-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Unwrapping Bacteria
article has not abstract
Published in the journal: Unwrapping Bacteria. PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004054
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004054Summary
article has not abstract
Imagine you must wrap a present—from the inside. You (as the “gift”) must first cover yourself with one or more sheets of light tissue paper, to keep from getting smudged or rattling around. Next, without leaving or tearing this papery cocoon, you must construct a rugged box to encase both gift and tissue, to protect against mishaps and external onslaughts. Finally, from deep within, using no tools that would damage or mar these previous bits of handiwork, you must overlay the whole parcel with a thin film of wrapping paper, colorfully patterned on its outer side but plain on its inner. If you are even more exuberant you may add ribbons, bows, and baubles to spruce up the completed package, but all the while you must remain embedded at center of these nested shells.
Pretty impressive trick, huh?
This lighthearted metaphor describes something very close to what gram-negative bacteria do on a moment-to-moment basis as they create the envelope that surrounds and defends them. The cytoplasm (the “gift”) is surrounded by the inner membrane (the “tissue paper”), the peptidoglycan cell wall (the “box”), and the outer membrane (the “wrapping paper”) (see Figure 1). In truth, the task is even more difficult than suggested by this analogy because the components must grow as the cell grows, divide when the mass doubles, allow some materials to cross while excluding others, and protect against a high internal turgor pressure, and none of these activities must compromise the integrity of any other element. In short, to create a cell “from the inside” requires the coordination of a remarkable suite of strategies and competing biochemical reactions. How all this is accomplished is the core concern of a new technique introduced by Paradis-Bleau et al. in this issue of PLOS Genetics [1].
Fig. 1. This schematic illustrates some of the pathways that must be coordinated to create an intact gram-negative bacterial cell envelope. 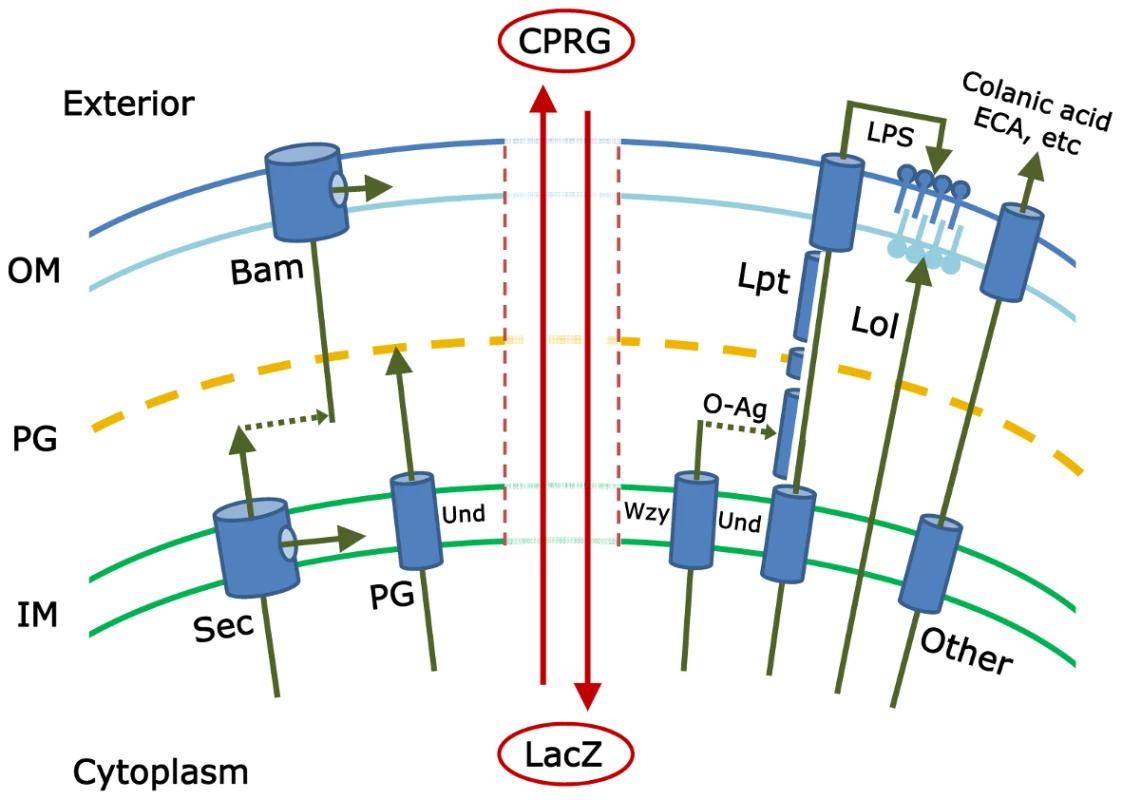
The inner membrane (IM), peptidoglycan (PG), and outer membrane (OM) form the principal layers into which components are inserted by pathways referenced in the text. The isoprenoid compound undecaprenyl-phosphate (Und) is an integral part of several of these pathways. The vertical central zone represents envelope damage that may accompany mutations in these pathways; breakdown of envelope integrity may allow the cytoplasmic LacZ protein to come into contact with and hydrolyze the extracellular compound CPRG (chlorophenyl red-β-D-galactopyranoside). The resulting red color marks the colonies of such mutants and is the basis for the genetic screen described by Paradis-Bleau et al. Note: the actual gram-negative