-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
RNA-directed DNA methylation (RdDM) is required for transcriptional silencing of transposons and other DNA repeats in Arabidopsis thaliana. Although previous research has demonstrated that the SET domain-containing SU(VAR)3–9 homologs SUVH2 and SUVH9 are involved in the RdDM pathway, the underlying mechanism remains unknown. Our results indicated that SUVH2 and/or SUVH9 not only interact with the chromatin-remodeling complex termed DDR (DMS3, DRD1, and RDM1) but also with the newly characterized complex composed of two conserved Microrchidia (MORC) family proteins, MORC1 and MORC6. The effect of suvh2suvh9 on Pol IV-dependent siRNA accumulation and DNA methylation is comparable to that of the Pol V mutant nrpe1 and the DDR complex mutant dms3, suggesting that SUVH2 and SUVH9 are functionally associated with RdDM. Our CHIP assay demonstrated that SUVH2 and SUVH9 are required for the occupancy of Pol V at RdDM loci and facilitate the production of Pol V-dependent noncoding RNAs. Moreover, SUVH2 and SUVH9 are also involved in the occupancy of DMS3 at RdDM loci. The putative catalytic active site in the SET domain of SUVH2 is dispensable for the function of SUVH2 in RdDM and H3K9 dimethylation. We propose that SUVH2 and SUVH9 bind to methylated DNA and facilitate the recruitment of Pol V to RdDM loci by associating with the DDR complex and the MORC complex.
Published in the journal: The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci. PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1003948
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003948Summary
RNA-directed DNA methylation (RdDM) is required for transcriptional silencing of transposons and other DNA repeats in Arabidopsis thaliana. Although previous research has demonstrated that the SET domain-containing SU(VAR)3–9 homologs SUVH2 and SUVH9 are involved in the RdDM pathway, the underlying mechanism remains unknown. Our results indicated that SUVH2 and/or SUVH9 not only interact with the chromatin-remodeling complex termed DDR (DMS3, DRD1, and RDM1) but also with the newly characterized complex composed of two conserved Microrchidia (MORC) family proteins, MORC1 and MORC6. The effect of suvh2suvh9 on Pol IV-dependent siRNA accumulation and DNA methylation is comparable to that of the Pol V mutant nrpe1 and the DDR complex mutant dms3, suggesting that SUVH2 and SUVH9 are functionally associated with RdDM. Our CHIP assay demonstrated that SUVH2 and SUVH9 are required for the occupancy of Pol V at RdDM loci and facilitate the production of Pol V-dependent noncoding RNAs. Moreover, SUVH2 and SUVH9 are also involved in the occupancy of DMS3 at RdDM loci. The putative catalytic active site in the SET domain of SUVH2 is dispensable for the function of SUVH2 in RdDM and H3K9 dimethylation. We propose that SUVH2 and SUVH9 bind to methylated DNA and facilitate the recruitment of Pol V to RdDM loci by associating with the DDR complex and the MORC complex.
Introduction
DNA methylation is an important epigenetic modification that contributes to transposable element silencing, genome stability, and gene regulation in plants and animals [1]–[3]. In Arabidopsis thaliana, DNA methylation can be established through a well-characterized RNA-directed DNA methylation (RdDM) pathway [1], [2]. The RdDM pathway is involved in the silencing of transposable elements and DNA repeats [1], [2], the stability of the genome [4], [5], the development of gametophytes and embryos [6]–[8], and the inheritance of stress-induced transcriptional silencing [9].
In the RdDM pathway, the multi-subunit DNA-dependent RNA polymerase IV (Pol IV) collaborates with RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) to produce double-stranded RNAs, which are subsequently cleaved into 24-nt siRNAs (small interfering RNAs) by DICER-LIKE 3 (DCL3) [10]–[13]. The recruitment of Pol IV to chromatin requires SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1)/DNA-BINDING TRANSCRIPTION FACTOR 1 (DTF1) [13], [14]. Pol IV-dependent siRNAs are loaded onto ARGONAUTE 4 (AGO4) in the cytoplasm and then transported into the nucleus to complete the assembly of the RdDM effector complex [15]. The multi-subunit DNA-dependent RNA polymerase V (Pol V) produces long noncoding RNAs, which act as scaffold RNAs for the assembly of the RdDM effector complex [16]–[18]. The DDR complex, which is composed of DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), and RNA-DIRECTED DNA MEETHYLATION 1 (RDM1), is required for the recruitment of Pol V to RdDM loci and for the accumulation of Pol V-produced noncoding RNAs [17]–[23]. RNA-DIRECTED DNA METHYLATION 4 (RDM4)/DEFECTIVE IN MERISTEM SILENCING 4 (DMS4), an IWR1-like protein, is likely to act as a general transcription factor that is shared by Pol II, Pol IV, and Pol V [24]–[26]. KOW-CONTAINING TRANSCRIPTION FACTOR 1 (KTF1)/SUPPRESSOR OF TY INSERTION 5-LIKE (SPT5L) can bind to Pol V-produced noncoding RNAs and is recruited to chromatin in parallel with AGO4 [27]–[29]. INVOLVED IN DE NOVO 2 (IDN2) and IDN2 PARALOG 1 and 2 (IDP1 and IDP2) can form a complex required for RdDM [30]–[32]. IDN2 physically associates with the SWI/SNF chromatin-remodeling complex, which is required for noncoding RNA-mediated transcriptional silencing [33]. DOMAIN REARRANGED METHYLTRANSFERASE 2 (DRM2) is the main DNA methyltransferase that catalyzes DNA methylation at RdDM loci [34], [35]. MORC1 and MORC6, two members of the conserved Microrchidia (MORC) adenosine triphosphatase (ATPase) family, were thought to be required for decondensation of pericentromeric heterochromatin in a DNA methylation-independent manner [36]. However, another two reports indicated that MORC6/DEFECTIVE IN MERISTEM SILENCING 11 (DMS11) is involved in RNA-directed DNA methylation and transcriptional silencing [37], [38]. Further study is required to clarify the functional mechanism of the MORC family proteins in transcriptional silencing.
In Arabidopsis, H3K9 methylation is catalyzed by the conserved SU(VAR)3–9 homologs (SUVHs) and related proteins (SUVRs) [2], [39]. The SUVHs contain a catalytic SET domain and an N-terminal SRA (SET - or RING-associated) domain [39]. The active histone H3K9 methyltransferases SUVH4/KRYPTONITE (KYP), SUVH5, and SUVH6 are not only required for H3K9 dimethylation (H3K9me2) but also for DNA methylation [40], [41]. The connection between DNA methylation and histone H3K9me2 is dependent on the SRA domain in the SUVHs that directly bind to methylated DNA [42], [43]. CHROMOMETHYLASE 3 (CMT3), a plant-specific DNA methyltransferase, can directly bind to H3K9me2-containing nucleosomes and connect the histone H3K9me2 with DNA methylation at CHG sites [44]. At RdDM loci, canonical RdDM components including Pol IV, Pol V, and AGO4 are required for both DNA methylation and histone H3K9me2 [17], [45], [46]. Previous reports demonstrated that SUVH2 and SUVH9 are involved in RNA-directed DNA methylation and transcriptional silencing [47], [48], but the underlying mechanism is unknown.
In this study, we found that SUVH2 and SUVH9 associate with the DDR complex and the newly characterized MORC1-MORC6 complex and act as adaptor proteins to mediate Pol V occupancy at RdDM loci. The conserved catalytic active site of SUVH2 is not required for the function of SUVH2 in RdDM and histone H3K9me2. The results revealed the functional mechanism of SUVH2 and SUVH9 in the RdDM pathway.
Results
SUVH2 and SUVH9 associate with DDR components and MORC family proteins in vivo
SUVH2 and SUVH9 were previously demonstrated to be involved in RNA-directed DNA methylation [47], [48], but the underlying mechanism is unclear. To understand the role of SUVH2 and SUVH9 in RdDM, we generated the native promoter-driven SUVH2-3xMyc and SUVH9-3xFlag transgenic plants. The stably expressed SUVH2-3xMyc and SUVH9-3xFlag transgenic lines were used for affinity purification of the proteins that associate with SUVH2 and SUVH9. Mass spectrometric assay revealed that SUVH2 and SUVH9 were the most abundant proteins in affinity purification of SUVH2-3xMyc and SUVH9-3xFlag, respectively (Table 1). Moreover, two RdDM components DMS3 and DRD1 were identified in affinity purification of SUVH2-3xMyc, whereas MORC6/DMS11 and DMS3 were identified in affinity purification of SUVH9-3xFlag (Table 1). The presence of RdDM components DMS3 and DRD1 in affinity purification of SUVH2-3xMyc and SUVH9-3xFlag suggests that SUVH2 and SUVH9 are involved in RdDM by physically associating with RdDM components. MORC6/DMS11 was recently demonstrated to be involved in RNA-directed DNA methylation and transcriptional silencing [37], [38]. The finding of MORC6 in affinity purification of SUVH9-3xMyc suggests that MORC6 interacts with SUVH9. We carried out affinity purification of MORC6-3xFlag in MORC6-3xFlag transgenic plants and identified SUVH9 by mass spectrometric assay, confirming that SUVH9 can interact with MORC6 in vivo (Table 1). To confirm the interaction between SUVH2 and DMS3, we performed co-immunoprecipitation (co-IP) in SUVH2-3xMyc transgenic plants using either anti-Myc antibody or anti-DMS3 antibody. The results indicated that SUVH2-3xMyc and DMS3 were co-precipitated not only by anti-DMS3 antibody but also by anti-Myc antibody (Figure 1A; Figure S1A). The interaction between SUVH9 and MORC6 was also demonstrated by co-IP in the transgenic plants harboring both SUVH9-Flag and MORC6-Myc transgenes (Figure 1B). We therefore concluded that SUVH2 and/or SUVH9 can physically associate with the canonical RdDM components in vivo. However, we could not detect the interaction between SUVH9 and DMS3 by co-IP (Figure S1B, S1C), suggesting that the interaction is either weak or highly locus-specific.
