-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
article has not abstract
Published in the journal: Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion. PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003829
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003829Summary
article has not abstract
Cohesion between sister chromosomes is a critical mechanism used by eukaryotic cells to accomplish accurate chromosome segregation. As an analogy, imagine that you are struck one day by the (inexplicable) urge to segregate all your socks into two equal piles. The task will be much easier if you previously took the time to pair them up before tossing them in your dresser drawer. Similarly, keeping sister chromosomes together following DNA replication allows them to be efficiently sorted during cell division. In mitosis, cohesion at centromeres promotes bi-orientation of sister kinetochores by counteracting the pulling forces of microtubules emanating from opposite spindle poles. In meiosis, cohesion between chromosome arms facilitates segregation of recombined homologues during meiosis I by stabilizing the physical linkages (chiasmata) between them, and cohesion between centromeres is essential for accurate segregation of sisters in meiosis II [1], [2]. A study by Moshkin and colleagues in this issue of PLOS Genetics [3] sheds new light on how these processes are regulated.
Cohesion is brought about by ring-shaped cohesin complexes, which contain Smc1, Smc3, a kleisin (mainly Rad21/Scc1in mitosis and Rec8 in meiosis), and an associated SA/Scc3 subunit. In many animals, cohesion removal in mitosis occurs in two steps (Figure 1A). First, in the “prophase pathway,” phosphorylation of SA by kinases such as Polo triggers non-proteolytic removal of cohesin from chromosome arms. This promotes removal of the bulk of cohesin from the arms but, importantly, does not dissolve cohesion at centromeres. Later, once chromosomes are bi-oriented and the spindle checkpoint is satisfied, a proteolytic cohesion removal system is let loose: Separase cleaves the Rad21/Scc1 subunit of the remaining chromosome-bound cohesin, triggering chromosome separation and allowing anaphase [1]. In meiosis, removal of cohesin also occurs by a two-step process but, in contrast to mitosis, both steps require separase activity (Figure 1B). During meiosis I, separase cleaves Rec8 on the arms, leading to resolution of chiasmata and disjunction of homologues. Rec8 at centromeres is not cleaved until meiosis II, when the sisters separate, finally giving rise to a haploid gamete [1], [2].
