-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Activation of the Immune System by Combinations of Common Alleles
Plants and their pathogens are engaged in an endless evolutionary battle. The invention of new strategies by pathogens pushes plants to continuously update their defenses. This in turn leads the pathogens to circumvent these new defenses, and so on. Given the abundance of potential enemies, it is therefore not surprising that genes involved in defense against pathogens are among the most variable in plants. A drawback of this extreme variation in pathogen-recognition mechanisms is that at times the plant mistakes itself for an enemy, leading to autonomous activation of defense responses in the absence of pathogens. Conventional models for this phenomenon, called hybrid necrosis, require the interaction between two different genes. Here we show instead that hybrid necrosis can be triggered by interactions between variants of a single gene, ACD6 (ACCELERATED CELL DEATH 6). Several of these variants are common in natural Arabidopsis thaliana populations and can interact to give different levels of activation of the immune system. Our results provide important information into the evolution and operation of the plant defense system. Moreover, the abundant presence of ACD6 functional variation suggests a major role for this gene in modulating plant defenses in nature.
Published in the journal: Activation of the Immune System by Combinations of Common Alleles. PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004459
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004459Summary
Plants and their pathogens are engaged in an endless evolutionary battle. The invention of new strategies by pathogens pushes plants to continuously update their defenses. This in turn leads the pathogens to circumvent these new defenses, and so on. Given the abundance of potential enemies, it is therefore not surprising that genes involved in defense against pathogens are among the most variable in plants. A drawback of this extreme variation in pathogen-recognition mechanisms is that at times the plant mistakes itself for an enemy, leading to autonomous activation of defense responses in the absence of pathogens. Conventional models for this phenomenon, called hybrid necrosis, require the interaction between two different genes. Here we show instead that hybrid necrosis can be triggered by interactions between variants of a single gene, ACD6 (ACCELERATED CELL DEATH 6). Several of these variants are common in natural Arabidopsis thaliana populations and can interact to give different levels of activation of the immune system. Our results provide important information into the evolution and operation of the plant defense system. Moreover, the abundant presence of ACD6 functional variation suggests a major role for this gene in modulating plant defenses in nature.
Introduction
Despite its inherent advantage, resistance to pathogens is highly variable in natural populations [1]. One explanation for this lies in fluctuating pathogen pressures, which are expected to result in fitness tradeoffs between maintaining continuous defenses and the metabolic costs incurred in the absence of enemies [2]–[4]. An alternative explanation for individual differences in disease resistance comes from the evershifting front in the evolutionary arms race between pathogens and their hosts. Accordingly, immunity loci are among the most variable genes in both animal and plant genomes [5]–[7]. Yet, too much variation can be dangerous, and lead to inadvertent self-recognition and autoimmunity.
Autoactivation of defenses in the absence of pathogens has been observed both in inbred strains and in hybrid progeny. One of the most visible outcomes of this phenomenon is widespread necrosis due to extensive cell death in leaves, mimicking the hypersensitive response (HR) that is often mounted upon pathogen attack [8]. The most severe cases are those reported in intra - and interspecific plant hybrids. Most cases of hybrid necrosis have been identified in controlled crosses, but some occur in nature [9], [10].
About 2% of random crosses between wild strains (accessions) of Arabidopsis thaliana (henceforth Arabidopsis) result in F1 plants that are smaller than their parents and that have overt signs of leaf necrosis [11]. As in other species [8], [12]–[14], the causal genes identified so far encode either immune receptors or regulators of the immune response [11], [15]. Similar, but weaker symptoms are seen in inbred strains that carry a naturally occurring hyper-active allele of the ACCELERATED CELL DEATH 6 (ACD6) gene; like necrotic hybrids [11], these plants show enhanced resistance to pathogens, but suffer from compromised growth [16].
Here we report a new phenomenon, single-locus hybrid necrosis. Several special alleles at the ACD6 locus interact to activate the immune system independently of the presence of pathogens. The increased immunity in these hybrids is associated with a temperature-dependent reduction in size and fertility. These alleles are responsible for several cases of hybrid necrosis observed in controlled crosses, and, unlike the causal alleles for other cases of hybrid necrosis in Arabidopsis [11], [15], they are common and co-occur in nature. The causal alleles themselves are heterogeneous, and interactions between different combinations elicit different levels of defense responses. Furthermore, the high frequencies of these alleles in the Costa Brava region of Northeastern Spain suggest that they play a role in local adaptation.
Results
Interactions between ACD6 alleles causing hybrid necrosis
Hybrid necrosis cases in Arabidopsis differ in their severity, with some hybrids dying, while others are merely dwarfed. One of the mildest examples is provided by a cross between the Mir-0 and Se-0 accessions from Miramare in Italy and San Elano in Spain [11]. At 16°C, necrosis appeared after three to four weeks in older leaves of Mir-0×Se-0 hybrids, and both final size and fertility were reduced compared to their parents. As with other cases of hybrid necrosis in Arabidopsis, these phenotypes largely disappeared at 23°C [11] (Figure 1A,B). Expression of the disease resistance marker gene PR1 was elevated in the hybrids (Figure 1C), consistent with their increased resistance to the pathogen Hyaloperonospora arabidopsidis [11]. Importantly, resistance has been observed at temperatures that suppress the morphological defects [11].
Fig. 1. Identification of ACD6 as causal for Mir-0×Se-0 temperature-sensitive hybrid necrosis. 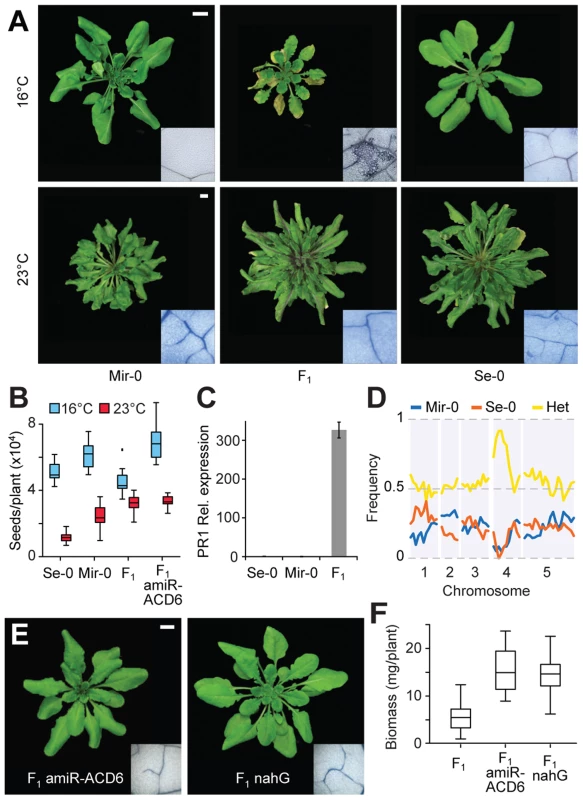
(A) Rosettes of a six-week-old Mir-0 and Se-0 plants and their F1 hybrid. Insets show leaves stained with Trypan blue for dead cells. (B) Seed set. Averages for 15 individuals are shown. Seed set is significantly different between F1 and F1 amiR-ACD6 plants at 16°C (p<0.001), but not at 23°C (p = 0.69). (C) Relative expression levels of PR1, as measured by quantitative RT-PCR (averages for five biological replicates). Expression values were normalized to those of Se-0 individuals. Differences in PR1 expression between Mir-0×Se-0 hybrids and the parental genotypes were significant at p<0.05. Error bars represent standard errors. (D) Genome-wide mapping of the hybrid necrosis phenotype in an F2 population. (E) Rosettes of six-week-old hybrids grown at 16°C, with necrosis suppressed by ACD6 knockdown or nahG-mediated depletion of SA (see the non-transgenic hybrid grown at 16°C in Figure 1A for comparison). Insets show leaves stained with Trypan blue for dead cells. (F) Aboveground biomass for six-week-old plants grown at 16°C. Averages for at least 10 individuals are shown. Size bars = 1 cm. Genetic mapping in the F2 generation established that the necrotic phenotype of Mir-0×Se-0 hybrids was linked to a single region of the genome (Figure 1D), in agreement with a 1∶1 segregation ratio of normal and F1-like plants in the F2 generation [11]. The final mapping interval of 290 kb (between 8.11 and 8.40 Mb on chromosome 4) included three immunity loci: At4g14370, which encodes an immune receptor of the TIR-NBS-LRR type; ACD6 (At4g14400); and the adjacent ACD6 paralog At4g14390. We knocked down each gene with artificial microRNAs (amiRNAs; [17]). Only the amiRNA against ACD6 suppressed leaf necrosis and increased plant size and seed production in hybrids (Figure 1B,E,F; Figure S1). ACD6 encodes an ankyrin repeat transmembrane protein that acts mainly through the hormone salicylate (SA) [16], [18]–[20]. In agreement, depletion of SA by expression of a bacterial salicylate hydroxylase, nahG [21], resulted in suppression of the hybrid phenotypes as well (Figure 1E,F).
