-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Engineering of a Novel Ligand in gH Confers to HSV an Expanded Tropism Independent of gD Activation by Its Receptors
To enter cells, all herpesviruses use the core fusion glycoproteins gH/gL and gB, in addition to species-specific glycoproteins responsible for specific tropism, etc. In HSV, the additional glycoprotein is the essential gD. We engineered in gH a heterologous ligand to the HER2 cancer receptor. The recombinant viruses entered cells through HER2, independently of gD activation by its receptors, or despite deletion of key residues that are part of the receptors’ binding sites in gD. The ligand activated gH in cis. Cumulatively, the receptor-binding and activating functions of gD were no longer essential and were replaced by the heterologous ligand in gH. Relevance to translational medicine rests in the fact that gH can serve as a tool to retarget HSV tropism to cancer-specific receptors. This expands the toolkit for the design of fully-virulent oncolytic-HSVs.
Published in the journal: The Engineering of a Novel Ligand in gH Confers to HSV an Expanded Tropism Independent of gD Activation by Its Receptors. PLoS Pathog 11(5): e32767. doi:10.1371/journal.ppat.1004907
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004907Summary
To enter cells, all herpesviruses use the core fusion glycoproteins gH/gL and gB, in addition to species-specific glycoproteins responsible for specific tropism, etc. In HSV, the additional glycoprotein is the essential gD. We engineered in gH a heterologous ligand to the HER2 cancer receptor. The recombinant viruses entered cells through HER2, independently of gD activation by its receptors, or despite deletion of key residues that are part of the receptors’ binding sites in gD. The ligand activated gH in cis. Cumulatively, the receptor-binding and activating functions of gD were no longer essential and were replaced by the heterologous ligand in gH. Relevance to translational medicine rests in the fact that gH can serve as a tool to retarget HSV tropism to cancer-specific receptors. This expands the toolkit for the design of fully-virulent oncolytic-HSVs.
Introduction
Entry of herpes simplex virus (HSV) into the cell is a multistep process that involves four virion glycoproteins (gD, gH/gL, gB), all of which are required. gD is species-specific, and a major determinant of HSV tropism. gH/gL and gB constitute the conserved fusion apparatus across the Herpesviridae family; gB exhibits features typical of viral fusion glycoproteins [1–6]. Many steps in the HSV entry process remain to be elucidated and the overall model is partly speculative. Inasmuch as the process initiates with gD binding to one of its receptors, and culminates with gB-mediated virion-cell fusion, the commonly accepted model envisions that the four viral glycoproteins are activated in a cascade fashion by the receptor-bound gD through intermolecular signaling among the glycoproteins themselves [1]. Specifically, following virion attachment to cells, the interaction of gD with one of its alternative receptors—nectin1, HVEM, and modified heparan sulphates [7–10]—results in conformational modifications to gD, in particular in the dislodgement of the ectodomain C-terminus, which carries the profusion domain [11–15]. Since this domain can interact with the heterodimer gH/gL [16,17], most likely this step is a critical event in the activation cascade. Recently, we have shown that gH/gL interacts with two interchangeable receptors, αvβ6 - and αvβ8-integrins, which promote HSV endocytosis, and most likely participate in the process of gH/gL activation [18]. Evidence for the activation cascade and for intermolecular signaling among the glycoproteins is indirect and rests on three sets of data: interactions among the four glycoproteins [17,19,20]; the ability of soluble gD to rescue the infection of gD-/- non-infectious virions and to promote fusion in a cell-cell fusion assay; the ability of soluble gD receptor to mediate virus entry into receptor-negative cells [15,21–23].
There is intense interest in HSV as an oncolytic agent (o-HSV) [24–27]. In the first and second generations o-HSVs, now in clinical trials, safety was obtained at the expense of virulence through single or multiple deletions. The most successful example is T-VEC, a HSV recombinant deleted in both copies of the γ134.5 gene and of ICP47 gene, and encoding the GM-CSF cytokine to boost the host immune response against the tumor [28]. In a phase III clinical trial, T-VEC improved the outcome of patients carrying metastatic melanoma [29]. A drawback of attenuation is that it strongly reduces the range of tumors against which the o-HSVs are effective. Thus, deletion of the γ134.5 genes restricts o-HSVs replication to cells defective in the PKR-dependent innate response. To overcome these limitations, non-attenuated o-HSVs retargeted to cancer-specific receptors and detargeted from the natural receptors were designed. They preserve the killing ability of wt-viruses [30,31]. So far, retargeting strategies entailed genetic modifications to gD, in particular the insertion of novel ligands, coupled with appropriate deletions for detargeting purposes [30,32–38]. The heterologous ligands included the IL13 cytokine, urokinase-type plasminogen activator or single chain antibodies (scFvs). The retargeting through genetic modifications obtained in the above-mentioned studies has clear advantages over retargeting through coupling of appropriate moieties to virions, and even more so over non-replicating viruses (see, for example [39]). In the former case virions maintain the retargeted phenotype generation after generation, even during replication in the tumor. In the latter case, targeting occurs only for a single generation, and viruses are usually non-detargeted, hence they also infect non cancer cells. Furthermore, non-replicating virions fail to propagate the therapeutic effect beyond the initially infected tumor cells.
The cancer-specific receptor selected in our laboratory is HER2 (human epidermal growth factor receptor 2), a member of the EGFR (epidermal growth factor receptor) family, overexpressed in breast, ovary, gastric carcinomas, glioblastomas, etc [40]. Two fully retargeted o-HSVs were generated. They differ in the portions of gD that were deleted for detargeting purposes. R-LM113 carries the deletion of the AA 6–38 N-terminal region [32,33]. R-LM249 carries the deletion of the 61–218 core region [34]. In both viruses, the deleted sequences were replaced with the scFv to HER2 derived from trastuzumab, a humanized MAb now in clinical practice. The scFv binds HER2 at high affinity (29.3 nM) [41]. In preclinical studies R-LM113 and R-LM249 exerted therapeutic effects against human breast and ovary cancers, and against a murine model of HER2+ glioblastoma [32–35,42]. Intraperitoneally-administered R-LM249 exerted therapeutic effect against metastases of ovary and breast cancers diffuse to the peritoneum, or to the brain [35].
Here, we engineered a heterologous ligand in gH. The aims were twofold, i.e. to better elucidate the respective roles of gD and gH/gL in HSV entry, and define whether gD is an absolute requirement for HSV entry, and to explore novel avenues in the design of retargeted o-HSVs. We report that the engineering in gH of a scFv to HER2 confers to the recombinant viruses the ability to use HER2 as the sole receptor, in the absence of gD receptors, or upon deletion of residues that form the nectin1/HVEM binding sites in gD.
Results
Design of R-VG803 and R-VG809, and verification of chimeric gH
R-VG803 carries the insertion of the scFv to HER2 (herein named scFv-HER2) at the N-terminus of gH, the mCherry red fluorescent marker in the UL37–UL38 intergenic region, and the LoxP-bracketed BAC sequences between UL3 and UL4 (schematic representation in Fig 1A). R-VG809 carries the deletion of the AA 6–38 portion in gD, and is otherwise identical to R-VG803. The recombinant viruses were generated by transfection of the recombinant BAC-genomes into SK-OV-3 cells, a HER2+ cell line derived from human ovary carcinoma, and resistant to trastuzumab [35]. The presence of the scFv—gH chimera in R-VG803 and R-VG809 was verified by sequencing of the entire ORF, and by immunoblot of Vero cells infected with R-VG803, R-VG809, or R-LM5 [43]. The latter is essentially a wt-virus with genetic modifications similar to those of R-VG803 and R-GV809, i.e. it carries wt-gD, the LoxP-bracketed BAC sequences, and EGFP (Enhanced green fluorescence protein) instead of mCherry. The annotated scFv-gH sequence is reported in S1 Fig. For immunoblotting, infected cell lysates were subjected to SDS-PAGE (sodium dodecyl sulphate polyacrylamide gel electrophoresis), and the blots were immunoreacted to polyclonal antibody (PAb) to gH [44]. The chimeric scFv—gH migrated with a slower electrophoretic mobility than wt-gH from R-LM5, and an apparent Mr of 130 K (Fig 1B).
