-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
Growth factor independent 1 (Gfi1) is a transcriptional repressor originally identified as a gene activated in T-cell leukemias induced by Moloney-murine-leukemia virus infection. Notch1 is a transmembrane receptor that is frequently mutated in human T-cell acute lymphoblastic leukemia (T-ALL). Gfi1 is an important factor in the initiation and maintenance of lymphoid leukemias and its deficiency significantly impedes Notch dependent initiation of T-ALL in animal models. Here, we show that immature hematopoietic cells require Gfi1 to competently integrate Notch-activated signaling. Notch1 activation coupled with Gfi1 deficiency early in T-lineage specification leads to a dramatic loss of T-cells, whereas activation in later stages leaves development unaffected. In Gfi1 deficient multipotent precursors, Notch activation induces lethality and is cell autonomous. Further, without Gfi1, multipotent progenitors do not maintain Notch1-activated global expression profiles typical for T-lineage precursors. In agreement with this, we find that both lymphoid-primed multipotent progenitors (LMPP) and early T lineage progenitors (ETP) do not properly form or function in Gfi1−/− mice. These defects correlate with an inability of Gfi1−/− progenitors to activate lymphoid genes, including IL7R, Rag1, Flt3 and Notch1. Our data indicate that Gfi1 is required for hematopoietic precursors to withstand Notch1 activation and to maintain Notch1 dependent transcriptional programming to determine early T-lymphoid lineage identity.
Published in the journal: Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors. PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003713
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003713Summary
Growth factor independent 1 (Gfi1) is a transcriptional repressor originally identified as a gene activated in T-cell leukemias induced by Moloney-murine-leukemia virus infection. Notch1 is a transmembrane receptor that is frequently mutated in human T-cell acute lymphoblastic leukemia (T-ALL). Gfi1 is an important factor in the initiation and maintenance of lymphoid leukemias and its deficiency significantly impedes Notch dependent initiation of T-ALL in animal models. Here, we show that immature hematopoietic cells require Gfi1 to competently integrate Notch-activated signaling. Notch1 activation coupled with Gfi1 deficiency early in T-lineage specification leads to a dramatic loss of T-cells, whereas activation in later stages leaves development unaffected. In Gfi1 deficient multipotent precursors, Notch activation induces lethality and is cell autonomous. Further, without Gfi1, multipotent progenitors do not maintain Notch1-activated global expression profiles typical for T-lineage precursors. In agreement with this, we find that both lymphoid-primed multipotent progenitors (LMPP) and early T lineage progenitors (ETP) do not properly form or function in Gfi1−/− mice. These defects correlate with an inability of Gfi1−/− progenitors to activate lymphoid genes, including IL7R, Rag1, Flt3 and Notch1. Our data indicate that Gfi1 is required for hematopoietic precursors to withstand Notch1 activation and to maintain Notch1 dependent transcriptional programming to determine early T-lymphoid lineage identity.
Introduction
Growth factor independent-1 (Gfi1) is a transcriptional repressor originally identified as a common proviral insertion site of the murine Moloney leukemia virus (MMLV) that conferred IL-2 independent growth to IL-2 dependent T-cell lymphomas [1]. Subsequently, Gfi1 was identified as the most commonly activated gene in MMLV-induced lymphoid malignancies [2]. Gfi1 contains an N-terminal “SNAG” domain that is required for transcriptional repression and nuclear localization [3] and six zinc fingers of which, three, four and five are required for specific DNA-binding [4], [5]. Gfi1−/− mice display decreased HSC fitness, an accumulation of myeloid progenitors, and a lack of mature neutrophils [6], [7], [8]. Furthermore, germline deletion of Gfi1 results in a 4-fold decrease in thymic cellularity and modest increases in apoptotic cells [9]; whereas, mice with a CD4-promoter-driven Cre and floxed Gfi1 alleles (Gfi1f/f) demonstrate no defects in absolute thymocytes numbers[10]. Taken together, these data have been interpreted to mean that Gfi1−/− thymic phenotypes are largely due to Gfi1 anti-apoptotic functions during early thymopoiesis.
Notch1 is a transmembrane receptor that is critical throughout metazoan development acting as a molecular switch to determine cell fate. Similarly, during hematopoiesis, activation of Notch1 is required for proper T cell development [11], [12], [13], [14], [15]. T cells arise from circulating bone marrow progenitors that enter the thymus and encounter Notch1 ligands of the Delta-like and Jagged family [16], [17], [18]. Ligand-engagement of Notch receptors results in a conformational change exposing internal cleavage sites. A disintegrin and metalloprotease (ADAM) - and γ-secretase complex-mediated cleavage results in intracellular Notch (ICN) release from the membrane, nuclear translocation [19], [20], [21], and subsequent binding to CBF1/Suppressor of Hairless/Lag1 (CSL/Rbpj-κ) ultimately leading to Notch target gene activation. As Notch1 signal strength increases in early T lineage progenitors (ETP) through double negative (DN) 3 pro-T cells, transcriptional programs are upregulated which enforce T lymphoid identity at the expense of other lineages [22]. Notch1 signaling strength is highest leading up to TCRβ-selection, however, early progenitors in the BM may also require low level Notch signals as one component of the stimulus to proliferate and differentiate into lymphoid progenitors. Although Notch1 signaling may not be required for the maintenance of adult hematopoietic stem cells [23], [24], it functions as a tumor suppressor during myeloid development [25], and inhibition of Notch1 in progenitors dramatically reduces the formation of ETPs disrupting downstream stages of T-cell development in the thymus [26].
T cell acute lymphoblastic leukemia (T-ALL) is a subset of acute lymphoblastic leukemia, the most prevalent pediatric malignancy comprising nearly 25% of all childhood cancers [27]. Translocations placing NOTCH1 under control of the TCRb locus, t(7;9)(q34;q34.3) first implicated NOTCH1 in T-ALL [28]. Yet additional activating NOTCH1 mutations were found in more than 50% of T-ALL patients [29]. Moreover, mutations in NOTCH1 [30] and NOTCH1 regulatory proteins [31] have also been identified in T-ALL [32] . All of these mutations are thought to create constitutively active forms of ICN through ligand-independent activation and ICN nuclear translocation [33]. Mutations in GFI1 have not been detected in human T-ALL [34] [32]; however, transgenic overexpression of Gfi1 can accelerate oncogene-driven murine models of T-ALL [35], [36].
Recently, we identified Gfi1 as an important factor in the initiation and maintenance of lymphoid leukemias [37]. Interestingly, in human T-ALL patients with NOTCH1 mutations, or a transcriptional signature indicative of activated NOTCH1, GFI1 was highly expressed; while in mice, Gfi1 loss of function profoundly blocked Notch-initiated leukemia. To further investigate this unique relationship, we used genetic mouse models, which constitutively and inducibly delete Gfi1, to demonstrate that Gfi1 is required in a cell autonomous manner for early thymocytes and lymphoid progenitors in the bone marrow to competently receive Notch signals. Furthermore, we show that Gfi1−/− lymphoid progenitors cannot respond to endogenous levels of Notch1, potentially explaining the dramatic reduction in Gfi1−/− ETP and LMPP numbers. Thus, our findings identify Gfi1 as a critical factor in the response of immature hematopoietic cells to Notch1 signaling.
