-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
article has not abstract
Published in the journal: Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics. PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003950
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003950Summary
article has not abstract
Fungi belong amongst the severest pathogens of humans, animals, and plants. For example, Candida spp. and Aspergillus spp. account for most invasive mycoses, and such infections are associated with high rates of mortality in hematology and oncology patients. Worrisomely, it has been estimated that 4% of all patients who die in hospitals die of invasive aspergillosis and 2% die of candidiasis [1]. Moreover, in agriculture, filamentous fungi are mainly responsible for severe loss of crops worldwide, destroying over 125 million tons of rice, wheat, maize, potatoes, and soybeans each year. Calculations for 2011 predicted that prevention of these losses would be sufficient to feed 600 million people [2]. However, due to the common negative perception of fungi as pathogens, we often lose sight of the beneficial role of fungi as producers of a cornucopia of life-saving drugs.
Janus-Like Role of Fungi
Some fungi are at the same time a “friend” and “foe” of humans. A good example of a filamentous fungus with a Janus-like role is A. terreus, which has opposing effects on human health. As a pathogen, this fungus is the third most significant cause of invasive aspergillosis, having a reported frequency of 3–12.5%. In addition, it also produces several mycotoxins that can be the cause of food spoilage, especially of cereals and nuts in tropical and subtropical climates. However, this fungus can also produce a number of organic acids and secondary metabolites that, due to their clinical relevance, are of major biotechnological and pharmaceutical interest. Lovastatin, a polyketide derivative, is the most important one because of its cholesterol-lowering properties (Table 1) and has been used successfully for decades to treat coronary artery disease, a prevalent cause of premature death in the Western world [3]. Moreover, this fungus can also produce gliotoxin, which acts as an immunosuppressive drug (Table 1).
Tab. 1. Selected fungal products from different compound classes, their application, and producers. 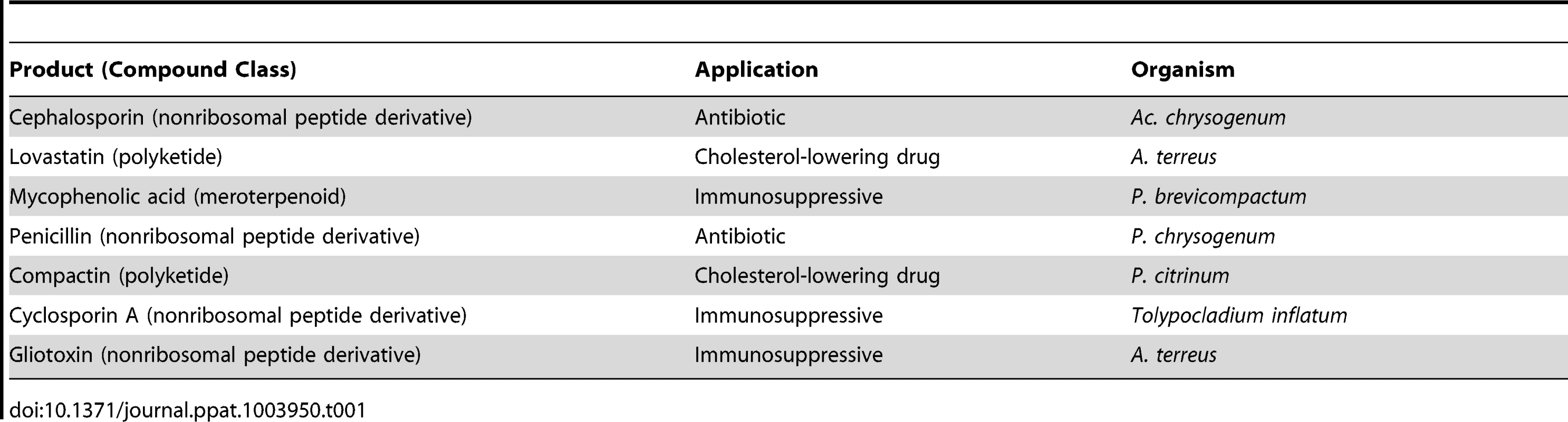
Two other representative examples of fungi with a Janus-like character are serious plant pathogens. Claviceps purpurea is the producer of ergot alkaloids, naturally occurring mycotoxins that contaminate grains. Consumption of contaminated food causes ergotism, a disease that resulted in more than 40,000 deaths in Europe in the Middle Ages. The toxic and therapeutic effects of ergot alkaloids have been known for centuries, and, nowadays, alkaloids form the basis of many synthetic drugs that treat the various symptoms of migraines [4]. A second example is Ashbya gossypii, a major plant pathogen that causes stigmatomycosis of cotton and citrus fruits in tropical and subtropical countries. In the 1940s, Ash. gossypii was found to be a high-level producer of riboflavin, also called vitamin B2. This essential vitamin cannot be synthesized by humans and must be supplied by food and dietary supplements. Nowadays, the industry uses Ash. gossypii as a producer of riboflavin [5], since microbial fermentation with a yearly production yield of about 9,000 tons is economically more feasible than chemical synthesis.
