-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
Genomic imprinting is a process that causes genes to be expressed from one allele only according to parental origin, the other allele being silent. Diseases can arise when the normally active alleles are not expressed. In this context, low level of expression of the normally silent alleles has been considered as genetic noise although such expression has never been further studied. Prader-Willi Syndrome (PWS) is a neurodevelopmental disease involving imprinted genes, including NDN, which are only expressed from the paternally inherited allele, with the maternally inherited allele silent. We present the first in-depth study of the low expression of a normally silent imprinted allele, in pathological context. Using a variety of qualitative and quantitative approaches and comparing wild-type, heterozygous and homozygous mice deleted for Ndn, we show that, in absence of the paternal Ndn allele, the maternal Ndn allele is expressed at an extremely low level with a high degree of non-genetic heterogeneity. The level of this expression is sex-dependent and shows transgenerational epigenetic inheritance. In about 50% of mutant mice, this expression reduces birth lethality and severity of the breathing deficiency, correlated with a reduction in the loss of serotonergic neurons. In wild-type brains, the maternal Ndn allele is never expressed. However, using several mouse models, we reveal a competition between non-imprinted Ndn promoters which results in monoallelic (paternal or maternal) Ndn expression, suggesting that Ndn allelic exclusion occurs in the absence of imprinting regulation. Importantly, specific expression of the maternal NDN allele is also detected in post-mortem brain samples of PWS individuals. Our data reveal an unexpected epigenetic flexibility of PWS imprinted genes that could be exploited to reactivate the functional but dormant maternal alleles in PWS. Overall our results reveal high non-genetic heterogeneity between genetically identical individuals that might underlie the variability of the phenotype.
Published in the journal: Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences. PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003752
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003752Summary
Genomic imprinting is a process that causes genes to be expressed from one allele only according to parental origin, the other allele being silent. Diseases can arise when the normally active alleles are not expressed. In this context, low level of expression of the normally silent alleles has been considered as genetic noise although such expression has never been further studied. Prader-Willi Syndrome (PWS) is a neurodevelopmental disease involving imprinted genes, including NDN, which are only expressed from the paternally inherited allele, with the maternally inherited allele silent. We present the first in-depth study of the low expression of a normally silent imprinted allele, in pathological context. Using a variety of qualitative and quantitative approaches and comparing wild-type, heterozygous and homozygous mice deleted for Ndn, we show that, in absence of the paternal Ndn allele, the maternal Ndn allele is expressed at an extremely low level with a high degree of non-genetic heterogeneity. The level of this expression is sex-dependent and shows transgenerational epigenetic inheritance. In about 50% of mutant mice, this expression reduces birth lethality and severity of the breathing deficiency, correlated with a reduction in the loss of serotonergic neurons. In wild-type brains, the maternal Ndn allele is never expressed. However, using several mouse models, we reveal a competition between non-imprinted Ndn promoters which results in monoallelic (paternal or maternal) Ndn expression, suggesting that Ndn allelic exclusion occurs in the absence of imprinting regulation. Importantly, specific expression of the maternal NDN allele is also detected in post-mortem brain samples of PWS individuals. Our data reveal an unexpected epigenetic flexibility of PWS imprinted genes that could be exploited to reactivate the functional but dormant maternal alleles in PWS. Overall our results reveal high non-genetic heterogeneity between genetically identical individuals that might underlie the variability of the phenotype.
Introduction
Imprinted genes are functionally mono-allelic in a parent-of-origin specific manner. Genomic imprinting is a non-Mendelian epigenetic form of gene regulation which is germline-inherited since the epigenetic marks are established in the parental gametes without altering the DNA sequence [1]. Compared to most other tissues the brain is enriched in genes showing an imprinted pattern of expression [2], vulnerable to environmental perturbation [3] and contributing to various neurodevelopmental diseases [4], [5]. This vulnerability, linked to a plasticity of gene regulation, might also allow a positive adaptation of an organism to a new external environment. It is important to examine situations in which partial loss of imprinting (LOI) rescues a mutant phenotype. Understanding the mechanisms underlying this positive effect could lead to therapeutic avenues that manipulate this rheostat function.
Necdin (Ndn) is an imprinted gene present in both human and mouse, and its maternally inherited allele is normally silenced [6]–[9]. The human NDN gene is located in a large imprinted domain. All the paternally expressed genes from this domain are candidate genes for some of the symptoms of Prader-Willi Syndrome (PWS), an orphan neurodevelopmental genetic disease [10] (OMIM 176270). The essential clinical diagnostic criteria include neonatal hypotonia and abnormal feeding behavior with a poor suck followed by a hyperphagia, resulting in severe obesity, and behavioral problems [11]–[14]. Breathing deficiency is a significant health concern for many patients and contributes to some cases of sudden death [15], [16]. Notably, there is considerable variability in symptom severity among patients [11].
Mouse strains with targeted inactivation of single PWS genes have been created, and heterozygous mice with a paternally inherited deficiency (+m/−p) are generally considered to be functionally null. Four independent Ndn-deficient mouse lines have been created [9], [17]–[19], three of which display PWS associated phenotypes [9], [17]–[21] including partial early post-natal lethality due to respiratory distress [20], [22].
In Muscatelli's Ndn-KO mouse model (named Ndntm1.1Mus), we observed a high level of phenotypic heterogeneity among the Ndn+m/−p mice within each litter; notably in the incidence and severity of apneas [22]. The stochastic nature of gene expression can generate pronounced phenotypic variations [23] and here we hypothesize that this inter-individual variability, among Ndn+m/−p mice, might result from a “stochastic” activation of the putatively silent maternal allele of Ndn.
In this study, we investigate this hypothesis by comparing homozygous mice deleted for both alleles of Ndn (Ndn−/−) with heterozygous Ndn (Ndn+m/−p) mice. We perform a comprehensive analysis of the in vivo expression and functional role of the Ndn maternal allele. We investigate the genetic context and the mechanism underlying this maternal expression. Finally, we show that the maternal allele of NDN is transcribed and that Necdin protein is present in human post-mortem Prader-Willi brains.
Results
Ndn−/− mutant mice present a more severe phenotype than Ndn+m/−p littermates
After 36 backcrosses on the C57Bl/6J genetic background, we measured the lethality of Ndn+m/−p mice versus Ndn+/+ mice, both derived from crosses between a wild-type (WT) female and a heterozygous male deleted for the Ndn maternal allele (−m/+p). As expected [17], Ndn+m/−p mice were significantly under-represented at weaning (28% reduction, 125 +/+ versus 91 Ndn+m/−p; CHI2 test, P<0.01). Furthermore, we confirmed that there was an equivalent number of Ndn+m/−p (118) versus Ndn+/+ (116) pups at birth, and 21% lethality between postnatal day (P) P0 and P3 in both sexes. However, in a cohort of 75 Ndn−/ − mutants, derived from crosses between a Ndn−/− female and a Ndn−/− male, we found a 43% lethality of the Ndn−/− pups (32/75), between P1 and P2, and these pups were visibly cyanotic. Altogether, these data suggest that, due presumably to respiratory deficiency, Ndn−/− newborns are twice as likely to die early compared to Ndn+m/−p newborns. This result is surprising because in theory there is no Ndn expression in either Ndn−/ − or Ndn+m/−p pups.
Next, we compared breathing pattern between Ndn−/ − and Ndn+m/−p mice. Previously, we demonstrated that newborn and young adult Ndn+m/−p mice present an irregular respiratory rhythm with frequent apneas [22]. Importantly, such apneas were more than twice as frequent in Ndn−/− compared to Ndn+m/−p mice (Table 1).
Tab. 1. Respiratory pattern and apnea in Ndn−/− young adult versus Ndn+m/−p mice. 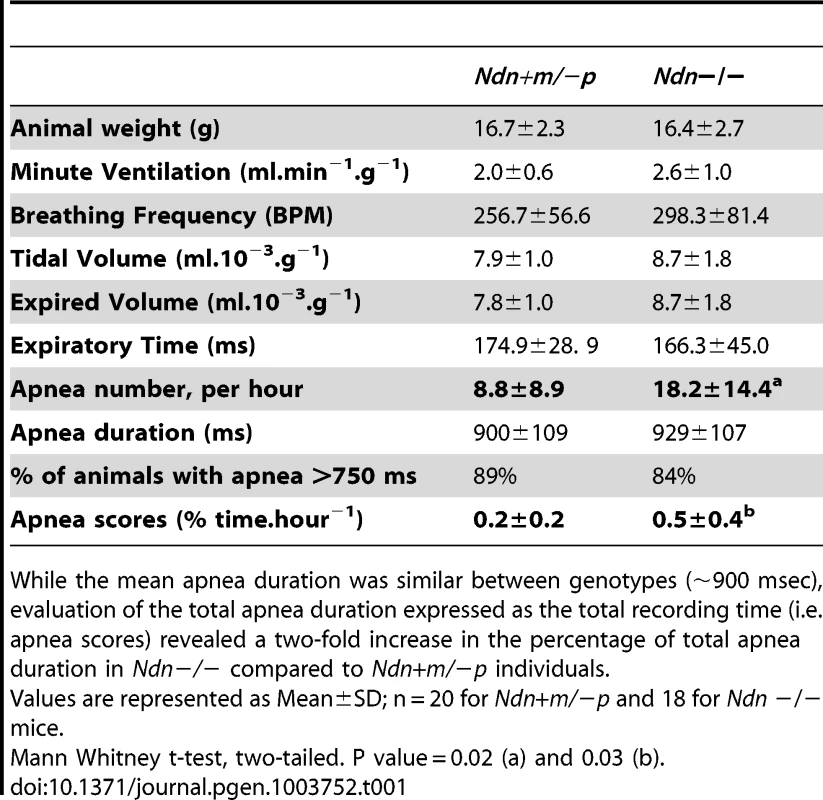
While the mean apnea duration was similar between genotypes (∼900 msec), evaluation of the total apnea duration expressed as the total recording time (i.e. apnea scores) revealed a two-fold increase in the percentage of total apnea duration in Ndn−/− compared to Ndn+m/−p individuals. In summary, the respiratory phenotype of Ndn−/ − homozygous mice is more severe than in Ndn+m/−p heterozygotes, suggesting a role for the maternally inherited Ndn allele.
