-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Molecular Mechanisms of Ethanol-Induced Pathogenesis Revealed by RNA-Sequencing
Acinetobacter baumannii is a common pathogen whose recent resistance to drugs has emerged as a major health problem. Ethanol has been found to increase the virulence of A. baumannii in Dictyostelium discoideum and Caenorhabditis elegans models of infection. To better understand the causes of this effect, we examined the transcriptional profile of A. baumannii grown in the presence or absence of ethanol using RNA-Seq. Using the Illumina/Solexa platform, a total of 43,453,960 reads (35 nt) were obtained, of which 3,596,474 mapped uniquely to the genome. Our analysis revealed that ethanol induces the expression of 49 genes that belong to different functional categories. A strong induction was observed for genes encoding metabolic enzymes, indicating that ethanol is efficiently assimilated. In addition, we detected the induction of genes encoding stress proteins, including upsA, hsp90, groEL and lon as well as permeases, efflux pumps and a secreted phospholipase C. In stationary phase, ethanol strongly induced several genes involved with iron assimilation and a high-affinity phosphate transport system, indicating that A. baumannii makes a better use of the iron and phosphate resources in the medium when ethanol is used as a carbon source. To evaluate the role of phospholipase C (Plc1) in virulence, we generated and analyzed a deletion mutant for plc1. This strain exhibits a modest, but reproducible, reduction in the cytotoxic effect caused by A. baumannii on epithelial cells, suggesting that phospholipase C is important for virulence. Overall, our results indicate the power of applying RNA-Seq to identify key modulators of bacterial pathogenesis. We suggest that the effect of ethanol on the virulence of A. baumannii is multifactorial and includes a general stress response and other specific components such as phospholipase C.
Published in the journal: Molecular Mechanisms of Ethanol-Induced Pathogenesis Revealed by RNA-Sequencing. PLoS Pathog 6(4): e32767. doi:10.1371/journal.ppat.1000834
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000834Summary
Acinetobacter baumannii is a common pathogen whose recent resistance to drugs has emerged as a major health problem. Ethanol has been found to increase the virulence of A. baumannii in Dictyostelium discoideum and Caenorhabditis elegans models of infection. To better understand the causes of this effect, we examined the transcriptional profile of A. baumannii grown in the presence or absence of ethanol using RNA-Seq. Using the Illumina/Solexa platform, a total of 43,453,960 reads (35 nt) were obtained, of which 3,596,474 mapped uniquely to the genome. Our analysis revealed that ethanol induces the expression of 49 genes that belong to different functional categories. A strong induction was observed for genes encoding metabolic enzymes, indicating that ethanol is efficiently assimilated. In addition, we detected the induction of genes encoding stress proteins, including upsA, hsp90, groEL and lon as well as permeases, efflux pumps and a secreted phospholipase C. In stationary phase, ethanol strongly induced several genes involved with iron assimilation and a high-affinity phosphate transport system, indicating that A. baumannii makes a better use of the iron and phosphate resources in the medium when ethanol is used as a carbon source. To evaluate the role of phospholipase C (Plc1) in virulence, we generated and analyzed a deletion mutant for plc1. This strain exhibits a modest, but reproducible, reduction in the cytotoxic effect caused by A. baumannii on epithelial cells, suggesting that phospholipase C is important for virulence. Overall, our results indicate the power of applying RNA-Seq to identify key modulators of bacterial pathogenesis. We suggest that the effect of ethanol on the virulence of A. baumannii is multifactorial and includes a general stress response and other specific components such as phospholipase C.
Introduction
Acinetobacters are Gram-negative bacteria that belong to the Moraxellaceae family [1]. The members of the Acinetobacter group are metabolically versatile since they can metabolize an important number of compounds such as aliphatic alcohols, some amino acids, decarboxylic and fatty acids, unbranched hydrocarbons, aromatic compounds, mandelate, and n-hexadecane. [2]. Moreover, accumulation of wax esters has been described for various Acinetobacter species [3]. These features have attracted attention toward several species of the genus given their potential use in the chemical industry.
Recently, A. baumannii has emerged as an opportunistic pathogen. Nosocomial and community acquired infections are associated with a wide spectrum of clinical manifestations, including pneumonia (the most frequent pathology associated with this microorganism), urinary tract infections, bacteremia and meningitis [4]-[7]. Furthermore, there has been a recent emergence of multidrug-resistant (MRD) isolates of A. baumannii strains resistant to a wide range of antimicrobial drugs such as aminopenicillins, ureidopenicillins, cephalosporins, chloramphenicol, and tetracycline [8],[9]. Indeed, 89% of Acinetobacter strains isolated from patients injured in Iraq and Afghanistan were resistant to at least two major classes of antibiotics [10],[11].
So far, lipopolysaccharide (LPS) [12],[13], an outer membrane protein named OmpA [14],[15], the pili [16], and two siderophore mediated iron-acquisition systems [17]–[19] have been proposed as determinants of A. baumannii pathogenicity. It is conceivable that additional elements could be involved in the pathogenesis of this bacterium. The complete genome sequences of several isolates of this species revealed the presence of homologues of virulence genes from other pathogens [20]–[23]. Examples include homologues of luxI and luxR that allow cell-cell communication, genes that encode two-component systems, genes that code for several hydrolytic enzymes, efflux pumps, and genes involved with resistance to antibiotics. However, in most cases, evidence regarding the contribution of each of these elements to Acinetobacter pathogenicity is lacking.
It was previously observed, that co incubation of yeast with A. baumannii promotes bacterial growth; the molecule responsible for this effect was shown to be ethanol. It was demonstrated that low concentrations of ethanol not only stimulated A. baumanni growth but also helped the ability of this bacteria to endure salt stress. Furthermore, in the presence of ethanol A. baumannii showed increased pathogenicity towards C. elegans [24]. It was subsequently reported that the increased pathogenicity of ethanol-fed A. baumannii was also observed when D. discoideum was used as a model host [25]. Furthermore, genes associated with A. baumannii virulence were identified by insertional mutagenesis; one of the genes identified by this strategy is homologous to pstC, which encodes a component of the high affinity phosphate transport system [25]. Interestingly, in some pathogenic bacteria the phosphate regulon appears to be part of a network that controls virulence and a particular stress response (recently reviewed by [26]). Another gene identified in the insertional screen was rpoH, which in many species, encodes the sigma factor that is responsible of the transcriptional induction of the genes that mediate the heat-stress response (HSR) [27]. The HSR has been shown to be required for full virulence in other pathogenic bacteria [28]–[31]. Thus, the potential contribution of the HSR to the virulence of A. baumannii remains as an attractive possibility.
To explore the underlying basis for the increased virulence of A. baumannii in the presence of ethanol, we characterized the transcriptional profile of A. baumannii grown in rich medium in the presence or absence of ethanol. Seventy genes whose expression is altered by the presence of ethanol in the growth medium were identified. Based on our results we suggest that the increased virulence of A. baumannii in the presence of ethanol is due to increased metabolic capacity, coupled with the expression of several key factors mostly related with stress responses that likely contribute to the virulence of this bacterium. In addition, ethanol promotes the expression of plc1, which encodes an A. baumannii-specific phospholipase C. We found that a plc mutant showed a reduction in A. baumannii-induced cytotoxicity of epithelial cells, suggesting that phospholipase C acts as a virulence factor in this bacterium.
Results/Discussion
Transcriptional profiling of A. baumannii by high throughput RNA sequencing
To identify genes important for an inducible virulence response in Acinetobacter, we first search for genes that were differentially expressed in the presence of ethanol. Total RNA was isolated from A. baumannii cells grown to mid-log phase in the absence or presence of ethanol (see Methods section). For the first set of samples, the mRNA was selectively enriched through a single step of rRNA depletion. These samples were fragmented and used to obtain cDNA libraries that were sequenced as described in Methods section. A total of 6,441,146 and 6,603,654 28 nt reads were obtained for each library (“no ethanol” and “ethanol”, respectively). Of these, 4,364,106, (no ethanol) and 4,824,047 (ethanol) reads mapped to multiple targets in the genome and only 312,266 (no ethanol) and 546,498 (ethanol) mapped uniquely. Analysis of 1% of the reads that mapped to the genome multiple locations revealed that most were derived from 23S, 16S, and 5S rRNA. In order to obtain a more representative sampling of the coding regions, the libraries were sequenced an additional two times. Additional RNA samples were prepared in duplicate experiments and subjected to three-cycles of rRNA depletion followed by size exclusion to remove small RNAs, but no substantial improvement in the number of unique reads was achieved with this procedure.
Combining all of the sequencing from both experiments produced a total of 3,596,474 unique reads that represent a total of 100,701,272 nucleotides (28 nt per read), a 25.3 fold average coverage of the A. baumannii genome. The sequences obtained from these experiments were mapped to the A. baumannii genome using the current annotation in Genbank (accession: CP000521 version CP000521.1). As expected, between 72 and 84% of the unique sequences correspond to previously annotated coding regions, whereas the remainder correspond to intercistronic regions, RNA molecules such as the tmRNA, ribonuclease P, 7S RNA, and possible regulatory RNAs, such as the TPP riboswitch. Two previously unannotated genes were also identified. These genes are a putative ferredoxine located downstream of A1S_0845, and a gene located upstream of A1S_2262 that has sequence similarity to SirA.
Two different approaches were used to demonstrate that the number of reads that map to a particular open reading frame correlate with the expression level of that gene. In the first approach, mRNA from two different tissues was analyzed using microarrays and RNA-Seq [32]. In the second one, microarray and RNA-seq data were compared with protein expression data obtained by shotgun mass spectroscopy [33]. In both cases, good levels of correlation were observed. Therefore, we computed the total number of reads for each gene, and this number was divided into windows of 250 bp to calculate the number of relative reads (NRR) per window. We observed that the highest NRR mapped to the loci corresponding to the tmRNA and RNase P, followed by the genes A1S_2840 (NRR 50,000), and A1S_2218 (NRR 41,000) that encode OmpA (Outer Membrane Protein A), and the pili subunit CsuA/B, respectively. The NRR was also high for genes encoding proteins related to transcription, translation and energy generation. We detected 163 genes with a NRR less than 1 (Table S1), suggesting that some genes are expressed at a low level or that these are not readily detected because of experimental bias. As shown in Figure 1, 50% of Acinetobacter genes have NRR values of approximately 150. Both technical and biological duplicates showed high reproducibility (Fig. 1).
