-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
Cell division in Escherichia coli begins with the assembly of FtsZ proteins into a ring-like structure, the Z-ring. Remarkably, the Z-ring localizes with very high precision at midcell. Currently, two molecular systems, nucleoid occlusion and the Min system, are known to localize the Z-ring. Here, we explore whether there are additional divisome localization systems in E. coli. Using quantitative fluorescence imaging, we show that slow growing cells lacking both known positioning systems continue to divide accurately at midcell. We find that the terminus region of the chromosome moves first to mid-cell where it functions as a positional landmark for the subsequent localization of the Z-ring. Furthermore, we provide evidence that this divisome positioning system involves MatP, ZapB, and ZapA proteins. Our work shows that E. coli can divide without the canonical mechanisms for localizing its cytokinetic ring. In particular, we identify that the Ter macrodomain acts as a landmark for the Z-ring in the presence of MatP, ZapB and ZapA proteins.
Published in the journal: Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in. PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004504
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004504Summary
Cell division in Escherichia coli begins with the assembly of FtsZ proteins into a ring-like structure, the Z-ring. Remarkably, the Z-ring localizes with very high precision at midcell. Currently, two molecular systems, nucleoid occlusion and the Min system, are known to localize the Z-ring. Here, we explore whether there are additional divisome localization systems in E. coli. Using quantitative fluorescence imaging, we show that slow growing cells lacking both known positioning systems continue to divide accurately at midcell. We find that the terminus region of the chromosome moves first to mid-cell where it functions as a positional landmark for the subsequent localization of the Z-ring. Furthermore, we provide evidence that this divisome positioning system involves MatP, ZapB, and ZapA proteins. Our work shows that E. coli can divide without the canonical mechanisms for localizing its cytokinetic ring. In particular, we identify that the Ter macrodomain acts as a landmark for the Z-ring in the presence of MatP, ZapB and ZapA proteins.
Introduction
Cell division is an essential cellular process that requires accurate spatial and temporal positioning of cytokinetic proteins. Assembly of the cell division apparatus, the divisome, must be coordinated closely with replication and segregation of chromosomes to ensure that each daughter cell receives an integral genome from the mother. The assembly of the divisome in Escherichia coli starts with the formation of a macromolecular structure, called the Z-ring, which encircles the rod-shaped cell in its geometric middle [1]–[4]. The Z-ring consists of filaments of FtsZ proteins, which are anchored to the cell membrane through the FtsA and ZipA linker proteins. The Z-ring serves as a scaffold for more than a dozen other divisome proteins, which build the cell envelope between the two daughters and mediate partitioning of the chromosomes into newly forming compartments [5].
In E. coli, the divisome is positioned at the cell center with remarkable accuracy [6]–[9]. How do nanometer-scale FtsZ proteins recognize the center of the cell with such high accuracy and at the same time provide faithful coordination between the divisome and the chromosome? The current view holds that Z-ring localization is governed by two independent mechanisms in E. coli, the Min system and nucleoid occlusion [2]–[4], [10] that both negatively regulate Z-ring polymerization. The Min system is composed of the MinC, MinD, and MinE proteins that together exhibit dynamic pole-to-pole oscillation [11]. While the MinD and MinE proteins are essential for such oscillation, MinC acts as the sole inhibitor of Z-ring formation by binding to FtsZ [12]. Considering that the minimum of the time-averaged concentration of MinC occurs at midcell, the Min system protects cell poles from developing septa and guides localization of the Z-ring to the center of the cell [13].
The nucleoid occlusion mechanism was first proposed on a phenomenological level to account for a lack of division septa from forming over the nucleoid [14], [15]. It has been established that the SlmA protein mediates nucleoid occlusion in E. coli [16], while a similar factor, the Noc protein, was found in Bacillus subtilis [17]. The two proteins do not share sequence similarity but they apparently function in a similar manner. Both SlmA [18]–[20] and Noc [21] are DNA-binding proteins that are capable of inhibiting Z-ring formation in their DNA-bound form. SlmA and Noc lack binding sites in the vicinity of the replication terminus (Ter). Such positioning assures that their Z-ring inhibiting activity is relieved at midcell when two daughter chromosomes segregate.
E. coli cells that lack both the Min system and SlmA are not capable of dividing in rich LB medium, instead forming long filamentous cells [16]. Although this finding could imply that the Min system and SlmA are the only localization systems for the divisome in E. coli, it was found that the same cells can grow and divide in nutrient poor M9 medium, and even in LB when FtsZ levels were artificially upregulated [16]. It was also found that deletion of SlmA alone did not cause any loss in cell division accuracy, and the correlations between the divisome and chromosome localizations remained the same in these cells compared to wild type [6]. These findings imply that there exists a SlmA-independent mechanism that localizes cell division proteins relative to chromosomes in E. coli. Similar to E. coli, evidence of Noc-independent nucleoid occlusion exists in B. subtilis [22], [23]. Moreover, it was found that B. subtilis cells were capable of positioning the Z-rings precisely at midcell in the complete absence of any nucleoid occlusion and the Min system [24]. These findings warrant revisiting the canonical model that the Min system and SlmA/Noc mediated nucleoid occlusion together are the sole factors coordinating the localization of cell division proteins in bacteria, and raise the question of what additional mechanisms bacterial cells use to position their divisome.
Here, we study cell division in E. coli strains lacking both the Min system and nucleoid occlusion factor SlmA to identify new mechanisms involved in Z-ring localization. We use high-resolution quantitative fluorescence imaging to resolve nanometer-scale changes in positions of the Z-rings and cell division planes. We show that Min and SlmA double deletion cells are capable of accurately localizing their division planes in slow growth conditions. In this process, E. coli frequently positions its Z-ring initially over the nucleoid center instead of at nucleoid-free regions. We determine that during the formation of the Z-ring, the nucleoid center is occupied by the Ter macrodomain region of the chromosome. MatP, ZapB, and ZapA proteins, which have been implicated in linking the Ter macrodomain and the Z-ring [25], affect the accuracy and the precision of the Z-ring positioning relative to the nucleoid center. However, E. coli ΔslmA Δmin cells without MatP, ZapB, and ZapA are still capable of positioning their Z-rings close to the cell centers, albeit with lower precision.
Results
ΔslmA Δmin cells divide at well-defined locations relative to cell poles
Details about how cell division occurs in E. coli ΔslmA Δmin strain has not yet been described in slow growth conditions where cells are capable of dividing and propagating. One would expect that if SlmA-mediated nucleoid occlusion and the Min system are the only two positioning systems in E. coli, then the division planes in these double mutant cells should be localized completely randomly. Surprisingly, we found this not to be the case. The majority of ΔslmA ΔminC cells appeared to divide about the cell center, and all the cells retained normal morphology in minimal M9 medium. To quantify the accuracy of division plane placement in these cells, we determined the relative volume fractions of two daughter cells that still adhere together by their poles after the division and compiled these ratios into a histogram (Figure 1). To calculate the volume fractions, we used fluorescent images of cells, which carried a cytosolic GFP label, and applied a quantitative image analysis procedure as described earlier [6]. As a reference, we determined the volume fraction distributions for the parental strain/wild type BW25113 (Figure 1A) and strain JW1165 having only a minC deletion (Figure 1B). For all the strains used in this work, see Table S1. As expected, the distribution of volume fractions for the parental strain consisted of a pronounced single peak at a value of 1/2, showing that upon division, the volume of each daughter cell is approximately equal. Note that all histograms are symmetric relative to 1/2 because both daughter cells are counted in these histograms. Also as expected, the distribution of volume fractions for the ΔminC strain showed distinct peaks at 1/4, 1/3, 2/3 and 3/4 values, in addition to the main peak at 1/2. All these peaks arise because of underlying nucleoid structure. Peaks at 1/4 and 3/4 values correspond to divisions where a mother cell distributes one of its nucleoids to one daughter cell and three to the other. Smaller peaks at 1/3 and 2/3 values correspond to division of cells with three nucleoids. The ΔminC strain also showed minicelling divisions which appeared as broad peaks on the tails of the volume fraction histogram.
