-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
Transposable elements are major components of eukaryotic genomes, comprising a large portion of the genome in some species. Miniature inverted-repeat transposable elements (MITEs), which belong to the class II DNA transposable elements, are abundant in gene-rich regions, and their copy numbers are very high; therefore, they have been considered to contribute to genome evolution. Because MITEs are short and have no coding capacity, they cannot transpose their positions without the aid of transposase, provided in trans by their autonomous element(s). It has been unknown how MITEs amplify themselves to high copy numbers in the genome. Our results demonstrate that the rice active MITE mPing is mobilized in the embryo by the developmental stage-specific up-regulation of an autonomous element, Ping, and thereby successfully amplifies itself to a high copy number in the genome. The short-term expression of Ping is thought to be a strategy of the mPing family for amplifying mPing by escaping the silencing mechanism of the host genome.
Published in the journal: Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE. PLoS Genet 10(6): e32767. doi:10.1371/journal.pgen.1004396
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004396Summary
Transposable elements are major components of eukaryotic genomes, comprising a large portion of the genome in some species. Miniature inverted-repeat transposable elements (MITEs), which belong to the class II DNA transposable elements, are abundant in gene-rich regions, and their copy numbers are very high; therefore, they have been considered to contribute to genome evolution. Because MITEs are short and have no coding capacity, they cannot transpose their positions without the aid of transposase, provided in trans by their autonomous element(s). It has been unknown how MITEs amplify themselves to high copy numbers in the genome. Our results demonstrate that the rice active MITE mPing is mobilized in the embryo by the developmental stage-specific up-regulation of an autonomous element, Ping, and thereby successfully amplifies itself to a high copy number in the genome. The short-term expression of Ping is thought to be a strategy of the mPing family for amplifying mPing by escaping the silencing mechanism of the host genome.
Introduction
Transposable elements (TEs) are DNA sequences that are capable of jumping from one genomic locus to another and make up a large fraction of eukaryotic genomes. More than 80% of the maize (Zea mays) and barley (Hordeum vulgare) genomes are composed of TEs [1], [2], and they constitute 35% and 14% of the genomes of rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana), respectively [3], [4]. TEs are harmful to the host because their mobilities perturb genome stability, whereas they play greatly generative roles in genome evolution such as alternation of gene structure, change of expression pattern, and rearrangement of chromosome structure [5], [6].
TEs are classified into two groups according to their transposition mechanisms: class I elements (retrotransposons) that transpose through a copy-and-paste mechanism via an RNA intermediate, and class II elements (transposons) that transpose through a cut-and-paste mechanism without undergoing an RNA intermediate. Class I elements easily attain tens of thousands of copies, whereas the majority of class II elements cannot amplify themselves to 50 copies at most. Unlike other class II elements, miniature inverted-repeat transposable elements (MITEs) have the capacity to amplify themselves to high copy numbers (hundreds or thousands) [7]–[9]. In the rice genome, MITEs are numerically predominant TEs [10], constituting 8.6% of the genome [11]. Because MITEs are too short (<600 bp) to encode any protein, their transpositions must depend on the proteins encoded by the autonomous elements. Well-studied MITEs are classified into the Stowaway and Tourist families, which belong to the Tc1/mariner and PIF/Harbinger superfamilies, respectively. Because MITEs are mainly deployed in gene-rich regions [10], [12] and affect adjacent gene expression [13], they are considered to play an important role in genome evolution. However, little is known about how MITEs attain high copy numbers.
Miniature Ping (mPing) is the first active MITE identified in the rice genome [14]–[16]. Although MITEs are deployed in the genome at a high copy number, the copy number of mPing exceptionally remains at a low level in most rice cultivars: indica and tropical japonica cultivars have fewer than 10 copies, and temperate japonica cultivars including Nipponbare have approximately 50 copies [14]. The transposition of mPing is suppressed in most rice cultivars, but, like other TEs, mPing is activated by exposure to various stress conditions such as gamma-ray irradiation [16], hydrostatic pressurization [17], cell culture [14], anther culture [15], and inhibition of topoisomerase II [18]. Introgression of distantly related genomes also causes mPing transposition [19], [20]. However, mPing is actively transposing without such stresses in the temperate japonica rice strain EG4 (cultivar Gimbozu) under natural growth conditions, and its copy number is approximately 1000 copies [21]. This indicates that mPing has overcome the silencing mechanism or established a novel strategy for its amplification in the EG4 genome. In this sense, mPing in EG4 is an appropriate material to study the amplification of MITEs in plant genomes.
The autonomous element Ping and its distantly related element Pong, which both belong to the PIF/Harbinger superfamily, provide two proteins required for mPing transposition. Both Ping and Pong have two open reading frames (ORFs), ORF1 and ORF2 [22], [23]. The former encodes a Myb-like DNA-binding protein, and the latter encodes a transposase lacking DNA binding domain. Transposase of most class II elements contains a conserved catalytic domain (DDE motif) and a DNA-binding domain [23], [24], whereas these domains are encoded separately by two ORFs in both Ping and Pong [22], [23]. The study of other members of the PIF/Harbinger superfamily suggested that the Myb-like DNA-binding protein directly binds to the subterminal regions of the transposon in order to recruit the transposase [25]. Both Myb-like protein and transposase of either Ping or Pong or both elements are necessary for mPing transposition [22], [23].
In this study, we demonstrate that mPing is actively transposing in the embryo of EG4 during the period from the regionalization of shoot apical meristem (SAM) and radicle to the formation of the first leaf primordium (3 to 5 days after pollination, DAP) with the aid of developmental stage-specific expression of Ping. Our results provide important evidence for the amplification mechanism not only of mPing but also of other MITEs.
Results
Transpositions of mPing during gametogenesis
Plants have acquired the silencing mechanism of TEs in germ cells. In Arabidopsis, for example, TEs are activated specifically in the vegetative nucleus of the pollen, and siRNAs from the activated TEs accumulate in the sperm cells [26]. On the basis of these results, Slotkin and colleagues proposed that siRNAs derived from TEs activated in the vegetative nucleus silence TEs in the sperm cells [26]. We conceived that mPing might overcome such a silencing mechanism in EG4. To confirm this hypothesis, we developed two F1 populations from reciprocal crosses between the mPing-active strain EG4 and the mPing-inactive cultivar Nipponbare, and investigated the transposition activity of mPing by transposon display (TD) analysis. Success of reciprocal crosses was confirmed by PCR analysis using locus-specific primers (Figure S1A). One of the results of TD analysis using two selective bases is shown in Figure 1A; all 16 possible primer combinations were analyzed. The banding patterns of F1 plants were almost the same as those of EG4. The bands that appeared in all F1 plants but not in the parental EG4 plant were derived from another parental Nipponbare plant (Figure S1B). Furthermore, the bands that appeared in only one of eight F1 plants but not in the parental EG4 plant are herein referred as de novo insertions. These bands were confirmed not to be PCR artifacts by sequence and locus-specific PCR analysis (Table S1 and Figure S2). We detected 15.5 de novo insertions per plant in the selfed progenies of EG4, whereas Nipponbare yielded no de novo insertions in the selfed progenies (Figure 1B). This confirmed that mPing is active in EG4 under natural growth conditions but inactive in Nipponbare. If mPing was specifically activated in the pollen of EG4, we could obtain de novo insertions only in the F1 plants from the Nipponbare/EG4 cross. However, we obtained de novo insertions in both Nipponbare/EG4 and EG4/Nipponbare populations (Figure 1B). Moreover, there was no significant difference in the number of de novo insertions per plant between the two F1 populations. This indicates that the activating factor(s) for the mPing transposition is present in both male and female gametes of EG4.
