-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Comparative Study between Transcriptionally- and Translationally-Acting Adenine Riboswitches Reveals Key Differences in Riboswitch Regulatory Mechanisms
Many bacterial mRNAs are regulated at the transcriptional or translational level by ligand-binding elements called riboswitches. Although they both bind adenine, the adenine riboswitches of Bacillus subtilis and Vibrio vulnificus differ by controlling transcription and translation, respectively. Here, we demonstrate that, beyond the obvious difference in transcriptional and translational modulation, both adenine riboswitches exhibit different ligand binding properties and appear to operate under different regulation regimes (kinetic versus thermodynamic). While the B. subtilis pbuE riboswitch fully depends on co-transcriptional binding of adenine to function, the V. vulnificus add riboswitch can bind to adenine after transcription is completed and still perform translation regulation. Further investigation demonstrates that the rate of transcription is critical for the B. subtilis pbuE riboswitch to perform efficiently, which is in agreement with a co-transcriptional regulation. Our results suggest that the nature of gene regulation control, that is transcription or translation, may have a high importance in riboswitch regulatory mechanisms.
Published in the journal: Comparative Study between Transcriptionally- and Translationally-Acting Adenine Riboswitches Reveals Key Differences in Riboswitch Regulatory Mechanisms. PLoS Genet 7(1): e32767. doi:10.1371/journal.pgen.1001278
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001278Summary
Many bacterial mRNAs are regulated at the transcriptional or translational level by ligand-binding elements called riboswitches. Although they both bind adenine, the adenine riboswitches of Bacillus subtilis and Vibrio vulnificus differ by controlling transcription and translation, respectively. Here, we demonstrate that, beyond the obvious difference in transcriptional and translational modulation, both adenine riboswitches exhibit different ligand binding properties and appear to operate under different regulation regimes (kinetic versus thermodynamic). While the B. subtilis pbuE riboswitch fully depends on co-transcriptional binding of adenine to function, the V. vulnificus add riboswitch can bind to adenine after transcription is completed and still perform translation regulation. Further investigation demonstrates that the rate of transcription is critical for the B. subtilis pbuE riboswitch to perform efficiently, which is in agreement with a co-transcriptional regulation. Our results suggest that the nature of gene regulation control, that is transcription or translation, may have a high importance in riboswitch regulatory mechanisms.
Introduction
For decades, genetic expression has generally been thought to be mostly regulated at the promoter level. Nevertheless, the description of many new mechanisms, such as small regulatory RNAs and ribozymes, clearly indicates that post-transcriptional regulation is as important as transcription initiation [1], [2]. Among the newly characterized mechanisms of post-transcriptional regulation are riboswitches, which are genetic modulators located in untranslated regions of mRNAs. Riboswitches are cellular sensors that modulate gene expression through their ability to alter their conformation in response to cellular changes [3]–[5]. These RNA switches, which have been observed in all kingdoms of life, can regulate transcription, translation, mRNA processing and mRNA splicing [3]. Riboswitches use various factors to control gene expression [3], such as metal ions [6], [7], temperature [8], [9], small metabolites [1], [3], [10]–[12], or uncharged tRNAs [13], [14], and mostly employ structural rearrangement to achieve gene expression regulation [5]. Recently, riboswitches have been found to regulate in trans the expression of the virulence factor PrfA in Listeria monocytogenes [15], suggesting that riboswitches may use an even wider range of regulation mechanisms than previously thought.
Riboswitches are composed of two modular domains consisting of an aptamer and an expression platform. The aptamer is the most conserved domain of the riboswitch and is involved in the binding of a specific cellular metabolite. The second domain, varying widely in sequence and structure, is the expression platform, which modulates gene expression mostly by altering the mRNA structure. Among the smallest riboswitches known to date, the purine-specific class comprises the adenine and the guanine riboswitches which are remarkably similar but exhibit a very high specificity and affinity toward their cognate ligands, adenine and guanine, respectively [16], [17]. Although guanine riboswitches negatively regulate expression by attenuating transcription [18], adenine-specific switches activate expression at the level of transcription [19], and presumably also at the level of translation [20]. For instance, while the Bacillus subtilis (B. subtilis) pbuE adenine riboswitch was predicted to control gene expression by modulating the formation of a transcription attenuator, the Vibrio vulnificus (V. vulnificus) add riboswitch was anticipated to modulate the expression by controlling the formation of a translation sequestrator (Figure 1A and 1B). Notably, previous work suggested that the add and pbuE adenine riboswitches behave differently in their ligand binding properties [20]–[22], suggesting that they possess significant differences in their respective mechanisms.
Fig. 1. pbuE and add adenine riboswitches exhibit different ligand binding properties. 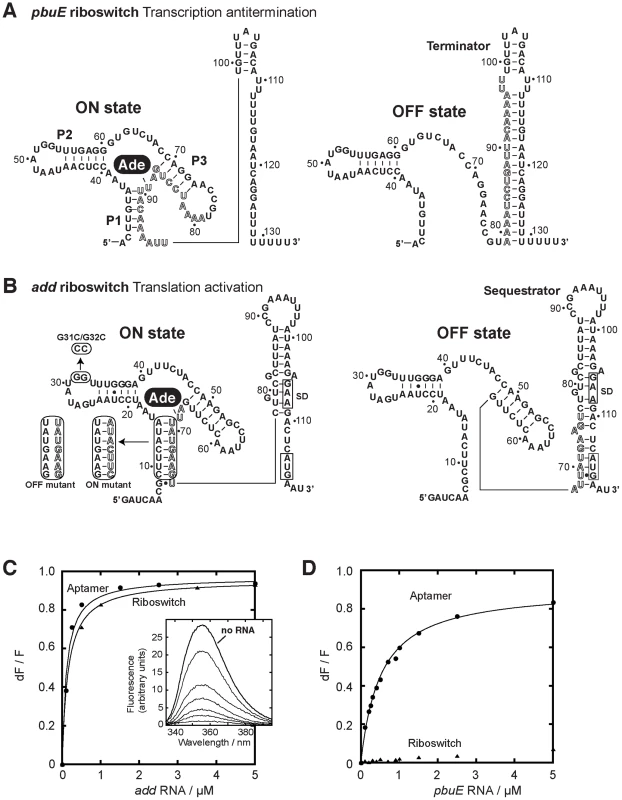
(A) Secondary structures of the pbuE adenine riboswitch associated with the ON and OFF states. The adenine ligand (Ade) is represented by a black oval. The adoption of the OFF state results in the folding of a terminator structure that promotes premature transcription termination while the presence of adenine favors the ON state and antitermination. Outlined letters represent nucleotides that are involved in the formation of both the terminator and the aptamer structures. The nucleotide numbering is derived from the present study. (B) Secondary structures of the add riboswitch associated with the ON and OFF states. The presence of adenine promotes translational activation. The Shine-Dalgarno (SD) and the AUG initiation codon are both boxed. Mutants used in this study are indicated in rounded rectangles. ON and OFF state mutants are indicated. The P1-3′ mutant corresponds to the wild-type P1 stem but in which the 3′ strand was mutated for the sequence corresponding to the ON state mutant. The nucleotide numbering is derived from this study. (C) Normalized 2AP fluorescence intensity plotted as a function of add aptamer (circles) and riboswitch (triangles) molecules. Changes in fluorescence (dF) were normalized to the maximum fluorescence (F) measured in the absence of RNA. Lines show the best fit to a simple binding model, yielding KDapp of 115±15 nM and 156±7 nM for the aptamer and the riboswitch, respectively. The insert shows fluorescence emission spectra for each add aptamer concentration. The indicated line represents 2AP fluorescence in absence of RNA. (D) Normalized 2AP fluorescence intensity plotted as a function of pbuE aptamer (circles) and riboswitch (triangles) molecules. An apparent binding affinity (KDapp) of 518±27 nM was calculated for the pbuE aptamer. No value was determined for the riboswitch due to the absence of significant fluorescence change. Most riboswitch studies are carried out in vitro by using renatured RNA molecules obtained from T7 RNA polymerase (RNAP) transcription systems. In various cases, however, transcription renaturation of RNA molecules is much longer in vitro than in vivo, suggesting that the transcription process dictates the RNA folding pathway and kinetic traps [23]–[27]. Indeed, during transcription elongation, because the upstream RNA section folds first, this will influence the folding pathway of the downstream RNA section [23]. Recently, the transcription process was shown to have an important role for the regulatory activity of an FMN-responsive riboswitch from B. subtilis [28]. In this elegant work, Breaker and coworkers observed that the riboswitch and the FMN ligand do not achieve thermodynamic equilibrium by the time the RNA polymerase reaches the decision point between transcription elongation or termination [28]. This indicates why higher FMN concentrations are required to trigger riboswitch regulation (T50) relative to the dissociation constant (KD). Because this mode of regulation primarily relies on the rates of ligand binding and riboswitch transcription, it was concluded that the riboswitch operates under a kinetic regime. Additional factors such as transcriptional pause sites were also observed to provide more time for the ligand to bind before the genetic decision is made. This is in contrast to a riboswitch operating under a thermodynamic regime in which the time needed to attain an RNA-ligand equilibrium is short compared to the transcriptional time scale and where the KD should be determinant for riboswitch activation. In principle, a “mixed regime” differing from a purely kinetic or thermodynamic regime may occur depending on cellular conditions [21]. For example, in the context of a normally kinetically-driven riboswitch, a change in cellular conditions favoring slower transcription (i.e., low NTP concentrations) could provide more time for the ligand to bind to the aptamer resulting in lower ligand concentrations to trigger riboswitch activity. As a result, this riboswitch would exhibit a more thermodynamic character in its regulation regime and thus the attribution of the riboswitch regulation regime (kinetic vs thermodynamic) should be achieved by taking in account biochemical parameters as well the cellular context. Because there are currently few examples of riboswitch regulation mechanism that have been characterized, it is crucial to examine in details the mechanisms of additional riboswitch representatives.
Herein we report the characterization and the comparison of the regulatory mechanisms of add and pbuE adenine riboswitches. Even though they bind the same ligand, we find that both add and pbuE representatives employ different regulation mechanisms to positively modulate gene expression. Our results with the add riboswitch are consistent with a thermodynamic model in which ligand binding and riboswitch regulation can occur post-transcriptionally, and where transcription-translation coupling is not required for efficient genetic control. In contrast, a kinetic regime is proposed for the pbuE riboswitch that needs to fold and to bind adenine co-transcriptionally to activate transcription. Our results show that the pbuE riboswitch regulation not only depends on the transcription elongation rate and transcriptional pausing but on the NusA elongation factor too. NusA positively affects riboswitch regulation most probably by reducing the transcription rate. Our findings provide the first evidences suggesting that transcriptional and translational riboswitches exhibit mechanistic regulatory differences.