Fig. 1. Analysis of the RNA-Seq short sequences (reads) mapped to the genome of A. baumannii. 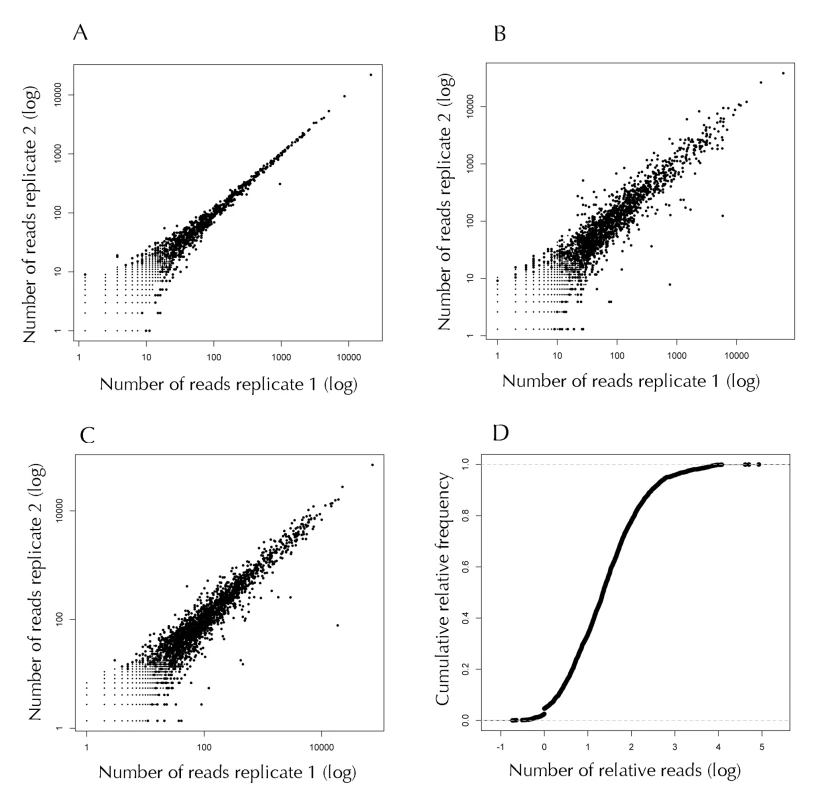
(A) Reproducibility between technical replicates. The total reads obtained from independent lanes of one flow cell were mapped to the genome of A. baumannii. The number of reads was normalized, and the absolute number of reads mapping to each coding region is compared. (B and C) Reproducibility between biological replicates. The reads obtained from independent libraries were mapped to the genome of A. baumannii, the number of reads between libraries was normalized and the absolute number of reads mapping to each coding region is compared. Panel B: Comparison between the libraries obtained from the cultures grown in the absence of ethanol. Panel C: Comparison between the libraries obtained from cultures grown in the presence of 1.1% ethanol. The R-squared values are: panel A, 0.99; panel B, 0.87; panel C, 0.94. (D) Cumulative relative frequency of the number of relative reads (NRR). To identify the genes that are induced or repressed by ethanol, we normalized the number of reads for each pair of libraries, and the number of reads for each gene was compared. The genes that reproducibly showed a ratio larger than 1.9 or below 0.5 in both biological replicates and a P value of 0.05 or less were considered as regulated by ethanol. From 101 genes that passed the first criterion, only 70 showed a P-value below 0.05. From these, 21 genes were repressed and 49 were induced by ethanol (Table 1).
Tab. 1. Up-regulated and down-regulated genes in A. baumannii (Ab) by ethanol. 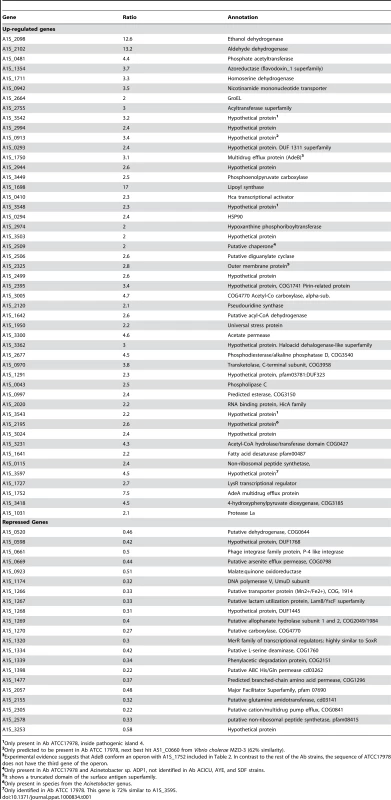
1Only present in Ab ATCC17978, inside pathogenic island 4. We also extracted total RNA from cultures grown to stationary phase in the presence of ethanol and performed a more limited RNA-Seq experiment that produced 146,170 unique reads. The NRR for each gene was calculated and arranged by rank order (Table S2). RNA from stationary phase cells with ethanol exhibited a large number of reads for genes related with the synthesis of siderophores and iron uptake. In fact, in this sample, 33 genes related with iron acquisition are among the 10% of the genes that showed the highest NRR values (Table S2). Interestingly, a putative operon encoding four proteins similar to the high-affinity phosphate transport system were also found among the 10% of the genes with highest NRR. Given the relevance of Fe and phosphate acquisition to bacterial pathogenesis we further explored the expression of these genes by qRT-PCR or using lacZ as reporter gene (see below).
Metabolic effect of ethanol
We examined in detail the genes that were induced by growth in the presence of ethanol during exponential phase. The most strongly induced genes encode proteins related to central metabolism or with ethanol/acetate assimilation. These included ethanol dehydrogenase (A1S_2098) and aldehyde dehydrogenase (A1S_2102), which showed an average induction of 12.6 and 13.2-fold, respectively. A. baumannii has two other genes that potentially encode a Fe-dependent ethanol dehydrogenase, i.e., A1S_2053, and A1S_2702; these genes are transcriptionally active since they showed a NRR of 28 and 194. However, their expression is not induced by ethanol. Other genes that may encode additional ethanol dehydrogenases are A1S_1788 and A1S_3436. We observed a slight induction of A1S_1788 by ethanol, but it showed a p-value higher than the selected cutoff (Table 2). Therefore, under our experimental conditions, A. baumannii seems to be oxidizing ethanol to acetate by the activity of the enzymes encoded by A1S_2098, A1S_2102 and perhaps A1S_1788 (Fig. 2).
Fig. 2. Metabolic pathways affected by the presence of ethanol. 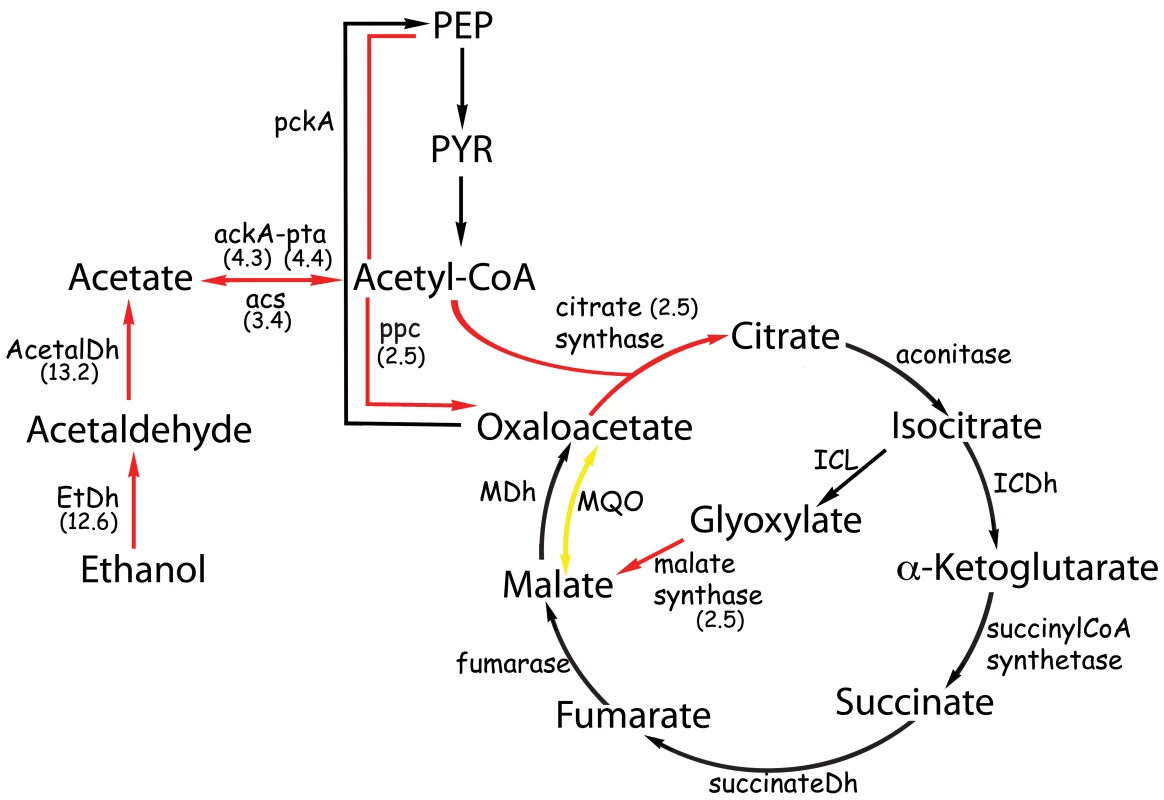
The numbers in parenthesis represent the ratio of the number of gene-specific mapped reads from the libraries obtained from cultures grown in ethanol and in the absence of ethanol. EtDh, ethanol dehydrogenase; acetalDh acetaldehyde dehydrogenase; ackA, acetate kinase; pta, phosphate transacetylase; acs, acetyl-CoA synthetase; ICDh, isocitrate dehydrogenase; ICL, isocitrate lyase; MDh, malate dehydrogenase; MQO, malate:quinone oxidoreductase; pckA, phosphoenolpyruvate carboxykinase; ppc, phosphoenolpyruvate carboxylase. Red and yellow arrows represent induced or repressed genes, respectively. Tab. 2. Genes with a P-value above the threshold but showing an induction of two fold or more in two independent experiments. 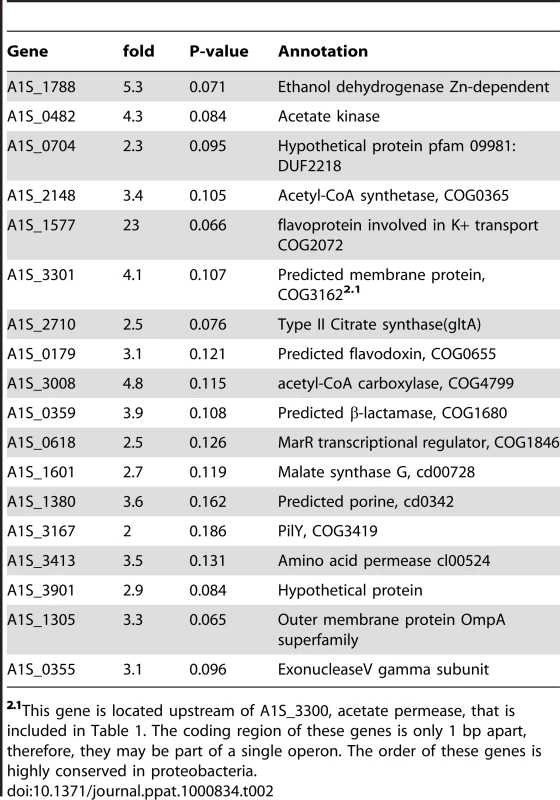
2.1This gene is located upstream of A1S_3300, acetate permease, that is included in Table 1. The coding region of these genes is only 1 bp apart, therefore, they may be part of a single operon. The order of these genes is highly conserved in proteobacteria. In many bacteria, after conversion to acetate, ethanol is further metabolized into acetyl-CoA and then assimilated through the glyoxylate cycle [34],[35]. Acetyl-CoA synthesis occurs by the action of ackA and pta (acetate kinase and phosphate acetyltransferase, respectively) as has been demonstrated for Corynebacterium glutamicum [36],[37] or through acs (acetyl-CoA synthetase) as has been demonstrated for Escherichia coli [38],[39]. In these bacteria, ackA and pta are arranged in a bicistronic operon [40],[41]. In A. baumannii, these genes are contiguous in the chromosome and therefore may be part of a single operon. From our data, we found that the genes coding for pta (A1S_0481) and ackA (A1S_0482) showed a 4.4 (Table 1) and 5.3 (Table 2) fold induction in the presence of ethanol. The same situation was found for the gene A1S_2148, which encodes acs which was induced 3.4 fold. Both acs and ackA showed a p-value higher than the threshold (Table 2), but nonetheless, evidence obtained by quantitative RT-PCR corroborated the induction of these two genes by ethanol (see below).
