-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
The Chordate phylum groups vertebrates, tunicates (including ascidians) and cephalochordates (amphioxus). These animals share a typical body plan characterized by the presence during embryonic life of a notochord and a dorsal neural tube. Ascidians, however, took a significantly different evolutionary path from other chordates resulting in divergent morphological, embryological and genomic features. Their development is fast and stereotyped with very few cells and ascidian genomes have undergone compaction and extensive rearrangements when compared to vertebrates, but also between ascidian species. This raises the question of whether developmental mechanisms controlling typical chordate structure formation are conserved between ascidians and vertebrates. Here, we have studied the set of ascidian genes which control the formation of the posterior part of the nervous system. We uncovered original usages of the signaling molecule Nodal and the transcription factor Otx. For example, Otx, which is a specific determinant of anterior identity in most metazoans, has been co-opted for the formation of the ascidian posterior nervous system. These two factors define a regulatory signature found in enhancers of posterior neural genes in two genomically divergent ascidian species.
Published in the journal: An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians. PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004548
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004548Summary
The Chordate phylum groups vertebrates, tunicates (including ascidians) and cephalochordates (amphioxus). These animals share a typical body plan characterized by the presence during embryonic life of a notochord and a dorsal neural tube. Ascidians, however, took a significantly different evolutionary path from other chordates resulting in divergent morphological, embryological and genomic features. Their development is fast and stereotyped with very few cells and ascidian genomes have undergone compaction and extensive rearrangements when compared to vertebrates, but also between ascidian species. This raises the question of whether developmental mechanisms controlling typical chordate structure formation are conserved between ascidians and vertebrates. Here, we have studied the set of ascidian genes which control the formation of the posterior part of the nervous system. We uncovered original usages of the signaling molecule Nodal and the transcription factor Otx. For example, Otx, which is a specific determinant of anterior identity in most metazoans, has been co-opted for the formation of the ascidian posterior nervous system. These two factors define a regulatory signature found in enhancers of posterior neural genes in two genomically divergent ascidian species.
Introduction
Neural tissue formation is a multi-step process through which embryonic cells acquire a neural phenotype. In vertebrate central nervous system (CNS) development, the first step is called neural induction. Naive ectodermal cells undergo a binary fate decision between epidermis and neural tissue in response to endomesodermal signals that modulate the FGF, BMP and Wnt signaling pathways [1]–[3]. While there may be variations between species, BMP inhibition together with FGF signaling activation are key events in neural induction. Concomitantly or following neural induction, neural tissue is patterned along the antero-posterior and medio-lateral axes. Acquisition of a differentiated neural phenotype involves further processes such as stabilization and reinforcement of the neural fate, specification of cellular identity and progression towards final differentiation. Each of these steps is controlled by complex mechanisms involving a variety of molecular players [4]–[6].
Non-vertebrate chordates include ascidians (tunicates) and amphioxus (cephalochordates). They form prototypical tadpole-like larvae with a dorsal hollow neural tube patterned similarly to vertebrates [7], [8]. The embryological process of neural induction also takes place in these animals but our current knowledge does not provide a unified view. In amphioxus, BMP activation represses neural tissue formation but FGF inhibition does not abolish neural tissue formation [9], [10]. In ascidians by contrast, FGF is essential for neural induction while BMP inhibition does not seem to be involved [11], [12].
Comparative embryology within each of these groups and with vertebrates provides an outstanding opportunity to assess the diversity of regulatory strategies leading to a common shared body plan and to test models of gene regulatory network evolution proposed in other bilaterian groups [13], [14]. In this context, ascidians can be regarded as interesting chordate evolutionary outliers with unique developmental and genomic features. Their mode of development, based on small cell numbers and invariant cell lineages, diverges markedly from that found in vertebrates and amphioxus [15]. In addition, ascidians also display a fast rate of evolution with extensive genome rearrangements and compaction as well as gene losses [16], [17]. Ascidian genomes are thus very different from other chordate genomes (for example, synteny and ultra conserved elements conserved between vertebrates and amphioxus are not found in ascidians) [18], [19]. Finally, the high conservation of ascidian cell lineages throughout ascidian groups allows the comparison of genomically divergent ascidian embryos with a cellular level of resolution [20]–[22].
The dorsal hollow neural tube of the ascidian larva is composed of three morphologically distinct regions: the sensory vesicle anteriorly, the visceral ganglion and the tail nerve cord posteriorly (Figure 1). While there are still debates on their precise homology to vertebrate CNS domains, they are thought to be equivalent to fore/midbrain, hindbrain and spinal cord respectively [23], [24]. The ascidian CNS has a dual origin and specification logic (reviewed in [25]). Three separate lineages, named according to the founding blastomeres of the 8-cell stage embryo, form the ascidian CNS (Figure 1). The A-line neural lineage originates from vegetal blastomeres and gives rise to the posterior part of the sensory vesicle and to the ventral and lateral parts of both visceral ganglion and tail nerve cord. Ectodermal blastomeres give rise to the anterior part of the sensory vesicle (a-line) and to the dorsal part of the visceral ganglion and tail nerve cord (b-line). While A-line CNS is specified autonomously [26], a - and b-line are specified through neural induction by FGF9/16/20 secreted from the vegetal hemisphere at the 16 - to 32-cell stage transition [11], [12], [27], [28]. Early target genes including Otx, Nodal, Elk and Erf are expressed at the 32-cell stage in all or part of the neural precursors (a6.5 and b6.5 blastomeres; Figures 1 and S2) where ERK signaling is active [11], [29], [30]. Interestingly, each of these precursors also contributes to the peripheral nervous system (PNS) following FGF9/16/20 induction [31], [32]. For example, the b6.5 blastomere gives rise to the dorsal midline of the tail epidermis, a neurogenic territory from which the epidermal sensory neurons of the PNS form (Figure 2A). Beside the requirement of Otx for anterior neural tissue formation [33] and the key role of Nodal in A-line CNS patterning and formation of the b6.5 derivatives [23], [29], [32], [34], [35], little is known for the function of these immediate target genes in neural fate acquisition or stabilization.