Results
Determination of the transcriptional start sites of add and pbuE riboswitch mRNAs
Previous studies on adenine riboswitches were performed using truncated versions of either the aptamer or the riboswitch domains, due to lack of information on promoter locations. Because this could lead to biased results, we determined the transcriptional +1 by performing primer extension analyses for both the add and the pbuE transcripts. Total RNAs were extracted either from V. vulnificus or B. subtilis and the +1 transcription start sites were determined for add and pbuE mRNAs, respectively (Figure S1A and S1B). The deduced RNA sequences differ from previously used truncated versions and new numbering nomenclature taking into account these variations were thus employed (Figure 1A and 1B). The newly determined transcription start site of the pbuE riboswitch differs from a previous report, which may result from different cellular genetic background [29].
Adenine riboswitches show distinct ligand binding properties
The fluorescent nucleobase 2-aminopurine (2AP) is strongly quenched when stacked upon adjacent nucleotides indicating changes in its immediate environment [20]–[22], [30]–[37]. The adenine riboswitch recognizes both 2AP and adenine in a similar manner, as previously shown using in-line probing and ß-galactosidase assays [22]. To investigate the ligand binding activity of the add and pbuE riboswitches, we took advantage of 2AP fluorescence to monitor the RNA-ligand interaction occurring in both riboswitches, using either the aptamer or the complete riboswitch (aptamer and platform). The fluorescence intensity of 2AP (at 50 nM) was first measured as a function of increasing concentrations of the add aptamer (from 0 to 5 µM). As shown in Figure 1C (insert), the 2AP fluorescence signal progressively decreased until near total quenching at 5 µM aptamer. The fluorescence data were well-fitted by a simple two-state binding model and an apparent dissociation constant KDapp of 115±15 nM was obtained. Furthermore, when the same experiment was repeated using the complete riboswitch sequence (Figure 1C), a very similar KDapp of 156±7 nM was obtained. This indicates that, for add, both the aptamer and the riboswitch bind equally well to the ligand, which is consistent with previous results obtained using truncated RNA molecules [20], [30].
The ligand binding activity of the pbuE adenine riboswitch was also monitored using 2AP assays (Figure 1D). Upon titrating the pbuE aptamer, a very efficient 2AP binding similar to that of add was observed (KDapp of 518±27 nM). In contrast to the aptamer, the pbuE riboswitch had very little effect on 2AP fluorescence, indicating that the expression platform has a negative influence on the ligand binding activity. These results are in agreement with previous fluorescence and probing data obtained using truncated riboswitch molecules [20]–[22], indicating that the natural pbuE adenine riboswitch inefficiently binds the ligand in vitro.
The add riboswitch exhibits structural changes upon ligand binding
Our data indicate that the add and pbuE riboswitches do not share similar ligand binding properties (Figure 1C and 1D). Indeed, in contrast to the pbuE riboswitch, both the add aptamer and riboswitch sequences perform ligand binding in vitro, suggesting that the unbound add riboswitch is competent to bind adenine and to fold upon ligand binding (Figure 2A). However, an alternative explanation is that the in vitro add structure is intrinsically adopting the ON structure even in absence of the ligand, which would favor constitutive ligand binding. To investigate whether the add riboswitch acts as a reversible switch and undergoes secondary structure rearrangement upon ligand binding, we used selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) to provide information about the folding of the riboswitch [38]. This technique is particularly useful to discriminate local nucleotide flexibility, where 2′OH groups are more reactive in flexible regions to electrophiles like N-methylisatoic anhydride (NMIA). When subjected to NMIA reaction in absence of adenine, the add riboswitch showed modifications throughout the entire riboswitch sequence where various single-stranded regions were reactive (Figure S2). Upon addition of adenine, clear changes were observed both in the aptamer and in the expression platform, where the Shine Dalgarno (SD) and AUG start codon were more reactive to NMIA (Figure 2B and Figure S2). Thus, these changes are consistent with the ligand-dependent increased accessibility of both the SD and the AUG codon sequences, which is required for the translation activation to take place.
Fig. 2. The add riboswitch secondary structure is modulated upon ligand binding. 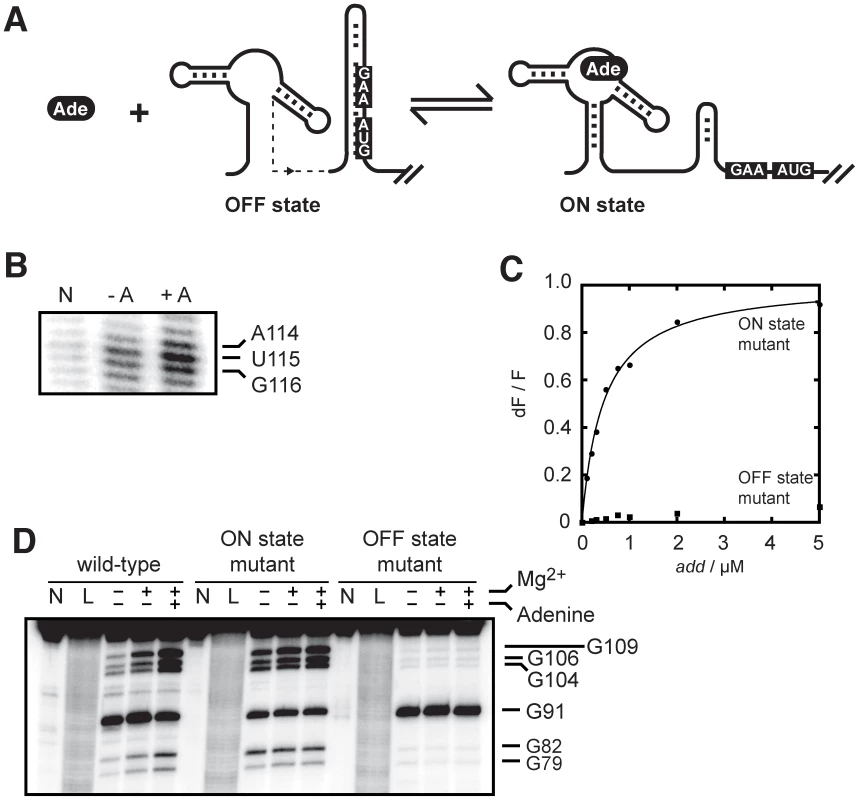
(A) Schematic showing control of gene expression by the add riboswitch upon adenine (Ade) binding. The accessibility of both the Shine-Dalgarno (GAA) and AUG sequences is increased in the ON state. (B) SHAPE modifications of the add riboswitch as a function of adenine concentration. The lane N denotes a reaction in which NMIA was omitted. SHAPE reactions were performed in absence (−A) and in presence of 10 µM adenine (+A). The AUG sequence showing increased accessibility in presence of adenine is indicated on the right. The complete gel is shown in Figure S2. (C) Normalized 2AP fluorescence intensity plotted as a function of add riboswitch ON (circles) and OFF (squares) state mutants. The sequence changes are shown in Figure 1B. An apparent binding affinity (KDapp) of 458±33 nM was obtained for the ON state mutant. No value was determined for the OFF state mutant due to the absence of significant fluorescence change. (D) RNase T1 partial cleavage of the add riboswitch showing the structural change of the expression platform in presence of adenine. Digestions were also done for the ON and OFF state mutants. Lanes N and L represent samples that were not reacted and that were subjected to partial alkaline digestion, respectively. Nuclease digestions were performed as a function of 10 mM magnesium ions and 10 µM adenine. Substantial cleavage sites are indicated on the right. The complete gel is shown in Figure S3. Because ligand binding reorganizes the riboswitch secondary structure, we speculated that the riboswitch conformation would be important for ligand binding. To test this, we introduced mutations in add to favor either the OFF or ON state (Figure 1B) and monitored the 2AP binding activity (Figure 2C). When we interconverted the 5′ and 3′ P1 stem sequences to prevent the formation of the sequestering stem while still allowing P1 stem formation (ON state mutant), we measured a 2AP binding activity (KDapp of 458±33 nM) very similar to the value obtained for the wild-type add riboswitch. However, when the 5′ sequence of the P1 stem was mutated to prevent P1 formation, thus making an OFF state conformer, very little binding activity was observed (Figure 2C). Because the mutated sequences are not directly involved in ligand binding, these results demonstrate that the add riboswitch binding is dependent on the adoption of the ON state. The ligand-dependent structural change of the add riboswitch was further characterized using a partial nuclease digestion assay using the single-stranded guanine-specific ribonuclease T1 (Figure 2D and Figure S3). A partial RNase T1 cleavage assay was first performed on the natural add riboswitch as a function of magnesium ions and adenine, where the increased exposition of the SD sequence (G104–G109 region) could be observed as a function of both magnesium ions and adenine (Figure 2D). Nuclease reaction sites were also determined for the complete riboswitch sequence which agreed well with previously obtained data (Figure S3) [22]. The comparison of the cleavage pattern between the wild-type and the two mutants confirmed that the wild-type riboswitch can readily switch from the OFF to the ON state upon adenine binding, consistent with our SHAPE data (Figure 2B). Together, our results show that the add riboswitch binds adenine in vitro and undergoes structural changes that are consistent with the SD and the start codon sequences being more accessible in the ligand-bound state.
The add riboswitch performs translational control in vitro
Our data demonstrate that the add riboswitch changes conformation in vitro upon ligand binding. To further investigate the riboswitch gene regulation mechanism, we developed an in vitro translation assay using different constructs. We hypothesized that if add undergoes structural changes upon ligand binding (Figure 2B and 2D), it should be able to efficiently control translation initiation as a function of adenine. In bacteria, the processes of transcription and translation are coupled events so that most bacterial genes initiate translation soon after the SD sequence has been transcribed. Because the add riboswitch regulates gene expression at the translational level, we investigated whether the transcription-translation coupling was important in the regulation control. We thus developed an in vitro translation assay where the coupling between transcription and translation is either allowed or disrupted by using a DNA or mRNA template, respectively. When performing in vitro translation assays where transcription and translation are coupled, the presence of adenine increased the level of synthesized protein by 2-fold after 15 minutes (Figure 3A, lower panel). In addition, when the add riboswitch was transcribed before translation (Figure 3A, upper panel), such that transcription and translation are uncoupled, the addition of adenine increased by 3-fold the expression of the protein. We then used both ON and OFF riboswitch mutants to demonstrate that the riboswitch conformations are responsible for the adenine-dependent modulation of translation. When we disrupted the P1 stem of the riboswitch (OFF state mutant), the level of synthesized protein became barely detectable (Figure 3B). However, when we interchanged both strands of P1 to prevent the formation of the SD sequestering stem (ON state mutant), the translation was constitutively activated (Figure 3B), and this independently of adenine. Taken together, these results indicate that the add riboswitch controls translation through conformational changes and that the coupling between transcription and translation is not required to efficiently perform gene regulation.
