-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
Plants, as sessile and photosynthetic organisms, have to constantly adjust their growth and development in response to the environment. While light and temperature are recognized as the most prominent environmental factors modulating plant photosynthetic metabolism, how the seasonal and daily adjustments are achieved is not understood. Global climate alterations will bring together the combination of light and temperature changes and will require an understanding of signal convergence. If we are to mitigate the impact of variable weather patterns on agriculture, it is critical to advance our understanding of the basis of plant responses to environmental variations. In our study we show that the antagonistic activity of key plant transcription factors involved in phytochrome red light photoreceptors signaling (PIFs and HY5) optimize photosynthetic pigment production in response to environmental cues. These light and temperature responsive transcription factors operate in cooperation with the circadian clock to regulate photosynthetic pigment production through a common gene promoter element.
Published in the journal: The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription. PLoS Genet 10(6): e32767. doi:10.1371/journal.pgen.1004416
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004416Summary
Plants, as sessile and photosynthetic organisms, have to constantly adjust their growth and development in response to the environment. While light and temperature are recognized as the most prominent environmental factors modulating plant photosynthetic metabolism, how the seasonal and daily adjustments are achieved is not understood. Global climate alterations will bring together the combination of light and temperature changes and will require an understanding of signal convergence. If we are to mitigate the impact of variable weather patterns on agriculture, it is critical to advance our understanding of the basis of plant responses to environmental variations. In our study we show that the antagonistic activity of key plant transcription factors involved in phytochrome red light photoreceptors signaling (PIFs and HY5) optimize photosynthetic pigment production in response to environmental cues. These light and temperature responsive transcription factors operate in cooperation with the circadian clock to regulate photosynthetic pigment production through a common gene promoter element.
Introduction
Light and temperature are prominent cues that signify seasonal and climatic change, as well as the phase of the daily light/dark cycle. The ability to sense and integrate these external signals is essential for plant life cycle progression and ultimately, survival. Central to this process is organization of photosynthetic machinery that captures energy from light. This vital process provides energy to fix carbon into organic matter that is essential for plant growth and development.
Light quality and quantity is sensed by a suite of specialized photoreceptors. An established class of these receptors are the red (R) and far-red (FR) light absorbing phytochromes that have an important role driving the switch from skotomorphogenic to photoautotrophic growth during seedling de-etiolation (reviewed by [1]). The phytochromes exist in two interconvertible forms. When the inactive-Pr form absorbs R light, it converts to the active, Pfr form and relocates from the cytoplasm into the nucleus. Once in the nucleus, phytochromes induce major changes in gene expression that modify the developmental program [2]. Amongst the phytochrome-mediated changes during this critical time is the promotion of the photosynthetic apparatus assembly, and the production of the photosynthetic pigments, chlorophylls and carotenoids [3]–[5] While chlorophyll production is required for photoautotrophic growth, excessive accumulation of the chlorophyll precursor protochlorophyllide can lead to harmful photo-oxidative damage [6]–[8]. It is therefore critical that this process is stringently regulated. Carotenoids are integral accessory pigments in the light absorbing antenna. In the greening process, carotenoids play an essential photoprotective role minimizing the potentially damaging effects of light on the photosynthetic machinery [9]–[11]. Therefore, to maximize light capture but curtail the potentially damaging effects of light over the emerging photosynthetic machinery, the production of carotenoids and chlorophylls has to take place in a tightly controlled and interdependent manner. This might be achieved in part by the circadian gating of isoprenoid pathway genes, that produce both carotenoids and chlorophylls, to ensure coordinated pathway gene expression [12]. However, we do not currently understand how this regulation is influenced by either the regular daily or unexpected variations in external light and temperature.
Earlier studies have shown that bHLH-Phytochrome Interacting Factors (PIFs) not only prevent the over-accumulation of protochlorophyllide in etiolated seedlings [8] but have a broader role in the seedling as repressors of photomorphogenic development [1], [13]. In accordance, a PIF quadruple mutant (pif1-1;pif3-3;pif4-2;pif5-3) (pifQ) exhibits a constitutive photomorphogenic phenotype in darkness and a global gene expression pattern that resembles R light-grown seedlings [1], [14]–[16]. Concurring with these observations, we previously showed that PIFs repress carotenoid accumulation by down-regulating the expression of the Arabidopsis thaliana gene encoding PHYTOENE SYNTHASE (PSY), the main rate-determining enzyme in the carotenoid pathway [17]. PIF1, a potent PSY gene repressor, effects control through direct binding to at least one of the the two G-box (CACGTG) elements found in the PSY promoter. Indeed, PIFs have been shown to preferentially target G-box motifs in a range of genes [8], [18]–[21]. Suppression of PSY transcription is strongest in etiolated seedlings when PIF are abundant, while exposure to light leads to a depletion in PIF levels and concomitant de-repression in PSY expression [17].
PIFs are strongly regulated by light and therefore their activity is conditional on the external light environment. In de-etiolated seedlings growing in light/dark cycles phytochromes control the degradation and re-accumulation of PIF proteins (PIF3, PIF4 and PIF5) generating a daily alternation in the abundance of these transcription factors [22]–[24]. For PIF4 and PIF5, additional temporal control is delivered by the circadian clock that regulates the phase of expression [23], [25]–[27]. As these PIFs are potent regulators of cell expansion in the seedling hypocotyl, this translates to a diurnal rhythm in hypocotyl elongation rate [23], [25]). Recent studies have shown that PIFs also participate in temperature signalling. PIF4, and to a lesser extent PIF5, are required for the promotion of growth in warmer conditions [28], [29]. Elevated temperatures promote the accumulation of phosphorylated PIF4 and enhance PIF4 binding at target promoters [30]–[32]. Indeed, PIF4 occupation at the FLOWERING TIME (FT) promoter increases with the eviction of H2A.Z nucleosomes by temperature [32].
For a broad range of responses PIFs act antagonistically with the bZIP transcription factor, HY5 [1], [33], [34]. HY5 drives photomorphogenic development by activating genes that promote photosynthetic machinery assembly, photopigment production, chloroplast development, and seedling cotyledon expansion [21], [35], while PIF suppression of these responses is required to maintain skotomorphogenesis [1], [33]. HY5 has been shown to regulate gene expression directly through interaction with “ACE” motifs (ACGT containing elements) that include the Z-box (ATACGTGT), C-boxes (GTCANN), G-boxes, and hybrid C/G (G) and C/A boxes [35]–[38]. Like PIFs, HY5 protein levels are light regulated – but in contrast to PIFs that are light labile, HY5 protein is stabilized by light [1], [39], [40]. Multiple photoreceptors participate in the accumulation of HY5 protein in the light, in part by reducing the nuclear levels of CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase that targets HY5 for proteasome mediated degradation in the dark [39]–[41]. The differential regulation of HY5 and PIF proteins by light means that they exhibit opposing diurnal expression patterns [23], [24], [35], [42].
Whereas PIF4 mediates warm temperature signalling, HY5 has been shown to have a role in cold temperature responses [43]. Here HY5 has been implicated in cold acclimation where it promotes anthocyanin accumulation, ROS production, and regulates a large swathe (∼10%) of cold inducible genes [43]–[45]. HY5 abundance is regulated by temperature. A shift in temperature from 20°C to 4°C elevates HY5 transcript levels and stabilises the HY5 protein through nuclear depletion of COP1 [43].