The ACD6 locus of Mir-0 had a similar organization as the reference Col-0 allele (Figure 2A), and a 7.2 kb genomic fragment spanning ACD6 reproduced the hybrid phenotype when transformed into Se-0 plants (Figure 2B, Table S1). In Se-0, there were two tandem copies of ACD6 (ACD6A and ACD6B; Figure 2A); only transformation of ACD6A into Mir-0 caused necrosis and reduced growth (Figure 2B, Table S1). Similar transgenic experiments with Col-0 and its ACD6 allele confirmed the specificity of the interaction between the Mir-0 and Se-0 alleles of ACD6 (Table S1).
Fig. 2. Mapping of causal regions in ACD6 alleles. 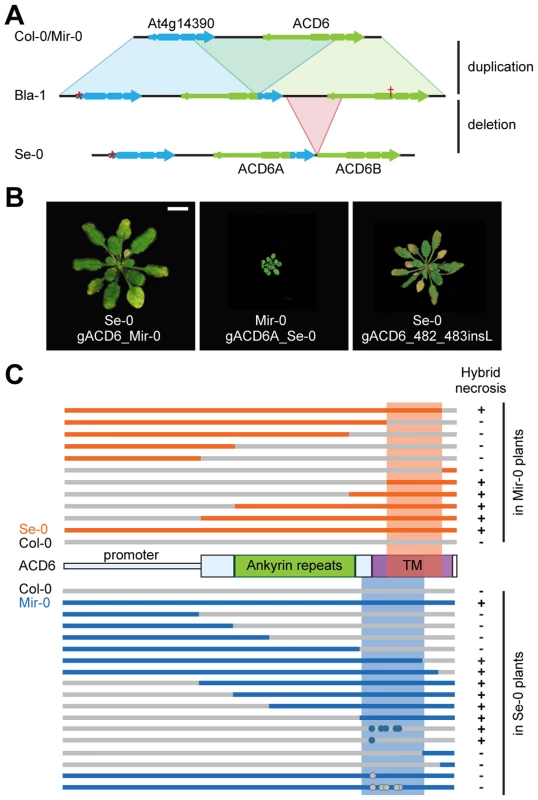
(A) Organization of the ACD6 locus in Col-0, Bla-1 and Se-0. Col-0 likely represents the ancestral configuration of the ACD6 region. A partial duplication of At4g14390 and ACD6 followed by a deletion of the 5′ portion of ACD6B likely produced first the Bla-1 and then the Se-0 arrangement. Asterisk indicates missing start codon in At4g14390, the cross a premature stop codon in the Bla-1 allele of ACD6B. Size bar = 1 cm. (B) Rosettes of six-week-old plants in which transgenic introduction of ACD6 alleles recapitulated the phenotype of hybrids. The gACD6_482_483insL transgene is the Col-0 allele of ACD6 with a single amino acid insertion between position 482 and 483. (C) Schematic representation of chimeric constructs used to map ACD6 sequences causal for hybrid necrosis. Dots indicate single amino acid changes. Experiments with chimeric transgenes showed that promoter activity did not account for differences between Mir-0, Se-0 and Col-0 alleles (Figure 2C). Domain swaps and site-directed mutagenesis further localized residues responsible for hybrid necrosis to the transmembrane domain for both the Mir-0 and Se-0 alleles (Figure 2C). These experiments point to the interallelic interaction occurring at the protein level.
Since the first report of hybrid necrosis in Arabidopsis [11], we have identified additional examples of hybrid necrosis, including several other Mir-0×Se-0-like cases (Table 1; Tables S2, S3). Using test crosses, segregation analyses, amiRNA knockdowns, and transformation with genomic fragments from Mir-0 and Se-0, we confirmed Mir-0 - and Se-0-like alleles of ACD6 as causal for several independent hybrid cases (Table S1, S2; Figure 3). We compared ACD6 sequences between these and other Arabidopsis strains to gain a better understanding of the activity and evolutionary background of these alleles. Notably, while Mir-0-like strains had a broad distribution that included much of the native range of the species throughout Eurasia, Se-0-like strains were only found in the Northeast of Spain, along the Costa Brava (Table 1).