Interestingly, A1S_3300, which encodes an acetate permease, was induced 4.6 fold (Table 1). In E. coli, acetate is excreted into the culture medium during exponential phase when the cells are grown in the presence of a high concentration of acetogenic sugars. However, when the culture begins the transition into stationary phase, acetate assimilation begins. In exponential phase, pyruvate is converted to acetate through the action of ackA and pta, whereas in stationary phase, acetate is assimilated by acs [38]. This does not seem to be the case in A. baumannii, since our results indicate that during exponential phase all these genes are simultaneously expressed. Therefore, a balance between assimilation and excretion may be occurring.
In many organisms, acetate is metabolized through the glyoxylate cycle, which in conjunction with other reactions of the citric acid cycle (TCA) allows the net synthesis of succinate from two molecules of acetyl-CoA. At the first step of this cycle, acetyl-CoA and oxaloacetate are used to form citrate. Isocitrate is then converted to glyoxylate and succinate, and finally malate is formed from glyoxylate and acetyl-CoA (Fig. 2). The enzymes that carry out these reactions are citrate synthase, aconitase, isocitrate lyase, and malate synthase [34],[35],[42],[43]. We observed that the genes encoding citrate synthase (A1S_2710), and malate synthase G (A1S_1601) were induced 2.5 and 2.7 fold respectively but with a p-value above the threshold (Table 2). qRT-PCR experiments verified that both genes are indeed induced by ethanol (see below). Therefore, acetate assimilation appears to take place through the glyoxylate cycle in A. baumannii. In E. coli there are two different malate synthases (i.e., malate synthase A and G encoded by aceB and glcB, respectively) [44]–[46]. This latter gene, glcB, codes for a secondary malate synthase that can replace the malate synthase A when aceB is mutated [44]. In contrast, only the malate synthase G is present in A. baumannii, similar to the situation found in C. glutamicum [37].
Unexpectedly, we did not observe an induction of the gene encoding isocitrate lyase (A1S_1008). This may be due to the presence of glucose in the media we used in these experiments, which may have repressed the expression of the isocitrate lyase gene. This result suggests that under these conditions, control of the isocitrate dehydrogenase by phosphorylation is sufficient for activation of the glyoxylate cycle. In A. baumannii the genes encoding isocitrate lyase, malate synthase and isocitrate kinase are not encoded by the same operon. Therefore, it is conceivable that these genes are differentially regulated.
We also detected the induction (2.5 fold) of A1S_3449, a gene encoding a putative phosphoenolpyruvate carboxylase (PEPCx) (Table 1). In other bacteria, this enzyme converts phosphoenolpyruvate to oxaloacetate accomplishing an anaplerotic function (Fig. 2) [47]. Another ethanol-induced enzyme related to the metabolism of acetate is a putative acetyl-CoA hydrolase/transferase, encoded by A1S_3231. The product of this gene is highly similar to proteins found in other bacteria, and it is 62% similar to Ach1 (acetyl-CoA hydrolase) from Saccharomyces cerevisiae. In S. cerevisiae, it has been shown that ach1 is not involved in the hydrolysis of acetyl-CoA, as originally thought. Instead, Ach1 transfers CoASH from succinyl-CoA to acetate, and this activity is required to support growth in acetate [48]. Due to the high similarity between CoA tranferases and hydrolases the actual activity of A1S_3231 in A. baumannii remains to be determined.
We detected a two-fold repression of A1S_3025 which encodes a putative malate∶quinone reductase (MQO). A. baumannii, like many other bacteria, possesses two genes that encode for a malate dehydrogenase, a membrane-associated malate∶quinone oxidoreductase (MQO) (A1S_0923), and a cytoplasmic malate dehydrogenase (MDH) (A1S_3025). Our results indicate that the expression level of A1S_3025 did not change in response to ethanol whereas the putative MQO A1S_0923 was down regulated by 50%. Given that MQO is repressed, MDH should be the main enzyme responsible for malate oxidation in this condition (Fig. 2). Similarly, MQO does not seem to play a significant role in malate oxidation in E. coli [49], but for C. glutamicum it has been shown that the malate∶quinone reductase (MQO) is the main enzyme catalyzing the oxidation of malate to oxalacetate [50]. A 2-fold reduction was also observed for A1S_1334 encoding L-serine deaminase. The reaction catalyzed by this enzyme yields pyruvate and ammonia. It is possible that this reduction helps prevent a further increase in the availability of acetyl-CoA above the level that ethanol catabolism produces.
Overall these results indicate that a variety of metabolic genes are affected by the presence of ethanol and show for the first time the metabolic pathways involved in ethanol assimilation in this bacterium.
Ethanol induces genes involved in the stress response and pathogenesis
In exponential phase, ethanol elicits the induction of 11 genes that encode hypothetical proteins that do not belong to any pfam or COG already described (Table 1). Five of these genes are unique to A. baumannii ATCC17978, and three of them are located in pathogenicity island 4 [20]. Of the remaining six, A1S_2195 is only present in organisms that belong to the Acinetobacter genus, and the other five have homologues in many other bacteria (Table 1). Of particular interest, A1S_2509 is present only in A. baumannii ATCC17978 and the non-pathogenic A. baylyi ADP1. The proteins encoded by these organisms are 40% identical; the first 50 residues of this protein showed high similarity with DjlC (DnaJ - containing domain protein) from E. coli and its homologues in other bacteria [28]. Interestingly, A1S_2509 is adjacent to a gene encoding an HSP70-like protein, and this arrangement is conserved among several bacteria [51]. It has been shown that DjlC produces a 10-fold activation of the ATPase activity of the HSP70-like protein [52], and it was proposed that DjlC and HSP70-like were required to respond to certain stress conditions. Therefore, it is possible that A1S_2509 may help to resist the ethanol stress together with A1S_2510 (HSP70-like). Consistent with this hypothesis, we observed that A1S_2510 (HSP70-like) is mildly induced by ethanol (1.4 fold, p-value 0.006) (data not shown).
The A1S_1641 gene, which encodes a fatty acid desaturase, was induced 2.2 fold (Table 1). In other microorganisms, it has been shown that an increase in the amount of unsaturated fatty acids facilitates adaptation to stressful conditions such as acid pH, ionic stress and ethanol [53]–[56]. Other genes induced by ethanol are A1S_1750 and A1S_1752, that likely are part of an operon and encode for an RND-type efflux pump that confers resistance to various antibiotics in A. baumannii BM4454 [57]. Interestingly, it has been shown that an RND efflux pump contributes to drug resistance and virulence of Francisella tularensis in mice [58]. We also observed the induction of A1S_1950, which encodes a protein that belongs to the universal stress protein family. It is known that members of this family are induced when the cell is exposed to agents that induce stress [59]–[61]. Interestingly, A. baumannii has five proteins that belong to this family A1S_1950, A1S_2692, A1S_2072, A1S_0214 and A1S_1246, but only A1S_1950 was induced by ethanol.
Recently, it was shown that overexpression of rpoH in E. coli, induces a set of genes that were not originally considered as a part of the heat-shock response (HSR); acpD is one of these genes [62]. The physiological role of AcpD is still uncertain since its original assignment was as an ACP-phosphohydrolase. AcpD was subsequently shown to be an azoreductase [63]. The induction of acpD (A1S_1354) by ethanol supports the idea that this gene is part of a stress response. A reduction of two-fold was detected for a cluster of five genes that could form an operon from A1S_1266 to A1S_1270; unfortunately, the function of these genes is unknown.