Quantitative expression of the Ndn maternal allele
Since we suspected a role of the maternal Ndn allele in the phenotype of Ndn+m/−p mice, we further investigated the expression of this allele using a specific anti-Necdin antibody in immunoblot analyses of protein extracts from different P1 brains (Figure S1A) or from individual E12.5 embryos (Figure S1B). We detected a specific signal at the expected size for WT animals but also a fainter signal (10–20 fold less intense) in four of the eight Ndn+m/−p animals, with no signal in Ndn−/− mice.
Consistent with our previous study [17], we did not detect maternal Ndn allele expression by RT-PCR (Figure S1C). We therefore increased the experimental sensitivity using RT-qPCR. We focused on different developmental stages (E12.5, P1 and adult) [24], using total brain tissue as well as brain structures known to highly express Ndn (hypothalamus) or to be involved in respiratory function (pons). A total of 258 individual animals on a C57Bl/6J genetic background, including 72 WT, 57 Ndn−/ − and 129 Ndn+m/−p, were analyzed.
In Ndn−/− mutant mice, no Ndn transcripts were detected irrespective of the brain structures or stages analyzed (data not shown). In WT individuals (Figure 1A), as expected [17], [24], we observed higher Ndn expression in P1 brains compared with expression in E12.5 embryos. In Ndn+m/−p individuals, maternal Ndn transcripts were detected, but the transcript level was reduced 800 (P1 brain) to 1500 (adult hypothalamus)-fold compared to WT individuals (comparing the medians, Figure 1B). Interestingly, there was a huge inter-individual variability (×100 to ×1000 between the extreme values) for all Ndn+m/−p mice, irrespective of the stages and tissues tested.
Fig. 1. Ndn expression analyzed by RT-qPCR in Ndn+m/−p mice. 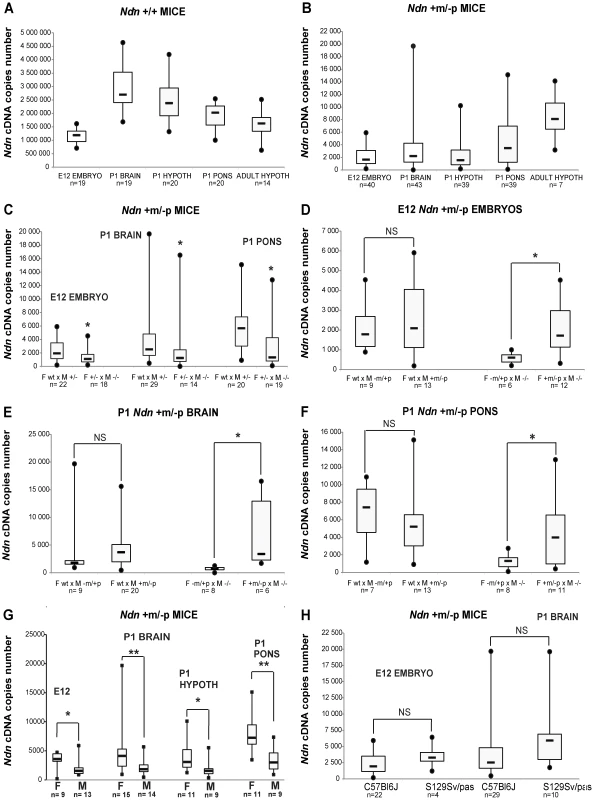
RT-qPCR analysis of Ndn transcripts. A) Ndn transcripts in E12 WT embryos and selected brain tissues from P1 (whole brain, hypothalamus, pons) or adult (hypothalamus) WT mice. B) Ndn transcripts in Ndn+m/−p E12 embryos and brain tissues from P1 (whole brain, hypothalamus, pons) or adult (hypothalamus) Ndn+m/−p mice. C–F) Quantification of Ndn transcripts in Ndn+m/−p E12 embryos (C,D) and whole brain (C,E) and pons (C,F) from P1 mice with respect to parental genotype (C) and to the maternal or paternal genotype contribution indicating a grandparental influence (D–F). G) Quantification of Ndn transcripts in Ndn+m/−p E12 embryos, P1 and adult Ndn+m/−p brain tissues in male (M) and female (F) mice. H) Quantification of Ndn transcripts in Ndn+m/−p E12 embryos and Ndn+m/−p P1 mice in C57Bl/6J and S129Sv/Pas mouse strains. The Ndn transcript copy number in Ndn+m/−p offspring (E12, P1 brain, P1 pons; n = 71) issued from a cross between a WT female and a Ndn+/− male is significantly more than 2 fold higher than the Ndn copy number in Ndn+m/−p offspring (n = 51) issued from a Ndn+/− female crossed with a Ndn−/− male (C). Considering separately the effect of the maternal or paternal genotype, we showed that when the mother is WT and the father is (+/−), with a maternal Ndn mutant allele (−m/+p) or a paternal Ndn mutant allele (+m/−p), then there is no difference in the copy number of Ndn maternal transcripts between the Ndn+m/−p individuals of the offspring of both types of crosses (n = 25 and n = 46, respectively) (D,E,F). However, we observed an effect of the maternal genotype, with a significant difference in the level of Ndn maternal transcripts between the Ndn+m/−p individuals (n = 22) issued from a (−m/+p female X −/− male) compared with the Ndn+m/−p individuals (n = 29) issued from a (+m/−p female X −/− male) (D,E,F); the +/+ or Ndn+m/−p maternal genotype is correlated with a significant three times higher level of Ndn maternal expression in the Ndn+m/−p offspring. Values are represented as Median (Q1, Q3). WMW test, two-tailed. * P value<0.05 and ** <0.01. We searched for factors that influence the level of transcripts of the Ndn maternal allele. While our results show an absence of a significant effect of the paternal genotype (Figure 1D,E,F), the maternal genotype clearly influences maternal Ndn expression in the Ndn+m/−p offspring. Although in all cases the offspring (+m/−p) have inherited a wild-type (+m) allele from the mother, offspring from the Ndn+/+ or Ndn+m/−p maternal genotype had a significant three-fold higher level of Ndn maternal expression compared to those from a Ndn−m/+p maternal genotype (Figure 1D,E,F). Importantly, the extensive variability of Ndn maternal expression is also positively correlated with both those maternal genotypes. In contrast, litters issued from Ndn−m/+p mothers showed both a lower level of Ndn expression and an absence of variability in the Ndn+m/−p offspring (Figure 1D,E,F). In addition, there is a gender-specific effect on the Ndn maternal expression in Ndn+m/−p offspring, with females expressing two-fold more Ndn expression compared to males (Figure 1G).
Finally we compared this maternal expression in Ndn+m/−p offspring from a C57Bl/6J or a 129Sv/Pas genetic background. In both mouse strains, a similar level of Ndn maternal expression was observed (Figure 1H).
We conclude that an extremely low but specific transcription of the maternally inherited Ndn allele in Ndn+m/−p individuals is found in at least two mouse strains (C57Bl/6J, 129Sv/Pas). Transcript numbers are highly variable irrespective of the developmental stage or the brain structure analyzed, even among littermates. Finally, the quantity of maternal Ndn transcripts depends significantly on the maternal genotype and on the gender.
Qualitative expression of the Ndn maternal allele at different developmental stages
We asked whether maternal Ndn expression was due to: 1) low but homogeneous expression in all tissues and/or 2) reduced but focal expression in specific structures and cell types.
