-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The PhoP-Dependent ncRNA Mcr7 Modulates the TAT Secretion System in
One of the best characterized two-component systems in Mycobacterium tuberculosis is represented by the PhoPR pair, with PhoR being the transmembrane sensor kinase and PhoP playing an essential part in controlling expression of virulence-associated genes, such as those encoding the ESX-1 secretion apparatus. Previous studies showed that mutations in phoP resulted in attenuation in the mouse model of infection, thus providing the basis for the development of a novel live attenuated Mycobacterium tuberculosis vaccine carrying a deletion in phoP which is today in clinical trials. To thoroughly investigate the role of PhoP in M. tuberculosis, we undertook a systems biology approach comprising ChIP-seq and RNA-seq technologies. We demonstrated binding of PhoP to at least 35 targets on the M. tuberculosis chromosome and direct impact on expression of 30 genes, while further amplification of the signal is provided by regulators acting downstream. The strongest binding site was located between rv2395 and PE_PGRS41, where transcription of the non-coding RNA (ncRNA) Mcr7 was demonstrated. Expression of Mcr7 was found to be restricted to M. tuberculosis species and totally silenced in a phoP mutant. Genetics and proteomics approaches proved that Mcr7 controls activity of the Twin Arginine (Tat) secretion system, thus modulating secretion of the immunodominant antigen Ag85 complex and the BlaC beta-lactamase.
Published in the journal: The PhoP-Dependent ncRNA Mcr7 Modulates the TAT Secretion System in. PLoS Pathog 10(5): e32767. doi:10.1371/journal.ppat.1004183
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004183Summary
One of the best characterized two-component systems in Mycobacterium tuberculosis is represented by the PhoPR pair, with PhoR being the transmembrane sensor kinase and PhoP playing an essential part in controlling expression of virulence-associated genes, such as those encoding the ESX-1 secretion apparatus. Previous studies showed that mutations in phoP resulted in attenuation in the mouse model of infection, thus providing the basis for the development of a novel live attenuated Mycobacterium tuberculosis vaccine carrying a deletion in phoP which is today in clinical trials. To thoroughly investigate the role of PhoP in M. tuberculosis, we undertook a systems biology approach comprising ChIP-seq and RNA-seq technologies. We demonstrated binding of PhoP to at least 35 targets on the M. tuberculosis chromosome and direct impact on expression of 30 genes, while further amplification of the signal is provided by regulators acting downstream. The strongest binding site was located between rv2395 and PE_PGRS41, where transcription of the non-coding RNA (ncRNA) Mcr7 was demonstrated. Expression of Mcr7 was found to be restricted to M. tuberculosis species and totally silenced in a phoP mutant. Genetics and proteomics approaches proved that Mcr7 controls activity of the Twin Arginine (Tat) secretion system, thus modulating secretion of the immunodominant antigen Ag85 complex and the BlaC beta-lactamase.
Introduction
Mycobacterium tuberculosis, the etiologic agent of tuberculosis in humans, is arguably the world's most important intracellular pathogen. The bacterium disseminates via the aerosol route from open pulmonary lesions of an infectious case and on reaching the alveoli of a susceptible individual is phagocytosed by resident macrophages. After infection, the tubercle bacillus resides asymptomatically for years in 95% of the infected persons. This latent state can be life-long but disease develops when the immune system weakens as a consequence of HIV co-infection, aging or malnutrition [1]. M. tuberculosis encounters a variety of environmental conditions and has to adapt to both the extracellular milieu and to the intracellular niche in order to survive [2]. Improved understanding of how this pathogen fine-tunes gene expression to support active growth and non-replicating persistence, and of how it copes with stresses encountered within the host would not only shed light on bacterial pathogenesis and biology but also aid the design of novel intervention strategies.
Well-known adaptation mechanisms in bacteria include two-component signal transduction systems (TCSS). These consist of a sensor protein that, upon reception of specific signal(s), activates its cognate transcription factor resulting in transcriptional regulation of a defined set of genes. The number of TCSS in M. tuberculosis is lower than typically found in bacteria of similar genome size, possibly reflecting the evolution of the bacillus as a human pathogen adapted to a predominantly intracellular environment [3]. Of the 11 TCSS present in M. tuberculosis H37Rv, the PhoPR TCSS is essential for virulence [4], as demonstrated in ex vivo and in vivo infection models, where inactivation of phoP led to greatly impaired growth [4], [5]. Consistent with these observations, a single nucleotide polymorphism (S219L) in the DNA binding domain of PhoP, which affected the ability of the regulator to control gene expression of the ESX-1 secretion system, was responsible for the reduced virulence of the attenuated strain H37Ra [6], [7]. Biochemical analyses conducted on the phoP mutant revealed that the synthesis of the cell wall components diacyltrehaloses, polyacyltrehaloses and sulfolipids, specific to pathogenic mycobacterial species, was also diminished as compared to wild type M. tuberculosis, thus providing an additional mechanism for attenuation [8]. Loss of these phenotypes is the basis of candidate vaccine strains carrying deletions in phoP, such as the recently developed MTBVAC, which showed great promise upon preclinical evaluation [9].
The PhoP protein is part of the PhoB/OmpR subfamily of transcription factors, characterized by an N-terminal regulatory domain and a DNA binding domain at the C-terminus. The crystal structure revealed the dimeric nature of the regulator and predicted PhoP to bind to direct repeats [10], in agreement with in vitro investigations of the PhoP-DNA interactions at a few operator regions [11], [12], [13].
Microarray-based transcriptomic studies of wild type and phoP mutant strains have been performed to identify the regulon controlled by PhoP. Genes belonging to lipid and intermediary metabolism, to the PE, PPE and PE_PRGS families and to the transcriptional regulator categories were found to be deregulated in phoP-deficient bacteria [5], [14], while a very recent publication [15] identified the genes under direct control of PhoP by means of ChIP-seq experiments on a PhoP-overexpressing strain.
Despite this body of knowledge, several questions remain unanswered. These include defining the external stimulus sensed by the PhoPR TCSS, performing ChIP-seq experiments in physiological conditions, obtaining a single-nucleotide resolution transcriptomic map, and identifying transcriptional regulators acting downstream of PhoP.
In this study, we used a systems biology approach, combining ChIP-seq, RNA-seq and in-depth proteomics, to thoroughly investigate the role of PhoP in the biology of M. tuberculosis. We identified a ncRNA, encoded by the mcr7 gene, as a major target of PhoP and showed its involvement in controlling secretion of Twin Arginine Translocation (Tat) substrates.
Results
Genome-wide identification of PhoP binding sites in M. tuberculosis by ChIP-seq
The PhoP regulon has been characterized previously by transcription profiling using microarrays in both the H37Rv [5] and MT103 strains [14] of M. tuberculosis. Those studies indicated that approximately 2% of genes are regulated by PhoP at the transcriptional level. However, little is known about the biophysical interactions between PhoP and the promoter regions of the genes controlled with the exception of a few well-characterized promoters [13]. Here, we applied ChIP-seq (chromatin immunoprecipitation with anti-PhoP antibodies followed by ultra-high throughput DNA sequencing) to locate PhoP binding sites across the M. tuberculosis H37Rv [16] chromosome. To avoid false positive signals we included an isogenic phoP mutant [11], which served as a control and reference sample in all experiments.
ChIP-seq analysis of cultures grown to exponential phase led to the identification of 35 significantly enriched (p<0.0001, FDR 0.00%) regions in H37Rv compared to the phoP mutant (Table 1). Several of these peaks were localized between divergently transcribed open reading frames (ORF) or upstream of validated or predicted operons [17], [18], [19], thus increasing the number of genes potentially affected by PhoP binding directly. These targets were randomly distributed along the M. tuberculosis genome (Figure 1A) and predominantly located upstream of ORF (Figure 1B). However, we also observed PhoP binding sites in the 3′-end of ORF, as shown for the hddA-ldtA genes (Figure 1B). All of the functional categories in which the M. tuberculosis ORFs have been grouped were represented in the ChIP-seq results, although clear prevalence of regulatory proteins was observed (12% of the total number of signals as compared to 5% representation in the genome). Remarkably, of the 35 regions detected by ChIP-seq, a number of them had not been described previously [5] as being associated with PhoP-regulated genes (i.e. mcr7, PE27, PPE43, PE31, Rv3778c, lpdA).
Fig. 1. Mapping of PhoP binding sites in the M. tuberculosis chromosome by ChIP-seq. 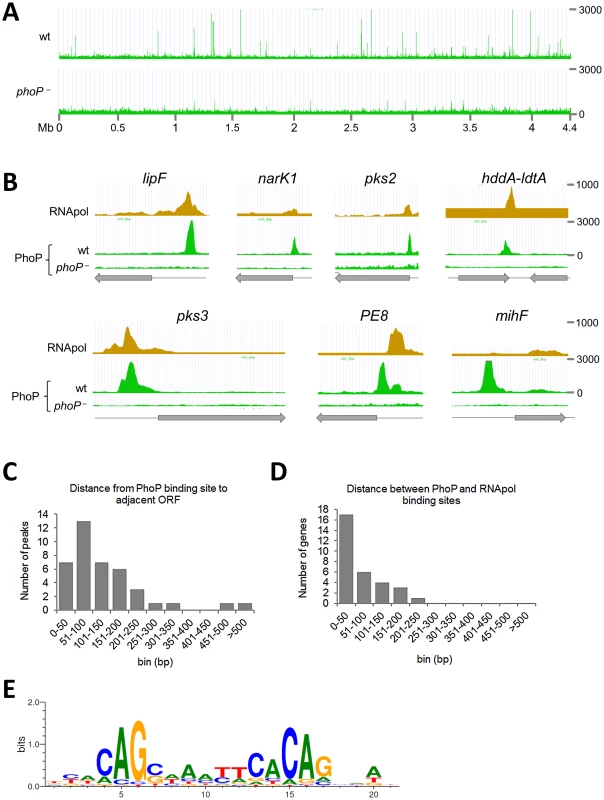
A. UCSC genome browser view showing PhoP binding regions across the M. tuberculosis genome. Peak height correlates with sequence reads and consequently with PhoP binding affinity. The use of a phoP mutant in this experiment (bottom panel) allows identification of false positive signals. B. PhoP and RNA polymerase (RNApol) ChIP-seq profiles for selected genes. Note that the PhoP binding site lies upstream that of RNApol for lipF, narK1, pks2 and pks3, known to be transcriptionally activated by PhoP. The position of the PhoP binding region lies downstream that of RNApol for the PE8 gene. PhoP binding at the 3′-end of hddA is shown as an example of non-canonical target. C. Bar diagram showing distance (in bp) between the PhoP binding sites and the start codon of adjacent ORFs. D. Bar diagram showing distance between the position of the PhoP binding site and the summit of the RNApol peak calculated from ChIP-seq data. E. Sequence logo showing the consensus sequence derived from the PhoP binding sites identified by ChIP-seq. Tab. 1. Summary of ChIP-seq results showing localization of PhoP binding sites in the M. tuberculosis genome. 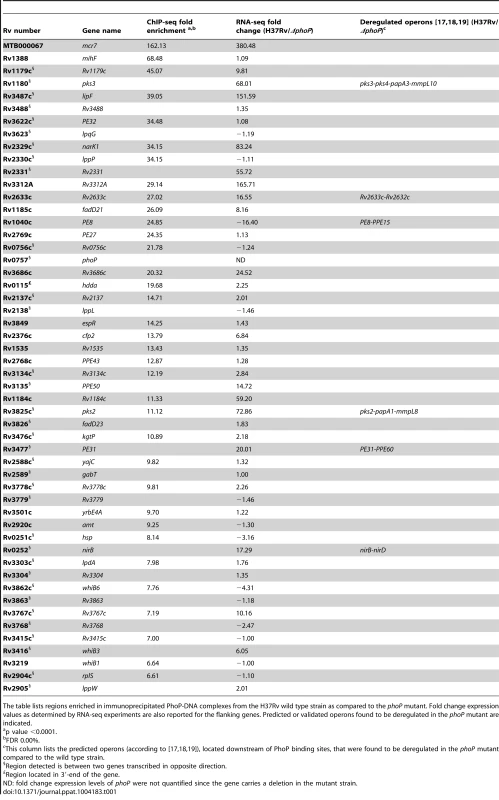
The table lists regions enriched in immunoprecipitated PhoP-DNA complexes from the H37Rv wild type strain as compared to the phoP mutant. Fold change expression values as determined by RNA-seq experiments are also reported for the flanking genes. Predicted or validated operons found to be deregulated in the phoP mutant are indicated. The distance between the PhoP peak and the ORF start site was calculated for each gene and plotted as reported in Figure 1C. The majority (83%) of the PhoP peaks were between 0 and 200 bp upstream of the ORF start site, with 50% of them within the first 100 bp. Two binding sites were considerably further away from the closer ORF: these were the cases of lipF (>500 bp) and rv1535 (472 bp). Only one case was observed with the PhoP peak lying within the ORF: rv2137c, where the summit was located 97 bp downstream of the ATG start codon.
