-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Investigating Fungal Outbreaks in the 21st Century
article has not abstract
Published in the journal: Investigating Fungal Outbreaks in the 21st Century. PLoS Pathog 11(5): e32767. doi:10.1371/journal.ppat.1004804
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004804Summary
article has not abstract
What Causes Fungal Outbreaks?
Public attention has been drawn to recent high-profile outbreaks of mycotic diseases, such as those of fungal meningitis and other infections linked to contaminated steroids [1] and an outbreak of necrotizing cutaneous mucormycosis linked to a tornado [2]. However, fungal outbreaks are more common than most people appreciate, and reports of outbreaks caused by unusual fungal pathogens are increasing. The Mycotic Diseases Branch at the Centers for Disease Control and Prevention (CDC) investigates 3–6 fungal outbreaks per year, many of which are caused by rare fungi with limited diagnostic and treatment options. This is a considerable increase from the 1990s, when the Branch investigated 1–2 hospital-based outbreaks per year, generally caused by yeast and traced to a single source. Although the exact reasons for this increase are unknown, the increased number of patients with impaired immune system may have contributed to this trend.
The majority of fungal outbreaks can be attributed to either environmental exposure or a contaminated product (Table 1). For example, two recent outbreaks were linked to contaminated medications. In 2012, two medications produced by a single compounding pharmacy in Florida were contaminated with Fusarium sp. and Bipolaris sp., respectively, shipped to 15 states, and injected into the eyes of patients undergoing vitrectomies. As a result, 47 patients developed endophthalmitis, and most lost vision [3]. In the 2012 fungal meningitis outbreak, methylprednisolone acetate (MPA) contaminated with Exserohilum rostratum and several other microorganisms was shipped to 23 states, potentially exposing nearly 14,000 individuals to this contaminated medication. As a result, 752 people developed meningitis, arachnoiditis, or spinal/paraspinal abscesses, and 64 patients died, making this the deadliest fungal outbreak to date [1].
Tab. 1. Examples of fungal outbreaks. 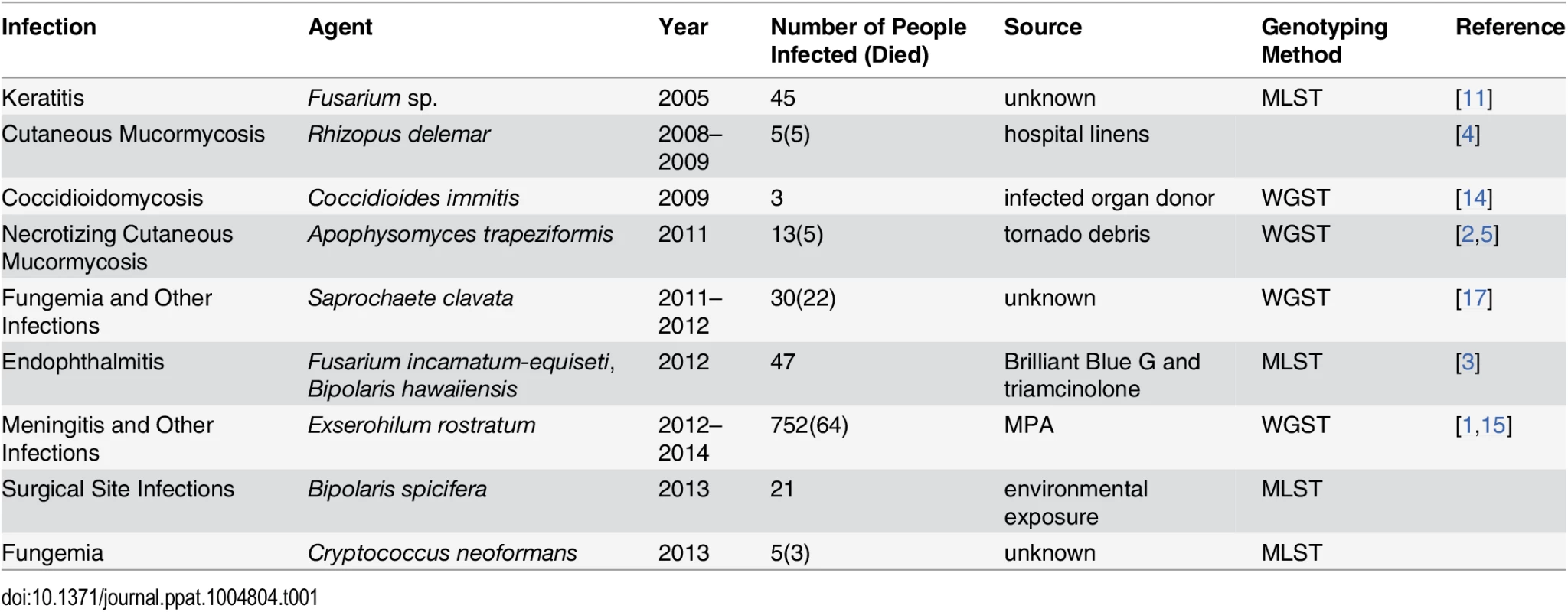
Environmental exposure is the other common cause of fungal outbreaks [2,4]. For example, a recent cluster of Rhizopus delemar infections in a children’s hospital in New Orleans was linked to contaminated linens [4]. In this outbreak, five children died from cutaneous mucormycosis, and R. delemar was isolated from linens, linen shelves, and bins at the hospital [4]. In another example, 13 people developed necrotizing cutaneous mucormycosis caused by Apophysomyces trapeziformis after receiving puncture wounds caused by flying debris during a tornado [2]. Although A. trapeziformis was not recovered from the local environment, whole genome sequence typing (WGST) of the isolates showed that at least three different strains were involved, suggesting environmental exposure [2,5]. In addition, many other outbreaks and clusters caused by dimorphic and other fungal pathogens have been linked to environmental exposure [6–8].