Fig. 1. 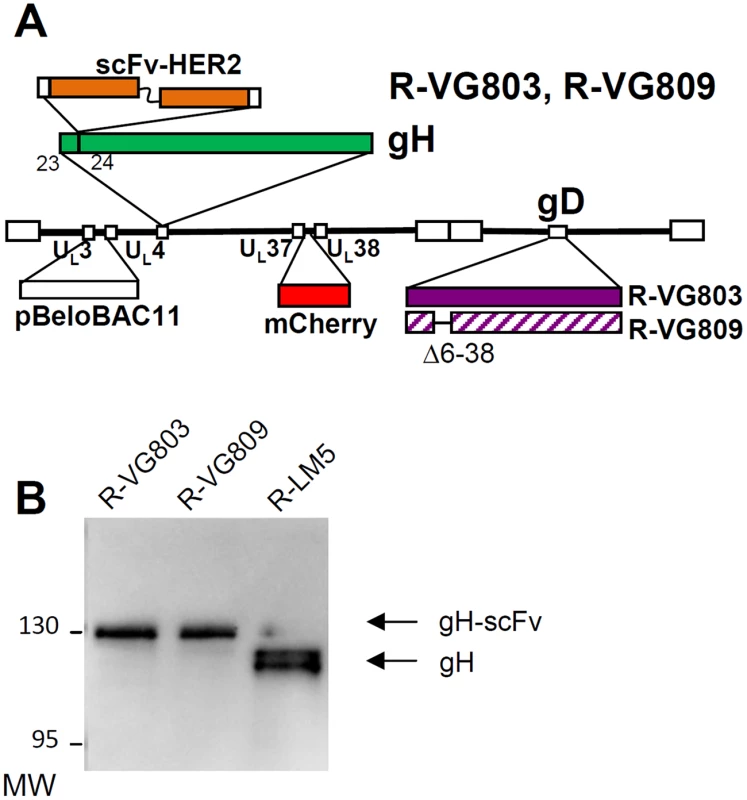
(A) Schematic drawing of R-VG803 and R-VG809 genomes. Sequence arrangement of HSV-1 genome shows the inverted repeat sequences as rectangular boxes. The scFv-HER2 sequence (VL-linker-VH) is inserted between AA 23 and 24 of gH, bracketed by upstream and downstream Gly-Ser linkers, 8 and 12 AA long, respectively. LOX-P-bracketed p-Belo-BAC and mCherry sequences are inserted between UL3-UL4, and between UL37-UL38 regions, respectively. The gD sequence is wt in R-VG803, and carries the deletion of the AA 6–38 region in R-VG809. (B) R-VG803 and R-VG809 express the chimeric scFv—gH glycoprotein. Lysates of Vero cells infected with R-VG803, R-VG809 or R-LM5 were subjected to PAGE. gH was detected by immunoblot [<em class="ref">44</em>]. Figures on the left represent the migration position of the 130K and 95K MW markers. R-VG803 infects cells that express HER2 as the sole receptor, in the absence of a gD receptor
Initially, we engineered R-VG803. To test whether it can use HER2 as an entry receptor, we made use of J-HER2 cells. The parental J cells express no receptor for gD, hence cannot activate gD, and are not infected by wt-HSV [7]. J-HER2 cells transgenically express HER2 as the sole receptor [43]. As controls, we included J-nectin and J-HVEM cells, which transgenically express nectin1 or HVEM as receptors and are infected by wt-HSV [7], and a panel of human and animal cells, which express the human or animal nectin1/HVEM. The panel included CHO, BHK, keratinocytic HaCaT, human fibroblastic HFF14, epithelial HeLa, the neuronal SK-N-SH cells, and the HER2-positive SK-OV-3 cancer cells. As shown in Fig 2A, R-VG803 infected J-HER2 cells. The infection of J-nectin1, J-HVEM, and of the animal and human cells (Fig 2A) was not surprising, given that R-VG803 encodes a wt-gD. Furthermore, R-VG803 could perform cell-to-cell spread in J-HER2 cells. Cells were infected at 0.01 PFU/cell, overlaid with medium containing MAb 52S (ascites fluid 1 : 10,000). At day 1 infection involved single cells. In the following day infection involved clusters of cells (Fig 2B).
Fig. 2. R-VG803 infects cells that express HER2 as the sole receptor (J-HER2 cells) in a HER2-dependent manner, and progeny virus spreads in these cells. 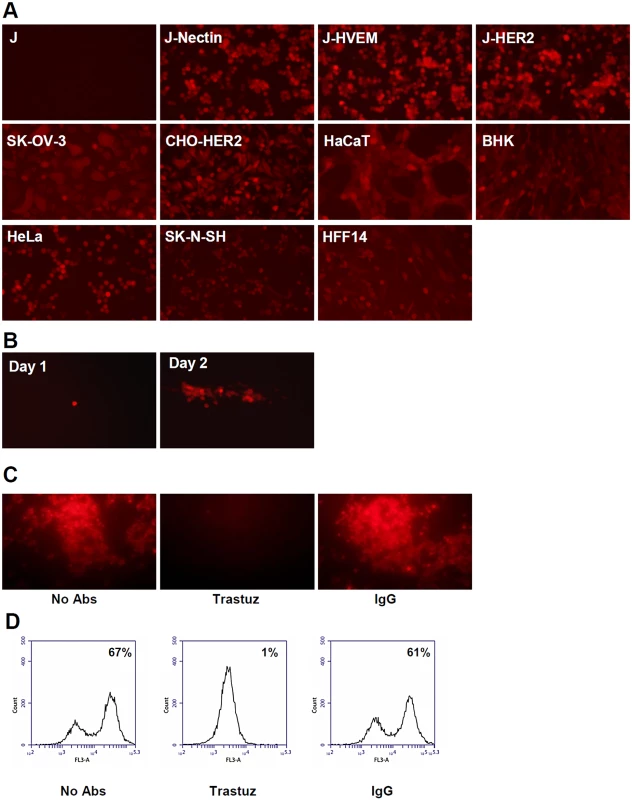
J cells express no receptor for wt-HSV. J-HER2, J-Nectin, J-HVEM only express the indicated receptor. (A) The indicated cells were infected with R-VG803 (2 PFU/cell as titrated in SK-OV-3), and monitored for red fluorescence microscopy. (B) J-HER2 cells were infected with R-VG803 (0.01 PFU/cell), overlaid with medium containing the neutralizing MAb 52S (ascites fluid 1:10,000) [45], and monitored daily for red fluorescence. Pictures of a same plaque are shown. (C, D) Trastuzumab inhibits R-VG803 infection of J-HER2 cells. J-HER2 cells were infected with R-VG803 in the presence of trastuzumab (trastuz) (28 μg/ml, final concentration) or control IgGs (28 μg/ml, final concentration). Infection was monitored by fluorescence microscopy (C), or flow cytometry (D). All pictures in panel A were taken with an exposure time of 0.6 sec. The whole pictures showing SK-OV-3- and HFF14-infected cells were adjusted as follows; increase in brightness 25%, increase in contrast 50%. Panel B pictures were adjusted as follows, increase in brightness 20%, increase in contrast 30%. We confirmed that R-VG803 infection occurs through the HER2 receptor, by blocking the infection with trastuzumab, in fluorescence microscopy (Fig 2C), and flow cytometry (Fig 2D) assays. The results validate the inference that R-VG803 uses HER2 as the portal of entry in J-HER2 cells. This finding supports two fundamental conclusions. First, infection with a gH-retargeted HSV can take place in the absence of a gD receptor. Under these conditions, gD is physically present but functionally ablated as receptor-binding glycoprotein, as it can not be activated by its cognate receptor(s) and can not transmit the activation to gH. Second, the tropism of HSV can be modified by engineering a heterologous ligand in gH.
Receptor usage in cells that harbour both HER2 and nectin1/HVEM
We analysed the receptor usage in cells that express both sets of receptors, HER2 and nectin1/HVEM, exemplified by SK-OV-3 cells. The question was whether one receptor was preferentially used over the other, or each one was used alternatively. In the latter case, we expected that a block in the access to one of the two sets of receptors—e.g. HER2—should result in low extent of inhibition, whereas the simultaneous block to both sets of receptors should result in strong inhibition. The latter was indeed the case. As controls, we included the two retargeted viruses R-LM113 and R-LM249, and wt R-LM5. R-LM113 is detargeted from natural gD receptors [33,43], even though the AA 6–38 deletion in gD removes only some residues implicated in the nectin1-binding site, in addition to the entire HVEM binding site. The nectin1 binding site is widespread in the molecule, and includes the Ig-folded core and portions located between AA 35–38, 199–201, 214–217, 219–221 [12,13,34]. SK-OV-3 cells were infected with the indicated viruses, in the presence of trastuzumab, MAb HD1 to gD, or both. Fig 3A shows that trastuzumab or HD1 exerted almost no inhibition on R-VG803 when given singly, but practically abolished infection when given together. In contrast, R-LM113 and R-LM249 were inhibited by trastuzumab alone. Thus, R-VG803 can use alternatively HER2 or nectin1/HVEM to infect SK-OV-3 cells. Usage of one or the other portals of entry by R-VG803 depends on the spectrum of receptors displayed by the cells.
