-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
Reprogramming into a naïve, pluripotent state during the oocyte-to-embryo transition is directed by the oocyte cytoplasm. To understand how this reprogramming is controlled, we searched for C. elegans mutants in which the activation of embryonic genome, a landmark event demarcating the switch from a germline - to embryo-specific transcription, is initiated precociously in germ cells. This screen identified a novel function for LIN-41, a member of the TRIM-NHL protein family, in preventing a premature onset of embryonic-like differentiation and teratoma formation in developing oocytes, thus ensuring a successful passage between generations. This is the first example of such a regulator in cells that are poised for embryonic development. Interestingly, the majority of molecular “roadblocks” to reprograming that have been identified so far are epigenetic regulators. However, we propose that, at least in germ cells, LIN-41-like regulators may fulfill an analogous role in the cytoplasm, which has possible implications for the generation of human pluripotent stem cells.
Published in the journal: The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in. PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004533
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004533Summary
Reprogramming into a naïve, pluripotent state during the oocyte-to-embryo transition is directed by the oocyte cytoplasm. To understand how this reprogramming is controlled, we searched for C. elegans mutants in which the activation of embryonic genome, a landmark event demarcating the switch from a germline - to embryo-specific transcription, is initiated precociously in germ cells. This screen identified a novel function for LIN-41, a member of the TRIM-NHL protein family, in preventing a premature onset of embryonic-like differentiation and teratoma formation in developing oocytes, thus ensuring a successful passage between generations. This is the first example of such a regulator in cells that are poised for embryonic development. Interestingly, the majority of molecular “roadblocks” to reprograming that have been identified so far are epigenetic regulators. However, we propose that, at least in germ cells, LIN-41-like regulators may fulfill an analogous role in the cytoplasm, which has possible implications for the generation of human pluripotent stem cells.
Introduction
There is a special relationship between germ cells and pluripotency, i.e,. the ability to adopt alternative cell fates. First, germ cells transmit the pluripotent potential to recreate all types of cells in a new individual. Second, germ cells give rise to pluripotent cell lines such as embryonic germ or carcinoma cells and oocyte cytoplasm has the capacity to reprogram somatic nuclei [1], [2]. Finally, in disease, germ cells can abnormally differentiate into diverse somatic cell types, forming teratomas. However, during normal development, the ability to differentiate into all three embryonic germ layers is restricted to the cells of the early embryo. Combined, these observations suggest that the reprogramming potential of germ cells is kept at bay by repressive mechanisms. Depletion of several chromatin modifiers, either alone or combined with an ectopic overexpression of somatic cell fate-specifying transcription factors, can induce reprogramming of C. elegans germ cells into somatic cells [3]–[5]. The loss of these factors appears to primarily impact proliferating (pre-meiotic) germ cells and affects chromatin-based regulation. In contrast, our previous work in the same animal demonstrated that a conserved RNA-binding protein, GLD-1/Quaking, prevents teratomatous differentiation of post-mitotic germ cells [6], [7]. Importantly, in gld-1 mutants, the germline-to-soma transition is accompanied by a precocious onset of embryonic (or zygotic) genome activation (EGA), suggesting a causal connection between EGA and pluripotency. In other animals, the connection between EGA and pluripotency has been also postulated based on the temporal correlation between EGA and the acquisition of a pluripotent chromatin landscape [8], [9].
These observations prompted us to examine whether new regulators of pluripotency can be identified based on a precocious onset of EGA in the germline. Here, we report the discovery of one such novel regulator of pluripotency, LIN-41/TRIM71. LIN-41 belongs to the TRIM-NHL protein family [10]. These proteins contain a TRIpartite Motif (TRIM) consisting of a RING finger domain (commonly endowing a protein with E3 ubiquitin ligase activity, for example [11]–[13]), two B-Box motifs and a coiled-coil domain. Additionally, they also carry six so-called NHL repeats (named after NCL-1, HT2A and LIN-41) and may contain a filamin domain, which have been implicated in both protein-protein and protein-RNA interactions [13]–[17]. Consistently, different molecular functions have been attributed to LIN-41-like proteins, but many questions remain open; for example, it is not clear whether all the domains function together and/or are used in a tissue context-dependent manner [11], [14], [18]–[20]. The TRIM-NHL family includes well-known regulators of self-renewal and differentiation. For example, in Drosophila melanogaster, Brat inhibits neuroblast self-renewal, cell growth and ribosome synthesis in the larval brain [21]–[24] and Mei-P26 restricts growth and proliferation in the ovarian stem cell lineage [25]. Defects in TRIM-NHL proteins have also been associated with human pathologies, for example TRIM32 has been implicated in the Bardet–Biedl Syndrome and the Limb-Girdle Muscular Dystrophy [12], [26], [27]. Recently, human LIN-41 has been shown to promote reprogramming of differentiated cells into induced pluripotent stem cells (iPSCs) [28]. Here, we demonstrate a role for LIN-41 in controlling pluripotency during development of an animal. In C. elegans, LIN-41 is a well-known component of the somatic heterochronic pathway, which temporally controls the transition from larval to adult cell fates [29], [30]. The lin-41 germline phenotype described here indicates that, by preventing the onset of embryonic events in developing oocytes, LIN-41 also ensures a successful transition between generations. However, based on our analyses on both existing and newly created LIN-41 mutations, LIN-41 appears to function in the germline and the soma via two distinct molecular mechanisms. Our study identifies the first cytoplasmic “molecular roadblock” to reprogramming in developing oocytes and we propose it to be required to delay the onset of embryonic differentiation until after fertilization.
Results
To understand how the onset of pluripotency is controlled during C. elegans development, we executed a genetic screen to identify factors that prevent EGA in the adult germline. To monitor EGA, we created a strain expressing GFP from an early embryonic promoter, vet-4 (very early transcript 4) [31], [32]. Thus, to identify novel regulators of developmental plasticity, we searched for mutants expressing the EGA-GFP in the adult germline (Figure 1A). In addition to a new allele of gld-1, this screen yielded two mutants that, in contrast to the embryo-specific EGA-GFP expression in wild-type animals, expressed EGA-GFP within the gonads (Figure 1B). Several lines of evidence suggested that the phenotype of the two mutant strains was caused by alterations in the same gene, lin-41 (Figures 1C and S1A–D). In these mutants (alleles rrr3 and rrr4, Figure 1C), the EGA-GFP expression was restricted to the proximal region of the oogenic germline (Figure 1B). Consistent with this, RNAi-mediated depletion of lin-41 resulted in a similar expression of EGA-GFP in the gonad (Figure S1C) and a transgenic construct expressing LIN-41 fully rescued the germline defects of lin-41(rrr3) animals (Figure S1D). To further examine the role of LIN-41 in controlling EGA, we verified that the endogenous vet-4 is also abnormally transcribed in lin-41(rrr3) gonads. Indeed, by in situ hybridization, we could detect vet-4 to be expressed in the proximal gonads of lin-41(rrr3), but not wild-type animals (Figure 1D). Next, to examine the extent of embryonic-like transcription in lin-41(rrr3) gonads, we monitored the levels of vet-4 and other additional early embryonic transcripts by reverse transcription and quantitative PCR (RT-qPCR) (Text S1). We found that these transcripts were expressed in mutant, but not wild-type gonads (Figure 1E), further demonstrating that, in lin-41 mutants, embryonic transcription is prematurely activated in the germline. Importantly, we detected no obvious changes in levels or expression pattern of GLD-1 in lin-41(rrr3) gonads (Figure S2), suggesting that the gonadal phenotype of lin-41 mutants is not caused by defective expression of GLD-1.