What Role Does Epidemiology Play in Investigating Fungal Outbreaks?
Descriptive epidemiology (i.e., detailed assessment of patients’ demographic characteristics, clinical histories, and the geographic and temporal distribution of cases) is essential for generating hypotheses about the potential source of infection. For example, cases occurring over several weeks to months and scattered geographically suggest a common source outbreak that involves a widely distributed product, especially when case patients have undergone similar medical treatments [1,3]. Conversely, cases occurring among persons with exposure to a common location suggest environmental transmission. Specifically, environmental transmission may be likely among patients with invasive mold infections cared for in the same hospital [4,9,10] or among people with dimorphic fungal infections who participated together in outdoor activities [6,8]. Hypotheses generated through descriptive epidemiology can be tested through analytical epidemiological studies and by microbiological testing of suspected sources (Fig 1).
Fig. 1. CDC epidemiologists are collecting environmental samples in a histoplasmosis outbreak investigation. 
Why Is Molecular Genotyping Important?
Molecular genotyping complements epidemiological findings. Whereas at least some isolates from common source outbreaks are expected to be genetically identical [1,3], environmental transmission is often associated with multiple strains or species [5,11]. Thus, molecular genotyping is a powerful tool to support or refute epidemiologically generated hypotheses. For example, results of molecular typing directly influenced the response to a cluster of Bipolaris sp. infections detected among patients recovering from cardiothoracic surgeries in three hospitals in Texas, Arkansas, and Florida (United States) that was recently investigated by the CDC. Because of the similarity of the patient population, a common product was suspected but could not be identified. However, multilocus sequence typing (MLST) revealed that none of the patients shared strains with the same genotype and that at least two species were involved, strongly supporting environmental exposure rather than a common source. These molecular results provided critical evidence during an early stage of the investigation and focused investigators on the environment rather than on medications and devices used by these patients.
Similar results were obtained when molecular typing was used to investigate a multistate outbreak of Fusarium keratitis, in which a case-control study implicated a specific type of contact lens solution and suggested a common source outbreak, although Fusarium was not isolated from intact product or the production facility. However, the identification of 19 distinct genotypes of Fusarium from case patients provided evidence for independent contamination events in case patients’ local environments [11].
