-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
Prolific sheep have proven to be a valuable model to identify genes and mutations implicated in female fertility. In the Lacaune sheep breed, large variation in litter size is genetically determined by the segregation of a fecundity major gene influencing ovulation rate, named FecL and its prolific allele FecLL. Our previous work localized FecL on sheep chromosome 11 within a locus of 1.1 Mb encompassing 20 genes. With the aim to identify the FecL gene, we developed a high throughput sequencing strategy of long-range PCR fragments spanning the locus of FecLL carrier and non-carrier ewes. Resulting informative markers defined a new 194.6 kb minimal interval. The reduced FecL locus contained only two genes, insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) and beta-1,4-N-acetyl-galactosaminyl transferase 2 (B4GALNT2), and we identified two SNP in complete linkage disequilibrium with FecLL. B4GALNT2 appeared as the best positional and expressional candidate for FecL, since it showed an ectopic expression in the ovarian follicles of FecLL/FecLL ewes at mRNA and protein levels. In FecLL carrier ewes only, B4GALNT2 transferase activity was localized in granulosa cells and specifically glycosylated proteins were detected in granulosa cell extracts and follicular fluids. The identification of these glycoproteins by mass spectrometry revealed at least 10 proteins, including inhibin alpha and betaA subunits, as potential targets of B4GALNT2 activity. Specific ovarian protein glycosylation by B4GALNT2 is proposed as a new mechanism of ovulation rate regulation in sheep, and could contribute to open new fields of investigation to understand female infertility pathogenesis.
Published in the journal: The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary. PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003809
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003809Summary
Prolific sheep have proven to be a valuable model to identify genes and mutations implicated in female fertility. In the Lacaune sheep breed, large variation in litter size is genetically determined by the segregation of a fecundity major gene influencing ovulation rate, named FecL and its prolific allele FecLL. Our previous work localized FecL on sheep chromosome 11 within a locus of 1.1 Mb encompassing 20 genes. With the aim to identify the FecL gene, we developed a high throughput sequencing strategy of long-range PCR fragments spanning the locus of FecLL carrier and non-carrier ewes. Resulting informative markers defined a new 194.6 kb minimal interval. The reduced FecL locus contained only two genes, insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) and beta-1,4-N-acetyl-galactosaminyl transferase 2 (B4GALNT2), and we identified two SNP in complete linkage disequilibrium with FecLL. B4GALNT2 appeared as the best positional and expressional candidate for FecL, since it showed an ectopic expression in the ovarian follicles of FecLL/FecLL ewes at mRNA and protein levels. In FecLL carrier ewes only, B4GALNT2 transferase activity was localized in granulosa cells and specifically glycosylated proteins were detected in granulosa cell extracts and follicular fluids. The identification of these glycoproteins by mass spectrometry revealed at least 10 proteins, including inhibin alpha and betaA subunits, as potential targets of B4GALNT2 activity. Specific ovarian protein glycosylation by B4GALNT2 is proposed as a new mechanism of ovulation rate regulation in sheep, and could contribute to open new fields of investigation to understand female infertility pathogenesis.
Introduction
Women, cattle, goats and ewes have generally one or two offspring, whereas other mammals, such as sows, rodents and dogs are prolific and produce more than three offspring. It relies on the number of ovulations at each estrus cycle i.e. the ovulation rate (OR), for which the underlying genetic mechanism was puzzling until the identification of fecundity genes in sheep, bone morphogenetic protein-15 (BMP15), growth and differentiation factor-9 (GDF9) and BMP receptor-1B (BMPR1B) [1]. Following the discovery of sheep fecundity genes, several research groups have focused on BMP15 and GDF9 and they have found numerous mutations associated with human ovarian pathologies such as premature ovarian failure or polycystic ovary syndrome [2]. Thus, prolific sheep are now considered as valuable models for identifying genes and mutations involved in mechanisms controlling the ovarian function, for agronomical purposes such as genetic selection of prolificacy, and for clinical purposes in the case of female infertility or subfertility.
In the meat strain of the Lacaune sheep breed, large variation in litter size has been observed and genetic studies explained this variation by the segregation of at least two major genes influencing OR and prolificacy, one being X-linked and named FecX, the second being autosomal and named FecL [3], [4]. FecX is known as BMP15 and in the Lacaune breed, the mutant allele (FecXL) associated with high prolificacy was identified as a pCys321Tyr substitution altering the BMP15 protein function [3]. Heterozygous FecXL mutation is associated with a twofold increase in OR, but homozygous FecXL/FecXL ewes are sterile, thus mimicking the phenotype observed for the other 5 mutations described in the ovine BMP15 gene [5]–[7].
The influence of the autosomal FecLL mutation on OR is additive with one copy increasing OR by about 1.5 and two copies by about 3.0 [4], [8]. We have recently established that the FecL locus influences both the ovarian activity and the endocrine profiles [9]. Indeed, increased OR in homozygous FecLL/FecLL (thereafter named L/L) ewes is associated with an increased number of gonadotropin-dependent follicles with a diameter greater than 3 mm, an increase in plasma estradiol concentrations, and an increase in the frequency of Luteinizing Hormone (LH) pulsatility during the follicular phase, leading to a precocious LH surge. In contrast, plasma concentrations of Follicle Stimulating Hormone (FSH) were not different compared to wild-type ewes. Based on ovarian phenotype and endocrine profiles, these findings suggest that the FecLL mutation affects ovarian function in a different way compared to other known hyperprolificacy-associated mutations, all affecting genes of the bone morphogenetic protein signaling system, BMP15, GDF9 and BMPR1B [1], [10].
The FecLL mutation associated with increased OR has not yet been identified. In a previous work, a full genome scan localized the FecL locus on sheep chromosome 11 (OAR11). Fine mapping reduced the interval containing FecL to markers BM17132 and FAM117A, corresponding to a synteny block of 1.1 megabases on human chromosome 17 (HSA17), which encompasses 20 genes [8]. With the aim to identify the FecL gene and its hyperprolificacy-associated mutation, we combined different approaches based on genetic fine mapping (both classical development of genetic markers and high throughput Roche 454 sequencing strategy), gene expression analysis, histochemistry and protein identification by mass spectrometry. From our results, we propose B4GALNT2, encoding the glycosylation enzyme beta-1,4-N-acetyl-galactosaminyl transferase 2, as the FecL gene. Finally, the FecL sheep model of prolificacy-associated mutation leads to the discovery of a new pathway involved in the regulation of folliculogenesis and ovulation rate.
Results
Fine mapping
The interval of localization previously published corresponded to a synteny block of 1.1 megabases on HSA17 [8]. Genotyping additional markers further reduced it across the whole experimental Lacaune pedigree (F1, BC and F1xBC representing 189 animals). This reduced interval was comprised between markers GNGT2-M2 (OAR11 : 36899194) and Ms162 (OAR11 : 37387389) encompassing 488 kb on the ovine chromosome 11 (OAR11, ovine genome version 3.1 released October 2012) and corresponded to a block of synteny of 479 kb on bovine chromosome 19 (BTA19, bovine genome version 4.6.1 released October 2011). This entire region was sequenced with the Roche 454 sequencing technology, using long-range PCR, in one heterozygous L/+ animal and two homozygous +/+ and L/L animals. Sixty-two polymorphisms were evidenced and an appropriate subset was genotyped on the recombinant animals allowing the reduction of the locus. This new interval of localization, comprised between two SNP markers on OAR11 g.36910171T>C (recombinant ewe n°990855) and g.37107627G>C (recombinant ewe n°60718), was estimated at 197 kb based on ovine genome OARv3.1 (Figure 1). This region encompasses 3 predicted protein-coding genes on the ovine genome, named B4GALNT2 (beta-1,4-N-acetyl-galactosaminyl transferase 2), EZR (ezrin) and IGF2BP1 (insulin-like growth factor 2 mRNA binding protein 1).
Fig. 1. Map of the FecL locus on ovine chromosome 11. 
The genes are indicated above the line, markers are indicated by points under the line. The FecL locus (197 kb on OARv3.1, or 194.6 kb, our own sequencing) is flanked by the two closest recombinant markers, g.36910171T>C and g.37107627G>C. Recombinants: white box, zero-recombinant zone; gray boxes, zone with one recombinant; black boxes, at least two recombinants with FecL. N: no Allele sharing between wild-type and carrier animals for the FecLL allele. Screening of the polymorphisms fully associated with the FecLL mutation
In order to identify all the polymorphisms contained within this 197 kb interval, we analyzed separately the sequences of the two homozygous L/L and +/+ animals. Sequence information coming from the Roche 454 sequencing technology was contained within 13 and 17 independent sequence contigs in L/L and +/+ animals, respectively. We have then completed these sequences by Sanger sequencing to link contigs with each other. By comparing the two sequences, we identified 49 polymorphisms (43 SNP, 4 microsatellites and 2 Insertion/Deletion, Table 1). None of the detected variants affected the coding sequence of the annotated genes. Polymorphisms were first tested on a set of one L/L, one L/+ and 2 to 6 +/+ animals for allele sharing. If the allele associated with the L-haplotype was not found on a wild chromosome, then the marker was tested on successive subsets of wild chromosomes from the Lacaune families and other sheep populations, as described in the materials and methods section. There were only two polymorphisms segregating as the FecLL mutation, i.e. fully associated with the hyperprolific phenotype: the SNP g.36938224T>A, localized in the intron 7 of B4GALNT2, and the SNP g.37034573A>G localized in the intergenic sequence between B4GALNT2 and EZR, 10.4 kb upstream of EZR (Figure 1).