Results
Loss of Gfi1 and activation of intracellular Notch1 results in thymic hypoplasia
To further elucidate the mechanisms that protect Gfi1 deficient T cells from T-ALL transformation, we investigated the requirement for Gfi1 in developing T cells exposed to Notch1 activation. To do so, we bred mice in which Cre recombinase expression is driven by the T-cell-specific proximal-Lck promoter [38] with both Gfi1fex4–5 (Gfi1f) mice and germline Gfi1Δex2–3 (Gfi1−) or Gfi1 Δex4–5 deficient mice (Gfi1 Δ) resulting in LckCre+Gfi1f/− (or LckCre+Gfi1f/Δ) animals. Notably, we observed a similar 3–4-fold reduction in total thymocytes as previously published in Gfi1 germline deleted mice [9] (Figure S1). Next, we bred the LckCre+Gfi1f/Δ model with a Rosa26-driven intracellular-Notch1 (ICN) transgene, in which ICN-IRES-eGFP expression is prevented by a floxed “stop” cassette (ROSAlslICN) [39]. In the LckCre+Gfi1f/Δ ROSAlslICN mice, Cre expression should activate ICN and eGFP expression while simultaneously deleting Gfi1 (Figure 1A). As previously reported [40], we find that ICN activation, in the presence of Gfi1, leads to an accumulation of DP and CD8+ T cells at the expense of CD4+ cells (Figure 1B, GFP Positive LckCre+Gfi1+/+ROSAlslICN). In contrast, when activation of ICN is coupled with Gfi1 deletion, the majority of GFP+ cells are CD4 or CD8 single positive cells (Figure 1B, GFP positive, LckCre+Gfi1f/Δ ROSAlslICN). Moreover, ICN expression coupled with Gfi1 deletion led to a dramatic reduction in thymus size (Figure 1C). Further analysis of total thymocyte numbers revealed a 17-fold decrease in total cellularity when activation of ICN was combined with loss of Gfi1 (Figure 1D, p<0.05). Notably, this phenotype was not observed in control LckCre+ROSAlslICN or in LckCre+Gfi1f/Δ thymocytes where activation of ICN or deletion of Gfi1 occurs separately (Figure 1D and Figure S1). The few remaining thymocytes present in the LckCre+Gfi1f/Δ ROSAlslICN mice either lacked equivalent ICN transgene activation, as measured by eGFP (Figure 1E, p<0.01,) or failed to delete the floxed allele of Gfi1 (Figure 1F). Moreover, the significant decrease in the percentage of GFP+ cells in LckCre+Gfi1f/Δ ROSAlslICN mice (Figure 1E) is underrepresented by the flow cytometric plots shown. For example, the absolute number of GFP+ thymocytes in LckCre+ROSAlslICN mice is 49.8×106 versus 0.35×106 GFP+ thymocytes in LckCre+Gfi1f/Δ ROSAlslICN mice, a 142-fold decrease in the total number of GFP+ thymocytes between ICN-signaled Gfi1-sufficient versus ICN-signaled Gfi1-deficient cells. Taken together, these data demonstrate that Gfi1 is required to withstand chronic ICN signaling during the stages of development in which T cell malignant transformation occurs [41], [42].
Fig. 1. Loss of Gfi1 and activation of intracellular Notch1 results in thymic hypoplasia. 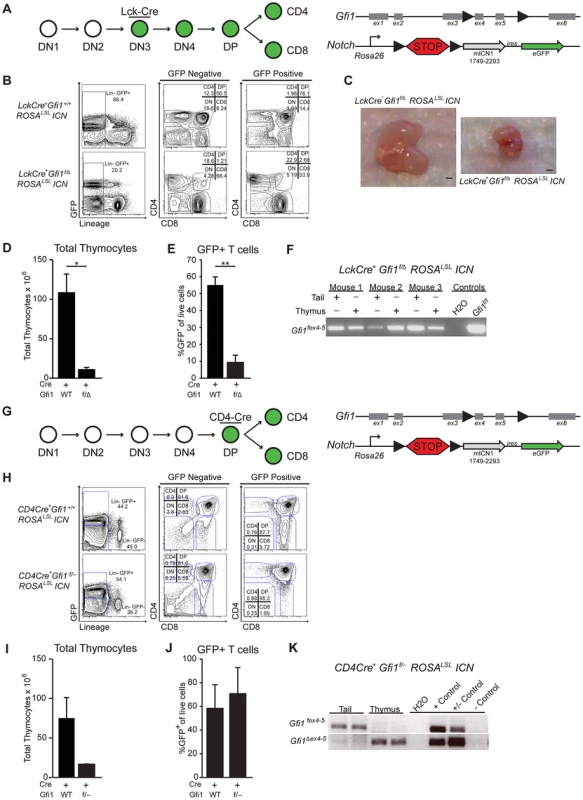
(A) Top left: Schematic of T cell development demonstrating that the proximal Lck-driven Cre activity is off during early stages (DN1-2) but is activated later during the DN3 stage of T cell development. Top right: Schematic of the floxed Gfi1 locus as well as the ICN transgene which has a floxed “STOP” cassette (lsl) preventing activation of ICN and GFP. (B) Example plots of flow cytometric analysis of thymic T cell populations for both GFP negative (left) and GFP positive (right) cells from LckCre+Gfi1+/+ROSALSLICN and LckCre+Gfi1f/ΔROSALSLICN mice. (C) Photograph of thymi from indicated mice. Scale bar is 1 mm. (D) Total thymocyte numbers from LckCre+ Gfi1+/+ROSALSLICN (n = 20) and LckCre+Gfi1f/ΔROSALSLICN (n = 4) mice. (E) Percentage of live, eGFP-expressing (a marker of Notch activation) thymocytes as determined by flow cytometric analysis. (F) PCR analysis of 3 separate mice for floxed alleles of Gfi1 (Gfi1fex4–5). (G) Top left: Schematic of T cell development demonstrating the CD4-driven Cre activity is off in early stages (DN1–4) but activates later during the DP stage of T cell development. Top right: Schematic of the floxed Gfi1 locus as well as the ICN transgene which has a floxed “STOP” cassette (lsl) preventing activation of ICN and eGFP. (H) Example plots of flow cytometric analysis of thymic T cell populations for both GFP negative (left) and GFP positive (right) cells from CD4Cre+Gfi1+/+ROSALSLICN and CD4Cre+Gfi1f/− ROSALSLICN mice. (I) Total thymocyte numbers from CD4Cre+Gfi1+/+ROSALSLICN (n = 9) and CD4Cre+Gfi1f/− ROSALSLICN (n = 2) mice. (J) Percentage of live, eGFP-expressing (a marker of Notch) thymocytes as determined by flow cytometric analysis. (K) PCR analysis of two mice for floxed alleles (top) or deleted alleles of Gfi1 (bottom). Representative FACS plots and pictures are shown. Experiments were repeated 2–3 times. Averages with SEM are shown in bar graphs. Students T-test were performed, *p≤0.05, **p≤0.01. To determine whether this apparent synthetic lethal relationship was dependent upon the stage of transgene activation or whether Notch-signaled pre-leukemic T cells generally require Gfi1, we utilized CD4Cre transgenic mice and repeated the above experiments. Notably, CD4Cre is expressed in DP thymocytes, and deletion of floxed Gfi1, Notch1, or Rbpj-κ by CD4Cre does not result in a reduction of thymocytes [10], [43], [44]. Therefore, any lethality caused by deleting Gfi1 and activating Notch should not be due to a specific developmental requirement for these factors alone, but instead would reflect a synergistic phenotype. Thus, we bred CD4Cre transgenic mice to Gfi1f/− ROSAlslICN mice (Figure 1G) and examined the effects on thymocyte development. Similar to LckCre-mediated activation, CD4Cre activation of ICN lead to an accumulation of DP and CD8 SP T cells at the expense of other populations (Figure 1H, CD4Cre+ROSAlslICN, GFP Positive). Comparatively, deletion of Gfi1 led solely to the development of DP T cells (Figure 1H, CD4Cre+ Gfi1f/− ROSAlslICN, GFP Positive). However, in contrast to the published CD4CreGfi1fex4–5/fex4–5 mice [10] or CD4Cre+ROSAlslICN mice, the CD4Cre+Gfi1f/− ROSAlslICN mice displayed a dramatic decrease in total cellularity similar to LckCre+Gfi1f/−ROSAlslICN mice (Figure 1I). Despite the decrease in total number of thymocytes, the percentage of CD4CreGfi1f/−ROSAlslICN thymocytes able to activate the ICN transgene was equivalent in CD4CreROSAlslICN signaled cells with or without Gfi1 as measured by eGFP (Figure 1J). Furthermore, CD4CreGfi1f/− ROSAlslICN thymocytes were able to efficiently delete the floxed allele of Gfi1 in thymocytes where Cre is active, but not in control Cre inactive tail tissue (Figure 1K). Thus, the presence of eGFP-expressing Gfi1Δ/− cells in this model suggests that the DP and SP T-cells do not absolutely require Gfi1 to express activated ICN, even though this combination results in dramatically decreased thymic cellularity.
