTPRC), SPN (CD43), IL2RA, IL13RA1. This group of genes is highly relevant; CD43−/− mice show a pronounced Th2 phenotype and exhibit increased in inflammation in two allergic mouse models [10] and association studies have identified both IL2RA and TGFBR2 as susceptibility loci for allergy and asthma [11]. CD143 is a regulator of CD4+ T-cell memory and CD45 isoforms are key markers of memory CD4+ T-cells, though it is not possible to determine the exact isoform mis-expressed due to the positioning of the gene expression probe in the 5′ UTR of CD45 [12]. To determine the relationship between the observed changes in gene expression and DNA methylation we determined the average promoter methylation for all genes represented on the 450K methylation array. Changes in gene expression and associated promoter DNA methylation between patients and controls were not significantly negatively correlated (Pearson's r = 0.14, P = 0.07), as would be expected if DNA methylation were acting to silence transcription (Figure S3B).Thus, although it was not possible to separate patients from controls by gene expression profile, a small set-of disease-relevant miss-expressed genes were identified.
Fig. 2. In vivo CD4+ T-cell methylation separates patients from controls during and outside the pollen season. 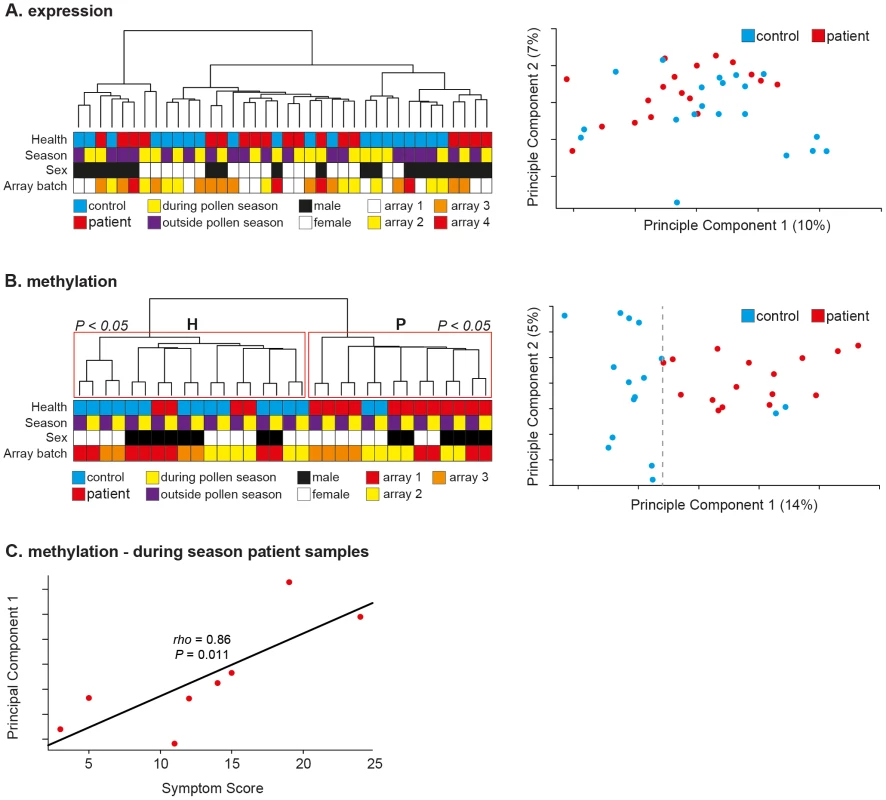
(A) Unsupervised hierarchical clustering of gene expression data of CD4+ T-cells from SAR patients (N = 9) and healthy control subjects (N = 10) both during and outside the pollen season (left panel). Sample annotation is illustrated by colored boxes below the dendrogram. Principle components analysis of the same gene expression data failed to cluster samples by disease status (right panel). Array batch indicates which samples were analyzed on the same microarray slide. (B) Unsupervised hierarchical clustering of quantitative genome-wide DNA methylation data of CD4+ T-cells from SAR patients (N = 8) and healthy control subjects (N = 8) both during and outside the pollen season (left panel). Repeated bootstrap resampling of the data to calculate p-values for each cluster revealed that the two main clusters (H & P) were significantly supported by the data (P<0.05). Principle components analysis of the same DNA methylation data also revealed clear separation by disease state along the main principle component (right panel). Array batch indicates which samples were analyzed on the same microarray slide. (C) Plot of patient symptom score during season with PCA1 value revealed a highly significant correlation (Spearman's rho = 0.86, P = 0.01). In contrast, unsupervised hierarchical clustering of all probes by DNA methylation profile resulted in clear separation of the samples by disease state, regardless of season (during or outside pollen season); seven of eight healthy controls grouped in cluster ‘H’, and six of eight patient samples grouped in cluster ‘P’ (Figure 2B, left panel). Consensus clustering identified clusters ‘H’ and ‘P’ with high statistical confidence (Multiscale Bootstrap Resampling; P<0.05). Interestingly, the paired measurements (during and outside season) of each sample also clustered neatly together; again implicating disease status as the major modifier of DNA methylation profile between patients and controls samples, while also highlighting the reproducibility and accuracy of the data (paired samples were collected and processed at least five months apart) (Figure 2, left panel). PCA resulted in even clearer separation of samples along the main principle component, with only one healthy sample clustering among the patient samples (Figure 2, right panel). LOO analysis correctly classified samples as patient or healthy control in all samples collected outside season (χ2;P<0.0001), and all but two healthy samples collected during season (χ2;P = 0.01). PCA1 explained approximately 14% of the variation in the data and 3-times the variation explained by PCA2 (5%). Although our findings indicated that disease status was the major mediator of DNA methylation differences between patients and healthy controls an effect of pollen season was also evident. With the exception of one outlier, healthy samples clustered tightly together along PCA1, whereas patient samples showed greater spread along PCA1 (Figure 2B, right panel). Given this observation, we tested if the position along PCA1 of patient samples was associated with patient symptom score during season. Symptom scores for each patient are listed in Supplemental Table S1. Significantly, we found that symptom score explained 74% of the variation in patient sample variation along PCA1, a strong and highly significant correlation (Spearman's rho = 0.86, P = 0.011) (Figure 2C). To our knowledge, such a strong association between individual or genome-scale markers and symptoms has not been previously been described in allergy, nor in other inflammatory diseases. However, given the small sample size (Npatients = 8) of the study presented here, the use of DNA methylation as a marker of disease severity needs to be tested in a much larger cohort. The association with disease severity may also explain the difficulty in identifying an epigenetic component to other immune-related diseases in which the disease course is more variable and complex to assess. However, as patient symptoms can vary dramatically during the pollen season it is important that the observed correlation between DNA methylation and symptom severity is tested at several time-points during a pollen season to validate the robustness of the preliminary observation reported here. If our findings are applicable to other inflammatory diseases, an important implication is that DNA methylation may help to stratify such diseases.
The observed differences in DNA methylation between patients and controls were small (mean absolute change = 1.2%±2.3 SD), bi-directional, and genome-wide, 12,000 probes (3.5% of all probes) were found to have changed significantly (Mann-Whitney U-test; P<0.01; unadjusted) (Figure S4A). Indeed, the 1,000 most significantly altered probes changed by only ±10% (Figure S4B). Given the small size of the observed methylation differences and the known technical variation associated with 450K methylation arrays [13], we selected five CpG loci for validation by pyrosequencing in 4 SAR patients and 4 healthy controls. We selected three CpGs that showed significant methylation differences between patients and healthy controls (among top 50 altered probes) which were also located in annotated gene promoters (PIEZO1 promoter CpG, RPP21 promoter CpG, HLA-DMA promoter CpG) and 2 control CpG loci (unmethylated CpG; GAPDH promoter & methylated CpG; CD74 promoter). Pyrosequencing primers are listed in Table S2. Array methylation was highly significantly correlated with pyrosequencing methylation for all CpG loci (Spearman's rho = 0.98, P<10−6) (Figure S4C), and the absolute difference in methylation measurements between the array and pyrosequencing across all CpGs was small (median difference = 2.41%±3.2 MAD; mean difference = 4.1%±4 SD). The methylated (Figure S4D) and unmethylated (Figure S4E) control CpGs were also validated by pyrosequencing, although agreement between array and pyrosequencing was slightly better for the unmethylated CpG site. Critically, pyrosequencing confirmed the direction and scale of DNA methylation change observed between patients and controls by 450K methylation array at the three disease-associated test loci (Figure S4F–S4H).