Fig. 1. Cell lineages of the ascidian central nervous system. 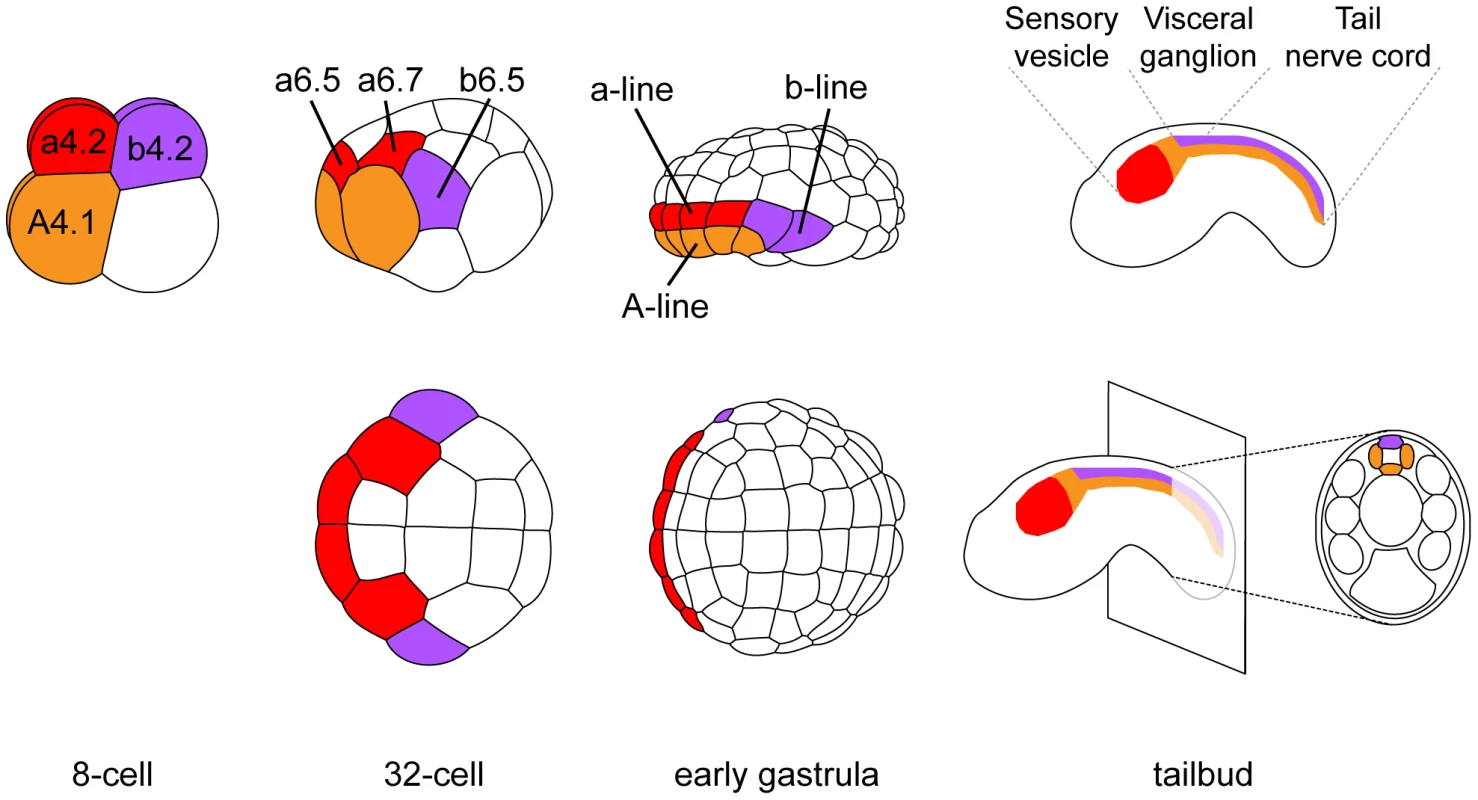
At each developmental stage, cells contributing to the central nervous system are colored according to their origin in the 8-cell stage embryo. a-line CNS (red) originates from anterior animal blastomeres (a4.2 pair) and forms the anterior sensory vesicle. A-line CNS (orange) originates from anterior vegetal blastomeres (A4.1 pair) and forms the posterior sensory vesicle, the visceral ganglion and the tail nerve cord (only the ventral and lateral parts for the latter two regions). b-line CNS (purple) originates from posterior animal blastomeres (b4.2 pair) and forms the dorsal part of visceral ganglion and tail nerve cord. Drawings for 8-cell to early gastrula stages: lateral view with animal to the top and anterior to the left (top row) and animal view with anterior to the left (bottom row). Drawings for tailbuds are lateral views with dorsal to the top and anterior to the left and a cross-section through the tail showing the four cells originating from two distinct lineages (A- and b-line). Fig. 2. FGF and Nodal signaling are required for posterior neural tissue formation. 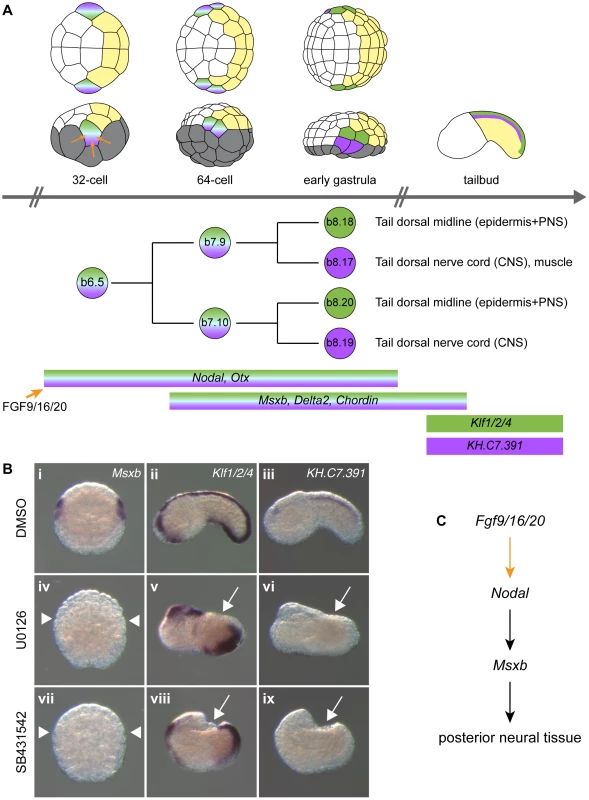
A) Schematic representation of b6.5 lineage history with representation of embryos, cell lineage and gene expression at different stages. The different tissues and precursors are color coded: vegetal cells in grey, anterior (a-line) ectoderm in white, posterior (b-line) ectoderm in yellow, dorsal tail epidermis in green and dorsal tail nerve cord in purple. Embryos are in animal view (top row) or lateral view (bottom row) with anterior to the left. B) Expression of early and late b6.5 lineage markers when FGF-Erk and Nodal signaling pathways are disrupted. Msxb which is normally expressed in the four daughter cells of the b6.5 blastomere (Bi) is not expressed in U0126-treated (Biv) and SB431542-treated (Bvii) embryos. Animal views of Msxb at early gastrula stages (stages 10/11) (Bi, iv, vii). Schematic animal views of stage 10 embryos are depicted as insets in Msxb panels: anterior ectoderm in white, posterior ectoderm in yellow and gene expression in blue. Expression of Klf1/2/4 is lost in tail dorsal midline for both treatments (Bv and Bviii). The dorsal tail nerve cord marker KH.C7.391 is suppressed (Bvi and Bix). Lateral view with dorsal to the top and anterior to the left (Bii, iii, v, vi, viii and ix) at stage 19. Control DMSO-treated embryos (Bi-iii), U0126-treated embryos (Biv-vi) and SB431542-treated embryos (Bvii-ix). White arrows and arrowheads indicate sites with a loss of expression. C) Gene interactions revealed by loss-of-function data. In order to gain insights into post-neural induction events, we focused our attention on the regulation of Msxb and Delta2, markers of the progeny of the b6.5 blastomeres. Both genes are expressed from the 64-cell stage (after neural induction) in the b6.5 progeny (b7.9 and b7.10 blastomere pairs; Figure 2A and [36], [37]) and are required for further specification and differentiation of these progenitors. Msxb is a marker of the entire b6.5 lineage until neurula stages, and is required for tail dorsal epidermal midline and dorsal nerve cord formation [23], [35]. Delta2 is involved in the specification of epidermal sensory neurons within the epidermal midline [32], [38].
In this study, we show that FGF signaling is necessary and sufficient for b6.5 fate acquisition in posterior ectoderm. Downstream of FGF, Nodal is necessary for b6.5 fate. Although it cannot induce neural tissue on its own, it is sufficient to posteriorize FGF-induced neural tissue. This led us to search for other factors acting with Nodal downstream of FGF. We uncovered a critical function for the transient expression of Otx in posterior neural fate acquisition. Using this simple model of regulation, we were able to isolate b6.5 lineage specific enhancers for both Msxb and Delta2. We further show that this mode of regulation is shared with the distantly related ascidian Phallusia mammillata, strengthening our proposal that Otx, a well known regulator of anterior neural tissues in many metazoans, has been co-opted in ascidians for posterior nervous system formation.
Results
FGF signaling is necessary and sufficient for posterior ectodermal cells to adopt a b6.5 fate
Previous reports indicated that induced b6.5 fates are lost after abolition of FGF signaling [11], [28], [35]. We extended these results using a pharmacological inhibitor of FGF/MEK signaling (U0126), three early markers of b6.5 progeny (Msxb, Delta2 and Chordin) and two tailbud markers of dorsal tail epidermis midline and dorsal nerve cord, Klf1/2/4 and KH.C7.391 respectively (Figures 2 and S1). MEK inhibition led to a conversion of neural b6.5 progenitors into epidermis as demonstrated by the loss of expression of all neural markers, coupled to the ectopic expression of the epidermal marker Ap2-like2 at gastrula stages (Figure S1).
Previous reports indicated that activation of the FGF pathway in explanted ectodermal precursors leads to the induction of neural fate in cells normally fated to form epidermis, with different neural fates achieved in a-line and b-line blastomeres [11], [12], [27], [32]. We confirmed that this was also the case in whole embryos. We treated whole embryos either with recombinant FGF protein from the 16-cell stage or overexpressed FGF9/16/20 by electroporation using the pFOG driver (expressed from the 16-cell stage throughout the entire ectoderm [39]). As expected, the epidermis marker Ap2-like2 was strongly down-regulated throughout the ectoderm (data not shown). The posterior neural markers Nodal, Msxb and Delta2 were ectopically expressed throughout the posterior ectoderm (b4.2 lineage or b-line ectoderm), and the anterior neural marker Dmrt1 was activated throughout the anterior ectoderm (a4.2 lineage or a-line ectoderm) (Figures 3A-H and S2). Chordin, which is normally expressed in the progeny of b6.5 as well as in a8.26 and a8.28 blastomere pairs (Figure 3C), was expressed throughout the posterior ectoderm and in part of the anterior ectoderm in response to ectopic FGF treatment (Figures 3C and 3G).