How Can WGST Help?
When available, conventional methods of molecular genotyping, such as MLST, can rapidly generate results; however, identical patterns obtained by these methods are often difficult to interpret, because they can be attributed to both common origin as well as to the low discriminatory power of the typing method. In particular, conventional genotyping methods are often unable to differentiate among strains from clonal populations with low genetic diversity, such as Cryptococcus neoformans or Cryptococcus gattii [12,13]. For example, in a recent cluster of C. neoformans infections in an Arkansas hospital investigated by the CDC, three of five cases of cryptococcosis were caused by isolates with identical ST5 (A5/M5) genotypes, which is also one of the most common MLST genotypes among environmental and clinical strains [13], making it impossible to determine whether these strains were likely to have been acquired independently or from a common source. In addition, certain genotyping methods, such as microsatellite typing, may generate homoplasy, patterns that look indistinguishable but are not related by descent. Furthermore, for the majority of fungi, conventional molecular genotyping methods are simply not available.
WGST provides a highly sensitive tool for molecular genotyping that can be applied to any pathogen without prior knowledge of the genome, which is especially useful for investigating outbreaks caused by rare fungal pathogens for which population structure information may be unavailable. This method has recently been applied to investigate several fungal outbreaks: (i) to confirm molecular identity of Coccidioides immitis from three organ recipients who shared the same donor [14], (ii) to investigate genetic relationships among isolates of E. rostratum from patients and contaminated methylprednisolone [15], (iii) to confirm genetic identity between isolates of C. immitis from soil in Washington state and a case patient with coccidioidomycosis acquired in that state [16], (iv) to demonstrate multiple origins of the rare mold A. trapeziformis in the tornado-associated cluster of necrotizing cutaneous mucormycosis [5], and (v) to demonstrate genetic identity among strains of Saprochaete clavata, a highly unusual fungal pathogen, associated with a multicenter outbreak in France [17].
The discriminatory power of WGST allows estimation of genetic relatedness among strains of pathogens without prior knowledge of the underlying population structure. For example, WGST analysis of E. rostratum isolates from the fungal meningitis outbreak indicated that 19 isolates from patients and contaminated medication lots had identical genomes of 33.8 Mb and no more than two single nucleotide polymorphisms (SNPs) differentiated any two isolates, confirming a likely single origin of the outbreak strains. By contrast, more than 20,000 SNPs were detected between any two control strains of E. rostratum, confirming the genetic diversity among unrelated strains [1,15]. Conversely, genetically identical isolates of A. trapeziformis as well as isolates separated by thousands of SNPs were identified when WGST was used to investigate the etiology of necrotizing cutaneous mucormycosis, which was consistent with environmentally acquired infections [2,5].
What Are the Limitations of WGST and What Is the Future of Fungal Outbreak Investigations?
Although the results of WGST can significantly enhance epidemiological investigations, this method is still unacceptably slow for most real-time investigations. For example, WGST results for the Exserohilum outbreak were generated 6 months after the initial investigation was completed and therefore provided mostly confirmatory data [15]. In order for WGST to become applicable for real-time investigations, time and cost of generating and analyzing WGST data need to be reduced. WGST methods are already widely used for investigating bacterial and viral outbreaks. However, most fungal genomes are at least ten times larger than those of bacteria and viruses; therefore, considerably more time and resources are needed to generate and process fungal genomes.
Development of a curated public database containing assembled genomes of major fungal pathogens will significantly accelerate analyses and implementation of WGST into public health by providing reference genomes and control strains for assessing genetic diversity in a population as well as facilitate data sharing among institutions. It is also possible that for some fungi, WGST can be substituted with a high-density MLST or a targeted SNP-based typing system that can provide as much information as WGST. As population genomic data for fungal pathogens accumulate, targeted SNPs, MLST loci, or both can be selected for high-resolution targeted genotyping for population genetic studies and molecular epidemiological investigations.