Peripheral T cells do not require Gfi1 to survive activated Notch signaling
To more precisely define the developmental stages susceptible to ICN activation and Gfi1 deletion, we mated the ROSAlslICN or Gfi1f/− ROSAlslICN transgenic mice to transgenic mice that activate Cre expression after TCR positive selection (distal-LckCre = DLC) [45]. Similar to published reports, we found that less than 5% of the thymocytes in DLC+ROSAlslICN or DLC+Gfi1f/− ROSAlslICN expressed eGFP, and only at very late stages of T cell development (Figures 2A–C, S2). As such, we examined peripheral splenic T cells and found no statistical differences in total cellularity (Figure 2D) or in the percentages of GFP+ T cells between DLC+ROSAlslICN and DLC+Gfi1f/−ROSAlslICN mice (Figure 2E). Furthermore, FACS sorted GFP+ T cells displayed complete excision of the floxed allele of Gfi1 (Gfi1fex4–5) and had detectable levels of the deleted allele of Gfi1 (Gfi1Δ ex4–5, Figure 2F). These cells still demonstrated a partial phenocopy of Gfi1 deficiency in that they have an increase in the frequency of the CD8+ population (Figure 2G); however, no differences were observed in the immunophenotype of ICN-activated T cells, with or without Gfi1. These data provide strong evidence to suggest that the ICN+Gfi1Δ/Δ -induced hypocellularity phenotype is limited to a window during development in which T cells are susceptible to transformation (i.e. after TCRβ-selection). However, as that window closes and developmental transcriptional programs turn off, they are no longer susceptible to phenotypes caused by ICN activation and Gfi1 deletion.
Fig. 2. Peripheral T cells do not require Gfi1 for Notch activation. 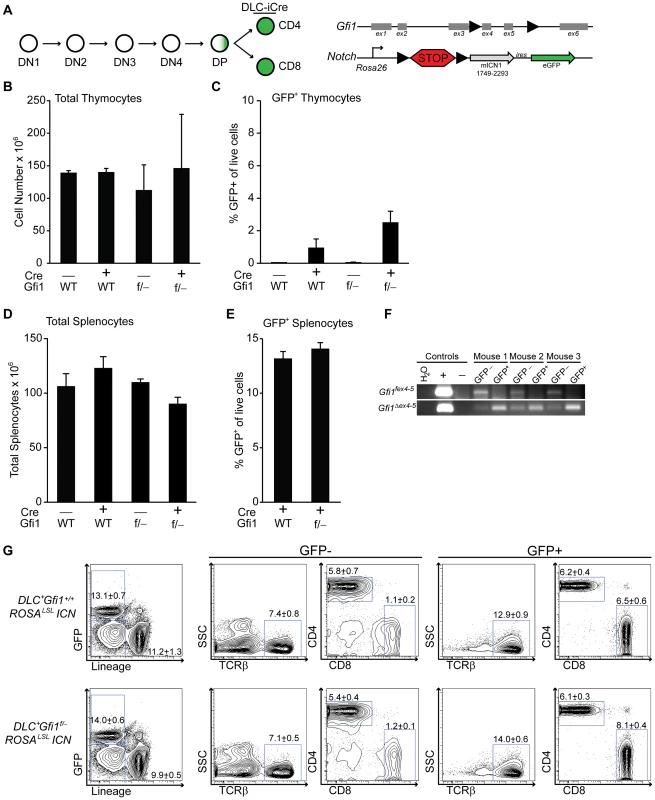
(A) Top left: Schematic of T cell development demonstrating the distal Lck-driven Cre (DLC-iCre) activity is off during most stages of T cell development, but activates in single-positive cells. Top right: Schematic of the floxed Gfi1 locus as well as the ICN transgene which has a floxed “STOP” cassette (lsl) preventing activation of ICN and GFP. (B–C) Total thymocyte numbers (B) and percentage of live, GFP-expressing thymocytes (C) as determined by flow cytometric analysis of Gfi1+/+ ROSALSLICN (n = 4), DLC-iCre+Gfi1+/+ROSALSLICN (n = 2), Gfi1f/−ROSALSLICN (n = 2), and DLC-iCre+Gfi1f/−ROSALSLICN (n = 4) mice. Averages with SD are shown (B–C). (D) Total splenocyte numbers of Gfi1+/+ ROSALSLICN (n = 8), DLC-iCre+Gfi1+/+ROSALSLICN (n = 10), Gfi1f/−ROSALSLICN (n = 5), and DLC-iCre+Gfi1f/−ROSALSLICN (n = 16) mice. (E) Percentage of live, GFP-expressing splenocytes as determined by flow cytometric analysis. (F) PCR analysis of splenic T cells from three DLC-iCre+Gfi1f/−ROSALSLICN mice independently FACS-sorted for TCRβ+ GFP− or GFP+ cells and genotyped for floxed (top) and deleted (bottom) alleles of Gfi1. (G) Example plots of flow cytometric analysis of splenic T cell populations for both GFP negative (left) and GFP positive (right) cells. Representative FACS plots and pictures are shown. Experiments were repeated 2–3 times. Averages with SEM are shown in bar graphs (D–E). One-way ANOVAs and student T tests were performed but no significant differences were found. Gfi1 is required for lymphoid lineage priming
Having established that deletion of Gfi1 early in T cell development mimics the phenotype of Gfi1 germline deletion, (LckCre+Gfi1f/Δ, Figure S1A–E) and that overexpression of intracellular Notch1 does not rescue this defect (LckCre+Gfi1f/Δ ROSAlslICN, Figure 1A–F) we further observed a direct relationship between the stage of lymphoid developmental and the synthetic lethal combination of deleting Gfi1 and activating ICN. This combination was most profound in early stages of T cell development (LckCre) in that GFP+ Gfi1Δ/Δ cells were not detectable. In contrast, at later developmental stages (CD4Cre) the absolute requirement for Gfi1 was lost (albeit with hypocellularity) and GFP+ Gfi1 Δ/Δ T cells could be detected. However, at very late stages of T cell development (DLCre) GFP+ Gfi1Δ/Δ T cells could be detected with no obvious defect in the numbers of peripheral or thymic T cells. Thus, we hypothesized that Gfi1 must be most critical during the earliest stages of lymphoid development where progenitors first experience lymphoid transcriptional programming, which includes Notch1 signaling. However, these data do not delineate between a selective event in which cells without Gfi1 die, versus an instructive event in which cells without Gfi1 fail to undergo proper lineage commitment and lymphoid gene expression changes.
To clarify this, we next performed a series of in vitro assays to concisely test the cell autonomous requirement for Gfi1 in lymphoid priming by inducibly deleting Gfi1 in the context of chronic ICN expression. First, we isolated Lin− BM from RosaCreERT2Gfi1fex4–5 and control Gfi1fex4–5 mice, and retrovirally transduced the stem and progenitor cells with GFP-marked ICN. GFP+ cells were FACS-sorted and plated in methylcellulose as previously described [46], [47] in the presence of 4-hydroxy tamoxifen (4-OHT, to induce Cre activity and delete Gfi1fex4–5) or vehicle control (EtOH). After one week in culture, CFU were enumerated, methylcellulose was disrupted and CFU were re-plated into 4-OHT or control-containing methylcellulose. This process was repeated for three weeks of plating (Figure 3A, diagram left). Untransformed progenitor cells generally produce 100–200 CFU within the first week, but fail to produce robust CFU in subsequent replatings [5], [46], [47]. In the absence of Cre expression, 4-OHT had no effect on CFU number or replating ability (Figure 3A, middle, Cre Neg: EtOH vs. OHT). However, in the presence of Cre, 4-OHT treatment dramatically reduced the number of CFU (Fig. 3A middle, Week 1, Cre Pos: EtOH vs. OHT: 363 to 156, p<0.01). Replating of Gfi1f/f, or RosaCre+Gfi1f/f vehicle-treated CFU led to similar numbers of CFU seven days later, whereas replating of RosaCre+Gfi1Δ/Δ resulted in an additional three-fold reduction in total CFU (Figure 3A middle, Week 1 vs. 2 : 156 to 57, p<0.01). Moreover, the CFU that did form in the absence of Gfi1 displayed substantially fewer cells per CFU demonstrating their inability to respond to ICN overexpression in the same manner as Gfi1f/f controls (Figure 3A, right).
