-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
Deciphering the specific contribution of individual motifs within cis-regulatory modules (CRMs) is crucial to understanding how gene expression is regulated and how this process is affected by sequence variation. But despite vast improvements in the ability to identify where transcription factors (TFs) bind throughout the genome, we are limited in our ability to relate information on motif occupancy to function from sequence alone. Here, we engineered 63 synthetic CRMs to systematically assess the relationship between variation in the content and spacing of motifs within CRMs to CRM activity during development using Drosophila transgenic embryos. In over half the cases, very simple elements containing only one or two types of TF binding motifs were capable of driving specific spatio-temporal patterns during development. Different motif organizations provide different degrees of robustness to enhancer activity, ranging from binary on-off responses to more subtle effects including embryo-to-embryo and within-embryo variation. By quantifying the effects of subtle changes in motif organization, we were able to model biophysical rules that explain CRM behavior and may contribute to the spatial positioning of CRM activity in vivo. For the same enhancer, the effects of small differences in motif positions varied in developmentally related tissues, suggesting that gene expression may be more susceptible to sequence variation in one tissue compared to another. This result has important implications for human eQTL studies in which many associated mutations are found in cis-regulatory regions, though the mechanism for how they affect tissue-specific gene expression is often not understood.
Published in the journal: Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity. PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004060
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004060Summary
Deciphering the specific contribution of individual motifs within cis-regulatory modules (CRMs) is crucial to understanding how gene expression is regulated and how this process is affected by sequence variation. But despite vast improvements in the ability to identify where transcription factors (TFs) bind throughout the genome, we are limited in our ability to relate information on motif occupancy to function from sequence alone. Here, we engineered 63 synthetic CRMs to systematically assess the relationship between variation in the content and spacing of motifs within CRMs to CRM activity during development using Drosophila transgenic embryos. In over half the cases, very simple elements containing only one or two types of TF binding motifs were capable of driving specific spatio-temporal patterns during development. Different motif organizations provide different degrees of robustness to enhancer activity, ranging from binary on-off responses to more subtle effects including embryo-to-embryo and within-embryo variation. By quantifying the effects of subtle changes in motif organization, we were able to model biophysical rules that explain CRM behavior and may contribute to the spatial positioning of CRM activity in vivo. For the same enhancer, the effects of small differences in motif positions varied in developmentally related tissues, suggesting that gene expression may be more susceptible to sequence variation in one tissue compared to another. This result has important implications for human eQTL studies in which many associated mutations are found in cis-regulatory regions, though the mechanism for how they affect tissue-specific gene expression is often not understood.
Introduction
Gene expression is initiated by the binding of transcription factors (TFs) to cis-regulatory modules (CRMs) such as enhancer elements, which give rise to specific patterns of temporal and spatial activity [1]. Recent years have seen a dramatic increase in the ability to identify the location of regulatory elements using genome-wide information on TF occupancy [2], [3], [4], [5], [6], [7], cofactor recruitment [8], [9] and chromatin modifications [10], [11], [12], [13], [14], [15]. These studies have identified thousands to hundreds-of-thousands of regulatory elements that could be potentially active at a given time and in a given cell-type during development. Given this extensive regulatory landscape, it has become an enormous challenge to decode how CRMs function in terms of their spatio-temporal activity. Detailed dissection of individual elements has revealed developmental enhancers whose function is dependent on the presence of individual TF motifs [16], [17], combinations of motifs [18], [19], [20], and the specific arrangement [21], [22], [23], [24], [25], [26] or not [2], [27], [28], [29] of those motifs. These examples have inspired much debate over the relative importance of each of these variables to enhancer function, but systematic rules to understand their contribution have not emerged.
While dissection of endogenous enhancers has proved to be an extremely powerful approach to understand enhancer function [19], [20], [29], [30], [31], [32], individual enhancers instantiate only one of many possible solutions that can give rise to a specific gene expression pattern [7], [28], [33], thereby limiting the range of functional rules that are generally explored. Synthetic elements offer the possibility to examine a wide range of motif compositions and motif positioning rules while ensuring, as much as possible, the neutrality of non-motif (i.e. spacer) sequences. Synthetic promoter-YFP libraries in yeast, for example, were used to quantify and model the effect of different promoter motif configurations on the levels of YFP expression [34], [35]. Similarly, synthetic constructs combined with massively parallel sequencing were used to dissect the relationship between DNA sequence and activity of constructs transiently transfected into cell lines [36] or injected into mouse tail veins [37]. These approaches offer the clear advantage of scale, as thousands of elements with different DNA sequence combinations can be examined simultaneously. However, they are limited by the simplicity of the read-out, which is a relative measure of the levels of the CRM's activity at a single time point or condition. For most developmental enhancers, the impact of sequence changes on the timing or tissue-specificity of gene expression is equally important. It is also not clear to what extent episomal DNA from transient transfection or tail-vein injections recapitulates the impact of chromatin context and nucleosome positioning on gene expression. In these respects, reporter assays in transgenic animals provide invaluable information. Although generally difficult to scale, multi-stage assays using stable transgenic embryos yield precise information about when and where an enhancer is active in an in vivo chromatinized context.
In this study, we systematically engineered 63 synthetic elements that differ with respect to the number and kinds of TF motifs they contain, as well as the relative spacing and orientation of these motifs. These elements were specifically designed to assess three properties of enhancer motif organisation relevant to metazoan development: (1) the ability of homotypic clusters of individual motifs to function as developmental enhancers, (2) the ability of combinations of different kinds of motifs (heterotypic motif clustering) to generate new emergent activity, and (3) the effect of changes in motif organisation, such as number, spacing and orientation, on the robustness of enhancer activity in both space and time. While many studies have focused on the effect of point mutations on enhancer activity, there is an enormous amount of structural variation within natural populations, including the sequence of Drosophila [38], [39] and humans [40]. To assess the influence of small insertions or deletions, we have systematically changed the spacing between motifs for four heterotypic pairs of TF motifs.
To examine these properties, we focused on ten motifs recognized by TFs that form part of a highly studied regulatory network that governs Drosophila mesoderm development [41], [42], including the downstream effector TFs of Wingless (known as Wnt in vertebrates) and Dpp (BMP in vertebrates) signaling pathways. We generated very simple elements consisting of six motifs in either a homotypic or heterotypic combination, which were stably integrated into the Drosophila genome. The resulting patterns of expression driven by these elements suggest a number of interesting features governing CRM function. First, despite the known importance of combinatorial activity to refine spatial patterns of expression, elements with multiple copies of an individual motif can drive complex patterns of expression. For example, although pMad is known to act cooperatively with lineage TFs in both Drosophila and vertebrates, it is sufficient to transduce Dpp activity when its motifs are in the appropriate configuration. Second, combining motifs for as few as two TFs can lead to novel emergent expression. Although the importance of cooperative DNA binding in the regulation of development has long been supported by other studies, the sufficiency of so few sites speaks to the extent to which even minor changes in cis-regulatory sequence can lead to the evolution of novel expression profiles. Third, the spacing and orientation of motifs is not only essential for enhancer activity in terms of binary on-off effects, but also has more quantitative effects on the robustness of gene expression, including inter - and intra - embryo variability. Fourth, these effects of motif organization, often referred to as motif grammar, are tissue-specific. The same ‘two-TF’ enhancer can function using very flexible motif spacing in one tissue, yet have rigid constraints in another, demonstrating an additional way in which the organization and function of CRMs acts to reduce the constraints of pleiotropy for regulatory mutations.
Results
Design of synthetic developmental enhancers
For each factor, synthetic CRMs were generated by combining six motifs, separated by a spacer sequence of defined length. Therefore, there were two aspects to the design of the synthetic elements; the choice of motif instance used for each TF and the sequence of the spacer. First, for the TF motifs, we selected high affinity motifs for each factor, as much as possible. TFs often recognize the same or highly similar sequences. This includes not only members of the same family of TFs, e.g. GATA factors (such as Pnr), but also TFs with apparently unrelated DNA binding domains, e.g. the bHLH factor Twist and the zinc finger TF Snail. While we cannot change this inherent property of TFs, we did try to increase the potential specificity of the motif (or word instance) used here for the particular TFs we are interested in by using motifs derived from in vivo occupancy data for nine of the ten factors [2], [7]. For clarity, we refer to each construct by the name of the TF from which ChIP data was used to learn the motif. However, the sequence of all motif instances used, as well as their similarity to other TF motifs is provided in Table S1.
Second, we tried to select a neutral spacer sequence that does not include known TFBS, based on our current knowledge. This is not trivial, as in addition to the spacer sequence itself, the border sequence bridging the spacer and the known motif (red bar, Figure 1A) can in itself create an additional binding site. To minimize this possibility, rather than using a common spacer, we computed an optimal spacer sequence for each combination of motifs to minimize the chance of inadvertently generating additional binding sites, based on current information (Supplemental Methods (Text S1)). During the course of this study, our results show that the spatio-temporal activity of the designed elements is primarily driven by the intended TF motifs and not from the spacer sequence, as indicated by two lines of evidence: (1) The concordance between the activities of homotypic and heterotypic elements for the same TFs, which were designed with different spacer sequences (described below) and 2) the very similar spatio-temporal activity of two heterotypic CRMs, which were specifically designed with identical TF motifs but with two different spacer sequences (pMad-Tin, see below).
