-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
Lichtheimia species are ubiquitous saprophytic fungi, which cause life-threating infections in humans. In contrast to the mucoralean pathogen R. oryzae, Lichtheimia species belong to the ancient mucoralean lineages. We determined the genome of L. corymbifera (formerly Mycocladus corymbifer ex Absidia corymbifera) and found high dissimilarities between L. corymbifera and other sequenced mucoralean fungi in terms of gene families and syntenies. A highly elevated number of gene duplications and expansions was observed, which comprises virulence-associated genes like proteases, transporters and iron uptake genes but also transcription factors and genes involved in signal transduction. In contrast to R. oryzae, we did not find evidence for a recent whole genome duplication in Lichtheimia. However, gene duplications create functionally diverse paralogs in L. corymbifera, which are differentially expressed in virulence-related compared to standard conditions. In addition, new potential virulence factors could be identified which may play a role in the regulation of the adaptation to iron-limitation. The L. corymbifera genome and the phylome will advance further research and better understanding of virulence mechanisms of these medically important pathogens at the level of genome architecture and evolution.
Published in the journal: Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina). PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004496
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004496Summary
Lichtheimia species are ubiquitous saprophytic fungi, which cause life-threating infections in humans. In contrast to the mucoralean pathogen R. oryzae, Lichtheimia species belong to the ancient mucoralean lineages. We determined the genome of L. corymbifera (formerly Mycocladus corymbifer ex Absidia corymbifera) and found high dissimilarities between L. corymbifera and other sequenced mucoralean fungi in terms of gene families and syntenies. A highly elevated number of gene duplications and expansions was observed, which comprises virulence-associated genes like proteases, transporters and iron uptake genes but also transcription factors and genes involved in signal transduction. In contrast to R. oryzae, we did not find evidence for a recent whole genome duplication in Lichtheimia. However, gene duplications create functionally diverse paralogs in L. corymbifera, which are differentially expressed in virulence-related compared to standard conditions. In addition, new potential virulence factors could be identified which may play a role in the regulation of the adaptation to iron-limitation. The L. corymbifera genome and the phylome will advance further research and better understanding of virulence mechanisms of these medically important pathogens at the level of genome architecture and evolution.
Introduction
The basal lineages of terrestrial fungi, formerly Zygomycota, were recently shown to be polyphyletic and were therefore separated into four separate subphyla [1]. Especially the order Mucorales of the Mucoromycotina encompasses several human pathogenic species. Although infections with mucoralean fungi (mucormycosis) are less common as compared to aspergilloses or candidioses, these fungi are increasingly recognized as the source of infection in immunocompromised patients [2]. Mucormycoses are associated with rapid blood vessel invasion and massive destruction of tissue (necrosis) [3], [4]. Mortality rates are high (∼50%) and treatment mainly includes a combination of antifungals and extensive surgery [2], [5]–[7]. In addition, mucoralean pathogens are resistant to a variety of antifungals including voriconazole which makes treatment even more complicated [8].
The order Mucorales comprises 240 described species, of which at least 20 have been found to be involved in mucormycosis. Genome sequences have been published for only two important pathogenic species within the Mucorales, namely Rhizopus oryzae ( = R. arrhizus) and Mucor circinelloides. These species are closely related and represent derived lineages within the group. However, a large proportion of pathogenic Mucorales (10 species) belong to more basal groups including the genera Lichtheimia, Rhizomucor, Apophysomyces, Saksenaea and Syncephalastrum. Recently, the first report of the involvement of Thamnostylum lucknowense, an ancient mucoralean fungus, in human infections has been published [9]. To date, almost nothing is known about the genomic structure and pathogenicity mechanisms of these basal groups.
Lichtheimia species are ubiquitous saprophytic molds and represent the second and third most common cause of mucormycosis in Europe and worldwide, respectively [2], [7], [10], [11]. The genus Lichtheimia was formerly included in the genus Absidia based on morphological similarities [12]. However, based on the higher growth optimum as well as morphological and molecular data Lichtheimia species were separated from the mesophilic Absidia species [13]. Today the genus encompasses five thermotolerant species, of which three are known to be clinically relevant, namely L. corymbifera, L. ramosa and L. ornata [14]. In addition to the distinct phylogenetic position at the base of mucoralean fungi, Lichtheimia species exhibit differences in physiology compared to the sequenced pathogens M. circinelloides and R. oryzae, including a higher maximum growth temperature (48–52°C vs 37°C and <45°C) and differences in susceptibility to certain antifungals [8], [15]. Moreover, filamentously growing Mucor and Rhizopus species have been shown to be able to form yeast cells which were also found in patient material and thus might be of relevance during infection [16]–[18]. In contrast, no yeast-like growth forms of Lichtheimia species have been observed to date. In addition, pulmonary Lichtheimia infections following solid organ transplantation seem to be associated with a higher risk to develop disseminated disease [19]. Besides its role in human infections, L. corymbifera is also believed to be associated with Farmer's lung disease (FLD), a hypersensitivity disorder resulting from frequent contact of mouldy material in agriculture [20]. Nothing comparable has been described for other mucoralean species. In addition to their pathogenicity towards humans, several Lichtheimia species are known as contaminants of several food products (e.g. cocoa, peanuts, olive products) [21]–[23]. However, despite the known role of Lichtheimia species in infection and diseases, several Lichtheimia species play an important role in the fermentation of soy products in Asian cuisine [24].The large evolutionary distance and notable differences in infection strategies between Lichtheimia and the two sequenced mucoralean pathogens indicate that they independently evolved their ability to infect humans by developing specific pathogenesis mechanisms. To gain insight into the genomic differences between these groups of pathogens, here we report the genome sequence of the type-strain of L. corymbifera (FSU 9682, CBS 429.75, ATCC 46771) which has been shown to be a typical strain in terms of virulence and physiology for this species [25] and compare it to published genomes of mucoralean fungi and other fungal phyla.
Results/Discussion
Genome assembly and structure
The genome of the type-strain of L. corymbifera (FSU 9682, CBS 429.75, ATCC 46771) was sequenced by a combination of 454 sequencing of a shotgun and 8 kb paired-end library in combination with Illumina sequencing of a paired-end read library (Materials and methods, Table S1). The final assembly comprises 209 scaffolds with a N50 scaffold size of 367,562 nt and a total length of 33.6 Mb (Table 1), which is comparable to the genome size of other zygomycetous fungi [26]. Mucoralean genomes are generally believed to contain large amounts of repetitive elements representing around 35% of the genome [27]. However, analysis of the L. corymbifera genome shows a much smaller content of repetitive elements, with only 4.7% of the assembly representing repetitive elements including DNA transposons, LTR and non-LTR retrotransposons (Table S2). This finding is consistent with the results of the k-mer analyses on the Illumina reads where only low amounts of potential repetitive regions were found. Of note, all previous estimates of repetitive elements in mucoraleans correspond to species with large genomes such as R. oryzae (46 Mb; 20% repetitive elements), Absidia glauca (52 Mb; 35%) and P. blakesleeanus (54 Mb; 35% repetitive elements) [27], [28]. Interestingly, a Lichtheimia-specific gene expansion in the heterokaryon incompatibility genes was discovered (see section gene expansion) which are involved in the recognition of non-self DNA and may contribute to the low amount of repetitive elements. Another mechanism of protection against transposons and viruses is RNA interference resulting in sequence specific RNA degradation [29]. Several predicted proteins with functional domains associated with this mechanism were found including a dicer-like protein, one argonaute-2 protein and a translation initiation factor 2C homolog. However, the exact effects of these mechanisms on the amount of repetitive elements remain to be determined.
Tab. 1. Statistics of the <i>L. corymbifera</i> genome. 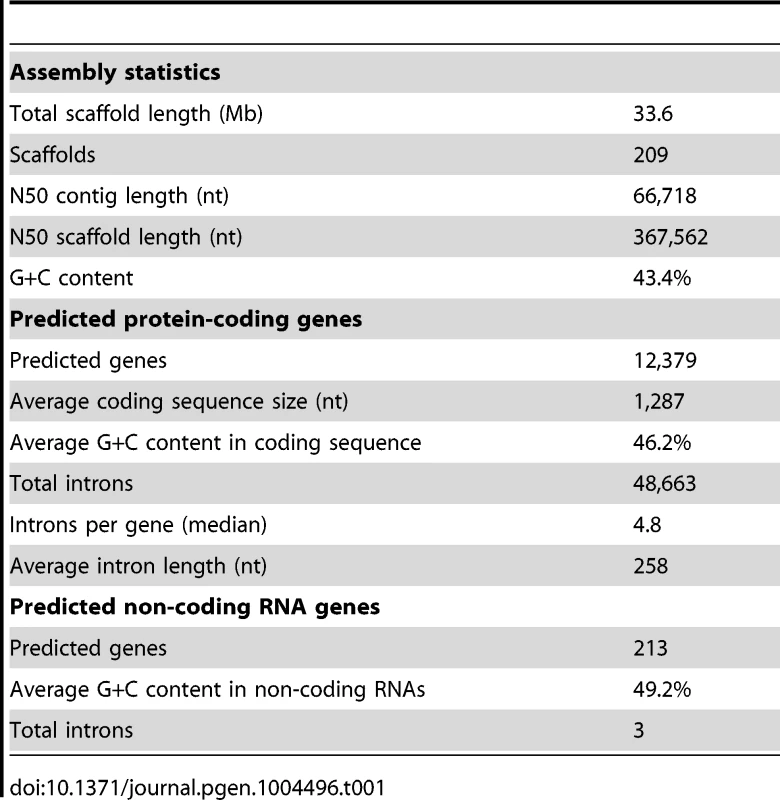
Heterozygosity was shown for several fungi including the basal lineage fungus Batrachochytrium dendrobatidis [30]. In order to test for potential heterozygous regions and estimate the genome size of L. corymbifera, k-mer analyses based on the Illumina reads were performed using an algorithm described previously [31] (Material and Methods). Analysis resulted in a relatively clear single peak with a slightly trailing left flank for all k-values (Figure S1). The distribution could be dissected into three components, each showing a normal distribution with similar variance, but different means and different proportions. The main component represents the potential homozygous part of the genome (94%), whereas two small components represent the potential heterozygous part of the genome (4%), and most likely some repeat regions that occur at relatively low frequency (2%). It has to be noted that the potential heterozygous part is rather small and could as well be explained e.g. by regions that are difficult to sequence and therefore have lower k-mer coverage. The lack of heterozygocity is in accordance with the general assumption that mucoralean fungi are haploid during vegetative growth. Based on the k-mer analysis for different k-mer lengths (41, 59, 69, and 79 nt) a total genome size of around 35 Mb was predicted which is close to our total scaffold length of 33.6 Mb (96% of k-mer predicted size).