At E12.5, using immunohistochemistry (IHC) and in situ hybridization (ISH) on frozen serial sections, Necdin protein and transcripts were detected in the same structures of four out of nine Ndn+m/−p embryos (Figure 2). Importantly, no protein or transcripts were detected in Ndn−/− individuals (Figure S2). Expression of the maternally inherited Ndn allele was detected in a restricted number of cells of specific nervous structures in which the paternally inherited Ndn allele is normally expressed in WT animals (Figure 2 A,B,C,D,E). Interestingly, the cerebral cortex, the tongue and the myotome, which show expression of the paternal allele in WT, do not express the maternal Ndn allele (data not shown). In contrast to WT E10.5 embryos, no maternal Ndn expression was detected in Ndn+m/−p E10.5 embryos (n = 9) (Figure S3). At P1, a stage when expression normally peaks [24], we detected Necdin protein by IHC in the brain of Ndn+m/−p newborns (n = 6) (Figure 3). Necdin presented a similar expression pattern in Ndn+m/−p adults (Figure S4). At both developmental stages, this expression was restricted to a limited number of cells in several, but not all nuclei that express Ndn in WT animals, such as the hypothalamic (Figure 3B) and the raphe nuclei (Figure 3C,D).
Fig. 2. Ndn expression in Ndn+m/−p E12.5 embryos. 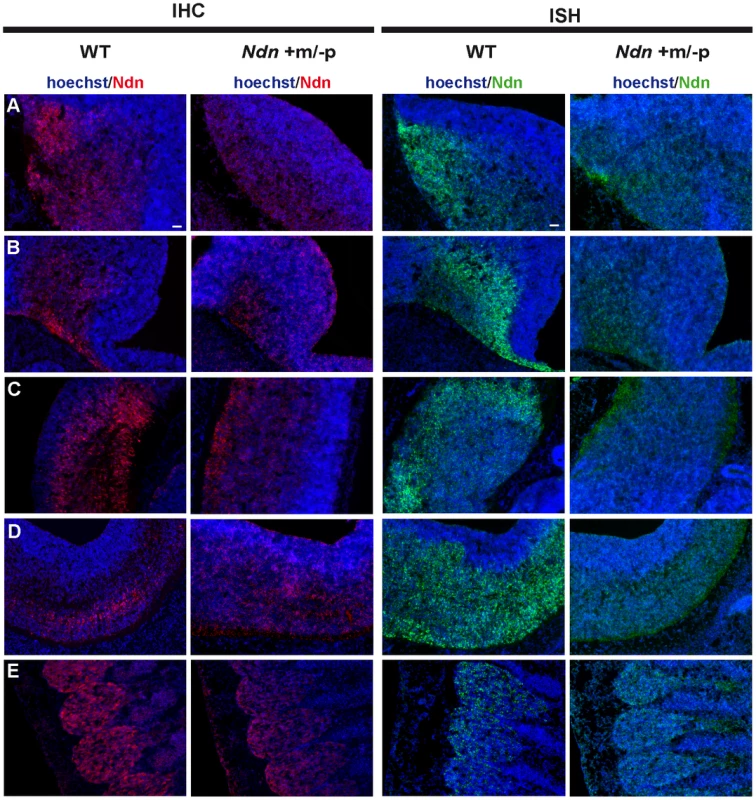
Expression of Ndn in the nervous system of WT and Ndn+m/−p embryos at E12.5 revealed by IHC or ISH on frozen sections using a Necdin specific antibody (Ndn,red) or Ndn RNA probe (green). Tissue sections are visualized using a Hoechst labeling (blue). Expression is detected in both genotypes, at the protein and transcript levels, in the preoptic area (A), supraoptic area (B), thalamus (C), pons (D) and in the dorsal root ganglia (E). Note that the level of expression is weaker and more restricted in Ndn+m/−p embryos. Other structures, like the tegmentum, the subthalamus and the spinal cord also express the Ndn maternal allele. Scale bar: 50 µm. Fig. 3. Ndn expression in Ndn+m/−p brains of neonates. 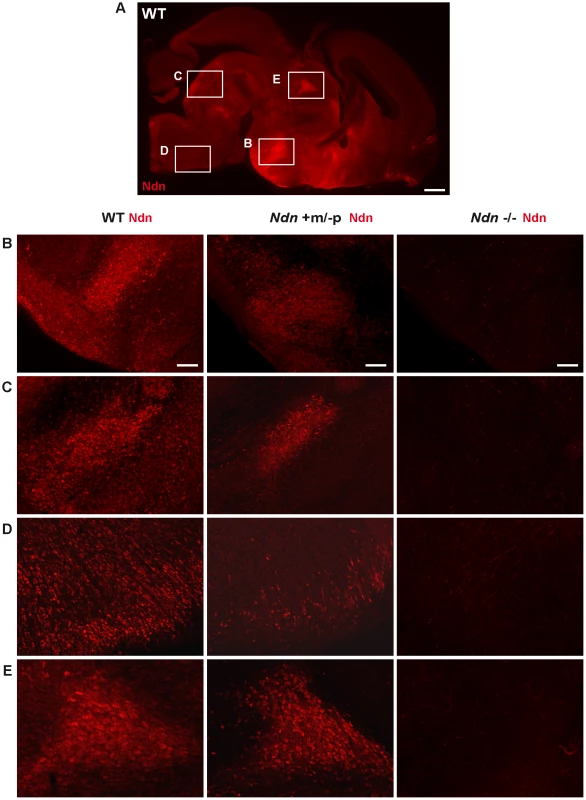
Presence of Necdin in WT, Ndn+m/−p and Ndn−/− brains of neonates (P0) revealed by IHC on sagittal sections using an anti-Necdin specific antibody (red). Immunolabeling on a WT whole brain sagittal section illustrates the ventral expression of Necdin (A). Such expression is compared between the three genotypes at the dorsal medial nucleus of the hypothalamus (B), the dorsal and magnus raphe nuclei (C and D) respectively and the paraventricular thalamus nuclei (E). Scale bar: 500 µm (A) and 100 µm (B,C,D,E). Thus, at the anatomical level, using ISH and IHC, we conclude that the Ndn maternal allele is expressed in a subset of Ndn+m/−p individuals and, compared to WT mice, is restricted to a limited population of cells in specific nervous system structures. Noticeably, there is considerable inter-individual variability, even between littermates (Figure 1 and data not shown).
We addressed the question of intra-individual variation by studying the raphe nuclei, a structure defined by 5HT-expressing neurons, all of which express Necdin in WT mice [22]. We double immunostained P1 brains using anti-Necdin and anti-5HT antibodies and determined the number of 5HT/Necdin positive neurons in the different raphe nuclei (B1 to B9) of WT, Ndn+m/−p and Ndn−/− newborns (Figure 4A). We confirmed both inter-individual and intra-individual variation in the number of 5HT/Necdin double positive neurons in Ndn+m/−p raphe nuclei. For instance, in the same individual, 78% of 5HT positive neurons in B1/B2 raphe nuclei were Necdin positive although in other raphe nuclei no Necdin expression was detected (Figure 4A). We conclude that there is also intra-individual variation in the expression of maternal Ndn allele in the raphe nuclei.
Fig. 4. Ndn expression in the 5HT raphe nuclei of Ndn+m/−p individuals. 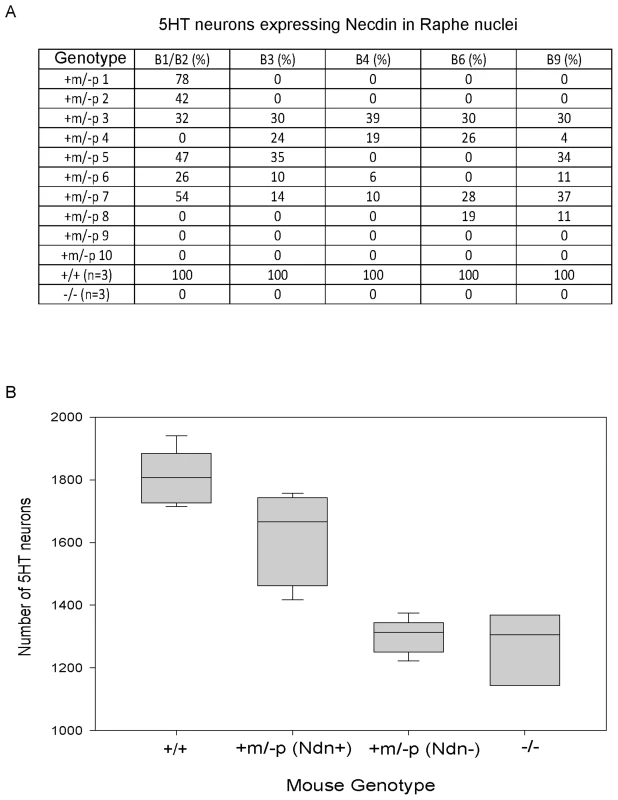
(A) % of Necdin immunoreactive cells among 5HT positive cells located in the B1 to B9 raphe nuclei. Wild-type (n = 3), Ndn−/− (n = 1) and Ndn+m/−p (n = 10) newborn mice were analyzed. (B) Number of 5HT-expressing neurons in the B1/B2 raphe nuclei of +/+ (n = 9), Ndn +m/−p (n = 18) and Ndn−/− (n = 8) individuals. Ndn+m/−p (n = 18) individuals are divided in two populations: a population in which Necdin immunolabeling is detected in a mean of 46% of 5HT neurons of Ndn+m/− (Ndn+, n = 9) individuals and a population in which no Necdin/5HT colabelling is detected in the B1/B2 raphe nuclei of Ndn+m/−p (Ndn−, n = 9) individuals. Scale bar: 10 µm. Ndn maternal expression plays a functional role at the cellular level
Previously, we observed alterations in the 5HT system [22] in Ndn+m/−p mice. Here, we analyzed the cellular defects in Ndn−/− newborn mice (n = 8) in comparison with Ndn+m/−p (n = 18) and WT newborns (n = 9). Using 5HT immunolabelling, we counted the number of 5HT neurons in the B1/B2 raphe nuclei (Fig. 4B). We found a significant 28% reduction (WMW test, P<0.001) in the number of 5HT-expressing neurons between Ndn−/− (1306 (1204,1337); n = 8) and WT newborns (1807 (1738, 1882); n = 9). Interestingly, in the B1/B2 raphe nuclei compared to WT mice, the Ndn+m/−p individuals that expressed Necdin, with a mean of 46% of 5HT neurons Necdin-positive (Ndn+m/−p (Ndn+), Figure 4B), had only a 8% reduction (WMW test, P<0.001) in the number of 5HT-expressing neurons (1666 (1489,1733); n = 9). In contrast, the Ndn+m/−p individuals (Ndn+m/−p (Ndn−), Figure 4B) that do not show Necdin expression had a significant 28% reduction (WMW test, P<0.001) in the number of 5HT-expressing neurons (1313 (1258,1335); n = 9) similar to the results observed in Ndn−/− P0 mice. Thus expression of the maternal Ndn allele in Ndn+m/−p individuals correlates with an increased number of 5HT-expressing neurons.