Fig. 1. Simple synthetic elements containing homotypic TF motifs can generate complex spatio-temporal activity. 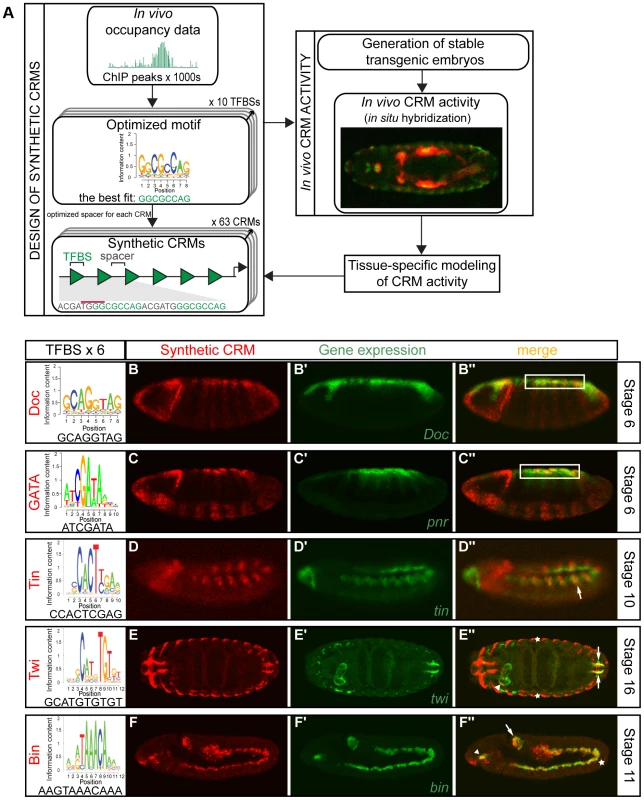
(A) Schematic representation of flow from design of synthetic CRMs, assessment of in vivo activity to tissue-specific modeling. Optimized TFBSs (from ChIP experiments) were separated by spacers, with minimal affinity to known TF binding models, and placed in front of a minimal promoter and lacZ reporter and integrated into the Drosophila genome. (B–F) Homotypic synthetic CRMs containing six TFBSs from the represented sequence logo for each TF, separated by a 6 bp spacer. CRM activity was assessed by double fluorescent in situ hybridization of the lacZ reporter gene driven by synthetic CRM (red) and the corresponding endogenous gene (green). Synthetic CRMs composed from GATA (B–B″) and Doc (C–C″) motifs drive expression in the presumptive amnioserosa (white box). Tin synthetic CRM (D–D″) is expressed in the dorsal mesoderm (arrow), while Twi CRM (E–E″) is expressed in the foregut weakly (white arrowhead), hindgut (white arrows) visceral mesoderm and ectoderm (asterisks). Bin synthetic CRM (F–F″) is active in the foregut (arrowhead), midgut (asterisk) and hindgut (arrow) visceral mesoderm (VM). All embryos are shown laterally with anterior to the left, except for (E) which is a dorsal view. Each synthetic CRM, which were on average 105 bp in length, was cloned into a common minimal lacZ reporter vector and stably integrated into the same location in the Drosophila genome using the phiC31 system [43], to allow for a direct comparison of enhancer activities. The ability of each CRM to drive spatio-temporal lacZ expression was assessed during all stages of embryogenesis by fluorescent in situ hybridization to examine the full regulatory potential of the DNA sequence.
The regulatory potential of homotypic CRMs for different types of TFs
The combinatorial binding of TFs provides complexity to regulate refined spatial patterns of expression, and forms the basis for logical operations within Gene Regulatory Networks driving development [44]. However, in addition to combinatorial activity, homotypic clusters of an individual TF's motif are also present in regulatory regions in the vicinity of developmental genes in both Drosophila [45] and vertebrates [46]. Although prevalent in vivo, the role of these clusters in regulating gene expression, and the properties governing how they function, is currently not clear. Studies examining the ability of clusters of motifs to regulate gene expression in transgenic reporter assays have had varied success: this includes (although not exhaustive) homotypic clusters of motifs that were sufficient to drive activity [47], [48], [49], [50], [51], [52] and those that were not [48], [53], [54], [55], [56]. However, as the elements in each of these individual studies were designed and tested in different ways, it is impossible to deduce any functional inference across studies.
We initiated this study by systematically examining the activity of elements composed of a cluster of six identical motifs. In total we tested homotypic CRMs with motifs for ten essential factors, encompassing multiple types of DNA binding domains: bHLH (Twist (Twi)), homeobox (Tinman (Tin), Bagpipe (Bap)), T-box (Dorsocross (Doc)), GATA zinc finger (Pannier (Pnr)), MADS box (Myocyte enhancer factor 2 (Mef2)), FoxF (Biniou (Bin)), HMG-domain (T-cell factor (TCF)), Ets-domain (Pointed (Pnt)) and the MH1 domain (pMad). Seven out of ten of these very simple elements were able to drive sequence-specific spatio-temporal expression (Figures 1B–F, 2, S1). In six cases, the expression profiles driven by clusters of a single motif were sufficient to partially (and in the case of two, almost completely) recapitulate the expression of the TF whose in vivo occupied motif was used to construct the CRM. For example, the synthetic CRMs containing Doc enriched and GATA motifs drove expression in the presumptive amnioserosa, cells where the endogenous Doc and pnr genes are expressed (white boxes, Figure 1B″,C″). Similarly the synthetic CRM containing Tin motifs drove expression in a subset of the dorsal mesoderm, colocalizing with the expression of the endogenous tin gene (Figure 1D″, arrow). Although multimerizing the preferred Twist E-box motif was not sufficient to drive mesoderm expression at early stages of development, this synthetic CRM could activate expression in the hindgut visceral mesoderm (VM) at later stages, overlapping the endogenous twist gene's expression (Figure 1E″, arrow).
Fig. 2. Altering the number of pMad motifs affects visceral mesoderm activity. 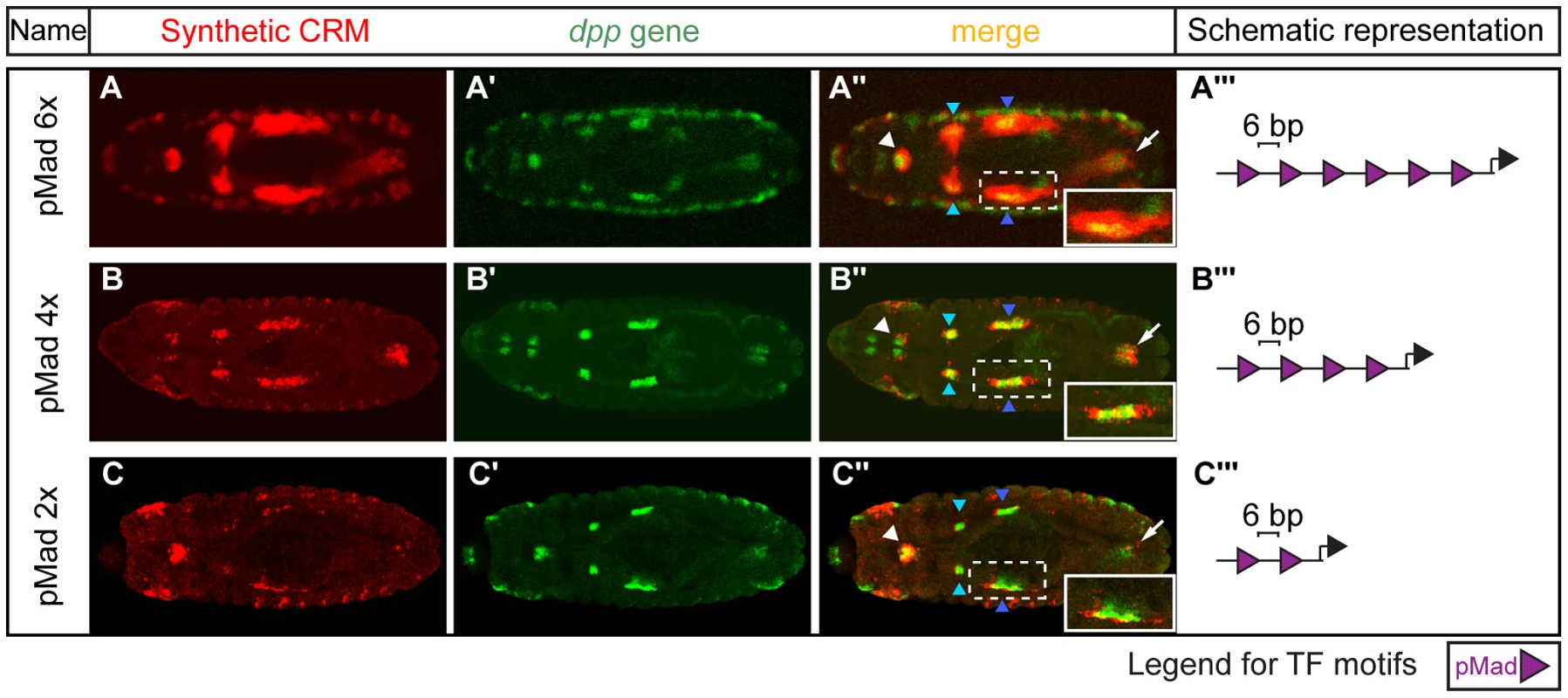
Double in situ hybridizations against (A–C) the lacZ reporter gene driven by the pMad homotypic synthetic CRMs (red) and (A′–C′) the endogenous dpp gene (green). (A″–C″) Expression patterns in the foregut (white arrowhead); hindgut (arrow); parasegment 3 (PS3) (light blue arrowhead) and 7 (PS7) (dark blue arrowhead) of the midgut visceral mesoderm (VM). Dashed box is enlarged in the inset (white box). Embryos are dorsally oriented, with anterior to the left, stage 13/14. (A′″–C′″) Schematic representations of the CRM composition, where purple triangles depict the number and orientation of pMad sites with indicated spacing (bp) between adjacent sites. The activity of two homotypic CRMs, containing motifs for either pMad (discussed below) or Bin, stood out as they were sufficient to recapitulate almost the entire domain of the TF's activity. For the Bin CRM, this included the foregut, midgut and hindgut VM (Figures 1F″, S2) through multiple stages of development. This result suggests that motifs for this FoxF factor are sufficient to regulate expression throughout the VM at multiple stages of development, consistent with Bin's essential requirement for VM development [57] and extensive enhancer occupancy [7], [58]. In contrast to the six CRMs that gave ‘expected’ activity, the activity of the homotypic dTCF CRM only partially overlaps wingless expression, (the ligand that activates the cascade leading to dTCF activation). The CRM's activity is restricted to segmental groups of cells that are in close proximity to, but not always adjacent to, the wg stripes (Figure S1A″), which may reflect more ‘long’-range signaling from Wg, or alternatively the activity of an additional TF that can occupy this dTCF motif.
