-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
Chromatin insulators are eukaryotic genome elements that upon binding of specific proteins display barrier and/or enhancer-blocking activity. Although several insulators have been described throughout various metazoans, much less is known about proteins that mediate their functions. This article deals with the identification and functional characterization in Paracentrotus lividus of COMPASS-like (CMPl), a novel echinoderm insulator binding protein. Phylogenetic analysis shows that the CMPl factor, encoded by the alternative spliced Cmp/Cmpl transcript, is the founder of a novel ambulacrarian-specific family of Homeodomain proteins containing the Compass domain. Specific association of CMPl with the boxB cis-element of the sns5 chromatin insulator is demonstrated by using a yeast one-hybrid system, and further corroborated by ChIP-qPCR and trans-activation assays in developing sea urchin embryos. The sns5 insulator lies within the early histone gene cluster, basically between the H2A enhancer and H1 promoter. To assess the functional role of CMPl within this locus, we challenged the activity of CMPl by two distinct experimental strategies. First we expressed in the developing embryo a chimeric protein, containing the DNA-binding domain of CMPl, which efficiently compete with the endogenous CMPl for the binding to the boxB sequence. Second, to titrate the embryonic CMPl protein, we microinjected an affinity-purified CMPl antibody. In both the experimental assays we congruently observed the loss of the enhancer-blocking function of sns5, as indicated by the specific increase of the H1 expression level. Furthermore, microinjection of the CMPl antiserum in combination with a synthetic mRNA encoding a forced repressor of the H2A enhancer-bound MBF1 factor restores the normal H1 mRNA abundance. Altogether, these results strongly support the conclusion that the recruitment of CMPl on sns5 is required for buffering the H1 promoter from the H2A enhancer activity, and this, in turn, accounts for the different level of accumulation of early linker and nucleosomal transcripts.
Published in the journal: The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo. PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003847
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003847Summary
Chromatin insulators are eukaryotic genome elements that upon binding of specific proteins display barrier and/or enhancer-blocking activity. Although several insulators have been described throughout various metazoans, much less is known about proteins that mediate their functions. This article deals with the identification and functional characterization in Paracentrotus lividus of COMPASS-like (CMPl), a novel echinoderm insulator binding protein. Phylogenetic analysis shows that the CMPl factor, encoded by the alternative spliced Cmp/Cmpl transcript, is the founder of a novel ambulacrarian-specific family of Homeodomain proteins containing the Compass domain. Specific association of CMPl with the boxB cis-element of the sns5 chromatin insulator is demonstrated by using a yeast one-hybrid system, and further corroborated by ChIP-qPCR and trans-activation assays in developing sea urchin embryos. The sns5 insulator lies within the early histone gene cluster, basically between the H2A enhancer and H1 promoter. To assess the functional role of CMPl within this locus, we challenged the activity of CMPl by two distinct experimental strategies. First we expressed in the developing embryo a chimeric protein, containing the DNA-binding domain of CMPl, which efficiently compete with the endogenous CMPl for the binding to the boxB sequence. Second, to titrate the embryonic CMPl protein, we microinjected an affinity-purified CMPl antibody. In both the experimental assays we congruently observed the loss of the enhancer-blocking function of sns5, as indicated by the specific increase of the H1 expression level. Furthermore, microinjection of the CMPl antiserum in combination with a synthetic mRNA encoding a forced repressor of the H2A enhancer-bound MBF1 factor restores the normal H1 mRNA abundance. Altogether, these results strongly support the conclusion that the recruitment of CMPl on sns5 is required for buffering the H1 promoter from the H2A enhancer activity, and this, in turn, accounts for the different level of accumulation of early linker and nucleosomal transcripts.
Introduction
Chromatin insulators are specialized DNA elements that upon binding of specific proteins display barrier and/or directional enhancer-blocking activity. The analysis of the genome-wide localization of insulator binding proteins (IBPs) in vertebrates and Drosophila suggests that insulators partition the eukaryotic genome in autonomous functional domains by promoting the formation of physical loop structures and/or mediate tethering of the chromatin fiber to structural elements within the nucleus [1], [2]. In vertebrates, CCCTC-binding factor (CTCF) is the only IBP that has been well characterized. Mechanistically, CTCF and its associated co-factors, most notably cohesin, are important in establishing long range chromatin interaction [3], [4]. This is illustrated by the CTCF-dependent intra - and inter-chromosomal interaction necessary for allele specific transcription within the mouse β-globin locus and at the imprinting control region in the H19/Igf2 locus [5]–[7]. Similarly, upon binding near the ins and syt8 promoters, located more that 300 kb away, CTCF stabilizes their interaction and affects gene expression at the human insulin locus [8].
Distinct families of insulators, defined by the IBPs necessary for their activity, have been described in drosophila. The best characterized IBPs are Zeste-white5 (Zw5) and Boundary Element Associated Factor 32 (BEAF-32), that bind to the first identified enhancer-blocking insulators scs and scs′ [9], [10], Suppressor of Hair-wing [Su(Hw)] of the gypsy retrotransposon [11], and dCTCF [12]. The functions of all Drosophila insulators converge as chromatin organizer into that of CTCF in vertebrates. Zw5 and BEAF-32 interact with each other to generate a chromosomal loop that include the 87A7 hsp70 locus [13]. Su(Hw) and dCTCF colocalize at several insulator bodies of diploid nuclei, but not in polytene chromosomes, with the Centrosomal Protein 190 (CP190) which is necessary for both insulator body formation and enhancer-blocking activity [14], [15]. BEAF-32 has also been shown to recruit CP190 to specific DNA sites [16], suggesting that loop formation mediated by CP190 might be a common mechanism for insulator function in drosophila.
A DNA element displaying features common to other chromatin insulators has been found at the 3′ end of the sea urchin P. lividus H2A gene, within the tandem repeat of the early histone unit. As reported, the 462 bp sns5 fragment is required for regulation of histone gene expression in the early embryo as well as for H2A silencing at gastrula stage [17], [18]. A physically separable sns fragment of 265 bp, displaying directional enhancer-blocking function in both sea urchin and mammalian cells [19]–[21], was previously identified in sns5. Most importantly, sns5, but not the enhancer-blocker sns, placed in flanking location of a γ-retrovirus vector prevents position effect variegation, improves transgene expression at randomly integration sites in erythroid cells, and by binding erythroid and ubiquitous transcription factors modifies nucleosomal histones to maintain a euchromatic state at the provirus locus [22]. Four protein binding sites have been identified by DNaseI footprinting in the sns5 element, namely -A, -B, -CT, and -D box, all required for the enhancer-blocking and silencing functions, and none of them resemble the CTCF binding-site consensus sequence [23]. Also the ArsI element, the only other insulator so far characterized in sea urchins, does not belong to the CTCF type [24]. It follows that the identification of sea urchin IBPs is of some importance to unravel the mechanism of action of insulators in chromosome organization and gene expression in this species.
There is at least an additional reason to identify sns5 IBPs, that is, the mechanism of function of sns5 can be studied within the natural histone gene context. We have in fact presented compelling evidence that its role is to attenuate the H2A enhancer in the interaction with the downstream H1 promoter in order to assure the different level of accumulation of nucleosomal and linker transcripts during sea urchin embryogenesis [25].
In this paper, we describe the identification and functional characterization of a novel homeodomain-containing IBP encoded by the Compass/Compass-like locus that is exclusive to the ambulacrarians.