Fig. 1. Models for regulation of chromosome cohesion in mitosis and meiosis. 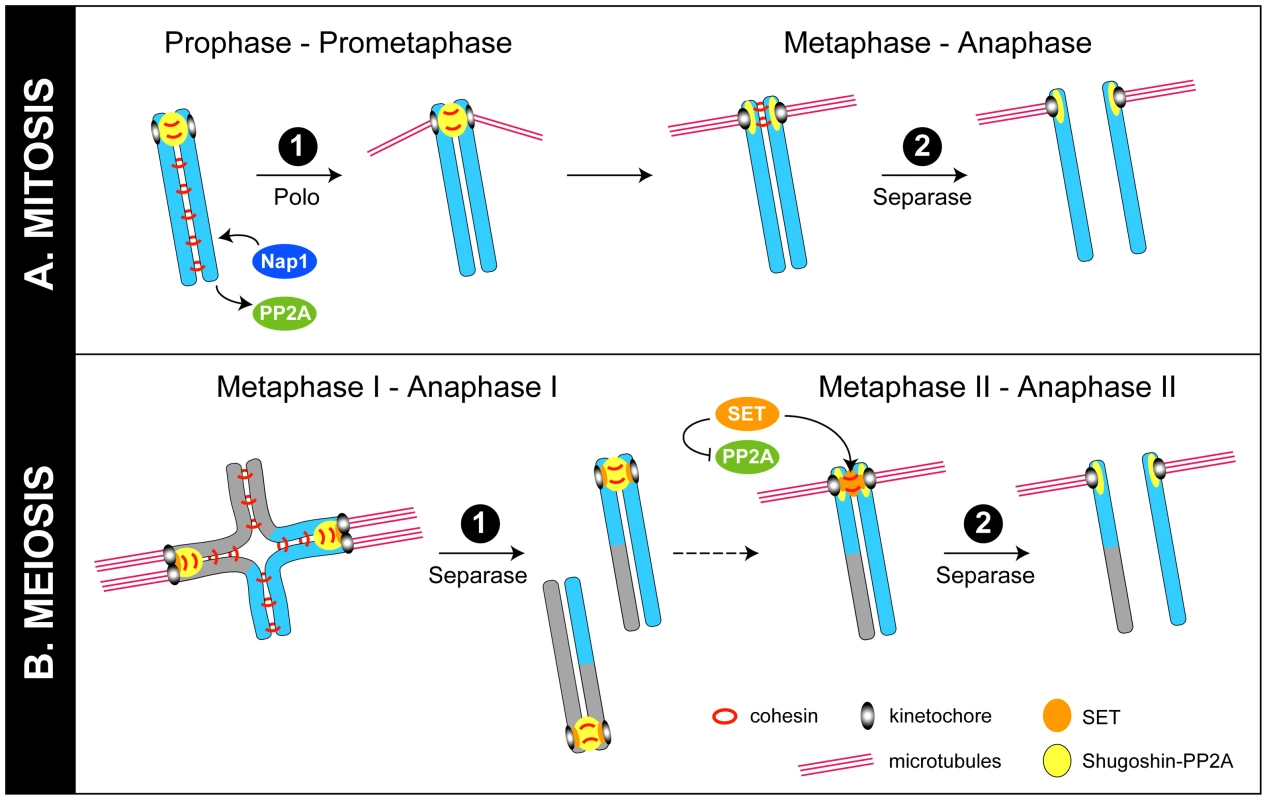
(A) In mitosis, cohesin (red rings) is phosphorylated and removed from chromosome arms by the “prophase pathway.” In this issue, Moshkin et al. provide evidence that Nap1 (blue) can displace PP2A (green) from cohesin to further promote cohesin release from chromosomes [3]. Cohesin at centromeres is protected by shugoshin-PP2A (yellow) until the metaphase–anaphase transition, when tension across bi-oriented sister kinetochores leads to the movement of shugoshin-PP2A away from cohesin at inner centromeres, allowing cohesin to be cleaved by separase [6]. It is not known if Nap1 has a role at this stage. (B) In meiosis I, phosphorylated cohesin linking sister chromosome arms is cleaved by separase, allowing recombined homologues to segregate. Cohesin at centromeres, however, is protected by shugoshin-PP2A, so that sister chromosomes remain together. In meiosis II, as in mitosis, the movement of shugoshin-PP2A away from inner centromeres on bi-oriented chromosomes allows cohesin between sisters to be phosphorylated and cleaved by separase [5], [6]. Recent work from Chambon et al. and Qi et al. is consistent with the view that the Nap1-related protein SET (orange) relocates to inner centromeres in meiosis II to inhibit PP2A and provide an additional means to encourage cohesin phosphorylation and cleavage by separase [8], [9]. Although the two-step removal systems in mitosis and meiosis are distinct, a common protein complex is implicated in protecting centromeric cohesion during the first step in both cases. Shugoshin/MEI-S332 family proteins collaborate with the phosphatase PP2A to prevent cohesin removal at centromeres [2], [4]. In mitosis, shugoshin-PP2A complexes antagonize SA phosphorylation by mitotic kinases, preventing removal by the prophase pathway (Figure 1A). In meiosis, shugoshin-PP2A antagonizes phosphorylation of Rec8, preventing cleavage by separase (Figure 1B) [2], [4]. A key question has been: What subsequently allows centromeric cohesion to be cleaved by separase in the second step? One proposed model is that, in response to tension across bi-oriented sister kinetochores, shugoshin-PP2A complexes move away from cohesin complexes at inner centromeres, making cohesin susceptible to removal by separase [5], [6]. Newly published studies, described below, propose two additional (related) mechanisms that target PP2A to make cohesin sensitive to removal.
