-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
Spinal Muscular Atrophy (SMA) is a prevalent childhood neuromuscular disease, which in its most common form causes death by the age of two. One in fifty Americans is a carrier for SMA, making this genetic disease a serious health concern. SMA is caused by loss of function mutations in the survival motor neuron 1 (SMN1) gene. SMN is an essential protein and has a well-characterized function in the assembly of small nuclear ribonucleoproteins (snRNPs), which are core components of the spliceosome. To elucidate the phenotypic consequences of disrupting specific SMN protein interactions, we have generated a series of SMA-causing point mutations, modeled in Drosophila melanogaster. Using this system, we have shown that key aspects of SMN structure and function are conserved between humans and flies. Intragenic complementation analyses reveal the potential for dominant negative interactions between wild-type and mutant SMN subunits, highlighting the essential nature of the YG box in formation of higher-order SMN multimers. These results provide a basis for future studies investigating therapy targeted at restoration of functional SMN oligomers.
Published in the journal: SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in. PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004489
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004489Summary
Spinal Muscular Atrophy (SMA) is a prevalent childhood neuromuscular disease, which in its most common form causes death by the age of two. One in fifty Americans is a carrier for SMA, making this genetic disease a serious health concern. SMA is caused by loss of function mutations in the survival motor neuron 1 (SMN1) gene. SMN is an essential protein and has a well-characterized function in the assembly of small nuclear ribonucleoproteins (snRNPs), which are core components of the spliceosome. To elucidate the phenotypic consequences of disrupting specific SMN protein interactions, we have generated a series of SMA-causing point mutations, modeled in Drosophila melanogaster. Using this system, we have shown that key aspects of SMN structure and function are conserved between humans and flies. Intragenic complementation analyses reveal the potential for dominant negative interactions between wild-type and mutant SMN subunits, highlighting the essential nature of the YG box in formation of higher-order SMN multimers. These results provide a basis for future studies investigating therapy targeted at restoration of functional SMN oligomers.
Introduction
Proximal spinal muscular atrophy (SMA) is a common neuromuscular disorder, recognized as the most prevalent genetic cause of early childhood mortality [1]. SMA is characterized by degeneration of motor neurons in the anterior horn of the lower spinal cord, and progressive symmetrical paralysis. Coupled with this loss of motor function, SMA patients display severe atrophy of the proximal muscles. The onset of symptoms and their severity can vary, leading to an historical classification of SMA into three distinct subtypes [1]. More recently, clinicians have recognized that SMA is better characterized as a continuous spectrum disorder, ranging from severe (prenatal onset) to nearly asymptomatic [2].
Almost two decades ago, mutations in the survival motor neuron 1 (SMN1) gene were shown to be causative for SMA [3]. The disease typically results from homozygous deletion of SMN1; however, a small fraction of SMA patients have lost one copy of SMN1 and the remaining copy contains a point mutation [4]. The best characterized function for the ubiquitously expressed SMN protein is in the biogenesis of Sm-class small nuclear ribonucleoproteins (snRNPs), core factors of the spliceosome [5], [6]. In addition, SMN has been implicated in numerous other cellular activities, including axonal transport, neuronal pathfinding, formation and function of neuromuscular junctions, myoblast fusion and maintenance of muscle architecture [4], [7]–[11]. Despite this multitude of putative functions attributed to SMN, or perhaps because of it, the precise pathophysiological mechanisms that give rise to SMA are the subject of considerable debate. Seemingly straightforward questions of disease pathogenesis have yet to be definitively answered. For example, is SMA caused by a cell-autonomous reduction of SMN protein levels in motor neurons [12]–[16] or is it a more systemic defect involving other cell types [17]–[24]?
Irrespective of the question of cellular autonomy, the molecular etiology of SMA also remains unclear. Is the neuromuscular dysfunction seen in SMA patients caused by a loss of Sm-class spliceosomal snRNPs, ultimately leading to defects in pre-mRNA splicing? Or is it due to some non-canonical function of SMN and/or snRNPs? Experiments using animal models of severe SMA suggest that the pre-mRNA splicing defects observed in late-stage SMA animals are indeed tissue-specific [25], [26]. However, such deficits are only detectable later in the disease course, after the onset of neuromuscular dysfunction [27]–[29]. Complicating matters, SMN-deficient animals are developmentally delayed or arrested [22], [29]–[34], making the selection of properly staged control animals critical to the comparison of phenotypes. Moreover, alternative splicing of the SMN gene duplicate (SMN2) in humans or mouse models is another variable, creating a feedback loop [35], [36] that can negatively regulate SMN expression. In the fruitfly, the vast majority of Smn pre-mRNA transcripts are intronless (Flybase; [37]). Therefore, we set out to create an allelic series of Drosophila SMA models wherein we could specifically focus on SMN protein function, in the absence of other complicating factors.
Results and Discussion
Generation of Drosophila models of SMA patient mutations
To identify which functions of SMN are critical to the pathology of SMA, we aimed to create disease-relevant models that disrupt subsets of SMN interactions. Point mutations are useful in this context, as they can disrupt specific functions of multi-domain proteins, leaving other functions unaffected. To date, twenty-five different SMN1 point mutations have been identified in SMA patients [4]. Many of these mutations are located at residues that are conserved between humans and insects (Fig. 1A). In this report, transgenic Drosophila bearing twelve of these SMA-causing point mutations were generated (Fig. 1B). The salient features of the Smn transgenic cassette have been previously described [28], including a 3X FLAG tag that was inserted immediately downstream of the ATG start codon. The transgenes were also expressed under control of the native Smn promoter and 3′ flanking sequences [28] and inserted at a landing site located within band 86Fb on chromosome 3R using the PhiC31 integrase system [38]. Note that all of the constructs (including an SmnWT control) were injected directly into embryos heterozygous for the SmnX7 microdeletion, a null mutation [39] that was recombined with the appropriate PhiC31 landing site prior to injection (see Methods for details). Note that the SmnX7 deletion removes the promoter region and the entire Smn open reading frame, leaving behind only 44 bp of the 3′ UTR [39]. Thus, this mutant produces no Smn mRNA.