The Treasure Trove: Secondary Metabolites Produced by Filamentous Fungi
From the numerous above-mentioned examples, it is clear that filamentous fungi have an extensive metabolism and produce a wealth of bioactive compounds. Many of their secondary metabolites contribute substantially to the pathogenicity of fungi and to the toxicity of contaminated food and crops [2], [6]. Notably, secondary metabolites differ from primary metabolites in that they (1) are not directly derived from any intermediary metabolism, (2) are often produced during a specific morphogenetic program, and (3) are not essential for survival of the producing organisms [7]. However, secondary metabolites may provide abiotic (melanins) and biotic properties as antifungal, antibacterial, and insecticidal agents [7]–[9]. Importantly, the content and composition profile of secondary metabolites differs from species to species and even within strains of a single species.
Secondary metabolites are highly diverse, and their biosynthesis genes are often organized in clusters and controlled by transcriptional regulation and chromatin remodeling [10]. Currently, they can be classified into different compound classes [7], including polyketides, fatty acid derivatives, and nonribosomal peptides that are produced by large modular enzymes, as well as terpenes and alkaloids. Moreover, several hybrid secondary metabolites are also known, namely meroterpenoids, which consist of polyketides and isoprenes, as well as polyketide-nonribosomal peptide hybrids. A selection of secondary metabolites with pharmaceutical relevance is provided in Table 1.
Traditionally, bioactive secondary metabolites were discovered by screening single organisms or complex samples. For example, the immunosuppressive cyclosporine A was discovered 45 years ago from a Norwegian soil sample [11]. Today, genome mining studies have revealed a potentially large number of unknown secondary metabolite genes that are often not expressed under standard laboratory conditions. Thus far, activation of “silent” gene clusters has been achieved by overexpression of cluster-encoded or global regulators, epigenetic modification, and cocultivation experiments [reviewed in 9]. For example, cocultivation of A. nidulans with an actinomycete triggered production of orsellinic acid, and induced expression of the A. nidulans transcriptional regulator AfoA triggered production of the polyketide asperfuranone [12], [13]. These novel approaches will be of great value for identifying and engineering new secondary metabolites for biotechnological and pharmaceutical applications.