The Ndn C57Bl/6J maternal allele is never expressed in wild-type mice
We next asked whether the low level of maternal Ndn expression was also present in WT mice. In order to discriminate between paternal and maternal allele-specific Ndn expression in WT mice, we identified mouse strains carrying transcribed polymorphisms in the Ndn gene. Three such polymorphisms (two SNPs in the 3′-untranslated region (UTR) and one 5bp indel in the 5′-UTR) were identified between Mus musculus (C57BL/6J) and Mus spretus strains.
First, to analyze the SNPs, we performed two quantifications of allele-specific expression by pyrosequencing (QUASEP) assays on RT-PCR products from F1 brains of six pups with a C57BL/6J mother and Mus spretus father, and did not detect expression of the maternal (C57BL/6J) Ndn allele in these brain samples (data not shown). To further increase the sensitivity for detection of maternal Ndn transcripts, we designed specific TaqMan probes distinguishing between the presence and absence of the 5 bp indel in the 5′-UTR and used RT-qPCR for allele-specific quantification. However, this assay also did not reveal any Ndn transcripts from the C57BL/6J maternal allele in (C57BL/6J×Mus spretus) F1 brains from 32 pups (Figure S5). We conclude that in this wild-type mixed genetic context, we do not detect any expression of the maternal C57Bl/6J Ndn allele.
Competition between the promoters of the Ndn alleles
The Ndn+m/−p heterozygous mice described by Gerard et al [9] (named Ndntm2Stw) present a more severe phenotype with in particular a higher lethality at birth compared to Ndntm1.1Mus+m/−p mice. We failed to detect expression of the Ndn maternal allele in Ndntm2Stw+m/−p embryos (n = 8) at two developmental stages (E12.5 and E14.5), using the IHC and ISH approaches (Figure S6 and data not shown). Importantly, in Ndntm2Stw mice, the Ndn coding sequence has been replaced by the β-Galactosidase sequence, and the Ndn promoter and regulatory sequences have been retained allowing β.Gal expression from the paternal allele [9]. In contrast, in Ndntm1.1Mus mice, the promoter and the first two thirds of the Ndn coding sequence were replaced with a loxP site. The complete lack of Ndn maternal expression in the Ndntm2Stw+m/−p mice could suggest that presence of the active paternal Ndn promoter suppresses expression of the Ndn maternal allele. We propose that the Ndn maternal allele is expressed only when the paternal promoter is absent or silenced, consistent with the absence of expression of the maternally inherited Ndn allele in WT mice (Figure S10A).
To further explore this question, we created a transgenic mouse line (TG45 named TG) containing a modified Bacterial Artificial Chromosome (BAC) in which the Ndn coding sequence was replaced by the eGFP sequence under the control of the Ndn promotert; this BAC transgene is present in one or two copies and its expression is not regulated by imprinting mechanism (Figure S7 and data not shown). In these mice, the expression of eGFP was restricted to the brain regions in which Ndn is normally expressed (Figure 5 and data not shown). We then studied this eGFP expression in newborn WT, Ndn+m/−p, Ndn−/− hypothalamus (using the Ndntm1.1Mus strain) and in the hypothalamus from a mouse line (Ndn++) in which Ndn is over-expressed (Figure 5 and Figure S10B). We performed colabelling and observed an inverse correlation between the intensity of Necdin immunolabeling and eGFP fluorescence on coronal brain sections (Figure 5). In the absence of Ndn expression (Ndn−/−, TG+) the number of eGFP-positive cells was the highest compared to eGFP-positive cells when Ndn is over-expressed (Ndn++, TG+) (Figure 5). This result was quantified by immunoblotting (Figure S8A). Finally, in (WT, TG+) mice, we independently quantified for each cell the eGFP and Necdin signals in two hypothalamic nuclei (Figure S8 B,C). For both structures, we observed two distinct populations of cells and conclude that approximately half of the cells expressed Necdin while the other half expressed eGFP; only very few cells co-expressed eGFP and Necdin (Figure S8D). Thus, at the cellular level, the eGFP expression level is inversely correlated with Necdin expression level.
Fig. 5. Necdin expression and eGFP expression in different mouse genetic backgrounds with or without the BAC Ndn-eGFP transgene (TG+ or TG−). 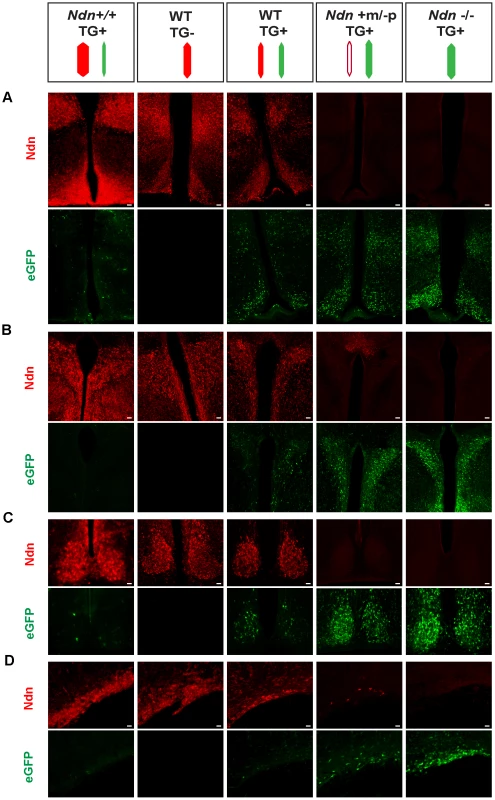
WT(TG−), WT(TG+), Ndn++(TG+), Ndn+m/−p(TG+) and Ndn−/−(TG+) mice are analyzed by IHC to detect Necdin or eGFP expression. The coloured dashes indicate: Ndn maternal allele (white), Ndn paternal allele (red) and BAC TG (green); the width of the dashes indicates the level of expression of Ndn or eGFP. Immunostaining for Necdin (red) and eGFP fluorescence (green) was performed on coronal cryosections of brains at P0. The expression was studied in several hypothalamic nuclei: dorsomedial hypothalamus and arcuate (A), paraventricular (B), suprachiasmatic (C) and supraoptic (D). Interestingly, in a Ndn+m/−p (TG+) hypothalamus with a faint expression of the maternal allele, we also detect fewer eGFP-positive cells than in a Ndn−/− (TG+) hypothalamus. Scale bar: 20 µm. Our results are consistent with a model whereby two Ndn alleles in the same cell, both of which include at least the Ndn promoter and regulatory sequences and neither of which is silenced by imprinting, triggers allelic exclusion at the transcriptional level favouring the expression of one allele only per cell (Figure S10).
Analysis of the methylation profile at the Ndn DMR
Variation in DNA methylation at the DMRs of imprinted genes has been reported in different tissues, importantly in brain, and might be a source of gene expression and phenotypic variations [25]. We therefore studied DNA methylation in a secondary DMR (42 CpGs), previously shown to be correlated with imprinted regulation of Ndn expression [7], [26] (Figure S9). We found no major changes in methylation on the Ndn maternal allele in Ndn+m/−p brains. However, methylation of this DMR occurs after the blastula stage and our failure to detect modifications to methylation in this DMR could be because only a few neurons express the maternal Ndn allele, and this escapes our global brain analysis.