Tab. 1. Polymorphisms within the minimal FecL locus and allele sharing. 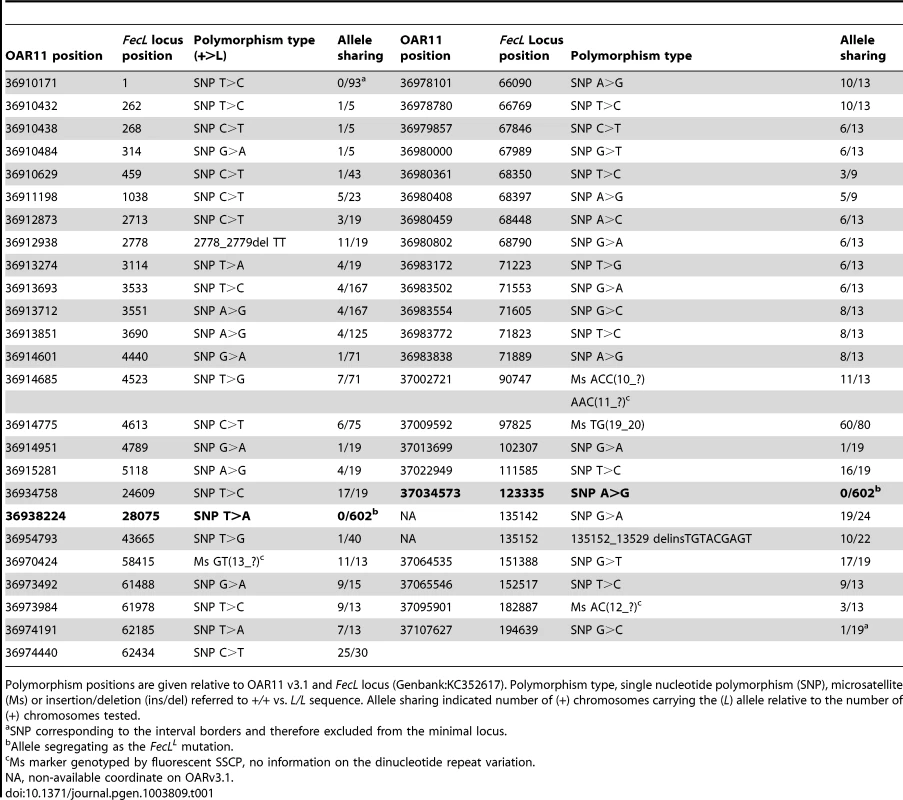
Polymorphism positions are given relative to OAR11 v3.1 and FecL locus (Genbank:KC352617). Polymorphism type, single nucleotide polymorphism (SNP), microsatellite (Ms) or insertion/deletion (ins/del) referred to +/+ vs. L/L sequence. Allele sharing indicated number of (+) chromosomes carrying the (L) allele relative to the number of (+) chromosomes tested. The sequencing of the locus indicated a real interval of 194.6 kb (GenBank:KC352617) and allowed to correct and complete the ovine reference genome sequence in this region. The locus effectively contained the full sequence of the B4GALNT2 gene based on the ovine B4GALNT2 mRNA (sequenced from Lacaune sheep, GenBank:KC175557), the full sequence of the IGF2BP1 gene based on the bovine IGF2BP1 mRNA (GenBank:NM_001192454) and a BLAST hit with the bovine EZR mRNA (GenBank:NM_174217) that we assumed to be a pseudogene. Indeed, the bovine EZR gene is located in a region of BTA9 that was not syntenic of the ovine FecL locus on OAR11 and the predicted sequence of this EZR annotation on OAR11 carried a premature STOP codon limiting the predicted protein to 223 amino-acids instead of 581. Consequently, only B4GALNT2 and IGF2BP1 were considered as positional candidate genes for FecL.
Gene expression analysis
The mRNA expression of the 2 positional candidate genes within the FecL locus was checked by real-time quantitative PCR in different tissues of the reproductive axis (hypothalamus, pituitary gland and ovarian granulosa and theca cells) isolated from +/+ and L/L ewes. In the hypothalamus and the pituitary gland, B4GALNT2 and IGF2BP1 mRNA amounts were similar between genotypes, but they were significantly higher in the ovarian cells of the L/L, compared to +/+ ewes (Figure 2). Interestingly, B4GALNT2 exhibited a 1000-fold higher expression level in the L/L granulosa and theca cells, whereas IGF2BP1 expression was enhanced 6-fold only in granulosa cells and was found unchanged in theca cells. To check if this over-expression might concern other genes of the locus, the expression of the flanking genes within the “one recombinant zone” (Figure 1), i.e. PHOSPHO1, ABI3 and GNGT2 on the one side and GIP, SNF8, UBE2Z, ATP5G1 and CALCOCO2 on the other side, was also studied. None of these genes showed an altered expression in the different L/L tissues of the reproductive axis. Given that GIP was not expressed in those tissues, its expression was checked in intestine and no difference was found between genotypes (supplemental figure S1). Moreover, as IGF2BP1 is a RNA binding protein controlling the steady-state level of the MYC oncogene mRNA [11] and the ß-actin (ACTB) gene translation [12] we checked for MYC mRNA expression and ACTB protein accumulation in Lacaune granulosa cells as a possible consequence of IGF2BP1 overexpression (supplemental figure S2). Real-time PCR analysis showed a large decreased expression of the MYC gene in large follicles compared to small ones (18-fold, p<0.001), but no alteration associated with elevated IGF2BP1 mRNA level in L/L granulosa cells. ACTB accumulation checked by western blotting was not affected by the L/L genotype either.
Fig. 2. Expression of genes within the minimal FecL locus. 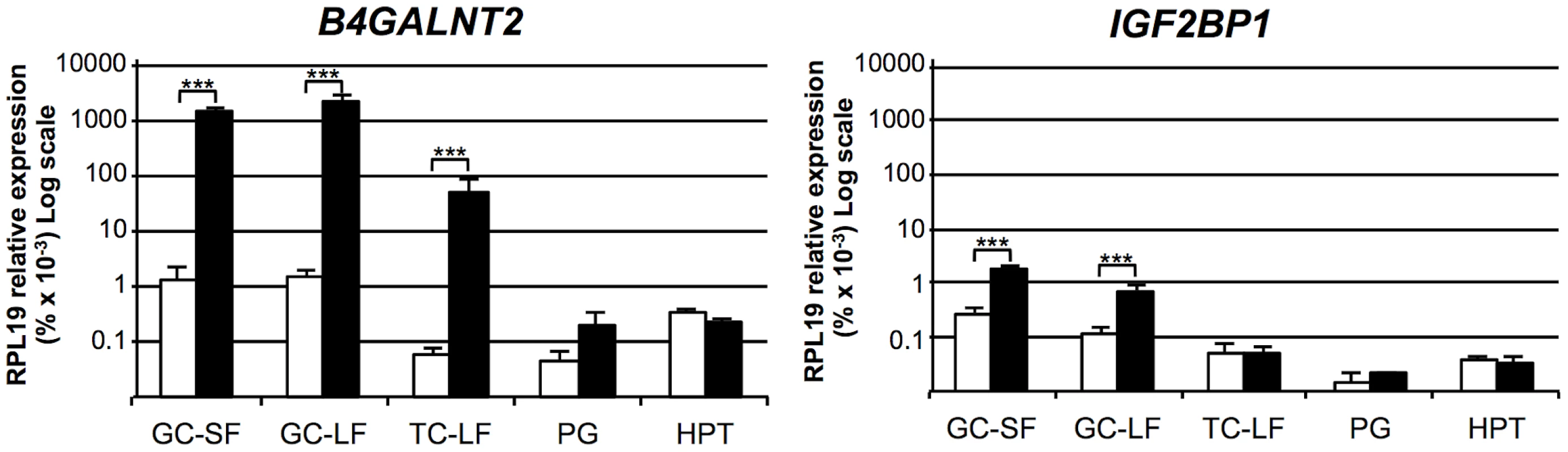
Total RNA from granulosa cells (GC) from small (SF, 1–3 mm), granulosa cells and theca cells (TC) from large (LF, ≥6 mm) follicles, pituitary gland (PG) and hypothalamus (HPT) were reverse-transcribed and submitted to real-time PCR analysis for quantification of B4GALNT2 and IGF2BP1 gene expression. Data are means ± SEM of relative expression to the reference gene RPL19 showed on a log scale. Asterisk indicates significant difference between means (n = 5) from non-carriers (+/+) and homozygous carriers of the FecLL mutation (L/L), **: p<0.01; ***: p<0.001. Thus, the localization of the B4GALNT2 gene within the minimal locus, the presence of the SNP g.36938224T>A on OAR11, localized in the intron 7 of this gene as the possible causal mutation, and the presence of a very high overexpression of the gene in the ovarian cells of L/L ewes led us to further investigate B4GALNT2 ectopic expression and activity in the ovary of the FecLL carrier animals.
Localization of B4GALNT2 protein and its activity in the FecLL ovary
B4GALNT2 is a β1,4-N-acetylgalactosaminyltransferase which was previously shown to be involved in the synthesis of the Sd(a) antigen, a carbohydrate expressed on erythrocytes, colonic mucosa and other tissues. B4GALNT2 transfers a beta-1,4-linked GalNAc to the galactose residue of an alpha-2,3-sialylated chain found on both N - and O-linked glycans [13]. Immunohistochemistry experiments using an antibody raised against B4GALNT2 showed that the enzyme was detectable in the granulosa cells of L/L ovarian follicles only (Figure 3). The use of the Dolichos Biflorus Agglutinin (DBA) lectin and the KM694 antibody raised against the Sd(a) antigen, both specific of the glycosylation activity of B4GALNT2, clearly localized the targets of the enzyme only in the granulosa cells and the antral follicular fluid of L/L ovaries (Figure 4). To confirm the glycosylation activity of B4GALNT2, +/+ ovine granulosa cells were transiently transfected by a B4GALNT2 expressing construct. Specific staining with DBA was observed in B4GALNT2-transfected +/+ granulosa cells (Figure 5), indicating that overexpression of the B4GALNT2 gene is directly related to positive DBA staining.