Fig. 3. Nodal acts downstream of FGF to posteriorize induced neural tissue. 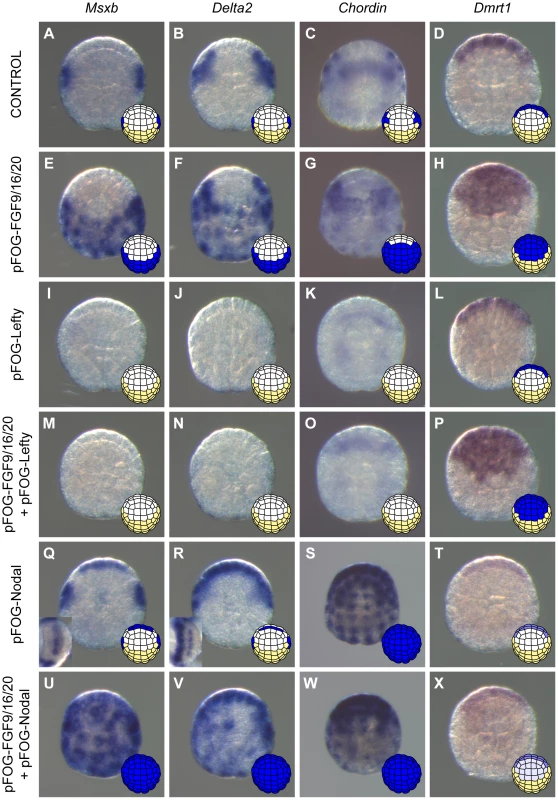
Expression of posterior neural markers (Msxb (A), Delta2 (B), and Chordin (C)) and of the anterior neural marker Dmrt1 (D) in control embryos. FGF9/16/20 overexpression using the pFOG promoter via electroporation led to ectopic expression of Msxb (E) and Delta2 (F) throughout posterior ectoderm, of Chordin (G) through most of the ectoderm except the anterior-most part and of Dmrt1 (H) throughout anterior ectoderm at early gastrula stages (st. 10/11). These effects were suppressed by inhibition of Nodal signaling through Lefty overexpression (M-O) except for Dmrt1 (P). Overexpression of Lefty alone inhibited posterior marker expression (I-K) but did not affect expression of the anterior marker Dmrt1 (L). Overexpression of Nodal using the pFOG driver was sufficient to activate Msxb (Q) and Delta2 (R) in the neural plate, and Chordin (S) throughout the ectoderm. Ectopic Chordin expression was stronger in anterior ectoderm than in posterior ectoderm possibly reflecting the difference in expression levels between anterior and posterior expressing cells in control embryos. Dmrt1expression was downregulated (T). Combined overexpression of FGF9/16/20 and Nodal led to ectopic activation of Msxb (U) and Delta2 (V) in both anterior and posterior ectoderm. Under these conditions, Chordin was still expressed throughout the ectoderm but at weaker levels (W). Overexpression of Nodal downregulated ectopic activation in anterior ectoderm of Dmrt1 induced by FGF9/16/20 (X). Animal view with anterior to the top for all except insets in Q and R that show neural plate view with vegetal side to the left. For each panel a schematic animal view of stage 10 embryos depicts anterior ectoderm in white, posterior ectoderm in yellow and gene expression in blue. Nodal activation at the 32-cell stage was a likely direct consequence of FGF signaling. FGF treatment activated Nodal ectopic expression in the presence of protein synthesis inhibitor (Figures S2), suggesting the absence of a transcriptional relay. In addition, a previously identified b6.5-specific Nodal enhancer has the same regulatory logic as the FGF-responsive enhancer of the direct FGF target gene Otx [30]. Msxb, Delta2 and Chordin are more likely to be indirect targets of FGF as they are activated later at the 64-cell stage.
In the following sections, we will precisely define the regulatory interactions between FGF, Nodal, Otx, Msxb, Delta2 and Chordin in the b6.5 lineage.
Nodal signaling posteriorizes FGF-induced neural tissue
To determine the function of Nodal during b6.5 fate acquisition, we blocked the function of its receptor with the pharmacological inhibitor SB431542 or overexpressed the Nodal antagonist Lefty in the ectoderm using electroporation. Both perturbations led to a loss of expression of Msxb, Delta2 and Chordin in b-line neural lineage at gastrula stages (Figures 2B, 3I-K and S1). At later stages, expression of the dorsal tail nerve cord marker KH.C7.391 was lost, as was the dorsal expression of the tail midline marker Klf1/2/4 (Figure 2B). This altered genetic program was similar to that obtained in response to FGF inhibition, suggesting that Nodal acts downstream of Fgf9/16/20 in b-line neural specification (Figure 2C). Consistent with this, FGF-induced ectopic activation of Msxb, Delta2 and Chordin was suppressed by Lefty overexpression (Figure 3M-O). Nodal was however not the sole mediator of FGF action, as its inhibition was not sufficient to convert the b6.5 progeny into epidermis, marked by Ap2-like2 expression (Figure S1).
We next overexpressed Nodal throughout the ectoderm using the pFOG driver and analyzed marker expression in the a - and b-line ectoderm. Ectopic expression of Chordin was observed throughout the ectoderm (Figure 3S), independently of the FGF induction status of the cells. Ectopic Chordin expression was stronger in a-line ectoderm, possibly reflecting the stronger levels detected in a8.26 and a8.28 blastomeres compared to b6.5 progeny in control embryos (Figure 3C). By contrast, we did not detect ectopic activation of Msxb and Delta2 in posterior (b-line) ectoderm (Figure 3Q,R). However, anterior neural tissue precursors (a6.5 lineage) ectopically expressed these two genes (Figure 3Q,R) and had reduced Dmrt1 expression (Figure 3T). These data indicate that anterior neural precursors adopted a posterior identity in response to Nodal expression. Consistent with these observations, co-electroporation of pFOG-FGF9/16/20 and pFOG-Nodal, led to the induction of posterior neural tissue in anterior ectoderm, demarcated by the ectopic activation of both Msxb and Delta2 and by the repression of Dmrt1 (Figure 3U,V and X).
The results of this section indicate that Nodal alone is required, though not sufficient, to induce neural tissue and that it can posteriorize FGF-induced neural tissue. Interestingly, expansion of the anterior neural marker Dmrt1 to posterior b-line territories was not observed following Nodal signaling inhibition in either wild type or FGF-induced contexts (Figure 3L, P). These results are consistent with the presence of a Nodal-independent factor necessary for Dmrt1 expression and anterior neural fate acquisition in a-line ectoderm [40], [41] (see discussion).
In summary, three genes expressed downstream of FGF in the b6.5 progeny show different requirements regarding Nodal signaling: Chordin can be activated in the entire ectoderm while Msxb and Delta2 are positive targets of Nodal solely in FGF-induced neural cells.
Otx is required for posterior neural tissue formation
The conversion of a6.5 anterior neural precursors into posterior neural fate upon ectopic activation of Nodal signaling (Figure 3 Q, R) suggests that posterior neural fates may result from the cooperation of Nodal with another FGF-target. Otx is a conspicuous candidate since it is expressed in all neural precursors downstream of FGF signaling (Figure S2) and is coexpressed with Nodal in posterior neural precursors marked by Msxb and Delta2 expression [11], [27] (Figures 2, 3 and S2).
We first tested the requirement of Otx in b6.5 fate acquisition by injecting a specific translation-blocking morpholino antisense oligonucleotide (MO). Otx morpholino injection led a full loss of Msxb and Delta2 expression at stage 10 (Figure 4C, F). The resulting embryos displayed gastrulation and neurulation defects reminiscent of FGF or Nodal signaling inhibition. The tail midline marker Klf1/2/4 was strongly affected (Figure 4I). Dorsal tail epidermis midline staining originating from b6.5 was abolished while posterior-most staining (originating from b6.6 lineage) was maintained. Ventral midline expression was also kept but the domain of expression appeared reduced in size. Dorsal tail nerve cord did not form either as revealed by the loss of the marker KH.C7.391 (Figure 4J). We obtained similar results by overexpressing a dominant negative form of Otx, OtxHDenR (a fusion protein between the Otx homeodomain and the repressor domain of Engrailed) [42] in the ectoderm (Figure S4). The phenotypes appeared milder probably because OtxHDenR was only targeted to the ectoderm and because of the mosaic inheritance of the transgene introduced by electroporation. In addition, we observed that expression of the epidermal marker Ap2-like2 was unchanged following overexpression of OtxHDenR (Figure S4). Similarly to what has been observed for Nodal inhibition, b-line neural lineage did not form neural tissue upon Otx loss-of-function but did not form epidermis either.