Fig. 3. Lesions and reduced biomass in different hybrids. 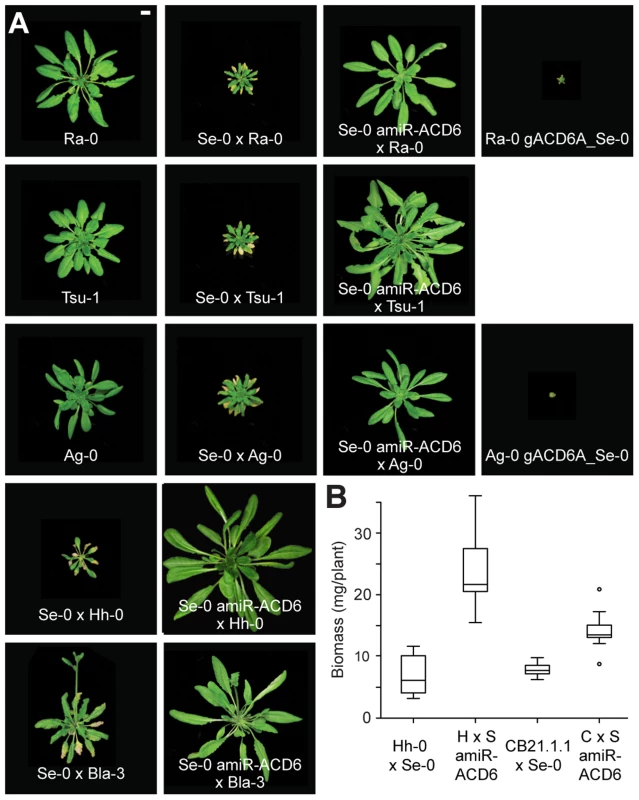
(A) Rosettes of six-week-old plants. Size bar = 1 cm. (B) Aerial biomass for six-week-old plants grown at 16°C. Averages for at least eight individuals are shown. CB21.1-1 carries a Mir-0-like class III allele of ACD6, and crosses with Se-0 resulted in milder necrosis than for accessions carrying a Mir-0-like class I or II allele. Accordingly, the effect of knocking down ACD6 in these hybrids had a smaller effect on biomass production. Differences between transgenic lines and the respective hybrids were significant at p<0.001. Tab. 1. Accessions with <i>ACD6</i> hybrid necrosis alleles (see also <em class="ref">Tables S1</em> and <em class="ref">S2</em>). 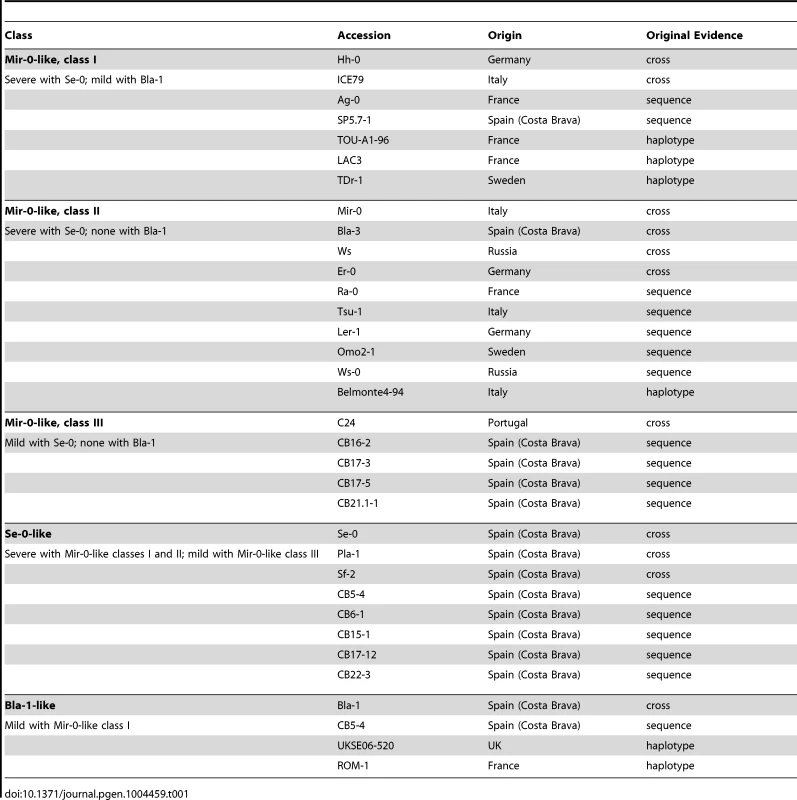
A complex evolutionary history for Se-0-like alleles
As described above, the ACD6 locus in Se-0 contained an additional ACD6 copy. This organization is most likely derived, since we did not find it in other Arabidopsis strains or in Arabidopsis lyrata. The ACD6 paralog At4g14390, located immediately upstream of ACD6, lacked a start codon in Se-0, while a fragment from the promoter through part of the first intron was missing in ACD6B, indicating that only one of the three genes, ACD6A, was functional. ACD6A appeared to be a chimeric gene that formed recently through intralocus duplication and recombination, deduced from the 3′ portion being almost identical to that of the upstream At4g14390 gene, and the 5′ region from the first intron on being almost identical to that of the downstream ACD6B gene (Figure 2A). Thus, the ACD6A sequences causal for hybrid necrosis were derived from the At4g14390 pseudogene, Se-0-like alleles of At4g14390 were found in other strains with the ancestral two-gene organization of the ACD6 locus (Figure 4A, Figure S2A), suggesting that the pseudogenized state of At4g14390 preceded the duplication event.
Fig. 4. Functional diversity of the ACD6 alleles. 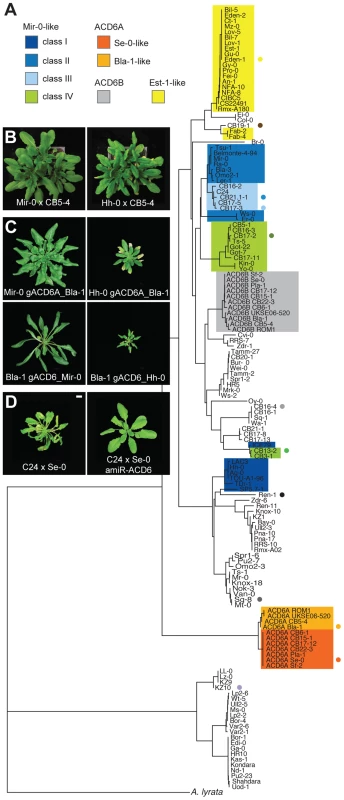
(A) Hierarchical clustering of ACD6 alleles from 131 Arabidopsis accessions and the Arabidopsis lyrata strain MN47. Classes are described in the text and in Table 1. Dots indicate genotypes found at the CB16 collection site near Llagostera, Spain (see Fig. 5B). The deeply diverged group above the A. lyrata MN47 allele is the KZ-10 group (described in [16]). (B) Rosettes of six-week-old plants grown at 16°C. The CB5-4 accession carries a Bla-1-like allele of ACD6, and produced mild necrosis only when crossed to Hh-0-like accessions. Crosses with CB5-4 instead of Bla-1 are shown because an additional, genetically independent mechanism of incompatibility between Bla-1 and Hh-0 complicated observation of the necrosis phenotype in these crosses. (C) Rosettes of six-week-old plants transformed with different genomic constructs and grown at 16°C. (D) Rosettes of six-week-old plants grown at 16°C. C24 has a Mir-0-like class III ACD6 allele; interaction with the Se-0 allele resulted in mild hybrid necrosis. Size bars = 1 cm. Across seven Se-0-like strains examined in detail, the entire 14 kb ACD6 locus lacked any polymorphisms, supporting a very recent origin of this allele in Northeastern Spain or a recent selective sweep in this region. The Bla-1 strain, also from this region, had an arrangement that likely reflects the ancestral state with respect to the Se-0 allele, lacking the partial deletion of ACD6B (Figure 2A). Bla-1 crosses, however, produced hybrid necrosis only in combination with two of the Mir-0-like strains, Hh-0 and ICE79 (Figure 4B; Table 1; Table S2). Although ACD6B was expressed in Bla-1 (Figure S2B), a premature stop codon was predicted to truncate the open reading frame. The encoded protein lacks therefore most of the ankyrin repeats along with the transmembrane domain required for ACD6 activity [22] (Figure 2A). Few additional derived polymorphisms distinguished the Se-0-like from the Bla-1-like alleles. Transgenic experiments confirmed that ACD6A was causal for Bla-1×Hh-0 hybrid necrosis. They also showed that the five non-synonymous SNPs distinguishing the Se-0 and Bla-1 alleles of ACD6A were responsible for the failure of the Bla-1 allele to interact with the Mir-0 allele (Figure 4C; Table S1).