Fig. 3. Immunostaining for B4GALNT2 in Lacaune sheep ovary. 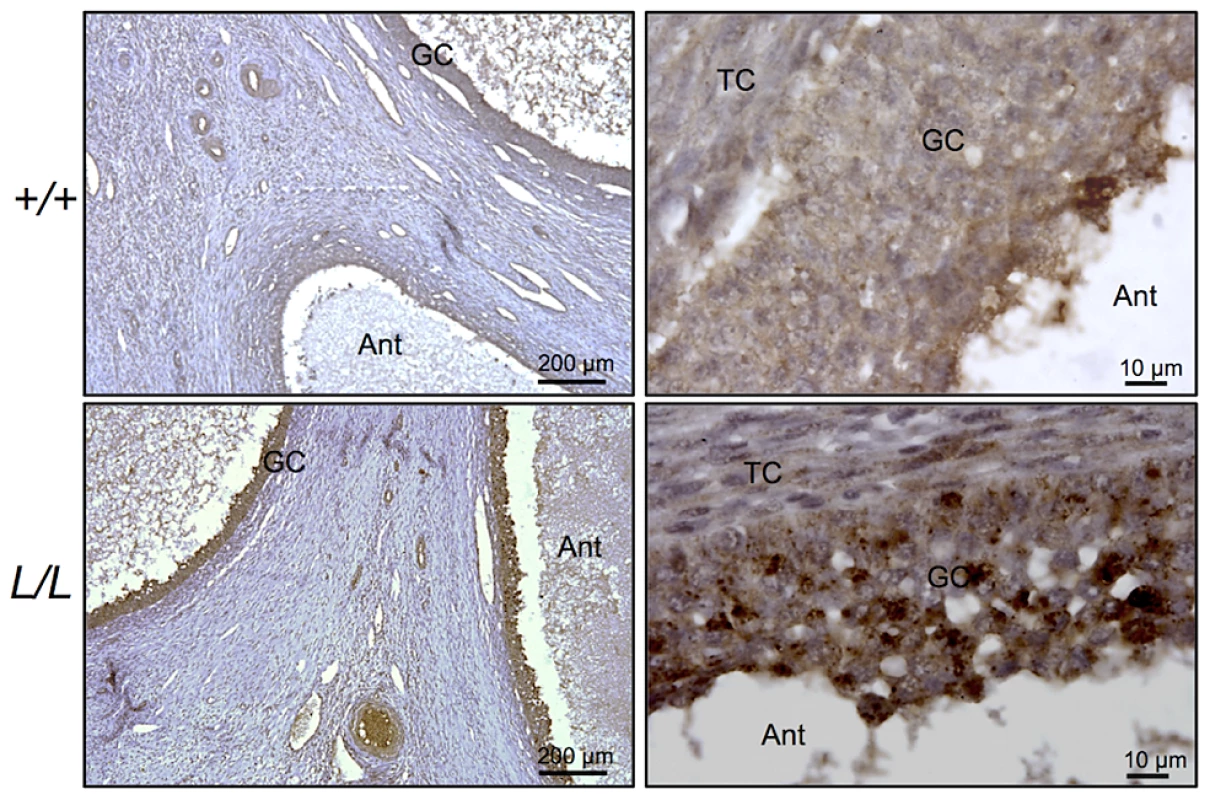
Photomicrographs of ovarian sections from +/+ and L/L ewes stained with anti-B4GALNT2 rabbit polyclonal antibody (1/50 dilution). Sections were counterstained with hematoxylin. A black segment indicates the microscopy magnification scale. GC, granulosa cell layer; TC, theca cell layer; Ant, antral cavity. Fig. 4. B4GALNT2 transferase activity revealed by DBA lectin and KM694 antibody staining in Lacaune sheep ovary. 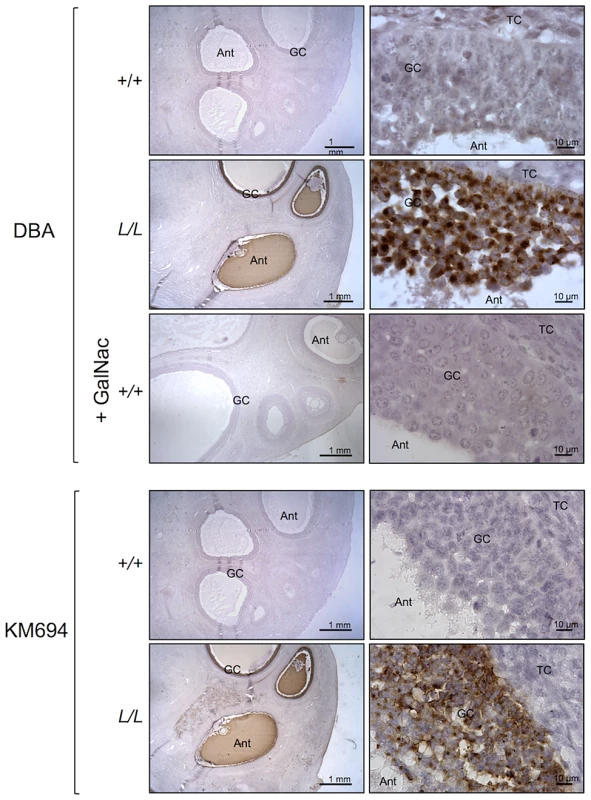
Photomicrographs of ovarian sections from +/+ and L/L ewes and stained either with biotinylated-DBA lectin (500 ng/ml) or KM694 mouse monoclonal antibody (1/1000 dilution). A GalNac treatment (200 µM) was used to compete for DBA staining as specificity control. Sections were counterstained with hematoxylin. A black segment indicates the microscopy magnification scale. GC, granulosa cell layer; TC, theca cell layer; Ant, antral cavity. Fig. 5. B4GALNT2 transferase activity revealed by DBA lectin after in vitro overexpression of B4GALNT2 in ovine granulosa cells. 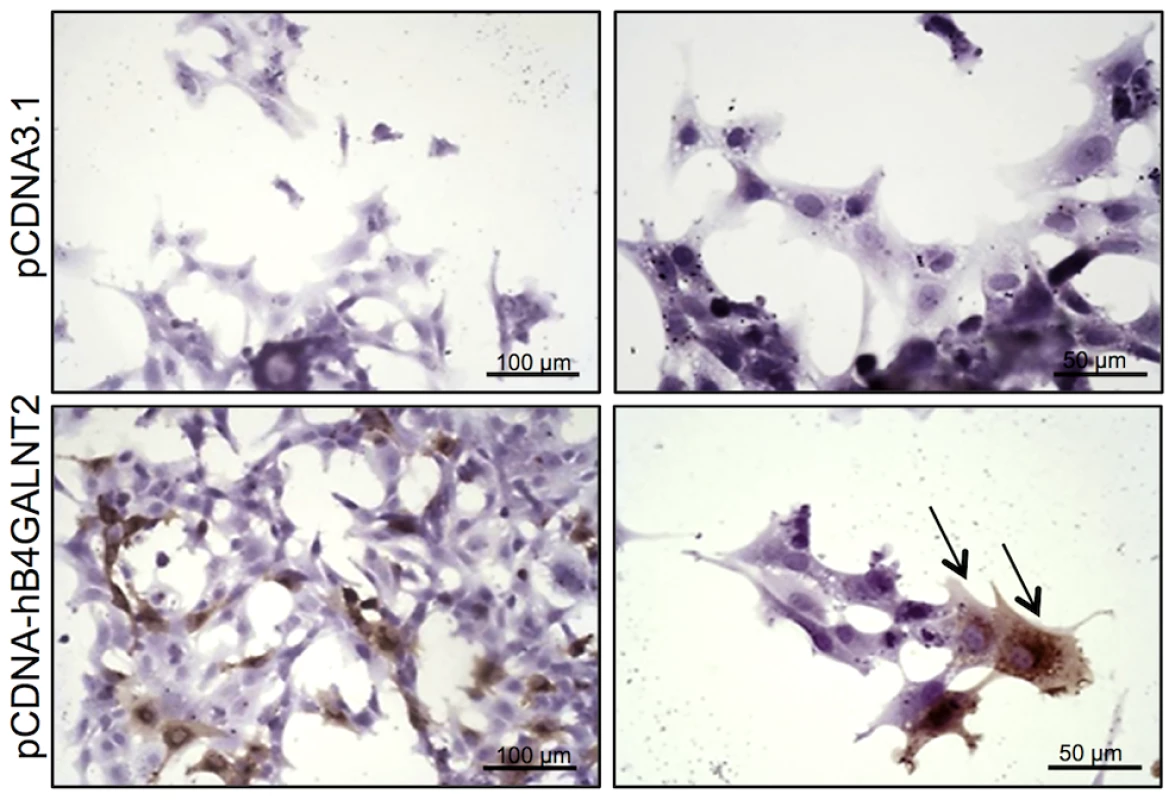
Primary ovine granulosa cells from +/+ small antral follicles were transiently transfected with either the pCDNA-hB4GALNT2 expressing the human form of B4GALNT2 or the empty pCDNA3.1 vector. Twenty-four hours after transfection, cells were stained with biotinylated-DBA lectin (500 ng/ml). Arrows indicated DBA positive staining only in B4GALNT2 transfected cells. Cells were counterstained with hematoxylin. A black bar indicates the microscopy magnification scale. The presence of a B4GALNT2 activity in the granulosa cells of the FecLL carrier ewes was confirmed by the results of DBA lectin and KM694 antibody precipitation and subsequent western-blot experiments using L/L and +/+ granulosa cell extracts and follicular fluids (Figure 6). Indeed, DBA or KM694 staining revealed very different glycoprotein profiles between L/L and +/+ ewes. In both granulosa cells and follicular fluids, at least 7 glycoprotein forms of various molecular weights (ranging from 40 to over 250 kDa) were retained by DBA lectin precipitation in the L/L ewes and were absent in non-carrier ewes. The KM694 immunoprecipitation of the proteins contained in the follicular fluids of the FecLL carrier ewes evidenced a subset of these glycoproteins, with a high molecular weight.
Fig. 6. Western immunoblotting analysis of B4GALNT2 transferase activity in Lacaune sheep granulosa cells and follicular fluids. 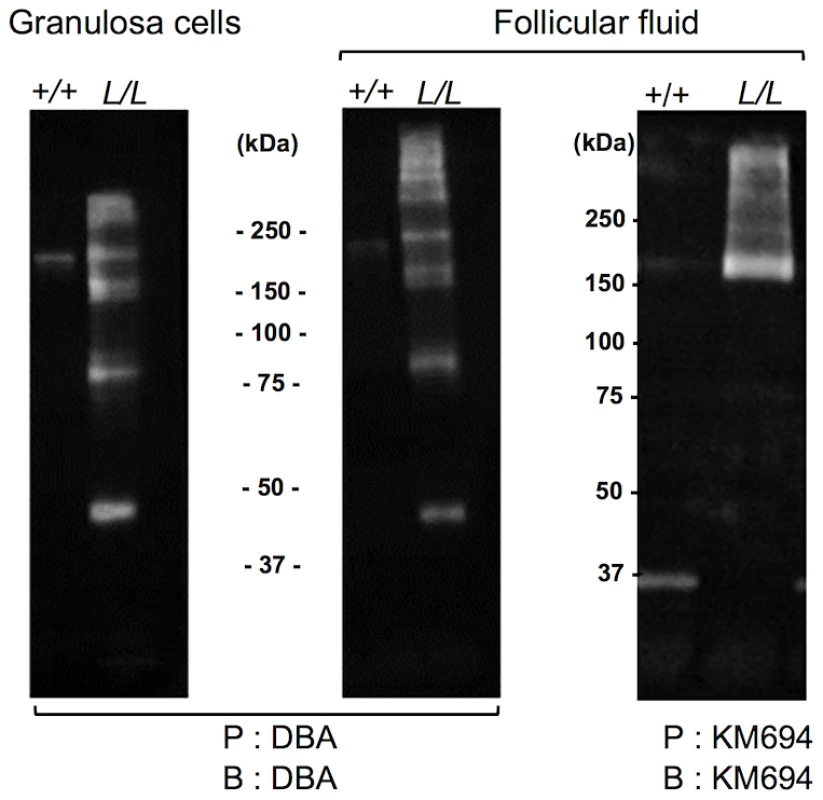
Granulosa cell protein extracts (50 µg) and follicular fluids (200 µg) from +/+ and L/L large antral follicles were precipitated (P) by agarose-DBA lectin or sepharose-protein A-KM694 monoclonal antibody. The resulting purified glycoproteins were separated on SDS-PAGE, transferred on nitrocellulose membrane and revealed after blotting (B) using biotinylated-DBA lectin or KM694 monoclonal antibody. Mass spectrometry identification of the glycosylated targets of B4GALNT2
We aimed at identifying the glycoproteins which are the targets of B4GALNT2 in ovary after their purification from L/L follicular fluids using DBA lectin affinity, followed by high-resolution mass spectrometry. Peptides and proteins purified from L/L and +/+ follicular fluids were identified with at least four independent peptides and a high probability threshold (>95%). This allowed identifying, on two repetitions of the experiment, 10 glycoproteins only present in FecLL follicular fluids, and then suspected to be the main substrates of B4GALNT2 activity (Table 2). Identified proteins were of a large molecular weight range (ranging from 39 to 613 kDa) and most of them were extracellular matrix or membrane-associated proteins. Interestingly, the inhibin α and βA subunits, leading to the production of Activin A and Inhibin A, and the chondroitin sulfate proteoglycan Versican are well known to be directly involved in female reproductive function [14]–[16]. These proteins represent promising physiological candidates to understand the mechanism by which the FecLL mutation affecting B4GALNT2 expression increases the ovulation rate in Lacaune sheep population.