Fig. 4. Otx is required for posterior neural tissue formation. 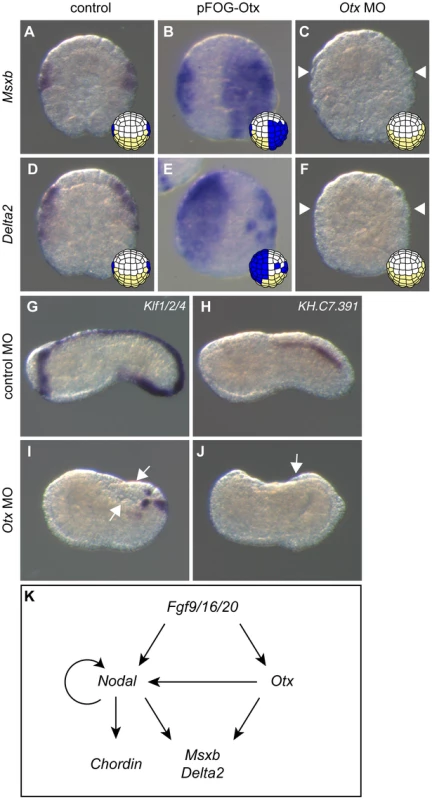
Overexpressing Otx in the ectoderm using the pFOG driver is sufficient to activate Msxb (B) and Delta2 (E) compared to control embryos (A, D) at stage 10. Upon injection of the Otx MO, Msxb (C) and Delta2 (F) expression is abolished at stage 10. The dorsal expression of the tail midline marker Klf1/2/4 is lost except in the posterior-most and ventral regions (I). The dorsal nerve cord marker KH.C7.391 expression is also suppressed (J). Control MO-injected embryos at stage 10 (A, D) and stage 19 (G, H). Animal view with anterior to the top (B, D-F). Vegetal view with anterior to the top (A and C). Lateral view with dorsal to the top, anterior to the left (G-J). White arrows and arrowheads indicate sites with a loss of expression. (K) Summary of gene interactions reported in this study and from previous studies [11], [29], [30], [35]. We next tested the effect of Otx overexpression using the pFOG driver. Although we expected that Otx would need to cooperate with Nodal to activate Msxb and Delta2, Otx overexpression was sufficient to activate both of these latter genes throughout the ectoderm (Figure 4B, E). When we overexpressed simultaneously Otx and Nodal throughout the ectoderm, we simply observed an addition of each molecule effect with no increase in the number of embryos ectopically expressing Msxb and Delta2 in the ectoderm (data not shown). To better understand these results, we further explored possible transcriptional interactions between Nodal and Otx that may control maintenance of their expression following the initial induction by FGF (Figure S2). We detected robust activation of Nodal expression at the 64-cell stage when Otx was ectopically expressed (Figure S3Aii). Accordingly, Nodal expression was repressed by the overexpression of OtxHDenR (Figure S3Aiii). This interaction between Otx and Nodal was not reciprocal, since Otx expression was not changed upon modulation of Nodal signaling (Figure S3Avi, vii). Nodal signaling inhibition also prevented Nodal expression (Figure S3Aiv), suggesting the existence of an autoregulatory loop on Nodal similarly to what has been described in vertebrates [43]. The ectopic activation of Msxb and Delta2 in the ectoderm by Otx overexpression did not require the activation of Nodal, as overexpression of Lefty did not significantly block Otx effect (Figure S3B). By contrast, Nodal-mediated ectopic expression of Msxb and Delta2 in anterior neural precursors was inhibited by OtxHDenR overexpression (Figure S3C).
These data demonstrate that Otx is an essential regulator of b6.5 lineage derived posterior neural tissue formation. Figure 4K provides a schematic representation of the gene regulatory network acting downstream of FGF in b-line ectoderm.
The genomic hardwiring of Msxb and Delta2 regulation
We next used the above functional evidence to isolate cis-regulatory DNA regions responsible for neural marker expression in the b6.5 lineage. We reasoned that the enhancer responsible for b6.5 lineage expression should integrate both Otx and Nodal inputs. Nodal is a ligand which controls gene expression through the activation of the Smad2/3 nuclear effector. A Smad2/3/Smad4 complex can directly bind DNA with low affinity through poorly defined GC rich regions or through (C)AGAC Smad Binding Element (SBE) consensus sequences [44]. However, high affinity binding is usually achieved through association with a DNA binding cofactor. In several instances, Fox transcription factors have been shown to fulfill this function [44]–[46]. We consequently searched the Msxb locus for the co-occurrence of Otx and Fox/Smad binding sites. We selected the core consensus sequences GGATTA for Otx, TGTTT for Fox from the Jaspar database [47], and AGAC for Smad [44]. We searched for regions enriched in Otx-, Fox - and Smad - core binding site motifs by first scanning, in Ciona intestinalis type A [48], the 50 kb genomic region that includes Msxb up to its two flanking genes. We arbitrarily chose a 300 bp window and found 15 regions that contained at least one of each motif. To reduce the number of candidates we increased the stringency by increasing the number of the least frequent site, which is Otx. We chose a more degenerate site for this additional motif, GATTA, as in [42]. Adding one or two GATTA motifs yielded 7 and 4 candidate regions, respectively. We focused on the latter 4 regions and searched whether the Ciona savignyi orthologous regions harbored a similar combination of binding sites using Vista suite [49]. A single region matched this criterion and was named “msxb-b6.5 line” according to its enhancer activity (see below) (Figure 5). This region is located just upstream of Msxb on a peak of conservation and contains 6 putative Otx, 5 putative Fox binding sites and 6 putative SBEs (Figures 5A, B and S5). This region falls within a region bound in vivo by Otx at early gastrula stages as revealed by ChIP-on-Chip experiment (Figure 5B) [50].
Fig. 5. The b6.5 line enhancers of Msxb and Delta2. 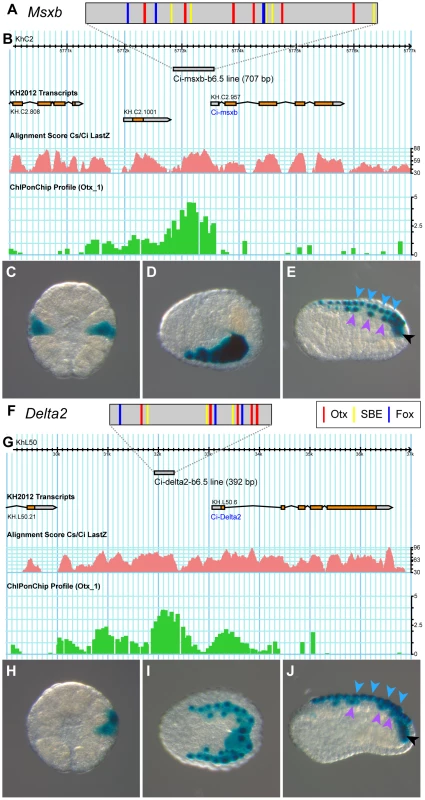
Schematic organization of tested enhancers depicting putative Otx (GATTA) (red bars), Fox (AAACA) (blue bars) binding sites and SBEs (AGAC) for Msxb (A) and Delta2 (F). Genomic browser view of gene loci with gene models (Msxb: KH.C2.957, Delta2: KH.L50.6), tested enhancers (grey bar), alignment profile of C. intestinalis and C. savignyi genomic sequences (pink) and ChiP-on-Chip data (green) [50] for Msxb (B) and Delta2 (G) (extracted from the Aniseed genome browser: http://www.aniseed.cnrs.fr/fgb2/gbrowse/ciona_intestinalis/ [70], and from the Ghost genome browser http://ghost.zool.kyoto-u.ac.jp/cgi-bin/gb2/gbrowse/kh/ [71]). Representative pictures for X-gal staining of electroporated embryos with the “msxb-b6.5 line” enhancer at stage 10 (C), stage 14 (D) and stage 16 (E), and with the “delta2-b6.5 line” enhancer at stage 10 (H), stage 14 (I) and stage 18 (J). Arrowheads indicate dorsal midline epidermis (blue), dorsal nerve cord (purple) and secondary muscle (black). Vegetal view, anterior to the top (C, H). Dorsal view, anterior to the left (D, I). Lateral view, dorsal to the top and anterior to the left (E, J). Additional staining was also observed in mesenchymal cells, a tissue highly permissive to transcriptional assays in Ciona [42]. We amplified this 707 bp fragment from C. intestinalis type B genomic DNA. The sequence obtained is very similar to the reference type A sequence but contains only 4 Fox binding sites and 5 SBEs (Figure S5). Placed upstream of the minimal promoter of Fog and the reporter gene LacZ [39], [51], this fragment drove transcription throughout b6.5 derivatives from the early gastrula stage (Figure 5C-E and Table S1). Thus, searching for enrichment in Otx, Fox and Smad putative binding sites in conserved non-coding genomic DNA was sufficient to isolate a region, which binds Otx in vivo at the early gastrula stage and is transcriptionally active in posterior neural precursors.
The same logic led to the identification of a Delta2 enhancer active in the b6.5 lineage. A single genomic region at the Delta2 locus harbored a combination of Otx, Fox and SBE sites within 300 bp in both C. intestinalis and C. savignyi and was named “delta2-b6.5 line” (Figure 5). This 392 bp long region is located within 2 kb upstream of Delta2, harbors a strong level of conservation, contains 5 Otx sites, 3 Fox sites and 3 SBEs; and is bound in vivo by Otx (Figures 5F-G and S6). When electroporated in C. intestinalis embryos it drove expression in b6.5 derivatives from early gastrula stages (Figure 5H-J and Table S1).
Overall, these results indicate that Msxb and Delta2 share similar regulatory motifs in their enhancers.
Msxb enhancer activity relies on Otx, Fox and Smad binding sites
We next assayed the relative contribution of Otx, Fox and Smad binding motifs to enhancer activity in the b6.5 lineage, focusing on the “msxb-b6.5 line” enhancer. Progressive shortening of this region on both sides (Figure S7 and Table S1) identified an active 273 bp long fragment (msxb-B) containing 3 Otx binding motifs, 2 overlapping Fox binding motifs and 4 Smad motifs (Figure 6A-B). This fragment was still active in inverted orientation (Msxb-B-inv), as expected from an enhancer (Figure S8B). Msxb-B enhancer activity was abolished when the Otx morpholino was injected and when Lefty was overexpressed (Figure 6B-D).