Non-coding RNA prediction and annotation
We annotated 174 tRNAs in L. corymbifera. Although R. oryzae (239) comprises many more tRNAs, we found unique anticodons among the basal fungi in Lichtheimia: CCC (Gly), AAA (Phe) and GAT (Ile). In contrast, only L. corymbifera misses the anticodons CAC (Val), CCT (Arg) and TAT (Ile). Three GTA (Tyr) tRNAs were predicted with introns in L. corymbifera, while the number was higher in other mucoralean fungi (up to 10). No selenocysteine and possible suppressor tRNAs were predicted. We found the downstream half of 28S rRNA only, but no 18S rRNA in the current assembly. We expect at least two operons (18S – 5.8S – 28S rRNA) as found in R. oryzae. In addition to 5S rRNAs located close to the operons, we were able to identify several independent 5S rRNA copies (Table S3). Another housekeeping ncRNA, present in all kingdoms of life, is the ribozyme RNase P, which processes tRNAs by cleaving off nucleotides on the 3′ end of tRNAs [32]. We detected this gene as expected in a single copy per genome, but two identical copies are apparently present in the genome of R. oryzae, which may result from whole genome duplication in R. oryzae [28]. The pseudoknot in the centre of the molecule is accredited with the catalytic function and highly conserved in evolution [33]. However, the L. corymbifera candidate varies exceptionally in sequence, while the secondary structure is maintained. Whether the function of the molecule is affected has to be analyzed. The evolutionary related RNase MRP was invented at the origin of eukaryots with dual function: (a) initiation of mitochondrial replication and (b) separation of 18S rRNA from 5.8S rRNA [34]. One copy per basal fungal genome was detected. The signal recognition particle containing a ncRNA part (SRP RNA) guides proteins to the endoplasmatic reticulum [35]. One copy was detected in Lichtheimia, whereas two copies were identified in the genome of R. oryzae. Surprisingly, the covariance model of mucoralean fungi, in agreement with Rhizopus, Batrachochytrium and Monosiga (RF00017) is much closer related to metazoans than to other known fungi SRP RNAs. We detected the RNA components of the major spliceosome and collected indications for a functional minor spliceosome. Except for U4 snRNA all five RNAs involved in U2-splicing were detected in Lichtheimia. U4 snRNA was not part of the assembly; however an U4-candidate was identified in the originally sequenced read data. Additionally, four of five RNAs involved in AT-AC-splicing were found. However, several special secondary structures were discovered, which may alter the functionality of the minor spliceosome: (i) The third stem of U12 snRNA is atrophied and the last stem is shorter than expected for all basal fungi. (ii) U4atac is not detected in Lichtheimia. The other basal fungi show one inconspicuous copy, which is assumed to to be an assembly mistake. However, no similar homologous gene was detected in reads either. (iii) The second half of U6atac is highly divergent (Figure S2 and supplemental material: http://www.rna.uni-jena.de/supplements/lichtheimia/index.html). Eleven CD-box snoRNAs and 3 H/ACA snoRNAs were identified, which are mainly conserved in sequence and structure among basal fungi. For further details we refer to the supplemental material (www.rna.uni-jena.de/supplements/lichtheimia/index.html). Additionally, several ncRNA candidates could be proposed, which have to be functionally characterized in future experiments. A riboswitch, binding to thiamine pyrophosphate (TPP) was found in all basal fungi. For Lichtheimia a potential telomerase RNA is suggested, which is surprisingly closely related to the shortest known telomerase RNAs in ciliates (150 nt Tetrahymena paravorax). This is unexpected, since the longest telomerase is known from the fungus Saccharomyces cerevisiae (1,220 nt). Although the alignment of the usually extremely divergent telomerase RNA is very convincing in sequence and secondary structure (see supplemental material), no homologs in another basal fungus and no interacting ciliate protein homolog were found in our current assembly. U7 snRNA is known to interact with the downstream region of histone mRNA for inhibition of degradation. Four similar candidates for this short ncRNA were identified. In eukaryotes, polymerase III transcripts (e.g. U6 snRNA, RNase P, RNase MRP, SRP RNA, U6atac snRNA) usually display a typical promoter region: −10 nt TATA box, PSE element, Oct region [36]. Therefore, a search for conserved motifs was conducted in Lichtheimia promoter regions. However, we were not able to identify even one of these motifs. This highlights a possible modified polymerase III activity for basal fungi and has to be investigated in detail in further work. A phylogeny among basal fungi and Schizosaccharomyces pombe as outgroup based on ncRNAs (except 18S and 28S rRNA) was reconstructed, see Figure S2 (B and C). In accordance with protein and traditional rRNA phylogeny Lichtheimia groups basal to P. blakesleeanus and the other two investigated fungi.
Protein-coding gene prediction and annotation
To aid prediction of protein-coding genes, RNA-seq analyses were performed for three different growth conditions in three biological replicates (see Material and Methods). The use of different conditions should ensure a higher number of expressed genes, thereby allowing evidence-based gene predictions for many gene models. On average, each replicate has a 70-fold genome coverage, which sums up to a 630-fold genome coverage (Table S4). Prediction of protein-coding genes was performed using AUGUSTUS [37], resulting in 12,379 predicted genes. Genes were functionally annotated by comparing to GenBank sequences using BLASTp (E-value≤10−25), and by scanning for the presence of conserved domains using the InterProScan function of BLAST2GO [38]. BLAST hits were obtained for 7,917 genes, InterProScan results were found in 10,066 genes and at least one Gene Ontology (GO) term was assigned to 7,435 genes based on the union of BLAST and InterProScan results. The raw reads of the DNA - and RNA-seq experiments, the final genome assembly, the structural and functional gene prediction are available at http://www.ebi.ac.uk/ena/data/view/PRJEB3978). The genome data are also accessible via HKI Genome Resource (http://www.genome-resource.de/).
Comparison of protein-coding genes between L. corymbifera and other completely sequenced genomes
An exhaustive comparison of L. corymbifera genome with other 24 completely sequenced genomes including the major fungal groups (Chytridiomycota, Mucoromycotina, Asco - and Basidiomycota) was performed. This comparison included the reconstruction of L. corymbifera phylome, which encompasses the complete set of evolutionary histories of L. corymbifera genes (Material and Methods). It was carried out using the previously described PhylomeDB pipeline [39]. In brief, for each L. corymbifera protein-coding gene we searched for homologs, and multiple sequence alignments were built, and Maximum Likelihood analyses were performed to reconstruct a phylogenetic tree. The phylome is available through phylomeDB (http://phylomedb.org), with the phylome ID 245. The phylome was used to establish phylogeny-based orthology and paralogy relationships among genes in the species considered [40], and to detect gene expansions (see below). In addition, we used two complementary approaches, gene concatenation and super-tree [41], to reconstruct the species tree that represents the evolution of the 25 species considered. In the first approach, 58 genes were selected that were present in 21 out of 25 in single copy. Their corresponding alignments were then concatenated and a maximum-likelihood species tree was reconstructed (Material and Methods). In the second approach, 9,478 trees present in the phylome were used to build a super-tree using a gene tree parsimony approach, a method which finds the topology that minimizes the total number of duplications in the phylome [42]. Both resulting trees presented a similar topology, which placed L. corymbifera at the base of the other Mucorales species (Figure 1). The only difference found between the trees by the complementary approaches was the position of Schizosaccharomyces pombe, which appeared at the base of Ascomycota in the super-tree tree while in the concatenated tree it grouped with S. cerevisiae. To assess the level of overlap in genetic content between the different species an all-against-all comparison of the 25 genomes was performed. The results indicate between 50% and 75% of the proteins encoded in the other three Mucorales species had homologs in L. corymbifera (Figure 1). Surprisingly, this percentage of shared gene content with L. corymbifera was similar to that of Schizosaccharomyces pombe (60.6%), which is higher than that found in the more closely related chytrid B dendrobatidis (an average of 41.1%). Figure 1 also shows how these homologs are distributed in differently defined groups. Most interestingly, the fraction of species-specific proteins (grey bars in the figure) is particularly high in large genomes (e.g., over half of the largest genomes Laccaria bicolor and Puccinia graminis). Over 25% of the proteins apparently are specific for L. corymbifera.
Fig. 1. Species tree including the 25 species used during phylome reconstruction. 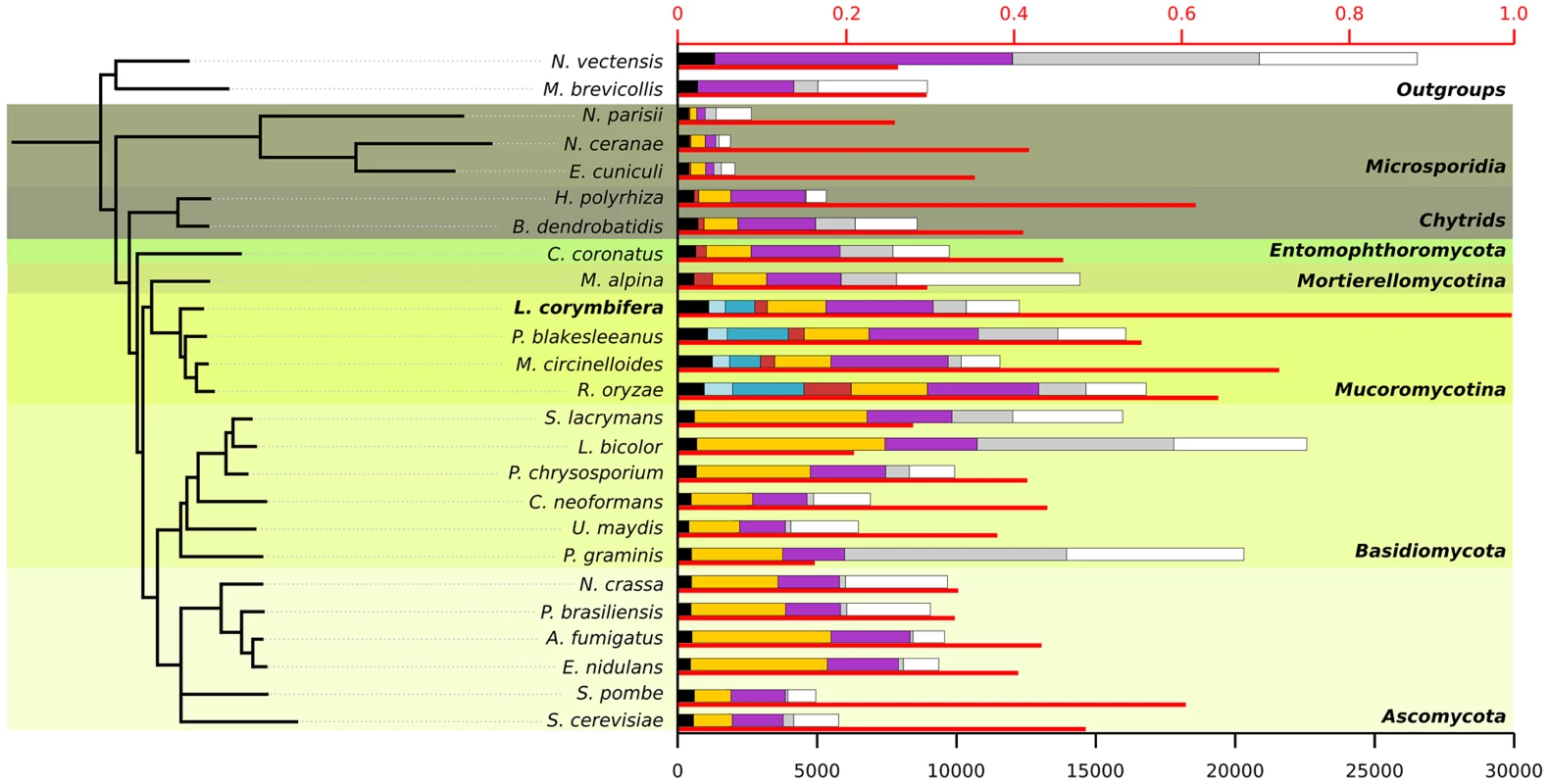
For each species the thin red bar represents the proportion of proteins that have a homolog in L. corymbifera (upper axis). The coloured bars represent the number of proteins found in different ranges of species (lower axis): black: wide-spread proteins found in at least 23 of the 25 species, light blue: proteins found exclusively in all four Mucorales species, darker blue: proteins found only in Mucorales species, red: proteins found in early diverging fungi, yellow: proteins found in fungi, purple: proteins found in fungi and at least one of the outgroups, grey: species-specific proteins without orthologs in other species but with paralogs within the genome, white: proteins with no homologs. All nodes in the tree have a bootstrap support of 100. Conserved gene regions in L. corymbifera and other mucoralean genomes
Since the Lichtheimia lineage separated early in mucoralean evolution we can expect that severe genomic re-arrangements have taken place during evolution, causing substantial differences between the genome structures of L. corymbifera and other Mucorales. Only 57.7% of the gene families present in Lichtheimia are also present in at least one of the other mucoralean genomes while only 36.7% were found in all four genomes representing 70.4% and 53.7% of the total L. corymbifera genes, respectively (Figure 2 A). Conserved regions, in terms of gene order, between mucoralean genomes were examined and evaluated with respect to the amount of conserved genes of these regions. A total of 230 regions with a minimum of 3 conserved genes of L. corymbifera were found that were present in at least one of the other genomes. These regions were interspersed over 41.1% of the scaffolds but covered only 7.6% of the L. corymbifera genome reflecting the high dissimilarity between the mucoralean genomes (Figure 2 B). Only 6 regions were shared with all species. The total number of shared clusters was found to be consistent with the phylogenetic distance between the species (Figure 2 B+C). Genes in the conserved regions are members of different gene families and contain a variety of functional domains.