Fig. 1. Transposition of mPing in reciprocal crosses between EG4 and Nipponbare. 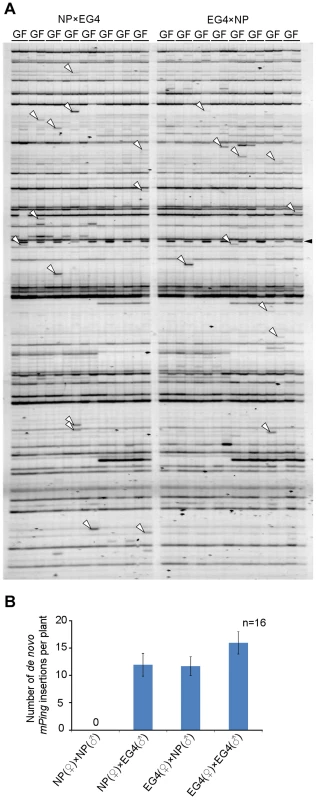
(A) Transposon display (TD) for mPing of the F1 population from reciprocal crosses between EG4 and Nipponbare. One of the results of TD analysis using two selective bases is shown. The cross combinations are indicated at the top of the profiles, respectively. G and F indicate parental EG4 and the F1 plants, respectively. White and black arrowheads indicate the bands representing the de novo mPing insertion and the band derived from Nipponbare genome, respectively. (B) Mean numbers of de novo mPing insertions in a single F1 plant and in a self-pollinated plant. The cross combinations are indicated at the bottom of the profile. All 16 possible primer combinations were analyzed, and mean values were calculated using 16 individuals (n = 16). Bars indicate SE. Transpositions of mPing during ontogeny of EG4 plants
We performed TD analysis of mPing using genomic DNA samples extracted from endosperm, radicle, and leaf blades of eight progenies (S1) derived from a single parental EG4 plant (S0), and investigated the mPing transposition during ontogeny of rice plants (Figure 2A). One of the results of TD analysis using two selective bases is shown in Figure S3; all 16 possible primer combinations were analyzed. We examined de novo insertions in the same way as described above. Consequently, a total of 228 de novo insertions were detected. These insertions were divided into five groups (Figure 2B): (1) endosperm-specific insertions that appeared only in the endosperm sample, (2) radicle-specific insertions that appeared only in the radicle sample, (3) leaf-specific insertions that appeared only in one sample from the 1st to 3rd leaf blades, but not in the 4th and 5th leaf blades, (4) shoot-specific insertions that appeared in at least one sample of 1st, 2nd, and 3rd leaf blades, and in at least one sample of 4th and 5th leaf blades, and (5) radicle/shoot-specific insertions that appeared in both radicle and leaf blade samples. These de novo insertions were confirmed by sequence and locus-specific PCR analysis (Table S2 and Figure S4). Numbers of each insertion obtained in this study are summarized in Figure 2C. Plant development is divided roughly into three successive phases: embryogenesis, vegetative phase, and reproductive phase. If mPing transposed in the SAM of the S0 plant during vegetative and/or reproductive phases, the de novo insertions would segregate according to Mendel's law among the S1 progenies. We obtained no band that appeared in at least two S1 progenies and was not seen in the S0 plant. This indicates that the transmissible insertion of mPing to the next generation seldom (or never) arises during the vegetative and reproductive phases.
Fig. 2. De novo mPing insertions during rice ontogeny. 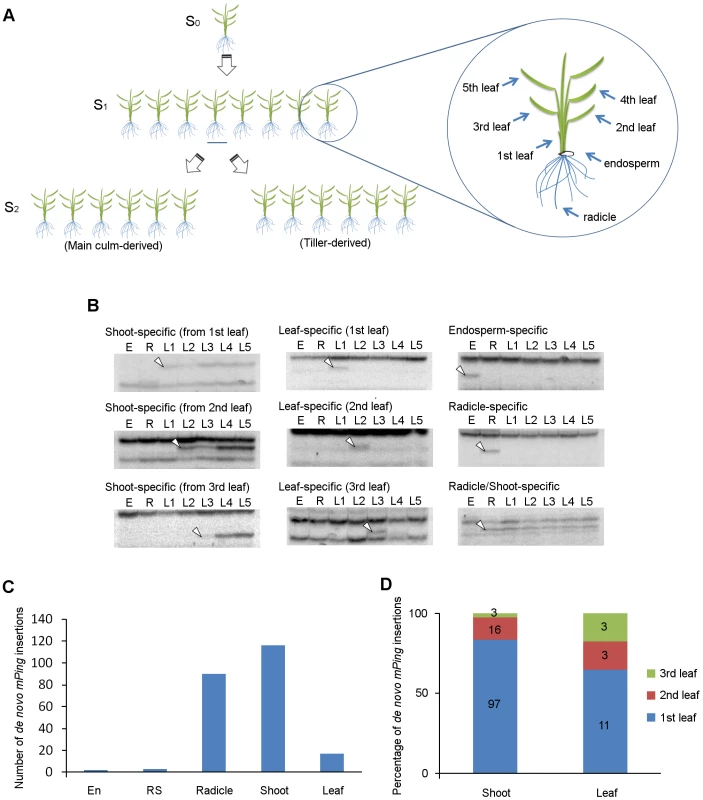
(A) Experimental setup for the ontogenical analysis to determine the timing of mPing transposition in EG4. Eight progenies (S1) derived from a single parental EG4 plant (S0) were grown in a greenhouse. Endosperm, radicle, and leaf blades (1st to 5th) of each S1 plant were sampled for DNA extraction. S2 seeds were harvested from the main culm and the primary tiller of each S1 plant to investigate the inheritance of de novo mPing insertions. The 2nd leaf blade of S0 and S2 plants was also sampled. Detailed information is provided in the ‘Materials and Methods’. (B) mPing insertions were detected by transposon display. Representative images of shoot-, leaf-, endosperm-, radicle-, and radicle/shoot-specific insertions are shown. White arrowheads indicate the bands representing the de novo mPing insertion. E: endosperm, R: radicle, L1–L5: 1st to 5th leaf blades. (C) The number of organ-specific de novo insertions in EG4. All 16 possible primer combinations were analyzed. En: endosperm-specific insertion, RS: radicle/shoot-specific insertion, R: radicle-specific insertion, Shoot: shoot-specific insertion, Leaf: leaf-specific insertion. (D) Percentage of leaf positions where the first de novo mPing insertion was found. Shoot: shoot-specific insertion, Leaf: leaf-specific insertion. Flowering plants have evolved a unique reproductive process called double fertilization. In this process, either of two sperm cells in pollen fuses with either an egg cell or a central cell in the ovule, and then the egg cell fertilized with the sperm cell initiates embryogenesis [27]. In rice, the SAM and radicle are regionalized in the embryo 3 DAP, and three leaves and the radicle are already present in the mature embryo [28]. We detected only three radicle/shoot-specific insertions (Figure 2C), indicating that mPing scarcely transposes during the period from the onset of gametogenesis to the early stage (until 3 DAP) of embryogenesis. Among the 228 de novo insertions, 116 and 17 were shoot-specific and leaf-specific insertions, respectively (Figure 2C). This indicates that mPing actively transposes in the embryo during the period from the regionalization of SAM and radicle (at 3 DAP) to the formation of the 3rd leaf primordia (at 8 DAP). Of the 133 shoot - and leaf-specific insertions, 108 were of the 1st leaf blade (Figure 2D). Since the 1st leaf primordium is formed at 5 DAP, the most active phase of the mPing transposition was considered to be from 3 to 5 DAP. We detected a large number of radicle-specific insertions as well as shoot-specific insertions, and the sum of these insertions accounted for 90% of all insertions detected in this study (Figure 2C). Taken together, we concluded that mPing in EG4 most actively transposes in the 3 to 5 DAP embryo.
