-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Phosphatidic Acid Produced by Phospholipase D Promotes RNA Replication of a Plant RNA Virus
All characterized eukaryotic positive-strand RNA [(+)RNA] viruses replicate their genomes using the viral replication complexes (VRCs), which contain multiple viral and host components, on intracellular membranes. Phospholipids are major constituents of cellular membranes; however, the function(s) of phospholipids in genome replication of (+)RNA viruses remains largely unknown. Here, we show that Red clover necrotic mosaic virus (RCNMV), a plant (+)RNA virus, induces a high accumulation of phosphatidic acid (PA) in infected plant leaves. PA-producing enzymes, phospholipase Dα (PLDα) and PLDβ, are associated with RCNMV VRCs. PA interacts with the viral replication protein and enhances the viral replication by upregulating the activity/assembly of the VRCs in vitro. In summary, RCNMV alters cellular lipid metabolism via PLD to establish a suitable environment for viral replication.
Published in the journal: Phosphatidic Acid Produced by Phospholipase D Promotes RNA Replication of a Plant RNA Virus. PLoS Pathog 11(5): e32767. doi:10.1371/journal.ppat.1004909
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004909Summary
All characterized eukaryotic positive-strand RNA [(+)RNA] viruses replicate their genomes using the viral replication complexes (VRCs), which contain multiple viral and host components, on intracellular membranes. Phospholipids are major constituents of cellular membranes; however, the function(s) of phospholipids in genome replication of (+)RNA viruses remains largely unknown. Here, we show that Red clover necrotic mosaic virus (RCNMV), a plant (+)RNA virus, induces a high accumulation of phosphatidic acid (PA) in infected plant leaves. PA-producing enzymes, phospholipase Dα (PLDα) and PLDβ, are associated with RCNMV VRCs. PA interacts with the viral replication protein and enhances the viral replication by upregulating the activity/assembly of the VRCs in vitro. In summary, RCNMV alters cellular lipid metabolism via PLD to establish a suitable environment for viral replication.
Introduction
Positive-strand RNA [(+)RNA] viruses are the most abundant plant viruses, and include many viruses economically important in agriculture. (+)RNA plant viruses have a limited coding capacity. To replicate and achieve successful infection in their hosts, they need to use host proteins, membranes, lipids, and metabolites. All characterized eukaryotic (+)RNA viruses replicate their genomes using viral replication complexes (VRCs), which contain multiple viral and host components on intracellular membranes [1–6]. A growing number of studies have suggested that plant viruses have evolved ways to hijack plant host factors and reprogram host cell metabolism for their successful infection [6, 7]. Conversely, plants have evolved the ability to recognize viruses through specific interaction with viral proteins or viral double-stranded RNA intermediates for restricting virus infection [8, 9]. Viruses must circumvent or suppress such surveillance systems and host defense mechanisms. Thus, viruses must be evolved to achieve a good balance between hijacking/reprogramming host factors for efficient viral replication and avoiding the danger of stimulating antiviral defense responses.
Red clover necrotic mosaic virus (RCNMV) is a (+)RNA plant virus and a member of the genus Dianthovirus in the family Tombusviridae. The genome of RCNMV consists of RNA1 and RNA2. RNA1 encodes a p27 auxiliary replication protein, p88pol RNA-dependent RNA polymerase (RdRp), and a coat protein [10]. RNA2 encodes a movement protein that is required for viral cell-to-cell movement [10, 11]. p27, p88pol, and host proteins form a 480-kDa replicase complex, which is a key player in the viral RNA replication [12]. p27 and p88pol colocalize at the endoplasmic reticulum (ER) [13, 14], where RCNMV replication takes place [15]. Our previous studies showed that RCNMV uses host heat shock proteins (HSPs), HSP70 and HSP90 [16], and ADP-ribosylation factor 1 (Arf1) [15] for the formation of the 480-kDa replicase complex and p27-induced ER membrane alterations. Arf1 is a small GTPase that regulates COPI vesicle formation. Sar1, another small GTPase that regulates COPII vesicle-mediated trafficking, and Arf1 are recruited from their original subcellular locations to RCNMV replication sites via p27, and p27 interferes with host membrane trafficking pathway in plant cells [15, 17]. Mammalian and yeast Arf1 recruits and/or stimulates its effector proteins, including a coatomer, phosphatidylinositol 4 kinase III β (PI4KIIIβ), and phospholipase D (PLD) [18]. Arf1 can activate mammalian PLD1 and PLD2 directly. PLD hydrolyses structural phospholipids such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE) to produce phosphatidic acid (PA) and remaining headgroups. PA production resulting from Arf1-mediated PLD activation has been proposed to be associated with vesicle formation [19].
The 12 different PLD isoforms encoded in the Arabidopsis thaliana genome are classified into six groups (α, β, γ, δ, ε, and ζ) based on sequence similarity and in vitro activity [20]. PLDζ1 and ζ2 have N-terminal phox homology (PX) and pleckstrin homology (PH) domains and share high sequence similarities to two PX/PH-PLDs in mammals. The remaining PLDs contain the Ca2+-dependent phospholipid-binding C2 domain and are unique to plants.
PA is normally present in small amounts (less than 1% of total phospholipids), but rapidly and transiently accumulates in lipid bilayers in response to different abiotic stresses such as dehydration, salt, and osmotic stress [20–22]. PA also accumulates in response to several microbe-associated molecular patterns (MAMPs) in plant cells and positively regulates salicylic acid (SA)-mediated defense signaling [23–27]. Moreover, effector proteins of bacterial and fungal pathogens, such as Cladosporium fulvum Avr4 and Pseudomonas syringae AvrRpm1 and AvrRpt2, trigger PA accumulation in their host cells, and multiple PLD isoforms contribute to AvrRpm1-triggered resistance in Arabidopsis thaliana [28–30]. PLDδ plays a positive role in MAMPs-triggered cell wall based immunity and nonhost resistance against Blumeria graminis f. sp. hordei [31]. Moreover, overexpression of rice diacylglycerol (DAG) kinase, which catalyzes the conversion of DAG to PA, enhances resistance against tobacco mosaic virus and Phytophthora parasitica infections in tobacco plants [32]. In accordance with this, direct application of PA to leaves has been shown to induce the expression of pathogenesis-related (PR) genes and cell death [28,33]. These findings indicate that PA is a positive regulator in plant defense against pathogens. In contrast, PLDβ1 acts like a negative regulator of the generation of reactive oxygen species (ROS), the expression of PR genes, and plant defenses against biotrophic pathogens in rice and Arabidopsis [34–36].
In this study, using two-step affinity purification and liquid chromatography–tandem mass spectrometry (LC/MS/MS) analysis, we identified Nicotiana benthamiana PLDα and PLDβ as interaction partners of RCNMV replication protein, p88pol. Gene-silencing and pharmacological inhibition approaches show that PLDs-derived PA plays a positive role in viral RNA replication. Consistent with this role, direct application of PA to plant cells or plant-derived cell-free extracts enhanced RCNMV RNA replication and negative-strand RNA synthesis, respectively. We found that p27 auxiliary replication protein interacted with PA in vitro and that the accumulation of PA increased in RCNMV-infected plant leaves. Together, our findings suggest that RCNMV hijacks host PA-producing enzymes to achieve successful RNA replication.
Results
To identify putative host proteins associated with the RCNMV replicase complex, we expressed six-His/FLAG-tagged p27 and p88pol replication proteins (p27-HF and p88pol-HF, respectively) together with an RNA2 replication template via agroinfiltration in N. benthamiana. Two-days after infiltration (dai), RCNMV replication proteins were purified via sequential affinity purification using nickel-agarose beads and FLAG-affinity resins as described in the Materials and Methods section. Note that p27-HF and p88pol-HF replication proteins can support RNA2 replication in N. benthamiana plants, although their activities were low compared with those of non-tagged p27 and p88 pol (S1 Fig).
The purified fraction was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis to separate the proteins that copurified with RCNMV replication proteins. A silver-stained gel showed the presence of many protein bands that were absent in the control fraction prepared from Agrobacterium-infiltrated N. benthamiana leaves expressing non-tagged p27 and p88pol together with RNA2 (Fig 1). LC/MS/MS analysis of the isolated proteins excised from gels led to the identification of many host proteins (Fig 1 and S1 Table). These proteins included PLDα and PLDβ, and several Arf1 effector proteins, such as coatomer subunits and clathrin heavy chain, in addition to previously identified host factors, HSP70 and HSP90 [16], and Arf1 [15]. It is known that the activities of yeast and mammalian PLDs are stimulated by Arf1 [37] and Arf1 is an essential host factor in RCNMV RNA replication [15]. Therefore, we investigated whether PLDα and PLDβ are also required for RCNMV RNA replication.
Fig. 1. Identification of proteins copurified with RCNMV replication proteins. 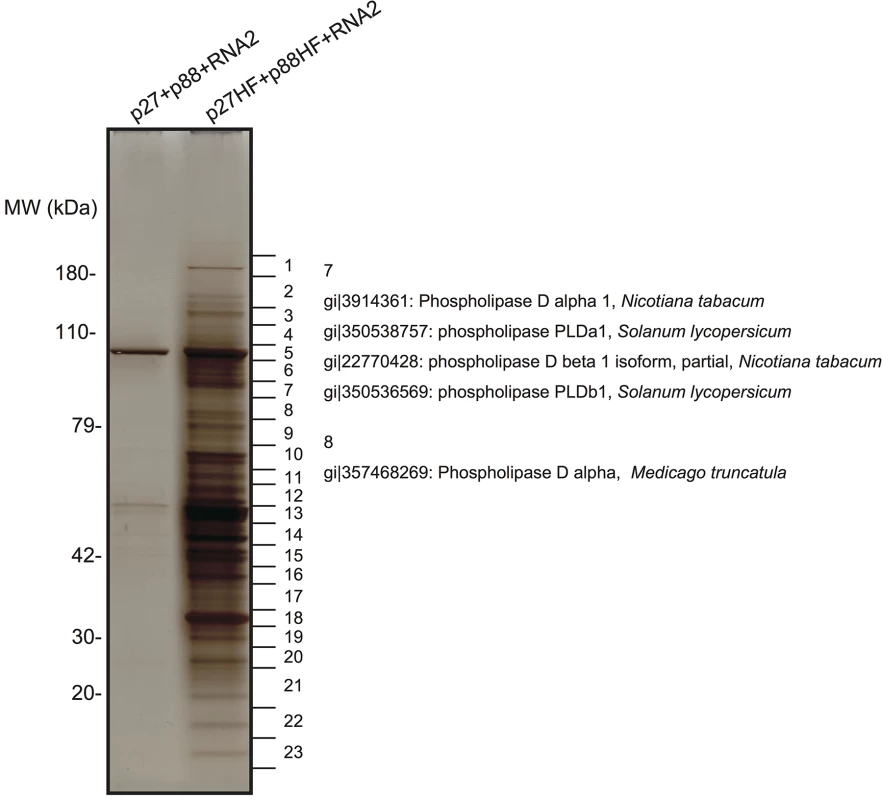
The solubilized total fractions prepared from Agrobacterium-infiltrated leaves expressing p27 plus p88 plus RNA2 or p27-His-FLAG (p27-HF) plus p88-His-FLAG (p88-HF) plus RNA2 were subjected to affinity purification with Ni-NTA agarose beads, followed by anti-FLAG affinity resins. The affinity-purified fractions were subjected to SDS-PAGE and stained using MS-compatible silver staining. Protein bands were excised, subjected to in-gel digestion, and analyzed by tandem mass spectrometry. Some of the proteins that were identified in LC/MS/MS analysis are listed in S1 Table. We isolated cDNAs encoding PLDα and PLDβ from N. benthamiana leaves. Deduced amino acid sequences and peptide sequences identified from LC/MS/MS analysis are presented in S2 Fig. To confirm the association of N. benthamiana (Nb)PLDα and NbPLDβ with RCNMV replication proteins, we performed a co-immunoprecipitation (co-IP) assay in N. benthamiana plants using green fluorescent protein (GFP)-fused NbPLDα or NbPLDβ as bait proteins. We co-expressed p88pol-HF together with p27 and RNA2, because p88pol can be detected by immunoblot in N. benthamiana only when viral RNA replication takes place [12]. Both p27 and p88pol-HF were co-immunoprecipitated with GFP-NbPLDα or GFP-NbPLDβ, but not with GFP (Fig 2A), confirming the association of these PLDs with RCNMV replication proteins.