One of the aims of this work was to identify potential virulence factors whose expression is induced in presence of ethanol. In this regard, two general traits were observed. First, there was the mild induction of Hsp90, GroEL, and Lon. In many bacterial species, these genes are part of the heat-shock stress response (HSR) [64]–[69]. Second, several genes known to be important for survival under diverse stress conditions exhibited increased expression (Table 1). Members of the HSR are chaperones that refold or prevent aggregation of misfolded proteins [27],[70], and Lon is a protease that hydrolyzes proteins with unstructured regions [71]. In many bacteria, the control of the HSR is mediated by the sigma factor σ32, encoded by the rpoH gene [27]. It has been shown that the HSR is required for full virulence in some pathogenic bacteria. For instance, DnaJ-like (HSP40) from Vibrio tapetis is required for cytotoxicity of hemocytes [28], σ32 is required for the invasion of epithelial cells by Neisseria gonorrhoeae [29] and the chaperons HSP90 and GroEL of many pathogenic bacteria induce the production of interleukin-8, modulating the immune response [30],[31],[72]. Consistent with these reports, a strain of A. baumannii carrying a transposon insertion in the gene encoding for σ32 (rpoH) was shown previously to be avirulent in the presence of ethanol towards C. elegans and D. discoideum [20]. Therefore, it is conceivable that ethanol could exacerbate the virulence of A. baumannii through the induction of heat-shock proteins, such as Hsp90, GroEL and Lon. The products of other genes listed in Table 1 may also help A. baumannii to tolerate stress conditions. Since it has been shown that one stress response might help bacteria to contend with other stress conditions [73]–[76], it is attractive to hypothesize that ethanol could improve the ability of A. baumannii to survive in the host since that several pathways of stress responses are activated.
Among the genes that we detected as induced by ethanol (Table 1), A1S_0043 encoding a phospholipase C deserves particular attention. This protein has been recognized as a virulence factor in other pathogenic bacteria [77]–[79]. For this reason, we further analyzed this gene as described below.
Quantitative RT-PCR of selected targets
To validate the induction of some of the genes identified by RNA-Seq (Table 1), we carried out qRT-PCR experiments. The expression of A1S_2664, A1S_0294, and A1S_0043 that encode for GroEL, HSP90 and phospholipase C, respectively, was measured, along with several genes whose expression was induced by ethanol but showed a p-value above the threshold (Table 2). These latter genes were A1S_0482, A1S_2148, A1S_2710, and A1S_1601 which encode acetate kinase, acetyl_CoA synthetase, citrate synthase, and malate dehydrogenase G, respectively. Ethanol dehydrogenase (A1S_2098) was used as a positive control. The down-regulated gene A1S_1266 was also included in this analysis. A1S_2846 and A1S_0880 were used as internal controls to calculate the fold-change after treatment with ethanol (see methods section). To test the expression of the genes above mentioned, we used total RNA isolated from exponential cultures of A. baumannii grown in the absence or presence of ethanol.
As shown in Fig. 3A, qRT-PCR experiments confirmed that the expression of all these genes was regulated by ethanol. Moreover, the fold change detected for each gene was similar to the ratio of induction and repression observed by RNA-Seq (Fig. 3B). These results support the conclusions outlined in the previous section regarding ethanol metabolism.
Fig. 3. Fold change of selected genes determined by qRT-PCR. 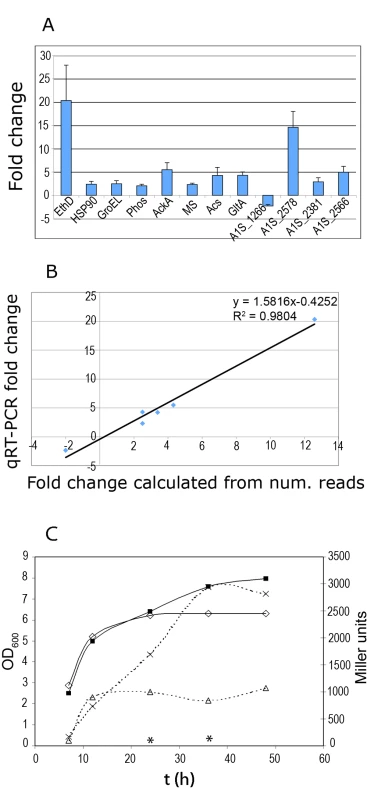
(A) Fold change of the expression levels of genes A1S_2098 encoding ethanol dehydrogenase (fold change 20.3); A1S_0294, HSP90 (2.38); A1S_2664, GroEL (2.46); A1S_0043, phospholipase C (2.02); A1S_0482, acetate kinase (5.5); A1S_1601, malate synthase G (2.3); A1S_2148 acetyl-CoA synthetase (4.25); A1S_2710, citrate synthase (4.3); A1S_1266, hypothetical protein (−2.3). These genes were tested using RNA obtained from cultures grown in 1.1% ethanol and without ethanol. The genes tested using RNA obtained from stationary phase cultures with and without ethanol are: A1S_2578, A1S_2381, and A1S_2566, encoding a putative non-ribosomal peptide synthetase (similar to the subunit F of the enterobactin synthetase), acinetobactin synthetase subunit E, and a siderophore receptor protein, respectively. (B) Correlation between the expression ratios of selected genes determined by qRT-PCR and RNA-Seq. (C) Time course of expression of the ptsS-lacZ fusion along the growth curve. A. baumannii cells carrying the plasmid expressing pstS-lacZ were grown in YPDA medium in the presence or absence of ethanol. At the indicated times, cell growth was monitored as culture turbidity at 600nm (continuous lines) from cultures grown in the absence (open squares) or presence (filled squares) of ethanol. β-Galactosidase activity (dashed lines) was determined from cultures grown in the absence (open triangles) or presence (multiplication symbol) of ethanol. Asterisks indicate the β-galactosidase activity from cultures grown in the presence of 10 mM phosphate buffer. These time points showed an OD600 of 6 and 6.2, respectively. All results are the mean of three experiments, with standard deviations of less than 15%. To validate the high expression level of the genes detected in the samples obtained from stationary phase cultures grown in the presence of ethanol, we also measured the expression levels of A1S_2381, A1S_2566, and A1S_2578 by qRT-PCR. These genes were randomly chosen among the genes that are related to Fe uptake and showed a high number of reads (NRR) (Table S2). A1S_2381 is required for acinetobactin synthesis and is located within a locus of 13 genes that are involved in the synthesis and transport of acinetobactin in A. baumannii ATCC19606 and A. baumannii ATCC17978. In contrast, A1S_2566 and A1S_2578 (encoding a protein required for siderophore synthesis and a siderophore receptor, respectively) are part of a second locus for Fe acquisition that is present in A. baumannii ATCC17978 but not in A. baumannii ATCC19606 [17]–[19]. To test the expression of these genes, we used total RNA from stationary phase cells that were grown in the presence or absence of ethanol (see methods). As shown in Fig. 3B, the expression of these genes was induced by the presence of ethanol in the culture medium, although this effect could be indirect given that a higher O.D.600 is reached when ethanol is included in the culture medium [24] (see panel C in Figure 3).
We also tested the induction of expression for the genes that belong to the putative pts operon (A1S_2448 to A1S_2445) which encode a high-affinity phosphate transport system. For this study, we used lacZ as reporter gene. The promoter region upstream of A1S_2448 (homologous to pstS) was cloned into pMP220. The amount of β-galactosidase produced by A. baumannii transformed with this construction was low when 10 mM PO4 was included in the culture medium (Fig. 3C, measures indicated by asterisks). The expression of lacZ was evaluated in A. baumannii cells grown in the absence or presence of ethanol at different time points of incubation. As shown in Fig. 3C, a low level of β-galactosidase activity was detected for both cultures at OD600 <3, but higher activities were observed as the cultures approached stationary phase. At 24 h of incubation, in the presence of ethanol higher amounts of β-galactosidase were detected nevertheless both cultures showed a similar OD indicating that increased β-galactosidase was not due to higher cell numbers (Fig. 3C). To ensure that plasmid integrity was intact through these experiments, we rescued the plasmids from both ethanol treated and untreated cultures at the end of the experiment and found that in each case the plasmid were functional and without detectable rearrangements. Thus, these results indicate that the high-affinity phosphate transport system of A. baumannii is highly expressed at high cell densities and that A1S_2448 is induced after incubation with ethanol.
The induction of the transport genes A1S_2448-45 is of particular significance since it has been demonstrated previously that a strain of A. baumannii carrying an insertion in A1S_2447 (homologue of ptsC) was avirulent towards C. elegans and D. discoideum [20]. Therefore, it is conceivable that ethanol could exacerbate the virulence of A. baumannii taking advantage of the induction of these uptake systems (Fe and phosphate) that in other bacteria have also been related with virulence [26].
It has been reported that community-acquired Acinetobacter infections are associated with underlying conditions such as alcoholism, smoking, chronic obstructive pulmonary disease and diabetes [80]–[82]. Our results provide mechanistic insight into how ethanol may modulate A. baumannii infections. Furthermore, the molecular mechanism underlying this effect could be multifactorial, given that ethanol up regulates TLR2 causing inflammation of the airway epithelium [83]. Ethanol also induces a delay of viability loss in stationary-phase cultures of bacteria [84], and we demonstrate that ethanol induces a stress response that may give the pathogen a better fitness to survive in the host.
Generation of a plc1 mutant strain
One of the genes that was induced by ethanol encoded a phospholipase C (plc; A1S_0043). A. baumannii has another gene encoding a phospholipase C (A1S_2055). Both genes are absent in the non-pathogenic A. baylyi ADP1 but present in other strains of A. baumannii that have been sequenced [23]. The proteins encoded by A1S_0043 and A1S_2055, show a similarity of 72%, and both are highly similar (75% similarity) to the phospholipases reported for Burkholderia pseudomallei. As is the case for phospholipase C proteins in other bacteria [85],[86], the N-terminal region contains the conserved residues that are recognized by the twin-arginine secretion system; therefore both phospholipases may be secreted. In addition, from the gene arrangement it can be proposed that A1S_0043 is expressed as a monocistronic mRNA since no other coding region is predicted to be located in the adjacent 404 bp downstream A1S_0043, and the upstream ORF, A1S_3479, is transcribed in opposite direction.
To evaluate the contribution of the phospholipase C (encoded by A1S_0043, from here on referred to as plc1) to A. baumannii virulence, we isolated a mutant strain carrying the insertion of a kanamycin cassette in the coding region of plc1 (see methods). This mutant strain did not show any apparent growth defects upon growth in liquid medium (data not shown). The ability of this strain to produce cellular damage on a monolayer of epithelial cells was tested as described below.