Fig. 1. LIN-41 prevents activation of embryonic transcription in the germline. 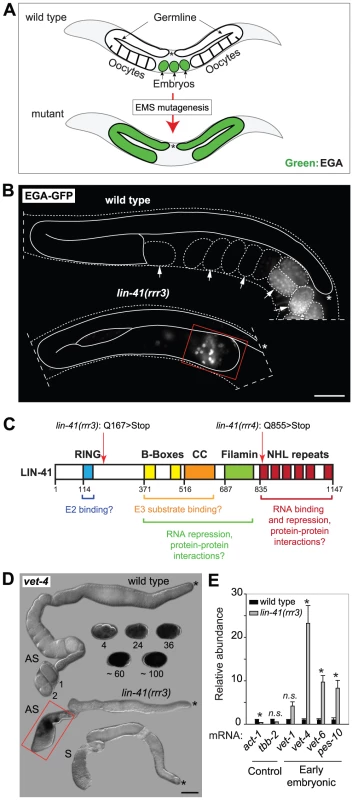
A. Summary of a genetic screen to identify mutants inducing EGA in the adult germline. In wild-type, the EGA-GFP reporter (green, marking embryonic transcription) is expressed in embryos. In mutants, this reporter is abnormally expressed in the germline. Asterisks here and in the subsequent figures mark the distal end of the gonad. B. Fluorescent micrographs of live animals expressing EGA-GFP. The gonads are outlined with a continuous line, the embryos and animals with dashed lines. Arrows point to selected embryos, the older of which express EGA-GFP. The lin-41(rrr3) mutants express EGA-GFP abnormally in the proximal gonad (boxed in red). This phenotype was observed in all examined animals (n>100). Scale bar: 25 µm. C. LIN-41 protein domains and their putative functions. Mutations identified in this study are indicated. D. In situ hybridization against an endogenous EGA transcript, vet-4. Shown are light micrographs of gonads and wild-type embryos (at the indicated stages), which were hybridized with antisense (AS) or sense (S) probes for the vet-4 mRNA. In contrast to the wild-type gonads, vet-4 mRNA was detected in the proximal region of all lin-41(rrr3) gonads (boxed in red; n = 20). Scale bar: 20 µm. E. Detection of additional EGA transcripts by RT-qPCR. “Early embryonic”: mRNAs normally expressed in the early embryo following EGA. Each bar represents the mean of three independent biological replicates, the error bars represent the standard error of the mean (SEM) and the significance of the differences has been calculated with the Student's t-test (symbols: “*”, p<0.05; “n.s.”, not significant). In wild-type animals, Pol II-dependent transcription is repressed in oocytes, which is seemingly at odds with the embryonic-like transcription in the proximal gonads of lin-41 animals. To investigate this potential discrepancy, we examined the transcription-initiating phosphorylation of serine 5 (Ser5P) within the C-terminal domain (CTD) of Pol II [33]. In contrast to wild-type gonads, Ser5P was detected in the majority of the cells in the proximal gonads of lin-41(rrr3) animals (Figure 2A), indicating ongoing Pol II-dependent transcription. Apart from EGA, the onset of embryonic development is marked by the degradation of germline mRNAs and proteins [34]. To examine this aspect of the germline-to-soma transition in lin-41 animals, we followed the expression levels of RME-2, a yolk receptor present in oocytes [35], and PGL-1, a constitutive component of germ cell-specific RNA/protein granules [36]. In contrast to wild-type animals, which express RME-2 in developing oocytes and PGL-1 throughout the germline, we found that both proteins were absent from the proximal lin-41(rrr3) gonads (Figure 2B–C), indicating that cells in this gonadal region lose germline identity. To test this further, we monitored expression of several transcripts that are normally expressed in somatic lineages. By RT-qPCR (Text S1), we found that several of these transcripts (for example the myogenic hlh-1/MyoD) were abnormally expressed in lin-41(rrr3) gonads (Figure 2D). Additionally, we examined the expression of several hox genes, which control the positional identities of cells during animal body formation [37]. While the hox transcripts were not expressed in wild-type gonads, they were strongly expressed in lin-41(rrr3) gonads (Figure 2D). Finally, we analyzed the expression of the muscle lineage markers UNC-120 and muscle myosin, the intestine lineage marker ELT-2 and a GFP reporter driven from a pan-neuronal unc-119 promoter (nGFP). We observed that lin-41(rrr3) gonads contained numerous cells expressing muscle and neuronal markers (Figures 2E and S3; 44/45 examined gonads contained cells expressing UNC-120, 10/18 cells expressing muscle myosin, and 57/57 cells expressing the nGFP). Only few gonads contained ELT-2-expressing cells (3/35 gonads and only in few cells), which might reflect a competitive advantage of some differentiation programs in the lin-41 teratoma. During embryogenesis, most body-wall muscles of an adult animal are specified by the transcription factor PAL-1/CDX [38]. The PAL-1-dependent transcription is relatively well understood and involves the activation of its direct targets, such as HLH-1/MyoD and UNC-120/SRF [39]. In wild-type oocytes, expression of PAL-1 is insufficient for the induction of its target genes (Figure S4A–B) [40]. Nevertheless, we observed that the numbers of UNC-120-expressing cells in lin-41(rrr3) gonads were significantly reduced upon pal-1 RNAi (Figure S4B). Thus, the differentiation into muscles in lin-41 gonads appears, at least partly, to mimic the pathway driving muscle formation in embryos. Together, these findings indicate that lin-41 germ cells in the proximal gonad undergo a dramatic reprogramming, which results in the acquisition of an embryonic-like state and teratomatous differentiation.
Fig. 2. Cells in the proximal regions of lin-41 gonads lose germline characteristics and differentiate into somatic cells, forming a teratoma. 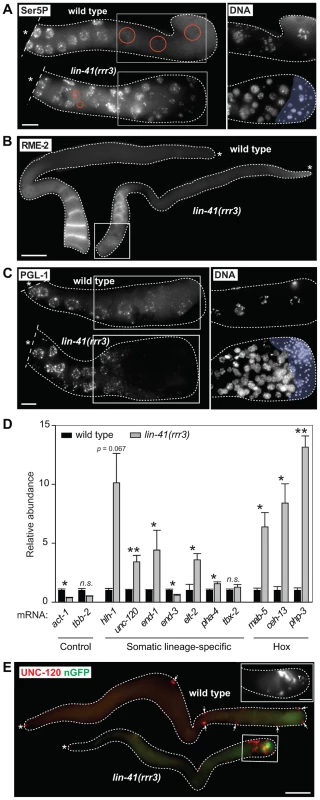
A. Fluorescent maximum intensity projections of gonads immunostained for the transcription-activating phosphorylation of Ser5 within the Pol II CTD (Ser5P). This phosphorylation was absent in the most-proximal wild-type gonads, but was present in all corresponding lin-41(rrr3) gonads (n>40). Encircled in red are nuclei containing low or no Ser5P. The corresponding DAPI-stained nuclei, from the boxed areas, are on the right. The proximal-most lin-41 gonad contains sperm (lightly colored in A and C), which explains the lack of Ser5P in this region. Scale bar: 10 µm. B. Fluorescent maximum intensity projections of gonads immunostained for an oocyte-expressed protein, the yolk receptor RME-2. In wild-type gonads, RME-2 is expressed in developing oocytes. In lin-41(rrr3) gonads, RME-2 is expressed in oocyte-like cells but is absent from the most proximal cells (boxed). Scale bar: 25 µm. C. Fluorescent maximum intensity projections of gonads immunostained for a germline-specific protein, PGL-1. PGL-1 (concentrated in RNA/protein granules) is present throughout the wild-type gonad but is eliminated in the proximal lin-41(rrr3) gonad (all gonads, n = 25). The corresponding DAPI-stained nuclei from the boxed areas are on the right. Scale bar: 10 µm. D. Detection of somatic cell-specific transcripts by RT-qPCR. “Somatic lineage-specific” indicates mRNAs expressed in somatic cell lineages: muscle (hlh-1, unc-120) or pharynx/gut (end-1, end-3, elt-2, pha-4 and tbx-2). “Hox” genes direct various aspects of somatic development. Each bar represents the mean of three independent biological replicates, the error bars represent the SEM and the significance of the differences has been calculated with the Student's t-test (“*”, p<0.05; “**”, p<0.01; “n.s.”, not significant). E. Fluorescent maximum intensity projections of gonads immunostained for the muscle-lineage marker UNC-120 and for a neuronal GFP reporter (nGFP). Arrows point to UNC-120-containing cells of the somatic gonad. In contrast to wild-type, UNC-120 and nGFP-expressing cells (boxed) are present within the lin-41(rrr3) germline. The inset shows nGFP-expressing cells from a different lin-41(rrr3) gonad, which extend long, neuronal-like processes (arrowhead). Scale bar: 50 µm. To better understand the germline-to-soma transition in lin-41 animals, we examined cells in lin-41(rrr3) gonads in a time-course experiment (Figure 3A). Until immediately after the end of spermatogenesis, the morphology and numbers of germ cells in lin-41 and wild-type gonads appeared similar. However, concomitantly with the onset of oogenesis, differences between the lin-41 and wild-type germlines began to emerge. The proximal region of wild-type gonads contained fully-grown oocytes harboring chromosomes arrested at the diakinesis stage of meiosis I. In stark contrast, the proximal region of lin-41 gonads contained oocyte-like cells that were about to divide, as evidenced by the presence of highly condensed chromosomes (marked by the phosphorylation of histone H3 on serine 10, Ser10P [41], and microtubule spindles (Figure 3A–B). Consistent with entering a mitotic cell cycle, cells in the proximal lin-41(rrr3) gonads did not express HIM-3 (Figure 3C), a synaptonemal complex component [42]. Wild-type oocytes eliminate centrosomes, presumably to ensure the correct ploidy in embryos [31], [43], [44]. In contrast, by monitoring a constitutive centrosome component, SPD-2 [45], we found that centrosomes were present in the proximal lin-41(rrr3) gonads (Figure S5). These centrosomes could duplicate (Figures 3D and S5) and were able to nucleate microtubule spindles (Figure 3D). Finally, in addition to the cell cycle markers, we monitored expression of an EGA reporter (EGA-mCherry) and the muscle-lineage marker UNC-120 and observed that their expression followed the onset of mitosis (Figure 3A). Taken together, the absence of LIN-41 leads to the elimination of germline proteins, induction of EGA, a change from the meiotic to the mitotic cell cycle and somatic-like differentiation. Thus, rather than completing oogenesis, cells in the proximal lin-41 gonads execute events that, in wild-type development, only occur during embryogenesis.