Our findings agree with a recent quantitative study of genome-wide methylation in CD4+ T-cells from monozygotic twins discordant for the autoimmune disease, psoriasis, in which affected and unaffected siblings were distinguished by numerous, small, bi-directional changes in DNA methylation, with no one CpG exhibiting a significant change in DNA methylation level [2]. Moreover, and very recent study of DNA methylation in CD4+ T-cells from patients with the autoimmune disease Systemic Lupus Erythematosus (SLE) also reported widespread small changes in DNA methylation [14]. Interestingly, we found that the genic location of significantly altered probes was enriched for gene-bodies and non-genic regions (χ2; P<0.0001) (Figure S5A). This highlights the importance of using unbiased genomic technologies; as assays targeted towards detection of large changes in DNA methylation in promoter regions may miss subtle but informative changes in other genomic compartments. To further dissect the genomic compartmentalization of the observed changes in DNA methylation we focused on regulatory elements whose function is known to be modified by DNA methylation, namely promoters, DNaseI hypersensitive sites (DHS) and enhancer elements. Interestingly, differentially methylated probes appeared enriched in annotated enhancers compared to those located in DHSs and promoters (χ2; P<10−5) (Figure S5B). This enrichment was observed both during and outside the pollen season (Figure S5B). Enhancer probes differentially methylated during and outside the pollen season showed a significant overlap (Fisher's exact test; P<0.001) (Figure S5C). Significantly, those enhancer probes differentially methylated both during and outside the pollen season (N = 960) showed a clear tendency towards loss of methylation in patients in contrast to the bi-directional changes observed for all differentially methylated probes (Figure S5D). As the activity of many enhancer elements and consequently their associated genes, is affected by DNA methylation, this finding may reflect the epigenetic re-modeling of enhancer elements in CD4+ T-cells. However, as most enhancers are only active in a small number of tissues we cross-referenced the observed differentially methylated enhancers with a recently published experimentally determined set of Th1-, Th2 - and Th0-specific enhancers [15], [16]. Only 1 of the differentially methylated enhancer probes (N = 960) mapped to the Th1/Th2/Th0 enhancers. Thus, is seems unlikely that the observed changes in enhancer methylation have function gene expression consequences in Th2 cells, the key pathogenic cell-type in allergy. However, as Th2 cells constitute <3% of the total CD4+ T-cell population, we cannot exclude large functionally relevant changes in DNA methylation at enhancers in other CD4+ T-cell subsets.