To gain insight into the interplay between PhoP and the transcriptional complex, we compared previous ChIP-seq data of RNA polymerase (RNApol) [17] with the PhoP profile obtained here. We observed that PhoP distribution mirrored that of RNApol at the putative promoter regions (Figure 1B). Closer examination indicated that PhoP binding sites precede those of RNApol. Additional confirmation came from calculation of the distance between the PhoP and RNApol signals, which was between 0 and 100 bp for most of the genes (Figure 1D). This might indicate a role of PhoP in positioning RNApol as a prerequisite for transcriptional control. Exceptionally, the PhoP binding region upstream of PE8 lies downstream of the RNApol binding site (Figure 1B). Curiously, we noticed that some strong PhoP peaks lacked a concomitant RNApol signal as illustrated by mihF (Figure 1B).
In order to confirm the ChIP-seq data independently, we quantified a selection of PhoP binding sites from the immunoprecipitated DNA of H37Rv, its phoP mutant and a control experiment performed without antibody, obtained from biological replicates. The results validated those obtained in ChIP-seq experiments (Figure S1).
Identification of the PhoP consensus sequence
We used the MEME suite to identify the PhoP consensus sequence from ChIP-seq signals. Two hundred bp surrounding the summit of the peaks were scanned and a motif was found in 83% of the instances (p-value between 5.88e-09 and 4.40e-05, Figure 1E and Worksheet 1 in Table S1). Additional, though more divergent, copies of the same consensus sequence were found in 11 peaks (31%) (Worksheet 1 in Table S1). Overall, six ChIP-seq signals (rv1535, PPE43, lpdA, rv3767, whiB1 and the region between yajC and gabT) were not found to be associated with the identified motif, suggesting either higher divergence of the sequences or indirect PhoP binding, i.e. mediated by other proteins. While the 5′ to 3′ orientation of the motif generally corresponded to the orientation of the gene associated with the ChIP-seq signal (22 out of 29 cases), we observed 7 exceptions, the most notable being the well-known PhoP-regulated gene pks3 [13], [14]. In this case the motif was localized on the opposite strand as compared to the direction of transcription of pks3.
Correlation between PhoP binding and transcriptional regulation
To explore the relationship between binding of PhoP and transcriptional control on the target genes, we performed deep transcriptomic analysis by RNA-seq under the same in vitro conditions employed for ChIP-seq. We compared exponentially growing H37Rv wild type to the isogenic phoP mutant and quantified gene expression according to the functional categories in the TubercuList database (http://tuberculist.epfl.ch), generating results reported in Worksheet 1 in Table S2. An arbitrary 3-fold threshold was applied to the dataset for further analysis.
Integration of the ChIP-seq and RNA-seq data revealed that 19 PhoP binding sites were associated with altered expression of the flanking gene(s) (Table 1). Since some of these ORFs are part of predicted operons [17], [18], [19], the total number of genes under direct control of PhoP was found to be at least 30 in the experimental conditions tested (Table 1). The region showing the most remarkable affinity for PhoP (162-fold enrichment in ChIP-seq) lay between rv2395 and PE_PGRS41. This signal correlated with the expression of a small transcript (Mcr7) in the intergenic region that was severely affected by deletion of PhoP. This small transcript is further characterized later in this work. Other examples are represented by the operons composed of pks3-pks4-papA3-mmpL10 and rv2633c-rv2632c, which were more expressed in the wild type strain, whereas the PE8-PPE15 transcriptional unit was induced upon deletion of phoP, thus demonstrating the dual role of the regulator. On the contrary, 15 ChIP-seq peaks did not correlate with the presence of deregulated transcripts in their vicinity. The most striking signal in this group was the one upstream of mihF.
Deeper inspection of the RNA-seq results uncovered 140 transcripts whose expression underwent changes in the phoP mutant (Worksheet 1 in Table S2). Since 30 of these were part of the aforementioned operons, the remaining 110 were likely to be indirectly controlled by PhoP through regulatory cascades. It is worth recalling that PhoP binds upstream of genes encoding several transcriptional regulators (espR, whiB1, whiB3, whiB6) that may act downstream. Interestingly, an almost equal proportion of genes was found to be activated (68) or repressed (72) as a consequence of the mutation. Upon clustering these transcripts into functional categories, we observed significant enrichment for the “lipid metabolism” group among the up-regulated genes (p = 0.0016, Fisher's Exact test) and for the “PE/PPE” category among the down-regulated genes (p = 0.0024, Fisher's Exact test). Further discussion of the PhoP regulon will be presented elsewhere.
Independent validation of the RNA-seq data was obtained for a subset of genes by quantitative reverse transcription PCR (qRT-PCR), which confirmed the excellent correlation between high-throughput results and targeted quantification (Figure S1).
Identification of a small ncRNA as the major PhoP-regulated target
The most prominent PhoP binding site in the genome lay between genes rv2395 and PE_PGRS41 (Figure 2A) but, surprisingly, transcription of neither gene differed between strain H37Rv and its phoP mutant (Worksheet 1 in Table S2). However, in a previous study of ncRNA in M. bovis BCG, the mcr7 gene encoding a 350 nt transcript, was located within this region [20].
Fig. 2. Characterization of mcr7 as the major PhoP-regulated target. 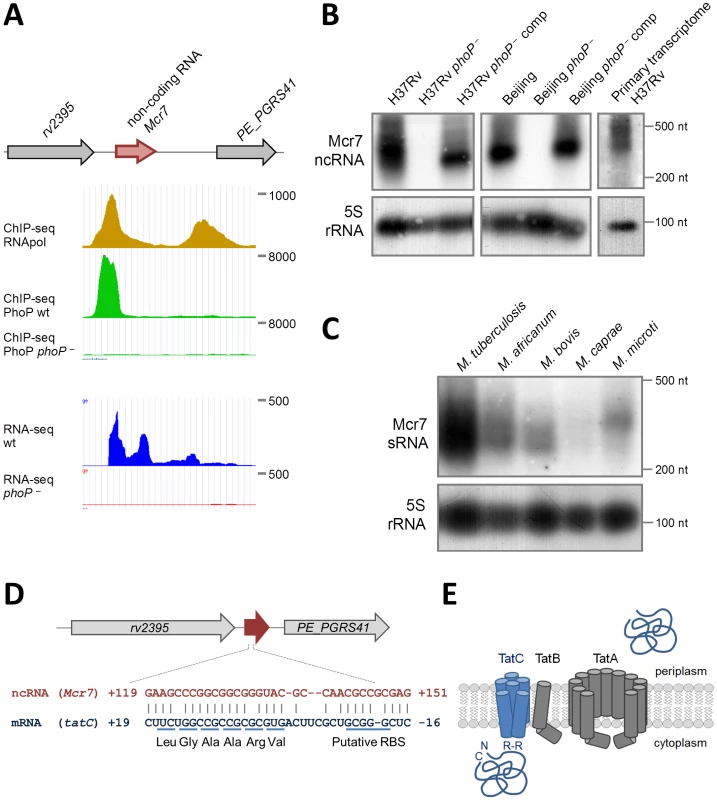
A. Diagram showing chromosomal location of mcr7 in the rv2395-PE_PGRS41 intergenic region. ChIP-seq and RNA-seq profiles of the mcr7 locus showing PhoP binding and transcriptional signals. Note the absence of transcription in the phoP mutant that correlates with lack of PhoP-binding in this strain. B. Northern blot analysis using a mcr7 antisense probe in wild type, phoP mutant and phoP-complemented strains in H37Rv and Beijing GC1237 genetic backgrounds. Detection of the Mcr7 transcript in the primary, unprocessed transcriptome from H37Rv is also shown. Expression of the 5S rRNA is used as a control of RNA loaded in each lane. C. Northern blot analysis of Mcr7 in representative species of the M. tuberculosis complex. Note the higher expression level of this non-coding RNA in M. tuberculosis H37Rv relative to other members of the M. tuberculosis complex. Expression of the 5S rRNA is used as a control of RNA loaded in each lane. D. Complementarity between Mcr7 and the 5′-end of the tatC mRNA. The predicted ribosome binding site (RBS) and positions of the first 6 codons of tatC are indicated. E. Schematic representation of the Tat system involved in protein translocation from the cytoplasm to the extracellular environment. TatC is involved in recognition of Arg-Arg (RR) motifs within the signal peptide of secreted proteins. Northern blot analysis was performed on RNA extracted from wild type M. tuberculosis, phoP mutants and complemented strains in two different genetic backgrounds: the H37Rv laboratory strain and GC1237, a clinical isolate belonging to the Beijing family [21]. We detected an RNA of approximately 350 nt in length in wild type and complemented strains (Figure 2B), whose 5′-end could be mapped from the RNA-seq profile to coordinate 2,692,165 in the H37Rv genome. In contrast, we were unable to identify this RNA in the phoP mutants even when 10-times more RNA was used in Northern blot experiments (Figure S2). These results were also confirmed by qRT-PCR showing barely detectable levels of Mcr7 in the M. tuberculosis phoP mutant (Figure S2). The complete lack of expression of this ncRNA in the M. tuberculosis phoP mutant validates the findings obtained by ChIP-seq and RNA-seq and confirms the phoP mutant as an Mcr7-deficient strain as well. Next, we sought to establish whether Mcr7 is a primary transcript or conversely processed from a longer RNA. Detection of an mcr7 transcript of approximately 350 nt in length in the primary transcriptome of H37Rv (Figure 2B) ruled out the latter possibility and indicated that Mcr7 is a primary, unprocessed RNA.