Fig. 1. An allelic series of SMN missense mutations in Drosophila. 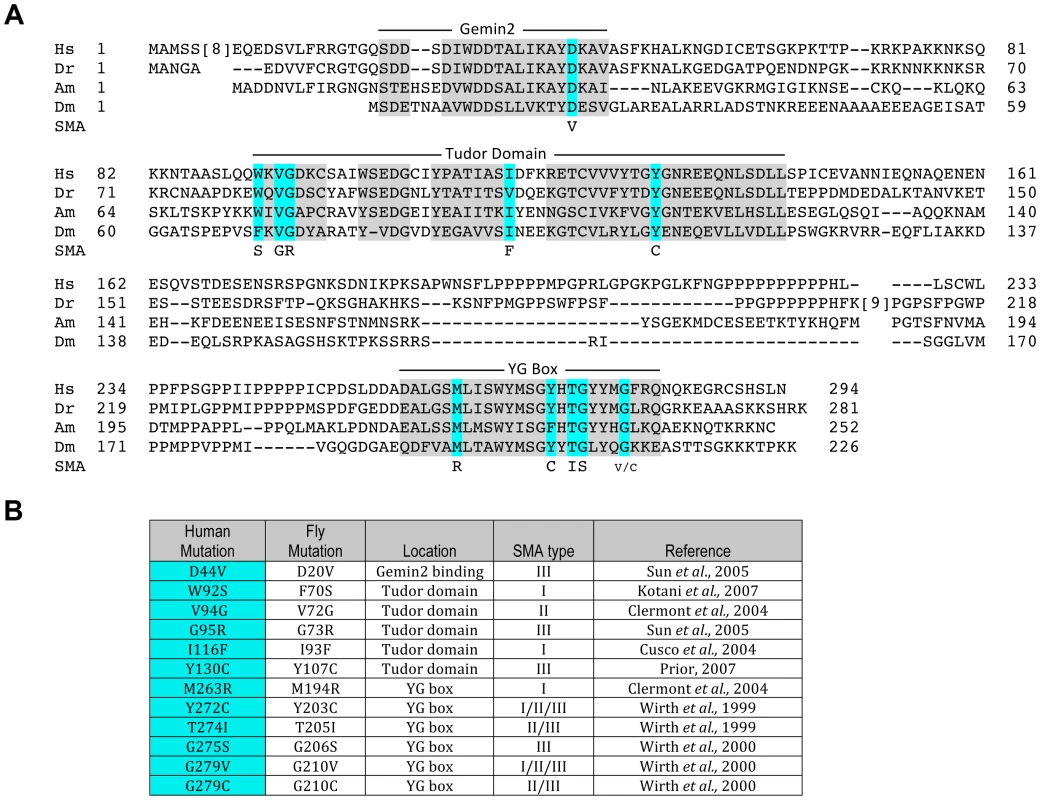
(A) Alignment of SMN orthologues from Homo sapiens (Hs), Danio Rerio (Dr), Apis Mellifera (Am), and Drosophila melanogaster (Dm). Conserved regions, including the Gemin2 binding domain, the Tudor domain and the YG box of SMN, are shaded in gray. Residues known to be mutated in SMA patients are highlighted in aqua. SMA-causing missense residues (SMA) are listed below the highlighted residues. (B) Summary of the human SMA patient mutations that were modeled in Drosophila. These mutations affect all major domains of SMN, and encompass all three classes of SMA severity, with Type I being the most severe and Type III being the mildest form. When multiple SMA types are listed for a given mutation, this indicates that differing phenotypic severities were observed in patients bearing the same mutation. As shown in Fig. 1, one of the mutations lies in the region responsible for Gemin2 binding, five are located in the Tudor domain, and six in the YG box oligomerization domain of SMN, thus mirroring the distribution of point mutations identified in SMA patients. For phenotypic analysis of the point mutants in our model, we crossed the transgenic flies (SmnX7, Flag-SmnTg) with an SmnX7 null allele to obtain flies that are homozygous null for endogenous Smn and hemizygous for the Flag-Smn transgene. We adopted this approach because human SMA patients that express SMN1 missense mutations are also typically hemizygous and because we observed an improvement in the viability of the transgenic mutants when crossed to SmnX7, as compared to the self-cross. This latter finding indicates the presence of recessive alleles in the transgenic background that contribute to the phenotype of the homozygotes.
SMA patient-derived mutations show a range of phenotypic severities in Drosophila
When expressed in an SmnX7/X7 null background, the hemizygous Flag-SmnWT construct showed robust rescue of adult viability (∼67%, see Fig. 2A), consistent with our previous findings using an SmnX7/D null background [28]. The degree of rescue achieved by expressing each of the different SMA point mutations varied (Fig. 2A). Three of the transgenes (SmnM194R, SmnY203C and SmnG206S) failed to rescue the larval lethality of the null animals (Fig. 2A). In contrast, a second group of point mutants rescued adult viability to a degree roughly similar to that of the SmnWT transgene, including: SmnD20V, SmnF70S and SmnG73R and SmnI93F. A third group of mutants can be characterized as having intermediate phenotypes: SmnV72G, SmnY107C, SmnT205I, SmnG210C and SmnG210V. This last category can be categorized as pupal-lethal, as the majority of these animals die prior to eclosion. The SmnG210C mutation produced a much milder phenotype (∼45% rescue to adulthood) compared to the SmnG210V mutation (∼5% rescue) despite the fact that they are located at the same residue. Interestingly, SmnV72G was the only mutation that rescued the larval lethality of the null animals but then also resulted in complete pupal lethality (Fig. 2A). Thus, when expressed in Drosophila, disease-causing Smn point mutations display a range in the age of symptomatic onset and in life-expectancy.
Fig. 2. SMA patient-derived mutations show a range of life expectancies in Drosophila. 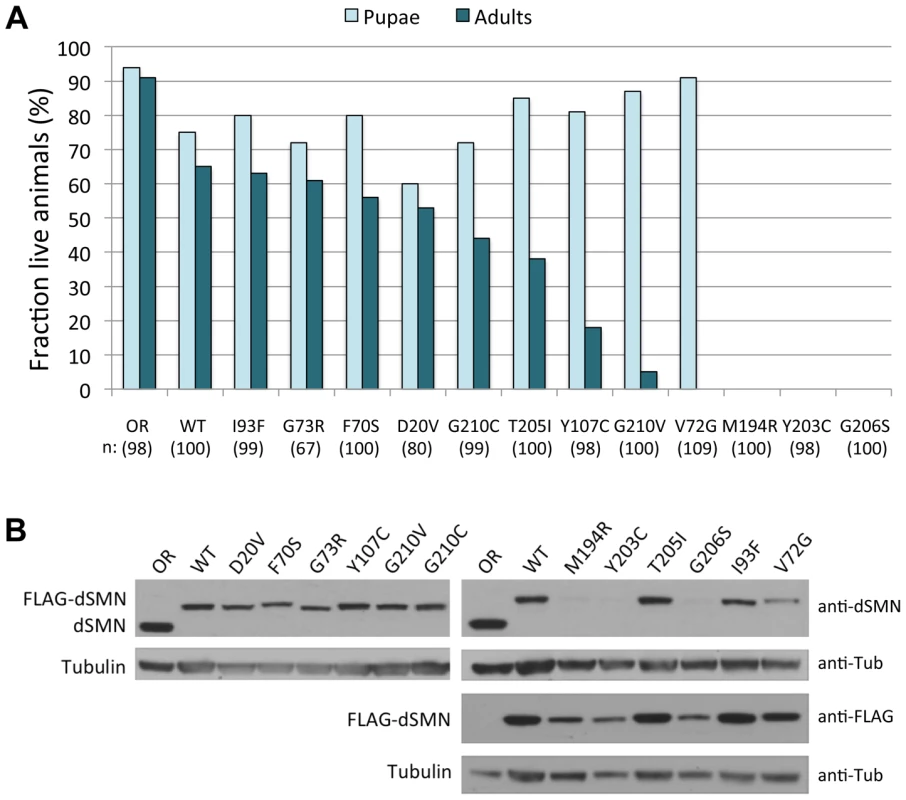
(A) Viability analyses of SMA point mutations in Drosophila. SmnX7/TM6.tb GFP flies were crossed with each mutant line (SmnX7,SmnTg/TM6.tb GFP) and hemizygous first instar mutant larvae (SmnX7/X7, SmnTg/− where Tg stands for transgene) were collected and followed through development. Oregon-R (OR) animals served as controls. The data for each genotype are expressed as a fraction of pupae or adults over the total number of starting larvae (n), listed below each genotype. Expression of the WT transgene (SmnX7/X7,SmnWT/−) shows robust rescue of the null phenotype (∼67% adults). The mutants show a range in severity, from very severe (larval lethal), to intermediate (pupal lethal), to mild (adults). (B) Expression levels of Smn transgenes were examined by western blotting. Larval lysates from homozygous mutant lines (SmnX7/X7,SmnTg/Tg), were probed with anti-dSMN or anti-FLAG antibodies, as indicated. The slower migrating bands represent the FLAG-tagged transgenic proteins and the faster migrating band corresponds to endogenous dSMN, present only in the OR controls. The mutant transgenes show similar levels of dSMN protein compared to SmnWT animals, with the exception of SmnM194R, SmnY203C and SmnG206S, which show a significant reduction. Tubulin was used as a loading control. As shown previously [28], the amount of dSMN protein expressed from transgenes driven by the native Smn promoter and integrated at site 86Fb is several-fold lower than that of the endogenous gene (Fig. 2B). However, we observed robust rescue of the null phenotype in the presence of the SmnWT transgene, as these animals are both viable and fertile. A majority of the SMA alleles expressed equivalent levels of transgenic protein when compared to SmnWT. The exceptions were SmnM194R, SmnY203C and SmnG206S, which showed a significant reduction in dSMN levels (Fig. 2B). Because the constructs were all inserted into the identical genomic location, and this same level of expression was observed in multiple independent transformants (Fig. S1), the reduction is most likely due to instability of the mutant proteins rather than differences in mRNA transcription. Furthermore, the very low levels of dSMN protein in the SmnM194R, SmnY203C and SmnG206S animals are consistent with their severe, larval-lethal phenotypes.