Fig. 2. RCNMV replication proteins interact with NbPLDα and NbPLDβ. 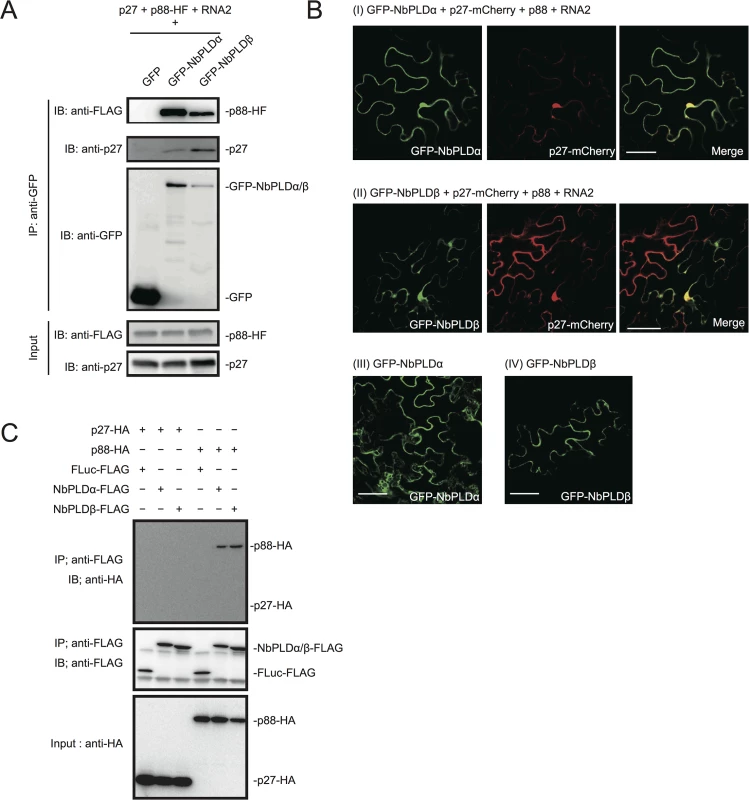
(A) RCNMV replication proteins interacted with NbPLDα and NbPLDβ in a co-IP assay in N. benthamiana. Green fluorescent protein (GFP)-fused NbPLDα (GFP-NbPLDα) or GFP-NbPLDβ was coexpressed with p27, six-His/FLAG-tagged p88pol (p88pol-HF), RNA2, and p19 of RNA silencing suppressor of Tomato bushy stunt virus in N. benthamiana leaves. Total protein was extracted and subjected to immunoprecipitation of GFP-fused proteins by anti-GFP antibody. Proteins were analyzed by immunoblot using antibodies as indicated. (B) NbPLDα and NbPLDβ were colocalized with p27 during viral replication. GFP-NbPLDα or GFP-NbPLDβ was coexpressed with C-terminally mCherry-fused p27 (p27-mCherry), p88pol, RNA2, and p19 (panel I and II) or coexpressed with empty vector and p19 (panel III and IV) in N. benthamiana leaves by Agrobacterium infiltration. Fluorescence was visualized by confocal microscopy at 4 days after infiltration. The merging of the green and red fluorescence is shown as a yellow color. Bars, 50 μm. (C) p88pol interacted with NbPLDα and NbPLDβ in a co-IP assay in BYL. Appropriate combinations of capped transcripts were added to BYL. After in vitro translation at 25°C for 2 hours, the extract was solubilized and subjected to immunoprecipitation of FLAG-tagged proteins by anti-FLAG antibody. Proteins were analyzed by immunoblot using antibodies as indicated. To investigate whether PLDs localize at VRCs, GFP-NbPLDα or GFP-NbPLDβ was co-expressed with mCherry-fused p27 (as a marker of VRC), p88pol, and RNA2 via agroinfiltration in N. benthamiana. At 4 dai, the fluorescence of GFP-NbPLDα and GFP-NbPLDβ was partially merged with the fluorescence of p27-mCherry in large aggregate structures (Fig 2B panels I and II), which are induced by p27 and are thought to be the site of RCNMV RNA replication [13,15,16]. The expression of GFP-NbPLDα or GFP-NbPLDβ alone did not induce such aggregated structures (Fig 2B panels III and IV), suggesting that both NbPLDα and NbPLDβ are recruited to viral replication sites during RCNMV replication.
To investigate whether p27 or p88pol interact with NbPLDα and NbPLDβ, we performed a co-IP assay in cell lysates prepared from evacuolated tobacco BY-2 protoplasts (BYL) [38]. C-terminally HA-tagged viral replication protein (p27-HA or p88pol-HA) was coexpressed with C-terminally FLAG-tagged PLD (NbPLDα-FLAG or NbPLDβ-FLAG) from capped transcripts in BYL. In BYL, p88pol-HA was easily detected by immunoblot using anti-HA antibody in the absence of other viral components after 2 hour of in vitro translation (Fig 2C). After in vitro translation, the extracts were subjected to immunoprecipitation using FLAG-affinity resin. Immunoblot analysis showed that p88pol-HA, but not p27-HA, was copurified with both NbPLDα-FLAG and NbPLDβ-FLAG (Fig 2C). p88pol-HA was not copurified with C-terminally FLAG-tagged firefly luciferase (FLuc-FLAG) that was used as a negative control, excluding the possibility that p88pol-HA binds to FLAG-affinity resin nonspecifically. In reciprocal co-IP experiments using HA antibody, we also found copurification of NbPLDα-FLAG with p88pol-HA, but not with FLuc-HA or p27-HA (S3 Fig). However, as NbPLDβ-FLAG was co-purified not only with p88pol-HA but also with FLuc-HA or p27-HA (S3 Fig), we could not confirm the specific interaction of p88pol with NbPLDβ in BYL.
Both PLDα and PLDβ are required for RCNMV infection
To test whether NbPLDα and NbPLDβ are required for infection of host plants with RCNMV, endogenous transcript levels of NbPLDα or NbPLDβ were downregulated using Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) in N. benthamiana plants. A TRV vector harboring a partial fragment of NbPLDα (TRV:NbPLDα) or NbPLDβ (TRV:NbPLDβ) was expressed via Agrobacterium-mediated expression. An empty TRV vector (TRV:00) was expressed as a control. Newly developed leaves were inoculated with RCNMV RNA1 and RNA2 at 18 dai. Two days after RCNMV inoculation, three inoculated leaves from three different plants were harvested and mixed, and total RNA was extracted. No morphological defects such as chlorotic and stunted phenotypes were observed at this stage (S4 Fig). Semiquantitative reverse-transcription-PCR (RT-PCR) or quantitative RT-PCR (RT-qPCR) analyses confirmed the specific reduction of NbPLDα or NbPLDβ mRNAs in plants infiltrated with TRV:NbPLDα or TRV:NbPLDβ, respectively (Figs 3 and S5). Northern blot analyses using ribonucleotide probes specifically recognizing RCNMV RNA1 or RNA2 showed that the accumulation of RCNMV RNAs was dramatically reduced in both NbPLDα - and NbPLDβ-knockdown plants compared with control plants (Fig 3).
Fig. 3. Knockdown of NbPLDα and NbPLDβ mRNAs levels via gene silencing inhibits the accumulation of RCNMV RNAs in N. benthamiana. 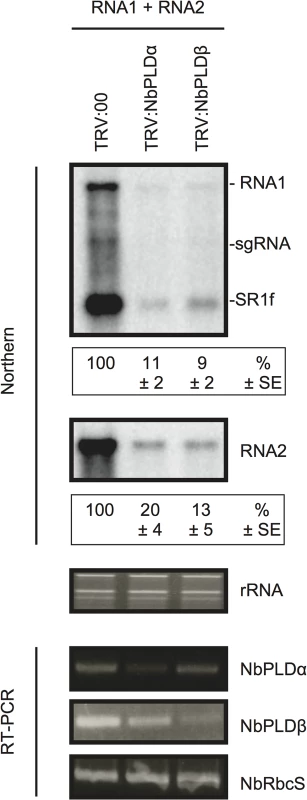
The tobacco rattle virus (TRV) vector harboring a partial fragment of N. benthamiana NbPLDα and NbPLDβ mRNAs (TRV:NbPLDα and TRV:NbPLDβ, respectively) was expressed in N. benthamiana via Agrobacterium infiltration. The empty TRV vector (TRV:00) was used as a control. The newly developed leaves were inoculated with RCNMV RNA1 and RNA2 at 18 days after agroinfiltration. Total RNA was extracted from the mixture of three independent inoculated leaves 2 days after virus inoculation. Accumulation of RCNMV RNA was analyzed by northern blotting. Ethidium bromide-stained ribosomal RNAs (rRNAs) are shown below the northern blots, as loading controls. The numbers below the images represent the relative accumulation levels (means ± standard error) of viral RNAs (RNA1 and RNA2, respectively) using the Image Gauge program (Fuji Film), which were calculated based on three independent experiments. NbPLDα and NbPLDβ mRNA levels were assessed by RT-PCR using primers that allow the amplification of the region of coding regions that are not present in TRV vectors. RT-PCR results for the RbcS gene demonstrated that equal amounts of total RNA were used for the RT and showed equivalent efficiency of the RT reaction in the samples. sgRNA, subgenomic RNA; SR1f, a small RNA fragment that derived from non-coding region of RNA1 [47]. It is known that, in PLDβ1 knockdown transgenic rice plants, the generation of ROS and the expression of defense-related genes are induced even in the absence of pathogen infection [35]. Therefore, it is possible that the poor viral infection in NbPLDβ-knokedown plants was due to activated defense responses. To address this possibility, we tested the effects of NbPLDα - or NbPLDβ-knockdown by TRV-mediated VIGS on the expression of defense-related genes by RT-qPCR analysis. The defense-related genes analyzed here were SA-signaling marker genes (PR-1, PR-2, and PR-5) [39,40], jasmonic acid (JA)-signaling marker genes (LOX1, PR-4, and PDF1.2.) [39,41], ROS-detoxification enzymes (APX, GST, and SOD) [39], MAMP-triggered immunity marker genes (CYP71D20 and ACRE132) [42], and mitogen-activated protein kinases (WIPK and SIPK) [42]. The expression levels of these defense-related genes were not significantly increased in NbPLDα - or NbPLDβ-knockdown plants compared with those in TRV control plants (S5 Fig), excluding the possibility that the reduced viral infection in NbPLDα- or NbPLDβ-knockdown plants are due to activated defense responses in these plants. Some genes including PR-1, PR-2, and CYP71D20 genes were even repressed in NbPLDα - or NbPLDβ-knockdown plants (S5 Fig), consistent with the positive roles of PLDs and PLD-derived PA on plant defense signaling [24–30]. Altogether, these results suggest that both NbPLDα and NbPLDβ play a positive role in RCNMV infection.
PA promotes viral RNA replication
To test the possible contribution of PLD-derived PA to viral RNA replication, we exploited the transphosphatidylation activity of PLDs, which uses primary alcohols as substrates to form an artificial phosphatidyl alcohol. The preferential formation of this compound impairs PA production [43]. Thus, we tested the effect of n-butanol that inhibits PA production by PLDs on RCNMV RNA replication. N. benthamiana protoplasts were inoculated with RCNMV RNA1 and RNA2 and incubated with n-butanol or tert-butanol, an alcohol with no inhibitory effect on PA production, and viral RNA accumulation was determined by northern blot analysis. Increasing n-butanol concentration caused a progressive reduction of viral RNA accumulation (Fig 4A). By contrast, viral RNA accumulation was only moderately reduced in protoplasts treated with tert-butanol compared with the water control. Note that n-butanol did not affect the accumulation of rRNA (Fig 4A). The inhibitory effect of n-butanol on RCNMV replication was also observed in tobacco BY-2 protoplasts (S6 Fig).