The maternally inherited allele of NDN is expressed in hypothalamus of PWS patients
We assessed whether the expression of the maternal Ndn allele observed in heterozygous Ndn+m/−p mice also occurs in PWS patients. Using a specific human NDN RNA probe and an anti-Necdin antibody, we performed an ISH and IHC on hypothalamic sections obtained from brains from two adult PWS patients (one with a deletion and one with a maternal disomy) and one PWS infant (9 months old with a deletion); age and sex matched control individuals were included as positive controls (Table S1). In all patients, we found NDN transcripts and protein in the paraventricular and supra optic nuclei (Figure 6). We confirmed NDN mRNA expression in five more adult PWS patients (25–64 years of age) and one PWS child (6 months old). Expression of NDN also occurred in the cortex of PWS patients (data not shown). The results contradict the widely accepted assumption that in PWS patients the maternal allele is totally silenced in the brain. These findings are in full agreement with the results obtained in our heterozygous Ndn+m/−p mice.
Fig. 6. NDN expression in PWS patients. 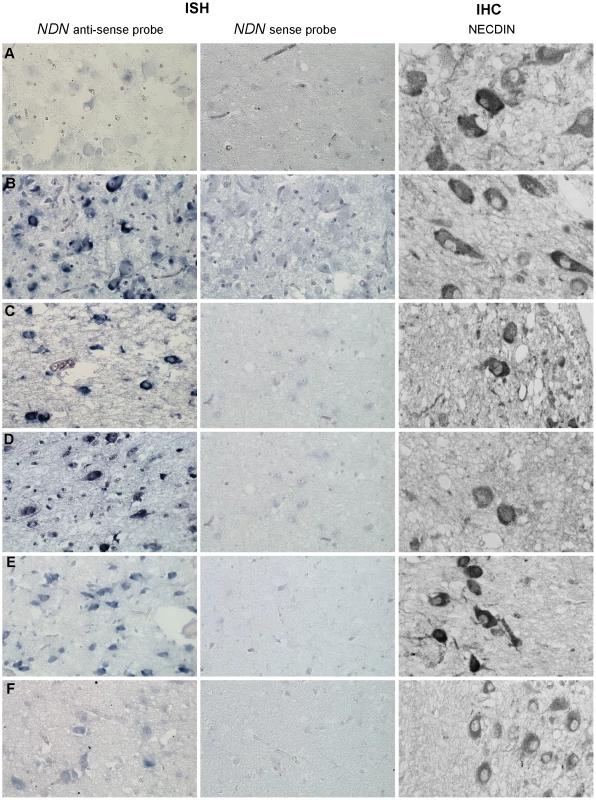
Detection of NDN transcripts revealed by ISH, and using a NDN anti-sense probe, on PVN brain sections from control individuals (A,C,E) and PWS patients (B,D,F). A NDN-sense probe was used as a negative control. IHC on SON brain sections, using a NECDIN specific antibody, was performed on the same control and PWS patients. The expression was studied in the 94-118 adult control male (A) and the 95104 adult PWS patient with a maternal uniparental disomy (B), in the 88-017 adult control male (C) and the 00-028 PWS adult patient with a deletion (D), in the 97-153 control infant (E) and the 99-079 PWS infant with a deletion (F). Scale bar: 20 µm. Discussion
In this article we report a stochastic expression of the maternally inherited allele of the Ndn gene in mice where the paternal gene has been inactivated. We showed an extremely low and very variable number of transcripts but nevertheless confirmed that these transcripts are translated into Necdin protein. Furthermore a comparison between Ndn−/− and Ndn+m/−p pups showed that the lethality, due to respiratory distress, is decreased two-fold in Ndn+m/−p compared to Ndn−/−. In agreement with this decreased lethality, surviving Ndn+m/−p adult mice present two-fold fewer apneas and more 5HT-expressing neurons, in a manner that is positively correlated with maternal Ndn expression. This confirms the functional importance of the extremely weak expression of the Ndn maternal allele. Finally NDN transcripts and protein were also detected in brain tissue from human PWS patients.
Furthermore, our results strongly suggest that expression of the maternal allele of Ndn only occurs in the absence of expression of the paternal Ndn allele. In addition, they are consistent with a model where, without an imprinting regulation, competition between two Ndn promoters results in a monoallelic expression. In this model, imprinting mechanisms create an allelic exclusion that dictates that the maternal allele is inactivated.
Loss of imprinting in mouse models for PWS and in PWS patients
Prior to this study, it was widely accepted that only the paternal alleles of PWS candidate genes are expressed, the maternal alleles being totally silenced. However, in brains of mice, with a deletion of the imprinting center, an incomplete silencing of paternally inherited PWS genes as well as a low level of expression of maternal alleles of PWS genes, was reported but not investigated [27]. LOI was also observed in lymphoblasts of two PWS patients with a deletion and two atypical PWS patients with a maternal disomy [28], [29], but these studies were not extended to include expression profiles in the brain. LOI has been described in other contexts, particularly in some cancers [30]. Our study addresses for the first time the robustness of silencing of the maternal alleles of PWS candidate genes in brain. Our results show that in both mice and humans, in the absence of the paternally inherited Ndn gene, the maternal Ndn allele is expressed in the brain at very low level but sufficiently to allow Necdin protein production. A similar mechanism might be hypothesized for any of the PWS genes in PWS patients. For example, the imprinted Magel2/MAGEL2 PWS gene, showed a similar loss of imprinting in Magel2+m/−p heterozygous mice and PWS human brains (F.M. and D.S. unpublished data). Significantly, the high variability of expression of maternal alleles of these genes might explain the large degree of heterogeneity in the severity of PWS symptoms.
The relevance of Ndn maternal allele expression
The two-fold reduction of post-natal mortality in Ndn+m/−p mice compared to Ndn−/− mice, suggests that even the low level of maternal Necdin protein is sufficient to rescue 50% of the mice, in comparison with the Ndn−/− mice. Nevertheless 50% of Ndn−/− individuals survive suggesting that another compensatory system is activated when the level of Ndn expression is null or very low in Ndn+m/−p mice.
A surprising degree of inter-individual variability was observed in the number of Ndn transcripts amongst Ndn+m/−p mice. The degree of maternal Ndn expression is correlated with the severity of the phenotype, in that the number of apneas is significantly increased in Ndn−/− mice compared to Ndn+m/−p mice. Previously, we published that those apneas might be correlated with an alteration of the 5HT system [22]. Here we showed that the number of 5HT-expressing neurons is reduced by 28% in Ndn−/− compared to WT mice while Ndn+m/−p mice are divided in two distinct populations with 30% and 10% fewer 5HT-neurons respectively. The lowest reduction (10%) of 5HT-expressing neurons is observed in those Ndn+m/−p individuals co-expressing Necdin in 5HT neurons. These data support a link between the expression of the Ndn maternal allele and the degree of survival, the severity of apneas and the number of 5HT neurons in the B1/B2 raphe nuclei.
Variability of Ndn maternal expression
Ndn maternal expression presents a high inter-individual variability (1 to 3 orders of magnitude), even among Ndn+m/−p individuals from the same litter, irrespective of the age and brain structure analyzed. Intra-individual variability of Ndn expression was also detected in the brain structures. This expression is limited to some, but not all, of the brain regions that normally express Ndn with no evidence of ectopic expression. In those brain regions the number of neurons expressing Ndn is clearly less than in wild-type animals and variability of Ndn expression amongst the Ndn+m/−p offspring was linked to both maternal genotype and gender, being additive factors. A Ndn+m/−p mouse with a +/+ or Ndn+m/−p mother (a mouse who has inherited a wild-type Ndn allele from her grandmother) is predisposed to the highest level of expression and to a greater inter-individual variability, a phenomenon referred to as transgenerational epigenetic inheritance [31]. In contrast, paternal genotype has no impact. Furthermore maternal Ndn allele expression was two-fold higher and more variable in female mice compared to male mice. This may reflect the increased genetic variability in females: some genes escaping X inactivation, such as Jarid1c, which codes for a histone demethylase [32], showing higher expression in females. This could explain our observations concerning maternal allele Ndn expression. Alternatively or additionally, female-specific hormones could be involved.
An interesting observation resulting from transcriptome profiling is the very high variability between individuals in steady state levels of a range of mRNAs, often reaching an order of magnitude [33]. This might explain why the penetrance of a given genotype is often incomplete [23], [31], [33]. This type of epigenetic phenomenon might also be involved in the variable expression of the maternal Ndn gene and consequently might lead to survival of some Ndn+m/−p mice. Nevertheless, even in Ndn−/ − mice the penetrance of the phenotype (postnatal lethality and apneas) is not complete, suggesting that another mechanism involving a “compensatory pathway” takes place. This compensatory pathway might result from an increase of a gene expression linked to the lack of Ndn expression or might also result from the stochastic variability in gene expression described above [33] that occurs independent of the state of Ndn expression.