Infection of epithelial cells
Incubating a monolayer of epithelial cells in the presence of A. baumannii has been reported to elicit several morphological and physiological changes, such as loss of viability as revealed by trypan blue staining, detachment from the culture plate, and a general shrinking of the cells [14],[87]. Consistent with these reports we found that after infection, FaDu epithelial cells became permeable to trypan blue indicating that A. baumannii compromises the membrane permeability. Furthermore, after 18 h of infection we observed extensive detachment of the cell monolayer and cellular death in many of the remaining cells (Fig. 4B and C; stained non-infected controls are shown in Fig. S1). It has been shown that the intracellular enzyme lactate dehydrogenase (LDH) is released into the culture medium after any insult that compromises the integrity of the plasma membrane. Therefore, we assessed cell damage by measuring LDH release upon infection with A. baumannii. For this assay, a monolayer of FaDu cells was infected with A. baumannii and the amount of LDH released was measured after 22 h of incubation. The amount of damage produced by the different strains was estimated as percent of the amount LDH released when the cells were infected with wild-type A. baumannii. As shown in Fig. 4D, only live bacteria triggered LDH release indicating that cell damage is a consequence of the bacterial infection.
Fig. 4. Cell damage associated with A. baumannii infection. 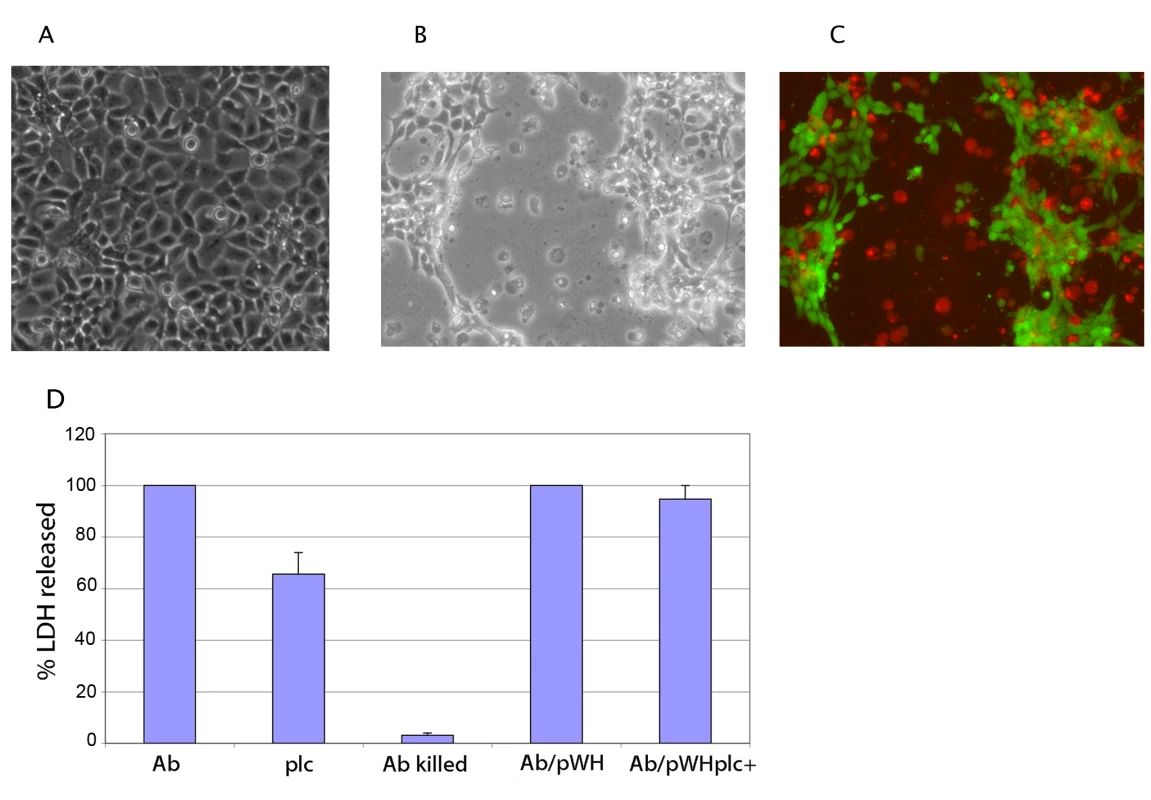
(A) Uninfected monolayer of FaDu cells observed at 150X with phase-contrast microscopy; (B) FaDu cells infected with A. baumannii after 18 h of incubation and observed at 150X with phase-contrast microscopy; (C) the same monolayer shown in panel B, stained with LIVE/DEAD reagent (Invitrogen) and observed by fluorescence microscopy. The live cells show green fluorescence whereas dead cells are observed as red. (D) Percentage of LDH released to the culture medium after 22 h of infection with A. baumannii wild-type, (Ab); plc1Δ::kan mutant, (plc); A. baumannii wild-type carrying pWH1266-Gm (Ab/pWH); plc1Δ::kan mutant carrying pWH1266-Gm/plc1+ (plc/pWHplc+); A. baumannii cells killed with formaldehyde, (Ab killed). Using this assay, the cytotoxic effect produced by the strain carrying the plc1Δ::kan allele was determined. A reduction in the amount of LDH released into the culture medium upon infection with this strain was observed (Fig. 4D). To further validate this result an infection was carried out with either the wild-type or the plc1 mutant transformed with pWH1266-Gm (empty vector) and pWH1266-Gm containing the plc1 gene, respectively. Cells infected with wild-type and plc mutant strain containing the plc1+ gene released a similar amount of LDH (Fig. 4D), indicating that phospholipase C contributes to cause cellular damage. Bacteria recovered from the infection plate were used to confirm that the complementing plasmid was stably maintained without detectable rearrangements (data not shown).
Phospholipase C has been reported as a virulence factor in many bacteria, such as Pseudomonas aeruginosa, Legionella monocytogenes and in the Gram-positive bacteria Clostridium perfringens [77],[88]. P. aeruginosa has an acidic phospholipase that shows strong hemolytic activity, and contributes to its ability to cause cellular damage [89]. Recently it has been shown that L. monocytogenes uses a phosphatidylinositol-specific phospholipase C to escape efficiently from the phagosome in macrophages [79],[90], and it also has a phosphatidylcholine-prefering phospholipase C that is involved in the escape from the phagosome in epithelial cells [91],[92]. B. pseudomallei has two phospholipases C that hydrolyze phosphatidylcholine and sphingomyelin and neither is hemolytic for human erythrocytes. However, it was reported that Plc-2 has a significant role in the virulence of this pathogen towards HeLa cells, whereas Plc-1 seems to have a minor one [78]. Thus, our results demonstrating that A. baumannii phospholipase C is important for virulence is consistent with finding with several other organisms.
Overall our results demonstrate a number of important conclusions:
First, RNA-Seq provides a comprehensive, detailed overview of the bacterial transcriptome. Even though contaminating rDNA reads can be present in large numbers, large numbers of unique reads can still be obtained by deep sequencing.
Second, in A. baumannii, ethanol is efficiently assimilated as a carbon source through the glyoxylate cycle that is a pathway required for full virulence in many pathogens.
Third, ethanol induces the expression of many proteins related to stress, including UspA, Hsp90, GroEL, Lon. Specific stress responses may help bacteria to contend with adverse condition pathogens face during infection.
Fourth, ethanol induces the expression of a phospholipase C that contributes to A. baumannii cytotoxicity.
Fifth, ethanol promotes the growth of A. baumannii, and presumably during stationary phase the resources of the medium are more efficiently utilized by the induction of the two Fe uptake systems and the high-affinity phosphate transporter system. Thus, overall these studies contribute a wealth of new information into the pathogenic response of Acinetobacter baumannii.
Methods
Bacterial strains, plasmids, oligonucleotides, and growth conditions
A. baumannii ATCC17978 was grown in YPDA culture medium (1% yeast extract, 2% peptone, 2% dextrose, and 0.012% adenine sulfate) at room temperature, or at the temperature indicated. When indicated, 1.1% ethanol was added to the culture medium. In this study 1.1% ethanol was used instead of 1% ethanol as previously published [20] because 1.1% ethanol caused more reproducible effects previous reported on A. baumannii growth. Bacterial strains were also grown in LB. When required, antibiotics were added at the following concentrations: ampicillin (100 µg/ml), kanamycin (50 µg/ml), gentamicin (20 µg/ml), tetracycline (10 µg/ml). Cloning were performed using the plasmids pCR2.1-TOPO (Invitrogen) and pUC19R (Invitrogen). pUC4K was used as source of the kanamycin cassette (GE Healtcare Life Sciences). pJQ200 was used as suicide plasmid [93]. pJQ200 confers GmR and carries the sacB gene from Bacillus subtilis to counterselect the presence of the plasmid in the presence of sucrose. This plasmid was also used as a source of the GmR cassette. pWH1266 was used as a shuttle vector for E. coli and A. baumannii [94]. pMP220 was used to construct transcriptional fusions to the promoter-less lacZ [95]. The sequences of the oligonucleotides used in this work are in Table S3.
Recombinant DNA techniques
Chromosomal DNA was obtained using the GenElute Bacterial Genomic DNA kit from Sigma-Aldrich. Plasmids were purified using the plasmid purification kit from Qiagen. DNA was amplified using PrimeStar HS Taq polymerase or LA Taq polymerase (Takara) according to the recommendations of the manufacturer. Transformation of E. coli was carried out using CaCl2 competent cells [96]. Electroporation was used to transform A. baumannii cells, following the protocols previously reported [97].
RNA isolation
A baumannii cultures were grown to mid-log phase (OD600 nm = 1.8) in YPDA or YPDA with 1.1% ethanol, at room temperature and shaking of 150 rpm. The cells were collected at 4°C and the RNA was isolated using the RiboPure-Bacteria kit (Ambion) according to the manufacturer's instructions. Residual DNA in the samples was removed using DNaseI. The integrity of the RNA was analyzed using an Agilent bioanalyzer (Agilent technologies). The MICROBExpress kit (Ambion) was used to remove the 23S and 16S rRNA from the total RNA samples. When indicated, the samples of enriched mRNA were subject to a final step of purification using the Megaclear kit from Ambion. To evaluate the degree of rRNA depletion the samples were analyzed on an Agilent bioanalyzer. The same protocol was used to isolate RNA from A. baumannii cultures from stationary phase. In this case, the cells were collected three hours after the OD600 of the cultures did not show any further increase.
cDNA synthesis and preparation of the library for high-throughput sequencing
The first two libraries (corresponding to the cultures with or without ethanol) were obtained using 2 µg of enriched mRNA that was depleted only once with the ribo-minus beads included in the MICROBExpress kit. For the samples corresponding to the biological duplicates, we performed two steps of depletion with ribo-minus beads and an additional purification step using the Megaclear kit. Double-stranded cDNA was obtained using hexameric random primers and the Super-script double-stranded cDNA synthesis kit from Invitrogen. The cDNA was purified using the Qiaquick PCR Purification kit from Qiagen, and subject to a partial digestion with DNaseI in order to obtain a substantial enrichment of fragments between 100 and 300 bp. After digestion, the sample was loaded on a 1.2% agarose gel, and the fragments between 100 and 300 bp were purified using the Qiaquick Gel Extraction kit from Qiagen. The ends of the fragments were end-repaired and A-tailed. For this, the End-It kit (Epicenter) was used according to the manufacturer's instructions. The repaired cDNA was purified using the Qiaquick PCR purification kit and A-tailed using the Klenow fragment of the DNase polymerase (NewEngland-BioLabs) and dATP. The sample was purified and ligated to the genomic adapters provided by Illumina. After ligation, the sample was loaded on a 2% agarose E-gel (Invitrogen), and the fragments between 150 and 350 bp were excised from the gel and purified using the Qiaquick gel extraction kit. A PCR reaction of the gel-purified cDNA was performed in 50 µl using the 1X master mix Phusion-High Fidelity DNA polymerase, and the primers 1.1 and 2.1 provided by Ilumina. The reaction was amplified with 17 cycles, and the sample loaded on a 1.2% agarose gel and the fragments between 150 and 350 bp were excised from the gel and purified. The sample was quantified spectrophotometrically using a nanodrop (Thermo) and sequenced in a Genome analyzer II (Illumina).