Endosperm is a triploid tissue that is produced by fusing a central cell containing two polar nuclei with one of two sperm cells in no particular order. The endosperm formation occurs in parallel with embryogenesis. The endosperm-specific insertions result from the mPing transposition occurred in either gametogenesis or endosperm formation. We observed only two endosperm-specific insertions (Figure 2C), supporting that mPing scarcely transposes during the period from the onset of gametogenesis to the early stage of embryogenesis. The relationship between the banding patterns obtained in TD analysis and the timing of mPing transposition is summarized in Figure S5.
Inheritance of de novo mPing insertions to the next generation
In order for mPing to amplify, the de novo insertions must be transmitted to the next generation. We performed TD analysis using 12 progenies (S2) derived from the main culm and the primary tiller of a single selfed parent (S1) to investigate whether the de novo insertions detected in ontogenical analysis are inheritable (Figure S6). Both radicle-specific and leaf-specific insertions in the S1 plants were not detected in the S2 progenies (0 of 15, 0 of 2, respectively). In contrast, 85% (11 of 13) of the shoot-specific insertions that were detected in the S1 plants also appeared in the S2 progenies. This value (85%) is consistent with the estimated number of inheritable de novo insertions in our previous report [21]. Thus most of the de novo insertions that arose in the 3 to 5 DAP embryo were successfully inherited to the next generation.
Excisions of mPing during ontogeny of EG4 plants
We have already determined the sites of all mPing insertions (1163 in total) in the EG4 genome [13], and have investigated mPing excisions in a small EG4 population using locus-specific primer pairs [29], [30]. Here we examined the timing of the mPing excision with locus-specific PCR using the genomic DNA samples that were used for the ontogenical analysis of the de novo insertion. We randomly chose 48 markers for this study (Table S3). We divided the mPing excisions into five types with the same criteria as those used for the de novo insertions: endosperm-, radicle-, leaf-, shoot-, and radicle/shoot-specific excisions (Figure S7). There were no endosperm-specific and radicle/shoot-specific excisions, indicating that no mPing transposition occurs during the period from the onset of gametogenesis to the early stage of embryogenesis. We detected seven radicle-specific, six leaf-specific, and three shoot-specific excisions. All shoot-specific excisions were detected from the 1st leaf blade sample. These results indicate that, like the de novo insertion, the mPing excision also occurs during the period from the regionalization of the SAM and radicle to the formation of the first leaf primordium, although we cannot completely rule out the possibility that these excisions occur also in somatic cells of mature tissues. Thus, in addition to the experimental results of the de novo insertion, we concluded that mPing of EG4 was most actively transposing in the 3 to 5 DAP embryo.
Expression pattern of Ping in EG4
Both Ping and Pong provide a Myb-like protein and a transposase, which are encoded by their ORF1 and ORF2, respectively (Figure 3A), and have been considered as autonomous elements responsible for the mPing transposition. We investigated the expression of Ping-ORF1, Ping-ORF2, Pong-ORF1, and Pong-ORF2 during embryogenesis to evaluate which autonomous element plays a predominant role in driving the mPing transposition in EG4. Reverse transcription-PCR analysis revealed that Ping-ORF1 and Ping-ORF2 constitutively expressed in the ovary during embryogenesis (Figure 3B). On the other hand, no transcriptions of Pong-ORF1 and Pong-ORF2 (Figure 3B) were observed. This strongly suggests that Ping predominantly controls the mPing transposition in EG4.
Fig. 3. Ping expression during seed development. 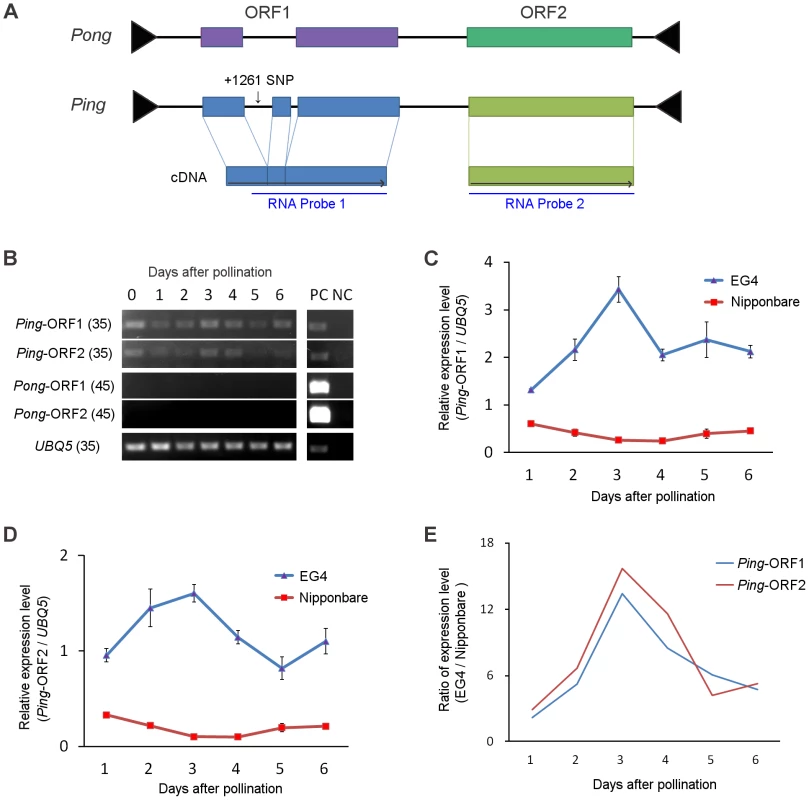
(A) Structure of the Ping and Pong elements. Terminal inverted repeats are indicated by black triangles. Boxes represent ORF1 and ORF2, respectively. Gray horizontal arrows indicate the direction of transcription. RNA probes used are indicated below the ORFs. (B) Reverse-transcription PCR analysis of Ping-ORF1, Ping-ORF2, Pong-ORF1, and Pong-ORF2. Numbers in parentheses are PCR cycle numbers. PC: positive control (0.1 ng genomic DNA), NC: negative control (non-reverse-transcribed RNA). (C) Real-time quantitative PCR of Ping-ORF1 and (D) Ping-ORF2. The expression level in the Nipponbare ovary just after pollination was set as 1. The results are presented as means of three biological replicates. Bars indicate SE. (E) The ratio of Ping expression level of EG4 to that of Nipponbare. The means in (C) and (D) were used for calculation. We performed real-time quantitative PCR (qPCR) analysis to compare the expression level of Ping-ORF1 and -ORF2 between EG4 and Nipponbare during embryogenesis. In all developmental stages from 1 to 6 DAP, the expression levels of both Ping-ORF1 and -ORF2 were higher in EG4 than in Nipponbare (Figure 3C, D). Since EG4 harbors seven copies of Ping, whereas Nipponbare has only one copy (Table S4), the difference in the expression levels between EG4 and Nipponbare is considered to be attributable to the different copy number of Ping. However, we found that Ping of EG4 showed different expression patterns from that of Nipponbare. In Nipponbare, the expression level of Ping-ORF1 and -ORF2 gradually declined until 3 DAP, and restored to the basal level at 6 DAP. In contrast, in EG4, the expression levels of both Ping-ORF1 and -ORF2 rapidly increased, with a peak at 3 DAP (Figure 3C, D). The ratio of relative expression level (EG4/Nipponbare) clearly demonstrated that Ping might be up-regulated in a developmental stage-specific manner in the ovary of EG4 (Figure 3E). Since mPing transposed during the period from 3 to 5 DAP, the rapid increase in Ping expression most likely drive the mPing transposition.
