-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Histone H4 Lysine 12 Acetylation Regulates Telomeric Heterochromatin Plasticity in
Recent studies have established that the highly condensed and transcriptionally silent heterochromatic domains in budding yeast are virtually dynamic structures. The underlying mechanisms for heterochromatin dynamics, however, remain obscure. In this study, we show that histones are dynamically acetylated on H4K12 at telomeric heterochromatin, and this acetylation regulates several of the dynamic telomere properties. Using a de novo heterochromatin formation assay, we surprisingly found that acetylated H4K12 survived the formation of telomeric heterochromatin. Consistently, the histone acetyltransferase complex NuA4 bound to silenced telomeric regions and acetylated H4K12. H4K12 acetylation prevented the over-accumulation of Sir proteins at telomeric heterochromatin and elimination of this acetylation caused defects in multiple telomere-related processes, including transcription, telomere replication, and recombination. Together, these data shed light on a potential histone acetylation mark within telomeric heterochromatin that contributes to telomere plasticity.
Published in the journal: Histone H4 Lysine 12 Acetylation Regulates Telomeric Heterochromatin Plasticity in. PLoS Genet 7(1): e32767. doi:10.1371/journal.pgen.1001272
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001272Summary
Recent studies have established that the highly condensed and transcriptionally silent heterochromatic domains in budding yeast are virtually dynamic structures. The underlying mechanisms for heterochromatin dynamics, however, remain obscure. In this study, we show that histones are dynamically acetylated on H4K12 at telomeric heterochromatin, and this acetylation regulates several of the dynamic telomere properties. Using a de novo heterochromatin formation assay, we surprisingly found that acetylated H4K12 survived the formation of telomeric heterochromatin. Consistently, the histone acetyltransferase complex NuA4 bound to silenced telomeric regions and acetylated H4K12. H4K12 acetylation prevented the over-accumulation of Sir proteins at telomeric heterochromatin and elimination of this acetylation caused defects in multiple telomere-related processes, including transcription, telomere replication, and recombination. Together, these data shed light on a potential histone acetylation mark within telomeric heterochromatin that contributes to telomere plasticity.
Introduction
Saccharomyces cerevisiae telomeres and mating type loci (HMR and HML) are well-characterized transcriptionally silenced domains. Heterochromatin tends to initiate at a specific DNA element and propagates along a chromatin fiber to repress the expression of nearby genes [1]. The major structural components of heterochromatin in yeast are known as Sir (silent information regulators) proteins, including Sir2, Sir3 and Sir4 [1]. For telomeric heterochromatin, the double-stranded telomeric DNA-binding protein Rap1 interacts with Sir4 and thereby recruits the whole Sir complex to the telomeres [2]. Once recruited to telomeres, Sir2 deacetylates a critical K16 acetyl mark on histone H4 [3], a process required for Sir proteins to bind throughout the subtelomeric regions of ∼3 kb proximal to the terminal ∼350-bp TG-tracts [4]–[8].
The spreading of heterochromatin must be restricted by boundaries between silent and active chromatin [9]. Since histone H4 amino acid residues 16 to 29 are primarily required for Sir3 binding [4], histone H4K16 acetylation dominantly prevents Sir-mediated heterochromatin spreading [6], [7]. Genetic and biochemical data also show that unacetylated H4 K5, 8, 12 together can substitute for the mutation of H4K16 in Sir3 binding [4], [5]. Therefore, it has been generally believed that the acetylation of histone H4 at K5, 8 and 12, catalyzed by NuA4 complex [6], [10], cumulatively antagonizes heterochromatin spreading. However, it remains unclear whether H4 K5, 8 and 12 contribute equally to this process.
Yeast heterochromatin possesses the ability to exchange chromatin-bound Sir3 for soluble unbound protein throughout the cell cycle [11]. In addition, heterochromatin is surprisingly permissive to activators, co-activators and transcriptional preinitiation-complex (PIC) as well [12]–[14]. Moreover, heterochromatin doesn't prohibit active base-pair substitutions [15]. These lines of evidence have revealed the dynamic aspect of silent - and hetero-chromatin. One outstanding question of concern is what contributes to the dynamics of heterochromatin. Accordingly, it would be beneficial to know if altering the dynamics of heterochromatin structure has biological consequences.
In this study, we show that the heterochromatin in yeast harbors a dynamic H4K12 acetylation mark which suppresses Sir-mediated aberrant condensation of telomeric heterochromatin and promotes telomere-related processes, including telomere transcription, replication and recombination. Thus, histone acetylation may provide a scheme for yeast cells to maintain partially flexible telomeric heterochromatin that allows for normal changes in DNA metabolism.
Results
A distinct system for studying de novo telomeric heterochromatin formation in yeast
To better understand the orchestrated events that are involved in establishing telomeric heterochromatin, we developed a new telomeric heterochromatin formation assay, building upon the de novo telomere addition system originally established by Gottschling's lab [16]. This new assay allowed us to study telomeric heterochromatin formation under conditions where global gene expression, especially the expression of the silencing-related genes, was not affected. Technically, we used a galactose-inducible HO endonuclease to cut a chromosome to expose a pre-inserted 81-bp telomeric “seed” for further telomere addition [16] (Figure 1A). Chromosome “healing” was hardly observed in the first 6 hours upon HO induction. Later, the telomere seed was gradually extended in a telomerase dependent manner ([16] and Figure 1B).
Fig. 1. De novo telomeric heterochromatin assembly on a short telomere. 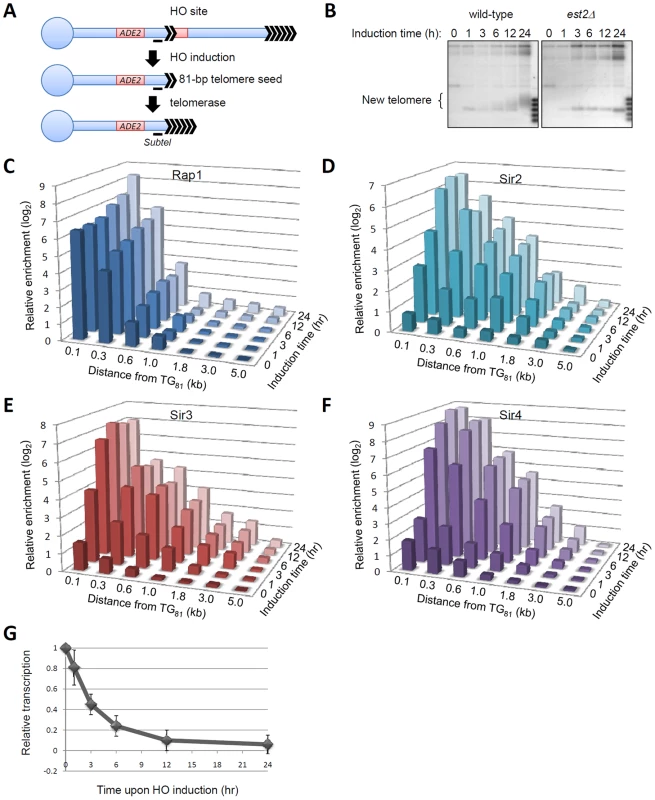
(A) Schematic representation of the de novo telomeric heterochromatin formation system [16]. The ADH4 locus was replaced in a haploid yeast strain with a 6-kb fragment consisting of the ADE2 selection marker, 81-bp telomeric DNA sequence and the recognition site for the HO endonuclease. The LYS2 gene was placed approximately 10 kb from the natural telomere VII-L. HO was induced by the addition of galactose to the media. ADE2 was a gene adjacent to the telomere seed. The black line indicated the primer set (SubTel) used for ChIP analysis. (B) De novo telomere addition analysis [16]. After HO induction, yeast cells were harvested at time points as indicated. Genomic DNA was isolated, digested with BstXI and subjected to Southern-blot analysis. The blots were hybrid with a telomere seed-proximal probe. (C–F) ChIP analyses of Rap1 (C), Sir2-myc (D), Sir3-myc (E) and Sir4-myc (F) in the time course of HO induction. Shown were the real-time Q-PCR results of the ChIP products. X axis indicated the distance from the 81-bp telomere seed. Y axis indicated the timed intervals after HO induction. Z axis indicated the enrichment values (IP/Input) at indicated regions relative to ACT1. Values that were greater than 1 indicated more enrichment at indicated regions than background. (G) mRNA analysis of ADE2 upon HO induction. The mRNA level of ADE2 was normalized by ACT1. The relative mRNA before galactose addition was set as 1. The recruitment of Rap1 and Sir proteins onto regions 100-bp to 5.0-kb from the telomere seed upon HO induction was then monitored by chromatin immunoprecipitation (ChIP). High occupancy of Rap1 was observed at the 81-bp telomere seed prior to HO induction (Figure 1C), indicating that the 81-bp telomeric DNA tract is sufficient for Rap1 recruitment. Upon HO induction, the Rap1 occupancy at the newly formed telomere was barely altered in the first 6 hours and then gradually increased along with telomere elongation (Figure 1B and 1C). Notably, upon HO induction, Sir proteins were recruited to the new telomere within the first 6 hours in an extremely efficient manner (Figure 1D–1F). Once occupying a newly formed telomere, Sir proteins began to spread into the lateral regions up to 3 kb proximal to the newly formed telomeres (Figure 1D–1F). Consistently, the expression of ADE2, a gene near the telomere seed, was gradually reduced upon HO induction (Figure 1G). Therefore, telomeric heterochromatin was quickly assembled at the 81-bp telomeric seed after HO induction.