Fig. 4. The effects of n-butanol and exogenously supplied PA on RCNMV RNA replication in N. benthamiana protoplasts. 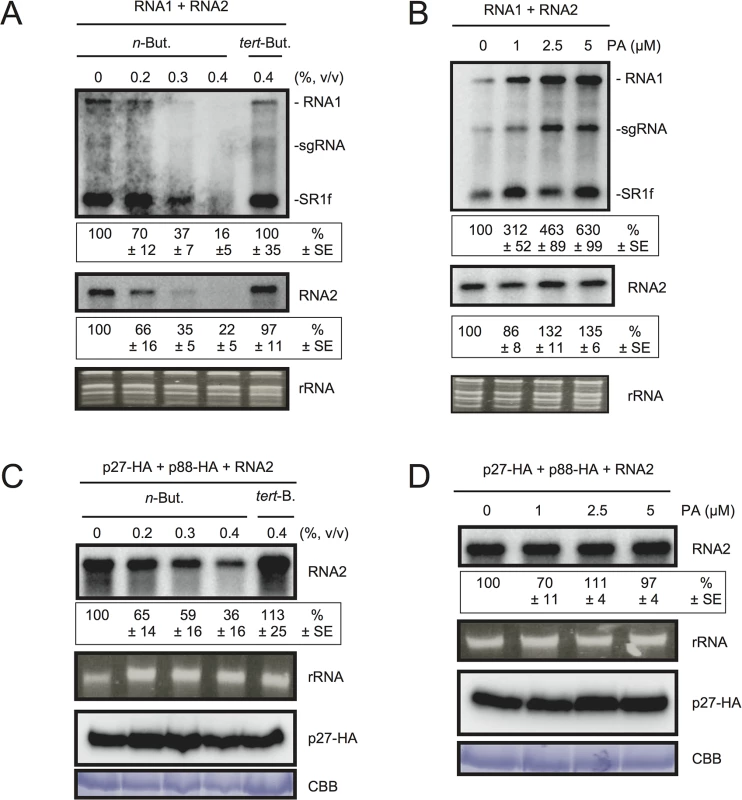
(A) An inhibitor of PLDs impairs RCNMV RNA replication in a single cell. N. benthamiana protoplasts were inoculated with in vitro transcribed RNA1 and RNA2. The inoculated protoplasts were incubated at 20°C for 18 hours in the presence of n-butanol. (B) An exogenously supplied phosphatidic acid (PA) enhances RCNMV RNA replication. N. benthamiana protoplasts were inoculated with in vitro transcribed RNA1 and RNA2. The inoculated protoplasts were incubated at 20°C for 18 hours in the presence of progressively increasing concentrations of PA. (C) An inhibitor of PLDs impairs replication of RNA2 in a single cell. N. benthamiana protoplasts were inoculated with in vitro transcribed RNA2 together with plasmids expressing p27-HA and p88-HA. The inoculated protoplasts were incubated at 20°C for 18 hours in the presence of n-butanol. (D) The effect of an exogenously supplied PA on the replication of RNA2. N. benthamiana protoplasts were inoculated with in vitro transcribed RNA2 together with plasmids expressing p27-HA and p88-HA. The inoculated protoplasts were incubated at 20°C for 18 hours in the presence of progressively increasing concentrations of PA. Total RNA was analyzed by northern blotting using ribonucleotide probes that recognize specifically RCNMV RNA1 and RNA2, respectively. Ethidium bromide-stained rRNAs were used as loading controls and are shown below the northern blots. The numbers below the images represent the relative accumulation levels (means ± standard error) of viral RNAs (RNA1 and RNA2, respectively) using the Image Gauge program, which were calculated based on three independent experiments. In (C) and (D), total protein was analyzed by Immunoblot using anti-HA antibody. Coomassie Brilliant Blue (CBB) staining served as a loading control. sgRNA, subgenomic RNA; SR1f, a small RNA fragment that derived from non-coding region of RNA1 [47]. We also tested the effect of n-butanol and tert-butanol on defense-related gene expressions in N. benthamiana protoplasts. n-butanol did not increase the expression of defense-related genes (S7 Fig). This result corresponds well with the finding that n-butanol has no effects on the basal transcription of the PR-1 gene in Arabidopsis seedlings [27]. In contrast, tert-butanol treatment caused the induction of CYP71D20, ACRE132, and WIPK genes (S7 Fig). This may explain weak negative effect of tert-butanol on RCNMV replication (Figs 4A and S6). Altogether, these results suggest that PLD-derived PA plays a positive role in viral RNA replication.
To verify further the importance of PA in viral RNA replication, commercially available soy-derived PA was supplied to RCNMV-inoculated N. benthamiana protoplasts and viral RNA accumulation was determined by northern blot analysis. Exogenously added PA enhanced the accumulation of RNA1 in a dose-dependent manner (up to 6-fold increase by 5 μM PA), whereas the effect of PA on the accumulation of RNA2 was negligible (Fig 4B). Neither exogenously supplied PC nor PE significantly affected the accumulation of viral RNA in N. benthamiana protoplasts (S8 Fig). These results suggest that PA plays an important role in RCNMV RNA replication, and that the requirement for PA may differ between RNA1 and RNA2.
Replication of RNA2 depends entirely on RNA1 simply because RNA2 uses replication proteins supplied from RNA1. The negative impact of n-butanol on the accumulation of RNA2 (Fig 4A) may be the indirect action of this primary alcohol through inhibition of RNA1 replication. Therefore, it is possible that RNA2 replication does not require any PLD-derived PA. To investigate this possibility, we inoculated N. benthamiana protoplasts with RNA2 and plasmids expressing p27 and p88pol, as suppliers of the replication proteins. The accumulation of RNA2 was decreased by n-butanol in a dose-dependent manner (Fig 4C). Note that in this experiment, the accumulation of p27 replication protein was not significantly changed (Fig 4C). These results indicate that PLD-derived PA was also required for the replication of RNA2 as in the case of RNA1. However, the replication of RNA2 was not enhanced by exogenously supplied PA (Fig 4D), suggesting that the threshold of PA requirement for RNA2 replication is lower than that for RNA1. The differential requirement for PA in the replication of RNA1 and RNA2 is discussed later.
Next, we investigated the effect of n-butanol on RNA replication of Brome mosaic virus (BMV), another plant (+)RNA virus, which is unrelated to RCNMV. Increasing n-butanol concentration caused progressive reduction in the accumulation of BMV RNA (Fig 5A). Co-IP experiments showed interactions between BMV replication proteins and NbPLDβ-FLAG (Fig 5B and 5C). These results suggest that PLD-derived PA is also important for BMV RNA replication. However, exogenously added PA did not affect the accumulation of BMV RNA (Fig 5D) as similarly seen for RCNMV RNA2.
Fig. 5. BMV RNA replication requires PLD-derived PA in N. benthamiana protoplasts. 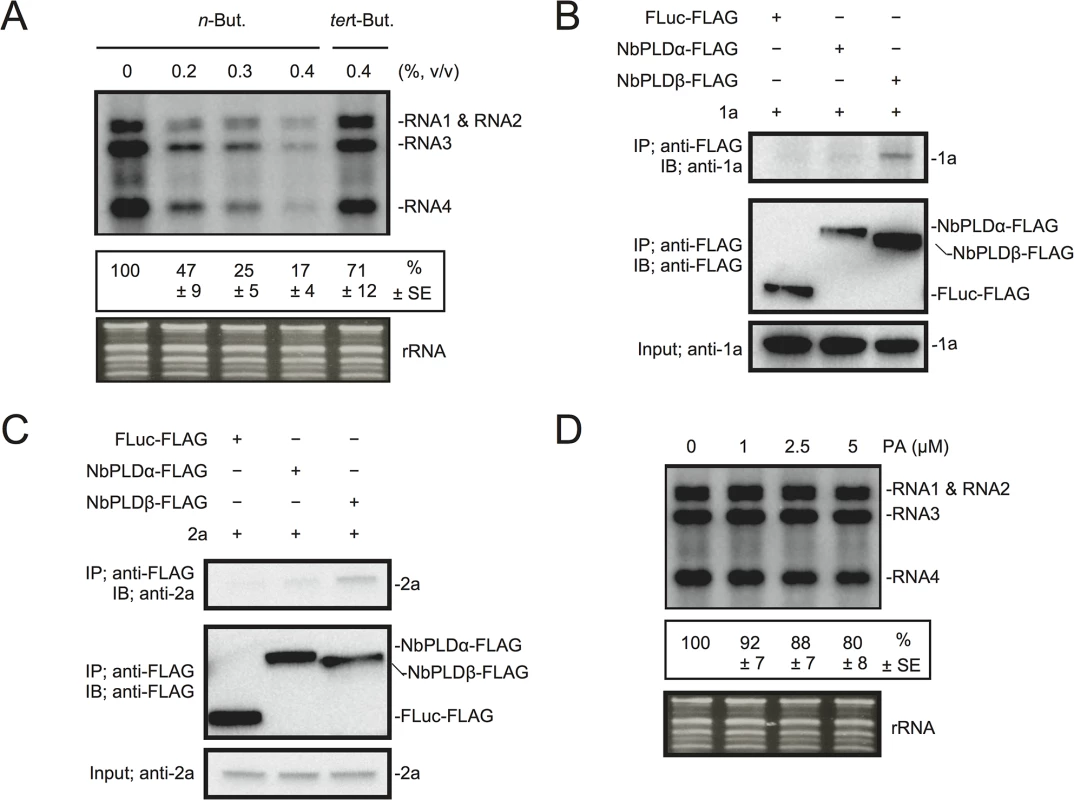
(A) An inhibitor of PLDs impairs BMV RNA replication in a single cell. N. benthamiana protoplasts were inoculated with in vitro transcribed BMV RNA1, RNA2, and RNA3. The inoculated protoplasts were incubated at 20°C for 18 hours in the presence of n-butanol. (B)-(C) Brome mosaic virus replication proteins 1a (B) and 2apol (C) interact with NbPLDβ. Appropriate combinations of capped transcripts were added to BYL. After in vitro translation at 25°C for 2 h, the extract was solubilized and mixed with a 10 μl bed volume of anti-FLAG M2 antibody agarose and incubated further at 4°C for 1 hours. After washing, proteins copurified with FLAG-tagged proteins were analyzed by immunoblotting with appropriate antibodies. (D) The effect of an exogenously supplied PA on the replication of BMV. N. benthamiana protoplasts were inoculated with BMV RNA1, RNA2, and RNA3 transcribed in vitro. The inoculated protoplasts were incubated at 20°C for 18 hours in the presence of progressively increasing concentrations of PA. Total RNA was analyzed by northern blotting using ribonucleotide probes that specifically recognize the 3’-UTR of BMV RNAs. Ethidium bromide-stained rRNA was used as a loading control and is shown below the northern blots. The numbers below the images represent the relative accumulation levels (means ± standard error) of viral RNAs (RNA1 + RNA2 + RNA3 + RNA4) using the Image Gauge program, which were calculated based on three independent experiments. PA stimulates VRC activity in vitro
PA acts as a second messenger in signal transduction during multiple biotic and abiotic stress responses and plays multiple roles including that for transcriptional reprogramming [20–22]. To investigate whether PA has a direct role in viral RNA replication, we took advantage of BYL, a nucleus-depleted (therefore, the effects of PA on transcriptional reprogramming are negligible) in vitro translation/replication system that has been used successfully to recapitulate the negative-strand RNA synthesis of RCNMV [12, 44–49]. Addition of PA into BYL stimulated the accumulation of newly synthesized negative-strand RNA (Fig 6A), and moderately enhanced the accumulation of the 480-kDa replicase complex (Fig 6B), suggesting that PA stimulates the activity and/or assembly of the viral replicase complex in a direct manner. The stimulation effects of PA on the accumulation of the 480-kDa replicase complex was less obvious than that on the accumulation of newly-synthesized viral RNA at the highest concentration of PA used in this experiment (50 μM). This result may also support a direct role for PA in the enhancement of RdRP activity rather than the formation of the replicase complex.