This study examines the impact of light and temperature on the control of photopigment production by HY5 and PIFs. ChIP, EMSA and transcript analysis established that HY5 and PIF1/PIF4 impart antagonistic regulation to common gene targets through direct binding to the same G-box cis element. This dynamic activation-suppression transcriptional module does not act alone, but with the circadian clock to modify the level of rhythmic gene expression. However, abrupt changes in either light or temperature adjusts the equilibrium of HY5 and PIF bound to target promoters altering the transcriptional response. In this way the expression of photopigment biosynthesis genes can be revised by external signals.
Results
HY5 is required for light induction of PSY expression, and photopigment synthesis
Our earlier work showed that PIFs restrict the accumulation of carotenoids during de-etiolation, in part by negatively regulating PSY, a rate limiting step in the carotenoid biosynthetic pathway [17]. We demonstrated that sequential removal of PIFs leads to incremental rises in PSY mRNA levels, illustrating that PIFs act redundantly to suppress PSY expression. This data also suggested the existence of a PSY activator that becomes more effective with PIF depletion. As HY5 is known to act in opposition to PIFs for a number of photomorphogenic responses [7], [14], [37], [46], we tested whether HY5 fulfilled this role by quantifying PSY gene expression in the hy5 mutant (hy5-215) during red light-triggered de-etiolation (Figure 1A). Our data confirmed that hy5 perturbs PSY accumulation following exposure to light. Likewise, red light induction of carotenoid and chlorophyll levels is severely attenuated in the hy5 mutant (Figure 1B–C). This response contrasts with that in pif1 and pifQ mutants, where PSY mRNA and photopigment levels are significantly elevated in dark and red light illuminated seedlings [17]. Because mutants defective in the HY5 homologue HYH did not exhibit photopigment accumulation defects under our conditions, we therefore focussed our analysis on HY5 (Figure S1). Our data illustrate that HY5 and PIFs have opposing roles in the regulation of PSY expression and carotenoid and chlorophyll biosynthesis during seedling de-etiolation.
Fig. 1. HY5 is a positive regulator of carotenoid and chlorophyll biosynthesis and controls PSY gene expression by promoter binding. 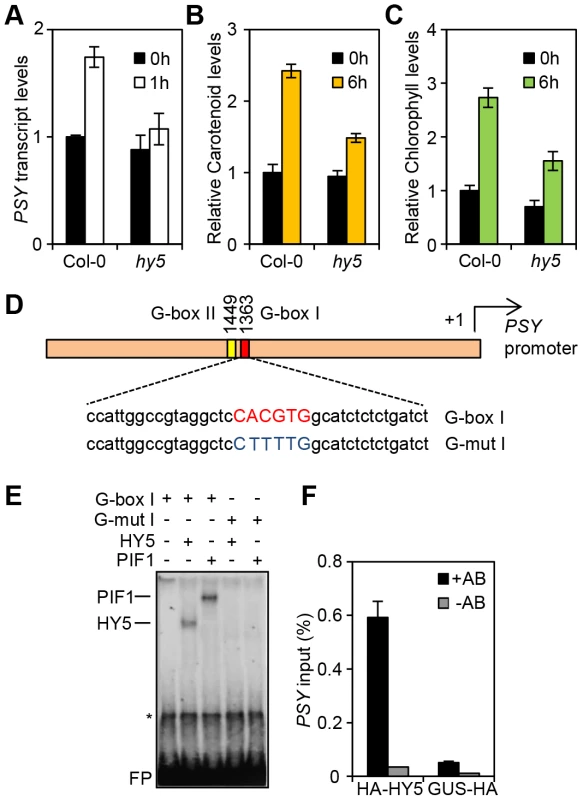
(A) PSY expression in 3-day old Col-0 and hy5-215 seedlings grown in the dark (0 h, black columns) or after 1 h Red light illumination (1 h, white columns). Expression was measured in biological triplicates by qPCR. PSY expression levels were normalized against APT1 levels and expressed relative to Col-0 dark (0 h). Error bars represent± Standard Error (SE). (B–C) Carotenoid and chlorophyll content in 3-day old Col-0 and hy5-215 seedlings. Photopigment content was measured in the Dark (0 h) or after 6 h R light illumination (40 µmol m−2 s−1). Error bars represent ±SE of biological triplicate sets. Levels (µg/g fresh weight) are expressed relative to Col-0 Dark sample. (D) Schematic representation of the PSY promoter with the location of the two G-box motifs. Under the diagram, the sequence of the G-box I probe and the mutant G-box I probe (G-mutI) labelled with 32P for Electrophoretic Mobility Shift Assays (EMSA) is indicated. (E) HY5 and PIF1 bind in-vitro to the G-box I in the PSY promoter in EMSA assays. For the assay, radiolabelled probes were incubated with TnT produced PIF1 and HY5 proteins. G-box I or G-mut I probes were used as indicated. Asterisk (*) corresponds to a TnT non-specific band and FP stands for free probe. (F) HY5 binds in-vivo in Chromatin immunoprecipitation (ChIP) assays to the G-box region in the PSY promoter. ChIP assays were carried out using a 35S:: HA-HY5 line with antibodies against the HA-tag. A 35S::GUS-HA line was used as a control (+HA antibody samples). A second set of samples were processed without antibody and used as negative controls (-HA antibody). Immunoprecipitated DNA was amplified by qPCR using primers against the G-box region in the PSY promoter (see Table S1). For the assay, plants were grown for 1 week at 22°C in white light (80 µmol m−2 s−1) and then transferred to 1 week growth under Red light (40 µmol m−2 s−1). Error bars indicate ± SE of biological triplicates. HY5 binds to the PSY promoter through a G-box element
Since HY5 and PIFs confer antagonistic regulation to PSY, we wanted to establish whether this was mediated at the PSY promoter. Previous chromatin immunoprecipitation (ChIP)-chip analysis indicated that HY5 binds to promoter regions that carry different types of the generic ACE motif (CACGT) including CA hybrid (GACGTA), CG hybrid (GACGTG), Z-boxes (ATACTGTGT), as well as G-boxes (CACGTG), the main recognition motif for PIFs [17], [21], [35], [37]. The PSY promoter contains two G-boxes (I and II), separated by 80 base pairs, but not CG, CA or Z type motifs (Figure S2 and Figure 1D). We therefore reasoned that the opposing actions of HY5 and PIF1 on PSY expression could be mediated through the same regulatory G-box element. This we tested using an in-vitro electrophoretic mobility shift assay (EMSA). Previously we showed that PIF1 did not bind in-vitro to G-box II [17], therefore our analysis is focussed exclusively on G-box I. Consistent with our previous results we detected PIF1 binding to a template encompassing the PSY G-box I (Figure 1D, E) [17]. In the same assay, we were able to demonstrate that like PIF1, HY5 bound to a labelled probe containing this G-box, but not a mutated version (G-mut I) where the G-box had been disrupted (Figure 1E).