Histone H4K5 and K12 acetylation partially survived the formation of telomeric heterochromatin
Since histone H4 hypoacetylation was required for the onset of telomere silencing [1], [4], [5] and K16 seemed to be the unique deacetylation target for Sir2 on H4 [3], we were curious about the deacetylation kinetics of different lysines on H4 (K5, K8, K12 and K16) during heterochromatin formation. To avoid recombination-induced histone acetylation, we deleted the RAD52 gene in our strains [17]. The result showed that, prior to HO cleavage, all lysine acetylation was robust at the subtelomeric region (indicated as SubTel in Figure 1) (Figure 2A). Upon HO induction, H4K8 and K16 acetylation levels decreased most substantially and were largely eliminated by 6 hours (Figure 2A), a time point when Sir proteins saturated SubTel (Figure 1D–1F). In contrast, H4K5 and K12 acetylation, especially K12 acetylation, decayed more slowly at SubTel during HO induction, and were still detectable after 24-hour HO induction (Figure 2A). These observations raise a possibility that H4K5 and K12 acetylation may partially survive the formation of new telomeric heterochromatin.
Fig. 2. Histone H4K12 acetylation survives heterochromatin formation. 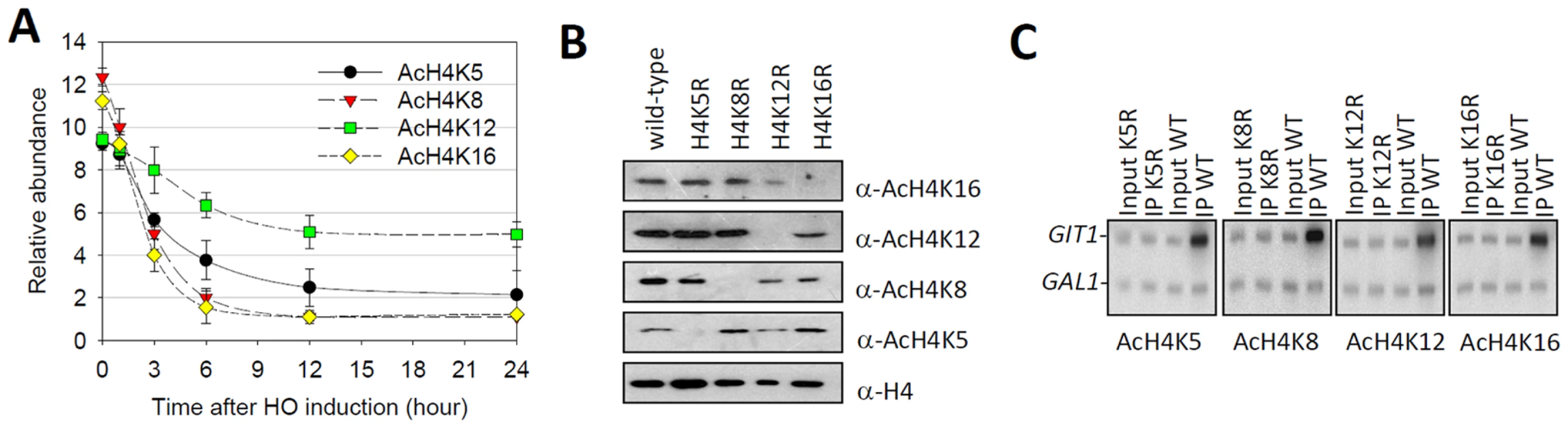
(A) ChIP analyses of H4K5 acetylation, K8 acetylation, K12 acetylation and K16 acetylation in the time course of HO induction. Shown were the real-time Q-PCR results of the ChIP products. Error bars represented standard error of the mean for three independent experiments. The relative abundance indicated the enrichment value (IP/Input) at SubTel relative to HMR. Values that were greater than 1 indicated more enrichment at the SubTel than background (HMR). (B) Whole-cell extracts from NSY429 strains expressing wild-type histone H4 (lane 1) and arginine substitution mutations of H4 Lys5 (lane 2), Lys8 (lane 3), Lys12 (lane 4) or Lys16 (lane 5) were subjected to western blot analysis with the specific antibodies indicated on the right. (C) ChIP assays were performed as in (A) except that chromatin from strains that contained either wild-type or H4 lysine substituted mutants was used. Q-PCR with 32P-ATP incorporation was done with primer pairs directed against GIT1 and GAL regions. PCR products were separated by 6% TBE gel and product abundance was detected using a PhosphorImager. To exclude the possibility that anti-acetyl-lysine antibodies cross-reacted with other subtelomeric factors, we compared the levels of H4 lysine acetylation in a wild-type strain to that of “unacetylated strains” where the corresponding lysines were mutated to arginines, respectively, thereby preventing anti-acetyl lysine antibodies from recognizing corresponding residues. In the western-blot experiment, we found that chromatin derived from strains with the lysine to arginine mutation was not detected by the corresponding anti-acetyl lysine antibodies (Figure 2B). In the ChIP experiment, we found that in the wild-type strain, all antibodies enriched GIT1, a typical hyperacetylated region [18], [19] (Figure 2C). In contrast, lysine to arginine mutations eliminated corresponding anti-acetyl lysine antibodies from enriching any chromatin fragments (Figure 2C). Additionally, increasing the amount of anti-acetyl-H4K12 antibody in the immunoprecipitation (IP) procedure couldn't recover chromatin in the H4K12R strain (Figure S1). These data are consistent with previous report [20], and indicate that all anti-acetyl-H4 lysine antibodies perform appropriately in our ChIP experiments.
Histone H4K12 is modestly acetylated at telomeric heterochromatin
To determine if the acetylation of H4K5 and K12 was generally associated with native telomeres that were not undergoing a double-strand break response [21], we performed ChIP to examine the acetylation profile of H4 lysines at all yeast telomeres. Sir2 binding was firstly mapped (Figure 3, pink bars) so as to mark the silencing status of the individual telomeres. We found that the abundance of Sir2 at different telomeres varied (Figure 3). Low amounts of Sir2 were mostly observed at telomeres that contain Y' elements (http://www.yeastgenome.org/ and Figure 3), supporting an anti-silencing role for these Y' elements [22]. The acetylation levels at subtelomeric regions were normalized to an HMR region, where we detected lowest level of acetylation (Figure S3, region D). Figure 3 showed that H4K12 acetylation was observed at nearly all telomeres in a manner that was independent of Sir2 binding. In contrast, the relative amount of H4K5, K8 or K16 acetylation at telomeres seemed to correlate in an opposing manner to the level of Sir2 binding (Figure 3). For example, at the end of left arm of chromosome II where there was little Sir2 binding and all lysine acetylations were robust. In contrast, at the end of right arm of chromosome II where there was abundant Sir2, only H4K12 acetylation was detectable (Figure 3). Essentially identical results were also obtained when yeast were grown in galactose culture (Figure S2). Furthermore, H4K12 acetylation was also detected at some regions of yeast HM heterochromatin (Figure S3). Taken together, these data suggest that H4K12 acetylation exists within yeast heterochromatin.
