-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
Botrytis cinerea is the causal agent of gray mold diseases in a range of dicotyledonous plant species. The fungus can reproduce asexually by forming macroconidia for dispersal and sclerotia for survival; the latter also participate in sexual reproduction by bearing the apothecia after fertilization by microconidia. Light induces the differentiation of conidia and apothecia, while sclerotia are exclusively formed in the absence of light. The relevance of light for virulence of the fungus is not obvious, but infections are observed under natural illumination as well as in constant darkness. By a random mutagenesis approach, we identified a novel virulence-related gene encoding a GATA transcription factor (BcLTF1 for light-responsive TF1) with characterized homologues in Aspergillus nidulans (NsdD) and Neurospora crassa (SUB-1). By deletion and over-expression of bcltf1, we confirmed the predicted role of the transcription factor in virulence, and discovered furthermore its functions in regulation of light-dependent differentiation, the equilibrium between production and scavenging of reactive oxygen species (ROS), and secondary metabolism. Microarray analyses revealed 293 light-responsive genes, and that the expression levels of the majority of these genes (66%) are modulated by BcLTF1. In addition, the deletion of bcltf1 affects the expression of 1,539 genes irrespective of the light conditions, including the overexpression of known and so far uncharacterized secondary metabolism-related genes. Increased expression of genes encoding alternative respiration enzymes, such as the alternative oxidase (AOX), suggest a mitochondrial dysfunction in the absence of bcltf1. The hypersensitivity of Δbctlf1 mutants to exogenously applied oxidative stress - even in the absence of light - and the restoration of virulence and growth rates in continuous light by antioxidants, indicate that BcLTF1 is required to cope with oxidative stress that is caused either by exposure to light or arising during host infection.
Published in the journal: The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen. PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004040
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004040Summary
Botrytis cinerea is the causal agent of gray mold diseases in a range of dicotyledonous plant species. The fungus can reproduce asexually by forming macroconidia for dispersal and sclerotia for survival; the latter also participate in sexual reproduction by bearing the apothecia after fertilization by microconidia. Light induces the differentiation of conidia and apothecia, while sclerotia are exclusively formed in the absence of light. The relevance of light for virulence of the fungus is not obvious, but infections are observed under natural illumination as well as in constant darkness. By a random mutagenesis approach, we identified a novel virulence-related gene encoding a GATA transcription factor (BcLTF1 for light-responsive TF1) with characterized homologues in Aspergillus nidulans (NsdD) and Neurospora crassa (SUB-1). By deletion and over-expression of bcltf1, we confirmed the predicted role of the transcription factor in virulence, and discovered furthermore its functions in regulation of light-dependent differentiation, the equilibrium between production and scavenging of reactive oxygen species (ROS), and secondary metabolism. Microarray analyses revealed 293 light-responsive genes, and that the expression levels of the majority of these genes (66%) are modulated by BcLTF1. In addition, the deletion of bcltf1 affects the expression of 1,539 genes irrespective of the light conditions, including the overexpression of known and so far uncharacterized secondary metabolism-related genes. Increased expression of genes encoding alternative respiration enzymes, such as the alternative oxidase (AOX), suggest a mitochondrial dysfunction in the absence of bcltf1. The hypersensitivity of Δbctlf1 mutants to exogenously applied oxidative stress - even in the absence of light - and the restoration of virulence and growth rates in continuous light by antioxidants, indicate that BcLTF1 is required to cope with oxidative stress that is caused either by exposure to light or arising during host infection.
Introduction
Botrytis cinerea Persoon: Fries (teleomorph Botryotinia fuckeliana (de Bary) Whetzel) is a necrotrophic pathogen with a broad host range causing gray mold disease in several economically important plants including grape vine and strawberry [1]–[2]. Sources of infection by B. cinerea are the conidia that are ubiquitously distributed in the air. After landing on the plant surface, the conidia germinate and form short germ tubes which directly penetrate. Infection may also occur from already established mycelia, in this particular case, multicellular infection structures (infection cushions) are formed. Usually, after penetration the epidermal and underlying cells die and B. cinerea establishes a primary (restricted) infection. Then, the fungus starts a massive outgrowth (spreading) that results finally in the maceration of the plant tissue (soft rot) and formation of conidia for new infections. During the interaction, the fungus produces phytotoxic metabolites e.g. botrydial (BOT) and botcinic acid (BOA), necrosis-inducing proteins, reactive oxygen species (ROS), cell wall-degrading enzymes, peptidases and phytohormones [1], [3]–[5]. Gene replacement approaches have identified only a few virulence factors to date, probably because of their redundancy [6], [2]. Hence, the importance of the toxic secondary metabolites BOT and BOA for infection becomes apparent only when strains are lacking both toxins [7]–[9].
The asexual macroconidia formed by branched conidiophores are undisputedly the main source of primary inoculum under given environmental conditions, such as high humidity and moderate temperatures. The dark pigmented sclerotia, however, serve as survival structures e.g. for over-wintering. They may germinate vegetatively to yield new mycelia and conidia, or they can act as the female parent in sexual reproduction when microconidia (male parent) carrying the opposite mating type are present. Apothecia, containing the linear asci with the ascospores, develop on the fertilized sclerotia [10]. It is difficult to estimate how frequently sexual reproduction happens in nature; nevertheless, it is assumed that it significantly contributes to genetic variation of B. cinerea [11]. If isolates produce sclerotia and microconidia they can function as female and male partners in reciprocal crosses [12]; however, not all B. cinerea isolates form sclerotia. As early as 1929, a comparative morphological study reported on three groups of B. cinerea isolates predominantly forming sclerotia, sterile mycelia or conidia [13]. When isolates are able to produce both conidia and sclerotia such as the sequenced strain B05.10 [14], [2], it happens in a light-dependent fashion: full-spectrum/white light induces the formation of conidia while its absence results in formation of sclerotia, and even limited exposures to light are sufficient to suppress sclerotial development [15]. B. cinerea was found to respond to different wavelengths of light: UV light results in conidiation, red light promotes sclerotial development and blue light leads to the accumulation of undifferentiated aerial mycelia [15]–[16]. Based on these observations it was hypothesized in 1975 that two receptors sensing UV/blue light and red/far-red light, respectively, are involved in regulation of asexual reproduction in B. cinerea [17]. In accordance with this model, the inspection of the genome sequence has revealed the existence of two UV light-sensing cryptochromes, two blue light sensors, two opsin-like proteins that may sense green light as well as three red/far-red light-sensing phytochromes. Notably, the number of phytochromes has been expanded in B. cinerea in comparison to Aspergillus nidulans and Neurospora crassa with one and two representatives, respectively, suggesting a special role of red light in its life cycle [18], [2].
The best studied fungus in terms of light signaling is N. crassa that mainly responds to blue light in order to regulate mycelial carotenoid biosynthesis, conidiation, the circadian clock, and formation of protoperithecia [19]. Most of these responses are mediated via the WHITE COLLAR transcription factors WC-1 and WC-2 that form a complex (WCC) in response to blue light leading to the activation of gene expression [20]. In accordance with the fact that blue light is the most active wavelength in N. crassa, deletions of the cryptochromes, opsins and phytochromes did not result in obvious phenotypes [21]–[24]. In contrast, and similarly to B. cinerea, A. nidulans does respond to UV, blue and red light and forms asexual spores in the light, but preferentially undergoes sexual reproduction (cleistothecia) in the dark [25]–[26]. It was demonstrated that the single cryptochrome CryA represses the formation of cleistothecia in the light [27], and that blue light sensed via the WCC complex (LreA/LreB) and red light sensed via the single phytochrome FphA act in concert to trigger the formation of conidia in the light [28]. However, unlike A. nidulans, B. cinerea possesses homologues of VVD-1 and FRQ-1 that are involved in photoadaptation and the circadian clock in N. crassa [18], [20], [29].
In B. cinerea, the photoreceptors, with the exception of the opsin BOP1 whose deletion affected neither affect light-dependent differentiation nor virulence [30], have not been functionally studied so far. However, “blind” B. cinerea mutants will be helpful to study the role of light in the fungus-host interaction without interfering with the host's metabolism. Recently, the reason for the “blind: always conidia” phenotype of two natural isolates (T4, 1750) has been identified: different single nucleotide polymorphisms (SNPs) in bcvel1 encoding a VELVET family protein result in truncated proteins and consequently in the deregulation of the light-dependent differentiation and in severely reduced virulence suggesting an interrelationship between light perception and virulence in B. cinerea [31]–[32].
This study describes the identification of a novel virulence-associated gene by a random mutagenesis approach using Agrobacterium tumefaciens-mediated transformation (ATMT). The identified gene bcltf1 encodes a GATA-type transcription factor homologous to A. nidulans and A. fumigatus NsdD, N. crassa SUB-1, and Sordaria macrospora PRO44 for whom the involvement in differentiation programs were demonstrated [33]–[38]. To our knowledge, the functions of this GATA transcription factor are described for the first time in a plant pathogen and reveal some overlapping features with the characterized homologues with regard to regulation of reproduction. Additionally, we report on specific functions of this transcription factor with regard to maintenance of ROS homoeostasis (production vs. scavenging of ROS), secondary metabolism and virulence.
Results
Identification of the GATA-type transcription factor BcLTF1
To identify genes in B. cinerea that are associated with the infection process, 2,367 hygromycin-resistant mutants obtained through A. tumefaciens-mediated transformation (ATMT) were screened for virulence on detached tomato leaves. The assay revealed 560 mutants that were either avirulent or less virulent than the parental wild-type strain B05.10 [39]. So far, phenotypes have been confirmed for 193 mutants on primary leaves of living bean plants (Phaseolus vulgaris), including the mutants PA31, PA2411, and PA3417 (Fig. 1A). These mutants are impaired in their ability to penetrate (PA3417) or to colonize the host tissue (PA31, PA2411), and are furthermore affected in light-dependent differentiation (data not shown). TAIL-(thermal asymmetric interlaced) PCR analyses revealed that the T-DNA integrations in the genomes of the mutants have occurred in the same genomic region (Fig. 1B). Identical integration events occurred for the RB flanks of the T-DNAs in all mutants i.e. 1.608 kb upstream of the annotated open reading frame (ORF) B0510_3555 (Broad Institute; for details see Materials and Methods), while different situations were found for the LB-flanking regions. In PA3417, the T-DNA insertion caused the deletion of four base pairs, in the other two mutants TAIL-PCR failed because no amplicons were obtained (PA31), or the remaining vector sequence was amplified (PA2411).
Fig. 1. Identification of bcltf1 as putative virulence factor by an ATMT approach. 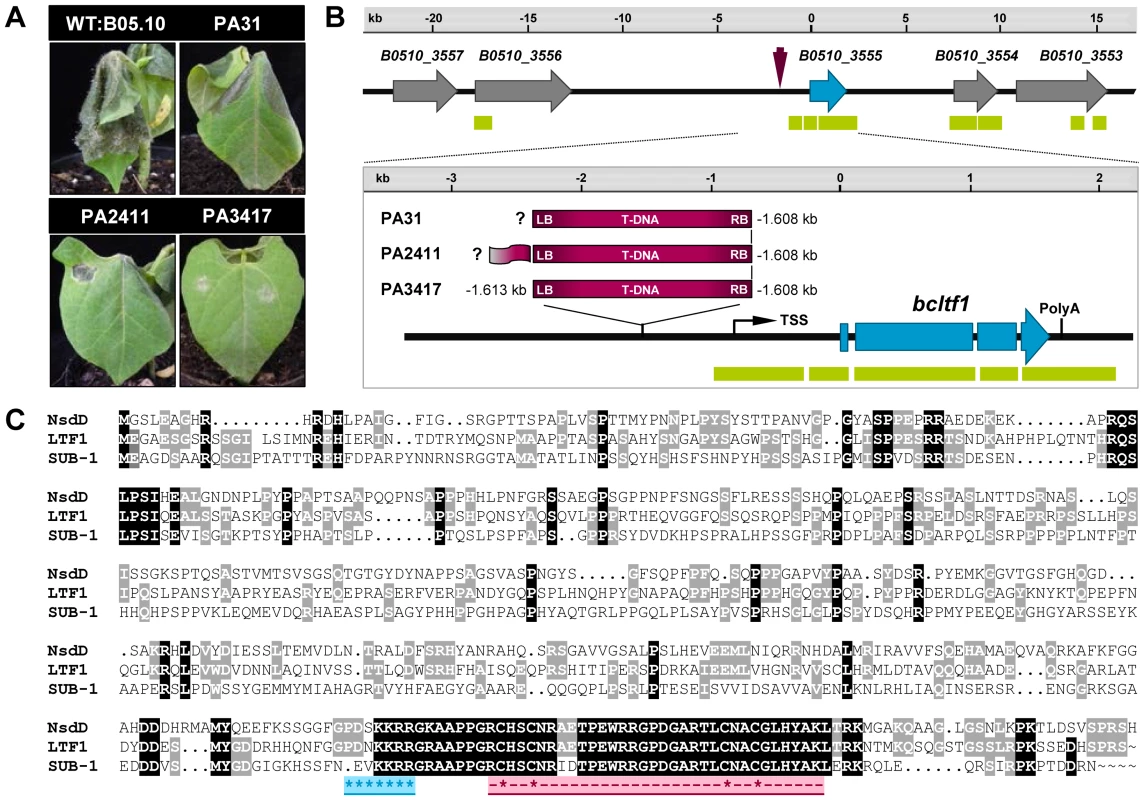
(A) Virulence phenotypes of the recipient strain B05.10 and the ATMT mutants PA31, PA2411, and PA3411. Primary leaves of Phaseolus vulgaris were inoculated with conidial suspensions and incubated for 5 d. (B) T-DNA insertion sites in the ATMT mutants were identified by TAIL-PCR analyses. In all three mutants, the insertion of the T-DNA (purple arrow) took place 1.608 kb upstream of an ORF hereinafter referred to as bcltf1 (blue arrows). The genomic region in strain B05.10 (B. cinerea Database, Broad Institute) with annotated genes and matching EST sequences (green bars) is shown. (C) Sequence alignment of BcLTF1 with homologous proteins from A. nidulans and N. crassa. The alignment was generated using ClustalW (http://genius.embnet.dkfz-heidelberg.de/) and the following protein sequences: B. cinerea LTF1 (481 aa; B0510_3555), A. nidulans NsdD (461 aa; AAB16914), and N. crassa SUB-1 (466 aa; NCU01154.5). Amino acids that are identical in all sequences are shaded black, amino acids that are shared by B. cinerea and N. crassa or A. nidulans are shaded in gray. The nuclear localization signal (NLS) is highlighted in blue, the GATA zinc finger motif (PF00320) with the four conserved cysteine residues (indicated by asterisks) is highlighted in pink. BlastP analyses using the sequence of B0510_3555 as query revealed the GATA-type transcription factors NsdD and SUB-1 from A. nidulans (e-value 6e-46) and N. crassa (e-value 9e-40), respectively, as putative orthologues. The alignment of protein sequences of B0510_3555, NsdD and SUB-1 showed low levels of sequence similarity (48% and 35%, respectively), except for the highly conserved GATA zinc finger domains in the C-terminal regions (Fig. 1C). Due to its expression profile and functions in B. cinerea - as described in the following - the protein is referred to as BcLTF1 (B. cinerea light-responsive transcription factor 1).
To confirm the linkage between the identified T-DNA integrations upstream of bcltf1 and the observed phenotypes of PA31, PA2411, and PA3417, bcltf1 was deleted and over-expressed in the B05.10 genomic background. Two different replacement mutants exhibiting identical phenotypes were generated by deleting either the ORF only (Δbcltf1-A6) or the ORF including 1.7-kb of the 5′-noncoding region (Δbcltf1-B1). Expression of bcltf1 under control of a constitutive promoter (PoliC) in the wild-type background resulted in two over-expressing mutants (OE::bcltf1-T6 and -T7). Complementation of deletion mutants was done by re-introduction of bcltf1 into the native gene locus (bcltf1COM) (for details, see Materials and Methods, Table 1, Fig. S1, Table S1).