Fig. 2. Conserved regions of the L. corymbifera genome with other mucoralean genomes. 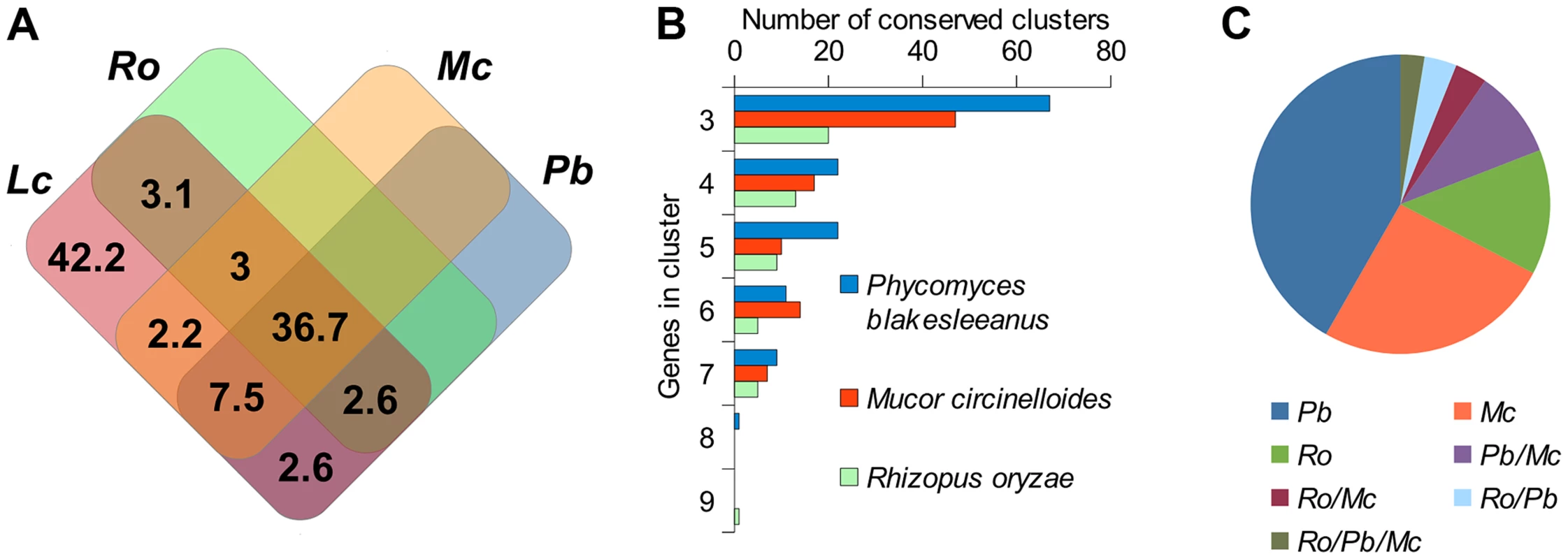
(A) Venn diagram of shared gene families between L. corymbifera and other mucoralean fungi based on GhostFam gene families. Numbers indicate percentage of L. corymbifera gene families. (B) Number and size of conserved clusters of L. corymbifera with other mucoralean genomes. (C) Proportions of conserved clusters of L. corymbifera shared with different mucoralean genomes. Occurrences in more than one of the genomes are indicated by a slash between the species. Gene duplications in L. corymbifera
In accordance with former results [28] a higher number of gene families with two members were detected for R. oryzae but also for L. corymbifera as compared to other fungi (6.99%±0.58%) (Figure 3 A). Whole genome duplication has been previously described for R. oryzae based on the presence of gene duplications and duplication of large genomic regions (segmental duplications) [28]. To investigate whether segmental duplications and thus a potential WGD also occur in L. corymbifera, the genomes were scanned for the presence of duplicated regions using GECKO2 [43], [44]. Consistent with the former findings, our analysis showed a high number of segmental duplications in R. oryzae covering more than 10% of the genome [28] while fewer duplicated regions were found in L. corymbifera covering less than 4% of its genome (Figure 3 B). Thus, the gene duplications seem not to result from recent WGD as in R. oryzae but may result from an ancient genome duplication in mucoralean fungi as suggested by Marcet-Houben et al. [45] which is no longer detectable in the duplicated gene clusters. The possibility of ancient WGD in mucoralean genomes is currently investigated in more detail (Corrochano et al., pers. comm.). The genome of L. corymbifera also shows increased numbers of gene families with a higher number of genes indicating that gene duplication and the preservation of the gene copies seem to be a common process in mucoralean genomes and may be independent from WGD. This will be further addressed in the next section.
Fig. 3. Gene duplication and duplication of genomic regions within mucoralean genomes in comparison to the genome of A. fumigatus, which (i) inhabits the same natural habitats and (ii) causes similar symptomatology in human like L. corymbifera and (iii) serves as model organism for causative agents of invasive mycoses [56], [75]. ![Gene duplication and duplication of genomic regions within mucoralean genomes in comparison to the genome of <i>A. fumigatus</i>, which (i) inhabits the same natural habitats and (ii) causes similar symptomatology in human like <i>L. corymbifera</i> and (iii) serves as model organism for causative agents of invasive mycoses <em class="ref">[56]</em>, <em class="ref">[75]</em>.](https://www.prelekara.sk/media/cache/resolve/media_object_image_small/media/image/79f8ecf0cd8b185a79a206e12e82c873.png)
The genome of A. fumigatus serves as measure for low incidences of singular and segmental gene duplications [115], [116]. (A) Comparison of gene families between L. corymbifera, R. oryzae and non-mucoralean fungi. Gene families are based on GhostFam homology families. Values for L. corymbifera and R. oryzae are excluded from the “mean fungi” value. (B) Regions with a minimum of 3 genes were tested for multiple occurrences within the genomes by GECKO2. Gene expansions in the L. corymbifera genome
In addition to gene duplications shared by all mucoralean fungi a high amount of species-specific duplications was detected. Therefore, the phylome was scanned in search of expansions of protein families that occurred specifically in L. corymbifera. For each tree, ETE [46] was used to find nodes that contained at least five L. corymbifera sequences and no other fungal sequence (Figure 4 A and B). Overlapping expansions were fused when they shared more than half of their members. We found 75 expansions that fulfilled those requirements. Five expansions contained more than 30 members, with the largest containing 331 paralogous genes. In contrast, the large genome of R. oryzae contains approximately twice the number of expansions, with the largest encompassing 1,888 members. As some of those expansions are likely the result of the presence of transposons, we scanned them for the presence of transposon-linked domains using the Pfam database and HMMER3 [47], [48]. If the expansions that contain transposons were excluded, 66 groups of paralogous proteins will be left in L. corymbifera comprising a total of 820 genes (Figure 4 B).
Fig. 4. Gene expansions and tandem duplications found in L. corymbifera. 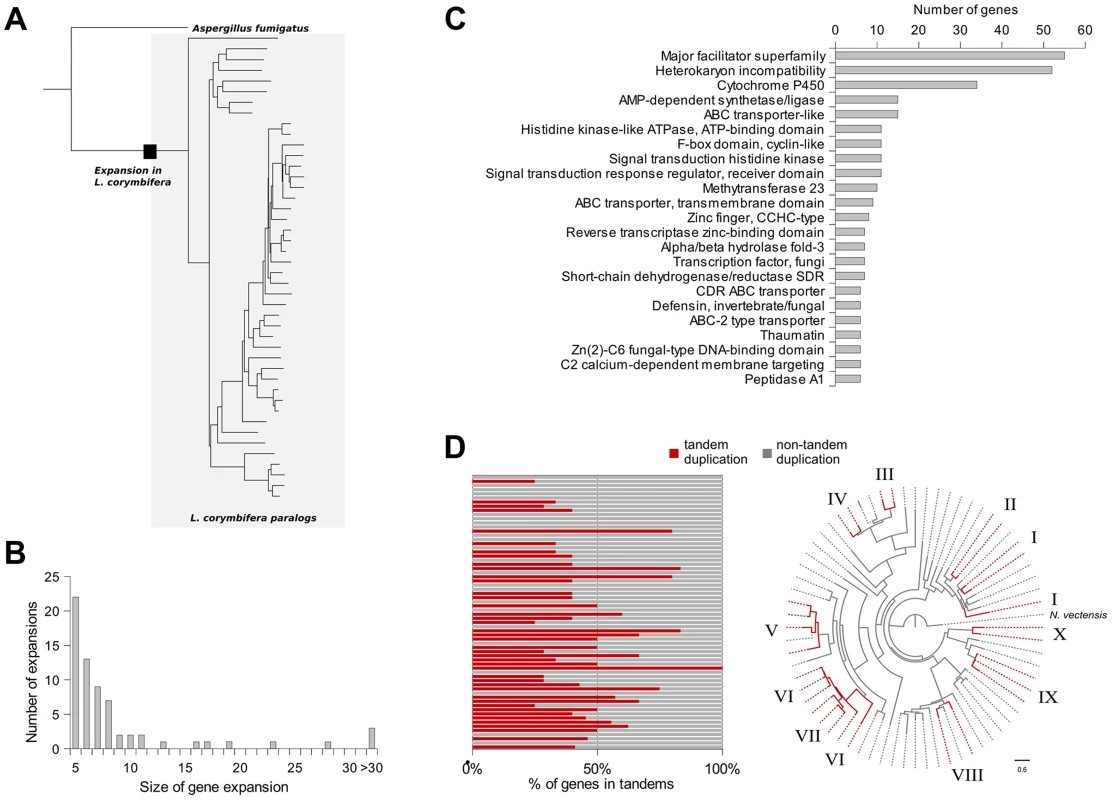
(A) Tree representing an expansion of HET proteins in L. corymbifera. Branches enclosed in the grey shaded area represent paralogs of L. corymbifera. The black square represents the point where the expansion started. (B) Number and size of gene expansions in the L. corymbifera genome. (C) Main functional domains of gene expansions based on PFAM annotation. The numbers of genes with the different functional domains were combined if a domain was present in more than one expansion. (D) Proportion of genes within gene expansions which are arranged in tandem duplications. Each bar represents an expansion with the red part as the percentage of tandem duplicated genes (left). Clustering of tandem duplicated genes of cytochrome P450 genes in L. corymbifera (based on reconstruction with RaxML [130]). Red branches represent tandem duplicated genes. Numbers at the branch tips indicate different tandems. The most abundant expanded groups (with 331 and 242 members) are rather heterogeneous in terms of functional domains, thus there is no particular function that could be assigned to them. The largest group with a dominating domain contains in total 56 members, of which 52 possess a heterokaryon incompatibility protein (HET) domain (PF06985) (Figure 4 C). Interestingly, this domain was so far attributed nearly exclusively to ascomycetes (with only one exception for the basidiomycete Moniliophthora perniciosa), where the HET proteins control somatic allorecognition (non-self-recognition) during the formation of heterokaryons [49]. However, Mucorales, opposite to ascomycetes, do not form heterokaryons by fusion of somatic cells but only during sexual reproduction and zygospore-formation. Since HET domain proteins are absent in all other sequenced zygomycetous genomes it is unlikely that they play a general role in the sexual reproduction but seem to be specific for Lichtheimia. Interestingly, these HET genes were differentially regulated under stress conditions. Several copies of the HET domain proteins were down-regulated under iron-depletion and hypoxia. Since these genes are absent in all other mucoralean fungi it is unclear which functions they serve in L. corymbifera. In addition it is unclear where these genes originate since they do not occur in other basal fungi and show only very weak similarity with the HET proteins of dikaryan fungi.
Several expansions contain transporters: major facilitator superfamily (MFS, PF07690, PF12832, PF05977, PF13347), ABC transporters (PF00005, PF00501, PF01061, PF00664, PF06422), sugar (and other) transporters (PF00083). In addition, some interesting expansions are connected to the transcription regulation function, which is discussed in more detail in a separate section and signal transduction pathways (see supplemental Material, Table S5 and S6). Four expanded groups are characterized by the cytochrome P450 (PF00067) domain (Figure 4 C). Interestingly, mucoralean pathogens like L. corymbifera have been shown to be resistant to several antifungals including voriconazole [8], [50] which could be explained by high copy numbers and isoforms of the target genes. Thus, gene duplication and expansion might be important for the success of L. corymbifera in human infections.
Strikingly, these domains (MFS transporters, HET and cytochrome P450) were also the dominant domains in genes which were localized in tandem duplications (Figure S3). Tandem duplicated genes were found to be present in 42 of the 66 gene expansion groups covering 38% of all genes in the expanded groups (Figure 4 D). However, additional smaller tandems were found which did not fit the criteria of gene expansions. A total of 701 genes are organized in such tandem repeats (Figure S3). In addition, duplicated genes were frequently found to be located on the same scaffold which may result from older tandem duplications.