Fig. 3. The add riboswitch positively regulates gene expression in vitro and in vivo. 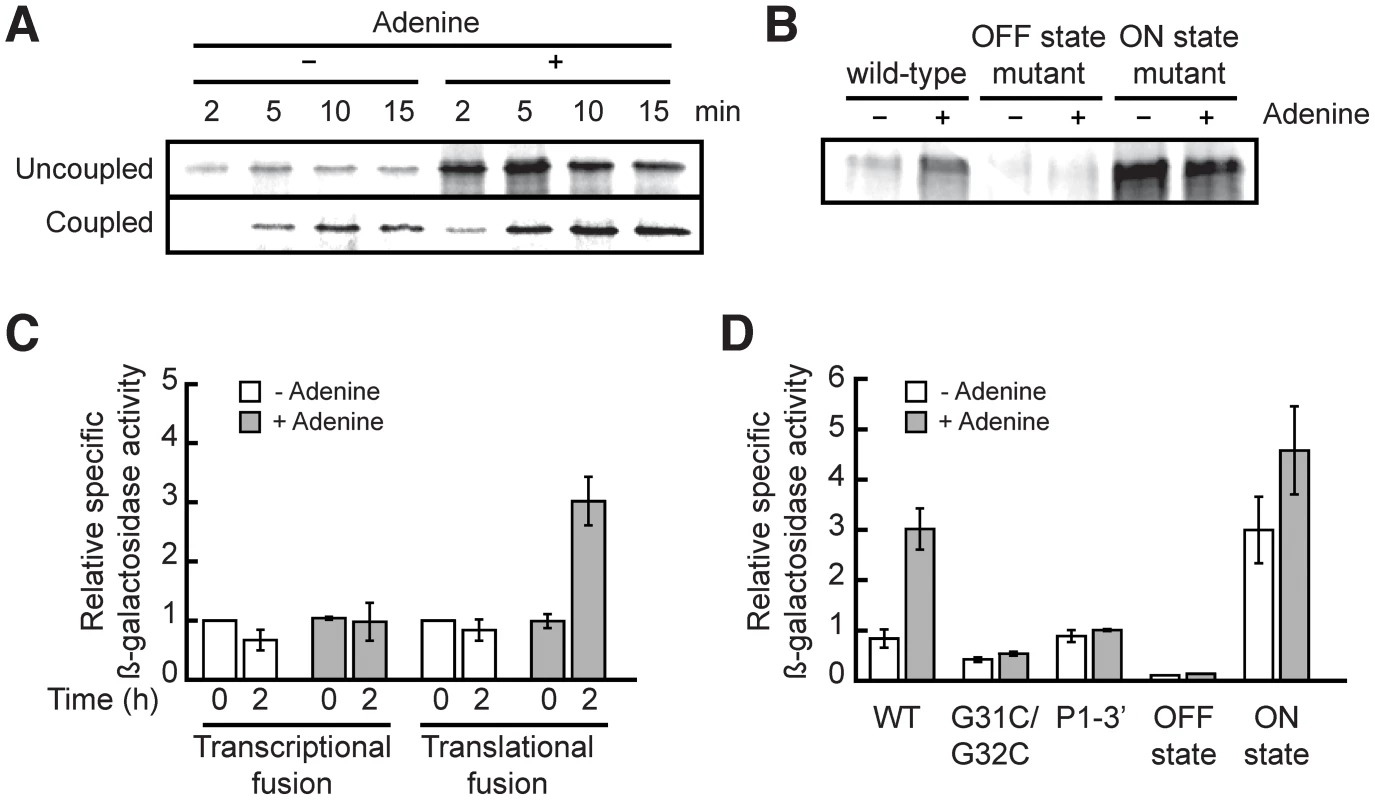
(A) In vitro translation assays of the add riboswitch using pre-transcribed mRNA (uncoupled) or DNA (coupled) as template. (B) In vitro translation assays of different mutations of the add riboswitch using pre-transcribed mRNA as template. (C) In vivo β-galactosidase assays with transcriptional and translational fusions of the add riboswitch to the lacZ gene. (D) In vivo β-galactosidase assays with mutants of the add'-'lacZ translational fusion. In all β-galactosidase assays, adenine was added at an OD600 of 0.3 (time = 0) to a final concentration of 500 µM. The add riboswitch performs translational control in vivo
Although the add riboswitch has been characterized in vitro, no in vivo data are available to assess the riboswitch regulation mechanism. To address this, we engineered transcriptional and translational constructs of the add riboswitch fused to the reporter gene lacZ in Escherichia coli (E. coli). Using primer extension assays, we confirmed that the transcription start site of our constructs in E. coli is identical to that of V. vulnificus (Figure S1).
We tested our constructs by growing cells containing wild-type riboswitches in minimal medium in absence and in presence of adenine. As seen in Figure 3C, the addition of adenine had no significant effect on the transcriptional fusion, which indicates that the transcript level was not affected by adenine. In contrast, the β-galactosidase activity of the translational fusion was increased by 3-fold by the addition of adenine, in agreement with the adenine-dependent translational activation mechanism of the add riboswitch. We also determined that the optimal concentration of adenine needed for translation activation is 500 µM (data not shown). Because the intracellular adenine concentration in bacteria is around 1.5 µM [39], this suggests that adenine does not penetrate the cell freely, or is rapidly used by the metabolism to reach homeostasis.
Additionally, we generated constructs of the add riboswitch translational fusion to confirm in vivo our fluorescence and probing data obtained in vitro. We first mutated two nucleotides in the loop L2 (G31C/G32C, Figure 1B) to prevent the formation of the loop-loop interaction. This interaction is critical for the folding of the riboswitch in presence of adenine [22]. As shown in Figure 3D, this mutation makes the riboswitch non-responsive to adenine. We then mutated both strands of the P1 stem independently or additionally to shift the equilibrium toward ON or OFF states. For instance, when the 5′ strand of the P1 stem was mutated to prevent P1 stem formation (OFF state mutant), all activity of the translational fusion was lost, suggesting that the sequestering stem is formed constitutively in absence of P1. On the other hand, when we mutated the 3′ strand of P1 to disrupt both the P1 and sequestering stems (P1-3′ mutant), we reestablished the same basal level than in the wild-type fusion, but lost the adenine effect on translation. Finally, by swapping the sequence of the two strands of P1 to favor the formation of the ON state mutant, we observed a constitutively active translation suggesting that the presence of the P1 stem correlates with translation activation. The results obtained with these mutants confirm that the add riboswitch controls translation in vivo by sequestering the SD region specifically under low adenine concentration. Overall, although riboswitch-driven genetic repression was previously demonstrated for several riboswitches [4], [18], [40]–[45], our study provides novel insights about ligand-dependent translation activation in vivo occurring through the action of a riboswitch.
The transcription process is important for the pbuE riboswitch ligand binding activity
Although most riboswitches can bind their ligand in vitro, we and others have reported that the pbuE riboswitch exhibits very poor adenine binding activity (Figure 1D) [20]–[22]. Nevertheless, the pbuE adenine riboswitch can modulate the expression of a lacZ reporter gene [19]. Thus, we speculated that the in vivo transcriptional context might be essential for the ligand binding activity of the pbuE riboswitch. To investigate this, we developed an in vitro assay where full-length transcription depends on the binding of the ligand. However, in the absence of the ligand, a prematurely terminated transcript should be produced. Since 2,6-diaminopurine (DAP) has been recently crystallized in complex with an adenine riboswitch aptamer [32], showing a very similar structure compared to the adenine:aptamer complex [17], we used DAP which exhibits ∼30-fold higher affinity compared to adenine [19]. The assay was performed using single-round transcription reactions [28], [46], which were carried out by using B. subtilis RNA polymerase (RNAP) in presence of increasing concentrations of the ligand. As shown in Figure 4A, a significant increase of readthrough transcripts was observed as a function of DAP concentration, which occurred concomitantly with the reduction of the prematurely terminated transcript. The fraction of readthrough transcript for each reaction was calculated and the DAP concentration required to obtain half of the change in transcription elongation, defined as T50 [28], was determined to be 2.1±0.2 µM (Figure 4A). These results suggest that the pbuE adenine riboswitch requires a transcriptional context to efficiently bind DAP.