Taken together, these results indicate that homotypic clusters of an individual motif are often sufficient to regulate specific patterns of spatio-temporal activity, analogous to the more commonly studied combinatorial elements. Multimerizing single TF motifs can provide remarkable specificity, as demonstrated by the non-random overlap of CRM activity with part or all of the TF's expression in six out of ten cases tested. Conversely, it is interesting that not all activator clusters are sufficient to drive expression, in contrast to what might be assumed from the activity of yeast GAL4 sites. For example, three synthetic CRMs (multimers of Pnt, Mef2 and Bap motifs) yielded no activity (Figure S1B–D). In the case of Mef2 and Twist, the lack of general mesoderm activity is surprising given that both TFs have very well characterised DNA binding specificities in both Drosophila and vertebrates. This diversity in CRM output may reflect inherent differences in the regulatory potential among different TFs or in their ability to act cooperatively in a homotypic manner to potentiate their activity.
The extent of CRM activity scales with the number of TF motifs
The dpp gene, coding for the ligand of the Dpp signaling cascade, is expressed in the foregut, hindgut and midgut VM in parasegment 3 (PS3) and 7 (PS7) at stage 13/14 of embryogenesis [59]. dpp expression is restricted to these VM domains through the integration of activating inputs from Ubx and Bin [57], [59], and repression via Wg signaling [60]. The downstream effector TF of Dpp signaling is the phosphorylated form of Mad (pMad). Our synthetic CRM containing six pMad motifs could recapitulate almost the entire expression of dpp in the VM during these stages of embryogenesis (Figure 2A). Therefore, despite the complexity in the network of upstream factors regulating dpp expression, once dpp is expressed, sites for its downstream effector (pMad) alone are sufficient to activate enhancer activity in these sub-tissue domains. The spatial boundaries of the enhancer's activity are most likely refined through the action of Brinker (Brk). Brk is a transcriptional repressor whose expression is negatively regulated by Dpp signaling [61]. This results in cells with high levels of pMad and low Brk (Dpp responding cells, where our synthetic CRM is active) and those with high Brk and low pMad (neighbouring cells outside the Dpp signaling domain, where our CRM is inactive). As Brk and pMad can recognize the same motif [62], there is often direct competition between the two TFs to regulate enhancer activity, where the relative levels of both TFs serve to limit the spatial domain of Dpp target gene expression, as nicely demonstrated for the endogenous Ubx enhancer [62].
The sufficiency of pMad motifs alone to activate enhancer activity was unexpected given previous studies in both Drosophila [56] and vertebrates [63], [64], indicating that Mad proteins bind to enhancers cooperatively with other tissue-specific ‘lineage’ factors, and have little activity alone. We therefore further investigated the regulatory potential of pMad sites in isolation by directly comparing the activity of elements containing six, four or two motifs, in the same orientation and with the same spacer sequence (Figures 2, S3). The expression of lacZ in PS7 of the midgut VM (Figure 2) is particularly informative of pMad activity: the Dpp signaling cascade activates pMad in an autocrine [65] and paracrine [66] fashion, which likely results in the highest levels of pMad activation in VM cells in PS7 and lower levels in adjacent parasegments. The homotypic CRM containing six pMad motifs reflects this paracrine signaling, having a larger domain of expression covering neighboring cells compared to the dpp expressing cells (Figure 2A). In contrast, the synthetic CRM containing four motifs had more restricted activity to a narrow domain anterior and posterior to the dpp expressing cells (Figure 2B), while the CRM containing only two motifs was active only in the Dpp producing cells (autocrine signaling) (Figure 2C). Therefore, pMad sites alone, when present in close proximity, are sufficient to activate enhancer activity in the absence of specific lineage TFs. However, the extent of the activity is dependent on the number of available pMad motifs and on the balance between pMad and Brk concentration.
In addition to the number of cells in which the enhancer was active, we also observed a strong correlation between the number of sites and the strength of the CRM. In the amnioserosa, for example, the CRM with six pMad sites drove robust expression at stage 11 (Figure S3A″), while four sites resulted in reduced activity, and the CRM with two sites drove only very weak expression in the amnioserosa (Figure S3B,C). This trend was even more dramatic at stage 14, when the pMad concentration in the amnioserosa decreases [67]. While the CRM with six pMad sites drove strong activity at stage 14 (Figure S3D), the CRMs with either four or two sites were inactive in the amnioserosa at this stage. The number of pMad sites, thus, appears to be able to compensate for a decrease in the amount of accessible pMad protein at stage 14, presumably by providing a larger platform for cooperative binding.
Changing motif organisation within a CRM affects its activity in a tissue-specific manner
Cooperative interactions between TFs can sometimes occur through direct protein-protein interactions (PPI), which may introduce constraints in the organization of the TFs' motifs within CRMs [1]. Taking advantage of the design flexibility of synthetic elements, we generated heterotypic CRMs composed of motifs for TFs known to exhibit protein-protein or genetic interactions: namely motifs for Tin with either Pnr, Doc, dTCF, or pMad [68], [69], [70], [71], to systematically explore two properties: 1) The influence of changes in the relative spacing and orientation of motifs on the robustness of CRM activity in different tissues, and 2) The ability of different combinations of TF motifs to generate emergent expression patterns in a particular tissue, not observed for CRMs containing only one kind of motif.
Each heterotypic CRM contained three pairs of TF motifs for either Doc-Tin, GATA-Tin, dTCF-Tin, or pMad-Tin. For each TF pair, on average nine constructs were tested in transgenic animals, in which the spacing and/or orientation of the Tin motif was systematically altered (Table S2). Heterotypic CRMs containing GATA-Tin or Doc-Tin combinations resulted in a pattern of expression that was largely the sum of the activity of each of the corresponding homotypic CRMs alone (data not shown). Changing the spacing and orientation of the sites (Table S2) had little or no effect on CRM activity, indicating a minimal role for motif positioning, or grammar, in these instances. The dTCF-Tin heterotypic CRMs did not have any activity, despite the fact that the homotypic CRMs containing six dTCF or Tin sites drove specific activity in segmental groups of cells in the ectoderm and mesoderm, respectively (Figures 1D, S1A). This lack of activity in the heterotypic context most likely reflects the reduction in the number of motifs used, from six in the homotypic constructs to three motifs for each factor in the heterotypic CRM. Correspondingly, homotypic construct containing four dTCF sites [53], [54] had no embryonic activity.
In contrast to the GATA-Tin and Doc-Tin heterotypic pairs, whose activity was robust to changes in motif organisation, the activity of the pMad-Tin heterotypic CRMs changed depending on the motif spacing. As discussed above, pMad motifs alone are sufficient to regulate expression in PS3 and PS7 of the midgut VM. In these cases, the strength of activity was dependent on the number of motifs present, with a dramatic reduction in expression observed going from four to two sites (Figure 2). A homotypic CRM containing three pMad sites separated by 13 bp also showed a dramatic reduction in VM activity (Figure 3A), a result that may stem from either or both the reduction in sites or the increase in spacing (from 6 bp to 13 bp). Interestingly, when a Tin motif is inserted within each 13 bp ‘spacer’ sequence, this pMad-Tin heterotypic design can restore activity in the midgut VM (Figure 3B, white square), giving a pattern of expression similar to the pMad homotypic CRM with 6 bp spacing. The ameliorative effects of the Tin sites were limited by distance. Increasing the spacing between adjacent pMad and Tin motifs from 2 to 8 bp, which increases the spacing between pMad motifs from 13 to 25 bp, caused a progressive reduction in lacZ expression in the midgut VM (Figure 3). This VM activity is primarily driven through the pMad and Tin motifs and not the spacer sequence, as changing the spacer sequence had a minimal effect on VM enhancer activity, while a similar distance effect was observed by altering the length of spacer between the two sites (compare Figures 3 and S5). These results suggest that the occupancy of Tin sites acts either to bridge the distance between pMad sites (up to ∼20 bp) facilitating cooperative pMad regulation or alternatively to facilitate pMad-Tin combinatorial regulation of VM expression.
Fig. 3. Changes in motif organisation have a graded effect on activity of CRMs in the visceral mesoderm. 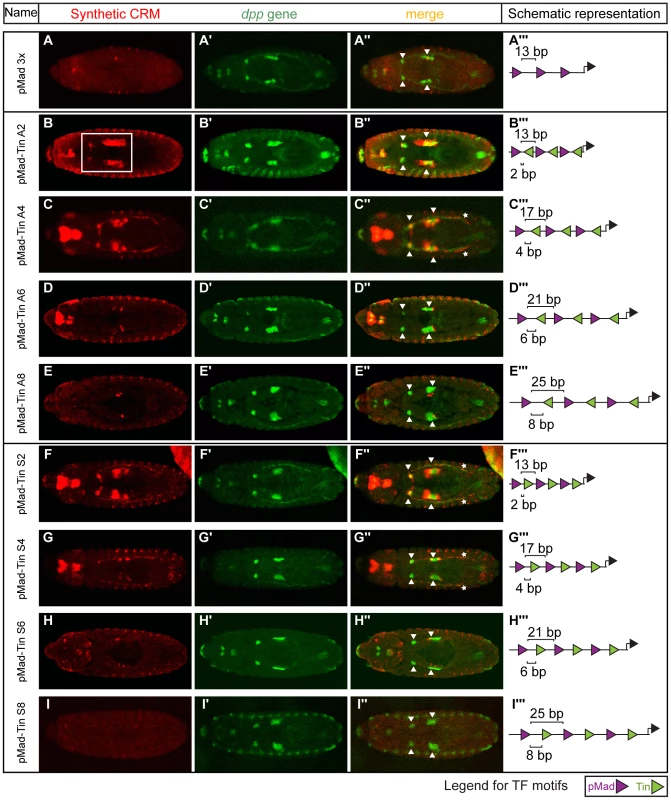
Double in situ hybridization against the (A–I) lacZ reporter gene driven by the synthetic CRMs (red) and (A′–I′) the endogenous dpp gene (green). (A″–I″) CRM activity in midgut visceral mesoderm is indicated with arrowheads or white box (B) and in heart with the asterisks (C″, F″, G″). Embryos are dorsally oriented, with anterior to the left, stage 13/14. (A′″–I′″) CRM composition, where triangles (pMad – purple, Tin – green) depict the number and orientation of sites. Spacing between adjacent pMad-Tin sites (below) and pMad-pMad sites (above) is indicated. To distinguish between these two possibilities, we examined the effect of altering the relative orientation of the sites, reasoning that if Tin occupancy acts indirectly to help pMad recruitment altering the orientation of the Tin site should have little effect on activity. However, we observed that changes in motif orientation did influence CRM activity: one orientation of the Tin motif (arbitrarily referred to as antisense (A)) typically drove stronger VM expression compared to CRMs with the motif in the opposite orientation (referred to as sense (S)) (Figure 3, compare C to G). This result indicates that the Tin motifs are not just neutral sequences bridging the spacing between homotypic pMad motifs, but rather suggest a specific mechanism of cooperative Tin-pMad DNA binding, and therefore transcriptional regulation.