Horizontal Gene Transfer Contributes to the Diversity of Fungal Metabolism
The immense diversity of secondary metabolites found in fungal species raises the intriguing question of their evolutionary origin. Lateral or horizontal gene transfer (HGT) across species barriers has long been known for bacteria but was thought to play a minor role in the evolution of eukaryotic species. However, recent data from genome sequencing studies have indicated that HGT has contributed considerably to this richness in metabolites [14], [15]. Investigation of 60 completely sequenced fungal genomes using strict phylogenomic criteria showed that 713 bacterial genes had been transferred to fungal genomes [16]. Several of these genes are involved in carbohydrate metabolism, allowing fungi to propagate under extreme environmental conditions. Early evidence that fungi acquired bacterial genes came from sequence comparison of beta-lactam antibiotic biosynthesis genes. The first two steps of beta-lactam antibiotic biosynthesis are catalyzed by a nonribosomal peptide synthetase and isopenicillin N synthase, leading to the intermediate isopenicillin N, which is common to all penicillins and their derivatives. Sequencing of the corresponding genes (pcbAB and pcbC) has provided evidence that these two genes are clustered in both bacteria and fungi (Figure 1). Furthermore, the clustered organization and the lack of any intronic sequences led to the assumption that bacterial beta-lactam antibiotic genes were transferred to fungal species such as A. nidulans, Penicillium chrysogenum, and Acremonium chrysogenum. The transfer of bacterial genes to a eukaryotic nucleus has a wide range of consequences. For example, the foreign genes have to adapt to the eukaryotic host system and its expression machinery. In prokaryotes, pathway-specific regulators commonly determine gene expression, while in eukaryotes, expression of secondary biosynthesis genes is often governed by global regulators. Good examples of such eukaryotic global regulators are LaeA and VeA, both part of a multi-subunit protein complex that controls the expression of genes, including those that encode beta-lactams, that are involved in fungal secondary metabolite processes [17], [18].
Fig. 1. Horizontal gene transfer of beta-lactam biosynthesis genes from prokaryotes to eukaryotes. 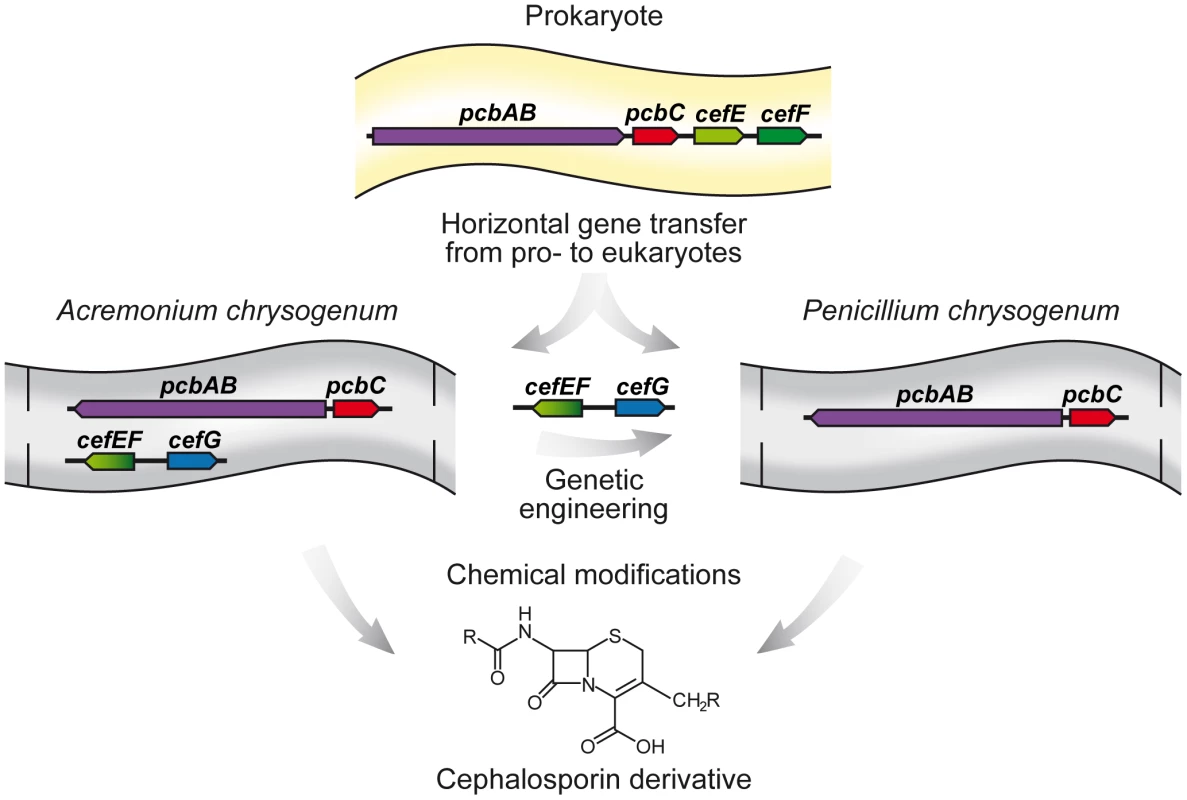
The antibiotic biosynthesis genes are derived either from gram-positive (e.g. Streptomyces spp.) or gram-negative (e.g. Lysobacter spp.) bacteria. The first two steps of both penicillin and cephalosporin C biosynthesis are catalyzed by the gene products of pcbAB and pcbC. P. chrysogenum harbors one additional gene, penDE (not shown), to perform the last step of penicillin biosynthesis, whereas Ac. chrysogenum has obtained several additional genes for production of cephalosporin C. Genetic engineering approaches have been used to introduce these genes into P. chrysogenum. Chemical engineering approaches have enabled the precursors of both the Ac. chrysogenum and P. chrysogenum biosynthesis pathway to be used for the production of new cephalosporin derivatives. Fungal Beta-lactam Antibiotics as a Key for Current and Future Treatment of Patients