Fig. 6. PA enhances accumulation of negative-strand RNA1 and the 480-kDa replicase complex in BYL. 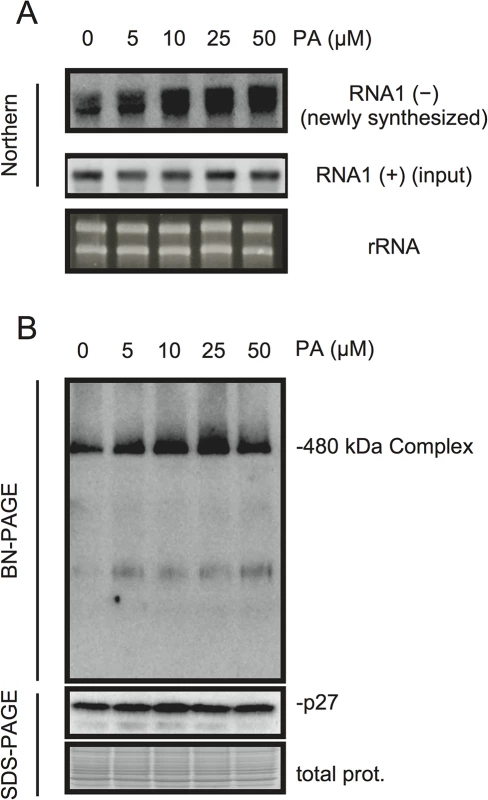
BYL was incubated for 4 h at 17°C with in vitro-transcribed RNA1 (300 ng) in the presence of various concentrations of PA as indicated. Accumulation of negative-strand RNA1 was detected by northern blotting (A). Total protein solubilized from BYL was subjected to blue native (BN)- and SDS-PAGE analyses, followed by immunoblotting with anti-p27 antisera (B). EtBr-stained rRNA and Coomassie Brilliant Blue (CBB)-stained cellular proteins are shown as loading controls. p27 replication protein binds to PA in vitro
Because PA directly stimulated the viral negative-strand RNA synthesis in BYL, we hypothesized that p27, the multifunctional RCNMV replication protein, has an affinity for PA. To investigate this possibility, we conducted a lipid overlay assay using bacterially expressed, purified C-terminally FLAG-tagged p27 (p27-FLAG) [49]. Purified p27-FLAG protein was incubated with phospholipid-spotted nitrocellulose membranes, and the interaction between p27-FLAG and phospholipids was detected using anti-FLAG antibody. p27 gave strong PA binding signals on the blot (Fig 7A and 7B). p27 also exhibited weak binding to phosphatidylinositol-4-phosphate (PI4P), phosphatidylinositol (3,5)-bisphosphate, phosphatidylinositol (4,5)-bisphosphate, and phosphatidylinositol (3,4,5)-trisphosphate, but negligible binding to other lipids, including PC and PE (Fig 7A and 7B). These results were consistent with the findings that neither exogenously supplied PC nor PE promoted RCNMV replication in N. benthamiana protoplasts (S8 Fig). N - or C-terminal halves of p27 fragments did not give PA binding signals (Fig 7C and 7D), suggesting that overall protein conformation may be important for p27–PA binding.
Fig. 7. p27 interacts with PA in vitro. 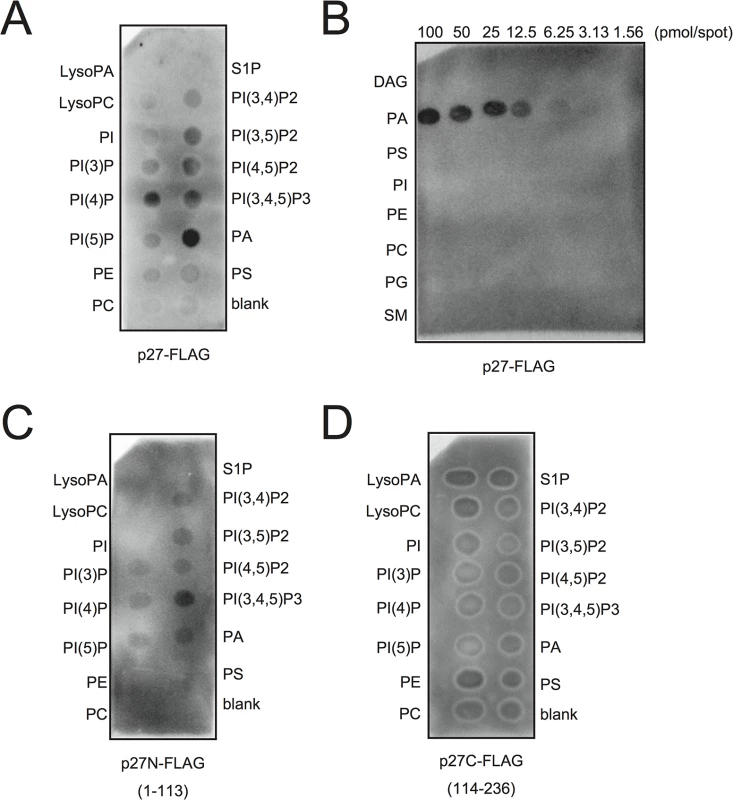
p27-FLAG or its variant proteins expressed in E. coli BL21 (DE3) were purified by Ni-NTA agarose beads. Lipid overlay analysis of various lipids with full-length p27-FLAG (A)-(B), N-terminal half of p27-FLAG (p27N-FLAG) (C), or C-terminal half of p27-FLAG (p27C-FLAG) (D). Purified protein (500 ng) was used, followed by immunoblotting with anti-FLAG antibody. LysoPA, lysophosphatidic acid; LysoPC, lysophosphochorine; PI, phosphatidylinositol; PI(3)P, phosphatidylinositol 3-phosphate; PI(4)P, phosphatidylinositol 4-phosphate; PI(5)P, phosphatidylinositol 5-phosphate; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PI(3,4)P2, phosphatidylinositol (3,4)-bisphosphate; PI(3,5)P2, phosphatidylinositol (3,5)-bisphosphate; PI(4,5)P2, phosphatidylinositol (4,5)-bisphosphate; PI(3,4,5)P3, phosphatidylinositol (3,4,5)-trisphosphate; PA, phosphatidic acid; PS, phosphatidylserine; DAG, diacylglycerol; PG, phosphatidylglycerol; SM, sphingomyelin. RCNMV promotes PA accumulation in planta
Next we investigated whether RCNMV infection affects the amount of PA in plant leaves. N. benthamiana leaves were inoculated with RCNMV via agroinfiltration. At 2 dai, lipids were extracted and the amount of PA was analyzed by thin layer chromatography (TLC). Compared with control plant leaves infiltrated with Agrobacterium harboring an empty vector, the signal intensity of a lipid spot that showed a migration similar to that of soy-PA was increased in RCNMV-infected plant leaves (about 3-fold higher) (Fig 8A and 8B). To identify the lipid species of the spot, the same samples were again subjected to TLC, and the spot was scratched out from Coomassie Brilliant Blue R-250 stained TLC plates and subjected to LC/MS analysis. The lipid was identified as PA (Fig 8C–8E). We concluded that RCNMV infection upregulated PA accumulation in plants. It is known that expressions of PLD genes were induced by pathogen infection [50]. Therefore, the enhanced accumulation of PA in RCNMV-infected plants could be due to elevated accumulation of PLD through induction of PLD gene expression. To examine this possibility, we investigated whether mRNA levels of NbPLDα and NbPLDβ were upregulated in RCNMV-infected plants by RT-qPCR analysis. The accumulation levels of NbPLDα and NbPLDβ transcripts in RCNMV-infected plants were about 1.2 - and 1.9-fold higher, respectively, than those in control plants, although the increase in NbPLDα transcripts was insignificant by the Student’s t-test (S9 Fig).
Fig. 8. RCNMV stimulates the accumulation of PA in a plant tissue. 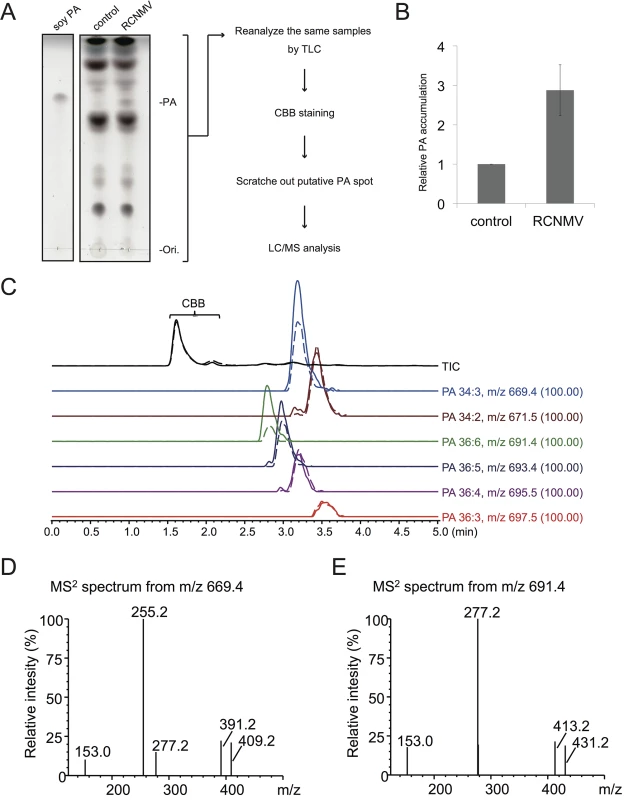
(A) The extracted lipids from Agrobacterium-infiltrated leaves expressing empty vector or RCNMV RNA1 plus RNA2 were subjected to thin layer chromatography (TLC) and phospholipids were visualized by CuSO4 staining. The same samples were again subjected to TLC, and the spot was scratched out from Coomassie Brilliant Blue R-250 (CBB) stained TLC plates and subjected to LC/MS analysis. (B) Relative PA accumulation was quantified using ImageJ software. Bars represent means and standard error of values obtained from three independent biological samples. (C) Total ion and major extracted ion chromatograms of the TLC separated bands. The broken lines indicate the extracts from empty vector-expressed leaves, and the solid lines indicate the ones from the leaves expressing RCNMV RNA1 plus RNA2. (D)-(E), MS2 spectra produced by CID of m/z 669.4 (D) and m/z 691.4 (E). The signal at m/z 153.0, 255.2, and 277.2 indicate the dehydrated glycerol phosphate, carboxylate anion of C16:0 and C18:3, respectively. Ions of m/z 391.2 and 413.2 arose from the neutral loss of C18:3, and m/z 409.2 and 431.2 arose from the C18:3 neutral loss as ketene from the each precursor ion. The other ions, m/z 671.5, 693.4, 695.5, and 697.5 also produced the similar MS2 spectra as described above, and were confirmed as phosphatidic acid. NbPLDα - or NbPLDβ-knockdown plants exhibit reduced PA accumulation
We compared the accumulation of endogenous PA in NbPLDα or NbPLDβ knockdown plants with that in TRV control plants by TLC analysis. As expected from their predicted function, NbPLDα - or NbPLDβ-knockdown plants showed reduced accumulation of PA compared with TRV control plants (S10 Fig), suggesting that these PLDs contribute to PA production in N. benthamiana. These results further supported the idea that RCNMV-induced PLDs-derived PA plays an important role in RCNMV replication.