To establish if we could verify our findings in-vivo we conducted a ChIP assay using the 35S::HA-HY5 lines used in a previous study that identified genome wide the targets of HY5 regulation [35]. After immunoprecipitation of protein-DNA complexes using the HA antibody, enriched DNA sequences were amplified by qPCR using primers bordering the PSY promoter G-box region. As shown in Figure 1F, in contrast to the 35S::GUS-HA and the no-antibody controls, we detected significant enrichment of PSY promoter sequences containing G-box I in 35S::HA-HY5 samples. These results together with our in vitro analysis suggest that HY5 regulates PSY transcription through direct binding to the G-box region of the promoter.
HY5 enhances carotenoid and chlorophyll pigment synthesis at cooler ambient temperatures
As PIFs (e.g. PIF4 and PIF5) and HY5 have been implicated in temperature signalling we wanted to assess whether this action extended to carotenoid and chlorophyll regulation [28]–[31], [43]–[45], [47]–[51]. We found that in both etiolated and red light exposed wild type seedlings carotenoid and chlorophyll levels rose incrementally with temperature (17°C, 22°C and 27°C) (Figure 2A–C). Compared to the wild type, the pifQ mutant had constitutively elevated carotenoid and chlorophyll levels under all conditions, but still remained responsive to temperature (Figure 2A, B). This suggests that while specific PIFs (e.g. PIF4) may operate at warmer temperatures, the collective action of PIFs maintain control of carotenoid and chlorophyll levels over a temperature range [28], [30]–[32], [47], [49] In contrast, hy5 suppressed red light-induction of carotenoid and chlorophyll levels (Figure 2A–C). While the effects of hy5 were apparent across temperatures, its impact was most marked in the cooler 17°C conditions.
Fig. 2. Photosynthetic pigment accumulation is temperature sensitive and dependent on HY5 and the PIFs. 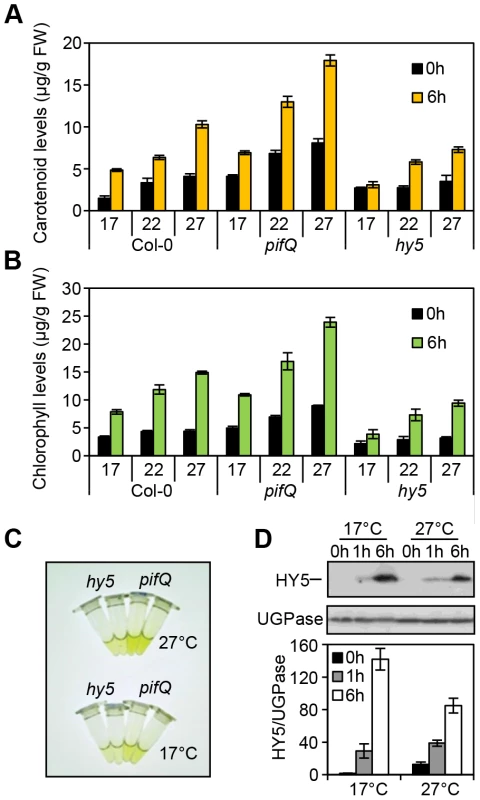
(A–B) Carotenoid and chlorophyll accumulation in Col-0, hy5-215 and pifQ (pif1-1 pif3-3 pif4-2 pif5-3) 3 day-old seedlings grown at different temperatures (17, 22 and 27°C) in the dark (black columns) or after 6 h Red light illumination (40 µmol m−2 s−1) (yellow or green columns). For measurements seedlings were kept for two days at 22°C and 1 day at the indicated temperature in darkness. On day 3 they were subjected to red light treatment (for 6 h). The control set was kept in darkness (0 h time point). Graphs represent the results for biological triplicates sets. Error bars indicate ± SE. (C) hy5-215 and pifQ accumulate different levels of pigments. Illustrative picture of the chlorophyll accumulation response in hy5-215 compared to the pifQ at 17°C and 27°C. Plant material and chlorophyll extraction was conducted as indicated in (A–B). (D) HY5 protein accumulates to moderately higher levels at 17°C than at 27°C in response to light. Immunoblot of HY5 protein in 35S::HA-HY5 seedlings grown in darkness for 5 days at 17°C and 27°C before illuminating with Red light (40 µmol m−2 s−1) for 1 h and 6 h. Quantification of protein levels was conducted relative to the UGPase signal. Error bars represent ± SE of three biological repeats. We next measured HY5 protein levels in etiolated and de-etiolating seedlings at 17°C and at 27°C. Concurring with published work, our data show that HY5 transcript and protein levels increase steadily following exposure to 1 h then 6 h red light (Figure 2D, Figure S3) [39]. In the dark, HY5 transcript and protein levels were low and not particularly affected by temperature in our study range. However, we detected higher levels of HY5 transcripts and protein at 17°C when compared to 27°C, after exposure to -red light (Figure 2D, Figure S3). Thus, HY5 - abundance correlates well with both the light and the temperature requirement for HY5 control of photopigment levels (Figure 2A–C).
HY5 and PIFs regulate a subset of photosynthetic pigment genes through a common mechanism
To establish whether G-box element convergence represented a generic mechanism through which HY5 and PIF operate, we tested the impact of pifQ and hy5 mutations on the expression of other genes central to carotenoid or chlorophyll biosynthesis and function with G-box elements in their promoters. These genes were chosen from genome wide transcriptional analyses specific to PIFs or HY5 [14], [21], [35], [44]. To rule out binding to other promoter elements, the selected genes did not possess any of the alternative high affinity binding sites for HY5: C, Z or C/A, G/A or G/A boxes (Figure S1). For carotenoid biosynthesis, in addition to PSY, we selected VIOLAXANTIN DE-EPOXIDASE (VDE) that is involved in the conversion of violaxanthin to zeaxanthin for optimal photoprotection in the xanthophyll cycle [52]. Chlorophyll biosynthesis genes are represented by PROTOCHLOROPHYLLIDE OXIDOREDUCTASE C (PORC), that phototransforms endogenous protochlorophyllide to chlorophyllide, GENOMES UNCOUPLED 5 (GUN5) that encodes the ChlH subunit of Mg-chelatase, a key enzyme in the chlorophyll branch of the tetrapyrrole pathway [53], [54], and the Photosystem I LIGHT-HARVESTING COMPLEX 4 (LHCA4) [55], [56].
The transcript levels of these genes tightly correlated with the carotenoid and chlorophyll accumulation data (compare Figure 3 with Figure 2A, B). Consistently, mRNA levels were elevated in dark or light-grown pifQ at 17°C and 27°C, while hy5 led to reduced red light induction of each target gene, particularly at 17°C. To establish whether HY5 and PIFs regulate this suite of genes through common G-box elements we conducted ChIP analysis with lines expressing 35S::HA-HY5, 35S::PIF1-TAP and 35S::PIF4-TAP. In these experiments seedlings were grown in more natural 12L:12D cycles sampling at 2 or 3 h post dawn (T2, T3) or 8 h post dusk (T20) (Figures 4 and Figure S2). Levels of binding to the VDE promoter, were very low which prevented detailed analysis (data not shown). However, we detected enrichment of HY5, PIF1 and PIF4 relative to controls, at G-box containing regions of PSY, LHCA4, PORC and GUN5 promoters. G-box cis element convergence therefore appears to represent a common mechanism through which HY5 and PIFs regulate some carotenoid and chlorophyll biosynthetic genes (Figure 4).