Fig. 3. Acetylation of H4K12 at native telomeric heterochromatin. 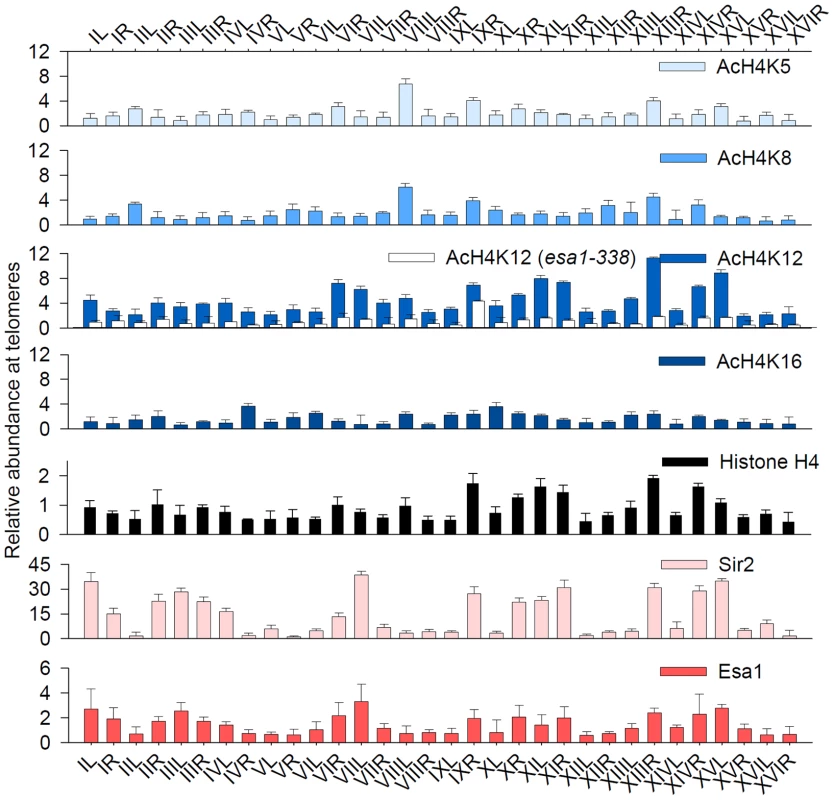
Relative abundance of H4K5 acetylation, K8 acetylation, K12 acetylation, K16 acetylation, histone H4, Sir2-myc and Esa1-myc at 32 telomeres. Real-time PCR was done with primer pairs against regions ∼0.5 kb from each telomere. PCR data for acetylations were normalized to an HMR region (region D in Figure S3); PCR data for Sir2-myc were normalized to ACT1; PCR data for Esa1-myc were normalized to a subtelomeric region ∼4 kb from the end of chromosome III-R (Stel). The relative abundance of lysine acetylation was normalized to H4 occupancy levels. Esa1 is responsible for H4K12 acetylation at telomeric heterochromatin
We next set out to ascertain how the subtelomeric H4K12 acetylation pattern was established. Genome-wide nucleosome H4K5, K8 and K12 acetylation in budding yeast is catalyzed by Piccolo NuA4 in a non-targeted manner [23]. We found that the subtelomeric H4K12 acetylation was sensitive to esa1-338 mutation (Figure 3, white bars). Since Esa1 is the catalytic subunit of NuA4 complex, these results support the idea that NuA4 is responsible for heterochromatic H4K12 acetylation. Interestingly, ChIP analysis revealed that Esa1 was specifically enriched at Sir2-abundant subtelomeres (Figure 3, red bars). Thus, heterochromatic H4K12 was acetylated by NuA4 in a targeted manner while the euchromatic H4K12, as the case of other H4 lysines, was acetylated by Piccolo NuA4 in a non-targeted manner [23]. We found that the binding of Esa1 at subtelomeric region was restricted to the distal region because there was no Esa1 binding in the subtelomeric domain 3.2 kb∼7.6 kb from the end of chromosome XIV-R (Figure S4A and S4B).
A previous study showed that Hat1, another lysine acetyltransferase in yeast, also possesses H4K12 acetylation activity [24], however it is predominantly localized to the cytoplasm and thought to specifically acetylate free histone H4 [25]. Western-blot results showed that HAT1 deletion did not affect chromatic H4K12 acetylation (Figure S4D). ChIP analyses revealed that the H4K12 acetylation level at the subtelomeric region of ChrXIV-R was not reduced, but rather modestly increased in hat1Δ cells (Figure S4E). Therefore, these data suggest that Hat1 is likely not needed for dynamic H4K12 acetylation at telomeric heterochromatin.
Histone H4K12 acetylation regulates telomere replication
Histone acetylation is thought to regulate chromatin assembly [26]. To address whether H4K12R mutation affects subtelomeric nucleosomal organization, the chromatin from wild-type and H4K12R mutant cells was digested with increasing concentrations of micrococcal nuclease (MNase) and the chromatin structure of subtelomere III-L was analyzed by Southern-blot as previously reported [27]. The MNase digestion of the subtelomeric region at specific sites indicated the presence of a regular array of nucleosomes (Figure 4A). There was little difference in subtelomeric nucleosome organization when comparing wild-type to H4K12R cells (Figure 4A), indicating that H4K12 does not influence the nucleosome organization of subtelomeric DNA on chromosome III-L. Surprisingly, the upper bands representing the undigested telomere-containing DNA, were much shorter in H4K12R cells when compared to wild-type cells (Figure. 4A), indicating that H4K12 acetylation affects the telomere length of chromosome III-L.
Fig. 4. Histone H4K12 acetylation regulates telomere replication. 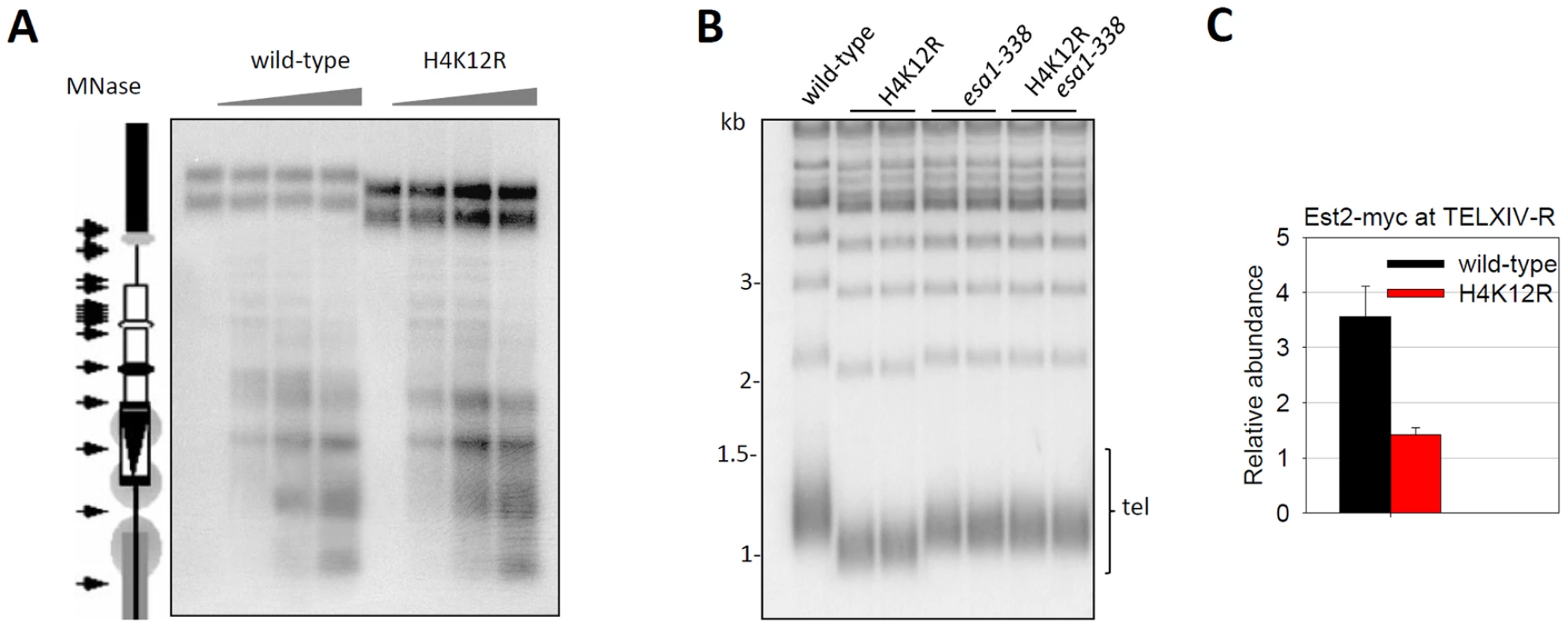
(A) Sensitivity of subtelomere III-L chromatin to MNase. Chromatin samples from wild-type and H4K12R cells were digested with increasing concentrations of MNase (0 U, 50 U, 150 U and 300 U) for 10 minutes and genomic DNA was purified, digested with BamHI and resolved in 1.2% agarose gel. Then the cutting profile was visualized after hybridization with an internal probe from Ty5-1. This probe abutted the BamHI site selected for the analyses and extended from position 1495 to 1725 of S. cerevisiae chromosome III. A scaled representation of the telomere III-L was shown on the left. Arrows pointed to the main cuts detected in chromatin. Gray circles represented translationally phased nucleosomes. The upper bands represented MNase-undigested telomere containing DNA. (B) Telomere-blot of wild-type and mutant cells. The esa1-338 mutation of ESA1 was a point mutation at position 338 (Glu to Gln), which is the catalytic site of Esa1. Genomic DNA was digested with XhoI, separated by 1.0% agarose gel, transferred to a nitrocellulose membrane and hybridized with a probe for yeast telomeric DNA. Tel: terminal TG-tracts-containing DNA fragments. (C) ChIP experiment of Est2-myc in wild-type and H4K12R cells. The enrichment value represented the ratio (IP/Input) at TELXIV-R relative to ACT1. To investigate whether H4K12 acetylation regulates telomere length in a universal manner, we examined the average telomere length of wild-type and H4K12R mutant cells. Strikingly, Southern-blot analysis revealed that telomeres in H4K12R mutant cells were much shorter than that in wild-type cells (Figure 4B). Consistently, the ESA1 mutant esa1-338, which lost telomeric H4K12 acetylation (Figure 3), also had shorter telomeres (Figure 4B). However, the telomere length in H4K12R esa1-338 double mutation cells resembled that in esa1-338 cells, rather than that in H4K12R cells (Figure 4B). Because H4K12R mutation didn't affect Esa1-telomere association (Figure S4B), it remains possible that the shorter telomeres observed in esa1-338 mutant could not be solely attributed to a defect in H4K12 acetylation.