Results
Identification and sequence characterization of the COMPASS-like protein family
To identify the trans-acting factors that interact with the sns5 insulator in P. lividus, we used a yeast one-hybrid genetic assay [26]. Briefly, a cDNA library of N-terminal fusions to the GAL4 activation domain was screened using as bait a yeast strain bearing a stably integrated pentamer of the boxB cis-element upstream of both the HIS3 and lacZ reporter genes. From this screening we isolated a ∼2.2 kb cDNA clone encoding a predicted protein of 396 amino acids, which contains a Compass domain followed by an atypical Homeodomain at the C-terminus (Figure 1A). The former domain is shared exclusively among members of the SATB and COMPASS (CMP) protein families [27], [28]. SATB proteins possess an atypical Homeodomain with phenylalanine, instead of tryptophan, at the 48th residue and a single glycine insertion between the first and second helices, whereas CMP proteins contain two atypical Homeodomains with a ten amino-acid insertion between the second and third helices (Figure 1B) [28]. Differently from the above described proteins, the sea urchin predicted protein exhibits a unique atypical Homeodomain bearing an eleven amino acid long insertion between helices II and III (Figure 1B). For these reasons, we have named this newly identified factor COMPASS-like (CMPl).
Fig. 1. The CMPl protein family. 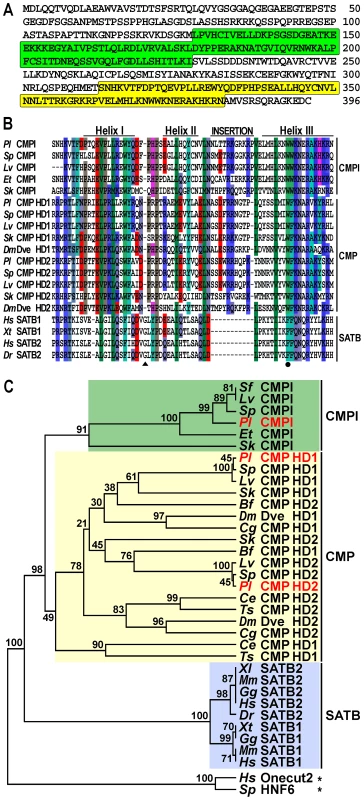
(A) Predicted amino-acid sequence of the P. lividus CMPl, with Compass- and Homeo- domain highlighted in green and yellow, respectively. (B) Comparison of the CMPl, CMP and SATB Homeodomains among various species. Differently coloured boxes highlight similarities; dashes represent the gaps inserted for maximal alignment; position 48 is marked by a filled circle, whereas the glycine insertion in SATB sequences is pointed by a triangle. (C) Rooted neighbor-joining tree constructed from the Homeodomains of representative CMPl, CMP and SATB family members. P. lividus sequence names are in red; numbers above nodes record percent bootstrap values, while asterisks indicate outgroups. Complete taxonomic names and accession numbers of all the sequences used to elaborate the tree are listed in Supplementary Table S1. By blasting the public databases with the sequence coding for the Homeodomain of the P. lividus CMPl protein, we show that the above mentioned differences are completely conserved in orthologs of various sea urchin species and in the hemichordate Saccoglossus kowalevskii (Figure 1B). Such a high degree of conservation suggests that these proteins play important role(s) in echinoderms and hemichordates, altogether forming the Ambulacraria group of deuterostome metazoans [29].
To clarify the phylogenetic relationship between SATB, CMP and CMPl, we built a neighbor-joining tree using set of Homeodomain sequences from various metazoans. As expected, in this analysis orthologs of the SATB family, which have only been identified in vertebrates [30], [31], comprised a monophyletic clade (Figure 1C). Orthologs of the CMP family, which instead have been described only in invertebrates [31], also formed a clade. Importantly, the ambulacrarian CMPl sequences formed a distinct clade supported by a high bootstrap value, suggesting that they constitute a novel family of proteins. In spite of extensive searches in the currently available databases of several metazoans, additional CMPl orthologs were not identified, indicating that the CMPl family probably exists only in ambulacrarians.
Alternative splicing and differential accumulation of the Cmp/Cmpl locus transcripts during sea urchin development
In order to obtain the nucleotide sequence of the Cmpl gene, we BLAST-searched the P. lividus genome database (whole genome shotgun assembly v1.0, http://octopus.obs-vlfr.fr/blast/oursin/blast_oursin.php) using the Cmpl full cDNA sequence as a query. Several overlapping scaffolds and contigs were isolated (Supplementary Table S1), from which the overall sequence was derived. The gene structure was inferred by aligning the genome sequence with that of the Cmpl cDNA and by the use of the Genscan software. We further coupled this analysis to the screening of the available P. lividus EST resources. By this approach we retrieved several hits of different size (Supplementary Figure S1). Collectively, these cDNAs harbor a nearly identical 5′-UTR and utilize the same translation initiation sequence, but only one of them almost entirely matched to Cmpl. Intriguingly, four of the remaining cDNAs appeared instead larger and highly divergent at the 3′-side compared to the query sequence (Supplementary Figure S1). We noticed that this fragment actually maps within the Cmpl gene, being partitioned in a couple of additional exons, namely e5 and e6, which are spliced out in the Cmpl mRNA being mutually exclusive with respect to e7–9 (Figure 2A and B).
Fig. 2. The Cmp/Cmpl locus and its products. 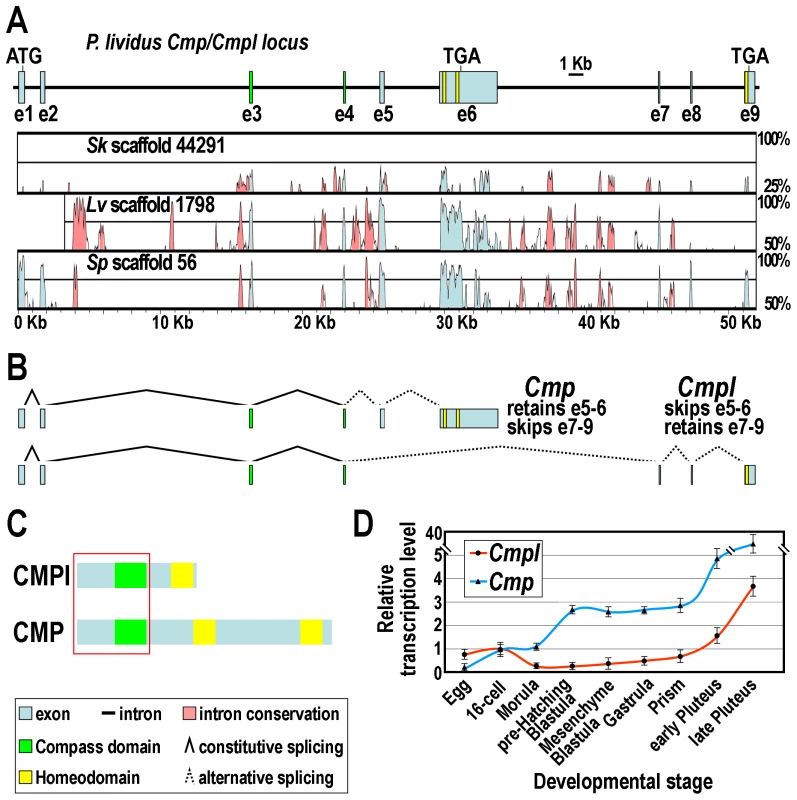
(A) Schematic drawing of the Cmp/Cmpl gene structure showing the positions of exons. Phylogenetic footprinting analysis is shown beneath the diagram. (B) Structure of the alternatively spliced mRNAs. (C) Diagrammatic representation of the CMPl and CMP protein domain organization. Red square indicates common amino acidic sequence at the N-terminal side. (D) qRT-PCR analysis of Cmpl and Cmp transcripts throughout P. lividus embryogenesis. Values at the various stages are shown as fold difference with respect to the 16-cell stage, which displays roughly equal amount of Cmpl and Cmp mRNAs. Bars are standard errors for the qPCR replicas. To assess the conservation of the Cmpl locus across ambulacrarians, we extended the BLAST searches to the public genomic databases of the Strongylocentrotus purpuratus and Lytechinus variegatus sea urchins, and to that of S. kowalevskii. From each of the mentioned database, a single genomic scaffold was retrieved (Figure 2A and Supplementary Table S1). Of importance, by phylogenetic footprinting performed by comparison of nucleotide sequences with the VISTA software, we established that the genomic organization of the P. lividus Cmpl locus is fully conserved in the two evolutionarily distant sea urchins, as well as in the hemichordate (Figure 2A). Furthermore, the retrieving of a single Cmpl hit from the fully completed genome sequence of S. purpuratus leads us to presume that Cmpl is most probably a single copy gene in sea urchins.