Fig. 3. Expression levels of genes related to photosynthetic pigment accumulation at 17°C and 27°C in Col-0, pifQ (pif1-1 pif3-3 pif4-2 pif5-3) and hy5-215. 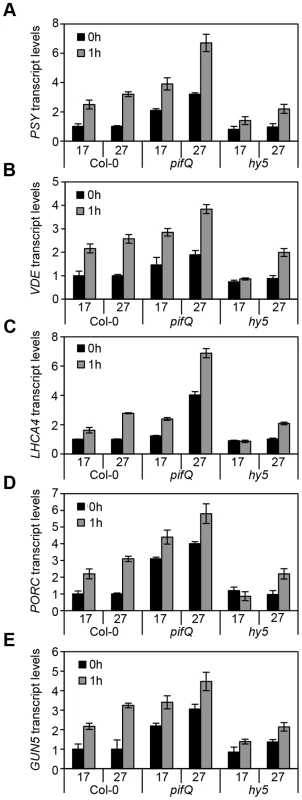
(A–E) Gene expression measured by qPCR for PSY, VDE, LHCA4, PORC and GUN5 in 3-day old etiolated seedlings. Samples were grown in the dark for 2 days at 22°C and then moved to 1 day at the indicated temperature, before illumination for 1 h with R light (1 h, grey columns) or maintained in darkness (0 h, black columns). Expression is represented relative to Actin7. Measurements were taken for biological triplicates. Error bars represent ± SE. Fig. 4. Chromatin immunoprecitation assays for 35S::HA-HY5, 35S::TAP-PIF1 and 35S::PIF4-HA grown in red-diurnals at 17°C and 27°C. 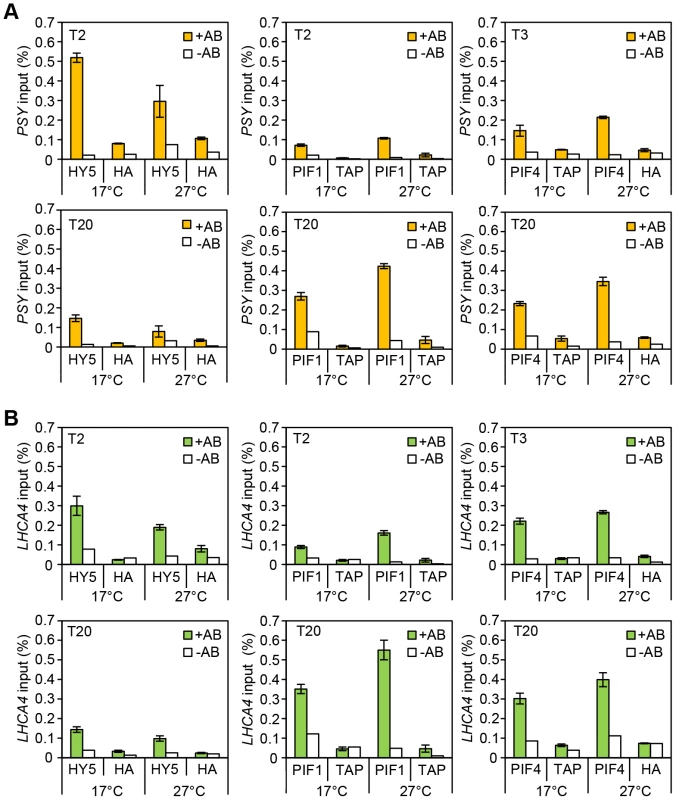
Two week old seedlings grown for one week in white diurnal cycles (12 h light/12 h dark, 80 µmol m−2 s−1) at 22°C, followed by one week under Red diurnal cycles (12 h light/12 h dark, 40 µmol m−2 s−1) were used for the experiment. On the last day, samples were taken 2 or 3 h after the lights came on (T2/T3) and 8 h after the lights were off (T20). ChIP was carried out using antibodies against the tag (anti-HA or anti-MYC). 35S::GUS-HA (labelled HA) or a 35S::GFP-TAP (TAP) lines were used as controls. A non-antibody control sample was processed in parallel in each case (white bars). Immunoprecipitated DNA was analysed by qPCR using specific primers covering the G-box containing region in the promoters for the indicated genes (see Table S1 for primer information). The assay was carried out in triplicates. Error Bars represent ±SE. (A) ChIP results for PSY. (B) ChIP results for LHCA4. PIF1 action is temperature regulated
Consistent with the more prominent role for HY5 at cooler temperatures (Figure 2) we detected an enrichment of HY5 at each promoter at 17°C compared to 27°C (Figures 4, and Figure S4). This may reflect a change in HY5 protein abundance that is detectable after exposure to 6 h of red light (Figure 2D). Previously we showed that heat leads to an accumulation of phosphorylated PIF4. This process appears to be extremely temperature sensitive as stepped increases in temperature lead to incremental rises of modified forms of PIF4 (Figure S5 A). We also observed increased binding of PIF4 to PSY, LHCA4 and GUN5 at 27°C compared to 17°C, but this was relatively modest [30] (Figures 4 and Figure S4). Interestingly, we detect slightly elevated protein levels and enhanced PIF1 promoter binding at 27°C suggesting that PIF1 is also temperature-regulated (Figures 4, Figure S4 B and Figure S5). In support of this notion our genetic data illustrates that like pif4pif5, pif1 has a greater impact on carotenoid and chlorophyll accumulation at 27°C compared to 17°C (Figure S6). Our results show the relative binding, particularly of HY5 and PIF1, at the G-box motifs in the promoters of several photopigment genes varies according to the ambient temperature regime.
HY5 and PIFs exhibit opposite diurnal shifts in promoter binding
Analysing binding during the daytime compared to the night allowed us to establish whether the documented diurnal changes in HY5, PIF4 and PIF1 abundance [16], [22]–[24], [35], [42] led to corresponding changes in binding. The levels and the proportion of HY5 bound to PSY, LHCA4, PORC and GUN5 promoters was greater at T2 than T20 (Figures 4, Figure S4 and Figure S5 C). PIF1 had the converse response with less bound at T2 (when PIF1 is less abundant) than T20 (Figures 4, Figure S4 and Figure S5 B). This trend was also evident, but less marked, for PIF4. This may reflect less dramatic diurnal changes in 35S::PIF4, the line used in this study, compared to 35S::HY5 and 35S::PIF1 (Figures 4, Figure S5 D and Figure S4). We then conducted EMSA to test whether a change in protein abundance is sufficient to drive a switch in binding to the G-box promoter fragment. In this assay the “primary” protein (either HY5 or PIF1) was incubated with a fixed amount of the G-box I probe and then the “challenge” protein was added at increasing concentrations (Figure S7). We could not detect a switch in binding when an equimolar amount of either “challenge” protein was added. However, binding was detected with increased concentrations of “challenge” protein, with a correlative decrease in “primary” protein binding. This indicates that when provided in excess HY5 or PIF1 can indeed prevent the other protein from binding.