Fig. 1. Relative volume fractions of daughter cells after division. 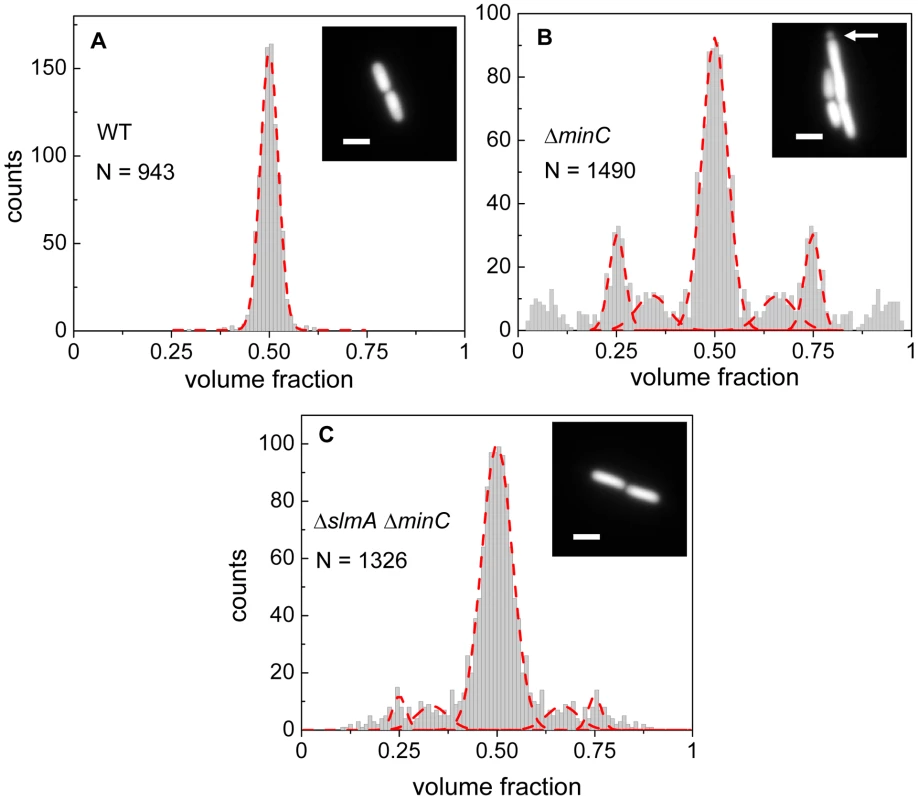
(A) Wild type (BW25113), (B) ΔminC (JW1165), (C) ΔslmA ΔminC (PB194) strains. Volume fractions are calculated as the ratio of one daughter cell's volume to the sum of both daughters' volumes. Red dashed lines in the histogram show fittings of different peaks with a Gaussian function. The centers of fitting lines are fixed to 1/4, 1/3, 1/2, 2/3 and 3/4 values. The insets in the histograms show fluorescent images of cells from the respective strains. The arrow in the inset of panel (B) points to a minicelling division. All scale bars correspond to 2 µm. The volume fraction distribution for the ΔslmA ΔminC strain (PB194) showed, qualitatively similar to the ΔminC strain, distinct peaks at 1/2, 1/4 and 3/4 positions with discernible peaks also at 1/3 and 2/3 values (Figure 1C). Gaussian fits to the peaks in the histogram showed that the majority of ΔslmA ΔminC cells divide at about midcell (75%) while 6.5% divided approximately at the quarter position, and 14% between the quarter and half-cell length from one of the poles. Interestingly, the frequency of central divisions for the ΔslmA ΔminC strain was higher than for the strain having only a minC deletion (50%) while the frequency of ΔminC cells dividing at a quarter (20%) and a third of the cell length from the poles (16%) was higher compared to the ΔslmA ΔminC strain. Additionally, the double mutant strain produced essentially no minicells (0.2% of total divisions), although they were noticeably present in the minC deletion strain (7% of total divisions). The presence of peaks at the 1/2, 1/4, and 3/4 positions indicates that, despite a lack of nucleoid occlusion factor SlmA in the double mutant strain, there remains a high level of coordination between nucleoids and the Z-rings in E. coli cells. Comparison between ΔminC ΔslmA double mutants and ΔminC single mutant strains further shows that removal of SlmA suppresses minicell production and biases cell division towards the cell center.
The cell division plane shifts from midcell to quarter-cell with increasing cell length
The length of the double mutant cells immediately following division, 2.83±1.54 µm, was 18% longer than that of the BW25113 parental strain, 2.41±0.36 µm (Figure S1). The main factor contributing to the length difference was long cells making up about 10% of the ΔslmA ΔminC population, whose lengths were about twice that of the majority of the population. It was noticeable that these longer double mutant cells divided with higher prevalence at 1/4 and 3/4 positions compared to shorter cells. To quantify this tendency, we plotted the frequency of central divisions and the frequency of divisions at the quarter cell length from the poles as a function of mother cell length (Figure 2). In this analysis, the central divisions were considered to be all divisions in which volume fractions were within 0.50±0.10. The divisions at quarter cell positions were considered when the corresponding volume fraction ratios were within 0.25±0.05. For the ΔslmA ΔminC cells, the frequency of cell divisions at the quarter cell length from the poles increased considerably as the cells reached a length of about 6 µm (Figure 2A). About 50% of cells longer than 6 µm preferentially divided at the quarter-cell length from the poles, while a smaller fraction, about 25% of cells, divided at the cell center. The data for the ΔminC cells showed a very similar sharp transition of the cell division plane from the center to the quarter locations as the cell length reached about 5.2 µm (Figure 2B). A marked increase in the frequency of 1/4 divisions indicates that some positional signal guides the cell division plane from midcell to its quarter positions as the cells reach a relatively well-defined length.
Fig. 2. Division frequency at the 1/4 and 1/2 cell positions with respect to mother cell length. 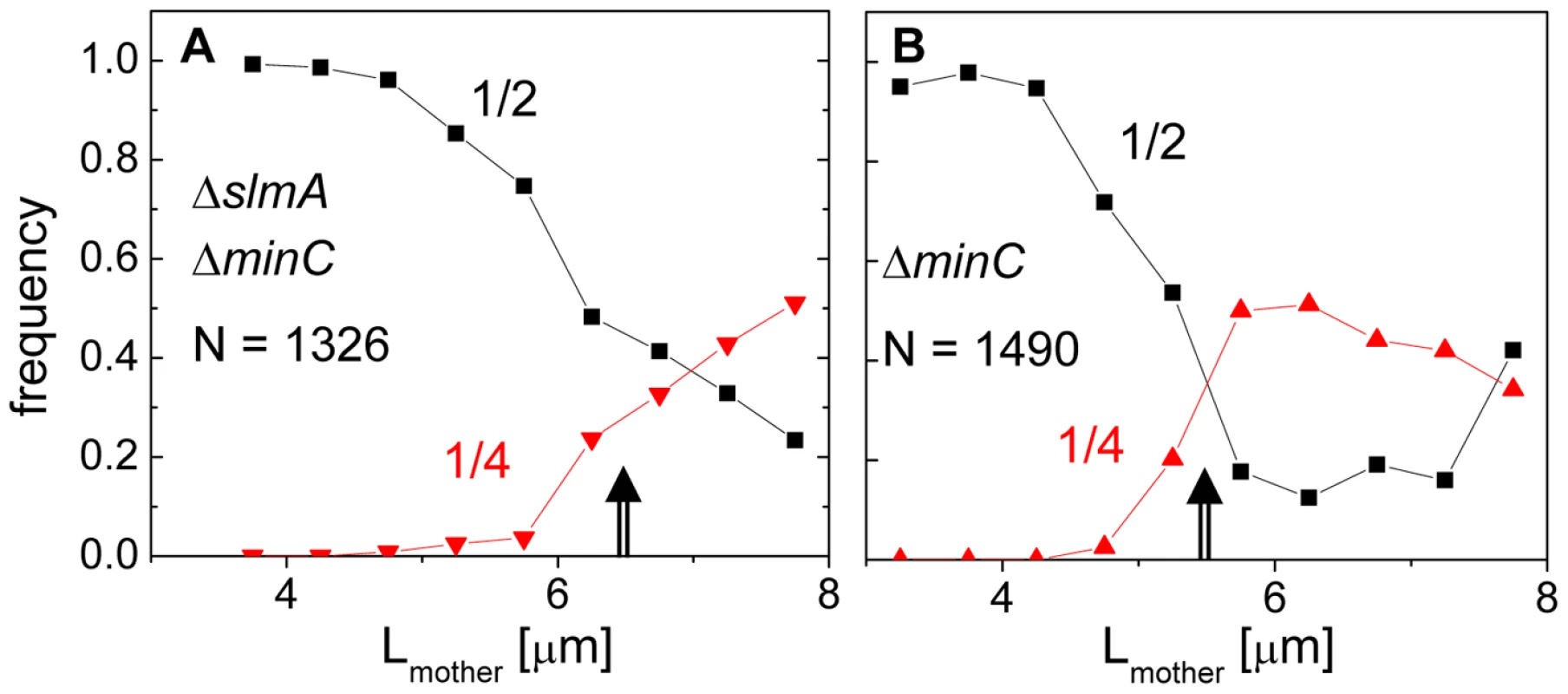
(A) Data for the ΔslmA ΔminC double mutant strain (PB194); (B) ΔminC strain (JW1165). Cell lengths are binned at 0.25 µm intervals. Arrows point to transition regions from centrally occurring divisions to divisions at cell quarters. The lengths of the mother cells are measured just before cell division. Note that only a few cells from both strains are longer than 8 µm, limiting analysis for longer cells. Z-rings localize to the centers of nucleoids
To investigate this positional signal, we determined the placement of the Z-ring relative to the nucleoid and cell centers using the previously described E. coli ΔslmA Δmin double mutant strains with ZipA-GFP (TB86 λCH151) and FtsZ-GFP (TB86 λDR120) labels [16]. In these measurements, the nucleoid was stained with DAPI. As a reference, the parental strain with the same labeling was also imaged. Representative cell images are shown in Figure S2. We noticed that in a few ΔslmA Δmin cells, the nucleoids were displaced noticeably from the cell center. In these cases, the positions of the Z-rings followed the centers of the nucleoids rather than the centers of the cytosolic volumes (Figure 3A). To quantify the tendency of the Z-ring to localize over the nucleoid center, we measured the distance between the Z-ring and cell center, ΔXz, as