We then investigated the presence of mcr7 in the Mycobacterium genus by bioinformatic analysis and found that it is predicted to be restricted to the M. tuberculosis complex. We therefore obtained expression data for mcr7 in five representative species. Surprisingly, the Mcr7 ncRNA is only weakly expressed in M. africanum, M. bovis, M. caprae and M. microti as compared to the high expression levels in M. tuberculosis (Figure 2C).
Biological function of the Mcr7 ncRNA
After demonstrating the strict PhoP regulation and the predominant expression of mcr7 in M. tuberculosis species, we tried to assign a biological role to this ncRNA. Most trans-acting ncRNA act by limited complementarity with their target mRNA, which results in post-transcriptional regulatory mechanisms [22]. A highly structured fold of Mcr7 with a 33-nt free loop (Figure S3) was predicted using the RNAfold server. A bioinformatic search for putative targets of Mcr7 resulted in 18 candidates with complementarity in their 5′-end portion (Figure S3). Given that some ncRNA exert their regulatory function through interaction with their loop structures [23], we focused on mRNAs that annealed with the 33-nt loop. As our previous unpublished results suggested that secretion of Tat-dependent substrates was affected in the phoP mutant strain, we focused on the predicted interaction between tatC and Mcr7. Interestingly, the 5′-end of the tatC mRNA is predicted to base pair with the major loop of Mcr7 (Figure 2D). The interacting region includes the putative ribosome binding site (RBS) and the first 6 codons of the tatC mRNA, suggesting that Mcr7 probably prevents ribosome loading and, consequently, translation of tatC mRNA. The tatC gene is essential for M. tuberculosis [24], and encodes a transmembrane protein that is part of the TatABC general secretory apparatus required for export of proteins with a twin arginine motif (RR) in their signal peptide [25] (Figure 2E). TatC recognizes the RR motif prior to protein translocation through the TatA channel (Figure 2E).
Proteomic analysis shows enrichment of Tat-dependent substrates in the secretome of M. tuberculosis phoP mutant lacking mcr7
Our prediction suggested that Mcr7 might regulate tatC at the post-transcriptional level by occlusion of the RBS and the consequent translational down-regulation (Figure 2D). Consequently, we studied the secretome from exponentially grown cultures of strain H37Rv, its phoP mutant and a phoP complemented mutant by in-depth proteomics. The enrichment ratio for each protein in the secreted fraction was calculated as the log2 of normalized peptide abundance between the desired strains. Results are presented in Worksheet 1 in Table S3. Upon applying a cutoff based on the Significance B value (B<0.05), 37 proteins were found to be more secreted in the phoP mutant compared to the wild type strain. Sixteen of these (43.24%) exhibited an RR motif within the first 50 aminoacids. On the contrary, 6 out of 35 proteins, that were more abundant in the wild type displayed the RR motif (17.14%).
These encouraging findings prompted us to compare the abundance of previously predicted Tat substrates [24], [26], [27] in our secretome experiments. Results indicated that these were significantly more present in the secreted fraction of the phoP mutant relative to wild type and complemented strains (Figure 3A). In addition, we compared the relative secretion levels of EsxA (ESAT-6), EsxB (CFP-10), EspA and EspC since these proteins are well-known PhoP-dependent ESX-1 secretion substrates [7] and thus serve as controls. As expected, the secretome of the phoP mutant contained very low amounts of EsxA, EsxB, EspA and EspC, thus showing the opposite trend to Tat-dependent substrates (Figure 3A). Next, we validated these results by Western blot analysis of Ag85C [26] and Rv2525c [24] as known Tat-dependent substrates and EsxA as a PhoP-dependent ESX-1 substrate. The results corroborated the proteomic studies: the secreted fraction of the phoP mutant showed higher levels of Ag85C and Rv2525c proteins compared to the wild type and complemented mutant strains. On the contrary, EsxA secretion was undetectable in the phoP mutant compared to the strains harboring a wild type phoP allele (Figure 3B). Taken together, these results are consistent with a regulatory model involving PhoP, Mcr7 and tatC mRNA since the absence of Mcr7 in the phoP mutant would result in more efficient TatC translation and therefore increased secretion (Figure 3C).
Fig. 3. Analysis of the M. tuberculosis secretome identifies a high proportion of Tat substrates enriched in the M. tuberculosis phoP mutant. 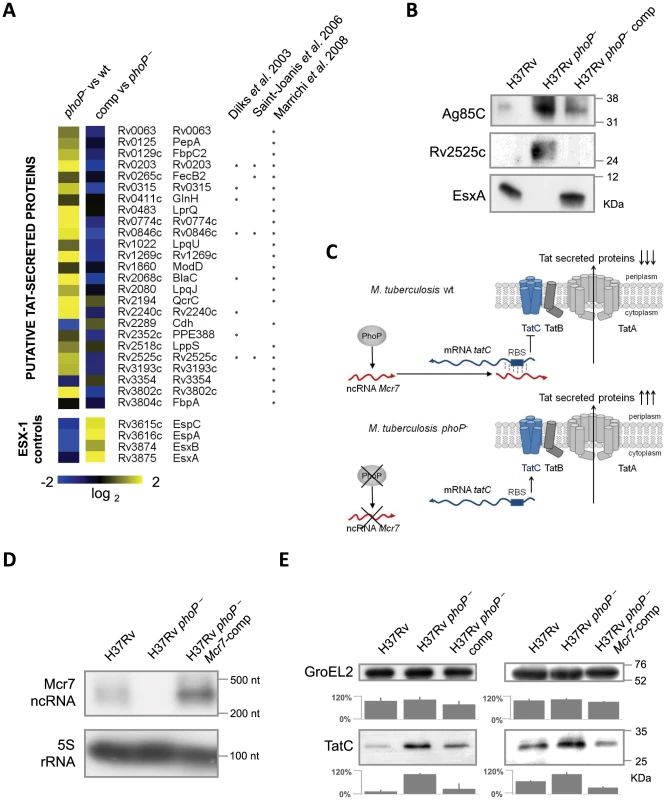
A. Heat map displaying values from in-depth proteomic quantifications of the secreted protein fraction. Values for EsxA, EsxB, EspA and EspC are displayed as internal controls since these proteins are barely detectable in the M. tuberculosis phoP mutant. The other proteins are representative examples of putative Tat-secreted substrates according to [24], [26], [27]. Note the opposite trend between EsxA/EsxB/EspA/EspC and Tat-secreted proteins. B. Western blot analysis of Ag85C, Rv2525c and EsxA in M. tuberculosis wild type, phoP mutant and complemented strains. Results are representative of three independent experiments. C. Proposed regulatory network involving PhoP, Mcr7 and tatC. PhoP controls transcription of Mcr7 (red transcript), which anneals to the 5′-end of the tatC mRNA (blue transcript) occluding the RBS (boxed) and consequently impairing translation of TatC. D. Northern blot analysis of Mcr7 in wild type, phoP mutant and mcr7-complemented strains of M. tuberculosis. Introduction of mcr7 under the 16S rRNA promoter in a phoP mutant efficiently restores production of this molecule. Expression of the 5S rRNA is used as a control of RNA loaded in each lane. E. Analysis of TatC expression by Western blot in wild type, phoP mutant, phoP-complemented mutant and mcr7-complemented strains of M. tuberculosis. Note the higher expression levels of TatC in the phoP mutant compared to the wild type. Complementation of a phoP mutant with phoP or mcr7 restored wild type levels of TatC. GroEL2 is used as a control of protein loaded in each lane. Signal intensity was quantified from three independent experiments and plotted in the graphs below the Western blot images. Reintroduction of mcr7 in an M. tuberculosis phoP mutant restores secretion of Tat-dependent proteins to wild type levels
In order to confirm that mcr7, but no other PhoP-dependent genes, influenced secretion of Tat substrates via post-transcriptional regulation of tatC mRNA, we restored Mcr7 production in the M. tuberculosis phoP mutant that we have previously demonstrated to be mcr7 deficient (Figure 2). The mcr7 gene was cloned downstream of the promoter for the 16S rRNA gene and the resultant construct was integrated into the chromosome of the H37Rv phoP mutant, thereby obtaining the mcr7-complemented strain. Northern blot experiments confirmed the authenticity and length of the mcr7 transcript (Figure 3D and Figure S4). Detection of TatC in whole-cell lysates of H37Rv, its phoP mutant, the phoP-complemented mutant and the mcr7-complemented strain demonstrated increased production of this protein in the phoP mutant in agreement with our proposed model (Figure 3E). Reintroduction of phoP complemented this phenotype as expected. Ectopic expression of mcr7 in the phoP mutant was sufficient to restore TatC levels to the wild type condition (Figure 3E), indicating that mcr7 per se was able to modulate expression of TatC, presumably by regulating translation of tatC mRNA.
To examine the phenotypic effect caused by reintroducing mcr7, we first showed by qRT-PCR that expression of the ncRNA from the surrogate promoter was only about 6-fold higher in the complemented mutant relative to the wild type strain (Figure 4A). Furthermore, we demonstrated that reintroduction of mcr7 in a phoP mutant did not influence the expression of the PhoP regulon that remained at undetectable levels in both the phoP mutant and mcr7-complemented strains (Figure 4A). Additionally, we proved that transcription of the tatC mRNA in the phoP mutant and in the mcr7-complemented strains showed no significant difference as compared to H37Rv (Figure 4A). Overall, these data ruled out a transcriptional impact of mcr7 on gene expression and supported the notion of a post-transcriptional effect exerted by the ncRNA on tatC.