Fig. 1. Detection of the protein interaction by co-IP and gel filtration analyses. 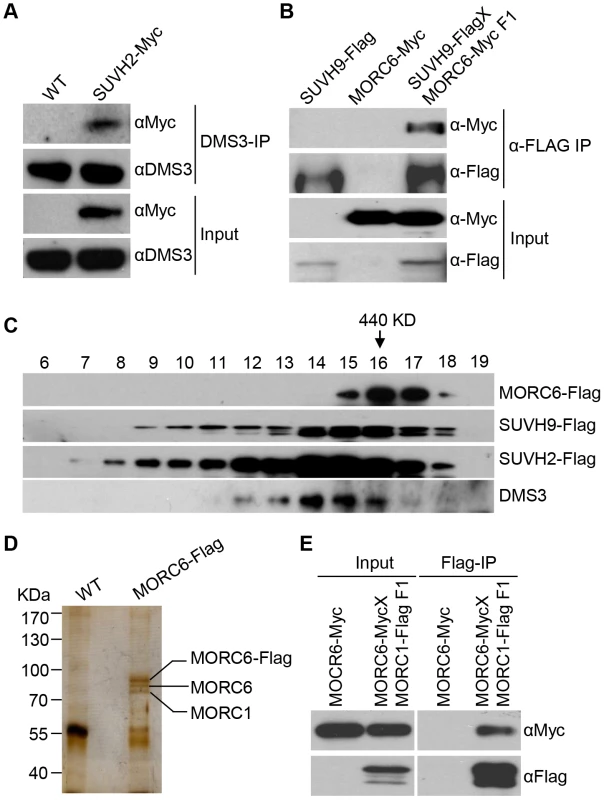
(A) Detection of the interaction between SUVH2-3xMyc and DMS3 by co-IP. The protein extract of SUVH2-3xMyc transgenic plants was immunoprecipitated by anti-Myc antibody. The precipitate was subjected to Western blotting with the antibodies against the Myc epitope and endogenous DMS3. (B) The interaction between SUVH9-3xFlag and MORC6-3xMyc as determined by co-IP. The F1 offspring generated from the cross between SUVH9-3xFlag and MORC6-3xMyc transgenic plants were used for co-IP. (C) The elution profile of MORC6-3xFlag, SUVH2-3xMyc, SUVH9-3xFlag, and DMS3 as determined by gel filtration. Antibodies specific for the Flag epitope and endogenous DMS3 were used for Western blotting. The arrow indicates the fraction of a 440-KDa standard protein. (D) The protein extracts from the wild type and MORC6-3xFlag transgenic plants were affinity purified by anti-Flag antibody. The purified proteins were run on an SDS-PAGE gel and subjected to silver staining. The three separated bands at ∼70–100 KDa were cut out for mass spectrometric analysis. (E) The interaction between MORC1 and MORC6 as determined by co-IP. Tab. 1. List of affinity co-purified proteins of SUVH2-Myc, SUVH9-Flag, and MORC6-Flag. 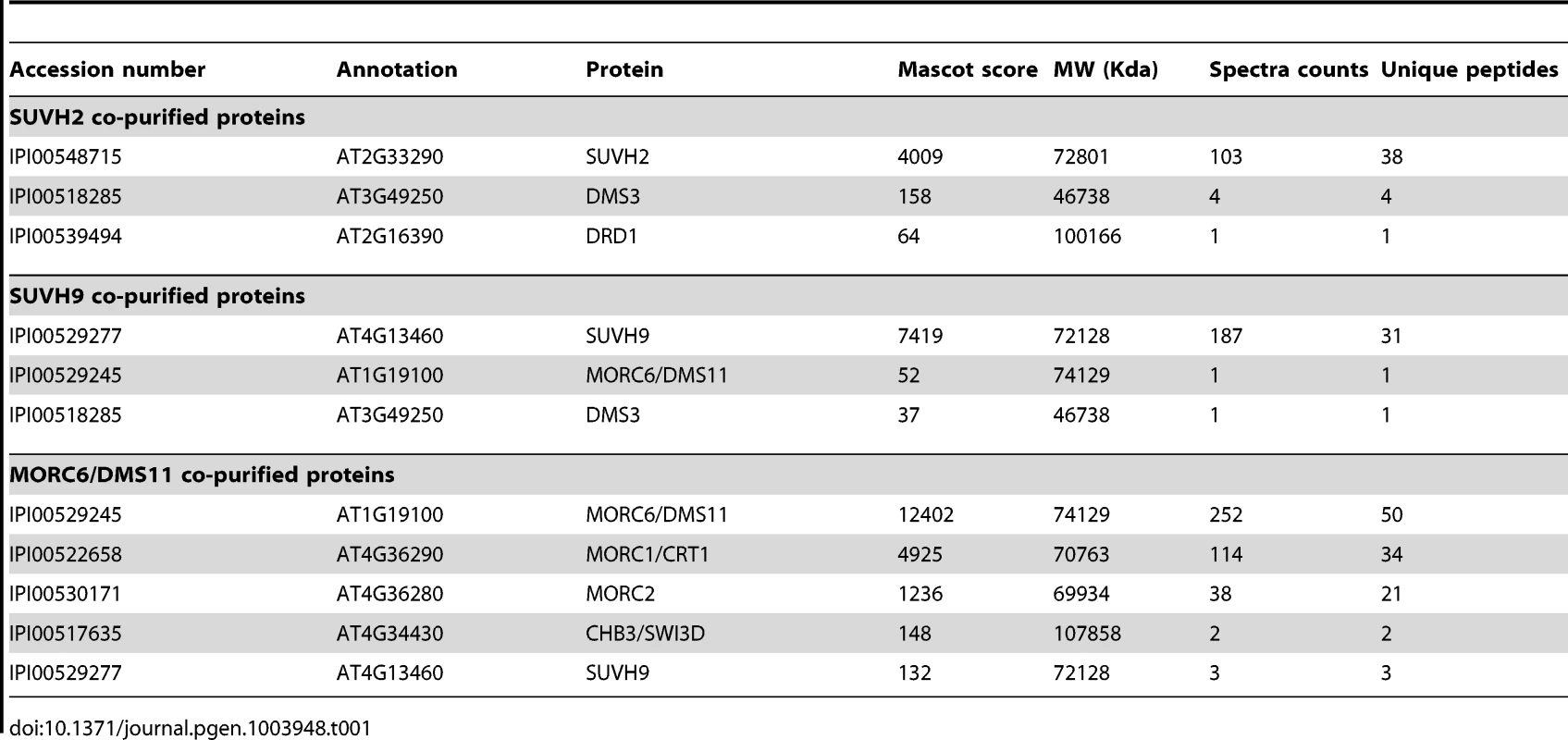
We carried out gel filtration to determine whether SUVH2, SUVH9, and related RdDM components act as protein complexes in vivo. For gel filtration, the protein extracts from the wild-type plants and SUVH2-3xMyc, SUVH9-3xFlag, and MORC6-3xFlag transgenic plants were separately eluted and then subjected to Western blotting (Figure 1C). The elution profile of these protein extracts revealed that each protein (SUVH2-3xMyc, SUVH9-3xFlag, and MORC6-3xFlag) was eluted in the form of a high-molecular-weight protein complex rather than as a monomer. SUVH2-3xMyc, SUVH9-3xFlag, and DMS3 were co-eluted with the peak at the size between 440 KDa and 669 KDa, which is consistent with the mass spectrometric analysis indicating that both SUVH2 and SUVH9 associated with DMS3 (Figure 1C). The elution peak of MORC6-3xFlag was <440 KDa, indicating that the complex containing MORC6 is smaller than the complex containing SUVH2, SUVH9, or DMS3 (Figure 1C). The small molecular size of the MORC6 elution peak indicated that MORC6 may form a different complex that does not include SUVH2, SUVH9, and DMS3 (Figure 1C).
Mass spectrometric analysis indicated that affinity purification of MORC6-3xFlag produced a large number of peptides corresponding to MORC1 and another MORC family protein, MORC2 (AT4G36280), in MORC6-3xFlag transgenic plants (Table 1). Therefore, MORC1, MORC2, and MORC6 may form a tight complex in vivo. Affinity purification of MORC6-3xFlag generated three specific silver-stained bands on SDS-PAGE gel (Figure 1D). The three bands were separately cut from the gel and subjected to mass spectrometric analysis. The results indicated the three bands from top to bottom correspond to the MORC6-3xFlag fusion protein, the endogenous MORC6 protein, and the endogenous MORC1 protein (Figure 1D). These results suggest that MORC6 not only forms a homodimer with another MORC6 but also forms a heterodimer with its homologs MORC1 and MORC2 in vivo. The formation of the MORC1-MORC6 heterodimer was confirmed by co-IP between MORC1-3xFlag and MORC6-3xMyc in the transgenic plants harboring both MORC1-3xFlag and MORC6-3xMyc transgenes (Figure 1E). Furthermore, the interaction between MORC6 and MORC1 or MORC2 was examined by yeast two-hybrid assay. The results demonstrated that MORC1, MORC2, and MORC6 can form a homodimer or heterodimer (Figure S2), which is consistent with the result of the gel filtration assay indicating that MORC6 acts in a distinct protein complex in vivo (Figure 1C).