Tab. 1. <i>B. cinerea</i> strains used in this study. 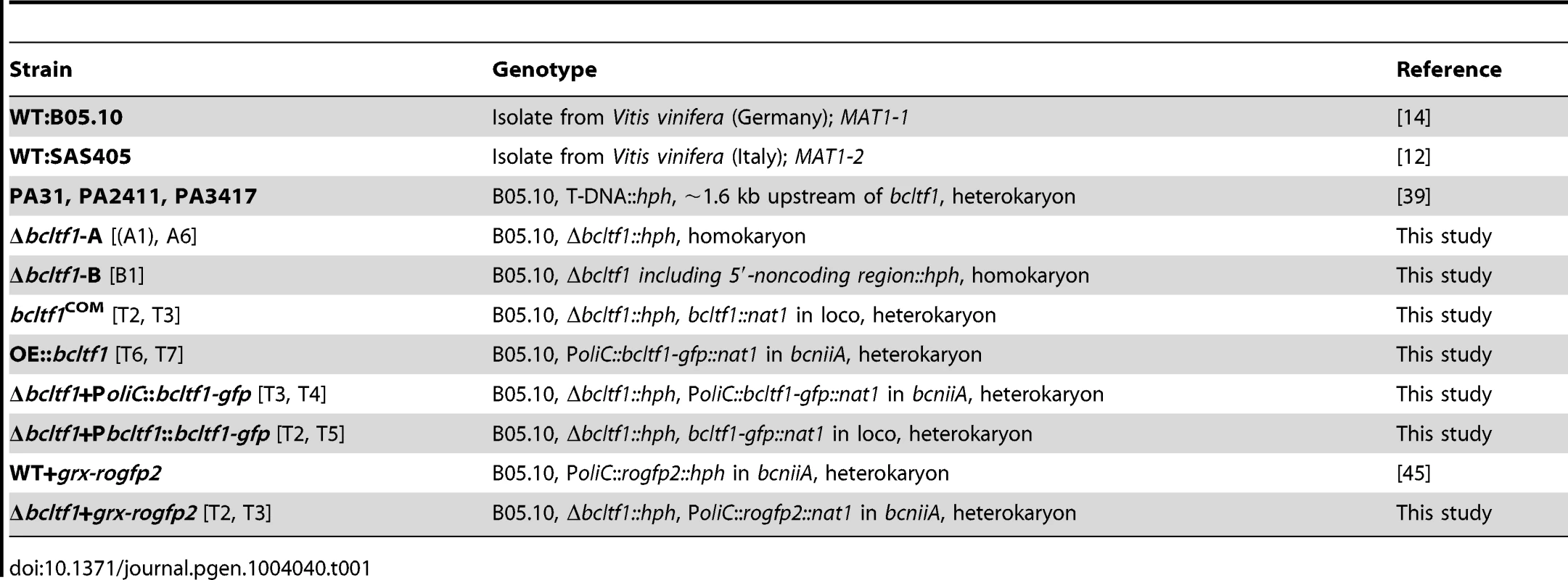
Bcltf1 expression is strongly induced by light
Given that BcLTF1 is implicated in regulation of light-dependent differentiation as suggested by the ATMT phenotypes, we studied its expression pattern during conditions inducing either conidiation (incubation in continuous light (LL) or in 12 h light/12 h dark rhythm (LD)) or sclerotia formation (incubation in continuous darkness (DD)) in the wild-type strain B05.10. Northern blot analyses revealed that bcltf1 was highly expressed during incubation in the light while only very weak expression occurred during sclerotial development in the dark. For comparison, expression of bcssp1 (homologue of S. sclerotiorum “sclerotia-specific protein 1” [40]) is restricted to developing sclerotia while expression of bcpks13 (polyketide synthase (PKS) putatively involved in melanin biosynthesis [41]) is restricted to conidiation in LL (Fig. 2A). To further analyze the effect of light on the expression of bcltf1, we exposed wild-type cultures that were grown for 2 d in DD for different periods to white light (5 to 300 min). High expression levels of bcltf1 became visible from 15 min light pulse (LP) while only low expression occurred in cultures that were kept in DD. In contrast to other light-induced genes such as bop1 (opsin-like protein [30]) and bcccg1 (homologue of N. crassa “clock-controlled gene-1” [42]) whose expression levels decreased again after prolonged exposure to light (from exposure times of 120 min and longer), expression levels of bcltf1 remained stable (Fig. 2B). Light induction of bcltf1, bop1, and bcccg1 was also observed during infection of P. vulgaris leaves (in stage of lesion spreading at 3 dpi), even though expression levels of bcltf1 were very low in comparison to those found in cultures grown on solid PDAB medium made from mashed bean leaves (Fig. 2C). Finally, dark-grown submerged cultures of B05.10 were exposed to ROS under dark conditions. Exposure to light, but neither the addition of hydrogen peroxide (H2O2), nor the presence of singlet oxygen (1O2), superoxide anions (O2−•) and hydroxyl radicals (OH•) affected the expression of bcltf1. Expression levels of bcccg1 increased in response to all treatments in contrast to bcgpx1 (encoding a glutathione peroxidase) that is specifically induced by H2O2 (Fig. 2D).
Fig. 2. Expression of bcltf1 depends on light. 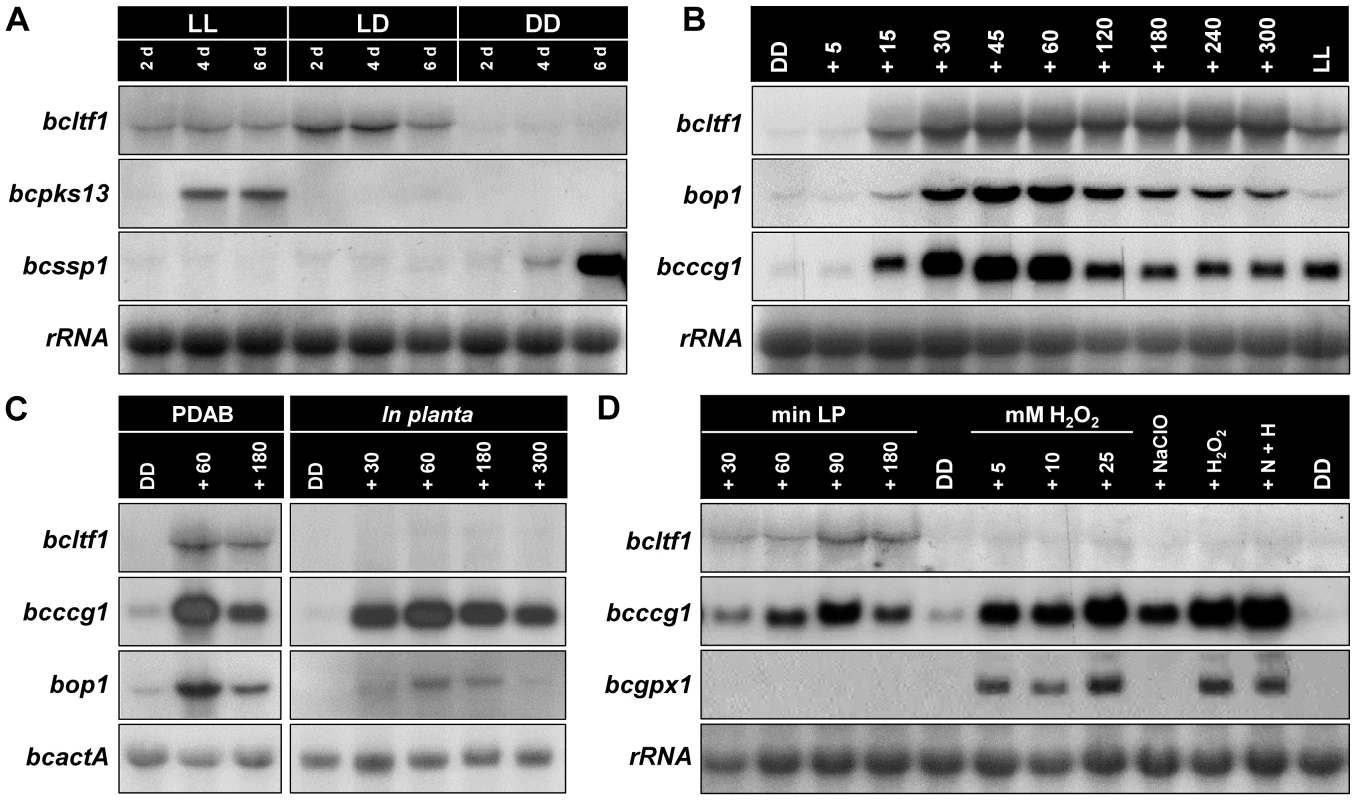
Gene expression was studied in wild-type strain B05.10 by northern blot analyses. rRNA or bcactA are shown as loading controls. For more details see Materials & Methods. (A) Bcltf1 is expressed during conidiation but not during sclerotial development. B05.10 was incubated on solid CM for the indicated time periods in continuous light (LL) and light-dark (LD) for induction of conidiation, and in continuous darkness (DD) for induction of sclerotium formation (sclerotia initials at 6 dpi). Genes exhibiting stage-specific expression profiles are shown: expression of bcpks13 is restricted to premature conidiation in LL, bcssp1 is exclusively expressed in developing sclerotia. (B) Expression of bcltf1 is induced by light. Wild type was incubated on solid CM for 52 h in DD or LL. DD-grown cultures were then exposed to white light for the indicated periods (5 to 300 min). (C) Bcltf1 is weakly expressed during infection of P. vulgaris. Primary leaves of living plants were inoculated with conidial suspensions of the wild type (four droplets per leaf) and incubated for 3 d in DD. Then plants were exposed to light for the indicated periods (30 to 300 min) or kept in DD. For comparison, light responses of B05.10 grown on solid PDAB (containing mashed bean leaves) are shown. (D) Expression of bcltf1 does not respond to oxidative stress. Liquid CM was inoculated with non-sporulating mycelia, and incubated for 3 d in DD at 170 rpm. Then, cultures were harvested (DD, controls), exposed to light (30 to 180 min), or exposed in darkness to oxidative stress induced by H2O2 (5 to 25 mM) or singlet oxygen (generated by simultaneous addition of 10 mM NaClO and 10 mM H2O2 (N+H)). All cultures that were exposed to oxidative stress were harvested after 30 min. Bcgpx1 is a known H2O2-induced gene. Taken together, among the conditions tested bcltf1 is exclusively highly expressed in response to light, and represents therefore an eligible candidate for controlling differentiation processes (conidiation vs. sclerotial development) and virulence in a light-dependent manner.
BcLTF1 is localized in the nuclei
To localize BcLTF1 in living hyphae, the coding sequence of bcltf1 was fused to the codon-optimized gene encoding the green fluorescent protein (GFP) [43] and expressed under control of a constitutive promoter in the bcltf1 deletion mutant (Δbcltf1+PoliC::bcltf1-gfp) (for details, see Materials and methods, Table 1, Fig. S1E). Merged images of GFP fluorescence and nucleic acid-specific Hoechst staining patterns showed that BcLTF1-GFP localizes to the nuclei in germinated conidia irrespective of the applied light conditions (Fig. 3A). Nuclear localization of BcLTF1-GFP was also observed in infectious hyphae during infection of onion epidermal cells (Fig. 3B). Similar results were obtained for BcLTF1-GFP that was expressed from its native promoter (Δbcltf1+Pbcltf1::bcltf1-gfp). GFP fluorescence, although weaker when compared to that of the constitutively expressed fusion protein, was detected in the nuclei independently of the illumination treatment (data not shown). Expression of BcLTF1-GFP under control of PoliC or its native promoter in the Δbcltf1 genomic background indicated the functionality of the fusion protein, as the native expression rescued all phenotypes of the deletion mutant while PoliC-driven expression resulted in phenotypes that are presumably due to the inappropriate expression of BcLFT1 especially in dark conditions (Fig. S2, Fig. S1F). Accordingly, mutants expressing PoliC::bcltf1-gfp in the wild-type background were generated as BcLTF1-overexpressing strains hereafter referred to OE::bcltf1 (T6, T7) (Fig. S1F, Table 1).
Fig. 3. BcLTF1-GFP fusion proteins are localized in the nuclei. 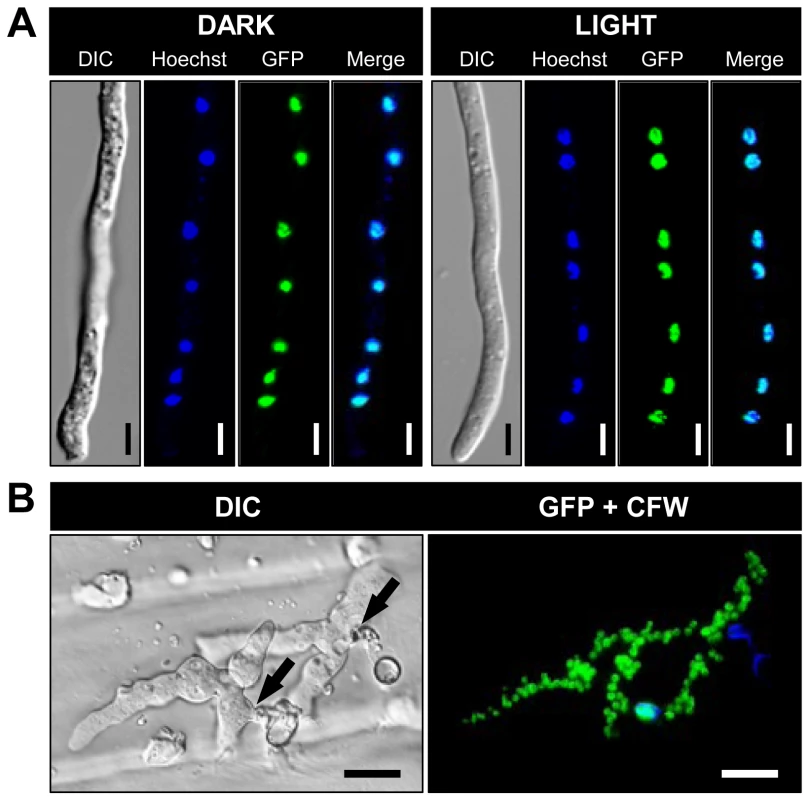
(A) Subcellular localization of BcLTF1-GFP does not depend on illumination conditions. The fusion protein was expressed from the constitutive A. nidulans oliC promoter in the Δbcltf1 background. Conidia were incubated on microscope slides for 18 h in DD (dark), or 12 h in DD followed by 6 h in LL (light). Nuclei were visualized using the fluorescent dye Hoechst 33342. Scale bars, 5 µm. (B) BcLTF1-GFP localizes to the numerous nuclei in infectious hyphae. Onion epidermis strips were inoculated with conidial suspensions and incubated for 24 h in DD. Conidia and hyphae on the surface were stained with the fluorescent dye calcofluor white (CFW). Sites of penetration are indicated by black arrows. Scale bars, 20 µm. Bcltf1 mutants are affected in light-dependent differentiation programs
The deletion as well as the over-expression of bcltf1 cause severe growth phenotypes that are related to the light treatment. Deletion mutants (Δbcltf1-A6, -B1) failed to grow on minimal medium (CD) in any light condition (Fig. 4A), and radial growth rates on synthetic complete medium (CM) correlated with the exposure times to light: while growth rates nearly equivalent to the wild type were observed in DD (92% of WT), incubation in LD and LL conditions significantly decreased growth rates to 43% and 27% of DD, respectively (Fig. 4B). The inhibitory effect of white light could be assigned to the blue light fraction, as growth rates were similarly decreased under incubation in continuous white and blue light, while growth rates in green, yellow and red light were comparable to those of Δbcltf1 cultures grown in DD (Fig. S3). Although Δbcltf1 mutants were severely impaired in growth in LD, they colonized the whole Petri dish after two weeks of incubation accompanied by reduced aerial hyphae formation and precocious and enhanced conidiation (155% of conidia produced by WT; data not shown). In contrast, the overexpressing mutants produced more aerial mycelia and reduced numbers of conidia giving the colonies a fluffy appearance. Light of different wavelengths (in LD rhythm) failed to alter the phenotypes of Δbcltf1 and OE::bcltf1 mutants. In contrast, the wild type formed more aerial hyphae and fewer conidia and sclerotia during incubation in blue and yellow/red light, respectively (data not shown). In general, phenotypes of the dark-grown cultures were more pronounced than those incubated in LD as both Δbcltf1 and OE::bcltf1 mutants failed to produce any sclerotia. Instead, the deletion mutants produced conidia and the overexpression mutants accumulated cotton ball-like mycelial aggregates in the dark (Fig. 4C).