Histone H4K12R mutation reduces the accessibility of telomeres to telomerase
The major structural components of telomere heterochromatin are Sir2, Sir3 and Sir4 [1]. Previous studies have shown that deletion of SIR3 or SIR4 caused telomere shortening, suggesting that disturbing telomere heterochromatin structure affects telomere replication [28]. Because the distal telomeres are devoid of nucleosome structure [29], we proposed that the telomere length defect observed in H4K12R mutant was attributable to a change in telomere heterochromatin structure. To test this possibility and to elucidate the molecular mechanism by which histone H4K12 acetylation affects telomere length, we examined the structural change(s) of telomere heterochromatin in H4K12R mutant cells. Immuno-staining analysis of myc-tagged Sir3 (Sir3-myc) with monoclonal anti-myc antibody revealed that H4K12R mutation did not affect the sub-cellular localization of telomeres (Figure S5), excluding the possibility that H4K12 affects the perinuclear localization of telomeres, which potentially regulates telomerase activity [30].
A distinctive feature of telomeric heterochromatin involves high-order structure, such as a fold-back loop-like structure [31]–[33] and such structure plays a role in telomere length homeostasis [28]. It was possible that H4K12R mutation affected the access of telomerase machinery to telomere end and thereby impaired telomere replication. To test this idea, we performed a ChIP experiment to compare the telomere binding of Est2, the catalytic subunit of telomerase, in wild-type and H4K12R cells. Interestingly, the telomere association of Est2 was greatly reduced by H4K12R mutation (Figure 4C), supporting our idea that H4K12 influences the recruitment of telomerase to telomeres.
Histone H4K12 acetylation suppresses the aberrant accumulation of Sir proteins at telomeric heterochromatin
The formation of heterochromatin in yeast is dependent on Sir proteins [1]. To determine if H4K12 acetylation directly modulated the binding of Sir proteins, we carried out ChIP to compare the chromatin binding of Sir2 and Sir3 in wild-type and histone mutant cells at regions 0.7 kb∼12 kb from the end of chromosome XIV-R (TELXIV-R) (Figure 5A–5C). We found that Sir proteins in wild-type cells were present only in telomeric heterochromatin as far as 3 kb from the end of the chromosome. In H4K16R mutant cells, the abundance of Sir proteins was reduced within heterochromatin but was greatly increased at heterochromatin adjacent regions (Figure 5B and 5C), a result that is representative of a typical heterochromatin over-spreading phenotype [6], [7]. Notably, compared to a H4K5R or H4K8R mutation that did not alter the binding of Sir proteins at TELXIV-R telomere (Figure 5B and 5C), the H4K12R mutation resulted in 2.52-fold more Sir2 binding and 3.09-fold more Sir3 binding 0.7 kb from the telomere end, but not at regions farther from the chromosome end. We also examined Sir2 binding at all other telomeres and found that the H4K12R mutation resulted in enhancement of Sir2 binding at most telomeres, especially at those where Sir2 was already abundant (Figure S6A). Because the H4K12R mutation does not perturb the transcriptional profile of the genome [34], and the expression and nuclear distribution of Sir proteins were not affected by the H4K12R mutation (Figure S5 and Figure S7), we therefore favor a model where the acetylation status of H4K12 directly affects Sir protein binding. We also analyzed the heterochromatin structure in H4K12Q mutant. Presumably, a lysine to glutamine mimicked a hyperacetylated state of the corresponding lysine. However, we found that the abundance and distribution of Sir2 and Sir3 in H4K12Q cells were largely similar to that in H4K12R cells (Figure S6B and S6C). We proposed that glutamine does not accurately represent the hyperacetylated state of H4K12 in this case. Together, these data suggest that the H4K12 acetylation suppresses the over-congregation of Sir proteins at telomeric heterochromatin.
Fig. 5. Regulation of telomere structure by histone H4K12 acetylation. 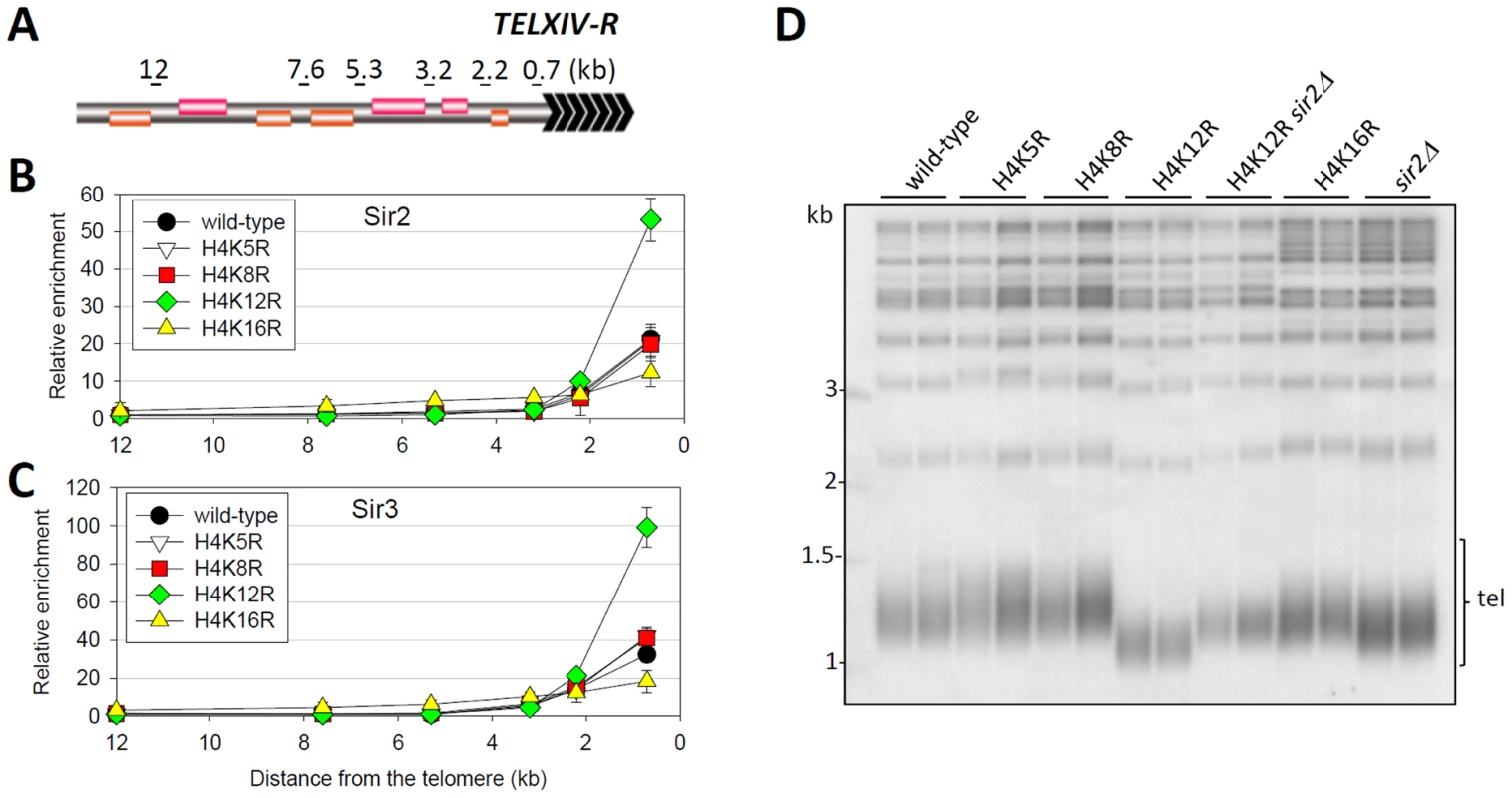
(A) Schematic diagram of the subtelomeric region of the right arm of chromosome XIV. Distance from each primer set to chromosome end was shown on the top. (B) and (C) ChIP analysis of Sir2-myc (B) and Sir3-myc (C) in wild-type and histone H4 mutant cells. The Q-PCR data were normalized to ACT1. (D) Telomere-blot of wild-type and histone mutant cells. To investigate if the regulation of telomere replication by H4K12 acetylation is dependent on the telomeric heterochromatin structure, we detected the effect of SIR2 deletion on the telomere length in H4K12R mutant cells. As shown in Figure 5D, sir2Δ cells exhibited a modest reduction of telomere length, which is much longer than that observed in H4K12R mutant cells. Interestingly, deletion of SIR2 efficiently rescued the severe telomere-shortening phenotype in H4K12R mutant cells (Figure 5D), indicating that H4K12 acetylation regulates telomere length though the Sir2 pathway. In addition, telomere length in H4K5R, K8R or K16R cells was indistinguishable from that in wild-type cells (Figure 5D), consistent with their already defined effect on the telomeric association of Sir proteins. Taken together, these data strongly supported the notion that H4K12 acetylation regulates telomere replication directly via modulating telomeric heterochromatin structure.