Sequence analysis revealed that, as expected, the protein encoded by the largest splice variant was identical to CMPl for 233 amino acids at the N-terminal side, including the Compass domain, but strongly diverged in the C-terminal region. Most notable is the presence of two atypical Homeodomains, with an insertion of ten amino acids between helices II and III (Figure 2C and Figure 1B). On the bases of these findings, coupled to the phylogenetic analysis based on Homeodomain sequences (Figure 1B and C), we designated this protein as the sea urchin CMP ortholog. Therefore, and unexpectedly, the genetic information for CMPl and CMP proteins partially overlaps in representative genomes of both the ambulacrarian taxa.
We then looked by qPCR at the time-course of accumulation of the two splice forms, utilizing primers that distinguish them. As shown in Figure 2D, both transcripts are maternally stored in the unfertilized egg and present at all stages of development. However, Cmpl mRNA is accumulated in the embryo at about a three - to ten-fold lower level than is the Cmp mRNA. After fertilization, Cmpl transcript abundance declines throughout the very early cleavage (up to morula stage), followed by a slight and steady increase until the prism stage. At this time, a later sharp burst in the message prevalence is detected through the pluteus larva.
The Cmp transcript is the most abundant and it is accumulated in the embryo following three main phases of expression. Just after fertilization, the mRNA level rapidly raises to peak at the 16-cell stage. A second increase in transcript level occurs approximately from the morula stage, to peak as the pre-hatching blastula is approached. The terminal phase of mRNA accumulation begins at the prism stage, by which time a dramatic climb in the transcript abundance is observed. Thus, these results clearly established distinct temporal expression patterns for the alternative splice products of the Cmp/Cmpl locus.
Altogether, our findings indicate that the genomic organization of the Cmp/Cmpl locus is evolutionary conserved across ambulacrarians, and that the mRNAs generated by the alternative spliced Cmp/Cmpl transcript exhibit distinct temporal expression profile in the sea urchin embryo.
CMPl, but not CMP, specifically binds the boxB cis-element in vivo
To ascertain the binding activity of CMPl to the sns5 chromatin in sea urchin embryos, we performed quantitative ChIP assays. To this end, we expressed different portions of the CMPl protein in E. coli. As the fragment corresponding to the N-terminal 98–270 amino acid residues gave the maximum yield of the protein in a soluble form, we have generated a polyclonal antibody against this peptide. Being the first 134 residues of this peptide shared by CMPl and CMP, we predicted that the anti-CMPl antibody should rather be able to react with both proteins. Indeed, in western blot assay, the antibody recognized two distinct protein bands at roughly 40 and 90 kDa in sea urchin nuclear extracts at morula and gastrula stages (Figure 3A). These molecular weights were congruent with those predicted for CMPl and CMP proteins, respectively, whereas no reaction occurred with the pre-immune serum (Figure 3A).
Fig. 3. Association of the CMPl protein to the boxB cis-element on sns5. 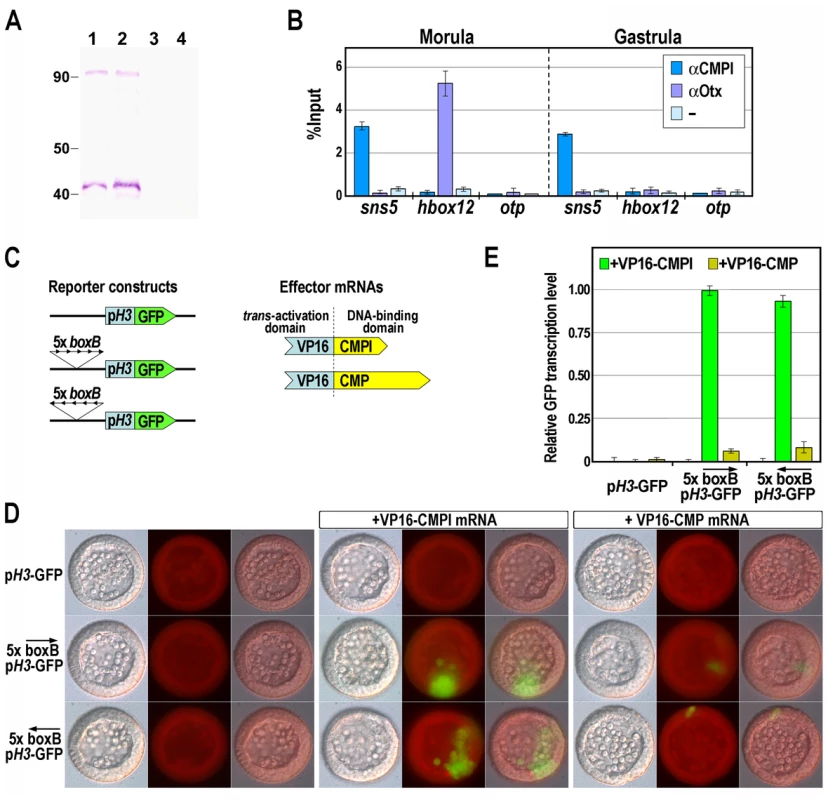
(A) Western blot analysis to test the specificity of the anti-CMPl antibody. Nuclear extracts from morula (lanes 1, 3) and gastrula (lanes 2, 4) embryos were fractioned by SDS-PAGE, blotted on nitrocellulose membrane and incubated with anti-CMPl antibody (lanes 1, 2) or pre-immune serum (lanes 3, 4). (B) ChIP-qPCR analysis of the sns5 occupancy by CMPl. ChIP assays were performed on chromatin extracted from embryos at the indicated stages and precipitated with antiserum against CMPl or Otx, or incubated without adding antibodies (−), followed by qPCR amplification of an sns5 fragment containing the boxB sequence, or hbox12 and otp promoter fragments. As described previously, the association of the Otx transcription factor to its binding site within the hbox12 promoter correlates with the hbox12 transcriptional state [32]. Data are normalized according to the percent of input method. Bars are as in Figure 2D. (C) Scheme of the reporter and effector constructs used in the trans-activation assay. (D) Trans-activation analysis in developing sea urchin embryos observed at the mesenchyme blastula stage. DIC, epifluorescence and merged images, respectively ordered from top to bottom, are shown for each embryo. (E) GFP expression levels assessed by qPCR in transgenic embryos at the mesenchyme blastula stage. Graphs show n-fold changes in mRNA expression level of GFP based on the normalized threshold cycle numbers, with respect to the 5×boxB-pH3-GFP/VP16-CMPl co-injected embryos. Bars are as in Figure 2D. Chromatin containing the sns5 region was consistently precipitated by the affinity-purified anti-CMPl antibody, in samples obtained from cultures of embryos at morula and gastrula stages (Figure 3B; see Materials and Methods). As a negative control, we selected two additional genes, hbox12 [32] and otp [33], [34], that do not share significant sequence similarity with sns5 in their promoters. As expected, both genes were clearly negative to CMPl occupancy from the same ChIP preparations (Figure 3B). Furthermore, only negligible amounts of sns5 sequences were precipitated from chromatin of both developmental stages by the unrelated antiserum from the same host species against the Otx regulator [32], used as control. Overall, these results point out the specific and constitutive association of CMPl to sns5 sequence in the native chromatin. However, as the antibody effectively recognizes epitopes common to both CMPl and CMP, these experiments may represent the full impact of both proteins on the sns5 chromatin.