Although fungi have most probably obtained genes for beta-lactam antibiotic biosynthesis through HGT from prokaryotes (Figure 1), they are more efficient producers of these secondary metabolites and, therefore, are preferentially chosen for industrial manufacturing. Currently, beta-lactam antibiotics are the major anti-infective agents worldwide, having an estimated world market of about 22 billion US dollars at the dosage form level [19]. In contrast to penicillin, which is mainly active against gram-positive bacteria, cephalosporin C is a broad-spectrum antibiotic that affects both gram-positive and gram-negative bacteria. Both antibiotics are produced by filamentous fungi, namely P. chrysogenum and Ac. chrysogenum. Both fungi share the first two steps of beta-lactam biosynthesis that are catalyzed by the gene products of pcbAB and pcbC, while the subsequent steps, resulting in either penicillin or cephalosporin C, are different. In P. chrysogenum, a single biosynthesis step is necessary to transform isopenicillin N into penicillin, and this is catalyzed by the penDE gene product. In contrast, several additional steps are needed in Ac. chrysogenum to generate cephalosporin C. The corresponding genes are part of a so-called “early” gene cluster, including pcbAB and pcbC, and a “late” gene cluster, comprising the genes cefEF and cefG [reviewed in 20]–[22]. Using genetic engineering approaches, genes from Ac. chrysogenum were transferred into P. chrysogenum, making this fungus an alternative producer of intermediates of cephalosporin C biosynthesis (Figure 1) [22].
Cephalosporin C as a natural product exerts only weak antibiotic activity. This activity has been gradually increased through the generation of semisynthetic derivatives, representing a good example of effective chemical modification of natural products [21]. Pharmaceutical companies have recently announced the development of cephalosporins that are particularly suitable for treating methicillin-resistant Staphylococcus aureus (MRSA), an increasing problem in hospitals. Recently, ceftobiprole became the first broad-spectrum cephalosporin with activity against MRSA to be assessed in late-stage clinical trials [23]. Furthermore, another cephalosporin derivative, ceftaroline, is one of only a few new antibiotics that have been recently launched [24], [25], indicating that cephalosporin derivatives remain extremely valuable antibiotics for current and future medical applications.
In summary, fungi can play a Janus-like role, being at the same time “friend” and “foe” to humans. In other words, they can not only act as serious pathogens themselves but can also produce a cornucopia of highly beneficial drugs and antibiotics. This unique ability of such fascinating filamentous fungal species thereby makes them of key interest in further biotechnological and pharmaceutical research.
Zdroje
1. MircusG, HagagS, LevdanskyE, SharonH, ShadkchanY, et al. (2009) Identification of novel cell wall destabilizing antifungal compounds using a conditional Aspergillus nidulans protein kinase C mutant. J Antimicrob Chemother 64 : 755–763.