Differences in DNA methylation are often assumed to reflect changes in DNA methylation in a given type of cell; however such differences can also result from changes in the proportions of cell types in samples. Indeed, a recent study reported that a significant proportion of the DNA methylation differences observed between breast tumours (N = 248) reflected the number of infiltrating T-cells, and was significantly associated with prognosis [17]. Thus, the observed, small, genome-wide and bi-directional changes in DNA methylation between SAR patients and controls may reflect changes in the proportions of CD4+ T-cell sub-populations between patients and controls. Given that DNA methylation profile clearly separates samples by disease-state both during and outside the pollen season we hypothesized that the observed differences may have been due to differences in memory CD4+ T-cell subsets. Indeed, we have previously observed a reduction in CD4+ central memory T-cells (TCM) in a small cohort of allergic patients [18]. Here we extended this study, using FACS analysis to quantify CD4+ T-cell subsets in an additional 26 subjects (Npatients = 35, Ncontrols = 12) (Figure S6). SAR patients were significantly depleted for CD4+ central memory T-cells (TCM) (11% of total CD4+ T-cells) relative to healthy individuals (18.7% of total CD4+ T cells), a reduction of 41% in the TCM population in patients relative to controls (Mann-Whitney; P = 0.001) (Figure 3). The values for TCM cells in healthy individuals reported here are similar to those reported in independent studies [19], affirming the accuracy of our methodology. Altered TCM proportions are being implicated in a growing number of immune-related diseases [19]–[21]. This finding was also consistent with enrichment for the Gene Ontology (GO) term, ‘negative regulation of memory T-cell differentiation’, found when genes containing the 100 most significantly changed probes were analyzed (Figure S5E, right panel). However, given the enrichment for altered probes in non-genic and non-promoter regions of genes, the results of such GO term analyses must be viewed with caution. As the quantification of CD4+ T-cell subsets and DNA methylation profiling were performed on different cohorts of patients and controls, conclusions drawn from these independent observations must be cautious. Determination of DNA methylation profile, CD4+ T-cell subset structure and symptom severity in a large cohort of patients and controls at multiple time-points during and outside the pollen season would allow a more robust analysis of the inter-dependence of these variables. Although our results suggest that differences in T-cell sub-populations may contribute to the observed differences in DNA methylation between SAR patients and controls, they do not exclude a direct role for DNA methylation changes within CD4+ T-cell subsets. Analysis of gene expression and DNA methylation in each memory CD4+ T-cell subset in a large cohort of patients and healthy controls is required to directly test the association between CD4+ T-cell subset changes and observed changes in total CD4+ T-cell methylation. It is technically challenging to obtain sufficient numbers of each subtype of CD4+ T-cell from standard (40 mL) blood samples. An alternate future strategy may be to determine reference genomes for each CD4+ T-cell subtype and use these subtype specific methylomes to estimate sub-type proportions from a total CD4+ T-cell methylome [22]. This approach has recently been successfully applied to the study of rheumatoid arthritis [23].
Fig. 3. CD4+ T-cell subsets differ between healthy controls and patients with seasonal allergic rhinitis. 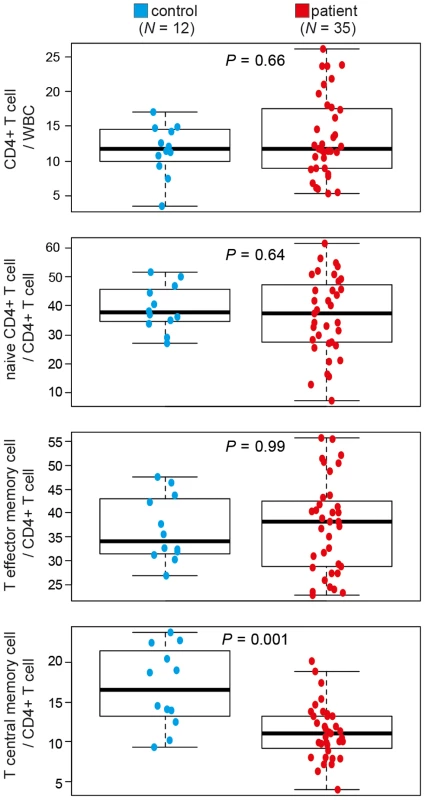
Total CD4+ T-cells, naïve CD4+ T-cells, CD4+ T central memory cells and T effector memory cell numbers in patients with seasonal allergic rhinitis (SAR) (N = 35) and healthy controls (N = 12) were determined by flow cytometry. WBC, white blood cells. Differences between samples were determined by Mann-Whitney U-test. The ability to separate patients and healthy controls by quantitative DNA methylation array, but not by gene expression array is interesting. Assuming little or no change in DNA methylation or gene expression profiles within CD4+ T-cell subtypes, small changes in the proportions of subtypes should result in very subtle changes in both the DNA methylation and gene expression profiles of total CD4+ T-cells. These subtle changes can be detected by the DNA arrays, given 1) their sensitivity – they are quantitative and accurate, and 2) their power – they assess 450,000 CpG sites. Gene expression arrays are not quantitative and have a restricted dynamic range of values which is often determined by probe design, not gene transcript levels. Moreover, whereas gene expression arrays typically have probes targeting ∼40,000 transcripts, only a small proportion of these are actually expressed (informative) in any given cell type, further reducing the power of the assay. Moreover, DNA methylation profiles are very stable in normal somatic cells, showing less inter-individual variation than RNA levels. Thus, whereas detecting small changes (<5%) in CD4+ T-cell substructure by gene expression microarray is theoretically possible, it would require a very large sample size; much greater than that used in this study (Npatients = 9, Ncontrols = 10; both during and outside the pollen season). Our results support the use of both DNA methylation arrays and gene expression arrays simultaneously, particularly, where cell-type composition may contribute to the molecular signature of the disease.