We addressed this question by performing a in vivo trans-activation assay. To this purpose, pentamer of the boxB cis-element was introduced, in both orientations, upstream of the H3 minimal promoter in the pH3-GFP vector, to obtain the 5×boxB-pH3-GFP reporter constructs (Figure 3C). As effectors we used synthetic mRNAs encoding for a forced transcription activator, in which the activation domain of the viral VP16 protein was joined to the DNA binding domain of either CMPl or CMP. Each transgene, alone or in combination with a chimeric effector mRNA, was then microinjected into sea urchin zygotes, embryos were allowed to develop and scored for GFP expression. As shown in Figure 3D, the control pH3-GFP vector and the 5×boxB-pH3-GFP constructs were not expressed in the absence of effectors in all of the injected embryos (n>300). As expected, and in agreement with the one-hybrid and ChIP results, a significant fraction of embryos (52%, n>300) injected with the reporter construct along with VP16-CMPl mRNA exhibited patches of clonal GFP-expression, irrespectively of the orientation of the cis-acting element on the transgene. By contrast, expression of the reporter was barely detectable in a minor fraction of embryos (5%, n>300) co-injected with equal amounts of the VP16-CMP mRNA (Figure 3D). qPCR measurements further confirmed that GFP expression was weakly evoked in these specimen, with respect to the VP16-CMPl co-injected embryos (Figure 3E). Altogether, these results support the contention that most, if not all, of the boxB binding sequences specifically recruits CMPl in vivo.
Further insights were obtained by examining the specificity of binding of CMPl versus CMP to the boxB element in the natural chromatin context of sns5, within the histone gene cluster. The DNA replication-dependent sea urchin early histone genes are organized in a single large cluster made up of almost 2000 tandem repeats of the 5′-H2B-H3-H2A-H1-H4-3′ unit [35]. Coordinate transcription of these genes is limited to the cleavage and reaches its maximum at the morula stage. The M30 cis-regulatory sequence, upstream the H2A promoter, upon binding of the MBF1 activator displays a bidirectional enhancer activity [25], [36]. Remarkably, as we previously shown [25], the H1 promoter is shielded by the M30 enhancer activity by the sns5 insulator, which is located at the 3′end of the H2A transcription unit (Figure 4A).
Fig. 4. Knock-down of the M30-enhancer and/or sns5-insulator functions and effect on the endogenous early histone gene transcription. 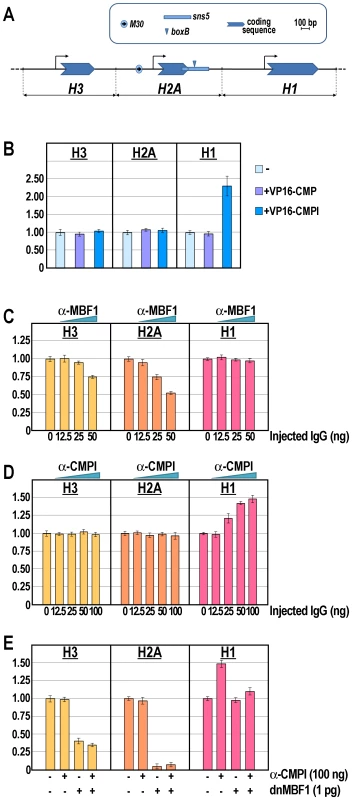
(A) Annotated map of the P. lividus early H3, H2A and H1 histone genes, highlighting the cis-regulatory sequence elements. The horizontal black line represents the genomic DNA, while the bent arrows denote the putative transcription start site. (B–E) qPCR analysis of histone gene transcription carried out in embryos at morula stage injected with the VP16-CMPl/VP16-CMP transcripts (B), or with increasing amounts of the α-MBF1 (C) or α-CMPl (D) antibodies, or the chimeric dnMBF1 mRNA together with saturating dosage of the α-CMPl antibody (E). Graphs show n-fold changes in mRNA expression level of histone genes based on the normalized threshold cycle number of injected embryos compared to that of the uninjected control embryos. Data were derived from two independent microinjection experiments and each bar represents the average of triplicate samples from the two batches of embryos. Sea urchin zygotes were microinjected with either the VP16-CMPl or the control VP16-CMP synthetic transcripts. Then, the expression of the H3, H2A and H1 genes was analyzed by qPCR at the morula stage. As expected, the injection of the VP16-CMP transcript had no detectable effect on histone genes expression (Figure 4B). Likewise, the mRNA levels of H3 and H2A did not show relevant change following the injection of the VP16-CMPl transcript. By contrast, the number of molecules of H1 mRNA was more than double in embryos expressing VP16-CMPl (Figure 4B). The most obvious explanation for the enhancement of H1 expression is that VP16-CMPl acted as a transcriptional activator on the H1 promoter. Alternatively, VP16-CMPl, by competing with the binding of the endogenous CMPl protein, impaired the enhancer-blocking activity of the sns5 element, thus allowing the H2A enhancer to act on the H1 promoter. This hypothesis is in line with our previous in vivo competition assays showing that inhibition of the sns5 led to up-regulation of only the H1 gene [25]. In conclusion, whatever is the mechanism, these experiments, as well as ChIP and trans-activation assays, strongly suggest that CMPl, but not CMP, associates to the boxB site in vivo.
CMPl binding to the boxB site mediates the sns5 enhancer-blocking activity
The finding that CMPl occupies the sns5 chromatin provided an opportunity to examine the function of an IBP in its natural gene context. To challenge CMPl activity, increasing amounts of the affinity-purified anti-CMPl antibodies, or control antibodies against the H2A enhancer binding factor MBF1, were injected into the sea urchin zygotes and histone gene expression analyzed by qPCR at the morula stage. In these experiments, injection of anti-MBF1 provoked a dose-dependent negative effect on the expression of the nucleosomal H2A and H3, but not the linker H1, genes (Figure 4C), excluding a unspecific effect of the injection. Also, it should be noted that H2A was more strongly affected compared to H3. These results are in agreement with those previously obtained by the in vivo inhibition of the H2A enhancer [25].
In strict accordance with previous findings [25 and this paper], both H2A and H3 mRNAs did not vary their abundance upon injection of different doses of anti-CMPl (Figure 4D), while the number of H1 mRNA molecules increased proportionally with the augmentation of the CMPl antiserum (Figure 4D). Taken together, these findings strongly point a role of the CMPl factor in mediating the sns5 insulator function by binding to the boxB cis-element.