Accumulation of Ping transcripts in the embryo triggers mPing transposition
We investigated the spatial expression pattern of Ping by in situ hybridization using Ping-specific probes. The probe positions were indicated in Figure 3A. The Ping transcripts were detected in all tissues, viz. embryo, endosperm, and ovary wall, in both EG4 and Nipponbare (Figure 4A–C, S8). Among the tissues, the 3 DAP embryo of EG4 yielded an exceptionally strong signal, indicating a high accumulation of Ping transcripts (Figure 4A), whereas the 5 DAP embryo showed a much lower accumulation of Ping transcripts in EG4 (Figure 4D–F). Such a drastic change in accumulation quantity of Ping transcripts with the advance of embryogenesis was consistent with the change in the expression quantity of Ping with the advance of embryogenesis, which was investigated by real-time qPCR (Figure 3C–E). These results suggest that the tissue - and developmental stage-specific accumulation of the Ping transcripts triggers mPing transposition at this stage in EG4. To confirm this hypothesis, we evaluated the spatial expression pattern of Ping in the SAM during the vegetative phase. As described above, mPing hardly transposes in the SAM during this phase. The Ping transcripts were detected in all tissues including the SAM, and, as expected, there was no obvious difference in the signal intensity between EG4 and Nipponbare (Figure 4G–I). Thus the Ping transcripts proved to accumulate developmental stage-specifically only in the tissue where mPing actively transposes. We therefore concluded that the high accumulation of Ping transcripts triggers the transposition of mPing in the 3 DAP embryo of EG4.
Fig. 4. Detection of Ping-ORF1 spatial expression patterns by in situ hybridization analysis. 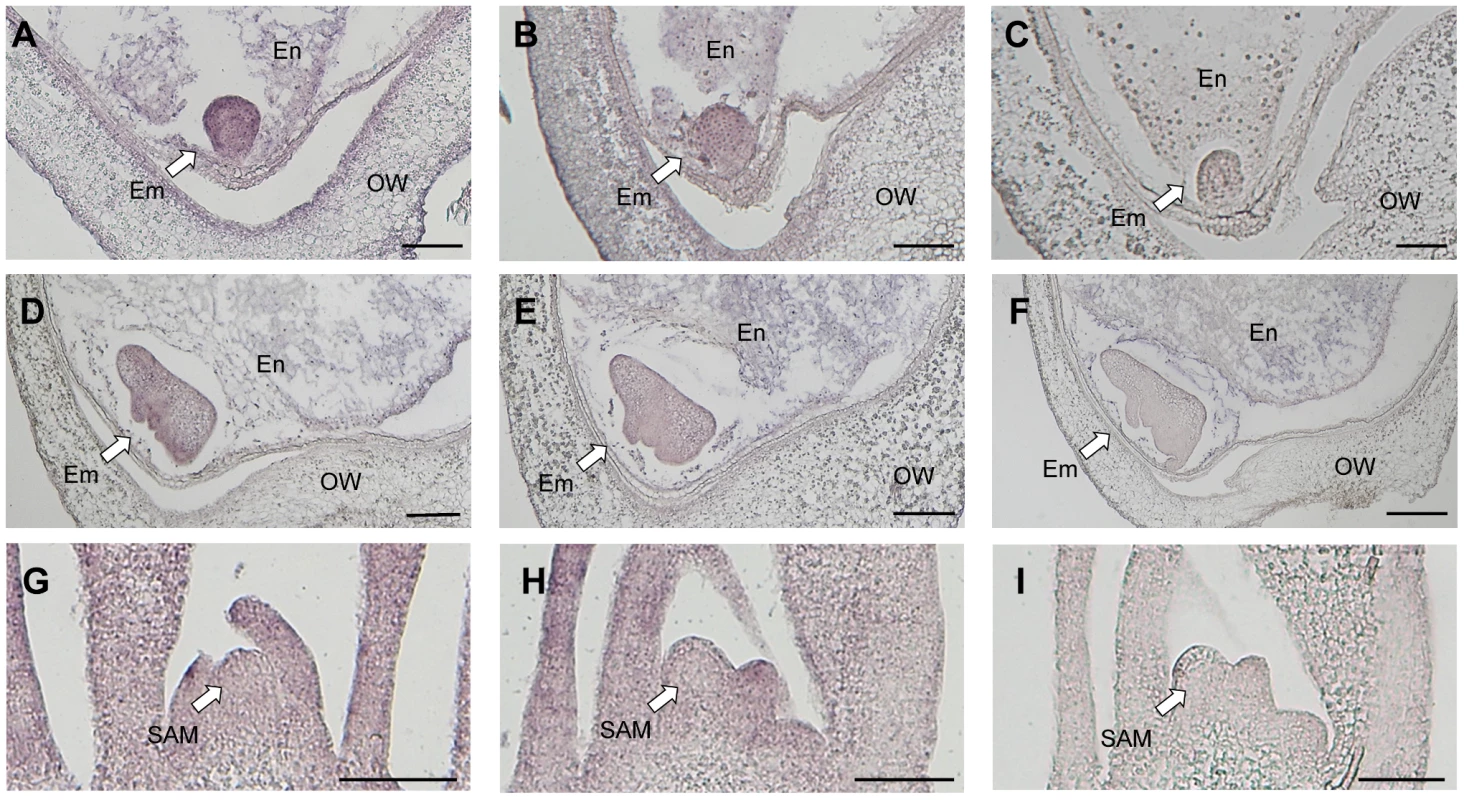
Longitudinal sections through the ovary 3 days after pollination of (A, C) EG4 and (B) Nipponbare; the ovary 5 days after pollination of (D, F) EG4 and (E) Nipponbare; and the shoot apical meristem of (G, I) EG4 and (H) Nipponbare seedlings were hybridized with antisense (A, B, D, E, G, H) or sense (C, F, I) RNA probes. Little staining was obtained with the sense probe (F). Em: embryo, En: endosperm, OW: ovary wall, SAM: shoot apical meristem. Scale bars represent 100 µm. SNP in an intronic region of Ping-ORF1
EG4 has seven Ping elements (Ping-1 to -7), whereas Nipponbare has only one (Ping-N) (Table S4). When we sequenced and compared all Ping elements, a single nucleotide polymorphism (SNP) in the first intronic region of Ping-ORF1 was detected between EG4 and Nipponbare (Figure 5A). Ping-N has a ‘T’ nucleotide on the SNP region, whereas all Ping elements in EG4 have a ‘C’ nucleotide. We named the former ‘T-type Ping’ and the latter ‘C-type Ping’.