Fig. 3. LIN-41 inhibits the transition from meiosis to mitosis in developing oocytes. 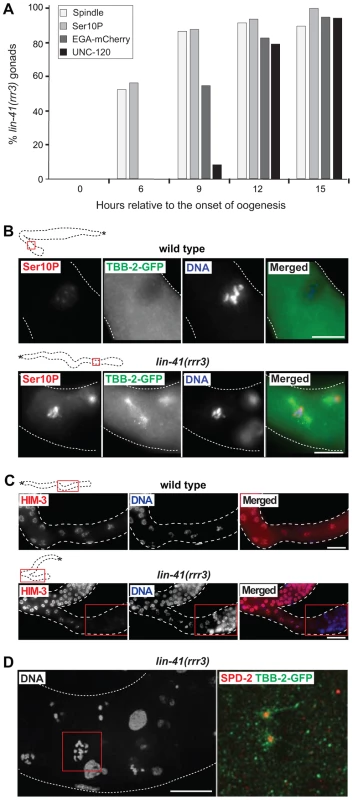
A. Time-course of EGA, mitotic chromosome condensation, spindle formation and somatic-like differentiation in lin-41(rrr3) gonads. The numbers indicate the fractions of lin-41(rrr3) gonads expressing EGA-mCherry (here the vet-4 promoter drives mCherry-tagged H2B) or assembling mitotic spindles (visualized with GFP-tagged β-tubulin; TBB-2-GFP), which was observed in live animals at the indicated time points. Additionally, these animals were immunostained for a mitotic marker (Ser10P) or UNC-120. At least 30 gonads were examined per each time-point/marker. B. Fluorescent maximum intensity projections of selected regions (boxed in red on the schematic gonads) of wild-type and lin-41(rrr3) gonads, immunostained for Ser10P and microtubule spindle (TBB-2-GFP), also stained by DAPI. Scale bars: 10 µm. C. Fluorescent maximum intensity projections of selected regions of wild-type and lin-41(rrr3) central-proximal gonads, immunostained for the meiotic marker HIM-3 and also stained by DAPI. Scale bars: 25 µm. D. Confocal images of maximum intensity projections of selected cells in the proximal lin-41(rrr3) gonad stained by DAPI, immunostained for the centrosomal component SPD-2 and for TBB-2-GFP, one day after the L4-to-adult molt. In contrast to wild-type gonads (not shown), cells in the proximal lin-41 gonads contained duplicated centrosomes (red), facilitating the assembly of microtubule spindles (green). Number of observed lin-41 cells forming a spindle with duplicated centrosomes: 60/60. Scale bar: 10 µm. In addition to the germline defects, lin-41(rrr3) animals displayed somatic abnormalities: decreased size (dumpy phenotype), appeared sick and occasionally bursted through the vulva. These phenotypes have been extensively described by Slack and colleagues and are caused, at least in part, by a precocious translation of the transcription factor LIN-29 [29]. To determine whether the gonadal phenotype reflects LIN-41 function in the germline, or it is indirectly caused by the loss of LIN-41 in the soma, we created a transgene driving lin-41 expression from a heat-shock promoter (hsp-16.41). Due to a general insensitivity of germ cells to the heat-shock promoter-driven expression [46], this transgene was not expressed in the germline but, when crossed into the lin-41(rrr3) mutant background and cultivated at an elevated temperature (24–25°C, which is apparently enough to drive sufficient expression of lin-41 in the soma), it rescued the somatic lin-41 defects (the transgenic animals no longer appeared sick or short; Figure 4A). Despite the somatic rescue, these animals still developed teratomas (Figure 4B, 50/50 examined animals), suggesting that, in controlling the germline-to-soma transition, LIN-41 functions autonomously in the germline. To examine this further, we immunostained gonads using antibodies raised against LIN-41 and found that, indeed, LIN-41 was present in the cytoplasm of germ cells starting from the late pachytene stage and culminating in the fully-grown oocytes (Figure 4C). Interestingly, LIN-41 was often absent from the most-proximal oocytes (Figures S1D and S6), suggesting a possible connection between oocyte maturation and/or ovulation and LIN-41 levels. LIN-41 expression was limited to the oogenic germline (i.e., it was absent in sperm, e.g., S1D), suggesting that the germline-to-soma transition in lin-41 gonads is caused by the loss of LIN-41 function in the developing oocytes.
Fig. 4. LIN-41 controls the germline-to-soma transition autonomously in the germline. 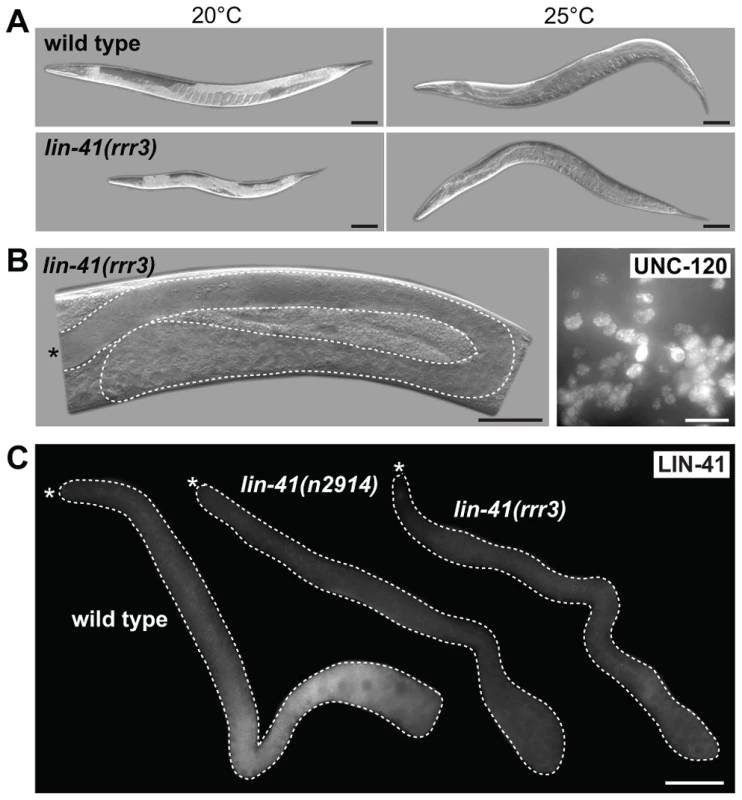
A. DIC micrographs of live animals, either wild-type (upper panels) or lin-41(rrr3) (lower panels), carrying a GFP-LIN-41 rescuing transgene driven by a heat-shock promoter (hsp-16.41). Control animals grown at 20°C are shown on the left and animals grown at 25°C (allowing leaky expression form the heat-shock promoter) on the right. Scale bars: 50 µm. B. Left panel: a DIC micrograph of a live lin-41(rrr3) animal grown at 25°C. The gonad is outlined with a dashed line. Note the absence of oocytes and the presence of a proximal germline tumor. Scale bar: 50 µm. Right panel: fluorescent maximum intensity projections of a small area from a proximal gonad immunostained for the muscle-lineage marker UNC-120, indicating teratoma formation in lin-41(rrr3) animals grown at 25°C. Scale bar: 10 µm. C. Gonads of the indicated genotypes immunostained for LIN-41. Scale bar: 50 µm. In the soma, LIN-41 is thought to associate with and repress the mRNA encoding a transcription factor, LIN-29, and LIN-29 depletion suppresses the somatic defects of lin-41 mutants [29]. In contrast to these observations in the soma, lin-29 mRNA appears to be either poorly or not at all expressed in the germline (our unpublished results and [47]). Consistently, we found that RNAi-mediated depletion of LIN-29 did not suppress the germline defects of lin-41(rrr3) mutants (though, it suppressed the somatic defects, as expected [29]). We obtained similar results in lin-41; lin-29 double mutants (Figure S7). Thus, LIN-41 may function in the germline and soma via distinct targets and/or mechanisms.