Furthermore, we predicted that the block of the H2A enhancer function might counteract the rise of H1 expression due to inhibition of CMPl binding to sns5. Indeed, the results showed in Figure 4E fulfilled this assumption, as co-injection of dnMBF1 mRNA, the dominant-negative version of MBF1 [25], along with saturating amounts of anti-CMPl caused a significant drop in the prevalence of H1 transcripts. Remarkably, in these embryos the number of mRNA molecules for the linker histone was comparable to that of the control uninjected embryos, confirming the independence of H1 transcription from the MBF1/enhancer positive input. Once again, the mRNA abundance of H2A and H3 (although to a lesser extent than H2A) decreased with the microinjection of dnMBF1, irrespectively of the anti-CMPl presence (Figure 4E).
On the basis of these results, we conclude that recruiting of the CMPl factor to the boxB cis-element is critical for the enhancer-blocking activity of the sns5 insulator during the early embryogenesis of the sea urchin.
Discussion
Chromatin insulator functions rely upon the assembly of protein complexes initiated by insulator DNA-binding proteins (IBPs). Although few IBPs have been identified so far, recognition motifs for IBPs represent one of the most conserved non-coding DNA elements in metazoan genomes [37], [38], indicating that these proteins have a critical role in transcriptional regulation. The functions of the vast majority of known IBPs apparently converge as chromatin organizer into that of the paradigmatic CTCF protein. Nevertheless, CTCF orthologs are apparently absent in organisms such as yeast and plants [39], which do have insulator elements embedded in their genome. As mentioned, two insulators have been characterized in sea urchins, namely sns5 and ArsI, and none of them do contain a CTCF binding consensus sequence, raising the question of whether or not a sea urchin CTCF ortholog is involved in enhancer-blocking and chromatin organization as it does in Drosophila and vertebrates. Of importance, it should be emphasized that a DNA sequence erroneously annotated in the S. purpuratus database as a sea urchin CTCF-related gene rather encodes the well characterized Zinc-finger MBF1 transcription activator [36]. Hence, it remains as a possibility that the CTCF ortholog has been lost during evolution in some echinoderms, as occurred in several nematodes [39].
CMPl is a compass - and homeo-domain containing regulator of a novel family
Previous studies in the sea urchin model led to the identification of several candidate proteins (such as ISWI, Sin3A, PARP1, and more) that are recruited on the ArsI insulator [40]. Although all these factors are probably part of protein complexes required for enhancer-blocking activity of ArsI, none of them directly binds the insulator DNA sequence and therefore should not be considered a sensu stricto IBP. Here we have described the identification and functional characterization of CMPl which, to our knowledge, represents the first IBP of a non-model organism. Our molecular analyses indicate that although Cmpl is a single copy gene in the sea urchins, at least two distinct transcripts exist, due to alternative RNA splicing. They encode the CMPl and CMP proteins that are identical in their N-terminal sequences, where the Compass domain is located. Such a domain is followed by one or two atypical Homeodomains, respectively in CMPl and CMP. The sea urchin CMP and CMPl proteins belong to distinct families. Indeed, the former, and likely the most abundant, protein displays features of the COMPASS family, containing two 48W-type Homeodomains, each embedding a decapeptide insertion between helices II and III. Although displaying a 48W signature, the single Homeodomain of CMPl exhibits instead an unusual K/R-rich insertion of eleven residues which is not found in any other known Homeodomain protein but is conserved in sea urchins and S. kowalevskii. As for most of the Drosophila IBPs, apparently, there are no sequence homologs of CMPl in vertebrates. Thus CMPls constitute an additional, previously not described, ambulacrarian-specific family of proteins within those containing both the Compass - and Homeo-domain.
Cmp/Cmpl locus and the monophyly of the Ambulacraria
Although the CMP protein is rather conserved among invertebrates, our phylogenetic analysis strongly suggests that hemichordates and echinoderms share a unique genomic organization at the Cmp/Cmpl locus. Consequently, the CMPl protein is found neither in chordates nor in protostomes. The monophyly of Ambulacraria is supported by three morphological characters: a trimeric arrangement of the adult coeloms, an axial complex with hydropore, and a dipleurula larva with neotroch [41]. At the molecular level, monophyly is also supported by 18S rDNA gene analyses [41]–[44], a unique mitochondrial gene code [45]–[46], the presence of three distinct posterior Hox genes [47], and now the possession of the Cmp/Cmpl locus.
The functional significance of the emergence and loss of CMPl, respectively in ambulacrarians and chordates, is not clear at the moment. On the other hand, the Cmp gene is thought to have a highly flexible behavior during evolution. According to a current model, Cmp genes had emerged from a common ancestor with the POU class homeobox genes, acquiring the insertions in the COMPASS - and Homeo-domain [31]. In the lineage to vertebrates, SATB genes emerged and the genomic structure changed after the divergence of the amphioxus and vertebrate ancestors. SATB gene may have arisen from the Cmp gene by domain shuffling between Cmp and Onecut genes and eventually the Cmp gene was lost from the ancestral vertebrate [31]. In light of this scenario, the genomic configuration of the ambulacrarian Cmp/Cmpl locus could be emerged by similar mechanisms.
Last, but not least, many insulator proteins are known to have multiple functions, and a wider functional analysis of the possible CMPl functional contribution to the sea urchin embryogenesis, outside the sns5 region, should help in the elucidation of the matter. In any case, this study provides an additional step towards an understanding of the evolution of Cmp, Cmpl and SATB genes in lower deuterostomes.
sns5-directed binding activity and functional role of CMPl
The specific binding of CMPl to the boxB cis-element of the sns5 chromatin insulator is substantiated by several lines of evidence. First, a cDNA encoding the CMPl protein was recovered following a yeast one-hybrid assay, using the boxB sequence as bait. Second, constitutive occupancy of the sns5 chromatin by the CMPl protein was demonstrated by ChIP assay, using a CMPl antiserum. Third, specific interaction between the CMPl Homeodomain and the boxB element was further verified by a trans-activation assay in which boxB was placed upstream of the GFP reporter and the resulting transgene injected together with an mRNA encoding the CMPl Homeodomain fused to VP16. Fourth and most important, the expression of the VP16-CMPl chimeric protein, by competing with the endogenous CMPl, acts as either a trans-activator of the H1 promoter or as inhibitor of the enhancer-blocking activity of sns5, or both. Whatever is the mechanism, such a result definitively proved the specific association of the CMPl Homeodomain with the sns5 native chromatin.
Intriguingly, this result clearly indicates that the Homeodomain alone is sufficient for CMPl effective DNA binding, in apparent contrast to the Compass domain-mediated oligomerization required for SATB1 to bind specific DNA sequences [48]. Although the Compass domain has been initially assimilated to a PDZ-like domain, a recent structural study indicated that it rather resembles a classic ubiquitin domain, even in the absence of sequence homology [49]. According to this study, the Compass domain mediates homo-tetramerization of SATB1 in solution. In particular, two dimers are joined together through multiple hydrogen bonds and hydrophobic interactions within their interfaces. Among these reciprocal interactions, the 97E98F162H residues are necessary for the formation of a hemi-dimer, and the 136K137W138N triad is important for the formation of the other dimer.