Fig. 4. Mutations of bcltf1 alter light-dependent growth and differentiation programs. 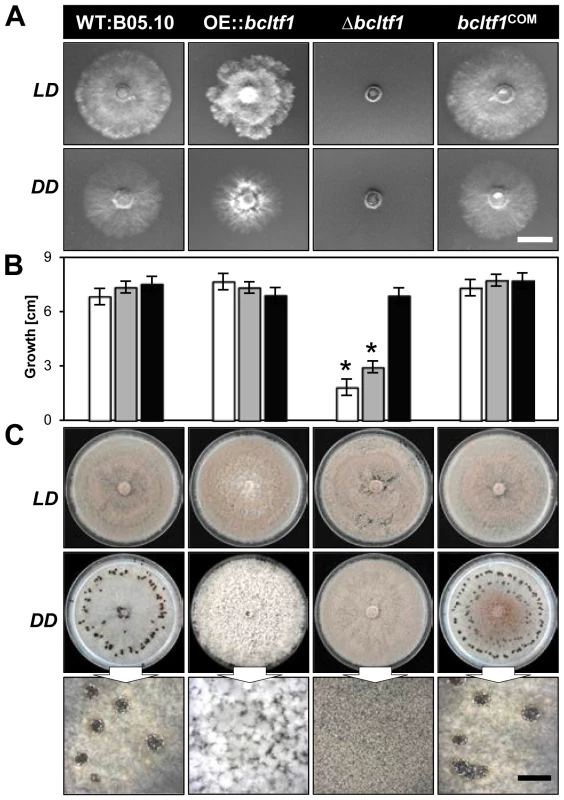
(A) BcLTF1 is essential for allowing growth on minimal medium. Indicated strains were incubated for 2 d on solid CD. Scale bar, 1 cm. (B) Exposure to light negatively affects radial growth rates of Δbcltf1 mutants on synthetic complete medium. Strains were incubated on solid CM in LL (white bars), LD (gray bars), and DD (black bars). Colony diameters (growth in cm) were determined after 3 d of incubation. Mean values and standard deviations were calculated from five colonies per strain and light condition. Asterisks indicate significant differences compared to WT:B05.10 in each condition (p<0.001). (C) Both overexpression and deletion of BcLTF1 modulate the differentiation program in DD. Strains were incubated for 14 d on solid CM. Close-up views of the cultures incubated in DD are shown in the lower panel. Scale bar, 5 mm. Evidently, light and BcLTF1 have an impact on colony morphology and the mode of reproduction, however, other differentiation programs in B. cinerea are not affected by light and mutations of bclft1. Conidial germination is induced by nutrients or hydrophobic surfaces in a similar fashion in wild type, OE::bcltf1 and Δbcltf1 mutants in LL and DD, and germling fusions via conidial anastomosis tubes (CATs) were observed for all strains (Fig. S4A; data not shown). Nevertheless, the forced expression of bcltf1 that is marked by bright nuclear BcLTF1-GFP signals, results in malformed germ tubes (Fig. S4B). The lack of the GFP signal accompanied by a wild-type-like morphology in a percentage of germ tubes suggests that the overexpression construct might become lost during conidiogenesis, so the outcome of studies involving OE::bcltf1 conidia needs to be carefully considered.
Moderate expression levels of bcltf1 are crucial for virulence
Though all T-DNA insertions upstream of bcltf1 impair virulence, differences exist between mutant PA3417, that is not able to penetrate resulting in the accumulation of mycelia on the top of the leaf, and mutants PA31 and PA2411 that are impaired in their ability to colonize the host tissue (Fig. 1A). However, the penetration defect of PA3417 is restricted to the conidia as infections from mycelia have been observed (data not shown). To examine how BcLTF1 is implicated in the infection process, the defined mutants (Δbcltf1, OE::bcltf1) were tested for their capabilities to penetrate and to proliferate inside a living host. To monitor the penetration events, onion epidermal strips were inoculated with conidial suspensions or agar plugs with non-sporulating mycelia (Fig. 5A). At 24 hpi, all strains had formed branched infection structures (infection cushions) originated from the already established mycelia on the agar plugs. Presumably, OE::bcltf1 mutants produced them more frequently than the wild type due to the significantly increased proliferation of aerial hyphae. In contrast, infection cushions were less often observed for Δbcltf1 mutants known to form fewer aerial hyphae (data not shown). Conidia derived from wild-type and Δbcltf1 strains formed short germ tubes that immediately penetrated the host cells indicating that the transcription factor is dispensable for entering the host. However, the majority of conidia with elevated bcltf1 expression levels produced elongated and branched germ tubes that failed to penetrate.
Fig. 5. BcLTF1 is required for full virulence. 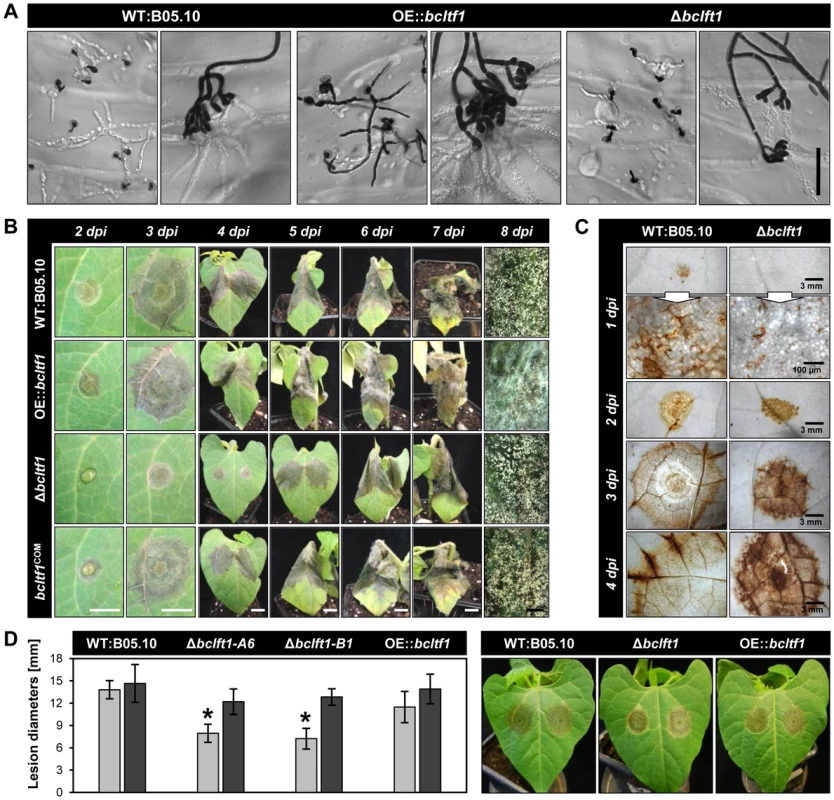
(A) Overexpression of bcltf1 affects the formation of infection structures. Onion epidermal strips were inoculated with conidial suspensions (on the left) or non-sporulating mycelia (on the right), and incubated for 24 h in DD. Conidia and hyphae on the surface were stained with lactophenol blue whereas invasively growing hyphae remain colorless. Scale bars, 50 µm. (B) Deletion of bcltf1 impairs the capability to colonize primary leaves of P. vulgaris. Leaves of living plants were inoculated with conidial suspensions and incubated in LD. White scale bars, 1 cm; black scale bars, 2 mm. (C) Spreading lesions provoked by Δbcltf1 mutants are characterized by increased H2O2 accumulation. Primary leaves were inoculated with conidial suspensions, detached 1 to 4 dpi, and incubated for 2 h in DAB solution. Plant tissues were decolorized prior to microscopy. (D) Addition of ascorbic acid restores virulence of Δbcltf1 mutants. Conidia were suspended in GB5 (gray) or in GB5 supplemented with 5 g/l ascorbic acid (black). Mean values and standard deviations of lesion diameters were calculated from eight lesions per strain and condition at 3 dpi. Asterisks indicate significant differences compared to WT:B05.10 in each condition (p<0.001). Pictures of lesions derived from ascorbic acid-supplemented conidial suspensions were taken 3 dpi. Primary leaves of living bean plants (P. vulgaris) were inoculated with conidial suspensions or mycelia-containing agar plugs derived from wild-type and mutant strains, and incubated in LD and DD to study the impact of light on infection. As can be seen from the time course experiments, Δbcltf1 mutants were significantly impaired in the infection process irrespective of the inoculation method and incubation conditions (in DD or LD) because in all cases distinct primary lesions emerged one day later than for the wild type and the OE::bcltf1 mutants (Fig. 5B, Fig. S5A,B). The reduced proliferation of Δbcltf1 hyphae inside the host became visible by 1 dpi in onion cells, and was likewise visualized in living bean tissues by trypan blue staining at 2 and 3 dpi (Fig. S5C). However, infection by the deletion mutant progressed until the whole leaf was colonized (9 dpi) and conidia were formed. Although germ tubes of OE::bcltf1 were severely impaired in their capacity to penetrate onion epidermal cells, infections of P. vulgaris derived from conidia and mycelia were comparable to those caused by the wild type with regard to lesion expansion. In contrast, colonization of the host tissue by OE::bcltf1 was accompanied by increased accumulation of aerial hyphae and reduced conidiation (Fig. 5B, Fig. S5A). In summary, light or its absence did not affect the infection process by wild type and bcltf1 mutant strains, and elevated and reduced expression levels of bcltf1 resulted in reduced penetration rates via germ tubes (OE::bcltf1) and retarded infection (Δbcltf1), respectively.
Increased ROS accumulation accounts for reduced virulence of Δbcltf1 mutants
B. cinerea induces an oxidative burst and hypersensitive cell death in the host, and it was shown that the degree of virulence on Arabidopsis correlates with the levels of superoxide (O2−) and hydrogen peroxide (H2O2) [44]. To monitor the accumulation of H2O2 in wild type - and Δbcltf1-infected plant tissues, inoculated bean leaves were sampled at 1 to 4 dpi and treated with 3,3′-diaminobenzidine (DAB); a compound that is oxidized by H2O2 in the presence of peroxidases yielding a dark-brown product. Microscopic observations revealed H2O2 accumulation in anticlinal walls of epidermal cells that were in close contact with wild-type germlings at 1 dpi. In contrast, no H2O2 accumulation was observed beneath the Δbcltf1 infection droplets at this time point even though conidia had germinated and entered the plant tissue (data not shown). However, after a further two days, significantly increased H2O2 accumulation in spreading lesions of the bcltf1 deletion mutant was observed (Fig. 5C). Therefore, in this particular case, increased ROS accumulation does not positively correlate with virulence. To explore the possibility that the elevated ROS levels may negatively affect the ability of the Δbcltf1 mutant to colonize the host tissue, we added ascorbic acid as an antioxidant to the conidial suspensions of wild type and bcltf1 mutants prior to inoculation. Whereas no differences were observed for infections by wild type and OE::bcltf1 strains, the addition of the antioxidant restored virulence of Δbcltf1 mutants as deduced from lesion sizes at 3 dpi (Fig. 5D). Consequently, the abnormal ROS accumulation may account for the virulence defect of the Δbcltf1 mutants.
BcLTF1 is required for maintaining ROS homoeostasis
For reasons thus far un-established, BcLTF1 is required to tolerate light during saprophytic growth on solid media, but not during plant colonization. To unravel the mechanism of light sensitivity, we modified the standard growth medium with different supplements and monitored colony growth and morphology during incubation in LL, LD and DD. Osmotic stabilization of the medium by adding 0.7 M sorbitol elevated growth rates of Δbcltf1 mutants in LL and LD conditions almost to those found in DD (66% and 94% of DD) (Fig. 6A) suggesting that the mutants have difficulty to cope with hypo-osmotic conditions, especially during illumination. On the other hand, the addition of 0.7 M NaCl appeared to cause ionic stress and prevented growth of Δbcltf1 mutants also during incubation in DD (25% of DD). These findings may imply that cell wall integrity and/or the osmotic stress response is impaired when bcltf1 is absent.
Fig. 6. BcLTF1 is needed to cope with oxidative stress and regulates the generation of ROS. 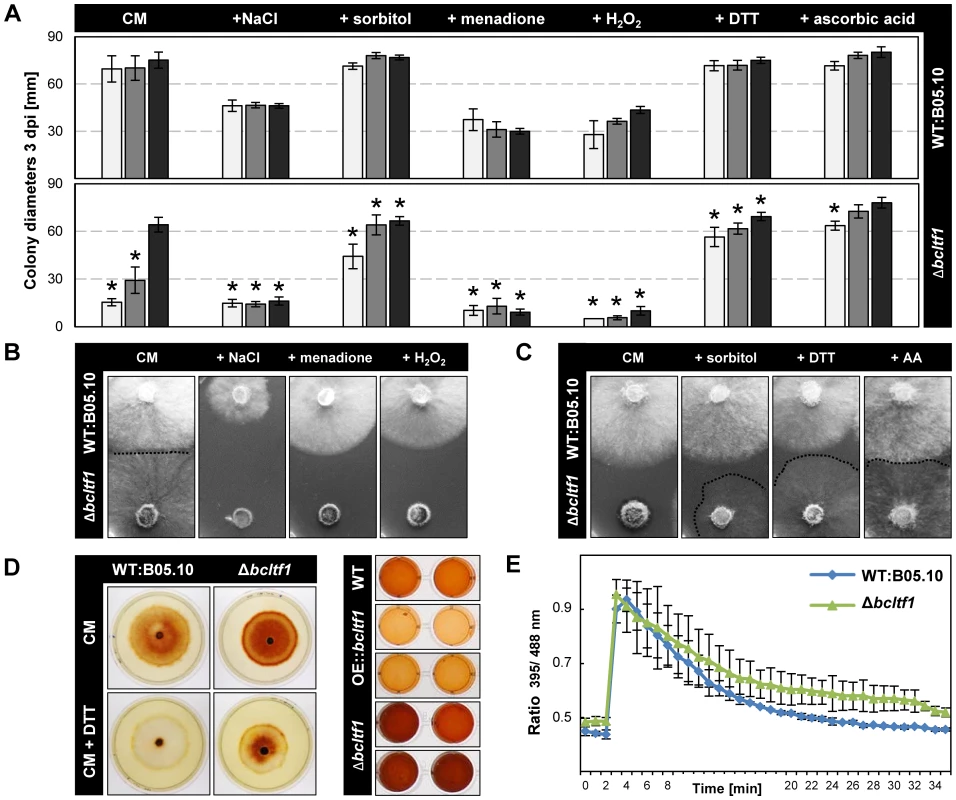
(A) Radial growth rates of Δbcltf1 mutants are affected by supplements. Strains were incubated in LL (white), LD (gray), or DD (black) on solid CM (control), and CM with 0.7 M NaCl, 0.7 M sorbitol, 300 µM menadione, 7.5 mM H2O2, 800 µM DTT (dithiothreitol) or 28 mM AA (ascorbic acid). Mean values and standard deviations were calculated from five colonies per strain and condition. Asterisks indicate significant differences compared to WT:B05.10 in each condition (p<0.001). (B) Salt and oxidative stress prevent growth of Δbcltf1 mutants even in the absence of light. Pictures were taken after 2 d in DD. (C) Ascorbic acid and DTT restore light tolerance but not the proliferation of aerial hyphae. Pictures were taken after 2 d in LL. Hardly visible Δbcltf1 colonies are highlighted by dashed lines. (D) Bcltf1 mutants are affected in generation of H2O2. Strains were incubated for 2 d in DD on CM or CM supplemented with 750 µM DTT. Then, colonies were flooded with DAB solution. DTT prevents oxidation of DAB in the range of wild-type colonies but not in the range of Δbcltf1 colonies (on the left). Identical quantities of fresh mycelia of B05.10, Δbcltf1 (A6, B1) and OE::bcltf1 (T6, T7) were incubated in DAB solution (on the right). (E) The cytosolic glutathione redox potential is not significantly affected in the absence of bcltf1. Reactions of redox-sensitive reporter roGFP2 expressed in Δbcltf1 and wild type were compared. Reduction of roGFP2 is displayed after an oxidation event provoked by addition of 10 mM H2O2 after 2 min. Indicated values are the means of triplicates; standard deviations are shown. For details, see Materials and Methods. Considering the finding of the altered ROS accumulation in Δbcltf1-infected plant tissues and the compensating effect of ascorbic acid in virulence, we pursued two strategies to study whether ROS play a role in the light-dependent growth defect of the deletion mutants. Firstly, the effect of oxidative stress caused by addition of 300 µM menadione (artificial source of superoxide radicals (O−2•)) and 7.5 µM H2O2 (a source of highly reactive hydroxyl radicals (OH•) upon reaction with metal ions), respectively, was monitored revealing that the deletion mutants were not able to cope with these conditions irrespective of the light conditions (Fig. 6A,B). Secondly, the use of antioxidants was intended to protect fungal strains against ROS. Both the addition of 800 µM dithiothreitol (DTT) and 28 mM ascorbic acid restored radial growth rates of Δbcltf1 mutants in LL and LD, but not the proliferation of aerial hyphae (Fig. 6C). To study whether the growth defect of the Δbcltf1 mutants on minimal medium (CD) can be rescued by reducing oxidative stress, we supplemented CD medium containing either nitrate or ammonium a single nitrogen source with ascorbic acid. In fact, the supplementation increased the growth rates of the mutants to certain extents in LD and DD, but failed to restore wild-type growth rates (Fig. S6). The fact that Δbcltf1 mutants are hypersensitive to exogenously applied oxidative stress even in the absence of light, and that on the contrary, the supplementation with antioxidants facilitated growth during incubation in LL indicate that the deletion mutants are unable to cope with oxidative stress that arises during the exposure to light.