Data analysis
The raw reads of 35 bp were truncated as 28-mers and remapped with the Efficient Local Alignment of Nucleotide Data (ELAND) allowing for 1 and 2 nt mismatches. The output file containing only the sequences that mapped once in the genome was further analyzed to ascertain genome coverage and to assign the number of reads per locus (orf or intercistronic region). To identify the genes regulated by ethanol, the libraries were initially compared by pairs; for this, the number of reads for each coding region was determined, the number of total reads was normalized between these libraries and the ratio of reads between ethanol and no ethanol was calculated. The genes that showed a ratio larger than 1.9 and lower that 0.5 were considered potential candidates. Finally, the number of reads for the four libraries was normalized and the Student's t-test was applied for each gene. Those genes that showed a P-value lower or equal to 0.05 were considered as genes regulated by ethanol. To obtain information regarding the level of expression among the genes, we calculate the number of relative reads (NRR) per coding region using a window of 250 bp.
Quantitative RT-PCR
The RNA was isolated as described in the previous section. As templates for this assay we used the same RNA samples that were used for the synthesis of first Illumina libraries and two additional pair of samples that were independently obtained. The reverse transcription step was carried out using the iScript Select cDNA Synthesis Kit from Bio-Rad, according to the manufacturer's instructions. The primer3 software [98] was used to select primers that would amplify a product of approximately 200 bp. The quantitative real-time PCR assay was performed with SYBR-Green I master mix (Applied Biosystems) in a LightCycler 480 system. Reactions were set up according to the manufacturer's instructions, and three technical replicates for each sample were included. The amplification conditions were: 95°C, 5 min (ramp/rate of 4.8°C/s), followed by 45 cycles of 95°C 10 sec (ramp/rate 4.8°C/s), 55°C 20 sec (ramp/rate 2.5°C/s), and 72°C 30 sec (ramp/rate 4.8°C/s). The specificity of the reaction was confirmed by obtaining a melting curve from 95 to 55°C and visualizing the amplified product in a 5% polyacrylamide/TAE gel. The absence of product using only RNA in the PCR reaction (without reverse transcriptase) was also verified. The Cp value was defined as the cycle in which the fluorescence value was above the background. The efficiency of the amplifications for each pair of primers was determined obtaining a standard curve using serial dilutions of DNA. The efficiency was calculated using the formula E = 10(1/-s)X100 where s is the slope of the curve. The fold change was calculated using the 2−ΔΔCt (2−ΔΔCp) method [99]. A1S_2846 encoding a putative sulfite reductase was used as internal control. Similar results were obtained if A1S_0880 encoding MinC was used as internal control instead of A1S_2846.
Isolation of mutant strains and recombinant plasmids
To obtain the A. baumannii mutant strain in the plc1 gene, a fragment of 2579 bp carrying the plc1 gene was amplified by PCR using the oligonucleotides A1S_0043.1 and A1S_0043.2 and cloned into pCR2.1-TOPO plasmid. An internal fragment of 519 bp from the coding region of plc1 was removed by inverse PCR using the oligonucleotides A1S_0043.A and A1S_0043.B and substituted with a kanamycin resistance cassette. The DNA fragment carrying the plc1Δ::kan allele was subcloned into pJQ200. The resultant plasmid was used to electroporate A. baumannii cells. Single recombinants appeared after overnight incubation on LB plates in the presence of gentamicin. Double recombinants were selected plating serial dilutions of different GmR colonies on LB plates with kanamycin and 3% sucrose. The proper replacement was confirmed by PCR. Plasmid pWH1266 (ApR TcR) that is stable in A. baumannii was used to carry the plc1 gene. To generate this construct a PCR fragment carrying plc1 (oligonucleotides A1S_0043.1 and A1S_0043.2) was cloned into pWH1266 using the BamH1 and SalI sites. It has been shown that a gene cloned in these sites is expressed under control of the Tet promoter. The resulting plasmid does not confer any Tc resistance and given that A. baumannii ATCC17978 is ApR, we proceeded to construct the plasmids pWH1266-Kan and pWH1266-Gm, in which a kanamycin or gentamicin resistance cassette was cloned into the EcoRI site of pWH1266. The gene conferring GmR resistance was obtained by PCR using the oligonucleotides acc3 and acc4. The gene conferring KanR was obtained by PCR using the oligonucleotides Kanfw1 and Kanrev1. The plc1 gene was cloned in pWH1266-KanR and pMH266-GmR. As a control the gfp gene was cloned in pWH1266-Kan and introduced to A. baumannii; as expected, green-fluorescent cells were observed. The pMP220/2248p plasmid carrying the transcriptional fusion of lacZ to the control region of A1S_2448 was constructed by cloning a PCR fragment of 913 bp obtained by PCR using the oligonucleotides AB2448up2 and AB2448dw5. This fragment carries the promoter region located upstream of A1S_2448.
β-Galactosidase assay
A. baumannii cells carrying pMP220/2248p were grown in YPDA-Tc, or YPDA-Tc supplemented with 1% ethanol, or 10 mM PO4 buffer pH 7. The cultures were grown aerobically at 30°C and aliquots were assay at different time points. β-galactosidase activity was determined in Chloroform/SDS-permeabilised cells. Hydrolysis of o-nitrophenyl-β-D-galactopyranoside was carried out at 37°C. Activities are expressed in terms of cell density using the formula of Miller [100].
Lactate dehydrogenase (LDH) assay
The FaDu cell line originating from a hypopharyngeal carcinoma was obtained from ATCC (ATCC HTB-43). The cell line was grown under 5% CO2 at 37°C in Eagle's minimum essential medium with Earle's balanced salt solution (ATCC 30–203) supplemented with 10% heat-inactivated fetal bovine serum (Gibco 16140) and 1% of a solution containing penicillin/streptomycin at 10,000 U/ml and 10 mg/ml, respectively (Gibco 15140). The cells were seeded in 12 well-plates and infected when they reached 5×105–7×105 cells per well. Before infection, the monolayer of epithelial cells was carefully washed with PBS, and fresh medium without antibiotics was added. Bacterial strains were grown on plates of LB or LB with the appropriate antibiotic and incubated overnight at 37°C. The next day a suspension of bacterial cells was prepared in PBS, and the OD600 was registered and adjusted (2 OD600 nm = 1×109 cells/ml). Formaldehyde-fixed bacteria were prepared by incubation of the suspension in 1% formaldehyde for 4 h at 4°C as described [101]. The epithelial cells were infected at an MOI of 100 with no more than 10 µl of bacterial suspension. Mock-infections and infections were done in duplicate. The plates were centrifuged at 1,500 rpm for 5 min and then incubated for 22 hrs at 37°C and 5% CO2. The amount of LDH released into the culture medium was determined according to the manufacturer's instructions (BioVision Research Products. Mountain View, CA). Each set of experiments was performed in triplicate. Bacteria recovered from the infection plate were used to determine the number of colony forming units on plates with and without gentamicin. The restriction pattern of the plasmid obtained from these cells was analyzed by double digestions with EcoRI and SalI or EcoRI and BamHI.
Supporting Information
Zdroje
1. BaumannP
DoudoroffM
StanierRY
1968 A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter). J Bacteriol 95 1520 1541
2. JuniE
1978 Genetics and physiology of Acinetobacter. Annu Rev Microbiol 32 349 371
3. WaltermannM
StovekenT
SteinbuchelA
2007 Key enzymes for biosynthesis of neutral lipid storage compounds in prokaryotes: properties, function and occurrence of wax ester synthases/acyl-CoA: diacylglycerol acyltransferases. Biochimie 89 230 242
4. FagonJY
ChastreJ
DomartY
TrouilletJL
GibertC
1996 Mortality due to ventilator-associated pneumonia or colonization with Pseudomonas or Acinetobacter species: assessment by quantitative culture of samples obtained by a protected specimen brush. Clin Infect Dis 23 538 542
5. MetanG
AlpE
AygenB
SumerkanB
2007 Acinetobacter baumannii meningitis in post-neurosurgical patients: clinical outcome and impact of carbapenem resistance. J Antimicrob Chemother 60 197 199
6. WisplinghoffH
BischoffT
TallentSM
SeifertH
WenzelRP
2004 Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39 309 317
7. SeifertH
StrateA
PulvererG
1995 Nosocomial bacteremia due to Acinetobacter baumannii. Clinical features, epidemiology, and predictors of mortality. Medicine (Baltimore) 74 340 349
8. PoirelL
LebessiE
HeritierC
PatsouraA
FoustoukouM
2006 Nosocomial spread of OXA-58-positive carbapenem-resistant Acinetobacter baumannii isolates in a paediatric hospital in Greece. Clin Microbiol Infect 12 1138 1141
9. DijkshoornL
NemecA
SeifertH
2007 An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5 939 951
10. HujerKM
HujerAM
HultenEA
BajaksouzianS
AdamsJM
2006 Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50 4114 4123
11. TienHC
BattadA
BryceEA
FullerJ
MulveyM
2007 Multi-drug resistant Acinetobacter infections in critically injured Canadian forces soldiers. BMC Infect Dis 7 95
12. KnappS
WielandCW
FlorquinS
PantophletR
DijkshoornL
2006 Differential roles of CD14 and toll-like receptors 4 and 2 in murine Acinetobacter pneumonia. Am J Respir Crit Care Med 173 122 129
13. ErridgeC
Moncayo-NietoOL
MorganR
YoungM
PoxtonIR
2007 Acinetobacter baumannii lipopolysaccharides are potent stimulators of human monocyte activation via Toll-like receptor 4 signalling. J Med Microbiol 56 165 171
14. ChoiCH
LeeEY
LeeYC
ParkTI
KimHJ
2005 Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol 7 1127 1138
15. ChoiCH
HyunSH
LeeJY
LeeJS
LeeYS
2008 Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol 10 309 319
16. TomarasAP
DorseyCW
EdelmannRE
ActisLA
2003 Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149 3473 3484
17. ZimblerDL
PenwellWF
GaddyJA
MenkeSM
TomarasAP
2009 Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 22 23 32
18. DorseyCW
TomarasAP
ConnerlyPL
TolmaskyME
CrosaJH
2004 The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology 150 3657 3667
19. MiharaK
TanabeT
YamakawaY
FunahashiT
NakaoH
2004 Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 150 2587 2597
20. SmithMG
GianoulisTA
PukatzkiS
MekalanosJJ
OrnstonLN
2007 New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21 601 614
21. VallenetD
NordmannP
BarbeV
PoirelL
MangenotS
2008 Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS ONE 3 e1805 doi:10.1371/journal.pone.0001805