SMA-causing mutations are biochemically similar in human and Drosophila
The ability of SMN to interact with itself [40], [41] is important for its function in RNP assembly [42], and mutations that disrupt SMN self-oligomerization are unstable in vivo [43]. We have shown previously that dSMN(Y203C) and dSMN(G206S) proteins are oligomerization defective [28]. We therefore tested dSMN(M194R) for its ability to self-interact. The mutant protein was N-terminally tagged with a 3X FLAG tag to distinguish it from the endogenous dSMN, and co-transfected into S2 cells (a Drosophila embryonic cell line) along with either a Myc-tagged dSMN(M194R) construct or a wild-type control. Immunoprecipitations were performed with anti-FLAG antibodies and the amount of co-precipitating protein was visualized by western blotting. As shown in Fig. 3A, dSMN(M194R) protein is also severely defective in its ability to self-oligomerize. Thus, the low levels of dSMN in SmnM194R, SmnY203C and SmnG206S animals can be explained by the inability of these proteins to form higher-order SMN complexes. These observations are consistent with findings that the equivalent mutations in human SMN (SMNM263R, SMNY272C and SMNG275S) are defective in self-oligomerization in vitro [40], [44], suggesting a high level of conservation between the structure-function relationships of Drosophila and human SMN.
Fig. 3. Interaction of SMN mutants with proteins involved in RNP assembly. 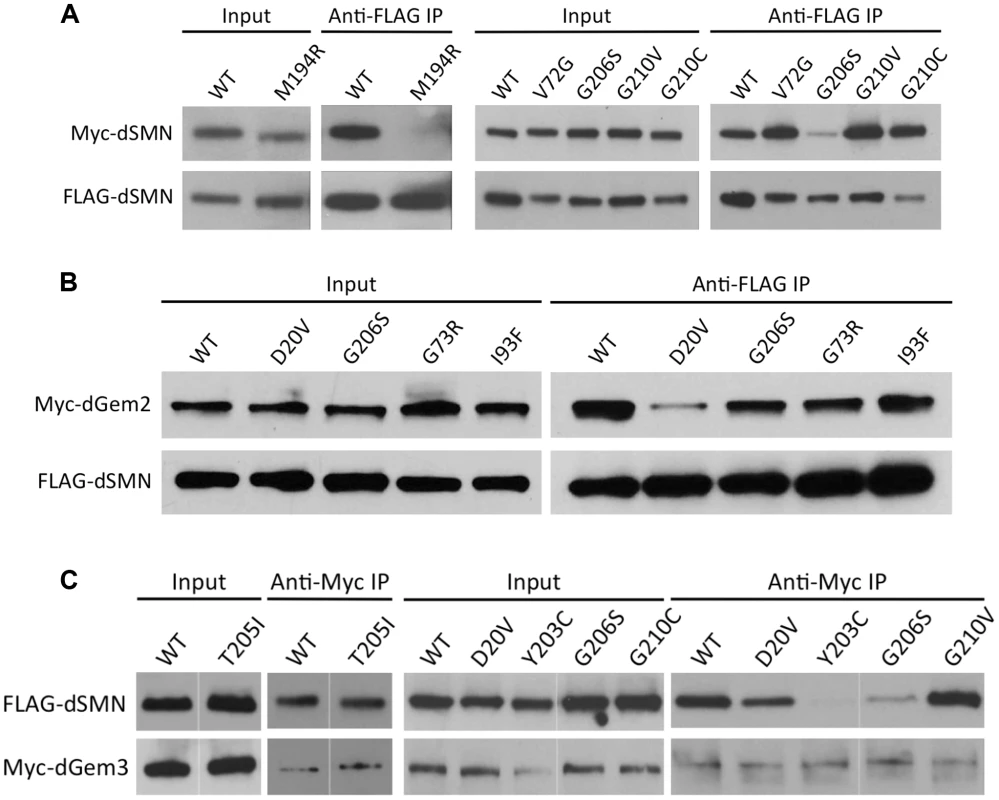
(A) SmnM194R and SmnG206S are defective in a self-oligomerization assay. Note that oligomerization of SmnY203C and SmnT205I was tested previously [28]. Lysates were prepared from cells co-expressing FLAG- and Myc-tagged versions of dSMN mutants or WT controls. Co-immunoprecipitation was performed with anti-FLAG antibody followed by western analysis with anti-Myc antibody to visualize the amount of Myc-tagged dSMN that co-precipitated with the FLAG-tagged protein. (B) dSMN(D20V) protein shows defective binding to dGem2. Immunoprecipitation was performed with anti-FLAG antibody followed by western analysis with anti-Myc antibody to visualize the amount of Myc-tagged dGem2. (C) dSMN(Y203C) and dSMN(G206S) are defective in their ability to bind dGem3. Immunoprecipitation of Myc-dGem3 was followed by probing with anti-FLAG antibody to visualize Flag-dSMN. Among the proteins that comprise the SMN complex in humans, collectively known as ‘Gemins,’ orthologues for Gemins2, 3, and 5 are the only genes that have been identified and validated in Drosophila. We and others [33], [45] have thus far failed to detect a biochemically stable interaction between dSMN and dGemin5 (dGem5), although dGem5 reportedly co-localizes with dSMN in various subcellular organelles [46]. To further establish whether dSMN can recapitulate the biochemical characteristics of SMA patient mutations, and to understand how these characteristics correlate with animal phenotypes, we analyzed the mutant dSMN proteins for their ability to bind dGem2 and dGem3 using a co-transfection assay.
The binding of dGem2 was reduced by a single Smn point mutation. As shown in Fig. 3B, dGem2 binding to dSMN(D20V) was reduced as compared to wild-type, but not eliminated. None of the other dSMN mutants showed any decrease in binding to dGem2 (Fig. 3B). This was not a surprising finding, as residue D20 lies within the known Gemin2 binding domain of SMN (Fig. 1A), and is consistent with experiments on its human counterpart, SMN(D44V) [47], [48]. Interestingly, SmnD20V flies display a mild phenotype, suggesting that the observed residual interaction with dGem2 is sufficient for its function in vivo. Indeed, SMA patients with the D44V mutation have been diagnosed with the mild form of SMA, type III [49].