To detect ROS production by the different strains, colonies of wild type and bclft1 mutants grown on CM and DTT-supplemented CM were stained with DAB solution. The intensive brown coloration of mutant colonies, even in presence of DTT that creates a reducing environment, indicated an increased accumulation of H2O2 in the range of the Δbcltf1 colonies in comparison to the wild type (Fig. 6D). In a second approach, the H2O2 production by equal amounts of biomasses of wild type, bcltf1 deletion and overexpressing mutants was studied, confirming the increased H2O2 production by the deletion mutants and revealing furthermore the decreased production by mutants overexpressing BcLTF1 (Fig. 6D). Formation of O−2• was monitored by treating wild type and mutant mycelia with nitro blue tetrazolium (NBT). Blue precipitates were found in hyphal tip segments of all strains (data not shown) demonstrating that mutations of bcltf1 do not alter the net accumulation of O2−. The fact that Δbcltf1 mutants accumulate higher amounts of H2O2 under all light conditions, either caused by its inappropriate production and/or detoxification, may explain the general hypersensitivity of the mutants to (additional) exogenously applied oxidative stresses such as the exposure to menadione, H2O2 or light.
To see whether the cellular redox status (glutathione pool) is altered in Δbcltf1 mutants, we expressed the redox-sensitive green fluorescent protein (roGFP2) in the mutant background and compared the ratio of fluorescence intensities (395 nm = oxidized state/488 nm = reduced state) with those of the wild type expressing roGFP2 [45]. Similar initial redox statuses were found in Δbcltf1 and wild-type hyphae, and roGFP2 was oxidized and subsequently reduced in a similar fashion in both strains after addition of 10 mM H2O2 (Fig. 6E). Thus, the altered ROS homoeostasis of the mutants that is characterized by the overproduction of H2O2 and the hypersensitivity to oxidative stress and light is not accompanied by an altered cytosolic redox status.
BcLTF1 modulates the transcriptional responses to light
To gain broader insight into the functions of BcLTF1 especially in those that are related to illumination, a genome-wide approach was initiated to compare gene expression profiles in the wild type and the Δbcltf1 mutant in response to a light stimulus. For these experiments, strains were cultivated in DD, and then exposed to white light for 60 min (WT:B05.10+LP; Δbcltf1+LP) or kept in darkness for additional 60 min (WT:B05.10-D; Δbcltf1-D) (for details, see Material and Methods). RNA from four biological replicates was extracted, labeled and hybridized to NimbleGen microarrays containing oligonucleotides representing all predicted B. cinerea genes as well as non-matching expressed sequence tags (ESTs) [2].
Statistical tests were performed to detect differentially expressed genes: light responses in wild type (+LP/D in WT) and mutant (+LP/D in Δbcltf1), and Δbcltf1 effects in darkness (Δbcltf1/WT in D) and light (Δbcltf1/WT in LP). In total, 2,156 differentially expressed spotted genes corresponding to 2,074 distinct genes were identified and could be assigned to 11 expression profiles (Fig. 7, Table S2). 293 genes showed a light-responsive pattern in the wild-type background: the expression of 244 genes was induced by light (referred to as light-induced genes; bclig1-244), and the expression of 49 genes was repressed by light (referred to as light-repressed genes; bclrg1-49). The expression levels of the majority of light-responsive genes were altered in the absence of BcLTF1, i.e. light induction/repression did not happen (genes in profiles 5 and 6) or just disappeared as genes are already de-regulated in Δbcltf1 in the absence of light (genes in profiles 7 and 8). Only 34% of light-responsive genes (in profiles 3 and 4) exhibited similar light responses in both genomic backgrounds, demonstrating that the absence of bcltf1, which itself is responsive to light, severely affects the expression pattern of other light-responsive genes. Interestingly, some genes were found whose expression responded to light exclusively in the absence of BcLTF1; they were either induced (12 genes in profile 9) or repressed by light (44 genes in profile 10). Nevertheless, the loss of the BcLTF1 had a more pronounced effect on genome-wide gene expression than the light stimulus. In total, the expression levels of 1,972 spotted genes corresponding to 1,905 distinct genes were changed in the Δbcltf1 mutant and most of them (1,539 genes; 81%) were not related to a light response: 748 genes were overexpressed (profile 1) and 791 genes were underexpressed (profile 2) irrespective of the illumination condition (Fig. 7, Table S2).
Fig. 7. BcLTF1 modulates the transcriptional responses to light. 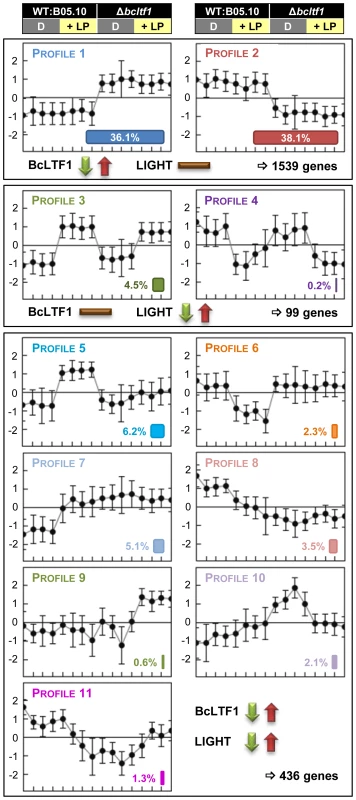
Wild type B05.10 and the Δbcltf1 mutant were grown on solid CM for 52 h in DD; then, cultures were exposed for 60 min to light (+LP) or kept for additional 60 min in darkness (D). Four biological replicates were performed. RNA samples were labeled and hybridized to microarrays with all predicted genes of B. cinerea. Statistical analysis revealed 2,074 differentially expressed genes. Relative expressions of the genes (i.e. log2-normalized intensities scaled by gene) in the four conditions (16 hybridizations) were clustered, and 11 of the 14 theoretically possible expression profiles were found (for more details, see Materials and Methods). This figure represents the centroid views of the 11 expression profiles (x = hybridization and y = log2-normalized intensity scaled by gene). The percentages among the 2,074 genes are indicated for each profile. Genes whose expression is only affected by the deletion of bcltf1 belong to profiles 1 (748 genes) and 2 (791 genes). Profiles 3 (94 genes) and 4 (5 genes) comprise genes whose expression is affected by light in a similar manner in both genomic backgrounds (WT, Δbcltf1). Genes (436) belonging to the other profiles are affected by light treatment and the deletion of bcltf1. Functional enrichment analyses using the GSEA (Gene Set Enrichment Analyses [46]–[47]) method and defined sets of genes were performed to reveal biological processes that are affected by light and/or in the absence of BcLTF1. Hence, the group of light-induced genes in wild type is enriched for genes implicated in the oxidative stress response (OSR) or light perception, while amino acid-transporter-encoding genes are overrepresented in the group of light-repressed genes. Even though a majority of bcltf1-dependent genes display similar expression profiles in darkness and light, the group of overexpressed genes in the Δbcltf1 mutant in darkness is significantly enriched in genes involved in light perception and respiration, and the group of overexpressed genes in the light is enriched in secondary metabolism-related genes. In both light conditions, the group of underexpressed genes in Δbcltf1 is enriched in transporter-encoding genes (Table 2).
Tab. 2. Functional categories that are enriched due to light treatment or deletion of BcLTF1. 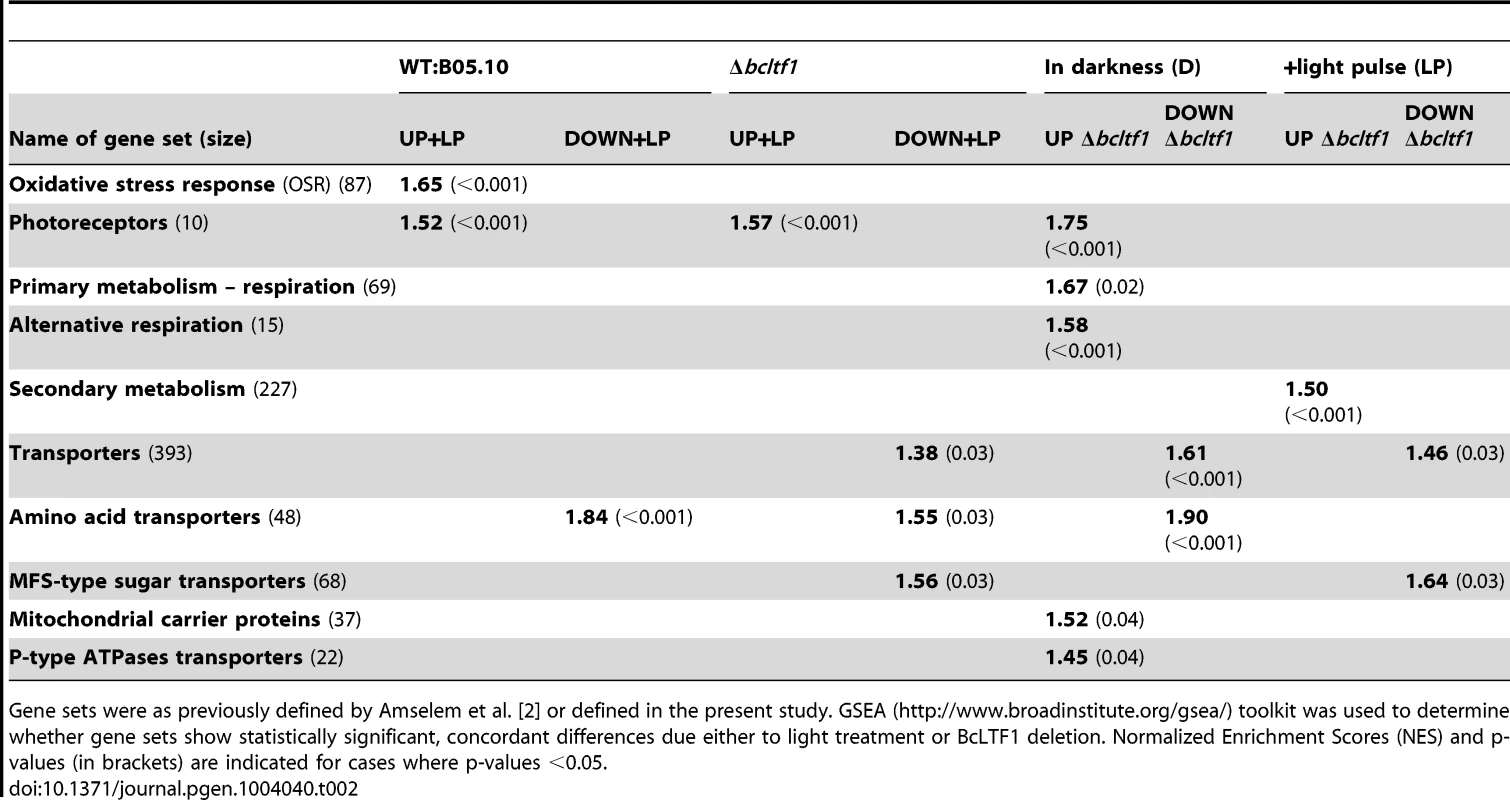
Gene sets were as previously defined by Amselem et al. [2] or defined in the present study. GSEA (http://www.broadinstitute.org/gsea/) toolkit was used to determine whether gene sets show statistically significant, concordant differences due either to light treatment or BcLTF1 deletion. Normalized Enrichment Scores (NES) and p-values (in brackets) are indicated for cases where p-values <0.05. Identification of putative regulators of light-dependent differentiation
Currently, it is not known how light and its absence, trigger differentiation in B. cinerea. Because young undifferentiated mycelia of the wild type, that are able to form conidia or sclerotia dependent on the illumination conditions, were subjected to the transcriptomic analysis, two sets of “developmental genes” among the light-responsive genes can be expected, those implicated in induction of conidiation or in suppression of sclerotial development.
Light responses are based on the perception of light as a signal and the induction of subsequent signaling events. Though it is not clear why photoreceptors should be regulated on the level of gene expression, their light-responsive expression is a known phenomenon in N. crassa [37], [48]. Accordingly, expression of most of the predicted photoreceptor-encoding genes of B. cinerea was induced by light (Fig. 8A, Table S3). Bcvvd1 encoding a putative blue light receptor was the strongest up-regulated gene with a 37-fold increase. Further light-induced genes are those encoding cryptochromes (bccry1, bccry2), opsins (bop1, bop2) and one of three phytochromes (bcphy2). Whereas similar intensities were found for bcvvd1 in the wild type and the Δbcltf1 mutant, amplitudes of light induction of the other photoreceptor-encoding genes were much lower in the mutant than in the wild-type background as the genes in Δbcltf1 were already higher expressed in the dark (bccry1, bccry2, bop2, bcphy2) or less induced by light treatment (bop1, bop2) than in the wild type. Bcwcl1 encoding a putative blue light-sensing transcription factor was not found among the light-induced genes in this microarray experiment. However, further analysis showed that bcwcl1 is expressed higher during incubation in LL than in DD (data not shown), indicating that light also affects the expression of this gene. Among the light-induced genes bclov3 (PAS/LOV protein 3) might be related to light perception as the deduced protein sequence shares similarities with N-terminal parts of plant phototropins. These proteins are autophosphorylating protein kinases harboring N-terminal blue light-sensory (LOV) and C-terminal kinase domains [49].