Non-additive activity by combining two different motifs: Emergence of heart activity
Combining pMad and Tin sites resulted in CRM activity in the dorsal mesoderm, VM and amnioserosa –representing essentially the summation of the expression profiles of the respective homotypic CRMs (Figures 1D, 2A, S4). However, in contrast to GATA-Tin and Doc-Tin CRMs, whose joint expression profiles were largely additive, the heterotypic pMad-Tin CRMs were also sufficient to drive expression in a new domain, the developing heart (Figure 4). Cardioblast specification requires the action of a large number of TFs and signaling cascades (for review, see [72]). Tin, Pnr (GATA factor), and Doc together with Dpp and Wg signaling are essential for heart development [73], [74], [75], [76], [77], [78], a tissue in which these factors regulate each others' expression [70], [75], [76], [77], [78], [79], [80], and act as a collective unit to regulate enhancer activity [2], [27]. Given this complex regulation, we did not anticipate that a simple element containing only pMad and Tin sites would be able to drive expression in the heart. Importantly, the emergence of this pattern was highly dependent on the CRM architecture: Among our constructs, heart activity was only observed in CRMs containing three pMad and three Tin motifs placed in close proximity (within 2–4 bp) to one another (Figure 4B–E). In contrast, no heart activity was observed in CRMs where the spacing between motifs was increased to 6 bp or 8 bp.
Fig. 4. Emergence of heart expression by combining two TFs' motifs in limited configurations. 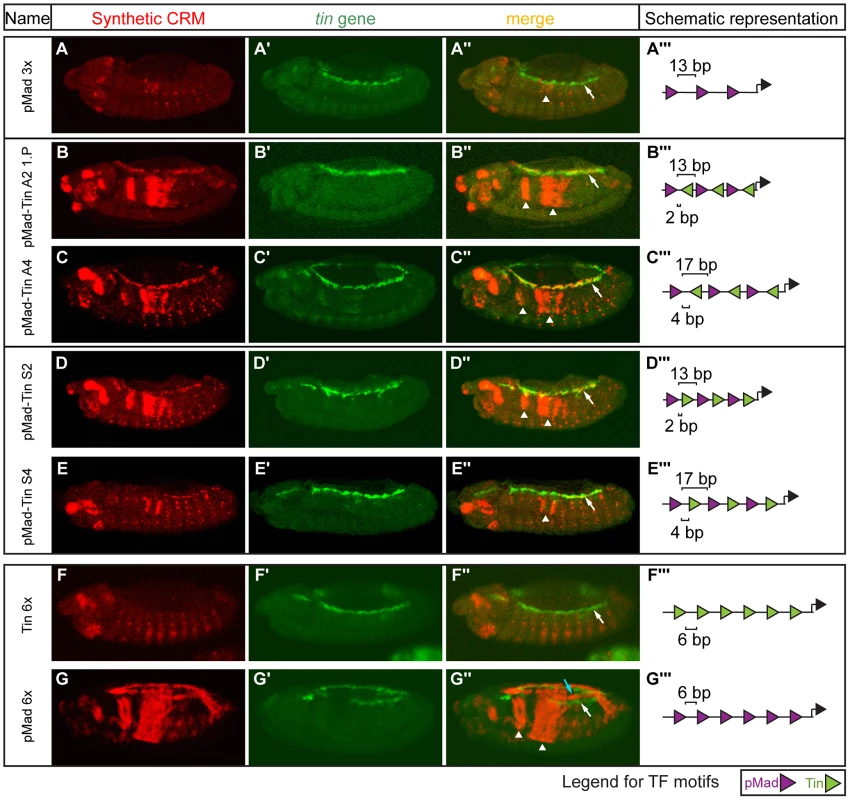
Double in situ hybridization against the (A–G) lacZ reporter gene driven by the synthetic CRMs (red) and (A′–G′) the endogenous tin gene (green). (A–A″) pMad 3× CRM with 13 bp spacer has no heart activity (white arrow) and a severe reduction in visceral mesoderm activity (arrowhead). (B″–E″) pMad-Tin CRMs containing 2 and 4 bp spacers were active in the heart (white arrows) as indicated by colocalization with tin gene. (F″,G″) CRMs composed of only Tin or only pMad sites were unable to drive expression in heart. Expression in the midgut visceral mesoderm is indicated with arrowheads (A″–E″, G″) and in amnioserosa with light blue arrow (G″). Embryos are laterally oriented, with anterior to the left, stage 13/14. (A′″–G′″) CRM composition, where triangles (pMad – purple, Tin – green) depict the number and orientation of sites. Spacing between adjacent pMad-Tin sites (below) and pMad-pMad sites (above) is indicated. Even in the ‘optimal’ motif configuration, we observed embryo-to-embryo variability, indicating that expression in the heart is significantly less robust (i.e. more influenced by stochastic events) than is expression driven by pMad-Tin constructs in the VM and other tissues. To better assess the robustness of these constructs activity, we made use of P-element transgenesis to integrate our CRMs into random locations in the genome. We reasoned that non-robust CRM activity would be more highly influenced by variation in chromatin context at different genomic positions compared to more robust expression profiles. Across these random-insertion sites, the pMad-Tin CRMs drove highly consistent expression in the VM, amnioserosa, and dorsal mesoderm, while heart activity in those same embryos varied dramatically as a function of genomic location (Table S3). In fact, the pMad-Tin A2 CRM was only active in the heart in the context of a transgenic fly line obtained using a P-element (Figure 4B), and not with the phiC31-mediated integration (Figure 3B). These results indicate that the heart activity of pMad-Tin CRMs is teetering on the edge of activation, being highly sensitive to both the motif context within the enhancer and the enhancer context within its chromatin environment. Thus, while the combined activity of these two TFs can give rise to emergent activity in the developing heart, this is not a robust mechanism to generate heart expression.
Sensitivity to motif organisation varies in a tissue-specific manner
Examining CRM activity in the heart revealed that some CRMs exhibit varied activity among embryos, and even within an embryo, as demonstrated by the pMad-Tin S4 construct, which drove expression throughout the entire heart in some embryos (Figure 3G), but only in a posterior portion of the heart in others (Figure 4E). To assess this variability in a systematic manner, and to provide quantitative data to which we could fit a model explaining CRM activity (see below), we applied two measures of the robustness of CRM activity: (1) Penetrance, defined as the fraction of embryos within a population that show CRM activity in the relevant tissue (in this case the VM or heart), and determined by any spatial overlap of lacZ expression with dpp (VM) or tin (heart) at a defined developmental stage (Figures 5A, S6A). (2) CRM expressivity, defined as the fraction of tissue-specific regions within an embryo (i.e. the proportion of the VM or heart) that display CRM expression (Figures 5A, S6A). In the midgut VM, for example, there are four domains of CRM activity (Figure 5A). If the CRM is active in all four domains, it has an expressivity of 1, while activity in two out of four domains has an expressivity of 0.5. To minimize systematic error, we implemented automated image analysis (see Materials and Methods) of embryos at a consistent stage of development (Figures 5A, S6A). In using penetrance and expressivity as quantitative metrics of CRM activity, we avoided issues arising from directly comparing non-linearly amplified signals from standard in situ hybridization of lacZ levels between CRMs. Penetrance provides a reliable measure of enhancer embryo-to-embryo variability, while expressivity provides a readout of intra-embryo enhancer variability – both of which we exploit to assess the effect of motif organization on the robustness of activity in two tissues.