Fig. 4. The pbuE riboswitch requires a transcriptional context to bind adenine. 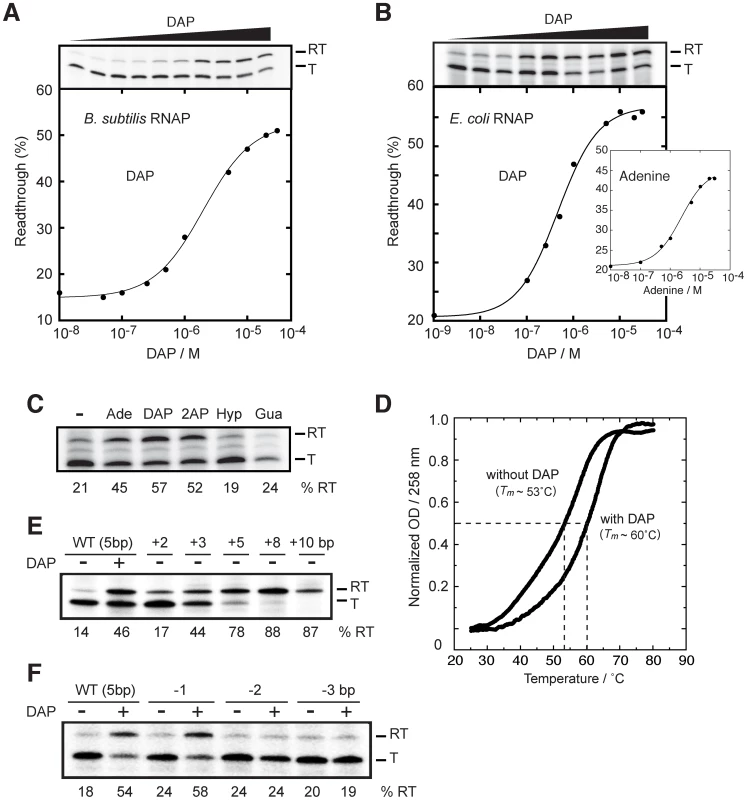
(A) Single-round in vitro transcriptions of the pbuE riboswitch using the B. subtilis RNA polymerase (RNAP). Top, transcription reactions performed as a function of 2,6-diaminopurine (DAP). Readthrough (RT) and terminated (T) products are indicated on the right. Bottom, percentage of readthrough products plotted as a function of DAP concentration. The line shows a two-state model from which a T50 value of 2.1±0.2 µM DAP was calculated. (B) Single-round in vitro transcriptions of the pbuE riboswitch using the E. coli RNAP. Top, transcription reactions performed as a function of DAP. Bottom, percentage of readthrough products plotted as a function of DAP concentration. A T50 value of 0.5±0.1 µM was obtained from the fitting. The insert shows transcription reactions performed as a function of adenine concentration. A T50 value of 2.3±0.3 µM was calculated for adenine. (C) Single-round in vitro transcriptions performed in presence of various ligands. Reactions were performed in absence (−) of ligand and in presence of 10 µM adenine (Ade), 2,6-diaminopurine (DAP), 2-aminopurine (2AP), hypoxanthine (Hyp) and guanine (Gua). Percentages of readthrough (RT) products are indicated below the gel. (D) Thermal denaturation of the pbuE aptamer in absence and in presence of 10 µM DAP. Melting temperatures (Tm) were evaluated by determining the temperature required to obtain half of the melting transition (normalized OD = 0.5) [79], [80]. (E) Single-round in vitro transcriptions performed as a function of the P1 stem elongation. The P1 stem was elongated by 2, 3, 5, 8 and 10 bp and resulting constructs were assayed using single-round in vitro transcriptions. The wild-type pbuE riboswitch contains a P1 stem of 5 bp. (F) Single-round in vitro transcriptions performed as a function of P1 stem destabilization. The P1 stem was shortened by 1, 2 and 3 bp and resulting constructs were assayed using single-round in vitro conditions. Next, to establish whether the riboswitch activity is polymerase dependent, we repeated the experiment using the E. coli RNAP and obtained a T50 value of 0.5±0.1 µM. Moreover, when we substituted DAP for the natural ligand adenine, we observed a transcription modulation that was characterized by a higher value of T50 (2.3±0.3 µM), consistent with the lower affinity of adenine for the riboswitch aptamer (Figure 4B, insert) [19]. No ligand-dependent transcription modulation was observed when using the bacteriophage T7 RNA polymerase (data not shown), suggesting that specific elements to bacterial polymerase (e.g., pause sites) may be important for riboswitch activity. Together, these results show that the pbuE riboswitch depends on transcription to perform ligand binding.
Transcription elongation depends on adenine-related ligands and requires aptamer stabilization
It has been previously shown that the adenine aptamer exhibits efficient ligand binding in presence of adenine, 2AP and DAP, but not with guanine-related compounds [19]. In presence of 10 µM ligand, efficient transcription readthrough was observed for adenine, 2AP and DAP, the latter resulting in the highest transcription readthrough (57%, as shown in Figure 4C). However, hypoxanthine and guanine failed to support transcription readthrough, also consistent with in-line probing data, showing their inefficiency to bind the adenine riboswitch aptamer domain [19]. Our results indicate that transcription readthrough is only observed in presence of ligands known to bind the adenine riboswitch aptamer, suggesting that transcription elongation is achieved via a riboswitch-mediated control mechanism. The inability of the pbuE riboswitch to efficiently bind adenine post-transcriptionally was suggested to result from the formation of a highly stable terminator stem (Figure 1D) [20]–[22]. Accordingly, we speculated that the binding of the ligand to the aptamer domain stabilizes the aptamer structure and prevents formation of the terminator. We thus carried out thermal denaturation experiments (TDE) of the pbuE aptamer to determine whether ligand-binding induces aptamer stabilization. TDE monitors the heat-induced unfolding of the RNA as a function of temperature by observing absorbance changes [47]. Using TDE, we followed the absorption of the aptamer at 258 nm in absence and in presence of DAP (Figure 4D). After normalizing the data, melting temperatures corresponding to half change in absorbance were determined to be ∼53°C and ∼60°C in absence and presence of the ligand, respectively. Thus, our results are consistent with the idea that ligand binding to the RNA promotes aptamer stabilization, which is in agreement with previous studies on the pbuE aptamer using optical-trapping assays [48]. The aptamer stabilization is central for the disruption of the highly stable terminator stem and for the transcription elongation.
Since our results show that ligand binding promotes pbuE transcription elongation and thermal stability of the riboswitch aptamer (Figure 4C and 4D), we carried out in vitro transcription experiments to determine to which extent aptamer stabilization is important for the riboswitch regulation (Figure 4E). To do so, we modulated the stability of the P1 stem that is directly involved in the switching mechanism (Figure 1A). By altering the sequence located to the 5′ side of the P1 stem as a way to extend the P1 stem by 2, 3 and 5 bp, we determined transcription readthrough efficiencies of 17%, 44% and 78%, respectively (Figure 4E). These results indicate that transcription elongation can be modulated solely by altering the stability of the P1 stem, suggesting that aptamer-ligand interactions are not strictly required for transcription elongation. Even higher readthrough efficiencies were observed when the P1 stem was extended by 8 and 10 bp, suggesting that the ON conformer was further stabilized (Figure 4E).
The transcriptional control of the pbuE riboswitch was also studied by removing base pairs from the P1 stem (Figure 4F). By destabilizing the latter, it is predicted that the OFF conformer is favored, which should inhibit the ligand-induced production of the readthrough transcript. While the removal of 1 bp did not significantly alter the transcription control, the removal of 2 or 3 bp completely abolished the ligand-induced response (Figure 4F), suggesting that a P1 stem of at least 4 bp is required for efficient riboswitch regulation. These results correlate well with our previous study showing that aptamers with reduced P1 stem do not exhibit efficient ligand binding activity [30].
The pbuE riboswitch regulation is consistent with a kinetic regime
It has been previously hypothesized that the pbuE adenine riboswitch is driven by a kinetic regulation mechanism, in which not only ligand binding but also RNAP transcription rates are important to establish the riboswitch activity [21]. Under this control regime, it is expected that ligand binding is highly dependent on a “temporal window” defined by the RNAP sequence position. For instance, while RNAP must have transcribed the aptamer region for ligand binding, the presence of the downstream terminator domain strongly precludes this binding [20]–[22]. Thus, it is expected that further transcription of the terminator sequence should reduce ligand binding. If this is true, high transcription rates should reduce ligand binding, and inversely, low transcription rates should improve ligand binding. To determine whether the pbuE riboswitch operates under such a kinetic regime, we conducted single-round transcription assays in presence of various rNTP concentrations and monitored transcriptional control using a range of DAP concentrations. When analyzing transcription reactions performed using rNTP concentrations of 20 and 150 µM, we calculated T50 values of 0.8±0.1 µM and 1.9±0.3 µM, respectively (Figure 5A), showing that the rNTP concentration is proportional to the ligand concentration (T50) required to trigger riboswitch activity. In addition, a ∼2-fold decrease in the transcription readthrough was observed in presence of 20 µM rNTP, consistent with the influence of the latter on transcription termination [49]. We observed no difference for experiments performed at 65 µM or 150 µM rNTP, most probably because transcription rates are optimal at 65 µM. However, clear differences in T50 were observed when using the E. coli RNAP, which is consistent with transcription elongation being modulated by altering rNTP concentrations (Figure S4). Thus, our results support the idea that RNAP transcription rates can influence riboswitch activity, in agreement with a kinetically-driven regulation regime as determined for the FMN riboswitch [28].