The domain structure of LIN-41 reflects the diversity of functions that have been associated with TRIM-NHL proteins (Figure 1C). Several of these proteins function as E3 ubiquitin ligases, which require a functional RING domain [10], [30]. However, a sequence alignment of the C. elegans LIN-41 RING domain with those of other Caenorhabditis species indicates that a highly conserved proline, critical for canonical E3-E2 interactions [48], is not found in the nematode LIN-41 RING domains (Figure 5A). Moreover, mutating five cysteine residues that are critical for the RING domain zinc finger structure (C114S, C117S, C130S, C151S, C154S; Figure 5A) resulted in a protein that rescued both somatic and germline defects of lin-41(rrr3) animals (Figure 5B). Although we cannot rule out the possibility that LIN-41 associates with additional factors to regulate ubiquitination, these results suggest that the nematode LIN-41 does not function as a direct E3 ubiquitin ligase.
Fig. 5. LIN-41 does not function as an E3 ubiquitin ligase. 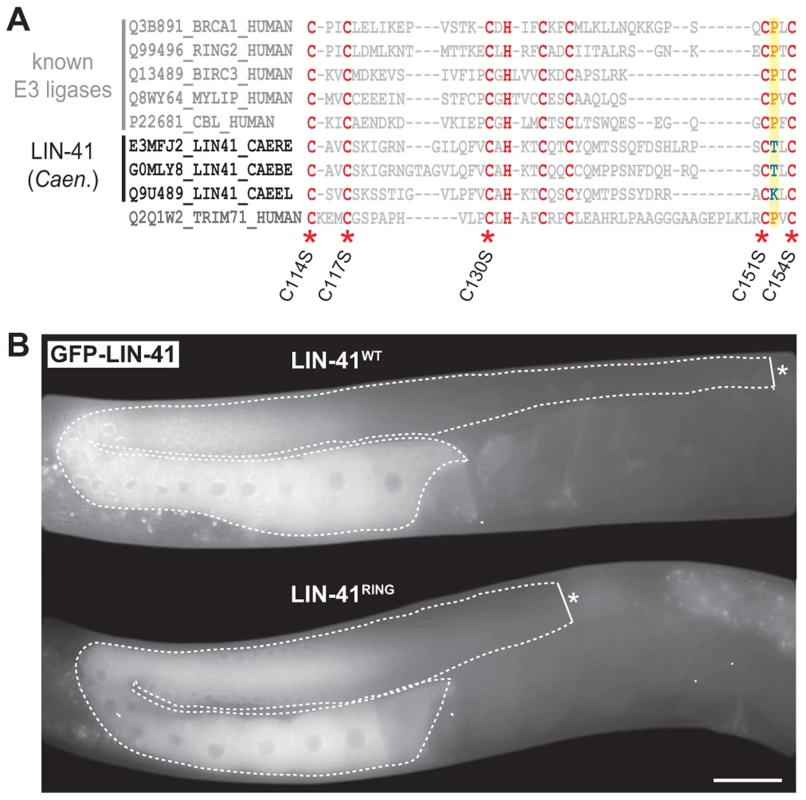
A. Sequence alignment of the mammalian TRIM71/LIN-41 RING domain with those from Caenorhabditis species and different known E3 ubiquitin ligases. Asterisks indicate the position of conserved cysteines (C. elegans: C114, C117, C130, C151, C154), which have been mutated to serines (S) to disrupt the domain (LIN-41RING). A highly conserved proline residue (P, highlighted in yellow), which is critical for canonical E3-E2 interactions, is absent in the nematode LIN-41 proteins. B. Fluorescent micrographs of live animals expressing wild-type (LIN-41WT) and mutated (LIN-41RING) GFP-tagged LIN-41 in the lin-41(rrr3) mutant background. The gonads are outlined with a dashed line. No differences in the distribution or in the levels of the two proteins have been observed (n>50). Scale bar: 20 µm. The coiled-coiled, filamin and NHL domains of the human homolog of LIN-41, TRIM71, constitute the minimal region responsible for binding mRNA and inhibiting translation [19], which is the function attributed to the C. elegans LIN-41 in the soma [29]. One previously isolated lin-41 allele, ma104, is a transposon insertion into the sequence coding for the filamin domain [29]. Intriguingly, lin-41(ma104) mutants display the somatic defects, but the animals are apparently fertile [29]. We confirmed this and, although the brood size in lin-41(ma104) animals is decreased [29], we found no obvious differences in the levels or localization of the germline LIN-41ma104 (Figure 6A) and no evidence for a precocious EGA in the gonads (0/35 worms were expressing the EGA reporter in their gonads). To understand the effect of the ma104 mutation on the LIN-41 protein, we examined the cDNA product of the lin-41(ma104) allele. We found that the ma104 mutation resulted in an insertion of 16 amino acids into the filamin domain (Figure 6B). To gain a mechanistic insight into the ma104 mutation, the filamin domain (residues 691–821) was subcloned, overexpressed in bacteria, and the protein purified to homogeneity. The protein was crystallized and the structure determined at high resolution (1.68 Å; for data collection and refinement statistics, see Table S1). The final crystallographic model encompasses residues 691–729 and 758–820, whereas a long insert (730–757) that is only found in Caenorhabditis LIN-41 protein sequences could not be built due to high flexibility. We found that the LIN-41 filamin structure exhibits a classical immunoglobulin (IG)-like domain fold consisting of seven β-strands arranged in two antiparallel β-sheets (Figure 6C) [49]. A structural search with the LIN-41 filamin domain against the Protein Data Bank (PDB), using DALI, identified the filamin domains most structurally similar to that of LIN-41, which yielded the filamin domains from the Dictyostelium discoideum gelation factor, the human TRIM45 and Filamin-A (PDB IDs 1QFH, 1WLH, 2DS4 and 3RGH, respectively) with root-mean-square deviation values for Cα positions between 1.6 and 2.0 Å [50]. Both crystal packing analysis using PISA [51] and SEC MALS experiments of the protein in solution reveal the oligomeric state of this protein domain as monomeric. Importantly, the 16-residue insertion present in the ma104 allele maps to the mid-section of the second β-strand and is very likely perturbing the filamin IG-like fold (Figure 6B–C). Specifically, the 16-residue insert will prevent completion of one of the β-sheets resulting in solvent access to the hydrophobic protein core of the IG-like β-sandwich and thereby severely destabilizing the fold. This makes it very unlikely that the filamin domain of the ma104 allele is properly folded to exert its biological function.