Epigenetic regulation plays a key role in Th differentiation. Pronounced changes in DNA methylation patterns are observed at several key loci during helper T cell differentiation [24]. Up-regulation of Il4, IL5, and IL13 gene expression in Th2 cells is accompanied by a pronounced loss of DNA methylation and gain of permissive histone marks across the locus [25], [26]. Similarly, the promoter of the Th1 gene, Ifng, is unmethylated in Th1 cells, but hypermethylated in Th2 cells, reinforcing helper T-cell identity. DNA demethylation also occurs at regulatory regions of the FOXP3 gene in Treg cells [27], and at the IL17A promoter in Th17 cells [28]. Allergy involves an inappropriate Th2 response to a benign allergen such as pollen [29], and several observations point to a key role for epigenetics in the pathogenesis of SAR. Murine studies have established that a diet rich in methyl donors, such as folic acid enhances allergic airway disease in progeny [30], and knock-out studies of the DNA methyltransferase, Dnmt3a, resulted in dysregulation of important Th2 cytokines, including Il13, and increased inflammation in a mouse model of asthma [31]. In humans, large meta-analysis of GWAS identified few loci associated with SAR [32], and those loci identified did not contain genes encoding Th2 genes or other genes of known relevance for SAR. A recent study identified stable and functional DNA demethylation at the key regulatory gene FOXP3 in patients cured by specific immunotherapy (SIT) [33], directly linking disease reversal with DNA demethylation. Recent studies have reported differences in DNA methylation in airway epithelial cells between asthmatic children and healthy controls [34] and reported that methylation levels at the key cytokine gene, IL2, in cord blood was associated with asthma exacerbations in childhood [35]. However, systematic clinical studies of the genome-wide distribution and role of DNA modifications in Th differentiation in SAR are lacking.
Analysis of the transcriptome of CD4+ T-cell subsets have failed to identify clear and reproducible differences between patients and healthy controls in several immune-diseases [6], [8], [9]. The circulating total CD4+ T-cell population is complex, comprised of several subsets that vary markedly between individuals. This complexity renders identification of subtle allergy-specific transcriptional signals challenging with current approaches. Unlike gene expression microarrays which typically assay 20,000–40,000 transcripts and have a small dynamic range, the DNA methylation microarrays employed here assay ∼450,000 individual CpG sites with quantitative accuracy. We suggest that quantitative DNA methylation microarrays can act as a proxy measure of the cell-population structure of samples, and as such, may be powerful analytical tools for diseases in which the proportions of different cell sub-types is likely to be of pathogenic significance such as immune-diseases and cancer [2], [17], [19]. Indeed, a very recent publication of genome-wide methylation in CD4+ T-cells from patients with Systemic Lupus Erythematosus (SLE), identified similar, small widespread changes in DNA methylation and associated these epigenetic changes with changes to CD4+ T-cell populations in SLE patients [14]. This exciting finding is completely consistent with the preliminary results reported here and suggests that alterations in CD4+ T-cell populations may be a general feature of many immune diseases. As CD4+ T-cell population structure and DNA methylation profiles reported here were determined in different cohorts, simultaneous analysis of the DNA methylation and gene expression profiles of CD4+ T-cells in the same cohort of patients and controls is required to directly determine the contribution of changes in subtype structure between patients and controls to the observed difference in total CD4+ T-cell methylation. Our findings highlight the potential to stratify immune diseases with DNA methylation.