Fig. 5. SNP in the first intronic region of Ping-ORF1. 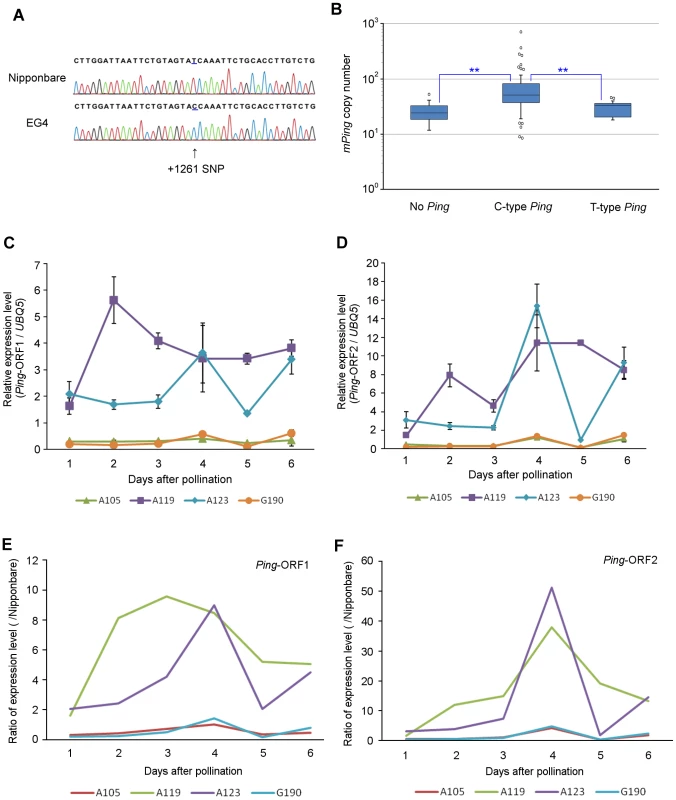
(A) Determination of the SNP sequence in the first intronic region of Ping-ORF1. The arrowhead indicates the position of the SNP. The number indicates the position of the Ping element. Ping harboring +1261C SNP and +1261T are named ‘C-type’ and ‘T-type’ Ping, respectively. (B) Box plots of mPing copy number in AG lines. The top and bottom of the boxes mark the first and third quartiles, respectively. The center line represents the median, and the whiskers show the range of observed values within 1.5 times the interquartile range from the hinges. Values beyond 1.5 times the interquartile range from the nearest hinge are marked by open circles. ‘No Ping’, ‘C-type Ping,’ and ‘T-type Ping’ indicate the groups having no Ping, C-type Ping, and T-type Ping, respectively. Expression of (C) Ping-ORF1 and (D) Ping-ORF2 during embryogenesis in mPing-active strains (A119 and A123) and mPing-inactive strains (A105 and G190). The results are presented as means of three biological replicates. Bars indicate SE. The ratio of (E) Ping-ORF1 and (F) -ORF2 expression level of A105, A119, A123, and G190 to that of Nipponbare. The means in (Fig. 3C) and (Fig. 3D) were used for calculation. In addition to EG4, several Aikoku and Gimbozu landraces (hereafter AG strains) are known to exhibit high mPing activity [21]. We investigated the SNP-type of Ping and the copy number of Ping and mPing in 93 AG strains, and evaluated the effect of C-type Ping on the mPing activity. These 93 AG strains were divided into three groups according to the SNP-type of the Ping allele (Table S4): strains harboring C-type Ping; strains harboring T-type Ping; and strains harboring no Ping. The strains with C-type Ping had more mPing copies than those with T-type Ping or no Ping (Figure 5B, Steel–Dwass test, p<0.01). This implies that the C-type Ping could drive the mPing transposition. We further investigated the expression patterns of Ping-ORF1 and -ORF2 in two mPing-active strains (A119 and A123) and two mPing-inactive strains (A105 and G190) during embryogenesis (from 1 to 6 DAP). A119 and A123 have six and ten copies of C-type Ping, respectively, and both A105 and G190 have one copy of T-type Ping (Table S4). Expression analysis revealed that A105 and G190 kept low expression levels of Ping-ORF1 and -ORF2, whereas A119 and A123 showed high expression levels with a peak around 3 DAP (Figure 5C–F). This indicates that, in EG4, A119, and A123, the developmental stage-specific expression of Ping is controlled by the same factor(s) described in the Discussion.
Discussion
Our final goal was to elucidate how MITEs attain their high copy numbers in the genome. To this end, we chose mPing, which is the only active MITE identified in rice, as a material and analyzed the timing of mPing transposition in the mPing-active strain EG4. Consequently, we successfully found one mechanism of the mPing amplification; mPing most actively transposes during the period from the regionalization of the SAM and radicle to the formation of the first leaf primordium (3 to 5 DAP) by the developmental stage-specific up-regulation of the autonomous element Ping.
The transpositions of TEs are categorized into germinal and somatic types according to the type of cells where the transposition takes place. LORE1a in Lotus japonicus is activated in plants regenerated from de-differentiated cells and transposes in male germ cells by the pollen grain-specific LORE1a transcription, resulting in the asymmetric transposition of LORE1a in the reciprocal crosses between the active and non-active lines [31]. Tag1 in Arabidopsis shows germinal transposition activity in both male and female germ cells. Consequently, the reciprocal crosses show symmetric transposition of Tag1 [32]. These results demonstrate that the transposition activity in reciprocal crosses reflects the tissue specificity of germinal transposition. In this study, reciprocal crosses between EG4 and Nipponbare showed the same mPing transposition activity, which may suggest that mPing in EG4 transposes in both male and female germ cells. However, we obtained only a few de novo endosperm-specific and radicle/shoot-specific insertions in the ontogenical analysis, although we detected a number of de novo shoot-specific and radicle-specific insertions. We therefore concluded that most mPing transposes not in germ cells but in somatic cells after pollination. Somatic transposition that occurs at the late stage of plant development often produces spotted and striped segments in tissues, such as maize seed coat variegation caused by Mutator excision from the bz2 gene [33], [34] and rice leaf color variegation by nDart1excision from the OsClpP5 gene [35]. In animals, somatic transposition is seldom transmitted to the next generation because germ cells are set aside from somatic cells at the early stage of embryogenesis. On the other hand, in plants, germ cells are generated from somatic cells at the reproductive stage. In rice, gametes are generated in the SAM; therefore, somatic transposition that occurred in the SAM can be transmitted to the next generation via gametes. In this study, we revealed that most mPing elements transposed in somatic cells of the embryo during the period from 3 to 5 DAP. Being a class II TE that transposes by a cut-and-paste mechanism, mPing is expected to be eliminated from genomic DNA with a certain frequency. However, a previous report demonstrated that the mPing excision sites would be repaired by utilizing a copy of mPing from either the sister chromatid or from the homologous chromosome [29]. The mPing excision site cannot be repaired if mPing transposes in germ cells, which are haploid. It is therefore considered that the somatic transposition of mPing is an important factor for mPing amplification in the genome.
The autonomous elements Ping and Pong mediate mPing transposition in the rice genome. Many japonica cultivars, including EG4 and Nipponbare, have both Ping and Pong. This study demonstrated that Ping plays a predominant role in mPing transposition in EG4. However, a heterologous expression assay using Arabidopsis and yeast showed that Pong had a higher catalytic capacity for mPing transposition than Ping [22], [23]. Furthermore, mPing transposition was observed under stress conditions in several rice cultivars harboring only Pong [14], [17], [19]. In this study, however, we detected very low expression of Pong through the development of rice plants, indicating that Pong would be epigenetically silenced at the transcriptional level in EG4. In contrast, Ping constitutively expressed in all organs including the SAM and embryo. Nevertheless, mPing could be transposing most actively in the embryo during the period from 3 to 5 DAP. Since the stage-specific up-regulation of Ping was observed during the period of mPing transposition, we hypothesized that the expression level of Ping needed to exceed a certain threshold of mPing transposition.
All mPing-active strains (EG4, A119, and A123) showed higher expression of Ping with a peak around 3 DAP than the mPing-inactive strains (Nipponbare, A105, and G190). Although further experiments are needed to elucidate the mechanism of developmental stage-specific up-regulation of Ping expression, we propose two hypotheses: (1) position - and dosage-effect, and (2) effect of SNP. The details of the hypotheses are as follows.
Position - and dosage-effect
Chromosomal position and copy number of TE often affect the transposition activity. The former is known as ‘position effect’ and the latter as ‘dosage effect’. Eight independent Tam3 copies residing in the Antirrhinum majus genome show different transposition activities from each other [36]. In Arabidopsis, germinal reversion frequency of Tag1 increases in proportion to its copy number [32]. The mPing-inactive strains Nipponbare, A105, and G190 have only one Ping at the same locus, whereas the mPing-active strains EG4, A119, and A123 have respectively seven, six, and ten copies of Ping at different loci except for the Ping-1 locus. Furthermore, the expression pattern of Ping showed slight variation among the mPing-active strains harboring only C-type Ping. These results suggest that the developmental stage-specific up-regulation of Ping expression is probably regulated by the position-effect and/or the dosage-effect.