The other significant limitation of current WGST is the difficulty in detecting and identifying fungal DNA directly in human clinical samples against the human DNA background. Proteomic and metagenomic methods have been used for culture-independent detection and typing of viral and bacterial pathogens [18–20], and these methods are being adapted for fungi [21]. Accumulation of fungal genomic data and the development of a WGST database for fungal pathogens will provide a necessary framework for developing metagenomic tools for detection and typing of fungi in clinical samples. In addition, genomic data will facilitate basic research aimed at understanding pathogenesis and improving antifungal therapies. As vulnerable patient populations increase and exposure to pathogenic fungi continues, the number of fungal outbreaks is also likely to increase. We anticipate that novel molecular tools coupled with thorough epidemiological investigation will continue to assume greater importance in recognizing, stopping, understanding, and preventing fungal outbreaks in the future.
Zdroje
1. Smith RM, Schaefer MK, Kainer MA, Wise M, Finks J, et al. (2013) Fungal infections associated with contaminated methylprednisolone injections. N Engl J Med 369 : 1598–1609. doi: 10.1056/NEJMoa1213978 23252499
2. Neblett Fanfair R, Benedict K, Bos J, Bennett SD, Lo YC, et al. (2012) Necrotizing cutaneous mucormycosis after a tornado in Joplin, Missouri, in 2011. N Engl J Med 367 : 2214–2225. doi: 10.1056/NEJMoa1204781 23215557
3. Mikosz CA, Smith RM, Kim M, Tyson C, Lee EH, et al. (2014) Fungal endophthalmitis associated with compounded products. Emerg Infect Dis 20 : 248–256. doi: 10.3201/eid2002.131257 24447640
4. Duffy J, Harris J, Gade L, Sehulster L, Newhouse E, et al. (2014) Mucormycosis outbreak associated with hospital linens. Pediatr Infect Dis J 33 : 472–476. doi: 10.1097/INF.0000000000000261 24667485
5. Etienne KA, Gillece J, Hilsabeck R, Schupp JM, Colman R, et al. (2012) Whole genome sequence typing to investigate the Apophysomyces outbreak following a tornado in Joplin, Missouri, 2011. PLoS One 7: e49989. doi: 10.1371/journal.pone.0049989 23209631
6. Lyon GM, Bravo AV, Espino A, Lindsley MD, Gutierrez RE, et al. (2004) Histoplasmosis associated with exploring a bat-inhabited cave in Costa Rica, 1998–1999. Am J Trop Med Hyg 70 : 438–442. 15100461
7. Guinea J, Garcia de Viedma D, Pelaez T, Escribano P, Munoz P, et al. (2011) Molecular epidemiology of Aspergillus fumigatus: an in-depth genotypic analysis of isolates involved in an outbreak of invasive aspergillosis. J Clin Microbiol 49 : 3498–3503. doi: 10.1128/JCM.01159-11 21832010
8. Cummings KC, McDowell A, Wheeler C, McNary J, Das R, et al. (2010) Point-source outbreak of coccidioidomycosis in construction workers. Epidemiol Infect 138 : 507–511. doi: 10.1017/S0950268809990999 19845993
9. Kainer MA, Keshavarz H, Jensen BJ, Arduino MJ, Brandt ME, et al. (2005) Saline-filled breast implant contamination with Curvularia species among women who underwent cosmetic breast augmentation. J Infect Dis 192 : 170–177. 15942908
10. Lutz BD, Jin J, Rinaldi MG, Wickes BL, Huycke MM (2003) Outbreak of invasive Aspergillus infection in surgical patients, associated with a contaminated air-handling system. Clin Infect Dis 37 : 786–793. 12955639
11. Chang DC, Grant GB, O'Donnell K, Wannemuehler KA, Noble-Wang J, et al. (2006) Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296 : 953–963. 16926355
12. Engelthaler DM, Hicks ND, Gillece JD, Roe CC, Schupp JM, et al. (2014) Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio 5: e01464–01414. doi: 10.1128/mBio.01464-14 25028429
13. Litvintseva AP, Kestenbaum L, Vilgalys R, Mitchell TG (2005) Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol 43 : 556–564. 15695645
14. Engelthaler DM, Chiller T, Schupp JA, Colvin J, Beckstrom-Sternberg SM, et al. (2011) Next-generation sequencing of Coccidioides immitis isolated during cluster investigation. Emerg Infect Dis 17 : 227–232. doi: 10.3201/eid1702.100620 21291593