We also analyzed the interaction of various Flag-tagged dSMN mutants with Myc-dGem3 by co-immunoprecipitation with anti-Myc antibodies. As shown in Fig. 3C, we found that binding of dGem3 was severely disrupted by two point mutations, SmnY203C (SMNY272C in humans) and SmnG206S (SMNG275S). Consistent with our findings, the human SMNY272C mutation was previously shown to disrupt Gemin3 binding in vitro [40]. Notably, we found that the SmnT205I (SMNT274I) mutation, which lies between residues Y203 and G206, did not affect dSMN's ability to interact with dGem3 (Fig. 3C). This finding is consistent with previous observations that SMNT274I is active in snRNP assembly, an activity that also requires Gemin3 [42]. Strikingly, the phenotype of the SmnT205I animals was much less severe than that of either the SmnY203C or SmnG206S mutants (Fig. 2A). Given that dSMN(T205I) and dSMN(G206S) proteins are both moderately defective in self-oligomerization ([28] and Fig. 3A), we attribute the more severe phenotype of the SmnG206S mutants to the inability of dSMN(G206S) to interact with dGem3. This interpretation is bolstered by the fact that null mutations in Gemin3 were shown to destabilize dSMN in vivo [33]. In summary, these results further demonstrate that important biochemical properties of SMN are conserved between Drosophila and humans [24], [28], [45].
Smn missense mutations display incomplete dominance
To further characterize the Smn point mutants, we crossed each of them to the wild-type (WT) rescue line, SmnWT. Since SMA patients display a recessive mode of inheritance, we expected that the SmnWT transgene would rescue organismal viability in the missense mutant backgrounds (genotype: SmnX7/X7,Flag-SmnWT/Mut) to roughly the same extent as it did in the hemizygous rescue line (SmnX7/X7,Flag-SmnWT/−). However, for many mutations we observed an intermediate level of rescue, between that observed when expressing the wild-type transgene alone versus that of the mutant transgenes alone (Fig. 4A). This dominant phenotype was most evident when the wild-type transgene was expressed in combination with the three most severe YG box alleles (M194R, Y203C and G206S). For example SmnX7/X7,Flag-SmnWT/Y203C animals displayed a much lower eclosion frequency (Fig. 4A) than did either of the two controls, SmnX7/X7,Flag-SmnWT/− (hemizygotes) or SmnX7/X7,Flag-SmnWT/WT (homozygotes). Importantly, expression of the wild-type Smn transgene in combination with the M194R, Y203C or G206S point mutants rescued the larval lethality associated with expression these alleles on their own (Fig. 4A). Thus, these dominant phenotypes are intermediate in their severity.
Fig. 4. Incomplete dominance of Smn mutants. 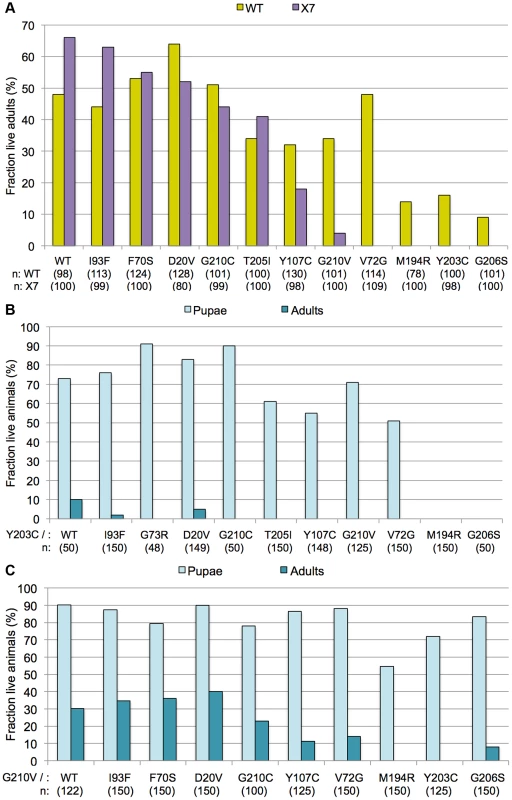
(A) Each of the Smn point mutant lines (SmnX7,SmnTg/TM6-tb.GFP) was crossed to the wild-type (WT) rescue line (SmnWT,SmnWT/TM6-tb.GFP). For most mutations, this resulted in an intermediate level of rescue, between that observed when expressing the wild-type transgene alone versus that of the mutant transgenes alone. SmnX7/X7,SmnTg/WT flies (WT) are shown in yellow and SmnX7/X7,SmnTg/− flies (X7) are shown in purple. Genotypes of the various transgenes are listed along the X-axis. Larval progeny were followed through development and the number of eclosing adult flies was measured and expressed as a fraction of the total number of starting animals (n) for each genotype. (B) The SmnX7,SmnY203C mutants were crossed with each of the other transgenic lines, both mutant (SmnX7,SmnTg) and wild-type (SmnX7,SmnWT). Larval progeny were followed through development and the number of pupae and adults was measured and expressed as a fraction of the total number of starting animals (n) for each genotype. (C) The SmnX7,SmnG210V mutants were crossed to the other alleles, as described in panel B. We note that SmnWT/WT homozygotes (obtained by an intercross of two different founder lines) showed a somewhat lower eclosion frequency (∼50%) than did the hemizygotes (∼70%). These findings suggest that second-site recessive alleles may contribute to the decreased fitness of the homozygotes and that outcrossing to a ‘clean’ SmnX7 chromosome ameliorates these effects. Moreover, the presence of second site recessives may also affect the results of the intragenic complementation analyses (Fig. 4A), as the transgenes were all inserted into the same genetic background. However, when co-expressed with SmnWT, the phenotypic severities observed for the point mutant lines correlated with the degree of rescue when expressed alone. For example, SmnG210V had a less severe phenotype (pupal lethal) than SmnG206S (larval lethal), when expressed alone. When co-expressed with the wild-type transgene, SmnG210V/WT flies also showed a milder phenotype than did SmnG206S/WT (Fig. 4A). There was one exception to this rule; SmnV72G displayed an early pupal-lethal phenotype when expressed alone, but had an eclosion frequency comparable to that of the mild mutations when co-expressed with SmnWT. In summary, complementation crosses with the mutant and wild-type Smn transgene display intermediate phenotypes.
Using SmnY203C and SmnG210V as test cases, we carried out additional complementation crosses to each of the other point mutant lines. As shown in Fig. 4B, co-expression of SmnY203C with the other transgenes had a negative effect on adult viability. When crossed to Y203C, alleles that showed strong rescue of the null phenotype when expressed alone (e.g. WT, D20V, G73R, or I93F) either completely failed to eclose as adults or did so at very low frequencies. However, this dominant effect was not fully penetrant, as a majority of the trans-heterozygous animals developed beyond larval stages and died as pupae (Fig. 4B). The only exceptions were crosses between Y203C and the other two severe mutations (M194R and G206S), which continued to die as larvae. SmnG210V co-expression with the other transgenes had a positive effect on pupation of flies expressing the severe mutations (Fig. 4C). Overall, when crossed to G210V, adult viability decreased. Crosses between G210V and either V72G or G206S were the only exceptions, with a small fraction of flies eclosing. This is in contrast to the pupal lethality observed when V72G was expressed alone and the larval lethality of G206S. Thus the intermediate phenotypes observed in these complementation crosses provide a clear example of the genetic principle of incomplete dominance.
Severe YG box mutants retain the ability to interact with wild-type SMN and display a more pronounced phenotype than Smn null animals
The most severe Smn point mutations in our collection (M194R, Y203C and G206S) all map to the YG box self-oligomerization domain (Fig. 1A), which is the most well-conserved region of SMN. Consistent with their YG box location, these three alleles are defective in self-interaction assays (Fig. 3A) and do not form stable SMN complexes in vivo (Fig. 2B). By such criteria, these three alleles would normally be considered to be protein nulls, yet their phenotype is actually more severe than that of the null allele, SmnX7/X7. Indeed, these larvae all died by ∼8–9 days after hatching, whereas ∼20% of the null mutants were still alive at this time point (Fig. 5A). The M194R, Y203C and G206S larvae are similar in size to Smn null mutants and are noticeably smaller and less active their SmnWT counterparts (Figs. 5B and 5C). These latter observations are consistent with previous analyses of Smn null mutants [19], [31], [33]. The long-lived larval phenotype displayed by the Smn null animals [28], [33], but not by the severe point mutants (Fig. 5A), suggests that zygotic expression of the oligomerization-defective point mutations inhibits the function of the endogenous Gemins and/or the maternally contributed (wild-type) dSMN.