Fig. 6. The filamin domain of LIN-41 is not essential for the germline function. 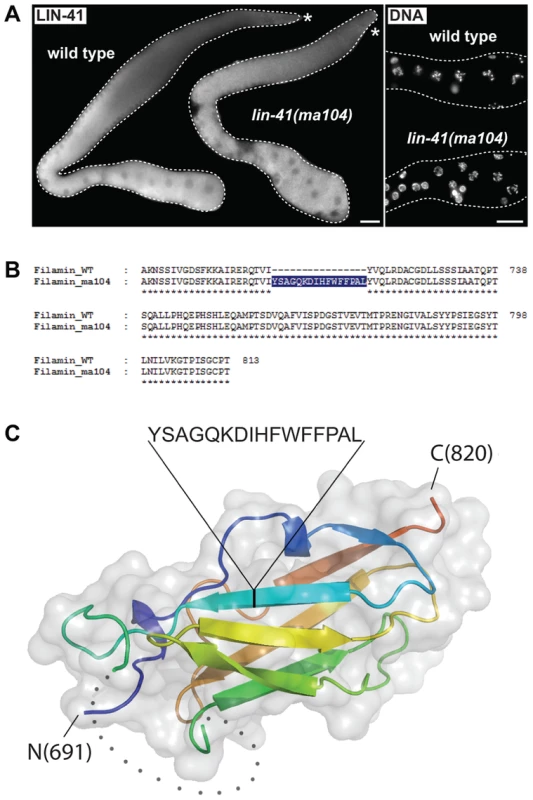
A. Left: fluorescent micrographs of gonads of the indicated genotypes immunostained for LIN-41. Right: corresponding maximum intensity projections of DAPI staining of the proximal-most region of the gonads shown on the left. Scale bars: 25 µm. B. Amino acid sequences of the filamin domain for wild-type and lin-41(ma104) mutant (derived from a cDNA sequence). C. Crystal structure of the LIN-41 filamin domain. The domain is presented as a cartoon model in rainbow colors from blue (N-terminus) to red (C-terminus) with a transparent surface. The position and sequence of the 16-residue insert present in the lin-41(ma104) allele in the second β-strand is highlighted. The disordered sequence stretch 730–757 which is not included in the model is displayed as grey dots. In the fly Brat and the mammalian TRIM71/LIN-41, the NHL domain is essential for mRNA regulation [14], [19], [52]. One of the lin-41 alleles reported here, rrr4, introduces a premature stop codon within the first NHL repeat (Figure 1C), potentially triggering mRNA degradation via nonsense-mediated mRNA decay (NMD). Indeed, inhibiting NMD (by depleting an NMD component, SMG-2 [53]), restored the wild-type expression pattern and levels of LIN-41rrr4 (Figure 7A). LIN-41rrr4 is expected to lack the NHL domain and we found that the gonads expressing this LIN-41 variant displayed lin-41-like germline and somatic defects (Figure 7A). Thus, the NHL domain appears to be essential for LIN-41 functions in both germ and somatic cells.
Fig. 7. LIN-41 may control the germline-to-soma transition independently from its role in mRNA regulation. 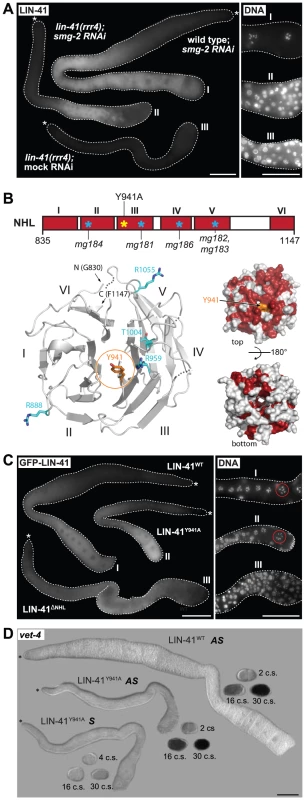
A. Fluorescence micrographs of lin-41(rrr4) gonads stained for LIN-41. Left: suppressing nonsense-mediated mRNA decay (by smg-2 RNAi) restores the expression of LIN-41rrr4. Right: by maximum intensity projections of DAPI staining, LIN-41rrr4 does not rescue the oocyte defects observed in LIN-41-depleted gonads. In contrast to the wild-type gonad containing oocytes in the proximal-most region (I), the gonad expressing LIN-41rrr4 (II) (n = 15) accumulates smaller nuclei, which are similar to those in the LIN-41-depleted gonad (III). Scale bars: 50 µm. B. Top: a schematic view of the LIN-41 NHL domain and its six β-propellers marked in red (I–VI). Previously identified mutations [29] and our point mutant “LIN-41Y941A” are indicated. Bottom left: a homology model of the LIN-41 NHL domain viewed from the electropositive side. The NHL propeller blades are numbered (I–VI) and N-/C-termini are indicated. The residue Y941 is shown in orange atom colors. Known substitution mutations [29] are displayed as sticks in cyan (atom colors). Loops that could not be modeled due to lack of homology are shown as dotted lines. Bottom right: surface representation of the LIN-41 NHL domain homology model in two orientations (rotated by 180° along a horizontal axis). Fully conserved surface-exposed residues are marked in red (see alignment in Figure S8). Y941 (in orange) is in the center of the highly conserved surface patch on the electropositive side of the NHL domain. C. Left: gonads expressing the indicated GFP-tagged LIN-41 variants in otherwise lin-41(rrr3) gonads. Right: by maximum intensity projections of DAPI staining, LIN-41ΔNHL did not rescue the oocyte defects, as evident by the accumulation of smaller nuclei similar to those in the LIN-41-depleted gonad. In contrast, gonads expressing LIN-41Y941A contained overall normal oocytes and LIN-41Y941A rescued the sterility of lin-41(rrr3) animals. At least 50 gonads per strain were examined. Scale bars: 50 µm. D. In situ hybridization against an endogenous EGA transcript, vet-4. Shown are light micrographs of gonads and embryos (at the indicated cell stages, “c.s.”), which were hybridized with antisense (AS) or sense (S) probes for the vet-4 mRNA. Similarly to the gonads expressing the rescuing LIN-41WT, vet-4 mRNA was absent from the gonads expressing LIN-41Y941A. Scale bar: 50 µm. The NHL domain structure of Brat forms a six-bladed β-propeller [52]. Several point mutations in Brat and TRIM71 that disrupt mRNA regulation affect residues on the electropositive side of the NHL domain [19], [23], [52], [54] (Figures S8, S9), highlighting the importance of this surface for mRNA regulation. Point mutations in the NHL domain have been also reported in LIN-41 (Figures 7B and S9A–B) [29]. Importantly, although these mutations display defects in the soma, the animals are fertile, suggesting that the mutant proteins fulfill the gonadal functions [29]. To better interpret these mutations, we initially attempted, unsuccessfully, to express the LIN-41 filamin-NHL or a NHL-only domain constructs for protein structure determination. Thus, we created a homology model of the LIN-41 NHL domain based on the crystal structure of the Brat NHL domain. Interestingly, we found that most of the existing point mutations in the LIN-41 NHL domain also affect amino acids residing on the electropositive surface of the NHL domain (Figure 7B). These observations suggest that i) the electropositive surface of the NHL domain plays a conserved function in mRNA regulation and ii) the germline and somatic functions of the NHL domain involve different mechanisms. To explore this further, we introduced an additional mutation (Y941A) on the electropositive surface of the NHL domain (Figures 7B and S9). Potentially, this mutation is more informative than the other existing NHL mutations because mutation of the corresponding residue (Y702A) in TRIM71 is known to abolish mRNA regulation [19]. In contrast to the deletion of the whole NHL domain from otherwise rescuing (FLAG - and GFP-tagged) LIN-41 protein, which, as expected, caused defects in both the soma and the germline, we found that the LIN-41Y941A variant largely suppressed the germline defects and sterility of lin-41 animals (Figure 7C), including the precocious expression of the endogenous vet-4 transcript (Figure 7D), though it continued to display the somatic defects. Thus, if the same domains/residues determine mRNA regulation in LIN-41 as in TRIM71, the germline function of LIN-41 might be independent from mRNA binding.
Discussion
Cytoplasmic regulators of pluripotency
In contrast to the much-publicized regulation of pluripotency via DNA and chromatin modifications, the potential for cytoplasmic regulation has largely been neglected. Our findings suggest that proteins like LIN-41 and GLD-1 can function in the cytoplasm as molecular “roadblocks” to reprogramming, analogous to the nuclear factors. Similarly, components of P-granules (germline-expressed RNPs) have been recently reported to facilitate maintenance of germline identity in proliferating germ cells, i.e., at the stage prior to GLD-1 expression [55]. Whether teratomatous differentiation in the absence of P-granules reflects precocious activation of embryonic transcription, or has a different etiology, remains to be determined. However, in contrast to P-granules, that impact multiple aspects of RNA metabolism, GLD-1 and LIN-41 are expected to have more specific functions. Intriguingly, the germline-to-soma transition in the absence of LIN-41 or GLD-1 involves similar events: loss of germline proteins, retention of centrosomes, execution of mitosis and activation of the embryonic genome. Two GLD-1 mRNA targets, important for the germline-to-soma transition, encode the CDK-2 partner protein CYE-1/cyclin E and the transcription factor PAL-1/Cdx [6], [31]. However, cyclin E and PAL-1 are co-expressed with LIN-41 in the developing wild-type oocytes [56], suggesting that their expression is not regulated by LIN-41. Thus, GLD-1 and LIN-41 may regulate pluripotency via different targets and/or mechanisms. While GLD-1 directly binds and regulates the expression of its mRNA targets, the molecular function of LIN-41 remains elusive. Our analysis suggests that the germline function of LIN-41 may be independent from mRNA binding, though it does not exclude its role in posttranscriptional regulation, for example as a component of a regulatory RNP. In addition to binding RNA, several TRIM-NHL proteins have been shown to modulate functions of other proteins, for example, by their sequestration (e.g., TRIM3 appears to regulate p21 [13], [17]) or by linking structural proteins (e.g., Wech bridges Talin and ILK for proper embryonic muscle attachment [16]). These interactions depend, at least in part, on the NHL domains of both proteins. Thus, LIN-41 could regulate the germline-to-soma transition by associating, via its NHL domain, with another protein.