Fig. 3. Gfi1 is required to enforce ICN1 activation of lymphoid genes. 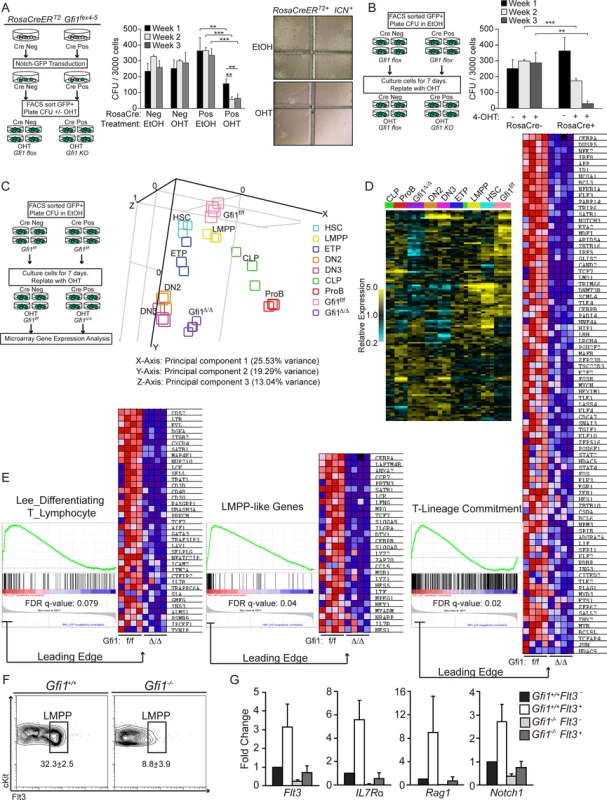
(A) Schematic for ICN colony forming unit (CFU) assay +/− Gfi1. CFU from FACS sorted ICN+eGFP expressing Lin− cells from either Gfi1f/f or RosaCreERT2+ Gfi1f/f were plated at 3000 cells/plate in 1 µM 4-OHT or EtOH control and cultured for 7 days (left). CFU/plate were then enumerated, disrupted and replated at 3000 cells/plate for additional two platings (middle). Representative pictures of RosaCreERT2+Gfi1f/f in EtOH or 4-OHT are shown (right). (B) Schematic of FACS-sorted ICN+eGFP expressing Lin− cells from either Gfi1f/f or RosaCreERT2+Gfi1f/f plated at 3000 cells/plate in EtOH and cultured for 7 days, then replated in 1 µM 4-OHT for an additional two platings (left). Enumeration of CFU (right). (C) Schematic of CFU conditions: similar to (B), but after 7 days in 1 µM 4-OHT, CFU were disrupted and RNA was isolated for microarray analysis (left). Principal component analysis of CFU with FACS-sorted HSC and lymphoid progenitor populations is shown. Note that Gfi1f/f CFU, but not Gfi1Δ/Δ CFU, cluster with LMPP. (D) Heatmap of 125 statistically different T-lineage commitment genes between Gfi1f/f CFU and Gfi1Δ/Δ CFU in indicated HSC and progenitor populations. Note that although Gfi1Δ/Δ CFU cluster with DN2 & DN3 cells, many of the 125 genes show differential expression suggesting Gfi1 Δ/Δ CFU are not characteristic of normal DN2 or DN3 cells. (E) GSEA enrichment plots and leading edge heatmaps demonstrate Gfi1f/f CFU transcriptionally mimic the expression of the indicated genesets, specific for lymphoid progenitors, while Gfi1Δ/Δ CFU fail to induce these genes. (F) Flow cytometric plots of Flt3+ LSK (LMPP) in BM progenitors from Gfi1+/+ and Gfi1−/− mice (N = 6/genotype); averages with SEM are shown. (G) Relative gene expression of indicated genes from FACS sorted Flt3− and Flt3+ LSK from Gfi1+/+ and Gfi1−/− mice. Averages with SD are displayed from triplicates (N = 2). In the absence of ICN overexpression, interruption of Gfi1 function promotes monocytic over granulocytic CFU formation [5]. Activation of ICN in myeloid lineages has recently been suggested to be lethal [25]. To avoid potential confounding factors of ICN activation in Gfi1-deficient myeloid progenitors, we next repeated the above assay (Figure 3A), but after FACS-sorting GFP+ ICN-transduced Lin− cells, we plated them for one week in the absence of 4-OHT in order to promote lymphoid priming and differentiation by ICN overexpression. After seven days in culture, CFU were enumerated, disrupted and plated in 4-OHT containing methylcellulose for an additional seven days for two rounds of replating (Figure 3B, left). Lymphoid-primed Gfi1f/f CFU were again unaffected by addition of 4-OHT through subsequent replatings. Although cells from RosaCre+Gfi1f/f generated equivalent CFU to cells from Gfi1f/f mice while cultured without 4-OHT, upon addition of 4-OHT, these cells again demonstrated a significant reduction in total CFU and cells per CFU compared to Gfi1f/f controls (Figure 3B right, Week 2 : 300 to 174, p<0.001). These data suggest that lymphoid-primed CFU also require Gfi1 to competently respond to ICN signaling.
To verify that this in vitro model truly reflects the characteristics of lymphoid progenitors, we repeated the experiment and examined global gene expression patterns. ICN-transduced Gfi1f/f and RosaCre+Gfi1Δ/Δ lineage-negative bone marrow cells were cultured for seven days without 4-OHT (to induce lymphoid-priming,) and then an additional seven days in 4-OHT (to induce deletion of Gfi1f/f alleles) before RNA was isolated and microarray expression analysis was performed (Figure 3C, left). Recently, global RNA-seq and ChIP-seq analyses defined a subset of genes that definitively distinguish FACS-sorted early lymphoid populations based upon their transcriptional networks [48]. Restricting our analysis to these genes, we first questioned whether they demonstrated statistically significant gene expression differences with or without Gfi1. Of the 378 tested, 125 genes displayed p-values <0.05 and were then used to cluster the expression signatures from both ICN-transduced CFU as well as normal FACS sorted lymphoid progenitors (Table S1) [49]. Principal component analysis (PCA) clustered Gfi1f/f CFU closest to LMPP populations demonstrating that the CFU partially mimic important transcriptional programs of in vivo lymphoid progenitors (Figure 3C, right). However, upon loss of Gfi1, PCA revealed that Gfi1Δ/Δ CFU no longer cluster with LMPP (Figure 3C–D), demonstrating a global inability to maintain lymphoid progenitor priming.
We next used an unbiased approach and applied gene set enrichment analysis (GSEA) [50] to our entire dataset. GSEA showed enrichment of published lymphoid progenitor signatures in Gfi1f/f CFU, whereas Gfi1Δ/Δ CFU showed no such enrichment (Figure 3E, “Lee_differentiating T_lymphocyte”). The same enrichment pattern was observed using more recently published LMPP-like and T-lineage commitment gene lists not yet curated in the MSigDB (Figure 3E “LMPP-like Genes” & “T-Lineage Commitment”). Indeed, further analysis [51] of gene expression differences between Gfi1f/f and Gfi1Δ/Δ ICN CFU, demonstrated significant changes in cell surface markers (Table S2, GO Cellular Component GO:0009986, p<2.49×10−16) and CD antigen genes (Table S3, HUGO Genenames.org, p<3.40×10−30). These data suggest that much (but not all) of the ICN-instructed lymphoid progenitor programs are dependent upon Gfi1. Taken together, we conclude that i) ICN-transduced Gfi1+/+ CFU share critical transcriptional programs with lymphoid bone marrow progenitors; ii) loss of Gfi1 results in a subsequent loss of key elements of those ICN-regulated transcriptional networks necessary for proper lymphoid lineage identity; and iii) Gfi1 is required in ICN-signaled (FACS sorted) cells in a cell autonomous fashion.