We conducted yeast two-hybrid assay to determine whether SUVH2 and SUVH9 interact with RdDM components and found that both SUVH2 and SUVH9 can interact with DMS3 (Figure 2A, 2B), which is consistent with the results from mass spectrometric analysis (Table 1). Moreover, in yeast two-hybrid assay, SUVH2 weakly interacts with MORC1 rather than with MORC2 and MORC6, whereas SUVH9 interacts with all the three MORC family proteins (Figure 2C, 2D). The interaction of SUVH9 is weaker with MORC2 than with MORC1 and MORC6 (Figure 2D). The interaction between SUVH9 and MORC6 detected by yeast two-hybrid assay is consistent with the results from the affinity purification of both SUVH9-3xFlag and MORC6-3xFlag (Table 1). However, the interaction between SUVH9 and the other two MORC family proteins MORC1 and MORC2 was not found by mass spectrometric assay of SUVH9-3xFlag affinity purification (Table 1). Similarly, the interaction between SUVH2 and MORC1 was also not detected by mass spectrometric assay (Table 1). The failure to detect these interactions is likely due to the low expression levels of these proteins as well as the weak interactions. We constructed truncated SUVH2 and SUVH9 sequences for yeast two-hybrid assay to determine the key domains of SUVH2 and SUVH9 that are required for the interaction with the RdDM components (Figure S3A). Yeast two-hybrid assay indicated that the truncated SUVH2 sequence without its C-terminal SET domain (SUVH2-c) can still interact with DMS3, whereas it cannot interact with MORC1 (Figure S3B). The other truncated versions of SUVH2 (SUVH2-a, SUVH2-b) interact with neither DMS3 nor MORC1 (Figure S3B). Likely, the truncated SUVH9 version without its SET domain (SUVH9-c) can interact with DMS3 but not with MORC1 and MORC6, and the other SUVH9 truncated versions (SUVH9-b, SUVH9-c) can interact with none of the three proteins (Figure S3C). The results suggest that the full length of SUVH2 and SUVH9 is required for their interaction with MORC1 and MORC6, whereas the sequence containing SRA and Pre-SET domains is sufficient for SUVH2 and SUVH9 to interact with DMS3.
Fig. 2. Detection of the interaction of SUVH2 and SUVH9 with RdDM components by yeast two-hybrid assay. 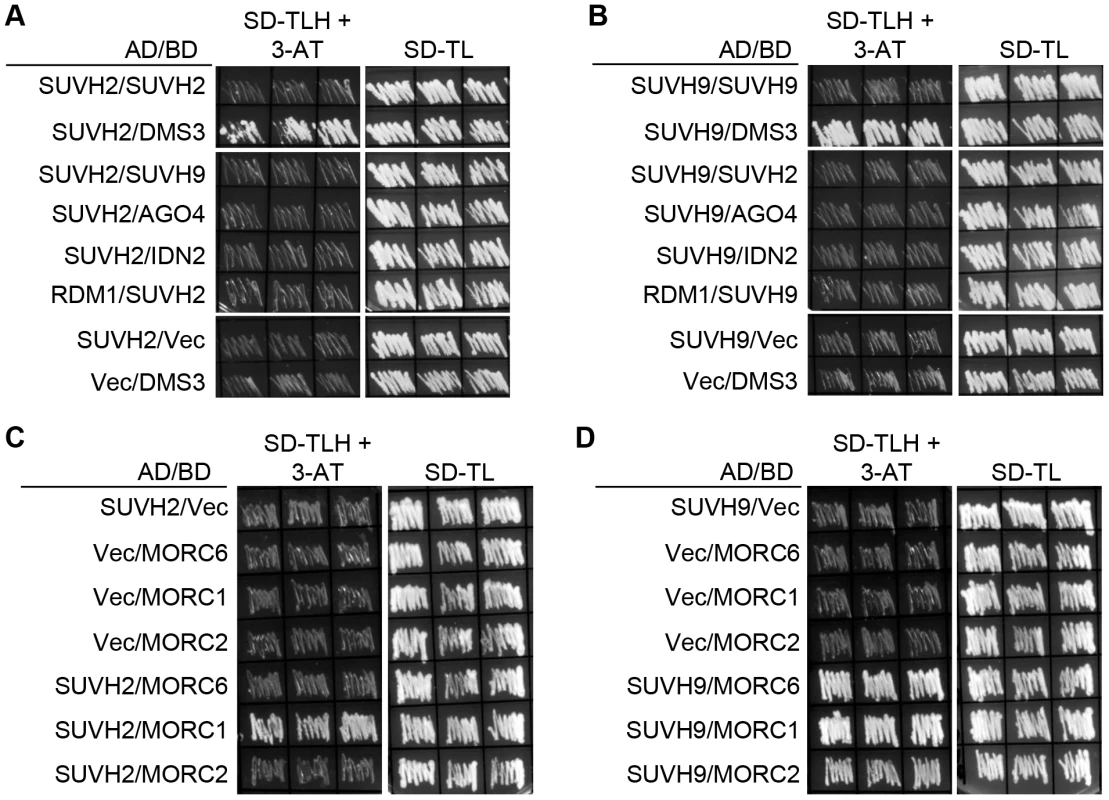
The yeast strains harboring GAL4-AD and GAL4-BD fusion constructs were streaked on plates with indicated media. SD-TLH, the synthetic dropout medium without Trp, Leu, and His. SD-TL, the synthetic dropout medium without Trp and Leu. “Vec” represents the empty pGADT7 or pGBKT7 vector. (A) The interaction of SUVH2 with SUVH9, AGO4, IDN2, and RDM1. (B) The interaction of SUVH9 with SUVH2, DMS3, AGO4, IDN2, and RDM1. (C) The interaction of SUVH2 with the MORC family proteins MORC1, MORC2, and MORC6. (D) The interaction of SUVH9 with the MORC family proteins. Effect of suvh2 and suvh9 on accumulation of Pol IV-dependent siRNAs
We carried out small RNA deep sequencing to compare the global effect of suvh2, suvh9, and suvh2suvh9 relative to that of the canonical RdDM mutants nrpd1, nrpe1, and dms3 on the accumulation of Pol IV-dependent siRNAs. We obtained at least 2.6 million uniquely genome-matched small RNA reads for the small RNA libraries of the wild type, suvh2, suvh9, suvh2suvh9, nrpd1, nrpe1, and dms3 (Table S1). Pol IV-dependent 24-nt-siRNA regions were defined as the genomic regions in which the abundance of 24-nt siRNAs was significantly reduced in nrpd1 relative to the wild type. Consistent with previous reports [49]–[51], we found that Pol IV-dependent siRNAs are predominantly enriched in centrometric regions where highly repetitive DNA sequences exist, and are also present in intergenic regions of each chromosome, especially when the regions contain transposable elements (Figure 3A; Table S2). Both NRPE1 and DMS3 are required for the production of Pol V-dependent noncoding RNAs and act at the downstream step of the RdDM pathway [17], [18]. Our small RNA data indicated that NRPE1 and DMS3 predominantly contribute to accumulation of the Pol IV-dependent siRNAs produced from intergenic regions at two arms of each chromosome, but that both proteins have little impact on the Pol IV-dependent siRNAs produced from highly duplicated DNA repeats in centromeric regions (Figure 3A; Table S2), which is consistent with previous reports [50], [51]. Accumulation of Pol IV-dependent siRNAs was only marginally reduced in the suvh2 and suvh9 single mutants and the reduction was significantly enhanced in the suvh2suvh9 double mutant (Figure 3A). The effect of suvh2suvh9 on accumulation of Pol IV-dependent siRNAs is comparable to that of nrpe1 and dms3 at the whole-genome level (Figure 3A), suggesting that SUVH2 and SUVH9 function redundantly at the downstream step of the RdDM pathway.
Fig. 3. Small RNA analyses by small RNA Northern blotting and small RNA deep sequencing. 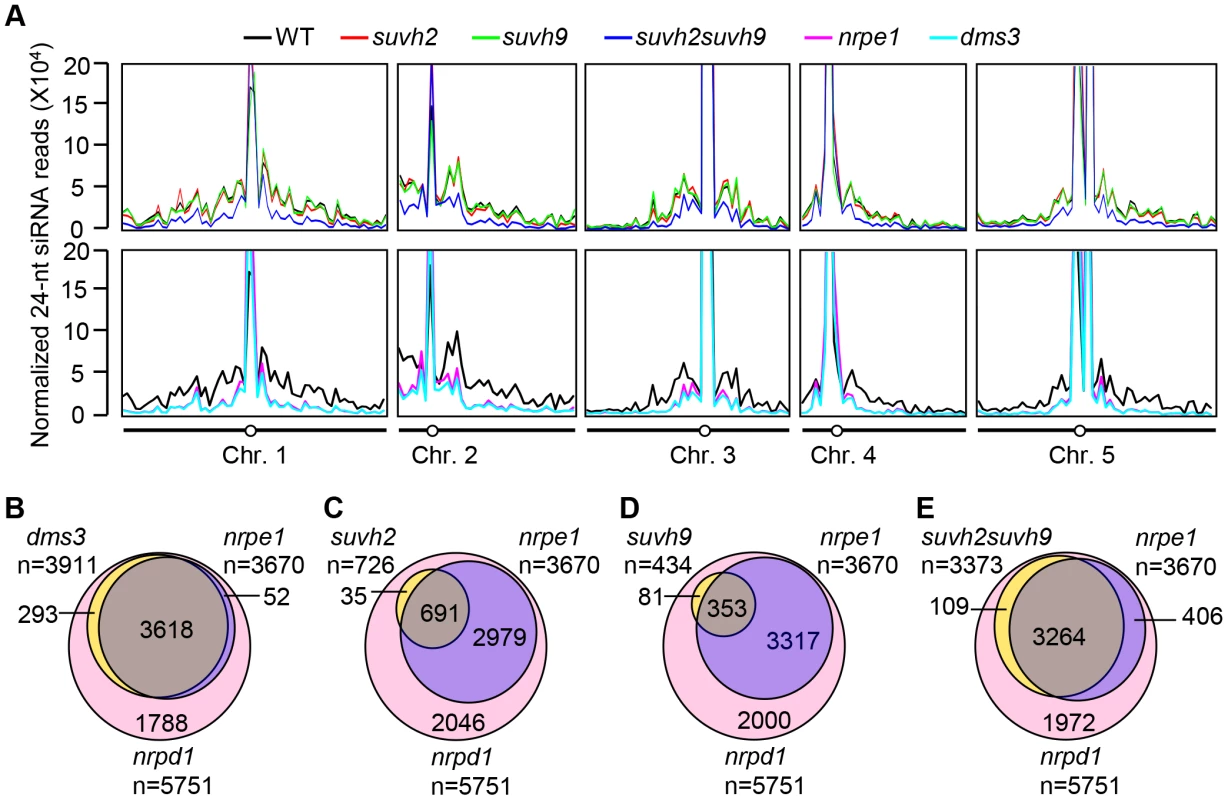
(A) Diagrams show the distribution of Pol IV-dependent 24-nt siRNAs across all five Arabidopsis chromosomes in the wild type, suvh2, suvh9, suvh2suvh9, nrpe1, and dms3. Normalized 24-nt siRNA reads per ten million in consecutive 500-Kb genomic regions of each chromosome are indicated by the Y coordinate, whereas all five Arabidopsis chromosomes are shown along the X coordinate. (B–D) Venn diagrams indicate the numbers and overlaps of siRNA regions that show decreased siRNAs in dms3 (B), suvh2 (C), suvh9 (D), and suvh2suvh9. (E) compared to nrpd1 and nrpe1. The results are from the small RNA deep sequencing data for the wild type, nrpd1, nrpe1, dms3, suvh2, suvh9, and suvh2suvh9. In total, we identified 5751 siRNA regions where the abundance of 24-nt siRNAs is strongly decreased (>5-fold decrease) in nrpd1 compared to the wild type (Figure 3B, 3C, 3D, 3E). In 63.8% (3670/5751) of the Pol IV-dependent siRNA regions, 24-nt siRNAs are significantly decreased in nrpe1 (Figure 3B, 3C, 3D, 3E). Thus, Pol IV-dependent siRNA regions were divided into two groups: Pol V-independent siRNA regions and Pol V-dependent siRNA regions. Of the 3670 Pol V-dependent siRNA regions, 98.6% (3618/3670) have 24-nt siRNAs that are also decreased in dms3, whereas only 14.1% (293/2081) of Pol IV-dependent and Pol V-independent siRNA regions are significantly decreased in 24-nt siRNAs in dms3 (Figure 3B). The highly similar effect of nrpe1 and dms3 on 24-nt siRNAs supported the inference that DMS3 facilitates the recruitment of Pol V and mediates the production of Pol V-dependent noncoding RNAs at a later step in the RdDM pathway.