A sequence alignment of Compass domain across species shows that these residues are all highly conserved among SATB proteins (Supplementary Figure S2), indicating that the Compass domain may have similar biological functions in vertebrates. By contrast, residues at positions 97, 98, and 162 are divergent in ambulacrarian sequences, and the 136K137W138N tripeptide exhibits an inverted orientation in the primary sequence (Supplementary Figure S2). The observed changes in key residues of the Compass domain could explain the different behaviour in DNA binding activity between CMPl and SATB1.
Expression of a CMP Homoeodomain-VP16 chimera in the developing embryo only modestly affected GFP expression in the trans-activation assay, and failed to prejudice sns5 enhancer-blocking function. Altogether, these findings lead us to conclude that no, or maybe very weak, interaction occurs between CMP and the boxB cis-element. It is known that the transcription factor LFB1/HNF1 contains a twenty-one amino-acids insertion between helices II and III which forms a large extra-loop that does not affect the overall structure of the Homeodomain [50]. On this basis, we speculate that the different-size insertions in CMPl and CMP Homeodomains do not participate in DNA interactions. Rather, we reckon that substitution of specific amino acids in Homeodomain sequences may account for differences in DNA binding affinity. Indeed, the Homeodomain of CMPl has a certain sequence similarity with those of CMP (58% and 66%, respectively; Figure 1B) but, notably, significant differences are detectable even in the helix III, which is known to be responsible for the discrimination of binding sequences [51]–[52].
The identification of CMPl in the sea urchin is of some importance to unravel the mechanism of action of insulators in chromosome organization and gene expression in this species. A principal criticism on studying insulators is that, often, the assays are performed by using artificial constructs in a chromosome context different than that of theirs, and it is known that the enhancer/promoter combination influences the activity [53]. In sharp contrast, our experiments provide a clear demonstration of the CMPl-dependent anti-enhancer function of the sns5 insulator in its natural chromatin context. Indeed, we have shown that the inhibition of the CMPl/boxB interaction, either by expressing the VP16-CMPl chimera or by titrating the CMPl factor through injection of specific antibodies, allowed an up-regulation of only the endogenous H1 gene. Most importantly, impairment of the H2A enhancer activity by expressing the forced repressor dnMBF1 restores the normal level of H1 transcripts in embryos in which CMPl is titrated by saturating amounts of the specific antiserum.
Altogether, the results presented in this article strongly suggest that the recruitment of the CMPl protein on the sns5 insulator is absolutely required for buffering the H1 promoter from the H2A enhancer activity, and this, in turn, accounts for the different level of accumulation of early nucleosomal and linker transcripts.
We have previously published several manuscripts analyzing the properties of the sns5 insulator in mammalian cells [20]–[22]. The finding that CMPl is an ambulacrarian-specific factor raises the paradoxical question of how sns5 works as an insulator in cells in which CMPl is absent. For completeness, it should be also remarked that neither the sns5 sequence does exist in the normal vertebrate genome but, if introduced by means of specific vectors, it is bound by several nuclear factors that are normally not recruited in sea urchin cells [21], [22]. In particular, through EMSA and ChIP assays we have demonstrated the occupancy of the boxB element by the Oct1 Homeodomain-containing regulator [21], [22]. In light of this evidence, an explanation to the above mentioned paradox could be that the unconventional recruitment of Oct1 on sns5 in mammalian cells somehow compensates for the lack of CMPl. A pertinent study in transgenic plants documents the enhancer-blocking activity of BEAD-1, a CTCF-dependent human-derived insulator [54], [55]. As reported above, no functional equivalents of CTCF factor and correspondent binding sites have been identified in plants. However, a large number of zinc-finger factors exhibit at least some degree of similarity at the amino acid level with the zinc-fingers of the vertebrate CTCF proteins [56] and may provide a similar function. In any case, the fact that insulators retain their activity in a unnatural host organism implies that at least a proportion of the insulator machinery in eukaryotes may be evolutionarily conserved.
Materials and Methods
Yeast one-hybrid screening
poly(A)+ RNA was extracted from total RNA of P. lividus embryos at the gastrula stage and cDNA library was made by using the Matchmaker One-Hybrid System kit (Clontech). Briefly, poly(A)+ RNA was reverse transcribed and the cDNA products were inserted into a shuttle vector, pGAD10, containing the GAL4 activation domain. Transformation of E. coli DH5α cells yielded >6×105 independent colonies. Pentamer of the 38 bp boxB sequence was used as the bait to select DNA-binding domains encoded in the library. The bait was inserted into pHISi-1 and pLacZi reporter vectors, and the recombinant plasmids were introduced sequentially into the genome of the yeast strain YM4271. Transformants were tested for growth on medium lacking histidine (SD/−His) in the presence of increasing concentrations of 3-aminotriazol (3-AT). Cells whose growth was inhibited by 5 mM 3-AT were selected as the host for the library screen. Transformation with the library was carried out by using LiCl-PEG, and transformants grown on SD/−His and SD/−Leu selective medium were tested for β-galactosidase activity. Plasmids from positive yeast clones were isolated by homogenization with glass beads and then individually transferred into DH5α cells for amplification. To eliminate false positives, these plasmids were separately introduced into yeast cells containing either the boxB bait or the p53 binding site, and transformants were tested for β-galactosidase activity. In this assay, a single plasmid conferred expression only in the boxB host. Sequence analysis revealed that such a plasmid harbored a ∼2.2 kb cDNA insert encoding for CMPl (Genbank accession number: KF421245).
Sequence analysis
BLASTP, BLASTX, and TBLASTN searches in public sequence databases (http://blast.hgsc.bcm.tmc.edu/blast.hgsc; http://www.molgen.mpg.de/~ag_seaurchin) with P. lividus CMPl and CMP as queries were performed and yielded candidates for many phyla. In some cases, a CMP coding region was deduced by Genscan analysis of genomic contigs. Protein domain architecture was defined via use of the Pfam (http://pfam.sanger.ac.uk) and SMART (http://smart.embl-heidelberg.de) databases. Multiple sequence alignments were generated using ClustalW, the output file was formatted using BioEdit, and neighbor-joining tree was constructed using the MEGA software package (http://www.megasoftware.net). The reliability of branching points was inferred by bootstrapping using 1000 replicates. Genomic sequence comparisons for phylogenetic footprinting were performed with the VISTA platform (http://genome.lbl.gov/vista/index.shtml), using window size varied between 50 and 100 bp, with 70% and 50% conservation respectively for sea urchins and S. kowalevskii.
Antibody production, ChIP and qPCR
A DNA fragment corresponding to amino acids 98–270 of CMPl was subcloned into the pGEX-4T-1 expression vector to create a glutathione-S-transferase (GST) fusion protein. This protein was affinity purified from bacterial extracts and then used to generate a rabbit polyclonal antibody (Eurogentec, BE). After serum preincubation with a sepharose-4B column bound to the GST protein, the anti-CMPl antibody was purified by sepharose-4B affinity columns bound to the GST-CMPl protein. The specificity of the antibody was assayed by western blot detection against GST-CMPl and Thioredoxin-CMPl proteins.