Discussion
A growing number of studies have suggested that PLD and PLD-derived PA play vital roles in environmental responses in plants [19–22,50]. The properties of PA (i.e., normally present in small amounts, and rapidly and transiently accumulates in response to various environmental cues) seem to be suitable for its function in biotic and abiotic responses in which plants need to rapidly accommodate their surrounding environments. Indeed, PA has been shown to accumulate in response to several MAMPs and pathogen effector proteins, or SA that is a key hormone involved in plant resistance against biotrophic pathogens [23–29]. PA induces PR gene expression and cell death [28,33,34], and has been proposed to act as an important component in resistance to biotrophic pathogens such as tobacco mosaic virus and Phytophthora parasitica. Plants have diverse numbers of PLD isoforms and they appear to have distinct but somewhat overlapping functions in cellular responses [50]. In Arabidopsis, it is proposed that multiple PLD isoforms cooperatively contribute to AvrRpm1-triggered resistance [30]. In rice, PLDβ1 acts like a negative regulator of defense signaling because PLDβ1-knockdown rice plants exhibit constitutive ROS production, expression of PR genes, and enhanced resistance against pathogens [35]. In this study, we showed that PLD and PA are essential for and play a key role in RCNMV replication. Poor infection of RCNMV to NbPLDα - or NbPLDβ-knockdown plants was not due to constitutively activated defense responses, indicating that both NbPLDα and NbPLDβ act as essential host factors in RCNMV replication. The findings reveal novel aspects of PLD and PA in their roles during biotic stress responses in plants.
The replication of Tomato bushy stunt virus is enhanced by deletion of the PAH1 gene encoding a PA phosphatase, which converts PA into DAG in yeast [51]. Moreover, ectopic expression of Arabidopsis PA phosphatase, Pah2 in N. benthamiana results in the inhibition of tombusviruses and RCNMV infection [51]. These findings suggest that PA is positively involved in the life cycles of these viruses. However, whether viruses manipulate PA production for viral replication has been unknown. In the current study, we showed that RCNMV replication proteins interacted with PA-producing enzymes, NbPLDα and NbPLDβ. RCNMV infection induced a high accumulation of PA in plant tissues, suggesting that RCNMV alters cellular lipid metabolism to establish a suitable environment for viral replication. It is known that transcription of PLD genes is induced by pathogen infection [50]. RCNMV infection increased the accumulation of NbPLDα and NbPLDβ transcripts (S9 Fig). Currently, whether the enhancement of PA accumulation observed in RCNMV-infected plants is due to the upregulation of PLD gene expression remains unknown. PLDs may be activated through direct or indirect interaction with RCNMV replication proteins. Although we failed to show the interaction between p88pol and NbPLDα or NbPLDβ in vivo because p88pol accumulates below detection limits in the absence of viral RNA replication in N. benthamiana [12], p88pol interacted with PLDs, at least with NbPLDα in a co-IP assay in BYL. The interaction of p88pol with PLDs seems to make sense for viral replication strategy because it brings them to the sites of replication. This strategy could increase PA only at VRCs and not at other cellular membranes where PA might affect cellular metabolism or activate the SA-mediated defense responses that is detrimental for successful viral infection. This is critical for the viral life cycle because PA interacts with p27 auxiliary replication protein (Fig 7) and enhances viral replication through upregulating the activity and/or assembly of the 480-kDa replicase complex (Fig 6). To our knowledge, this is the first demonstration of a functional role for PA in (+)RNA virus replication. It is likely that PA is involved in RNA replication of many (+)RNA viruses. Indeed, the replication proteins, 1a and 2apol of BMV, a virus unrelated to RCNMV, also interacted with NbPLDβ, and BMV RNA replication was sensitive to n-butanol, which is an inhibitor of PLDs-derived PA production (Fig 5). However, exogenously added PA did not affect the accumulation of BMV RNA (Fig 5), implying that the threshold of PA requirement for BMV RNA replication seems to be lower than that for RCNMV. The differential PA requirement may be explained by very weak affinity of BMV replication proteins with NbPLDα, which is more active in PA production than NbPLDβ (S10 Fig). Moreover, Dengue virus induces the accumulation of several lipids in infected mosquito cells, including PA [52]. It has been reported that Coxsackievirus B3 and mouse hepatitis coronavirus replication is insensitive to n-butanol [53,54]. However, because PA can also be formed through the combined action of phospholipase C and DAG kinase [55], it is unknown whether replication of these viruses depends on PA or not.
How does PA affect viral RNA replication? PA binding to proteins modulates the catalytic activity of target proteins, tethers proteins to the membranes, and promotes the formation and/or stability of protein complexes [55]. We found that exogenous PA enhanced the accumulation of newly synthesized viral RNA and the formation of 480-kDa replicase complexes in BYL in vitro translation/replication systems, and that p27 had affinity for PA in vitro (Figs 6 and 7). Therefore, PA could promote viral replicase activity and/or assembly directly. The 480-kDa replicase complexes contain p27 oligomer. Therefore, it is possible that PA-binding to p27 in the replicase complexes assists the conformational change of p27 that is suitable for RNA synthesis. Alternatively, PA could serve as an assembly platform for host PA-binding proteins. PA binds to various proteins, including transcription factors, kinases, phosphatases, enzymes involved in central metabolism, and proteins involved in vesicular trafficking and cytoskeletal rearrangements [20,21]. Several known cellular PA-binding proteins were also identified in our LC/MS/MS analysis (S1 Table). These include NADPH oxidase [56], glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [57,58], and SNF1-related kinase [58]. RCNMV-induced accumulation of high PA levels may facilitate the recruitment of these PA-binding proteins to viral replication sites. One of the candidate proteins is GAPDH-A. Although GAPDH-A is not required for RCNMV replication, it recruits RCNMV movement protein to viral replication sites and plays an important role in virus cell-to-cell movement [59]. Therefore, PA may also function in bridging viral replication and cell-to-cell movement. RCNMV induces large ER aggregates in infected cells, which are thought to be viral RNA replication factories [13–16]. PA may play a direct role in the formation of RNA replication factories. In support of this hypothesis, deletion of pah1 in yeast causes the expanded ER membranes and leads to enhanced TBSV replication on these membranes, although TBSV replicates normally at peroxisomes [51]. In addition, it is also possible that PA might affect unknown cellular factors that are involved in viral RNA replication. Further studies are needed to elucidate the molecular functions of PA in viral RNA replication.
Our results suggest that RNA1 and RNA2 have differential PA requirements in RNA replication. How does this difference contribute to RCNMV infection? RNA1 requires a larger amount of PA for maximum efficiency of its replication than that required by RNA2. This may reflect differences in the translation and replication mechanisms between RNA1 and RNA2. Translation of MP from RNA2 couples with RNA replication: only progeny RNA2 generated de novo through the RNA replication pathway could function as mRNA [60]. Poor enhancement of RNA2 replication by PA may be beneficial for switching from replication to translation. By contrast, because RNA1 has a cap-independent translation enhancer that is an effective recruiter of translation factors [61], newly synthesized RNA1 serves as a template for further translation of the replication proteins that would activate PA production in infected cells. Moreover, RNA1 also serves as a template for transcription of subgenomic RNA, which encodes CP. Therefore, PA-mediated enhancement of RNA1 replication may be suitable for the production of CP subgenomic RNA during the late stage of viral infection. High PA requirement for maximum RNA1 replication may explain the use of both NbPLDα and NbPLDβ as essential host factors.
A growing number of studies have suggested that multiple lipid species affect virus replication and that (+)RNA viruses employ a multifaceted strategy to rewire host machinery involved in lipid transport and synthesis [62]. HCV infection stimulates the production of cellular PC, PE, and PI4P and HCV co-opts PI4KIIIα for replication [63,64]. In addition, HCV infection stimulates the accumulation of cellular sphingomyelin, which binds to and activates HCV NS5B polymerase [65,66]. Enterovirus utilizes PI4KIIIβ for RNA replication and viral RdRP 3Dpol binds to PI4P [67]. Enteroviruses also upregulate cellular uptake of fatty acids, which are channeled toward highly upregulated PC synthesis in infected cells [68]. The elevated PC has been proposed to serve as a building block for the formations of the viral replication factory [68, 69]. Recently, it has been shown that PE and PC stimulate TBSV RdRP activity in vitro [70]. RCNMV p27 showed affinity, not only for PA but also for other phospholipids such as PI4P in vitro. However, our LC/MS/MS analysis failed to detect any PI4P-producing enzymes, such as PI4K. Combined lipidomics, proteomics, and transcriptome analysis will be helpful for a comprehensive understanding of lipid species involved in viral RNA replication.
Materials and Methods
Gene cloning and plasmid construction
Plasmids given the prefix ‘‘pBIC” were used for Agrobacterium infiltration, ‘‘pUC”, ‘‘pRC” and ‘‘pR” were used for in vitro transcription, ‘‘pCold” was used for protein expression in Escherichia coli. pUCR1 [71] and pRC2_G [72] are full-length cDNA clones of RNA1 and RNA2 of the RCNMV Australian strain, respectively. pB1TP3, pB2TP5, and pB3TP8 are full-length cDNA clones of RNA1, RNA2, and RNA3 of the BMV M1 strain, respectively [73] (generous gift from Paul Ahlquist). The constructs described previously used in this study include pBICp27 [71], pBICp88 [71], pBICR2 [71], pBICR1R2 [71], pBICp19 [71], pCOLDIp27-FLAG [49], pCOLDIp27N-FLAG [49], and pCOLDIp27C-FLAG [49]. pUC118 was purchased from Takara Bio Inc. (Shiga, Japan). Escherichia coli DH5α was used for the construction of all plasmids. All plasmids constructed in this study were verified by sequencing.
RNA extraction from Nicotiana benthamiana leaves was performed using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Reverse-transcription-PCR (RT-PCR) was catalyzed by Superscript III reverse transcriptase (Invitrogen) using oligo (dT) [16]. Primers to amplify coding sequences of NbPLDα or NbPLDβ were designed based on the N. benthamiana RNA seq data (Transcriptome version 5: http://sydney.edu.au/science/molecular_bioscience/sites/benthamiana/) [74].
pBICp27-His-FLAG
A PCR fragment from pBICp27 was amplified using primers 5'-CGGGGATCCATGGGTTTTATAAATCTTTCG-3' and 5'-CGGGGATCCCTACTTGTCATCGTCGTCCTTGTAATCATGATGATGATGATGATGAAAATCCTCAAGGGATTTG-3'. The amplified fragment was digested with BamHI and inserted into the corresponding region of pBICP35.
pBICp88-His-FLAG
A PCR fragment from pBICp88 was amplified using primers 5'-CCGGGTACCATGGGTTTTATAAATCTTTCG-3' and 5' - CGGGGTACCTTACTTGTCATCGTCGTCCTTGTAATCATGATGATGATGATGATGTCGGGCTTTGATTAGATCTTTG-3'. The amplified fragment was digested with KpnI and inserted into the corresponding region of pBICP35.
pBYLp27-HA
A PCR fragment from pUBp27-HA [16] was amplified using primers 5' - CTGGCGCGCCATGGGTTTTATAAATCTTTCGCTTTTTGATGTGG-3' and 5'-CTGGCGCGCCTTAAGCGTAATCTGGAACATCGTATGGGTA-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBYL2 [12].
pBYLp88-HA
A PCR fragment from pUBp88-HA [16] was amplified using primers 5' - CTGGCGCGCCATGGGTTTTATAAATCTTTCGCTTTTTGATGTGG-3' and 5'-CTGGCGCGCCTTAAGCGTAATCTGGAACATCGTATGGGTA-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBYL2.
pBYLNbPLDα-FLAG
A PCR fragment from cDNA derived from N. benthamiana was amplified using primers 5'-CTGGCGCGCCATGGCTCAGATTCTGCTTCATGGAACTCTCC-3' and 5'-CTGGCGCGCCCTACTTGTCATCGTCGTCCTTGTAATCTGTAGTGAGGATAGGAGGAAGG-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBYL2.