Fig. 5. Quantifying the penetrance and expressivity of CRM activity. 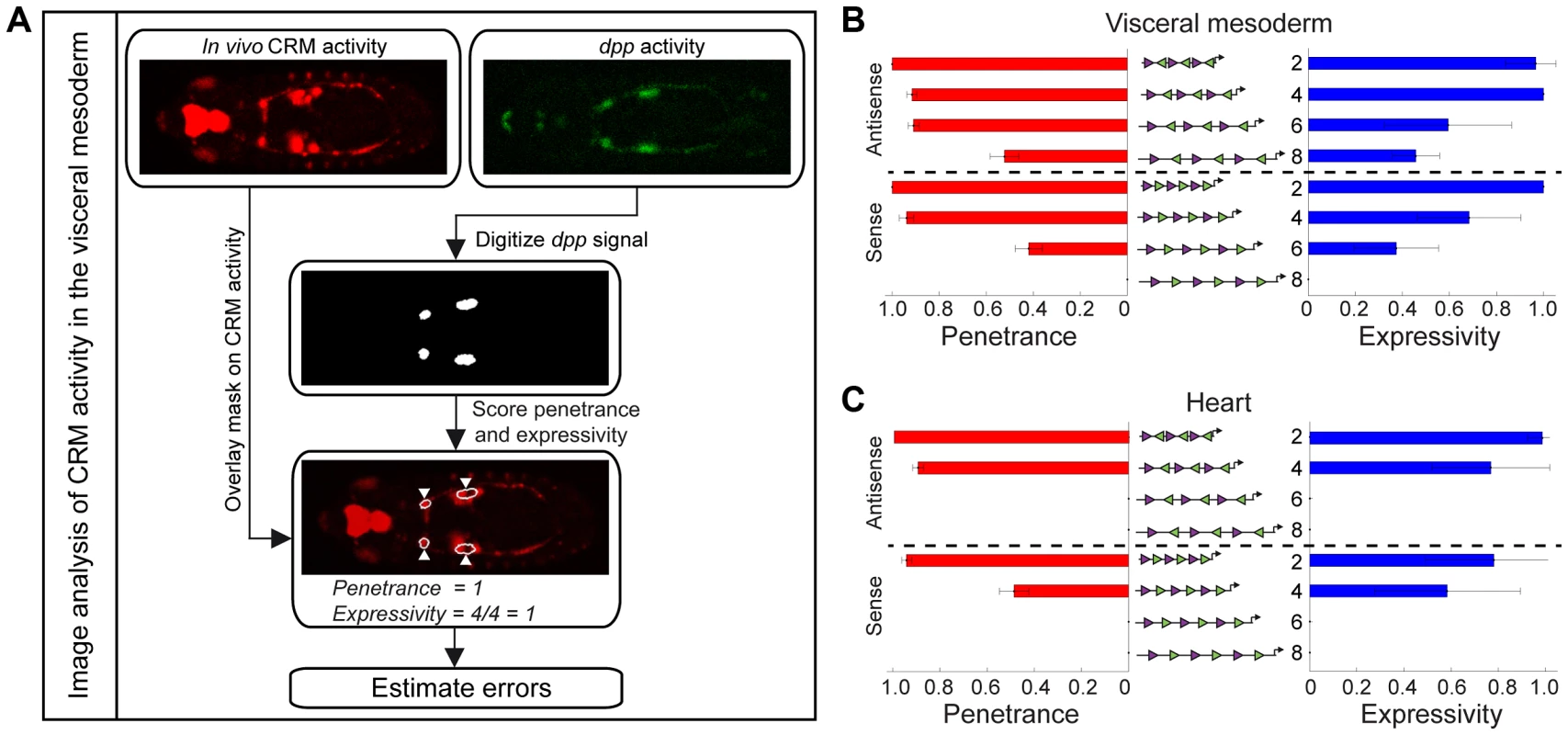
(A) Automated image analysis workflow. The gene expression pattern was digitized to create a mask for the tissue of interest. Comparing the mask with CRM activity enabled rapid and reliable scoring of both penetrance and expressivity. Errors in penetrance and expressivity were estimated as described in Supplemental Methods (Text S1). Penetrance (red bars) and expressivity (blue bars) of pMad-Tin heterotypic CRMs in the VM (B) and the heart (C). Note that P-element pMad-Tin A2 line was used to quantify heart activity. The CRM penetrance of around one hundred embryos was measured for each of the 8 pMad-Tin synthetic CRMs (Table S4). With this quantification, differences in activity became more striking in several regards. First, within a tissue, subtle differences in activity between CRMs with different motif configurations become clear. For example, in the antisense orientation, changing the motif spacing from 2 bp to either 4 bp or 6 bp spacing resulted in only a slight decrease in penetrance, from 1 to 0.91 in the VM. However, the expressivity of these CRMs was quite distinct. The CRM with a 4 bp spacing had an expressivity of 1, while this number was nearly halved (0.59) for the CRM with a 6 bp spacing (Figure 5B; Table S5). The extreme effect of a small (2 bp) change in motif spacing suggests that direct, and potentially cooperative, interactions among bound factors have been disrupted, leaving the enhancer's activity more prone to variation.
A second striking observation is the extent to which changes in CRM architecture impacts activity in a tissue-specific manner. This is made clear by relative differences in CRM penetrance between the VM and heart. In the antisense orientation, for example, CRM penetrance remains almost unchanged in the VM when the motif spacing between the pMad and Tin sites is changed from 2 to 6 bp (Figure 5B). In contrast, the penetrance in the heart drops from 0.89 at a 4 bp spacing to zero when the motifs are spaced by 6 bp. A similar dramatic effect was observed by flipping the orientation of the Tin motif from antisense to sense, at a 4 bp spacing, which caused the penetrance of heart activity to drop from 0.89 to 0.48 (Figure 5C). These switch-like transitions in penetrance indicate that heart activity can only occur with a very restricted motif organization, which relies on close proximity of all TF motifs.
Taken together these results highlight an important property of cis-regulatory activity in multicellular organisms: An enhancer element (e.g. composed of only two types of motifs as described here) can require a very restricted motif configuration to regulate expression in one tissue (heart), but yet be much more flexible in its motif organisation to drive robust activity in another (VM).
Modeling TF occupancy reveals tissue-specific rules of TF cooperativity
The extent to which cooperative interactions, including higher-order interactions across multiple proteins, contribute to enhancer activity is difficult to assess by simply visualizing expression patterns. Here, computational models can be extremely helpful to explore the effect of potential interactions on CRM function [33], [81]. To better understand the contribution of higher-order interactions among TFs on our synthetic CRMs, we used fractional site occupancy modeling (Figures S7; Methods (Text S1)), which describe DNA-protein and protein-protein interactions as thermodynamic processes, an approach that has been successfully used to understand other regulatory elements [82], [83], [84], [85], [86], including Drosophila enhancers [26], [87].
As fractional site occupancy models analyze the probability of every possible configuration of binding events, the complexity of the models increases exponentially with the number of TFBSs. To avoid these complications, we aimed to identify the simplest model that recapitulates the observed CRM activity. In line with our observations (as seen in Figures 3 and 5), and with research in another tissue [71], the model assumes the presence of direct cooperative interactions between neighbouring pMad and Tin proteins. Protein-protein interactions were modeled as the extent of overlap between spheres of “interaction space” around each bound protein, with sense and antisense orientations having different effective spherical radii (Figure S7, and includes parameters for the strength of possible cooperativity between bound TFs (Supplemental Methods (Text S1)). The model specifically explores whether additive interactions between Tin-pMad pairs are sufficient to recapitulate the observed experimental data or if potential ‘higher order’ cooperativity among pMad-Tin pairs with nearby pMad or Tin bound proteins are required to drive robust CRM activity.
For VM activity driven by the pMad-Tin heterotypic CRMs, a model that includes an additional degree of cooperativity beyond pMad-Tin pairs fit the data better compared to a model in which pMad-Tin pairs act in an independent additive manner (Figures 6A, S8A). A key difference between the models lies in their predictions of the robustness of shorter CRMs: if higher order interactions are central for robust CRM activity then shorter CRMs will have a sharper decrease in robustness compared to pMad-Tin interactions alone. To experimentally test this, we halved the size of two heterotypic CRMs, generating CRMs with pMad-Tin-pMad motifs in S2 and A4 configuration. Such small CRMs could drive expression in the midgut VM (Figure 6B,C), albeit at a reduced level. In contrast, CRMs with only one pMad and Tin site, representing the smallest possible cooperative binding configuration (a pMad-Tin pair), drastically reduced all VM activity (Figure 6D,E). Incorporating higher level cooperativity into the model, without any further fitting, significantly improved the quality of the prediction of the shortened CRMs (Figures 6F,G, S8B). This suggests that pMad-Tin-pMad is the minimal configuration essential for robust VM expression. Importantly, this model was robust to a ‘leave-two-out’ cross validation iterated over all possible orderings of the 12 CRMs, arguing against over-fitting (Figure S8C). Finally, to test the model's prediction that additive interactions between pMad-Tin pairs with other bound pMad or Tin proteins are insufficient to drive robust VM expression, we tested the activity of a CRM with the motif configuration Tin-pMad-Tin. This CRM had very weak VM activity (Figure S8D), consistent with a requirement for higher order interactions between multiple pMad-Tin pairs for robust CRM activity (Figure 6G). In summary, increasing the complexity of TF cooperativity resulted in significantly improved consistency with experiment compared to considering only independent pMad-Tin cooperative pairs (Figure 6G).
Fig. 6. A fractional occupancy model learned different cooperative rules of TF binding that explain CRM penetrance and expressivity. 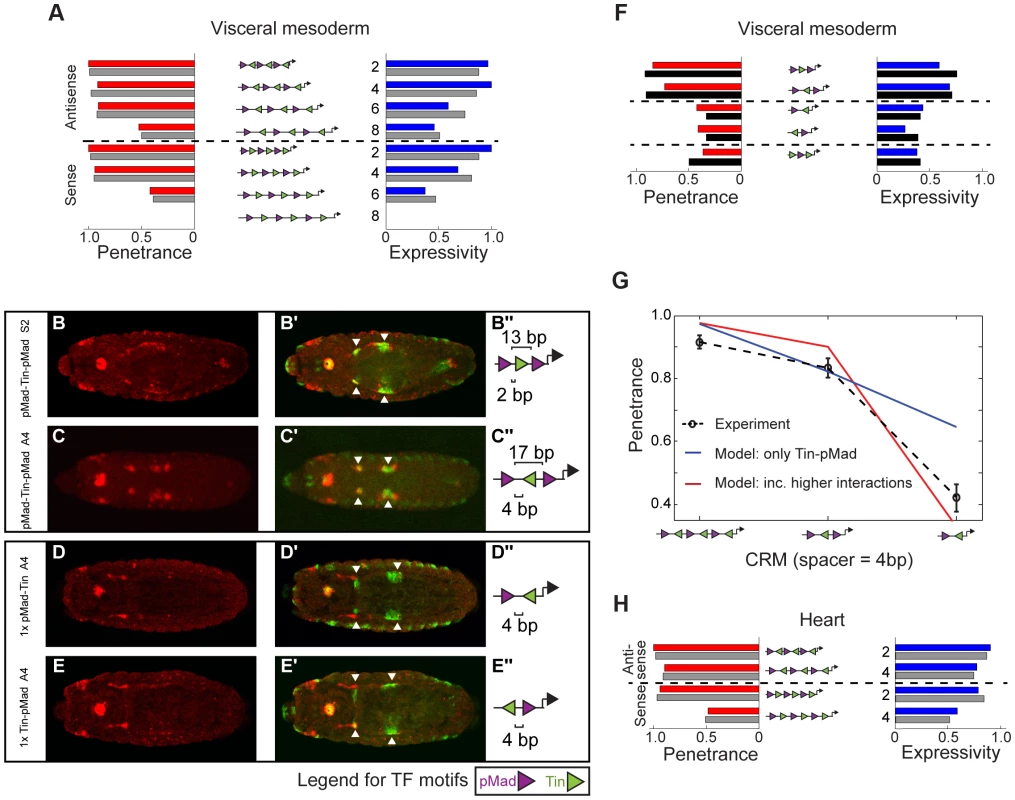
(A) Red and blue bars represent the experimentally measured penetrance and expressivity, respectively, of the six-motif pMad-Tin heterotypic CRMs in the VM. Gray bars represent the model's fitted results when pMad-Tin-pMad cooperative interactions were included. (B–E) CRM activity for short pMad-Tin heterotypic CRMs. Double in situ hybridization against the lacZ reporter gene driven by the synthetic CRMs (B–E, red) and the endogenous dpp gene (B′–E′) green), where arrowheads indicate the midgut visceral mesoderm (VM). Embryos are dorsally oriented, with anterior to the left, stage 13/14. (B″–E″) Schematic representations of the CRM composition, where purple and green triangles depict the number and orientation of pMad or Tin sites, respectively. Spacing between adjacent pMad-Tin sites (below) and pMad-pMad sites (above) is indicated. (F) Penetrance (red bars) and expressivity (blue bars) of the short pMad-Tin heterotypic CRMs in the VM with the model's prediction (black bars) when pMad-Tin-pMad cooperative interactions were included. (G) Model predictions for pMad-Tin enhancers of different motif number with antisense orientation of the Tin site and 4 bp separation between sites. Red (blue) curve corresponds to the model prediction with (or without) pMad-Tin-pMad interactions. (H) As (A) but for the heart, where only non-zero results are shown. Note that we fit the heart data including the penetrance and expressivity results from the P-element pMad-Tin A2 line. Gray bars represent the model fit when considering Tin-pMad-Tin as the minimal cooperative TF configuration. Next, we addressed how heterotypic pMad-Tin CRMs lead to activity in the heart. Simple models with cooperativity between bound Tin and pMad can recapitulate the observed penetrance and expressivity in the heart for CRMs (Figures 6H, S8E,F). However, only the model including higher-order cooperative interactions between three neighboring units of Tin-pMad-Tin was consistent with both the shorter constructs and the six TF motif CRMs (Figures 6H, S8E,F). We note that the true minimal motif arrangement to generate robust heart activity is likely to be much more complex. In line with this, the Tin-pMad-Tin CRM has no heart activity. Interestingly, the effective range of cooperative TF interactions learned by the model for the heart was considerably lower (<5 bp) than for the VM (<9 bp) (Table S6).