a function of the distance between the nucleoid center and the cell center, ΔXn, for all cells in a population having a single nucleoid (Figure 3B, C). The numerical procedure to determine the centers of the cell, nucleoid, and Z-ring is described in the Text S1. As can be seen from Figure 3B, displacements of nucleoids away from cell centers were associated with correlated displacements of Z-rings. To further analyze the extent of co-localization between the nucleoid center and the Z-ring, we determined the standard deviations of distances between the Z-rings and nucleoid centers, , and between the Z-rings and cell centers . We separated the data into two distinct groups – polar Z-rings and centrally located ones. The precision of central Z-ring placement relative to nucleoid centers, (Figure 3E), was more than two times higher than the positioning of Z-rings relative to cell centers, , in ΔslmA Δmin cells with ZipA-GFP label (Figure S3A). We found very similar co-localization characteristics for central Z-rings (, ) in FtsZ-GFP labeled ΔslmA Δmin cells (Figure S3B, S4), confirming that the co-localization effect is not related to a specific Z-ring label. The collection of distribution statistics for all measured strains can be found in Table S2. Interestingly, for wild type cells (Figure 3D, F) co-localization between Z-rings and nucleoid centers () was somewhat lower than in ΔslmA Δmin cells (Ansari-Bradley test ; F-test ), while the precision of Z-ring placement in the vicinity of the cell centers () was significantly higher when compared to ΔslmA Δmin cells (Ansari-Bradley test ; F-test ). Similar values of and as in wild type cells were found also for Δmin and ΔslmA single deletion strains (Figure S5; Table S2).
Fig. 3. Localization of ZipA-GFP labeled Z-rings relative to cell center and the center of nucleoids. 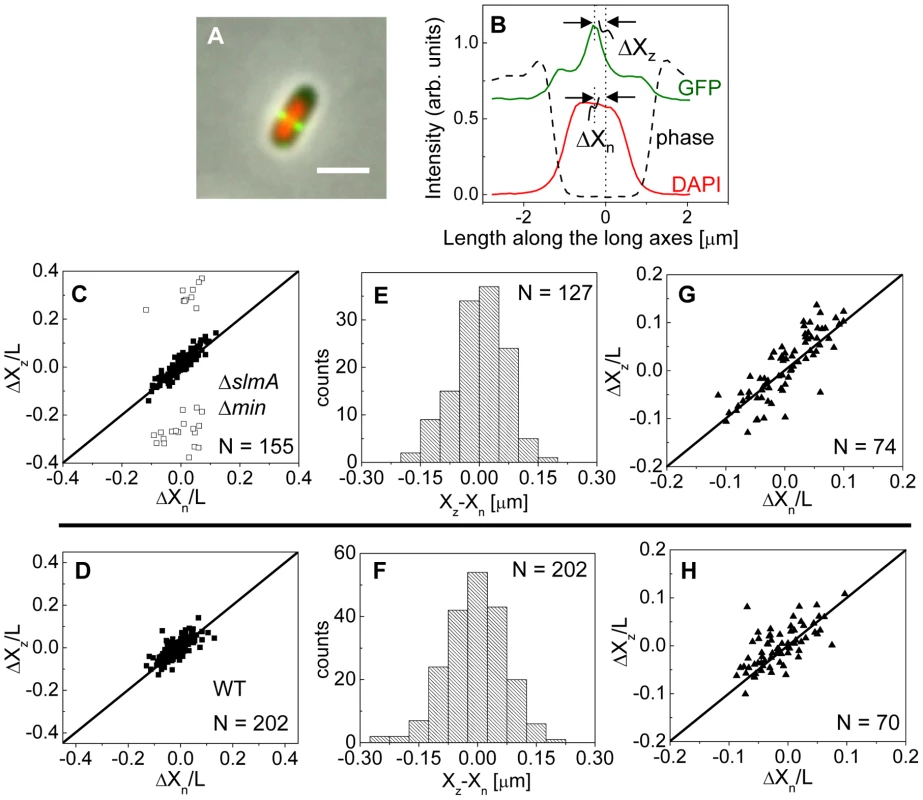
(A) A composite of ZipA-GFP (green), DAPI stained nucleoid (red) and phase contrast images (grey) of a ΔslmA Δmin cell with a distinctly off-center placed nucleoid. The scale bar is 2 µm. (B) The intensity line profiles of each image plane along the long axis of the cell for the cell shown in panel A. The displacement of the nucleoid relative to the cell center is ΔXn, and the displacement of the ZipA-GFP labeled Z-ring is ΔXz. (C) ΔXz vs. ΔXn for ΔslmA Δmin cells (strain TB86) scaled by cell length L. Solid rectangles mark central and open rectangles mark polar Z-rings. The solid line corresponds to . Data are shown only for cells with a single nucleoid. (D) ΔXz vs. ΔXn for the parental strain (strain JMBW5). (E), (F) Distribution of distances between the Z-ring center and nucleoid center for ΔslmA Δmin strain and parental strain, respectively. Data for central Z-rings are shown. (G), (H) ΔXz vs. ΔXn for cells that show a Z-ring over a compact nucleoid in ΔslmA Δmin and in parental strain, respectively. Co-localization of the Z-ring to nucleoid centers was present already in the early stages of chromosomal replication before a distinct bi-lobed morphology appeared in nucleoid images (Figure 3G, H). To distinguish bi-lobed nucleoids from compact nucleoids, we inspected intensity line profiles taken over DAPI stained nucleoids. We considered a nucleoid to be compact if its DAPI intensity line profile near the nucleoid center lacked any discernable dips (e.g. DAPI profile in Figure 3B). In ΔslmA Δmin cells with a compact nucleoid, the level of co-localization between the nucleoid and the Z-ring, , was comparable to the value characterizing the whole cell population, (F-test, ; Ansari Bradley test ). A Similar conclusion can be drawn also for the wild type cells where (F-test ; Ansari Bradley test ) and for Δmin and ΔslmA single deletion strains (Figure S5; Table S2). These comparisons indicate that nucleoid centers and Z-rings can co-localize in early stages of replication when the nucleoid morphology is compact both in wild type cells and in cells where one or both of the known Z-ring positioning systems have been removed.
The bias in localization of the Z-rings to the centers of nucleoids was even more visually striking in longer ΔslmA Δmin cells that had two or more well-separated nucleoids (Figure 4A). We found a strong preference for the Z-ring to position over the centers of nucleoids as compared to regions between fully segregated nucleoids (Figure 4B). We refer to the former as the new division sites (N) and the latter as the old division sites (O). The probability of finding a Z-ring over the center of nucleoids (N sites) was 98±1%, while the probability of finding a Z-ring in the inter-nucleoid space between fully segregated nucleoids (O sites) decreased to 59±8% (Figure 4C). Note that Z-rings can be present in both division sites at the same time. The tendency of the Z-rings to preferentially localize at ¼ positions from the cell pole in longer cells, i.e. in new sites, is consistent with our earlier observation that in longer ΔslmA Δmin cells, divisions occur preferentially at ¼ positions from the cell pole (Figure 2). Taken together, the analysis of the placement of the Z-rings and nucleoid centers in multi-nucleoid ΔslmA Δmin cells further supports the hypothesis that a positional signal guides the Z-rings to the nucleoid centers.