Thus, tandem duplications and a high amount of gene retentions may give an additional explanation for the high amounts of duplicated genes in L. corymbifera comparable to the observations in plant genomes where segmental duplications (resulting from WGD) and tandem duplications play different roles in the enrichment of genes of several gene families [51], [52]. Tandem duplication and the retention of duplicated genes would be an explanation for the severe differences in the size of gene families between mucoralean fungi with only 53% of gene families with the same size in L. corymbifera and R. oryzae (see Figure S3 C).
To investigate if the different gene copies may have different functions and thus may contribute to rapid adaptations to different environmental conditions we analysed the expression of tandem duplicated genes under infection-associated stress conditions (iron depletion and hypoxia; see Table S7). Differential expression of at least one gene of the tandem clusters under at least one of the conditions was found for 71 tandems. Strikingly, only 7 tandems were co-regulated while in 64 cases expression of the copies was different including six cases were copies were antithetically regulated (Figure S4). These results are consistent with the hypothesis that the high prevalence and maintenance of duplicated genes leads to diversification of gene functions.
Alternative splicing
Duplicated genes can lead to the diversification of gene functions of the two copies which has been discussed in the section above. In addition, alternative splicing (AS) can increase the functional diversity. Gene prediction resulted in 841 alternative splicing events in a total of 683 genes (5.5% of total genes) comparable to the situation in S. cerevisiae [53]. Based on the analysis of the RNAseq data alternative splicing could be verified for 273 genes (2.2% of total genes) (Figure S5 A and Table S8). Alternative donor and acceptor are the dominant groups of alternative splicing events (>75% of the total events) which is similar to the situation in several higher eukaryotes [54] (Figure S5 A). Comparison of alternatively spliced genes with genes in tandem duplications and gene expansions showed that only 12 (4.4% of genes with AS) in these groups are also alternatively spliced. If AS occurs in tandem duplicated genes, it occurs in only one of the copies except in one case. This is in accordance with recent results in S. cerevisiae which show that duplicated genes can replace one alternatively spliced gene and that alternative splicing is often lost after gene duplication [55]. To test if AS plays a role in the stress adaptation of L. corymbifera we analysed the potential alteration in alternative splicing pattern during stress adaptation. Significant changes were only detected for 16 and 23 genes under iron depletion and hypoxia (<0.2% of the total genes), respectively (Figure S5 B). Based on the high incidence of gene duplication and the differential expression of the copies as well as the comparably low number of alternatively spliced genes, maintenance of duplicated genes seems to play a more important role for the generation of functionally distinct paralogs than alternative splicing.
Identification and expression of potential virulence factors under infection-related conditions
a) Iron uptake genes
Iron is an essential trace element for all organisms and plays a crucial role in fungal pathogenicity [56]–[58]. Elevated host iron levels are an important prerequisite for mucormycosis and the iron permease FTR1 has been shown to be crucial for virulence in R. oryzae [59]–[62]. The genome of L. corymbifera contains four copies of FTR1, of which three are located next to a multicopper oxidase and may share the same promoter. Both FTR1 and multicopper oxidase are important players of the reductive pathway and have been shown to form functional complexes in C. albicans [63]. Thus, co-expression of both genes and maintenance of their proximity may contribute to effective iron uptake and, hence, to the virulence of L. corymbifera. The higher copy number of FTR1 in L. corymbifera compared to R. oryzae (one copy) suggests more efficient employment of this pathway and, probably, optimisation of the different copies to different environmental conditions in L. corymbifera. To investigate the expression of iron-uptake genes under iron limited conditions we added the iron chelator bathophenanthrolinedisulfonic acid (BPS) to overnight cultures of L. corymbifera (200 µM final concentration) and analysed gene expression via RNA sequencing (see Material and Methods for details). Consistent with this hypothesis only one of the copies of FTR1 (LCor01036.1) was up-regulated under iron limitation in the gene expression experiments while another copy (LCor06326.1) was moderately down-regulated (Figure 5 A). In addition, all multicopper oxidases which are co-localized with FTR1 were regulated in the same manner as the corresponding FTR1 gene and expressed at comparable levels (Figure 5 A). The two remaining copies (LCor00518.1, LCor04103.1) were expressed constitutively at low levels and may be either specific for other conditions or generally silenced. In addition to FTR1 and multicopper oxidases, ferric reductases are key elements in the reductive pathway of iron acquisition. Two of the three ferric reductases were up-regulated under iron limitation (LCor07115.1, LCor11373.1), whereas the third one was constitutively expressed (Figure 5 B).
Fig. 5. Expression of iron uptake genes under iron limited conditions. 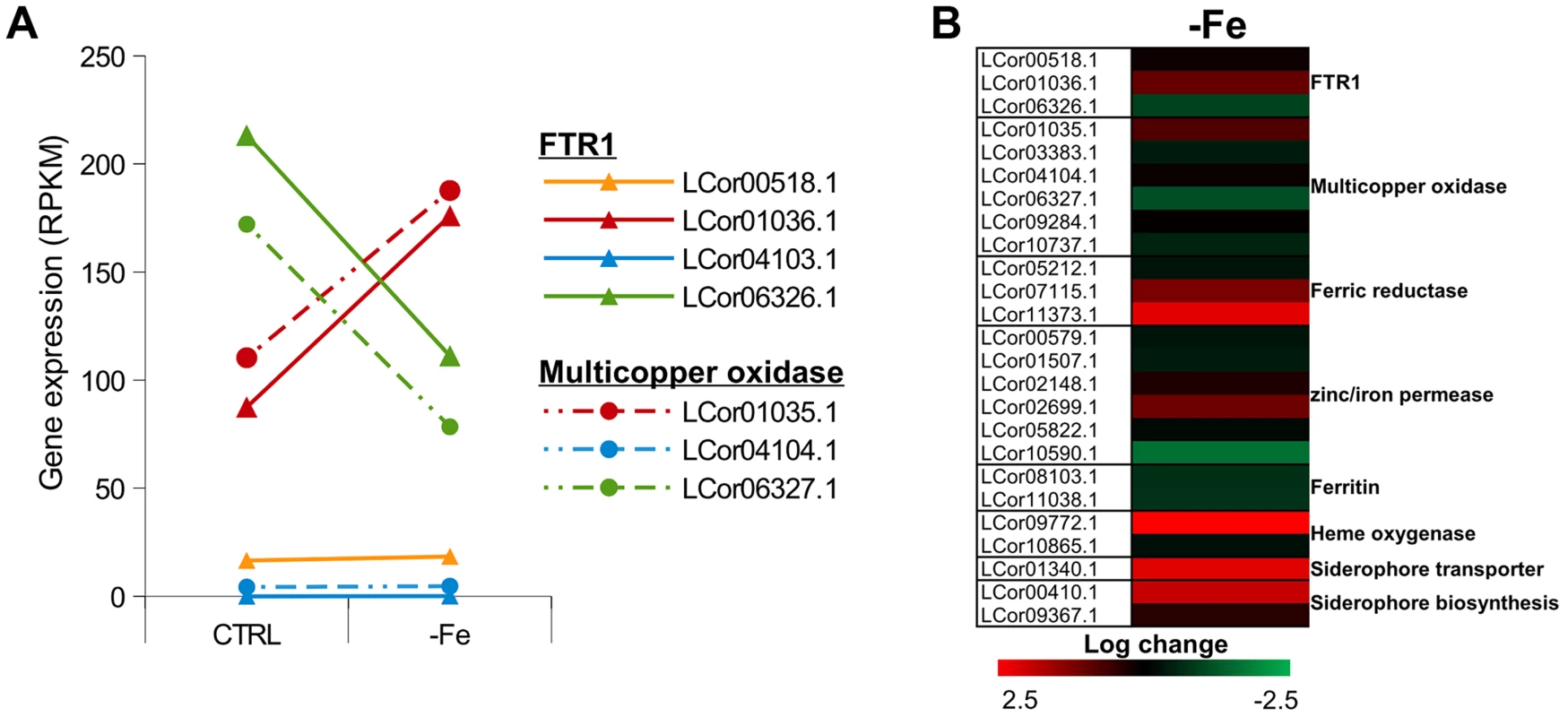
(A) Expression levels of FTR1 domain genes and their corresponding multicopper oxidases under standard conditions and iron limitation. (B) Heat map showing the regulation of iron uptake genes under iron-limited conditions. Besides the role of the reductive pathway little is known about the iron uptake systems in mucoralean pathogens. Our analysis uncovered the presence of additional genes involved in iron uptake including zinc/iron permeases, heme oxygenases and siderophore transporters (Table 2). Heme utilization may contribute to growth within the host, since mucoralean pathogens rapidly invade blood vessels [3] and may use hemoglobin as iron source [60]. Accordingly, one of the heme oxygenases (LCor09772.1) was strongly up-regulated under iron limitation (Figure 5 B). All available mucoralean genomes, including L. corymbifera, lack non-ribosomal peptide synthetases (NRPSs) and are therefore unable to produce hydroxamate siderophores. Instead they produce polycarboxylate siderophores (rhizoferrin), which have a much weaker binding activity compared to hydroxamate siderophores and are produced by direct fermentation [64], [65]. In addition, mucoralean fungi are also able to utilize deferoxamine, a bacterial siderophore which is used as an iron-chelator in human therapy [66], [67]. Interestingly, zygomycetous species have been shown to live in close relationship with bacteria, including cases containing bacterial endosymbionts indicating that xenosiderophores might play a role in the development of siderophore uptake systems [68], [69]. However, L. corymbifera has also been shown to be a potent producer of siderophores itself [70]. Under iron-limitation the putative siderophore transporter of L. corymbifera (LCor01340.1) was up-regulated (Figure 5 B) supporting the role of siderophores in the iron acquisition of mucoralean fungi. Interestingly, based on the expression data we found an additional gene (LCor00410.1) that may be involved in the siderophore metabolism of L. corymbifera, containing functional domains which are typical for genes involved in regulation and synthesis of siderophores in bacteria (lucA/lucC PF04183 and FhuF PF06276). Thus, the gene may encode a novel candidate for a regulator of siderophore biosynthesis in mucoralean fungi.
Tab. 2. Iron uptake genes in the <i>L. corymbifera</i> genome. 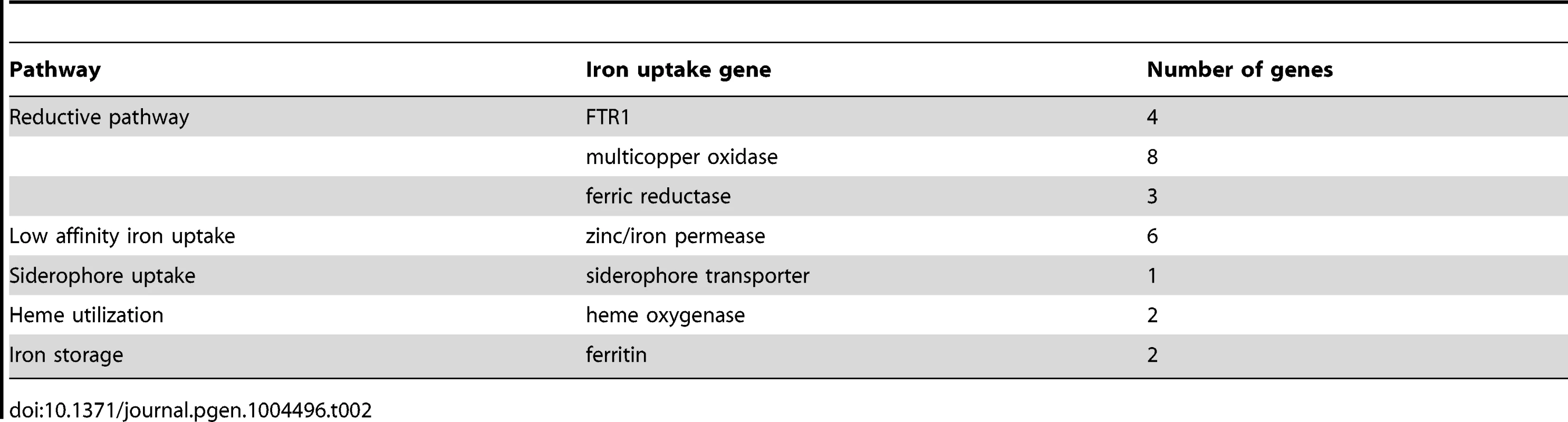
Although fungi generally lack ferritin as intracellular iron storage, ferritin has been found in several mucoralean species [62], [71]. Based on orthology searches, ferritin genes could be identified in all mucoralean genomes. In addition, domain search in Spizellomyces punctatus and Allomyces macrogynus (Origins of Multicellularity, BROAD) revealed also the presence of potential ferritin genes. Since these two groups represent the most basal fungal lineages, apart from the highly derived microsporidians [72], ferritin seems to have been lost in some microsporidians and the higher fungi. Interestingly, in higher fungi, the loss of ferritin coincides with the appearance of sidA, an important gene in hydroxamate siderophore production (Figure S6). Siderophores are known to serve as intracellular iron storages in several Asco - and Basdiomycota species [73], [74]. Thus, a plausible hypothesis is that maintenance of ferritin was not necessary in derived fungi due to the gain of importance of siderophores. The expression of the two ferritin genes (LCor08103.1, LCor11038.1) was slightly decreased under iron-limitation (∼1.6×, Fig. 5B). Nothing is known about the dynamics and functions of fungal ferritins. The slight decrease may be sufficient to stabilize free iron concentrations in the cytoplasm.