Fig. 4. Reintroduction of mcr7 in an M. tuberculosis phoP mutant restores secretion of Tat-dependent substrates to wild type levels. 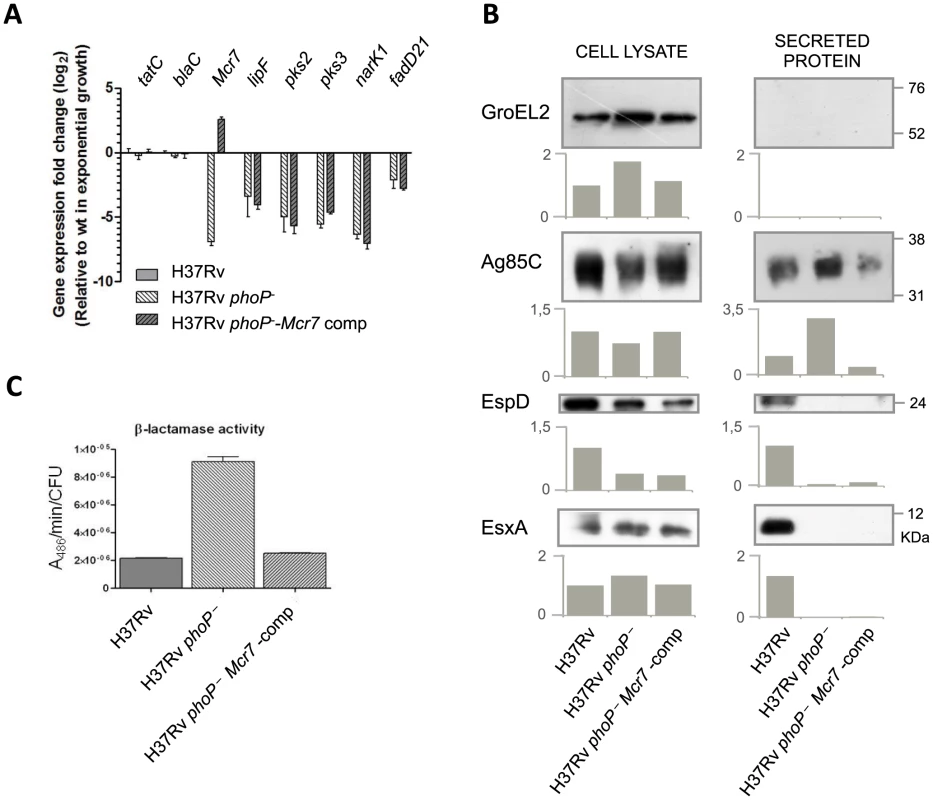
A. qRT-PCR analysis of tatC, mcr7 and other genes from the PhoP regulon (lipF, pks2, pks3, narK1 and fadD21). Bars represent fold changes in the expression levels in the phoP mutant and mcr7-complemented strains relative to M. tuberculosis wild type grown exponentially in 7H9 medium. Expression of the tatC mRNA is independent of phoP mutation and mcr7 expression. Note that complementation with mcr7 does not affect expression of the PhoP regulon. Results are representative of three independent experiments and error bars are the standard deviation of the mean. B. Western blot experiments of GroEL2, Ag85C, EspD and EsxA in the whole cell and secreted fractions of M. tuberculosis wild type, its phoP mutant and the mcr7-complemented strains. Equal protein amounts were loaded per well. GroEL2 is used as a control of bacterial integrity in each sample. Signal intensity was quantified and plotted in the graphs below the Western blot images. Results are representative of three independent experiments. C. Measure of β-lactamase activity of the BlaC protein by nitrocefin chromogenic assay. Activity is calculated relative to the CFU/ml in each strain. We then investigated whether reintroduction of mcr7 in the phoP mutant restored secretion of Tat-dependent substrates to wild type levels. Western blot analysis of Ag85C in the whole-cell lysate and secreted fractions showed that while Ag85C is produced at very similar levels in all strains, secretion of this protein was more pronounced in the phoP mutant compared to the wild type and mcr7 complemented strains (Figure 4B). By contrast, inspection of ESX-1 substrates showed no detectable secretion of EsxA and EspD in either the phoP mutant or the mcr7-complemented mutant strains (Figure 4B). Therefore, the Mcr7 ncRNA did not impact the activity of the ESX-1 secretion apparatus whereas it did affect protein secretion through the Tat system.
Finally, we measured the enzymatic activity of a Tat-dependent substrate, the well-characterized BlaC [28] beta-lactamase using the chromogenic cephalosporin substrate nitrocefin. The results indicated faster reaction kinetics in the phoP mutant relative to wild type (Figure 4C), a finding correlated with the protein secretion levels observed in proteomic studies. Again, complementation with mcr7 was sufficient to successfully restore BlaC activity to wild type levels (Figure 4C). Since no deregulation of blaC transcription was observed in the phoP mutant (Worksheet 1 in Table S2) and in the mcr7-complemented strain (Figure 4A), we attributed this effect to post-transcriptional regulation of TatC by Mcr7.
Discussion
High-resolution systems biology is helping greatly to unravel the complexities of the M. tuberculosis “regulome”. Recent works have uncovered a plethora of ncRNA [29] and reconstructed the hypoxia regulatory network [15] in this pathogen. In this study we integrated data from complementary high-throughput sequencing technologies and obtained extensive knowledge on PhoP-dependent transcriptional regulation in the tubercle bacillus. Specifically, ChIP-seq identified the PhoP binding sites along the M. tuberculosis chromosome (Figure 1), whereas strand-specific, single-nucleotide resolution transcriptomic analyses revealed previously unknown features of the PhoP regulatory network in vitro. Although good overlap was observed between RNA-seq data and published transcriptomic analyses ([5], see Worksheet 2 in Table S2 for comparison), major progress has been made as compared to traditional microarray-based approaches as indirect regulatory effects present in former studies [5], [14] have been unmasked. Importantly, we found many genes that were deregulated in the phoP mutant despite the absence of a PhoP binding signal in the respective promoter regions. In this regard, PhoP was found to control expression of several other regulatory proteins (e.g. EspR, WhiB1, WhiB3, WhiB6), which act in downstream regulatory cascades [30], [31]. Independent confirmation for this conclusion was presented recently in a regulatory model predicting production of acyltrehalose-derived lipids to be coordinated by a PhoP-WhiB3 network via regulation of pks2 and pks3 [15]. Additional proof was obtained upon comparing the transcriptome of the phoP mutant with the predicted binding sites of WhiB1, WhiB3 and WhiB6 (information available at http://www.tbdb.org/). Indeed, the overlap was found to include rv0996, rv1004c, rv1040c, rv2274c and rv3289c for WhiB1, rv1040c for WhiB3, and rv2396 for WhiB6. Concerning EspR, deregulation of the espACD operon was reported in an espR knockout strain [32], where a binding site for EspR was demonstrated [30]. Interestingly, PhoP was shown to bind upstream of lipF and of lppL, where EspR is also present [30], thereby increasing the complexity of the regulatory machinery at these loci. The small ncRNA Mcr7 can also be considered as an intermediate regulator in the PhoP global network, although it likely exerts its function at the post-transcriptional level. We will come back to Mcr7 later in the discussion.
Comparison of ChIP-seq and RNA-seq profiles uncovered several genes associated with a PhoP binding site but whose expression was not altered in a PhoP-deficient strain. We hypothesize that these genes may be subjected to additional layers of regulation or may respond to yet unexplored environmental conditions. This is exemplified by the mihF gene, which, despite its upstream PhoP binding site, was not found to be deregulated. Since the signal sensed and the downstream components of the PhoPR two-component system have not yet been completely elucidated, it is conceivable that signal transduction originating from PhoR may involve other factors than PhoP, thereby fine-tuning gene expression in M. tuberculosis in different conditions.
Galagan and colleagues recently mapped the binding sites of 50 transcription factors, including PhoP, in M. tuberculosis [15] by exploiting a tetracycline-inducible promoter system to overexpress the FLAG-tagged version of the protein of interest and using anti-FLAG antibodies in ChIP-seq experiments. Contrary to their approach, we worked in physiological conditions and performed immunoprecipitation assays using antibodies directed against native PhoP, thus avoiding artifacts due to abnormal expression levels or to biased protein-antibody interaction. In addition, use of the phoP mutant allowed false positive signals to be avoided. Interestingly, the number of peaks pinpointed in our work (35) was considerably smaller than that reported in Galagan et al. [15], where several signals were detected in intergenic as well as in intragenic regions. This could reflect the different methods employed, since artificial expression of PhoP may have increased binding to low affinity sites. Head-to-head comparison revealed that all but two of the peaks (yajC-gabT and lpdA-rv3304) identified here were also present in the other study (see Worksheet 2 in Table S1 and Worksheet 3 in Table S1 for detailed comparison).
The position of the PhoP peak with respect to the ORF start site merits discussion. We noticed that in the case of lipF and rv1535, the binding site was located >400 bp upstream of the translation start codon. This is consistent with previous results of footprinting assays for lipF [13] and with the presence of long 5′-UTRs, with presumptive regulatory roles, for lipF and rv1535 in the respective RNA-seq profiles. Interesting observations were made upon alignment of the PhoP and RNApol ChIP-seq profiles. PhoP was located upstream of the enzyme in most cases, suggesting a role as a positive regulator, later confirmed by RNA-seq data. On the contrary, PE8 was the only gene associated with a PhoP signal downstream of the RNApol peak, indicating potential steric hindrance and thus prevention of RNApol progression throughout the coding sequence. ChIP-seq analysis can therefore provide clues as to the role fulfilled by a transcription factor depending on the position of its binding site with respect to RNApol.
Inspection of the PhoP targets uncovered unusual binding sites in the 3′-end of ORF such as the one between hddA and ldtA. Since this peak is at the end of two convergent genes, it likely corresponds to an unmapped small RNA. Closer inspection of this intergenic region revealed the presence of a novel small transcript downregulated in the phoP mutant. Another case is represented by rv2137c, where the PhoP interacting region was mapped within the ORF, suggesting an alternative, PhoP-dependent start codon. Indeed, a polypeptide starting at the ATG codon at nucleotide 106, in frame with the currently annotated start site, shows more than 85% identity with the corresponding proteins in all other mycobacteria whose genomes have been sequenced.
The bipartite PhoP consensus sequence derived from ChIP-seq analysis is consistent with the crystal structure of the dimeric PhoP regulator that is predicted to bind to direct repeats [10]. It also agrees with previous footprinting experiments demonstrating binding of PhoP upstream of its own gene [11], [13] and in the promoter regions of lipF, fadD21, pks2 [13]. On the other hand, the divergent orientation of the PhoP binding motif relative to the pks3 gene can be subjected to different interpretations. It could be that the adjacent rv1179c is the gene directly controlled by PhoP while pks3 undergoes indirect regulation. Alternatively, transcriptional regulation in that locus might be independent of directional positioning of the transcription factor.
The last years have witnessed increased attention to ncRNA in prokaryotic organisms, including Salmonella enterica [33], Legionella pneumophila [34], Listeria monocytogenes [35] and M. tuberculosis [29]. These molecules have been predicted to exert their function at the post-transcriptional level by modulating translation of RNAs [36]. This process has important implications when bacteria face environmental stresses since it allows faster responses than classical transcriptional regulation. In this study we disclose the Mcr7 ncRNA encoded by the mcr7 gene, located between rv2395 and PE_PGRS41 (Figure 2). The latter was described as highly repressed in a phoP mutant from microarray experiments by Walters and co-workers [5]. Thanks to the increased resolution provided by RNA-seq, we can now identify the heavily deregulated gene as mcr7 rather than PE_PGRS41. The position of the probes in the microarray assay probably did not allow such precision. A similar observation can be made for a study that characterized the transcriptional differences between the avirulent strain H37Ra and H37Rv [6]. The gene encoding PE_PGRS41 (and likely the associated intergenic region carrying mcr7) was found to be the most highly deregulated. Importantly, H37Ra is a natural mutant in the phoP gene since it carries a polymorphism in the DNA binding domain [7], [11], thus indicating that reduced virulence is associated with lack of PhoP activity and impaired expression of the locus encoding mcr7. We confirmed this prediction by measuring the expression levels of mcr7 in H37Ra by qRT-PCR. The ncRNA was found to be poorly detectable as compared to H37Rv (Figure S5).