Temporal regulation of the germline-to-soma transition by LIN-41
LIN-41-mediated regulation of cell fate transition between generations is somewhat reminiscent of LIN-41 function in the hetrochronic pathway in the soma. However, specific mutations within the NHL and filamin domains of LIN-41 result mainly in somatic but not germline defects (this study and [29]). In addition, LIN-41-dependent repression of LIN-29 appears to be restricted to the soma. Thus, although LIN-41 regulates developmental transitions in both germ - and somatic cells, it may do so through different molecular mechanisms and/or targets.
In the soma, down-regulation of LIN-41, which is mediated by the let-7 miRNA, allows terminal differentiation [57], [58]. In the germline, LIN-41 levels decrease in the most-proximal oocytes, so that LIN-41 is absent from the early embryos. An interesting possibility is that the absence of LIN-41 is a trigger for the onset of embryonic differentiation. In order to test this hypothesis, we attempted to over-express LIN-41 from the heat shock promoter in very early embryos. However, LIN-41 was efficiently expressed only after gastrulation (our unpublished observation), i.e., several cell divisions after EGA, making the experiment inconclusive. The down-regulation of LIN-41 occurs while Pol II-dependent transcription is globally repressed in the oocytes, suggesting that the regulation occurs at the mRNA or the protein level. To test for possible regulation at the mRNA level, we expressed a rescuing LIN-41 under the control of a truncated 3′UTR missing most of the sequence, including the let-7 binding sites [58]. While the expression of this LIN-41 protein started earlier (more distally) in the germline, suggesting posttranscriptional regulation of lin-41 mRNA in this part of the gonad, LIN-41 was still down-regulated in the oocytes and embryos (our unpublished observation), hinting at a possible regulation at the protein level. If so, testing the functional significance of LIN-41 degradation will require the dissection of regulatory motifs in the protein (for example phosphorylation).
Mammalian LIN-41/TRIM71 proteins and pluripotency
TRIM-NHL proteins are known to control the proliferation versus differentiation decision in germ - and neuronal stem cell lineages [23], [25], [59] and, intriguingly, C. elegans LIN-41 has been recently reported to control the regenerative ability of neurons [60]. While these examples highlight the importance of TRIM-NHL proteins for maintaining homeostasis in self-renewing tissues, these proteins have not previously been implicated in controlling pluripotency during development. In our study, we describe LIN-41 as a critical component of the timing mechanism controlling the oocyte reprogramming capacity. To our knowledge, this is the first example of such a regulator in cells that are ready for embryonic development, providing the initial glimpse into a pathway controlling one of the most fundamental developmental transitions. While the in vivo roles of the mammalian LIN-41/TRIM71 are poorly understood, the murine TRIM71 is expressed and functions in developing embryos [61], [62]. TRIM71 is also preferentially expressed in embryonic stem (ES) cells [18], which are derived from pluripotent embryonic cells. In ES cells, TRIM71 represses the expression of Cdkn1, an inhibitor of the cell cycle progression, thereby promoting proliferation [18]. While this role appears opposite to LIN-41 function in the C. elegans germline, TRIM71 presumably associates with many mRNAs, making additional roles likely. Intriguingly, the human LIN-41 has been recently shown to facilitate reprogramming of fibroblasts into iPSCs [28]. In this context, LIN-41, combined with several “pluripotency” transcription factors, can circumvent the requirement for c-Myc in reprogramming [28]. c-Myc facilitates reprogramming in several ways, including by inhibiting differentiation [63], and LIN-41 appears to play a similar role by repressing mRNAs encoding pro-differentiation factors [28]. Although the targets and, perhaps, the mechanisms may differ, it is striking that the C. elegans LIN-41 appears to fulfill an analogous function in the germline. Thus, dissecting LIN-41 targets and the mechanism are exciting objectives for the future research.
Materials and Methods
Nematode culture, mutants, RNAi and transgenic lines
N2 animals were maintained as previously described [64] and were grown at 20°C unless stated otherwise. For alleles and transgenic lines, see Supplemental Material. For RNAi, L1 larvae (L4 for fog-2), grown at 25°C, were fed with bacteria expressing dsRNAs (targeting lin-41, pal-1, fog-2 or smg-2 from the Open Biosystem library or lin-29 from the Ahringer library) and screened one day after the L4-to-adult molt in the same (lin-41, pal-1 and lin-29) or in the second generation (fog-2 and smg-2). A bacterial strain carrying an “empty” vector was used as a negative control (mock RNAi).
Mutagenesis and whole genome sequencing
EMS mutagenesis [64] was performed on a strain (# 1284, see Text S1) carrying the EGA-GFP (integrated at two chromosomal locations to increase GFP fluorescence). F2 animals derived from ≈10.000 F1s were screened. Candidate mutations were identified as previously described [65]. Each mutant was back-crossed four times against the parental strain before genome sequencing. Genomic DNAs (gDNAs) were isolated using Gentra Puregene Tissue Kit 4 g (Qiagen). DNA libraries were created from 50 ng of gDNA (Nextera DNA kit from Illumina). The sequencing data were generated using Hi Seq 2000 (Illumina).
Processing of sequence data and detection of sequence variants
Sequence reads were aligned to the May 2008 C. elegans assembly (obtained from http://hgdownload.soe.ucsc.edu/goldenPath/ce6/chromosomes/) using “bwa” [66]; version 0.6.1-r104) with default parameters, but only retaining single-hit alignments (“bwa samse -n 1” and selecting alignments with “X0:i:1”). The resulting alignments were converted to BAM format, sorted and indexed using “samtools” [67]; version 0.1.18). In order to quantify contamination by Escherichia coli, reads were similarly aligned to a collection of E. coli genomes (NCBI accession numbers NC_008253, NC_008563, NC_010468, NC_004431, NC_009801, NC_009800, NC_002655, NC_002695, NC_010498, NC_007946, NC_010473, NC_000913 and AC_000091), which typically resulted in less than 1% aligned reads. Sequence variants were identified using GATK [68]; version 1.5.31) indel realignment and base quality score recalibration, followed by SNP and INDEL discovery and genotyping for each individual strain using standard hard filtering parameters, resulting in a total of six to eight thousand sequence variations in each strain compared to the reference genome. Finally, the number of high quality (score > = 500) single nucleotide substitutions of EMS-type (G/C→A/T transitions [69], not found in other any other mutant strain or in the parent strain (typically less than 1% of the total number of variants per strain) were counted in sequential windows of 1 Mb to identify regions of increased variant density.
Real-time quantitative PCR on dissected gonads
RNA was isolated from gonads dissected from one day-old (after the L4-to-adult molt) animals. cDNA was synthesized with oligo(dT) primers using the ImProm II Reverse transcription system from Promega according to manufacturer's instructions. cDNA was used for qPCR with the Absolute QPCR SYBR green ROX mix (AbGene) on an ABI PRISM 7700 system (Applied Biosystems). qPCR reactions were performed as previously described [31]. At least one primer in each pair is specific for an exon-exon junction. Human carrier RNA was added to each sample before RNA extraction, allowing normalization to hGAPDH. Standard curves for quantification were generated from a serial dilution of input cDNA for each primer pair. The amount of target present in each replicate was derived from a standard curve; an average was calculated for the triplicates. To compare total mRNA levels, the qPCR results were normalized to human GAPDH and to the wild-type values for each primer pair and fold enrichments were calculated. For primers used, see Text S1.