Only three transcription factors were up-regulated under iron depleted conditions (Figure S7). Interestingly, one of these genes (LCor08192.1) shows similarities to GATA type regulators which are known to be involved in the adaptation to iron limitation in higher fungal pathogens (e.g. A. fumigatus and Histoplasma capsulatum) [75], [76]. Thus, this transcription factor may represent a key regulator in iron acquisition, and therefore an important virulence factor of L. corymbifera. A second transcription factor (LCor01236.1) resembles CRZ1, which is a calcineurin regulated TF and indicates a possible involvement of the calcineurin pathway in the adaption to iron limitation.
b) Secreted proteases
Besides the iron uptake systems, hydrolytic enzymes like proteases are known as important virulence factors in fungal pathogens, e. g., R. microsporus and C. albicans [77]–[79]. In addition, gene expansion of secreted proteases was observed in the R. oryzae genome [28]. The genome of L. corymbifera contains a total of 413 predicted proteases representing 3.3% of all genes comparable to the situation in R. oryzae which contains 630 proteases (3.6% of all genes). However, the number of secreted proteases differs between the two species, moreover, in L. corymbifera the relative amount of secreted proteases is nearly twice as high as in R. oryzae: in L. corymbifera 13% (53) of the proteases are predicted to be secreted while in R. oryzae this number reaches only 7% (44). The most important classes of secreted proteases are serine and aspartate proteases representing 55% and 36% of total secreted proteases, respectively (Table 3). Comparison of secreted aspartic proteases (SAP) of R. oryzae revealed an enrichment of SAPs compared to other fungal genomes [28]. However, the number of SAPs is comparable in L. corymbifera (24) and R. oryzae (28) indicating that the presence of a higher number of these enzymes is a general feature of mucoralean pathogens. Several secreted proteases were activated under infection-related stress conditions (iron depletion, hypoxia). While iron depletion affected mainly the expression of aspartic and serine proteases, hypoxic conditions induced the expression of serine-, metallo - and some aspartic proteases.
Tab. 3. Protease families in the <i>L. corymbifera</i> genome. 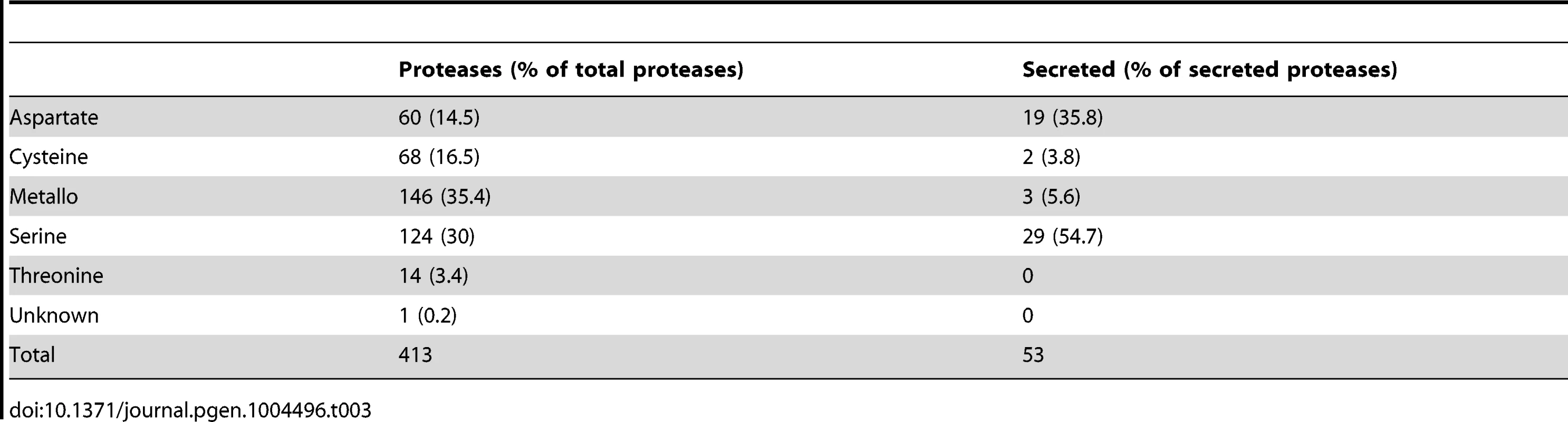
c) Transcription factors
The Lichtheimia genome encodes 768 putative transcription factors (TFs) representing 6.2% of total genes. This amount is comparable to the situation in R. oryzae (6.4% of total genes) but higher than the average content of TFs in other fungi (4.5%) [80]. Basic BLASTp analyses showed that 37 of the TFs (4.8% of total TFs) are specific for L. corymbifera.
The TFs were assigned to 53 families of DNA-binding domains (based on the InterProScan predictions). The great majority of these families have been described previously in fungal species [81]. However, 4 families have not been found in true fungal species before (putative representatives of 2 families, PF01167 (Tubby) and PF02319 (TDP), were predicted in microsporidia; the other 2 were described in plants and bacteria). Of these “new” TFs one (PF03106, DNA-binding WRKY domain), represented in 7 Lichtheimia proteins, has been previously described only in plants. Interestingly, in plants these TFs are numerous and have diverse functions, including pathogen defense [82]. In L. corymbifera we found three of the seven members differentially regulated under hypoxic conditions (down: LCor09690.1; up: LCor02851.1, LCor08197.1) indicating a function in the stress response of this species (Figure S7).
Of 6 fungal-specific TF families [81], 2 families are not predicted in L. corymbifera genome, namely PF04769 (mating-type protein MAT a1) and PF02292 (APSES domain). The lack of MAT gene is expected because mating in Mucorales is regulated via sex plus and sex minus HMG transcription factors [83]. The absence of APSES domain may be compensated by the APSES-type DNA binding domain PF04383.
It is also noteworthy that the traditional proportion of the Zn fingers Zn(2)Cys(6) (Zn cluster) and Cys(2)His(2) is inverted in Lichtheimia. In all fungi observed so far, the Zn clusters are more abundant than C2H2 TFs, in fact they are normally the most numerous in the fungal genomes. In Lichtheimia, on the contrary, the number of C2H2 Zn fingers is nearly 1.5 times larger than the number of Zn clusters.
Comparative phylome-based analysis reveals several expanded TF families in L. corymbifera including MADS box TFs with 11 representatives instead of the usual 1–4 members. MADS box genes are known to play a role in a variety of functions (e.g, cell cycle, stress response, development [84]). Presumably, the expansion of the MADS box genes in Lichtheimia was accompanied by the delegation of some functions from other TFs or even neofunctionalization. The functional basis for such expansion as well as the potential roles of these TFs cannot be elucidated from their primary structure, because MADS box genes are not conserved except for the MADS domains. But at the epxressional level, we could find significant up-regulation of two of the 11 MADS box TFs (LCor03918.1, LCor08105.1). Thus, the copies do not seem to have completely overlapping functions.
Analysis of the phylomes allowed us to detect another exciting expansion, which is evidently characteristic for all Mucorales: the duplication of TBP, TATA binding protein (PF00352). As it has been recently shown for higher eukaryotes, core promoter recognition factors can be involved in modulating gene - and cell-type-specific programs of transcription, such as tissue differentiation, development, etc. [85]. These new functions are associated with a gene duplication of the TBP, resulting in TRF2 (and other) factors, which are highly similar to TBP but do not bind the TATA box. In fungi, the event of TBP duplication is exceptionally rare. A survey of all so far sequenced genomes revealed only 4 examples of such duplication: 3 in Ascomycetes/Sordariomycetes (Chaetomium globosum, Grosmannia clavigera and Podospora anserina) and 1 in the basidiomycete Laccaria bicolor. In contrast, in Mucorales all 4 considered species possess 2 copies of the TBP gene. It can be supposed that the duplicated TBP-like factors may play an additional role in condition-specific responses and thus may be of interest as potential virulence factors.
Temperature tolerance is an essential prerequisite for the infection of warm-blooded animals and was shown to be connected to the virulence of Lichtheimia species [25], [86]. The genome was surveyed for the presence of heat shock transcription factors (HSF). The total number of these TFs in L. corymbifera genome is 24, which is the highest number among all so far investigated fungi. This is in accordance with the known tolerance of L. corymbifera to high temperatures [13], [14]. However, it seems that this family expansion is not a specific trait of Lichtheimia but is characteristic for all Mucorales. Interestingly, HSF genes were also up-regulated under hypoxic conditions indicating additional functions of the different members of the HSF family in the response to different stresses and growth conditions.
It is curious that additionally to the abundant heat shock factors also a cold shock TF (PF00313) was found, which was not previously described in fungi. This can explain why Lichtheimia, although it does not grow at low temperatures, can tolerate cold as it was shown to survive periods of more than 5,000 years in ice [87].
The importance of the L. corymbifera genome for studying the infection biology of mucoralean pathogens: Concluding remarks
The genome of L. corymbifera represents the first insight into the genome structure of basal mucoralean pathogens. Despite the growing recognition of Mucorales as life-threatening clinically important human pathogens, little is known about the virulence traits of these fungi. The high dissimilarity between L. corymbifera and the other sequenced mucoralean pathogens R. oryzae and M. circinelloides in both evolutionary and functional sense underlines the importance of additional genome projects.
This study revealed a high proportion of duplicated and expanded genes in the L. corymbifera genome comparable to the situation in R. oryzae. However, clear evidence for a WGD can be detected only for R. oryzae, but not for L. corymbifera indicating that additional mechanisms contribute to the higher incidence of duplicated genes in mucoralean fungi. Tandem repeats seem to be an important source for gene duplication in L. corymbifera and may explain the rapid development of lineage-specific gene duplication and expansion in mucoralean fungi. Several species-specific gene duplications point at potential virulence traits including iron uptake genes, hydrolytic enzymes and genes which may contribute to resistance against antifungal agents like azoles (cytochrome P450 gene expansion). In contrast, alternative splicing does not seem to play an important role in the generation of orthologs and the adaption to stress conditions.
Based on these results, we postulate a relationship between genome fluidity by the generation and retention of additional gene copies and dynamics of adaptation to new environments. Higher genome flexibility results in a higher likelihood for a saprobic zygomycete to become a pathogen.
In addition we were able to shed light on the genes involved in iron uptake, which is a crucial step for virulence and thus for the development of an infection. We could identify additional genes which might be involved in iron-uptake besides the known virulence factor FTR1 L. corymbifera including transcription factors, siderophore transporters and a potential regulator involved in siderophore biosynthesis that has not been described in mucoralean fungi.
Our data represent a valuable resource for future research and the understanding of infection-associated mechanisms of mucoralean pathogens.