In the same genomic region of the CDC1551 strain, Abramovitch and colleagues postulated the existence of the aprABC locus with aprC corresponding to PE_PGRS41 and aprA and aprB corresponding to ORFs MT2466 and MT2467 [37]. These ORFs were not predicted in strain H37Rv as neither their codon usage nor positional base composition are typical of true protein coding sequences [16], [38]. The mcr7 gene completely overlaps the hypothetical aprA. A major PhoP binding site precedes the mcr7 gene but there is none immediately upstream of PE_PGRS41. Our results from Northern blot experiments clearly showed one prominent band of approximately 350 nt in length corresponding to Mcr7, that was first described in M. bovis BCG [20]. This genomic locus is restricted to species belonging to the M. tuberculosis complex, including M. canettii, and was not identified in M. kansasii and in M. marinum, although expression of mcr7 was found to be particularly prominent in M. tuberculosis only. The expression pattern of mcr7 tallies with the proposed evolutionary pathway of the tubercle bacilli [39]. Indeed, those lineages that evolved from a common M. tuberculosis-like ancestor by multiple deletions (M. africanum, M. microti, M. caprae and M. bovis) express low-levels of the ncRNA as compared to the M. tuberculosis strain. In light of our findings, it is tempting to speculate that modulation of the activity of the Tat secretion system by means of a small RNA has played a role in shaping the adaptation of tubercle bacilli and/or in restricting their host spectrum. We investigated the potential role played by Mcr7 in virulence by performing ex vivo and in vivo infections. Complementation of the phoP mutant with mcr7 alone did not restore the wild type virulence and the strain was more attenuated than the phoP mutant (Figure S6). This phenotype may be related to the ectopic overexpression of Mcr7, which was indeed associated with small colony size (data not shown).
The predicted folding model of Mcr7 revealed the presence of a 33-nt loop with the potential to anneal to three candidate mRNAs: rv2767c, rv2053c and tatC (Figure S3). Since our results provided convincing evidence for increased secretion of the Tat substrates, Ag85A and Ag85C, in phoP mutants, we prioritized the study of Tat-dependent secretion. However, we cannot exclude a post-transcriptional impact of Mcr7 on expression of the hypothetical membrane proteins Rv2767c and Rv2053c, although their role in M. tuberculosis physiology is questionable since previous proteomic experiments failed to detect them in the total proteome or in cellular subfractions [40], [41], [42], [43]). Proteomic analysis demonstrated that proteins secreted through the Tat system are more abundant in the extracellular fraction of the PhoP-deficient strain (Figure 3). A genetic approach relying on complementation of the phoP mutant with mcr7 proved the involvement of the ncRNA in the regulation of Tat-dependent secretion at the post-transcriptional level while no impact on the amount of mRNA was observed (Figures 3 and 4). This is the first report describing the function of a ncRNA in M. tuberculosis. Notably, M. tuberculosis phoP mutants display pleiotropic phenotypic effects including impaired secretion of ESX-1 substrates [7], compromised production of sulphatides (SL), diacyltrehaloses (DAT) and polyacyltrehaloses (PAT) [8] and reduced virulence in the macrophage and mouse models of infection [4], [5]. Mcr7 was found to be sufficient to re-establish the wild type phenotype with respect to secretion of Tat substrates whereas ESX-1 substrates were unaffected, thus evoking a specific regulatory cascade where Mcr7 acts downstream of PhoP.
Overall, this work refined the role played by PhoP in control of gene expression in M. tuberculosis. A previous study reported that PhoP is involved in the regulation of the ESX-1 secretion system [7] but no direct evidence had been provided so far. Here we uncovered the existence of a novel regulatory cascade composed of at least two regulatory factors, PhoP and EspR, that ultimately controls ESX-1 functions, such as secretion of EsxA, via regulation of the espACD locus [7], [30]. In addition, we demonstrated a role for the PhoP-dependent ncRNA Mcr7 in Tat-dependent secretion of well-known M. tuberculosis antigens, namely the immunodominant Ag85 complex. PhoP could therefore also mediate antigenicity and pathogenesis via the Ag85 complex itself and/or through trehalose 6,6-dimycolate, an abundant glycolipid in the mycobacterial cell wall whose biosynthesis is catalyzed by Ag85 proteins. The Ag85 complex is also involved in binding to human fibronectin, important for cell adhesion and invasion [44], [45]. Mcr7 could therefore represent the missing link between PhoP and the downstream processes required for successful infection of the host. Finally, our findings provided a new molecular basis to explain the better protection against tuberculosis conferred by the candidate vaccine strain MTBVAC, which carries a deletion in phoP [9]. While the reduced virulence results mainly from abrogation of the ESX-1 secretion system and possibly from lack of complex lipids, its efficacy may be ascribed to improved antigenicity properties following silencing of Mcr7 and the ensuing increase in secretion of Tat substrates such as the Ag85 proteins.
Materials and Methods
Ethics statement
All animal work has been conducted according to the national and international guidelines. The protocols for animal handling were previously approved by University of Zaragoza Animal Ethics Committee (protocol number PI43/10).
Bacterial strains and culture conditions
Mycobacterium tuberculosis H37Rv [16], GC1237 [21] wild type strains and their isogenic phoP mutants were previously described [11]. The growth rate of the wild type and of the mutant strains were similar (Figure S7). M. bovis AF2122/97 [46], M. caprae M57 [39], M. microti 15496 and M. africanum MAF419 [47] were used as representative strains of the M. tuberculosis complex. Mycobacterial strains were grown at 37°C in 7H9 medium (Difco) supplemented with 0.05% Tween 80 and 10% albumin-dextrose-catalase (ADC, Middlebrook) or on 7H10 plates supplemented with 10% ADC. For M. tuberculosis complex strains different from M. tuberculosis, 40 mM sodium pyruvate was added to the medium. Escherichia coli DH5α used for cloning procedures was grown at 37°C in LB broth or on LB agar plates. Kanamycin (20 µg/ml) and hygromycin (20 µg/ml) were used as appropriate.
Chemicals, antibodies and oligonucleotides used in this study
All chemicals were purchased from Sigma-Aldrich, unless otherwise stated. Immunoblotting was performed with mouse monoclonal anti-EsxA antibodies (Hyb 076-08, Abcam), mouse monoclonal anti-Ag85C antibodies (HYT27, Abcam), mouse monoclonal anti-GroEL2 antibodies (BDI578, Abcam), rabbit polyclonal anti-Rv2525c antibodies [24], rat polyclonal anti-EspD antibodies (kindly provided by Jeffrey Chen) and rabbit polyclonal anti-TatC (Eurogentec) antibodies. Polyclonal antibodies to the transcriptional regulator PhoP of M. tuberculosis were obtained from rabbits that received five doses of PhoP (0.5 mg), at weeks 0, 4, 8, 12 and 16, respectively. These anti-PhoP antibodies were validated by ELISA (ZEU-Immunotec Zaragoza, Spain). Sequences of the oligonucleotides used in this study will be provided upon request.
Plasmid construction
The pAZ31 plasmid was kindly provided by Ainhoa Arbues. The pWM222 plasmid was used for phoPR complementation in northern blot experiments and was constructed as follows. A 2.7 kbp region spanning the phoPR operon was amplified by PCR using primers (5′-ATACTAGTGGCATCACCCAACGCTTGTT-3′) and (5′-ATACTAGTGGTGAGCCAGCTGATCGG-3′). This PCR product was digested with SpeI and subsequently transferred into a pMV361 [48] derivative deleted from the phsp60 promoter. In this construct, the phoPR operon is expressed from its native promoter. Plasmid pLZ11 used for mcr7 expression was constructed by inserting a transcriptional fusion of the rrs (16S rRNA) promoter with the mcr7 transcript following a similar strategy to that described in [29]. This transcriptional fusion was accomplished using an overlapping two-step PCR strategy. Briefly, the rrs promoter was PCR amplified using primers rrsOV Fw: GACGTCCCGCAGCTGTCGAGCGCT and rrsOV Rv: GGGCCGCCGGCCCTGCCAGTCTAATACAAATCC. The mcr7 region was amplified using primers mcr7Ov Fw: GACTGGCAGGGCCGGCGGCCCGACACA and mcr7Ov Rv: AAGCTTCCACCTTCTCGTTACCCGCCTCTG. Both PCR products overlap in 21 bp (underlined nucleotides) and were used as self-templates in a PCR reaction. The entire transcriptional fusion was amplified by PCR using the flanking primers rrsOV Fw and mcr7Ov Rv, digested with HindIII and EcoRI and introduced between the HindIII and EcoRI sites of pMV361. The resulting construct was introduced in mycobacteria by electroporation and colonies carrying a chromosome-integrated vector were checked by PCR.
Chromatin immunoprecipitation experiments
Chromatin immunoprecipitation experiments were performed as previously described [49] with the following modifications. We performed two independent ChIP-seq experiments with the wild type strain H37Rv and one experiment with the control phoP mutant. Briefly, M. tuberculosis cultures were grown to exponential phase (optical density at 600 nm of 0.4) and cross-linked with 1% formaldehyde for ten minutes at 37°C. Cross-linking was quenched by addition of glycine (125 mM). Cells were then washed twice with Tris-buffered saline (TBS, 20 mM Tris-HCl pH 7.5, 150 mM NaCl), resuspended in 4 ml immunoprecipitation (IP) buffer (50 mM Hepes-KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail from Roche) and sonicated to shear DNA using Bioruptor (Diagenode). Cell debris was removed by centrifugation and the supernatant used in IP experiments. Nucleo-protein extracts were incubated with 50 µl of rabbit polyclonal anti-PhoP antibodies at 4°C for 2 days on a rotating wheel. Complexes were subsequently precipitated with Dynabeads (Dynal, anti-rabbit, Invitrogen) for three hours at 4°C. Beads were washed twice with IP buffer, once with IP buffer plus 500 mM NaCl, once with buffer III (10 mM Tris-HCl pH 8, 250 mM LiCl, 1 mM EDTA, 0.5% Nonidet-P40, 0.5% sodium deoxycholate), once with Tris-EDTA buffer pH 7.5. Elution was performed in 50 mM Tris-HCl pH 7.5, 10 mM EDTA, 1% SDS for 40 minutes at 65°C. Samples were finally treated with RNAse A for one hour at 37°C and cross-links were reversed by incubation for two hours at 50°C and for eight hours at 65°C in 0.5× elution buffer with 50 µg Proteinase K (Eurogentec). DNA was purified by phenol-chloroform extraction and quantified by Nanodrop and Qubit fluorometer according to the manufacturer's recommendations (Invitrogen).