Extensive sequence and functional variation in Mir-0-like alleles
In contrast to the Se-0-like alleles, there was considerable sequence variation among Mir-0-like alleles from Bla-3, Er-0, Hh-0, ICE79, and Ws-0 (Figure 4A), suggesting an older origin. ACD6 alleles from these strains shared four characteristic amino acid substitutions and a single amino acid insertion. An ACD6 transgene with the single amino acid insertion, a leucine between positions 482 and 483 (482_483insL), introduced into the Col-0 reference allele was sufficient to induce hybrid necrosis-like symptoms in Se-0 plants (Figure 2B, C; Figure S3), but only the Hh-0, and not the Mir-0 ACD6 transgene recapitulated the hybrid phenotype in the Bla-1 background (Figure 4C), indicating that variation in the sequence of ACD6 is responsible for the differences in the behavior of Mir-0-like accessions.
In agreement with Mir-0-like alleles being more broadly distributed, we found several additional strains with the Mir-0 causal polymorphism among a commonly used reference set of 96 strains [23] (Table 1; Figure 4A). Based on the phenotype of hybrids with Se-0 and Bla-1, we could divide these accessions into four classes. The first two were defined by the alleles described above: class I alleles, such as Hh-0, produced severely affected hybrids regardless of the crossing partner, while class II alleles, including Mir-0, interacted only with Se-0. Class III alleles also interacted only with Se-0, but produced milder symptoms compared to class II alleles. Finally, class IV alleles did not result in any necrosis (Figure 4A, D; Table 1; Tables S1, S2; Figure S4). The class III and IV alleles were distinguished from each other and from class I and II alleles by unique polymorphisms, suggesting that the reason for their different behavior resided in the ACD6 gene itself. F2 progeny involving class III and IV alleles did not produce additional phenotypic classes either, confirming the absence of independently segregating, extragenic suppressors of necrosis (Table S3), and supporting functional differentiation among ACD6 alleles.
Local co-occurrence of hybrid necrosis ACD6 alleles in Northeast Spain
Based on shared single nucleotide polymorphisms (SNPs) across the ACD6 region, we identified additional Mir-0-like strains in a set of 1,307 unique accessions that had been genotyped at high density [24]. This set included eight known Mir-0-like strains and four known Se-0/Bla-1-like strains. Sixty-eight additional accessions had patterns of polymorphism consistent with Mir-0-like class I, II or III alleles. Sequence analysis of 25 that were representative for different subgroups indicated that all carried a hybrid necrosis-inducing ACD6 allele. Test crosses with four of the new accessions confirmed that these alleles could induce hybrid necrosis in combination with Se-0 or Bla-1. At the same time, we found only four additional accessions with Se-0/Bla-1-like ACD6 alleles in this global sample. All four came from outside Spain and had Bla-1-like sequence and activity, interacting only with Hh-0 and not Mir-0 (Table 1, Tables S1, S2 and S4).
Because Mir-0 - and Se-0/Bla-1-like alleles co-occurred in the Northeast of Spain, we investigated the distribution of ACD6 alleles in this region in more detail. We first screened a set of 54 accessions with unique multi-locus genotypes collected in 2007 from the Costa Brava region (Table S5). We found that five each carried Mir-0 - and Se-0/Bla-1-like ACD6 alleles (Table 1). Crosses between these strains and with Mir-0, Hh-0, Se-0 and Bla-1 produced the expected phenotypes, as did ACD6 knockdowns (Figure S4; Table S2). To further determine whether ACD6 alleles responsible for hybrid necrosis co-occurred in local populations, we collected over 2,000 individuals representing 13 populations in March 2012 (Table S6). Sequencing of the ACD6 locus in 1,751 of these samples [25] confirmed that these populations harbored both Mir-0 - and Se-0/Bla-1-like alleles (Figure 5A). We focused on one population, CB16, with 640 individuals, in an uncultivated field on the immediate outskirts of Llagostera. Sequencing identified 12 different ACD6 alleles in this population, including two distinct Mir-0-like class III alleles (combined frequency 31%) and an Se-0-like allele (frequency 13%) (Figure 5B). Despite Arabidopsis being predominantly selfing, moderate levels of outcrossing are frequently observed in natural populations [26], and 12 plants were heterozygous at the ACD6 locus. Two of these hybrids were heterozygous for Mir-0 - and Se-0-like alleles (Figure 5B).
Fig. 5. Co-occurrence of different ACD6 alleles in the Costa Brava region. 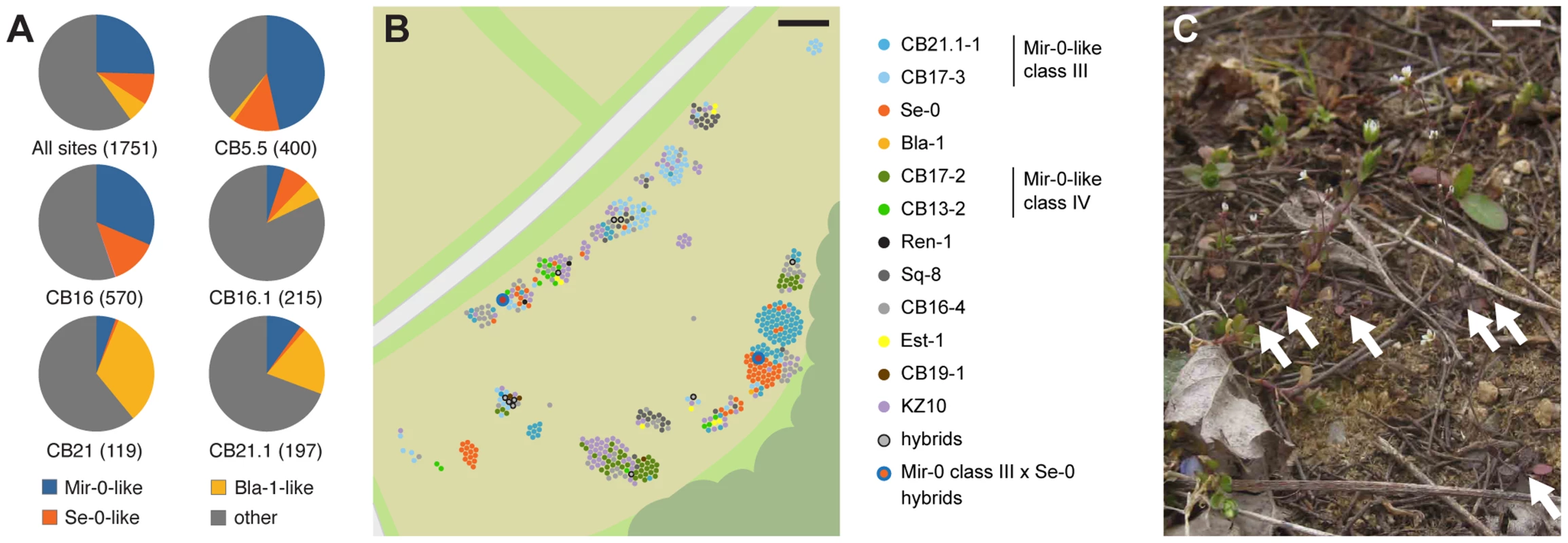
(A) ACD6 allele frequencies in different Costa Brava populations. The numbers of genotyped individuals are given in parentheses. “Other” alleles include Mir-0-like class IV alleles, which do not induce hybrid necrosis. (B) Distribution of individuals carrying different ACD6 alleles at the CB16 collection site near Llagostera, Spain. Alleles are named according to their similarity to those reported in Fig. 4A. (C) Example of Arabidopsis plants growing at site CB16. Arrows point to rosettes. Size bars = 10 m in B, 1 cm in C. We then analyzed 3,641 restriction site associated DNA (RAD) markers in 1,688 individuals from the Costa Brava region [25]. These included 147 individuals that shared the Se-0 allele at ACD6: despite the lack of diversity at ACD6 in these individuals, there was substantial genome-wide diversity in this group (Figure 6), indicating that the Se-0 allele had not merely spread as a clonal lineage, but also through outcrossing. While linkage disequilibrium (LD) around ACD6 tended to be generally lower than the genome-wide average, this was not the case for the Se-0 group, in agreement with a recent origin of the Se-0 allele (Figure S5). Mir-0-like alleles were found in Northeast Spain at a high frequency as well; out of 958 individuals with a unique multi-locus genotype, 22% (207) carried Mir-0-like alleles, while 9% (81) had an Se-0 allele and 7%(70) a Bla-1 allele. These percentages are significantly higher than in the global sample of Arabidopsis accessions (Mir-0-like = 6%; Se-0 = ND; Bla-1 = 0.3%; p<0.01).