In the suvh2 and suvh9 single mutants, 12.6% (726/5751) and 7.5% (434/5751) of the Pol IV-dependent siRNA regions show a significant decrease in 24-nt siRNAs, respectively (Figure 3C, 3D). The low effect of suvh2 and suvh9 on Pol IV-dependent siRNAs may account for the functional redundancy between SUVH2 and SUVH9. Although the effect of the suvh2 and suvh9 single mutants is quite low, we still find that most of these SUVH2 - and SUVH9-dependent siRNA regions (691/726 for SUVH2 and 353/434 for SUVH9) belong to Pol IV - and Pol V-dependent siRNA regions (Figure 3C, 3D). The results suggest that like Pol V, SUVH2 and SUVH9 may act downstream of the primary siRNA biogenesis in the RdDM pathway. In the suvh2suvh9 double mutant, a high percentage of Pol IV - and Pol V-dependent siRNAs regions (88.9%, 3264/3670) show a decrease in 24-siRNAs, whereas only 5.2% (109/2081) of Pol IV-dependent and Pol V-independent siRNAs regions have decreased 24-nt siRNAs (Figure 3E). The effect of suvh2suvh9 is comparable to that of the Pol V mutant nrpe1 and of the DDR complex mutant dms3 (Figure 3B, 3E)
Based on the small RNA deep sequencing data, we tested the effect of suvh2 and suvh9 on well-known Pol IV-dependent siRNAs, which include siRNAs from 180 bp centromeric repeats (180 bp CEN), AtGP1, AtSN1, and MEA-ISR loci. As expected, all these Pol IV-dependent siRNAs were blocked in the Pol IV mutant nrpd1, whereas AtSN1 siRNA and MEA-ISR siRNA but not 180 bp CEN siRNA and AtGP1 siRNA were reduced in the Pol V mutant nrpe1 and the DDR complex mutant dms3 (Figure S4). We found that AtSN1 siRNA and MEA-ISR siRNA were weakly reduced in suvh2 but not in suvh9. In the suvh2suvh9 double mutant, AtSN1 siRNA and MEA-ISR siRNA were reduced to much more extent than that in suvh2 (Figure S4). The effect of suvh2suvh9 on accumulation of AtSN1 siRNA and MEA-ISR siRNA is similar to that of nrpe1 and dms3 (Figure S4), suggesting that the function of SUVH2 and SUVH9 in Pol V-dependent siRNA accumulation is partially redundant. Neither suvh2 and suvh9 single mutants nor suvh2suvh9 double mutant reduced the Pol V-independent siRNAs, 180 bp CEN siRNA and AtGP1 siRNA (Figure S4). These results support the inference that SUVH2 and SUVH9 act together with Pol V and the DDR complex at a later step in the RdDM pathway.
SUVH2 and SUVH9 are required for Pol V-dependent noncoding RNAs and transcriptional silencing
We carried out quantitative RT-PCR to determine the effect of suvh2 and suvh9 on transcriptional silencing at RdDM loci. The results indicated that the RNA transcript levels of solo LTR, AtGP1, SDC, and AtSN1 are markedly induced in the canonical RdDM mutants including nrpe1, drd1, dms3, and nrpd1 (Figure 4A). The RNA transcript levels of solo LTR, AtGP1, and AtSN1 are either not affected or are weakly induced in either the suvh2 or suvh9 single mutant (Figure 4A). In the suvh2suvh9 double mutant, however, the induction of the RNA transcript levels of the three loci are significantly enhanced compared to the levels in the single mutants (Figure 4A). The SDC transcript is intermediately induced in suvh9 rather than in suvh2 and the transcript is drastically enhanced in the suvh2suvh9 double mutant (Figure 4A). These results suggest that SUVH2 and SUVH9 function redundantly in transcriptional silencing.
Fig. 4. RNA transcript analysis by semi-quantitative RT-PCR and RNA deep sequencing. 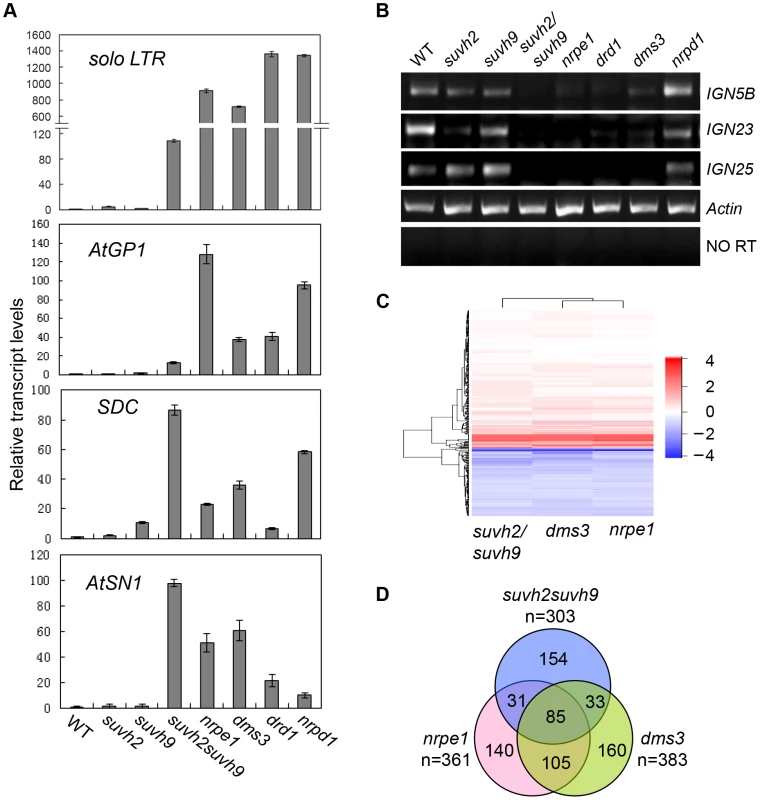
(A) The effect of suvh2, suvh9, and svuh2suvh9 on transcriptional silencing as determined by quantitative RT-PCR. The transcript levels of RdDM loci solo LTR, AtGP1, SDC, AtSN1 was tested. The actin gene was amplified as an internal control. (B) Pol V-dependent noncoding RNAs from IGN5B, IGN23, and IGN25 loci were measured by semiquantitative RT-PCR. No RT indicates the amplification of the actin gene using total RNAs as template without reverse transcription. (C) The genome-wide effect of suvh2suvh9, nrpe1, and dms3 on gene expression as determined by RNA deep sequencing. Heat map of log2 (Mutant/WT) was shown for each mutant. (D) The numbers and overlaps of the upregulated genes in suvh2suvh9, nrpe1, and dms3 relative to the wild type are shown in Venn diagram. Small RNA analysis indicated that the effect of suvh2suvh9 on Pol IV-dependent siRNAs is similar to that of the Pol V mutant, nrpe1 (Figure 3A, 3E). Moreover, both SUVH2 and SUVH9 can interact with DMS3 (Table 1; Figure 1A, 2A, 2B), which acts as a subunit of the DDR complex and is required for Pol V chromatin association at RdDM loci [22], [23]. Therefore, it is possible that SUVH2 and SUVH9 are involved in the accumulation of Pol V-dependent noncoding RNAs. The effect of suvh2 and suvh9 on the Pol V-dependent transcript AtSN1B was previously noted [48]. We determined the effect of suvh2, suvh9, and suvh2suvh9 on the Pol V-dependent noncoding RNAs at IGN5B, IGN23, and IGN25 by RT-PCR (Figure 4B; Figure S5). The results showed that the RNA transcript levels of the three tested loci were markedly reduced in nrpe1, drd1, and dms3 (Figure 4B; Figure S5), suggesting that the RNA transcripts of these loci are dependent on Pol V. The Pol V-dependent transcripts IGN5B, IGN23, and IGN25 were either not affected or marginally reduced in the suvh2 and suvh9 single mutants but were markedly reduced in the suvh2suvh9 double mutant (Figure 4B; Figure S5), which demonstrated that SUVH2 and SUVH9 are required for the production of Pol V-dependent noncoding RNAs at RdDM loci. SUVH2 and SUVH9 are functionally redundant in Pol V-dependent RNA accumulation.