Fig. 5. The rate of transcription is important for the pbuE riboswitch activity. 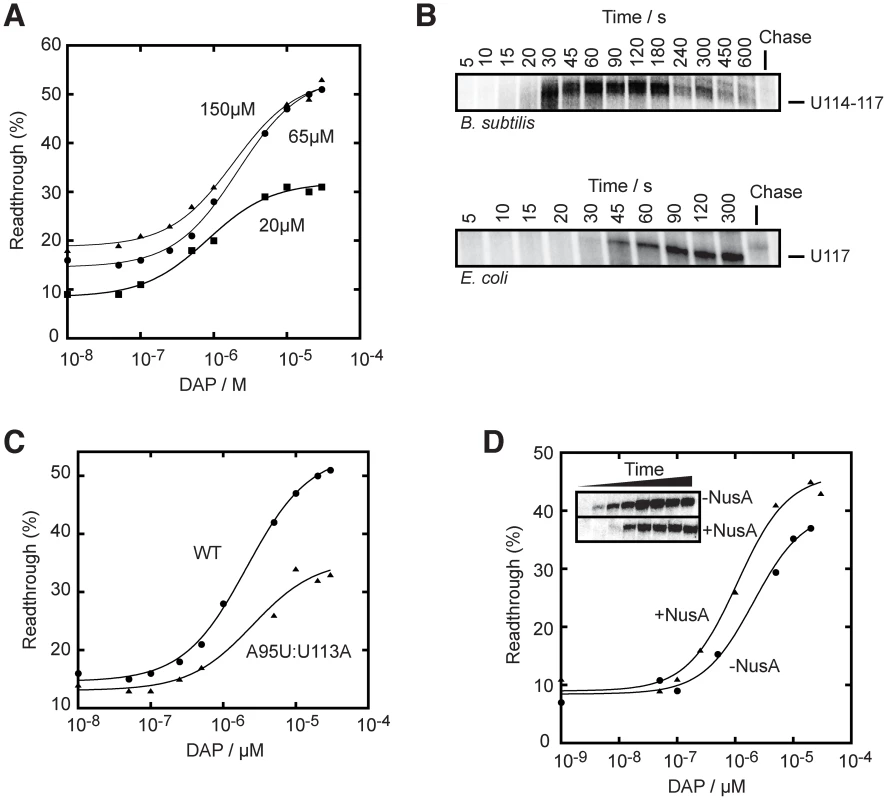
(A) Single-round in vitro transcriptions were performed using either 20 µM (squares), 65 µM (circles) or 150 µM (triangles) rNTP. T50 values of 0.8±0.1 µM, 2.1±0.2 µM and 1.9±0.3 µM were obtained for reactions using 20 µM, 65 µM and 150 µM rNTP, respectively. (B) Single-round transcription kinetics were performed and stopped at various time intervals. The complete gel is shown in Figure S5. Top, transcription time course showing a pausing site in the region U114–U117 that is observed when using the B. subtilis RNAP. Bottom, transcription time course performed with the E. coli RNAP also reveals a pausing site at position U117. Chase reactions were performed using 150 µM rNTP. (C) Single-round transcriptions using an A95U:U113A mutant that disrupts the riboswitch regulation. A T50 value of 2.6±0.1 µM was calculated. The readthrough efficiency is also significantly decreased in the context of the mutant (triangles). (D) Single-round experiments were performed in absence (circles) and in presence (triangles) of 1 µM NusA. T50 values of 2.1±0.7 µM and 1.1±0.3 µM were obtained in absence and in presence of NusA, respectively. The insert shows transcription kinetics done for 5, 10, 15, 20, 30, 45, 60 and 300 s in absence and in presence of 1 µM NusA. It can be observed that the presence of NusA delays the appearance of the full length species (15 s to 20 s), consistent with its role in slowing down transcription. Transcriptional pausing has been previously shown to be important in the transcriptional folding of ribozymes [23]–[25], and also for riboswitch activity [28], [50]. To determine potential transcriptional pause sites on the pbuE riboswitch, we performed transcription time courses (Figure 5B and Figure S5). We observed prominent transcriptional intermediates paused in the region U114–U117, which largely disappeared over incubation time and chase reaction (Figure 5B, upper panel, and Figure S5). Our data show a pause lifetime of ∼60 s (see Materials and Methods), which is similar to what has been found for the FMN riboswitch [28]. Notably, we observed a very similar pause site (U117) and half-life when using E. coli RNAP (Figure 5B, lower panel). The identified pause site corroborates a region (U110–U115) that was previously speculated to be part of a pause site [21]. To further investigate this pause site, we introduced mutations in the terminator domain that did not alter the base pairing potential of the stem but that modified the pause site sequence (Figure 5C). When performing in vitro transcription using the A95U:U113A mutant, we found that a higher ligand concentration was required to activate the pbuE riboswitch (T50 = 2.6±0.1 µM) and that the lifetime was decreased to ∼22 s. These results are consistent with the hypothesis that the disruption of the pause site decreases the time for the ligand to bind to the riboswitch, which resulted in a greater ligand concentration to promote riboswitch activity. In addition, the extent of transcription elongation was also decreased by a factor of ∼2-fold, consistent with a faster transcription rate reducing ligand binding and transcription elongation (Figure 5C).
The transcription elongation factor NusA positively affects pbuE riboswitch regulation
The transcription factor NusA is an RNA-binding protein known to modulate termination by increasing the E. coli RNAP pausing time and to reduce the rate of transcription [23], [51]. It has also been shown to assist the FMN riboswitch activity by reducing the transcription rate [28]. To verify if NusA could affect the pbuE riboswitch regulation mechanism, we performed in vitro transcription kinetics in absence and in presence of the B. subtilis NusA and observed a significant decrease in transcription rate in presence of NusA (∼15 s difference for significant full length formation, see Figure 5D, insert). When performing transcription reactions as a function of DAP concentrations, the ligand requirement was decreased in presence of NusA where values of T50 of 2.1±0.7 µM and 1.1±0.3 µM were obtained in absence and in presence of NusA, respectively (Figure 5D). No significant change of lifetime at pause sites was observed in presence of NusA. This suggests that, at least in our experimental conditions, NusA modulates riboswitch activity by decreasing the general transcription reaction [23].
Discussion
Our study on the regulation mechanisms of adenine riboswitches has demonstrated two major findings. First, we show that although the transcriptionally - and translationally-acting pbuE and add adenine riboswitches recognize the same ligand, they use different regulatory mechanisms to modulate gene expression. Second, we provide biochemical evidence about both the structural reversibility and the non-requirement of a transcription-translation coupling for the translationally-acting add riboswitch, all of which are consistent with a thermodynamic control. This regulation mechanism is in contrast to what we have observed for the transcriptionally-regulating pbuE riboswitch, which performs co-transcriptional binding and exhibits similar features to those found in kinetically-controlled riboswitches.
The translationally-regulating add riboswitch is a true riboswitch regulator
Our analysis of the add riboswitch confirms and extends prior reports suggesting that add undergoes structural changes to control translation initiation [20], [52]. However, a very different view emerges for the add riboswitch regulation regime when compared to the pbuE riboswitch for which our results suggest a kinetic regime. For instance, 2AP fluorescence showed that the presence of the add expression platform does not inhibit ligand binding (Figure 1C). Also, structural probing and mutagenesis studies of the add riboswitch revealed that the Shine-Dalgarno and the AUG codon sequences are more accessible in presence of adenine (Figure 2). These results are consistent with add modulating translation initiation in a ligand-dependent manner, which has been observed both in vitro and in vivo (Figure 3). Moreover, given that adenine can bind to the add riboswitch post-transcriptionally, it suggests that, unlike the pbuE variant, add is a reversible switch that can adopt either the OFF or ON structure at the equilibrium. A secondary structure analysis performed using the program mfold predicts similar free energies of −23.8 kcal/mol and −23.1 kcal/mol for the OFF and ON structures, respectively. This supports the idea that the add riboswitch can fluctuate readily between the ON and OFF states when compared to pbuE [22]. The structural reversibility of add is consistent with the riboswitch activity not requiring a coupling between transcription and translation. Indeed, both the structural reversibility and the absence of coupling for riboswitch regulation suggest that add may benefit of an extended time compared to pbuE because ligand binding can occur post-transcriptionally (after the riboswitch portion of the mRNA is transcribed). For the add riboswitch to operate under a purely dictating thermodynamic regime, it would require that the T50 value approximates the KD of the riboswitch-ligand complex. It may be difficult to ascertain whether this is the case as it would demand to determine the KD of the riboswitch-adenine complex in vivo. However, the quantitative determination of metabolite concentration in vivo is becoming increasingly widely applied to map relative concentration changes induced by environmental alterations [39]. Nevertheless, our results are consistent with add operating under a thermodynamic regime that may exhibit a mixed character depending of various cellular conditions, such as the concentration of adenine [21]. The intracellular ligand concentrations of kinetically-controlled riboswitches are typically significantly higher than KD values of corresponding riboswitch-ligand complexes. As observed for the pbuE adenine riboswitch, the adenine concentration in B. subtilis was found to be ∼30 µM [53]. Although the adenine concentration is not known in V. vulnificus, it is possible to provide an estimation about its intracellular concentration (∼1.5 µM) if we consider that a similar adenine concentration exists in both gammaproteobacteria V. vulnificus and E. coli [39]. In such a case, the add riboswitch could be in presence of adenine concentrations in the low micromolar range which would be closer to the determined KD values [20], [21], [30], [32], providing an additional indication about the thermodynamic character of the add riboswitch regulation regime. Together with these observations, the structural reversibility of the riboswitch and the absence of coupling requirement for the transcription-translation suggest that the add riboswitch regulates translation initiation using a thermodynamic regime that may be modulated depending on cellular conditions. Other translationally-regulating riboswitches responding to S-adenosylmethionine (SAM), adenosylcobalamin, thiamine pyrophosphate (TPP) and riboflavin have also been shown to exhibit ligand binding, structural rearrangement, and ribosome binding in vitro [44], [45], [54]–[59]. Whether all translationally-regulating ligand-binding riboswitches need to benefit from a thermodynamic regime to regulate gene expression will require further investigation.
Additional findings provided indications that other cellular players could be involved in bacterial translation initiation control. Recently, Burmann et al. have observed using NMR spectroscopy that the transcription factor NusG can associate alternatively with NusE or Rho [60]. Interestingly, because NusG contacts RNAP and that NusE is identical to the ribosomal protein S10, it was concluded that NusG may act as a functional link between transcription and translation. In a cellular context where translation initiation is inhibited, NusG is expected to be available to interact with Rho, which stimulates a Rho-dependent transcription termination. However, in conditions promoting translation initiation, the formation of a NusG-NusE complex should prevent Rho binding and transcription antitermination is predicted to take place. The molecular details about these mechanisms and to which extent they are linked to riboswitch translation initiation regulation will be a fertile subject for future research.
The pbuE riboswitch operates under a kinetic regime
In contrast to what we have observed for add, the pbuE aptamer shows a remarkable inefficiency to bind 2AP in presence of the expression platform (Figure 1D) [20]–[22]. However, in vitro ligand binding is attained when occurring in a transcriptional context (Figure 4A and 4B). While not as drastic as our observations with pbuE, ligand-binding inhibitory effects have often been observed for transcriptionally-regulating riboswitches when in presence of their expression platform, suggesting that the transcriptional context is critical for the ligand binding activity of some riboswitches [22], [28], [40], [41]. It is likely that the additional sequence downstream of the aptamer allows structures that are incompatible with ligand binding. Accordingly, the presence of Rho-independent terminators, which are GC-rich helical domains, may disrupt ligand-binding structures as seen for the pbuE adenine riboswitch [20]–[22]. Moreover, in the case of negative regulation such as the FMN riboswitch, the antiterminator domain must be very stable as it competes with both the terminator structure and the aptamer domain. However, during transcription, because the aptamer domain is synthesized before the terminator, it may fold without competing with the terminator, which allows ligand binding. This is indeed what is observed in the cases of pbuE and ribD riboswitches. Because riboswitch folding is strongly influenced by the transcriptional process, co-transcriptional ligand binding is critical for the “genetic decision” to take place. More work will be required to draw general rules about riboswitch transcription regulation mechanisms and will very likely reveal additional factors allowing greater flexibility in riboswitch genetic regulation.