HY5 and PIFs regulate photopigment biosynthetic genes in cooperation with the circadian clock
Through a red/dark cycle at 17°C, wild type expression profiles of PSY, LHCA4, GUN5 and PORC are rhythmic with differing phases of expression, suggesting underlying regulation by the circadian oscillator (Figure 5). The hy5 and pifQ mRNA profiles appear to approach or on occasion converge with the wild type at distinct phases in the cycle, suggesting that HY5 and PIF control of this gene set is gated by the clock (Figure 5). The contrasting day/night shifts in HY5 vs PIF1 binding to target promoters (Figure 4, Figure S4) inferred that the hy5 mutation should be more effective during the daytime and pifQ more potent at night. Unexpectedly, we found that this is not the case, as both hy5 and pifQ cause a shift in transcript levels during the day and the night (Figure 5). This indicates that during a diurnal cycle there is not a simple relationship between our recorded changes in promoter binding and the transcriptional response. It also indicates that HY5 and PIFs operate through the light/dark cycle to regulate this gene set. Interestingly, HY5 transcript levels are elevated in the pifQ mutant, while PIF4 and PIF5 (but not PIF1 or PIF3) mRNA levels are raised in hy5 at T2 and T20 (Figure S8). Such cross-regulation is predicted to increase the amplitude of expression in pifQ and conversely, reduce the amplitude in hy5 (Figure 5). In support of this notion, at least for PSY, LHCA4 and GUN5 the data show amplitude differences in pifQ compared to hy5 (Figure 5).
Fig. 5. Red diurnal expression patterns of photopigment genes targets of PIFs and HY5. 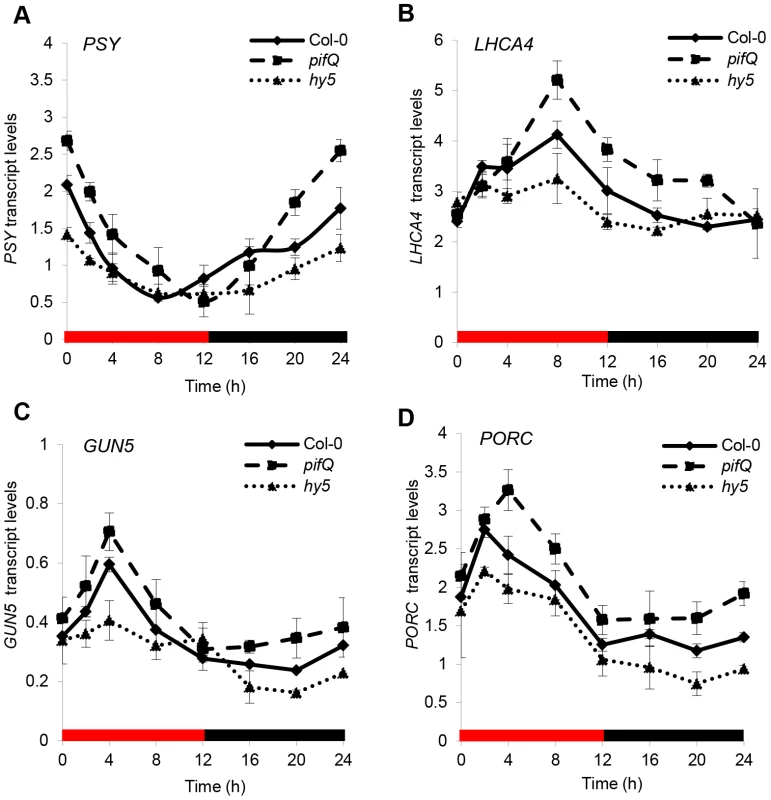
(A–D) Expression by qPCR of PSY (A), LHCA4 (B), GUN5 (C) and PORC (D) in Col-0, pifQ (pif1-1 pif3-3 pif4-2 pif5-3) and hy5-215 backgrounds. Plants were grown for one week under white diurnals (12 h light/12 h dark, 100 µmol m−2 s−1) at 22°C and then were transferred for 7 days to Red diurnal cycles at 17°C before sample collection at the indicated time points. Gene expression was analysed by qPCR. Black bars represent the dark period and red ones the illuminated times. Error bars represent ± SE of biological triplicates. The data presented suggest that HY5 and PIFs regulate gene expression in cooperation with the clock. This is further supported by the observation that the cross-regulated genes, HY5, PIF4 and PIF5 exhibit strong diurnal regulation, whereas PIF1 and PIF3 are not rhythmic and are not subject to cross-control (Figure S8 B) [22], [24]. Collectively, our analysis shows that HY5 and PIFs do not exhibit strong diurnal shifts in activity, but operate with the circadian clock to moderate rhythmic gene expression through the light/dark cycle.
Abrupt changes in HY5 and PIF protein abundance control binding to target gene promoters
The observation that the relative action of HY5 and PIF did not change during a diurnal cycle appeared to be at odds with our finding that light or temperature stabilisation of HY5 led to concomitant rises in binding activity and gene regulation (Figures 1, 3, 4 and Figure S4). This suggested that a sudden or sizeable increase in HY5 levels relative to PIFs may activate gene expression. To test this hypothesis we next manipulated HY5 and PIF1 levels in vivo to establish whether we observed corresponding changes in promoter binding and gene regulation. In these experiments plants were grown in 17°C diurnal cycles and protein levels were measured 3 h into the darkness in controls or following an end-of-day FR (EOD-FR) pulse which deactivates phytochrome [57]. This treatment led to a concurrent rise in PIF1 protein levels and fall in HY5 protein levels (Figure 6 A). In response to EOD-FR we observed correlative alterations in HY5 and PIF1 binding to the PSY, LHCA4, GUN5 and PORC promoters (Figure 6 B–E). We then tested whether EOD-FR led to predictable changes in gene regulation. PSY and GUN5 were unresponsive to the EOD-FR treatment (Figure S9). This result was unsurprising for PSY, as the EOD-FR timing coincided with the phase in which PIFs and HY5 are not fully engaged in transcriptional regulation. Likewise, the contribution of PIFs and HY5 to GUN5 transcriptional regulation was relatively low when compared to the other genes (Figure 5). Nonetheless, for LHCA4, PORC and VDE, the EOD-FR treatment elicited a decrease in transcript levels (Figure 7), a response that is consistent with the observed changes in PIF vs HY5 protein abundance and promoter binding.