envelope is much more complicated and includes, for example, the Tat secretory pathway; other specialized protein secretion systems; and additional envelope proteins, extracellular components, and pathways. Any of these may contribute to envelope integrity and therefore may be subjects of study by the screen introduced by Paradis-Bleau et al. [1]. To date, investigators have pieced together the major mechanisms by which each envelope subcomponent is synthesized and directed to one of four destinations (inner membrane, periplasm, or to one of the two faces of the outer membrane) (see Figure 1). Proteins are directed to the inner membrane and periplasm (the space between the inner and outer membranes) via the Sec or Tat secretory pathways [2], [3]; lipopolysaccharides and lipoproteins are directed to the outer and inner leaflets of the outer membrane via the Lpt and Lol pathways, respectively [4], [5]; proteins are inserted into the outer membrane by way of the Bam pathway [5]–[7]; the cell wall peptidoglycan is polymerized in the periplasm [8]; and carbohydrate polymers, such as colanic acid, enterobacterial common antigen (ECA), or capsule, are delivered to the cell surface or extracellular space [9]–[11]. And yet, despite all we know, at least two large questions remain. First, have all the pertinent biochemical pathways been described? Probably not, because little or nothing is known about the function of nearly one-third of the ∼400 predicted envelope proteins in Escherichia coli [12]. The second, more mysterious and difficult question is this: how are all these pathways choreographed so that the cell grows smoothly? The identities of these crucial regulatory processes are deeply enigmatic.
Paradis-Bleau and colleagues describe a genetic approach that promises to move us closer to finding the answers to these lingering questions [1]. They begin by assuming that mutations in individual biochemical steps or in overarching regulatory pathways will produce cells with defective envelopes. The simplest phenotype of such an envelope is that it becomes porous to substances that are otherwise contained within the cell or excluded from it. Alternately, envelope function may be so severely impaired that a fraction of the bacterial culture dies and releases cytoplasmic material. Paradis-Bleau et al. devised a simple screen to detect mutants that meet either of these criteria. The small molecule CPRG (chlorophenyl red-β-D-galactopyranoside) is incorporated into an agar plate, onto which mutagenized bacteria are spread. Wild-type E. coli cells form white colonies on plates containing this compound because CPRG cannot enter intact bacteria. However, cells with an impaired envelope may admit CPRG to the cytoplasm, where the LacZ protein hydrolyzes it to form a red product. Alternatively, if the mutants lyse, LacZ is released into the medium to contact CPRG (see Figure 1). In either case the colonies become visibly red and can be isolated for further study.