Fig. 3. Receptor usage by R-VG803 and R-VG809 in SK-OV-3 cells, detected through inhibition of infection by trastuzumab, MAb HD1, or combination thereof. 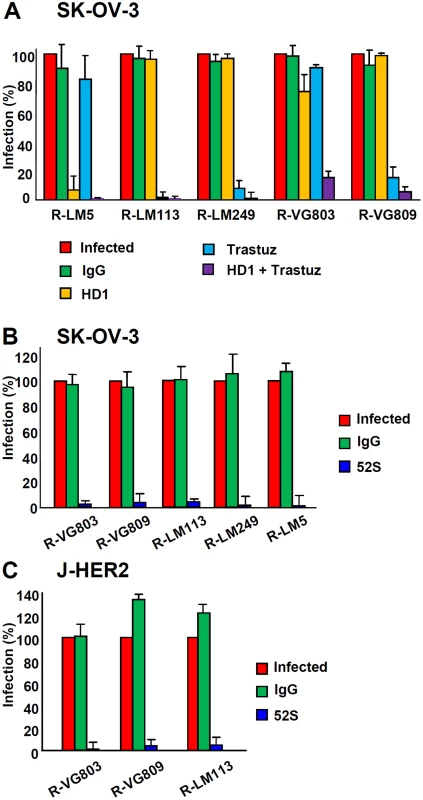
The indicated viruses were preincubated with HD1 (1.5 μg IgG/ml, final concentration) and then allowed to infect SK-OV-3 cells. When indicated, cells were pretreated with trastuzumab (28 μg/ml, final concentration), or control IgGs. Extent of infection was quantified 24 h later by means of flow cytometry, and expressed as percentage relative to cells infected with untreated virus, or untreated cells. Each value represents the average of three independent experiments ± S.D. (B, C) R-VG803 and R-VG809 infection of SK-OV-3 (B) and J-HER2 (C) cells is inhibited by MAb 52S to gH. The indicated virions were preincubated with MAb 52S (ascites fluid 1:25), or mouse IgGs for 1 h, prior to infection of SK-OV-3 or J-HER2 cells (2 or 0.3 PFU/cell, respectively), until harvesting at 24 h after infection. Infection was quantified by flow cytometry and expressed as % of cells infected with untreated virions. Each value represents the average of three independent experiments ± S.D. To characterize further the scFv—gH chimera we asked whether infection can be blocked by the neutralizing MAb 52S to gH. This MAb recognizes a continuous epitope, independent of gL, with critical residues at S536 and A537 [45,46]. R-VG803 infection of both SK-OV-3 and J-HER2 cells was abolished by MAb 52S (ascites fluid 1 : 25) (Fig 3B and 3C), indicating that a key functional domain in wt-gH was preserved in the chimera.
Deletion of AA 6–38 from R-VG803 gD results in a recombinant retargeted to HER2 via gH and detargeted from gD receptors
Inasmuch as R-VG803 infects J-HER2 cells independently of gD receptors and of neutralizing MAb to gD, we reasoned that it might be possible to engineer a recombinant carrying the scFv-HER2 in gH and the deletion of receptors’ binding sites from gD. We deleted the AA 6–38 region. R-VG809 failed to infect not only J-HVEM cells, but also J-nectin cells, and did not infect or infected very little the panel of animal and human cell lines employed above. It infected efficiently J-HER2, CHO-HER2 and SK-OV-3 cells (Fig 4A). In summary, R-VG809 exhibited a redirected tropism, strikingly different from that of R-VG803 (compare Fig 4A with Fig 2A). R-VG809 was also capable of cell-to-cell spread in J-HER2 cells (Fig 4B). Further validation that R-VG809 uses HER2 as portal of entry was provided by inhibition with trastuzumab (Fig 4C and 4D).
Fig. 4. R-VG809 infects cells that express HER2, fails to infect cells via gD receptors, and progeny virus spreads in J-HER2 cells. 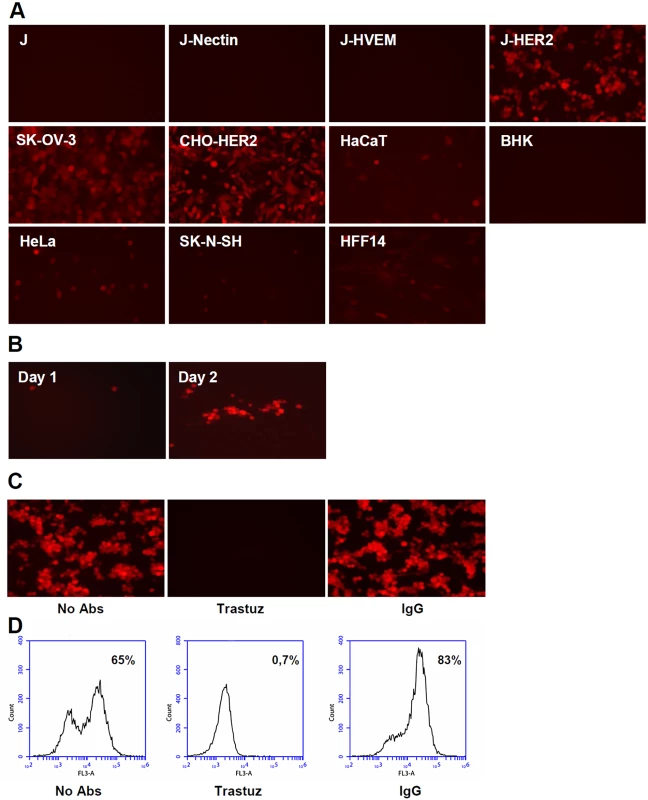
(A) The indicated cells were infected with R-VG809 (20 PFU/cell as titrated in SK-OV-3), and monitored for red fluorescence microscopy at 24 h after infection. (B) J-HER2 cells were infected with R-VG809 (0.01 PFU/cell), overlaid with medium containing the neutralizing MAb 52S to gH (ascites fluid, 1:10,000), and monitored daily for red fluorescence. Daily pictures of a same plaque are shown. (C, D) Trastuzumab inhibits R-VG809 infection of J-HER2 cells. J-HER2 cells were infected with R-VG809 in the presence of trastuzumab (trastuz) (28 μg/ml, final concentration) or control IgGs (28 μg/ml, final concentration). Infection was monitored by fluorescence microscopy (C), or flow cytometry (D). Panel B pictures were adjusted as follows, increase in brightness 20%, increase in contrast 30%. Analysis of the inhibitory effect of trastuzumab and MAb HD1 in SK-OV-3 cells shows that R-VG809 infection was inhibited by trastuzumab, even in the absence of MAb HD1 (Fig 3A), confirming that HER2 is the only portal for R-VG809. Infection of R-VG809 was blocked by MAb 52S, in agreement with the fact that R-VG809 and R-VG803 carry the same gH chimera (Fig 3B and 3C). We conclude that R-VG809 infection via the HER2-retargeted gH does not require the receptors’ binding sites in gD, and the receptor-mediated gD activation. Inasmuch R-VG809 does not carry the deletion of the entire gD open reading frame, we cannot formally rule out that gD serves a hypothetical, additional, so-to-say structural function, i.e. it facilitates the formation and/or stabilization of complexes among the glycoproteins.
Characterization of the gD AA 6–38 deletion
We characterized the detargeting effect exerted by the AA 6–38 deletion in gD. The retargeted phenotype exhibited by R-LM113 may result from the deletion per se or from the combined deletion-insertion. For example, the scFv insert, which is ~ 270 AA long, is likely to induce distortions in gD N-terminus, such that it can not any longer interact with the core of the molecule. Moreover, the insert may mask part of the nectin1 binding site in gD. To discriminate among these possibilities we measured R-VG809 replication in J-nectin1 and Vero cells. We included R-VG803 and R-LM5 for comparison. Fig 5A and 5B shows that R-VG809, but not R-VG803 and R-LM5, failed to replicate in J-nectin and Vero cells. Also the cytolytic effect of R-VG809 was strikingly different from those of R-VG803 and R-LM5, in that R-VG809 failed to kill J-nectin cells (Fig 5C). Parenthetically, the increase in cells viability exhibited by R-VG809 and R-LM5 between day 1 and 4 may be consequent to fact that some cells were not infected at day 0, and they replicated in the time interval of the assay. We conclude that the of AA 6–38 deletion in gD suffices to achieve full detargeting from both HVEM and nectin1, even in the absence of any insert.