Histone H4K12 acetylation regulates telomere recombination
Telomeres are hotspots for recombination when they are deprotected. Telomerase-negative yeast cells could undergo homologous recombination on Y' or TG1–3 telomeric sequences, thus generating Type I or Type II post-senescence survivors, respectively [35]. Since Type II survivors grew much faster than Type I, liquid culturing of post-senescent est2Δ cells yielded primarily Type II survivor cells [35] (Figure 6A). Interestingly, deletion of the catalytic subunit of telomerase EST2 in the H4K12R mutant background eventually led to the generation of a population of cells that contained amplified Y' telomeric sequence (Figure 6A), a hallmark of Type I survivor cells. Therefore, we conclude that H4K12 acetylation suppresses homologous recombination of TG1–3 tracts during the creation of telomerase-null post-senescent survivors.
Fig. 6. Histone H4K12 acetylation regulates telomere recombination. 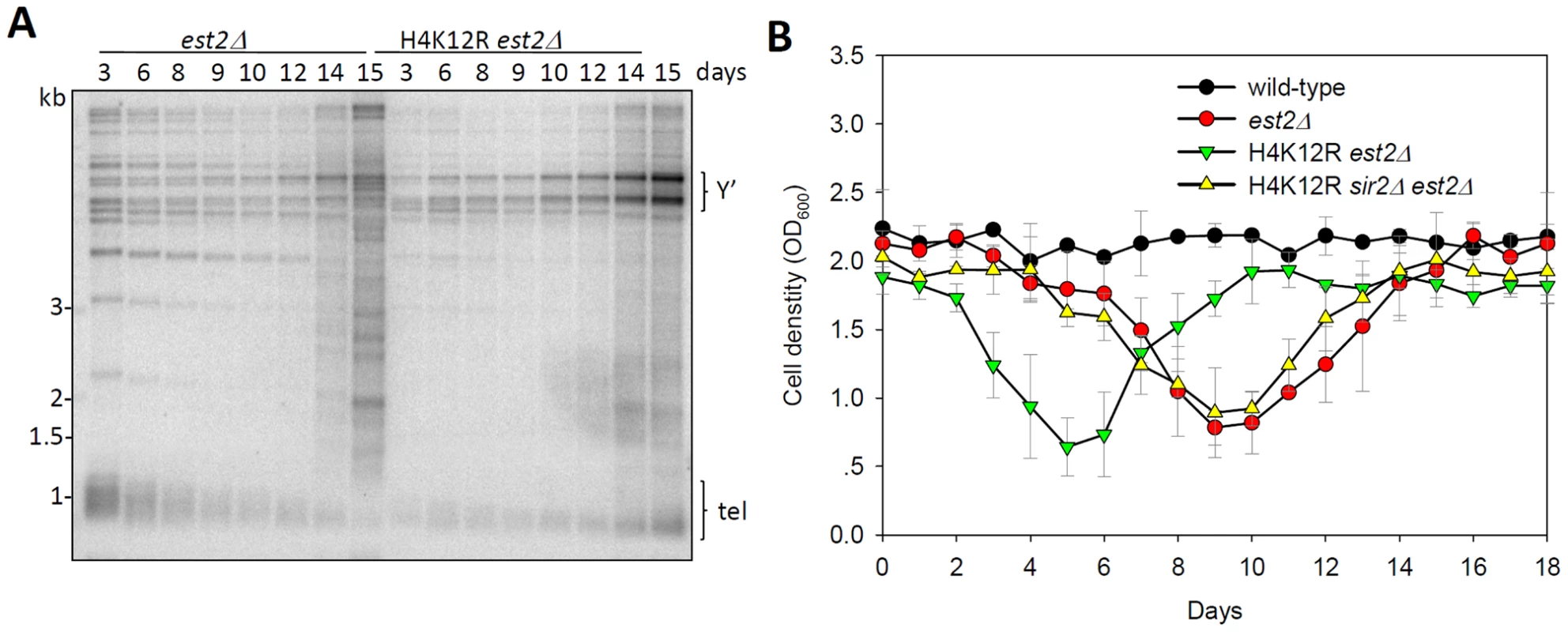
(A) Telomere-blot of est2Δ and est2Δ H4K12R cells. Cells were continuously passaged for the indicated number of days and the genomic DNA was subjected to telomere-blot. Y': Y' elements. (B) Senescence rates were measured in liquid culture by serially passaging strains of the indicated genotypes. Since histone H4K12 acetylation affected both telomere length and recombination, we wondered whether it regulated the senescence rate of telomerase inactive cells. Therefore, we compared the growth potential of est2Δand H4K12R est2Δ cells. The result showed that H4K12R mutation greatly accelerated the senescence rate of est2Δ cells (Figure 6B). Further deletion of SIR2 could suppress the accelerated senescence rate of H4K12R est2Δ cells (Figure 6B). These data suggested that H4K12 acetylation delays senescence driven by Sir-dependent telomere dysfunction.
Histone H4K12 acetylation facilitates basal transcription at telomeric heterochromatin
Previous work had shown that histone H4 mutations that led to increased telomere position effect (TPE) were usually associated with a dramatic decrease of K12 acetylation [36]. To further address this point, we carried out TPE assay [37] to evaluate the effects of histone mutations on the transcriptional state at heterochromatin. A URA3 gene was inserted into a locus that was proximal to the right telomeric TG-tracts of chromosome XIV. Strains were then tested for the relative URA3 expression in histone mutant strains compared with wild-type level. In agreement with a previous report [38], H4K16R mutation markedly reduced telomere silencing (Figure 7). Notably, H4K12R, but not K5R or K8R cells, had less URA3 expression than wild-type cells (Figure 7), suggesting that H4K12R mutation enhances telomere silencing. H4K12R mutation in sir2Δ background had similar URA3 expression to that in sir2Δcells (Figure 7), indicating that the increased silencing by H4K12R mutation is Sir-dependent. Therefore, we concluded that H4K12 acetylation contributes to the basal transcription within telomeric heterochromatin.