Fig. 5. Phenotypic characterization of three severe YG box mutants. 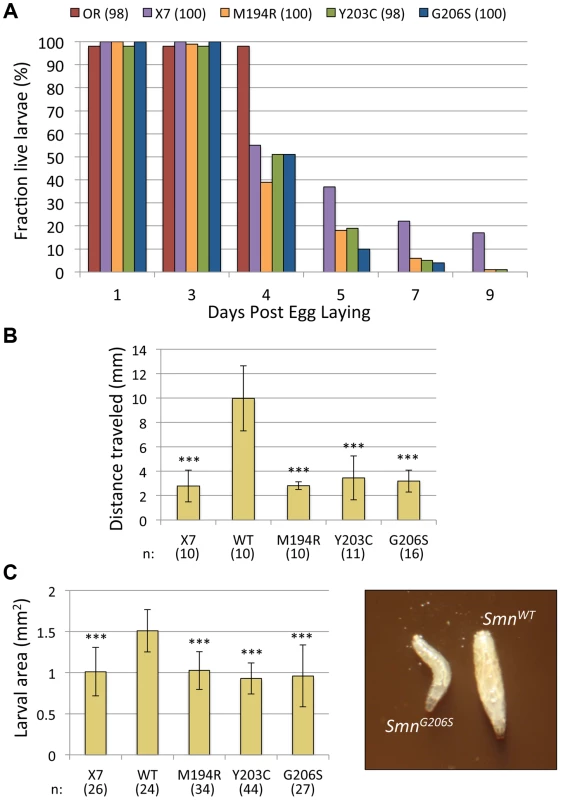
(A) Viability of larvae hemizygous for the SmnM194R, SmnG206S and SmnY203C mutations was assayed. The graph tracks survival of approximately100 larvae (n) for each genotype (SmnX7/X7,SmnTg/−), along with homozygous null mutant (SmnX7/SmnX7) and Oregon-R (OR) controls, over time. All three missense mutants die earlier than the null animals, suggesting a dominant negative effect. Note that by day 5 all of the wild-type control larvae have pupated. (B) Graph of average distance traveled over 20 s after stimulation with a needle (n>10 larvae were scored for each genotype). Larval movement was impaired for M194R, Y203C and G206S larvae, relative to WT transgene controls (P<1.3×10−5). (C) Graph of overall larval size, as measured by area (in mm2). For each genotype, n>24 larvae were scored (P<1.8×10−7). For illustration, an image of two representative larvae (SmnWT and SmnG206S) is shown. Despite the fact that the severe YG box mutants are unstable and fail to self-interact, we reasoned that the dominant negative phenotype seen upon co-expression of wild-type and mutant dSMN suggests that these two proteins might form heterodimers. We tested this idea by co-transfecting differentially tagged wild-type and mutant dSMN constructs and measuring the amount of co-precipitated protein. We assayed Y203C, G206S, G210V (Fig. 6A) along with a number of other mutant Flag-dSMN constructs, and found that they were able to pull down wild-type Myc-dSMN. Consistent with these findings, Martin et al. [44] recently showed that human SMN YG box mutants M263R, Y272C and G275S (corresponding to M194R, Y203C and G206S in flies) were completely unable to form dimers or higher-order multimers in vitro, as measured by multi-angle light scattering (SEC-MALS). In contrast, a relatively mild YG box mutation, T274I (T205I in flies), was only partially defective in self-interaction [28], [40], [44], [50]. We conclude that the dominant negative interaction of Smn alleles is not limited to zygotically expressed protein, but extends to the maternal contribution as well. Moreover, these findings show that important structural and functional features of the SMN YG box are conserved between vertebrates and invertebrates.
Fig. 6. Severe YG box mutants retain the ability to interact with wild-type SMN and overexpression of WT Flag-dSMN rescues their dominant negative effect. 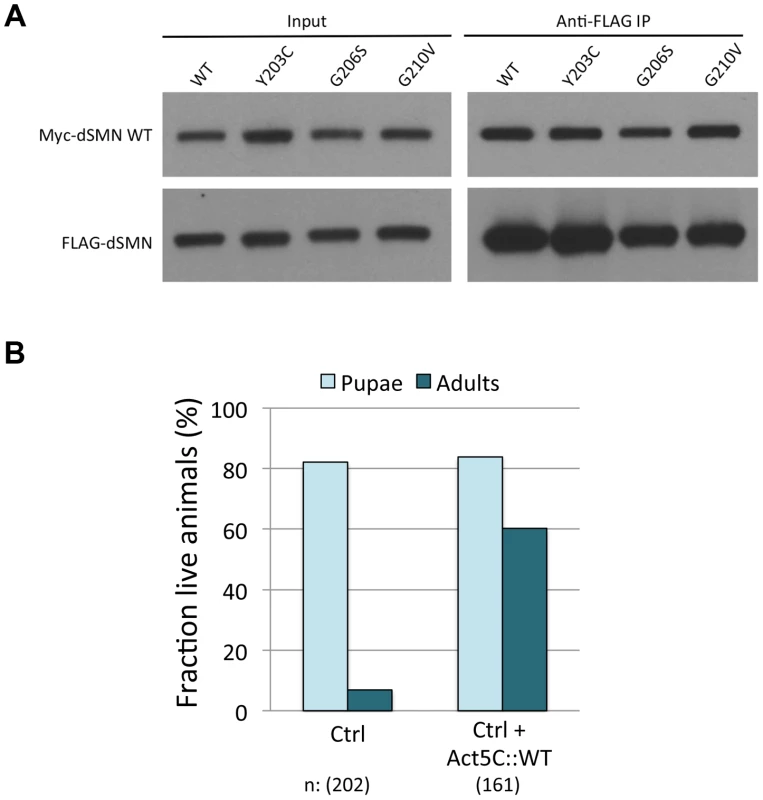
(A) Self-oligomerization defective YG box mutants are still able to interact with WT dSMN. Lysate from cells co-transfected with Myc tagged WT Smn and FLAG tagged mutant protein were immunoprecipitated with anti-FLAG antibody. The amount of co-precipitating Myc-WT dSMN was visualized by western blotting. A variety of FLAG-tagged dSMN mutants including Y203C, G206S and G210V were able to pull down Myc-WT dSMN. (B) The wild-type dSMN protein was overexpressed in an SmnY203C/WT background using a UAS::Flag-SmnWT transgene driven by Actin-5C::GAL4. Control larvae (UAS::SmnWT/CyoGFP; SmnX7,SmnWT/SmnX7,SmnY203C, labeled Ctrl) and overexpression larvae (UAS::SmnWT/Actin5C::Gal4; SmnX7,SmnWT/SmnX7,SmnY203C, labeled Ctrl+Act5C::WT) were followed over development and the numbers of pupae and adults were measured. Overexpression of dSMN ameliorates the dominant negative effect of the SmnY203C transgene by increasing the number of eclosing adults from ∼6% to ∼60%. Overexpression of Flag-SmnWT suppresses SMN dominant negative interactions
It is important to note that the dominant effect of expressing the M194R, Y203C and G206S transgenes was not observed when these alleles were carried over a balancer or a wild-type third chromosome. Smn transgenes integrated at the 86Fb landing site express relatively low levels of Flag-dSMN, compared to the endogenous gene located at 73A9 (Fig. 2B). One interpretation of these results is that high levels of wild-type dSMN expression from the endogenous gene can squelch the activity of the mutant transgenic protein. Alternatively, the observed decrease in viability of the heteroallelic Smn combinations could be due to second site recessive mutations, as the transgenes were all inserted into the same genetic background. To explicitly address this question, we over-expressed wild-type Flag-dSMN from a transgene located on the second chromosome, using the UAS-Gal4 system [51]. Next, we measured eclosion frequencies of UAS-Flag-Smn,act5C-Gal4; SmnX7/X7,Flag-SmnWT/Y203C transgenic rescue animals versus those that lack the act5C-Gal4 driver. As shown in Fig. 6B, overexpression of WT Flag-dSMN increased the eclosion frequency from ∼6% to ∼60%. Equivalent results were obtained with a tubulin-Gal4 driver. Thus, we can fully rescue the dominant negative effect of the heteroallelic SmnY203C/WT transgenes by overexpressing WT dSMN, demonstrating that the decrease in viability of these animals is not due to second site recessives. We therefore conclude that severe Smn YG box mutations can act in a dominant fashion, given that our transgenic alleles are expressed at relatively low (but equivalent) levels, the observed dominant behavior of the severe YG box mutants could also be viewed as creating a haploinsufficient condition, titrating away wildtype SMN monomers and/or Gemin components. In either event, the ‘squelching’ experiment (Fig. 6B) is important because it suggests that overexpression of full-length SMN protein (e.g. from SMN2) may be an effective therapy for SMA patients bearing severe SMN1 point mutations.