Immunostaining, antibodies, RNA in situ hybridization and microscopy
Immunostaining experiments were performed as previously described [70] with the following antibodies: PGL-1 [36] (dilution 1∶1000); SPD-2 [45] (“969LA”, 1∶800); GFP (Roche, 1∶700); phospo-Histone H3 Ser10 (“Ser10P”, Millipore, 1∶200); muscle myosin [71] (“5–6”, 1∶2.500); and UNC-120 (courtesy of Michael Krause, 1∶500). Immunostainings against RME-2 [35] (“INT”, dilution 1∶100), GLD-1 [72] (dilution 1∶5) and LIN-41 (courtesy of Helge Grosshans, “4796”, 1∶2.000) were performed as previously described [73] and against the Ser5P of Pol II CTD [74] (“3E8”, 1∶5) according to Seydoux and Dunn [33]. Immunostainings against HIM-3 [42] (courtesy of Monique Zetka, dilution 1∶500) were performed as previously described [75]. Secondary antibodies used in this study: goat anti-mouse IgG alexa-488 (Molecular Probes, 1∶600,), goat anti-rabbit IgG alexa-568 (Invitrogen, 1∶750) and goat anti-rat IgG alexa-568 (Molecular Probes, 1∶500). In situ hybridizations against the vet-4 mRNA were performed as previously described [31]. Unless indicated otherwise, the gonads were dissected from 1 day-old adults. Zeiss AxioImager Z1 microscope equipped with an Axiocam MRm REV 2 CCD camera was used for capturing pictures. Images were then exported into Adobe Photoshop CS4 and processed in an identical manner. A spinning disk multipoint confocal microscope equipped with an EM-CCD Cascade II camera (Photometrics) was used for capturing images for Figure 3D. Pictures were, then, deconvolved with the Huygens software and then processed in Imaris XP 7.1.1.
LIN-41 antibody
The affinity-purified (ELISA) rabbit anti-LIN-41 antibody (“4796”) was provided by Helge Grosshans (Magdalene Rausch & Helge Grosshans, unpublished data) and created against the VKNLKLSVLISQAESLQSKQIDLQQAIQTATKLMDSSDCDEMVLRQVFEKLASCQMGNEGTEPNNNILNVLMLACQVNEDDRLKFTAPQDGILLNKARQF sequence (residues 587–686). The rabbit was raised by SDIX in Newark, DE, USA.
LIN-41 variants
The LIN-41 point mutant transgene constructs “RING” and “Y941A” were created from the wild-type LIN-41 transgenic template by site-directed mutagenesis (Stratagene QuikChange method), whereas the deletion construct “ΔNHL” was created via two-step PCR. In any case, Phusion High-Fidelity DNA Polymerase (Fermentas) was used. For primers used see Text S1.
Cloning, expression and purification of LIN-41
The filamin domain (residues 691–821) of C. elegans LIN-41 (isoform B of Q9U489) was cloned into pOPINF [76] using In-Fusion (Clontech Laboratories Inc). The resulting expression construct was transformed into BL21 DE3 cells and the protein expressed via auto-induction at 20°C for 20 hours. Cells were harvested, then resuspended in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 20 mM imidazole, 0.2% Tween-20) and frozen at −80°C. The cell suspension was thawed and freshly supplemented with Complete EDTA-free protease inhibitors (Roche Diagnostics) and 3 U/ml Benzonase (Sigma) before passing through an Avestin EmulsiFlex-C3 cell disruptor. The clarified lysate was incubated with NiNTA affinity resin (Qiagen) in batch mode and the bound protein eluted in 50 mM Tris, pH 7.5, 500 mM NaCl, 125 mM imidazole. The protein was fractionated on a Superdex 75 HiLoad 16/60 (GE Healthcare) gel filtration column in GF buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 2 mM TCEP and 0.02% NaN3). The single peak fraction was pooled and digested overnight at 4°C with 3C protease to remove the N-terminal histidine tag. The released protein tag and 3C protease were removed by a second nickel-affinity step and the untagged filamin domain was further purified over a Superdex 75 column in GF buffer and concentrated to 7.5 mg/ml.
Crystallization, data collection and structure solution
All crystallization experiments were performed at 20°C using the sitting-drop vapour diffusion method via a Phoenix robot (Art Robbins) dispensing 100 nl drops. Removal of the N-terminal histidine tag from the filamin domain was needed to obtain crystals. The untagged filamin domain readily crystallized in many conditions. Crystals grown in 1.1 M sodium malonate, 0.1 M HEPES, pH 7.0, 0.5% v/v Jeffamine ED-2001, were harvested and cryoprotected in mother liquor containing 25% ethylene glycol. These crystals diffracted to 1.68 Å resolution at the SLS PX-III beamline and belonged to space group C2221 with one molecule per asymmetric unit. Diffraction data were integrated and scaled using XDS [77] and the structure was solved by the molecular replacement method using PHASER [78]. Phases from this solution were calculated and used for automatic model building with BUCCANEER [79]. The LIN-41 filamin structure was further improved by the crystallographic simulated annealing routine followed by individual B-factor refinement in PHENIX [80] and several rounds of manual rebuilding in COOT [81] and refinement in BUSTER [82]. The final structure was validated using COOT. Structural images for figures were prepared with PyMOL (http://pymol.sourceforge.net/). Atomic coordinates and structure factors for the LIN-41 filamin domain have been deposited in the PDB with entry code 4UMG.
Homology modeling
Amino acid sequences of the C. elegans LIN-41 (Uniprot Q9U489, 830–1147) and Homo sapiens TRIM71 (Uniprot Q2Q1W2, 591–868) NHL domains were submitted to the HHPRED server for homology detection and structure prediction [83]. The structure of the D. melanogaster Brat NHL domain (PDB 1Q7F) was the top hit in both searches resulting in very high scores for the LIN-41 NHL domain (Score = 241.22, E-value = 1e-33, 28% sequence identity) and the TRIM71 NHL domain (score = 22.54, E-value = 4.5e-35, 31% identity). The top alignments were edited for minimal local corrections and sent to the HHPRED MODELLER pipeline for modeling. Loops lacking template information for both the C. elegans and the human NHL domain models were removed in the final models. Structural figures were prepared using PyMOL (www.pymol.org).
Supporting Information
Zdroje
1. GurdonJB, MeltonDA (2008) Nuclear reprogramming in cells. Science 322 : 1811–1815.
2. YuJ, ThomsonJA (2008) Pluripotent stem cell lines. Genes Dev 22 : 1987–1997.
3. PatelT, TursunB, RaheDP, HobertO (2012) Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep 2 : 1178–1186.
4. TursunB, PatelT, KratsiosP, HobertO (2011) Direct conversion of C. elegans germ cells into specific neuron types. Science 331 : 304–308.
5. Kaser-PebernardS, MullerF, WickyC (2014) LET-418/Mi2 and SPR-5/LSD1 Cooperatively Prevent Somatic Reprogramming of C. elegans Germline Stem Cells. Stem Cell Reports 2 : 547–559.
6. CioskR, DePalmaM, PriessJR (2006) Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311 : 851–853.
7. WrightJE, CioskR (2013) RNA-based regulation of pluripotency. Trends Genet 29 : 99–107.
8. AkkersRC, van HeeringenSJ, JacobiUG, Janssen-MegensEM, FrancoijsKJ, et al. (2009) A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell 17 : 425–434.
9. VastenhouwNL, ZhangY, WoodsIG, ImamF, RegevA, et al. (2010) Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464 : 922–926.
10. WulczynFG, CuevasE, FranzoniE, RybakA (2011) miRNAs Need a Trim : Regulation of miRNA Activity by Trim-NHL Proteins. Adv Exp Med Biol 700 : 85–105.
11. RybakA, FuchsH, HadianK, SmirnovaL, WulczynEA, et al. (2009) The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 11 : 1411–1420.
12. KudryashovaE, KudryashovD, KramerovaI, SpencerMJ (2005) Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol 354 : 413–424.
13. RahejaR, LiuY, HukkelhovenE, YehN, KoffA (2014) The ability of TRIM3 to induce growth arrest depends on RING-dependent E3 ligase activity. Biochem J 458 : 537–545.
14. KwonSC, YiH, EichelbaumK, FohrS, FischerB, et al. (2013) The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20 : 1122–1130.