Materials and Methods
Ethics statement
In vitro and in vivo array studies (cohorts 1–3, Figure S1) were approved by the ethics board of University of Gothenburg and all participants provided written consent for participation. The quantification of CD4+ T-cell subtypes study (cohort 4, Figure S1), was approved by the ethics board of Linkoping University and all participants provided written consent for participation.
Study subjects
We recruited patients with SAR and matched healthy controls of Swedish origin at The Queen Silvia Children's Hospital, Gothenburg (cohorts 1–3, Figure S1). We recruited patients with SAR and matched healthy controls of Swedish origin at Linkoping University Hospital, Linkoping (cohort 4, Figure S1). SAR was defined by a positive seasonal history and a positive skin prick test or by a positive ImmunoCap Rapid (Phadia, Uppsala, Sweden) to birch and/or grass pollen. Patients with perennial symptoms or asthma were not included. The healthy subjects did not have any history for SAR and had negative ImmunoCap Rapid tests. Supplemental Figure S1 provides an overview of the experiments performed on each cohort used in the research presented here. Severity of patient symptoms (itchiness of the eyes, block sinus, running nose) during season were self-assessed using a visual analogue scale (1–10). The score for each symptom was summed to give a single symptom severity score for each patient (Supplemental Table S1).
A detailed description of all methods employed in this study can be found in Supplemental Text S1.
Supporting Information
Zdroje
1. BaranziniSE, MudgeJ, van VelkinburghJC, KhankhanianP, KhrebtukovaI, et al. (2010) Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature 464 : 1351–1356.
2. GervinK, VigelandMD, MattingsdalM, HammeroM, NygardH, et al. (2012) DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: identification of epigenetically dysregulated genes. PLoS Genet 8: e1002454.
3. GreerJM, McCombePA (2012) The role of epigenetic mechanisms and processes in autoimmune disorders. Biologics 6 : 307–327.
4. WangH, BarrenasF, BruhnS, MobiniR, BensonM (2009) Increased IFN-gamma activity in seasonal allergic rhinitis is decreased by corticosteroid treatment. J Allergy Clin Immunol 124 : 1360–1362.
5. Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. The International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee. Lancet 351 : 1225–1232.
6. SjogrenAK, BarrenasF, MuraroA, GustafssonM, SaetromP, et al. (2012) Monozygotic twins discordant for intermittent allergic rhinitis differ in mRNA and protein levels. Allergy 67 : 831–833.
7. KannoY, VahediG, HiraharaK, SingletonK, O'SheaJJ (2012) Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol 30 : 707–731.
8. LeeJC, LyonsPA, McKinneyEF, SowerbyJM, CarrEJ, et al. (2011) Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 121 : 4170–4179.
9. McKinneyEF, LyonsPA, CarrEJ, HollisJL, JayneDR, et al. (2010) A CD8+ T cell transcription signature predicts prognosis in autoimmune disease. Nat Med 16 : 586–591, 581p following 591.
10. CannonJL, CollinsA, ModyPD, BalachandranD, HenriksenKJ, et al. (2008) CD43 regulates Th2 differentiation and inflammation. J Immunol 180 : 7385–7393.
11. BottemaRW, KerkhofM, ReijmerinkNE, ThijsC, SmitHA, et al. (2010) Gene-gene interaction in regulatory T-cell function in atopy and asthma development in childhood. J Allergy Clin Immunol 126 : 338–346, 346 e331–310.
12. Salek-ArdakaniS, CroftM (2006) Regulation of CD4 T cell memory by OX40 (CD134). Vaccine 24 : 872–883.
13. BibikovaM, BarnesB, TsanC, HoV, KlotzleB, et al. (2011) High density DNA methylation array with single CpG site resolution. Genomics 98 : 288–295.