Fig. 8. Differentially expressed genes may encode regulators of light-dependent differentiation. 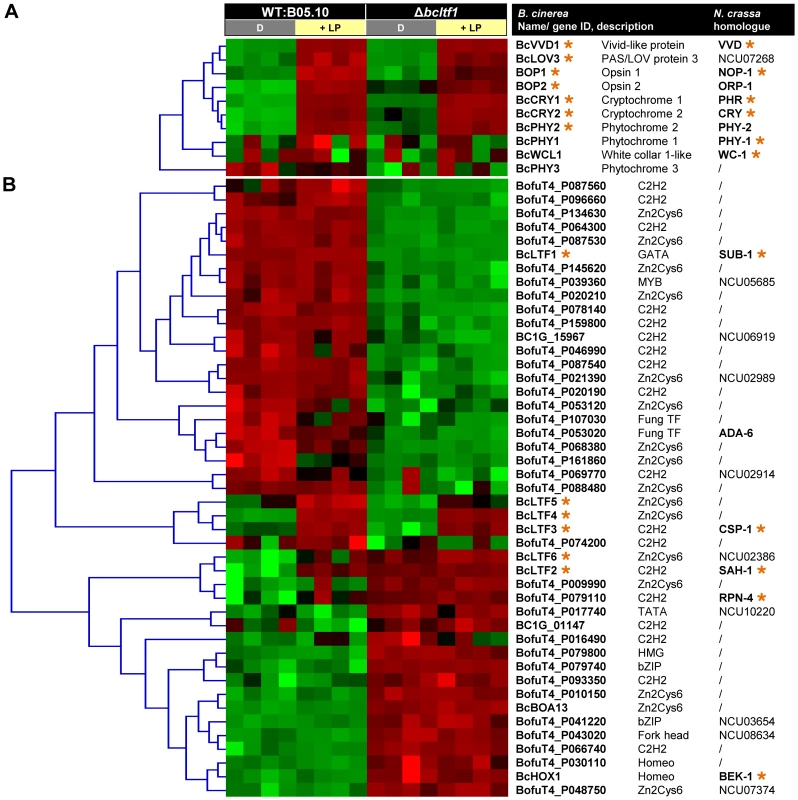
Relative expressions of the genes (i.e. log2-normalized intensities scaled by gene) in the four conditions (16 hybridizations) were clustered and depicted by color scale, where shades of green and red represent under- and overexpressed genes, respectively. Homologues in N. crassa were identified by a bidirectional best-hit (BDBH) approach. Light-induced genes in B. cinerea and N. crassa are marked with orange asterisks. For more details see Table S3. (A) Expression of most photoreceptor-encoding genes is similarly induced by light in wild type and Δbcltf1 strains. All predicted photoreceptors were clustered. (B) Expression levels of 45 TF-encoding genes are altered in response to light or deletion of bcltf1. TFs that were differentially expressed in at least one condition were subjected to clustering. Regulators that function downstream of signaling cascades are transcription factors (TFs) whose activities might be transcriptionally and/or post-transcriptionally modified. The genome of B. cinerea contains 406 genes encoding TFs of different families [2], [50], 45 TF-encoding genes were differentially expressed in at least one of the four conditions, i.e. displaying a light response in wild type or Δbcltf1 mutant, or a Δbcltf1 effect in darkness or in response to light (Fig. 8B, Table S3). Hierarchical clustering of the selected TFs across the 16 hybridizations highlights that the differential expression is mainly due to the bcltf1 mutation (43 TF-encoding genes) and to a smaller extent to the light condition (seven TF-encoding genes). In addition to bcltf1, the expression of five further TF-encoding genes was induced by light and the genes were accordingly named bclft2-bclft6. BcLTF2 and BcLTF3 are C2H2-TFs homologous to the light-responsive TFs SAH-1 and CSP-1 of N. crassa [37], [48]. BcLTF4 to BcLTF6 belong to the family of Zn2Cys6-TFs, and only a homologue of BcLTF6 is present in N. crassa. While the expression of bcltf3, bcltf4 and bcltf5 was similarly induced by light in both genomic backgrounds, the light-dependent expression pattern of the other two TF-encoding genes are affected in different ways in the absence of BcLTF1. Both bcltf2 and bcltf6 were overexpressed in the Δbcltf1 mutant in darkness, but only the expression level of bcltf6 was further increased in response to the light stimulus. Therefore, BcLTF1 is involved in repressing the transcription of bcltf2 and bcltf6 in the dark. Moreover, it can be assumed that BcLTF1 represses the expression of 16 other TF-encoding genes in a light-independent manner, including bcboa13 that belongs to the botcinic acid (BOA) gene cluster (see next section). The expression of one TF-encoding gene (BofuT4_P161860) was repressed by light in the wild type and the effect was found to be absent in the Δbcltf1 background, as the gene was no longer expressed (even in darkness). The gene encodes a member of the Zn2Cys6-TF family and is physically linked with genes of unknown functions displaying a similar expression pattern (Bclft1g1499-1505; Table S2).
Absence of BcLTF1 affects expression of secondary metabolism-related genes
Fungal secondary metabolite (SM)-biosynthetic genes are typically located adjacent to each other and exhibit similar patterns of expression. These clusters usually include a gene encoding the key enzyme responsible for synthesis of the raw product, genes encoding enzymes for modifications and the transport of the compound, and genes whose products regulate cluster gene expression [51]. The genome of B. cinerea comprises 43 genes encoding SM key enzymes including 21 polyketide synthases (PKSs), nine non-ribosomal peptide synthetases (NRPSs), six sesquiterpene cyclases (STCs), three diterpene cyclases (DTCs) and one dimethylallyl tryptophan synthase (DMATS) [2]. In our transcriptomic approach, 38 of the 43 genes were significantly expressed in at least one condition (see Materials and Methods). The expression patterns of 17 of these genes (45%) are significantly altered in the absence of BcLTF1, including 9 PKS-, 3 NRPS-, 4 STC - and the single DMATS-encoding gene (Fig. S7A). Remarkably, the majority (15) of these BcLTF1-dependent genes are not responsive to light. This group includes bcbot2 (bcstc1) that is part of the BOT-biosynthetic cluster (bcbot1-5) as well as bcboa6 and bcboa9 (PKSs) that belong to the two gene clusters (bcboa1-6, bcboa7-17) required for BOA biosynthesis. The other cluster genes were overexpressed as the key enzymes (Fig. S7B), suggesting that the mutants may produce more toxins under the tested conditions. Additionally, the two PKS-encoding genes bcpks12 and bcpks13 were under - and overexpressed in the Δbcltf1 mutant, respectively, and are considered to be associated with dihydroxynaphthalene (DHN)-melanin biosynthesis [41]. Beside the PKS yielding the polyketidic precursor, probably three other enzymes encoded by bcbrn1, bcbrn2 and bcsd1 are involved in melanin biosynthesis (Fig. S8). Increased expression levels of bcpks13, bcbrn1, bcbrn2, and bcscd1 in Δbcltf1 mutants were associated with the dark pigmentation of the culture medium, while expression of bcpks12 was exclusively detected in developing sclerotia of the wild type. Therefore, bcpks13 and bcpks12 are differentially expressed in Δbcltf1 mutants as well as during the life cycle of the wild type.
The expression patterns of only three key enzyme-encoding genes (bcnrps2, bcphs1, bcstc5) were significantly affected by light. Notably, one of them corresponds to a cluster of light-induced genes (bcphs1, bcphd1, bccao1) that encode homologues of enzymes involved in the biosynthesis of retinal, the chromophore for opsin, in Fusarium fujikuroi (phytoene synthase, phytoene dehydrogenase, and carotenoid oxygenase). As in F. fujikuroi, the cluster contains a forth co-regulated gene encoding an opsin (bop2) [52]–[54] (Fig. S9). Though genes were still induced by light in the absence of BcLTF1, the absolute expression levels were much lower than those found in the wild type in response to light, suggesting decreased retinal production and an altered function of BOP2 in the absence of BcLTF1.
Light and BcLTF1 affect the expression of ROS-related genes
Given the finding that mutations of bcltf1 affect the accumulation of H2O2, we inspected the transcriptomic data for genes involved in ROS metabolism to gain insight whether the net over-accumulation is due to an inappropriate detoxification and/or generation of ROS. Oxidative stress response (OSR) systems include superoxide dismutases (SODs) that convert O−2• into the less toxic H2O2, and catalases (CATs), peroxidases (PRDs), and peroxiredoxins (PRXs) that detoxify H2O2 and thereby prevent the formation of OH• via the Fenton reaction. Important non-enzymatic mechanisms include the oxidation of compounds such as glutathione, ascorbate and carotenoids. Reduced glutathione (GSH) provided by glutathione reductases (GLRs) is used by glutathione peroxidase (GPX), glutaredoxins (GRXs) as well as by glutathione S-transferases (GSTs) that conjugate GSH with various substrates such as peroxides [55]. In B. cinerea, several enzyme activities are represented by multiple genes, thus, four SODs, eight CATs, 16 PRDs, nine PRXs, one TRX, five GRXs and 26 GSTs have been identified (Table S4). The set of light-induced genes in the wild type is significantly enriched for OSR-related genes (Table 2), among them are two CATs (bccat3, bccat4), two PRDs (bcprd1, bcprd2) and three GSTs (bcgst3, bcgst8, bcgst15) (Table S4). These genes are not induced by light in the absence of bcltf1 suggesting that their products may contribute to coping with light-related oxidative stress. Nevertheless, twenty genes among them SOD-, CAT-, PRD-, PRX - and GST-encoding genes exhibited increased or decreased expression levels in the mutant background, reflecting a general deregulation of OSR-related genes.
Several enzymes of B. cinerea may generate ROS either as secondary messenger (e.g. formation of O−2• by the NADPH oxidase (NOX)) or as side products of metabolic processes (e.g. oxidation of carbohydrates, fatty acids and amino acids yielding H2O2). Forty-eight putative oxidase-encoding genes were identified in the genome, and expression levels of twenty-five genes were affected by either the light stimulus and/or the deletion of bcltf1. Four oxidase-encoding genes were induced by light in both genomic backgrounds, and a further 11 oxidase-encoding genes exhibited increased expression levels in the mutant background. Seven out of the 11 identified DAO (D-amino acid oxidase)-encoding genes are less expressed in the bclft1 mutant than in the wild type, irrespective of the illumination condition, a fact that might be related with the weak expression of amino acid transporter-encoding genes (Table 2). Additionally, two out of the four identified putative NADH:flavin oxidoreductase/NADH oxidase-encoding genes showed increased expression levels in the absence of BcLTF1 while the genes encoding the catalytic subunits of the NOX complex (bcnoxA and bcnoxB) were slightly underexpressed (Table S4).
Absence of BcLTF1 results in increased alternative respiration
Beside the mentioned ROS-generating enzymes, the mitochondrial electron transport chain (ETC) massively contributes to intracellular ROS levels. Complexes I and III are forming O2− that is in turn dismutated to H2O2 to prevent the formation of OH• [56]. Though expression levels of nuclear-encoded subunits of the different protein complexes of the respiratory chain were not affected in the Δbctlf1 mutant (data not shown), strongly increased expression levels were observed for the gene encoding the alternative oxidase (BcAOX1) (Table S4, Fig. S10). This enzyme accepts electrons from the ubiquinone pool and reduces oxygen directly, hence bypassing the electron flux through complexes III and IV (cytochrome c oxidase, COX), and related ROS generation [57]. In addition to complex I (NADH:ubiquinone oxidoreductase), B. cinerea possesses five putative NADH dehydrogenases that may deliver electrons to the ubiquinone pool thereby bypassing complex I. In fact, bcnde2 and bcndi1 encoding external and internal NADH dehydrogenases, respectively, were overexpressed in the deletion mutants. Additionally, genes encoding cytochrome c (bccyc1), a cytochrome c peroxidase (bcccp2), an uncoupling protein (bcucp1) and several other mitochondrial carrier proteins were more highly expressed in the absence of BcLTF1, further supporting the deregulation of the ETC in the mutant background. CCP2 may accept electrons from complex III/cytochrome c for reduction of H2O2, hence avoiding the electron flux through COX [58], and uncoupling proteins are considered to decrease ROS production by facilitating the transport of ions across the membrane [56], [59]. Taken together, the observed transcriptional changes in the Δbcltf1 mutant suggest that alternative enzymes compete with the ETC complexes for electrons to decrease the generation of mitochondrial ROS.
Discussion
Random mutagenesis via ATMT is successfully used in plants and filamentous fungi to generate new phenotypes and to identify the corresponding genes. In a recent study, we reported on an ATMT approach in B. cinerea yielding 2,367 mutants with 560 exhibiting reduced virulence in a first screening. For two out of four randomly chosen candidates tested, the genes could be linked to the ATMT phenotype by deletion approaches. As reported for other fungal ATMT libraries, T-DNA integrations were mostly random, single copy, and occurred preferentially in noncoding (regulatory) regions [39]. The latter facts hold true for the ATMT mutants we are describing in this study, however, the fact that three independent mutants out of 77 virulence-attenuated mutants analyzed so far were found to contain T-DNA insertions at the same gene locus, indicates that “hot spots” for T-DNA insertions exist in the genome of B. cinerea.
While the deletion of bcltf1 mainly affects the advanced stages of infection, the overexpression of bcltf1 impairs specifically the penetration process by germ tubes which exhibit an abnormal branching pattern. In contrast, penetration via infection cushions from already established mycelia is not impaired indicating that these modes of penetration are independently regulated processes. This is in agreement with previous reports on mutants that are impaired in mycelia - but not in conidia-derived infection [60]. The retarded colonization of the host tissue by Δbcltf1 mutants is accompanied and likely caused by increased accumulation of ROS as the addition of antioxidants to the inoculation droplets restored virulence to wild-type levels. Since the deletion mutants accumulate more H2O2 than the wild type under axenic conditions, the increased H2O2 levels in Δbcltf1-infected plant tissues probably are the consequence of inappropriate ROS production by the fungus rather than by the plant. However, the increased accumulation has a negative impact on invasion of the host tissue by the mutant, whereas the application of antioxidants does not interfere with virulence of the wild type, raising the question to which extent ROS produced by the host (in terms of an oxidative burst) or the fungus are contributing to the outcome of the interaction.
Homologues of BcLTF1 have been characterized so far only in very few fungal species, revealing their requirement for sexual reproduction in Sordariomycetes (N. crassa, S. macrospora), and Eurotiomycetes (A. nidulans and A. fumigatus) [33], [35], [36], [38]. Due to the lack of sclerotia, Δbcltf1 as well as OE::bcltf1 mutants are female sterile, and for crossings of Δbcltf1 microconidia (MAT1-1) with sclerotia of the reference strain SAS405 (MAT1-2) no apothecia were obtained (data not shown) indicating that the TF is required for sexual reproduction in B. cinerea as well. However, significant differences also exist that may reflect the requirement to adapt to different ecological niches. NsdD in A. flavus clearly regulates the mode of asexual reproduction, either via formation of conidia or sclerotia in a light-independent manner [61].
Regardless of the fact that the mentioned fungi are distantly related and exhibit very different modes of reproduction including different asexual and sexual structures, reproduction is controlled by different environmental cues. Therefore, protoperithecia formation in N. crassa requires nutrient limitation and light [19], and limited air exchange favors sexual reproduction in A. nidulans while nutrient limitation, light and other stresses shift the ratio from sexual to asexual reproduction [25]–[26]. On the contrary, light alone controls asexual reproduction in B. cinerea by inducing conidiation and suppressing sclerotial development preventing thereby the simultaneous formation of both structures. In contrast, the formation of microconidia is induced by nutrient limitation [62], and also the formation of apothecia in the laboratory takes place in the absence of nutrients and requires a photoperiod [10]. Hence, light has an outstanding function in B. cinerea because it determines the mode of reproduction.
In accordance with the findings that B. cinerea undergoes photomorphogenesis and that its germ tubes, conidiophores and apothecia stipes exhibit phototropic responses [63], a number of light-responsive genes (293) could be identified by genome-wide expression analysis. Similar studies have revealed 314, 533, and 250 light-responsive genes in N. crassa, A. nidulans, and A. fumigatus, respectively [37], [64]–[65]. Fifteen min of white light was sufficient to induce the transcription of bcltf1 and two other tested light-responsive genes (bop1, bcccg1), and maximal induction was achieved at exposure times of 45–60 min. BcLTF1 is an important modulator of the transcriptional responses to light as it influences the expression patterns of the majority of light-responsive genes (66%). Similar observations have been made for SUB-1 in N. crassa which acts in a hierarchical light-sensing cascade, being – as an early light-responsive gene – involved in induction of the majority of late light-responsive genes [37]. Noteworthy, transcript levels of both bcltf1 and sub-1 increase in response to light but no light responsiveness of A. nidulans nsdD has been reported [64], indicating that NsdD is unlikely to be involved in light signaling in the same fashion as its homologues in B. cinerea and N. crassa.
White light is composed of different wavelengths; therefore different photoreceptors are assumed to be involved in perception and subsequent signaling. The inducible effect of the blue light fraction on expression of bcltf1 is likely mediated via a WHITE COLLAR-like transcriptional complex (WCC) formed by the GATA transcription factors BcWCL1 and BcWCL2 that were reported to interact in the nuclei [66]. In fact, induction of bcltf1, bop1 and bcccg1 by white light requires BcWCL1 because in its absence the expression levels are much lower than in the wild type [P. Canessa, J. Schumacher, M. Hevia, P. Tudzynski, L. Larrondo, unpublished]. Nevertheless, an induction is still visible suggesting the involvement of other photoreceptors/wavelengths in transcriptional activation of bcltf1 and other light-induced genes. Δbcltf1 mutants exhibit similarly decreased radial growth rates in blue and white light suggesting that the blue light fraction accounts for the toxicity of white light, possibly by provoking greater oxidative stress than other wavelengths. Remarkably, the light-protecting function seems to be unique to BcLTF1 because N. crassa Δsub-1 and S. macrospora Δpro44 mutants exhibit comparable vegetative growth in light and darkness [L. Larrondo, pers. commun., 38].