Fig. 6. Otx, Fox and Smad putative binding sites control msxb-B enhancer activity. 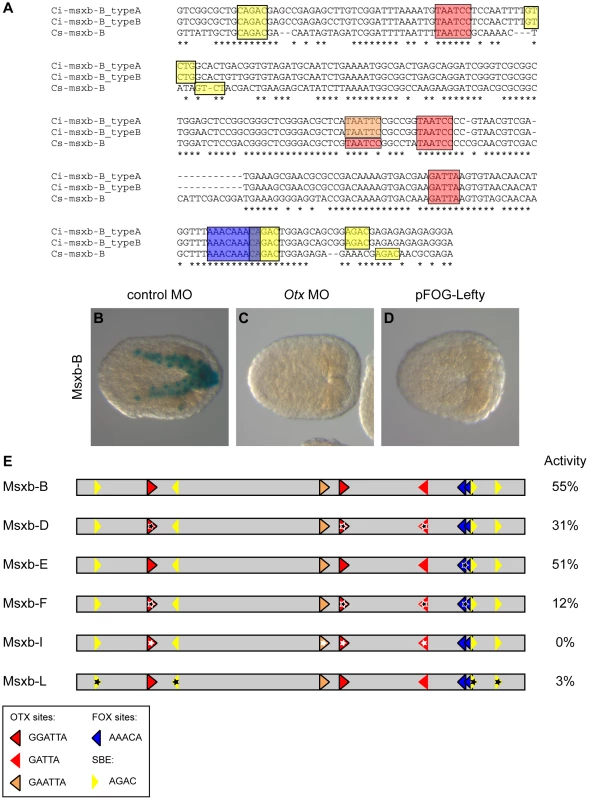
A) Alignment of msxb-B sequences from C. intestinalis type A, C. intestinalis type B and C. savignyi. Putative transcription factor binding sites are in colored boxes as follows: canonical Fox (AAACA) in dark blue, canonical Otx (GATTA) in red, non-canonical Otx (GAATTA) in orange and SBE (AGAC) in yellow. B) The msxb-B enhancer is active in b6.5 derivatives as revealed by X-gal staining on late gastrula. Its activity is abolished upon injection of the Otx MO (C) or overexpression of Lefty (D). E) Schematic view of msxb-B enhancer and its mutated versions. Putative transcription factor binding sites position and orientation are represented by colored arrows with the same color code as in (A). Mutations are depicted by stars. The precise mutations are described in the main text and in figure S8. Transcriptional activity of the different enhancers was measured as the percentage of embryos with staining in the b6.5 derivatives at late gastrula stages (stage 14) (Table S1). Simultaneous mutation of the 3 Otx sites through a single nucleotide modification in the core (GATTA = >GcTTA) (construct Msxb-D) led to a partial loss of activity (Figure 6E). Since activity was not completely suppressed, we looked for potential Otx binding motifs with altered core sequence. Interestingly, we found a GAATTA motif that corresponds to a canonical GGATTA sequence in Ciona savignyi (Figure 6A). Simultaneous mutation of this and the 3 canonical Otx sites (GNATTA = >GNcgTA) (construct Msxb-I) led to a complete loss of activity.
We next mutated the 4 conserved Smad Binding Elements (AGAC = >ctAC) and found these sites to be essential for Msxb-B activity (Msxb-L construct; Figure 6A, E).
We finally mutated the Fox sites. Two AAACA sites overlap in the AAACAAACA sequence (Figure 6A). We generated either a single nucleotide change that matches in the core of each Fox site (AAACgAACA, Msxb-E) or a single nucleotide change in each core (AAgCAAgCA, Msxb-H) (Figures 6E and S8). These mutations did not affect enhancer activity. Additional mutation (AACA = >AgCA, Msxb-G) of the three more degenerate AACA consensus found in the sequence, but not conserved in C. savignyi, also had no effect (Figure S8). We then tested the effect of mutating Fox sites in the sensitized context of the Msxb-D element where 3 Otx sites are mutated and where activity is decreased. The Msxb-F fragment (3 Otx sites mutated, 2 canonical Fox sites mutated) displayed a further reduction in activity (Figure 6E), suggesting that Otx and Fox sites may work together to control Msxb-B activity.
Mutational analysis indicates that Msxb regulation through the Msxb-B enhancer may involve putative Fox binding sites and requires the presence of putative Otx and Smad binding sites to be transcriptionally active in b6.5 derived cells.
A conserved regulatory logic across distantly related ascidian genera
We tested the transcriptional activity of the Ciona Msxb and Delta2 enhancers that we identified in a distantly related and genomically divergent ascidian, Phallusia mammillata. When each construct was electroporated in P. mammillata embryos, we detected LacZ activity in dorsal tail epidermis midline, dorsal nerve cord and secondary muscle, the same territories that are stained in C. intestinalis (Figure 7B, D and Table S2). These results suggest that the regulatory logic of these enhancers is interpreted in the same way in C. intestinalis and P. mammillata embryos. The similar enhancer activity between these two species possibly reflects conservation of the combination of transcription factors, the trans-regulatory logic, acting upstream of Msxb and Delta2. We further tested this possibility by determining the expression patterns of Msxb and Delta2 in P. mammillata by in situ hybridization (Figure S9). We observed that both genes are activated in the b6.5 lineage at the 64-cell stage (b7.9 and b7.19 blastomeres) like the C. intestinalis orthologous genes. This expression was abolished when inhibitors of the FGF/MEK (U0126) and Nodal (SB431542) signaling pathways were applied to the embryos (Figure 7G-L).
Fig. 7. A shared regulatory logic in Ciona intestinalis and Phallusia mammillata. 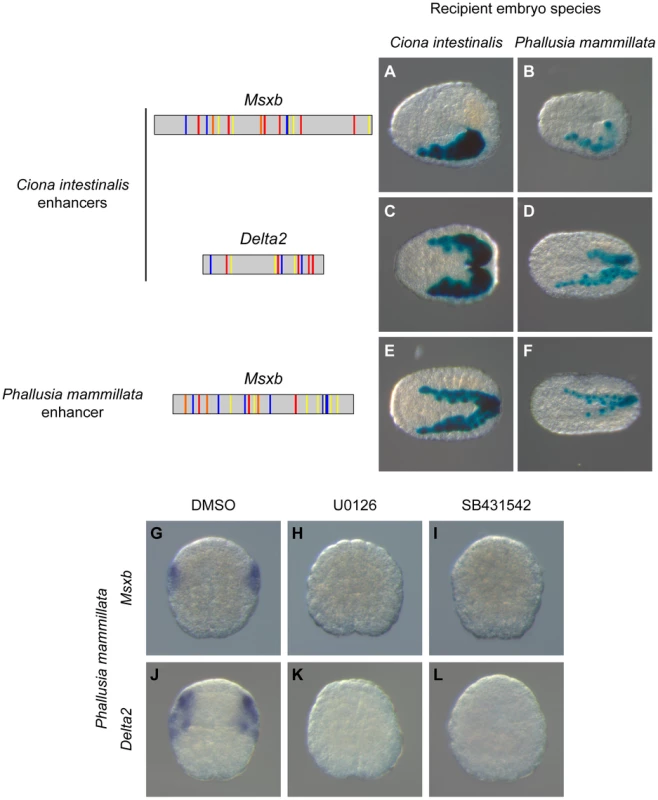
Schematic organization of tested enhancers with the same color code used in figures 4 and 5. Reporter gene activity is detected by X-gal staining after electroporation of “Ci-msxb-b6.5 line” enhancer (A, B), “Ci-delta2-b6.5 line” enhancer (C, D) and “Pm-msxb-b6.5 line” enhancer (E, F) into C. intestinalis (A, C, E) or P. mammillata (B, D, F) embryos. Transcriptional activity of the different enhancers, measured as the percentage of embryos with staining in the b6.5 derivatives, is detailed in Tables S1 and S2. In Phallusia mammillata embryos, Msxb (G) and Delta2 (J) are expressed in b6.5 derivatives. This expression is abolished upon inactivation of the FGF/MEK (H, K) or Nodal (I, L) signaling pathway. Dorsal view with anterior to the left (A-F). Animal view, anterior to the top (G-L). These results led us to search for enhancers regulating Msxb expression in P. mammillata. Employing the same strategy we used for C. intestinalis genes, we searched the Pm-Msxb locus for regions enriched in Otx, Fox and Smad binding motifs and conserved in the sister species Phallusia fumigata. We isolated a 587 bp fragment containing 6 Otx, 7 Fox binding motifs and 7 SBEs and located just upstream of Pm-Msxb (Figure S10). This fragment, “Pm-msxb-b6.5 line”, whose sequence could not be aligned with that of “Ci-msxb-b6.5 line”, was active in b6.5 derivatives when electroporated in P. mammillata (Figure 7F) or C. intestinalis (Figure 7E) embryos (Tables S1 and S2). Therefore, the functional knowledge acquired in C. intestinalis was sufficient to isolate an active enhancer with expected activity in another species, P. mammillata.