pBYLSP6NbPLDβ-FLAG
A PCR fragment from cDNA derived from N. benthamiana was amplified using primers 5'-CTGGCGCGCCATGGCTCATTTGTCTTATTCTGATTCTATTAG-3' and 5'-CTGGCGCGCCTCACTTGTCATCGTCGTCCTTGTAATCGATATCTGGAAAGGTTTCACAGCCAGGGAG-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBYL2 to generate pBYLNbPLDβ-FLAG. A PCR fragment from pBYLNbPLDβ-FLAG was amplified using primers 5'-GAGGATCCCCGATTTAGGTGACACTATAGAGACCCAAGCTGGCGCGCCATGGCTCATTTGT-3' and 5'-AGTCCCGGGCACACCCTTATACTCGTGATCTCCTCC-3'. The amplified fragment was digested with BamHI and XmaI, and inserted into the corresponding region of pBYLNbPLDβ-FLAG.
pBYLAtPAH1-HA
A PCR fragment from cDNA derived from A. thaliana was amplified using primers 5'-CTGGCGCGCCATGAGTTTGGTTGGAAGAGTTGGGAGTTTGATTT-3' and 5'-GCTGGCGCGCCTCAAGCGTAATCTGGAACATCGTATGGGTATTCAACCTCTTCTATTGGCAGTTTCC-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBYL2.
pBYLFLuc-HA
A PCR fragment from pLUCpA60 [75] was amplified using primers 5'-CTGGCGCGCCATGGAAGACGCCAAAAACATAAAGAAAGGCCC-3' and 5'-CTGGCGCGCCTTAAGCGTAATCTGGAACATCGTATGGGTACACGGCGATCTTTCCGCCCTTCTTGGCC-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBYL2.
pBYLFLuc-FLAG
A PCR fragment from pLUCpA60 [75] was amplified using primers 5'-CTGGCGCGCCATGGAAGACGCCAAAAACATAAAGAAAGGCCC-3' and 5'-CTGGCGCGCCTTACTTGTCATCGTCGTCCTTGTAATCCACGGCGATCTTTCCGCCCTTCTTGGCC-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBYL2.
pBICp27-mCherry
A PCR fragment from pUBp27-mCherry [15] was amplified using primers 5'-AGGATCCGGATGGGTTTTATAAATCTTTCGCTTTTTGATGTGG-3' and 5'-GGGGTACCCTACTTGTACAGCTCGTCCATGCCGCCGGTGG-3'. The amplified fragment was digested with BamHI and KpnI, and inserted into the corresponding region of pBICP35 [76].
pBICsGFP-NbPLDα
A PCR fragment from pBYLNbPLDα-FLAG was amplified using primers 5'-CTGGCGCGCCATGGCTCAGATTCTGCTTCATGGAACTCTCC-3' and 5'-CTGGCGCGCCCTATGTAGTGAGGATAGGAGGAAGGTAGTCAG-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBICsGFP [13].
pBICsGFP-NbPLDβ
A PCR fragment from pBYLNbPLDα-FLAG was amplified using primers 5'-CTGGCGCGCCATGGCTCATTTGTCTTATTCTGATTCTATTAG-3' and 5'-CTGGCGCGCCTCAGATATCTGGAAAGGTTTCACAGCCAGGGAG-3'. The amplified fragment was digested with AscI and inserted into the corresponding region of pBICsGFP.
pTVNbPLDα
A PLDα cDNA fragment was amplified from cDNA derived from N. benthamiana (NbPLDα) RNA using primers 5'-ATCCCCCGGGAGTTCCTTGTAGCTCTGTGACATCCCC-3' and 5'-GCAGCCCGGGTCAGCCAACATAAACCAGAGATCGATGGACGG-3'. The generated PCR product was then cloned in the antisense orientation into the XmaI site of pTV00 [77].
pTVNbPLDβ
A PLDβ cDNA fragment was amplified from cDNA derived from N. benthamiana (NbPLDβ) RNA using primers 5'-CCCCCGGGTCGACTTTTCTGACTGAGTTCCTGAGGTGTAGCAGG-3' and 5'-AGCCCGGGTTGATACCAATGGAGATTGCTCTAAAAATTGCC-3'. The generated PCR product was then cloned in the antisense orientation into the XmaI site of pTV00.
Plant growth conditions
N. benthamiana plants were grown on commercial soil (Tsuchi-Taro, Sumirin-Nosan-Kogyo Co. Ltd.) at 25 ± 2°C and 16 h illumination per day.
RNA preparation
RCNMV RNA1 and RNA2 were transcribed from SmaI-linearized pUCR1 and pRC2_G, respectively, using T7 RNA polymerase (TaKaRa Bio, Inc). BMV RNAs were transcribed from EcoRI-linearized pB plasmids using T7 RNA polymerase and capped with a ScriptCapm7G capping system (Epicentre Biotechnology). Capped mRNAs were transcribed from NotI-linearized pBYL plasmids using T7 or SP6 RNA polymerase (TaKaRa Bio, Inc) and capped with a ScriptCapm7G capping system (Epicentre Biotechnology). All transcripts were purified with a Sephadex G-50 fine column (Amersham Pharmacia Biotech). RNA concentration was determined spectrophotometrically, and its integrity was verified by agarose gel electrophoresis.
Tandem affinity purification
Four-week-old N. benthamiana plants were agroinfiltrated as described previously [15]. At 2 days postinfiltration (dpi), total proteins were extracted from 6 g of leaves in 10 ml of buffer A (50 mM HEPES, 150 mM NaCl, 0.1% 2-mercaptoethanol, 0.5% Triton X-100, 5% glycerol, pH 7.5) containing 30 mM imidazole and a cocktail of protease inhibitors (Roche). Following the removal of cell debris by filtering the mixture through cheesecloth, the extract was centrifuged at 21,000g at 4°C for 10 min and the supernatant was mixed with Ni-NTA beads (400 μl) (Qiagen, Hilden, Germany) and incubated for 1 h at 4°C with gentle mixing. The beads were washed three times with 1 ml of buffer A containing 100 mM imidazole. The bound proteins were eluted with 1 ml of buffer A containing 500 mM imidazole. The eluted proteins were mixed with 50 μl of ANTI-FLAG M2-Agarose Affinity Gel (Sigma-Aldrich) and incubated for overnight at 4°C with gentle mixing. The beads were washed 3 times with 1 ml of buffer A. The bound proteins were eluted with 300 μl of buffer A containing 150 μg/ml 3 × FLAG peptides (Sigma-Aldrich). The eluted proteins were concentrated by acetone precipitation and dissolved in 1 × NuPAGE sample buffer (Invitrogen). The purified proteins were separated by sodium dodecyl sulfate (SDS)-PAGE (NuPAGE 3%–12% bis-Tris gel: Invitrogen) and visualized by silver staining (Wako, Osaka, Japan). Proteins in excised gel pieces were subjected to digestion with trypsin, LC–MS/MS analysis, and MASCOT searching as described previously [12].
Silencing of PLDα and PLDß in N. benthamiana
Appropriate combinations of silencing vectors were expressed via Agrobacterium infiltration in 3 - to 4-week-old N. benthamiana plants as described previously [16]. At 18 dpi, the leaves located above the infiltrated leaves were inoculated with in vitro transcribed RNA1 and RNA2 (500 ng each). At 2 days after inoculation, three inoculated leaves from three different plants infected with the same inoculum were pooled, and total RNA was extracted using RNA extraction reagent (Invitrogen), treated with RQ1 RNase-free DNase (Promega, Madison, WI), purified by phenol–chloroform and chloroform extractions, and precipitated with ethanol. Viral RNAs were detected by northern blotting, as described previously [16]. The mRNA levels of NbPLDα and NbPLDß were examined by RT-PCR using primer pairs 5′ -TATCAAGGTAGAGGAGATAGGTGC-3′ and 5′-TACATCATCTCCATCGTTCTCCTC-3′, and 5′-GAAGGCTTCAAAGCGCCATG-3′ and 5′-CTTAGGCAAGGGACATCAGC-3′, respectively. As a control to show the equal amounts of cDNA templates in each reaction mixture, the ribulose 1,5-biphosphate carboxylase small subunit gene (RbcS), a gene that is constitutively expressed, was amplified by RT-PCR as described previously [16].
Protoplasts experiments
N. benthamiana protoplasts were prepared according to Kaido et al. (2014) [59]. N. benthamiana protoplasts were inoculated with RCNMV RNA1 (1.5 μg) and RNA2 (0.5 μg) and incubated with various concentrations of n-butanol (Sigma-Aldrich), tert-butanol (Sigma-Aldrich), phosphatidic acid (PA) (Soy-derived; Avanti Polar Lipid), phosphatidyl choline (PC) (Soy-derived; Avanti Polar Lipid) or phosphatidyl ethanolamine (PE) (Soy-derived; Avanti Polar Lipid) at 20°C for 18 h. Phospholipids were dissolved in dimethylsulfoxide. Total RNA was extracted and subjected to northern blotting, as described previously [15]. Each experiment was repeated at least three times using different batches of protoplasts.
BYL experiments
The preparation of BYL was as described previously [38,46]. The BYL translation/replication assay was performed essentially as described previously [46]. Briefly, 300 ng of RNA1 was added to 30 μL of BYL translation/replication mixture in the presence of various concentrations of PA. The mixture was incubated at 17°C for 240 min. Aliquots of the reaction mixture were subjected to northern and immunoblotting analyses, as described previously [45,46,48].
Coimmunopurification assay
Four-week-old N. benthamiana plants were agroinfiltrated as described previously [15]. At 4 days postinfiltration (dpi), total proteins were extracted from 0.33 g of leaves in 1 ml of buffer A containing a cocktail of protease inhibitors (Roche). Following the removal of cell debris by centrifugation at 21,000g at 4°C for 10 min, the supernatant was mixed with GFP-Trap agarose beads (10 μl) (ChromoTek) and incubated for 1 h at 4°C with gentle mixing. The beads were washed 3 times with 1 ml of buffer A. The bound proteins were eluted by addition of 1 × SDS gel loading buffer, followed by incubation for 3 min at 95°C. Protein samples were subjected to SDS-PAGE, followed by immunoblotting with appropriate antibodies.
FLAG - or HA-tagged proteins were expressed in BYL by adding an in vitro transcript. After incubation at 25°C for 120 min, a 10-μl bed volume of anti-HA Affinity Matrix (Roche) or ANTI-FLAG M2-Agarose Affinity Gel (Sigma-Aldrich) was added to the BYL and further incubated for 60 min with gentle mixing. The resin was washed three times with 200 μl of TR buffer [38] supplemented with 150 mM NaCl and 0.5% Triton X-100. The bound proteins were eluted by addition of 1 × SDS gel loading buffer, followed by incubation for 3 min at 95°C. Protein samples were subjected to SDS-PAGE, followed by immunoblotting with appropriate antibodies.
Subcellular localization assays
Appropriate combinations of fluorescent protein-fused proteins were expressed in N. benthamiana leaves by Agrobacterium infiltration. Fluorescence of GFP and mCherry was visualized with confocal microscopy at 4 dai as described previously [59].
Protein purification and lipid overlay assay
Protein expression in E. coli BL21 (DE3) and subsequent purification were done as described previously [15]. The concentration of purified protein was measured using a Coomassie Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). The purified protein was subjected to SDS-PAGE and visualized with Coomassie brilliant blue R-250 to check its purity.
Lipid overlay assays were conducted as recommended by the manufacture’s protocol. Briefly, the membrane (PIP Strips or Membrane Lipid Arrays; Echelon Bioscience Inc) was incubated in 3% fatty acid free BSA (Sigma-Aldrich) in a mixture of phosphate-buffered saline and 0.1% Tween 20 (PBST) for 1 h at room temperature (RT) and then incubated in the same solution containing 500 ng of purified recombinant protein for 1 h at RT. After washing three times with PBST, the membrane was incubated with a mouse anti-FLAG antibody (1 : 10000; Sigma-Aldrich) for 1 h at RT, followed by three washes with PBST. An anti-mouse IgG conjugated with horseradish peroxidase (1 : 10000; KPL) was used as a secondary antibody. Binding of proteins to phospholipids was visualized by incubation with a chemiluminescent substrate.