Recently, ligand binding parameters were studied for riboswitches responding to c-di-GMP and preQ1 where additional regulation strategies were characterized [61]–[63]. For instance, the transcriptionally-regulating PreQ1 riboswitch was found to exhibit two different coexisting stem-loop structures in the expression platform [61]. Upon PreQ1 binding to the riboswitch, it was observed that the equilibrium of the competing hairpins becomes significantly altered. By studying the riboswitch mechanism, the authors provided a model for how a riboswitch presenting no obvious overlap between aptamer and terminator domains may regulate gene expression by employing bistable sequence elements [61]. In addition, the structural basis of ligand binding by a c-di-GMP riboswitch was obtained [62], [63]. It was found that the affinity of the complex is very strong, exhibiting a KD of ∼10 pM [63]. When comparing to the adenine and FMN riboswitches, it was observed that although the on-rate of c-di-GMP is similar to that of both riboswitches, the off-rate is significantly slower by ∼5 orders of magnitude [63]. Thus, it is expected that the complex has a very slow approach to equilibrium so that ligand binding is effectively irreversible on a biological time scale, consistent with the riboswitch operating under a kinetic regulation regime. Thus, together with previously characterized riboswitches, these findings show that riboswitches may use various regulation strategies to achieve gene modulation.
In the present study, our findings are consistent with pbuE operating under a kinetic regime where the rate of ligand binding, more than the dissociation constant itself, is crucial in the decision of the transcription outcome. Our results corroborate with previous findings showing that fine-tuning of the transcription elongation is central not only for riboswitch regulation, but for RNA folding in general. As such, transcription is central for the Tetrahymena group I intron folding and splicing and for the bacterial RNase P RNA catalytic activity [64]. Because of the uneven elongation rate of the RNAP, which is modulated by several factors such as pause sites and transcription factors, many RNA structures will accumulate even though they are not the most optimally stable. For instance, pause sites in riboswitches play important roles in the case of the FMN riboswitch [28], and more recently for a pH-responsive riboregulator [50]. In both cases, the presence of the NusA transcription factor helps to decrease the polymerase rate, which increases the decision time frame. Additionally, both the pbuE adenine and FMN riboswitches demonstrate similarities as they carry a pause site in a U-rich sequence that is important for the riboswitch regulation (Figure 5B and 5C). Even though pbuE and FMN riboswitches possess a pause site, NusA does not appear to increase RNAP pausing. Instead, NusA generally reduces the rate of transcription, as previously observed in the case of the RNase P [23]. In principle, transcriptional pausing can be affected by cellular conditions to yield longer pause lifetimes. Because of this, lower adenine concentrations may be required to bind the aptamer and to trigger gene regulation. Thus, because changes in cellular conditions are inherently involved in the riboswitch regulation regime, it is likely that the pbuE riboswitch regime may exhibit a more thermodynamic character depending upon transcription time scale, rate of ligand binding and protein factors involved at the transcriptional level [21].
General considerations for riboswitch regulation mechanisms
While they bind to the same ligand, transcriptionally - and translationally-regulating adenine riboswitches employ regulatory mechanisms that differ in several aspects (Figure 6). For instance, although the add riboswitch could in principle bind adenine only during transcription, and could require coupling between transcription and translation, it appears as we have found here that this is not the case (Figure 3A). To occur after transcription, ligand binding requires similar free energies for the adoption of both ON and OFF state structures, and may thus impose an additional selection pressure on the riboswitch. Similar free energies for both structures also suggest that such regulatory systems are more prone to “leakiness” from their OFF state, i.e., adoption of the ON state even in absence of adenine. Leakiness from the pbuE riboswitch is expected to be very low given the presence of a very stable terminator structure [22], which is important given that the regulated gene is a purine efflux pump [53], [65]. Because the terminator stem is highly stable and disrupts the aptamer domain in absence of adenine [22], it is thus important that the riboswitch binds the ligand in a co-transcriptional manner (Figure 6), as this allows the binding to occur before the terminator is transcribed. The formation of the RNA-ligand complex increases the stability of aptamer domain and this process is aided by the presence of a pause site in the expression platform that gives more time for the ligand to bind. Thus, transcriptionally-regulating adenine riboswitches may rely on different regulatory mechanisms, compared to translationally-regulating ones, to increase the “window of decision” for gene regulation. However, because reversible switches such as add can perform ligand binding post-transcriptionally, there is no obvious need for these switches to contain pause sites such as found in pbuE (Figure 6). Given that add operates at the level of translation, it will be important to experimentally determine if such a structural reversibility is a pre-requisite to control ribosome accessibility, and if so, to assess whether a non-required coupling between transcription and translation is always needed. Additional work will be required to determine if our findings may be expanded to other classes of ligand-dependent bacterial riboswitches.
Fig. 6. Schematic showing proposed regulation mechanisms for add and pbuE adenine riboswitches. 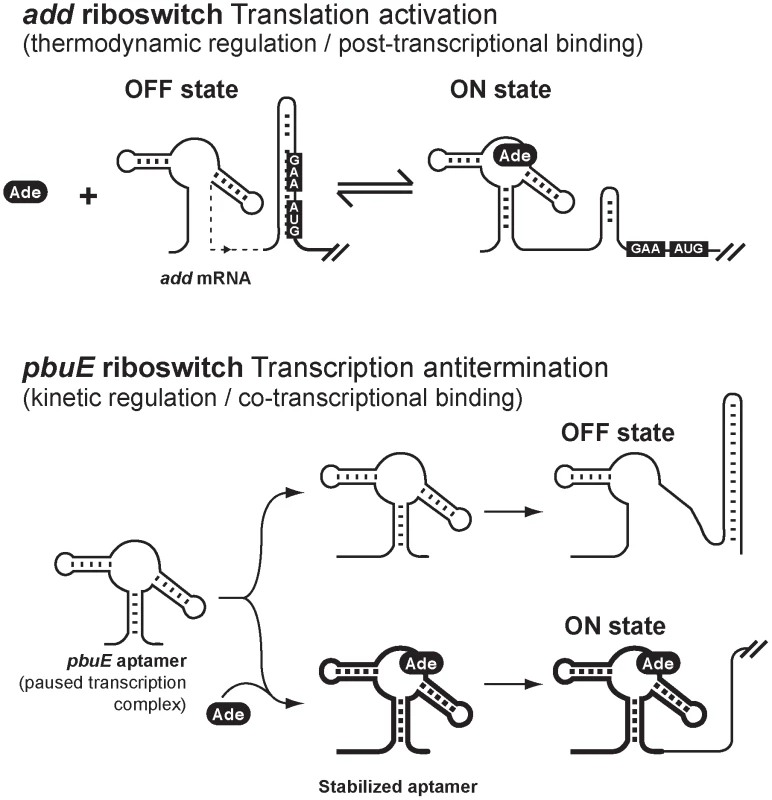
Top, regulation mechanism of the add adenine riboswitch. The OFF state is represented with the Shine-Dalgarno (GAA) and AUG start codon sequences base paired in the sequestrator helix. Upon adenine (Ade) binding, the ON state is formed which increases the accessibility of both GAA and AUG sequences. The structural reversibility and the lack of requirement of a transcription-translation coupling for the regulatory activity of the riboswitch are consistent with a thermodynamic regulation regime. Bottom, regulation mechanism of the pbuE adenine riboswitch. In this regulation mechanism, a low intracellular adenine concentration leads to the formation of the OFF state. However, an elevated adenine concentration may co-transcriptionally bind to the riboswitch aptamer on a paused transcription complex, thereby stabilizing the aptamer, which will then ultimately lead to the formation of the ON state and the expression of pbuE. Because adenine binding occurs co-transcriptionally and is largely dependent on the rate of transcription, the regulation mechanism is consistent with a kinetic regime. Aptamers having thick lines represent stabilized complexes in presence of adenine. Materials and Methods
Strains
Derivatives of E. coli MG1655 were used in all experiments. DH5α strain was used for cloning procedures. Transcriptional and translational fusions of V. vulnificus add gene were constructed by inserting a PCR product (using chromosomal DNA of V. vulnificus YJ016 as template) into the plasmids pFRΔ [66] and pRS1551 [67], respectively. Table 1 describes the oligonucleotides used in this study. PCR fragment containing −237 to +18 relative to add start codon (oligonucleotides EM620–EM621) was digested by EcoRI and BamHI and ligated into EcoRI/BamHI digested pFRΔ and pRS1551 to generate the transcriptional add'-lacZ and translational add'-'lacZ fusions. Other constructs were generated by the three-step PCR mutagenesis as described previously [68]. Briefly, the add'-lacZ construct was used as a template for two independent PCR reactions. Oligonucleotides EM194–EM644 and EM195–EM643 were used to generate the G31C/G32C construct. Oligonucleotides EM194–EM827 and EM195–EM826 were used to construct the OFF state mutant, and oligonucleotides EM194–EM1012 and EM195–EM1011 were used to generate the P1-3′ mutant construct. The two PCR products were mixed to serve as a template for a third PCR reaction using oligonucleotides EM194–EM195. The resulting PCR products were digested by EcoRI and BamHI and ligated into EcoRI/BamHI-digested pFRΔ and pRS1551. The ON state mutant was generated by adding the P1-3′ mutation to the OFF state mutant using the same three-step PCR method. Transcriptional and translational fusions were inserted in single copy into the bacterial chromosome of wild-type strain EM1055 (described in [69]) at the λ att site as described previously [67]. Stable lysogens were screened for single insertion of recombinant λ by PCR as described [70].
Tab. 1. Oligonucleotides used in this study. 
In vitro RNA synthesis
RNA was transcribed by the T7 RNA polymerase (Roche, Germany) using a PCR product as a template. Transcription reactions were performed in T7 transcription buffer (40 mM Tris-HCl at pH 8.0, 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine), with 400 µM NTPs (A, C, U and G), 20 units RNA guard, 20 units T7 RNA polymerase and 0.5 µg DNA template. After 4 h of incubation at 37°C, the mixture was treated with 2 units of Turbo DNase (Ambion) and extracted once with phenol-chloroform. RNA transcripts were purified on denaturing acrylamide gel. The primers used for generating DNA templates for in vitro RNA synthesis were EM821–EM886 (wild-type add) and EM890–E886 (add OFF state and add ON state mutants). To generate the transcription templates, the genomic DNA of EM1005 strains harboring either λadd'-'lacZ, λaddOFF'-'lacZ, or λaddON'-'lacZ fusions were used as template for PCR reactions. The aptamer sequences used in this study are based on the genomic sequence to which a GCG sequence was added to the 5′ side to allow high transcription yield and to minimize 5′ heterogeneity [71].
Fluorescence spectroscopy
Fluorescence was performed on a Quanta Master fluorometer. Data were collected at 10°C in 10 mM MgCl2, 50 mM Tris-HCl (pH 8.0) and 100 mM KCl. Spectra were corrected for background and intensities were determined by integrating data collected over the range 365–475 nm. 2AP was excited at 300 nm to obtain a good separation between the Raman and the fluorescence peak.