Effect of SNP
Intronic SNPs are known to cause drastic effects on gene expression. In humans, an intronic SNP in SLC22A4 affects transcriptional efficiency in vitro, owing to an allelic difference in affinity to the transcriptional factor RUNX1 [37]. Furthermore, a SNP located in the intronic enhancer region of the thyroid hormone receptor β gene enhances pituitary cell-specific transcriptional activity [38]. In this study, we demonstrated that a SNP is present in the intronic region of Ping-ORF1, and Ping elements in the AG strains were categorized into either T-type or C-type Ping according to the SNP-type. Since all strains that showed a peak in the expression analysis had only C-type Ping, the intronic SNP might influence the developmental stage-specific up-regulation of Ping expression. T-type Ping was present in 14 AG strains as one copy, and its chromosomal location did not differ among strains. In contrast, the copy number of C-type Ping varied from one to ten, and their chromosomal locations, except for Ping-1, differed from each other. These results indicate that T-type Ping has lost its activity, whereas C-type Ping may be still active in the rice genome. Furthermore, we found that the copy number of mPing was significantly larger in strains harboring C-type Ping than in strains harboring T-type Ping. This strongly supports that C-type SNPs in the intronic region of Ping contribute to the amplification of mPing, presumably by the developmental stage-specific up-regulation of Ping expression.
Since the transposition of TEs often damages the host genome, TEs with high transposition activity are targeted by the silencing mechanisms. Nevertheless, MITEs amplify to very high copy numbers not only in plant genomes but also in animal genomes. Very little is known about how MITEs attain their high copy numbers by escaping the silencing mechanism. The transposition of mPing is transiently induced by various stresses [14]–[18], indicating that the activity of mPing is suppressed by the silencing mechanisms in many cultivars. Thus, mPing must overcome the silencing mechanism in order to maintain the transposition activity under natural growth conditions. Our results revealed that mPing in EG4 was mobilized by the sufficient supply of Ping transcripts produced only during the period of mPing transposition. This stage-specific activation is thought to be a strategy of the mPing family to amplify mPing by escaping from the silencing mechanism of the host genome. Since no active MITEs other than mPing so far have been identified, it is very difficult to elucidate if the other MITEs also attain their high copy numbers in the same way as mPing amplifies. Given that the other active MITEs are identified, however, our study will help to understand their amplification mechanisms. Our previous study documented the generation of new regulatory networks by a subset of mPing insertions that render adjacent genes stress inducible [13]. In addition to mPing, other MITEs also contribute to gene and genome evolution via providing new promoter regulatory sequences, transcriptional termination elements, and new alternative exons [39], suggesting that the amplification of MITEs causes gene and genome evolution. Our results provide clues to further understand not only the amplification mechanism of MITEs but also the co-evolution of MITEs and the host genome.
Materials and Methods
Plant materials and sampling
EG4 (cultivar Gimbozu), Nipponbare, and 94 Aikoku/Gimbozu landraces were used in this study (Table S4). Aikoku/Gimbozu landraces were provided from the GenBank project of the National Institute of Agrobiological Science, Ibaraki, Japan. Reciprocal crosses between EG4 and Nipponbare were made in a green house. Before pollination, all anthers were removed from the flowers of maternal plants. The pollinated flowers were covered with protective bags to prevent outcrossing until harvest. After harvesting, success of crosses was checked with the molecular markers. For ontogenical analysis, eight progenies of EG4 (S1) derived from a single parental plant (S0) were grown in a greenhouse, and all S2 seeds were harvested. For S1 plants, each seed was cut into two halves, and the half including the embryo was germinated and the other was sampled. After germination, the radicle and the 1st, 2nd, 3rd, 4th, and 5th leaf blades were sampled. The second leaf was collected from S0 and S2 plants. For estimation of Ping and mPing copy numbers, eight bulked plants were sampled. For RNA extraction, ovaries before pollination and ovaries at 1, 2, 3, 4, 5, and 6 DAP were collected. All samples were immediately frozen in liquid nitrogen and stored at −80°C until use.
DNA extraction and transposon display
DNA extraction and transposon display was performed following a published protocol [30]. For DNA extraction from endosperm, we used GM quicker 2 (Nippon Gene).
Locus-specific PCR
Sequencing of mPing-flanking fragments excised from transposon display gels and primer design were performed following a published protocol [30]. The genomic locations of the mPing insertion sites were forecasted by a BLAST search in the Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp/) [40], [41] using mPing flanking sequences as queries. To prepare enough templates for PCR, whole genome amplification was performed using an illustra GenomiPhi V2 Kit (GE Healthcare). mPing excision was detected by PCR with mPing-sequence characterized amplified region (SCAR) markers [29]. PCR was performed in 10-µl reaction volumes containing 10 ng of the template DNA, 5 µl of GoTaq Green Master mix (Promega), 5% DMSO, and 0.25 µM of each primer (Table S3). PCR conditions were as follows: 94°C for 3 min; 40 cycles of 98°C for 10 s, 57°C for 30 s, and 72°C for 45 s; and 72°C for 5 min. To detect the presence of Ping-N, -1, -2, -3, -4, -5, -6, and -7, eight Ping-SCAR markers were used. The genomic locations of the Ping insertion sites were referred from a previous report [42]. For detection of the Ping-1 allele, PCR was performed in 10-µl reaction volumes containing 10 ng of template DNA, 0.2 U of KOD FX Neo (Toyobo), 1×PCR buffer for KOD FX Neo (Toyobo), and 0.2 µM of each primer (Table S5). PCR conditions were as follows: 94°C for 3 min; 35 cycles of 98°C for 10 s, 60°C for 30 s, and 68°C for 90 s; and 72°C for 1 min. For detection of Ping-N, -2, -3, -4, -5, -6, and -7 alleles, PCR was performed in 10-µl reaction volumes containing 10 ng of template DNA, 5 µl of GoTaq Green Master mix (Promega), 5% DMSO, and 0.25 µM of each primer (Table S5). PCR conditions were as follows: 94°C for 3 min; 35 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 45 s; and 72°C for 1 min.
RNA isolation and expression analysis
Total RNA was isolated using TriPure isolation reagent (Roche) and digested using RNase-free DNase (TaKaRa). First strand cDNA was synthesized using a Transcriptor first strand cDNA synthesis kit (Roche). For reverse transcription PCR, PCR was performed in 10 µl reaction volumes containing cDNA generated from 4 ng total RNA, 0.2 U of KOD FX Neo (Toyobo), 1×PCR buffer for KOD FX Neo (Toyobo), and 0.5 µM of each primer. PCR conditions were as follows: 94°C for 3 min; 35 or 45 cycles of 98°C for 10 s, 60°C for 10 s, and 68°C for 10 s. Relative quantification of Ping-ORF1 and Ping-ORF2 were calculated by the 2−ΔΔCT method [43] using Light cycler 1.5 (Roche). The UBQ5 gene was used as the calibrator gene. The thermal profile consisted of 10 min at 95°C; and 45 cycles of 4 s at 95°C, 10 s at 60°C, and 1 s at 72°C. Amplification data were collected at the end of each extension step. The primer pairs used in this study are listed in Table S6.
Paraffin sectioning and in situ hybridization
Plant samples were fixed with 4% (w/v) paraformaldehyde and 1% Triton X in 0.1M sodium phosphate buffer for 48 h at 4°C. They were then dehydrated in a graded ethanol series, substituted with 1-butanol, and embedded in Paraplast Plus. The samples were sectioned at 8-µm thickness using a rotary microtome. Fragments of Ping-ORF1 (1091 bp) and Ping-ORF2 (1368 bp) were cloned into pBlueScript SK+ (Stratagene) and sequenced. For digoxigenin-labeled antisense/sense RNA probe synthesis, in vitro transcription was performed using T7 RNA polymerase and T3 RNA polymerase. In situ hybridization and immunological detection with alkaline phosphatase were performed according to Kouchi and Hata [44].