Tab. 2. DBA-purified proteins present in L/L follicular fluid identified by mass spectrometry. 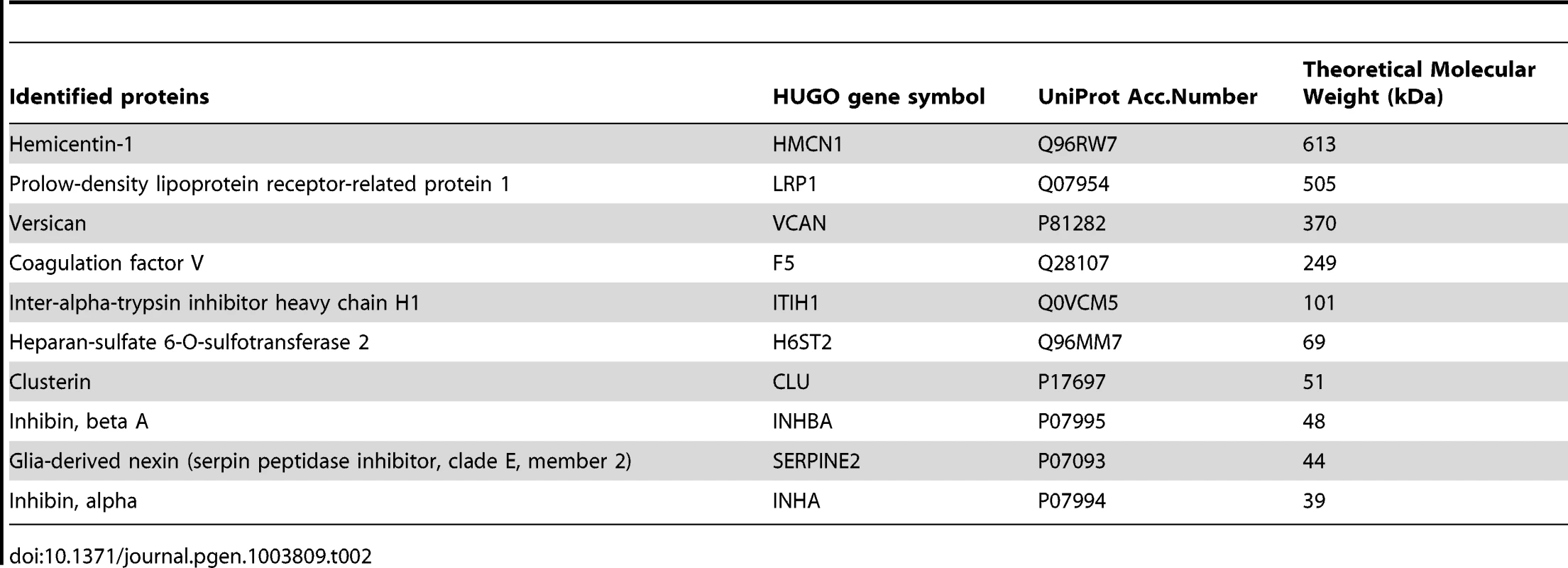
Discussion
Genetic characterization of the FecL locus
The fine mapping strategy combining marker development through high-throughput sequencing and genotyping of selected recombinant animals allowed the delimitation of the FecL locus on OAR11 between SNP markers g.36910171T>C and g.37107627G>C. With the double aim to densify the locus with more informative polymorphisms and to directly identify the causal polymorphism, we developed a systematic high-throughput sequencing of the locus with the Roche 454 technology on finely chosen animals. The targeting of the locus was done by overlapping long-range PCR fragments (around 10 kb), a method successfully used to detect genomic variations in BRCA1/BRCA2 locus in human [17]. We experienced some bias in detecting polymorphisms, especially SNP, in the reads depth of the L/+ animal due non-independent reads [18] and PCR-dependent allele disequilibrium. Nevertheless the use of the two independent sequences L/L and +/+ allowed the elimination of this latter bias. The polymorphisms evidenced were further studied for allele sharing. The different appropriate subsets of markers that were genotyped on recombinant ewes led to the reduction of the minimal interval to 194.6 kb. Within this interval of localization, 2 SNPs have been found fully associated with the FecLL mutation, namely SNP g.36938224T>A and SNP g.37034573A>G. The SNP g.36938224T>A was localized in the intron 7 of B4GALNT2, in a sequence portion that was only conserved in the bovine orthologous gene, and not in other species. The SNP g.37034573A>G was localized in the intergenic sequence between B4GALNT2 and IGFBP1 in a non-conserved region enriched with LINES and SINES repetitive elements.
The finding of non-coding SNPs as causal mutations contrasts with other known mutations affecting ovulation rate through alteration of the coding sequence of BMP15, GDF9 and BMPR1B genes impairing the protein function [10]. To go further in the fine mapping study, different strategies can be proposed. The first one is to find additional recombinant animals within the genetic families. A second one is to find recombinant animals at the population level, i.e. animals carriers of a shorter L-haplotype and finely phenotyped. However the probability to find such animal recombining within an interval of 195 kb is very low. If only one of these two SNP is the causal mutation, one way to prove it is to find an animal carrying a recombination between the two markers. However, the 2 SNPs are 96 kb apart and the probability to find such animal is nearly null. A third possibility is to increase the number of wild chromosomes (i.e. wild haplotypes) tested for allele sharing. Animals coming from 9 different breeds have been genotyped (representing 180 wild haplotypes) but did not allow the elimination of one or the other putative causal mutation.
Expressional candidates of the FecL gene and effect of the SNPs
Annotation available on the last release of the sheep genome assembly (OARv3.1) indicated that the minimal 194.6 kb interval, that we entirely sequenced, contained only 3 potential protein encoding genes, B4GALNT2, EZR and IGF2BP1. The EZR annotation is very recent (October 2012) and this gene was not considered as an expressional candidate for FecL at the beginning of the work. Moreover, if the localizations of B4GALNT2 and IGF2BP1 on OAR11 were coherent with their syntenic location on BTA19 and HSA17, the presence of the EZR gene annotation is intriguing. Indeed, in bovine (UMD3.1) and human (GRCh37) genomes, this gene is positioned on BTA9 and HSA6, respectively (www.Ensembl.org). Moreover, Blastn analysis (www.livestockgenomics.csiro.au) of the bovine EZR cDNA (NM_174217) indicated sequences producing significant alignments on Ovis aries breed Texel contig_127582, OAR11 and OAR8. Only matched sequences on OAR8 presented an exon/intron structure in a syntenic region of BTA9 and HSA6, and then should correspond to the ovine EZR gene. The high identity match we found between bovine EZR cDNA and both the ovine reference genome on OAR11 and our own FecL locus sequence (98% coverage, 95% identity) evidenced the presence of an EZR spliced pseudogene with a premature stop codon impairing the translation of an EZR protein. For those reasons, only IGF2BP1 and B4GALNT2 were considered as positional candidate genes for FecL in this study.
Due to the absence of polymorphism in the coding sequence of IGF2BP1 and B4GALNT2 genes, we searched for an expressional candidate to discriminate between the two genes. Real-time PCR analysis showed that both IGF2BP1 and B4GALNT2 expression was significantly affected by FecLL, specifically in ovarian cells indicating a tissue-specific regulation by the FecLL mutation. Interestingly, no differential expression was observed for genes outside of the minimal locus, reinforcing the genetic fine mapping result. For follicles collected at the same size class, the differential expression observed between +/+ and L/L follicles could not be attributed to the existence of a difference in follicle maturity as already observed for the hyperprolificacy-associated Booroola mutation [19]. Indeed, IGF2BP1 and B4GALNT2 expression in granulosa cells was not different between small and large antral follicles that were at different maturity stages as attested by marker expression, such as LHR, INHA, CYP19A1 and CYP11A1 as previously shown [9] or MYC (present study).
SNP g.36938224T>A, localized in the intron 7 of B4GALNT2, and SNP g. 37034573A>G in the inter-genic region, were the only two polymorphisms segregating as the FecLL mutation. They were then thought to be directly the cause of the huge overexpression of B4GALNT2 and in a lesser extent of IGF2BP1 in the ovaries of FecLL carrier ewes, through a molecular mechanism that remains to be determined. In order to explain this differential expression we searched in silico for transcriptional factors able to bind these SNP locations. However, we failed to find matches with consensus sequences binding known transcriptional factors at each site. Experimentally, electromobility shift assays with ovine granulosa cell nuclear extracts failed also to discriminate between SNPs and between prolific and wild type alleles. As a second in silico approach, we also searched matches in Patrocles, the database of polymorphic microRNA-target interactions (www.patrocles.org). Polymorphism in miRNA target sites might be important effectors of phenotypic variation as attested by the hypermuscularity phenotype in Texel sheep [20]. Interestingly, the B4GALNT2 intronic SNP g.36938224T>A was found to create a motif GTGTGAGA in FecLL which is a recognition site for MIR342 that is conserved among bovine and human. However, we cannot reconcile this finding with the current knowledge on gene regulation by microRNAs, indicating that those small noncoding RNAs usually targeted matured mRNA in the 3′-untranslated regions within cytoplasmic protein complexes to repress protein synthesis [21]. Another hypothesis is that the intronic location of the SNP g.36938224T>A may be associated with alternative splicing of the B4GALNT2 mRNA, but analysis by RT-PCR amplification between exon 6 and terminal exon 11 failed to evidence such alternative splicing between L/L and +/+ mRNA. In human gastrointestinal cancer cells the expression of B4GALNT2 is dependent on the promoter methylation status [22]. As in other species, a large CpG island is present in the vicinity of the ovine B4GALNT2 promoter. By bisulfite sequencing or restriction site analysis by methyl sensitive enzymes of granulosa cell derived DNA from +/+ and L/L animals, we failed to detect any differential methylation status. Thus, it remains to be determined how the g.36938224T>A and/or g.37034573A>G SNPs are able to distantly regulate B4GALNT2 and IGF2BP1 expression at the mRNA level specifically in ovarian cells, maybe through yet unknown tissue-specific DNA-protein interactions.