15. LoedigeI, StotzM, QamarS, KramerK, HennigJ, et al. (2014) The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev 28 : 749–764.
16. LoerB, BauerR, BornheimR, GrellJ, KremmerE, et al. (2008) The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat Cell Biol 10 : 422–428.
17. LiuY, RahejaR, YehN, CiznadijaD, PedrazaAM, et al. (2014) TRIM3, a tumor suppressor linked to regulation of p21(Waf1/Cip1.). Oncogene 33 : 308–315.
18. ChangHM, MartinezNJ, ThorntonJE, HaganJP, NguyenKD, et al. (2012) Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun 3 : 923.
19. LoedigeI, GaidatzisD, SackR, MeisterG, FilipowiczW (2013) The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res 41 : 518–532.
20. LeeSH, ChoS, Sun KimM, ChoiK, ChoJY, et al. (2014) The ubiquitin ligase human TRIM71 regulates let-7 microRNA biogenesis via modulation of Lin28B protein. Biochim Biophys Acta 1839 : 374–386.
21. AramaE, DickmanD, KimchieZ, ShearnA, LevZ (2000) Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 19 : 3706–3716.
22. FrankDJ, EdgarBA, RothMB (2002) The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129 : 399–407.
23. BetschingerJ, MechtlerK, KnoblichJA (2006) Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124 : 1241–1253.
24. LeeCY, WilkinsonBD, SiegristSE, WhartonRP, DoeCQ (2006) Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell 10 : 441–449.
25. NeumullerRA, BetschingerJ, FischerA, BushatiN, PoernbacherI, et al. (2008) Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454 : 241–245.
26. FroskP, WeilerT, NylenE, SudhaT, GreenbergCR, et al. (2002) Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet 70 : 663–672.
27. ChiangAP, BeckJS, YenHJ, TayehMK, ScheetzTE, et al. (2006) Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci U S A 103 : 6287–6292.
28. WorringerKA, RandTA, HayashiY, SamiS, TakahashiK, et al. (2014) The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14 : 40–52.
29. SlackFJ, BassonM, LiuZ, AmbrosV, HorvitzHR, et al. (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5 : 659–669.
30. EcsediM, GrosshansH (2013) LIN-41/TRIM71: emancipation of a miRNA target. Genes Dev 27 : 581–589.
31. BiedermannB, WrightJ, SenftenM, KalchhauserI, SarathyG, et al. (2009) Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev Cell 17 : 355–364.
32. SeydouxG, MelloCC, PettittJ, WoodWB, PriessJR, et al. (1996) Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382 : 713–716.
33. SeydouxG, DunnMA (1997) Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 124 : 2191–2201.
34. StitzelML, SeydouxG (2007) Regulation of the oocyte-to-zygote transition. Science 316 : 407–408.
35. GrantB, HirshD (1999) Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell 10 : 4311–4326.
36. KawasakiI, ShimYH, KirchnerJ, KaminkerJ, WoodWB, et al. (1998) PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94 : 635–645.
37. McGinnisW, KrumlaufR (1992) Homeobox genes and axial patterning. Cell 68 : 283–302.
38. HunterCP, KenyonC (1996) Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell 87 : 217–226.
39. LeiH, LiuJ, FukushigeT, FireA, KrauseM (2009) Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development 136 : 1241–1249.
40. MootzD, HoDM, HunterCP (2004) The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 131 : 3263–3272.
41. HsuJY, SunZW, LiX, ReubenM, TatchellK, et al. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102 : 279–291.
42. ZetkaMC, KawasakiI, StromeS, MullerF (1999) Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13 : 2258–2270.
43. AlbertsonDG, ThomsonJN (1993) Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res 1 : 15–26.
44. Mikeladze-DvaliT, von TobelL, StrnadP, KnottG, LeonhardtH, et al. (2012) Analysis of centriole elimination during C. elegans oogenesis. Development 139 : 1670–1679.
45. KempCA, KopishKR, ZipperlenP, AhringerJ, O'ConnellKF (2004) Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell 6 : 511–523.
46. StringhamEG, DixonDK, JonesD, CandidoEP (1992) Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell 3 : 221–233.
47. ReinkeV, GilIS, WardS, KazmerK (2004) Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 : 311–323.
48. BudhidarmoR, NakataniY, DayCL (2012) RINGs hold the key to ubiquitin transfer. Trends Biochem Sci 37 : 58–65.
49. BorkP, HolmL, SanderC (1994) The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol 242 : 309–320.
50. HolmL, RosenstromP (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–549.
51. KrissinelE, HenrickK (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372 : 774–797.
52. EdwardsTA, WilkinsonBD, WhartonRP, AggarwalAK (2003) Model of the brain tumor-Pumilio translation repressor complex. Genes Dev 17 : 2508–2513.
53. PageMF, CarrB, AndersKR, GrimsonA, AndersonP (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19 : 5943–5951.
54. HarrisRE, PargettM, SutcliffeC, UmulisD, AsheHL (2011) Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell 20 : 72–83.
55. UpdikeDL, KnutsonAK, EgelhoferTA, CampbellAC, StromeS (2014) Germ-Granule Components Prevent Somatic Development in the C. elegans Germline. Curr Biol 24 : 970–975.
56. BrodiganTM, LiuJ, ParkM, KipreosET, KrauseM (2003) Cyclin E expression during development in Caenorhabditis elegans. Dev Biol 254 : 102–115.
57. ReinhartBJ, SlackFJ, BassonM, PasquinelliAE, BettingerJC, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 : 901–906.
58. VellaMC, ChoiEY, LinSY, ReinertK, SlackFJ (2004) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev 18 : 132–137.
59. SchwambornJC, BerezikovE, KnoblichJA (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136 : 913–925.
60. ZouY, ChiuH, ZinovyevaA, AmbrosV, ChuangCF, et al. (2013) Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 340 : 372–376.
61. Maller SchulmanBR, LiangX, StahlhutC, DelConteC, StefaniG, et al. (2008) The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7 : 3935–3942.
62. SchulmanBR, Esquela-KerscherA, SlackFJ (2005) Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn 234 : 1046–1054.
63. KnoepflerPS (2008) Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell 2 : 18–21.
64. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
65. ZurynS, Le GrasS, JametK, JarriaultS (2010) A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 186 : 427–430.
66. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
67. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
68. DePristoMA, BanksE, PoplinR, GarimellaKV, MaguireJR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43 : 491–498.
69. DrakeJW, BaltzRH (1976) The biochemistry of mutagenesis. Annu Rev Biochem 45 : 11–37.
70. LinR, HillRJ, PriessJR (1998) POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell 92 : 229–239.
71. PriessJR, ThomsonJN (1987) Cellular interactions in early C. elegans embryos. Cell 48 : 241–250.
72. ScheckelC, GaidatzisD, WrightJE, CioskR (2012) Genome-wide analysis of GLD-1-mediated mRNA regulation suggests a role in mRNA storage. PLoS Genet 8: e1002742.
73. NavarroRE, ShimEY, KoharaY, SingsonA, BlackwellTK (2001) cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development 128 : 3221–3232.
74. ChapmanRD, HeidemannM, AlbertTK, MailhammerR, FlatleyA, et al. (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318 : 1780–1782.
75. BurgerJ, MerletJ, TavernierN, RichaudeauB, ArnoldA, et al. (2013) CRL2(LRR-1) E3-ligase regulates proliferation and progression through meiosis in the Caenorhabditis elegans germline. PLoS Genet 9: e1003375.
76. BerrowNS, AldertonD, OwensRJ (2009) The precise engineering of expression vectors using high-throughput In-Fusion PCR cloning. Methods Mol Biol 498 : 75–90.
77. KabschW (2010) Xds. Acta Crystallogr D Biol Crystallogr 66 : 125–132.
78. McCoyAJ, Grosse-KunstleveRW, AdamsPD, WinnMD, StoroniLC, et al. (2007) Phaser crystallographic software. J Appl Crystallogr 40 : 658–674.
79. CowtanK (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62 : 1002–1011.
80. AfoninePV, Grosse-KunstleveRW, EcholsN, HeaddJJ, MoriartyNW, et al. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68 : 352–367.
81. EmsleyP, LohkampB, ScottWG, CowtanK (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66 : 486–501.
82. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P et al. (2011). BUSTER Version 2.11.4, Cambridge, United Kingdom: Global Phasing Ltd.
83. SodingJ, BiegertA, LupasAN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–248.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