Fig. 6. End of Day FR (EOD-FR) effect over 35S::HA-HY5 and 35S::TAP-PIF1 binding to G-box regions in the promoters of genes related to carotenoids and chlorophyll accumulation at 17°C. 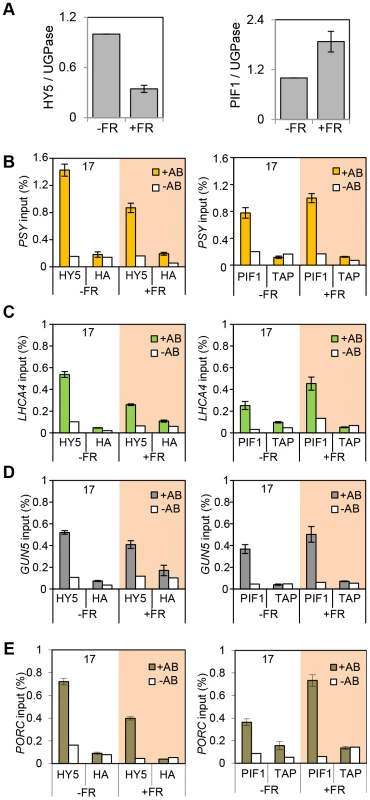
(A) Protein content quantification relative to the signal of UGPase for 35S::HA-HY5, 35S::PIF1-TAP at time 15 h (T15), in samples treated with (+FR) or without (-FR) an EOD saturating FR light pulse (3000 µmol). Triplicate Immunoblots were carried out using 2 week old seedlings. Seedlings were grown for one week in 12 h light/12 h dark white diurnal cycles at 22°C and then transferred to 17°C, 12 h light/12 h dark Red diurnal cycles. Protein was extracted at T15 and quantified against UGPase signal. (B–D) Chromatin Immunoprecipitation assay for PSY (B), LHCA4 (C), GUN5 (D) and PORC (E) G-box regions in 35S::HA-HY5 and 35S::PIF1-TAP plants treated with (+) or without(−) an EOD FR pulse at T15. +AB indicates samples treated with antibody, -AB stands for no antibody controls. Plants were grown as indicated in (A) and in material and methods. The HA- and MYC- controls and ChIP procedure were described in Figure 4. Error bars represent ± SE of biological triplicates. Fig. 7. Photopigment gene expression response to an End of Day (EOD)-Far Red (FR) light treatment in Col-0. 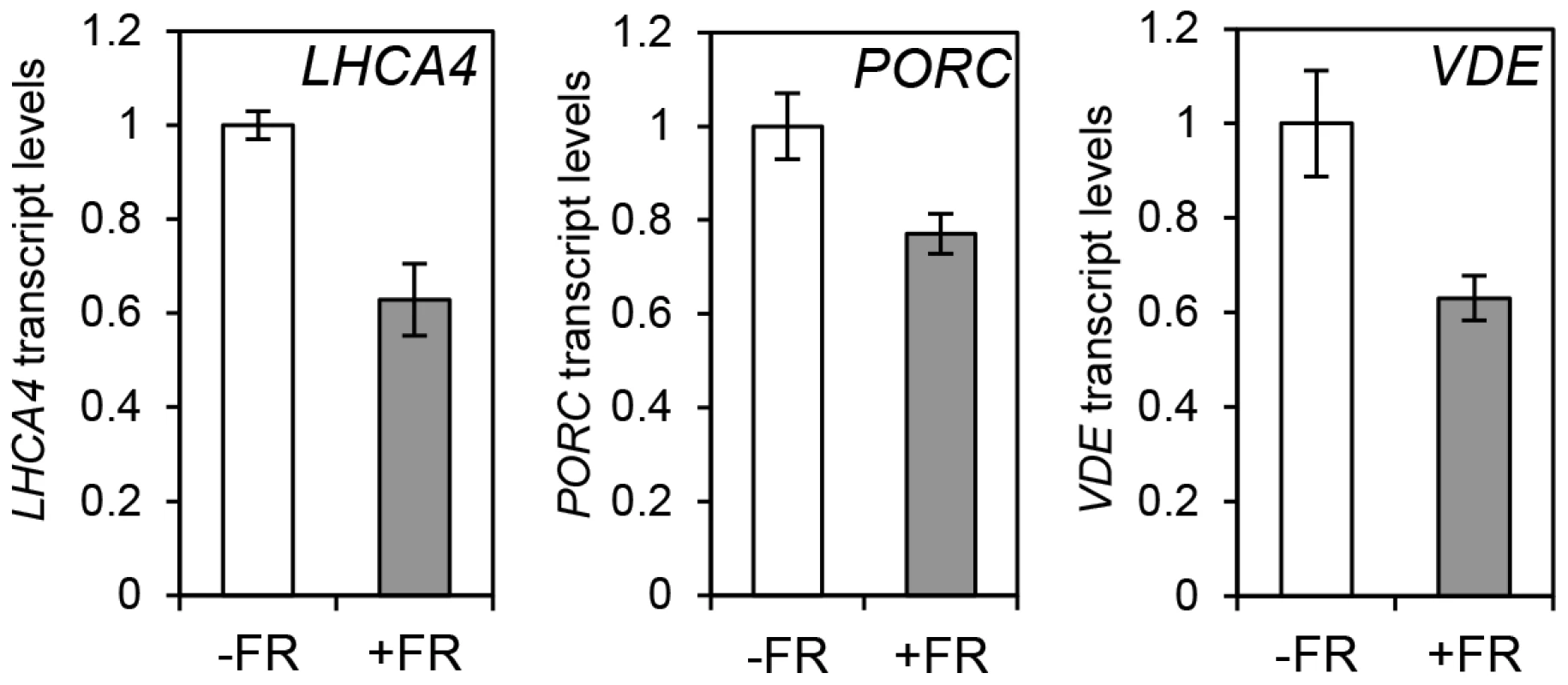
Expression levels for LHCA4, PORC and VDE in Col-0 plants treated without (−FR) or with (+FR) a saturating FR pulse (3000 µmol) at the end of the day (EOD) and collected at T15. Samples were grown as in Figure 6, and used for gene expression measurements by qPCR. Levels are expressed relative to Col-0 (-FR) T15 sample and normalized against ACT7 expression. Error bars represent ± SE of biological triplicates. HY5 regulates photosynthetic capacity at cooler temperatures
Our data illustrate that HY5 acts antagonistically with PIFs to control photosynthetic pigment genes through a common cis element, particularly at cooler temperatures. As this work was conducted in seedlings we wanted to establish the significance of this regulation in older plants. We tested this by measuring photosynthetic capacity in WT and hy5 adult plants (grown in 12L:12D) shifted to either 17°C or 27°C from 22°C. The hy5 mutant had lower chlorophyll levels and CO2 net flux m−2 s−1 compared to WT, and this difference is more marked at 17°C vs 27°C (Figure 8 A–B). Our previous studies of pifQ demonstrated that PIFs also influence the accumulation of carotenoids and chlorophylls under photoperiodic conditions [17]. It therefore appears that the HY5 and PIF driven transcriptional switch is important for controlling the production of photosynthetic pigments beyond the seedling stage. The temperature dependence of the hy5 mutation on chlorophyll levels and CO2 uptake was evident seven days following the transfer from 22°C to either 17°C or 27°C. This indicates the process is dynamic and that HY5 is required to actively maintain chlorophyll content in response to changing external light and temperature signals. The impact of hy5 on carbon assimilation may have consequences for growth as fresh weight is markedly reduced in hy5 mutants at 17°C but not at 27°C (Figures 8 C and D).