Given the similarity of gene signatures between ICN-CFU and LMPP and the reliance of these cells in vitro on Gfi1, we questioned whether endogenous levels of Notch1 signaling experienced in vivo by lymphoid progenitor cells of Gfi1−/− mice may engender the same phenotypes identified in the transgenic and retroviral overexpression systems. To answer this question, we examined the LMPP (Flt3 high, Lin−, cKit+, Sca1+) in the BM reasoning that: i) LMPP are the first lymphoid progenitors to respond to Notch1 signaling [52], ii) ICN+Gfi1f/f CFU clustered closest to FACS-sorted LMPP, and iii) differences in the expression of Flt3 have been reported in Gfi1−/− LSK [7], [8]. Similar to previous reports [7], [53], we observed a decrease in the percentage and total number of LMPPs in Gfi1−/− mice (Figure 3F). To determine whether Gfi1−/− phenotypic LMPP are functionally similar to wild type LMPP, we FACS sorted LSK and LMPP from Gfi1+/+ and Gfi1−/− mice and tested for the induction of lymphoid signature genes coincident with Flt3 expression in LMPP. Whereas Gfi1+/+ progenitors upregulated the expression of Flt3, IL7R, Rag1 and Notch1 3–10 fold during the transition from Flt3−LSK to LMPP, Gfi1−/− progenitors did not induce these genes to the same degree (Figure 3G). Furthermore, we found lower expression of each of these genes in Gfi1−/− Flt3−LSK, suggesting that phenotypically normal Gfi1−/− progenitors have a functional defect in their ability to prime lymphoid transcriptional programs. Taken together, these data indicate that loss of Gfi1 in lymphoid-primed progenitors results in a cell-autonomous inability to maintain a lymphoid specific transcriptional program.
Gfi1 deficiency results in loss of early thymic progenitors (ETP)
ETPs are thought to be the progeny of lymphoid-primed progenitors, which reside within the BM and have significant overlap in transcriptional signatures with LMPP [49]. Therefore, we hypothesized that the role of Gfi1 during lymphoid priming may be most pronounced in ETPs, in particular since these cells experience a dramatic increase in Notch1 signaling. Gfi1+/− lymphoid progenitors express less Gfi1 protein than wild type cells [54], therefore we examined the effect of Gfi1 deletion and haploinsufficiency upon ETP numbers. We found a Gfi1 dose-dependent reduction in ETP percentages and absolute numbers in Gfi1+/− and Gfi1−/− thymi compared to Gfi1+/+ controls (Figure 4A–B; 6.4×104, 1.7×104, 0.06×104 between Gfi1+/+, Gfi1+/− and Gfi1−/− respectively; p<0.05 and p<0.01). Thus, we conclude that Gfi1−/− mice have few phenotypically normal ETPs.
Fig. 4. Gfi1 deficiency results in reduced number and function of ETP. 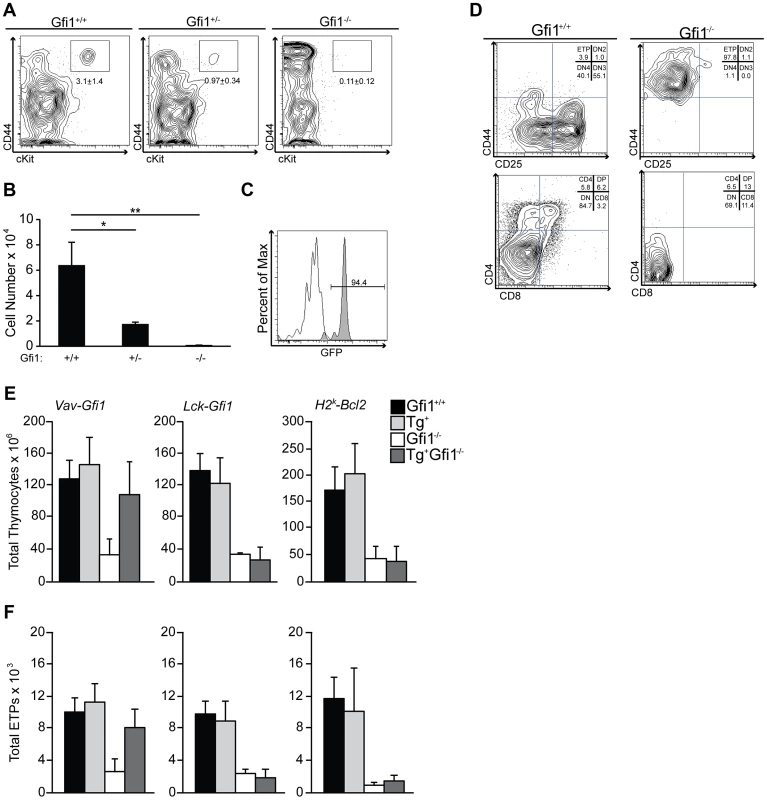
(A) Gfi1+/+ (n = 4), Gfi1+/− (n = 6) and Gfi1−/− (n = 3) thymocytes were analyzed for the percent of Lin−, CD25−, cKit+, CD44+ ETPs; Average ± SD. (B) Total cell number or ETPs from (A). (C) Histogram of GFP from Gfi1GFP/+ knock-in mice (grey shaded peak) compared to Gfi1+/+ littermate control (white peak,) demonstrates Gfi1 expression in ETPs (N = 5/genotype). (D) ETPs were FACS-sorted from Gfi1+/+ and Gfi1−/− mice directly onto OP9-DL1 stroma and cultured for 15 days. T cell development stages were assessed by flow cytometry. Experiments were repeated three times. One representative example is shown. (E–F) Transgenic rescue of Gfi1−/− T cell development using Vav-Gfi1 (N = 9/genotype), Lck-Gfi1 (N = 6/genotype), and H2K-Bcl2 (N = 6/genotype) in Gfi1+/+ and Gfi1−/− mice. Total thymocyte numbers (E) were calculated and flow cytometry was performed to identify ETPs (F). Next, we asked whether ETPs normally express Gfi1 and whether Gfi1−/− ETPs can respond to Notch1 signaling. To address the first question, we used Gfi1-GFP knock-in mice (Gfi1GFP/+) [55] in which eGFP replaces Gfi1 coding exons, and the expression of eGFP mirrors that of endogenous Gfi1. We found that Gfi1GFP/+ ETP are clearly eGFP+, demonstrating that Gfi1 is highly expressed in ETPs (Figure 4C). To address the latter question, we FACS-sorted Gfi1+/+ and Gfi1−/− ETP cells and exposed them to the Notch ligand, Delta-like 1 (DL1), by culturing the cells on OP9-DL1 stroma. Gfi1−/− ETP failed to respond and did not progress through T cell development whereas their Gfi1+/+ ETP controls began to express both CD4 and CD8 after 15 days in culture (Figure 4D). Thus, these data demonstrate that phenotypically defined Gfi1 deficient ETPs do not properly function in response to Notch ligands in vitro.
We next sought to genetically rescue Gfi1 expression both before and after lymphoid progenitors experience increases in basal Notch1 signaling. To examine whether endogenous levels of Notch1 signaling were correctly interpreted, we examined total thymocyte and ETP numbers, both of which are critically dependent on Notch1 [11], [26]. First, we mated Vav-Gfi1 transgenic mice, which express Gfi1 in all hematopoietic stem/progenitors and mature lineages [56], to germline Gfi1−/− mice. Gfi1 expression in this model occurs before endogenous increases in Notch1 signals [56]. We then analyzed the total number of thymocytes and the formation of ETPs by flow cytometry. Vav-mediated expression of Gfi1 rescued both the total thymocyte numbers (Figure 4E) and the total numbers of ETPs (Figure 4F) to the levels of Gfi1+/+ controls. Next, we mated germline Gfi1−/− mice to Lck-Gfi1 transgenic mice [57] to re-express Gfi1 at the height of Notch1 target gene expression in the thymus [57], [58], [59]. Transgenic Lck-Gfi1 expression failed to rescue germline Gfi1−/− defects in total thymocyte (Figure 4E) or ETP numbers (Figure 4F). These data corroborate that Gfi1 is required early during lymphoid progenitor development and further suggest that Gfi1 is required to properly respond to endogenous levels of Notch1 signaling.