Lipid extraction and TLC
Four-week-old N. benthamiana plants were inoculated with RCNMV via agroinfiltration. At 2 dai, 0.33 g of infiltrated leaves were ground in liquid nitrogen and extracted in 900 μl of water. Total lipids were extracted by adding 3 ml CHCl3/CH3OH (2 : 1, v/v) to each sample. The samples were vortexed and centrifuged at 1690g, at 4°C for 10 min. The organic phase was recovered and dried under nitrogen gas stream. Lipids were dissolved in 100 μl CHCl3/CH3OH (2 : 1, v/v). Subsequently, 5 μl of the samples were analyzed on TLC plates (Merck, Germany). The chromatography was performed using CHCl3/CH3OH/formic acid/acetic acid (12 : 6:0.6 : 0.4, v/v). Plates were air-dried, soaked in 10% CuSO4, and charred at 180°C for 10 min to visualize lipids.
To identify lipid species, the air-dried TLC plates were stained for an hour in a 0.03% Coomassie Brilliant Blue R-250 solution containing 20% of methanol and 0.5% of acetic acid. Destaining of the plates was performed with 20% methanol containing 0.5% acetic acid for 5 min. After drying the plates for a few minutes, the blue bands of interest were scratched out and transferred to glass tubes. The scratched silica gels were mixed with chloroform/methanol (3/7, v/v), followed by the centrifugation at 1,690g, at 4°C for 10 min. The supernatants were subjected to the LC-MS analysis using LCMS-IT-TOF mass spectrometer (Shimadzu, Kyoto, Japan). A TSK gel ODS-100Z column (2.0 × 150 mm, 5 μm, Tosoh, Tokyo, Japan) was eluted isocratically with acetonitrile/methanol/2-propanol/water (6/131/110/3, by volume) containing 19.6 mM of ammonium formate and 0.2% of formic acid at a flow rate of 0.2 mL/min. The MS was performed using an electrospray ionization interface operated in negative ion mode, under the following conditions: CDL temperature, 200°C; block heater temperature, 200°C; nebulizing gas (N2) flow, 1.5 L/min. The MS data were acquired in the range of m/z 600 to 1,000 using 10 msec ion accumulation time. The MS2 data were acquired in the range of m/z 125 to 500, using 50 msec ion accumulation time and automatic precursor ion selection in the range of m/z 650 to 750. CID parameters were follows: energy, 50%; collision gas (argon) 50%.
Quantitative RT-PCR analysis
Total RNA extracted from N. benthamina leaves or protoplasts were subjected to reverse transcription using PrimeScript RT reagent Kit (Takara) using oligo-dT and random primers according to manufacturer’s protocol. Real-time PCR was carried out using SYBR Premix Ex Taq (RR420A, Takara). Primers used for real-time PCR analysis were listed in S2 Table. Quantitative analysis of each mRNA was performed using a Thermal cycler Dice Real Time System TP800 (Takara).
Accession numbers
NbPLDα and NbPLDβ were registered through DDBJ and accession numbers LC033851 and LC033852, respectively, were given on March 11 2015.
Supporting Information
Zdroje
1. den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010; 64 : 241–256. doi: 10.1146/annurev.micro.112408.134012 20825348
2. Hyodo K, Kaido M, Okuno T. Host and viral RNA-binding proteins involved in membrane targeting, replication and intercellular movement of plant RNA virus genomes. Front Plant Sci. 2014; 5: Article 321
3. Laliberté JF, Zhang H. Viral manipulation of plant host membranes. Annu Rev Virol. 2014; 1 : 237–259.
4. Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008; 6 : 363–374. doi: 10.1038/nrmicro1890 18414501
5. Mine A, Okuno T. Composition of plant virus RNA replicase complexes. Curr Opin Virol. 2012; 2 : 669–675. doi: 10.1016/j.coviro.2012.09.014 23083891
6. Nagy PD, Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol. 2012; 10 : 137–149. doi: 10.1038/nrmicro2692 22183253
7. Hyodo K, Okuno T. Host factors used by positive-strand RNA plant viruses for genome replication. J. Gen. Plant Pathol. 2014; 80 : 123–135.
8. Mandadi KK, Scholthof KB. Plant immune responses against viruses: how does a virus cause disease? Plant Cell. 2013; 25 : 1489–1505 doi: 10.1105/tpc.113.111658 23709626
9. Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013; 11 : 745–760. doi: 10.1038/nrmicro3120 24129510
10. Okuno T, Hiruki C. Molecular biology and epidemiology of Dianthoviruses. Adv Virus Res. 2013; 87 : 36–71.
11. Kaido M, Tsuno Y, Mise K, Okuno T. Endoplasmic reticulum targeting of the Red clover necrotic mosaic virus movement protein is associated with the replication of viral RNA1 but not that of RNA2. Virology. 2009; 395 : 232–242. doi: 10.1016/j.virol.2009.09.022 19819513
12. Mine A, Takeda A, Taniguchi T, Taniguchi H, Kaido M, Mise K, et al. Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of Red clover necrotic mosaic virus. J Virol. 2010; 84 : 6070–6081. doi: 10.1128/JVI.00054-10 20375154
13. Kusumanegara K, Mine A, Hyodo K, Kaido M, Mise K, Okuno T. Identification of domains in p27 auxiliary replicase protein essential for its association with the endoplasmic reticulum membranes in Red clover necrotic mosaic virus. Virology. 2012; 433 : 131–141. doi: 10.1016/j.virol.2012.07.017 22898643
14. Turner KA, Sit TL, Callaway AS, Allen NS, Lommel SA. Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology. 2004; 320 : 276–290. 15016550
15. Hyodo K, Mine A, Taniguchi T, Kaido M, Mise K, Taniguchi H, et al. ADP ribosylation factor 1 plays an essential role in the replication of a plant RNA virus. J Virol. 2013; 87 : 163–176. doi: 10.1128/JVI.02383-12 23097452
16. Mine A, Hyodo K, Tajima Y, Kusumanegara K, Taniguchi T, Kaido M, et al. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J Virol. 2012; 86 : 12091–12104. doi: 10.1128/JVI.01659-12 22933272
17. Hyodo K, Kaido M, Okuno T. Traffic jam on the cellular secretory pathway generated by a replication protein from a plant RNA virus. Plant Signal Behav. 2014; 9: e28644. 24714629
18. Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011; 12 : 362–375. doi: 10.1038/nrm3117 21587297
19. Jang JH, Lee CS, Hwang D, Ryu SH. Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog Lipid Res. 2012; 51 : 71–81 doi: 10.1016/j.plipres.2011.12.003 22212660
20. Li M, Hong Y, Wang X. Phospholipase D - and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta. 2009; 1791 : 927–935. doi: 10.1016/j.bbalip.2009.02.017 19289179
21. Testerink C, Munnik T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot. 2011; 62 : 2349–2361. doi: 10.1093/jxb/err079 21430291
22. Hong Y, Zhang W, Wang X. Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ. 2010; 33 : 627–635. doi: 10.1111/j.1365-3040.2009.02087.x 19968827
23. Bargmann BO, Laxalt AM, Riet BT, Schouten E, van Leeuwen W, Dekker HL, et al. LePLDbeta1 activation and relocalization in suspension-cultured tomato cells treated with xylanase. Plant J. 2006; 4 : 358–368.
24. Kirik A, Mudgett MB. SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc Natl Acad Sci U S A. 2009; 106 : 20532–20537. doi: 10.1073/pnas.0903859106 19918071
25. van der Luit AH, Piatti T, van Doorn A, Musgrave A, Felix G, Boller T, et al. Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 2000; 123 : 1507–1516. 10938366
26. Yamaguchi T, Minami E, Ueki J, Shibuya N. Elicitor-induced activation of phospholipases plays an important role for the induction of defense responses in suspension-cultured rice cells. Plant Cell Physiol. 2005; 46 : 579–587. 15695430
27. Janda M, Šašek V, Chmelařová H, Andrejch J, Nováková M, Hajšlová J, et al. Phospholipase D affects translocation of NPR1 to the nucleus in Arabidopsis thaliana. Front Plant Sci. 2015; 6: Article 59.
28. Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerström M. Phospholipase-dependent signalling during the AvrRpm1 - and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J. 2006; 47 : 947–959. 16925603
29. de Jong CF, Laxalt AM, Bargmann BO, de Wit PJ, Joosten MH, Munnik T. Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J. 2004; 39 : 1–12. 15200638
30. Johansson ON, Fahlberg P, Karimi E, Nilsson AK, Ellerström M, Andersson MX. Redundancy among phospholipase D isoforms in resistance triggered by recognition of the Pseudomonas syringae effector AvrRpm1 in Arabidopsis thaliana. Front Plant Sci. 2014; 5: Article 639.