SNP detection
PCR was performed in 10-µl reaction volumes containing 10 ng of template of DNA, 5 µl of GoTaq Green Master mix (Promega), 5% DMSO, and 0.25 µM of each primer. PCR conditions were as follows: 94°C for 3 min; 35 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s; and 72°C for 1 min. PCR primers used in this study are listed in Table S6. Because the original sequence contained an Afa I restriction site, one mutation was introduced into the reverse primer. The 5-µl PCR products were mixed with 5 µl restriction mixture containing 1 U Afa I (TaKaRa), 33 mM Tris-acetate (pH 7.9), 10 mM Mg-acetate, 0.5 mM Dithiothreitol, 66 mM K-acetate, and 0.01% (w/v) bovine serum albumin. After 16 h incubation at 37°C, DNA gel electrophoresis was performed. PCR products (502 bp) including +1261T SNP were not digested, whereas PCR products including +1261C SNP were digested into two fragments (352 bp and 150 bp).
Estimation of Ping and mPing copy number
To determine the copy number of Ping by Southern blot analysis, genomic DNA samples were digested with Eco RI restriction enzyme. These samples were loaded onto an agarose gel, separated by electrophoresis, blotted onto a nylon membrane, and probed with the Ping fragment. The mPing copy number was determined by real-time quantitative PCR as described previously [45] with little modification. Quantitative PCR was performed using the LightCycler 480 system (Roche). PCR was performed in 20 µl reaction volumes containing 5 µl genomic DNA (0.4 ng/µl), 1×LightCycler 480 SYBR Green I Master mix (Roche), and 0.5 µM of each primer. Specificity of the amplified PCR product was assessed by performing a melting curve analysis on the LightCycler 480 system.
Supporting Information
Zdroje
1. SchnablePS, WareD, FultonRS, SteinJC, WeiF, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326 : 1112–1115 doi: 10.1126/science.1178534
2. WickerT, ZimmermannW, PerovicD, PatersonAH, GanalM, et al. (2005) A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv-eIF4E locus: recombination, rearrangements and repeats. Plant J 41 : 184–194 doi: 10.1111/j.1365-313X.2004.02285.x
3. International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 : 793–800 doi: 10.1038/nature03895
4. Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 : 796–815 doi: 10.1038/35048692
5. FedoroffNV (2012) Transposable Elements, Epigenetics, and Genome Evolution. Science 338 : 758–767 doi: 10.1126/science.338.6108.758
6. CowleyM, OakeyRJ (2013) Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet 9: e1003234 doi: 10.1371/journal.pgen.1003234
7. BureauTE, WesslerSR (1992) Tourist: A Large Family of Small Inverted Repeat Elements Frequently Associated with Maize Genes. Plant Cell 4 : 1283–1294.
8. BureauTE, WesslerSR (1994) Stowaway: A New Family of Inverted Repeat Elements Associated with the Genes of both Monocotyledonous and Dicotyledonous Plants. Plant Cell 6 : 907–916 doi: 10.1105/tpc.6.6.907
9. WesslerSR, BureauTE, WhiteSE (1995) LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev 5 : 814–821 doi:10.1016/0959-437x(95)80016-x
10. OkiN, YanoK, OkumotoY, TsukiyamaT, TeraishiM, et al. (2008) A genome-wide view of miniature inverted-repeat transposable elements (MITEs) in rice, Oryza sativa ssp. japonica. Genes Genet Syst 83 : 321–329 doi: 10.1266/ggs.83.321
11. ChenJ, LuC, ZhangY, KuangH (2012) Miniature inverted-repeat transposable elements (MITEs) in rice were originated and amplified predominantly after the divergence of Oryza and Brachypodium and contributed considerable diversity to the species. Mob Genet Elements 2 : 127–132 doi: 10.4161/mge.20773
12. HanY, QinS, WesslerSR (2013) Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genomics 14 : 71 doi:10.1186/1471-2164-14-71
13. NaitoK, ZhangF, TsukiyamaT, SaitoH, HancockCN, et al. (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461 : 1130–1134 doi:10.1038/nature08479
14. JiangN, BaoZ, ZhangX, HirochikaH, EddySR, et al. (2003) An active DNA transposon family in rice. Nature 421 : 163–167 doi:10.1038/nature01214
15. KikuchiK, TerauchiK, WadaM, HiranoH-Y (2003) The plant MITE mPing is mobilized in anther culture. Nature 421 : 167–170 doi:10.1038/nature01218
16. NakazakiT, OkumotoY, HoribataA, YamahiraS, TeraishiM, et al. (2003) Mobilization of a transposon in the rice genome. Nature 421 : 170–172 doi:10.1038/nature01219
17. LinX, LongL, ShanX, ZhangS, ShenS, et al. (2006) In planta mobilization of mPing and its putative autonomous element Pong in rice by hydrostatic pressurization. J Exp Bot 57 : 2313–2323 doi: 10.1093/jxb/erj203
18. YangX, YuY, JiangL, LinX, ZhangC, et al. (2012) Changes in DNA methylation and transgenerational mobilization of a transposable element (mPing) by the Topoisomerase II inhibitor, Etoposide, in rice. BMC Plant Biol 12 : 48 doi:10.1186/1471-2229-12-48
19. ShanX, LiuZ, DongZ, WangY, ChenY, et al. (2005) Mobilization of the Active MITE Transposons mPing and Pong in Rice by Introgression from Wild Rice (Zizania latifolia Griseb.). Mol Biol Evol 22 : 976–990 doi:10.1093/molbev/msi082
20. YasudaK, TsukiyamaT, KarkiS, OkumotoY, TeraishiM, et al. (2012) Mobilization of the active transposon mPing in interspecific hybrid rice between Oryza sativa and O. glaberrima. Euphytica 192 : 17–24 doi: 10.1007/s10681-012-0810-1
21. NaitoK, ChoE, YangG, CampbellMA, YanoK, et al. (2006) Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA 103 : 17620–17625 doi: 10.1073/pnas.0605421103
22. YangG, ZhangF, HancockCN, WesslerSR (2007) Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci USA 104 : 10962–10967 doi: 10.1073/pnas.0702080104
23. HancockCN, ZhangF, WesslerSR (2010) Transposition of the Tourist-MITE mPing in yeast: an assay that retains key features of catalysis by the class 2 PIF/Harbinger superfamily. Mob DNA 1 : 5 doi:10.1186/1759-8753-1-5
24. HarenL, Ton-HoangB, ChandlerM (1999) INTEGRATING DNA: Transposases and Retroviral Integrases. Annu Rev Microbiol 53 : 245–281 doi: 10.1146/annurev.micro.53.1.245
25. SinzelleL, KapitonovVV, GrzelaDP, JurschT, JurkaJ, et al. (2008) Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc Natl Acad Sci USA 105 : 4715–4720 doi: 10.1073/pnas.0707746105
26. SlotkinRK, VaughnM, BorgesF, TanurdzićM, BeckerJD, et al. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136 : 461–472 doi: 10.1016/j.cell.2008.12.038
27. HamamuraY, SaitoC, AwaiC, KuriharaD, MiyawakiA, et al. (2011) Live-cell imaging reveals the dynamics of two sperm cells during double fertilization in Arabidopsis thaliana. Curr Biol 21 : 497–502 doi: 10.1016/j.cub.2011.02.013
28. ItohJ-I, NonomuraK-I, IkedaK, YamakiS, InukaiY, et al. (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46 : 23–47 doi: 10.1093/pcp/pci501
29. MondenY, NaitoK, OkumotoY, SaitoH, OkiN, et al. (2009) High potential of a transposon mPing as a marker system in japonica × japonica cross in rice. DNA Res 16 : 131–140 doi: 10.1093/dnares/dsp004
30. TsukiyamaT, TeramotoS, YasudaK, HoribataA, MoriN, et al. (2013) Loss-of-Function of a Ubiquitin-Related Modifier Promotes the Mobilization of the Active MITE mPing. Mol Plant 6 : 790–801 doi: 10.1093/mp/sst042
31. FukaiE, UmeharaY, SatoS, EndoM, KouchiH, et al. (2010) Derepression of the Plant Chromovirus LORE1 Induces Germline Transposition in Regenerated Plants. PLoS Genet 6 doi:10.1371/journal.pgen.1000868
32. LiuD, CrawfordNM (1998) Characterization of the germinal and somatic activity of the Arabidopsis transposable element Tag1. Genetics 148 : 445–456.