Fig. 5. Detargeting conferred to R-VG809 by the AA 6–38 deletion in gD. 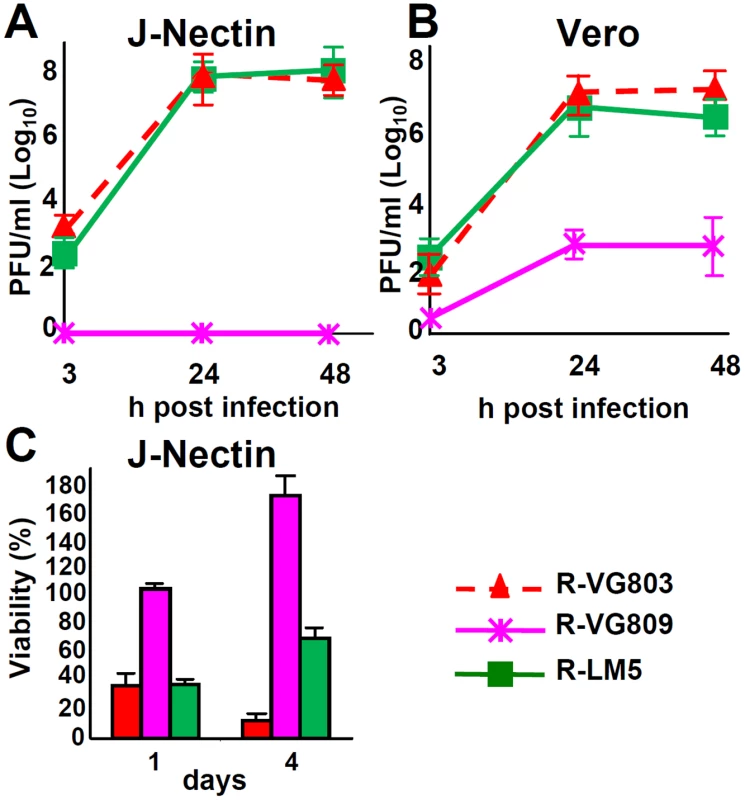
(A, B) Growth curves of R-VG803, R-VG809 and R-LM5 in J-nectin (A) and Vero (B) cells, infected at an input multiplicity of infection of 0.1 or 1 PFU/cell, respectively, and harvested at the indicated times (h) after infection. Progeny viruses were titrated in SK-OV-3 cells. (C) Cell killing ability of R-VG803, R-VG809 and R-LM5 for J-nectin cells. Cells were infected with the indicated viruses at 2 PFU/cell. Cell viability was determined by AlamarBlue, in triplicate monolayers, at the indicated days after infection. Each point or column represents the mean of triplicates ± S.D. Replication and cell killing ability of R-VG803 and R-VG809
Replication efficiency and cell killing are key properties for any candidate o-HSV. We verified the replication efficiency of R-VG803 and R-VG809 in J-HER2 cells, in comparison to that of R-LM113, R-LM249 and R-LM5. Fig 6A shows that the yields of R-VG803 and R-LM113 in cells infected at 0.1 PFU/cell were practically undistinguishable, implying that the extent of replication in J-HER2 cells is independent of whether the retargeting is achieved through gH or gD. Fig 6B compares the yields of R-VG803, R-VG809 and R-LM5 in J-HER2 cells infected at 0.01 PFU/cell. R-VG809 was somewhat hampered relative to R-VG803. R-VG809 was capable of cell-to-cell spread in J-HER2 cells; the decrease relative to R-VG803 likely reflected the lower extent of replication than the spread per se (Fig 6C).
Fig. 6. Replication of R-VG803 and R-VG809 in J-HER2 and SK-OV-3 cells. 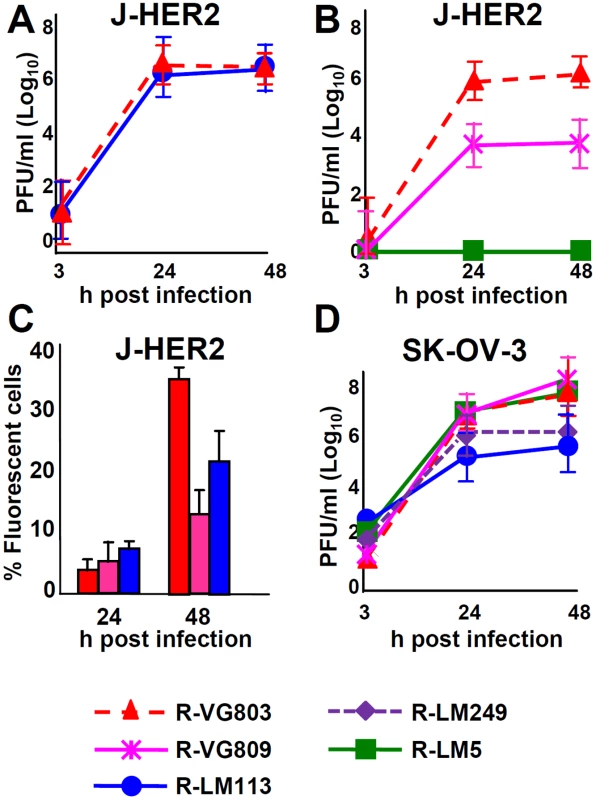
(A, B) Growth curves of R-VG803, R-VG809, R-LM113, R-LM5 in J-HER2. J-HER-2 cells were infected at 0.1 PFU/cell (A), or 0.01 PFU/cell (B). The viral titres of the input viruses were determined in the same cell line. Progeny virus was harvested at the indicated times and titrated in J-HER2 cells. (C) Comparison of cell-to-cell spread by R-VG803, R-VG809 and R-LM113 in J-HER2 cells. J-HER2 cell monolayers were infected with the indicated viruses at 0.01 PFU/cell. The percentage of fluorescent cells at 24 and 48 h after infection was determined by flow cytometry. (D) Growth curves of the indicated viruses (0.1 PFU/cell) in SK-OV-3. Input and progeny viruses were titrated in SK-OV-3 cells. Results are the average of at least two independent experiments ± S.D. Of interest was the growth in SK-OV-3 cells, as these are cancer cells, resistant to trastuzumab [35]. R-VG803 and R-VG809 replicated equally well, could not be differentiated from the wt R-LM5, and replicated somewhat better than R-LM113 and R-LM249 (Fig 6D).
Lastly, we analyzed the ability of R-VG803, R-VG809 to kill the SK-OV-3 tumor cells, in comparison to R-LM113, R-LM249 and R-LM5. Cytotoxicity caused by R-VG803, R-VG809, R-LM113 and R-LM249 were very similar one to the other, and much higher than that of R-LM5 (Fig 7).
Fig. 7. Cell killing ability of R-VG803 and R-VG809 for SK-OV-3 cells. 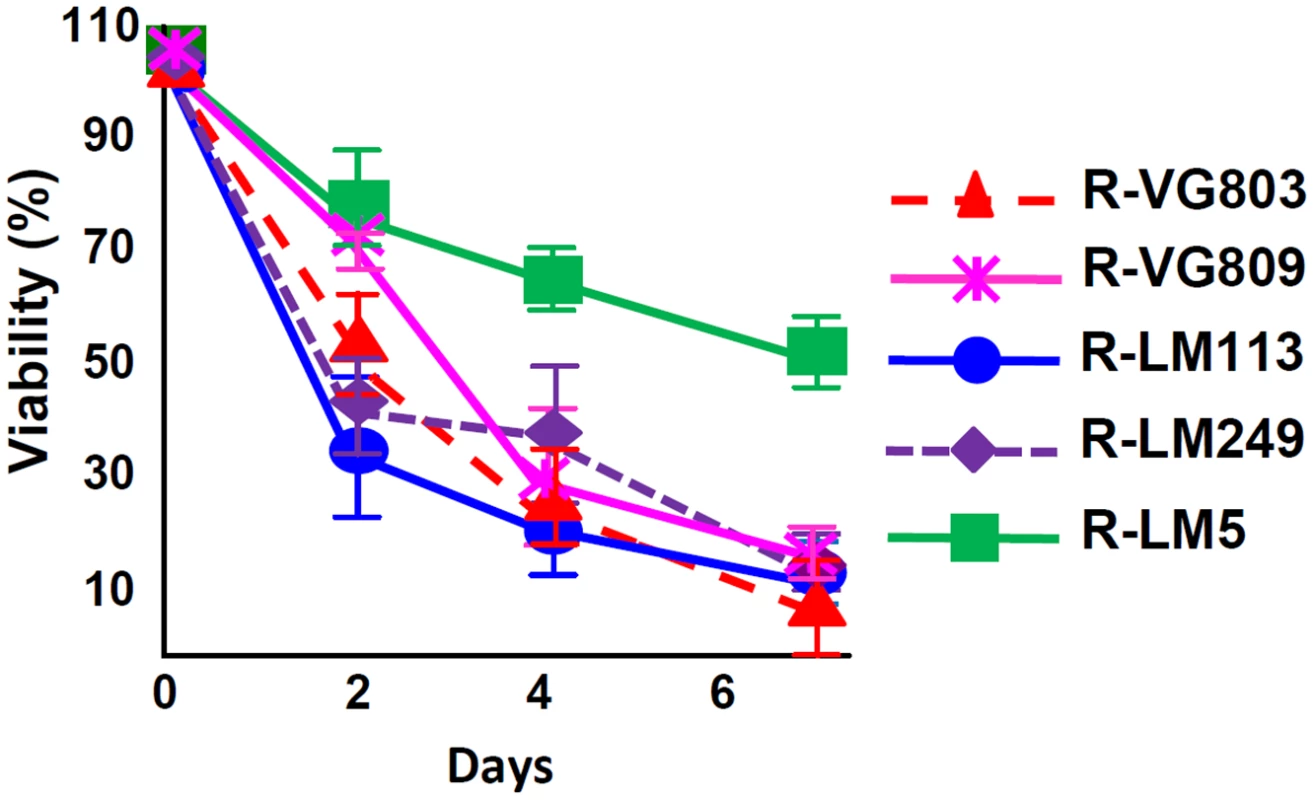
Cells were infected with the indicated viruses at 2 PFU/cell. Cell viability was determined by AlamarBlue, in triplicate monolayers. Each figure represents the average of triplicates ± S.D. Discussion
The engineering of a novel ligand—a single chain antibody (scFv) directed to HER2—in gH conferred to HSV an expanded tropism for cells which express HER2 as the sole receptor. Virus entry mediated by the chimeric scFv-HER2–gH could occur in the absence of gD receptors, despite deletion of the receptor-binding sites in gD, or presence of gD-neutralizing MAbs. Basically, the key functions of gD are no longer essential, and can be replaced by a ligand in gH. This finding impacts on current view of how HSV enters cells, and on the strategies for retargeting the HSV tropism to receptors of choice.