Fig. 4. Positioning of Z-rings relative to nucleoids in multi-nucleoid cells. 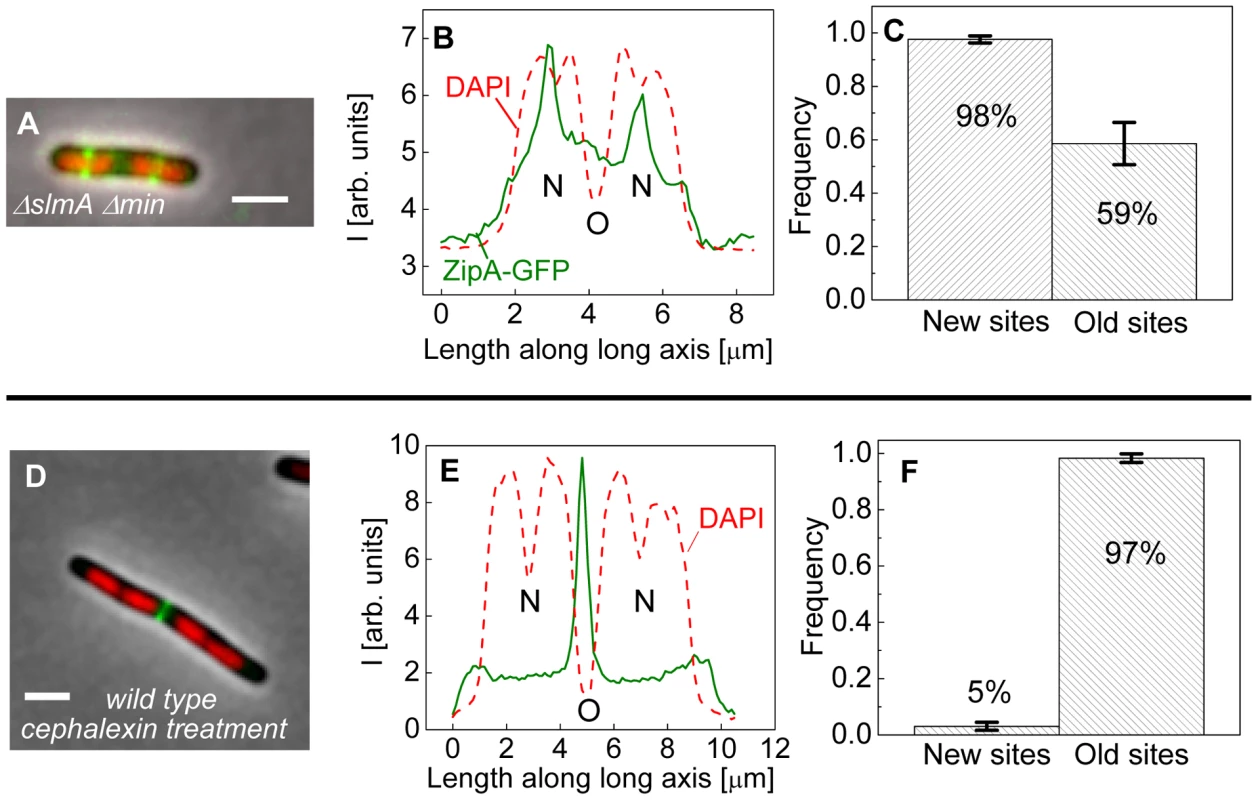
(A) A composite image of longer ΔslmA Δmin cell. ZipA-GFP (green), DAPI stained nucleoid (red), and phase contrast images (grey) have been overlaid. Scale bar is 2 µm. (B) Nucleoid and ZipA-GFP density distributions along the long axis of the cell for the cell shown in panel (A). The positions marked by “N” correspond to the new division sites at the centers of the nucleoids and the position marked by “O” to old division site between fully segregated nucleoids. (C) Frequency of Z-rings in the double mutant cells at the new and old replication sites. Only cells that have two or more distinct nucleoids have been analyzed. Error bars represent standard deviations over three independent measurements each involving about 50 cells. (D)–(F) the same for wild type cells that have been treated for 2 hours with 20 µg/ml cephalexin. To determine if wild type cells would display the same behavior as multi-nucleoid ΔslmA Δmin cells we induced an elongated, multi-nucleoid cell morphology by treating cells with cephalexin. Cephalexin does not inhibit Z-ring assembly but prevents Z-ring constriction by inhibiting the downstream protein FtsI (PBP3). Interestingly, in elongated wild type cells the Z-rings appeared essentially only at midcell even when new sites were present (Figure 4 D–F). Z-rings in cephalexin treated ΔslmA ΔminC cells still showed a preference to the new division sites as did their untreated counterparts (Figure S6 A–C). We also analyzed ΔslmA and ΔminC single deletion cells after cephalexin treatment. ΔslmA cells behaved as wild type cells (Figure S6 D–F) while Z-rings in the ΔminC cells showed a preference to the new division sites as in ΔslmA Δmin cells (Figure S6 G–I). These comparisons show that the putative positioning signal only manifests itself when it is not conflicting the regulation due to the Min system. It is important to note that such conflict does not occur in wild type cells in normal growth conditions because in this case the nucleoid center and the concentration minimum for MinC coincide. As Figures 3 D–H show, the localization signal emanating from the nucleoid center is important in Z-ring localization in wild type cells under normal growth conditions.
Co-localization between the Ter macrodomain and the Z-ring
Approximately at the time of Z-ring formation, the center of the nucleoid is known to be occupied by the Ter region of the chromosome [26], [27], which forms a well-defined unit – the Ter macrodomain [28], [29]. In E. coli, MatP is a dispensable protein that defines the Ter macrodomain by connecting 23 specific sites in a chromosomal region that spans about 800 kb [28]. Based on previous works [26], [27], it appeared plausible that the Z-rings might position over the Ter macrodomain. To investigate if this hypothesis is correct, we labeled the Ter region of the chromosome with a MatP-mCherry construct that was expressed from its endogenous matP locus and we labeled the Z-ring with ZipA-GFP (Figure 5A, B). The measurements revealed a very strong correlation in the placement of the MatP-labeled Ter macrodomain and the Z-ring in ΔslmA Δmin cells (Figure 5C). Notably, the Z-ring co-localized with the MatP focus in all cases, even including cases when the MatP focus was at the nucleoid periphery close to the cell pole. In wild type cells, correlations were also strongly present although in a few cases (4 out of 166) the Z-ring could be observed to localize at the center of the cell when the MatP locus was close to the cell pole (Figure 5D). These events were also present in the ΔslmA single deletion strain but were absent from the ΔminC strain (Figure S7) indicating that the Min system reduces correlations between the Z-ring and the Ter macrodomain. The measured co-localization precision between the MatP-labeled Ter foci and Z-ring centers was σXz-XMatP = 56 nm for ΔslmA Δmin (Figure 5E) and σXz-XMatP = 66 nm for wild type cells (Figure 5F). Similar values for σXz-XMatP also were found for ΔslmA and Δmin single deletion strains (Figure S7). All the measurements of co-localization precision σXz-XMatP were close to our resolution limit and thus consistent with the hypothesis that the Ter macrodomain and Z-ring co-localize in E. coli unless the Min system prevents such co-localization from happening.