In summary, while our experimental data is suggestive of cooperative TF interactions being likely necessary for CRM activity, the modeling has formalized this and systematically identified the range of interactions and the likely minimum level of higher-order TF cooperativity required for activity in both the VM and heart. Taken together, the modeling provides regulatory rules that explain how the same two TF motifs can give rise to activity in two different tissues (VM and heart) depending on the motif organization within the CRM.
Discussion
In this study, we stably integrated simple synthetic CRMs into transgenic Drosophila embryos, and then combined a quantitative analysis of enhancer activity with fractional site occupancy modeling to determine the contribution of motif organization to activity in two tissues. The results of these analyses highlight a number of organizational features that contribute to an enhancer's activity during development.
Tissue-specific sensitivity to motif organisation
While quantifying the activity of a simple ‘two-TF motif’ CRM (pMad-Tin), our results show that enhancer activity can exhibit very different sensitivity to motif organization in one tissue compared to another (Figure 7). Several mechanisms could account for this interesting effect, including different concentrations of the TF (i.e. pMad or Tin) in the different tissues, the availability of tissue-specific co-factors, or tissue-specific priming of the enhancer, which may increase the ease by which the enhancer is activated.
Fig. 7. Motif organisation affects the ability of two TF motifs to elicit activity in a tissue-specific manner. 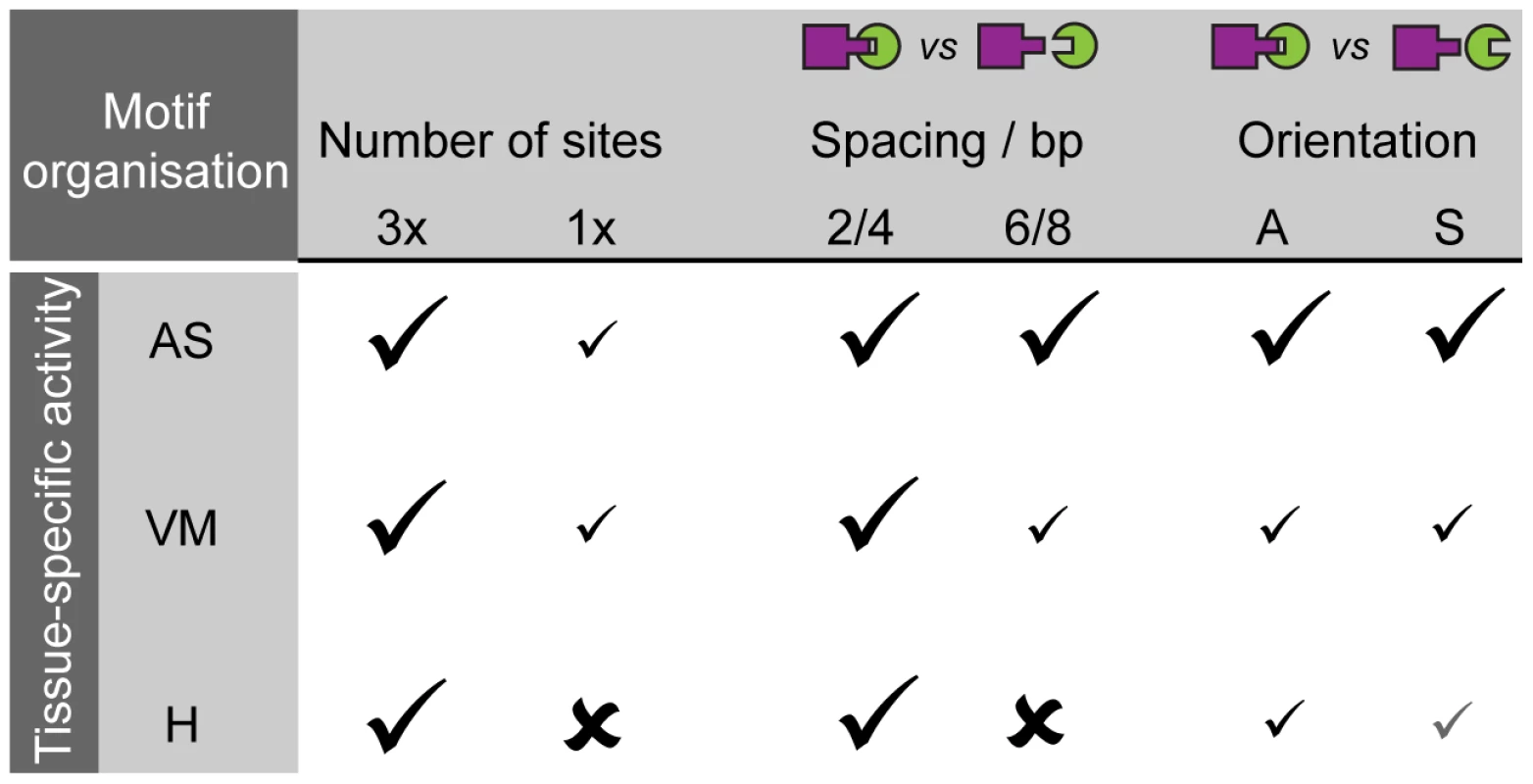
Schematic table summarizing our experimental data of CRM activity for pMad-Tin CRMs, which display tissue-specific sensitivity to motif organisation (spacing, orientation) in different tissues: amnioserosa (AS), visceral mesoderm (VM) and heart (H). The effect is not always an on-off switch, but can produce a range of responses (depicted with differential strength of check marks), which affects robustness of CRM activity. An elegant dissection of the endogenous spa enhancer demonstrated that completely rearranging the relative order and spacing of TF binding sites could switch its cell type-specific activity from cone cells to photoreceptors in the eye [20]. In comparison, the changes in motif organisation introduced in our study were much more subtle such that the relative order of motifs was completely preserved. Yet only changing the spacing or orientation of motifs altered the robustness of enhancer activity in a tissue-specific manner. This result indicates that small insertions or deletions in CRMs, that do not affect the TF motifs themselves, could still have significant effects on gene expression in one tissue while having no effect in another. A study examining the activity of neuroectoderm enhancers between Drosophila species supports this model, where reduced spacing between Dorsal and Twist sites results in broader neuroectodermal stripes of CRM activity, while increased motif spacing resulted in progressively narrower stripes [88]. Studies of both endogenous enhancers and the synthetic CRMs described here provide compelling evidence that the exact positioning of motifs within CRMs is crucial for the robustness of their activity in one tissue, while it may be largely dispensable in another. Different cell types can therefore interpret the same motif content of a given enhancer in different manners.
Non-additivity of CRM motifs: Variabile heart expression
The Drosophila heart is composed of two cell types, cardioblasts and pericardial cells, each of which requires the integration of many regulatory proteins for proper specification and diversification [72]. A characterized pericardial enhancer, eve MHE, for example, contains pMad and Tin binding sites in addition to sites for dTCF, Twi, Ets proteins, and Zfh1 [17], [89]. Given this complexity, it was surprising that a simple element built from pMad and Tin sites alone was sufficient to drive expression in the heart, albeit at a later developmental stage. Our analyses indicate that this activity is due to cooperativity binding between Tin and pMad, facilitated by a very specific motif arrangement. Using crystal structure data from close homologues of pMad [90] and Tin [91], we modelled the two TFs interaction on DNA, using a similar range of motif spacing (Figure S9). This 3D structural model indicates that it is possible for the DNA binding domains of these two proteins to both bind to DNA at a 2 bp spacing and to physically interact at a 2 bp and 4 bp spacing, but not at 6 bp spacing. Although done by homologue mapping, this structural data is consistent with our functional analyses of CRM activity, and further supports direct DNA binding cooperativity between these two TFs.