When wt-HSV enters target cells, gD serves two major functions. It serves as major receptor binding glycoprotein, and determinant of the viral tropism, i.e. it dictates which cells HSV will or will not infect. Secondly, the encounter of HSV with a cell carrying a gD receptor is signaled to gH/gL and gB, to trigger fusion. In essence, the receptor-bound gD initiates the cascade of activation of the entry glycoproteins [1–3,47,48] The control exerted by gD on virion-cell fusion ensures that the activation of the viral fusion machinery occurs only when HSV has reached a receptor-positive cell. In contrast to wt-virus, when the gH-retargeted viruses infect J-HER2 cells the activation of the chimeric scFv—gH does not require gD activation by its receptor, or receptor-binding sites in gD. gD is functionally ablated as receptor-binding glycoprotein and as activator of the downstream glycoproteins. gD is no longer a requirement to trigger fusion. Its functions have been taken over by the scFv in gH.
In wt virus, the activation exerted by the receptor-bound gD on gH/gL necessarily occurs through intermolecular signaling. We refer to it as trans-signaling, as opposed to a signaling that occurs intramolecularly, herein referred to as cis-signaling. A novelty of our results is that the activation of gH can occur in cis, i.e. the scFv activates the gH moiety in the chimera. In the past, Klupp and Mettenleiter generated a non-viable PrV recombinant, carrying a deletion in gL [49]. Upon serial blind passages, a viable recombinant carrying a gD-gH fusion was isolated [50]. Subsequently, Cairns et al. constructed a similar HSV gD-gH chimera, in which the entire ectodomain of gD was fused to the N-terminus of gH (named chimera 22 in their work) [51]. In complementation assays, the HSV chimera rescued the infection of a gD-/- gH+ virus, or of a gH-/- gD+ virus. It was not tested for complementation of a double deletion gD-/- gH-/- virus. There are two key differences between the previous report [51] and our finding. First, in the complementation assays, the wt-gD in the gH-/- gD+ virus had the possibility to activate in trans the gH moiety in the gD-gH chimera. Conversely, the gD moiety in the chimera had the possibility to activate in trans the wt-gH present in the gD-/- gH+ virions. In either case, the activation may have taken place in-trans, as concluded by the Authors. Formal evidence for cis-activation of the gD-gH chimera was not provided [51]. Secondly, irrespective of the activation mechanism, in the complementing system the gH activation was mediated by gD, which has a binding site on gH [14–16,52], and not by a heterologous ligand. The latter was indeed an unexpected possibility. Previous attempts to develop systems for HSV-mediated cell fusion, or HSV infection independent of gD led to partial indications as follows. In the cell-cell fusion assay, a partial deletion in the N-terminus of gH was reported to induce low, constitutive levels of fusion by gB, in the absence of gD or gD receptors [48]. Whether, once present in the virion, the same deletion will lead to a constitutive, low level gD-independent entry, or will lead to an exhausted fusion/entry machinery has not been ascertained. Uchida and collaborators reported on mutations in virion gB, or virion gH, that render these glycoproteins independent of gD activation by its major receptor nectin1, but are still dependent on the activation by so-called unconventional gD receptors (e.g. nectin3 present in J cells [14]), or receptors to a retargeted gD (e.g. EGFR for a EGFR-retargeted gD). The ability of the mutant forms of gB or gH to carry out entry independently of any form of gD activation, or with a form of gD deleted in receptor-binding sites was not established [53]. Hence, previous studies are strikingly different from current study, where the receptor-binding activity of gD for nectin1/HVEM was ablated by deletion of key residues, and a heterologous receptor-binding activity was implanted in gH.
As regards the field of oncolytic HSVs, our data show that gH accepted the insertion of a hetelogous ligand and became a tool for the retargeting of HSV tropism to a heterologous receptor. The ligand may be at least 270 AA in size, i.e. about 1/3 of gH ectodomain. The gH-mediated retargeting could be combined with detargeting, through a suitable deletion in gD. This ensued in the fully retargeted R-VG809, whose replication and killing capacity for SK-OV-3 cells did not substantially differ, or were even better than those of the gD-retargeted R-LM113 and R-LM249. In essence, changes in tropism through modifications in gH or in gD yield o-HSVs with substantially similar growth and lytic properties. Remarkably, both the gH - and the gD-retargeted o-HSVs grew almost as efficiently as the wt R-LM5. They represent an improvement over the first generation retargeted o-HSVs, which were marred by a relatively low replication capacity [30,54]. We highlight that so far, gD was the only glycoprotein that successfully enabled the retargeting of HSV [30,33–38,54]. Earlier efforts to use glycoproteins other than gD, e.g. gC, did not meet with success [55].
Current findings expand the toolkit for generation of non attenuated retargeted o-HSVs. Two prospective applications are worth noting. The anti-HER2 huMAbs and small molecule inhibitors of HER2 signaling now in clinical trials have non-overlapping mechanisms of action, and patients clearly benefit from combinations [56,57]. However, a fraction of patients does not respond. The responders develop resistance, often within a year of treatment [58]. In the resistant cancer cells, the HER2 ectodomain is preserved, and the modifications affect the signaling portions of the receptor. This type of resistance is recapitulated in SK-OV-3 cells, which are HER2+ and trastuzumab-resistant [35]. The observation that R-VG809, as well as R-LM249 [34,35], can grow and kill SK-OV-3 cells raises the possibility that treatment with HER2-retargeted o-HSVs could be applied to patients who developed resistance to the anti-HER2 specific therapeutics. Secondly, the heterogeneity in cancers cells represents a limit to numerous therapeutic approaches. Heterogeneity is observed also in the extent of expression of cancer receptors. The possibility to retarget o-HSV tropism to cancer receptors via gH and via gD opens the way to the design of double-retargeted o-HSVs, which may be better suited to counteract cancer cell heterogeneity than singly-retargeted o-HSVs.
Materials and Methods
Cells and viruses
The receptor negative J cells, their counterparts expressing HER2, nectin1, HVEM and CHO-HER2 were described [7,43]. HFF14 cells were received by Dr. Frank Neipel (University of Erlangen). Vero, RS, SK-OV-3, HaCaT, BHK, HeLa and SK-N-SH cells were received by ATCC. The wt - HSV-1(F), R-LM113, R-LM249 and R-LM5 were described [33,34,43,59].