The observation that expression levels of light-induced genes (bop1, bcccg1) decreased after prolonged exposure to light (from 120 min on) indicates the existence of photoadaptation processes in B. cinerea. In N. crassa, photoadaptation involves a further blue light-sensing protein (VIVID), whose expression is induced by light in a WCC-dependent manner, and that then negatively acts on the WCC to prevent further activation of light-responsive genes [67]–[68]. The homologue of B. cinerea (bcvvd1) is highly induced by light in both wild-type and Δbcltf1 strains, however, whether BcVVD1 impacts light-induced gene expression and is required for light tolerance like its counterpart in N. crassa needs to be studied. VIVID, the WCC and the FREQUENCY (FRQ) protein are furthermore implicated in running the circadian clock in N. crassa. Like vvd, expression of frq is turned on by the WCC in response to light, and subsequently FRQ blocks its own transcription by physically interacting with the WCC [20]. B. cinerea possesses a homologue of frq whose expression is induced by light in wild type and the bcltf1 deletion mutants to a similar extent, suggesting that the circadian clock is set by light in a BcLTF1-independent manner, as it was described for SUB-1 in N. crassa [37]. The fact that expression of bcltf1 in contrast to its homologue in N. crassa is not subjected to photoadaptation underlines the importance of the transcription factor for mediating the tolerance towards light in B. cinerea.
Based on the finding that the deletion of BcLTF1 results in hyperconidiation, it can be concluded that BcLTF1 acts as a general repressor of conidiation. This result was unexpected because the light-responsiveness of bcltf1 may suggest a stimulatory role of the TF on light-dependent differentiation processes. Additionally, a direct role for BcLTF1 in the induction of sclerotia development appears unlikely as its overexpression results in the accumulation of undifferentiated aerial mycelia instead of sclerotia. This fluffy phenotype resembles those of wild type colonies that have been illuminated with blue light [15]–[16] supporting the role of BcLTF1 in mediating blue light responses. Hence, it is concluded that conidiation is initiated by the differentiation of aerial hyphae (triggered by blue light, involving WCC and BcLTF1) followed by their differentiation to conidiophores (possibly triggered by the other light wavelengths/photoreceptors).
In addition to BcLTF1 that represses conidiation in light and darkness, five other light-responsive TFs were identified that may regulate differentiation in a light-dependent fashion. Homologues of bcltf2 and bcltf3 in N. crassa (sah-1/short aerial hyphae and csp-1/conidial separation) are inducible by light and control asexual development [36]–[37]. Increased expression levels of bcltf2 in the Δbcltf1 background in darkness correlate with the hyper-conidiation phenotype of the mutant, suggesting that BcLTF2 indeed may act as a positive regulator of conidiation. Light-responsive TFs may be also implicated in suppressing sclerotial development in the light, for instance by induction of transcriptional repressor(s) of sclerotia-related gene expression. Further studies on the light-responsive TFs are envisaged to reveal whether and how they are involved in differentiation of reproductive structures. Likewise these TFs may be involved in regulation of DHN-melanin biosynthesis, a pigment that is incorporated in the reproductive structures giving conidiophores, conidia and sclerotia of B. cinerea their characteristic gray to black color [69]–[70]. Accordingly, Δbcltf1 and Δbcvel1 mutants [31]–[32] exhibiting a hyper-conidiation phenotype produce melanin in excess leading to a dark coloration of the culture broths. In contrast to other DHN-melanin-producing fungi, B. cinerea possesses two highly similar PKS-encoding genes (bcpks12 and bcpks13) [41], while single copies for the other melanogenic genes exist. The relevance of having two copies of the PKS appears obscure when both enzymes are functional. However, considering the differential expression pattern of bcpks12 and bcpks13 in the Δbcltf1 background and during the life cycle of the wild type and the fact that B. cinerea produces, in contrast to other fungi, the same pigment under very different conditions, it is conceivable that BcPKS12 and BcPKS13 are responsible for sclerotia - and conidiophore/conidia-specific melanin biosynthesis, respectively. The observation that non-sporulating B. cinerea mutants produce an orange pigment during intensive illumination suggests furthermore that the fungus is able to produce carotenoids, a group of pigments that is produced by plants and fungi to protect cells from free radicals, ROS and singlet oxygen [71]. Consequently, increased carotenoid production may result in extended life spans as demonstrated in Podospora anserina [72]. Genes putatively involved in retinal biosynthesis in B. cinerea are inducible by light as their homologues in N. crassa and F. fujikuroi, but unlike the situation in N. crassa, genes in the two other fungi are organized together with the opsin-encoding gene in a gene cluster [37], [54]. Nevertheless, as described in N. crassa, light-induction of carotenogenesis in B. cinerea relies on the WCC [P. Canessa, J. Schumacher, M. Hevia, P. Tudzynski, L. Larrondo, unpublished]. Given the protective function of the carotenoids, their decreased production by Δbcltf1 mutants, as concluded from lower gene expression levels, may contribute to the hypersensitivity of the mutant to light and oxidative stress.
Several observations suggest that BcLTF1 is essential for maintaining the ROS homoeostasis. Expression levels of many ROS-related genes are altered in the absence of BcLTF1, though it is still ambiguous which of the changes (decreased scavenging, increased generation in the peroxisomes and/or mitochondria) cause the significantly increased net accumulation of H2O2 and therefore rendering Δbcltf1 mutants more sensitive to oxidative stress arising from exogenous ROS, light, and during infection. Some but not all phenotypes of Δbcltf1 mutants can be restored by antioxidants illustrating that the unbalanced ROS homoeostasis accounts for decreased virulence and mediates light toxicity, but it does not cause the observed developmental defects. Although clear evidences suggest an altered cellular redox status in the deletion mutants, the use of roGFP2 as sensor failed to confirm that, because the initial cytosolic glutathione pool is reduced and recovers after a short-term oxidation event caused by H2O2 in a manner almost as in the wild-type. However, given that the equilibrium of the glutathione pool is limited between different cellular compartments, as it was reported for plant cellular compartments [73], the mutant may experience oxidative stress in another compartment other than the cytosol. The overexpression of nuclear genes encoding enzymes of the alternative respiration pathway in Δbcltf1 mutants indicates the reprogramming of the mitochondria probably as a consequence of their dysfunction (retrograde signaling) [74]. The key component of the alternative pathway is the AOX that functions as the terminal oxidase (instead of the copper-containing COX in cytochrome ETC) transferring electrons to oxygen. Studies in different fungi reported an increased aox expression level in response to the inhibition of later steps of cytochrome ETC [75]–[78], to oxidative stress [79]–[82], and to copper deficiency causing COX assembly defects [83]–[84]. AOX activity has been reported to be dispensable for virulence in M. oryzae, B. cinerea and A. fumigatus, however, in the latter it contributes to oxidative stress resistance [85]–[86], [77; C. Levis, pers. commun.]. In P. anserina, AOX activity in complex III-deficient mutants leads to an extended lifespan associated with a decreased ROS production [87], [76]. On the contrary, the long-lived phenotype of mutants with a nonfunctional COX (deletion of the copper chaperone PaCOX17) is accompanied by increased ROS production [88]. Accordingly, studies in plant cells demonstrated the oxidation of the mitochondrial glutathione pool in response to the inhibition of COX (without affecting the cytosolic redox status), but not in response to inhibition of complex III [89]. From the present data it is difficult to define the reason for increased alternative respiration in Δbcltf1 mutants, i.e. whether increased overall ROS production or an impairment of the cytochrome ETC causes the expression of the corresponding genes. However, the observation that the homologues of the copper chaperone COX17 and the high-affinity copper transporter CTR3 are significantly overexpressed in the Δbcltf1 mutant may indicate an altered copper homoeostasis in the mutants which in turn may affect the activity of COX and other enzymes requiring copper as cofactor.
Regardless of how increased alternative respiration is achieved, the altered mode of respiration is expected to reduce mitochondrial ROS production and to cause further global changes in the metabolism of the fungus, because the electron flux through the alternative enzymes is not coupled with the translocation of protons across the membrane and therefore does not allow for proton gradient-driven ATP synthesis. Retrograde signaling in yeast cells was shown to affect glycolysis, the tricarboxylic acid cycle, peroxisomal fatty acid oxidation as well as the glyoxylate cycle [90]. We suggest therefore that the significantly altered secondary metabolism in Δbcltf1 mutants is due to an altered primary metabolism rather than to a direct involvement of BcLTF1 in regulation of secondary metabolism-related genes.
Taken together, it can be assumed that light affects B. cinerea in two ways (Fig. 9); directly via generation of singlet oxygen that damage macromolecules, and indirectly via its sensing by fungal photoreceptors altering gene expression. Products of light-responsive genes are implicated in ROS homoeostasis (via generation or scavenging), photoadaptation, the circadian clock, in promoting conidiation and apothecia formation and in preventing sclerotial development. Many, but not all, responses are altered in the absence of BcLTF1; and a hallmark of the deletion mutants is the unbalanced ROS homoeostasis (net ROS overproduction) accompanied by a retrograde response (alternative respiration) accounting for reduced virulence, light toxicity and potentially for altered secondary metabolism. Phenotypic and expression data suggest that BcLTF1 exerts repressing as well as activating functions in constant darkness and in response to light, though its transcription level is very low in darkness.
Fig. 9. Photoresponses in B. cinerea are modulated by the light-responsive transcription factor BcLTF1. 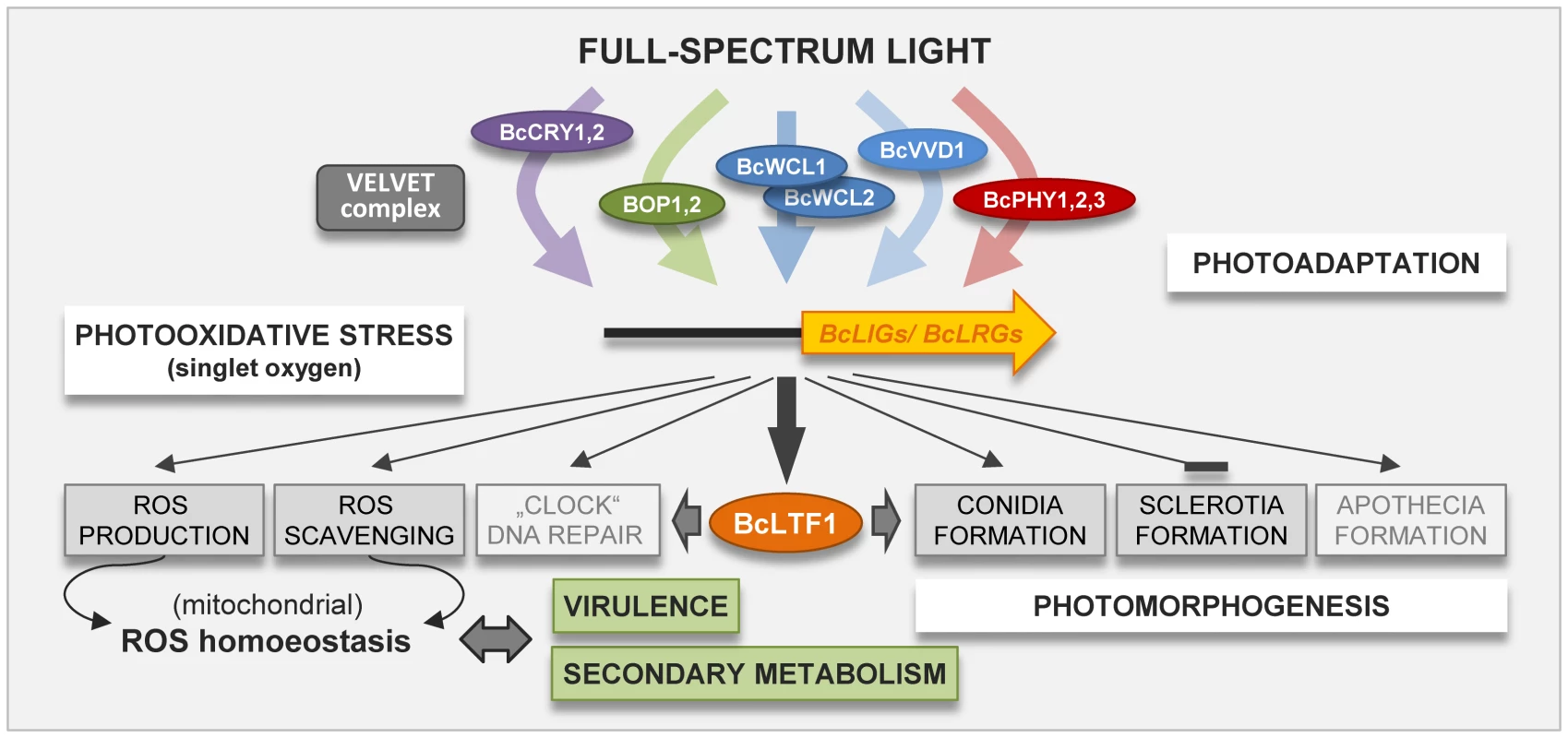
Light determines the mode of asexual reproduction (conidia vs. sclerotia), is required for differentiation of the fruiting bodies (apothecia) and is further considered to set the circadian clock. The cellular redox status is affected by emerging singlet oxygen and other ROS; compensation is achieved by induction of antioxidant systems including enzymes and carotenoids. Bcltf1 represents one of the 244 light-induced genes, and its absence affects several light-dependent features such as the formation of reproductive structures. The deletion of BcLTF1 leads to an unbalanced ROS status (net overproduction of H2O2), possibly in the mitochondria, as suggested by the upregulation of the alternative respiration pathway. The altered ROS status is considered to cause the inability of the deletion mutant to tolerate additional oxidative stress that arises during illumination (singlet oxygen), exposure to ROS (H2O2, menadione) and during host infection (oxidative burst). Materials and Methods
B. cinerea strains and growth conditions
Strain B05.10 of B. cinerea Pers. Fr. [B. fuckeliana (de Bary) Whetzel] is an isolate from Vitis (Table 1) and is used as the recipient strain for genetic modifications. Genome sequences of B05.10 and T4 were recently published [2], [91]. Strains were cultivated on plates containing solid synthetic complete medium (CM) [92], modified Czapek-Dox (CD) as minimal medium (2% sucrose, 0.1% KH2PO4, 0.3% NaNO3, 0.05% KCl, 0.05% MgSO4×7 H2O, 0.002% FeSO4×7 H2O, pH 5.0), or solid potato dextrose medium (Sigma-Aldrich, Germany) supplemented with 10% homogenized bean leaves. Acidification of the culture medium (solid CM pH 8) was monitored using the pH indicator bromothymol blue (0.1%) (Sigma-Aldrich, Germany). Cultures were incubated at 20°C under white light (12 h light/12 h darkness (LD) or continuous light (LL)) for conidiation, and in continuous darkness for induction of sclerotial formation. White light (9000 lx at culture level) was generated by using a set of Sylvania Standard F18W/29-530 “warm white” and F36W/33-640 “cool white” fluorescent bulbs. Wavelengths of transmitted light were controlled using Petri dish chambers covered with Roscolux polyester filters (#381 Baldassari Blue, #389 Chroma Green, #312 Canary, #27 Med Red; http://www.rosco.com/filters/roscolux.cfm); transmission spectra of the filters are shown in Fig. S3. Crossings using SAS405 (MAT1-2) as reference strain were performed as described previously [31]. For DNA and RNA isolation, mycelia were grown on solid CM medium with cellophane overlays.