15. Litvintseva AP, Hurst S, Gade L, Frace MA, Hilsabeck R, et al. (2014) Whole-genome analysis of Exserohilum rostratum from an outbreak of fungal meningitis and other infections. J Clin Microbiol 52 : 3216–3222. doi: 10.1128/JCM.00936-14 24951807
16. Litvintseva AP, Marsden-Haug N, Hurst S, Hill H, Gade L, et al. (2015) Valley Fever: Finding New Places for an Old Disease: Coccidioides immitis Found in Washington State Soil Associated With Recent Human Infection. Clin Infect Dis 60: e1–3. doi: 10.1093/cid/ciu681 25165087
17. Vaux S, Criscuolo A, Desnos-Ollivier M, Diancourt L, Tarnaud C, et al. (2014) Multicenter Outbreak of Infections by Saprochaete clavata, an Unrecognized Opportunistic Fungal Pathogen. MBio 5: e02309–14. doi: 10.1128/mBio.02309-14 25516620
18. Wendt JM, Kaul D, Limbago BM, Ramesh M, Cohle S, et al. (2014) Transmission of methicillin-resistant Staphylococcus aureus infection through solid organ transplantation: confirmation via whole genome sequencing. Am J Transplant 14 : 2633–2639. doi: 10.1111/ajt.12898 25250717
19. Rawat A, Engelthaler DM, Driebe EM, Keim P, Foster JT (2014) MetaGeniE: characterizing human clinical samples using deep metagenomic sequencing. PLoS One 9: e110915. doi: 10.1371/journal.pone.0110915 25365329
20. Calderaro A, Arcangeletti MC, Rodighiero I, Buttrini M, Gorrini C, et al. (2014) Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry applied to virus identification. Sci Rep 4 : 6803. doi: 10.1038/srep06803 25354905
21. Cushion MT, Keely SP (2013) Assembly and annotation of Pneumocystis jirovecii from the human lung microbiome. MBio 4: e00224. doi: 10.1128/mBio.00224-13 23592264
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary TuberculosisČlánek Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic PlagueČlánek Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host SpeciesČlánek Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR SelectionČlánek Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus GenomesČlánek Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from FerrioxamineČlánek Remembering MumpsČlánek Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling PathwayČlánek Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Parasites and Their Heterophagic Appetite for Disease
- The Elusive Role of the Prion Protein and the Mechanism of Toxicity in Prion Disease
- Intestinal Colonization Dynamics of
- Activation of Typhi-Specific Regulatory T Cells in Typhoid Disease in a Wild-Type . Typhi Challenge Model
- The Engineering of a Novel Ligand in gH Confers to HSV an Expanded Tropism Independent of gD Activation by Its Receptors
- Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary Tuberculosis
- Group Selection and Contribution of Minority Variants during Virus Adaptation Determines Virus Fitness and Phenotype
- Phosphatidic Acid Produced by Phospholipase D Promotes RNA Replication of a Plant RNA Virus
- A Ribonucleoprotein Complex Protects the Interleukin-6 mRNA from Degradation by Distinct Herpesviral Endonucleases
- Characterization of Transcriptional Responses to Different Aphid Species Reveals Genes that Contribute to Host Susceptibility and Non-host Resistance
- Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic Plague
- Natural Killer Cell Sensing of Infected Cells Compensates for MyD88 Deficiency but Not IFN-I Activity in Resistance to Mouse Cytomegalovirus
- Manipulation of the Xanthophyll Cycle Increases Plant Susceptibility to
- Ly6C Monocytes Regulate Parasite-Induced Liver Inflammation by Inducing the Differentiation of Pathogenic Ly6C Monocytes into Macrophages
- Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host Species
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness
- Inhibition and Reversal of Microbial Attachment by an Antibody with Parasteric Activity against the FimH Adhesin of Uropathogenic .