Engineering of R-VG803 and R-VG809
First, we engineered R-VG801, by insertion of the sequence encoding the trastuzumab scFv between AA 23 and 24 of gH. Subsequently we engineered R-VG803 by insertion of mCherry sequences into the UL37-UL38 intergenic region of R-VG801. To generate R-VG801, the starting viral genome was pYEBac102, which carries LOX-P-bracketed p-BeloBAC sequences inserted between UL3 and UL4 of HSV-1 genome [60]. All engineering procedures were performed by means of galK recombineering [61]. Briefly, the GalK cassette, with homology arms to gH was amplified by means of primers gH6_galK_f ATGCGGTCCATGCCCAGGCCATCCAAAAACCATGGGTCTGTCTGCTCAGTCCTGTTGACAATTAATCATCGGCA and gH5_galK_r TCGTGGGGGTTATTATTTTGGGCGTTGCGTGGGGTCAGGTCCACGACTGGTCAGCACTGTCCTGCTCCTT. This cassette was electroporated in SW102 bacteria carrying pYEBac102. The recombinant clones carrying the galK cassette were selected on M63 plates (15 mM (NH4)2SO4, 100 mM KH2PO4, 1.8 μg FeSO4·7H2O, adjusted to pH 7) supplemented with 1 mg/L D-biotin, 0.2% galactose, 45 mg/L L-leucine, 1 mM MgSO4·7H2O and 12 μg/ml chloramphenicol. To exclude galK false positive colonies, the recombinant clones were plated on McConkey agar base plates, supplemented with 1% galactose and 12 μg/ml chloramphenicol, and checked by colony PCR with primer galK_129_f ACAATCTCTGTTTGCCAACGCATTTGG and galK_417_r CATTGCCGCTGATCACCATGTCCACGC. Next, the trastuzumab scFv cassette, bracketed by Ser-Gly linkers and by upstream and downstream homology arms to gH, was amplified using pSG-ScFvHER2-SG (a gift from Alfredo Nicosia) as template. pSG-ScFvHER2-SG was obtained by inserting the synthetic antiHER2 scFv cassette, designed on the basis of published information [62]; Sequence 18 from Patent WO2004065416 (Genbank CQ877234); Sequence 7 (pS2072a) from Patent WO2005100399 (Genbank CS276173) into an appropriate vector. The scFv cassette was bracketed by the Ser-Gly linkers detailed below. Relative to sequence 18 from Patent WO2004065416, nucleotides 769–771 were mutated in pSG-ScFvHER2-SG to generate a XhoI restriction site. Using pSG-ScFvHER2-SG as template, two separate fragments (# 1 and # 2) were PCR-amplified by means of oligonucleotides which contained homology arms to gH. Specifically, fragment # 1 was amplified by means of primers gH23_8SG_scFv4D5_f TCGTGGGGGTTATTATTTTGGGCGTTGCGTGGGGTCAGG TCCACGACTGGCATAGTAGTGGCGGTGGCTCTGGATCCG and scFv4D5_358_r GGAAACGGTTCGGATCAGCCATCGG, using pSG-ScFvHER2-SG as template. Fragment # 2 was amplified by means of gH24_12SG_scFv4D5r ATGCGGTCCATGCCCAGGCCATCCAAAAACCATGGGTCTGTCTGCTCAGTACCG GATCCACCGGAACCAGAGCC and scFv4D5_315_f GGAGATCAAATCGGATATGCCGATGG using pSG-ScFvHER2-SG as template. Thereafter, fragments # 1 and # 2 were annealed and extended to generate the entire scFv-HER2 cassette, bracketed by the Ser-Gly linkers and the homology arms to gH. The sequence of the upstream and downstream Ser-Gly linkers were HSSGGGSG, and SSGGGSGSGGSG, respectively. The linker between VL and VH had the sequence SDMPMADPNRFRGKNLVFHS. The recombinant bacterial clones carried the scFv-HER2 cassette in place of the galK cassette. They were selected on M63 plates, supplemented with 1 mg/L D-biotin, 0.2% deoxy-2-galactose, 0.2% glycerol, 45 mg/L L-leucine, 1 mM MgSO4·7H2O and 12 μg/ml chloramphenicol. Bacterial colonies were checked for the presence of inserted sequence by colony PCR.
The mCherry red fluorescent protein, under the CMV promoter, was inserted in the UL37-UL38 intergenic region of R-VG801 (coordinates 84156–84157), to generate R-VG803, following the two step procedure outlined above. Briefly, we first inserted the galK cassette, amplified by means of oligonucleotides UL37/38_galK_f CCGCAGGCGTTGCGAGTACCCCGCGTCTTCGCGGGGTGTTATACGGCCACCCTGTTGACAATTAATCATCGGCA and UL37/38_galK_r TCCGGACAATCCCCCGGGCCTGGGTCCGCGAACGGGATGCCGGGACTTAATCAGCACTGTCCTGCTCCTT. Subsequently, the galK sequence was replaced with the promoter-mCherry cassette, amplified by means of oligonucleotides UL37/38_CMV_mcherry_f CCGCAGGCGTTGCGAGTACCCCGCGTCTTCGCGGGGTGTTATACGGCCACCGATGTACGGGCCAGATATACG and UL37/38_pA_mcherry_1958_r TCCGGACAATCCCCCGGGCCTGGGTCCGCGAACGGGATGCCGGGACTTAACCATAGAGCCCACCGCATCC. The starting material for R-VG809 was the R-VG803 BAC genome. To generate the AA 6–38 deletion in gD, a galK cassette flanked by homology arms to gD was amplified by means of primers gD5_galK_f TTGTCGTCATAGTGGGCCTCCATGGGGTCCGCGGCAAATATGCCTTGGCGCCTGTTGACAATTAATCATCGGCA and gD39_galK_r ATCGGGAGGCTGGGGGGCTGGAACGGGTCCGGTAGGCCCGCCTGGATGTGTCAGCACTGTCCTGCTCCTT. Next, we replaced the galK sequence with a synthetic double stranded oligonucleotide gD_aa5_39_f_r TTGTCGTCATAGTGGGCCTCCATGGGGTCCGCGGCAAATATGCCTTGGCGCACATCCAGGCGGGCCTACCGGACCCGTTCCAGCCCCCCAGCCTCCCGAT. In all cases, the recombinant viruses were generated by transfection of SK-OV-3 cells with the appropriate recombinant BAC DNA (500 ng) by means of Lipofectamine 2000 (Life Technologies). Virus growth was monitored by red fluorescence. The structure of the viral recombinants was verified by sequencing the gH and mCherry ORFs, and gD ORF for R-VG809. Virus stocks were generated and titrated in SK-OV-3 cells, or in J-HER2 cells, as specified.
Expression of chimeric gH from R-VG803 and R-VG809
Lysates of Vero cells infected with R-VG803, R-VG809 or R-LM5 (3 PFU/cell) were subjected to PAGE, transferred to PVDF membranes. Immunoblot reactivity to polyclonal antibody (PAb) to gH was assayed as detailed [44].
Tropism of R-VG803 and R-VG809
The indicated cells were infected with R-VG803, or R-VG809 at 2 and 20 PFU/cell, respectively. Red fluorescence was monitored by fluorescence microscopy.
Block of R-VG803 and R-VG809 infection by MAbs to HER2 and gD
Replicate monolayers of J-HER2 or SK-OV-3 cells in 12 well plates were preincubated with trastuzumab or non-immune mouse IgG (28 μg/ml, final concentration) for 1 h, and then infected with R-VG803, R-VG809, R-LM113, R-LM249 or R-LM5 (0.3 or 2 PFU/cell for J-HER2 or SK-OV-3 cells, respectively), in the same medium. Alternatively, virions were preincubated with MAbs HD1 (1.5 μg/ml, final concentration), or MAb 52 S (ascites fluid 1 : 25) for 1 h at 37°C, and then allowed to absorb to cells for 90 min, in the absence or presence of trastuzumab, as indicated. Viral inocula were then removed, and cells were overlaid with medium containing the indicated antibodies. Virus replication was monitored at 24 h after infection by BD Accuri C6 flow cytometer. Results are expressed ad the mean of three independent experiments ± SD.
Virus growth determinations
To determine R-VG803 and R-VG809 growth in J-HER2, SK-OV-3, J-nectin, and Vero cells, the cells were infected with R-VG803, R-LM113, R-LM249, R-LM5 at the indicated MOI. Unabsorbed virus was inactivated by rinsing cells with pH 3 solution (40 mM citric acid, 10 mM KCl, 135 mM NaCl). Cells were harvested at 3 (0 time), 24 and 48 h after infection and progeny virus (intracellular plus extracellular) was titrated in J-HER2 or SK-OV-3 cells, as indicated.
Cytotoxity assay
SK-OV-3 and J-nectin cells were seeded in 96 well plates at 8x103 cell/well, and infected with R-VG803, R-VG809, R-LM113, R-LM249 and R-LM5 (2 PFU/cell) or mock-infected. AlamarBlue (10 μl/well, Life Technologies) was added to the culture media at indicated times after infection and incubated for 4 h at 37°C. Plates were read at 560 and 600 nm with GloMax Discover System (Promega). For each time point, cell viability was expressed as the percentage of AlamarBlue reduction in infected versus uninfected cells, excluding for each set of samples the contribution of medium alone. Each point represents the average of at least triplicate samples ± SD.