Role of the IGF2BP1 gene in the ovarian function
The IGF2BP1 gene was part of the minimal FecL locus and its expression was increased under the influence of the FecLL prolific allele (being g.36938224A and/or g.37034573G) in granulosa cells. This expression in ovine ovary is consistent with the IGF2BP1 expression already observed in mouse and human ovaries [23]. Elevated IGF2BP1 expression was also associated with ovarian carcinoma and proliferation deregulation through a c-myc (MYC) dependent mechanism [24]. Indeed, IGF2BP1 possesses RNA binding motifs and can stabilize the MYC mRNA [11] and enhance the translation of several other genes such as IGF2 and ß-actin (ACTB) by binding to their mRNA [25]. Interestingly, all these known target genes of IGF2BP1 action play an important role in follicle function. IGF2 is present in follicular fluid [26], expressed by granulosa [27] and theca cells [28] and participates in the maturation of ovarian follicles [29]. Actin is associated with granulosa cell shape changes along antral folliculogenesis [30] that may consequently affect steroid synthesis and proliferation [31]. MYC participates in the control of granulosa cell proliferation under gonadotropin and insulin stimulation [32], [33]. However, checked by MYC mRNA and ACTB protein accumulation, it seemed that the 6-fold overexpression of IGF2BP1 had no biological consequences in the granulosa cells of L/L ovaries.
Role of the B4GALNT2 gene in the increased ovulation rate of the FecLL ewes
The FecLL prolific allele was clearly associated with an ectopic overexpression (at least 1000-fold compared to wild-type) of B4GALNT2 mRNA in ovarian granulosa and theca cells from antral follicles. This ectopic expression seemed to occur specifically in the ovary, since expression checked by real-time qPCR in pituitary, hypothalamus (Figure 2), intestine (Figure S1) and adrenals (data not shown) showed no difference between genotypes, indicating a tissue-specific mechanism for B4GALNT2 expression regulation by the FecLL mutation. Such ectopic expression of B4GALNT2 has never been demonstrated in the ovary before. However, ectopic expression of B4galnt2 is the molecular basis for the action of the Mvwf locus, a major modifier of plasma von Willebrand factor (VWF) level in RIIIS/J mice [34]. Indeed a switch of B4galnt2 gene expression from intestine epithelial cells to vascular endothelial cells, resulting in aberrant VWF glysosylation, explained the phenotypic characteristics of the RIIIS/J mice similar to human type 1 von Willebrand disease. The region responsible for the Mvwf locus regulatory switch lies within a 30-kb genomic interval upstream to the B4galnt2 gene [35]. In the present study, one of the potential causal SNP (g.37034573A>G) lies 42 kb upstream to B4GALNT2 in a region which is not conserved between sheep and mouse.
The mRNA huge overexpression of B4GALNT2 observed in the L/L ovaries was accompanied by increased protein expression level as shown by specific immunohistochemistry, mainly in granulosa cells. As previously stated, B4GALNT2 is involved in the synthesis of the Sd(a) antigen on various protein targets [13] with the transfer of a terminal GalNAc, specifically recognized by DBA lectin [34] and the KM694 antibody [36], [37]. Using DBA lectin staining, we evidenced the GalNac transferase activity exclusively in the granulosa cells of the FecLL ovaries. The glycosylated targets of B4GALNT2 were mainly secreted in the follicular fluid as shown by histochemistry and western-blotting experiments. However, the pattern of target proteins revealed by DBA lectin and KM694 antibody was different. This discrepancy could be explained by the strict recognition specificity of the Sd(a) antigen (GalNac transfer to terminal α2,3-sialylated galactose residue in the ß1,4 linkage) by the KM694 antibody but a wider spectrum of terminal GalNac recognition by DBA lectin, indicating that B4GALNT2 could create other carbohydrate structures than the Sd(a) antigen. Moreover, we demonstrated that the in vitro overexpression of B4GALNT2 was responsible for the DBA lectin staining. Regarding these results, we assume that an atypical glycosylation of proteins within the granulosa cells of FecLL ewes, which does not occur in wild-type cells, is due to the overexpression of B4GALNT2. This could represent the initiating mechanism of the increased ovulation rate which characterizes the FecLL Lacaune sheep. In transgenic mice, manipulating glycosylation at the oocyte level led to slight increased fecundity or primary ovarian insufficiency depending on the glycosyltransferase being invalidated [38], [39]. Anyway, it proves the importance of glycosylation in the control of ovarian function.
In order to go further in the role of B4GALNT2 in the FecLL ovaries, we tried to identify the target proteins recognized by the DBA lectin using lectin affinity purification. Through comparative mass spectrometry analysis, we identified several atypically glycosylated proteins secreted in the follicular fluid of L/L ewes. Among the glycoproteins identified, were the inhibin subunits (INHA and INHBA) participating in the inhibin A and the activin A hormone formation [16]. Inhibin A and activin A are dimeric glycoproteins belonging to the transforming growth factor-beta (TGFβ) superfamily. They are produced by granulosa cells and can accumulate in high concentrations in follicular fluid. Activin A is considered to act mainly through auto/paracrine signaling in granulosa and theca cells [16], while inhibin A acts mainly through endocrine negative feedback regulation of pituitary FSH secretion. However, inhibin A has also been shown to exert a blocking action on the activin A and other TGFß member dependent regulation of steroidogenesis and proliferation within the ovary [16]. Immunization against inhibin can promote an increase in ovulation rate and prolificacy [40] through enhanced follicular development in sheep [41], goat [42] and water buffalo [43]. It has been suggested that inhibin antibodies may act primarily by an intraovarian paracrine action rather than by reducing the suppressive action of inhibin on pituitary FSH release [43], [44]. Inhibin A is a heterodimer of α - (INHA) and ßA - (INHBA) subunits, whereas activin A is a homodimer of ßA subunits. Interestingly, the differential subunit association (α-ßA or ßA-ßA) was dependent on glycosylation events implicating N-linked oligosaccharide [45]. The B4GALNT2 atypical glycosylation of INHA and INHBA could alter the subunits association, and then change the biological activity or the physiological ratio of activin A and inhibin A produced by the FecLL ovaries. Given that no difference in FSH plasmatic concentration during the follicular phase was observed between +/+ and L/L Lacaune ewes [9], one might suspect a more direct consequence on activin A signaling within the ovary. Experiments are ongoing to demonstrate the direct effect of B4GALNT2-dependent glycosylation on inhibin A and activin A biological activity at the auto/paracrine and endocrine levels.
Other good physiological candidates to explain increased OR in Lacaune sheep were the proteoglycans versican (VCAN) and inter-alpha trypsin inhibitor (heavy chain H1, ITIH1), implicated in follicular fluid formation and osmotic gradient, cumulus expansion, follicular remodeling and finally fertility [15], [46]. The coagulation factor F5, the serine protease inhibitor of the serpin family SERPINE2 and the heparan sulfate proteoglycan sulfotransferase (HS6ST2) would be other important regulators of coagulation, controlling antithrombin, plaminogen or fibrinogen activities present in follicular fluid [47]–[51]. Clusterin (CLU), a sulfated glycoprotein can also act on the ovarian function through its protective effect on granulosa cells against apoptosis during follicular atresia [52], and it is well known that a reduction in atresia is associated with increased ovulation rate [53]. Clusterin is a binding protein of the low-density lipoprotein receptor-related protein-2 (LRP2) [54], and may participate in cholesterol delivery to steroidogenic cells as LRP8 does in the bovine ovary [55]. Interestingly, in Lacaune sheep follicular fluids, we evidenced the presence of LRP1 that could have the same function on cholesterol uptake but can also act as a TGFß receptor (type 5) and regulate its signaling in ovarian cells [56]. Hemicentins are extracellular matrix proteins implicated in cell contacts, adhesion and migration [57]. Hemicentin-1 (HMCN1) carries a von Willebrand A domain that may explain its glycosylation by B4GALNT2. Hemicentins interact with fibulins that are estrogen regulated and overexpressed in ovarian cancer cells [58], [59], but no direct role of hemicentins has been described in the ovarian function. Finally, all the glycoproteins identified could have a role in the ovarian follicle function, may be through the control of intra-follicular activity of hormones or growth factors and/or their transfer outside of the follicle to the general blood circulation.
In conclusion, the present study reports strong evidence for the B4GALNT2 gene to be the FecL fecundity gene in Lacaune sheep. We propose that its overexpression in granulosa cells under the influence of only 1 or 2 non-coding regulatory SNP can induce an atypical glycosylation of follicular target proteins such as inhibin subunits and that is the starting point of the mechanism explaining increased ovulation rate and prolificacy in this breed. For the first time a fecundity gene in sheep does not belong to the TGFß/BMP signaling genes and it opens new fields of investigation regarding ovarian glycosylation and the pathogenesis of fertility disorders in women.
Materials and Methods
Animals
The presence of the Lacaune autosomal fecundity locus and its prolific allele FecLL was checked in our experimental Lacaune meat strain flock (n = 189) as described [8]. The three genotypes at the FecL locus are called +/+, L/+ and L/L representing FecL+/FecL+, FecLL/FecL+ and FecLL/FecLL, respectively. Briefly, as a unique haplotype is associated with the FecLL mutation, the presence of this particular mutant haplotype was established by the genotyping of four close markers, including the DLX3:c.*803A>G SNP that alone provides accurate classification of animals (99.5%) as carriers or non-carriers of the mutation. The absence of the FecXL mutation was checked in the studied Lacaune ewes by direct genotyping of the mutation [3]. The estrus cycles of all adult Lacaune ewes were synchronized with intravaginal sponges impregnated with fluorogestone acetate (FGA, 40 mg, Intervet, Angers, France) for 14 days. All procedures were approved by the “Direction Départementale des Services Vétérinaires de Haute-Garonne” (approval number C31-429-01) for the agricultural and scientific research agency INRA (French National Institute for Agricultural Research), and conducted in accordance with the Guide for the Care and Use of Agricultural Animals in Research and Teaching.