Fig. 8. HY5 modulates photosynthetic acclimation efficiency at low temperatures. 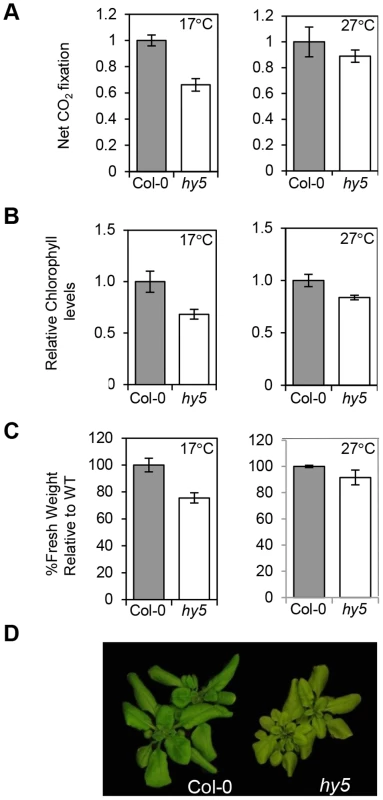
(A) CO2 net flux measurements for Col-0 and hy5-215 acclimated to 17°C or 27°C. Plants were grown for 3 weeks under 12 h light/12 h dark white light cycles at 22°C and transferred for 7 days to same photoperiod cycles at 17°C or 27°C. CO2 assimilation was measured in parts per million (ppm). Flux was calculated per unit area (m2) and results expressed relative to Col-0. Measurements were conducted in duplicates for two independent sets of 48 plants. Error bars represent ± SE. (B) Chlorophyll content for plants used in the CO2 assimilation experiment. Chlorophyll was extracted for 12 randomly selected plants. Plants were grown as described in (A). Values were calculated in µg/g fresh weight and expressed relative to Col-0. Error bars represent ± SE. (C) Fresh weight difference for Col-0 and hy5-215 17°C and 27°C acclimated plants. Weight was measured in grams and result is expressed as a percentage of Col-0 (assigned value of 100%). Bars represent SE of two sets of 48 plants. (D) Pictures of representative Col-0 and hy5-215 plants acclimated to 17°C and used in the experiments described (A–C). Discussion
HY5 and PIFs co-regulate photosynthetic genes at common G-box motifs
Despite the knowledge that light and temperature are among the most relevant environmental signals modulating photosynthetic pigment production, there is no integrative view on how these adjustments are achieved. We have shown that by responding to external light and temperature signals, HY5 opposes PIF action to deliver environmental control of common target genes involved in photosynthesis and photoprotection. This dual control system appears to operate in conjunction with the circadian oscillator to adjust levels of rhythmic photosynthetic gene expression.
Our previous work, and that of others, identified the PIFs as negative regulators of chlorophyll and carotenoid accumulation [8], [17], [51], [58]. However, optimization of these essential responses would require an additional regulator that acts antagonistically to PIFs. Here we have shown that HY5 fulfils this role as a positive regulator of photosynthetic pigment synthesis (Figure 1). Our earlier research demonstrated that PIFs control the expression of PSY, a rate limiting step in carotenogenesis. This is achieved by directly binding to G-box motifs in the PSY promoter [17]. Previous genome wide gene expression and ChIP-CHIP analyses highlighted an overlap in PIFs and HY5 gene targets and identified G-boxes as potential HY5 targets [7], [35], [36], [44], [59]. We have shown in-vitro and in-vivo that HY5 binds to the same G-box containing region targeted by PIF1 in the PSY promoter (Figure 1). Our data indicates that HY5 and PIFs act antagonistically to control PSY gene expression via a common cis element. This regulation is not confined to PSY, as we presented evidence that other central carotenoid and chlorophyll pathway genes are regulated through the same dual input mechanism. HY5 and PIFs do not always act antagonistically. In the case of anthocyanin biosynthesis, PIF3 and HY5 both positively regulate the pathway by activating transcription of the same biosynthetic genes by binding to distinct cis-promoter elements (G-box and another ACE motif respectively) [19]. In this instance PIF3 binding is facilitated by the presence of HY5, suggesting cooperativity of action. However, our analysis of photopigment gene regulation, together with a recent study focussing on REACTIVE OXYGEN SPECIES genes, indicates that when HY5 and PIF act in opposition, G-box convergence is a common mechanism [7].
Coaction of HY5 and PIF antagonists fine tune rhythmic photopigment gene expression
Light has opposing effects on HY5 and PIF levels and/or activity [1], [34], [60], [61]. HY5 protein accumulates in the light and is degraded in the dark [39], [41]. In contrast, PIFs accumulate in the dark to promote dark development, and light induces rapid phytochrome dependent phosphorylation, degradation and deactivation [22], [60], [62]–[64]. Accordingly we established that HY5 promotes the expression of PSY, VDE, PORC, LHCA4 and GUN5 genes following exposure to light, while PIFs strongly suppress the transcription of these genes in etiolated seedlings (Figure 3).
In diurnal cycles we showed that the levels of HY5, PIF1 and PIF4 bound to common G-box elements correlated with times that these proteins are abundant (Figures 4, Figure S4 and Figure S5). Furthermore, in vitro EMSA assays demonstrated that binding of either HY5 or PIF1 to the G-box region of the PSY promoter was reduced when the opposing challenge protein was provided in excess (Figure S7). This, analysis illustrated that the change in relative HY5:PIF promoter binding could at least partly be driven by alterations in protein abundance. However, when we analysed the transcript profiles of wild type plants compared to hy5 and pifQ mutants, there was no obvious correlation between the shift in binding dominance and the relative effectiveness of HY5 and PIFs through the light/dark cycle (Figure 5). A potential explanation is that cross-regulation between HY5 and PIFs may dampen the diurnal swings in response. Such a mechanism has been recently reported by Chen and coworkers[7]. This study showed that PIF1 and PIF3 physically interact with HY5 and its homologue HYH. Through this interaction these antagonistic pairs of transcription factors, PIF1/PIF3 and HY5/HYH, moderate each other's control of ROS-responsive genes. It is therefore possible that this cross-control is a more broadly utilised “buffering” mechanism that dampens antagonistic HY5-PIF responses. Alternatively, an independent regulator such as the circadian clock may be modulating the level or activity of HY5 or PIFs at the promoter. Indeed, we showed that HY5/PIF control of PSY, PORC, LHCA4 and GUN5 expression is gated by the circadian clock (Figure 5). These findings are consistent with earlier work that shows that the HY5 protein physically interacts with CCA1, which in turn enhances the binding of HY5 to the LHCB1*1 promoter [65]. It is not yet known how PIFs interface with the oscillator. However, an in vitro interaction between PIF3 and TOC1/PRR1 has been reported [66], [67], and GUN5, a gene identified as a HY5/PIF target in this study, was previously shown to be directly and negatively regulated by TOC1 [68]–[70]. PRR9, PRR7 and PRR5 are also reported to be negative regulators of chlorophyll and carotenoid biosynthesis [71]. Thus, PIFs may regulate specific gene targets in concert with TOC1/PRR1, other PRRs or their regulators. Further analysis will be required to evaluate these and other possible links between HY5, PIFs and the clock.
Changes in protein abundance alter HY5/PIF promoter binding
The diurnal switch between HY5 - and PIF - dominant promoter binding did not lead to correlative diurnal shifts in transcriptional regulation (Figures 4, 5 and Figure S4). However, a sudden change in HY5 or PIF protein abundance in-vivo did elicit a matching alteration in gene expression in LHCA4, PORC and VDE (Figure 7). Supplying an EOD-FR pulse, which deactivates phyB, induced a simultaneous fall in HY5 and rise in PIF1 protein abundance, with correlative alterations in promoter binding and transcript regulation (Figures 6 and 7). These results illustrate that abrupt changes in HY5 or PIF levels can lead to corresponding adjustments in signalling. Sizeable adjustments in HY5 and PIF levels and activity are known to occur in etiolated seedlings following exposure to light, and are predicted in plants exposed to abrupt changes in temperature [1], [28], [29], [35], [41], [43], [61], [62], [72], [73].