Fig. 7. Histone H4K12 acetylation regulates basal transcription at telomeric heterochromatin. 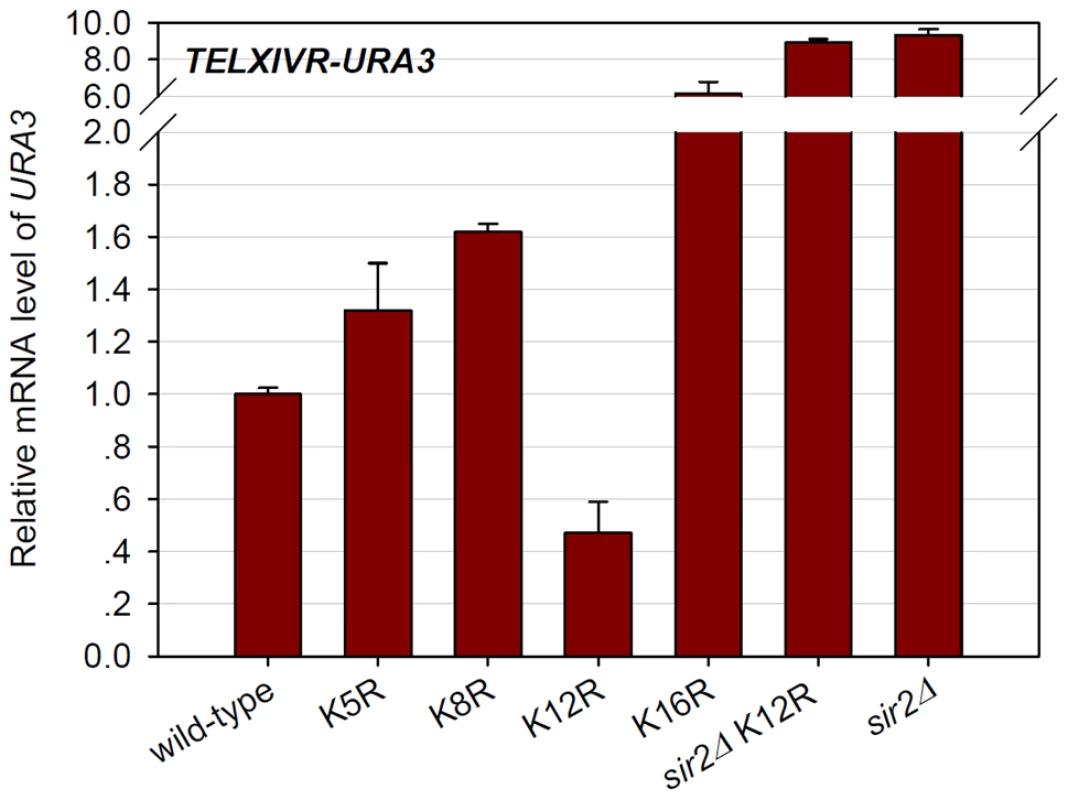
URA3 gene was artificially inserted into a locus that is proximal to the telomeric TG-tracts of chromosome XIV-R. The mRNA of URA3 in wild-type and mutant strains were measured by real-time PCR and normalized to ACT1. The relative mRNA level of URA3/ACT1 in wild-type strain was set as 1. Therefore, values above “1” indicated higher URA3 expression. Discussion
In this study, we have employed the de novo telomere addition assay [16] as a de novo telomeric heterochromatin formation system, to monitor chromatin dynamics occurred on a newly-formed telomere (Figure 1). Compared with the traditional Sir3-induction system [39]–[41], this system works in a more physiological condition, with normal Sir proteins level and much less transcriptional changes across the genome. Hence, the kinetics of multiple events in the course of telomeric heterochromatin formation can be more accurately followed.
The experimental observations we have made in this study establish histone H4K12 acetylation as an important component of yeast telomeric heterochromatin. We have provided phenotypic, genetic and mechanistic evidence to support the presence of H4K12 acetylation inside telomeric heterochromatin. By using stringently controlled ChIP analyses, we detected H4K12 acetylation at most telomeres (Figure 3). Compared with euchromatic H4K12 acetylation, the level of heterochromatic H4K12 acetylation was relatively low but was greatly elevated by SIR2 deletion (Figure S3), a phenomenon also observed in earlier reports [20], [42]. Therefore, H4K12 acetylation, as is the case of acetylation of other H4 lysines, is suppressed by heterochromatin structure. Genetically, the H4K12R mutation, which mimicked an unacetylated state of K12, increased Sir protein binding at telomeric heterochromatin and altered several dynamic telomere-related chromosomal processes (Figure 4, Figure 5, Figure 6, Figure 7). Heterochromatic H4K12 acetylation coincides with H4K12 as a memory mark for the heritable chromatin structure in yeast [36]. Mechanistically, Esa1, the catalytic subunit of NuA4 HAT, bound to silenced telomeres and was responsible for H4K12 acetylation (Figure 3). Since Arp4, another subunit of NuA4 complex, is also enriched at heterochromatic domains [43], it is possible that the whole NuA4 complex binds to telomeric heterochromatin and plays a direct role in the acetylation of H4K12. Earlier work suggested a phosphorylated H2A-dependent mechanism for the recruitment of NuA4 during DNA damage repair [44]. However, due to the fact that normal telomeres are protected from being recognized as double-strand DNA break and phosphorylated H2A is absent from normal telomeres, we propose that yeast cells take a distinct strategy such as a Rap1-dependent mechanism to recruit NuA4 onto telomeric heterochromatin [45].
Esa1 preferentially acetylated nucleosome on both H4K5 and K12 in vivo [46] and in vitro (Figure S4C). This raised a question of why H4K5 was hypoacetylated at telomeric heterochromatin. We suspected that another histone deacetylase besides Sir2 was involved in the establishment of the acetylation pattern at telomeric heterochromatin. Two candidates are Rpd3 and Hda1, which displayed the highest in vivo activity toward acetylated H4K5 from among H4K5, K8, K12 and K16 [47]. Indeed, our recent study did reveal a genetic interaction between Rpd3L and H4K5 at subtelomeric regions [48]. Moreover, analysis of the CIDMS/MS spectra shows K12 to be the most highly acetylated site (54%) from among H4K5, K8 and K12, followed by K5 (32%), and K8 (24%) [49], suggesting that K12 acetylation covers a much wider range of yeast chromatin than that of K5 or K8 acetylation.
NuA4 complex also functions to prevent the Sir complex from spreading out of heterochromatic domains [19], [50]–[52]. Therefore, mutation of ESA1 or other key subunits of the NuA4 complex resulted in gene silencing near heterochromatin [19], [50], [51] and a modest reduction of silencing within telomeric heterochromatin [53], [54]. The histone H4K12 acetylation per se has little effect on heterochromatin boundary activity (Figure 5B and 5C), however, since it is the uniquely acetylated site within heterochromatin, abolishment of histone H4K12 caused a increase of heterochromatin silencing (Figure 7 and [36]).
Chromatin modifications have been implicated in telomere elongation in several organisms [55], [56]. Recent study on H4K16 demonstrated the relationship between histone acetylation and telomere regulation [57]. The natural presence of H4K12 acetylation at telomeres and Sir-dependent regulation of telomere replication via H4K12 have provided additional direct evidence supporting the proposal that chromatin modifications affect telomere homeostasis. Elimination of H4K12 acetylation increased subtelomeric binding of Sir proteins (Figure 5B and 5C), accelerated senescence in est2Δ cells (Figure 6B) and suppressed homologous recombination within TG-tracts (Figure 6A). By contrast, inactivation of Sas2, the acetyltransferase of H4K16 [38], decreased Sir3 binding at telomere ends and thereby delayed senescence in tlc1Δ cells through homologous recombination-dependent mechanism [57]. Therefore, H4K12 acetylation and K16 acetylation seem to play opposite roles in the Sir-dependent regulation of homologous recombination at telomeric heterochromatin. It is quite interesting that H4K12 and K16 are in close proximity to each other but have opposing roles in regulating telomere dynamics through Sir-dependent mechanisms. Finally, a recent paper showed that Esa1 and Rpd3L controlled H4K12 acetylation, which is necessary for cell growth and viability [46]. Although H4K12R does not change the genome-wide transcription profile [34], it is still possible that H4K12 also fine-tunes the chromatin structure at sequences other than the telomeric heterochromatin.
In conclusion, heterochromatin is known as a highly condensed chromatin domain that is transcriptionally silent [1]. However, pioneering studies have recently revealed the dynamic aspect of yeast heterochromatin [11]–[13], [15]. In this study, we have built on these pioneering studies and shown that the H4K12R mutation led to a more condensed telomeric heterochromatin structure (Figure 5) and more static telomere metabolism (Figure 4, Figure 5, Figure 6, Figure 7). Therefore, we propose that H4K12 provides a mechanism for yeast cells to maintain partial plasticity of their telomeric heterochromatin (Figure 8). Interestingly, histone H4K12 acetylation has also been observed at the chromocenter in fly [58]. Mst1, the orthologue of Esa1 in fission yeast, acetylates H3K4 at pericentric heterochromatin to regulate heterochromatin reassembly [59]. TIP60, the orthologue of NuA4 in mammals, physically interacts with Sirt1, the mammalian homologue of Sir2 [60], and associates with tri-methylated H3K9, a hallmark of heterochromatin [61]. Hence, it will be of great interest to determine if histone acetylation also plays a general role in heterochromatin dynamics in other eukaryotes.