Structural basis for the YG box dominant negative phenotype
Within the SMN YG box dimer [44], conserved tyrosine and glycine residues are thought to form a network of intersubunit interactions, wherein each tyrosine side chain packs against the main chain atoms of the of the i+3 glycine on the opposing helix. As illustrated in Fig. 7, molecular modeling of Drosophila SMN dimers reveals that the Y203C mutation is predicted to disrupt this intermolecular Y-G contact. Wild-type homodimers can form two such interactions, whereas Y203C:WT heterodimers can form only one of them; Y203C homodimers are incapable of making these contacts (Fig. 7). Similarly, mutation of G206 to a bulkier serine is predicted to have a disruptive effect. Most important, the Y203-G206 contact occurs precisely at the point at which the two helices cross, so this interaction is expected to be most critical for stability of the SMN dimer.
Fig. 7. SMNY203C is predicted to disrupt an intermolecular Y-G contact, weakening or preventing oligomerization. 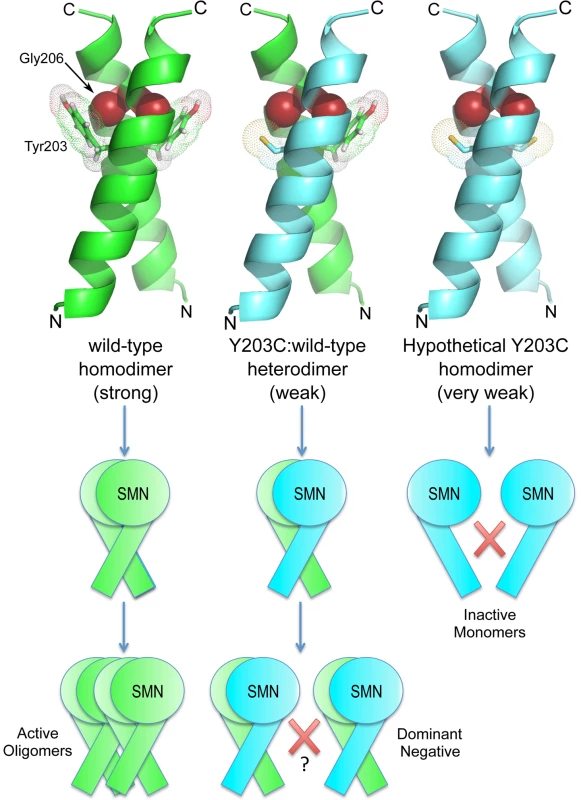
A combination of experimental analysis and structural modeling of Drosophila SMN dimers reveals that wild-type homodimers (left column) can make two Y-G contacts and are able to form higher-order structures (active SMN oligomers). In contrast, Y203C homodimers (right) are unable to make these contacts and do not dimerize (inactive SMN monomers). Interestingly, Y203C:WT heterodimers (middle) make only one Y-G contact but the loss of one interaction does not prevent dimerization. These findings suggest that the dominant negative activity of Y203C is due to an inability of the heterodimers to form active, higher-order oligomers. Interestingly, the highly conserved threonine residue present within the 203YxTG206 motif does not participate directly in the dimer interface [44]. Consistent with this observation, we found that the T205I mutant interacts with dGem3, whereas the Y203C and G206S constructs do not. These results suggest that dGem3 interacts with a multimeric form of SMN.
Because the human SMN crystal structure [44] does not contain any information regarding residues that are located upstream of M263 (M194 in the fruitfly), we are currently unable to create an accurate molecular model of the M194R mutation. However, it is interesting to note that constructs containing the proximal N-terminal region of human SMN (i.e. residues 252–294 instead of 263–294) have a strong tendency to form higher-order structures, primarily tetramers and octamers [44]. Taken together with the relative instability of dSMN(M194R) protein (Fig. 2B), its inability to self-interact in cultured cells (Fig. 3A), and the severe larval-lethal phenotype of this mutation (Fig. 4), these results support the view that formation of higher-order multimers of SMN is important for its function in vivo (Fig. 7).
Using the Drosophila system to assay SMN protein function
Gaining crucial insight from simpler model organisms is a proven strategy for unraveling complicated biological questions. Using a single point mutation (SmnT205I), we recently showed that the larval locomotion and adult viability defects associated with loss of SMN can be uncoupled from the snRNP assembly function of SMN [28]. More specifically, we found that complete loss of SMN did not result in appreciable changes in the splicing of minor intron genes [28], [29], thus arguing for a non-splicing mechanism for SMA etiology. In this report, we have conducted genetic and biochemical characterization of eleven additional Smn mutants. We show that this allelic series captures a range of phenotypic severities, further suggesting a high level of conservation between the human and fruitfly SMN proteins and related pathways.
The complex organization and polymorphic nature of the two human SMN genes complicates the analysis of SMA patient phenotypes. Indeed, SMN2 copy number variation is the best known modifier of SMA, potentially masking the phenotype of SMN1 point mutants [4]. However, SMN2 copy number is not always predictive of SMA disease severity [52]. Thus, developing an animal model that is an accurate predictor of SMN protein function is an important goal.
We find good correlation between the biochemical properties of the YG box mutants and their phenotypic severities. Patients bearing point mutations in SMN1 are relatively rare, and genotypic information (SMN2 copy number) is often not available. Such is the case for the only patient reported to bear a G275S mutation (G206S in flies), who presented with a mild form of SMA but SMN2 copy number was not determined [53]. Biochemically speaking, G275S should be a severe mutation, as this mutant is unable to form oligomers [44]. Consistent with these findings, G206S is a severe mutation in the fly and fails to bind Gemin3 (this work). Similarly, four human patients bearing the T274I mutation presented with intermediate forms of SMA, yet they had only a single copy of SMN2 [53], [54]. Given that the T274I mutant was shown to be active in Sm core assembly [42], the milder human phenotype was perhaps to be expected. Concordantly, the corresponding fly mutant, T205I, also displays an intermediate phenotype ([28]; this work). The same is true for D44V (D20V), which is located in the Gemin2 binding domain, and presents with a relatively mild phenotype in both humans and flies.