- The EBNA-2 N-Terminal Transactivation Domain Folds into a Dimeric Structure Required for Target Gene Activation
- Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection
- The NLRP3 Inflammasome Is a Pathogen Sensor for Invasive via Activation of α5β1 Integrin at the Macrophage-Amebae Intercellular Junction
- Sequential Conformational Changes in the Morbillivirus Attachment Protein Initiate the Membrane Fusion Process
- A Two-Component DNA-Prime/Protein-Boost Vaccination Strategy for Eliciting Long-Term, Protective T Cell Immunity against
- cAMP-Signalling Regulates Gametocyte-Infected Erythrocyte Deformability Required for Malaria Parasite Transmission
- Response Regulator VxrB Controls Colonization and Regulates the Type VI Secretion System
- Evidence for a Novel Mechanism of Influenza Virus-Induced Type I Interferon Expression by a Defective RNA-Encoded Protein
- Dust Devil: The Life and Times of the Fungus That Causes Valley Fever
- TNF-α Induced by Hepatitis C Virus via TLR7 and TLR8 in Hepatocytes Supports Interferon Signaling via an Autocrine Mechanism
- The Recent Evolution of a Maternally-Inherited Endosymbiont of Ticks Led to the Emergence of the Q Fever Pathogen,
- L-Rhamnosylation of Wall Teichoic Acids Promotes Resistance to Antimicrobial Peptides by Delaying Interaction with the Membrane
- Rapid Sequestration of by Neutrophils Contributes to the Development of Chronic Lesion
- Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus Genomes
- The Expression of Functional Vpx during Pathogenic SIVmac Infections of Rhesus Macaques Suppresses SAMHD1 in CD4 Memory T Cells
- Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from Ferrioxamine
- TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation
- Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site
- Remembering Mumps
- The Role of Horizontal Gene Transfer in the Evolution of the Oomycetes
- Advances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi
- Investigating Fungal Outbreaks in the 21st Century
- Systems Biology for Biologists
- How Does the Dinoflagellate Parasite Outsmart the Immune System of Its Crustacean Hosts?
- FCRL5 Delineates Functionally Impaired Memory B Cells Associated with Exposure
- Phospholipase D1 Couples CD4 T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication
- Influenza A Virus on Oceanic Islands: Host and Viral Diversity in Seabirds in the Western Indian Ocean
- Geometric Constraints Dominate the Antigenic Evolution of Influenza H3N2 Hemagglutinin
- Widespread Recombination, Reassortment, and Transmission of Unbalanced Compound Viral Genotypes in Natural Arenavirus Infections
- Gammaherpesvirus Co-infection with Malaria Suppresses Anti-parasitic Humoral Immunity
- A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
- Mechanisms of Stage-Transcending Protection Following Immunization of Mice with Late Liver Stage-Arresting Genetically Attenuated Malaria Parasites
- The Myelin and Lymphocyte Protein MAL Is Required for Binding and Activity of ε-Toxin
- Genome-Wide Identification of the Target Genes of AP2-O, a AP2-Family Transcription Factor
- An Atypical Mitochondrial Carrier That Mediates Drug Action in
- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Helminth Infection and Commensal Microbiota Drive Early IL-10 Production in the Skin by CD4 T Cells That Are Functionally Suppressive
- Circulating Pneumolysin Is a Potent Inducer of Cardiac Injury during Pneumococcal Infection
- ExoT Induces Atypical Anoikis Apoptosis in Target Host Cells by Transforming Crk Adaptor Protein into a Cytotoxin
- Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement
- Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
- Varicella Viruses Inhibit Interferon-Stimulated JAK-STAT Signaling through Multiple Mechanisms
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