Fig. 8. Model for the regulation of telomeric heterochromatin plasticity by histone H4K12 acetylation. 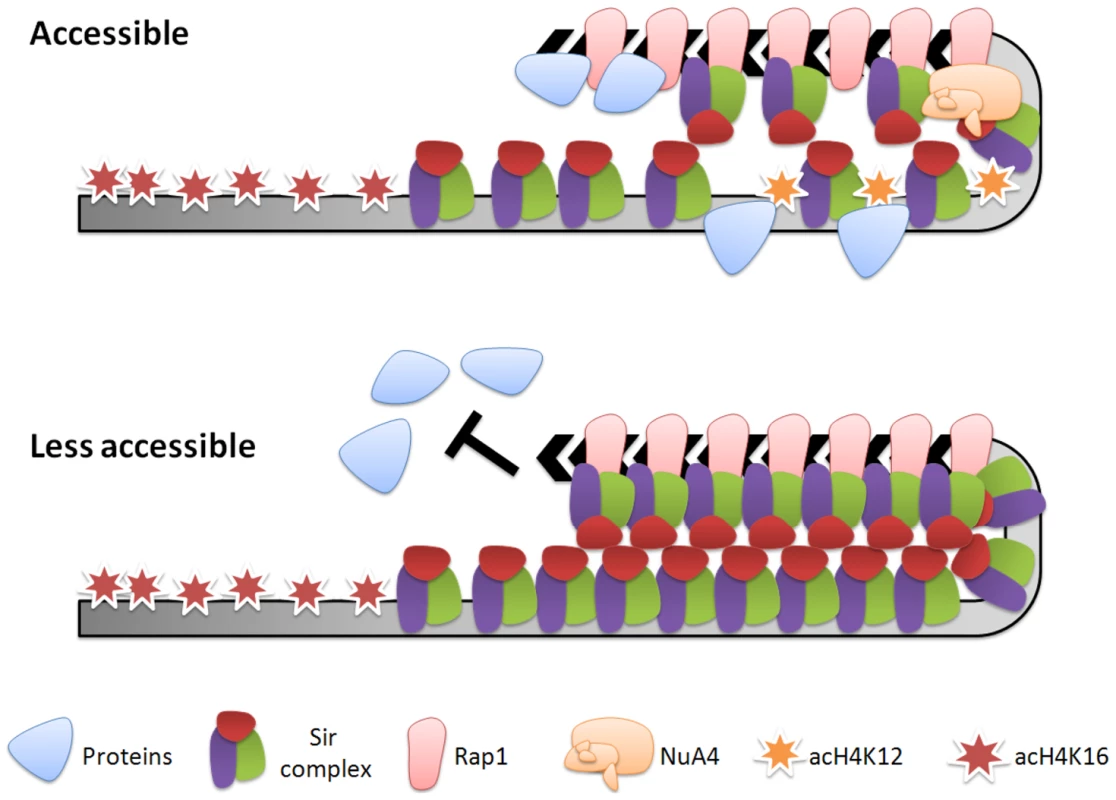
In wild-type cells where subtelomeric histone H4K12 is partially acetylated, heterochromatin structure is plastic and accessible to proteins, such as the RNA polymerase machinery and the telomerase machinery. In H4K12 deacetylated strain, heterochromatin becomes aberrantly condensed and less accessible to proteins, thereby inhibiting chromosomal processes. Materials and Methods
Yeast strains and reagents
Antibodies, yeast strains and primers used in this study are listed in Tables S1, S2, S3, respectively.
Chromatin immuno-precipitation
ChIP assays were performed as described [18], [19]. Most ChIP products were directly analyzed by real-time Q-PCR using SYBR green as a label (TOYOBO). Alternatively, pellet and whole-cell extract DNAs were analyzed by Q-PCR performed in a linear range with 32P-dATP, electrophoresis through 6% PAGE in Tris-Borate-EDTA buffer, and phosphorimager quantification of radioactive bands in dried gels. The relative enrichment value represented the ratio (IPs/Input) at indicated loci relative to internal control. All ChIP experiments were performed in triplicate on paired isogenic wild-type and mutant strains.
HPLC analysis of DNA methylation status
After protein expression was induced by galactose for indicated time, total DNA was isolated from yeast cells by glass beads lysis, proteinase K digestion and extraction with QIAGEN Genomic DNA Kit, followed by RNaseA and RNaseT digestion, phenol/chloroform extraction and re-precipitation. Finally, purified genomic DNA was digested into mononucleoside 5′-monophosphates with Nuclease P1 (Sigma). Nucleosides were separated by an AQ-C18 column (5 µm, 4.6×250 mm, Welch Materials Inc.) guarded by a precolumn (Phenomenex, Security Guard), using a Beckman device (SYSTEM GOLD 125 Solvent Module and SYSTEM GOLD 166 Detector). The eluate was obtained using a flow rate of 1 ml/min and 100% buffer A (10 mM KH2PO4/H3PO4, pH 3.7) for 50 minutes, followed by a shift to 70% A/30% methanol for 10 minutes, and then to 100% buffer A for 10 minutes. Peaks were quantified by measuring their heights with a 32 Karat software V7.0 after identity confirmation by comparison of spikes with different combinations of dCMP and 5′-me-dCMP (Hongene Biotechnology Ltd. Shanghai). At least three independent yeast clones were assayed for each construct.
Southern blot
Southern blot was performed as described [62]. Yeast genomic DNAs were digested with restriction enzymes as indicated, separated by 1.2% agarose gel electrophoresis, and transferred to Hybond-N membrane (Amersham). The blot was hybridized with a 32P-dCTP incorporated probe as indicated. The radioactive signal was detected by phosphorimager.
Micrococcal nuclease sensitivity assay
Micrococcal nuclease (MNase) sensitivity assay was performed as described [27]. Briefly, Cells from 50-ml cultures were collected by centrifugation, treated with zymolyase, and digested with micrococcal nuclease (MNase). genomic DNA samples were purified, digested with BamHI, resolved in 1.2% agarose gels. Then the cutting profiles were visualized after hybridization with an internal probe from Ty5-1.
Immunofluorescence of yeast cells
Cells were grown in YPD medium overnight to a density of ∼1–2×107 cells/ml and were fixed for 30 min by incubation with 3.7% formaldehyde. Next, cells were washed with 0.1 M potassium phosphate (pH 6.5) and P solution (1.2 M sorbitol, 1 M K2PO4), and re-suspended in P solution. Cells were subsequently treated with 0.1 mg/ml Zymolyase (20T, MP Biomedicals) for 10 min, washed with P solution, spotted on Poly-L-Lysine pre-treated slides. After rinsing in PBS-T buffer (PBS containing 0.1% Triton X-100 and 1% BSA), slides were incubated overnight with anti-Myc, anti-Rap1 and anti-Nop1 antibody diluted in PBS containing 1% BSA. Slides were then washed with PBS-T and incubated with the appropriate secondary antibodies conjugated to Cy3 or fluorescein isothiocyanate (FITC). The DNA fluorescence signal was detected by DAPI (1 µg/ml in Phosphate Buffered Saline (PBS) solution) staining. Slides were mounted with PBS containing 1 mg/ml p-phenylenediamine, 2.5 µM NaOH, and 90% glycerol.
Confocal microscopy was performed on a Leica TCS SP2 microscope with a 63× lamda blue objective (oil). Image processing including similar filtration and threshold levels was standardized for all images.
Purification of Esa1 protein
N terminal 6xHis tagged Esa1 protein was overexpressed and purified in E. coli according to the manufacturer's instructions (GE Healthcare).