ChIP experiments were performed essentially as described in [32], with minor modifications. Briefly, formaldehyde cross-linked sea urchin embryos at morula and gastrula stages were incubated in cell lysis buffer (10 mM HEPES pH 8.0 and 85 mM KCl, containing the following protease inhibitors: 0.5% NP40, 1 µg/ml leupeptin, 1 µg/ml aprotinin, 1 mM PMSF), for 10 min on ice. Nuclei were then resuspended in nuclear lysis buffer (50 mM Tris-HCl pH 8.1, 10 mM EDTA and 1% SDS, containing the same protease inhibitors as in cell lysis buffer) and incubated on ice for 10 min. Chromatin was sonicated using a Bandelin Sonopuls ultrasonic homogenizer to an average fragment size of 150 to 500 bp, as determined by agarose gel electrophoresis. To reduce non-specific background, the samples were diluted into five volumes of ChIP dilution buffer (16.7 mM Tris-HCl pH 8.1, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, plus proteinase inhibitors) and incubated with 100 µl of a salmon sperm DNA/protein A-sepharose slurry for 1 h at 4°C, with mixing. Ten percent of chromatin was withdrawn (Input) and processed as the immunoprecipitated chromatin. Aliquots of chromatin containing 25 µg of DNA were incubated in the absence of antibodies (as a negative control) or either with the anti-CMPl or the anti-Otx serum overnight at 4°C. The immune complexes were adsorbed to protein A-sepharose beads, which were sequentially washed with a low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 150 mM NaCl), a high salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 500 mM NaCl), a LiCl buffer (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0) and twice in 1×TE buffer (10 mM Tris-HCl, 0.1 mM EDTA pH 8.0). The immune complexes were eluted with the elution buffer (1% SDS, 0.1 M NaHCO3), digested with RNase at 37°C and treated with proteinase K in 0.3 M NaCl at 65°C for 4 h to reverse the cross-links. DNA from chromatin samples was extracted with phenol/chloroform, precipitated with ethanol and dissolved in 50 µl of water. DNA samples were then quantified by readings in a Qubit Fluorometer (Invitrogen) using the Quant-iT dsDNA HS assay kit (Invitrogen).
The enrichment of histone regulatory sequences in 100 pg aliquots of genomic DNA purified from the precipitated chromatin fractions was examined by qPCR as described previously [25], [32]. qPCR experiments were performed from two different batches and all reactions were run in triplicate on the 7300 Real-Time PCR system (Applied Biosystems) using SYBR Green detection chemistry. ROX was used as a measure of background fluorescence and, at the end of the amplification reactions, a ‘melting-curve analysis’ was run to confirm the homogeneity of all amplicons. Calculations from qPCR raw data were performed by the RQ Study software version 1.2.3 (Applied Biosystems), using the comparative Ct method. Primer efficiencies (i.e., the amplification factors for each cycle) were found to exceed 1.9. In every experiment, a no-template control was included for each primers set. Primers used in this study are listed in Supplementary Table S2.
The amounts of Cmp/Cmpl and histone gene transcription in control and injected embryos were evaluated as follows. Total RNA from batches of 150 embryos at the desired stage was extracted by using the Power SYBR Green Cells-to-CT kit (Ambion) and reverse transcribed following the manufacturer's recommendations. The resulting cDNA sample was further diluted and the equivalent amount corresponding to one embryo was used as template for qPCR analysis, using the primers indicated in Supplementary Table S2. A cytochrome oxidase or the mbf1 mRNA, which are known to be expressed at a constant level during development [25], [32], were used to normalize all data, in order to account for fluctuations among different preparations.
DNA constructs, synthetic transcripts and microinjection
The pH3-GFP DNA plasmid was constructed as follows. A PCR fragment harboring the H3 minimal promoter (spanning from −62 to +60 with respect to the transcription start site) was inserted in the plasmid polylinker of a pGL3 vector containing the GFP coding sequence. The 5×boxB-pH3-GFP reporter constructs were obtained by shotgun cloning of ligated double-stranded oligonucleotides, bearing the boxB cis-regulatory sequence, upstream of the H3 promoter of the pH3-GFP plasmid. The VP16-CMPl and VP16-CMP effector constructs were obtained by fusing the DNA-binding domain coding sequences of either CMPl or CMP to those of the VP16 activation domain cloned in the CS2+nls expression vector. All DNA clones were checked by sequencing.
Capped mRNAs were synthesized from the linearized pCS2-constructs using the mMessage-mMachine kit (Ambion). Purified RNAs were resuspended at 0.5 mg/ml and 2 pl were then microinjected either alone or in combination with the linearized pH3-GFP and 5×boxB-pH3-GFP constructs.
Microinjection was conducted as previously described [57]–[58]. In the trans-activation assay, more than 100 injected embryos per experiment were scored for GFP expression at the mesenchyme blastula stage and each experiment was repeated three times with different batches of eggs. As a control of the expression of the injected transgenes, we used an actively transcribed GFP construct driven by the full promoter of the hbox12 gene [32]. Images were captured with a Leica DC300F digital camera. As for in vivo titration assays with antibodies, affinity purified rabbit polyclonal IgG reacting with either CMPl or MBF1 were diluted to a final concentration of 12.5–100 ng/pl in ultrapure water containing 30% glycerol, and eventually injected into sea urchin zygotes.
Supporting Information
Zdroje
1. DixonJR, SelvarajS, YueF, KimA, LiY, et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485 : 376–380.
2. BusheyAM, DormanER, CorcesVG (2008) Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32 : 1–9.
3. HouC, ZhaoH, TanimotoK, DeanA (2008) CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci USA 105 : 20398–20403.
4. PhillipsJE, CorcesVG (2009) CTCF: master weaver of the genome. Cell 137 : 1194–1211.
5. MurrellA, HeesonS, ReikW (2004) Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36 : 889–893.
6. KurukutiS, TiwariVK, TavoosidanaG, PugachevaE, MurrellA, et al. (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103 : 10684–10689.
7. SplinterE, HeathH, KoorenJ, PalstraRJ, KlousP, et al. (2006) CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 20 : 2349–2354.
8. XuZ, WeiG, ChepelevI, ZhaoK, FelsenfeldG (2011) Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol 18 : 372–378.
9. GasznerM, VazquezJ, SchedlP (1999) The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev 13 : 2098–2107.
10. ZhaoK, HartCM, LaemmliUK (1995) Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81 : 879–889.
11. GeyerPK, CorcesVG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6 : 1865–1873.
12. MoonH, FilippovaG, LoukinovD, PugachevaE, ChenQ, et al. (2005) CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6 : 165–170.
13. BlantonJ, GasznerM, SchedlP (2003) Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev 17 : 664–675.
14. GerasimovaTI, LeiEP, BusheyAM, CorcesVG (2007) Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28 : 761–772.
15. MohanM, BartkuhnM, HeroldM, PhilippenA, HeinlN, et al. (2007) The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26 : 4203–4214.
16. BusheyAM, RamosE, CorcesVG (2009) Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23 : 1338–1350.
17. Di CaroD, MelfiR, AlessandroC, SerioG, Di CaroV, et al. (2004) Down-regulation of early sea urchin histone H2A gene relies on cis regulative sequences located in the 5′ and 3′ regions and including the enhancer blocker sns. J Mol Biol 342 : 1367–1377.
18. Di CaroV, CavalieriV, MelfiR, SpinelliG (2007) Constitutive promoter occupancy by the MBF-1 activator and chromatin modification of the developmental regulated sea urchin alpha-H2A histone gene. J Mol Biol 365 : 1285–1297.
19. PallaF, MelfiR, AnelloL, Di BernardoM, SpinelliG (1997) Enhancer blocking activity located near the 3′ end of the sea urchin early H2A histone gene. Proc Natl Acad Sci USA 94 : 2272–2277.