2. FisherMC, HenkDA, BriggsCJ, BrownsteinJS, MadoffLC, et al. (2012) Emerging fungal threats to animal, plant, and ecosystem health. Nature 484 : 186–194.
3. ManzoniM, RolliniM (2002) Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs. Appl Microbiol Biotechnol 58 : 555–564.
4. SchardlCL, PanaccioneDG, TudzynskiP (2006) Ergot alkaloids—Biology and molecular biology. Alkaloids Chem Biol 63 : 45–86.
5. KatoT, ParkEY (2012) Riboflavin production by Ashbya gossypii. Biotechnol Lett 34 : 611–618.
6. ScharfDH, HeinekampT, BrakhageAA (2014) Human and plant fungal pathogens: The role of secondary metabolites. PLOS Pathog 10: e1003859 doi:10.1371/journal.ppat.1003859
7. KellerNP, TurnerG, BennettJW (2005) Fungal secondary metabolism—From biochemistry to genomics. Nat Rev Microbiol 3 : 937–947.
8. KhajoA, BryanRA, FriedmanM, BurgerRM, LevitskyY, et al. (2011) Protection of melanized Cryptococcus neoformans from lethal dose gamma irradiation involves changes in melanin's chemical structure and paramagnetism. PLOS ONE 6: e25092 doi:10.1371/journal.pone.0025092
9. BrakhageAA, SchroeckhV (2011) Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet Biol 48 : 15–22.
10. BrakhageAA (2013) Regulation of fungal secondary metabolism. Nat Rev Microbiol 11 : 21–32.
11. DreyfussM, HärriE, HofmannH, KobelH, PacheW, et al. (1976) Cyclosporin A and C—New metabolites from Trichoderma polysporum (Link ex Pers.) Rifai. Eur J Appl Microbiol 3 : 125–133.
12. ChiangYM, SzewczykE, DavidsonAD, KellerN, OakleyBR, et al. (2009) A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc 131 : 2965–2970.
13. SchroeckhV, ScherlachK, NutzmannHW, ShelestE, Schmidt-HeckW, et al. (2009) Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA 106 : 14558–14563.
14. FitzpatrickDA (2012) Horizontal gene transfer in fungi. FEMS Microbiol Lett 329 : 1–8.
15. Teichert I, Nowrousian M (2011) Evolution of genes for secondary metabolism in fungi. In: Pöggeler S, Wöstemeyer J, editors. The Mycota XIV. Berlin, Heidelberg: Springer Verlag. pp. 231–255.
16. Marcet-HoubenM, GabaldonT (2010) Acquisition of prokaryotic genes by fungal genomes. Trends Genet 26 : 5–8.
17. BayramÖ, BrausGH (2012) Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol Rev 36 : 1–24.
18. JainS, KellerN (2013) Insights to fungal biology through LaeA sleuthing. Fungal Biol Rev 27 : 51–59.
19. DemainAL (2009) Antibiotics: Natural products essential to human health. Med Res Rev 29 : 821–842.
20. SchmittEK, HoffB, KückU (2004) Regulation of cephalosporin biosynthesis. Adv Biochem Eng Biotechnol 88 : 1–43.
21. OzcengizG, DemainAL (2013) Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation. Biotechnol Adv 31 : 287–311.
22. WeberSS, BovenbergRAL, DriessenAJM (2012) Biosynthetic concepts for the production of beta-lactam antibiotics in Penicillium chrysogenum. Biotechnol J 7 : 225–236.
23. NoelGJ, StraussRS, AmslerK, HeepM, PypstraR, et al. (2008) Results of a double-blind, randomized trial of ceftobiprole treatment of complicated skin and skin structure infections caused by gram-positive bacteria. Antimicrob Agents Chemother 52 : 37–44.
24. LaudanoJB (2011) Ceftaroline fosamil: A new broad-spectrum cephalosporin. J Antimicrob Chemother 66(Suppl 3): iii11–iii18.
25. ButlerMS, BlaskovichMA, CooperMA (2013) Antibiotics in the clinical pipeline in 2013. J Antibiot (Tokyo) 66 : 571–591.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 3- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