The fraction of quenched 2AP fluorescence was determined by monitoring the fluorescence data using a fixed concentration of 2AP (50 nM). Titrations were performed using an increasing concentration of a given aptamer or riboswitch molecule. Because the total RNA concentration is in large excess relative to 2AP, the binding can be described by the equationwhere dF and F are the change in fluorescence intensity and the maximum fluorescence intensity in the absence of RNA, respectively. KDapp is the apparent dissociation constant and the parameter a is a dimensionless constant proportional to the ratio of quantum yields of 2AP in the complex and in free solution [22]. The value of a is obtained with KDapp by nonlinear least-squares fitting following the Levenberg-Marquardt algorithm and typically corresponds to a value of 0.05. The reported errors are the standard uncertainties of the data from the best-fit theoretical curves. The standard uncertainty of the measurement is thus assumed to be approximated by the standard deviation of the points from the fitted curve [22], [72], [73]. For each experiment, at least three measurements have been performed and all exhibited very similar uncertainties from best-fit curves.
Partial RNase T1 cleavage
Radioactively 5′-labeled RNA was incubated in 50 mM Tris-HCl (pH 8.0) and 100 mM KCl in the presence of MgCl2 and/or adenine at the indicated concentrations for 5 min at 37°C. RNase T1 (1 U) was allowed to react for 2 min and reactions were quenched by adding an equal volume of a solution of 97% formamide and 10 mM EDTA. Products were separated on a denaturing polyacrylamide gel, which was subsequently dried and exposed to PhosphorImager screens.
SHAPE assays
The SHAPE reaction was prepared using 1 pmol of purified RNA that was resuspended in two volumes of 0.5× TE buffer, in which was added one volume of 3.3× folding buffer containing 333 mM K-HEPES (pH 8.0), 333 mM NaCl, 10 mM MgCl2 and the indicated concentration of adenine. The samples were heated to 65°C and allowed to cool down to 30°C before being pre-incubated 10 min at 37°C. N-methylisatoic anhydride (NMIA, Invitrogen) dissolved in dimethyl sulfoxide (DMSO) was then added and allowed to react for 80 min at 37°C. Modified RNA were precipitated, washed with 70% ethanol and resuspended in 0.5× TE buffer. Reverse transcription reactions were performed as previously described [38] and products were separated on a 5% denaturing polyacrylamide gel. Gels were dried and exposed to PhosphorImager screens. The RNA molecule used for SHAPE assays corresponds to nucleotides 1 to 165 of the transcribed mRNA. The region used for the primer corresponds to nucleotides 146 to 165.
Transcriptional and translational β-galactosidase assays
Kinetic assays for β-galactosidase activities were performed as described previously [74] using a SpectraMax 250 microtitre plate reader (Molecular Devices). Briefly, overnight bacterial cultures were incubated in LB media at 37°C and diluted 1000-fold into 50 ml of fresh LB media at 37°C. Cultures were grown with agitation to an OD600 of 0.3 before adding adenine at the indicated concentrations. Specific β-galactosidase activity was calculated using the formula Vmax/OD600. The reported results represent data of at least three experimental trials.
In vitro translation assays
Translation reactions were performed in E. coli S30 extracts (Promega, L1030). Coupled and uncoupled reactions were performed using either 25 nM of DNA or 250 nM RNA as template, respectively. Template RNAs were transcribed as described above with T7 RNA polymerase. Reactions contained the following (in a total volume of 15 µl): 0.4 ml [35S] methionine (588 Ci/mmol), 4.5 µl of S30 extract, 6 µl of S30 premix, 0.1 µM of amino acid mix without methionine, and the template. When indicated, adenine was added at a final concentration of 500 µM. Reaction mixtures were incubated at 37°C for 30 min and samples were analyzed on 15% SDS-PAGE, exposed on phosphor screen, and revealed on a Storm 860 (Molecular Dynamics).
Single-round transcription
E. coli RNAP was purchased from Epicentre Biotechnologies. B. subtilis RNAP and B. subtilis NusA were purified as previously described [28], [75]. The B. subtilis sigmaA transcription factor was purified as previously described [76]. DNA templates were produced by recursive PCR using oligonucleotides containing the xpt promoter sequence for transcriptions using the E. coli polymerase [77]. The GlyQS promoter was used for transcriptions using the B. subtilis polymerase [78]. A transcription start site was generated 51 nt upstream of the aptamer domain. The construct was engineered to allow transcription initiation using an ApC dinucleotide and a halt at position +1 by omission of CTP, and a readthrough transcript product terminating at 40 nt after the AUG start codon. P1 stem mutants were made by altering the required number of base pair to achieved the indicated P1 stem length. Thus, elongated P1 stem constructs were performed by mutating the P1 stem 5′ sequence to complement the corresponding 3′ sequence to generate a P1 stem of the indicated length. Shortened P1 stem constructs were made by changing the P1 5′ sequence to its Watson-Crick complement so that base pair formation with the P1 3′ sequence is inhibited. Single-round transcriptions were performed as previously described [77], using 300 fmol DNA templates and 0.6 µg of either E. coli or B. subtilis RNA polymerase together with an equivalent amount of the sigmaA factor. Transcriptions were initiated in a tube containing 1× transcription buffer including 150 µM ApC, 0.75 µM UTP, 0.25 µM [α-32P], 2.5 µM of ATP and GTP [28] by incubating at 37°C for 15 min, which was subsequently incubated on ice for 10 min. Transcription elongation was initiated by adding rNTP to a final concentration of 65 µM in presence or absence of ligand at 37°C for 15 min. Heparin (20 µg/mL) was added to prevent transcription re-initiation. The rNTP concentration was decreased to 20 µM when performing time course experiments. Sequencing transcription reactions were performed by including 120 µM of 3′-O-methyl-NTP. Reactions were stopped by adding one volume of 95% formamide. Transcription products were resolved using denaturing gel electrophoresis. Gels were dried and exposed to PhosphorImager screens. Experiments have been performed at least three times and all exhibited very similar uncertainties (<10%). The uncertainties on calculated T50 values were obtained as previously reported [46].
Thermal denaturation
300 nM RNA was incubated in 10 mM MOPS (pH 8.0), 25 mM NaCl and 2 mM MgCl2 in presence or absence of 10 µM DAP, and was degassed 5 min. Denaturation profiles were obtained at 258 nm using a Shimadzu UV2501 spectrophotometer equipped with a temperature controller. Samples were heated at a rate of ∼0.5°C/min over a range of 20°C to 80°C and an average of 3 s was used for each reading. Absorbance data was normalized by subtracting the pre - and post-transition to obtain the proportion of the unfolded state. Data was smoothed over a 3°C range and melting temperature values were determined by evaluating the temperature required to obtain half of the transition of the resulting profiles.
Primer extension
Transcriptional +1 of add and pbuE riboswitches were determined as previously described [74]. Briefly, 40 µg of total RNA were incubated in presence of 2 pmol of radioactively 5′-labeled DNA oligonucleotides and the reverse transcription reaction was allowed according to the Superscript II protocol (Invitrogen, Burlington, ON). Reactions were precipitated and migrated on denaturing polyacrylamide gels. PCR reactions were used as sequencing markers. Gels were dried and exposed to PhosphorImager screens.