Fig. 6. Genome-wide analysis of Costa Brava populations. 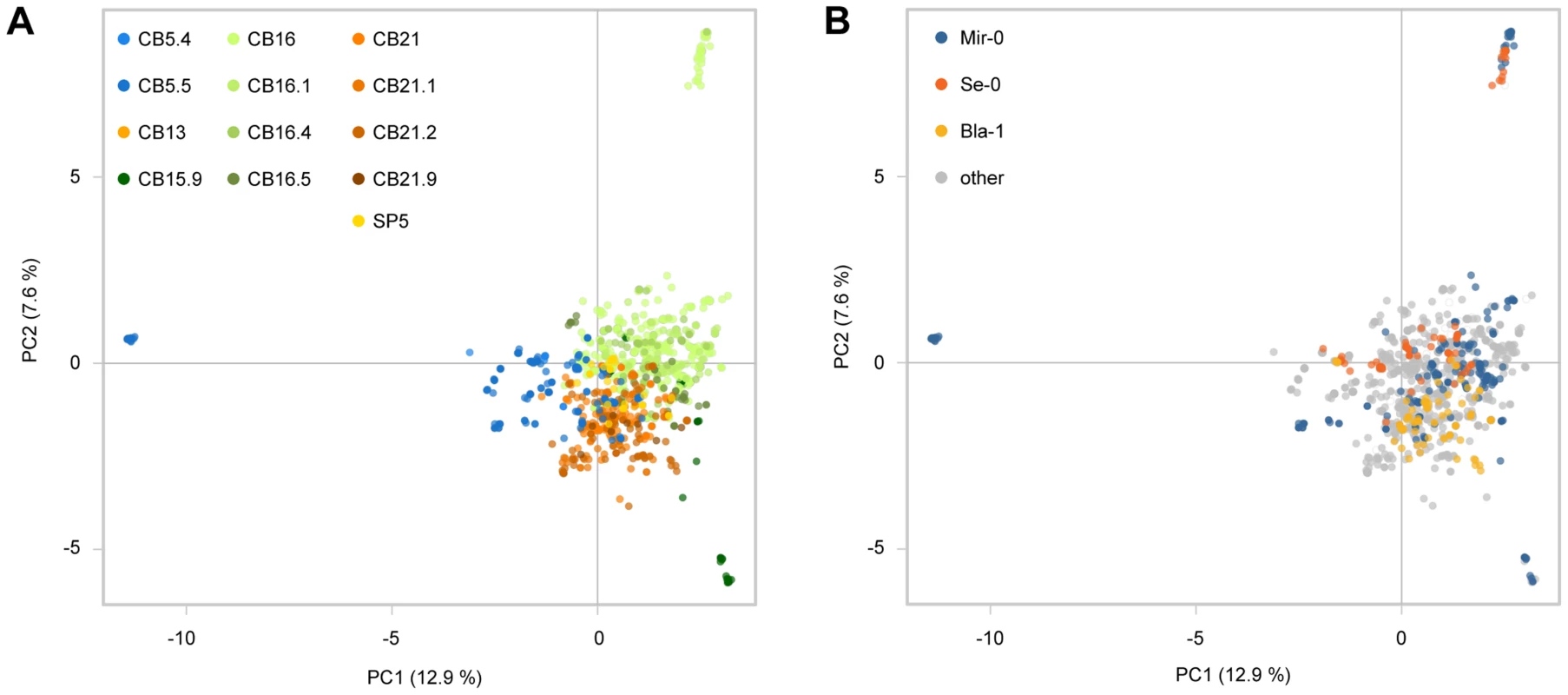
(A) Principal component analysis (PCA) of 1,688 Costa Brava individuals with 730 SNP markers with complete information. Populations are color coded. (B) Same as in (A), but individuals are color coded by ACD6 allele types. Discussion
Hybrid necrosis caused by allelic interactions at a single locus
Hybrid necrosis has attracted the attention of plant breeders and evolutionary biologists for decades [8], not least because they conform to the classical two-gene models of Bateson, Dobzhansky and Muller for the evolution of genetic incompatibilities [11]–[15]. We find that interactions between two alleles of a single gene, ACD6, are sufficient to induce similar hyper-activation of the plant immune system, making this the first described case of single-locus hybrid necrosis.
Insights into ACD6 function from hybrid necrosis alleles
ACD6 encodes a transmembrane protein with ankyrin repeats. In plants with the induced gain-of-function allele acd6-1 a single amino acid change in the transmembrane domain leads to constitutive activation of defense responses, resulting in increased pathogen resistance, extensive leaf necrosis and stunted growth [18], [19], [22]. We recently described a similarly hyper-active allele of ACD6 in natural strains of Arabidopsis [16]. This allele, dubbed ACD6-Est from the name of the strain in which it was initially discovered, segregates at intermediate frequencies in the global Arabidopsis population, and strains carrying it display similar phenotypes as the acd6-1 mutant. Although ACD6-Est carries many different non-synonymous substitutions compared to the reference allele, only two amino acid changes in the transmembrane domain are required for its gain-of-function activity (Figure S3, Table S7).