Previous reports suggest that disruption of the RdDM machinery leads to derepression not only of transposable elements but also of their flanking genes [2], [32], [33]. We performed RNA deep sequencing to compare the effect of suvh2suvh9 vs. nrpe1 and dms3 on gene expression at the whole-genome level. For each RNA library, we obtained at least 14.7 million RNA reads that were mapped to the Arabidopsis genome (Table S3). The Heat map results indicated that the genome-wide effect of suvh2suvh9 on gene expression is generally similar to that of nrpe1 and dms3 (Figure 4C). The genes upregulated in the RdDM mutants nrpe1 and dms3 are thought to be RdDM loci (Table S2). We found that 361, 383, and 303 genes that are significantly (log2 (Mutant/WT)>1; P<0.01) upregulated in nrpe1, dms3, and suvh2suvh9, respectively (Table S4; Figure 4D), suggesting that the three mutants have a similar effect on gene expression at the whole-genome level. Of the 383 genes upregulated in dms3, 49.6% (190/383) are also upregulated in nrpe1 (Figure 4D), supporting the inference that DMS3 acts together with NRPE1 in the RdDM pathway. Of the 303 genes upregulated in suvh2suvh9, nearly half (49.2%, 149/303) are upregulated in either nrpe1 or dms3 (Figure 4D). The data are consistent with the finding that SUVH2 and SUVH9 associate with DMS3 and mediate transcriptional silencing through the RdDM pathway.
SUVH2 and SUVH9 are involved in the association of Pol V and DMS3 with chromatin at RdDM loci
Given that Pol V-dependent noncoding RNAs are markedly decreased in the suvh2suvh9 double mutant, we determined whether suvh2suvh9 reduces Pol V chromatin association at RdDM loci. The NRPE1-Flag transgene was introduced and expressed in WT, dms3, and suvh2suvh9 equivalently (Figure S6A). By CHIP assay using anti-Flag antibody, we found that NRPE1 was enriched at the RdDM loci including solo LTR, IGN5, IGN22, IGN25, and IGN26, and that the enrichment was decreased in the DDR complex mutant dms3 (Figure 5A), which is consistent with previous reports [17], [18]. Interestingly, our results indicated that the enrichment of NRPE1 on chromatin at these RdDM loci was also significantly decreased in suvh2suvh9 (Figure 5A), suggesting that SUVH2 and SUVH9 are required for Pol V chromatin association. The results demonstrate that SUVH2 and SUVH9 not only physically associate with the DDR complex but also act together with the complex to facilitate Pol V occupancy at RdDM loci.
Fig. 5. Effect of suvh2 and suvh9 on the occupancy of Pol V and the DDR complex at RdDM loci. 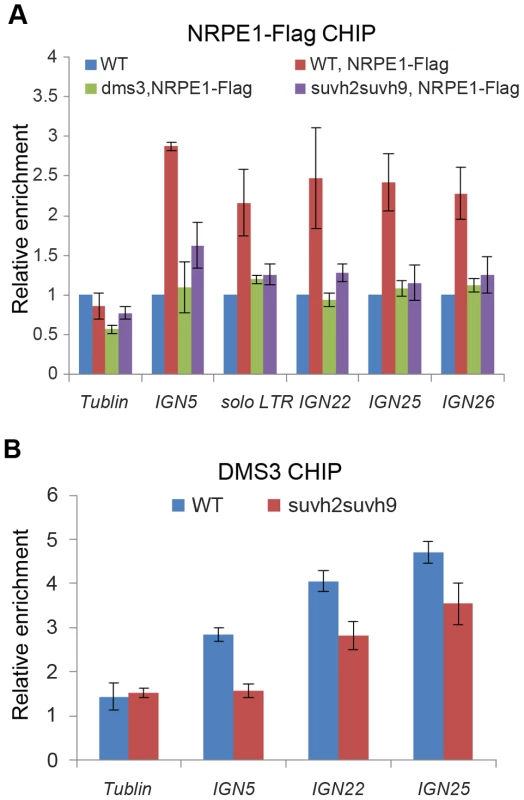
(A) The effect of suvh2suvh9 on Pol V occupancy at RdDM loci was determined by CHIP assay with anti-Flag antibody. The enrichment of the largest subunit of Pol V, NRPE1, was measured at IGN5, solo LTR, IGN22, IDN25, and IGN26. The abundance of NRPE1 on the actin gene was used as an internal negative control. Enrichment of NRPE1-Flag on RdDM loci and the tublin gene was normalized by the abundance of NRPE1-Flag on the actin gene. (B) The effect of suvh2suvh9 on the occupancy of the DDR complex at RdDM loci was determined by CHIP assay with anti-DMS3 antibody. Enrichment of the DDR complex component DMS3 was determined in the wild type and suvh2suvh9. Relative enrichment of DMS3 is shown. The DDR complex was previously demonstrated to be co-purified with Pol V [22], whereas no Pol V subunits were identified in SUVH2 and SUVH9 co-purified proteins (Table 1). The results suggest that the role of SUVH2 and SUVH9 in Pol V occupancy is probably indirectly through their action on the DDR complex. Thus, we determined whether suvh2suvh9 affects the association of the DDR complex with chromatin at RdDM loci. We carried out DMS3 CHIP assay in the wild type as well as in suvh2suvh9 by using anti-DMS3 antibody. The specificity of the antibody was confirmed by Western blotting assay (Figure S6B). The CHIP assay demonstrated that DMS3 occupancy at RdDM loci including IGN5, IGN22, and IGN25 is significantly reduced in suvh2suvh9 relative to the wild type (Figure 5B), suggesting that SUVH2 and SUVH9 enable the association of the DDR complex with chromatin, thereby facilitating the recruitment of Pol V to RdDM loci.
SUVH2 and SUVH9 are required for DNA methylation and H3K9me2 at RdDM loci
The effect of suvh2 and suvh9 on DNA methylation at RdDM loci was evaluated previously [47], [52]. At the genome-wide level, suvh2 reduces DNA methylation at a large number of RdDM loci, whereas suvh9 affects only a very small portion of RdDM loci [52], [53]. We determined the effect of suvh2 and suvh9 on DNA methylation by chop-PCR (Figure 6A). The results indicated that DNA methylation of AtSN1, IGN5, IGN25, and solo LTR was either not reduced or only marginally reduced in the suvh2 and suvh9 single mutants but the reduction was greatly enhanced in the suvh2suvh9 double mutant (Figure 6A). The reduction of DNA methylation in suvh2suvh9 was comparable to that in the canonical RdDM mutants nrpe1, dms3, and drd1 (Figure 6A). The results demonstrate that SUVH2 and SUVH9 are functionally redundant in DNA methylation at RdDM loci.
Fig. 6. Effect of suvh2 and suvh9 on DNA methylation and histone H3K9me2 at RdDM targets. 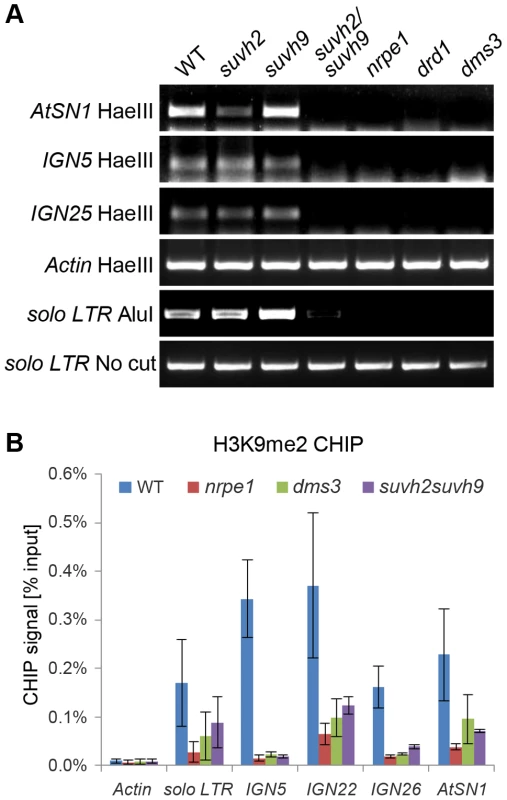
(A) The effect of suvh2, suvh9, and suvh2suvh9 on DNA methylation as indicated by chop-PCR. DNA methylation was determined for AtSN1, IGN5, IGN25, and solo LTR. Genomic DNA from the wild type, nrpd1, nrpe1, dms3, suvh2, suvh9, and suvh2suvh9 was cleaved by the DNA methylation-sensitive restriction enzyme HaeIII or AluI and subjected to semiquantitative PCR. The actin gene, which lacks HaeIII recognition site, was amplified as an internal control. The solo LTR locus was amplified using uncut genomic DNA as template for a control. (B) Effect of suvh2suvh9 on histone H3K9me2 was measured by CHIP assay with anti-H3K9me2 antibody. The RdDM mutants nrpe1 and dms3 were included as controls, in which the H3K9me2 levels at indicated RdDM loci were decreased. The actin gene, which lacks detectable H3K9me2, was used as a negative control. Previous studies revealed that disruption of canonical RdDM components can lead to reduction of repressive histone H3K9me2 at RdDM loci [17], [45], [46]. To determine whether H3K9me2 at these loci is related to the putative histone methyltransferases SUVH2 and SUVH9, we preformed chromatin immunoprecipitation (CHIP) assay using anti-H3K9me2 antibody. The results indicated that the H3K9me2 levels at solo LTR, IGN5, IGN22, IGN26, and AtSN1 loci are decreased in the Pol V mutant nrpe1 and the DDR complex mutant dms3 (Figure 6B), which is consistent with previous results [17], [46]. Moreover, we found that the H3K9me2 levels of these loci are markedly decreased in the suvh2suvh9 double mutant (Figure 6B), suggesting that SUVH2 and SUVH9 are required for H3K9me2 at RdDM loci.