14. AbsherDM, LiX, WaiteLL, GibsonA, RobertsK, et al. (2013) Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations. PLoS Genet 9: e1003678.
15. HawkinsRD, LarjoA, TripathiSK, WagnerU, LuuY, et al. (2013) Global chromatin state analysis reveals lineage-specific enhancers during the initiation of human T helper 1 and T helper 2 cell polarization. Immunity 38 : 1271–1284.
16. AranD, HellmanA (2013) DNA methylation of transcriptional enhancers and cancer predisposition. Cell 154 : 11–13.
17. DedeurwaerderS, DesmedtC, CalonneE, SinghalSK, Haibe-KainsB, et al. (2011) DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med 3 : 726–741.
18. ZhangH, CardellLO, BjorkanderJ, BensonM, WangH (2013) Comprehensive profiling of peripheral immune cells and subsets in patients with intermittent allergic rhinitis compared to healthy controls and after treatment with glucocorticoids. Inflammation 36 : 821–829.
19. NauschN, BourkeCD, ApplebyLJ, RujeniN, LantzO, et al. (2012) Proportions of CD4+ memory T cells are altered in individuals chronically infected with Schistosoma haematobium. Sci Rep 2 : 472.
20. SiegelAM, HeimallJ, FreemanAF, HsuAP, BrittainE, et al. (2011) A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 35 : 806–818.
21. PraksovaP, StouracP, BednarikJ, VlckovaE, MikulkovaZ, et al. (2012) Immunoregulatory T cells in multiple sclerosis and the effect of interferon beta and glatiramer acetate treatment on T cell subpopulations. J Neurol Sci 319 : 18–23.
22. HousemanEA, AccomandoWP, KoestlerDC, ChristensenBC, MarsitCJ, et al. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13 : 86.
23. LiuY, AryeeMJ, PadyukovL, FallinMD, HesselbergE, et al. (2013) Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 31 : 142–147.
24. NorthML, EllisAK (2011) The role of epigenetics in the developmental origins of allergic disease. Ann Allergy Asthma Immunol 106 : 355–361; quiz 362.
25. LeeDU, AgarwalS, RaoA (2002) Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 16 : 649–660.
26. LeeGR, KimST, SpilianakisCG, FieldsPE, FlavellRA (2006) T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 24 : 369–379.
27. HuehnJ, PolanskyJK, HamannA (2009) Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol 9 : 83–89.
28. JansonPC, LintonLB, BergmanEA, MaritsP, EberhardsonM, et al. (2011) Profiling of CD4+ T cells with epigenetic immune lineage analysis. J Immunol 186 : 92–102.
29. BarnesPJ (2011) Pathophysiology of allergic inflammation. Immunol Rev 242 : 31–50.
30. HollingsworthJW, MaruokaS, BoonK, GarantziotisS, LiZ, et al. (2008) In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest 118 : 3462–3469.
31. YuQ, ZhouB, ZhangY, NguyenET, DuJ, et al. (2012) DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proc Natl Acad Sci U S A 109 : 541–546.
32. RamasamyA, CurjuricI, CoinLJ, KumarA, McArdleWL, et al. (2011) A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol 128 : 996–1005.
33. SwamyRS, ReshamwalaN, HunterT, VissamsettiS, SantosCB, et al. (2012) Epigenetic modifications and improved regulatory T-cell function in subjects undergoing dual sublingual immunotherapy. J Allergy Clin Immunol 130 : 215–224 e217.
34. StefanowiczD, HackettTL, GarmaroudiFS, GuntherOP, NeumannS, et al. (2012) DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children. PLoS One 7: e44213.
35. CurtinJA, SimpsonA, BelgraveD, Semic-JusufagicA, CustovicA, et al. (2013) Methylation of IL-2 promoter at birth alters the risk of asthma exacerbations during childhood. Clin Exp Allergy 43 : 304–311.
Štítky
Genetika Reprodukčná medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