Standard molecular methods
Fungal genomic DNA was prepared according to Cenis [93]. For Southern blot analysis, fungal genomic DNA was digested with restriction enzymes (Fermentas, Germany), separated on 1% (w/v) agarose gels and transferred to Amersham Hybond-N+ filters (GE Healthcare Limited, UK). Blot hybridizations with random-primed α-32P-dCTP-labelled probes were performed as described previously [94]. PCR reactions were performed using the high-fidelity DNA polymerase Phusion (Finnzymes, Finland) for cloning purposes and the BioTherm Taq DNA Polymerase (GeneCraft, Germany) for diagnostic applications. Replacement fragments and expression vectors were assembled in Saccharomyces cerevisiae by exploiting its homologous recombination machinery [36], [66]. Sequencing of DNA fragments was performed with the Big Dye Terminator v3.1 sequencing kit (Applied Biosystems, USA) in an ABI Prism capillary sequencer (model 3730; Applied Biosystems). For sequence analysis, the program package DNAStar (Madison, USA) was used. Protocols for protoplast formation and transformation of B. cinerea were described by Schumacher [66]. Regenerated protoplasts were overlaid with SH agar containing 70 µg/ml hygromycin B (Invitrogen, The Netherlands) and 140 µg/ml nourseothricin (Werner-Bioagents, Germany), respectively. Resistant colonies were transferred to agar plates containing CM medium supplemented with the respective selection agent in a concentration of 70 µg/ml. Single conidial isolates were obtained by spreading conidial suspensions on selective media and transferring single colonies to new plates.
Northern blot analyses
Total RNA was isolated making use of the TRIzol reagent (Invitrogen, The Netherlands). Samples (25 µg) of total RNA were transferred to Hybond-N+ membranes after electrophoresis on a 1% (w/v) agarose gel containing formaldehyde, according to the method of Sambrook et al. [95]. Blot hybridizations were carried out by use of random-primed α-32P-dCTP-labelled probes [94]. Analyzed genes are (Fig. 2): Bcssp1 (BC1G_03185) encoding the homologue of S. sclerotiorum ssp1 [40]; bcpks12 (AY495617) and bcpks13 (AY495618) encoding polyketide synthases putatively involved in melanin biosynthesis [41]; bop1 (BC1G_02456) encoding an opsin-like protein, expression is induced upon H2O2 [30] and light treatment (this study); bcccg1 (BC1G_11685) representing the homologue of N. crassa ccg-1 [42]), expression is induced upon H2O2 and light treatment (this study); bcactA (AJ000335) encoding actin is used in this study to detect fungal RNA in mixed (in planta) samples; bcgpx1 (BC1G_02031) encoding the glutathione peroxidase, expression is induced upon H2O2 treatment [96].
Identification of bcltf1 as virulence-associated gene
Strains PA31, PA2411 and PA3417 belong to an insertional mutant library of B. cinerea B05.10 that was generated by A. tumefaciens-mediated transformation (ATMT). Southern blot analyses demonstrating the existence of single T-DNA copies in the three mutants (data not shown) were performed according to Giesbert et al. [39]. The T-DNA-flanking regions were identified by TAIL-(thermal asymmetric interlaced) PCR as described previously [39]. BlastN analyses using the amplified T-DNA flanking regions revealed discontinuous sequences at the respective locus in both genome sequences of T4 (1-kb gap upstream of BofuT4_P09344; 480 aa) and B05.10 (2-kb gap upstream of BC1G_10441; 133 aa) (B. cinerea genome project, URGI; http://urgi.versailles.inra.fr/Species/Botrytis/) [2]. Re-sequencing of both strains yielded continuous sequences (B. cinerea Database, Broad Institute; http://www.broadinstitute.org/annotation/genome/botrytis_cinerea/ [91], hence, the T-DNA insertion occurred 1.608 kb upstream of the ORF hereafter called bcltf1 (Fig. 1B). Bcltf1 of B05.10 encodes a protein of 481 aa (BcLTF1B05.10: B0510_3555), that of T4 a protein of 480 aa (BcLTF1T4: BofuT4_P09344, BcT4_9306). Putative transcriptional start (TSS) and PolyA signals (Fig. 1B) were predicted using the FGENESH tool (http://linux1.softberry.com/). The nuclear localization signal (NLS) and the GATA zinc finger motif were predicted by WoLF PSORT (http://wolfpsort.org/) and Pfam Search (http://pfam.sanger.ac.uk), respectively (Fig. 1C). No nuclear export signals (NES) were identified by using NetNES 1.1 (http://www.cbs.dtu.dk/services/NetNES/). Putative phosphorylation sites for protein kinase A (PKA: S78, S95, S184, S185) and protein kinase C (PKC: T91, S105, S124, S194, T453, T457, S467, S468) in BcLTF1B05.10 were identified by running NetPhosK (http://www.cbs.dtu.dk/services/NetPhosK/).
Construction of bcltf1 mutants
For generation of fragments mediating the replacement of bcltf1 (Δbcltf1-A) and bcltf1+1.7 kb of the 5′-noncoding region (Δbcltf1-B), respectively, different 5′ flanks (A, B) were assembled with the same 3′ flank and a hygromycin resistance cassette by yeast recombinational cloning [36] (Table S1, Fig. S1A). Replacement fragments were transformed into B. cinerea B05.10. Homologous integration events in hygromycin-resistant transformants were detected by diagnostic PCR using the primers pCSN44-trpC-T and pCSN44-trpC-P, binding in the hygromycin resistance cassette and the primers bcltf1-A-hi5F/B-hi5F and bcltf1-hi3R, binding upstream and downstream of the bcltf1-flanking regions, respectively (Table S1, Fig. S1B). Single spore isolates were screened for the absence of bcltf1 alleles using primers bcltf1-WT-F and bcltf1-WT-R matching the substituted coding region. For Southern blot analysis, genomic DNA of the mutants and the recipient strain B05.10 was digested with PstI, and the blot was hybridized with the 3′ flank of the replacement fragment. The hygromycin resistance cassette contains an additional PstI restriction site resulting in smaller hybridizing fragments in the replacement mutants (2.3 kb) than in the wild type (7.2 kb) (Fig. S1C). Strain Δbcltf1-A1 was discarded because a second ectopic integration event occurred. Taken together, two independent mutants were generated lacking either only bcltf1 (Δbcltf1-A6) or bcltf1 and its 5′-noncoding region (Δbcltf1-B1). The mutant Δbcltf1-A6 was complemented by targeted integration of bcltf1 or bcltf1-gfp at the native gene locus thereby replacing the hygromycin resistance cassette. For vector construction, all fragments indicated in Fig. S1A were amplified by PCR using oligonucleotides listed in Table S1, and assembled in one step in S. cerevisiae yielding plasmids pbcltf1-COM and pbcltf1-gfpin loco. Replacement constructs were transformed into Δbcltf1-A6, and diagnostic PCR confirmed the targeted integration in two transformants per construct (bcltf1COM:T2, T3; and Δbcltf1+Pbcltf1::bcltf1-gfp: T2, T5) (Fig. S1D). For over-expressing the BcLTF1-GFP fusion protein, the ORF was introduced in vector pNAN-OGG comprising gene flanks for targeted integration at bcniiA (locus of nitrite reductase), a nourseothricin resistance cassette and the expression cassette with gfp under control of the constitutive oliC promoter [66] (Fig. S1E). The construct was transformed into B05.10 yielding transformants WT+PoliC::bcltf1-gfp (T6, T7), and into Δbcltf1-B1 yielding transformants Δbcltf1+PoliC::bcltf1-gfp (T3, T4). Targeted integration at bcniiA was verified by diagnostic PCR as described previously [66] (data not shown). Northern blot analyses revealed the constitutive expression of bcltf1-gfp in the mutant background and the overexpression of bcltf1-(gfp) in the wild-type background, respectively (Fig. S1F).
Germination and germling fusion assays
Conidial germination was monitored according to Doehlemann et al. [97]. For testing nutrient - and hydrophobicity-induced germination, cleaned conidia were incubated in Gamborg B5+10 mM glucose on glass surfaces and in double distilled water on polypropylene foil, respectively, in LL or DD conditions. Germination rates were determined in triplicates after 4 h and 8 h of incubation for glucose-induced and after 24 h for polypropylene-induced germination. Germling fusions via conidial anastomosis tubes were monitored according to Roca et al. [98]. Briefly, conidia were plated on solid Vogel's medium and incubated for 17 h at 21°C in the dark. Samples were analyzed by using light microcopy.
Virulence assays
For penetration assays, onion epidermal layers were prepared as described [31]. Conidial suspensions or non-sporulating mycelia from 3-d-old cultures were used for inoculation. The staining with lactophenol aniline blue (Sigma-Aldrich, Germany) allowed the identification of the penetrations sites after 24 h of incubation. For infection assays on French bean (Phaseolus vulgaris cv. 90598), primary leaves of living plants were inoculated with 7.5-µl droplets of conidial suspensions (2×105 conidia/ml Gamborg B5+2% glucose) or non-sporulating mycelia [31]. Infected plants were incubated in plastic boxes at 22°C under natural illumination conditions (LD) and continuous darkness (DD), respectively. Trypan blue and DAB (3,3′-diaminobenzidine) staining of lesions were performed as described previously [31]. Bright field images were taken using the Zeiss SteREO Discovery V.20 stereomicroscope or the AxioScope.A1 microscope equipped with an AxioCamMRc camera and the AxiovisionRel 4.8 software package (Zeiss, Germany).
Detection of reactive oxygen species (ROS)
To detect accumulating H2O2, strains were grown on solid CM covered with cellophane for 3 d in DD. Fresh mycelia (25 mg) were weighed, placed in wells of a 24-well plate and flooded with 1 ml DAB solution (0.5 mg/ml DAB in 100 mM of citric acid buffer, pH 3.7). Samples were incubated for 2 h in darkness at room temperature. The colorless DAB is oxidized by H2O2 in the presence of fungal peroxidases and horseradish peroxidase (positive control) resulting in a brown coloration of the staining solution. To detect accumulating O2.−, germinated conidia were incubated for 2 h in NBT staining solution (0.5 mg/ml nitrotetrazolium blue chloride in 50 mM sodium phosphate buffer, pH 7.5). Samples were analyzed by using light microcopy. The colorless substrate NBT is converted to a blue precipitate in the presence of O2−.
Measurement of the cellular glutathione redox potential
Vector pNAN-GRX1-roGFP2 carrying the reduction-oxidation sensitive green fluorescent protein (roGFP) and a nourseothricin resistance cassette [45], was transformed into the mutant Δbcltf1-A6 (Table 1). The homologous integration of the construct at bcniiA in two independent transformants (T2, T3) was verified by diagnostic PCR as described previously [66]. Reactions of roGFP2 in wild type B05.10 (WT+grx-rogfp2) and bcltf1 deletion mutants (Δbcltf1+grx-rogfp2) were monitored using a fluorometer as described by Heller et al. [45]. Briefly, germinated conidia (conidia cultivated for 12 h in liquid CM) were washed twice and transferred to 96-well plates. Fluorescence was measured at the bottom with 3×3 reads per well and an excitation wavelength of 395±5 nm for the oxidized state and 488±5 nm for the reduced state of roGFP2 using a Tecan Safire fluorometer. Relative fluorescence units (RFU) were recorded to calculate the Em395/Em488 ratio.
Fluorescence microscopy
For microscopy, conidia were suspended in Gamborg B5 solution supplemented with 2% glucose and 0.02% ammonium phosphate, and incubated overnight under humid conditions on microscope slides or on onion epidermal strips. Nuclei were stained using the fluorescent dye Hoechst 33342 [66], and fungal cell walls using calcofluor white (CFW) as described previously [60]. Fluorescence and light microscopy was performed with a Zeiss AxioImager.M1 microscope equipped with the ApoTome.2 technology for optical sectioning with structured illumination. Differential interference microscopy (DIC) was used for bright field images. Specimens stained by Hoechst 33342 and CFW were examined using the filter set 49 DAPI shift free (excitation G 365, beam splitter FT 395, emission BP 445/50), GFP fluorescence using filter set 38 (excitation BP 470/40, beam splitter FT 495, emission BP 525/50). Images (optical sections and Z-stacks) were captured with an AxioCam MRm camera and analyzed using the Axiovision Rel 4.8 software package (Zeiss, Germany).
Microarrays analyses
Wild type B05.10 and the Δbcltf1 mutant were grown for 3 d on solid CM in LD before non-sporulating mycelia were transferred to solid CM covered with cellophane overlays. Cultures were incubated for 52 h in DD, and then half of the cultures were transferred to white light (+LP). After 60 min all cultures (WT-D; WT+LP; Δbcltf1-D; Δbcltf1+LP) were harvested. Material derived from four independent experiments was used for RNA isolation (Trizol procedure). Total RNA was treated with the DNA-free kit to remove any trace of DNA (Ambion - Applied Biosystems, France). Synthesis of double-stranded cDNA, Cy3-labeling and hybridization on microarrays were done by PartnerChip (http://www.partnerchip.fr/) using the procedures established by NimbleGen (Roche) and the reagents from Invitrogen (Life technologies, France). To study the complete transcriptome of B. cinerea, NimbleGen 4-plex arrays with 62,478 60-mer specific probes covering all the 20,885 predicted gene models and non-mapping ESTs of B. cinerea [2] were used. The arrays also include 9,559 random probes as negative controls. Data processing, quality controls and differential expression analysis were performed using ANAIS methods [99]. Probe hybridization signals were subjected to RMA-background correction, quantile normalization, and gene summarization [100]–[101]. Genes with a normalized intensity over the threshold (1.5×95th percentile of random probes) in at least one hybridization were considered as expressed and kept for further analyses. Differentially expressed genes (light response in wild type and mutant, Δbcltf1 effect in darkness and light) were then identified using a one-way ANOVA test. To deal with multiple testings, the ANOVA p-values were then submitted to a False Discovery Rate correction. Genes with a corrected p-value <0.05, and more than a 2-fold change in transcript level were considered as significantly differentially expressed. BlastN analyses for the 20,885 spotted genes [2; URGI database] were performed to identify the matching gene models derived from the re-sequencing approaches by Staats & van Kan [91; Broad Institute]. By this, the number of differentially expressed genes was reduced because incorrect gene models derived from discontinuous sequences were revised and furthermore previously non-matching ESTs could be mapped. The four gene lists were then pooled and cluster analyses were performed to highlight the different expression profiles encountered. For this purpose, the log2-normalized intensities scaled by gene across the 16 hybridizations were clustered using Genesis tools [102]. Gene enrichment analyses were further performed with GSEA toolkit [46]–[47] to highlight significantly enriched functions compared to the complete list of functionally annotated B. cinerea genes [2]. Details on the experiments, raw and normalized values are available at NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) (Accession: GPL17773). Furthermore, lists of differentially expressed genes are available at http://urgi.versailles.inra.fr/Data/Transcriptome.
Supporting Information
Zdroje
1. WilliamsonB, TudzynskiB, TudzynskiP, van KanJA (2007) Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol 8 : 561–580.
2. AmselemJ, CuomoCA, van KanJA, ViaudM, BenitoEP, et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7: e1002230.
3. Van KanJA (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci 11 : 247–253.
4. Sharon A, Elad Y, Barakat R, Tudzynski P (2004) Phytohormones in Botrytis-plant interactions. In: Elad Y, Williamson B, Tudzynski P, Delen N, editors. Botrytis: biology, pathology, control. Kluwer, Dordrecht. pp. 163–179.
5. ChoquerM, FournierE, KunzC, LevisC, PradierJM, et al. (2007) Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol Lett 277 : 1–10.
6. Tudzynski P, Kokkelink L (2009) Botrytis cinerea: Molecular aspects of a necrotrophic life style. In: Deising H, editor. The Mycota V, Plant Relationships, 2nd Edition. Springer-Verlag Berlin Heidelberg. pp. 29–50.
7. SiewersV, ViaudM, Jimenez-TejaD, ColladoIG, Schulze GronoverC, et al. (2005) Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol Plant-Microbe Interact 18 : 602–612.
8. PinedoC, WangCM, PradierJM, DalmaisB, ChoquerM, et al. (2008) Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem Biol 3 : 791–801.
9. DalmaisB, SchumacherJ, MoragaJ, LePêcheurP, TudzynskiB, et al. (2011) The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol Plant Pathol 12 : 564–579.
10. FaretraF, AntonacciE (1987) Production of apothecia of Botryotinia fuckeliana (de Bary) Whetz under controlled environmental conditions. Phytopathol Mediterr 26 : 29–35.
11. GiraudT, FortiniD, LevisC, LerouxP, BrygooY (1997) RFLP markers show genetic recombination in Botryotinia fuckeliana (Botrytis cinerea) and transposable elements reveal two sympatric species. Mol Biol Evol 14 : 1177–1185.
12. FaretraF, AntonacciE, PollastroS (1988) Sexual behaviour and mating system of Botryotinia fuckeliana, teleomorph of Botrytis cinerea. J Gen Microbiol 134 : 2543–2550.