Mechanism underlying the maternal expression
The lack of detection of expression of the C57Bl/6J maternal allele in WT mice (with a paternal M. Spretus allele) suggests that expression of the Ndn maternal allele is associated with the absence of an active paternal Ndn promoter, as confirmed by our study of the Ndn+m/−p Ndntm2Stw embryos that did not express the maternal allele of Ndn. Similarly, in a third Ndn-KO mouse model, in which the Ndn-promoter also drives β-gal expression [18], no maternal Ndn transcripts were detected by RT-qPCR [34] in Ndn+m/−p mice. Previously, Chamberlain et al. observed a low level of Ndn maternal expression only in the absence of an active paternal PWS-imprinting center, and suggested a trans effect where the paternal PWS-IC acts on the maternal allele [27]. Furthermore, our data suggest that, even in the absence of imprinted regulation of Ndn, as is the case for the Ndn-eGFP BAC transgene, it appears that there is a transcriptional regulation predisposing to a monoallelic expression of Ndn. This result might be explained by promoter competition for transcriptional activators, or a mechanism involving physical contact in trans between promoters [27], [35]–[37].
Expression levels of Ndn mRNA and protein
Given the extremely low level of Ndn transcripts in +m/−p mice as estimated by RT-qPCR it is surprising that the protein was detectable by immunohistochemistry and by Western blot. Importantly, the absence of antibody staining on samples from −/ − mice ruled out the possibility of cross-reactivity with proteins sharing epitopes with Necdin. Until relatively recently, it has been assumed that transcript abundance is the main, although not the only, determinant of protein abundance. Experiments aimed at addressing this question have lead to an emerging body of evidence changing this view and, in every organism that has been examined to date at a global level, steady-state transcript abundance only partially predicted protein abundance [38]. This lack of correlation suggests a strong regulatory role for all processes downstream of transcription. Furthermore, it has been shown that in many situations, transcription, translation and degradation are often extensively coupled and regulate each other through feedback loops. This coupling might enhance responsiveness to the environment and might help reduce inter-cellular variability in gene expression, which is by nature a stochastic event [39].
Collectively, these results suggest that very low level expression of PWS maternally silenced genes might be sufficient to alleviate specific PWS symptoms [23]. Importantly, we show that the quantity of Ndn transcripts is not, at least in neurons, a good indicator of its protein level and hence its functional importance [38].
An understanding of the context in which the Ndn maternal allele might be transcribed is an important step towards the development of a pharmacological therapy to trigger and/or increase the expression of this maternal allele in PWS patients. Furthermore, our results provide a further indication of the high non-genetic heterogeneity between genetically identical individuals that might, in this case, underlie LOI and contribute to variability in the phenotype [40].
Materials and Methods
Breeding of mice
Mice were handled and cared in accordance with the Guide for the Care and Use of Laboratory Animals (N.R.C., 1996) and the European Communities Council Directive of September 22th 2010 (2010/63/EU, 74). Experimental protocols were approved by the institutional Ethical Committee guidelines for animal research with the accreditation no. B13-055-19 from the French Ministry of Agriculture.
Ndn deficient mice were maintained on the C57BL/6J background and the paternal mutation was transmitted by crossing Ndn−m/+p males with C57BL/6J WT females (from Janvier Company). In parallel, since the Ndn-KO allele was created using a 129/SvPas ES cell line, we maintained the mutation via a maternal transmission on the 129/SvPas genetic background using Charles River male mice. All Ndn mice were genotyped by PCR as previously described [21]. Genotype of Ndn−/− mice was confirmed by a secondary intra-deletional PCR whose primers were: 5′-GATCCGAAGGCGCAGACATG-3′ and 5′-CTGCCCATGACCTCTTTCAC-3′ generating a 420 bp fragment indicating the presence of Ndn WT allele.
The Ndn ++ over-expressing mouse line is the Magel2 KO (+m/−p and −/−) mouse line created previously in our team. Consequently, it is an over-expression of the endogenous Ndn gene rather than being a transgenic mouse. Magel2 being imprinted, closed to Ndn and belonging to the MAGE family gene, as Ndn. We observed this overexpression at the transcript and protein level and we estimated, by western blot quantification, the level of overexpression (a factor of 1.7 fold). They were also maintained onto C57BL/6J background.
Importantly, all the mouse lines used in this study, excepted the Ndntm2Stw+m/−p mouse line, have been created in our laboratory and maintained on pure genetic background. The Ndntm2Stw+m/−p mouse have also been bred onto C57Bl/6 for over 30 generations in Wevrick's laboratory.”
Western blotting
P1 brain and E12 whole embryos were rapidly dissected and crushed in lysis buffer as previously [41]. For each sample, proteins (30 mg) were separated on a 12% SDS-PAGE and transferred onto nitrocellulose membranes (Protran Whatman, Dutscher). Membranes were incubated overnight with a rabbit polyclonal antibody against Necdin (Upstate; 1∶1000) or with a rabbit polyclonal antibody against GFP (Sigma, G1544) and subsequently with an anti-rabbit horseradish peroxydase antibody (GE Healthcare, Buckinghamshire,UK; 1∶3000). In both experiments, membranes were reprobed using a mouse anti-α-Tubulin antibody (Sigma, T6074). For Necdin immunolabeling was visualized by enhanced chemiluminescence. For GFP immunolabeling was visualized by Gbox (Syngen). Quantification was performed using ImageJ.
Plethysmography
We performed plethysmography in weight-matched littermate mice that were 6 weeks old, unrestrained and unanesthetized. Spontaneous breathing activities were recorded in normoxic conditions using whole-body plethysmograph (EMKA Technologies, Paris, France). After a 30 min period of stabilization in the apparatus, respiratory parameters were calculated breath-by-breath during a 30 min period of measurement. The mean of each parameter was automatically calculated from this 30 min period of measurement using EMKA technologies Datanalyst software. Apneas have been defined here as an absence of a respiratory signal during at least three respiratory cycles in resting conditions.
Reverse transcription and real-time quantitative PCR
Classical RT-PCR was performed as previously described [17].
For RT-qPCR, mice were sacrified at E12.5, P1 or as adults. Whole embryos, whole P1 brains, P1 pons and P1 or adult hypothalamus tissues were rapidly collected and frozen in liquid nitrogen prior to RNA isolation using standard conditions. Subsequently, total RNA samples were incubated with DNase (TURBO DNA-free; Ambion). Messenger RNAs from 1 µg of total RNAs were reverse-transcribed in a total volume of 20 µL using the M-MLV, reverse transcriptase RNAse H minus, point mutant (Promega) and oligod(T)15 in the presence of a synthetic external, heterologous and noncompetitive poly(A) Standard RNA (SmRNA) used to calibrate the reverse transcription [42] (patent WO2004.092414). At the end of the RT, total volume was brought up to 100 µL and real-time PCR was performed using the Rotorgene System (Qiagen) to determine the number of SmRNA and Ndn cDNA molecules in 5 µL of the RT product. The specific forward and reverse primers were designed using “Universal Probe Library” software (Roche Diagnostics) in the region deleted in the Ndn KO-allele. The sequences of the primer pair used were: Necdin-Forward 5′-AACAACCGTATGCCCATGA-3′, Necdin-Reverse 5′-CTTCACATAGATGAGGCTCAGGAT-3′ (60 bp). The primer sequences and the quantification conditions of calibrator cDNAs (Standard cDNAs) are protected by the patent WO2004.092414. To discriminate specific from nonspecific cDNA products, a melting curve was obtained at the end of each run, by a slow temperature elevation up to 98°C (0.1°C.s-1). Before RT, absence of traces of genomic DNA in the purified total RNA samples was ruled out by real-time PCR of the non-deleted Ndn sequence. Quantification cycles were converted into the number of cDNA copies using the quantification curve specific for each primer pair that had been previously established from serial dilutions of purified PCR products. The equation of the calibration curve for NDN cDNA was performed in four replicates for each dilution ranging from 10 to 1×109 copies : Ct = −3.3417 Log [cDNA]i+39.049, r2 = 0.9988. No amplification was obtained in Ndn−/ − individuals only. In the other mice, the lowest and highest copy numbers quantified were 92 and 4,638,062, respectively. For each sample, the number of Ndn cDNA copies was normalized according to relative efficiency of RT determined by the standard cDNA quantification. Finally, gene expression was expressed as the cDNA copy number quantified in 5 µL aliquot of RT product.
Immunohistochemistry
Specificity of Necdin protein detection was controlled on tissues from Ndn−/− animals. Fixed brains was dissected, cryopreserved and sectioned (14 µm) using a cryostat (Leica CM3050S). Embryos and post-natal mice (P1) were sacrificed and treated as previously [21]. Antibodies used were: rabbit polyclonal anti-Necdin (07-565; Millipore, Bedford, MA, USA; 1∶500), mouse monoclonal anti-GFP (Interchim, NB600-597; 1∶500), goat polyclonal anti-5HT (Immunostar, 20079; 1∶300).
Sections were washed twice in PBS and incubated with Hoechst (33258, Sigma; 1∶2000) and corresponding fluorochrome-conjugated secondary antibodies, goat anti-rabbit Alexa Fluor 488 or Alexa Fluor 555 (Molecular Probes, Invitrogen; 1/500), goat anti-mouse Alexa Fluor 488 (Molecular Probes, Invitrogen; 1/500), donkey anti-goat Cy3 (Chemicon, AP180C; 1/1000) diluted in the blocking buffer without BSA. Sections were examined on a Zeiss Axioplan 2 microscope with an Apotome module. Quantification of labeled cells was performed using ImageJ.