31. Pinosa F, Buhot N, Kwaaitaal M, Fahlberg P, Thordal-Christensen H, Ellerström M, et al. Arabidopsis phospholipase dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol. 2013; 163 : 896–906. doi: 10.1104/pp.113.223503 23979971
32. Zhang W, Chen J, Zhang H, Song F. Overexpression of a rice diacylglycerol kinase gene OsBIDK1 enhances disease resistance in transgenic tobacco. Mol Cells. 2008; 26 : 258–264. 18679055
33. Park J, Gu Y, Lee Y, Yang Z, Lee Y. Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 2004; 134 : 129–136. 14730067
34. Zhao J, Devaiah SP, Wang C, Li M, Welti R, Wang X. Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens. New Phytol. 2013; 199 : 228–240. doi: 10.1111/nph.12256 23577648
35. Yamaguchi T, Kuroda M, Yamakawa H, Ashizawa T, Hirayae K, Kurimoto L, et al. Suppression of a Phospholipase D gene, OsPLDβ1, activates defense responses and increases disease resistance in rice. Plant Physiol. 2009; 150 : 308–319. doi: 10.1104/pp.108.131979 19286937
36. Canonne J, Froidure-Nicolas S, Rivas S. Phospholipases in action during plant defense signaling. Plant Signal Behav. 2011; 6 : 13–18. 21248491
37. Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993; 75 : 1137–1144. 8261513
38. Komoda K, Naito S, Ishikawa M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc Natl Acad Sci U S A. 2004; 101 : 1863–1867. 14769932
39. Wang X, Yan Y, Li Y, Chu X, Wu C, Guo X. GhWRKY40, a Multiple Stress-Responsive Cotton WRKY Gene, Plays an Important Role in the Wounding Response and Enhances Susceptibility to Ralstonia solanacearum Infection in Transgenic Nicotiana benthamiana. PLoS One. 2014; 9: e93577. doi: 10.1371/journal.pone.0093577 24747610
40. Zhu F, Xi DH, Yuan S, Xu F, Zhang DW, Lin HH. Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol Plant Microbe Interact. 2014; 27 : 567–577. doi: 10.1094/MPMI-11-13-0349-R 24450774
41. Nakano M, Nishihara M, Yoshioka H, Takahashi H, Sawasaki T, Ohnishi K, et al. Suppression of DS1 phosphatidic acid phosphatase confirms resistance to Ralstonia solanacearum in Nicotiana benthamiana. PLoS One. 2013; 8: e75124. doi: 10.1371/journal.pone.0075124 24073238
42. Huang PY, Yeh YH, Liu AC, Cheng CP, Zimmerli L. The Arabidopsis LecRK-VI.2 associates with the pattern recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 2014; 79 : 243–255 doi: 10.1111/tpj.12557 24844677
43. Munnik T, Arisz SA, De Vrije T, Musgrave A. G Protein Activation Stimulates Phospholipase D Signaling in Plants. Plant Cell. 1995; 7 : 2197–2210. 12242371
44. An M, Iwakawa HO, Mine A, Kaido M, Mise K, Okuno T. A Y-shaped RNA structure in the 3' untranslated region together with the trans-activator and core promoter of Red clover necrotic mosaic virus RNA2 is required for its negative-strand RNA synthesis. Virology. 2010; 405 : 100–109. doi: 10.1016/j.virol.2010.05.022 20561661
45. Hyodo K, Mine A, Iwakawa HO, Kaido M, Mise K, Okuno T. Identification of amino acids in auxiliary replicase protein p27 critical for its RNA-binding activity and the assembly of the replicase complex in Red clover necrotic mosaic virus. Virology. 2011; 413 : 300–309. doi: 10.1016/j.virol.2011.02.017 21440279
46. Iwakawa HO, Kaido M, Mise K, Okuno T. cis-Acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3'-untranslated region of Red clover necrotic mosaic virus RNA1. Virology. 2007; 369 : 168–181. 17727911
47. Iwakawa HO, Mizumoto H, Nagano H, Imoto Y, Takigawa K, Sarawaneeyaruk S, et al. A viral noncoding RNA generated by cis-element-mediated protection against 5' - 3’ RNA decay represses both cap-independent and cap-dependent translation. J Virol. 2008; 82 : 10162–10174. doi: 10.1128/JVI.01027-08 18701589
48. Iwakawa HO, Mine A, Hyodo K, An M, Kaido M, Mise K, et al. Template recognition mechanisms by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J Virol. 2011; 85 : 497–509. doi: 10.1128/JVI.01754-10 20980498
49. Mine A, Hyodo K, Takeda A, Kaido M, Mise K, Okuno T. Interactions between p27 and p88 replicase proteins of Red clover necrotic mosaic virus play an essential role in viral RNA replication and suppression of RNA silencing via the 480-kDa viral replicase complex assembly. Virology. 2010; 407 : 213–224. doi: 10.1016/j.virol.2010.07.038 20828775
50. Zhao J. Phospholipase D and phosphatidic acid in plant defence response: from protein–protein and lipid–protein interactions to hormone signalling. J Exp Bot. 2015; 66 : 1721–1736. 25680793
51. Chuang C, Barajas D, Qin J, Nagy PD. Inactivation of the host lipin gene accelerates RNA virus replication through viral exploitation of the expanded endoplasmic reticulum membrane. PLoS Pathog. 2014; 10: e1003944. doi: 10.1371/journal.ppat.1003944 24586157
52. Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, et al. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012; 8: e1002584. doi: 10.1371/journal.ppat.1002584 22457619
53. Duijsings D, Wessels E, van Emst-de Vries SE Melchers WJ, Willems PH, van Kuppeveld FJ. Reduction of phospholipase D activity during coxsackievirus infection. J Gen Virol. 2007; 88 : 3027–3030. 17947526
54. Verheije MH, Raaben M, Mari M, Te Lintelo EG, Reggiori F, van Kuppeveld FJ, et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008; 4: e1000088. doi: 10.1371/journal.ppat.1000088 18551169
55. Guo L, Wang X. Crosstalk between Phospholipase D and Sphingosine Kinase in Plant Stress Signaling. Front Plant Sci. 2012; 3: Article 51.
56. Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, et al. Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell. 2009; 21 : 2357–2377. doi: 10.1105/tpc.108.062992 19690149
57. Kim SC, Guo L, Wang X. Phosphatidic acid binds to cytosolic glyceraldehyde-3-phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J Biol Chem. 2013; 288 : 11834–11844. doi: 10.1074/jbc.M112.427229 23504314
58. McLoughlin F, Arisz SA, Dekker HL, Kramer G, de Koster CG, Haring MA, et al. Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem J. 2013; 450 : 573–581. doi: 10.1042/BJ20121639 23323832
59. Kaido M, Abe K, Mine A, Hyodo K, Taniguchi T, Taniguchi H, et al. GAPDH-A recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement. PLoS Pathog. 2014; 10: e1004505. doi: 10.1371/journal.ppat.1004505 25411849
60. Mizumoto H, Iwakawa H, Kaido M, Mise K, Okuno T. Cap-independent translation mechanism of Red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J Virol. 2006; 80 : 3781–3791 16571795
61. Iwakawa HO, Tajima Y, Taniguchi T, Kaido M, Mise K, Tomari Y, et al. Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3' untranslated region. J Virol. 2012; 86 : 7836–7849. doi: 10.1128/JVI.00538-12 22593149
62. Konan KV, Sanchez-Felipe L. Lipids and RNA virus replication. Curr Opin Virol. 2014; 9 : 45–52. doi: 10.1016/j.coviro.2014.09.005 25262061
63. Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010; 6: e1000719. doi: 10.1371/journal.ppat.1000719 20062526
64. Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011; 9 : 32–45. doi: 10.1016/j.chom.2010.12.002 21238945
65. Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, et al. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 2012; 8: e1002860. doi: 10.1371/journal.ppat.1002860 22916015
66. Weng L, Hirata Y, Arai M, Kohara M, Wakita T, Watashi K, et al. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J Virol. 2010; 84 : 11761–11770. doi: 10.1128/JVI.00638-10 20844041
67. Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010; 141 : 799–811. doi: 10.1016/j.cell.2010.03.050 20510927
68. Nchoutmboube JA, Viktorova EG, Scott AJ, Ford LA, Pei Z, Watkins PA, et al. Increased long chain acyl-Coa synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog. 2013; 9: e1003401. doi: 10.1371/journal.ppat.1003401 23762027
69. Belov GA. Modulation of lipid synthesis and trafficking pathways by picornaviruses. Curr Opin Virol. 2014; 9 : 19–23. doi: 10.1016/j.coviro.2014.08.007 25240228
70. Pogany J, Nagy PD. Activation of Tomato bushy stunt virus RdRp by cellular Hsp70 is enhanced by phospholipids in vitro. J Virol. 2015; 89 : 5714–5723. doi: 10.1128/JVI.03711-14 25762742
71. Takeda A, Tsukuda M, Mizumoto H, Okamoto K, Kaido M, Mise K, et al. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005; 24 : 3147–3157. 16096641
72. Xiong ZG, Lommel SA. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology. 1991; 182 : 388–392. 2024474
73. Janda M, French R, Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5' extensions on transcript infectivity. Virology. 1987; 158 : 259–262. 18644564
74. Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse P. De Novo Transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS ONE. 2013; 8: e59534. doi: 10.1371/journal.pone.0059534 23555698
75. Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T. Capindependent translational enhancement by the 3' untranslated region of red clover necrotic mosaic virus RNA1. J Virol. 2003; 77 : 12113–12121. 14581548
76. Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, et al. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 2002; 532 : 75–79. 12459466
77. Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001; 25 : 237–245. 11169199
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary TuberculosisČlánek Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic PlagueČlánek Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host SpeciesČlánek Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR SelectionČlánek Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus GenomesČlánek Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from FerrioxamineČlánek Remembering MumpsČlánek Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling PathwayČlánek Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Parasites and Their Heterophagic Appetite for Disease
- The Elusive Role of the Prion Protein and the Mechanism of Toxicity in Prion Disease
- Intestinal Colonization Dynamics of
- Activation of Typhi-Specific Regulatory T Cells in Typhoid Disease in a Wild-Type . Typhi Challenge Model
- The Engineering of a Novel Ligand in gH Confers to HSV an Expanded Tropism Independent of gD Activation by Its Receptors
- Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary Tuberculosis
- Group Selection and Contribution of Minority Variants during Virus Adaptation Determines Virus Fitness and Phenotype
- Phosphatidic Acid Produced by Phospholipase D Promotes RNA Replication of a Plant RNA Virus
- A Ribonucleoprotein Complex Protects the Interleukin-6 mRNA from Degradation by Distinct Herpesviral Endonucleases
- Characterization of Transcriptional Responses to Different Aphid Species Reveals Genes that Contribute to Host Susceptibility and Non-host Resistance
- Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic Plague
- Natural Killer Cell Sensing of Infected Cells Compensates for MyD88 Deficiency but Not IFN-I Activity in Resistance to Mouse Cytomegalovirus
- Manipulation of the Xanthophyll Cycle Increases Plant Susceptibility to
- Ly6C Monocytes Regulate Parasite-Induced Liver Inflammation by Inducing the Differentiation of Pathogenic Ly6C Monocytes into Macrophages
- Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host Species
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness
- Inhibition and Reversal of Microbial Attachment by an Antibody with Parasteric Activity against the FimH Adhesin of Uropathogenic .
- The EBNA-2 N-Terminal Transactivation Domain Folds into a Dimeric Structure Required for Target Gene Activation
- Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection
- The NLRP3 Inflammasome Is a Pathogen Sensor for Invasive via Activation of α5β1 Integrin at the Macrophage-Amebae Intercellular Junction
- Sequential Conformational Changes in the Morbillivirus Attachment Protein Initiate the Membrane Fusion Process
- A Two-Component DNA-Prime/Protein-Boost Vaccination Strategy for Eliciting Long-Term, Protective T Cell Immunity against
- cAMP-Signalling Regulates Gametocyte-Infected Erythrocyte Deformability Required for Malaria Parasite Transmission
- Response Regulator VxrB Controls Colonization and Regulates the Type VI Secretion System
- Evidence for a Novel Mechanism of Influenza Virus-Induced Type I Interferon Expression by a Defective RNA-Encoded Protein
- Dust Devil: The Life and Times of the Fungus That Causes Valley Fever
- TNF-α Induced by Hepatitis C Virus via TLR7 and TLR8 in Hepatocytes Supports Interferon Signaling via an Autocrine Mechanism
- The Recent Evolution of a Maternally-Inherited Endosymbiont of Ticks Led to the Emergence of the Q Fever Pathogen,
- L-Rhamnosylation of Wall Teichoic Acids Promotes Resistance to Antimicrobial Peptides by Delaying Interaction with the Membrane
- Rapid Sequestration of by Neutrophils Contributes to the Development of Chronic Lesion
- Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus Genomes
- The Expression of Functional Vpx during Pathogenic SIVmac Infections of Rhesus Macaques Suppresses SAMHD1 in CD4 Memory T Cells
- Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from Ferrioxamine
- TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation
- Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site
- Remembering Mumps
- The Role of Horizontal Gene Transfer in the Evolution of the Oomycetes
- Advances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi
- Investigating Fungal Outbreaks in the 21st Century
- Systems Biology for Biologists
- How Does the Dinoflagellate Parasite Outsmart the Immune System of Its Crustacean Hosts?
- FCRL5 Delineates Functionally Impaired Memory B Cells Associated with Exposure
- Phospholipase D1 Couples CD4 T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication
- Influenza A Virus on Oceanic Islands: Host and Viral Diversity in Seabirds in the Western Indian Ocean
- Geometric Constraints Dominate the Antigenic Evolution of Influenza H3N2 Hemagglutinin
- Widespread Recombination, Reassortment, and Transmission of Unbalanced Compound Viral Genotypes in Natural Arenavirus Infections
- Gammaherpesvirus Co-infection with Malaria Suppresses Anti-parasitic Humoral Immunity
- A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
- Mechanisms of Stage-Transcending Protection Following Immunization of Mice with Late Liver Stage-Arresting Genetically Attenuated Malaria Parasites
- The Myelin and Lymphocyte Protein MAL Is Required for Binding and Activity of ε-Toxin
- Genome-Wide Identification of the Target Genes of AP2-O, a AP2-Family Transcription Factor
- An Atypical Mitochondrial Carrier That Mediates Drug Action in
- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Helminth Infection and Commensal Microbiota Drive Early IL-10 Production in the Skin by CD4 T Cells That Are Functionally Suppressive
- Circulating Pneumolysin Is a Potent Inducer of Cardiac Injury during Pneumococcal Infection
- ExoT Induces Atypical Anoikis Apoptosis in Target Host Cells by Transforming Crk Adaptor Protein into a Cytotoxin
- Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement
- Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
- Varicella Viruses Inhibit Interferon-Stimulated JAK-STAT Signaling through Multiple Mechanisms
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