It is interesting to note, that although pMad and Tin sites are sufficient to drive expression in the heart from stage 13 to 14 (when placed in a limited motif arrangement), nature appears to use other enhancer configurations to regulate this critical function. There are two important aspects to this finding. First, heart activity arising from CRMs containing pMad and Tin sites alone is not robust. The enhancers are on ‘the edge’ of activation, where subtle changes in motif positioning or enhancer location switch activity between embryos and within embryos. Second, endogenous enhancers that are bound only by pMad and Tin – with no known input from other factors – direct expression in the dorsal mesoderm and not in the heart, at stage 10 [2], [92]. In our synthetic situation, pMad and Tin sites also drive robust expression in the dorsal mesoderm, in addition to variable weak expression in the heart. Therefore, although pMad and Tin sites alone are sufficient to drive heart activity in limited motif contexts, this mechanism is most likely not robust enough to be generally used to drive heart expression in vivo. This is consistent with recent studies showing that heart enhancer activity is elicited by the collective action of many TFs, which can occupy enhancers with considerable flexibility in terms of their motif content and configuration [2], [27]. Our pMad-Tin synthetic elements uncovered a very simple, although not very robust, alternative mechanism to regulate heart activity, and represent a nice example of how combinatorial regulation can lead to emergent expression profiles more than the simple sum of its parts.
Robustness of CRM activity
The expression of key developmental genes is generally buffered against variation in genetic backgrounds and environmental conditions. This may occur at many levels including RNA polymerase II pausing [93], [94] and the presence of partially redundant enhancers [95], [96], [97], [98]. However, robust expression may also be buffered by the motif content within an enhancer to ensure a stable regulatory function. CRMs, for example, often include additional binding sites to those that are minimal and necessary [99]. In the context of the pMad-Tin synthetic CRMs, the motif organization can also act to ensure robust activity. Our results demonstrate that even in situations where the composition of motifs and their relative arrangement are maintained, subtle changes in the spacing between the motifs could have dramatic effects on enhancer output. Interestingly, this effect seems to be very tissue-specific, with some tissues maintaining robust activity whilst others lost all enhancer activity.
Taken together, the data presented in this study demonstrate that subtle alterations in motif organization can affect the ability of different tissues to ‘read’ an enhancer, which in turn may allow each tissue to fine-tune enhancer activity based on fluctuations in its molecular components.
Materials and Methods
Design of the sequence used in the synthetic CRMs
Binding affinity models (PWMs) for Twi, Tin, Mef2, Bap, Bin, dTCF, pMad, and Doc2 and GATA were derived from ChIP-chip data analyses [2], [7]. The model for Pnt was generated using published footprints (Supplemental Methods (Text S1)). PWMs were first trimmed on each side to remove positions with an information content (IC) of less than 0.4 (trimming stopped at the first IC position > = 0.4). The sequence that best fits the PWM model was then determined for each trimmed PWM and is referred to hereafter as “TFBS”. All TFBS sequences used to design the synthetic CRMs are available in Table S1. For each CRM, a ‘neutral’ spacer sequence (a linker sequence placed between motifs) was heuristically determined by minimizing the sequence affinity for known TF PWM models (Supplemental Methods (Text S1)).
Cloning and transgenesis of the synthetic CRMs
Synthetic CRMs were generated from long oligonucleotides synthesized by Eurofins MWG Operon with compatible cohesive ends upon annealing for cloning. The forward and reverse strand oligonucleotides were phosphorylated, annealed and subsequently ligated into the pDUO2n [7], to generate stable, transgenic Drosophila lines using the phiC31 site-specific integrase [43]. The pH-Pelican [100] vector was use to test the robustness of enhancer activity at different genomic locations by random P-element transgenesis. The sequence of each CRM was verified to ensure that there were no synthesis errors and is provided in Table S2.
In situ hybridization and microscopy
CRM activity was assessed in embryos from transgenic flies using fluorescent in situ hybridization as described previously [101]. The following ESTs or full length cDNAs from the Drosophila Gene Collection (DGC) were used to generate probes: RE13967 (bap), RE40937 (doc2), RE20611 (dpp), GM04312 (dTCF), SD02611 (pnr) and AT15089 (twi). cDNAs used for bin and tin, lacZ, Mef2 and pnt were generous gifts from M. Frasch, U. Elling and M. Taylor respectively. Double or triple in situ hybridizations were performed using anti-fluorescein-POD, anti-DIG-POD and anti-biotin-POD antibodies (Roche, 1∶2000 dilution) and were developed sequentially with Cy3, fluorescein, and Cy5 tyramide signal amplification reagents (Perkin Elmer TSA kit). The lacZ expression patterns were imaged using Zeiss LSM 510 FCS or LSM 510 META confocal microscopes with A-Plan 10×/0.25 PH1 objective.
Automated image analysis for the quantification of tissue expressivity
Background subtraction of both the CRM and tissue-specific channels was performed using a morphological opening with disk size greater than the largest relevant VM region (typical disk size of 25 pixel radius). The tissue-specific subset of images (e.g. dpp or tin in situs) were segmented using the Ilastik software package (www.ilastik.org). The segmented images were analysed using Matlab. The segmented regions in each image were smoothened by performing dilation (disk size of 5 pixel radius) followed by equivalent erosion. An area threshold (>200 pixels) was used to remove small, segmented regions. Finally, the perimeter of the segmented regions was calculated (using Matlab function bwperim) and overlaid onto the CRM expression data.
Calculation of penetrance and expressivity
The penetrance was calculated for approximately 100 embryos for each of the twelve lines (Table S4). Bootstrapping was used to estimate the error in the penetrance measurements. The expressivity was calculated from around 16 carefully staged and positioned embryos (based on morphology and markers for VM (dpp) and heart (tin) tissues) for each line (Table S5). Embryos aligned dorsally were imaged and four regions of midgut VM were assigned, as shown in Figure 5A. The observed heart expression (also viewed dorsally) occurs in two rows of cells along either side of the embryo. We separated each row into an anterior and posterior segment, resulting in four heart regions (Figure S6A). The posterior segments, to the right of the PS7 VM region, correspond roughly to the heart proper, while the defined anterior heart segments correspond roughly to the region often referred to as the ‘aorta’. The penetrance in both the VM and heart was therefore measured as signal in one to four different regions of the tissue.
Modeling of CRM activity
A fractional occupancy model was used to analyze the experimental data [102]. Our methodology was similar to other thermodynamic models used to understand CRM activity in Drosophila (e.g. [26], [87]). The model had at most four parameters: two parameters described the relevant protein-protein interactions; and two parameters were used to distinguish sense and antisense binding effects. Mathematical details are provided in the Supplemental Methods (Text S1).
Ethics statement
This work is carried out in Drosophila, and was conducted in compliance with EMBL's guidelines.
Supporting Information
Zdroje
1. SpitzF, FurlongEE (2012) Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13 : 613–626.
2. JunionG, SpivakovM, GirardotC, BraunM, GustafsonEH, et al. (2012) A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148 : 473–486.
3. LiXY, MacArthurS, BourgonR, NixD, PollardDA, et al. (2008) Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27.
4. MacArthurS, LiXY, LiJ, BrownJB, ChuHC, et al. (2009) Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80.
5. SandmannT, GirardotC, BrehmeM, TongprasitW, StolcV, et al. (2007) A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev 21 : 436–449.
6. ZeitlingerJ, ZinzenRP, StarkA, KellisM, ZhangH, et al. (2007) Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21 : 385–390.
7. ZinzenRP, GirardotC, GagneurJ, BraunM, FurlongEE (2009) Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462 : 65–70.
8. ViselA, BlowMJ, LiZ, ZhangT, AkiyamaJA, et al. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457 : 854–858.
9. BlowMJ, McCulleyDJ, LiZ, ZhangT, AkiyamaJA, et al. (2010) ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet 42 : 806–810.
10. BernsteinBE, BirneyE, DunhamI, GreenED, GunterC, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489 : 57–74.
11. BonnS, ZinzenRP, GirardotC, GustafsonEH, Perez-GonzalezA, et al. (2012) Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44 : 148–156.
12. ErnstJ, KheradpourP, MikkelsenTS, ShoreshN, WardLD, et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473 : 43–49.
13. HeintzmanND, HonGC, HawkinsRD, KheradpourP, StarkA, et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459 : 108–112.
14. HeintzmanND, StuartRK, HonG, FuY, ChingCW, et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39 : 311–318.
15. RoyS, ErnstJ, KharchenkoPV, KheradpourP, NegreN, et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330 : 1787–1797.
16. BusserBW, HuangD, RogackiKR, LaneEA, ShokriL, et al. (2012) Integrative analysis of the zinc finger transcription factor Lame duck in the Drosophila myogenic gene regulatory network. Proc Natl Acad Sci U S A 109 : 20768–20773.
17. HalfonMS, CarmenaA, GisselbrechtS, SackersonCM, JimenezF, et al. (2000) Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103 : 63–74.
18. IpYT, ParkRE, KosmanD, YazdanbakhshK, LevineM (1992) dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev 6 : 1518–1530.
19. StruffiP, CoradoM, KaplanL, YuD, RushlowC, et al. (2011) Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development 138 : 4291–4299.
20. SwansonCI, EvansNC, BaroloS (2010) Structural rules and complex regulatory circuitry constrain expression of a Notch - and EGFR-regulated eye enhancer. Dev Cell 18 : 359–370.
21. D'AlonzoRC, SelvamuruganN, KarsentyG, PartridgeNC (2002) Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem 277 : 816–822.
22. ErivesA, LevineM (2004) Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci U S A 101 : 3851–3856.
23. InoueJ, SatoR, MaedaM (1998) Multiple DNA elements for sterol regulatory element-binding protein and NF-Y are responsible for sterol-regulated transcription of the genes for human 3-hydroxy-3-methylglutaryl coenzyme A synthase and squalene synthase. J Biochem 123 : 1191–1198.
24. PanneD, ManiatisT, HarrisonSC (2007) An atomic model of the interferon-beta enhanceosome. Cell 129 : 1111–1123.
25. ThanosD, ManiatisT (1995) Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83 : 1091–1100.
26. ZinzenRP, SengerK, LevineM, PapatsenkoD (2006) Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol 16 : 1358–1365.
27. JinH, StojnicR, AdryanB, OzdemirA, StathopoulosA, et al. (2013) Genome-wide screens for in vivo Tinman binding sites identify cardiac enhancers with diverse functional architectures. PLoS Genet 9: e1003195.