13. PaulWRC (1929) A comparative morphological and physiological study of a number of strains of Botrytis cinerea Pers. with special reference to their virulence. Trans Br Mycol Soc 14 : 118–134.
14. QuiddeT, OsbournAE, TudzynskiP (1998) Detoxification of α-tomatine by Botrytis cinerea. Physiol Mol Plant Pathol 52 : 151–165.
15. TanKK, EptonHAS (1973) Effect of light on the growth and sporulation of Botrytis cinerea. Trans Br Mycol Soc 61 : 147–157.
16. TanKK (1974) Blue-light inhibition of sporulation in Botrytis cinerea. J Gen Microbiol 82 : 191–200.
17. TanKK (1975) Interaction of near-ultraviolet, blue, red, and far-red light in sporulation of Botrytis cinerea. Trans Br Mycol Soc 64 : 215–222.
18. Schumacher J, Tudzynski P (2012) Morphogenesis and infection in Botrytis cinerea. In: Pérez-Martín J, Di Pietro A, editors. Topics in Current Genetics, Vol. Morphogenesis and Pathogenicity in Fungi. Springer-Verlag Berlin, Heidelberg. pp. 225–241.
19. LindenH, BallarioP, MacinoG (1997) Blue light regulation in Neurospora crassa. Fungal Genet Biol 22 : 141–150.
20. ChenCH, DunlapJC, LorosJJ (2010) Neurospora illuminates fungal photoreception. Fungal Genet Biol 47 : 922–929.
21. ShimuraM, ItoY, IshiiC, YajimaH, LindenH, et al. (1999) Characterization of a Neurospora crassa photolyase-deficient mutant generated by repeat induced point mutation of the phr gene. Fungal Genet Biol 28 : 12–20.
22. FroehlichAC, ChenCH, BeldenWJ, MadetiC, RoennebergT, et al. (2010) Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. Eukaryot Cell 9 : 738–750.
23. BieszkeJA, BraunEL, BeanLE, KangS, NatvigDO, et al. (1999) The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc Natl Acad Sci USA 96 : 8034–8039.
24. FroehlichAC, NohB, VierstraRD, LorosJ, DunlapJC (2005) Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot Cell 4 : 2140–2152.
25. AdamsTH, WieserJK, YuJH (1998) Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev 62 : 35–54.
26. HanKH (2009) Molecular Genetics of Emericella nidulans sexual development. Mycobiol 37 : 171–182.
27. BayramO, BiesemannC, KrappmannS, GallandP, BrausGH (2008) More than a repair enzyme: Aspergillus nidulans photolyase-like CryA is a regulator of sexual development. Mol Biol Cell 19 : 3254–3262.
28. PurschwitzJ, MüllerS, KastnerC, SchöserM, HaasH, et al. (2008) Functional and physical interaction of blue - and red-light sensors in Aspergillus nidulans. Curr Biol 18 : 255–259.
29. Rodriguez-RomeroJ, HedtkeM, KastnerC, MüllerS, FischerR (2010) Fungi, hidden in soil or up in the air: light makes a difference. Annu Rev Microbiol 64 : 585–610.
30. HellerJ, RuhnkeN, EspinoJ, MassaroliM, ColladoIG, et al. (2012) The MAP kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol Plant Microbe Interact 25 : 802–816.
31. SchumacherJ, PradierJM, SimonA, TraegerS, MoragaJ, et al. (2012) Natural variation in the VELVET gene bcvel1 affects virulence and light-dependent differentiation in Botrytis cinerea. PLoS One 7: e47840.
32. SchumacherJ, GautierA, MorgantG, StudtL, DucrotPH, et al. (2013) A functional bikaverin biosynthesis gene cluster in rare strains of Botrytis cinerea is positively controlled by VELVET. PLoS One 8: e53729.
33. HanKH, HanKY, YuJH, ChaeKS, JahngKY, et al. (2001) The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol Microbiol 41 : 299–309.
34. GrosseV, KrappmannS (2008) The asexual pathogen Aspergillus fumigatus expresses functional determinants of Aspergillus nidulans sexual development. Eukaryot Cell 7 : 1724–1732.
35. SzewczykE, KrappmannS (2010) Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryot Cell 9 : 774–783.
36. ColotHV, ParkG, TurnerGE, RingelbergC, CrewCM, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103 : 10352–10357.
37. ChenCH, RingelbergCS, GrossRH, DunlapJC, LorosJJ (2009) Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J 28 : 1029–1042.
38. NowrousianM, TeichertI, MasloffS, KückU (2012) Whole-genome sequencing of Sordaria macrospora mutants identifies developmental genes. G3 (Bethesda) 2 : 261–270.
39. GiesbertS, SchumacherJ, KupasV, EspinoJ, SegmüllerN, et al. (2012) Identification of pathogenesis-associated genes by T-DNA-mediated insertional mutagenesis in Botrytis cinerea: A type 2A phosphoprotein phosphatase and a SPT3 transcription factor have significant impact on virulence. Mol Plant Microbe Interact 25 : 481–495.
40. LiM, RollinsJA (2009) The development-specific protein (Ssp1) from Sclerotinia sclerotiorum is encoded by a novel gene expressed exclusively in sclerotium tissues. Mycologia 101 : 34–43.
41. KrokenS, GlassNL, TaylorJW, YoderOC, TurgeonBG (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA 100 : 15670–15675.
42. LorosJJ, DenomeSA, DunlapJC (1989) Molecular cloning of genes under control of the circadian clock in Neurospora. Science 243 : 385–388.
43. LerochM, MernkeD, KoppenhoeferD, SchneiderP, MosbachA, et al. (2011) Living colors in the gray mold pathogen Botrytis cinerea: codon-optimized genes encoding green fluorescent protein and mCherry, which exhibit bright fluorescence. Appl Environ Microbiol 77 : 2887–2897.
44. GovrinEM, LevineA (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10 : 751–757.
45. HellerJ, MeyerAJ, TudzynskiP (2012) Redox-sensitive GFP2: use of the genetically encoded biosensor of the redox status in the filamentous fungus Botrytis cinerea. Mol Plant Pathol 13 : 935–947.
46. SubramanianA, KuehnH, GouldJ, TamayoP, MesirovJP (2007) GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23 : 3251–3253.
47. MoothaVK, LindgrenCM, ErikssonKF, SubramanianA, SihagS, et al. (2003) PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34 : 267–273.
48. SmithKM, SancarG, DekhangR, SullivanCM, LiS, et al. (2010) Transcription factors in light and circadian clock signaling networks revealed by genome-wide mapping of direct targets for Neurospora white collar complex. Eukaryot Cell 9 : 1549–1556.
49. ChristieJM (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58 : 21–45.
50. SimonA, DalmaisB, MorgantG, ViaudM (2013) Screening of a Botrytis cinerea one-hybrid library reveals a Cys(2)His(2) transcription factor involved in the regulation of secondary metabolism gene clusters. Fungal Genet Biol 52 : 9–19.
51. HoffmeisterD, KellerNP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24 : 393–416.
52. ThewesS, Prado-CabreroA, PradoMM, TudzynskiB, AvalosJ (2005) Characterization of a gene in the car cluster of Fusarium fujikuroi that codes for a protein of the carotenoid oxygenase family. Mol Genet Genomics 274 : 217–228.
53. Prado-CabreroA, ScherzingerD, AvalosJ, Al-BabiliS (2007) Retinal biosynthesis in fungi: characterization of the carotenoid oxygenase CarX from Fusarium fujikuroi. Eukaryot Cell 6 : 650–657.
54. AvalosJ, EstradaAF (2010) Regulation by light in Fusarium. Fungal Genet Biol 47 : 930–938.
55. HellerJ, TudzynskiP (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu Rev Phytopathol 49 : 369–390.
56. RhoadsDM, UmbachAL, SubbaiahCC, SiedowJN (2006) Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol 141 : 357–366.
57. Van AkenO, GiraudE, CliftonR, WhelanJ (2009) Alternative oxidase: a target and regulator of stress responses. Physiol Plant 137 : 354–361.
58. VolkovAN, NichollsP, WorrallJA (2011) The complex of cytochrome c and cytochrome c peroxidase: the end of the road? Biochim Biophys Acta 1807 : 1482–1503.
59. JarmuszkiewiczW, Woyda-PloszczycaA, Antos-KrzeminskaN, SluseFE (2010) Mitochondrial uncoupling proteins in unicellular eukaryotes. Biochim Biophys Acta 1797 : 792–799.
60. SchumacherJ, de LarrinoaIF, TudzynskiB (2008) Calcineurin-responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development, and full virulence on bean plants. Eukaryot Cell 7 : 584–601.
61. CaryJW, Harris-CowardPY, EhrlichKC, MackBM, KaleSP, et al. (2012) NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot Cell 11 : 1104–1111.
62. UrbaschI (1985) Ultrastructural studies on the microconidia of Botrytis cinerea Pers. and their phialoconidial development. Phytopath Z 112 : 229–237.
63. Jarvis WR (1977) Botryotinia and Botrytis Species: Taxonomy, Physiology and Pathogenicity. A Guide to the Literature. Canadian Department of Agriculture, Ottawa. pp. 38–39.
64. Ruger-HerrerosC, Rodríguez-RomeroJ, Fernández-BarrancoR, OlmedoM, FischerR, et al. (2011) Regulation of conidiation by light in Aspergillus nidulans. Genetics 188 : 809–822.
65. FullerKK, RingelbergCS, LorosJJ, DunlapJC (2013) The fungal pathogen Aspergillus fumigatus regulates growth, metabolism, and stress resistance in response to light. MBio 4: e00142–13.
66. SchumacherJ (2012) Tools for Botrytis cinerea: new expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet Biol 49 : 483–497.
67. HeintzenC, LorosJJ, DunlapJC (2001) The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104 : 453–464.
68. SchwerdtfegerC, LindenH (2003) VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J 22 : 4846–4855.
69. DossRP, DeisenhoferJ, Krug von NiddaHA, SoeldnerAH, McGuireRP (2003) Melanin in the extracellular matrix of germlings of Botrytis cinerea. Phytochemistry 63 : 687–691.
70. ZeunR, BuchenauerH (1985) Effect of tricyclazole on production and melanin contents of sclerotia of Botrytis cinerea. Phytopathol Z 112 : 259–267.
71. GaoQ, Garcia-PichelF (2011) Microbial ultraviolet sunscreens. Nat Rev Microbiol 9 : 791–802.
72. StrobelI, BreitenbachJ, ScheckhuberCQ, OsiewaczHD, SandmannG (2009) Carotenoids and carotenogenic genes in Podospora anserina: engineering of the carotenoid composition extends the life span of the mycelium. Curr Genet 55 : 175–184.
73. SchwarzländerM, FrickerMD, MüllerC, MartyL, BrachT, et al. (2008) Confocal imaging of glutathione redox potential in living plant cells. J Microsc 231 : 299–316.
74. RhoadsDM, SubbaiahCC (2007) Mitochondrial retrograde regulation in plants. Mitochondrion 7 : 177–194.
75. DuarteM, VideiraA (2009) Effects of mitochondrial complex III disruption in the respiratory chain of Neurospora crassa. Mol Microbiol 72 : 246–258.
76. SellemCH, MarsyS, BoivinA, LemaireC, Sainsard-ChanetA (2007) A mutation in the gene encoding cytochrome c1 leads to a decreased ROS content and to a long-lived phenotype in the filamentous fungus Podospora anserina. Fungal Genet Biol 44 : 648–658.
77. GrahlN, DinamarcoTM, WillgerSD, GoldmanGH, CramerRA (2012) Aspergillus fumigatus mitochondrial electron transport chain mediates oxidative stress homeostasis, hypoxia responses and fungal pathogenesis. Mol Microbiol 84 : 383–399.
78. DufourE, BoulayJ, RinchevalV, Sainsard-ChanetA (2000) A causal link between respiration and senescence in Podospora anserina. Proc Natl Acad Sci USA 97 : 4138–4143.
79. MagnaniT, SorianiFM, MartinsVP, NascimentoAM, TudellaVG, et al. (2007) Cloning and functional expression of the mitochondrial alternative oxidase of Aspergillus fumigatus and its induction by oxidative stress. FEMS Microbiol Lett 271 : 230–238.
80. HondaY, HattoriT, KirimuraK (2012) Visual expression analysis of the responses of the alternative oxidase gene (aox1) to heat shock, oxidative, and osmotic stresses in conidia of citric acid-producing Aspergillus niger. J Biosci Bioeng 113 : 338–342.
81. YukiokaH, InagakiS, TanakaR, KatohK, MikiN, et al. (1998) Transcriptional activation of the alternative oxidase gene of the fungus Magnaporthe grisea by a respiratory-inhibiting fungicide and hydrogen peroxide. Biochim Biophys Acta 1442 : 161–169.
82. MartinsVP, DinamarcoTM, SorianiFM, TudellaVG, OliveiraSC, et al. (2011) Involvement of an alternative oxidase in oxidative stress and mycelium-to-yeast differentiation in Paracoccidioides brasiliensis. Eukaryot Cell 10 : 237–248.
83. BorghoutsC, ScheckhuberCQ, StephanO, OsiewaczHD (2002) Copper homeostasis and aging in the fungal model system Podospora anserina: differential expression of PaCtr3 encoding a copper transporter. Int J Biochem Cell Biol 34 : 1355–1371.
84. StumpferlSW, StephanO, OsiewaczHD (2004) Impact of a disruption of a pathway delivering copper to mitochondria on Podospora anserina metabolism and life span. Eukaryot Cell 3 : 200–211.
85. Avila-AdameC, KöllerW (2002) Disruption of the alternative oxidase gene in Magnaporthe grisea and its impact on host infection. Mol Plant Microbe Interact 15 : 493–500.
86. MagnaniT, SorianiFM, Martins VdeP, PolicarpoAC, SorgiCA, et al. (2008) Silencing of mitochondrial alternative oxidase gene of Aspergillus fumigatus enhances reactive oxygen species production and killing of the fungus by macrophages. J Bioenerg Biomembr 40 : 631–636.
87. LorinS, DufourE, BoulayJ, BegelO, MarsyS, et al. (2001) Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol Microbiol 42 : 1259–1267.
88. ScheckhuberCQ, HouthoofdK, WeilAC, WernerA, De VreeseA, et al. (2011) Alternative oxidase dependent respiration leads to an increased mitochondrial content in two long-lived mutants of the aging model Podospora anserina. PLoS One 6: e16620.
89. SchwarzländerM, FrickerMD, SweetloveLJ (2009) Monitoring the in vivo redox state of plant mitochondria: effect of respiratory inhibitors, abiotic stress and assessment of recovery from oxidative challenge. Biochim Biophys Acta 1787 : 468–475.
90. ButowRA, AvadhaniNG (2004) Mitochondrial signaling: the retrograde response. Mol Cell 14 : 1–15.
91. StaatsM, van KanJA (2012) Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot Cell 11 : 1413–1414.
92. PontecorvoGV, RoperJA, HemmonsLM, MacDonaldKD, BuftonAWJ (1953) The genetics of Aspergillus nidulans. Adv Genet 5 : 141–238.
93. CenisJL (1992) Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res 20 : 2380.
94. SiewersV, SmedsgaardJ, TudzynskiP (2004) The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl Environ Microbiol 70 : 3868–3876.
95. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
96. TemmeN, TudzynskiP (2009) Does Botrytis cinerea ignore H(2)O(2)-induced oxidative stress during infection? Characterization of Botrytis activator protein 1. Mol Plant Microbe Interact 22 : 987–998.
97. DoehlemannG, BerndtP, HahnM (2006) Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol Microbiol 59 : 821–835.
98. RocaMG, WeichertM, SiegmundU, TudzynskiP, FleissnerA (2012) Germling fusion via conidial anastomosis tubes in the grey mould Botrytis cinerea requires NADPH oxidase activity. Fungal Biol 116 : 379–387.
99. SimonA, BiotE (2010) ANAIS: Analysis of NimbleGen Arrays Interface. Bioinformatics 26 : 2468–2469.
100. IrizarryRA, HobbsB, CollinF, Beazer-BarclayYD, AntonellisKJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 : 249–264.
101. BolstadBM, IrizarryRA, AstrandM, SpeedTP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 : 185–193.
102. SturnA, QuackenbushJ, TrajanoskiZ (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18 : 207–208.
Štítky
Genetika Reprodukčná medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