Mir-0×Se-0 hybrids present more extreme phenotypes than strains with the ACD6-Est allele. Interactions between ACD6 alleles in Mir-0×Se-0 hybrids occur at the protein level, which is consistent with ACD6 forming oligomeric complexes [27]. As with acd6-1 and ACD6-Est, the causal amino acid changes in the Mir-0 and Se-0 alleles map to the transmembrane domain (Figure S3, Table S7). These observations confirm the functional importance of the transmembrane domain and suggest a major role of multimerization in regulating ACD6 activity. However, despite these structural similarities, plants with hyperactive ACD6 alleles and necrotic hybrids differ in their responses to temperature. While the immune phenotypes of plants with the Est-1 allele are largely insensitive to changes in temperature, the phenotypes of Mir-0×Se-0 hybrids and the acd6-1 mutant are attenuated at higher temperature (Figure S6), as is typical for immune responses [28].
Complexity of hybrid necrosis in Mir-0×Se-0-like crosses
A notable feature of the Mir-0×Se-0 system is variation in the expression of hybrid necrosis. Variation in the degree of lethality has been documented in interspecfic Drosophila crosses as well [29], [30]. Different from these other systems, we have shown that phenotypic variation is primarily controlled by the strength and specificity of interactions between several sub-categories of Mir-0 - and Se-0-like alleles.
Se-0-like alleles have a unique evolutionary history. The ancestral organization for this locus is likely to be the one found in the reference Col-0 strain, with two paralogs, At4g14390 and ACD6, derived from an ancient duplication event. Next, At4g14390 became pseudogenized, a state found in several Arabidopsis strains (Fig. S1A). This was followed by a tandem duplication creating the chimeric ACD6A gene upstream of ACD6B, which corresponds to ACD6 in the reference genome (Figure 2A, Figure S3). This configuration is found in Bla-1, which has partial Se-0-like activity. Finally, the promoter and first exon of ACD6B was deleted, giving rise to the Se-0 allele, while the ACD6B copy of Bla-1 appears to have independently suffered a nonsense mutation. A more recent origin of the Se-0 allele compared to Bla-1 is also supported by their geographical distribution; while Bla-1 is broadly distributed at low frequency in Europe, Se-0 is found only in Northeast Spain, where it rose to high frequency.
It is interesting that the causal polymorphisms for hybrid necrosis in the ACD6A allele are derived from At4g14390. These polymorphisms could accumulate freely in At4g14390 once it became a pseudogene and therefore relieved from selective pressures. The duplication event that gave rise to ACD6A “resuscitated” part of this pseudogene and made these polymorphisms part of a functional gene again. The importance of pseudogenization and resurrection in determining the fate of duplicated genes has been proposed before [31], [32], but instances in which a contribution to functional divergence has been documented are rare ([33], [34]; see also [35]). Gene conversion involving pseudogenes is known to be a major source of immunoglobulin diversity in chicken [36], [37] and of antigenic variation in several human pathogens [38]
Co-occurrence of Mir-0 and Se-0-like alleles in natural populations
Different from other examples of hybrid necrosis in Arabidopsis [11], [15], the Mir-0 and Se-0-like alleles are both locally common and can be found at high frequencies in the same population. In addition, we found individuals heterozygous for the causal alleles in nature. The high frequency of Mir-0 - and Se-0-like alleles in the Costa Brava region is suggestive of a role in local adaptation. This interpretation is supported by the lack of polymorphisms found among Se-0-like alleles, possibly due to a recent selective sweep for this allele. The possibility that these alleles would have instead achieved high frequency in this area simply by genetic hitchhiking is not supported by our finding that both Mir-0 - and Se-0-like alleles are present in several different genetic backgrounds (Figure 6).
An alternative explanation for the patterns observed in Costa Brava is that Mir-0×Se-0 hybrids are conditionally advantageous or heterotic. Increased resistance to pathogens in these hybrids could compensate for their reduced fitness, especially in the mild climate of Northeast Spain, which would likely mitigate the severity of the necrosis (Figure S7). This hypothesis is partially supported by the observation that hybrids carrying both causal ACD6 alleles could not be readily distinguished from inbred individuals in natural populations. It should be noted, however, that most plants in such population were small, probably due to abiotic stress exposure, possibly limiting the expression of hybrid necrosis symptoms (Figure 5C). Moreover, Mir-0×Se-0 hybrids can withstand long period of cold exposure without further reduction in their fitness, meaning that they would be able survive winter (most plants in Costa Brava seem to germinate in autumn and to overwinter as rosettes) (Figure 7). This situation would have similarities with what we observed for hyperactive Est-1 alleles of ACD6, which are maintained at intermediate frequencies by balancing selection [16]. Additional surveys of genetic diversity in the Costa Brava region would be required to test this hypothesis.
Fig. 7. Effect of low temperature on hybrids. 
(A) Rosettes of plants grown for four weeks at 16°C in long days and then transferred to 4°C in short days for 16 weeks. Size bar = 1 cm. (B) Seed set for plants that were exposed to a six-week period of 4°C (short days) one week after sowing, and afterwards returned to the indicated temperatures. Averages from at least 15 individuals are shown. Seed set was significantly different between F1 and F1 amiR-ACD6 plants at 16°C (p<0.001), but not at 23°C (p = 0.60). In conclusion, we have discovered a complex, single-gene hybrid necrosis system in which interactions between alleles with a diverse evolutionary history lead to different degrees of activation of the immune system. We have identified the causal polymorphisms and described the population genetic dynamics of the causal alleles. The complex structure and extraordinary functional diversity at the ACD6 locus not only make it very similar to conventional immune receptors of the NLR class [7], [39], [40], but also point to a central role of ACD6 in fine-tuning immunity in natural populations of Arabidopsis. Further investigation of this system will provide additional insight into the mechanism of ACD6 action and will help to define the fuzzy boundary between beneficial priming of resistance and deleterious autoimmunity in plants.
Materials and Methods
Plant material and growth conditions
Seeds were obtained from the European Arabidopsis Stock Center (NASC). Some of the crosses have been described [11]. Plants were grown in growth rooms under long days (16 hours of light, 8 hours of dark) at 16°C with 65% humidity, unless stated otherwise.
Genetic mapping
Genomic DNA was isolated from 96 Mir-0×Se-0 F2 plants with an F1-like phenotype using a BioSprint 96 (Qiagen, Hilden, Germany). Plants were genotyped using a panel of 149 SNPs [41] (Sequenom, San Diego, CA, USA). Fine mapping was performed using DNA from 864 additional necrotic F2 plants, isolated using a modification of the CTAB method for 96-well plates [42]. Markers used for fine-mapping are reported in Table S8. To test linkage of the necrosis phenotype in Bla-1×Hh-0 and ICE79×Bla-1, 96 F2 plants were genotyped for two markers flanking the ACD6 region. In addition, 16 to 62 F2 plants each were grown from 36 crosses among accessions from the Costa Brava region, or between these accession and Se-0 or Mir-0, and presence of leaf necrosis was assessed (Table S3).
Histology
Trypan blue (Sigma-Aldrich, St. Louis, MO, USA) staining was performed as described [43].