22. IaconoM
VillaL
FortiniD
BordoniR
ImperiF
2008 Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52 2616 2625
23. AdamsMD
GoglinK
MolyneauxN
HujerKM
LavenderH
2008 Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190 8053 8064
24. SmithMG
Des EtagesSG
SnyderM
2004 Microbial synergy via an ethanol-triggered pathway. Mol Cell Biol 24 3874 3884
25. WannerBL
1987 Phosphate regulation of gene expression in Escherichia coli.
NeidhardtF
IngrahamJL
LowKB
MagasanikB
SchaechterM
UmbargerHE
Escherichia coli and Salmonella: cellular and molecular biology Washington, D.C. American Society for Microbiology 1326 1333
26. LamarcheMG
WannerBL
CrepinS
HarelJ
2008 The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev 32 461 473
27. YuraT
NagaiH
MoriH
1993 Regulation of the heat-shock response in bacteria. Annu Rev Microbiol 47 321 350
28. LakhalF
Bury-MoneS
NomaneY
Le GoicN
PaillardC
2008 DjlA, a membrane-anchored DnaJ-like protein, is required for cytotoxicity of clam pathogen Vibrio tapetis to hemocytes. Appl Environ Microbiol 74 5750 5758
29. DuY
LenzJ
ArvidsonCG
2005 Global gene expression and the role of sigma factors in Neisseria gonorrhoeae in interactions with epithelial cells. Infect Immun 73 4834 4845
30. LinSN
AyadaK
ZhaoY
YokotaK
TakenakaR
2005 Helicobacter pylori heat-shock protein 60 induces production of the pro-inflammatory cytokine IL8 in monocytic cells. J Med Microbiol 54 225 233
31. HinodeD
YoshiokaM
TanabeS
MikiO
MasudaK
1998 The GroEL-like protein from Campylobacter rectus: immunological characterization and interleukin-6 and -8 induction in human gingival fibroblast. FEMS MicrobiolLett 167 1 6
32. MarioniJC
MasonCE
ManeSM
StephensM
GiladY
2008 RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18 1509 1517
33. FuX
FuN
GuoS
YanZ
XuY
2009 Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 10 161
34. KornbergHL
1966 The role and control of the glyoxylate cycle in Escherichia coli. Biochem J 99 1 11
35. KornbergHL
KrebsHA
1957 Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179 988 991
36. ArndtA
AuchterM
IshigeT
WendischVF
EikmannsBJ
2008 Ethanol catabolism in Corynebacterium glutamicum. J Mol Microbiol Biotechnol 15 222 233
37. GerstmeirR
WendischVF
SchnickeS
RuanH
FarwickM
2003 Acetate metabolism and its regulation in Corynebacterium glutamicum. J Biotechnol 104 99 122
38. WolfeAJ
2005 The acetate switch. Microbiol Mol Biol Rev 69 12 50
39. OhMK
RohlinL
KaoKC
LiaoJC
2002 Global expression profiling of acetate-grown Escherichia coli. J Biol Chem 277 13175 13183
40. KakudaH
HosonoK
ShiroishiK
IchiharaS
1994 Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J Biochem 116 916 922
41. ReinscheidDJ
SchnickeS
RittmannD
ZahnowU
SahmH
1999 Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology 145(Pt 2) 503 513
42. SerranoJA
CamachoM
BoneteMJ
1998 Operation of glyoxylate cycle in halophilic archaea: presence of malate synthase and isocitrate lyase in Haloferax volcanii. FEBS Lett 434 13 16
43. LohWH
RandlesCI
SharpWR
MillerRH
1984 Intermediary carbon metabolism of Azospirillum brasilense. J Bacteriol 158 264 268
44. MolinaI
PellicerMT
BadiaJ
AguilarJ
BaldomaL
1994 Molecular characterization of Escherichia coli malate synthase G. Differentiation with the malate synthase A isoenzyme. Eur J Biochem 224 541 548
45. OrnstonLN
OrnstonMK
1969 Regulation of glyoxylate metabolism in Escherichia coli K-12. J Bacteriol 98 1098 1108
46. CozzoneAJ
1998 Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu Rev Microbiol 52 127 164
47. SauerU
EikmannsBJ
2005 The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev 29 765 794
48. FleckCB
BrockM
2009 Re-characterisation of Saccharomyces cerevisiae Ach1p: fungal CoA-transferases are involved in acetic acid detoxification. Fungal Genet Biol 46 473 485
49. van der RestME
FrankC
MolenaarD
2000 Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Escherichia coli. J Bacteriol 182 6892 6899
50. MolenaarD
van der RestME
DryschA
YucelR
2000 Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J Bacteriol 182 6884 6891
51. ItohT
MatsudaH
MoriH
1999 Phylogenetic analysis of the third hsp70 homolog in Escherichia coli; a novel member of the Hsc66 subfamily and its possible co-chaperone. DNA Res 6 299 305
52. KluckCJ
PatzeltH
GenevauxP
BrehmerD
RistW
2002 Structure-function analysis of HscC, the Escherichia coli member of a novel subfamily of specialized Hsp70 chaperones. J Biol Chem 277 41060 41069
53. CipakA
JaganjacM
TehlivetsO
KohlweinSD
ZarkovicN
2008 Adaptation to oxidative stress induced by polyunsaturated fatty acids in yeast. Biochim Biophys Acta 1781 283 287
54. AllakhverdievSI
KinoshitaM
InabaM
SuzukiI
MurataN
2001 Unsaturated fatty acids in membrane lipids protect the photosynthetic machinery against salt-induced damage in Synechococcus. Plant Physiol 125 1842 1853
55. AlexandreH
RousseauxI
CharpentierC
1994 Ethanol adaptation mechanisms in Saccharomyces cerevisiae. Biotechnol Appl Biochem 20(Pt 2) 173 183
56. ZhangYM
RockCO
2008 Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6 222 233
57. MarchandI
Damier-PiolleL
CourvalinP
LambertT
2004 Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48 3298 3304
58. BinaXR
LavineCL
MillerMA
BinaJE
2008 The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol Lett 279 226 233
59. O'TooleR
WilliamsHD
2003 Universal stress proteins and Mycobacterium tuberculosis. Res Microbiol 154 387 392
60. KvintK
NachinL
DiezA
NystromT
2003 The bacterial universal stress protein: function and regulation. Curr Opin Microbiol 6 140 145
61. NachinL
NannmarkU
NystromT
2005 Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol 187 6265 6272
62. NonakaG
BlankschienM
HermanC
GrossCA
RhodiusVA
2006 Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev 20 1776 1789
63. ThomasJ
CronanJE
2005 The enigmatic acyl carrier protein phosphodiesterase of Escherichia coli: genetic and enzymological characterization. J Biol Chem 280 34675 34683
64. AudiaJP
PattonMC
WinklerHH
2008 DNA microarray analysis of the heat shock transcriptome of the obligate intracytoplasmic pathogen Rickettsia prowazekii. Appl Environ Microbiol 74 7809 7812
65. SlamtiL
LivnyJ
WaldorMK
2007 Global gene expression and phenotypic analysis of a Vibrio cholerae rpoH deletion mutant. J Bacteriol 189 351 362
66. QinJH
ShengYY
ZhangZM
ShiYZ
HeP
2006 Genome-wide transcriptional analysis of temperature shift in L. interrogans serovar lai strain 56601. BMC Microbiol 6 51
67. GreenHA
DonohueTJ
2006 Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J Bacteriol 188 5712 5721
68. Asadulghani
SuzukiY
NakamotoH
2003 Light plays a key role in the modulation of heat shock response in the cyanobacterium Synechocystis sp PCC 6803. Biochem Biophys Res Commun 306 872 879
69. Martinez-SalazarJM
Sandoval-CalderonM
GuoX
Castillo-RamirezS
ReyesA
2009 The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology 155 386 397
70. GrossC
NeidhardtFC
CurtissRIII
IngrahamJL
LinECC
1996 Function and regulation of the heat shock proteins. Eschericha coli and Salmonella: cellular and Molecular Biology. Washington, D.C. ASM Press 1382 1399
71. GurE
SauerRT
2008 Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev 22 2267 2277
72. ShelburneCE
CoopamahMD
SweierDG
AnFY
LopatinDE
2007 HtpG, the Porphyromonas gingivalis HSP-90 homologue, induces the chemokine CXCL8 in human monocytic and microvascular vein endothelial cells. Cell Microbiol 9 1611 1619
73. XuH
LeeHY
AhnJ
2008 Cross-protective effect of acid-adapted Salmonella enterica on resistance to lethal acid and cold stress conditions. Lett Appl Microbiol 47 290 297
74. GunasekeraTS
CsonkaLN
PaliyO
2008 Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190 3712 3720
75. KogaT
SakamotoF
YamotoA
TakumiK
1999 Acid adaptation induces cross-protection against some environmental stresses in Vibrio parahaemolyticus. J Gen Appl Microbiol 45 155 161
76. VolkerU
MachH
SchmidR
HeckerM
1992 Stress proteins and cross-protection by heat shock and salt stress in Bacillus subtilis. J Gen Microbiol 138 2125 2135
77. SongerJG
1997 Bacterial phospholipases and their role in virulence. Trends Microbiol 5 156 161
78. KorbsrisateS
TomarasAP
DamninS
CkumdeeJ
SrinonV
2007 Characterization of two distinct phospholipase C enzymes from Burkholderia pseudomallei. Microbiology 153 1907 1915
79. PoussinMA
LeitgesM
GoldfineH
2009 The ability of Listeria monocytogenes PI-PLC to facilitate escape from the macrophage phagosome is dependent on host PKCbeta. Microb Pathog 46 1 5
80. AnsteyNM
CurrieBJ
HassellM
PalmerD
DwyerB
2002 Community-acquired bacteremic Acinetobacter pneumonia in tropical Australia is caused by diverse strains of Acinetobacter baumannii, with carriage in the throat in at-risk groups. J Clin Microbiol 40 685 686
81. ChenM-Z
HsuehPo-Ren
LeeLi-Na
YuC-J
YangP-C
LuhK-T
2001 Severe Community-Acquired Pneumonia due to Acinetobacter baumannii. Chest 120 1072 1077