Roche 454 sequencing and data mining
Genomic DNA was extracted from blood samples following a salt-based DNA extraction [60]. Firstly, 45 long-range PCR fragments spanning 488 kb were amplified on genomic DNA from the heterozygous L/+ ewe n°982140, chosen as a dam of a double-recombinant ewe able to reduce the locus and because of its high level of homozygosity within the FecL locus. On a second time, 25 long-range PCR fragments spanning 250 kb were amplified from a homozygous L/L ewe and a homozygous +/+ ewe. The L/L ewe n°991012 was chosen at random, as it exists only one L-haplotype in the studied population. The +/+ ewe n°011182 was chosen for its homozygosity all along the locus because several wild-type haplotypes segregated in the population. Long-range PCR fragments were amplified on an ABI 9700 thermocycler (Applied Biosystems) using the Long PCR Enzyme Mix (Fermentas). Independently for each animal, the resulting fragments were purified and pooled all together at equal concentrations. These samples were then sequenced using the Roche 454 Life Sciences Genome Sequencer FLX (454 Life Science, Roche), following the manufacturer's instructions. Three shotgun libraries were prepared with 1 µg of pooled PCR product DNA using the Titanium General Library Preparation Kit. Nebulized, purified, and adaptor-linked DNA fragments were amplified using the GS FLX Titanium LV emPCR Kit, and sequencing on the FLX Genome Sequencer was performed using the GS FLX Sequencing Kit, Titanium Reagents XLR70. L/+, L/L and +/+ sequencing data from fastq files generated by 454 sequencer were cleaned using an in-house algorithm. A total of 356 873 reads with an average length of 366 bases were aligned on the sheep genome (OAR v2.0 - released March 2011 - draft sheep reference genome) with bwa software (bwasw algorithm [61], indicating a mean sequencing deepness of 103X. The resulting SAM format files were processed using samtools view, sort and merge functions [62]. Screening of polymorphisms was done “manually” using IGV software [63]. The position of all described polymorphisms within this manuscript is given according the last release of the sheep genome (OAR v3.1 - released October 2012 - sheep reference genome [64]).
Marker analysis, genotyping and Sanger sequencing
Prior to 454 sequencing, the search for single nucleotide polymorphisms (SNPs) was performed from ovine ESTs and BAC end sequence information, on a set of 2 L/L and 8 +/+ animals by single strand conformation polymorphism (SSCP) and silver staining as described [8]. Identified polymorphic fragments, after SSCP or 454 sequencing, were amplified by PCR on ABI 9700 thermocycler (Applied Biosystems). Microsatellites genotyping through fluorescent SSCP was performed on an ABI 3100 sequencer (Applied Biosystems). Depending on markers, SNP genotyping was done by SSCP and silver staining, or SSCP with fluorescent primers as described in Applied Biosystems Publication 116AP01-02, or by direct Sanger sequencing using the ABI Prism BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) on ABI 3100 sequencer (Applied Biosystems). The particular FecXL mutation was genotyped by SSCP as described [3].
After identification, the allelic sharing of a given marker was tested first on a subset of at least 2 to 6 +/+ Lacaune ewes, then the marker was tested on successive subsets of wild chromosome providing from: the Lacaune genetic family (n = 103), the dairy Lacaune GEBRO population (n = 173) and the Blanche du Massif-Central (BMC) population (n = 148) that shared common ancestors. Additional wild chromosomes from 9 different breeds genetically unlinked to the Lacaune breed were also used (n = 180, Limousine, Bizet, Rava, Suffolk, Causse du Lot, Charmoise, Préalpes du Sud, Berrichon du Cher, Noire du Velay, 10 animals of each breed).
Tissue collection
Estrus-synchronized ewes (+/+, n = 13 and L/L, n = 14) were slaughtered during the follicular phase 36 h after FGA sponge removal. Ovaries, pituitary gland and hypothalamus were collected from each animal. Pituitary gland and hypothalamus were immediately frozen in liquid nitrogen and stored at −80°C for further RNA extraction. Some ovaries were immediately placed in fixative Bouin's solution and embedded in paraffin for further histochemistry. Other ovaries were finely dissected to isolate individual antral follicles >1 mm in diameter. Once dissected, follicles were classified according to their size, small (1–3 mm) and large (≥6 mm) and with respect to genotype, independent of atresia. Granulosa cells and follicular fluids were recovered from small and large follicles as described previously [65], and theca layer was gently detached from the follicular wall of large follicles with forceps and washed in PBS to eliminate residual granulosa cells. Pools of each category of cells were established per animal and stored at −80°C for further RNA or protein extraction.
RNA extraction, Reverse transcription and quantitative PCR
Total RNA from ovarian cells and from pituitary gland was isolated using Nucleospin RNA II or Nucleospin RNA L kit, respectively, according to the manufacturer's protocol (Macherey-Nagel). Total RNA from hypothalamus was isolated by urea/LiCl precipitation and phenol extraction [66]. All RNA samples were DNAse-treated to avoid genomic DNA contamination and diluted at 0.5 µg/µl in RNAse-free water. RNA (1 µg) was reverse-transcribed using Superscript II reverse transcriptase (Invitrogen). Real-time quantitative PCR was run on a LightCycler 480 system (Roche Diagnostic) using Power SYBR Green PCR Master Mix (Applied Biosystems) in 384 wells-plate as described [8]. The specific primer sequences (Table 3) used for each gene were designed using the Beacon Designer 7 software (Premier Biosoft International). For each primer pair, efficiency curves were generated using serial dilutions of cDNA in abscissa and the corresponding cycle threshold (Ct) in ordinate. The slope of the log-linear phase reflects the amplification efficiency (E) derived from the formula E = e(−1/slope). Amplification efficiency obtained for each primer pair is indicated in Table 3. For quantification analysis, the Ct of target gene was compared with the internal reference gene RPL19 encoding an ubiquitous ribosomal protein, according to the ratio R = [EL19CtL19/EtargetCt target] expressed in percentage.
Tab. 3. Quantitative PCR primer sequences and their efficiency. 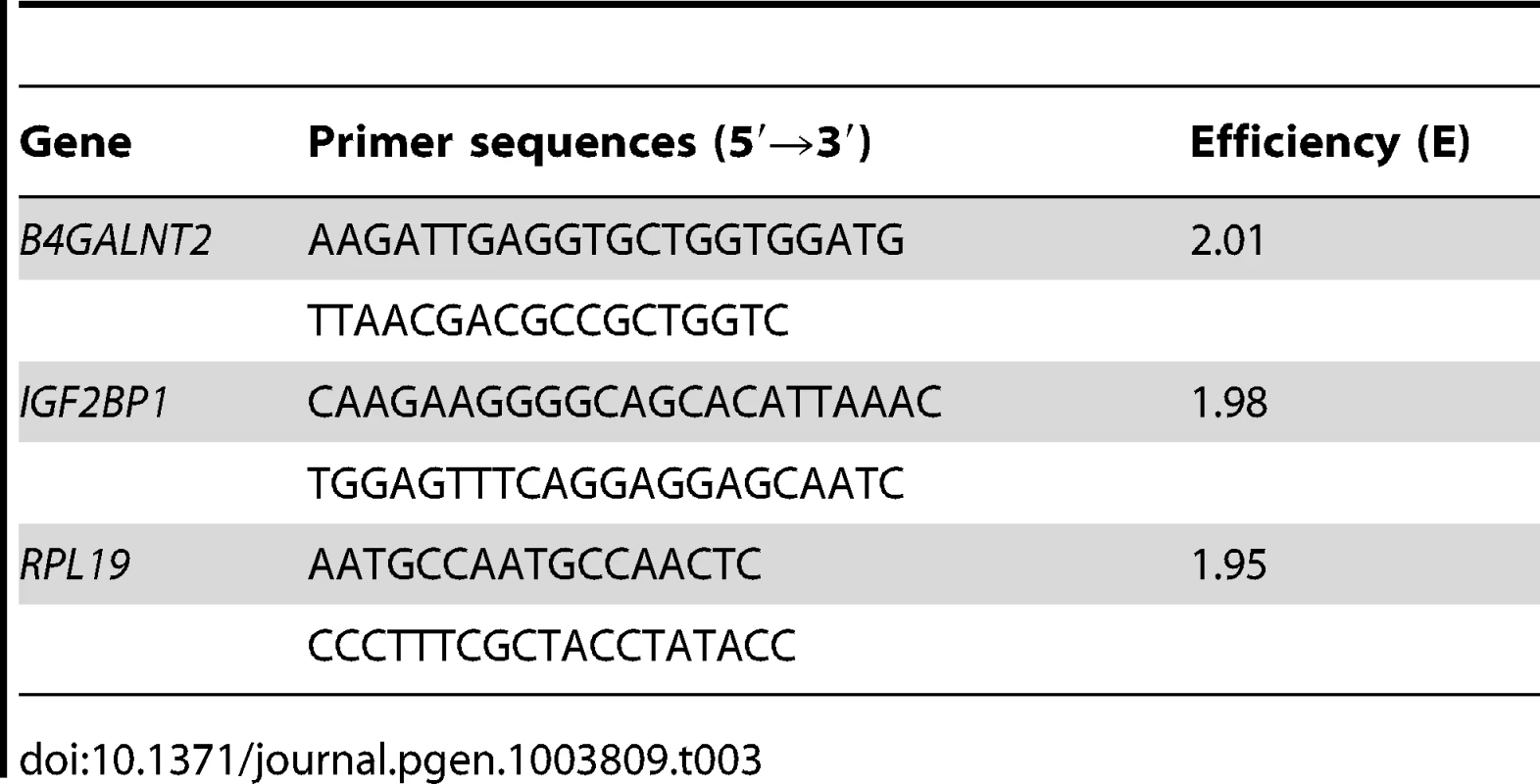
Histochemistry
Paraffin embedded ovaries were serially sectioned at a thickness of 7 µm. For immuno-histochemistry, deparaffinized sections were subjected to antigen unmasking solution (Vector Laboratories) boiled for 3 min. After two 5-min washes with 0.1% saponin in PBS, sections were incubated at 4°C for 30 min with PBS containing 0.1% saponin and 0.3% H2O2 to remove endogenous peroxidase activity. After three 5-min washes in 0.1% saponin in PBS, sections were incubated at 4°C overnight with goat polyclonal anti-B4GALNT2 antibody (sc-107334, Santa Cruz Biotechnology) diluted 1∶50 or mouse monoclonal KM694 (anti-Sda) antibody kindly provided by Shigeyuki Yamano (Kyowa Hakko Kirin Co., Japan) diluted 1∶1000 in PBS containing 0.1% saponin and 0.1% BSA. After three 5-min washes in PBS containing 0.1% saponin, sections were incubated for 2 hours at room temperature with the biotinylated secondary antibody (donkey anti-goat, Santa Cruz Biotechnology; donkey anti-mouse, Jackson ImmunoResearch Laboratories) diluted 1∶800 in PBS containing 0.1% saponin and 0.1% BSA, and washed thereafter. For lectin-histochemistry, deparaffinized sections were subjected to Carbo-Free blocking solution (Vector Laboratories) for 30 min. After a 5-min wash in PBS containing 0.05% Tween-20 (Sigma), sections were incubated with PBS containing 0.5 µg/mL of biotinylated DBA lectin (Dolichos Biflorus Agglutinin, Vector Laboratories) for 2 hours at room temperature, with or without 0.1 M of N-Acetyl-D-galactosamine (Sigma) to check for its specificity and then washed in PBS containing 0.05% Tween-20. For both approaches, stained sections were incubated with avidin-peroxidase conjugate from Vectastain Elite ABC kit (Vector Laboratories) and developed with 0.4 mg/ml DAB (3,3′-diaminobenzidine tetrahydrochloride dehydrate; Sigma) and 0.012% H2O2 in 50 mM Tris-HCl (pH 7.8) for 1 to 5 min at room temperature. Negative control sections involved omission of the primary antibody or the lectin from the procedure.