The catalytic active site of SUVH2 is dispensable for its function in RdDM and H3K9me2
The SET domain is responsible for histone methyltransferase activity and is conserved not only in plants but also in animals and fungi [39]. However, the SET domain-containing proteins SUVH2 and SUVH9 have no detectable histone methyltransferase activity by in vitro assay and the overall histone H3K9me2 level is not affected in the suvh2suvh9 double mutant [47]. It is necessary to determine whether the possible histone methyltransferase activity is required for the function of SUVH2 and SUVH9 in RdDM and H3K9me2 in vivo. We mutated two conserved residues (SUVH2-H596K and SUVH2-P600A) in the catalytic active site of the SET domain in SUVH2 and transformed the mutated SUVH2 sequences into suvh2suvh9 (Figure 7A). The transgenic plants harboring the stably expressed wild-type or mutant SUVH2 transgene were used for complementation assay. Previous studies suggest that the two residues SUVH2-H596 and SUVH2-P600 are highly conserved and are essential for the catalytic activity of the SET domain [54], [55]. If histone methyltransferase activity is required for the role of SUVH2 in RdDM and H3K9me2, disruption of the conserved catalytic active site in SUVH2 is expected to block its function in vivo. DNA methylation assay demonstrated that the DNA methylation defect in IGN5 and IGN23 caused by suvh2 was complemented by the two mutated SUVH2 transgene sequences as well as by the wild-type SUVH2 transgene (Figure 7B). Moreover, H3K9me2 CHIP assay indicated that the two mutations in SUVH2 also have no effect on the in vivo function of SUVH2 in H3K9me2 at the RdDM loci IGN5 and AtSN1 (Figure 7C). The results suggest that disruption of the putative histone methyltransferase activity of SUVH2 has no effect on the in vivo role of SUVH2 in RdDM and histone H3K9me2, supporting the finding that SUVH2 and SUVH9 are not active histone methyltransferases [47]. Histone H3K9me2 at RdDM loci is catalyzed by other histone H3K9 methyltransferases rather than SUVH2 and SUVH9.
Fig. 7. Mutation of the conserved SET domain in SUVH2 has no effect on DNA methylation and H3K9me2 at RdDM loci. 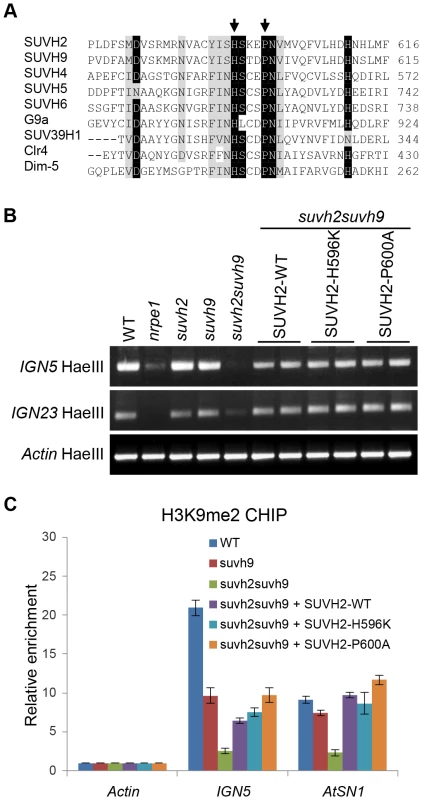
(A) Alignment of the catalytic active site in the Arabidopsis SET domain proteins SUVH2, SUVH9, SUVH4, SUVH5, and SUVH6, and their homologs including G9a and SUV39H1 in human, Clr4 in fission yeast, and Dim-5 in Neurospora. The conserved residues SUVH2-H596 and SUVH2-P600 that were subjected to point mutation are marked with arrows. (B) Site-directed mutagenesis was carried out to introduce the SUVH2-H596K and SUVH2P600A mutations in the SET domain of SUVH2. The construct harboring either wild-type or mutant SUVH2 sequence was transformed into suvh2suvh9 for complementation assay. For DNA methylation assay, genomic DNA was digested by the DNA methylation-sensitive restriction enzyme HaeIII followed by PCR. The DNA methylation levels at RdDM loci IGN5 and IGN23 were determined. (C) H3K9me2 CHIP assay was performed to test whether the mutant SUVH2 sequences complement the H3K9me2 defect caused by suvh2 in suvh2suvh9 at RdDM loci IGN5 and AtSN1. Enrichment of H3K9me2 was normalized by the abundance of H3K9me2 on the actin gene. Discussion
SUVH2 and SUVH9 function redundantly at a downstream step of the RdDM pathway [47], [48]. Previous reports suggest that disruption of the downstream RdDM components including NRPE1 and DMS3 indirectly affects accumulation of Pol IV-dependent 24-nt siRNAs [50], [51]. Our small RNA deep sequencing data indicate that the effect of suvh2suvh9 on 24-nt siRNA accumulation is comparable to that of nrpe1 and dms3 (Figure 3A, 3B, 3E; Figure S4), supporting the functional association of SUVH2 and SUVH9 with the downstream RdDM components NRPE1 and DMS3. We demonstrate that SUVH2 and SUVH9 can also physically associate with the components of the DDR complex and mediate Pol V occupancy at RdDM loci (Table 1; Figure 1, 2, 5A, 6). In the RdDM pathway, Pol V produces long noncoding RNAs that act as scaffold RNAs for the assembly of the RdDM effector complex [17]. The DDR complex is involved in the recruitment of Pol V to chromatin and facilitates the production of Pol V-dependent noncoding RNAs [18], [22], [23]. The association of SUVH2 and SUVH9 with the DDR complex is consistent with the function of SUVH2 and SUVH9 in Pol V occupancy at RdDM loci. The SRA domain of SUVH2 and SUVH9 can bind to methylated DNA [47], but the role of the methylated DNA-binding ability in RdDM remains elusive. Given the finding that SUVH2 and SUVH9 can physically associate with the DDR complex (Table 1; Figure 1, 2), the binding of SUVH2 and SUVH9 to methylated DNA is probably required for the occupancy of the DDR complex on chromatin. Our DMS3 CHIP assay indicates that the occupancy of DMS3 at RdDM sites is significantly reduced in suvh2suvh9 relative to the wild type (Figure 5B). Thus, reduced Pol V occupancy at RdDM sites in suvh2suvh9 is likely due to reduced occupancy of DMS3. We propose that the binding of SUVH2 and SUVH9 to methylated DNA mediates the occupancy of the DDR complex at RdDM loci, thereby recruiting Pol V to the loci (Figure 8). Noncoding RNAs produced by Pol V interact with 24-nt siRNAs bound by AGO4 and recruit the de novo DNA methyltransferase DRM2 to mediate DNA methylation [17], [18], [21]. Thus, SUVH2 and SUVH9 act as a linker between DNA methylation and Pol V transcription in the RdDM pathway (Figure 8).
Fig. 8. Model for the role of SUVH2, SUVH9 in RdDM. 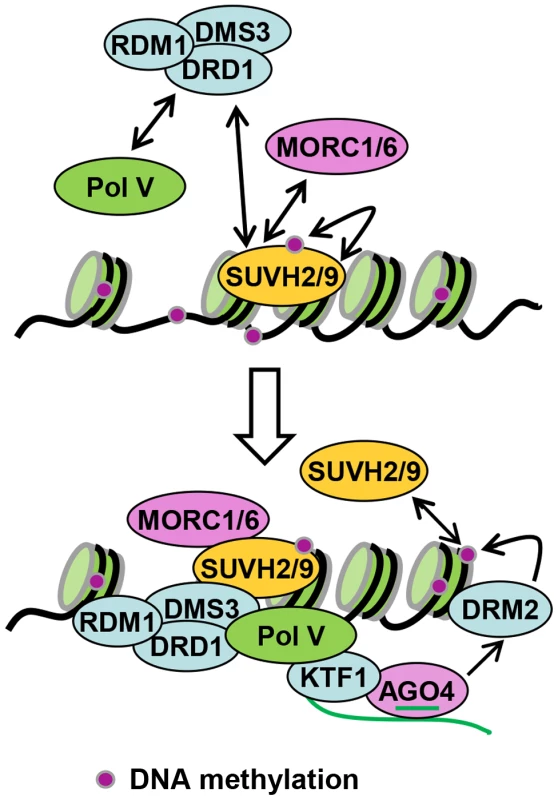
SUVH2 and SUVH9 bind to methylated DNA at RdDM loci and are at least partially required for the occupancy of the DDR complex at the loci. The DDR complex associates with Pol V and facilitates the recruitment of Pol V to RdDM loci. Pol V-produced noncoding RNAs interact with 24nt-siRNAs bound by AGO4, recruiting the de novo DNA methyltransferase DRM2 for DNA methylation. The methylated DNA is further bound by SUVH2 and SUVH9, resulting in a self-reinforcing loop that facilitates maintenance of DNA methylation at RdDM loci. Although SUVH2 and SUVH9 have been demonstrated to be required for H3K9me2 at RdDM sites (Figure 6B), SUVH2 and SUVH9 have no detectable histone methyltransferase activity by in vitro histone methyltransferase assay [47]. In Arabidopsis, another three SUVH proteins SUVH4/KYP, SUVH5, and SUVH6 have histone H3K9 methyltransferase activity and are required for maintenance of histone H3K9 methylation at the whole-genome level [2], [40], [41]. It is possible that H3K9me2 at RdDM sites is catalyzed by these active histone H3K9 methyltransferases. Our results indicate that disruption of the putative catalytic active site of SUVH2 has no effect on the function of SUVH2 in RdDM and H3K9me2 (Figure 7A, 7B, 7C), supporting the inference that SUVH2 and SUVH9 act as adaptor proteins that are involved in Pol V recruitment but not as active histone methyltransferases. H3K9me2 of RdDM loci requires not only SUVH2 and SUVH9 but also other RdDM components including NRPD1, NRPE1, and AGO4 [17], [45], [46], suggesting that SUVH2 and SUVH9 are implicated in H3K9me2 through the RdDM pathway.