For quantification of immunofluorescence, images were acquired using a confocal microscope (SP5-X, Leica), z stacks of 70 µm were performed for each image, and analyzed using ImageJ.
In Situ hybridization
All Ndn in situ hybridization experiments for the study of Ndn gene expression were performed on serial slices of those used for immunohistochemistry and performed as previously [43]. Specificity of Ndn mRNA detection was controlled on tissues from Ndn−/− animals and with the sense control riboprobes. A peroxidase-conjugated anti-digoxigenin-POD (1∶1250) antibody (Roche) was used to detect the Ndn hybridized riboprobe, visualized using a tyramide signal amplification (TSA-plus Biotin Kit, Perkin Elmer).
Quantification of allele-specific expression
We could identify two transcribed Single Nucleotide Polymorphisms (tSNPs) in the 3′-UTR of Ndn to discriminate between the C57BL/6J and Mus spretus alleles.
To determine allele-specific transcription levels, we performed QUASEP and RT-qPCR with allele-specific TaqMan probes on cDNA of C57BL/6J×Mus spretus F1 brains from P10–P14 pups. All RNA samples were treated with DNaseI (Agilent) to minimize any risk of contamination with genomic DNA. Subsequently, 2 µg of high-quality total RNA were reverse transcribed into cDNA (SuperScript III First Strand Synthesis System, Invitrogen) and oligo(dT)-priming according to manufacturer's instructions.
The QUASEP assays were designed using the PyroMark Assay Design Software 2.0 (Qiagen). PCR was performed with the FastStart High Fidelity PCR System (Roche) according to manufacturer's recommendations using the cDNA of C57BL/6J×Mus spretus F1 brains from 6 P10–P14 mice. Pyrosequencing was done on a PSQ 96MA Pyrosequencing System (Qiagen) with a sequencing primer (Table S2) and PyroGold SQA reagents (Qiagen). Data were analyzed with the PSQ 96MA 2.1.1 software (Qiagen) as previously described [44].
For allele-specific RT-qPCR, we used the 5 bp indel in the 5′-UTR of Ndn to design TaqMan probes specific for C57BL/6J and Mus spretus, respectively [45], [46]. Quantitative PCR was performed on an ABI 7500 Fast Real time PCR System using the cDNA of C57BL/6J×Mus spretus F1 brains from 32 P10–P14 mice. Briefly, the 20 µl reaction contained 10 µl TaqMan Fast Universal PCR Master Mix (2×), 3.6 µl 5 µM combined forward (C57BL/6J: 5′-CTTCCTCTGCTGGTCTCCAC-3′, Mus spretus: 5′-CTTCCTCTGCTGGTCTCCAC-3′) and reverse (C57BL/6J: 5′-GGGTCGCTCAGGTCCTTACT-3′, Mus spretus: 5′ - GGGTCGCTCAGGTCCTTACT-3′) primers (0.9 µM); 2 µl of each 2 µM TaqMan probe (C57BL/6J: FAM-CTCCAAGCCGCATCGGTCCTGCTC-BHQ1, Mus spretus: ATTO550-CTCCAAGCCGCATCGCATCGGTCC-BHQ2; 0.2 µM) and 2.4 µl cDNA. The qPCR thermal profile consisted of 95°C for 10 min, followed by 48 cycles of 95°C for 30 s and 60°C for 30 s. Real-time PCR data were analyzed with ABI SDS 2.0.6 software.
Methylation study
Unfertilized oocytes and blastocysts were collected from C57BL/6 superovulated females and directly embedded in agarose beads for bisulphite treatment as previously described [47]. Sperm was recovered from the epididymis. Adult brain and kidney were dissected from interspecific M. spretus X M. musculus F1 mice or from Ndn+m/−p mice and DNA extracted according to standard techniques.
Bisulphite treatment of HindIII-digested adult brain, kidney and sperm genomic DNAs was carried out as described [48]. Oocyte and blastocyst DNAs were treated as described [47]. A semi-nested PCR was used to amplify regions A (CpG sites 1 to 20) and B (CpG sites 21 to 42) from bisulphite treated DNA samples. Primers used to amplify region A were: 5′-TGTGTTATATAGGAGATTAGG-3′ (outside forward; first and second rounds), 5′-AAACTACCATAAAACCTT-3′ (outside reverse) and 5′-CTATCCTACATCTCACAA-3′ (inside reverse). Primers used to amplify region B were: 5′-ATTGTGAGATGTAGGATAG-3′ (outside forward; first and second rounds), 5′-CCATAACCTCTTTCACCATA-3′ (outside reverse), and 5′-AAACTACCATAAAACCTTC-3′ (inside reverse). PCRs were performed in 50 µl reactions containing 1.25% DMSO, 25 pmole of each primer, 0.2 mM dNTPs, 2.5 mM MgCl2, 1× PCR Buffer and 2.5 U Q-BioTaq DNA Polymerase (Quantum Appligene, Germany). Second round PCRs were performed using 1 µl of the purified primary PCR products. PCR cycles were 5 min at 94°C followed by 10 cycles of 30 s at 94°C, 30 s at 58°C, 25 s at 72°C, and by 25 cycles of 30 s at 94°C, 30 s at 58°C, 25 s plus 5 s at each cycle at 72°C, and 7 min at 72°C. Purified PCRs products were cloned into the pGEM-T Easy TA Vector (Promega) and sequenced using standard methods. For oocytes and blastocysts, PCRs were performed on samples prepared from at least three different batches of oocytes and blastocysts. Identical clones derived from oocytes and blastocysts secondary PCRs were considered as derived from one single allele and represented only once.
Generation of transgenic mice with Ndn-eGFP modified BAC
The BAC603M20 (Research Genetics; referred as BAC109 [43]) contains a 104 kb NotI insert including the Ndn gene. BAC109 was modified by homologous recombination in E.coli as described [49] in order to replace the Ndn open reading frame (ORF) by the eGFP ORF.
Cesium chloride gradient purified BAC DNA was microinjected in the pronucleus of C57BL/6× CBA mouse zygotes. Founders containing the BAC transgene were identified by amplifying an eGFP fragment by PCR. Transgenic founders were maintained on C57BL6 genetic background. Transgene copy numbers were determined by Southern blot of BglII digested genomic DNA hybridized with PCR probes.
Human material and studies
Hypothalamic material from 3 PWS patients and from controls, matched for age, sex, postmortem delay and fixation time were obtained through The Netherlands Brain Bank (NBB, Director Dr. I. Huitinga). Clinicopathological details are given in Supplementary Table S1.
Sections throughout the hypothalamus were collected at 1200 µm intervals and mounted and pretreated as previously [50], with 2 µg/ml of proteinase K.
For the detection of NDN mRNA we hybridized the sections with a 2000 ng/ml DIG-labeled RNA probe, complementary to bp1258–1578 of the human NDN mRNA (NM_002487.2). Hybridization and stringency washes were performed as previously described [51] at 60°C. Anti-DIG-Alkaline phosphatase-fab fragments (Roche), diluted 1∶3000 in buffer 1 (100 mM Tris, 150 mM NaCl pH 7.5) were used to detect DIG labeled RNA hybrids [50]. Specificity of the hybridization signal was verified by comparison with sections processed with sense probe under identical conditions.
For IHC, 6 µm sections of hypothalamus tissue, containing SON, PVN, and INF were collected, mounted and microwaved in Citrate Buffer as previously [50]. Necdin was detected with rabbit IgG, anti-Necdin (07-565; Millipore, Bedford, MA, USA) diluted 1∶500 in Supermix (SUMI: 0.25% gelatin (Merck) (w/v), 0.5% Triton X-100 in TBS, pH 7.6) for 1 hour at RT, followed by an overnight incubation at 4°C. Detection of Necdin immunoreactivity was performed according to the ABC method described before [52]. Antibody specificity was confirmed by the absence of ICC staining in the human hypothalamus after omission of the first antibody from the staining protocol.
Statistical analyses
Nonparametric statistical tools (Sigmastat software) or exact statistical tools (StatXact software) were used depending on the size of the sample (n). All tests are two-tailed tests. In the results, values are indicated as following: Mean±SD or (Q2 (Q1, Q3), n, P value) where Q2 is the median, Q1 is the first quartile and Q3 is the third quartile. Mann Whitney t-test or Wilcoxon-Mann-Whitney test (WMW in the text) are used. The level of significance was set at a P-value less than 0.05.
Supporting Information
Zdroje
1. ReikW, WalterJ (2001) Genomic imprintig: parental influence on the genome. Genetics 2 : 21–32.
2. PrickettAR, OakeyRJ (2012) A survey of tissue-specific genomic imprinting in mammals. Mol Genet Genomics 287 : 621–630.