Discussion
We have shown that Nodal and Otx, directly activated by the neural inducer FGF9/16/20 at the 32-cell stage, are required for posterior neural fate acquisition. We propose that these two genes act in concert to promote the activation of Msxb and Delta2 at the 64-cell stage. This simple model allowed us to isolate an enhancer for each gene containing Otx, Fox and Smad binding sites and active in the posterior neural lineage. We also showed that this regulatory logic is conserved, in a distantly related ascidian species Phallusia mammillata, in spite of extensive sequence disparity.
Molecular mechanisms downstream of neural induction for neural fate acquisition
FGF-triggered neural induction in Ciona appears, at first glance, to be a simple inductive process whereby two blastomeres (a6.5 and b6.5) receive a signal from the vegetal hemisphere and adopt a neural fate instead of an epidermal fate (Figures 1 and 2A). However, this event is tightly controlled: ectodermal cell competence is regulated [39], [40], embryo geometry [52] and various signaling pathways [41] also control the response of the ectoderm to the inducer.
We have shown that three FGF-dependent genes expressed in the b6.5 progeny from the 64-cell stage show differential regulation by Nodal signaling. Chordin is probably directly regulated by Nodal while Msxb and Delta2 need additional inputs from Otx. Our data provide additional connections and genomic hardwiring to a previously described network [35]. The network of genes regulating posterior neural fate is not linear and includes several regulatory loops (Figure 4K). FGF activates at least two direct target genes, Otx and Nodal, at the 32-cell stage, which collectively regulate secondary targets (i.e. Msxb and Delta2 at the 64-cell stage). Moreover, the regulation that we have uncovered involves a transcription factor and a signaling molecule that are expressed in the same cells. It is possible that this configuration allows very tight transcriptional control in a lineage-restricted manner using autocrine signaling. Finally, we have uncovered additional interactions that most likely maintain gene expression in a lineage-restricted manner following initial activation. For example, maintaining Nodal expression in the b6.5 progeny following FGF induction is apparently controlled both by Otx and Nodal itself (Figures S3 and 4K).
The actual mode of concerted regulation of Msxb and Delta2 by Otx and Nodal at the molecular level will need further investigation. We have proposed that the signaling molecule Nodal uses a Fox factor as a nuclear effector [44], [53]. This hypothesis led us to isolate three enhancers active in the b6.5 lineage. However, it is very likely that omitting Fox sites in our enhancer search would have led to the same outcome since Fox consensus sites (AAACA) are probably very abundant in the AT-rich ascidian genomes. Nevertheless, we observed that two overlapping Fox sites (AAACAAACA) are present in Msxb enhancers from both C. intestinalis and P. mammillata (Figures S5 and S10). However, mutation of these sites in “Ci-msxb b6.5 line” enhancer was silent unless some Otx sites were also mutated (Figure 6). The C. intestinalis genome encodes 29 predicted Fox factors whose expression pattern during early development has been determined [37], [54], but the number of candidate Fox factors (expressed in the b6.5 lineage or maternally provided) is beyond the scope of the current study. Although we cannot exclude the involvement of Fox factors in Msxb and Delta2 regulation, we would favor an alternative scenario explaining the concerted action of Otx and Nodal. We have shown that Smad Binding Elements (SBEs) are essential for msxb-B enhancer activity, and the active enhancers that we have isolated contain at least three SBEs. We could thus conceive that Otx itself serves as a co-factor for Nodal signaling and that it would interact directly with activated Smad2/3 on the enhancer to promote transcriptional activation.
Besides activating secondary FGF targets, the function of direct FGF targets is an opened question. Epidermal versus neural fate decision is primarily controlled by FGF signaling. We have shown that inhibition of FGF, Nodal or Otx function abolishes b-line neural fate. However, contrary to the inhibition of FGF, blocking Nodal or Otx function does not lead neural precursors to adopt the alternative epidermal fate (Figures S1 and S4). These observations can be explained by two non-exclusive hypotheses: epidermis fate inhibition is achieved directly upon reception of FGF signaling or several direct FGF targets contribute to epidermis repression. In particular, in addition to Otx and Nodal, genes such as Elk and Erf are expressed in neural progenitors and are likely direct FGF targets [30], but their function has not been determined.
Following their activation at the 64-cell stage in the b7.9/10 blastomeres, Msxb and Chordin remain expressed in all daughter cells (until mid-gastrula stages) but Delta2 expression becomes restricted in b8.18/20 blastomeres, precursors of the dorsal tail midline epidermis (Figure 2). This change in expression correlates with and may be involved in the fate restriction that occurs at early gastrula stages. This event is crucial since it separates central nervous system (dorsal nerve cord) and peripheral nervous system (dorsal tail midline epidermis) precursors. A similar CNS versus PNS segregation occurs at the same time in the anterior part of the embryo and involves FGF signaling [55]. While Msxb is essential for the formation of both dorsal tail epidermis midline and dorsal nerve cord [23], [35], the role of the two other genes remains to be investigated.
Otx and Nodal in chordate posterior neural tissue formation
Otx is a transcription factor expressed in the anterior nervous system, and which participates to anterior neural patterning in many bilaterians [56], [57]. In ascidians, a similar role has previously been ascribed to this gene in two distantly related species Ciona intestinalis and Halocynthia roretzi [21], [27], [33], [35], [42], [58]. The additional involvement of Otx in posterior neural tissue formation that we describe in the present study is rather unexpected. However, the function of Otx that we have addressed corresponds to a very early phase of its dynamic expression. Otx has been shown to be a direct target of FGF signaling at the 32-cell stage [11]. The expression is transient (from the 32-cell stage to the 112-cell stage) in both anterior (a6.5 lineage) and posterior (b6.5 lineage) neural tissue precursors and precedes a new and massive expression only in the anterior neural plate (from early gastrula stages). This early phase marks neural induction in both ascidian species studied [11], [59], [60]. While the onset of expression of Otx homologs in vertebrates may be broader than the prospective anterior central nervous system [61], there is no report, to our knowledge, of participation of Otx genes in posterior nervous system formation. We consequently propose that Otx has been co-opted in ascidians for posterior neural tissue specification. Whether this co-option is unique to ascidians will await more functional data in invertebrate deuterostomes.
We have shown that Nodal is required for posterior neural tissue formation and that Nodal can posteriorize FGF-induced neural tissue. Interestingly, Nodal signaling is also involved in posterior neural tissue formation in vertebrates [62]–[64]. However, this is most likely indirect through the control of mesoderm specification and patterning. Nodal signaling is rather thought to be an anti-neural pathway whose activity needs to be shut down for neural fate acquisition [65], [66]. Our study shows that the function of Nodal signaling in ascidians is different from vertebrates: Nodal is not incompatible with neural fate and it can directly posteriorize neural tissue.
In Ciona, Nodal expression in posterior neural precursors is the result of differential competence of animal blastomeres to respond to FGF. This competence is controlled by FoxA-a, expressed in anterior blastomeres [35], [40], [41]. When FoxA-a function is abolished, anterior neural ectoderm adopts a posterior identity and ectopically expresses Nodal and Delta2. A phenotype similar to what we observed for Nodal ectopic misexpression. However, Nodal is not the only factor involved in posterior identity definition. When we blocked Nodal function, posterior neural precursors did not adopt an anterior identity. This result suggests that either expression of FoxA-a is necessary for anterior identity definition and/or that additional factor(s) control posterior identity redundantly with Nodal. It will be interesting to probe the involvement of other signaling pathways (Wnt, FGF and retinoic acid) that are also major regulators of posterior neurectoderm formation in vertebrates [4].
A conserved regulatory signature in ascidians
Based on the combined regulation by Otx and Nodal, we were able to isolate enhancers containing putative Otx, Fox and Smad binding sites that control expression in the posterior neural lineage for two co-expressed genes. Interestingly, the “Ci-msxb-b6.5 line” enhancer is also active in anterior neurectoderm at tailbud stages (Figure S11) where several enhancers with an Otx signature have been described to be active [42]. This raises questions that will need further investigation. Are the same Otx-regulated enhancers re-used in different territories at different stages? Does the fragment we tested contain two distinct abutting or partially overlapping enhancers? These enhancers could consequently be the means for Otx co-option in posterior neural tissue. Finally, is Nodal signaling involved in later steps of anterior neurectoderm formation in C. intestinalis?