20. Di SimoneP, Di LeonardoA, CostanzoG, MelfiR, SpinelliG (2001) The sea urchin sns insulator blocks CMV enhancer following integration in human cells. Biochem Biophys Res Commun 284 : 987–992.
21. AcutoS, Di MarzoR, CalzolariR, BaiamonteE, MaggioA, et al. (2005) Functional characterization of the sea urchin sns chromatin insulator in erythroid cells. Blood Cells Mol Dis 35 : 339–344.
22. D'ApolitoD, BaiamonteE, BagliesiM, Di MarzoR, CalzolariR, et al. (2009) The sea urchin sns5 insulator protects retroviral vectors from chromosomal position effects by maintaining active chromatin structure. Mol Ther 17 : 1434–1441.
23. MelfiR, PallaF, Di SimoneP, AlessandroC, CaliL, et al. (2000) Functional characterization of the enhancer blocking element of the sea urchin early histone gene cluster reveals insulator properties and three essential cis-acting sequences. J Mol Biol 304 : 753–763.
24. HinoS, AkasakaK, MatsuokaM (2006) Sea urchin arylsulfatase insulator exerts its anti-silencing effect without interacting with the nuclear matrix. J Mol Biol 357 : 18–27.
25. CavalieriV, MelfiR, SpinelliG (2009) Promoter activity of the sea urchin (Paracentrotus lividus) nucleosomal H3 and H2A and linker H1 α-histone genes is modulated by enhancer and chromatin insulator. Nucleic Acids Res 37 : 7407–7415.
26. WangMM, ReedRR (1993) Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364 : 121–126.
27. FussB, HochM (1998) Drosophila endoderm development requires a novel homeobox gene which is a target of Wingless and Dpp signalling. Mech Dev 79 : 83–97.
28. BürglinTR, CassataG (2002) Loss and gain of domains during evolution of cut superclass homeobox genes. Int J Dev Biol 46 : 115–123.
29. SwallaBJ, SmithAB (2008) Deciphering deuterostome phylogeny: molecular, morphological and palaeontological perspectives. Philos Trans R Soc Lond B Biol Sci 363 : 1557–1568.
30. YasuiD, MiyanoM, CaiS, Varga-WeiszP, Kohwi-ShigematsuT (2002) SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419 : 641–645.
31. TakatoriN, SaigaH (2008) Evolution of CUT class homeobox genes: insights from the genome of the amphioxus, Branchiostoma floridae. Int J Dev Biol 52 : 969–977.
32. CavalieriV, Di BernardoM, AnelloL, SpinelliG (2008) cis-Regulatory sequences driving the expression of the Hbox12 homeobox-containing gene in the presumptive aboral ectoderm territory of the Paracentrotus lividus sea urchin embryo. Dev Biol 321 : 455–469.
33. CavalieriV, Di BernardoM, SpinelliG (2007) Regulatory sequences driving expression of the sea urchin Otp homeobox gene in oral ectoderm cells. Gene Expr Patterns 7 : 124–130.
34. CavalieriV, Di BernardoM, SpinelliG (2003) Impairing Otp homeodomain function in oral ectoderm cells affects skeletogenesis in sea urchin embryos. Dev Biol 262 : 107–118.
35. SpinelliG, BirnstielML (2002) The modulator is a constitutive enhancer of a developmentally regulated sea urchin H2A gene. Bioessays 24 : 850–857.
36. AlessandroC, Di SimoneP, BuscainoA, AnelloL, PallaF, et al. (2002) Identification of the enhancer binding protein MBF-1 of the sea urchin modulator alpha-H2A histone gene. Biochem Biophys Res Commun 295 : 519–525.
37. JungCH, MakuninIV, MattickJS (2010) Identification of conserved Drosophila specific euchromatin-restricted non-coding sequence motifs. Genomics 96 : 154–166.
38. KimTH, AbdullaevZK, SmithAD, ChingKA, LoukinovDI, et al. (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128 : 1231–1245.
39. HegerP, MarinB, SchierenbergE (2009) Loss of the insulator protein CTCF during nematode evolution. BMC Mol Biol 10 : 84.
40. YajimaM, FairbrotherWG, WesselGM (2012) ISWI contributes to ArsI insulator function in development of the sea urchin. Development 139 (19) 3613–3622.
41. PetersonKJ, EernisseDJ (2001) Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol Dev 3 : 170–205.
42. BromhamLD, DegnanBM (1999) Hemichordates and deuterostome evolution: robust molecular phylogenetic support for a hemichordate +echinoderm clade. Evol Dev 1 : 166–171.
43. FurlongRF, HollandPWH (2002) Bayesian phylogenetic analysis supports monophyly of Ambulacraria and of Cyclostomes. Zool Sci 19 : 593–599.
44. WadaH, SatohN (1994) Details of the evolutionary history from invertebrates to vertebrates, as deduced from the sequences of 18S rDNA. Proc Natl Acad Sci USA 91 : 1801–1804.
45. CastresanaJ, Feldmaier-FuchsG, PaaboS (1998) Codon reassignment and amino acid composition in hemichordate mitochondria. Proc Natl Acad Sci USA 95 : 3703–3707.
46. CastresanaJ, Feldmaier-FuchsG, YokoboriSI, SatohN, PaaboS (1998) The mitochondrial genome of the hemichordate Balanoglossus carnosus and the evolution of deuterostome mitochondria. Genetics 150 : 1115–1123.
47. PetersonKJ (2004) Isolation of Hox and Parahox genes in the hemichordate Ptychodera flava and the evolution of deuterostome Hox genes. Mol Phylogenet Evol 31 (3) 1208–1215.
48. PurbeyPK, SinghS, KumarPP, MehtaS, GaneshKN, et al. (2008) PDZ domain-mediated dimerization and homeodomain-directed specificity are required for high-affinity DNA binding by SATB1. Nucleic Acids Res 36 : 2107–2122.
49. WangZ, YangX, ChuX, ZhangJ, ZhouH, et al. (2012) The structural basis for the oligomerization of the N-terminal domain of SATB1. Nucleic Acids Res 40 : 4193–4202.
50. CeskaTA, LamersM, MonaciP, NicosiaA, CorteseR, et al. (1993) The X-ray structure of an atypical homeodomain present in the rat liver transcription factor LFB1/HNF1 and implications for DNA binding. EMBO J 12 (5) 1805–1810.
51. BürglinTR (2011) Homeodomain subtypes and functional diversity. Subcell Biochem 52 : 95–122.
52. GehringWJ, AffolterM, BürglinT (1994) Homeodomain proteins. Annu Rev Biochem 63 : 487–526.
53. ScottKC, TaubmanAD, GeyerPK (1999) Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153 : 787–798.
54. BellAC, WestAD, FelsenfeldG (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98 : 387–396.
55. Gudynaite-SavitchL, JohnsonDA, MikiBLA (2009) Strategies to mitigate transgene-promoter interactions. Plant Biotechnol J 7 : 472–485.
56. EngelbrechtCC, SchoofH, BöhmS (2004) Conservation, diversification and expression of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5 : 1–17.
57. CavalieriV, Di BernardoM, SpinelliG (2009) Functional studies of regulatory genes in the sea urchin embryo. Methods Mol Biol 518 : 175–188.
58. CavalieriV, GuarcelloR, SpinelliG (2011) Specific expression of a TRIM-containing factor in ectoderm cells affects the skeletal morphogenetic program of the sea urchin embryo. Development 138 : 4279–4290.
Štítky
Genetika Reprodukčná medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