Supporting Information
Zdroje
1. RuscheLN
KirchmaierAL
RineJ
2003 The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72 481 516
2. LuoK
Vega-PalasMA
GrunsteinM
2002 Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16 1528 1539
3. ImaiS
ArmstrongCM
KaeberleinM
GuarenteL
2000 Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403 795 800
4. HechtA
LarocheT
Strahl-BolsingerS
GasserSM
GrunsteinM
1995 Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80 583 592
5. CarmenAA
MilneL
GrunsteinM
2002 Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem 277 4778 4781
6. KimuraA
UmeharaT
HorikoshiM
2002 Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet 32 370 377
7. SukaN
LuoK
GrunsteinM
2002 Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32 378 383
8. Katan-KhaykovichY
StruhlK
2005 Heterochromatin formation involves changes in histone modifications over multiple cell generations. Embo J 24 2138 2149
9. BiX
BroachJR
2001 Chromosomal boundaries in S. cerevisiae. Curr Opin Genet Dev 11 199 204
10. SmithER
EisenA
GuW
SattahM
PannutiA
1998 ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A 95 3561 3565
11. ChengTH
GartenbergMR
2000 Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev 14 452 463
12. SekingerEA
GrossDS
2001 Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105 403 414
13. ChenL
WidomJ
2005 Mechanism of transcriptional silencing in yeast. Cell 120 37 48
14. AndrauJC
van de PaschL
LijnzaadP
BijmaT
KoerkampMG
2006 Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell 22 179 192
15. TeytelmanL
EisenMB
RineJ
2008 Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet 4 e1000247 doi:10.1371/journal.pgen.1000247
16. DiedeSJ
GottschlingDE
1999 Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99 723 733
17. TamburiniBA
TylerJK
2005 Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25 4903 4913
18. MeneghiniMD
WuM
MadhaniHD
2003 Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725 736
19. ZhouBO
WangSS
XuLX
MengFL
XuanYJ
2010 SWR1 complex poises heterochromatin boundaries for antisilencing activity propagation. Mol Cell Biol 30 2391 2400
20. SukaN
SukaY
CarmenAA
WuJ
GrunsteinM
2001 Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8 473 479
21. NegriniS
RibaudV
BianchiA
ShoreD
2007 DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev 21 292 302
22. ZhuX
GustafssonCM
2009 Distinct differences in chromatin structure at subtelomeric X and Y' elements in budding yeast. PLoS ONE 4 e6363 doi:10.1371/journal.pone.0006363
23. BoudreaultAA
CronierD
SelleckW
LacosteN
UtleyRT
2003 Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev 17 1415 1428
24. KleffS
AndrulisED
AndersonCW
SternglanzR
1995 Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem 270 24674 24677
25. Ruiz-GarciaAB
SendraR
GalianaM
PamblancoM
Perez-OrtinJE
1998 HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem 273 12599 12605
26. ShahbazianMD
GrunsteinM
2007 Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76 75 100
27. Vega-PalasMA
VendittiS
Di MauroE
1998 Heterochromatin organization of a natural yeast telomere. Changes of nucleosome distribution driven by the absence of Sir3p. J Biol Chem 273 9388 9392
28. PalladinoF
LarocheT
GilsonE
AxelrodA
PillusL
1993 SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75 543 555
29. WrightJH
GottschlingDE
ZakianVA
1992 Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev 6 197 210
30. SchoberH
FerreiraH
KalckV
GehlenLR
GasserSM
2009 Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23 928 938
31. de BruinD
KantrowSM
LiberatoreRA
ZakianVA
2000 Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol 20 7991 8000
32. Strahl-BolsingerS
HechtA
LuoK
GrunsteinM
1997 SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11 83 93
33. de BruinD
ZamanZ
LiberatoreRA
PtashneM
2001 Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409 109 113
34. DionMF
AltschulerSJ
WuLF
RandoOJ
2005 Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A 102 5501 5506
35. GrandinN
CharbonneauM
2007 Control of the yeast telomeric senescence survival pathways of recombination by the Mec1 and Mec3 DNA damage sensors and RPA. Nucleic Acids Res 35 822 838
36. SmithCM
HaimbergerZW
JohnsonCO
WolfAJ
GafkenPR
2002 Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc Natl Acad Sci U S A 99 Suppl 4 16454 16461
37. GottschlingDE
AparicioOM
BillingtonBL
ZakianVA
1990 Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63 751 762
38. MeijsingSH
Ehrenhofer-MurrayAE
2001 The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev 15 3169 3182
39. LiYC
ChengTH
GartenbergMR
2001 Establishment of transcriptional silencing in the absence of DNA replication. Science 291 650 653
40. KirchmaierAL
RineJ
2001 DNA replication-independent silencing in S. cerevisiae. Science 291 646 650
41. HoppeGJ
TannyJC
RudnerAD
GerberSA
DanaieS
2002 Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22 4167 4180
42. BraunsteinM
SobelRE
AllisCD
TurnerBM
BroachJR
1996 Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol 16 4349 4356
43. OgiwaraH
UiA
KawashimaS
KugouK
OnodaF
2007 Actin-related protein Arp4 functions in kinetochore assembly. Nucleic Acids Res 35 3109 3117
44. BirdAW
YuDY
Pray-GrantMG
QiuQ
HarmonKE
2002 Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419 411 415
45. ReidJL
IyerVR
BrownPO
StruhlK
2000 Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6 1297 1307
46. ChangCS
PillusL
2009 Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae. Genetics 183 149 160
47. RundlettSE
CarmenAA
KobayashiR
BavykinS
TurnerBM
1996 HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci U S A 93 14503 14508
48. ZhouJ
ZhouBO
LenzmeierBA
ZhouJQ
2009 Histone deacetylase Rpd3 antagonizes Sir2-dependent silent chromatin propagation. Nucleic Acids Res 37 3699 3713
49. SmithCM
GafkenPR
ZhangZ
GottschlingDE
SmithJB
2003 Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem 316 23 33
50. BabiarzJE
HalleyJE
RineJ
2006 Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev 20 700 710
51. ChiuYH
YuQ
SandmeierJJ
BiX
2003 A targeted histone acetyltransferase can create a sizable region of hyperacetylated chromatin and counteract the propagation of transcriptionally silent chromatin. Genetics 165 115 125
52. AltafM
AugerA
Monnet-SaksoukJ
BrodeurJ
PiquetS
2010 NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem 285 15966 15977
53. ClarkeAS
SamalE
PillusL
2006 Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell 17 1744 1757
54. AugerA
GalarneauL
AltafM
NouraniA
DoyonY
2008 Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol 28 2257 2270
55. BlascoMA
2007 The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8 299 309
56. YuEY
Steinberg-NeifachO
DandjinouAT
KangF
MorrisonAJ
2007 Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol Cell Biol 27 5639 5649
57. KozakML
ChavezA
DangW
BergerSL
AshokA
2010 Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. Embo J 29 158 170
58. TurnerBM
BirleyAJ
LavenderJ
1992 Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69 375 384
59. XhemalceB
KouzaridesT
2010 A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev 24 647 652
60. YamagataK
KitabayashiI
2009 Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem Biophys Res Commun 390 1355 1360
61. SunY
JiangX
XuY
AyrapetovMK
MoreauLA
2009 Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 11 1376 1382
62. MengFL
HuY
ShenN
TongXJ
WangJ
2009 Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. Embo J 28 1466 1478
Štítky
Genetika Reprodukčná medicína
Článek Composite Effects of Polymorphisms near Multiple Regulatory Elements Create a Major-Effect QTLČlánek Horizontal Transfer, Not Duplication, Drives the Expansion of Protein Families in ProkaryotesČlánek Segregating Variation in the Polycomb Group Gene Alters the Effect of Temperature on Multiple TraitsČlánek Global Analysis of the Impact of Environmental Perturbation on -Regulation of Gene ExpressionČlánek H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone DeacetylasesČlánek A Mutation in the Gene Encoding Mitochondrial Mg Channel MRS2 Results in Demyelination in the Rat
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Meta-Analysis of Genome-Wide Association Scans Identifies IL18RAP, PTPN2, TAGAP, and PUS10 As Shared Risk Loci for Crohn's Disease and Celiac Disease
- Composite Effects of Polymorphisms near Multiple Regulatory Elements Create a Major-Effect QTL
- Horizontal Transfer, Not Duplication, Drives the Expansion of Protein Families in Prokaryotes
- Genome-Wide Association Study SNPs in the Human Genome Diversity Project Populations: Does Selection Affect Unlinked SNPs with Shared Trait Associations?
- Friedreich's Ataxia (GAA)•(TTC) Repeats Strongly Stimulate Mitotic Crossovers in
- Zebrafish Mutation Leads to mRNA Splicing Defect and Pituitary Lineage Expansion
- Histone H4 Lysine 12 Acetylation Regulates Telomeric Heterochromatin Plasticity in
- Bub1-Mediated Adaptation of the Spindle Checkpoint
- Segregating Variation in the Polycomb Group Gene Alters the Effect of Temperature on Multiple Traits
- Signaling Role of Fructose Mediated by FINS1/FBP in
- RNF12 Activates and Is Essential for X Chromosome Inactivation
- Comparative Study between Transcriptionally- and Translationally-Acting Adenine Riboswitches Reveals Key Differences in Riboswitch Regulatory Mechanisms
- Global Analysis of the Impact of Environmental Perturbation on -Regulation of Gene Expression
- Application of a New Method for GWAS in a Related Case/Control Sample with Known Pedigree Structure: Identification of New Loci for Nephrolithiasis
- H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone Deacetylases
- A Mutation in the Gene Encoding Mitochondrial Mg Channel MRS2 Results in Demyelination in the Rat
- Transcription Initiation Patterns Indicate Divergent Strategies for Gene Regulation at the Chromatin Level
- The Transposon-Like Correia Elements Encode Numerous Strong Promoters and Provide a Potential New Mechanism for Phase Variation in the Meningococcus
- Proteins Encoded in Genomic Regions Associated with Immune-Mediated Disease Physically Interact and Suggest Underlying Biology
- A Novel RNA-Recognition-Motif Protein Is Required for Premeiotic G/S-Phase Transition in Rice ( L.)
- The Mucin-Like Protein OSM-8 Negatively Regulates Osmosensitive Physiology Via the Transmembrane Protein PTR-23
- Genome Sequencing and Comparative Transcriptomics of the Model Entomopathogenic Fungi and
- Rnf12—A Jack of All Trades in X Inactivation?
- Joint Genetic Analysis of Gene Expression Data with Inferred Cellular Phenotypes
- Evolutionary Conserved Regulation of HIF-1β by NF-κB
- Quaking Regulates Expression through Its 3′ UTR in Oligodendrocyte Precursor Cells
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone Deacetylases
- Evolutionary Conserved Regulation of HIF-1β by NF-κB
- Rnf12—A Jack of All Trades in X Inactivation?
- Joint Genetic Analysis of Gene Expression Data with Inferred Cellular Phenotypes
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