In contrast, the phenotypes of the Tudor domain mutants were harder to compare. Of the five mutations tested, two of them (F70S and I93F) were not as severe as one would have predicted from the human data (W92S, [55]; I116F, [56]). Using an S2 cell co-transfection assay similar to that in Fig. 3, we screened the entire panel of Smn mutants for their ability to bind to SmD1 and found no appreciable differences. Previous results suggest that Drosophila cells are less sensitive to the methylation status of Sm proteins than are human cells [57]–[59]. Given that the Tudor domain is thought to be a methyl-binding module, perhaps Drosophila are more tolerant to mutations in this region of SMN. However, we previously identified a synthetic lethal genetic interaction between Smn and Dart5 (the fruitfly orthologue of the arginine methyltransferase, PRMT5; see [59]), so methylation of Sm proteins (or other SMN interactors) may not be completely dispensable in flies. In the absence of more quantitative biochemical assays of SMN function (particularly for the Tudor domain mutants), we are currently unable to make good correlations between genotype and phenotype. Thus, additional efforts in this area will be needed.
Additional mutations, those that are patient-derived as well as those that are predicted by ultrastructural studies, should greatly aide future investigations of the SMN YG box oligomerization motif by providing an all-important organismal readout. Moreover, additional phenotypic analyses (longevity, locomotion, flight, neuromuscular development, etc.) particularly of adult animals, should provide quantitative measures of differences between the weaker Smn alleles. Correlating this information along with proteomic and RNomic analyses of these alleles will provide important data on SMN function and SMA etiology.
Materials and Methods
Fly stocks
All stocks were cultured on molasses and agar at room temperature (24±1°C) in half-pint bottles. Oregon-R was used as the wild-type allele. The SmnX7 microdeletion allele was a gift from S. Artavanis-Tsakonis (Harvard University, Cambridge, USA). This deficiency removes the promoter and the entire SMN coding region, leaving only the final 44 bp of the 3′ UTR [39].
Larval size and locomotion
Control and mutant larvae (73–77 hrs post egg-laying) were imaged at 10× magnification with a stereo dissection microscope (Leica) at 10× magnification. Images were captured with a digital camera and larval outlines were traced. Total pixel area was calculated using ImageJ software and measurements converted to square millimeters. For larval locomotion, Smn control and mutant larvae (73–77 hours post egg-laying) were placed on 1.5% agarose molasses plates and prodded with a needle to stimulate movement. After 20 s an image was recorded (7× magnification) and the tracks were traced and total distance traveled was measured using ImageJ software. P-values were generated using a 2-tailed student's t-test, assuming unequal variance.
Rescue constructs
As described in Praveen et al. [28], a ∼3 kb fragment containing the entire Smn coding region was inserted into the pAttB vector [38]. A 3× FLAG tag was inserted immediately downstream of the dSMN start codon. Point mutations were introduced into this construct using Quickchange (Invitrogen) site-directed mutagenesis according to manufacturer's instructions. The transgenes were injected directly into embryos heterozygous for the SmnX7 microdeletion [39] that was recombined prior to injection with the 86Fb PhiC31 landing site (Bloomington Stock Center, IN, USA). The injections were performed by BestGene Inc. (Chino Hills, CA).
Tissue culture and transfections
S2 cell lines were obtained from the Drosophila Genome Resource Center (Bloomington, IL). S2 cells were maintained in Express Five SFM (Gibco) supplemented with 1% penicillin/streptomycin and 9% L-glutamine. Cells were removed from the flask using a cell scraper and passaged to maintain a density of approximately 106–107 cells/mL. S2 cells were transferred to SF900 SFM (Gibco) prior to transfection with Cellfectin II (Invitrogen). Transfections were performed according to Cellfectin II protocol in a final volume of 3 mL in a T-25 flask containing 5×106 cells that were plated one hour before transfection. The total amount of DNA used in transfections was 2.5 ug.
Immunoprecipitation and western blotting
Larval lysates were prepared by crushing animals in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40) with 1× protease inhibitor cocktail (Invitrogen) and clearing the lysate by centrifugation at 13,000 RPM for 10 min at 4°C. S2 cell lysates were prepared by suspending cells in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40) with 10% glycerol and 1× protease inhibitor cocktail (Invitrogen) and disrupting cell membranes by pulling the suspension through a 25 gauge needle (Becton Dickinson). The lysate was then cleared by centrifugation at 13,000 RPM for 5 min at 4°C. Western blotting on lysates was performed using standard protocols. Rabbit anti-dSMN serum [28] was affinity purified. For Western blotting, dilutions of 1 in 2,500 for the affinity purified anti-dSMN, 1 in 10,000 for anti-α tubulin (Sigma), 1 in 10,000 for monoclonal anti-FLAG (Sigma), 1 in 10,000 for polyclonal anti-Myc and 1 in 5000 for monoclonal anti-Myc (Santa Cruz) were used. Anti-FLAG antibody crosslinked to agarose beads (EZview Red Anti-FLAG M2 affinity gel, Sigma) or anti-Myc antibody crosslinked to agarose beads (Sigma) were used to immunoprecipitate FLAG and Myc tagged proteins from cells.
Structural modeling
A structural model of a Drosophila SMN YG-box dimer was generated by replacement of side chains in the Martin et al. [44] structure (pdb code 4GLI) with those that differ in the Drosophila SMN sequence. For each substitution, the new side chain rotamer was adjusted to be identical to that found in the human structure. No changes to the main chain conformation were made and no steric clashes were observed in the final model.
Supporting Information
Zdroje
1. PearnJ (1980) Classification of spinal muscular atrophies. Lancet 1 : 919–922.
2. TizianoFD, MelkiJ, SimardLR (2013) Solving the puzzle of spinal muscular atrophy: what are the missing pieces? Am J Med Genet A 161A: 2836–2845.
3. LefebvreS, BurglenL, ReboulletS, ClermontO, BurletP, et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80 : 155–165.
4. BurghesAH, BeattieCE (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10 : 597–609.
5. FischerU, EnglbrechtC, ChariA (2011) Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev RNA 2 : 718–731.
6. MateraAG, WangZ (2014) A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15 : 108–121.
7. ShababiM, LorsonCL, Rudnik-SchonebornSS (2014) Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J Anat 224 : 15–28.
8. GouletBB, KotharyR, ParksRJ (2013) At the “junction” of spinal muscular atrophy pathogenesis: the role of neuromuscular junction dysfunction in SMA disease progression. Curr Mol Med 13 : 1160–1174.
9. FalliniC, BassellGJ, RossollW (2012) Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res 1462 : 81–92.
10. PraveenK, WenY, GrayKM, Van DuyneGD, MateraAG (2014) Point mutations in fruitfly Survival Motor Neuron (Smn) recapitulate the full range of phenotypic severity observed in human SMA patients. PLoS Genet in revision
11. HamiltonG, GillingwaterTH (2013) Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med 19 : 40–50.
12. GolicKG (2013) RNA-guided nucleases: a new era for engineering the genomes of model and nonmodel organisms. Genetics 195 : 303–308.
13. GogliottiRG, QuinlanKA, BarlowCB, HeierCR, HeckmanCJ, et al. (2012) Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J Neurosci 32 : 3818–3829.
14. ParkGH, Maeno-HikichiY, AwanoT, LandmesserLT, MonaniUR (2010) Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci 30 : 12005–12019.