When Paradis-Bleau et al. tested a library of random transposon insertions and an ordered set of gene deletions in E. coli, the screen identified envelope-damaging effects caused by ∼100 genes with no known function. For example, mutants lacking a functional elyC (ycbC) gene lysed as they approached stationary phase, a transition point whose regulation is poorly understood. This phenotype was suppressed by overexpressing enzymes involved in the synthesis of peptidoglycan or undecaprenyl pyrophosphate, this latter a critical isoprenoid that participates in the biosynthesis of several envelope components [10], [13], [14]. In addition, mutations in the ECA synthetic pathway exacerbated or suppressed this elyC-associated lethality. Thus, characterizing the behavior and genetic interactions of this single mutation has already identified a web of new envelope relationships. More are expected, since the technique allows investigators to bring a whole pantheon of genetic tools to bear on these questions, and because the screen can be adapted to organisms whose envelopes are even less well understood.
Of course, even such a significant advance does not solve all problems. First, the technique identifies only those mutants that are viable and that disable the envelope so that it leaks. Mutations that kill cells outright will not appear, nor will mutations that alter assembly without affecting envelope permeability. Second, the screen will probably return mutants that die or leak even if the affected processes have no direct bearing on the envelope (though these may be interesting in their own right). Third, there is an odd temperature limitation: the screen works well at 25°C, but a large background appears when cells are incubated at 37°C. The reason is not clear, but this, too, might be turned into a positive by inverting the approach to look for mutants that do not leak under these or other conditions, thereby expanding the screen's genetic reach. Thus, even its limitations presage the expansion of the technique for use in broader contexts.
In short, Paradis-Bleau et al. have performed a valuable service by developing this new tool for investigating the complexities of the bacterial cell envelope. And that's a gift for all of us, wrapped just right.
Zdroje
1. Paradis-BleauC, KritikosG, OrlovaK, TypasA, BernhardtT (2013) A genome-wide screen for bacterial envelope biogenesis mutants identifies a novel factor involved in cell wall precursor metabolism. PLoS Genet 10: e1004056 doi:10.1371/journal.pgen.e1004056
2. FaceySJ, KuhnA (2010) Biogenesis of bacterial inner-membrane proteins. Cell Mol Life Sci 67 : 2343–2362.
3. RobinsonC, MatosCF, BeckD, RenC, LawrenceJ, et al. (2011) Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim Biophys Acta 1808 : 876–884.
4. ZhangG, MeredithTC, KahneD (2013) On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol 16 : 1–7.
5. RicciDP, SilhavyTJ (2012) The Bam machine: a molecular cooper. Biochim Biophys Acta 1818 : 1067–1084.
6. HaganCL, KimS, KahneD (2010) Reconstitution of outer membrane protein assembly from purified components. Science 328 : 890–892.
7. StroudDA, MeisingerC, PfannerN, WiedemannN (2010) Biochemistry. Assembling the outer membrane. Science 328 : 831–832.
8. SobhanifarS, KingDT, StrynadkaNC (2013) Fortifying the wall: synthesis, regulation and degradation of bacterial peptidoglycan. Curr Opin Struct Biol S0959-440X(13)00129-2 [pii], 10.1016/j.sbi.2013.07.008.
9. WhitfieldC (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75 : 39–68.
10. KajimuraJ, RahmanA, RickPD (2005) Assembly of cyclic enterobacterial common antigen in Escherichia coli K-12. J Bacteriol 187 : 6917–6927.
11. IslamST, LamJS (2013) Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ Microbiol 15 : 1001–1015.
12. HuP, JangaSC, BabuM, Díaz-MejiaJJ, ButlandG, et al. (2009) Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol 7: e96 doi:10.1371/journal.pbio.1000096
13. LoveringAL, SafadiSS, StrynadkaNC (2012) Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem 81 : 451–478.
14. TengKH, LiangPH (2012) Structures, mechanisms and inhibitors of undecaprenyl diphosphate synthase: a cis-prenyltransferase for bacterial peptidoglycan biosynthesis. Bioorg Chem 43 : 51–57.
Štítky
Genetika Reprodukčná medicína
Článek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