Biomass and seed analysis
For dry weight analysis, the aerial parts of plants grown for six weeks at 16°C were dried at 85°C overnight, then weighed. For seed set analysis, 15 plants of each genotype were grown until the end of their life cycle in both 16°C and 23°C long days, with or without a six week vernalization treatment at 4°C in short days. Seeds were counted in six randomly chosen mature siliques per plant (at least 80 siliques per genotype under each condition). Total seed number for each plant was determined by multiplying seed averages per silique with the total number of siliques. The low seed production of Se-0 plants in 23°C conditions was due to a large fraction of aborted siliques. Tukey-Kramer tests were used to determine significance for multiple comparisons.
ACD6 sequence analysis
For Mir-0 and Hh-0, fosmid libraries were made using the Copy Control Fosmid Library production kit (Epicentre Biotechnologies, Madison, WI, USA) with 20 µg genomic DNA as starting material, following the manufacturer's instructions. The libraries were screened with two probes flanking the ACD6 locus. Fosmids were shotgun sequenced and individual sequences were assembled using SeqMan (DNAstar, Madison, WI, USA). PCR fragments covering the entire region in Se-0 were amplified and sequenced. A similar approach was used to sequence the full-length ACD6 gene from other accessions. Sequences were assembled using SeqMan and then aligned using CLUSTALW version 2 [44]. Neighbor joining trees were computed and plotted using MEGA5 [45].
Transgenes
The amiRNA against ACD6 has been described [16]. AmiRNAs targeting At4g14370 and At4g14390 were designed using the WMD online tool (http://wmd3.weigelworld.org; [17]; Table S9), and placed under control of the constitutive CaMV 35S promoter in pFK210 derived from [46]. All amiRNAs were transformed into Se-0 plants, and primary or later-generation transformants were crossed to Mir-0 or other accessions. The genomic fragments for Mir-0 and Hh-0 were PCR-amplified from fosmid clones. For the Se-0 genes in the ACD6 region, genomic DNA was used as template for PCR amplification; to ensure specific amplification, it was necessary to amplify two different fragments each for ACD6A and ACD6B, and then reassemble each gene using specific restriction enzyme sites. Other ACD6 alleles were similarly amplified from genomic DNA. Genomic constructs including the putative promoter region (from the 3′ end of the upstream gene) and several hundred base pairs of sequence beyond the putative 3′ UTR, were cloned into pFK202, a pGREEN-derived binary vector. Different regions were exchanged between alleles cloned into pFK202 using restriction enzymes; when restriction sites were not available, fragments were amplified from the two alleles, joined by overlap PCR and inserted into the appropriate genomic clone. Non-synonymous substitutions in the codons for amino acids 485, 486, 520 and 521 of the ACD6 protein, as well as a single leucine insertion between positions 482 and 483, were introduced by PCR-mediated mutagenesis into the Col-0 genomic construct [16]. Amino acid positions refer to the ACD6 protein sequence in the reference Col-0 strain, unless otherwise indicated. Constructs were introduced into plants by Agrobacterium tumefaciens-mediated transformation [47].
Haplotype analysis
Genome-wide genotype information for 1,307 accessions was obtained from ref. [24], and haplotype similarity was visually assessed for a 60 kb region centered around the ACD6 locus. The 120 accessions with the most similar haplotypes to known Mir-0-like alleles were divided into subgroups with identical or highly similar haplotypes, and one or two representative accessions from each subgroup were selected for further analysis. The sequence of the transmembrane region of ACD6 was obtained for these accessions and compared to the sequences of known Mir-0-like alleles (Table S4). Test crosses were made for four of these accessions (Table S2).
Expression assays
Quantitative reverse transcription PCR (RT-PCR) assays were performed as described [48], using RNA extracted from the 12th leaf of 6-week old plants. Expression levels were normalized against BETA-TUBULIN-2 (At5g62690). An experimentally quantified average amplification efficiency of 1.98 was used in the calculations [16]. Primers used for RT-PCR are given in Table S10.
Field collections
An initial screening of ACD6 sequences was performed on pooled leaf tissue collected from 27 sites in March 2012 in the Costa Brava region (Spain). One hundred seventy-nine pools (each from 6–10 plants) were assayed by PCR for the presence of Mir-0-like and Se-0-like sequences. Subsequently, approximately 2,200 samples were collected from the thirteen larger populations in the area. All samples were immediately frozen on dry ice to preserve their DNA. The region encoding the transmembrane domain of ACD6 (or ACD6B for Se-0-like accessions) was amplified by PCR and sequenced for all samples. Genotyping for the highly divergent KZ10-like alleles was performed using a previously described PCR-based assay [16].
Sequencing and genotyping of multiplexed RAD-tag sequencing
For RAD-tag sequencing [49], genomic DNA from 2,112 wild individuals was quantified using a Qubit (Life Technologies, Carlsbad, CA, USA) and all samples were normalized to 20 ng/µl. Eleven RAD-seq libraries were prepared following the modifications described in [50], with double digestions using PstI and MseI restriction enzymes (Thermo Fisher Scientific, Waltham, MA, USA). The final eight PCR reactions per library were pooled and 250–500 bp fragments were selected by gel extraction. Libraries were sequenced on a HiSeq2000 instrument (Illumina, San Diego, CA, USA) with single-end 101 bp reads.
Reads were processed with SHORE (ver. 0.9; [51]) and mapped to the reference sequence, Col-0, with BWA [52], using the default parameters and allowing 5% mismatches. All mapped reads were converted into BAM format by Samtools (ver. 0.1.18; [53]) for further analysis. Single nucleotide polymorphism (SNP) genotypes were generated using the subprogram UnifiedGenotyper implemented in GATK (ver. 2.3–6; [54]) using default parameters. Only polymorphisms present as non-singletons in 80 fully sequenced Arabidopsis accessions [55] were accepted, to reduce the possibility of erroneously mapped markers, especially heterozygous markers. We removed markers with over 10% heterozygous calls, more than 20% missing information, or minor allele frequency (MAF) below 0.01. To generate the data matrix for subsequent analysis we used individuals that had been successfully typed for ACD6 alleles, resulting in 1,688 samples and 3,641 markers.
Population genetic analysis
Principle component analysis (PCA) was carried out using the adegenet package with 730 SNPs that had complete information (ver. 1.3–6; [56]) in R (http://www.R-project.org). To determine the number of unique genotypes we calculated the genetic distance based on the pairwise difference among samples using MEGA version 5 [45]. A sliding window approach was used to calculate Fst using the program HBKpermute (see [57]) implemented in ‘analysis’ built by the C++ class library, ‘libsequence’ [58]. To calculate Fst, we used sliding windows of 10 SNPs, with 2 SNP steps. LD was estimated using the program ‘rsq’ implemented in ‘analysis’ [58]. LD decay was smoothened by estimating the least-square expectation of squared correlations (r2) from the nonlinear regression evaluation [59].
Accession numbers
Sequences have been deposited in GenBank under accession numbers KC019116 to KC019169.
Supporting Information
Zdroje
1. GilbertGS (2002) Evolutionary ecology of plant diseases in natural ecosystems. Annu Rev Phytopathol 40 : 13–43.
2. BurdonJJ, ThrallPH (2003) The fitness costs to plants of resistance to pathogens. Genome Biol 4 : 227.
3. HeilM, BaldwinIT (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7 : 61–67.
4. BrownJK (2002) Yield penalties of disease resistance in crops. Curr Opin Plant Biol 5 : 339–344.
5. SacktonTB, LazzaroBP, SchlenkeTA, EvansJD, HultmarkD, et al. (2007) Dynamic evolution of the innate immune system in Drosophila. Nat Genet 39 : 1461–1468.