Fig. 5. Positioning of the Z-ring relative to the MatP-labeled Ter macrodomain. 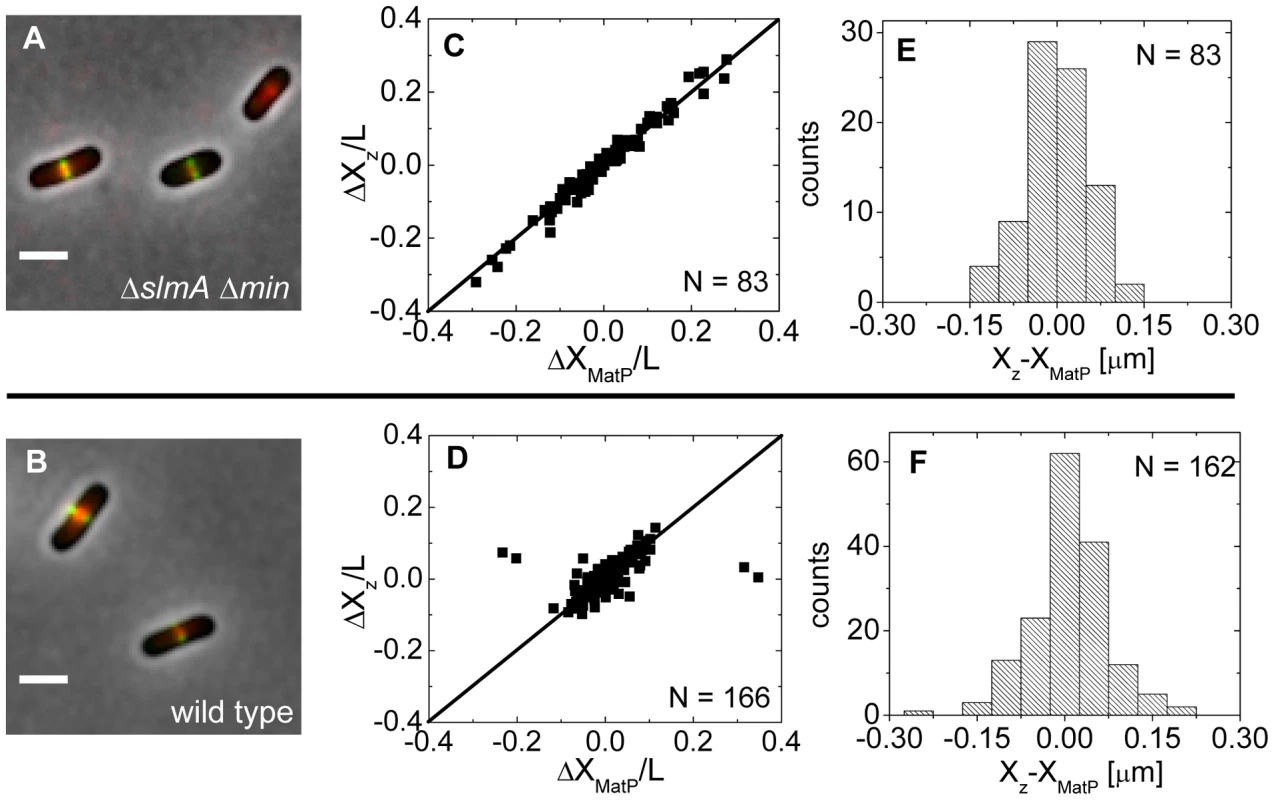
(A) A composite of ZipA-GFP (green), MatP-mCherry (red), and phase contrast image (grey) of ΔslmA Δmin cells (strain WD1). Scale bar is 2 µm. (B) The same for the wild type strain (strain WD2). (C) Location of ZipA-GFP labeled Z-ring (ΔXz) vs location of MatP-mCherry focus (ΔXMatP) in ΔslmA Δmin cells scaled by the cell length L. Both locations are referenced relative to the cell center. Solid symbols correspond to locations near the center of the nucleoid and open squares to locations near the poles. The straight line corresponds to . Only cells with a single MatP focus are analyzed. (D) ΔXz vs ΔXMatP for wild type cells. (E), (F) Distribution of distances between the Z-ring and the MatP focus along the long axes of the cell for ΔslmA Δmin and wild type cells, respectively. The Ter region arrives at the cell center before Z-ring formation
Previously, it was argued that the divisome anchors the Ter macrodomain to the cell center through a MatP-mediated link in which the divisome related proteins ZapA and ZapB participate [25]. The co-localization data (Figure 5) clearly supports the presence of this link, which we refer to as the Ter linkage. The data also raise the possibility that the Ter macrodomain may be important in positioning and stabilizing the location of the divisome. If the latter hypothesis is correct then there should be some time delay between the arrival of the Ter macrodomain at the cell center and the subsequent formation of the Z-ring. To test this hypothesis we followed the movement of the Ter macrodomain and the Z-ring in ΔslmA Δmin and wild type cells using MatP-mCherry and ZipA-GFP labels. Similar to an earlier report on wild type cells [25], [28], in ΔslmA Δmin cells under slow growth conditions the Ter macrodomain moved from the cell pole to the center of the cell at the beginning of the cell cycle (Figure 6 A–B, Figure S8, Movie M1, M2). During this movement, the Ter macrodomain either split into two distinct foci or displaced through the cell as a somewhat diffuse unit. The Ter region of the chromosome remained in the center of the nucleoid for the majority of the cell cycle before splitting into two foci during the late stage of cytokinesis. The Z-ring co-localized with the Ter macrodomain early in the cell cycle when the Ter region was positioned at the cell poles and during the majority of the cell cycle when the Ter region was localized as a single unit at midcell (Figure 6 A–B). However, our measurements showed that during the period in which the Ter macrodomain dislocated from the new pole to the cell center, the ZipA-GFP focus lagged behind the MatP-labeled Ter macrodomain. We measured the lag period to be (0.12±0.07)⋅Td for ΔslmA Δmin cells (Figure 6C). The doubling time, Td, was about 120 min in these growth conditions. In addition to the lag period, the accumulation of the Z-ring proteins and the Ter macrodomain in the center of the cell showed different time-dependent behaviors (Figure 6D). Following the beginning of the cell cycle, the MatP-mCherry labeled Ter macrodomain arrived at the cell center not only with a shorter delay but also accumulated in the center of the cell on average more rapidly than the ZipA-GFP marker for the Z-ring (Figure 6D). We observed a similar behavior for wild type cells (Figure S9) although the delay appeared somewhat smaller, (0.02±0.10) ⋅Td (Figure S10).
Fig. 6. Arrival of the MatP foci and the Z-ring at midcell. 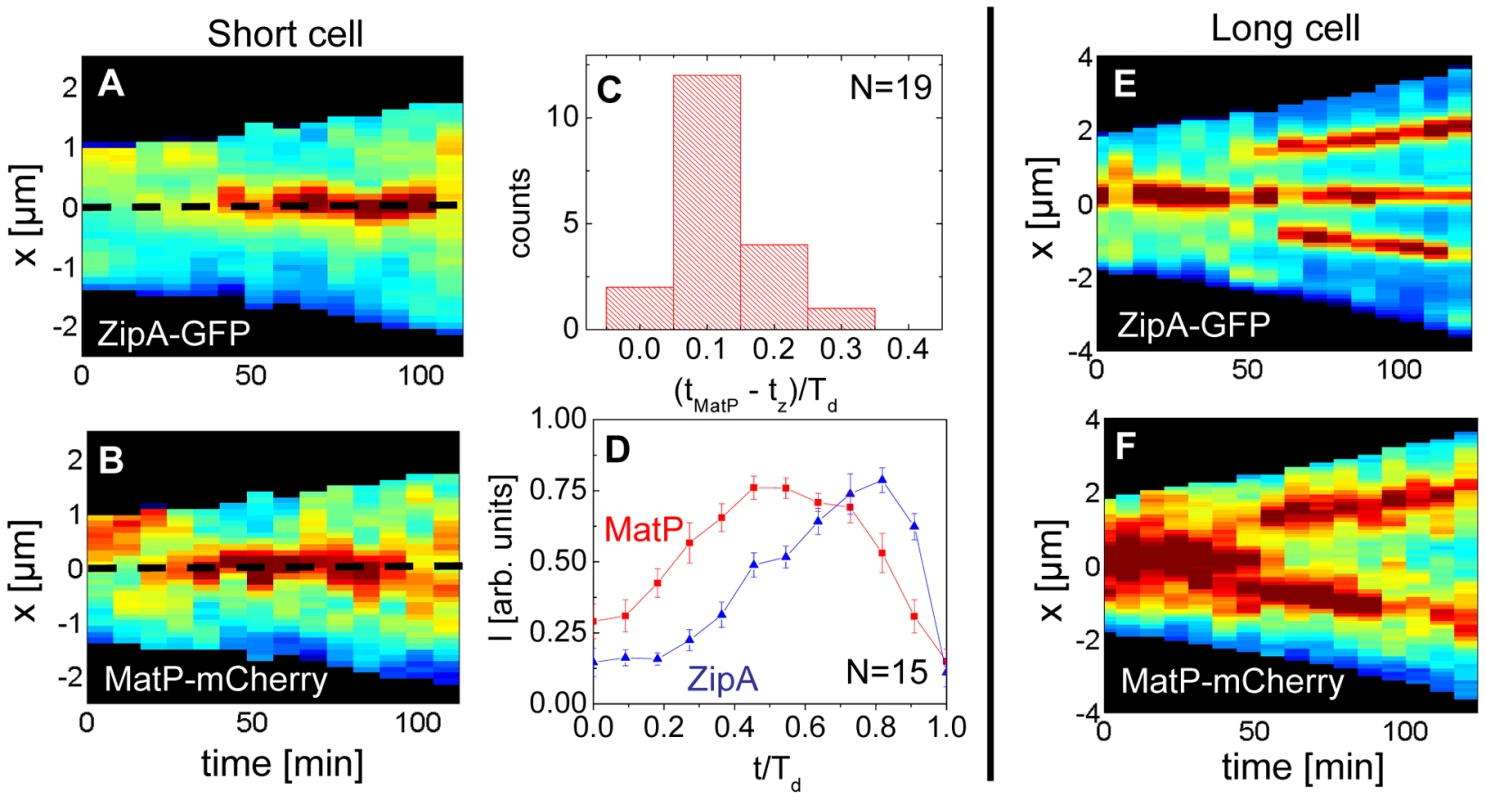
(A) Distribution of ZipA-GFP along the cell length as a function of time for a short ΔslmA Δmin cell (strain WD1). (B) Distribution of MatP-mCherry labeled Ter region for the same cell. In the heat maps blue corresponds to low and red to high intensity. The dashed black line approximately marks midcell. (C) Histogram of time differences between the arrival of MatP (tMatP) and ZipA (tz) at midcell. The times are expressed in doubling times. (D) Accumulation of ZipA-GFP (blue triangles) and MatP-mCherry (red rectangles) at midcell as a function of time. Each curve represents the average from measurements of 15 cells. Error bars represent standard errors. (E) Distribution of ZipA-GFP and (F) MatP-mCherry in a long ΔslmA Δmin cell. Time lapse measurements of the Ter macrodomain and the Z-ring