3. JirtleRL, SkinnerMK (2007) Environmental epigenomics and disease susceptibility. Nat Rev Genet 8 : 253–262.
4. MannMR, LeeSS, DohertyAS, VeronaRI, NolenLD, et al. (2004) Selective loss of imprinting in the placenta following preimplantation development in culture. Development 131 : 3727–3735.
5. LimAL, Ferguson-SmithAC (2010) Genomic imprinting effects in a compromised in utero environment: implications for a healthy pregnancy. Semin Cell Dev Biol 21 : 201–208.
6. JayP, RougeulleC, MassacrierA, MonclaA, MatteiMG, et al. (1997) The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet 17 : 357–361.
7. WatrinF, RoeckelN, LacroixL, MignonC, MatteiMG, et al. (1997) The mouse Necdin gene is expressed from the paternal allele only and lies in the 7C region of the mouse chromosome 7, a region of conserved synteny to the human Prader-Willi syndrome region. Eur J Hum Genet 5 : 324–332.
8. MacDonaldHR, WevrickR (1997) The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet 6 : 1873–1878.
9. GerardM, HernandezL, WevrickR, StewartCL (1999) Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet 23 : 199–202.
10. ChamberlainSJ, LalandeM (2010) Neurodevelopmental disorders involving genomic imprinting at human chromosome 15q11-q13. Neurobiol Dis 39 : 13–20.
11. CassidySB, SchwartzS, MillerJL, DriscollDJ (2012) Prader-Willi syndrome. Genet Med 14 : 10–26.
12. ButlerMG (2011) Prader-Willi Syndrome: Obesity due to Genomic Imprinting. Curr Genomics 12 : 204–215.
13. McAllisterCJ, WhittingtonJE, HollandAJ (2011) Development of the eating behaviour in Prader-Willi Syndrome: advances in our understanding. Int J Obes (Lond) 35 : 188–197.
14. DykensEM, LeeE, RoofE (2011) Prader-Willi syndrome and autism spectrum disorders: an evolving story. J Neurodev Disord 3 : 225–237.
15. FestenDA, de WeerdAW, van den BosscheRA, JoostenK, HoeveH, et al. (2006) Sleep-related breathing disorders in pre-pubertal children with Prader-Willi Syndrome and effects of growth hormone treatment. J Clin Endocrinol Metab 91 : 4911–5.
16. TauberM, DieneG, MolinasC, HebertM (2008) Review of 64 cases of death in children with Prader-Willi syndrome (PWS). Am J Med Genet A 146 : 881–887.
17. MuscatelliF, AbrousDN, MassacrierA, BoccaccioI, Le MoalM, et al. (2000) Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet 9 : 3101–3110.
18. TsaiTF, ArmstrongD, BeaudetAL (1999) Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome [letter]. Nat Genet 22 : 15–16.
19. KuwakoK, HosokawaA, NishimuraI, UetsukiT, YamadaM, et al. (2005) Disruption of the paternal necdin gene diminishes TrkA signaling for sensory neuron survival. J Neurosci 25 : 7090–7099.
20. RenJ, LeeS, PagliardiniS, GerardM, StewartCL, et al. (2003) Absence of Ndn, encoding the Prader-Willi syndrome-deleted gene necdin, results in congenital deficiency of central respiratory drive in neonatal mice. J Neurosci 23 : 1569–1573.
21. AndrieuD, MezianeH, MarlyF, AngelatsC, FernandezPA, et al. (2006) Sensory defects in Necdin deficient mice result from a loss of sensory neurons correlated within an increase of developmental programmed cell death. BMC Dev Biol 6 : 56.
22. ZanellaS, WatrinF, MebarekS, MarlyF, RousselM, et al. (2008) Necdin plays a role in the serotonergic modulation of the mouse respiratory network: implication for Prader-Willi syndrome. J Neurosci 28 : 1745–1755.
23. RajA, RifkinSA, AndersenE, van OudenaardenA (2010) Variability in gene expression underlies incomplete penetrance. Nature 463 : 913–918.
24. AndrieuD, WatrinF, NiinobeM, YoshikawaK, MuscatelliF, et al. (2003) Expression of the Prader-Willi gene Necdin during mouse nervous system development correlates with neuronal differentiation and p75NTR expression. Gene Expr Patterns 3 : 761–765.
25. SchneiderE, PliushchG, El HajjN, GaletzkaD, PuhlA, et al. (2010) Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res 38 : 3880–3890.
26. HanelML, WevrickR (2001) Establishment and maintenance of DNA methylation patterns in mouse Ndn: implications for maintenance of imprinting in target genes of the imprinting center. Mol Cell Biol 21 : 2384–2392.
27. ChamberlainSJ, JohnstoneKA, DuBoseAJ, SimonTA, BartolomeiMS, et al. (2004) Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice. Hum Mol Genet 13 : 2971–2977.
28. RoganPK, SeipJR, WhiteLM, WengerSL, SteeleMW, et al. (1998) Relaxation of imprinting in Prader-Willi syndrome [In Process Citation]. Hum Genet 103 : 694–701.
29. MuralidharB, MarneyA, ButlerMG (1999) Analysis of imprinted genes in subjects with Prader-Willi syndrome and chromosome 15 abnormalities. Genet Med 1 : 141–145.
30. JelinicP, ShawP (2007) Loss of imprinting and cancer. J Pathol 211 : 261–268.
31. ZucchiFC, YaoY, MetzGA (2012) The secret language of destiny: stress imprinting and transgenerational origins of disease. Front Genet 3 : 96.
32. XuJ, DengX, DistecheCM (2008) Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One 3: e2553.
33. TuranN, KatariS, CoutifarisC, SapienzaC (2010) Explaining inter-individual variability in phenotype: is epigenetics up to the challenge? Epigenetics 5 : 16–19.
34. WuN, LiZ, SuY (2012) The association between oxytocin receptor gene polymorphism (OXTR) and trait empathy. J Affect Disord 138 : 468–472.
35. XuN, TsaiCL, LeeJT (2006) Transient homologous chromosome pairing marks the onset of X inactivation. Science 311 : 1149–1152.
36. ThatcherKN, PeddadaS, YasuiDH, LasalleJM (2005) Homologous pairing of 15q11-13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum Mol Genet 14 : 785–797.
37. KruegerC, KingMR, KruegerF, BrancoMR, OsborneCS, et al. (2012) Pairing of homologous regions in the mouse genome is associated with transcription but not imprinting status. PLoS ONE 7: e38983.
38. VogelC, MarcotteEM (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13 : 227–232.
39. DahanO, GingoldH, PilpelY (2011) Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet 27 : 316–322.
40. PujadasE, FeinbergAP (2012) Regulated noise in the epigenetic landscape of development and disease. Cell 148 : 1123–1131.
41. AebischerJ, SturnyR, AndrieuD, RieussetA, SchallerF, et al. (2011) Necdin protects embryonic motoneurons from programmed cell death. PLoS One 6: e23764.
42. MoralesA, BonnetC, BourgoinN, TouvierT, NadamJ, et al. (2006) Unexpected expression of orexin-B in basal conditions and increased levels in the adult rat hippocampus during pilocarpine-induced epileptogenesis. Brain Res 1109 : 164–175.
43. WatrinF, Le MeurE, RoeckelN, RipocheMA, DandoloL, et al. (2005) The Prader-Willi syndrome murine imprinting center is not involved in the spatio-temporal transcriptional regulation of the Necdin gene. BMC Genet 6 : 1.
44. RufN, BahringS, GaletzkaD, PliushchG, LuftFC, et al. (2007) Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum Mol Genet 16 : 2591–2599.
45. HollandPM, AbramsonRD, WatsonR, GelfandDH (1991) Detection of specific polymerase chain reaction product by utilizing the 5′––3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A 88 : 7276–7280.
46. LivakKJ (1999) Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 14 : 143–149.
47. PrissetteM, El-MaarriO, ArnaudD, WalterJ, AvnerP (2001) Methylation profiles of DXPas34 during the onset of X-inactivation. Hum Mol Genet 10 : 31–38.
48. OlekA, OswaldJ, WalterJ (1996) A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res 24 : 5064–5066.
49. GongS, YangXW, LiC, HeintzN (2002) Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res 12 : 1992–1998.
50. ShanL, LiuCQ, BalesarR, HofmanMA, BaoAM, et al. (2012) Neuronal histamine production remains unaltered in Parkinson's disease despite the accumulation of Lewy bodies and Lewy neurites in the tuberomamillary nucleus. Neurobiol Aging 33 : 1343–1344.
51. PasterkampRJ, De WinterF, HoltmaatAJ, VerhaagenJ (1998) Evidence for a role of the chemorepellent semaphorin III and its receptor neuropilin-1 in the regeneration of primary olfactory axons. J Neurosci 18 : 9962–9976.
52. UnmehopaUA, van HeerikhuizeJJ, SpijkstraW, WoodsJW, HowardAD, et al. (2005) Increased melanin concentrating hormone receptor type I in the human hypothalamic infundibular nucleus in cachexia. J Clin Endocrinol Metab 90 : 2412–2419.
Štítky
Genetika Reprodukčná medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