6. NielsenR, BustamanteC, ClarkAG, GlanowskiS, SacktonTB, et al. (2005) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol 3: e170.
7. ClarkRM, SchweikertG, ToomajianC, OssowskiS, ZellerG, et al. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 : 338–342.
8. BombliesK, WeigelD (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8 : 382–393.
9. SalmonCE (1919) Papaver rhæas, P. dubium and the hybrid between them. New Phytol 18 : 111–118.
10. McNaughtonIH, HarperJL (1960) The comparative biology of closely related species living in the same area. II. Aberrant morphology and a virus-like syndrome in hybrids between Papaver rhoeas L. and P. dubium L. New Phytol 59 : 27–41.
11. BombliesK, LempeJ, EppleP, WarthmannN, LanzC, et al. (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236.
12. KrugerJ, ThomasCM, GolsteinC, DixonMS, SmokerM, et al. (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 : 744–747.
13. YamamotoE, TakashiT, MorinakaY, LinS, WuJ, et al. (2010) Gain of deleterious function causes an autoimmune response and Bateson–Dobzhansky–Muller incompatibility in rice. Mol Genet Genomics 283 : 305–315.
14. JeukenMJ, ZhangNW, McHaleLK, PelgromK, den BoerE, et al. (2009) Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21 : 3368–3378.
15. AlcázarR, GarcíaAV, KronholmI, de MeauxJ, KoornneefM, et al. (2010) Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat Genet 42 : 1135–1139.
16. TodescoM, BalasubramanianS, HuTT, TrawMB, HortonM, et al. (2010) Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465 : 632–636.
17. SchwabR, OssowskiS, RiesterM, WarthmannN, WeigelD (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 : 1121–1133.
18. LuH, RateDN, SongJT, GreenbergJT (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15 : 2408–2420.
19. RateDN, CuencaJV, BowmanGR, GuttmanDS, GreenbergJT (1999) The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11 : 1695–1708.
20. LuH, SalimianS, GamelinE, WangG, FedorowskiJ, et al. (2009) Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J 58 : 401–412.
21. GaffneyT, FriedrichL, VernooijB, NegrottoD, NyeG, et al. (1993) Requirement of salicylic Acid for the induction of systemic acquired resistance. Science 261 : 754–756.
22. LuH, LiuY, GreenbergJT (2005) Structure-function analysis of the plasma membrane - localized Arabidopsis defense component ACD6. Plant J 44 : 798–809.
23. NordborgM, HuTT, IshinoY, JhaveriJ, ToomajianC, et al. (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196.
24. HortonM, HancockAM, HuangYS, ToomajianC, AtwellS, et al. (2012) Genome-wide pattern of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44 : 212–216.
25. TodescoM, KimS-T, ChaeE, BombliesK, ZaidemM, et al. (2014) Data from: Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. Dryad Digital Repository http://dx.doi.org/10.5061/dryad.2nk64.
26. BombliesK, YantL, LaitinenR, KimS-T, HollisterJD, et al. (2010) Local-scale patterns of genetic variability, outcrossing and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet 6: e1000890.
27. Shrestha J (2010) Structural and functional study of ACCELERATED CELL DEATH 6 in Arabidopsis defense [Dissertation]. United States, Illinois: University of Chicago. 199 p.
28. HuaJ (2013) Modulation of plant immunity by light, circadian rhythm, and temperature. Curr Opin Plant Biol 16 : 406–413.
29. BarbashDA (2010) Genetic testing of the hypothesis that hybrid male lethality results from a failure in dosage compensation. Genetics 184 : 313–316.
30. PresgravesDC (2003) A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163 : 955–972.
31. Ohno S (1970) Evolution by Gene Duplication. New York: Springer Verlag. 160 p.
32. Lynch M (2007) The Origins of Genome Architecture. Sunderland, MA: Sinauer Associates. 389 p.
33. BekpenC, Marques-BonetT, AlkanC, AntonacciF, LeograndeMB, et al. (2009) Death and resurrection of the human IRGM gene. PLoS Genet 5: e1000403.
34. Trabesinger-RuefN, JermannT, ZankelT, DurrantB, FrankG, et al. (1996) Pseudogenes in ribonuclease evolution: a source of new biomacromolecular function? FEBS Lett 382 : 319–322.
35. SassiSO, BraunEL, BennerSA (2007) The evolution of seminal ribonuclease: pseudogene reactivation or multiple gene inactivation events? Mol Biol Evol 24 : 1012–1024.
36. ReynaudCA, AnquezV, GrimalH, WeillJC (1987) A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 48 : 379–388.
37. ThompsonCB, NeimanPE (1987) Somatic diversification of the chicken immunoglobulin light chain gene is limited to the rearranged variable gene segment. Cell 48 : 369–378.
38. BalakirevES, AyalaFJ (2003) Pseudogenes: are they “junk” or functional DNA? Annu Rev Genet 37 : 123–151.
39. BotellaMA, ParkerJE, FrostLN, Bittner-EddyPD, BeynonJL, et al. (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10 : 1847–1860.
40. BakkerEG, ToomajianC, KreitmanM, BergelsonJ (2006) A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18 : 1803–1818.
41. WarthmannN, FitzJ, WeigelD (2007) MSQT for choosing SNP assays from multiple DNA alignments. Bioinformatics 23 : 2784–2787.
42. DoyleJJ, DoyleJL (1987) A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull 19 : 11–15.
43. KochE, SlusarenkoA (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 : 437–445.
44. LarkinMA, BlackshieldsG, BrownNP, ChennaR, McGettiganPA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 : 2947–2948.
45. TamuraK, PetersonD, PetersonN, StecherG, NeiM, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 : 2731–2739.
46. HellensRP, EdwardsEA, LeylandNR, BeanS, MullineauxPM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 : 819–832.
47. Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 354 p.
48. LempeJ, BalasubramanianS, SureshkumarS, SinghA, SchmidM, et al. (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1: e6.
49. BairdNA, EtterPD, AtwoodTS, CurreyMC, ShiverAL, et al. (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3: e3376.
50. PolandJA, BrownPJ, SorrellsME, JanninkJL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253.
51. OssowskiS, SchneebergerK, ClarkRM, LanzC, WarthmannN, et al. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18 : 2024–2033.
52. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
53. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
54. McKennaA, HannaM, BanksE, SivachenkoA, CibulskisK, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20 : 1297–1303.
55. CaoJ, SchneebergerK, OssowskiS, GuntherT, BenderS, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43 : 956–963.
56. JombartT, AhmedI (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27 : 3070–3071.
57. HudsonRR, BoosDD, KaplanNL (1992) A statistical test for detecting geographic subdivision. Mol Biol Evol 9 : 138–151.
58. ThorntonK (2003) Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19 : 2325–2327.
59. RemingtonDL, ThornsberryJM, MatsuokaY, WilsonLM, WhittSR, et al. (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci U S A 98 : 11479–11484.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 7- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