28. BrownCD, JohnsonDS, SidowA (2007) Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science 317 : 1557–1560.
29. LibermanLM, StathopoulosA (2009) Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev Biol 327 : 578–589.
30. RastegarS, HessI, DickmeisT, NicodJC, ErtzerR, et al. (2008) The words of the regulatory code are arranged in a variable manner in highly conserved enhancers. Dev Biol 318 : 366–377.
31. SwansonCI, SchwimmerDB, BaroloS (2011) Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr Biol 21 : 1186–1196.
32. ParkerDS, WhiteMA, RamosAI, CohenBA, BaroloS (2011) The cis-regulatory logic of Hedgehog gradient responses: key roles for gli binding affinity, competition, and cooperativity. Sci Signal 4: ra38.
33. BullaugheyK (2011) Changes in selective effects over time facilitate turnover of enhancer sequences. Genetics 187 : 567–582.
34. GertzJ, SiggiaED, CohenBA (2009) Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457 : 215–218.
35. SharonE, KalmaY, SharpA, Raveh-SadkaT, LevoM, et al. (2012) Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 30 : 521–530.
36. MelnikovA, MuruganA, ZhangX, TesileanuT, WangL, et al. (2012) Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 30 : 271–277.
37. PatwardhanRP, HiattJB, WittenDM, KimMJ, SmithRP, et al. (2012) Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol 30 : 265–270.
38. ZichnerT, GarfieldDA, RauschT, StutzAM, CannavoE, et al. (2013) Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome research 23 : 568–579.
39. MassourasA, WaszakSM, Albarca-AguileraM, HensK, HolcombeW, et al. (2012) Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS genetics 8: e1003055.
40. WeischenfeldtJ, SymmonsO, SpitzF, KorbelJO (2013) Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet 14 : 125–138.
41. BonnS, FurlongEE (2008) cis-Regulatory networks during development: a view of Drosophila. Curr Opin Genet Dev 18 : 513–520.
42. CiglarL, FurlongEE (2009) Conservation and divergence in developmental networks: a view from Drosophila myogenesis. Curr Opin Cell Biol 21 : 754–760.
43. BischofJ, MaedaRK, HedigerM, KarchF, BaslerK (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 104 : 3312–3317.
44. DavidsonEH (2010) Emerging properties of animal gene regulatory networks. Nature 468 : 911–920.
45. LifanovAP, MakeevVJ, NazinaAG, PapatsenkoDA (2003) Homotypic regulatory clusters in Drosophila. Genome Res 13 : 579–588.
46. GoteaV, ViselA, WestlundJM, NobregaMA, PennacchioLA, et al. (2010) Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res 20 : 565–577.
47. HanesSD, RiddihoughG, Ish-HorowiczD, BrentR (1994) Specific DNA recognition and intersite spacing are critical for action of the bicoid morphogen. Mol Cell Biol 14 : 3364–3375.
48. Simpson-BroseM, TreismanJ, DesplanC (1994) Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell 78 : 855–865.
49. SzymanskiP, LevineM (1995) Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J 14 : 2229–2238.
50. FulkersonE, EstesPA (2010) Common motifs shared by conserved enhancers of Drosophila midline glial genes. J Exp Zool B Mol Dev Evol 316 : 61–75.
51. RothbacherU, BertrandV, LamyC, LemaireP (2007) A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development 134 : 4023–4032.
52. VincentJP, KassisJA, O'FarrellPH (1990) A synthetic homeodomain binding site acts as a cell type specific, promoter specific enhancer in Drosophila embryos. EMBO J 9 : 2573–2578.
53. BaroloS (2006) Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25 : 7505–7511.
54. RieseJ, YuX, MunnerlynA, EreshS, HsuSC, et al. (1997) LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell 88 : 777–787.
55. BaroloS, WalkerRG, PolyanovskyAD, FreschiG, KeilT, et al. (2000) A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell 103 : 957–969.
56. GussKA, NelsonCE, HudsonA, KrausME, CarrollSB (2001) Control of a genetic regulatory network by a selector gene. Science 292 : 1164–1167.
57. ZaffranS, KuchlerA, LeeHH, FraschM (2001) biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev 15 : 2900–2915.
58. JakobsenJS, BraunM, AstorgaJ, GustafsonEH, SandmannT, et al. (2007) Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network. Genes Dev 21 : 2448–2460.
59. SunB, HurshDA, JacksonD, BeachyPA (1995) Ultrabithorax protein is necessary but not sufficient for full activation of decapentaplegic expression in the visceral mesoderm. EMBO J 14 : 520–535.
60. YangX, van BeestM, CleversH, JonesT, HurshDA, et al. (2000) decapentaplegic is a direct target of dTcf repression in the Drosophila visceral mesoderm. Development 127 : 3695–3702.
61. MartyT, MullerB, BaslerK, AffolterM (2000) Schnurri mediates Dpp-dependent repression of brinker transcription. Nat Cell Biol 2 : 745–749.
62. SallerE, BienzM (2001) Direct competition between Brinker and Drosophila Mad in Dpp target gene transcription. EMBO Rep 2 : 298–305.
63. MullenAC, OrlandoDA, NewmanJJ, LovenJ, KumarRM, et al. (2011) Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147 : 565–576.
64. TrompoukiE, BowmanTV, LawtonLN, FanZP, WuDC, et al. (2011) Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147 : 577–589.
65. NewfeldSJ, ChartoffEH, GraffJM, MeltonDA, GelbartWM (1996) Mothers against dpp encodes a conserved cytoplasmic protein required in DPP/TGF-beta responsive cells. Development 122 : 2099–2108.
66. ImmergluckK, LawrencePA, BienzM (1990) Induction across germ layers in Drosophila mediated by a genetic cascade. Cell 62 : 261–268.
67. FernandezBG, AriasAM, JacintoA (2007) Dpp signalling orchestrates dorsal closure by regulating cell shape changes both in the amnioserosa and in the epidermis. Mech Dev 124 : 884–897.
68. BruneauBG, NemerG, SchmittJP, CharronF, RobitailleL, et al. (2001) A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106 : 709–721.
69. DurocherD, CharronF, WarrenR, SchwartzRJ, NemerM (1997) The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J 16 : 5687–5696.
70. GajewskiK, ZhangQ, ChoiCY, FossettN, DangA, et al. (2001) Pannier is a transcriptional target and partner of Tinman during Drosophila cardiogenesis. Dev Biol 233 : 425–436.
71. ZaffranS, XuX, LoPC, LeeHH, FraschM (2002) Cardiogenesis in the Drosophila model: control mechanisms during early induction and diversification of cardiac progenitors. Cold Spring Harb Symp Quant Biol 67 : 1–12.
72. CrippsRM, OlsonEN (2002) Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol 246 : 14–28.
73. BodmerR (1993) The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118 : 719–729.
74. AzpiazuN, FraschM (1993) tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev 7 : 1325–1340.
75. ReimI, FraschM (2005) The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 132 : 4911–4925.
76. GajewskiK, FossettN, MolkentinJD, SchulzRA (1999) The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development 126 : 5679–5688.
77. FraschM (1995) Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature 374 : 464–467.
78. LockwoodWK, BodmerR (2002) The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech Dev 114 : 13–26.
79. HerranzH, MorataG (2001) The functions of pannier during Drosophila embryogenesis. Development 128 : 4837–4846.
80. KlinedinstSL, BodmerR (2003) Gata factor Pannier is required to establish competence for heart progenitor formation. Development 130 : 3027–3038.
81. GibsonG (1996) Epistasis and pleiotropy as natural properties of transcriptional regulation. Theor Popul Biol 49 : 58–89.
82. AckersGK, JohnsonAD, SheaMA (1982) Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci U S A 79 : 1129–1133.
83. AmitR, GarciaHG, PhillipsR, FraserSE (2011) Building enhancers from the ground up: a synthetic biology approach. Cell 146 : 105–118.
84. LaikenSL, GrossCA, Von HippelPH (1972) Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J Mol Biol 66 : 143–155.
85. OppenheimAB, KobilerO, StavansJ, CourtDL, AdhyaS (2005) Switches in bacteriophage lambda development. Annu Rev Genet 39 : 409–429.
86. PtashneM (2005) Regulation of transcription: from lambda to eukaryotes. Trends Biochem Sci 30 : 275–279.
87. FakhouriWD, AyA, SayalR, DreschJ, DayringerE, et al. (2010) Deciphering a transcriptional regulatory code: modeling short-range repression in the Drosophila embryo. Mol Syst Biol 6 : 341.
88. CrockerJ, TamoriY, ErivesA (2008) Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol 6: e263.
89. KnirrS, FraschM (2001) Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol 238 : 13–26.
90. BabuRajendranN, PalasingamP, NarasimhanK, SunW, PrabhakarS, et al. (2010) Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors. Nucleic Acids Res 38 : 3477–3488.
91. LaRonde-LeBlancNA, WolbergerC (2003) Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev 17 : 2060–2072.
92. XuX, YinZ, HudsonJB, FergusonEL, FraschM (1998) Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev 12 : 2354–2370.
93. BoettigerAN, LevineM (2009) Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325 : 471–473.
94. LevineM (2011) Paused RNA polymerase II as a developmental checkpoint. Cell 145 : 502–511.
95. PerryMW, BoettigerAN, BothmaJP, LevineM (2010) Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol 20 : 1562–1567.
96. HongJW, HendrixDA, LevineMS (2008) Shadow enhancers as a source of evolutionary novelty. Science 321 : 1314.
97. PerryMW, CandeJD, BoettigerAN, LevineM (2009) Evolution of insect dorsoventral patterning mechanisms. Cold Spring Harb Symp Quant Biol 74 : 275–279.
98. FrankelN, DavisGK, VargasD, WangS, PayreF, et al. (2010) Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466 : 490–493.
99. LudwigMZ, Manu, KittlerR, WhiteKP, KreitmanM (2011) Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet 7: e1002364.
100. BaroloS, CarverLA, PosakonyJW (2000) GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29 : 726, 728, 730, 732.
101. FurlongEE, AndersenEC, NullB, WhiteKP, ScottMP (2001) Patterns of gene expression during Drosophila mesoderm development. Science 293 : 1629–1633.
102. AyA, ArnostiDN (2011) Mathematical modeling of gene expression: a guide for the perplexed biologist. Crit Rev Biochem Mol Biol 46 : 137–151.
Štítky
Genetika Reprodukčná medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