Gfi1−/− lymphoid progenitors are reduced in number, but also fail to induce genes normally downstream of Notch signals. To delineate a requirement for Gfi1 to integrate Notch signaling versus to survive an apoptotic selection event, we attempted to rescue the loss of ETPs and total thymocytes in Gfi1−/− mice by crossing them with Bcl2-transgenic mice (H2K-Bcl2), which would block apoptosis. Although Bcl2 overexpression was able to rescue most of the Gfi1 loss-of-function phenotypes in T-ALL [37], neither total thymocyte numbers or ETP numbers returned to Gfi1+/+ levels in Bcl2 transgenic Gfi1−/− mice (Figure 4E–F). Thus, forced expression of an anti-apoptotic molecule is insufficient to rescue Gfi1−/− T cell development defects.
Discussion
Notch1 is a central mediator of both T cell leukemogenesis and T cell development. ICN-target genes such as Myc [60], [61], [62], Hes1 [47], [63], Notch3 [64], [65] and IGF1R [66] are critical to T cell development and T-ALL, and Notch signaling directly controls expression of T-cell-lineage specific identity genes such as Tcf7 [67], [68] and Bcl11b [69]. Not surprisingly, interfering with the expression of Notch1 target genes disrupts Notch1 programing of developing T - or T-ALL cells. In contrast, we previously showed that Notch signaling does not directly regulate Gfi1 expression [37]. However, in this study we demonstrate that Gfi1 is still required to execute Notch1-driven developmental and pre-leukemic programs even though it is unlikely to be an ICN-downstream-target gene.
Previously, regulation of apoptosis was considered the dominant function of Gfi1 in developing T cells [9], [70]. In transformed lymphoid cells, loss of Gfi1 leads to induction of apoptosis through the exaggeration of p53-dependent target gene activation. Overexpression of Bcl2 or knockdown of p53 rescues Gfi1 loss of function phenotypes in T-ALL [37]. However, neither loss of p53 or overexpression of Bcl2 alters Gfi1−/− total thymocyte numbers (Figure 4 and data not shown). This may be due a lower threshold of DNA damage present in untransformed lymphoid precursors that is increased in T-ALL (due to oncogenic stress) resulting in hyperactivation of p53. Thus, a lack of Notch1-regulated gene expression observed in Gfi1−/− lymphoid precursors might previously have been ascribed to a selective event causing those cells that express lymphoid genes to die. Because loss of Gfi1 debilitates ICN-mediated lymphoid priming in a cell autonomous manner, we now conclude that repression of pro-apoptotic genes is only one of many biological functions that are integrated by Gfi1 during lymphoid priming and T lymphopoiesis. As expression of an anti-apoptotic effector was insufficient to rescue all of the defects associated with Gfi1 deficiency, we further conclude that Gfi1 is an obligate instructive factor that is critical to effectively maintain Notch1-dependent transcriptional programs necessary for lymphoid lineage commitment.
Previous studies have implicated Gfi1 at multiple stages of lymphoid development. For example, Gfi1 overexpression has been shown to partially rescue Lyl1 deficiency in LMPP [71]. Moreover, Gfi1 acts downstream of Ikaros in MPPs to mediate the differentiation choice between B cells and myeloid cells [72] by antagonizing Pu.1. Given previous data demonstrating that Pu.1 can restrain Notch1 signaling in pre-T cells [73] and that Pu.1 is a bona fide Gfi1 target gene [72], it is attractive to hypothesize that loss of Gfi1 leads to derepression of Pu.1 which in turn opposes Notch1. However, Notch1-signaled cells appear to have alternative mechanisms to antagonize Pu.1-responsive transcriptional circuits based on the observation that upregulation of Pu.1 or Nab2 (as seen in Gfi1−/− MPP [72], [74]) in Notch-activated Gfi1 deficient cells (GSE41162) was not observed. Instead, we find that Gfi1 is required to maintain critical lymphoid transcriptional programs activated by Notch1 such as Rag1, Dtx1, and Tcf7. It remains unclear how Gfi1 might maintain the activation of these genes; whether Gfi1 represses other transcriptional repressors, microRNAs, or whether loss of Gfi1 leads to alternative differentiation pathways for Notch-signaled cells has yet to be elucidated.
We discovered that the phenotypes associated with Notch1 activation and Gfi1 loss of function were most severe in early lymphoid precursors, while immature SP and peripheral T cells showed modest effects. This stage-specific phenomenon could be due to the fact that Notch1 and Gfi1 are both endogenously expressed and required for the normal development of T cells from lymphoid progenitors up to TCRβ selection [9], [15], [57]. Alternatively, once T cells have completed critical development checkpoints they may no longer be susceptible to manipulation of developmental transcriptional networks. For instance, during stages of development where activation of lymphoid-associated genes is critical to establishing a T-lineage identity, Gfi1 appears to be required to maintain the activation of Notch-driven lymphoid-restricted genes such as Tcf1. However, in a mature T cell, either the expression of these genes is maintained by other transcription networks, or an inability to maintain their expression does not result in phenotypic consequences because the cell's developmental potential has already been achieved. In either case, our work has uncovered an epistatic relationship between Notch1 and Gfi1 that is essential for proper lymphoid development.
Loss of Gfi1 phenocopies the loss of Notch1 and Tcf7 (Tcf1) with regard to the formation of ETPs, but unlike Notch1 and Tcf7, Gfi1 is also required for the survival or formation of lymphoid-primed progenitors upstream of the ETP [23], [67]. This suggests a unique role for Gfi1 in bridging lymphoid transcriptional programs from the earliest lymphoid-primed bone marrow progenitor to the thymic ETP before Notch1-regulated transcriptional programs become the dominant mechanism through which T lineage fate is enforced. Although our data do not exclude the possibility that Gfi1 participates in a shared, undiscovered, transcriptional network with other key “T cell-specific” transcription factors, it appears more likely that the phenocopy of Gfi1−/− ETP is due to the inability of Gfi1 deficient cells to integrate lymphoid progenitor transcriptional circuits, in particular those initiated by Notch1.
We have recently shown that Gfi1 deficient mice are protected from Notch1 mediated malignant transformation [37]. Here, we have uncovered a requirement for Gfi1 in Notch1 activated cells with implications for both normal lymphopoiesis as well as T cell transformation. Specifically, Gfi1 is required to maintain cellularity in Notch-signaled cells in a temporally regulated manner. These data help to clarify the almost absolute requirement for Gfi1 in Notch-mediated transformation. Gfi1 is required to maintain the pool of premalignant cells available for transformation, and to maintain Notch target genes essential for leukemogenesis. Thus, our data provide additional insight into the multiple mechanisms by which transcriptional networks may have evolved to protect developing lymphoid cells from transformation.
Materials and Methods
Mice
LckCre, CD4Cre [38], distal LckCre [45], Rosa26-lox-stop-lox-NotchIC [39], Lck-Gfi1 [75], Vav-Gfi1 [56], Gfi1fex4–5 [10], Gfi1Δex2–3 [76], Gfi1 Δex2–5 [6], RosaCreERT2 Gfi1fex4–5 [46] transgenic mice have all previously been described. Gfi1fex4–5 were bred to Gfi1Δex2–3 mice to generate Gfi1fex4–5, Δex2–3 mice to allow for more efficient deletion of the remaining floxed allele by LckCre, CD4Cre or distal LckCre transgenic mice. All mice were bred and housed in a specific-pathogen-free barrier facility at Cincinnati Children's Hospital Medical Center (CCHMC) Veterinary Services or at the Institut de recherches cliniques de Montréal (IRCM).
Ethics statement
The Institutional Animal Care and Use Committee at CCHMC and the Animal Care Committee at the IRCM reviewed and approved all animal experimentation protocols, certified animal technicians, regularly observed mice in all studies and took steps to maintain animal welfare and prevent undue suffering under protocol numbers 1D09075 and 2009-12 respectively.