Materials and Methods
Genome sequencing and assembly
A combination of Illumina and 454 sequencing was used for the L. corymbifera genome. A shotgun library and an 8 kb paired-end library were created and sequenced on a half plate on a Roche GS FLX Titanium each resulting in 1,168,226 shotgun reads (505,023,982 nt) and 519,989 paired-end reads (76,603,029 nt). In addition, a standard paired-end read library was prepared and sequenced in one channel Illumina HiSeq2000 (100 bp paired-end reads) resulting in 264,907,616 raw reads (26,490,761,600 nt) and 12,614,650 filtered and downsampled reads (1,261,465,000 nt). The 454 reads were separately assembled using Newbler (454 Life Sciences) and Mira [88] and both assemblies were unified using minimus2 [89]. The Illumina reads were used to solve homopolymeric regions using Nesoni (http://bioinformatics.net.au/software). This approach resulted in a total of 1,214 contigs (≥500 nt) with a total of 41,405,106 nt and a N50 of 66.718 nt. Finally, the contigs were mapped on Newbler predicted scaffolds using MUMmer [90] resulting in 209 scaffolds with a total length of 33.6 Mb (for statistics refer to Table 1). The raw DNA-seq reads and the resulting genome assembly is available at EMBL under the study accession number PRJEB3978 (http://www.ebi.ac.uk/ena/data/view/PRJEB3978).
Detection of transposable elements
Scaffolds of L. corymbifera were searched for repeats by Repbase and the server version of Censor [91], [92] (http://www.girinst.org/censor/index.php) using the eukaryotic repeat database.
K-mer analysis
The analysis was performed on the Ilumina reads with an algorithm described in the potato genome paper [31]. The algorithm was used to write a custom perl program. Based on the fastq data of the Illumina reads k-mers of 41, 59, 69, and 79 nt were detected and analyzed. Component estimation was done manually in R.
Non-coding RNA prediction, synteny and phylogeny
A local version of tRNAscan-SE v.1.23 [93] with parameters –omlfrF was used for the detection of tRNAs. Ouput files are supported in the supplemental material (http://www.rna.uni-jena.de/supplements/lichtheimia/index.html). With RNammer -S euk -m lsu,ssu,tsu -gff (v.2.1) [94] rRNAs were detected. The 1973 ncRNA classes currently available at RFAM (v.10.1) [95] were downloaded for homologous search. These classes were predicted with (I) BLAST (v.2.2.25) [96] with an E-value<10−4 (II) with infernal [97] using covariance models from RFAM and (III) by hand as indicated in main text. Genes discovered in the reads only were found with rnabob [98] in combination with various programs of the RNAViennaPackage v.2.0.2 (http://www.tbi.univie.ac.at/~ivo/RNA/). All ncRNA genes are available at the supplemental material in gff and fasta format (http://www.rna.uni-jena.de/supplements/lichtheimia/index.html). Additionally, sequence-structure-alignments for each RFAM-ncRNA class in stockholm format are provided. Motif search in promoter regions of polymerase III transcripts was performed with MEME (v.4.8.1) [99], rnabob and by hand. Synteny analysis: for all of our identified ncRNA positions in L. corymbifera and R. oryzae, five direct upstream and downstream located genes and their function were extracted, according to protein-annotation files. Pairwise alignments of syntenic proteins with a) -p blastn and b) -p tblastn and a minimum E-value of E<10−4 were performed. For ncRNA-phylogeny reconstruction the best scored ncRNA per ncRNA family was joined, which was identified in all species, except 18S and 28S rRNA, and S. pombe used as outgroup. A multiple alignment was created by Mafft with the L-INS-i method, 1000 iterations as module in the EPoS framework for phylogenetic analysis [100]. Out of this alignment we constructed a Neighbour Joining Tree (Kimura correction model, 1000 bootstrap replicates) and Mr. Bayes (v.3.1.2; two runs with each four chains and 5,000,000 generations).
Prediction of protein-coding genes and functional annotation
Evidence-driven gene prediction was performed using AUGUSTUS v2.7 [37] using the gene models from Rhizopus oryzae prediction was supported by the incorporate pooled Illumina RNA-seq data from three biological replicates of three different physiological conditions (control, hypoxia, iron depletion) sequenced on Illumina HiSeq 2000. After the raw RNA-Seq data were quality trimmed - using btrim [101], the data were pooled and mapped using the splice-junction mapper tophat2 [102]. From this mapping data the AUGUSTUS protocol (http://bioinf.uni-greifswald.de/bioinf/wiki/pmwiki.php?n=IncorporatingRNAseq.Tophat) was followed to create hints for gene structures in an iterative manner. Finally, the hints were incorporated during the AUGUSTUS predcition based on the Lichtheimia genome using the metaparameters of R. oryzae. For functional annotation predicted protein-coding genes were analyzed by BLASTp in BLAST2GO [38] with a minimum E-value of E≤10−25 and a HSP length cut-off of 33 amino acids. Conserved domains were identified using the InterProScan function of BLAST2GO and GO mapping was performed based on the BLAST and InterProScan results. Genome annotations are available at the ENA under the study accession number PRJEB3978 (http://www.ebi.ac.uk/ena/data/view/PRJEB3978).
Detection of differentially expressed genes under infection-related conditions
Paired-end RNA-seq data for three biological replicates of two infection-associated conditions (i.e., iron-depletion and hypoxia) and a control treatment was obtained.
L. corymbifera was grown on SUP agar [103] plates for 7 days at 37°C. Spores were washed off with sterile PBS, washed with PBS and counted using a Thoma chamber. Erlenmeyer flasks (500 ml) containing 100 ml of chemical defined medium (1.7 g/l YNB w/o amino acids and ammonium sulphate, 20 g/l Glucose, 5 g/l ammonium sulphate, 50 mg/l arginine, 80 mg/l aspartic acid, 20 mg/l histidine, 50 mg/l isoleucine, 100 mg/l leucine, 50 mg/l lysine, 20 mg/l methionine, 50 mg/l phenylalanine, 100 mg/l threonine, 50 mg/l tryptophane, 50 mg/l tyrosine, 20 mg/l valine) [60] were inoculated with 107 spores and grown for 16 h at 37°C under shaking. Afterwards (i) cultures were grown for additional 2 h under these conditions, (ii) the iron chelator bathophenanthrolinedisulfonic acid (BPS, Sigma) was added to a final concentration of 200 µm and cultures were incubated for additional 2 h under previous conditions or (iii) cultures were subjected to hypoxic conditions (1% oxygen, 5% CO2) and incubated for 2 h at 37°C under shaking. The mycelium was separated from the medium using a miracloth filter (Millipore) and immediately frozen in liquid nitrogen. For RNA isolation the mycelium was grounded using mortar and pestle under liquid nitrogen and total RNA was isolated using the RNAeasy Plant kit (Quiagen) according to the manufacturer's instructions.
Sequencing was performed using Illumina HiSeq 2000. Raw reads were quality-filtered using btrim [101] and mapped to the genome using tophat2 [102] (parameters: –no-discordant –no-mixed –b2-very-sensitive –max-intron-length 5000). Differentially expressed genes were identified with EdgeR [104] which also adjusted obtained p-Values for multiple testing. Transcripts with an absolute fold-change≥2 and an adjusted p-Value≤0.01 were considered differentially expressed. Results are available in Table S7.
Phylome reconstruction
The phylome, meaning the complete collection of phylogenetic trees for each gene in a genome, was reconstructed for the genome of L. corymbifera. 24 other fungal species were included in the reconstruction. A rough draft of the proteome of Mortierella alpina ([26]; PUBMED ID:22174787) was predicted using AUGUSTUS [32] due to the lack of a publicly available proteome. The phylome was reconstructed using an automated pipeline previously described in [39]. Briefly, for each protein in the L. corymbifera genome a Smith-Waterman search was performed against the fungal proteome database. Results were filtered using an e-value cut-off E<1e−5 and a continuous overlapping region of 0.5. At most 150 homologous sequences for each protein were accepted. Homologous sequences were then aligned using three different programs: MUSCLE v3.8 [105], MAFFT v6.712b [106], and kalign (http://www.biomedcentral.com/1471-2105/6/298/)]. Alignments were performed in forward and reverse direction (i.e. using the Head or Tail approach [107]), and the 6 resulting alignments were combined with M-COFFEE [108]. This combined alignment was trimmed with trimAl v1.3 [109] (consistency-score cut-off 0.1667, gap-score cut-off 0.9). Trees were reconstructed using the best-fitting evolutionary model. The selection of the model best fitting each alignment was performed as follows: a Neighbour Joining (NJ) tree was reconstructed as implemented in BioNJ [110]; the likelihood of this topology was computed, allowing branch-length optimization, using 7 different models (JTT, LG, WAG, Blosum62, MtREV, VT and Dayhoff), as implemented in PhyML v3.0 [111]; the model best fitting the data, as determined by the AIC criterion [112], was used to derive ML trees. Four rate categories were used and invariant positions were inferred from the data. Branch support was computed using an aLRT (approximate likelihood ratio test) based on a chi-square distribution. Resulting trees and alignments are stored in phylomeDB [39] (http://phylomedb.org), with the phylomeID 245. Trees were scanned using ETE v2 [46].
Orthology prediction
Orthologs between L. corymbifera and the other species included in the phylome were based on phylogenies obtained during phylome reconstruction. A species-overlap algorithm, as implemented in ETE v2 [46], was used to infer orthology and paralogy relationships. Briefly the algorithm decides whether a node in a tree is a speciation of a duplication node depending on the overlap of the species branching from the node. Overlap between those species will indicate a duplication node. Otherwise a speciation node will be considered.
Species tree reconstruction
The species tree was build using a concatenation method. 58 single-copy proteins that appeared in at least 21 of the 25 genomes were selected. After concatenation, the alignment was trimmed using trimAl [109]. Columns with more than 50% of gaps were removed. A conservation score of 50% of the alignment was used. The final alignment contained 46,793 positions. The tree was reconstructed using phyML [111]. LG model [113] was selected and a 4-categories GAMMA distribution was used. Bootstrap was obtained by creating 100 random sequences using SeqBoot from the phylip package. A tree was then reconstructed for each sequence and the consensus tree was inferred using phylip. All the nodes in the species tree had a bootstrap of 100. Additionally a species tree based on the super-tree reconstruction program DupTree [42] was reconstructed. The input contained the 9,478 trees obtained during phylome reconstruction. Both species trees showed a similar topology. The only difference pertained to the position of S. pombe. In the concatenated tree it appeared grouped with S. cerevisiae while in the super-tree it appeared in its correct position at the base of Ascomycota. This difference was collapsed into a multifurcation for the tree in figure 1.
Detection of conserved regions
For the detection of conserved regions, all genomes were modeled as strings of integers. BLAST analyses [96] were performed for all proteins in the four mucoralean genomes all-against-all, with an E-value threshold of 0.1. Homology families IDs were assigned to the protein-coding genes using GhostFam [114] with default parameters. Genomes were transformed into strings of gene IDs, which were then used as input for the reference gene cluster implementation in Gecko2 [43], [44]. The two parameters for the algorithm were the minimum size of the reference cluster/hypothetical conserved region “s” and the maximal distance “δ” (insertion or deletion of a gene). For every hypothetical gene cluster larger than s on the reference genome, all other genomes were tested for approximate occurrences of this reference gene cluster. The L. corymbifera genome was used as a reference genome and searched for gene clusters with parameters s = 3 (minimum size of the reference gene cluster) and δ = 0 (number of insertions and deletions), s = 4/δ = 1, s = 5/δ = 2, s = 6/δ = 3 and s = 7/δ = 4. Results of the different filter settings were combined and overlapping clusters were eliminated. Local rearrangements and duplications within the cluster occurrences were not punished. All regions that had approximate occurrences in at least one other genome were reported. If multiple occurrences did intersect, only the best scoring one was reported.
Detection of duplicated regions (segmental duplications)
To detect duplicated regions in the mucoralean species, each genome was analysed individually by using the single contigs as reference. As for the detection of conserved regions, the same homology assignment and parameters of s = 5 and δ = 2 were used. All regions with approximate occurrences in at least one other contig or the reference contig were reported, unless they intersected.
Detection of tandem duplications
Tandem duplications were defined by at least two genes assigned to the same GhostFam gene family and a maximum of three genes between the copies.
Prediction of alternative splicing
Predicted transcripts of the genomes were separated in alternative splicing events by Astalavista [24]. Events of predicted transcripts that contain splice-junctions have been confirmed by the number of split-mappings that confirm each of the exon-exon junctions (Table S8). For a read to support a splice-junction, the left part of the read was required to be included in one exon, and the right part had to be included in the other exon of a splice junction, with the first/last position before/after the split matching exactly the position of the predicted intron.