82. FalagasME
KarveliEA
KelesidisI
KelesidisT
2007 Community-acquired Acinetobacter infections. Eur J Clin Microbiol Infect Dis 26 857 868
83. BaileyKL
WyattTA
RombergerDJ
SissonJH
2009 Alcohol functionally upregulates Toll-like receptor 2 in airway epithelial cells. Alcohol Clin Exp Res 33 499 504
84. VulicM
KolterR
2002 Alcohol-induced delay of viability loss in stationary-phase cultures of Escherichia coli. J Bacteriol 184 2898 2905
85. OchsnerUA
SnyderA
VasilAI
VasilML
2002 Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc Natl Acad Sci USA 99 8312 8317
86. RossierO
CianciottoNP
2005 The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect Immun 73 2020 2032
87. LeeJC
OhJY
KimKS
JeongYW
ParkJC
2001 Apoptotic cell death induced by Acinetobacter baumannii in epithelial cells through caspase-3 activation. APMIS 109 679 684
88. TitballRW
1993 Bacterial phospholipases C. Microbiol Rev 57 347 366
89. VasilML
StonehouseMJ
VasilAI
WadsworthSJ
GoldfineH
2009 A complex extracellular sphingomyelinase of Pseudomonas aeruginosa inhibits angiogenesis by selective cytotoxicity to endothelial cells. PLoS Pathog 5 e1000420 doi:10.1371/journal.ppat.1000420
90. CamilliA
TilneyLG
PortnoyDA
1993 Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol 8 143 157
91. MarquisH
DoshiV
PortnoyDA
1995 The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun 63 4531 4534
92. GrundlingA
GonzalezMD
HigginsDE
2003 Requirement of the Listeria monocytogenes broad-range phospholipase PC-PLC during infection of human epithelial cells. J Bacteriol 185 6295 6307
93. QuandtJ
HynesMF
1993 Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127 15 21
94. HungerM
SchmuckerR
KishanV
HillenW
1990 Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87 45 51
95. SpainkHP
OkkerRJH
WijffelmanCA
PeesE
LugtenbergBJJ
1987 Promoters in the nodulation region of the Rhizobium leguminosarum symbiotic plasmid pRL1JI. Plant Mol Biol 9 27 39
96. AusubelFM
BrentR
KingstonRE
MooreDD
SeidmanJG
1987 Current Protocols in Molecular Biology. New York John Wiley
97. DorseyCW
TomarasAP
ActisLA
2002 Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl Environ Microbiol 68 6353 6360
98. RozenS
SkaletskyH
2000 Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132 365 386
99. LivakKJ
SchmittgenTD
2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402 408
100. MillerJ
1972 Experiments in molecular genetics. Cold Spring Harbor, N.Y. Cold Spring Harbor Laboratory
101. PatonJC
RogersTJ
MoronaR
PatonAW
2001 Oral administration of formaldehyde-killed recombinant bacteria expressing a mimic of the Shiga toxin receptor protects mice from fatal challenge with Shiga-toxigenic Escherichia coli. Infect Immun 69 1389 1393
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Suppression of mRNAs Encoding Tegument Tetraspanins from Results in Impaired Tegument TurnoverČlánek Novel Riboswitch Ligand Analogs as Selective Inhibitors of Guanine-Related Metabolic PathwaysČlánek The Physical Relationship between Infectivity and Prion Protein Aggregates Is Strain-DependentČlánek Rhomboid 4 (ROM4) Affects the Processing of Surface Adhesins and Facilitates Host Cell Invasion by
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 4- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Innate Recognition of Fungal Cell Walls
- Suppression of mRNAs Encoding Tegument Tetraspanins from Results in Impaired Tegument Turnover
- Junín Virus Infection of Human Hematopoietic Progenitors Impairs Proplatelet Formation and Platelet Release via a Bystander Effect Involving Type I IFN Signaling
- The Endosymbiotic Bacterium Induces Resistance to Dengue Virus in
- Natural Regulatory T Cells in Malaria: Host or Parasite Allies?
- Keratinocytes Determine Th1 Immunity during Early Experimental Leishmaniasis
- Spatial and Temporal Association of Outbreaks of H5N1 Influenza Virus Infection in Wild Birds with the 0°C Isotherm
- Novel Riboswitch Ligand Analogs as Selective Inhibitors of Guanine-Related Metabolic Pathways
- RNA Polymerase Activity and Specific RNA Structure Are Required for Efficient HCV Replication in Cultured Cells
- The Physical Relationship between Infectivity and Prion Protein Aggregates Is Strain-Dependent
- Inadequate Clearance of Translocated Bacterial Products in HIV-Infected Humanized Mice
- Topology and Organization of the Type III Secretion Needle Complex Components
- Temperature Modulates Plant Defense Responses through NB-LRR Proteins
- Peptide Inhibitors of Dengue-Virus Entry Target a Late-Stage Fusion Intermediate
- Identification of Host-Dependent Survival Factors for Intracellular through an siRNA Screen
- Exposure to HIV-1 Directly Impairs Mucosal Epithelial Barrier Integrity Allowing Microbial Translocation
- Increased Asymmetric Dimethylarginine in Severe Falciparum Malaria: Association with Impaired Nitric Oxide Bioavailability and Fatal Outcome
- Reconstitution of SARS-Coronavirus mRNA Cap Methylation
- Induces Brain Microvascular Endothelial Cell Detachment from the Matrix and Cleavage of Occludin: A Role for MMP-8
- Two Coregulated Efflux Transporters Modulate Intracellular Heme and Protoporphyrin IX Availability in
- The Type I NADH Dehydrogenase of Counters Phagosomal NOX2 Activity to Inhibit TNF-α-Mediated Host Cell Apoptosis
- Rhomboid 4 (ROM4) Affects the Processing of Surface Adhesins and Facilitates Host Cell Invasion by
- Increased Monocyte Turnover from Bone Marrow Correlates with Severity of SIV Encephalitis and CD163 Levels in Plasma
- The RING-CH Ligase K5 Antagonizes Restriction of KSHV and HIV-1 Particle Release by Mediating Ubiquitin-Dependent Endosomal Degradation of Tetherin
- Molecular Mechanisms of Ethanol-Induced Pathogenesis Revealed by RNA-Sequencing
- Highly Frequent Mutations in Negative Regulators of Multiple Virulence Genes in Group A Streptococcal Toxic Shock Syndrome Isolates
- Emergence and Pathogenicity of Highly Virulent Genotypes in the Northwest United States
- Structural and Functional Analysis of Viral siRNAs
- Prion Shedding from Olfactory Neurons into Nasal Secretions
- a GATA Transcription Factor That Directs Disparate Fates in Including Morphogenesis and Siderophore Biosynthesis
- Three Members of the 6-cys Protein Family of Play a Role in Gamete Fertility
- Complement as an Endogenous Adjuvant for Dendritic Cell-Mediated Induction of Retrovirus-Specific CTLs
- A Genomic Survey of Positive Selection in Provides Insights into the Evolution of Accidental Virulence
- Overcomes Stress of Azole Drugs by Formation of Disomy in Specific Multiple Chromosomes
- Blood Fluke Exploitation of Non-Cognate CD4 T Cell Help to Facilitate Parasite Development
- Antagonism of Tetherin Restriction of HIV-1 Release by Vpu Involves Binding and Sequestration of the Restriction Factor in a Perinuclear Compartment
- The Development of Therapeutic Antibodies That Neutralize Homologous and Heterologous Genotypes of Dengue Virus Type 1
- Deficiencies in Jasmonate-Mediated Plant Defense Reveal Quantitative Variation in Pathogenesis
- Interactions with Bacteria in the Context of Human Health and Disease
- Viral Capsid Is a Pathogen-Associated Molecular Pattern in Adenovirus Keratitis
- Electron Tomography Reveals the Steps in Filovirus Budding
- Selective Condensation Drives Partitioning and Sequential Secretion of Cyst Wall Proteins in Differentiating
- The Effect of Vaccination on the Evolution and Population Dynamics of Avian Paramyxovirus-1
- A Timescale for Evolution, Population Expansion, and Spatial Spread of an Emerging Clone of Methicillin-Resistant
- VacA Toxin/Subunit p34: Targeting of an Anion Channel to the Inner Mitochondrial Membrane
- Production of Extracellular Traps against and in Infected Lung Tissue Is Dependent on Invading Neutrophils and Influenced by Hydrophobin RodA
- A Differential Role for Macropinocytosis in Mediating Entry of the Two Forms of Vaccinia Virus into Dendritic Cells
- Impaired Innate Immunity in Mice but Preserved CD8 T Cell Responses against in -, -, - or -Deficient Mice
- SARS-CoV Pathogenesis Is Regulated by a STAT1 Dependent but a Type I, II and III Interferon Receptor Independent Mechanism
- Proteolysis of Human Thrombin Generates Novel Host Defense Peptides
- Multilayered Mechanism of CD4 Downregulation by HIV-1 Vpu Involving Distinct ER Retention and ERAD Targeting Steps
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Effect of Vaccination on the Evolution and Population Dynamics of Avian Paramyxovirus-1
- Reconstitution of SARS-Coronavirus mRNA Cap Methylation
- Deficiencies in Jasmonate-Mediated Plant Defense Reveal Quantitative Variation in Pathogenesis
- A Timescale for Evolution, Population Expansion, and Spatial Spread of an Emerging Clone of Methicillin-Resistant
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