Flow cytometry & FACS sorting
Thymi were harvested in Medium 199 (Invitrogen) and single cell suspensions were created. Bone marrow was flushed from femurs and tibias using Medium 199, spun down and RBC were lysed using ACK lysis buffer (Gibco). Cell counts were determined using a Coulter Counter (Beckman) and cells were then stained with various cocktails of monoclonal antibodies to the following antigens: Fc-block (2.4G2), CD4 (RM4–5), CD8a (53-6.7), CD44 (IM7), CD25 (PC61), cKit (2B8), Sca1 (D7), Flt3 (A2F10). Lineage cocktails for T cell development contained B220 (RA3-6B2), CD11b (M1/70), CD11c (N418), NK1.1, TCRγδ (UC7-13D5) and Ter119. Lineage cocktails for ETPs FACS plots contained B220 (RA3-6B2), CD3ε (145-2C11), CD8 (53-6.7), CD11b (M1/70), CD11c (N418), DX5, Gr1 (RB6-8C5), NK1.1, TCR γδ (UC7-13D5) and Ter119. Cells were stained at 4°C for 30 minutes before being washed and resuspended in PBS containing 2% FBS and 1 mM EDTA. Data was acquired on the BD LSRII, LSRFortessa or FACSCanto. Cells were FACS sorted on the BD FACSAriaII and recovered in PBS with 50% FBS.
PCR and gene expression analyses
Polymerase chain reaction (PCR) detection of the Gfi1fex4–5 allele was performed with primers 5′-CAGTCCGTGACCCTCCAGCAT-3′ and 5′-CTGGGAGTGCACTGCCTTGTGTT-3′, whereas detection of the Gfi1Δex4–5 allele was performed with primers 5′-CAGTCCGTGACCCTCCAGCAT-3′ and 5′-CCATCTCTCCTTGTGCTTAAGAT-3′. Gene expression analysis was performed on RNA isolated from TRI Reagent (Sigma) by phenol-chloroform extraction or by the RNeasy kit (QIAGEN). cDNA was synthesized from purified RNA using the cDNA High Capacity Archive Kit (Applied Biosystems) according to the manufacturer's instructions. Gene expression was assessed using Taqman probes (Applied Biosystems) or primers for cMyc (Mm03053277_s1, Mm00487803_m1), Dtx1 (Mm00492297_m1), Hes1 (Mm00468601_m1, Mm01342805_m1), Hey1 (Mm00468865_m1), Ptcra (Mn00478361_m1), Ccnd1(Mn00432359_m1), Notch1 (Mm00435245_m1, Mm00435249_m1), Notch3 (Mm01345646_m1) on an ABI Prism 7900. Threshold values were calculated and normalized to the endogenous control, Gapdh (Mm99999915_g1); then, the ΔΔCT method was used to calculate the fold change compared to Gfi1+/+ controls. Gene array data (GSE20282 or GSE41162) was analyzed using GeneSpring (version 12.0 Agilent Technologies) or the R software package.
Cell culture and in vitro differentiation
OP9-DL1 cells were cultured in 24-well plates at a concentration of 2×104cells/ml in α-MEM media supplemented with 20% FBS (charcoal stripped), β-mercapto ethanol, sodium pyruvate, and non-essential amino acids. OP9-DL1 cells were seeded 24 h before FACS sorted ETP were directly sorted onto the monolayer. Fresh IL7 (1 ng/mL) and Flt3L (5 ng/mL) were then added. Media was changed every 4–5 days and developing T cells were transferred onto a new monolayer of OP9-DL1 cells with fresh media and cytokines.
Retroviral transduction and CFU assays
Lineage negative cells were isolated from total BM using magnetic separation (Miltenyi) and then placed into StemSpan SF media (StemCell Technologies) containing IL-3, IL-6, IL-7, SCF, Flt3L and human IL-11 (Miltenyi) with 1% Glutamine and 1% Pen/Strep (Gibco). Cells were expanded for two days before being placed on Retronectin (Takara) coated plates preloaded with viral supernatants harvested from MigR1-ICN-ires-eGFP transfected 293T cells. Viral supernatants were spinfected at 1000 g at 4°C for 30 minutes. The process was repeated twice and the cells were expanded for 48 hours before FACS-sorting. eGFP+ cells were resuspended in MethoCult semi-solid media (StemCell Technologies) and allowed to grow for one week. CFU were enumerated, cells were then dissociated, counted and replated. 4-OHT was added at a final concentration of 1 µM to induce Cre activity. Gfi1 deletion was confirmed by PCR; any CFU demonstrating incomplete excision of floxed Gfi1 was excluded from gene expression array analysis.
Supporting Information
Zdroje
1. GilksCB, BearSE, GrimesHL, TsichlisPN (1993) Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol 13 : 1759–1768.
2. UrenAG, KoolJ, MatentzogluK, de RidderJ, MattisonJ, et al. (2008) Large-scale mutagenesis in p19(ARF) - and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 133 : 727–741.
3. GrimesHL, ChanTO, Zweidler-McKayPA, TongB, TsichlisPN (1996) The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol 16 : 6263–6272.
4. Zweidler-MckayPA, GrimesHL, FlubacherMM, TsichlisPN (1996) Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol 16 : 4024–4034.
5. ZarebskiA, VeluCS, BaktulaAM, BourdeauT, HormanSR, et al. (2008) Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity 28 : 370–380.
6. KarsunkyH, ZengH, SchmidtT, ZevnikB, KlugeR, et al. (2002) Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet 30 : 295–300.
7. HockH, HamblenMJ, RookeHM, SchindlerJW, SalequeS, et al. (2004) Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431 : 1002–1007.
8. ZengH, YucelR, KosanC, Klein-HitpassL, MoroyT (2004) Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 23 : 4116–4125.
9. YucelR, KarsunkyH, Klein-HitpassL, MoroyT (2003) The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med 197 : 831–844.
10. ZhuJ, JankovicD, GrinbergA, GuoL, PaulWE (2006) Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A 103 : 18214–18219.
11. RadtkeF, WilsonA, StarkG, BauerM, van MeerwijkJ, et al. (1999) Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10 : 547–558.
12. HadlandBK, ManleyNR, SuD, LongmoreGD, MooreCL, et al. (2001) Gamma -secretase inhibitors repress thymocyte development. Proc Natl Acad Sci U S A 98 : 7487–7491.
13. WilsonA, MacDonaldHR, RadtkeF (2001) Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med 194 : 1003–1012.
14. HanH, TanigakiK, YamamotoN, KurodaK, YoshimotoM, et al. (2002) Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol 14 : 637–645.
15. MaillardI, WengAP, CarpenterAC, RodriguezCG, SaiH, et al. (2004) Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood 104 : 1696–1702.
16. AndersonG, PongraczJ, ParnellS, JenkinsonEJ (2001) Notch ligand-bearing thymic epithelial cells initiate and sustain Notch signaling in thymocytes independently of T cell receptor signaling. Eur J Immunol 31 : 3349–3354.
17. SchmittTM, Zuniga-PfluckerJC (2002) Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17 : 749–756.
18. LeharSM, DooleyJ, FarrAG, BevanMJ (2005) Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood 105 : 1440–1447.
19. KopanR, SchroeterEH, WeintraubH, NyeJS (1996) Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A 93 : 1683–1688.
20. SchroeterEH, KisslingerJA, KopanR (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393 : 382–386.
21. StruhlG, AdachiA (1998) Nuclear access and action of notch in vivo. Cell 93 : 649–660.
22. RothenbergEV, ZhangJ, LiL (2010) Multilayered specification of the T-cell lineage fate. Immunol Rev 238 : 150–168.
23. MaillardI, KochU, DumortierA, ShestovaO, XuL, et al. (2008) Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell 2 : 356–366.
24. ChiangMY, ShestovaO, XuL, AsterJC, PearWS (2012) Divergent effects of supraphysiological Notch signals on leukemia stem cells and hematopoietic stem cells. Blood 121 (6) 905–17.