Genome resources
Genome data of Aspergillus fumigatus [115], Aspergillus nidulans [116], Batrachochydrium dendrobatidis, Cryptococcus neoformans, Encephalitozoon cuniculi [117], Rhizopus oryzae [28], Paracoccidioides brasiliensis, Schizosaccharomyces pombe [118], Nosema ceranae [119], Nematocida parisii [120], Puccinia graminis [121], Ustilago maydis and Coprinus cinerea [122] are genome sequencing projects of the Broad Institute of Harvard and MIT (http://www.broadinstitute.org/) (see Table S9 for detailed citations). Phycomyces blakesleeanus, Phanerochaete chrysosporium [123], Laccaria bicolor [124], Mucor circinelloides, Nematostella vectensis [125], Monosiga brevicollis [126] and Serpula lacrymans [127] genomic data were obtained from Joint Genome Institute (JGI). These sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community. The genomes of Homoloaphlyctis polyrhiza [128] and Mortierella alpina [26] were obtained from Genbank (Hp: PRJNA68115; Ma: PRJNA41211). The Neurospora crassa genome [129] was obtained from UniProt reference genomes. The Saccharomyces cerevisiae genome was obtained from Saccharomyces Genome database (SGD) (see Table S9) [53].
Supporting Information
Zdroje
1. HibbettDS, BinderM, BischoffJF, BlackwellM, CannonPF, et al. (2007) A higher-level phylogenetic classification of the Fungi. Mycol Res 111 : 509–547 doi:10.1016/j.mycres.2007.03.004
2. RodenMM, ZaoutisTE, BuchananWL, Knudsen Ta, Sarkisova Ta, et al. (2005) Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis 41 : 634–653 doi:10.1086/432579
3. Sugar AM (1992) Mucormycosis. Clin Infect Dis 14 Suppl 1: S126–9.
4. RibesJA, Vanover-SamsCL, BakerDJ (2000) Zygomycetes in human disease. Clin Microbiol Rev 13 : 236–301.
5. ChakrabartiA, DasA, MandalJ, ShivaprakashMR, GeorgeVK, et al. (2006) The rising trend of invasive zygomycosis in patients with uncontrolled diabetes mellitus. Med Mycol 44 : 335–342 doi:10.1080/13693780500464930
6. CornelyOA, VehreschildJJ, RüpingMJGT (2009) Current experience in treating invasive zygomycosis with posaconazole treatment options for zygomycosis. Clin Microbiol 15 : 77–81.
7. SkiadaA, PaganoL, GrollA, ZimmerliS, DupontB, et al. (2011) Zygomycosis in Europe: analysis of 230 cases accrued by the registry of the European Confederation of Medical Mycology (ECMM) Working Group on Zygomycosis between 2005 and 2007. Clin Microbiol Infect 17 : 1859–1867.
8. VitaleRG, de HoogGS, SchwarzP, DannaouiE, DengS, et al. (2012) Antifungal susceptibility and phylogeny of opportunistic members of the order mucorales. J Clin Microbiol 50 : 66–75 doi:10.1128/JCM.06133-11
9. XessI, MohapatraS, ShivaprakashMR, ChakrabartiA, BennyGL, et al. (2012) Evidence implicating Thamnostylum lucknowense as an etiological agent of rhino-orbital mucormycosis. J Clin Microbiol 50 : 1491–1494 doi:10.1128/JCM.06611-11
10. LanternierF, DannaouiE, MorizotG, ElieC, HuerreM, et al. (2012) A global analysis of mucormycosis in France: The RetroZygo Study (2005–2007). Clin Infect Dis 54 : 35–43 doi:10.1093/cid/cir880
11. AlvarezE, SuttonDa, CanoJ, Fothergill aW, Stchigela, et al. (2009) Spectrum of zygomycete species identified in clinically significant specimens in the United States. J Clin Microbiol 47 : 1650–1656 doi:10.1128/JCM.00036-09
12. Tieghem, VanP (1876) Troisiéme mémoire sur les Mucorinées. Ann des Sci Nat Bot 4 : 312–399.
13. HoffmannK, DischerS, VoigtK (2007) Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol Res 111 : 1169–1183 doi:10.1016/j.mycres.2007.07.002
14. Alastruey-IzquierdoA, HoffmannK, de HoogGS, Rodriguez-TudelaJL, VoigtK, et al. (2010) Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Absidia pro parte, Mycocladus). J Clin Microbiol 48 : 2154–2170 doi:10.1128/JCM.01744-09
15. De Hoog, GS, Guarro, J, Gene, J, Figueras M (2000) Atlas of Clinical Fungi. 2nd ed. Centraalbureau voor Schimmelcultures. p. 58–114.
16. HaidleCW, StorckR (1966) Control of dimorphism in Mucor rouxii. J Bacteriol 92 : 1236–1244.
17. CooperBH (1987) A case of pseudoparacoccidioidomycosis: Detection of the yeast phase of Mucor circinelloides in a clinical specimen Abstract. Mycopathologia 97 : 189–193.
18. HesseltineCW, FeatherstonC (1985) Anaerobic growth of molds isolated from fermentation starters used for foods in Asian countries. Mycologia 77 : 390–400.
19. SunH-Y, AguadoJM, BonattiH, ForrestG, GuptaKL, et al. (2009) Pulmonary zygomycosis in solid organ transplant recipients in the current era. Am J Transplant 9 : 2166–2171 doi:10.1111/j.1600-6143.2009.02754.x
20. BellangerA-P, RebouxG, BotterelF, CandidoC, RousselS, et al. (2010) New evidence of the involvement of Lichtheimia corymbifera in farmer's lung disease. Med Mycol 48 : 981–987 doi:10.3109/13693781003713711
21. CopettiMV, IamanakaBT, FrisvadJC, PereiraJL, TaniwakiMH (2011) Mycobiota of cocoa: from farm to chocolate. Food Microbiol 28 : 1499–1504 doi:10.1016/j.fm.2011.08.005
22. MphandeFA, SiameBA, TaylorJE (2004) Fungi, aflatoxins, and cyclopiazonic acid associated with peanut retailing in Botswana. J Food Prot 67 : 96–102.
23. BaffiMA, Romo-SánchezS, Ubeda-IranzoJ, Briones-PérezAI (2012) Fungi isolated from olive ecosystems and screening of their potential biotechnological use. N Biotechnol 29 : 451–456 doi:10.1016/j.nbt.2011.05.004
24. HongS, KimD, LeeM, BaekS, KwonS, et al. (2012) Zygomycota associated with traditional meju, a fermented soybean starting material for soy sauce and soybean paste. J Microbiol 50 : 386–393 doi:10.1007/s12275-012-1437-6
25. SchwartzeVU, HoffmannK, NyilasiI, PappT, VágvölgyiC, et al. (2012) Lichtheimia species exhibit differences in virulence potential. PLoS One 7: e40908 doi:10.1371/journal.pone.0040908
26. WangL, ChenW, FengY, RenY, GuZ, et al. (2011) Genome characterization of the oleaginous fungus Mortierella alpina. PLoS One 6: e28319 doi:10.1371/journal.pone.0028319
27. WöstemeyerJ, KreibichA (2002) Repetitive DNA elements in fungi (Mycota): impact on genomic architecture and evolution. Curr Genet 41 : 189–198 doi:10.1007/s00294-002-0306-y
28. MaL-J, IbrahimAS, SkoryC, GrabherrMG, BurgerG, et al. (2009) Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5: e1000549 doi:10.1371/journal.pgen.1000549
29. BuchonN, VauryC (2006) RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity (Edinb) 96 : 195–202 doi:10.1038/sj.hdy.6800789
30. JamesTY, LitvintsevaAP, VilgalysR, MorganJAT, TaylorJW, et al. (2009) Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog 5: e1000458 doi:10.1371/journal.ppat.1000458
31. XuX, PanS, ChengS, ZhangB, MuD, et al. (2011) Genome sequence and analysis of the tuber crop potato. Nature 475 : 189–195 doi:10.1038/nature10158
32. DeutscherMP (1984) Processing of tRNA in prokaryotes and eukaryotes. Crit Rev Biochem Mol Biol 17 : 45–71.
33. KachouriR, StribinskisV, ZhuY, RamosKS, WesthofE, et al. (2005) A surprisingly large RNase P RNA in Candida glabrata. RNA 11 : 1064–1072 doi:10.1261/rna.2130705
34. SchmittME, Clayton Da (1993) Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 13 : 7935–7941.
35. WalterP, IbrahimiI, BlobelG (1981) Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol 91 : 545–550.
36. DieciG, FiorinoG, CastelnuovoM, TeichmannM, PaganoA (2007) The expanding RNA polymerase III transcriptome. Trends Genet 23 : 614–622 doi:10.1016/j.tig.2007.09.001
37. StankeM, DiekhansM, BaertschR, HausslerD (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24 : 637–644 doi:10.1093/bioinformatics/btn013
38. ConesaA, GötzS, García-GómezJM, TerolJ, TalónM, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 : 3674–3676 doi:10.1093/bioinformatics/bti610
39. Huerta-CepasJ, Capella-GutierrezS, PryszczLP, DenisovI, KormesD, et al. (2011) PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res 39: D556–60 doi:10.1093/nar/gkq1109
40. GabaldónT (2008) Large-scale assignment of orthology: back to phylogenetics? Genome Biol 9 : 235 doi:10.1186/gb-2008-9-10-235
41. DelsucF, BrinkmannH, PhilippeH (2005) Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6 : 361–375 doi:10.1038/nrg1603
42. WeheA, BansalMS, BurleighJG, EulensteinO (2008) DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics 24 : 1540–1541 doi:10.1093/bioinformatics/btn230
43. BöckerS, JahnK, MixtackiJ, StoyeJ (2009) Computation of median gene clusters. J Comput Biol 16 : 1085–1099 doi:10.1089/cmb.2009.0098
44. JahnK (2011) Efficient computation of approximate gene clusters based on reference occurrences. J Comput Biol 18 : 1255–1274 doi:10.2144/000113833
45. Marcet-HoubenM, MarcedduG, GabaldónT (2009) Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evol Biol 9 : 295 doi:10.1186/1471-2148-9-295
46. Huerta-CepasJ, DopazoJ, GabaldónT (2010) ETE: a python Environment for Tree Exploration. BMC Bioinformatics 11 : 24 doi:10.1186/1471-2105-11-24
47. PuntaM, CoggillPC, EberhardtRY, MistryJ, TateJ, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301 doi:10.1093/nar/gkr1065
48. FinnRD, ClementsJ, EddySR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–37 doi:10.1093/nar/gkr367
49. PaolettiM, SaupeSJ, ClaveC (2007) Genesis of a Fungal Non-Self Recognition Repertoire. PlosOne e283 doi:10.1371/Citation
50. Alastruey-IzquierdoA, CuestaI, WaltherG, Cuenca-EstrellaM, Rodriguez-TudelaJL (2010) Antifungal susceptibility profile of human-pathogenic species of Lichtheimia. Antimicrob Agents Chemother 54 : 3058–3060 doi:10.1128/AAC.01270-09
51. CannonSB, MitraA, BaumgartenA, YoungND, MayG (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4 : 10 doi:10.1186/1471-2229-4-10
52. HanadaK, ZouC, Lehti-ShiuMD, ShinozakiK, ShiuS-H (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148 : 993–1003 doi:10.1104/pp.108.122457
53. HirschmanJE, BalakrishnanR, ChristieKR, CostanzoMC, DwightSS, et al. (2006) Genome Snapshot: a new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res 34: D442–5 doi:10.1093/nar/gkj117
54. KimE, MagenA, AstG (2007) Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35 : 125–131 doi:10.1093/nar/gkl924
55. MarshallAN, MontealegreMC, Jiménez-LópezC, LorenzMC, van HoofA (2013) Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet 9: e1003376 doi:10.1371/journal.pgen.1003376
56. SchrettlM, BignellE, KraglC, JoechlC, RogersT, et al. (2004) Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med 200 : 1213–1219 doi:10.1084/jem.20041242
57. RamananN (2000) A High-Affinity iron permease essential for Candida albicans virulence. Science (80-) 288 : 1062–1064 doi:10.1126/science.288.5468.1062
58. HowardDH (1999) Acquisition, transport, and storage of iron by pathogenic fungi. Clin Microbiol Rev 12 : 394–404.
59. FuY, LeeH, CollinsM, TsaiH-F, SpellbergB, et al. (2004) Cloning and functional characterization of the Rhizopus oryzae high affinity iron permease (rFTR1) gene. FEMS Microbiol Lett 235 : 169–176 doi:10.1016/j.femsle.2004.04.031
60. IbrahimAS, GebremariamT, LinL, LuoG, HusseinyMI, et al. (2010) The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol Microbiol 77 : 587–604 doi:10.1111/j.1365-2958.2010.07234.x
61. SymeonidisaS (2009) The role of iron and iron chelators in zygomycosis. Clin Microbiol Infect 15 Suppl 5 : 26–32 doi:10.1111/j.1469-0691.2009.02976.x
62. IbrahimAS, SpellbergB, WalshTJ, KontoyiannisDP (2012) Pathogenesis of Mucormycosis. Clin Infect Dis 54 : 1–7 doi:10.1093/cid/cir865
63. ZieglerL, TerzulliA, GaurR, McCarthyR, KosmanDJ (2011) Functional characterization of the ferroxidase, permease high-affinity iron transport complex from Candida albicans. Mol Microbiol 81 : 473–485 doi:10.1111/j.1365-2958.2011.07704.x
64. Thiekena, WinkelmannG (1992) Rhizoferrin: a complexone type siderophore of the Mucorales and entomophthorales (Zygomycetes). FEMS Microbiol Lett 73 : 37–41.