Supporting Information
Zdroje
1. WatersLS
StorzG
2009 Regulatory RNAs in bacteria. Cell 136 615 628
2. KeeneJD
2007 RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8 533 543
3. SerganovA
PatelDJ
2007 Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet 8 776 790
4. RothA
BreakerRR
2009 The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78 305 334
5. BlouinS
MulhbacherJ
PenedoJC
LafontaineDA
2009 Riboswitches: ancient and promising genetic regulators. Chembiochem 10 400 416
6. CromieMJ
ShiY
LatifiT
GroismanEA
2006 An RNA sensor for intracellular Mg(2+). Cell 125 71 84
7. DannCE3rd
WakemanCA
SielingCL
BakerSC
IrnovI
2007 Structure and mechanism of a metal-sensing regulatory RNA. Cell 130 878 892
8. MoritaMT
TanakaY
KodamaTS
KyogokuY
YanagiH
1999 Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev 13 655 665
9. MoritaM
KanemoriM
YanagiH
YuraT
1999 Heat-induced synthesis of sigma32 in Escherichia coli: structural and functional dissection of rpoH mRNA secondary structure. J Bacteriol 181 401 410
10. SerganovA
2009 The long and the short of riboswitches. Curr Opin Struct Biol 19 251 259
11. DambachMD
WinklerWC
2009 Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol 12 161 169
12. HenkinTM
2008 Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22 3383 3390
13. HenkinTM
2009 RNA-dependent RNA switches in bacteria. Methods Mol Biol 540 207 214
14. GrundyFJ
HenkinTM
1993 tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74 475 482
15. LohE
DussurgetO
GripenlandJ
VaitkeviciusK
TiensuuT
2009 A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139 770 779
16. BateyRT
GilbertSD
MontangeRK
2004 Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432 411 415
17. SerganovA
YuanYR
PikovskayaO
PolonskaiaA
MalininaL
2004 Structural Basis for Discriminative Regulation of Gene Expression by Adenine - and Guanine-Sensing mRNAs. Chem Biol 11 1729 1741
18. MandalM
BoeseB
BarrickJE
WinklerWC
BreakerRR
2003 Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113 577 586
19. MandalM
BreakerRR
2004 Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11 29 35
20. RiederR
LangK
GraberD
MicuraR
2007 Ligand-Induced Folding of the Adenosine Deaminase A-Riboswitch and Implications on Riboswitch Translational Control. Chembiochem 8 896 902
21. WickiserJK
CheahMT
BreakerRR
CrothersDM
2005 The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44 13404 13414
22. LemayJF
PenedoJC
TremblayR
LilleyDM
LafontaineDA
2006 Folding of the adenine riboswitch. Chem Biol 13 857 868
23. PanT
ArtsimovitchI
FangXW
LandickR
SosnickTR
1999 Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A 96 9545 9550
24. WongTN
SosnickTR
PanT
2007 Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci U S A 104 17995 18000
25. BrehmSL
CechTR
1983 Fate of an intervening sequence ribonucleic acid: excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry 22 2390 2397
26. ZarrinkarPP
WilliamsonJR
1994 Kinetic intermediates in RNA folding. Science 265 918 924
27. ZhangF
RamsayES
WoodsonSA
1995 In vivo facilitation of Tetrahymena group I intron splicing in Escherichia coli pre-ribosomal RNA. RNA 1 284 292
28. WickiserJK
WinklerWC
BreakerRR
CrothersDM
2005 The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18 49 60
29. LobanovKV
Korol'kovaNV
EreminaS
Errais LopesL
ProshkinSA
2007 [Mutations altering the specificity of the sensor RNA encoded by the Bacillus subtilis pbuE gene]. Genetika 43 859 864
30. LemayJF
LafontaineDA
2007 Core requirements of the adenine riboswitch aptamer for ligand binding. RNA 13 339 350
31. MulhbacherJ
LafontaineDA
2007 Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res 35 5568 5580
32. GilbertSD
StoddardCD
WiseSJ
BateyRT
2006 Thermodynamic and Kinetic Characterization of Ligand Binding to the Purine Riboswitch Aptamer Domain. J Mol Biol 359 754 768
33. EskandariS
PrychynaO
LeungJ
AvdicD
O'NeillMA
2007 Ligand-Directed Dynamics of Adenine Riboswitch Conformers. J Am Chem Soc
34. PrychynaO
DahabiehMS
ChaoJ
O'NeillMA
2009 Sequence-dependent folding and unfolding of ligand-bound purine riboswitches. Biopolymers 91 953 965
35. WardDC
ReichE
StryerL
1969 Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J Biol Chem 244 1228 1237
36. StiversJT
1998 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res 26 3837 3844
37. JeanJM
HallKB
2001 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc Natl Acad Sci U S A 98 37 41
38. MerinoEJ
WilkinsonKA
CoughlanJL
WeeksKM
2005 RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127 4223 4231
39. BennettBD
KimballEH
GaoM
OsterhoutR
Van DienSJ
2009 Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5 593 599
40. WinklerWC
NahviA
SudarsanN
BarrickJE
BreakerRR
2003 An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10 701 707
41. SudarsanN
WickiserJK
NakamuraS
EbertMS
BreakerRR
2003 An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev 17 2688 2697
42. GrundyFJ
LehmanSC
HenkinTM
2003 The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci U S A 100 12057 12062
43. MironovAS
GusarovI
RafikovR
LopezLE
ShatalinK
2002 Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111 747 756
44. WinklerWC
Cohen-ChalamishS
BreakerRR
2002 An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A 99 15908 15913
45. WinklerW
NahviA
BreakerRR
2002 Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419 952 956
46. BlouinS
LafontaineDA
2007 A loop-loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13 1256 1267
47. SchaakJE
BabitzkeP
BevilacquaPC
2003 Phylogenetic conservation of RNA secondary and tertiary structure in the trpEDCFBA operon leader transcript in Bacillus. Rna 9 1502 1515
48. GreenleafWJ
FriedaKL
FosterDA
WoodsideMT
BlockSM
2008 Direct observation of hierarchical folding in single riboswitch aptamers. Science 319 630 633
49. McDowellJC
RobertsJW
JinDJ
GrossC
1994 Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science 266 822 825
50. NechooshtanG
Elgrably-WeissM
SheafferA
WesthofE
AltuviaS
2009 A pH-responsive riboregulator. Genes Dev 23 2650 2662
51. FarnhamPJ
GreenblattJ
PlattT
1982 Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell 29 945 951
52. VerhounigA
KarcherD
BockR
2010 Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci U S A 107 6204 6209
53. NygaardP
SaxildHH
2005 The purine efflux pump PbuE in Bacillus subtilis modulates expression of the PurR and G-box (XptR) regulons by adjusting the purine base pool size. J Bacteriol 187 791 794
54. NouX
KadnerRJ
2000 Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci U S A 97 7190 7195
55. NahviA
SudarsanN
EbertMS
ZouX
BrownKL
2002 Genetic control by a metabolite binding mRNA. Chem Biol 9 1043
56. FuchsRT
GrundyFJ
HenkinTM
2006 The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol 13 226 233
57. BairdNJ
Ferre-D'AmareAR
Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16 598 609
58. LangK
RiederR
MicuraR
2007 Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res 35 5370 5378
59. RentmeisterA
MayerG
KuhnN
FamulokM
2007 Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res 35 3713 3722
60. BurmannBM
SchweimerK
LuoX
WahlMC
StittBL
2010 A NusE:NusG complex links transcription and translation. Science 328 501 504
61. RiederU
KreutzC
MicuraR
2010 Folding of a transcriptionally acting preQ1 riboswitch. Proc Natl Acad Sci U S A 107 10804 10809
62. KulshinaN
BairdNJ
Ferre-D'AmareAR
2009 Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol 16 1212 1217
63. SmithKD
LipchockSV
AmesTD
WangJ
BreakerRR
2009 Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16 1218 1223
64. PanT
SosnickT
2006 RNA folding during transcription. Annu Rev Biophys Biomol Struct 35 161 175
65. JohansenLE
NygaardP
LassenC
AgersoY
SaxildHH
2003 Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL). J Bacteriol 185 5200 5209
66. RepoilaF
MajdalaniN
GottesmanS
2003 Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol 48 855 861
67. SimonsRW
HoumanF
KlecknerN
1987 Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53 85 96
68. DesnoyersG
MorissetteA
PrevostK
MasseE
2009 Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J 28 1551 1561
69. MasseE
GottesmanS
2002 A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99 4620 4625
70. PowellBS
RivasMP
CourtDL
NakamuraY
TurnboughCLJr
1994 Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res 22 5765 5766
71. PleissJA
DerrickML
UhlenbeckOC
1998 T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. Rna 4 1313 1317
72. FlanneryBP
TeukolskySA
VetterlingWT
1992 Numerical Recipes in Fortran, 2nd Edn Cambridge Cambridge University Press, UK
73. RistM
MarinoJ
2001 Association of an RNA kissing complex analyzed using 2-aminopurine fluorescence. Nucleic Acids Res 29 2401 2408
74. PrevostK
SalvailH
DesnoyersG
JacquesJF
PhaneufE
2007 The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64 1260 1273
75. QiY
HulettFM
1998 PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol Microbiol 28 1187 1197
76. HelmannJD
2003 Purification of Bacillus subtilis RNA polymerase and associated factors. Methods Enzymol 370 10 24
77. MandalM
LeeM
BarrickJE
WeinbergZ
EmilssonGM
2004 A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306 275 279
78. GrundyFJ
WinklerWC
HenkinTM
2002 tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci U S A 99 11121 11126
79. PuglisiJD
TinocoIJr
1989 Absorbance melting curves of RNA. Methods Enzymol 180 304 325
80. AlbergoDD
MarkyLA
BreslauerKJ
TurnerDH
1981 Thermodynamics of (dG–dC)3 double-helix formation in water and deuterium oxide. Biochemistry 20 1409 1413
Štítky
Genetika Reprodukčná medicína
Článek Composite Effects of Polymorphisms near Multiple Regulatory Elements Create a Major-Effect QTLČlánek Horizontal Transfer, Not Duplication, Drives the Expansion of Protein Families in ProkaryotesČlánek Segregating Variation in the Polycomb Group Gene Alters the Effect of Temperature on Multiple TraitsČlánek Global Analysis of the Impact of Environmental Perturbation on -Regulation of Gene ExpressionČlánek H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone DeacetylasesČlánek A Mutation in the Gene Encoding Mitochondrial Mg Channel MRS2 Results in Demyelination in the Rat
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Meta-Analysis of Genome-Wide Association Scans Identifies IL18RAP, PTPN2, TAGAP, and PUS10 As Shared Risk Loci for Crohn's Disease and Celiac Disease
- Composite Effects of Polymorphisms near Multiple Regulatory Elements Create a Major-Effect QTL
- Horizontal Transfer, Not Duplication, Drives the Expansion of Protein Families in Prokaryotes
- Genome-Wide Association Study SNPs in the Human Genome Diversity Project Populations: Does Selection Affect Unlinked SNPs with Shared Trait Associations?
- Friedreich's Ataxia (GAA)•(TTC) Repeats Strongly Stimulate Mitotic Crossovers in
- Zebrafish Mutation Leads to mRNA Splicing Defect and Pituitary Lineage Expansion
- Histone H4 Lysine 12 Acetylation Regulates Telomeric Heterochromatin Plasticity in
- Bub1-Mediated Adaptation of the Spindle Checkpoint
- Segregating Variation in the Polycomb Group Gene Alters the Effect of Temperature on Multiple Traits
- Signaling Role of Fructose Mediated by FINS1/FBP in
- RNF12 Activates and Is Essential for X Chromosome Inactivation
- Comparative Study between Transcriptionally- and Translationally-Acting Adenine Riboswitches Reveals Key Differences in Riboswitch Regulatory Mechanisms
- Global Analysis of the Impact of Environmental Perturbation on -Regulation of Gene Expression
- Application of a New Method for GWAS in a Related Case/Control Sample with Known Pedigree Structure: Identification of New Loci for Nephrolithiasis
- H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone Deacetylases
- A Mutation in the Gene Encoding Mitochondrial Mg Channel MRS2 Results in Demyelination in the Rat
- Transcription Initiation Patterns Indicate Divergent Strategies for Gene Regulation at the Chromatin Level
- The Transposon-Like Correia Elements Encode Numerous Strong Promoters and Provide a Potential New Mechanism for Phase Variation in the Meningococcus
- Proteins Encoded in Genomic Regions Associated with Immune-Mediated Disease Physically Interact and Suggest Underlying Biology
- A Novel RNA-Recognition-Motif Protein Is Required for Premeiotic G/S-Phase Transition in Rice ( L.)
- The Mucin-Like Protein OSM-8 Negatively Regulates Osmosensitive Physiology Via the Transmembrane Protein PTR-23
- Genome Sequencing and Comparative Transcriptomics of the Model Entomopathogenic Fungi and
- Rnf12—A Jack of All Trades in X Inactivation?
- Joint Genetic Analysis of Gene Expression Data with Inferred Cellular Phenotypes
- Evolutionary Conserved Regulation of HIF-1β by NF-κB
- Quaking Regulates Expression through Its 3′ UTR in Oligodendrocyte Precursor Cells
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone Deacetylases
- Evolutionary Conserved Regulation of HIF-1β by NF-κB
- Rnf12—A Jack of All Trades in X Inactivation?
- Joint Genetic Analysis of Gene Expression Data with Inferred Cellular Phenotypes
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