Accession numbers
HSV-1 strain F, Genbank GU73477. gH coordinates 43783–46299
HSV-1 strain F, gD: Genbank L09242
scFv to HER2 Genbank CQ877234, CS276173
mCherry Genbank HM771696
Supporting Information
Zdroje
1. Campadelli-Fiume G, Menotti L, Avitabile E, Gianni T (2012) Viral and cellular contributions to herpes simplex virus entry into the cell. Curr Opin Virol 2 : 28–36. doi: 10.1016/j.coviro.2011.12.001 22440963
2. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R (2011) Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol 9 : 369–381. doi: 10.1038/nrmicro2548 21478902
3. Campadelli-Fiume G, Amasio M, Avitabile E, Cerretani A, Forghieri C, et al. (2007) The multipartite system that mediates entry of herpes simplex virus into the cell. Rev Med Virol 17 : 313–326. 17573668
4. Backovic M, DuBois RM, Cockburn JJ, Sharff AJ, Vaney MC, et al. (2010) Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc Natl Acad Sci U S A 107 : 22635–22640. doi: 10.1073/pnas.1011507107 21149698
5. Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, et al. (2010) Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol 17 : 882–888. doi: 10.1038/nsmb.1837 20601960
6. Rey FA (2006) Molecular gymnastics at the herpesvirus surface. EMBO Rep 7 : 1000–1005. 17016458
7. Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G (1998) The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol 72 : 9992–10002. 9811737
8. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG (1998) Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280 : 1618–1620. 9616127
9. Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, et al. (1999) A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99 : 13–22. 10520990
10. Montgomery RI, Warner MS, Lum BJ, Spear PG (1996) Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87 : 427–436. 8898196
11. Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, et al. (2001) Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell 8 : 169–179. 11511370
12. Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, et al. (2005) Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. Embo J 24 : 4144–4153. 16292345
13. Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, et al. (2011) Structure of herpes simplex virus glycoprotein d bound to the human receptor nectin-1. PLoS Pathog 7: e1002277. doi: 10.1371/journal.ppat.1002277 21980294
14. Cocchi F, Menotti L, Di Ninni V, Lopez M, Campadelli-Fiume G (2004) The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2. J Virol 78 : 4720–4729. 15078954
15. Fusco D, Forghieri C, Campadelli-Fiume G (2005) The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc Natl Acad Sci U S A 102 : 9323–9328. 15972328
16. Gianni T, Amasio M, Campadelli-Fiume G (2009) Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL through the C-terminal profusion. J Biol Chem 284 : 17370–17382. doi: 10.1074/jbc.M109.005728 19386594
17. Avitabile E, Forghieri C, Campadelli-Fiume G (2009) Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J Virol 83 : 10752–10760. doi: 10.1128/JVI.01287-09 19656900
18. Gianni T, Salvioli S, Chesnokova LS, Hutt-Fletcher LM, Campadelli-Fiume G (2013) alphavbeta6 - and alphavbeta8-integrins serve as interchangeable receptors for HSV gH/gL to promote endocytosis and activation of membrane fusion. PLoS Pathog 9: e1003806. doi: 10.1371/journal.ppat.1003806 24367260
19. Avitabile E, Forghieri C, Campadelli-Fiume G (2007) Complexes between Herpes Simplex Virus Glycoproteins gD, gB, and gH Detected in Cells by Complementation of Split Enhanced Green Fluorescent Protein. J Virol 81 : 11532–11537. 17670828
20. Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, et al. (2007) Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A 104 : 18718–18723. 18003913
21. Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, et al. (2004) The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc Natl Acad Sci U S A 101 : 7445–7450. 15123804
22. Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ (2010) Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol 84 : 12292–12299. doi: 10.1128/JVI.01700-10 20861251
23. Kwon H, Bai Q, Baek HJ, Felmet K, Burton EA, et al. (2006) Soluble V domain of Nectin-1/HveC enables entry of herpes simplex virus type 1 (HSV-1) into HSV-resistant cells by binding to viral glycoprotein D. J Virol 80 : 138–148. 16352538
24. Cattaneo R, Miest T, Shashkova EV, Barry MA (2008) Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat Rev Microbiol 6 : 529–540. doi: 10.1038/nrmicro1927 18552863
25. Lichty BD, Breitbach CJ, Stojdl DF, Bell JC (2014) Going viral with cancer immunotherapy. Nat Rev Cancer 14 : 559–567. doi: 10.1038/nrc3770 24990523
26. Campadelli-Fiume G, De Giovanni C, Gatta V, Nanni P, Lollini PL, et al. (2011) Rethinking herpes simplex virus: the way to oncolytic agents. Rev Med Virol 21 : 213–226. doi: 10.1002/rmv.691 21626603
27. Russell SJ, Peng KW, Bell JC (2012) Oncolytic virotherapy. Nat Biotechnol 30 : 658–670. doi: 10.1038/nbt.2287 22781695
28. Liu BL, Robinson M, Han ZQ, Branston RH, English C, et al. (2003) ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther 10 : 292–303. 12595888
29. Andtbacka RHI, Collichio FA, Amatruda T, Senzer NN, Chesney J, et al. OPTiM: A randomized phase III trial of talimogene laherparepvec (T-VEC) versus subcutaneous (SC) granulocyte-macrophage colony-stimulating factor (GM-CSF) for the treatment (tx) of unresected stage IIIB/C and IV melanoma; 2013.
30. Zhou G, Roizman B (2006) Construction and properties of a herpes simplex virus 1 designed to enter cells solely via the IL-13alpha2 receptor. Proc Natl Acad Sci U S A 103 : 5508–5513. 16554374
31. Zhou G, Ye GJ, Debinski W, Roizman B (2002) Engineered herpes simplex virus 1 is dependent on IL13Ralpha2 receptor for cell entry and independent of glycoprotein D receptor interaction. Proc Natl Acad Sci USA 99 : 15124–15129. 12417744
32. Menotti L, Cerretani A, Campadelli-Fiume G. A HSV recombinant exhibiting a single chain antibody to HER2/neu enters cells through the mammary tumor receptor, independently of the gD receptors; 2006; Seattle, USA. 16699034
33. Menotti L, Cerretani A, Hengel H, Campadelli-Fiume G (2008) Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J Virol 20 : 10153–10161. doi: 10.1128/JVI.01133-08 18684832
34. Menotti L, Nicoletti G, Gatta V, Croci S, Landuzzi L, et al. (2009) Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc Natl Acad Sci USA 106 : 9039–9044. doi: 10.1073/pnas.0812268106 19458262
35. Nanni P, Gatta V, Menotti L, De Giovanni C, Ianzano M, et al. (2013) Preclinical Therapy of Disseminated HER-2(+) Ovarian and Breast Carcinomas with a HER-2-Retargeted Oncolytic Herpesvirus. PLoS Pathog 9: e1003155. doi: 10.1371/journal.ppat.1003155 23382683
36. Zhou G, Roizman B (2007) Separation of receptor binding and pro-fusogenic domains of glycoprotein D of herpes simplex virus 1 into distinct interacting proteins. Proc Natl Acad Sci U S A 104 : 4142–4146. 17360490
37. Kamiyama H, Zhou G, Roizman B (2006) Herpes simplex virus 1 recombinant virions exhibiting the amino terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther 13 : 621–629. 16292350
38. Uchida H, Chan J, Goins WF, Grandi P, Kumagai I, et al. (2010) A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J Virol 84 : 12200–12209. doi: 10.1128/JVI.01633-10 20861246
39. Zhang KX, Kim C, Chow E, Chen IS, Jia W, et al. (2011) Targeting trastuzumab-resistant breast cancer cells with a lentivirus engineered to bind antibodies that recognize HER-2. Breast Cancer Res Treat 125 : 89–97. doi: 10.1007/s10549-010-0828-9 20232140
40. Jackson C, Browell D, Gautrey H, Tyson-Capper A (2013) Clinical Significance of HER-2 Splice Variants in Breast Cancer Progression and Drug Resistance. Int J Cell Biol 2013 : 973584. doi: 10.1155/2013/973584 23935627
41. Kubetzko S, Balic E, Waibel R, Zangemeister-Wittke U, Pluckthun A (2006) PEGylation and multimerization of the anti-p185HER-2 single chain Fv fragment 4D5: effects on tumor targeting. J Biol Chem 281 : 35186–35201. 16963450
42. Gambini E, Reisoli E, Appolloni I, Gatta V, Campadelli-Fiume G, et al. (2012) Replication-competent herpes simplex virus retargeted to HER2 as therapy for high-grade glioma. Mol Ther 20 : 994–1001. doi: 10.1038/mt.2012.22 22354378
43. Menotti L, Cerretani A, Campadelli-Fiume G (2006) A herpes simplex virus recombinant that exhibits a single-chain antibody to HER2/neu enters cells through the mammary tumor receptor, independently of the gD receptors. J Virol 80 : 5531–5539. 16699034
44. Gianni T, Cerretani A, Dubois R, Salvioli S, Blystone SS, et al. (2010) Herpes simplex virus glycoproteins H/L bind to cells independently of {alpha}V{beta}3 integrin and inhibit virus entry, and their constitutive expression restricts infection. J Virol 84 : 4013–4025. doi: 10.1128/JVI.02502-09 20147400
45. Showalter SD, Zweig M, Hampar B (1981) Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun 34 : 684–692. 6277788
46. Gompels UA, Carss AL, Saxby C, Hancock DC, Forrester A, et al. (1991) Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J Virol 65 : 2393–2401. 1707982
47. Heldwein EE, Krummenacher C (2008) Entry of herpesviruses into mammalian cells. Cell Mol Life Sci 65 : 1653–1668. doi: 10.1007/s00018-008-7570-z 18351291
48. Atanasiu D, Cairns TM, Whitbeck JC, Saw WT, Rao S, et al. (2013) Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors. MBio 4.