in longer (L>6 µm) ΔslmA Δmin cells indicate why these cells prefer divisions at the ¼ positions from the cell poles (cf. Figure 2A) and preferentially show Z-rings at the new division sites (cf. Figure 4). The measurements showed that a shift from the cell center to ¼ positions occurred when the Ter region moved from the center of the cell to ¼ positions from the cell poles (Figure 6 E–F, Movie M3). This was shortly accompanied by an appearance of the Z-rings in the same locations. In some cases we observed that the Z-ring completely disappeared from the central location, while in other cases, as shown in Figure 6 E–F, the Z-ring also persisted in the cell center and was able to complete division. Observations that the Z-ring follows the movement of the Ter macrodomain in a highly correlated manner for both single and multi-nucleoid ΔslmA Δmin cells are consistent with the hypothesis that the Ter macrodomain acts as a positional landmark for cell division proteins in these cells.
Localization of the Z-ring in the absence of the putative Ter linkage
If the MatP-ZapB-ZapA linkage is involved in the co-localization of the Ter macrodomain and the Z-ring in ΔslmA Δmin cells, then rendering the linkage dysfunctional by removal of any proteins of the linkage should make the placement of the Z-ring relative to the nucleoid center more random. To verify this prediction we constructed ΔslmA Δmin ΔmatP, ΔslmA Δmin ΔzapB, and ΔslmA Δmin ΔzapA triple deletion strains. The triple mutants were imaged using ZipA-GFP as a Z-ring label and DAPI as a stain for nucleoids (Figure 7 A–C). Indeed, the distributions of distances between the central Z-ring and nucleoid centers (Figure 7 D–F) were more than a factor of two wider after deletion of matP (; ), zapB (; ) and zapA (; ) from ΔslmA Δmin cells (). All p-values were calculated using single tailed Ansari-Bradley test. Note that the horizontal axes in Figure 7 D–F spans a distance that is three times larger than in the corresponding graphs for ΔslmA Δmin and the parental cells (Figure 3 E–F). Wider Xz−Xn distributions for the ΔslmA Δmin ΔzapA and ΔslmA Δmin ΔzapB strains compared to ΔslmA Δmin ΔmatP strain are likely caused by irregular Z-ring patterns in the former two strains. ZapA and ZapB have been identified as bundling agents for the FtsZ protofilaments [30], [31]. In the absence of these proteins aberrantly shaped Z-rings can be present at the division site which leads to higher uncertainty in Z-ring positions.
Fig. 7. Positioning of the Z-rings relative to the cell and nucleoid centers in triple deletion strains. 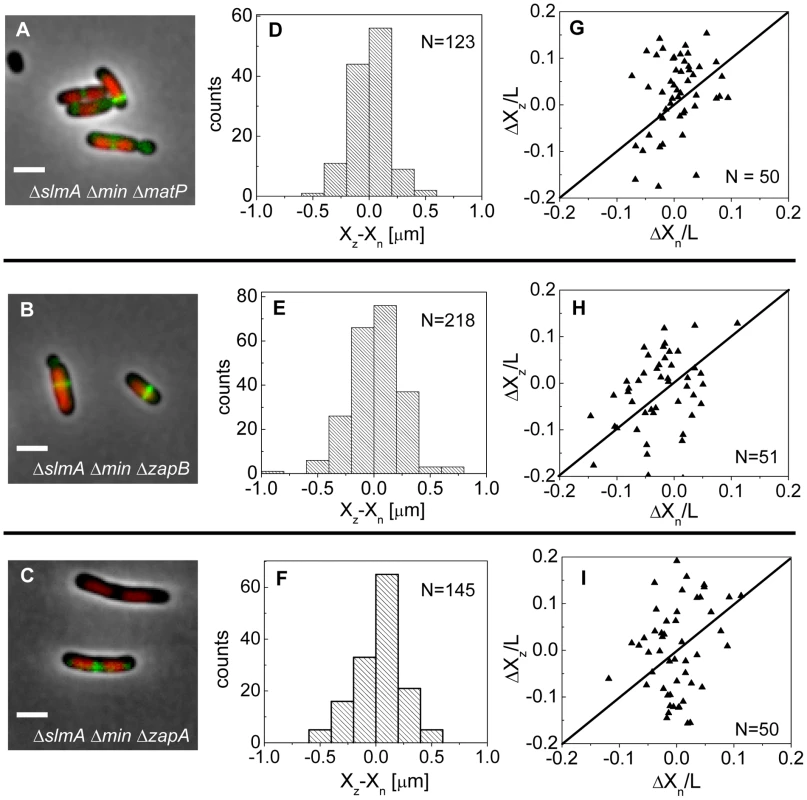
Composite of DAPI labelled nucleoid (red), ZipA-GFP (green) and phase contrast image in (A) ΔslmA Δmin ΔmatP, (B) ΔslmA Δmin ΔzapB, and (C) ΔslmA Δmin ΔzapA cells. Scale bar is 2 µm. (D)–(F) Distribution of distances between the Z-ring center and nucleoid center for ΔslmA Δmin ΔmatP, ΔslmA Δmin ΔzapB, and ΔslmA Δmin ΔzapA cells, respectively. (G)–(I) ΔXz vs. ΔXn in ΔslmA Δmin ΔmatP, ΔslmA Δmin ΔzapB, and ΔslmA Δmin ΔzapA cells, respectively. Data are from cells with a single compact nucleoid and a central Z-ring. Straight lines correspond to . We also observed a significantly higher percentage of polar Z-rings (Figure S11) and polar constrictions after deletion of matP, zapB, and zapA from ΔslmA Δmin background (Table S3). Note that only a fraction of polar Z-rings leads to polar constrictions. For example, in ΔslmA Δmin ΔmatP cells the frequency of polar Z-rings was 69% while the frequency of polar constrictions leading to minicelling divisions was 28%. Although the division planes were positioned much more randomly in triple deletion strains than in the ΔslmA Δmin strain, the cell length distributions (Figure S12) were not significantly affected except for ΔslmA Δmin ΔmatP strain which was longer. Constancy of cell length may indicate that the timing and duration of cell division are not affected by zapA and zapB deletions but may be affected by matP deletion. Considering MatP is also involved in organizing the Ter region of chromosome [28], it is conceivable that its deletion could affect cell length more so than a deletion of ZapA or ZapB. Altogether, these findings show that the Ter linkage strongly affects the accuracy and precision of division plane placement but it appears not to affect significantly the timing of cell division.