SUVH2 and/or SUVH9 can associate not only with the DDR complex but also with the MORC family proteins (Table 1; Figure 1, 2). A recent report suggests that involvement of MORC1 and MORC6/DMS11 in transcriptional silencing is not related to DNA methylation [36], whereas another two reports indicate that MORC6/DMS11 is a new component of the RdDM machinery [37], [38]. Our results demonstrate that MORC6/DMS11 and its homologs MORC1 and MORC2 associate with SUVH9 (Table 1; Figure 1B, 1C, 2D), which is required for Pol V occupancy in the RdDM pathway (Figure 5A). MORC6/DMS11 was previously demonstrated to be involved in the production of Pol V-dependent RNAs [37]. Our results suggest that involvement of MORC6/DMS11 in the production of Pol V-dependent RNAs is related to the physical association of SUVH2 and SUVH9 with MORC6/DMS11. SUVH2, SUVH9, the DDR complex, and the MORC family proteins act together and mediate Pol V occupancy at RdDM loci, thereby facilitating the production of Pol V-dependent noncoding RNAs (Figure 8). Moreover, the MORC family proteins can also mediate the change of chromatin superstructure and lead to transcriptional silencing without involvement of DNA methylation [36]. Our mass spectrometric assay has identified CHB3/SWI3D, a subunit of the SWI/SNF-type chromatin-remodeling complex, as a MORC6-associating protein in affinity purification of MORC6-3xFlag (Table 1). The SWI/SNF-type chromatin-remodeling complex was recently demonstrated to act in Pol V-dependent noncoding RNA-mediated transcriptional silencing by establishing positioned nucleosomes on specific genomic loci [33]. Therefore, in addition to the role of the MORC family proteins in the RdDM pathway, they may also act with the SWI/SNF-type chromatin-remodeling complex to contribute to RNA-mediated transcriptional silencing through a DNA methylation-independent manner.
Our results demonstrate that MORC1, MORC2, and MORC6 can form a homodimer or a heterodimer in vivo (Table 1; Figure 1D, 1E; Figure S2). A recent report suggests that the GHLK ATPase domain-containing MORC6/DMS11 can physically interact with the SMC hinge domain-containing protein DMS3 and form a functional analog of an authentic SMC protein, which functions in a dimer and binds complementary DNA strands during sister chromatid cohesion, chromatin condensation, and DNA repair [37], [56]. Given that DMS3 can form a homodimer as described previously [20], a MORC dimer and a DMS3 dimer may form a functional SMC analog in the RdDM pathway, which may help produce Pol V-dependent noncoding RNAs. However, no DMS3 peptide is identified by affinity purification of MORC6-3xFlag (Table 1), which does not support the tight in vivo interaction between the MORC dimer and the DMS3 dimer.
Overall, our results indicate that SUVH2 and SUVH9 are physically and functionally associated with the DDR complex and the MORC family proteins and facilitate Pol V occupancy at RdDM loci, concluding that SUVH2 and SUVH9 are canonical components of the RdDM machinery. SUVH2 and SUVH9 may bind to methylated DNA at RdDM loci and interact with the DDR complex at the loci, thereby recruiting Pol V to produce noncoding RNAs. Moreover, we demonstrate that the putative histone methyltransferase activity of SUVH2 is dispensable for the function of SUVH2 in RdDM and H3K9me2 at RdDM loci. Involvement of SUVH2 in H3K9me2 at RdDM loci is via its action in the RdDM pathway. Further study is required to elucidate how SUVH2, SUVH9, the DDR complex, and the MORC family proteins are coordinated to mediate Pol V occupancy at RdDM loci.
Materials and Methods
Plant materials, cloning, and generation of transgenic plants
The Arabidopsis materials included the wild-type Col-0, suvh2 (Gabi_kat_516A07), suvh9 (SALK_048033), suvh2suvh9 [48], nrpd1-3 (SALK_128428), nrpe1-11 (SALK_029919C), dms3 (Salk_125019C), drd1-6 [19], morc1 (Salk_008610), and morc6 (GK-599B06-023140). The seedlings were grown on MS medium plates under long-day conditions (16 h day time and 8 h night time) at 22°C. Two-week-old seedlings were directly used for the study or transplanted into soil when adult plants or flower tissues were needed.
The Flag- and/or Myc-tagged SUVH2, SUVH9, MORC1, and MORC6 genomic sequences were constructed into the modified binary pCAMBIA1305 vector or pRI909 vector. The DNA primers used for constructing were listed in Table S5. In these constructs, the genes were driven by their own native promoters, and the tags were fused to the C-terminal of the corresponding proteins. The constructs were transformed to wild-type plants by agrobacteria infection. T1 transgenic plants were grown on MS medium plates supplemented with 20 mg/ml hygromycin or 50 mg/ml kanamycin. The resistant seedlings were transplanted into soil and grown for the study. For complementation assay, the construct harboring the native promoter-driven SUVH2-3xMyc sequence was transformed into suvh2suvh9. The wild-type SUVH2 genomic sequence was subjected to site-directed mutagenesis, introducing the SUVH2-H596K and SUVH2-P600A mutations in the SUVH2-3xMyc construct.
Affinity purification and mass spectrometric analysis
A 3-g quantity of flower tissue from SUVH2-3xMyc, SUVH9-3xFlag, or MORC6-3xFlag transgenic plants was harvested for affinity purification by anti-Flag antibody. Flower tissue was ground in liquid nitrogen and suspended in Lysis buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 0.5 mM DTT, 1 mM PMSF, and 1 protease inhibitor cocktail tablet/50 ml [Roche]). The final supernatant was incubated with Anti-Flag M1 agarose (Sigma, A 4596) in Lysis buffer at 4°C for 2.5 h and then precipitated by centrifugation. The precipitant was washed four times, and the agarose-bound proteins were eluted with 3xFlag peptide (Sigma, F 4799).
The eluted proteins were run on a 10–12% SDS-PAGE gel and then subjected to silver staining with the ProteoSilver Silver Stain Kit (Sigma, PROT-SIL1). Total eluted proteins or specific target bands were cut from gels and extracted. Mass spectrometric analysis was performed as described previously [30]. Briefly, the peptides were eluted on a capillary column (50 µm×10 cm) packed with 5-µm spherical C18 reversed-phase material (YMC, Kyoyo, Japan). The eluted peptides were loaded onto a LTQ mass spectrometer (Thermo Fisher Scientific) for analysis. The peptide sequence data were searched against the IPI (International Protein Index) Arabidopsis protein database on the Mascot server (Matrix Science Ltd., London, UK).
Co-immunoprecipitation and gel filtration
SUVH2-3xMyc and SUVH9-3xFlag transgenic plants were used to determine whether SUVH2 and SUVH9 interact with DMS3 by co-immunoprecipitation. SUVH9-3xFlag transgenic plants were crossed to MORC6-3xMyc transgenic plants, and the offspring plants harboring both transgenes were used to determine whether SUVH9 interact with MORC6. For co-IP, protein extracts were incubated with the agarose-conjugated antibodies in Lysis buffer at 4°C for 2–3 h and precipitated by centrifugation. The precipitant was washed four times and then boiled for 5 min in 1×SDS sample buffer. After centrifugation, the supernatant was run on an SDS-PAGE gel, followed by Western blotting.
For gel filtration, total protein extracts from wild-type or transgenic plants were loaded onto a Superose 6 10/300 GL column (GE Healthcare, 17-5172-01) and the eluted solution was harvested once per 500 µl. The eluted fractions were run on a 10–12% SDS-PAGE gel and were then subjected to Western blotting. Standard proteins were used to determine the sizes of the eluted fractions.
Yeast two-hybrid assay
The cDNA sequences were cloned into pGADT7 and pGBKT7 vectors in frame to the C-termini of GAL4-AD and GAL4-BD, respectively. The DNA primers for cloning were listed in Table S5. The yeast strain PJ694a was co-transformed with pGADT7 and pGBKT7 constructs and grown on the synthetic dropout medium minus Trp and Leu (SD-TL). The positive yeast colonies were streaked on both SD-TL and SD-TLH (the synthetic dropout medium minus Trp, Leu, and His) for growth assay. Growth of transformed positive yeast strains on SD-TLH indicates the interaction between GAL-AD fusion protein and GAL4-BD fusion protein in corresponding yeast strains. 20 mM 3-AT was used to inhibit the background growth of transformed strains on SD-TLH medium.
Small RNA deep sequencing
Small RNA was extracted from two-week-old seedlings as described previously [57] and subjected to Illumina sequencing. Raw reads from small RNA libraries were processed to remove adapter sequences and were classified according to the barcodes. Small RNAs with 18 to 27 nt were mapped to the Arabidopsis genome (TAIR10) using the Bowtie program [58], and only perfectly matched reads were extracted for subsequent analysis. The whole Arabidopsis genome was split into 500-bp bins, and the appearances of 24-nt reads in each bin were counted. The read counts were then normalized by the library size. Reads per ten million (RPTM) were calculated for the adjusted small RNA abundance in each bin. The bins with low reads (<200 RPTM in all samples) were removed, and the regions were referred to as Pol IV-dependent siRNA regions if the RPTM value was five-time greater in the wild type than in the Pol IV mutant nrpd1. We also determine the RPTM value in Pol IV-dependent siRNA regions using 500-Kb windows to plot the profile of Pol IV-dependent siRNAs across chromosomes.