49. Klupp BG, Fuchs W, Weiland E, Mettenleiter TC (1997) Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol 71 : 7687–7695. 9311852
50. Klupp BG, Mettenleiter TC (1999) Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J Virol 73 : 3014–3022. 10074151
51. Cairns TM, Milne RS, Ponce-de-Leon M, Tobin DK, Cohen GH, et al. (2003) Structure-function analysis of herpes simplex virus type 1 gD and gH-gL: clues from gDgH chimeras. J Virol 77 : 6731–6742. 12767993
52. Fan Q, Longnecker R, Connolly SA (2014) Substitution of herpes simplex virus 1 entry glycoproteins with those of saimiriine herpesvirus 1 reveals a gD-gH/gL functional interaction and a region within the gD profusion domain that is critical for fusion. J Virol 88 : 6470–6482. doi: 10.1128/JVI.00465-14 24672037
53. Uchida H, Chan J, Shrivastava I, Reinhart B, Grandi P, et al. (2013) Novel mutations in gB and gH circumvent the requirement for known gD Receptors in herpes simplex virus 1 entry and cell-to-cell spread. J Virol 87 : 1430–1442. doi: 10.1128/JVI.02804-12 23152509
54. Zhou G, Roizman B (2005) Characterization of a recombinant herpes simplex virus 1 designed to enter cells via the IL13Ralpha2 receptor of malignant glioma cells. J Virol 79 : 5272–5277. 15827141
55. Laquerre S, Anderson DB, Stolz DB, Glorioso JC (1998) Recombinant herpes simplex virus type 1 engineered for targeted binding to erythropoietin receptor-bearing cells. J Virol 72 : 9683–9697. 9811702
56. Baselga J, Cortes J, Kim SB, Im SA, Hegg R, et al. (2012) Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med 366 : 109–119. doi: 10.1056/NEJMoa1113216 22149875
57. Baselga J, Bradbury I, Eidtmann H, Di Cosimo S, de Azambuja E, et al. (2012) Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet 379 : 633–640. doi: 10.1016/S0140-6736(11)61847-3 22257673
58. O'Sullivan CC, Connolly RM (2014) Pertuzumab and its accelerated approval: evolving treatment paradigms and new challenges in the management of HER2-positive breast cancer. Oncology (Williston Park) 28 : 186–194, 196. 24855725
59. Ejercito PM, Kieff ED, Roizman B (1968) Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2 : 357–364. 4300104
60. Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y (2003) Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol 77 : 1382–1391. 12502854
61. Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33: e36. 15731329
62. Sidhu SS, Li B, Chen Y, Fellouse FA, Eigenbrot C, et al. (2004) Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol 338 : 299–310. 15066433
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary TuberculosisČlánek Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic PlagueČlánek Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host SpeciesČlánek Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR SelectionČlánek Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus GenomesČlánek Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from FerrioxamineČlánek Remembering MumpsČlánek Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling PathwayČlánek Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Parasites and Their Heterophagic Appetite for Disease
- The Elusive Role of the Prion Protein and the Mechanism of Toxicity in Prion Disease
- Intestinal Colonization Dynamics of
- Activation of Typhi-Specific Regulatory T Cells in Typhoid Disease in a Wild-Type . Typhi Challenge Model
- The Engineering of a Novel Ligand in gH Confers to HSV an Expanded Tropism Independent of gD Activation by Its Receptors
- Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary Tuberculosis
- Group Selection and Contribution of Minority Variants during Virus Adaptation Determines Virus Fitness and Phenotype
- Phosphatidic Acid Produced by Phospholipase D Promotes RNA Replication of a Plant RNA Virus
- A Ribonucleoprotein Complex Protects the Interleukin-6 mRNA from Degradation by Distinct Herpesviral Endonucleases
- Characterization of Transcriptional Responses to Different Aphid Species Reveals Genes that Contribute to Host Susceptibility and Non-host Resistance
- Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic Plague
- Natural Killer Cell Sensing of Infected Cells Compensates for MyD88 Deficiency but Not IFN-I Activity in Resistance to Mouse Cytomegalovirus
- Manipulation of the Xanthophyll Cycle Increases Plant Susceptibility to
- Ly6C Monocytes Regulate Parasite-Induced Liver Inflammation by Inducing the Differentiation of Pathogenic Ly6C Monocytes into Macrophages
- Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host Species
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness
- Inhibition and Reversal of Microbial Attachment by an Antibody with Parasteric Activity against the FimH Adhesin of Uropathogenic .
- The EBNA-2 N-Terminal Transactivation Domain Folds into a Dimeric Structure Required for Target Gene Activation
- Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection
- The NLRP3 Inflammasome Is a Pathogen Sensor for Invasive via Activation of α5β1 Integrin at the Macrophage-Amebae Intercellular Junction
- Sequential Conformational Changes in the Morbillivirus Attachment Protein Initiate the Membrane Fusion Process
- A Two-Component DNA-Prime/Protein-Boost Vaccination Strategy for Eliciting Long-Term, Protective T Cell Immunity against
- cAMP-Signalling Regulates Gametocyte-Infected Erythrocyte Deformability Required for Malaria Parasite Transmission
- Response Regulator VxrB Controls Colonization and Regulates the Type VI Secretion System
- Evidence for a Novel Mechanism of Influenza Virus-Induced Type I Interferon Expression by a Defective RNA-Encoded Protein
- Dust Devil: The Life and Times of the Fungus That Causes Valley Fever
- TNF-α Induced by Hepatitis C Virus via TLR7 and TLR8 in Hepatocytes Supports Interferon Signaling via an Autocrine Mechanism
- The Recent Evolution of a Maternally-Inherited Endosymbiont of Ticks Led to the Emergence of the Q Fever Pathogen,
- L-Rhamnosylation of Wall Teichoic Acids Promotes Resistance to Antimicrobial Peptides by Delaying Interaction with the Membrane
- Rapid Sequestration of by Neutrophils Contributes to the Development of Chronic Lesion
- Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus Genomes
- The Expression of Functional Vpx during Pathogenic SIVmac Infections of Rhesus Macaques Suppresses SAMHD1 in CD4 Memory T Cells
- Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from Ferrioxamine
- TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation
- Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site
- Remembering Mumps
- The Role of Horizontal Gene Transfer in the Evolution of the Oomycetes
- Advances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi
- Investigating Fungal Outbreaks in the 21st Century
- Systems Biology for Biologists
- How Does the Dinoflagellate Parasite Outsmart the Immune System of Its Crustacean Hosts?
- FCRL5 Delineates Functionally Impaired Memory B Cells Associated with Exposure
- Phospholipase D1 Couples CD4 T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication
- Influenza A Virus on Oceanic Islands: Host and Viral Diversity in Seabirds in the Western Indian Ocean
- Geometric Constraints Dominate the Antigenic Evolution of Influenza H3N2 Hemagglutinin
- Widespread Recombination, Reassortment, and Transmission of Unbalanced Compound Viral Genotypes in Natural Arenavirus Infections
- Gammaherpesvirus Co-infection with Malaria Suppresses Anti-parasitic Humoral Immunity
- A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
- Mechanisms of Stage-Transcending Protection Following Immunization of Mice with Late Liver Stage-Arresting Genetically Attenuated Malaria Parasites
- The Myelin and Lymphocyte Protein MAL Is Required for Binding and Activity of ε-Toxin
- Genome-Wide Identification of the Target Genes of AP2-O, a AP2-Family Transcription Factor
- An Atypical Mitochondrial Carrier That Mediates Drug Action in
- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Helminth Infection and Commensal Microbiota Drive Early IL-10 Production in the Skin by CD4 T Cells That Are Functionally Suppressive
- Circulating Pneumolysin Is a Potent Inducer of Cardiac Injury during Pneumococcal Infection
- ExoT Induces Atypical Anoikis Apoptosis in Target Host Cells by Transforming Crk Adaptor Protein into a Cytotoxin
- Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement
- Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
- Varicella Viruses Inhibit Interferon-Stimulated JAK-STAT Signaling through Multiple Mechanisms
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