We have extended our study through cross-species transcriptional assay in two divergent ascidian species. Since Otx and Nodal display conserved expression in the b6.5 blastomeres in both C. intestinalis and H. roretzi [27], [29], [58], [67], it is very likely that they are also expressed in b6.5 in Phallusia mammillata, a species more closely related to C. intestinalis. This hypothesis can explain why we found conserved activity when C. intestinalis enhancers were tested in P. mammillata embryos. Importantly, we found that Msxb and Delta2 from P. mammillata are expressed under the control of FGF and Nodal signaling pathways in b-line neural precursors. Together with the isolation of an active enhancer for Pm-Msxb, these results strongly support that gene regulation is also conserved. We have tried to extend our comparison to Pm-Delta2 by testing several elements containing consensus Otx, Fox and Smad binding sites, but these elements were not active in posterior neural tissue precursors (data not shown). This can be explained by subtle changes in gene regulation or most likely by an incomplete understanding of the regulatory logic to be able to predict a functional enhancer (for example the tested elements had fewer Otx sites compared to the three active enhancers previously isolated). Interestingly, the Msxb enhancers that we isolated from each species do not show sequence conservation, they are not alignable. This is a general trend that has been observed by comparing ascidian genomes [21], [22]; mainly coding sequences retain sequence conservation and there is poor synteny conservation. This indicates that these genomes have largely diverged and underwent extensive reshuffling. This offers an excellent situation to probe enhancer evolution and transcription factor binding site turnover in genomes that control development of very similar embryos [20].
Materials and Methods
Embryo obtention and manipulation
Ciona intestinalis type B were provided by the Centre de Ressources Biologiques Marines in Roscoff. Phallusia mammillata were collected by diving in the Port-Vendres and Sète harbors, or collected from fishermen trawling in the Banyuls-sur-mer area. C. intestinalis embryology was performed as described in [32]. Staging was described according to [68]. P. mammillata embryos were handled the same way as Ciona except dechorionation was performed on unfertilized eggs for around 40 min with 0.1% trypsin and 0.5% sodium thioglycolate acid raised to basic pH by NaOH addition. Electroporation was performed as described [32] with the following modification: a single pulse of 25V for 32 ms (C. intestinalis) or 25 to 37V for 32 ms (P. mammillata).
Recombinant protein and inhibitor treatments were conducted as previously described [11], [27]–[29], [32]: bFGF (100 ng/ml) from the 16-cell stage, the protein synthesis inhibitor puromycin (200 µg/ml) from the 8-cell stage, the MEK inhibitor U0126 (4 µM) from the 8-cell stage and the TGFβ type 1 receptor inhibitor SB431542 (5 to 10 µM) from the 16-cell stage.
Standard control-MO (5'-CCTCTTACCTCAGTTACAATTTATA 3') and otx-MO (5′-ACATGTTAGGAATTGAACCCGTGGT-3′) were purchased from GeneTools LLC and were injected at 0.25 to 0.50 mM.
Gene model identifiers
The genes described in this study are represented by the following gene models in the KH2012 Ciona intestinalis assembly: Fgf9/16/20 (KH.C2.125), Otx (KH.C4.84), Nodal (KH.L106.16), Msxb (KH.C2.957), Delta2 (KH.L50.6), Chordin (KH.C6.145), Klf1/2/4 (KH.C5.154), KH.C7.391 (KH.C7.391), Dmrt1 (KH.S544.3), Lefty (KH.C3.411), Fog (KH.C10.574) and Ap2-like2 (KH.C7.43).
In situ hybridization, X-gal staining
Whole mount in situ hybridization and X-gal staining were performed as previously described [11]. Dig-labeled probes were synthesized from the following cDNAs for C. intestinalis: Msxb (cign067l18), Delta2 (cieg005o22), Chordin (cign055j01), Nodal (cicl090l02), Dmrt1 (ciad017d15), Klf1/2/4 (citb012d14), KH.C7.391 (cilv038e26) [69], Otx [27] and Ap2-like2 (cien223529) (Rothbächer et al., in preparation). For P. mammillata: Msxb (AHC0AAA214YL10RM1) and Delta2 (AHC0AAA62YG24RM1). While Msxb, Delta2 and Chordin expression in the b6.5 lineage starts at the 64-cell stage (st. 8), we analyzed early gastrula stages (st. 10/11) because expression is much stronger and more readily detectable by in situ hybridization.
Generation of electroporation constructs
Electroporation constructs for overexpression were generated using Gateway technology [51] with the promoter of Fiend of Gata (Fog) driving expression throughout ectoderm from the 16-cell stage [32], [39]. Constructs for Fgf9/16/20, Nodal and Lefty have already been described [32]. pFOG-Otx was generated by U. Rothbächer using the pENTRY clone cien28442 (Rothbächer et al., in preparation). A construct corresponding to the homeodomain of Otx fused to the Engrailed repressor domain has already been used [42] and was converted into a pENTRY clone using the following primers: attB1-OTXHD-Fw (5′-AAAAAGCAGGCTCAGAAAAAATGGTATACAGTTCGTCTAGAAAAC-3′) and attB2-EnR-Rev (5′-AGAAAGCTGGGTGAATTCTATACGTTCAGGTCCT-3′).
For transcriptional assay, genomic fragments were PCR amplified from sperm genomic DNA using AccuPrime Taq HiFi DNA polymerase (Invitrogen) and converted into pENTRY clones by a BP clonase reaction or TA cloning using the PCR8/GW/TOPO TA cloning kit (Invitrogen). The LR clonase reaction was performed to produce an expression clone with the genomic region in front of the minimal promoter of Fog and of nls-LacZ [51]. A detailed list of primers and vectors is described in Table S3. Enhancers msxb-A to -M (Figures 6, S7 and S8) were designed based on the msxb-OtxUP type B sequence and were synthesized as G-blocks Gene Fragments (Integrated DNA Technologies) flanked with AttB sequences (sequences listed in File S1). G-Blocks were shuffled into pDONR221 through BP reaction and through LR reaction into Rfa-bpFOG-nlsLacZ [51].
Supporting Information
Zdroje
1. LevineAJ, BrivanlouAH (2007) Proposal of a model of mammalian neural induction. Dev Biol 308 : 247–256 doi:10.1016/j.ydbio.2007.05.036
2. SternCD (2005) Neural induction: old problem, new findings, yet more questions. Development 132 : 2007–2021.
3. HarlandR (2000) Neural induction. Curr Opin Genet Dev 10 : 357–362.
4. AltmannCR, BrivanlouAH (2001) Neural patterning in the vertebrate embryo. Int Rev Cytol 203 : 447–482.
5. NiehrsC (2004) Regionally specific induction by the Spemann–Mangold organizer. Nat Rev Genet 5 : 425–434 doi:10.1038/nrg1347
6. RogersCD, MoodySA, CaseyES (2009) Neural induction and factors that stabilize a neural fate. Birth Defects Res Part C Embryo Today Rev 87 : 249–262 doi:10.1002/bdrc.20157
7. HollandLZ (2009) Chordate roots of the vertebrate nervous system: expanding the molecular toolkit. Nat Rev Neurosci 10 : 736–746.
8. Satoh N (2014) Developmental genomics of ascidians. Hoboken, New Jersey: John Wiley & Sons, Inc.
9. BertrandS, CamassesA, SomorjaiI, BelgacemMR, ChabrolO, et al. (2011) Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc Natl Acad Sci U S A 108 : 9160–9165 doi:10.1073/pnas.1014235108
10. YuJK, SatouY, HollandND, ShinIT, KoharaY, et al. (2007) Axial patterning in cephalochordates and the evolution of the organizer. Nature 445 : 613–617.
11. BertrandV, HudsonC, CaillolD, PopoviciC, LemaireP (2003) Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell 115 : 615–627.
12. DarrasS, NishidaH (2001) The BMP/CHORDIN antagonism controls sensory pigment cell specification and differentiation in the ascidian embryo. Dev Biol 236 : 271–288.
13. DavidsonEH, ErwinDH (2006) Gene regulatory networks and the evolution of animal body plans. Science 311 : 796–800 doi:10.1126/science.1113832
14. LemaireP (2006) Developmental biology. How many ways to make a chordate? Science 312 : 1145–1146.
15. Satoh N (1994) Developmental biology of ascidians. Cambridge: Cambridge University Press.
16. DehalP, SatouY, CampbellRK, ChapmanJ, DegnanB, et al. (2002) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298 : 2157–2167.
17. VoskoboynikA, NeffNF, SahooD, NewmanAM, PushkarevD, et al. (2013) The genome sequence of the colonial chordate, Botryllus schlosseri. eLife 2: e00569 doi:10.7554/eLife.00569
18. HollandLZ, AlbalatR, AzumiK, Benito-GutierrezE, BlowMJ, et al. (2008) The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res 18 : 1100–1111.
19. PutnamNH, ButtsT, FerrierDE, FurlongRF, HellstenU, et al. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453 : 1064–1071.
20. LemaireP (2011) Evolutionary crossroads in developmental biology: the tunicates. Development 138 : 2143–2152 doi:10.1242/dev.048975
21. Oda-IshiiI, BertrandV, MatsuoI, LemaireP, SaigaH (2005) Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development 132 : 1663–1674.