15. Paez-ColasanteX, SeabergB, MartinezTL, KongL, SumnerCJ, et al. (2013) Improvement of neuromuscular synaptic phenotypes without enhanced survival and motor function in severe spinal muscular atrophy mice selectively rescued in motor neurons. PLoS One 8: e75866.
16. FrugierT, TizianoFD, Cifuentes-DiazC, MiniouP, RoblotN, et al. (2000) Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet 9 : 849–858.
17. HuaY, SahashiK, RigoF, HungG, HorevG, et al. (2011) Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478 : 123–126.
18. MentisGZ, BlivisD, LiuW, DrobacE, CrowderME, et al. (2011) Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron 69 : 453–467.
19. ImlachWL, BeckES, ChoiBJ, LottiF, PellizzoniL, et al. (2012) SMN is required for sensory-motor circuit function in Drosophila. Cell 151 : 427–439.
20. HayhurstM, WagnerAK, CerlettiM, WagersAJ, RubinLL (2012) A cell-autonomous defect in skeletal muscle satellite cells expressing low levels of survival of motor neuron protein. Dev Biol 368 : 323–334.
21. SahashiK, LingKK, HuaY, WilkinsonJE, NomakuchiT, et al. (2013) Pathological impact of SMN2 mis-splicing in adult SMA mice. EMBO Mol Med 5 : 1586–1601.
22. BoyerJG, MurrayLM, ScottK, De RepentignyY, RenaudJM, et al. (2013) Early onset muscle weakness and disruption of muscle proteins in mouse models of spinal muscular atrophy. Skelet Muscle 3 : 24.
23. WalkerMP, RajendraTK, SaievaL, FuentesJL, PellizzoniL, et al. (2008) SMN complex localizes to the sarcomeric Z-disc and is a proteolytic target of calpain. Hum Mol Genet 17 : 3399–3410.
24. RajendraTK, GonsalvezGB, WalkerMP, ShpargelKB, SalzHK, et al. (2007) A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol 176 : 831–841.
25. ZhangZ, LottiF, DittmarK, YounisI, WanL, et al. (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133 : 585–600.
26. LottiF, ImlachWL, SaievaL, BeckES, Hao leT, et al. (2012) An SMN-dependent U12 splicing event essential for motor circuit function. Cell 151 : 440–454.
27. BaumerD, LeeS, NicholsonG, DaviesJL, ParkinsonNJ, et al. (2009) Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 5: e1000773.
28. PraveenK, WenY, MateraAG (2012) A Drosophila model of spinal muscular atrophy uncouples snRNP biogenesis functions of survival motor neuron from locomotion and viability defects. Cell Rep 1 : 624–631.
29. GarciaEL, LuZ, MeersMP, PraveenK, MateraAG (2013) Developmental arrest of Drosophila survival motor neuron (Smn) mutants accounts for differences in expression of minor intron-containing genes. RNA 19 : 1510–1516.
30. SchrankB, GotzR, GunnersenJM, UreJM, ToykaKV, et al. (1997) Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A 94 : 9920–9925.
31. ChanYB, Miguel-AliagaI, FranksC, ThomasN, TrulzschB, et al. (2003) Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet 12 : 1367–1376.
32. WinklerC, EggertC, GradlD, MeisterG, GiegerichM, et al. (2005) Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev 19 : 2320–2330.
33. ShpargelKB, PraveenK, RajendraTK, MateraAG (2009) Gemin3 is an essential gene required for larval motor function and pupation in Drosophila. Mol Biol Cell 20 : 90–101.
34. Bosch-MarceM, WeeCD, MartinezTL, LipkesCE, ChoeDW, et al. (2011) Increased IGF-1 in muscle modulates the phenotype of severe SMA mice. Hum Mol Genet 20 : 1844–1853.
35. JodelkaFM, EbertAD, DuelliDM, HastingsML (2010) A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum Mol Genet 19 : 4906–4917.
36. RuggiuM, McGovernVL, LottiF, SaievaL, LiDK, et al. (2012) A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol Cell Biol 32 : 126–138.
37. BrownJB, BoleyN, EismanR, MayGE, StoiberMH, et al. (2014) Diversity and dynamics of the Drosophila transcriptome. Nature
38. BischofJ, MaedaRK, HedigerM, KarchF, BaslerK (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 104 : 3312–3317.
39. ChangHC, DimlichDN, YokokuraT, MukherjeeA, KankelMW, et al. (2008) Modeling spinal muscular atrophy in Drosophila. PLoS One 3: e3209.
40. LorsonCL, StrasswimmerJ, YaoJM, BalejaJD, HahnenE, et al. (1998) SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet 19 : 63–66.
41. PellizzoniL, CharrouxB, DreyfussG (1999) SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A 96 : 11167–11172.
42. ShpargelKB, MateraAG (2005) Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A 102 : 17372–17377.
43. BurnettBG, MunozE, TandonA, KwonDY, SumnerCJ, et al. (2009) Regulation of SMN protein stability. Mol Cell Biol 29 : 1107–1115.
44. MartinR, GuptaK, NinanNS, PerryK, Van DuyneGD (2012) The survival motor neuron protein forms soluble glycine zipper oligomers. Structure 20 : 1929–1939.
45. KroissM, SchultzJ, WiesnerJ, ChariA, SickmannA, et al. (2008) Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc Natl Acad Sci U S A 105 : 10045–10050.
46. CauchiRJ, Sanchez-PulidoL, LiuJL (2010) Drosophila SMN complex proteins Gemin2, Gemin3, and Gemin5 are components of U bodies. Exp Cell Res 316 : 2354–2364.
47. OgawaC, UsuiK, AokiM, ItoF, ItohM, et al. (2007) Gemin2 plays an important role in stabilizing the survival of motor neuron complex. J Biol Chem 282 : 11122–11134.
48. ZhangR, SoBR, LiP, YongJ, GlisovicT, et al. (2011) Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly. Cell 146 : 384–395.
49. SunY, GrimmlerM, SchwarzerV, SchoenenF, FischerU, et al. (2005) Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum Mutat 25 : 64–71.
50. WirthB, HerzM, WetterA, MoskauS, HahnenE, et al. (1999) Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet 64 : 1340–1356.
51. BrandAH, PerrimonN (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 : 401–415.
52. YamamotoT, SatoH, LaiPS, NurputraDK, HarahapNI, et al. (2013) Intragenic mutations in SMN1 may contribute more significantly to clinical severity than SMN2 copy numbers in some spinal muscular atrophy (SMA) patients. Brain Dev
53. WirthB (2000) An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat 15 : 228–237.
54. HahnenE, SchonlingJ, Rudnik-SchonebornS, RaschkeH, ZerresK, et al. (1997) Missense mutations in exon 6 of the survival motor neuron gene in patients with spinal muscular atrophy (SMA). Hum Mol Genet 6 : 821–825.
55. KotaniT, SutomoR, SasongkoTH, SadewaAH, Gunadi, etal (2007) A novel mutation at the N-terminal of SMN Tudor domain inhibits its interaction with target proteins. J Neurol 254 : 624–630.
56. CuscoI, BarceloMJ, del RioE, BaigetM, TizzanoEF (2004) Detection of novel mutations in the SMN Tudor domain in type I SMA patients. Neurology 63 : 146–149.
57. GonsalvezGB, RajendraTK, TianL, MateraAG (2006) The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol 16 : 1077–1089.
58. GonsalvezGB, TianL, OspinaJK, BoisvertFM, LamondAI, et al. (2007) Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol 178 : 733–740.
59. GonsalvezGB, PraveenK, HicksAJ, TianL, MateraAG (2008) Sm protein methylation is dispensable for snRNP assembly in Drosophila melanogaster. RNA 14 : 878–887.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