Granulosa cell transient transfection
Granulosa cells from small antral +/+ follicles were seeded at 100 000 viable cells/chamber on Lab-Tek 8 chambers slide system (Thermo-Scientific) and cultured for 48 h at 37°C with 5% CO2, in McCoy's 5a medium (Sigma) supplemented with 3% fetal ovine serum (FOS). Cells were transiently transfected with 1 µg of empty pCDNA3.1 or pCDNA-hB4GALNT2 (kindly provided by A. Harduin-Lepers) using jetPEI transfection reagent (Polyplus Transfection) for 18 h with a DNA/JetPEI ratio of 1/2 (w/w) as specified by the manufacturer, and thereafter, medium was changed with fresh McCoy's 5a medium supplemented with 3% FOS for an extra 30 h. At the end of the culture period, cells were fixed in 4% paraformaldehyde for 10 min at 4°C. Slides were washed in PBS 0.1% saponin, and then stained with biotinylated DBA lectin (0.5 µg/µL) as described above.
Lectin precipitation and western blotting
Granulosa cell whole cell extracts were obtained by resuspension in RIPA lysis buffer as previously described [67]. Granulosa cell lysates and follicular fluids from large antral follicles were centrifuged at 15000 g for 20 min at 4°C, and the protein concentration in the supernatant was determined by a colorimetric assay (BC Assay kit; Uptima Interchim). Protein samples corresponding to 50 µg of granulosa cell extract or 200 µg of follicular fluid were incubated with 30 µl of agarose-coupled DBA lectin (Vector Laboratories) in 300 µL of RIPA buffer for 2 hours at room temperature on a rotating platform. Alternatively, 250 µg of protein from follicular fluid were incubated with 25 µL of protein A-sepharose (Vector Laboratories) and KM694 antibody (1∶300). After a brief centrifugation (10000 g, 2 min), supernatant was discarded and the precipitated protein complex linked to agarose or sepharose beads was washed 3 times with 1 mL RIPA buffer. After wash, the complex was resuspended in Laemmli sample buffer (Bio-Rad), fractionated using SDS-PAGE in 10% polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were either blocked for 30 min at room temperature in Carbo-Free solution, and stained with 0.5 µg/mL biotinylated-DBA lectin (Vector laboratories) for 1 h at room temperature in PBS, or blocked with 5% non-fat dry milk, 0.1% Tween 20 (Sigma) in PBS, stained for 2 h with KM694 antibody (1∶1000), followed by incubation with peroxidase-conjugated anti-mouse IgG (1∶2500; Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Glycoproteins precipitated by DBA lectin were revealed by ABC reaction (Vectastain, Vector Laboratories), followed by ECL Plus detection (GE Healthcare). Glycoproteins precipitated by KM694 were revealed by direct ECL Plus detection. Luminescence was captured using an Image Master VDS-CL box imager (Amersham Pharmacia Biotech).
Mass spectrometry analysis
For mass spectrometry identification of glycoproteins purified by DBA lectin affinity, proteins (1 mg) from a mix of follicular fluid of large and small follicles (one mix for each genotype) were incubated with 60 µL agarose-coupled DBA lectin in RIPA buffer, as indicated above. After the last wash step, purified glycoproteins were eluted from the precipitated complex through incubation with 0.2 M of N-Acetyl-D-galactosamine (Sigma) for 90 min at room temperature under stirring. After centrifugation (10000 g, 2 min), the supernatant was collected and frozen for subsequent analysis. Eluted samples were submitted to SDS-PAGE in 10% polyacrylamide gels (12 min at 90 V) and stained by Coomassie blue. Proteins were in-gel digested with trypsin as previously described [68]. Each peptide mixture from +/+ and L/L genotype was analyzed in triplicate by nanoflow liquid chromatography tandem mass spectrometry (nanoLC-MS/MS). All experiments were performed on a LTQ Orbitrap Velos Mass Spectrometer (Thermo Fisher Scientific, Bremen, Germany) coupled to an Ultimate 3000 RSLC chromatographer (Dionex, Amsterdam, The Netherlands). Samples were loaded on an LCPackings trap column (Acclaim PepMap 100 C18, 100 µm i.d×2 cm long, 3 µm particles) and desalted for 10 min at 5 µL/min with 4% solvent B. Mobile phases consisted of (A) 0.1% formic acid, 97.9% water, 2% acetonitrile (v/v/v) and (B) 0.1% formic acid, 15.9% water, 84% acetonitrile (v/v/v). Separation was conducted using a LCPackings nano-column (Acclaim PepMap C18, 75 µm i.d×25 cm long, 3 µm particles) at 300 nl/min by applying gradient consisted of 4–45% B for 120 min. The mass spectrometer was operated in data dependent scan mode. Survey full scan MS spectra (from 300–1800 m/z) were acquired in the Orbitrap analyser with R = 30 000. The 20 most intense ions were fragmented in the high-pressure linear ion trap by collision-induced dissociation. Dynamic exclusion was active during 30 s with a repeat count of 1. Polydimethylcyclosiloxane (m/z, 445.1200025) ions were used for internal calibration.
MS/MS ion searches were performed using Mascot search engine v 2.2 (Matrix Science, London, UK) against the mammalian section of Uniprot_sprot database (2012_07). The search parameters included trypsin as a protease with allowed two missed cleavages, carbamidomethylcysteine, methionine oxidation and acetylation of N-term protein as variable modifications. The tolerance of the ions was set to 10 ppm for parent and 0.8 Da for fragment ion matches. Mascot results obtained from the target and decoy databases searches were subjected to Scaffold 3 software (v 3.4.3, Proteome Software, Portland, USA). The raw data may be downloaded from ProteomeCommons.org linked to the Tranche data repository using the “follicular_fluid_sheep” keywords. Peptide and protein identification was done by the Peptide and Protein Prophet algorithms [69], [70] with a probability >95.0%. Only proteins with greater than four identified peptides were considered. Differentially expressed proteins were determined using the spectral counting quantitative module of Scaffold 3 Q+ software (version 3.4, Proteome Software, Portland, USA). To eliminate quantitative ambiguity into protein groups, we ignored all the spectra matching any peptide that is shared between proteins. Thereby, quantification performed with normalized spectral counts was carried out on distinct proteins identified from two biological replicates. T-test was performed to differentiate the significantly changed proteins with a p-value <0.05 between the two genotypes.
Data analysis
All experimental data are presented as means ± SEM. The genotype effect on gene expression was analyzed using t-test for comparisons between two means. For all analyses, differences with P>0.05 were considered as not significant.
Supporting Information
Zdroje
1. FabreS, PierreA, MulsantP, BodinL, Di PasqualeE, et al. (2006) Regulation of ovulation rate in mammals: contribution of sheep genetic models. Reprod Biol Endocrinol 4 : 20.
2. PersaniL, RossettiR, CacciatoreC, FabreS (2011) Genetic defects of ovarian TGF-β-like factors and premature ovarian failure. Journal of endocrinological investigation 34 : 244–251.
3. BodinL, Di PasqualeE, FabreS, BontouxM, MongetP, et al. (2007) A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep. Endocrinology 148 : 393–400.
4. BodinL, SanCristobalM, LecerfF, MulsantP, BibeB, et al. (2002) Segregation of a major gene influencing ovulation in progeny of Lacaune meat sheep. Genet Sel Evol 34 : 447–464.
5. GallowaySM, McNattyKP, CambridgeLM, LaitinenMP, JuengelJL, et al. (2000) Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet 25 : 279–283.
6. HanrahanJP, GreganSM, MulsantP, MullenM, DavisGH, et al. (2004) Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol Reprod 70 : 900–909.
7. Martinez-RoyoA, JuradoJJ, SmuldersJP, MartíJI, AlabartJL, et al. (2008) A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep. Animal Genetics 39 : 294–297.
8. DrouilhetL, LecerfF, BodinL, FabreS, MulsantP (2009) Fine mapping of the FecL locus influencing prolificacy in Lacaune sheep. Anim Genet 40 : 804–812.
9. DrouilhetL, TaragnatC, FontaineJ, DuittozA, MulsantP, et al. (2010) Endocrine Characterization of the Reproductive Axis in Highly Prolific Lacaune Sheep Homozygous for the FecLL Mutation. Biol Reprod 82 : 815–824.
10. VinetA, DrouilhetL, BodinL, MulsantP, FabreS, et al. (2012) Genetic control of multiple births in low ovulating mammalian species. Mamm Genome 23 : 727–740.
11. WeidensdorferD, StöhrN, BaudeA, LedererM, KöhnM, et al. (2009) Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15 : 104–115.
12. HüttelmaierS, ZenklusenD, LedererM, DictenbergJ, LorenzM, et al. (2005) Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438 : 512–515.
13. MontielM, Krzewinski-RecchiM, DelannoyP, Harduin-LepersA (2003) Molecular cloning, gene organization and expression of the human UDP-GalNAc:Neu5Acα2-3Galβ-R β1,4-N-acetylgalactosaminyltransferase responsible for the biosynthesis of the blood group Sda/Cad antigen: evidence for an unusual extended cytoplasmic domain. Biochem J 373 : 369.
14. RodgersRJ, Irving-RodgersHF (2010) Formation of the ovarian follicular antrum and follicular fluid. Biol Reprod 82 : 1021–1029.
15. RodgersRJ, Irving-RodgersHF (2010) The roles of the ovarian extracellular matrix in fertility. Soc Reprod Fertil Suppl 67 : 217–230.
16. KnightPG, SatchellL, GlisterC (2012) Intra-ovarian roles of activins and inhibins. Mol Cell Endocrinol 359 : 53–65.
17. HernanI, BorrasE, de Sousa DiasM, GamundiMJ, ManeB, et al. (2012) Detection of genomic variations in BRCA1 and BRCA2 genes by long-range PCR and next-generation sequencing. J Mol Diagn 14 : 286–293.