22. TakahashiH, MitaniY, SatohG, SatohN (1999) Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126 : 3725–3734.
23. ImaiKS, StolfiA, LevineM, SatouY (2009) Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136 : 285–293.
24. IkutaT, SaigaH (2007) Dynamic change in the expression of developmental genes in the ascidian central nervous system: Revisit to the tripartite model and the origin of the midbrain–hindbrain boundary region. Dev Biol 312 : 631–643 doi:10.1016/j.ydbio.2007.10.005
25. LemaireP, BertrandV, HudsonC (2002) Early steps in the formation of neural tissue in ascidian embryos. Dev Biol 252 : 151–169.
26. MinokawaT, YagiK, MakabeKW, NishidaH (2001) Binary specification of nerve cord and notochord cell fates in ascidian embryos. Development 128 : 2007–2017.
27. HudsonC, LemaireP (2001) Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev 100 : 189–203.
28. HudsonC, DarrasS, CaillolD, YasuoH, LemaireP (2003) A conserved role for the MEK signalling pathway in neural tissue specification and posteriorisation in the invertebrate chordate, the ascidian Ciona intestinalis. Development 130 : 147–159.
29. HudsonC, YasuoH (2005) Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development 132 : 1199–1210.
30. KhoueiryP, RothbacherU, OhtsukaY, DaianF, FrangulianE, et al. (2010) A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol 20 : 792–802 doi:10.1016/j.cub.2010.03.063
31. NishidaH (1987) Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev Biol 121 : 526–541.
32. PasiniA, AmielA, RothbacherU, RoureA, LemaireP, et al. (2006) Formation of the Ascidian Epidermal Sensory Neurons: Insights into the Origin of the Chordate Peripheral Nervous System. PLoS Biol 4: e225.
33. WadaS, SudouN, SaigaH (2004) Roles of Hroth, the ascidian otx gene, in the differentiation of the brain (sensory vesicle) and anterior trunk epidermis in the larval development of Halocynthia roretzi. Mech Dev 121 : 463–474.
34. HudsonC, LotitoS, YasuoH (2007) Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development 134 : 3527–3537.
35. ImaiKS, LevineM, SatohN, SatouY (2006) Regulatory blueprint for a chordate embryo. Science 312 : 1183–1187.
36. HudsonC, YasuoH (2006) A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development 133 : 2855–2864.
37. ImaiKS, HinoK, YagiK, SatohN, SatouY (2004) Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131 : 4047–4058.
38. Joyce TangW, ChenJS, ZellerRW (2013) Transcriptional regulation of the peripheral nervous system in Ciona intestinalis. Dev Biol 378 : 183–193 doi:10.1016/j.ydbio.2013.03.016
39. RothbacherU, BertrandV, LamyC, LemaireP (2007) A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development 134 : 4023–4032.
40. LamyC, RothbacherU, CaillolD, LemaireP (2006) Ci-FoxA-a is the earliest zygotic determinant of the ascidian anterior ectoderm and directly activates Ci-sFRP1/5. Development 133 : 2835–2844.
41. OhtaN, SatouY (2013) Multiple signaling pathways coordinate to induce a threshold response in a chordate embryo. PLoS Genet 9: e1003818 doi:10.1371/journal.pgen.1003818
42. HaeusslerM, JaszczyszynY, ChristiaenL, JolyJS (2010) A cis-regulatory signature for chordate anterior neuroectodermal genes. PLoS Genet 6: e1000912 doi:10.1371/journal.pgen.1000912
43. SchierAF (2003) Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol 19 : 589–621 doi:10.1146/annurev.cellbio.19.041603.094522
44. MassaguéJ, SeoaneJ, WottonD (2005) Smad transcription factors. Genes Dev 19 : 2783–2810 doi:10.1101/gad.1350705
45. GomisRR, AlarconC, HeW, WangQ, SeoaneJ, et al. (2006) A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci U A 103 : 12747–12752.
46. SilvestriC, NarimatsuM, von BothI, LiuY, TanNB, et al. (2008) Genome-wide identification of Smad/Foxh1 targets reveals a role for Foxh1 in retinoic acid regulation and forebrain development. Dev Cell 14 : 411–423.
47. Portales-CasamarE, ThongjueaS, KwonAT, ArenillasD, ZhaoX, et al. (2010) JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: D105–110 doi:10.1093/nar/gkp950
48. SatouY, MinetaK, OgasawaraM, SasakuraY, ShoguchiE, et al. (2008) Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol 9: R152.
49. FrazerKA, PachterL, PoliakovA, RubinEM, DubchakI (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–279 doi:10.1093/nar/gkh458
50. KuboA, SuzukiN, YuanX, NakaiK, SatohN, et al. (2010) Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development 137 : 1613–1623 doi:10.1242/dev.046789
51. RoureA, RothbacherU, RobinF, KalmarE, FeroneG, et al. (2007) A multicassette Gateway vector set for high throughput and comparative analyses in ciona and vertebrate embryos. PLoS ONE 2: e916.
52. TassyO, DaianF, HudsonC, BertrandV, LemaireP (2006) A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr Biol 16 : 345–358.
53. CarlssonP, MahlapuuM (2002) Forkhead transcription factors: key players in development and metabolism. Dev Biol 250 : 1–23.
54. YagiK, SatouY, MazetF, ShimeldSM, DegnanB, et al. (2003) A genomewide survey of developmentally relevant genes in Ciona intestinalis. III. Genes for Fox, ETS, nuclear receptors and NFkappaB. Dev Genes Evol 213 : 235–244.
55. WagnerE, LevineM (2012) FGF signaling establishes the anterior border of the Ciona neural tube. Dev Camb Engl 139 : 2351–2359 doi:10.1242/dev.078485
56. BoylPP, SignoreM, AnninoA, BarberaJP, AcamporaD, et al. (2001) Otx genes in the development and evolution of the vertebrate brain. Int J Dev Neurosci Off J Int Soc Dev Neurosci 19 : 353–363.
57. LichtneckertR, ReichertH (2005) Insights into the urbilaterian brain: conserved genetic patterning mechanisms in insect and vertebrate brain development. Heredity 94 : 465–477 doi:10.1038/sj.hdy.6800664
58. WadaS, KatsuyamaY, SatoY, ItohC, SaigaH (1996) Hroth an orthodenticle-related homeobox gene of the ascidian, Halocynthia roretzi: its expression and putative roles in the axis formation during embryogenesis. Mech Dev 60 : 59–71.
59. AkanumaT, NishidaH (2004) Ets-mediated brain induction in embryos of the ascidian Halocynthia roretzi. Dev Genes Evol 214 : 1–9 doi:10.1007/s00427-003-0368-y
60. MiyaT, NishidaH (2003) An Ets transcription factor, HrEts, is target of FGF signaling and involved in induction of notochord, mesenchyme, and brain in ascidian embryos. Dev Biol 261 : 25–38.
61. AngS-L, ConlonRA, JinO, RossantJ (1994) Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development 120 : 2979–2989.
62. FeldmanB, DouganST, SchierAF, TalbotWS (2000) Nodal-related signals establish mesendodermal fate and trunk neural identity in zebrafish. Curr Biol 10 : 531–534.
63. JiaS, WuD, XingC, MengA (2009) Smad2/3 activities are required for induction and patterning of the neuroectoderm in zebrafish. Dev Biol 333 : 273–284 doi:10.1016/j.ydbio.2009.06.037
64. ThisseB, WrightCV, ThisseC (2000) Activin - and Nodal-related factors control antero-posterior patterning of the zebrafish embryo. Nature 403 : 425–428 doi:10.1038/35000200
65. CamusA, Perea-GomezA, MoreauA, CollignonJ (2006) Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev Biol 295 : 743–755 doi:10.1016/j.ydbio.2006.03.047
66. ChangC, HarlandRM (2007) Neural induction requires continued suppression of both Smad1 and Smad2 signals during gastrulation. Development 134 : 3861–3872 doi:10.1242/dev.007179
67. MorokumaJ, UenoM, KawanishiH, SaigaH, NishidaH (2002) HrNodal, the ascidian nodal-related gene, is expressed in the left side of the epidermis, and lies upstream of HrPitx. Dev Genes Evol 212 : 439–446.
68. HottaK, MitsuharaK, TakahashiH, InabaK, OkaK, et al. (2007) A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn 236 : 1790–1805.
69. SatouY, YamadaL, MochizukiY, TakatoriN, KawashimaT, et al. (2002) A cDNA resource from the basal chordate Ciona intestinalis. Genesis 33 : 153–154.
70. TassyO, DaugaD, DaianF, SobralD, RobinF, et al. (2010) The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res 20 : 1459–1468 doi:10.1101/gr.108175.110
71. SatouY, KawashimaT, ShoguchiE, NakayamaA, SatohN (2005) An integrated database of the ascidian, Ciona intestinalis: towards functional genomics. Zool Sci 22 : 837–843.
72. ShiW, LevineM (2008) Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development 135 : 931–940.
73. MitaK, FujiwaraS (2007) Nodal regulates neural tube formation in the Ciona intestinalis embryo. Dev Genes Evol 217 : 593–601.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