Library preparation for ChIP-seq analysis and Illumina high-throughput sequencing
DNA fragments obtained from the immunoprecipitation procedure were used for library construction and sequencing with the ChIP-Seq Sample Preparation Kit (Illumina), according to the protocol provided by the manufacturer. One lane per library was sequenced on the Illumina Genome Analyzer IIx at the Lausanne Genomics Technologies Facility using the SR Cluster Generation Kit v2 and SBS 36 Cycle Kit v2. Data were processed with the Illumina Pipeline Software v1.40.
Genome annotation
All analyses in this study were carried out using the M. tuberculosis H37Rv annotation from the TubercuList database (http://tuberculist.epfl.ch/), which includes 4019 protein coding sequences (CDS), 73 genes encoding for stable RNAs, small RNAs and tRNAs. In order to quantify protein occupancy and transcription across the entire genome, 3080 intergenic regions (regions flanked by two non-overlapping CDS) were included, resulting in a total of 7172 features.
ChIP-seq data analysis
ChIP-seq analysis was performed using the HTSstation pipeline at EPFL (http://htsstation.epfl.ch/). Briefly, the single-ended sequence reads generated from ChIP-seq experiments were aligned to the M. tuberculosis H37Rv genome (NCBI accession NC_000962.2) using Bowtie [50] with options “-l 28 -best -strata”. Peaks were analysed using MACS v.1.4 [51] with parameters “-bw 200 -m 10100”. Alignment files were converted to bigWig format for visualization in the UCSC genome browser Mycobacterium tuberculosis H37Rv 06/20/1998 Assembly [52]. To determine the level of ChIP-seq enrichment for each feature, an enrichment ratio (ER) was calculated by dividing the read count for the ChIP-seq sample in the wild type strain by the read count for the mutant (control) sample.
PhoP binding site motifs were searched using the MEME Suite (http://meme.nbcr.net/meme/) in sequence regions encompassing 100 bp upstream and 100 bp downstream of the predicted peak summit. Motif sequence logo was obtained using WebLogo3 (http://weblogo.threeplusone.com/).
RNA extraction
Mycobacterial cultures (one for the wild type strain and one for the phoP mutant) were grown to exponential phase (OD600 = 0.5-0.6) and pelleted by centrifugation. To minimize RNA degradation bacteria were resuspended in 1 ml RNA Protect Bacteria Reagent (Qiagen), incubated for 5 min at room temperature and then centrifuged. Bacterial pellets were resuspended in 0.4 ml lysis buffer (0.5% SDS, 20 mM NaAc, 0.1 mM EDTA) and 1 ml phenol:chloroform (pH = 4.5) 1∶1. Suspensions were transferred to tubes containing glass beads (Qbiogene) and lysed using a ribolyser (Fast-prep instrument) with a three-cycle program (15 sec at speed 6.5 m) including cooling the samples on ice for 5 min between pulses. Samples were then centrifuged and the homogenate was removed from the beads and transferred to a tube containing chloroform:isoamylalcohol 24∶1. Tubes were inverted carefully before centrifugation and the upper (aqueous) phase was then transferred to a fresh tube containing 0.3 M Na-acetate (pH = 5.5) and isopropanol. Precipitated nucleic acids were collected by centrifugation. The pellets were rinsed with 70% ethanol and air dried before being re-dissolved in RNase-free water. DNA was removed from RNA samples using Turbo DNA free (Ambion) by incubation at 37°C for 1 h. RNA integrity was assessed by agarose gel electrophoresis and absence of contaminating DNA was checked by lack of amplification products after 30 PCR cycles.
Primary, unprocessed RNA from H37Rv was prepared as indicated in [53]. Briefly, 10 µg total RNA were treated with 10 U of Terminal 5′-phosphate dependent Exonuclease (Epicentre) for 24 h at 30°C followed by phenol extraction and isopropanol precipitation. Successful preparation of primary transcriptome was confirmed by lack of 23S/16S rRNA bands in agarose gels.
Library preparation for RNA-seq analysis and Illumina high-throughput sequencing
100 ng of total RNA were mixed with 5× Fragmentation buffer (Applied Biosystems), incubated for 4 minutes at 70°C and then transferred immediately on ice. RNA was purified using RNAClean XP beads (Beckman Coulter), according to the manufacturer's recommendations, and subsequently treated with Antarctic phosphatase (New England Biolabs). RNA was then re-phosphorylated at the 5′-end with polynucleotide kinase (New England Biolabs) and purified with Qiagen RNeasy MinElute columns. In order to ensure strand-specificity, v1.5 sRNA adapters (Illumina) were ligated at the 5′ - and 3′-ends using RNA ligase. Reverse transcription was carried out using SuperScript III Reverse Transcriptase (Invitrogen) and SRA RT primer (Illumina). Twelve cycles of PCR amplification using Phusion DNA polymerase were then performed and the library was finally purified with AMPure beads (Beckman Coulter) as per the manufacturer's instructions. A small aliquot (2.5 µl) was analyzed on Invitrogen Qubit and Agilent Bioanalyzer prior to sequencing on Illumina HiSeq 2000 using the TruSeq SR Cluster Generation Kit v3 and TruSeq SBS Kit v3. Data were processed with the Illumina Pipeline Software v1.82.
RNA-seq data analysis
The single-ended sequence reads generated from RNA-seq experiments were aligned to the M. tuberculosis H37Rv genome (NCBI accession NC_000962.2) using Bowtie2 with default parameters [54]. Read counts for all annotated features were obtained with htseq-count program (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html). Regions where genes overlapped were excluded from counting. Reads spanning more than one feature were counted for each feature. Since the RNA library was strand-specific, the orientation of sequence reads had to correspond to the orientation of annotated features to be counted. Analysis of differential gene expression was carried out using the DESeq package [55].
Quantitative PCR (qRT-PCR) for ChIP-seq and RNA-seq data validation
One microgram of M. tuberculosis RNA was converted to cDNA using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's recommendations. All PCR primers were designed using Primer Express software (Applied Biosystems). The 10 µl PCR reaction consisted of 1× Sybr Green PCR Master Mix (Applied Biosystems), 0.25 µM of each primer and 1 µl of 1∶10 diluted cDNA or IP DNA from immunoprecipitation reactions. Reactions were carried out in triplicate in an Applied Biosystems StepOnePlus Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. Melting curves were constructed to ensure that only one amplification product was obtained. In the case of qRT-PCR for RNA-seq data confirmation, normalization was obtained to the number of sigA molecules in each sample. Regarding the qPCR for ChIP-seq data validation, the number of target molecules was normalized to the mutant (control) sample, after subtraction of the background represented by the mock-IP (no antibody control).
Northern blot
Northern blot was performed using the DIG Northern starter kit (Roche) following the manufacturer's recommendations. Briefly, total RNA was separated using denaturing 1% agarose gels in 1× MOPS buffer containing 2% formaldehyde. RNA was transferred by capillary blotting to Hybond-N+ nylon membranes (Amersham) and UV-crosslinked prior to incubation with the desired probe. Digoxigenin (DIG)-labelled probes were synthesized to detect rrf (5S rRNA) and mcr7 transcripts using the primer pairs NB-5S-rRNA-fw (ttacggcggccacagcgg)/NB-T7-5S-rRNA-rv (taatacgactcactatagggtgtcctacttttccacccggagggg), NB-mcr7-fw (ccggcggcccgacacatg)/NB-T7-mcr7-rv (taatacgactcactatagggacccgctcaagcaggtcg) respectively. The T7 promoter used for in vitro transcription and labeling of RNA is underlined. RNA transcripts complementary to each probe were detected by Western-blot using an anti-DIG antibody conjugated to alkaline phosphatase and the chemiluminescent substrate CDP-Star.
Prediction of Mcr7 interactors
The secondary structure fold of mcr7 was predicted using the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Prediction of mcr7 putative targets was performed using TargetRNA (http://cs.wellesley.edu/~btjaden/TargetRNA2/index.html) allowing antisense complementarity from -80 to +20 relative to ORF translation start sites of M. tuberculosis H37Rv. A minimum hybridization seed of 7 nt and a p-value threshold of 0.05 were required for target transcripts.
Preparation of protein samples
In order to avoid albumin contamination in the secreted protein fraction, cultures were grown in 7H9 (Difco) 0.05% Tween 80 supplemented with 0.2% dextrose, 0.085% NaCl. After 2-3 weeks incubation at 37°C, cultures were pelleted by centrifugation. The supernatant containing secreted proteins was incubated with 10% trichloroacetic acid (TCA) for one hour in ice and then centrifuged at 4°C for 30 min. Pelleted proteins were rinsed with cold acetone and then resuspended in 150 mM TrisHCl pH 8. Protein integrity and absence of albumin contamination was checked by SDS-PAGE and Coomassie staining. The pelleted fraction of bacterial cultures was used for extraction of whole-cell proteins. The pellet was resuspended in PBS containing 1% triton ×100 and a cocktail of protease inhibitors (Roche) and sonicated for 30 minutes at 4°C using a Bioruptor (Diagenode). Samples were then centrifuged and the upper phase containing whole-cell lysate was used in downstream experiments. To prepare whole-cell extracts for detection of TatC by Western blot, proteins were further solubilized with 9 M urea, 70 mM DTT and 2% Triton X-100 followed by TCA precipitation and final resuspension in 150 mM TrisHCl pH 8.
Dimethyl labeling and SAX fractionation
Each sample (8 µg) was reconstituted in 50 µl of 4 M Urea, 10% acetonitrile and buffered with Tris-HCl pH 8.5 to a final concentration of 30 mM. Proteins were reduced using 10 mM dithioerythritol (DTE) at 37°C for 60 min. Cooled samples were subsequently incubated in 40 mM iodoacetamide at 37°C for 45 min in a light-protected environment. Reaction was quenched by addition of DTE to a final concentration of 10 mM. A two-step digestion was performed using Lys-C (1∶50 enzyme: protein) for 2 hours at 37°C. The lysates were first diluted 5-fold and samples were again digested overnight at 37°C using Mass Spectrometry grade trypsin gold (1∶50 enzyme: protein) and 10 mM CaCl2. Reaction was stopped by addition of 2 µl of pure formic acid (FA) and peptides were concentrated by vacuum centrifugation to a final volume of 70 µl. Samples were dimethyl-labeled as previously described [56]. The sample H37Rv phoP- was labeled with light dimethyl reactants (CH2O + NaBH3CN), the sample H37Rv was labeled with medium reactants (CD2O + NaBH3CN) and the sample H37Rv phoP- complemented was labeled with heavy methyl reactants (13CD2O + NABD3CN). As a final step of labeling procedure, samples were mixed in a 1∶1∶1 [(Light: Medium: Heavy) ratio and extensively lyophilized. Technical replicates were obtained. SAX fractionation was performed as previously described [57]. The eluted fractions were dried by vacuum centrifugation and used for LC-MS analysis.