RNA deep sequencing-based gene expression analysis
The mRNA extracted from each genotype was used for Illumina library construction. The libraries were subjected to single-end Illumina sequencing. For data analysis, 45-bp sequences were mapped to the Arabidopsis genome (TAIR10) using TopHat [59]. Only reads mapped uniquely to the genome with a maximum of two mismatches were used for further analysis. Cufflinks [60] was used to quantify changes in gene expression between genotypes. Genes with a significant differential expression (P<0.05) was chosen to plot Heat map using Gplots package in R.
Analyses of RNA transcripts by RT-PCR
Total RNA was extracted by the routine method with Trizol as described previously [57]. For RT-PCR, total RNA was treated with DNase to remove residual DNA contamination in each RNA sample. RT-PCR was performed with the TAKARA one-step RT-PCR kit. Either sequence-specific primers or oligo-dT were used for reverse transcription. The RNA transcript levels were determined by semi-quantitative RT-PCR or quantitative RT-PCR. All RT-PCR results were biologically repeated at least two times. The primers used for RT-PCR were listed in Table S5.
DNA methylation analysis
DNA methylation was tested by chop-PCR. Genomic DNA was cleaved with the methylation-sensitive restriction enzyme HaeIII or AluI, followed by PCR. The abundance of PCR products indicates the DNA methylation levels of the amplified loci. The actin gene sequence, which lacks an HaeIII recognition site, was amplified as an internal control. The DNA methylation analyses were repeated at least two times.
CHIP assay
The levels of histone H3K9me2, NRPE1-Flag and DMS3 on chromatin were determined by CHIP assay as described previously [17]. Two-week-old seedlings were soaked in 0.5% formaldehyde under vacuum for cross linking. After chromatin was extracted and sonicated, it was immunoprecipitated with agarose-conjugated antibodies. Anti-Flag M2 Affinity Gel (A2220, Sigma) is directly used for immunoprecipitation, whereas anti-H3K9me2 antibody (ab1220, Abcam) and anti-DMS3 antibody [30] are conjugated with Protein A agarose followed by immunoprecipitation. Precipitated DNA as well as input DNA was subjected to quantitative PCR. The actin gene that has no NRPE1 and H3K9me2 enrichment was used as an internal control. Enrichment of NRPE1-Flag, H3K9me2 and DMS3 on chromatin was normalized by that on the actin gene. The primers used for quantitative PCR were listed in Table S5. The indicated results were obtained from three independent replicates.
Supporting Information
Zdroje
1. MatzkeM, KannoT, DaxingerL, HuettelB, MatzkeAJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21 : 367–376.
2. LawJA, JacobsenSE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11 : 204–220.
3. CastelSE, MartienssenRA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14 : 100–112.
4. MirouzeM, ReindersJ, BucherE, NishimuraT, SchneebergerK, et al. (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461 : 427–430.
5. TsukaharaS, KobayashiA, KawabeA, MathieuO, MiuraA, et al. (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461 : 423–426.
6. SlotkinRK, VaughnM, BorgesF, TanurdzicM, BeckerJD, et al. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136 : 461–472.
7. MosherRA, MelnykCW, KellyKA, DunnRM, StudholmeDJ, et al. (2009) Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460 : 283–286.
8. Olmedo-MonfilV, Duran-FigueroaN, Arteaga-VazquezM, Demesa-ArevaloE, AutranD, et al. (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464 : 628–632.
9. ItoH, GaubertH, BucherE, MirouzeM, VaillantI, et al. (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472 : 115–119.
10. HerrAJ, JensenMB, DalmayT, BaulcombeDC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308 : 118–120.
11. XieZ, JohansenLK, GustafsonAM, KasschauKD, LellisAD, et al. (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104.
12. HaagJR, ReamTS, MarascoM, NicoraCD, NorbeckAD, et al. (2012) In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell 48 : 811–818.
13. LawJA, VashishtAA, WohlschlegelJA, JacobsenSE (2011) SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet 7: e1002195.
14. LiuJ, BaiG, ZhangC, ChenW, ZhouJ, et al. (2011) An atypical component of RNA-directed DNA methylation machinery has both DNA methylation-dependent and -independent roles in locus-specific transcriptional gene silencing. Cell Res 21 : 1691–1700.
15. YeR, WangW, IkiT, LiuC, WuY, et al. (2012) Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell 46 : 859–870.
16. ReamTS, HaagJR, WierzbickiAT, NicoraCD, NorbeckAD, et al. (2009) Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 33 : 192–203.
17. WierzbickiAT, HaagJR, PikaardCS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135 : 635–648.
18. WierzbickiAT, ReamTS, HaagJR, PikaardCS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41 : 630–634.
19. KannoT, MetteMF, KreilDP, AufsatzW, MatzkeM, et al. (2004) Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 14 : 801–805.
20. KannoT, BucherE, DaxingerL, HuettelB, BohmdorferG, et al. (2008) A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet 40 : 670–675.
21. GaoZ, LiuHL, DaxingerL, PontesO, HeX, et al. (2010) An RNA polymerase II - and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465 : 106–109.
22. LawJA, AusinI, JohnsonLM, VashishtAA, ZhuJK, et al. (2010) A protein complex required for polymerase V transcripts and RNA - directed DNA methylation in Arabidopsis. Curr Biol 20 : 951–956.
23. ZhongX, HaleCJ, LawJA, JohnsonLM, FengS, et al. (2012) DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19 : 870–875.
24. HeXJ, HsuYF, ZhuS, LiuHL, PontesO, et al. (2009) A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev 23 : 2717–2722.
25. KannoT, BucherE, DaxingerL, HuettelB, KreilDP, et al. (2010) RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep 11 : 65–71.
26. HaagJR, PikaardCS (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12 : 483–492.
27. Bies-EtheveN, PontierD, LahmyS, PicartC, VegaD, et al. (2009) RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep 10 : 649–654.
28. HeXJ, HsuYF, ZhuS, WierzbickiAT, PontesO, et al. (2009) An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4 - and RNA-binding protein. Cell 137 : 498–508.
29. RowleyMJ, AvrutskyMI, SifuentesCJ, PereiraL, WierzbickiAT (2011) Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet 7: e1002120.
30. ZhangCJ, NingYQ, ZhangSW, ChenQ, ShaoCR, et al. (2012) IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet 8: e1002693.
31. AusinI, GreenbergMV, SimanshuDK, HaleCJ, VashishtAA, et al. (2012) INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci U S A 109 : 8374–8381.
32. XieM, RenG, ZhangC, YuB (2012) The DNA - and RNA-binding protein FACTOR of DNA METHYLATION 1 requires XH domain-mediated complex formation for its function in RNA-directed DNA methylation. Plant J 72 : 491–500.
33. ZhuY, RowleyMJ, BohmdorferG, WierzbickiAT (2013) A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell 49 : 298–309.
34. CaoX, JacobsenSE (2002) Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12 : 1138–1144.
35. HendersonIR, DelerisA, WongW, ZhongX, ChinHG, et al. (2010) The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet 6: e1001182.
36. MoissiardG, CokusSJ, CaryJ, FengS, BilliAC, et al. (2012) MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336 : 1448–1451.
37. LorkovicZJ, NaumannU, MatzkeAJ, MatzkeM (2012) Involvement of a GHKL ATPase in RNA-directed DNA methylation in Arabidopsis thaliana. Curr Biol 22 : 933–938.
38. BrabbsTR, HeZ, HoggK, KamenskiA, LiY, et al. (2013) The stochastic silencing phenotype of Arabidopsis morc6 mutants reveals a role in efficient RNA-directed DNA methylation. Plant J 75(5): 836–46.
39. BaumbuschLO, ThorstensenT, KraussV, FischerA, NaumannK, et al. (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29 : 4319–4333.
40. JacksonJP, LindrothAM, CaoX, JacobsenSE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 : 556–560.
41. EbbsML, BenderJ (2006) Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18 : 1166–1176.
42. RajakumaraE, LawJA, SimanshuDK, VoigtP, JohnsonLM, et al. (2011) A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev 25 : 137–152.
43. JohnsonLM, BostickM, ZhangX, KraftE, HendersonI, et al. (2007) The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17 : 379–384.
44. DuJ, ZhongX, BernatavichuteYV, StroudH, FengS, et al. (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151 : 167–180.
45. ZilbermanD, CaoX, JacobsenSE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 : 716–719.
46. HuettelB, KannoT, DaxingerL, AufsatzW, MatzkeAJ, et al. (2006) Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 25 : 2828–2836.
47. JohnsonLM, LawJA, KhattarA, HendersonIR, JacobsenSE (2008) SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet 4: e1000280.
48. KuhlmannM, MetteMF (2012) Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol Biol 79 : 623–633.
49. ZhangX, HendersonIR, LuC, GreenPJ, JacobsenSE (2007) Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A 104 : 4536–4541.
50. MosherRA, SchwachF, StudholmeD, BaulcombeDC (2008) PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A 105 : 3145–3150.
51. LeeTF, GurazadaSG, ZhaiJ, LiS, SimonSA, et al. (2012) RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7 : 781–795.
52. StroudH, GreenbergMV, FengS, BernatavichuteYV, JacobsenSE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152 : 352–364.
53. ListerR, O'MalleyRC, Tonti-FilippiniJ, GregoryBD, BerryCC, et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 : 523–536.
54. ReaS, EisenhaberF, O'CarrollD, StrahlBD, SunZW, et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 : 593–599.
55. TrievelRC, BeachBM, DirkLM, HoutzRL, HurleyJH (2002) Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111 : 91–103.
56. JessbergerR (2002) The many functions of SMC proteins in chromosome dynamics. Nat Rev Mol Cell Biol 3 : 767–778.
57. HeXJ, HsuYF, PontesO, ZhuJ, LuJ, et al. (2009) NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 23 : 318–330.
58. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
59. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
60. TrapnellC, WilliamsBA, PerteaG, MortazaviA, KwanG, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28 : 511–515.
Štítky
Genetika Reprodukčná medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