65. DrechselH, TschierskeM, ThiekenA, JungG (1995) The carboxylate type siderophore rhizoferrin and its analogs produced by directed fermentation. J Ind Microbiol Microbiol 14 : 105–112.
66. BoelaertJR, de LochtM, Van CutsemJ, KerrelsV, CantinieauxB, et al. (1993) Mucormycosis during deferoxamine therapy is a siderophore-mediated infection: In vitro and in vivo animal studies. J Clin Invest 91 : 1979–1986 doi:10.1172/JCI116419
67. IbrahimAS, GebermariamT, FuY, LinL, HusseinyMI, et al. (2007) The iron chelator deferasirox protects mice from mucormycosis through iron starvation. J Clin Invest 117 : 2649–2657 doi:10.1172/JCI32338
68. SchmittI, Partida-MartinezLP, WinklerR, VoigtK, EinaxE, et al. (2008) Evolution of host resistance in a toxin-producing bacterial-fungal alliance. ISME J 2 : 632–641 doi:10.1038/ismej.2008.19
69. Partida-MartinezLP, de LoossCF, IshidaK, IshidaM, RothM, et al. (2007) Rhizonin, the first mycotoxin isolated from the zygomycota, is not a fungal metabolite but is produced by bacterial endosymbionts. Appl Environ Microbiol 73 : 793–797 doi:10.1128/AEM.01784-06
70. LarcherG, DiasM, RazafimandimbyB, BomalD, BoucharaJ-P (2013) Siderophore production by pathogenic Mucorales and uptake of deferoxamine B. Mycopathologia doi:10.1007/s11046-013-9693-5
71. CarranoCJ, BöhnkeR, MatzankeBF (1996) Fungal ferritins: the ferritin from mycelia of Absidia spinosa is a bacterioferritin. FEBS Lett 390 : 261–264.
72. Capella-GutierrezS, Marcet-HoubenM, GabaldonT (2012) Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol 10 : 47 doi:10.1186/1741-7007-10-47
73. HaasH, EisendleM, TurgeonBG (2008) Siderophores in fungal physiology and virulence. Annu Rev Phytopathol 46 : 149–187 doi:10.1146/annurev.phyto.45.062806.094338
74. SilvaMG, SchrankA, BailãoEFLC, BailãoAM, BorgesCL, et al. (2011) The homeostasis of iron, copper, and zinc in paracoccidioides brasiliensis, Cryptococcus neoformans var. grubii, and Cryptococcus gattii: a comparative analysis. Front Microbiol 2 : 49 doi:10.3389/fmicb.2011.00049
75. HaasH (2012) Iron - a key nexus in the virulence of Aspergillus fumigatus. Front Microbiol 3 : 28 doi:10.3389/fmicb.2012.00028
76. HwangLH, SethE, Gilmore Sa, SilA (2012) SRE1 regulates iron-dependent and -independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryot Cell 11 : 16–25 doi:10.1128/EC.05274-11
77. SchoenC, ReichardU, MonodM, KratzinHD, RüchelR (2002) Molecular cloning of an extracellular aspartic proteinase from Rhizopus microsporus and evidence for its expression during infection. Med Mycol 40 : 61–71.
78. SpreerA, RüchelR, ReichardU (2006) Characterization of an extracellular subtilisin protease of Rhizopus microsporus and evidence for its expression during invasive rhinoorbital mycosis. Med Mycol 44 : 723–731 doi:10.1080/13693780600936399
79. SchallerM, BorelliC, KortingHC, HubeB (2005) Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48 : 365–377 doi:10.1111/j.1439-0507.2005.01165.x
80. ParkS-Y, ChoiJ, LimS-E, LeeG-W, ParkJ, et al. (2013) Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog 9: e1003350 doi:10.1371/journal.ppat.1003350
81. ShelestE (2008) Transcription factors in fungi. FEMS Microbiol Lett 286 : 145–151 doi:10.1111/j.1574-6968.2008.01293.x
82. EulgemT, RushtonPJ, RobatzekS, SomssichIE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 : 199–206.
83. LeeSC, NiM, LiW, ShertzC, HeitmanJ (2010) The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev 74 : 298–340 doi:10.1128/MMBR.00005-10
84. MessenguyF, DuboisE (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316 : 1–21 doi:10.1016/S0378-1119(03)00747-9
85. GoodrichJA, TjianR (2011) Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet 11 : 549–558 doi:10.1038/nrg2847
86. LeachMD, CowenLE (2013) Surviving the heat of the moment: a fungal pathogens perspective. PLoS Pathog 9: e1003163 doi:10.1371/journal.ppat.1003163
87. HaselwandterK, EbnerMR (1994) Microorganisms surviving for 5300 years. FEMS Microbiol Lett 116 : 189–193.
88. ChevreuxB, WetterT, SuhaiS (1999) Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB) 45–56.
89. SommerDD, DelcherAL, SalzbergSL, PopM (2007) Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8 : 64 doi:10.1186/1471-2105-8-64
90. KurtzS, PhillippyA, DelcherAL, SmootM, ShumwayM, et al. (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12 doi:10.1186/gb-2004-5-2-r12
91. JurkaJ, KapitonovVV, Pavliceka, KlonowskiP, KohanyO, et al. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110 : 462–467 doi:10.1159/000084979
92. KohanyO, GentlesAJ, HankusL, JurkaJ (2006) Annotation, submission and screening of repetitive elements in Repbase: Repbase Submitter and Censor. BMC Bioinformatics 7 : 474 doi:10.1186/1471-2105-7-474
93. LoweTM, EddySR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25 : 955–964.
94. LagesenK, HallinP, RødlandEA, StaerfeldtH-H, RognesT, et al. (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35 : 3100–3108 doi:10.1093/nar/gkm160
95. GardnerPP, DaubJ, TateJ, MooreBL, OsuchIH, et al. (2011) Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 39: D141–5 doi:10.1093/nar/gkq1129
96. AltschulSF, GishW, MillerW, MyersEW, LipmanDJ (1990) Basic local alignment search tool. J Mol Biol 215 : 403–410 doi:10.1016/S0022-2836(05)80360-2
97. NawrockiEP, KolbeDL, EddySR (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics 25 : 1335–1337 doi:10.1093/bioinformatics/btp157
98. GautheretD, MajorF, CedergrenR (1990) Pattern searching/alignment with RNA primary and secondary structures: an effective descriptor for tRNA. CABIOS 6 : 325–331.
99. TimothyTL, ElkanC (1994) Fitting a mixture model by exceptation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology 28–36.
100. GriebelT, BrinkmeyerM, BöckerS (2008) EPoS: a modular software framework for phylogenetic analysis. Bioinformatics 24 : 2399–2400 doi:10.1093/bioinformatics/btn364
101. KongY (2011) Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98 : 152–153 doi:10.1016/j.ygeno.2011.05.009
102. KimD, PerteaG, TrapnellC, PimentelH, KelleyR, et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 doi:10.1186/gb-2013-14-4-r36
103. WöstemeyerJ (1985) Strain-dependent variation in ribosomal DNA arrangement in Absidia glauca. Eur J Biochem 146 : 443–448.
104. RobinsonMD, McCarthyDJ, SmythGK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140 doi:10.1093/bioinformatics/btp616
105. EdgarRC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 : 113 doi:10.1186/1471-2105-5-113
106. KatohK, KumaK, TohH, MiyataT (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33 : 511–518 doi:10.1093/nar/gki198
107. LandanG, GraurD (2007) Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol 24 : 1380–1383 doi:10.1093/molbev/msm060
108. WallaceIM, O'SullivanO, HigginsDG, NotredameC (2006) M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res 34 : 1692–1699 doi:10.1093/nar/gkl091
109. Capella-GutiérrezS, Silla-MartínezJM, GabaldónT (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 : 1972–1973 doi:10.1093/bioinformatics/btp348
110. GascuelO (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14 : 685–695.
111. GuindonS, DufayardJ-F, LefortV, AnisimovaM, HordijkW, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59 : 307–321 doi:10.1093/sysbio/syq010
112. AkaikeH (1973) Information theory and extension of the maximum likelihood principle. Proceedings of the 2nd international symposium on information theory 267–281.
113. LeSQ, GascuelO (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25 : 1307–1320 doi:10.1093/molbev/msn067
114. SchmidtT, StoyeJ (2007) Gecko and GhostFam. Methods in Molecular Biology Vol. 396 165–182 doi:_10.1007/978-1-59745-515-2_12
115. NiermanWC, PainA, AndersonMJ, WortmanJR, KimHS, et al. (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438 : 1151–1156 doi:10.1038/nature04332
116. GalaganJE, CalvoSE, CuomoC, MaL-J, WortmanJR, et al. (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438 : 1105–1115 doi:10.1038/nature04341
117. KatinkaMD, DupratS, CornillotE, MéténierG, ThomaratF, et al. (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414 : 450–453 doi:10.1038/35106579
118. WoodV, GwilliamR, RajandreamMA, LyneM, LyneR, et al. (2002) Genome sequence of Schizosaccharomyces pombe. Nature 415 : 871–880.
119. CornmanRS, ChenYP, SchatzMC, StreetC, ZhaoY, et al. (2009) Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog 5: e1000466 doi:10.1371/journal.ppat.1000466
120. CuomoCA, DesjardinsCA, BakowskiMA, GoldbergJ, MaAT, et al. (2012) Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth:. 2478–2488 doi:10.1101/gr.142802.112.2478
121. DuplessisS, CuomoCA, LinY, AertsA, TisserantE, et al. (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. doi: 10.1073/pnas.1019315108
122. StajichJE, WilkeSK, AhrénD, AuCH, BirrenBW, et al. (2010) Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci U S A 107 : 11889–11894 doi:10.1073/pnas.1003391107
123. MartinezD, LarrondoLF, PutnamN, GelpkeMDS, HuangK, et al. (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22 : 695–700 doi:10.1038/nbt967
124. MartinF, Aertsa, AhrénD, Bruna, DanchinEGJ, et al. (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452 : 88–92 doi:10.1038/nature06556
125. PutnamNH, SrivastavaM, HellstenU, DirksB, ChapmanJ, et al. (2007) Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317 : 86–94 doi:10.1126/science.1139158
126. KingN, WestbrookMJ, YoungSL, KuoA, AbedinM, et al. (2008) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451 : 783–788 doi:10.1038/nature06617
127. EastwoodDC, FloudasD, BinderM, MajcherczykA, SchneiderP, et al. (2011) The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333 : 762–765 doi:10.1126/science.1205411
128. JonesonS, StajichJE, ShiuS-H, RosenblumEB (2011) Genomic transition to pathogenicity in chytrid fungi. PLoS Pathog 7: e1002338 doi:10.1371/journal.ppat.1002338
129. GalaganJE, CalvoSE, BorkovichKa, SelkerEU, ReadND, et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 : 859–868 doi:10.1038/nature01554
130. Stamatakisa, LudwigT, MeierH (2005) RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21 : 456–463 doi:10.1093/bioinformatics/bti191
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