Mass spectrometry and data analysis
Each SAX fraction was resuspended in 2% acetonitrile, 0.1% FA for LC-MS/MS injections and then loaded on a homemade capillary pre-column (Magic AQ C18; 3 µm by 200 Å; 2 cm×100 µm ID) and separated on a C18 tip-capillary column (Nikkyo Technos Co; Magic AQ C18; 3 µm by 100 Å; 15 cm×75 µm). MS/MS data was acquired in data-dependent mode (over a 4 hr acetonitrile 2–42% gradient) on an Orbitrap Q exactive Mass spectrometer equipped with a Dionex Ultimate 3000 RSLC nano UPLC system and homemade nanoESI source. Acquired RAW files were processed using MaxQuant version 1.3.0.5 [58] and its internal search engine Andromeda [59]. MS/Ms spectra were searched against M. tuberculosis strain H37Rv database R23 (http://tuberculist.epfl.ch/) [60]. MaxQuant default identification settings were used in combination with dimethyl-labeling parameters. Search results were filtered with a false-discovery rate of 0.01. Known contaminants and reverse hits were removed before statistical analysis. Relative quantification within different conditions was obtained by calculating the significance B values for each of the identified proteins using Perseus [58].
Western blotting
Protein samples were quantified using the RC DC protein assay (BioRad) and equal amounts of protein preparations were loaded per well. Proteins were separated on SDS-PAGE 12–15% gels and transferred onto PVDF membranes using a semidry electrophoresis transfer apparatus (Bio-Rad). Membranes were incubated in TBS-T blocking buffer (25 mM Tris pH 7.5, 150 mM NaCl, 0.05% Tween 20) with 5% w/v skimmed milk powder for 30 min prior to overnight incubation with primary antibodies at the dilution indicated below. Membranes were washed in TBS-T three times, and then incubated with secondary antibodies for 1 h before washing. Antibodies were used at the following dilutions: 1∶2,000 for anti-EsxA, 1∶5,000 for anti-Ag85C, 1∶500 for anti-GroEL2, 1∶1,000 for anti-Rv2525, 1∶1,000 for anti-EspD and 1∶1,000 for anti-TatC. Horseradish peroxidase (HRP) conjugated IgG secondary antibodies (Sigma-Aldrich) were used at a 1∶20,000 dilution. Signals were detected using chemiluminescent substrates (GE Healthcare).
Beta-lactamase assay
Bacterial cultures were grown to OD 600 nm 0.6–0.8 and pelleted. Nitrocefin was added to culture supernatants at 50 mM final concentration and absorbance was measured at 486 nm (Synergy HT BioTEK) every 10 minutes for 3h. Slope of linear range was measured and normalized against total CFUs of the culture.
J774A.1 infection
Virulence of the different M. tuberculosis strains was evaluated in J774A.1 murine macrophages according to a previously published procedure [61], [62]. Briefly, cells were grown in DMEM medium containing 10% fetal bovine serum at 37°C under 5% CO2. 10,000 macrophages per well were seeded into a 384-well plate in a total volume of 45 µl and incubated at 37°C for 30 minutes before infection. Cells were infected at an MOI of 10 with titrated stocks of H37Rv, phoP mutant, phoP-complemented and mcr7-complemented strains. On day 3, macrophage survival was measured by exposing the infected cells to PrestoBlue Cell Viability Reagent (Life Technologies) for 1 hour. Fluorescence was read using a TECAN Infinite M200 microplate reader and statistical analysis was performed with the unpaired T-test method.
Mouse infection
C57BL/6 mice were infected intranasally with an inoculum of 2.5×104 cfu/ml (6 mice per group). Four weeks post-infection mice were euthanized and lungs were plated on 7H11 plates supplemented with 0.5% glycerol, 10% albumin-dextrose-catalase (ADC, Middlebrook), polymixin B 50 U/ml, trimethoprim 0.02 mg/ml and amphotericin B 0.01 mg/ml. Unpaired T-test was used for statistical analysis. The protocols for animal handling were previously approved by University of Zaragoza Animal Ethics Committee (protocol number PI43/10).
Data access
The ChIP-seq and RNA-seq datasets have been deposited in NCBI's Gene Expression Omnibus [63] under accession number GSE54241.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE (Proteomics Identification Database) partner repository [64] with the dataset identifier PXD000698.
Supporting Information
Zdroje
1. ErnstJD (2012) The immunological life cycle of tuberculosis. Nat Rev Immunol 12 : 581–591.
2. RussellDG (2011) Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev 240 : 252–268.
3. BretlDJ, DemetriadouC, ZahrtTC (2011) Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol Mol Biol Rev 75 : 566–582.
4. PerezE, SamperS, BordasY, GuilhotC, GicquelB, et al. (2001) An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol 41 : 179–187.
5. WaltersSB, DubnauE, KolesnikovaI, LavalF, DaffeM, et al. (2006) The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol 60 : 312–330.
6. LeeJS, KrauseR, SchreiberJ, MollenkopfHJ, KowallJ, et al. (2008) Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe 3 : 97–103.
7. FriguiW, BottaiD, MajlessiL, MonotM, JosselinE, et al. (2008) Control of M. tuberculosis ESAT-6 Secretion and Specific T Cell Recognition by PhoP. PLoS Pathogens 4: e33.
8. Gonzalo AsensioJ, MaiaC, FerrerNL, BariloneN, LavalF, et al. (2006) The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem 281 : 1313–1316.
9. ArbuesA, AguiloJI, Gonzalo-AsensioJ, MarinovaD, UrangaS, et al. (2013) Construction, characterization and preclinical evaluation of MTBVAC, the first live-attenuated M. tuberculosis-based vaccine to enter clinical trials. Vaccine 31 : 4867–4873.
10. MenonS, WangS (2011) Structure of the response regulator PhoP from Mycobacterium tuberculosis reveals a dimer through the receiver domain. Biochemistry 50 : 5948–5957.
11. Gonzalo-AsensioJ, SotoCY, ArbuesA, SanchoJ, del Carmen MenendezM, et al. (2008) The Mycobacterium tuberculosis phoPR Operon Is Positively Autoregulated in the Virulent Strain H37Rv. Journal of Bacteriology 190 : 7068–7078.
12. GuptaS, SinhaA, SarkarD (2006) Transcriptional autoregulation by Mycobacterium tuberculosis PhoP involves recognition of novel direct repeat sequences in the regulatory region of the promoter. FEBS Lett 580 : 5328–5338.
13. CiminoM, ThomasC, NamouchiA, DubracS, GicquelB, et al. (2012) Identification of DNA binding motifs of the Mycobacterium tuberculosis PhoP/PhoR two-component signal transduction system. PLoS One 7: e42876.
14. Gonzalo-AsensioJ, MostowyS, Harders-WesterveenJ, HuygenK, Hernandez-PandoR, et al. (2008) PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One 3: e3496.
15. GalaganJE, MinchK, PetersonM, LyubetskayaA, AziziE, et al. (2013) The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499 : 178–183.
16. ColeST, BroschR, ParkhillJ, GarnierT, ChurcherC, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 : 537–544.
17. UplekarS, RougemontJ, ColeST, SalaC (2013) High-resolution transcriptome and genome-wide dynamics of RNA polymerase and NusA in Mycobacterium tuberculosis. Nucleic Acids Res 41 : 961–977.
18. RobackP, BeardJ, BaumannD, GilleC, HenryK, et al. (2007) A predicted operon map for Mycobacterium tuberculosis. Nucleic Acids Res 35 : 5085–5095.
19. PriceMN, HuangKH, AlmEJ, ArkinAP (2005) A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res 33 : 880–892.
20. DiChiaraJM, Contreras-MartinezLM, LivnyJ, SmithD, McDonoughKA, et al. (2010) Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res 38 : 4067–4078.
21. CamineroJA, PenaMJ, Campos-HerreroMI, RodriguezJC, GarciaI, et al. (2001) Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med 164 : 1165–1170.
22. Gottesman S, Storz G (2011) Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3.
23. RomillyC, ChevalierC, MarziS, MasquidaB, GeissmannT, et al. (2012) Loop-loop interactions involved in antisense regulation are processed by the endoribonuclease III in Staphylococcus aureus. RNA Biol 9 : 1461–1472.
24. Saint-JoanisB, DemangelC, JacksonM, BrodinP, MarsollierL, et al. (2006) Inactivation of Rv2525c, a substrate of the twin arginine translocation (Tat) system of Mycobacterium tuberculosis, increases beta-lactam susceptibility and virulence. J Bacteriol 188 : 6669–6679.
25. PalmerT, BerksBC (2012) The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10 : 483–496.
26. MarrichiM, CamachoL, RussellDG, DeLisaMP (2008) Genetic toggling of alkaline phosphatase folding reveals signal peptides for all major modes of transport across the inner membrane of bacteria. J Biol Chem 283 : 35223–35235.
27. DilksK, RoseRW, HartmannE, PohlschroderM (2003) Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol 185 : 1478–1483.
28. McDonoughJA, HackerKE, FloresAR, PavelkaMSJr, BraunsteinM (2005) The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J Bacteriol 187 : 7667–7679.
29. ArnvigKB, ComasI, ThomsonNR, HoughtonJ, BoshoffHI, et al. (2011) Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog 7: e1002342.
30. BlascoB, ChenJM, HartkoornR, SalaC, UplekarS, et al. (2012) Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog 8: e1002621.
31. SinghA, CrossmanDK, MaiD, GuidryL, VoskuilMI, et al. (2009) Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog 5: e1000545.
32. RaghavanS, ManzanilloP, ChanK, DoveyC, CoxJS (2008) Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454 : 717–721.
33. KrogerC, DillonSC, CameronAD, PapenfortK, SivasankaranSK, et al. (2012) The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109: E1277–1286.
34. SahrT, BruggemannH, JulesM, LommaM, Albert-WeissenbergerC, et al. (2009) Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol 72 : 741–762.
35. Toledo-AranaA, DussurgetO, NikitasG, SestoN, Guet-RevilletH, et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 : 950–956.
36. GripenlandJ, NetterlingS, LohE, TiensuuT, Toledo-AranaA, et al. (2010) RNAs: regulators of bacterial virulence. Nat Rev Microbiol 8 : 857–866.
37. AbramovitchRB, RohdeKH, HsuFF, RussellDG (2011) aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol 80 : 678–694.
38. FleischmannRD, AllandD, EisenJA, CarpenterL, WhiteO, et al. (2002) Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol 184 : 5479–5490.
39. BroschR, GordonSV, MarmiesseM, BrodinP, BuchrieserC, et al. (2002) A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A 99 : 3684–3689.
40. GuS, ChenJ, DobosKM, BradburyEM, BelisleJT, et al. (2003) Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol Cell Proteomics 2 : 1284–1296.
41. MawuenyegaKG, ForstCV, DobosKM, BelisleJT, ChenJ, et al. (2005) Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol Biol Cell 16 : 396–404.
42. MalenH, PathakS, SoftelandT, de SouzaGA, WikerHG (2010) Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol 10 : 132.
43. SchubertOT, MouritsenJ, LudwigC, RostHL, RosenbergerG, et al. (2013) The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13 : 602–612.