Consistent with the role of ZapB and MatP in the Ter linkage, we observed a drastic loss of co-localization between the Z-rings and MatP foci in ΔslmA Δmin ΔzapB cells and in ΔslmA Δmin cells when the last 20 amino acids in the C-terminus of MatP were replaced by an mCherry fusion (matPΔC-mCherry) (Figure S13). The MatP C-terminal domain has been shown to be important for its interaction with ZapB [25]. While our data indicates that the Ter linkage determines the position of the Z-ring, it has been shown that the linkage is required to stabilize the position of the Ter macrodomain [25]. Our data do not contradict this finding. The MatP focus appeared more delocalized relative to the nucleoid center in the absence of the Ter link in the ΔslmA Δmin ΔzapB and ΔslmA Δmin ΔmatPΔC-mCherry strains compared to the ΔslmA Δmin and wild type strains (Figure S14). The Ter linkage thus appears to determine the position of the Z-ring and at the same time stabilize the position of the Ter macrodomain relative to the cell center once the Z-ring has formed.
In cells with compact nucleoids, representative of an early state of chromosome segregation, analysis of Z-ring positions relative to nucleoid-centers revealed essentially no co-localization (Figure 7G–I). The corresponding values for ΔslmA Δmin ΔmatP, ΔslmA Δmin ΔzapB, and ΔslmA Δmin ΔzapA strains were about a factor of 1.5 larger (230 nm, 344 nm, 280 nm, respectively) than these values for the whole cell population. These differences were statistically significant in both the F-test and in the Ansari-Bradley test. This evidence suggests that the Ter linkage is critical to the specific localization of the Z-ring with the chromosomal terminus at early states of chromosome segregation when the nucleoid morphology is compact. Once the bi-lobed nucleoid morphology emerges in the triple deletion strains, co-localization between the Z-ring and nucleoid center appears, though much more weakly than in the ΔslmA Δmin and parental strains.
Interestingly, the spatial distributions of those Z-rings that were not located at the poles still displayed a bias towards the cell center (Figure 7 D–F, Figure S11). The locations of constrictions in the triple deletion strains, which we measured from phase contrast images, showed an overall positioning bias towards cell centers as well (Figure S15). However, the corresponding distributions were significantly broader in triple deletion strains than in ΔslmA Δmin cells. The latter findings indicate that while triple deletion strains lack a mechanism to recognize centers of compact nucleoids, they still have a mechanism that can position Z-rings relative to cell center albeit with significantly lower precision and accuracy than the ΔslmA Δmin and parental strains.
Discussion
The Min system and SlmA-mediated nucleoid occlusion are the only two molecular systems responsible for positioning the cytokinetic ring in E. coli that have been identified thus far [2]–[5]. Here, we show that E. coli without these two known positioning systems is capable of coordinating cell division and chromosome segregation with high fidelity. The majority of ΔslmA Δmin cells position their division planes accurately relative to nucleoids in slow growth conditions and produce essentially no minicells. In searching for the mechanism responsible for the localization of the Z-ring in these double mutant cells, we found that the Z-rings have a strong tendency to co-localize with the nucleoid centers. Further investigation showed that the nucleoid centers were occupied by the Ter region of the chromosome at the time of Z-ring formation.
The Ter region of the E. coli chromosome is organized by MatP proteins [28]. MatP links the Ter macrodomain to the Z-ring through ZapB and ZapA proteins [25]. It was proposed earlier that the Z-ring acts as an anchor for the Ter macrodomain through this linkage [25]. Our time lapse measurements show a broader role of the Ter linkage. These measurements demonstrate that the MatP-decorated macrodomain arrives at the cell center a small fraction of the cell cycle before appreciable assembly of the Z-ring occurs in ΔslmA Δmin cells. This temporal relationship indicates that the Ter region of the chromosome through the Ter linkage localizes the cell division proteins in the early stage of cytokinesis. It is thus the Ter macrodomain that acts as an ‘anchor’ for cell division proteins during the formation of the divisome. However, the interactions between the Z-ring and the Ter macrodomain appear to stabilize the position of Ter macrodomain later in the cell cycle. During maturation of the divisome, especially when it becomes fixed to the cell wall, the divisome acts as a stabilizing element for the Ter macrodomain, holding it fixed in the cell center [25].
A positive regulation mechanism for cell division
The Ter linkage facilitates correct placement of the division plane relative to the chromosomes. Severing the linkage in ΔslmA Δmin cells leads to increased number of unviable minicells and less symmetric division of mother cells. Both outcomes limit the fitness of cells. Unlike the Min system and SlmA-mediated nucleoid occlusion, which are inhibitors of Z-ring formation, the Ter linkage represents a positive regulatory mechanism. The link guides cell division proteins to the location of the future division site and not away from the undesired locations in the cell as do the Min system and SlmA-mediated nucleoid occlusion.
The positive regulation by the Ter linkage is dynamic and it is likely not very strong. Time-lapse measurements show that the Ter linkage temporarily disassembles when the Ter region of the chromosome moves from the cell pole to its center. Also, the Ter region becomes disconnected from the divisome near the end of cytokinesis. The Ter linkage appears thus to provide a dynamic and reconfigurable connection, which biases assembly of cell division proteins towards the Ter region, but does not commit cells to division.
The Ter linkage and the Min system define two independent positioning systems for the divisome. The Min system is capable of positioning the Z-ring without any nucleoid in E. coli minicells albeit with somewhat lower precision than in wild type cells [32]. The position defined by the Min system may, however, not always match the position defined by the Ter macrodomain. In these conflicting cases, the Min system has the dominant effect over the Ter linkage. Consistent with this idea, we observed in long cephalexin treated wild-type cells that Z-rings localized only at the cell center rather than at the locations of MatP foci. Also, in the Min+ cells we observed no appreciable accumulations of the ZipA-GFP reporter near the cell poles although this location is favored by the Ter linkage at the early stages of the cell cycle.
Unlike the Min system, the effect of SlmA on the Ter linkage was less pronounced. The only observed consequence of deleting slmA in our measurements was the decrease in polar Z-rings and minicelling divisions in the ΔslmA Δmin strain compared to the ΔminC strain. We hypothesize that SlmA removal, i.e. removal of the negative regulator, effectively strengthens the positive regulation due to the Ter linkage. The stronger regulation due to the Ter linkage then leads to more abundant Z-rings in the vicinity of the Ter region(s) of the chromosome, which sequester more efficiently the Z-ring related proteins from other regions of the cell including cell poles. As a result, less polar Z-rings and minicelling divisions are present in the ΔslmA ΔminC than in the ΔminC cells. More work is needed to further test this hypothesis as well as to understand the exact mechanism of how SlmA regulates Z-ring assembly.
Positive regulation mechanisms in other bacteria
Evidence of positive control in localizing cell division proteins has been reported recently for several bacterial species including Streptomyces [33], Myxococcus xanthus [34] and Bacillus subtilis [22], [24]. In Streptomyces the positive control appears to be achieved by a combination of SsgA and SsgB proteins [33]. In M. xanthus, PomZ is shown to have a similar role [34]. Although these proteins arrive before FtsZ in both organisms, it remains unclear which molecular mechanisms are responsible for their own localization. PomZ appears to localize over the nucleoid although it has not been determined if it is linked to any specific chromosomal region [34]. Positioning of SsgA and SsgB relative to chromosome also is not clear yet.
A positive localization signal, or potentiaton as the authors refer to it, appears to be present also in B. subtilis [22]. However, the mechanism seems to be very different in B. subtlis in which the positive signal was reported to appear during the assembly of the replichore, i.e. much earlier than in E. coli. Moreover, it was observed that “some factor” attracted Z-ring assembly to the oldest division site in B. subtilis outgrowing spores that lacked Min and Noc proteins [24]. This is contrary to our observation in E. coli, where the Z-ring is biased towards sites between newly segregating nucleoids. Taking that B. subtilis is evolutionarily divergent from E. coli, differences are expected. It remains to be determined how widespread the Ter linkage is among other bacteria. MatP is conserved in enterobacteria [28], but taking its important functional role, structurally similar assemblies can be present more broadly.