25. KlinakisA, LobryC, Abdel-WahabO, OhP, HaenoH, et al. (2011) A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 473 : 230–233.
26. SambandamA, MaillardI, ZediakVP, XuL, GersteinRM, et al. (2005) Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol 6 : 663–670.
27. PuiCH (2000) Acute lymphoblastic leukemia in children. Curr Opin Oncol 12 : 3–12.
28. EllisenLW, BirdJ, WestDC, SorengAL, ReynoldsTC, et al. (1991) TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66 : 649–661.
29. WengAP, FerrandoAA, LeeW, MorrisJPt, SilvermanLB, et al. (2004) Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306 : 269–271.
30. SulisML, OW, ToselloV, PallippukamS, PalomeroT, et al. (2007) A Novel Class of Acitvating Mutations in NOTCH1 in T-ALL. Blood 110 : 213a.
31. ThompsonBJ, BuonamiciS, SulisML, PalomeroT, VilimasT, et al. (2007) The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 204 : 1825–1835.
32. De KeersmaeckerK, AtakZK, LiN, VicenteC, PatchettS, et al. (2013) Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 45 : 186–190.
33. MaleckiMJ, Sanchez-IrizarryC, MitchellJL, HistenG, XuML, et al. (2006) Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol 26 : 4642–4651.
34. ZhangJ, DingL, HolmfeldtL, WuG, HeatleySL, et al. (2012) The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481 : 157–163.
35. ZornigM, SchmidtT, KarsunkyH, GrzeschiczekA, MoroyT (1996) Zinc finger protein GFI-1 cooperates with myc and pim-1 in T-cell lymphomagenesis by reducing the requirements for IL-2. Oncogene 12 : 1789–1801.
36. SchmidtT, KarsunkyH, GauE, ZevnikB, ElsasserHP, et al. (1998) Zinc finger protein GFI-1 has low oncogenic potential but cooperates strongly with pim and myc genes in T-cell lymphomagenesis. Oncogene 17 : 2661–2667.
37. KhandanpourC, PhelanJD, VassenL, SchutteJ, ChenR, et al. (2013) Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell 23 : 200–214.
38. LeePP, FitzpatrickDR, BeardC, JessupHK, LeharS, et al. (2001) A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15 : 763–774.
39. MurtaughLC, StangerBZ, KwanKM, MeltonDA (2003) Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A 100 : 14920–14925.
40. RobeyE, ChangD, ItanoA, CadoD, AlexanderH, et al. (1996) An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell 87 : 483–492.
41. AllmanD, KarnellFG, PuntJA, BakkourS, XuL, et al. (2001) Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J Exp Med 194 : 99–106.
42. LiX, GounariF, ProtopopovA, KhazaieK, von BoehmerH (2008) Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med 205 : 2851–2861.
43. TanigakiK, TsujiM, YamamotoN, HanH, TsukadaJ, et al. (2004) Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20 : 611–622.
44. WolferA, BakkerT, WilsonA, NicolasM, IoannidisV, et al. (2001) Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat Immunol 2 : 235–241.
45. ZhangDJ, WangQ, WeiJ, BaimukanovaG, BuchholzF, et al. (2005) Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol 174 : 6725–6731.
46. HormanSR, VeluCS, ChaubeyA, BourdeauT, ZhuJ, et al. (2009) Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood 113 : 5466–5475.
47. EspinosaL, CathelinS, D'AltriT, TrimarchiT, StatnikovA, et al. (2010) The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell 18 : 268–281.
48. ZhangJA, MortazaviA, WilliamsBA, WoldBJ, RothenbergEV (2012) Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149 : 467–482.
49. LucS, LuisTC, BoukarabilaH, MacaulayIC, Buza-VidasN, et al. (2012) The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol 13 (4) 412–9.
50. SubramanianA, TamayoP, MoothaVK, MukherjeeS, EbertBL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102 : 15545–15550.
51. ChenJ, BardesEE, AronowBJ, JeggaAG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–311.
52. LaiAY, KondoM (2007) Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc Natl Acad Sci U S A 104 : 6311–6316.
53. ZhuangD, QiuY, KoganSC, DongF (2006) Increased CCAAT enhancer-binding protein epsilon (C/EBPepsilon) expression and premature apoptosis in myeloid cells expressing Gfi-1 N382S mutant associated with severe congenital neutropenia. J Biol Chem 281 : 10745–10751.
54. Ordonez-RuedaD, JonssonF, MancardiDA, ZhaoW, MalzacA, et al. (2012) A hypomorphic mutation in the Gfi1 transcriptional repressor results in a novel form of neutropenia. Eur J Immunol 42 : 2395–2408.
55. YucelR, KosanC, HeydF, MoroyT (2004) Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem 279 : 40906–40917.
56. PargmannD, YucelR, KosanC, SabaI, Klein-HitpassL, et al. (2007) Differential impact of the transcriptional repressor Gfi1 on mature CD4+ and CD8+ T lymphocyte function. Eur J Immunol 37 : 3551–3563.
57. SchmidtT, KarsunkyH, RodelB, ZevnikB, ElsasserHP, et al. (1998) Evidence implicating Gfi-1 and Pim-1 in pre-T-cell differentiation steps associated with beta-selection. EMBO J 17 : 5349–5359.
58. TaghonTN, DavidES, Zuniga-PfluckerJC, RothenbergEV (2005) Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev 19 : 965–978.
59. YuiMA, FengN, RothenbergEV (2010) Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol 185 : 284–293.
60. PalomeroT, LimWK, OdomDT, SulisML, RealPJ, et al. (2006) NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 103 : 18261–18266.
61. WengAP, MillhollandJM, Yashiro-OhtaniY, ArcangeliML, LauA, et al. (2006) c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 20 : 2096–2109.
62. SharmaVM, CalvoJA, DraheimKM, CunninghamLA, HermanceN, et al. (2006) Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol 26 : 8022–8031.
63. WendorffAA, KochU, WunderlichFT, WirthS, DubeyC, et al. (2010) Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 33 : 671–684.
64. MasieroM, MinuzzoS, PuscedduI, MoserleL, PersanoL, et al. (2011) Notch3-mediated regulation of MKP-1 levels promotes survival of T acute lymphoblastic leukemia cells. Leukemia 25 : 588–598.
65. SulimanS, TanJ, XuK, KousisPC, KowalskiPE, et al. (2011) Notch3 is dispensable for thymocyte beta-selection and Notch1-induced T cell leukemogenesis. PLoS One 6: e24937.
66. MedyoufH, GusscottS, WangH, TsengJC, WaiC, et al. (2011) High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J Exp Med 208 : 1809–1822.
67. WeberBN, ChiAW, ChavezA, Yashiro-OhtaniY, YangQ, et al. (2011) A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476 : 63–68.
68. GermarK, DoseM, KonstantinouT, ZhangJ, WangH, et al. (2011) T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A 108 : 20060–20065.
69. LiP, BurkeS, WangJ, ChenX, OrtizM, et al. (2010) Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329 : 85–89.
70. GrimesHL, GilksCB, ChanTO, PorterS, TsichlisPN (1996) The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc Natl Acad Sci U S A 93 : 14569–14573.
71. ZohrenF, SouroullasGP, LuoM, GerdemannU, ImperatoMR, et al. (2012) The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol 13 : 761–769.
72. SpoonerCJ, ChengJX, PujadasE, LasloP, SinghH (2009) A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity 31 : 576–586.
73. FrancoCB, Scripture-AdamsDD, ProektI, TaghonT, WeissAH, et al. (2006) Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A 103 : 11993–11998.
74. LasloP, SpoonerCJ, WarmflashA, LanckiDW, LeeHJ, et al. (2006) Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126 : 755–766.
75. DoanLL, KitayMK, YuQ, SingerA, HerblotS, et al. (2003) Growth factor independence-1B expression leads to defects in T cell activation, IL-7 receptor alpha expression, and T cell lineage commitment. J Immunol 170 : 2356–2366.
76. HockH, HamblenMJ, RookeHM, TraverD, BronsonRT, et al. (2003) Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18 : 109–120.
Štítky
Genetika Reprodukčná medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