Chambon et al. suggest that an inhibitor of PP2A, known as SET (or I2PP2A or TAF-I) [7], is required to inactivate shugoshin-PP2A [8]. They reported, as in previous proteomic studies, that SET is found in a complex with shugoshin, and that it more clearly co-localizes with shugoshin, PP2A, and cohesin at inner centromeres in meiosis II than in meiosis I. Morpholino-based depletion of SET from mouse oocytes caused some sister chromosomes to fail to separate in meiosis II [8]. Qi et al. reported similar findings [9]. In this case, SET depletion by RNAi did not detectably alter chromosome segregation in mouse oocytes, but overexpression of SET led to precocious sister separation in meiosis I. These results are broadly consistent with a model in which SET inhibits PP2A in meiosis II to allow Rec8 phosphorylation and cleavage by separase (Figure 1B).
SET is a member of a widely conserved family of proteins related to nucleosome assembly protein-1 (Nap1), which all form a distinctive “earmuff-like” structure. These include human and Drosophila SET, Vps75 in budding yeast, and the Nap1 proteins (e.g. six Nap1-like proteins in humans, Nap1 in Drosophila, and yNap1 in yeasts) [10], [11]. SET and Nap1 have both been extensively studied as histone chaperones; they can also associate with histone acetyltransferases (Vps75) and histone deacetylases (Nap1), and SET is a component of INHAT, an inhibitor of histone acetyltransferases [10]–[12]. It is notable that neither Chambon et al., nor Qi et al., formally showed that it is the ability of SET to inhibit PP2A activity that modulates cohesion. Therefore, a number of questions remain unanswered. Is SET really acting as a PP2A enzyme inhibitor? If so, does a similar mechanism exist in mitosis or in other organisms? Do the related Nap1 proteins play similar roles? The new study in this issue [3] expands on these points.
Moshkin et al. identified cohesin subunits in Drosophila Nap1 immunoprecipitates, and found that the genome binding sites of Nap1 resemble those of cohesin in ChIP-chip experiments [3]. Depletion or deletion of Nap1 caused increased chromosomal localization of PP2A, shugoshin/MEI-S332, and cohesin, and prevented the normal separation of chromosome arms in early mitosis. Based on this, and the movement of Nap1 into the nucleus in prophase, the ability of recombinant Nap1 to displace PP2A from cohesin complexes in vitro, and the ability of PP2A depletion or overexpression to reverse the cohesion defects caused by Nap1 depletion or overexpression, Moshkin et al. propose that Nap1 displaces PP2A from binding to cohesin. In this way, Nap1 increases cohesin phosphorylation and release by (presumably) the prophase pathway in mitosis, without necessarily inhibiting the enzyme activity of PP2A (Figure 1A).
Together, these three studies reveal that SET and Nap1 proteins regulate cohesion in meiosis and mitosis in more than one organism, and leave us with a variety of proposed ways in which the “guardian spirit” of cohesion (shugoshin-PP2A) can be relieved of its duties. It will now be interesting to determine if the different conclusions reached about the mechanism of SET and Nap1 action on PP2A reflect differences between the two proteins, between mice and flies, between meiosis and mitosis, or simply our currently incomplete understanding. Is counteracting PP2A action the only or main way in which these proteins regulate cohesion, or could their histone chaperone activity, or binding to histone-modifying enzymes, play a role? The future use of separation of function mutants of Nap1 and SET will likely help answer these questions. What normally restricts SET activity to meiosis II, and might cyclin A2–dependent kinases [13] contribute? Does SET function in mitosis? Does Nap1 play a role in meiosis, or influence centromeric cohesion in late mitosis? To what extent do tension-dependent shugoshin-PP2A relocation [5], [6] and PP2A inactivation [3], [8], [9] collaborate at centromeres? Because Nap1 can modulate gene expression, and cohesin itself may play a role in transcriptional regulation [1], [12], it will also be useful to exclude indirect effects on cell division.