Additional mechanisms for localization of cell division proteins
Deletion of any of the three proteins involved in the Ter linkage affects the midcell positioning of the Z-ring but does not lead to complete positioning randomness. Accordingly, a mechanism responsible for the localization of cell division proteins must exist in addition to the MatP-ZapB-ZapA mediated Ter linkage in ΔslmA Δmin cells. The mechanism does not appear to link Z-rings to nucleoid centers at early stages of chromosome segregation when there is no discernable bi-lobed nucleoid structure (compact nucleoids). Interestingly, later in chromosome segregation when a distinct bi-lobed morphology appears, stronger correlations between the Z-rings and nucleoid centers emerge. Two positioning mechanisms that link the nucleoid and divisome have been discussed in the past [35], [36] that can possibly explain such behavior. Both mechanisms rely on the transertional linkages that connect bacterial DNA through transcribed RNA and simultaneously translated membrane proteins to the plasma membrane of the cell [37]. In one hypothesis transertional linkages create local membrane crowding [35] that prevents Z-ring formation in the vicinity of the nucleoid. In another hypothesis, mechanical tension produced by the transertional linkages due to chromosomal segregation acts as a (positive) signal to guide localization of cell division proteins [36]. Further work can prove or disprove these ideas.
In conclusion, we have shown that E. coli lacking both the Min system and the nucleoid occlusion factor SlmA are able to localize their division planes at the centers of nucleoids as opposed to the nucleoid free regions in slow growth conditions. In this localization process, the Ter region of the chromosome acts as a landmark for the Z-ring. Removal of the Ter linkage, which involves MatP, ZapB, and ZapA proteins, significantly affects the accuracy and precision with which the Z-ring localizes over the nucleoid. Our data, however, is indicative that yet an unidentified, lower fidelity positioning system remains in E. coli ΔslmA Δmin cells even without the Ter linkage. Despite the lower fidelity, this unidentified positioning system still coordinates Z-ring localization relative to the cell center. Further studies are warranted to identify the molecular origins of this positioning mechanism.
Materials and Methods
Strains and growth conditions
All strains used in this study were derivatives of E. coli K-12. Descriptions of all strains and plasmids are given in Table S1. All bacteria were grown in M9 minimal medium (Sigma-Aldrich) supplemented with magnesium sulfate and either with 0.5% glucose or 0.3% glycerol. 20 µg/ml kanamycin, 35 µg/ml chloramphenicol, 20 µg/ml ampicillin was used to grow the strains with respective resistance markers. 50 µg/ml ampicillin was used to grow strains carrying pKen1-GFPμ2 plasmids. To grow long, multi-nucleoid cells, all strains were incubated with 20 µg/mL of cephalexin for approximately 2 hours. All bacteria were grown and imaged at 28°C.
Fluorescent microscopy
A Nikon Ti-E inverted fluorescence microscope with a 100X NA 1.40 oil immersion phase contrast objective was used for imaging the bacteria. Fluorescence was excited by a 200W Hg lamp through an ND4 or ND8 neutral density filter. Chroma 41004, 41001 and 31000v2 filtercubes were used to record mCherry, GFP and DAPI images, respectively. Images were captured by an Andor iXon DU897 camera and recorded using NIS-Elements software.
Cells were imaged on M9 agar pads for still imaging. For time lapse imaging home-made glass bottom dishes were used. Cells were pipetted to #1.5 glass coverslips on the bottom of the dish and covered with about 1 cm thick slab of M9 agar. No antibiotics were used in M9 agar during imaging. Agar was supplemented with IPTG (10–40 µM) for strains with ZipA-GFP constructs. For DAPI labeling cells were incubated in 0.2 µg/ml DAPI for 1/2 hour before spreading cells on the pads.
Image analysis
Matlab with the Image Analysis Toolbox and DipImage Toolbox (http://www.diplib.org/) were used for image analysis. In addition to Matlab, simpler image processing such as contrast and brightness adjustments were performed using ImageJ software (v1.41o). The procedures for finding volume fraction ratios is described in [6]. The procedure for finding nucleoid centers, centers of MatP foci, and Z-ring positions relative to cell center is given in Text S1.
Supporting Information
Zdroje
1. BiE, LutkenhausJ (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354 : 161–164.
2. MargolinW (2005) FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6 : 862–871.
3. LutkenhausJ (2007) Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76 : 539–562.
4. AdamsDW, ErringtonJ (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7 : 642–653.
5. de BoerPAJ (2010) Advances in understanding E. coli cell fission. Curr Opin Microbiol 13 : 730–737.
6. MännikJ, WuF, HolFJH, BissichiaP, SherrattDJ, et al. (2012) Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proc Natl Acad Sci U S A 109 : 6957–6962.
7. YuXC, MargolinW (1999) FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 32 : 315–326.
8. Den BlaauwenT, BuddelmeijerN, AarsmanMEG, HameeteCM, NanningaN (1999) Timing of FtsZ assembly in Escherichia coli. J Bacteriol 181 : 5167–5175.
9. GubermanJM, FayA, DworkinJ, WingreenNS, GitaiZ (2008) PSICIC: Noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput Biol 4: e1000233.
10. ShapiroL, McAdamsHH, LosickR (2009) Why and how bacteria localize proteins. Science 326 : 1225–1228.
11. RaskinDM, de BoerPAJ (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci U S A 96 : 4971–4976.
12. ShenB, LutkenhausJ (2010) Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol Microbiol 75 : 1285–1298.
13. HuangKC, MeirY, WingreenNS (2003) Dynamic structures in Escherichia coli: Spontaneous formation of MinE rings and MinD polar zones. Proc Natl Acad Sci U S A 100 : 12724–12728.
14. MulderE, WoldringhCL (1989) Actively replicating mucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol 171 : 4303–4314.
15. WoldringhCL, MulderE, HulsPG, VischerN (1991) Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol 142 : 309–320.
16. BernhardtTG, de BoerPAJ (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18 : 555–564.
17. WuLJ, ErringtonJ (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtillis. Cell 117 : 915–925.
18. TonthatNK, AroldST, PickeringBF, Van DykeMW, LiangSD, et al. (2011) Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J 30 : 154–164.
19. ChoHB, McManusHR, DoveSL, BernhardtTG (2011) Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci U S A 108 : 3773–3778.
20. ChoHB, BernhardtTG (2013) Identification of the SlmA active site responsible for blocking bacterial cytokinetic ring assembly over the chromosome. PLoS Genet 9: e1003304.
21. WuLJ, IshikawaS, KawaiY, OshimaT, OgasawaraN, et al. (2009) Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J 28 : 1940–1952.
22. MoriyaS, RashidRA, RodriguesCDA, HarryEJ (2010) Influence of the nucleoid and the early stages of DNA replication on positioning the division site in Bacillus subtilis. Mol Microbiol 76 : 634–647.
23. BernardR, MarquisKA, RudnerDZ (2010) Nucleoid occlusion prevents cell division during replication fork arrest in Bacillus subtilis. Mol Microbiol 78 : 866–882.
24. RodriguesCDA, HarryEJ (2012) The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet 8: e1002561.
25. EspeliO, BorneR, DupaigneP, ThielA, GigantE, et al. (2012) A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J 31 : 3198–3211.
26. WangXD, PossozC, SherrattDJ (2005) Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev 19 : 2367–2377.
27. FisherJK, BourniquelA, WitzG, WeinerB, PrentissM, et al. (2013) Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153 : 882–895.
28. MercierR, PetitM-A, SchbathS, RobinS, El KarouiM, et al. (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 : 475–485.
29. NikiH, YamaichiY, HiragaS (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14 : 212–223.
30. GalliE, GerdesK (2010) Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol 76 : 1514–1526.
31. BussJ, ColtharpC, HuangT, PohlmeyerC, WangS-C, et al. (2013) In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol 89 : 1099–1120.
32. SunQ, YuXC, MargolinW (1998) Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol 29 : 491–503.
33. WillemseJ, BorstJW, de WaalE, BisselingT, van WezelGP (2011) Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev 25 : 89–99.
34. Treuner-LangeA, AguiluzK, van der DoesC, Gomez-SantosN, HarmsA, et al. (2013) PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol Microbiol 87 : 235–253.
35. ZaritskyA, WoldringhCL (2003) Localizing cell division in spherical Escherichia coli by nucleoid occlusion. FEMS Microbiol Lett 226 : 209–214.
36. RabinovitchA, ZaritskyA, FeingoldM (2003) DNA-membrane interactions can localize bacterial cell center. Journal of Theoretical Biology 225 : 493–496.
37. NorrisV (1995) Hypothesis - chromosome separation in Escherichia coli involves autocatalytic gene expression, transertion and membrane-domain formation. Mol Microbiol 16 : 1051–1057.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