18. MarietteJ, NoirotC, KloppC (2011) Assessment of replicate bias in 454 pyrosequencing and a multi-purpose read-filtering tool. BMC Research Notes 4 : 149.
19. McNattyKP, JuengelJL, WilsonT, GallowaySM, DavisGH (2001) Genetic mutations influencing ovulation rate in sheep. Reprod Fertil Dev 13 : 549–555.
20. ClopA, MarcqF, TakedaH, PirottinD, TordoirX, et al. (2006) A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38 : 813–818.
21. FabianMR, SonenbergN, FilipowiczW (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79 : 351–379.
22. WangHR, HsiehCY, TwuYC, YuLC (2008) Expression of the human Sda beta-1,4-N-acetylgalactosaminyltransferase II gene is dependent on the promoter methylation status. Glycobiology 18 : 104–113.
23. HammerNA, HansenTvO, ByskovAG, Rajpert-De MeytsE, GrøndahlML, et al. (2005) Expression of IGF-II mRNA-binding proteins (IMPs) in gonads and testicular cancer. Reproduction 130 : 203–212.
24. KöbelM, WeidensdorferD, ReinkeC, LedererM, SchmittWD, et al. (2007) Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene 26 : 7584–7589.
25. RossAF, OleynikovY, KislauskisEH, TanejaKL, SingerRH (1997) Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol 17 : 2158–2165.
26. MongetP, MonniauxD, PisseletC, DurandP (1993) Changes in insulin-like growth factor-I (IGF-I), IGF-II, and their binding proteins during growth and atresia of ovine ovarian follicles. Endocrinology 132 : 1438–1446.
27. ZhouJ, AdesanyaOO, VatziasG, HammondJM, BondyCA (1996) Selective expression of insulin-like growth factor system components during porcine ovary follicular selection. Endocrinology 137 : 4893–4901.
28. PerksCM, Denning-KendallPA, GilmourRS, WathesDC (1995) Localization of messenger ribonucleic acids for insulin-like growth factor I (IGF-I), IGF-II, and the type 1 IGF receptor in the ovine ovary throughout the estrous cycle. Endocrinology 136 : 5266–5273.
29. MongetP, FabreS, MulsantP, LecerfF, ElsenJM, et al. (2002) Regulation of ovarian folliculogenesis by IGF and BMP system in domestic animals. Domest Anim Endocrinol 23 : 139–154.
30. BonnetA, Lê CaoKA, SancristobalM, BenneF, Robert-GraniéC, et al. (2008) In vivo gene expression in granulosa cells during pig terminal follicular development. Reproduction 136 : 211–224.
31. Le BellegoF, FabreS, PisseletC, MonniauxD (2005) Cytoskeleton reorganization mediates alpha6beta1 integrin-associated actions of laminin on proliferation and survival, but not on steroidogenesis of ovine granulosa cells. Reprod Biol Endocrinol 3 : 19.
32. DelidowBC, LynchJP, WhiteBA, PelusoJJ (1992) Regulation of proto-oncogene expression and deoxyribonucleic acid synthesis in granulosa cells of perifused immature rat ovaries. Biol Reprod 47 : 428–435.
33. DelidowBC, WhiteBA, PelusoJJ (1990) Gonadotropin induction of c-fos and c-myc expression and deoxyribonucleic acid synthesis in rat granulosa cells. Endocrinology 126 : 2302–2306.
34. MohlkeKL, PurkayasthaAA, WestrickRJ, SmithPL, PetryniakB, et al. (1999) Mvwf, a dominant modifier of murine von Willebrand factor, results from altered lineage-specific expression of a glycosyltransferase. Cell 96 : 111–120.
35. JohnsenJM, LevyGG, WestrickRJ, TuckerPK, GinsburgD (2008) The endothelial-specific regulatory mutation, Mvwf1, is a common mouse founder allele. Mamm Genome 19 : 32–40.
36. KawamuraYI, KawashimaR, FukunagaR, HiraiK, Toyama-SorimachiN, et al. (2005) Introduction of Sd(a) carbohydrate antigen in gastrointestinal cancer cells eliminates selectin ligands and inhibits metastasis. Cancer Res 65 : 6220–6227.
37. KawamuraYI, ToyotaM, KawashimaR, HagiwaraT, SuzukiH, et al. (2008) DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology 135 : 142–151.e143.
38. WilliamsSA, StanleyP (2008) Mouse fertility is enhanced by oocyte-specific loss of core 1-derived O-glycans. FASEB J 22 : 2273–2284.
39. WilliamsSA, StanleyP (2011) Premature ovarian failure in mice with oocytes lacking core 1-derived O-glycans and complex N-glycans. Endocrinology 152 : 1057–1066.
40. FrayMD, WrathallJH, KnightPG (1994) Active immunisation against inhibin promotes a recurrent increase in litter size in sheep. Vet Rec 134 : 19–20.
41. CampbellBK, GordonBM, TsonisCG, ScaramuzziRJ (1995) The effect of acute immuno-neutralisation of inhibin in ewes during the early luteal phase of the oestrous cycle on ovarian hormone secretion and follicular development. J Endocrinol 145 : 479–490.
42. MedanMS, WatanabeG, SasakiK, NaguraY, SakaimeH, et al. (2003) Effects of passive immunization of goats against inhibin on follicular development, hormone profile and ovulation rate. Reproduction 125 : 751–757.
43. LiDR, QinGS, WeiYM, LuFH, HuangQS, et al. (2011) Immunisation against inhibin enhances follicular development, oocyte maturation and superovulatory response in water buffaloes. Reprod Fertil Dev 23 : 788–797.
44. HoltzW, WangX, El-GayarM, KnightPG (2012) The effect of exogenous gonadotropins on ovarian function in goats actively immunized against inhibin. Theriogenology 77 : 253–259.
45. AntenosM, StemlerM, BoimeI, WoodruffTK (2007) N-Linked Oligosaccharides Direct the Differential Assembly and Secretion of Inhibin - and A-Subunit Dimers. Molecular Endocrinology 21 : 1670–1684.
46. BrownHM, DunningKR, RobkerRL, BoerboomD, PritchardM, et al. (2010) ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biology of Reproduction 83 : 549–557.
47. SuchanekE, Mujkic-KlaricA, GrizeljV, SimunicV, KopjarB (1990) Protein concentration in pre-ovulatory follicular fluid related to ovarian stimulation. Int J Gynaecol Obstet 32 : 53–59.
48. YamadaM, GentryPA (1995) Hemostatic profile of bovine ovarian follicular fluid. Can J Physiol Pharmacol 73 : 624–629.
49. YamadaM, GentryPA (1995) The hemostatic profile of equine ovarian follicular fluid. Thromb Res 77 : 45–54.
50. SemotokCA, JohnsonWH, LaMarreJ, GentryPA (2000) Amounts of selected coagulation factors in pre - and post-mortem follicular fluid are similar and do not correlate with molecular mass. Anim Reprod Sci 63 : 177–185.
51. HasanS, HosseiniG, PrincivalleM, DongJC, BirsanD, et al. (2002) Coordinate expression of anticoagulant heparan sulfate proteoglycans and serine protease inhibitors in the rat ovary: a potent system of proteolysis control. Biol Reprod 66 : 144–158.
52. ZwainI, AmatoP (2000) Clusterin protects granulosa cells from apoptotic cell death during follicular atresia. Exp Cell Res 257 : 101–110.
53. ScaramuzziRJ, BairdDT, CampbellBK, DriancourtM-A, DupontJ, et al. (2011) Regulation of folliculogenesis and the determination of ovulation rate in ruminants. Reprod Fertil Dev 23 : 444–467.
54. LakinsJN, PoonS, Easterbrook-SmithSB, CarverJA, TenniswoodMP, et al. (2002) Evidence that clusterin has discrete chaperone and ligand binding sites. Biochemistry 41 : 282–291.
55. ArgovN, SklanD (2004) Expression of mRNA of lipoprotein receptor related protein 8, low density lipoprotein receptor, and very low density lipoprotein receptor in bovine ovarian cells during follicular development and corpus luteum formation and regression. Mol Reprod Dev 68 : 169–175.
56. TsengWF, HuangSS, HuangJS (2004) LRP-1/TbetaR-V mediates TGF-beta1-induced growth inhibition in CHO cells. FEBS Lett 562 : 71–78.
57. VogelBE, MurielJM, DongC, XuX (2006) Hemicentins: what have we learned from worms? Cell Res 16 : 872–878.
58. RogerP, PujolP, LucasA, BaldetP, RochefortH (1998) Increased immunostaining of fibulin-1, an estrogen-regulated protein in the stroma of human ovarian epithelial tumors. Am J Pathol 153 : 1579–1588.
59. MollF, KatsarosD, LazennecG, HellioN, RogerP, et al. (2002) Estrogen induction and overexpression of fibulin-1C mRNA in ovarian cancer cells. Oncogene 21 : 1097–1107.
60. MontgomeryGW, SizeJA (1990) Extraction of DNA from sheep white blood cells. New Zealand Journal of Agricultural Research 33 : 437–441.
61. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
62. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
63. RobinsonJT, ThorvaldsdottirH, WincklerW, GuttmanM, LanderES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29 : 24–26.
64. ArchibaldAL, CockettNE, DalrympleBP, FarautT, KijasJW, et al. (2010) The sheep genome reference sequence: a work in progress. Anim Genet 41 : 449–453.
65. MonniauxD, PisseletC (1992) Control of proliferation and differentiation of ovine granulosa cells by insulin-like growth factor-I and follicle-stimulating hormone in vitro. Biol Reprod 46 : 109–119.
66. Hochereau-de ReviersMT, PerreauC, PisseletC, FontaineI, Monet-KuntzC (1990) Comparisons of endocrinological and testis parameters in 18-month-old Ile de France and Romanov rams. Domest Anim Endocrinol 7 : 63–73.
67. PierreA, PisseletC, DupontJ, Mandon-PépinB, MonniauxD, et al. (2004) Molecular basis of bone morphogenetic protein-4 inhibitory action on progesterone secretion by ovine granulosa cells. Journal of Molecular Endocrinology 33 : 805–817.
68. BourinM, GautronJ, BergesM, AttucciS, Le BlayG, et al. (2011) Antimicrobial potential of egg yolk ovoinhibitor, a multidomain Kazal-like inhibitor of chicken egg. J Agric Food Chem 59 : 12368–12374.
69. KellerA, NesvizhskiiAI, KolkerE, AebersoldR (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74 : 5383–5392.
70. NesvizhskiiAI, KellerA, KolkerE, AebersoldR (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75 : 4646–4658.
Štítky
Genetika Reprodukčná medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