33. LevyAA, BrittAB, LuehrsenKR, ChandlerVL, WarrenC, et al. (1989) Developmental and genetic aspects of Mutator excision in maize. Dev Genet 10 : 520–531 doi: 10.1002/dvg.1020100611
34. LevyA, WalbotV (1990) Regulation of the timing of transposable element excision during maize development. Science 248 : 1534–1537 doi: 10.1126/science.2163107
35. TsuganeK, MaekawaM, TakagiK, TakaharaH, QianQ, et al. (2006) An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. Plant J 45 : 46–57 doi: 10.1111/j.1365-313X.2005.02600.x
36. KitamuraK, HashidaSN, MikamiT, KishimaY (2001) Position effect of the excision frequency of the Antirrhinum transposon Tam3: implications for the degree of position-dependent methylation in the ends of the element. Plant Mol Biol 47 : 475–490 doi: 10.1023/A:1011892003996
37. TokuhiroS, YamadaR, ChangX, SuzukiA, KochiY, et al. (2003) An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet 35 : 341–348 doi:10.1038/ng1267
38. AlberobelloAT, CongedoV, LiuH, CochranC, SkarulisMC, et al. (2011) An intronic SNP in the thyroid hormone receptor β gene is associated with pituitary cell-specific over-expression of a mutant thyroid hormone receptor β2 (R338W) in the index case of pituitary-selective resistance to thyroid hormone. J Transl Med 9 : 144 doi: 10.1186/1479-5876-9-144
39. Guermonprez H, Henaff E, Cifuentes M, Casacuberta JM (2012) MITEs, Miniature Elements with a Major Role in Plant Genome Evolution. In: Grandbastien M-A, Casacuberta JM, editors. Plant Transposable Elements: Impact on Genome Structure and Function. Berlin, Heidelberg: Springer. pp. 113–124. doi: 10.1007/978-3-642-31842-9
40. KawaharaY, de la BastideM, HamiltonJP, KanamoriH, McCombieWR, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6 : 4.
41. SakaiH, LeeSS, TanakaT, NumaH, KimJ, et al. (2013) Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol 54: e6 doi: 10.1093/pcp/pcs183
42. OkiN, OkumotoY, TsukiyamaT, NaitoK, NakazakiT, et al. (2007) A Novel Transposon Pyong in the japonica Rice Variety Gimbozu. J Crop Res 52 : 39–43.
43. LivakKJ, SchmittgenTD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods 25 : 402–408 doi: 10.1006/meth.2001.1262
44. KouchiH, HataS (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238 : 106–119 doi: 10.1007/BF00279537
45. BaruchO, KashkushK (2012) Analysis of copy-number variation, insertional polymorphism, and methylation status of the tiniest class I (TRIM) and class II (MITE) transposable element families in various rice strains. Plant Cell Rep 31 : 885–893 doi: 10.1007/s00299-011-1209-5s
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Inflammation: Gone with Translation
- Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1
- Caspase Inhibition in Select Olfactory Neurons Restores Innate Attraction Behavior in Aged
- Accurate, Model-Based Tuning of Synthetic Gene Expression Using Introns in
- A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in
- Ancient DNA Analysis of 8000 B.C. Near Eastern Farmers Supports an Early Neolithic Pioneer Maritime Colonization of Mainland Europe through Cyprus and the Aegean Islands
- The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy
- Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
- Netrins and Wnts Function Redundantly to Regulate Antero-Posterior and Dorso-Ventral Guidance in
- Coordination of Wing and Whole-Body Development at Developmental Milestones Ensures Robustness against Environmental and Physiological Perturbations
- Phenotypic Dissection of Bone Mineral Density Reveals Skeletal Site Specificity and Facilitates the Identification of Novel Loci in the Genetic Regulation of Bone Mass Attainment
- Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of -Factors in Two Independent Origins of C Photosynthesis
- Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity
- Translational Regulation of Specific mRNAs Controls Feedback Inhibition and Survival during Macrophage Activation
- Rosa26-GFP Direct Repeat (RaDR-GFP) Mice Reveal Tissue- and Age-Dependence of Homologous Recombination in Mammals
- Abnormal Type I Collagen Post-translational Modification and Crosslinking in a Cyclophilin B KO Mouse Model of Recessive Osteogenesis Imperfecta
- : Clonal Reinforcement Drives Evolution of a Simple Microbial Community
- Reviving the Dead: History and Reactivation of an Extinct L1
- Defective iA37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA
- Early Back-to-Africa Migration into the Horn of Africa
- Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants
- Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia
- Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
- Palaeosymbiosis Revealed by Genomic Fossils of in a Strongyloidean Nematode
- Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- Genetic Background Drives Transcriptional Variation in Human Induced Pluripotent Stem Cells
- Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes
- N-WASP Is Required for Structural Integrity of the Blood-Testis Barrier
- The Transcription Factor TFII-I Promotes DNA Translesion Synthesis and Genomic Stability
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
- Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats
- ATRA-Induced Cellular Differentiation and CD38 Expression Inhibits Acquisition of BCR-ABL Mutations for CML Acquired Resistance
- The EJC Binding and Dissociating Activity of PYM Is Regulated in
- JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling
- Mouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
- Rasa3 Controls Megakaryocyte Rap1 Activation, Integrin Signaling and Differentiation into Proplatelet
- Transcriptional Control of Steroid Biosynthesis Genes in the Prothoracic Gland by Ventral Veins Lacking and Knirps
- Souffle/Spastizin Controls Secretory Vesicle Maturation during Zebrafish Oogenesis
- The POU Factor Ventral Veins Lacking/Drifter Directs the Timing of Metamorphosis through Ecdysteroid and Juvenile Hormone Signaling
- The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses
- Sequence of a Complete Chicken BG Haplotype Shows Dynamic Expansion and Contraction of Two Gene Lineages with Particular Expression Patterns
- Background Selection as Baseline for Nucleotide Variation across the Genome
- CPF-Associated Phosphatase Activity Opposes Condensin-Mediated Chromosome Condensation
- The Effects of Codon Context on Translation Speed
- Glycogen Synthase Kinase (GSK) 3β Phosphorylates and Protects Nuclear Myosin 1c from Proteasome-Mediated Degradation to Activate rDNA Transcription in Early G1 Cells
- Regulation of Gene Expression in Autoimmune Disease Loci and the Genetic Basis of Proliferation in CD4 Effector Memory T Cells
- Muscle Structure Influences Utrophin Expression in Mice
- BLMP-1/Blimp-1 Regulates the Spatiotemporal Cell Migration Pattern in
- Identification of Late Larval Stage Developmental Checkpoints in Regulated by Insulin/IGF and Steroid Hormone Signaling Pathways
- Transport of Magnesium by a Bacterial Nramp-Related Gene
- Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
- The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
- The Rim15-Endosulfine-PP2A Signalling Module Regulates Entry into Gametogenesis and Quiescence Distinct Mechanisms in Budding Yeast
- Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
- Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions
- Positive Feedback of Expression Ensures Irreversible Meiotic Commitment in Budding Yeast
- Hecate/Grip2a Acts to Reorganize the Cytoskeleton in the Symmetry-Breaking Event of Embryonic Axis Induction
- Regulatory Mechanisms That Prevent Re-initiation of DNA Replication Can Be Locally Modulated at Origins by Nearby Sequence Elements
- Speciation and Introgression between and
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Early Back-to-Africa Migration into the Horn of Africa
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