44. NaitoM, OharaN, MatsumotoS, YamadaT (1998) The novel fibronectin-binding motif and key residues of mycobacteria. J Biol Chem 273 : 2905–2909.
45. KuoCJ, BellH, HsiehCL, PtakCP, ChangYF (2012) Novel mycobacteria antigen 85 complex binding motif on fibronectin. J Biol Chem 287 : 1892–1902.
46. GarnierT, EiglmeierK, CamusJC, MedinaN, MansoorH, et al. (2003) The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci U S A 100 : 7877–7882.
47. MalagaW, ConstantP, EuphrasieD, CataldiA, DaffeM, et al. (2008) Deciphering the genetic bases of the structural diversity of phenolic glycolipids in strains of the Mycobacterium tuberculosis complex. J Biol Chem 283 : 15177–15184.
48. StoverCK, de la CruzVF, FuerstTR, BurleinJE, BensonLA, et al. (1991) New use of BCG for recombinant vaccines. Nature 351 : 456–460.
49. SalaC, HaouzA, SaulFA, MirasI, RosenkrandsI, et al. (2009) Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis. Mol Microbiol 71 : 1102–1116.
50. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
51. ZhangY, LiuT, MeyerCA, EeckhouteJ, JohnsonDS, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137.
52. DreszerTR, KarolchikD, ZweigAS, HinrichsAS, RaneyBJ, et al. (2012) The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res 40: D918–923.
53. SharmaCM, HoffmannS, DarfeuilleF, ReignierJ, FindeissS, et al. (2010) The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464 : 250–255.
54. LangmeadB, SalzbergSL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9 : 357–359.
55. AndersS, HuberW (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106.
56. BoersemaPJ, RaijmakersR, LemeerS, MohammedS, HeckAJ (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4 : 484–494.
57. WisniewskiJR, ZougmanA, MannM (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res 8 : 5674–5678.
58. CoxJ, MaticI, HilgerM, NagarajN, SelbachM, et al. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc 4 : 698–705.
59. CoxJ, NeuhauserN, MichalskiA, ScheltemaRA, OlsenJV, et al. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10 : 1794–1805.
60. LewJM, KapopoulouA, JonesLM, ColeST (2011) TubercuList—10 years after. Tuberculosis (Edinb) 91 : 1–7.
61. ChenJM, ZhangM, RybnikerJ, BasterraL, DharN, et al. (2013) Phenotypic profiling of Mycobacterium tuberculosis EspA point mutants reveals that blockage of ESAT-6 and CFP-10 secretion in vitro does not always correlate with attenuation of virulence. J Bacteriol 195 : 5421–5430.
62. KollyGS, BoldrinF, SalaC, DharN, HartkoornRC, et al. (2014) Assessing the essentiality of the decaprenyl-phospho-d-arabinofuranose pathway in Mycobacterium tuberculosis using conditional mutants. Mol Microbiol 92 : 194–211.
63. EdgarR, DomrachevM, LashAE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 : 207–210.
64. VizcainoJA, CoteRG, CsordasA, DianesJA, FabregatA, et al. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41: D1063–1069.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Combined Systems Approaches Reveal Highly Plastic Responses to Antimicrobial Peptide Challenge inČlánek Two Novel Human Cytomegalovirus NK Cell Evasion Functions Target MICA for Lysosomal Degradation
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Surveillance for Emerging Biodiversity Diseases of Wildlife
- The Emerging Role of Urease as a General Microbial Virulence Factor
- PARV4: An Emerging Tetraparvovirus
- Epigenetic Changes Modulate Schistosome Egg Formation and Are a Novel Target for Reducing Transmission of Schistosomiasis
- The Human Adenovirus E4-ORF1 Protein Subverts Discs Large 1 to Mediate Membrane Recruitment and Dysregulation of Phosphatidylinositol 3-Kinase
- A Multifactorial Role for Malaria in Endemic Burkitt's Lymphoma Pathogenesis
- Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating Activity of SARS-CoV Papain-Like Protease
- Cathepsin-L Can Resist Lysis by Human Serum in
- Epstein-Barr Virus Down-Regulates Tumor Suppressor Expression
- BCA2/Rabring7 Targets HIV-1 Gag for Lysosomal Degradation in a Tetherin-Independent Manner
- The Evolutionarily Conserved Mediator Subunit MDT-15/MED15 Links Protective Innate Immune Responses and Xenobiotic Detoxification
- Suppressor of Cytokine Signaling 4 (SOCS4) Protects against Severe Cytokine Storm and Enhances Viral Clearance during Influenza Infection
- T Cell Inactivation by Poxviral B22 Family Proteins Increases Viral Virulence
- Dynamics of HIV Latency and Reactivation in a Primary CD4+ T Cell Model
- HIV and HCV Activate the Inflammasome in Monocytes and Macrophages via Endosomal Toll-Like Receptors without Induction of Type 1 Interferon
- Virus and Autoantigen-Specific CD4+ T Cells Are Key Effectors in a SCID Mouse Model of EBV-Associated Post-Transplant Lymphoproliferative Disorders
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis
- Squalene Synthase As a Target for Chagas Disease Therapeutics
- The Contribution of Viral Genotype to Plasma Viral Set-Point in HIV Infection
- Combined Systems Approaches Reveal Highly Plastic Responses to Antimicrobial Peptide Challenge in
- Anthrax Lethal Factor as an Immune Target in Humans and Transgenic Mice and the Impact of HLA Polymorphism on CD4 T Cell Immunity
- Ly49C-Dependent Control of MCMV Infection by NK Cells Is -Regulated by MHC Class I Molecules
- Two Novel Human Cytomegalovirus NK Cell Evasion Functions Target MICA for Lysosomal Degradation
- A Large Family of Antivirulence Regulators Modulates the Effects of Transcriptional Activators in Gram-negative Pathogenic Bacteria
- Broad-Spectrum Anti-biofilm Peptide That Targets a Cellular Stress Response
- Malaria Parasite Infection Compromises Control of Concurrent Systemic Non-typhoidal Infection via IL-10-Mediated Alteration of Myeloid Cell Function
- A Role for in Higher Order Structure and Complement Binding of the Capsule
- Hip1 Modulates Macrophage Responses through Proteolysis of GroEL2
- CD8 T Cells from a Novel T Cell Receptor Transgenic Mouse Induce Liver-Stage Immunity That Can Be Boosted by Blood-Stage Infection in Rodent Malaria
- Phosphorylation of KasB Regulates Virulence and Acid-Fastness in
- HIV-Infected Individuals with Low CD4/CD8 Ratio despite Effective Antiretroviral Therapy Exhibit Altered T Cell Subsets, Heightened CD8+ T Cell Activation, and Increased Risk of Non-AIDS Morbidity and Mortality
- A Novel Mechanism Inducing Genome Instability in Kaposi's Sarcoma-Associated Herpesvirus Infected Cells
- Structural and Biochemical Characterization Reveals LysGH15 as an Unprecedented “EF-Hand-Like” Calcium-Binding Phage Lysin
- Hepatitis C Virus Cell-Cell Transmission and Resistance to Direct-Acting Antiviral Agents
- Different Modes of Retrovirus Restriction by Human APOBEC3A and APOBEC3G
- TNFα and IFNγ but Not Perforin Are Critical for CD8 T Cell-Mediated Protection against Pulmonary Infection
- Large Scale RNAi Reveals the Requirement of Nuclear Envelope Breakdown for Nuclear Import of Human Papillomaviruses
- The Cytoplasmic Domain of Varicella-Zoster Virus Glycoprotein H Regulates Syncytia Formation and Skin Pathogenesis
- A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase
- Are We There Yet? The Smallpox Research Agenda Using Variola Virus
- High-Efficiency Targeted Editing of Large Viral Genomes by RNA-Guided Nucleases
- Dynamic Functional Modulation of CD4 T Cell Recall Responses Is Dependent on the Inflammatory Environment of the Secondary Stimulus
- Bacterial Superantigens Promote Acute Nasopharyngeal Infection by in a Human MHC Class II-Dependent Manner
- Follicular Helper T Cells Promote Liver Pathology in Mice during Infection
- A Nasal Epithelial Receptor for WTA Governs Adhesion to Epithelial Cells and Modulates Nasal Colonization
- Unexpected Role for IL-17 in Protective Immunity against Hypervirulent HN878 Infection
- Human Cytomegalovirus Fcγ Binding Proteins gp34 and gp68 Antagonize Fcγ Receptors I, II and III
- Expansion of Murine Gammaherpesvirus Latently Infected B Cells Requires T Follicular Help
- Venus Kinase Receptors Control Reproduction in the Platyhelminth Parasite
- Molecular Signatures of Hemagglutinin Stem-Directed Heterosubtypic Human Neutralizing Antibodies against Influenza A Viruses
- The Downregulation of GFI1 by the EZH2-NDY1/KDM2B-JARID2 Axis and by Human Cytomegalovirus (HCMV) Associated Factors Allows the Activation of the HCMV Major IE Promoter and the Transition to Productive Infection
- Inactivation of Fructose-1,6-Bisphosphate Aldolase Prevents Optimal Co-catabolism of Glycolytic and Gluconeogenic Carbon Substrates in
- New Insights into Rotavirus Entry Machinery: Stabilization of Rotavirus Spike Conformation Is Independent of Trypsin Cleavage
- Prophenoloxidase Activation Is Required for Survival to Microbial Infections in
- SslE Elicits Functional Antibodies That Impair Mucinase Activity and Colonization by Both Intestinal and Extraintestinal Strains
- Timed Action of IL-27 Protects from Immunopathology while Preserving Defense in Influenza
- HIV-1 Envelope gp41 Broadly Neutralizing Antibodies: Hurdles for Vaccine Development
- The PhoP-Dependent ncRNA Mcr7 Modulates the TAT Secretion System in
- Cellular Superspreaders: An Epidemiological Perspective on HIV Infection inside the Body
- The Inflammasome Pyrin Contributes to Pertussis Toxin-Induced IL-1β Synthesis, Neutrophil Intravascular Crawling and Autoimmune Encephalomyelitis
- Papillomavirus Genomes Associate with BRD4 to Replicate at Fragile Sites in the Host Genome
- Integrative Functional Genomics of Hepatitis C Virus Infection Identifies Host Dependencies in Complete Viral Replication Cycle
- Co-assembly of Viral Envelope Glycoproteins Regulates Their Polarized Sorting in Neurons
- Targeting Membrane-Bound Viral RNA Synthesis Reveals Potent Inhibition of Diverse Coronaviruses Including the Middle East Respiratory Syndrome Virus
- Dual-Site Phosphorylation of the Control of Virulence Regulator Impacts Group A Streptococcal Global Gene Expression and Pathogenesis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Venus Kinase Receptors Control Reproduction in the Platyhelminth Parasite
- Dual-Site Phosphorylation of the Control of Virulence Regulator Impacts Group A Streptococcal Global Gene Expression and Pathogenesis
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis
- High-Efficiency Targeted Editing of Large Viral Genomes by RNA-Guided Nucleases
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