Chromosome segregation defects cause a range of problems, including chromosome instability and aneuploidy in cancer and, if they occur in meiosis, infertility, miscarriage, and birth defects [14], [15]. Cohesin gene mutations in cancer [16] and loss of sister chromosome cohesion in aged oocytes [15], [17], [18] may underlie some of these defects. Further understanding of cohesion regulation, including the newfound contribution of Nap1 and SET, may enhance our ability to prevent and treat these conditions.
Zdroje
1. NasmythK, HaeringCH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43 : 525–558 doi: 10.1146/annurev-genet-102108-134233
2. WatanabeY (2012) Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev Mol Cell Biol 13 : 370–382 doi: 10.1038/nrm3349
3. MoshkinYM, DoyenCM, KanTW, ChalkleyGE, SapK, et al. (2013) Histone chaperone NAP1 mediates sister chromatid resolution by counteracting protein phosphatase 2A. PLOS Genet 9: e1003719 doi: 10.1371/journal.pgen.1003719
4. CliftD, MarstonAL (2011) The role of shugoshin in meiotic chromosome segregation. Cytogenet Genome Res 133 : 234–242 doi: 10.1159/000323793
5. GomezR, ValdeolmillosA, ParraMT, VieraA, CarreiroC, et al. (2007) Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep 8 : 173–180 doi: 10.1038/sj.embor.7400877
6. LeeJ, KitajimaTS, TannoY, YoshidaK, MoritaT, et al. (2008) Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol 10 : 42–52 doi: 10.1038/ncb1667
7. LiM, MakkinjeA, DamuniZ (1996) The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem 271 : 11059–11062 doi: 10.1074/jbc.271.19.11059
8. ChambonJP, TouatiSA, BerneauS, CladiereD, HebrasC, et al. (2013) The PP2A inhibitor I2PP2A is essential for sister chromatid segregation in oocyte meiosis II. Curr Biol 23 : 485–490 doi: 10.1016/j.cub.2013.02.004
9. QiST, WangZB, OuyangYC, ZhangQH, HuMW, et al. (2013) Overexpression of SETβ, a protein localizing to centromeres, causes precocious separation of chromatids during the first meiosis of mouse oocytes. J Cell Sci 126 : 1595–1603 doi: 10.1242/jcs.116541
10. EitokuM, SatoL, SendaT, HorikoshiM (2008) Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci 65 : 414–444 doi: 10.1007/s00018-007-7305-6
11. DasC, TylerJK, ChurchillME (2010) The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci 35 : 476–489 doi: 10.1016/j.tibs.2010.04.001
12. MoshkinYM, KanTW, GoodfellowH, BezstarostiK, MaedaRK, et al. (2009) Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell 35 : 782–793 doi: 10.1016/j.molcel.2009.07.020
13. TouatiSA, CladiereD, ListerLM, LeontiouI, ChambonJP, et al. (2012) Cyclin A2 is required for sister chromatid segregation, but not separase control, in mouse oocyte meiosis. Cell Rep 2 : 1077–1087 doi: 10.1016/j.celrep.2012.10.002
14. BakhoumSF, ComptonDA (2012) Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest 122 : 1138–1143 doi: 10.1172/JCI59954
15. NagaokaSI, HassoldTJ, HuntPA (2012) Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 13 : 493–504 doi: 10.1038/nrg3245
16. SolomonDA, KimT, Diaz-MartinezLA, FairJ, ElkahlounAG, et al. (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333 : 1039–1043 doi: 10.1126/science.1203619
17. ChiangT, DuncanFE, SchindlerK, SchultzRM, LampsonMA (2010) Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 20 : 1522–1528 doi: 10.1016/j.cub.2010.06.069
18. ListerLM, KouznetsovaA, HyslopLA, KalleasD, PaceSL, et al. (2010) Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 20 : 1511–1521 doi: 10.1016/j.cub.2010.08.023
Štítky
Genetika Reprodukčná medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