Environmental temperature signals are delivered through HY5 and PIF1
Earlier work illustrated that HY5 is important for cold acclimation responses and enhancement of freezing tolerance [43]–[45]. At 4°C HY5 protein levels were shown to stabilise, particularly in the dark [43]. We have shown that HY5 also has a prominent role at the more moderate 17°C, but here its action depends on light. Exposure of etiolated seedlings to light leads to a rapid depletion in PIF levels and increase in HY5 (Figure 2D) [1], [16], [35], [41], [62]. The sudden change from PIF to HY5 dominance induces a switch from transcriptional repression to activation of target photopigment genes (Figure 3). This switch is more robust at 17°C compared to 27°C.
PIF4 is known to activate genes in a temperature dependent manner [30]–[32], [47], [50], [74]. PIF4 levels and its binding to target promoters are boosted by warm temperature [30]–[32]. In line with these studies we observed enhanced PIF4 enrichment at carotenoid and photosynthetic gene promoters under warmer conditions. Interestingly, PIF1 levels and binding to target promoters is also increased at 27°C compared to 17°C (Figures 4 and Figure S4). Our genetic data also show that PIF1 has a greater influence on carotenoid and chlorophyll levels at 27°C in etiolated seedlings (Figure S6). Thus, in the regulation of photosynthetic pigment synthesis, PIF1 action appears to be temperature dependent. The temperature response is particularly evident in etiolated seedlings as PIF protein levels are very high compared to HY5, and therefore PIF signalling predominates. Our data also show that the pifQ mutant has perturbed responses at 17°C and 27°C, suggesting that collectively, PIF1, PIF3, PIF4 and PIF5 operate over a broad temperature range (Figures 2 and 3).
HY5 maintains photosynthetic capacity at lower temperatures through development
We have shown that HY5 is not only important in developing seedling, but as for PIFs, it continues to regulate photopigment levels in adult plants (Figure 8) [17]. Our data demonstrate that HY5 plays a role in maintaining chlorophyll levels and CO2 uptake. Interestingly, our analysis indicates that HY5 has a greater impact in plants transferred (from 22°C) to 17°C compared to 27°C (Figure 8 A–C). This illustrates that maintenance of chlorophyll pool is very dynamic and that HY5 is required to augment chlorophyll synthesis and carbon uptake, particularly when temperatures fall. Furthermore at 17°C the fresh weight of hy5 is markedly reduced - compared to wild type plants at 17°C but mot at 27°C (Figure 8 C–D).HY5 has been shown to modulate several aspects of plant hormones pathways including auxins, giberellins and absicic acid signal transduction. Therefore, it is likely that alterations in hormone signalling will contribute to hy5 adult phenotype [34] However, the reduction in photosynthetic capacity observed in hy5 at 17°C (Figure 8) may also compromise growth.
This paper illustrates that signal convergence of antagonistic regulators HY5 and PIFs at a shared cis regulatory element provides an effective mechanism to integrate light and temperature signals. A similar type of control has been reported for the endoreduplication E2Fb and E2Fc transcription factors which antagonistically regulate DEL1 expression through a common cis-element [75], [76]. Differential regulation of E2Fb vs E2Fc protein levels by light, determines relative binding capacity and the level of DEL1 activation [77]. It is therefore possible that the single cis element activation-inactivation module is a prevalent signalling mechanism through which external signals can change or fine-tune transcriptional responses.
Materials and Methods
Plant material and growth conditions
Columbia-0 (Col-0) wild type and mutants were used for experiments. The mutant alleles corresponded to: hy5-215, hyh (GK-57200610-N323769), pif4pif5 (pif4-2, pif5-2), pif1-1, pifQ (pif1-2, pif3-3, pif4-2, pif5-2). Over expressing plants included 35S::PIF4-HA, 35S::HA-HY5 and 35S::TAP-PIF1. All have been previously described [14], [35], [58], [78], [79]. Seeds were surface sterilized, sown in GM-agar media and stratified in darkness for 3 days at 4°C before given a 3 h white light pulse to induce germination. For deetiolation experiments (qPCR and photosynthetic pigment measurements), 3 d-old seedlings were used. Seedlings were kept in the dark for 2 days at 22°C and transferred to the indicated temperature for 1d before exposure to red light (40 µmol m−2 s−1). For Red Diurnal experiments plants were grown for one week under white diurnals (12 h light/12 h dark, 100 µmol m−2 s−1) at 22°C and then were transferred for 7 days to Red diurnal cycles at 17°C before sample collection. In the case of ChIP experiments, samples were collected at T2 for PIF1 and HY5 and T3 for PIF4. Times were selected based on moments where the proteins have started to recover following phyB degradation (in the case of the PIFs, that starts at T0) and when levels were comparable among all proteins (in the case of HY5) ([80], [81],[22]).
Carotenoid and chlorophyll measurements
For the experiments, 3 d-old etiolated seedlings were used and treated as indicated above. Total carotenoids and chlorophylls were extracted and quantified spectrophotometrically as described by[17]. Concentration was expressed per sample fresh weight and measured in biological triplicates.
RNA isolation and transcript levels analysis by qPCR
For deetiolating experiments quantitative qPCR seedlings were prepared and sown as previously described, and grown for 2 days in the dark at 22°C and 1 day at the indicated temperature before red-light illumination (as indicated above). For diurnal expression analyses, 2 week old seedlings were used. Seedlings were grown under white light 12L:12D cycles (80 µmol m−2 s−1) at 22°C for one week before being transferred to 17°C red light 12∶12 dark/light cycles (40 µmol m−2 s−1) for one week and harvesting at the indicated time points. Samples were collected in RNAlater (Sigma). RNA was extracted with RNeasy Plant Mini kit (Qiagen). cDNA synthesis was performed using the SuperScript VILO cDNA synthesis kit (Invitrogen). The qPCR was set up with a liquid handling robot (Tecan Freedom EVO) and qPCR performed with a LightCycler 480 (Roche). All samples were processed in biological triplicates.
Electrophoretic Mobility Shift Assays (EMSA)
EMSA were conducted as described in [18]. In brief HY5 and PIF1 proteins were produced in an in-vitro transcription and translation system (TnT) (Promega) and incubated with a PSY promoter fragment generated by annealed oligonucleotides containing the G-box motif labelled with 32P-dCTP.
Chromatin Immunoprecipitation assays
ChIP assays were conducted according to [58]except that 2 week old plants were used for the assay. Unless otherwise stated, plants were grown for one week in 12∶12 white light diurnal cycles at 22°C and moved to one week growth in red 12∶12 diurnal cycles at the testing temperature. Samples were harvested at the indicated time points during the diurnal cycle. For the end-of-day FR experiment, a saturating FR pulse (3000 µmol total) was given at the end of the day and samples harvested 3 h after the pulse. A non-FR treated sample was harvested at the same time point. The sequence of the primers used in these experiments to amplify G-box containing promoter regions of individual genes is shown in Table S1.
Immunoblots
Total proteins were extracted from 100 mg of tissue in a buffer containing 100 mM Tris-HCl pH 8, 50 mM EDTA, 0.25 M NaCl, 0.7% SDS and 1 mM DTT. Samples were heated at 65°C for 10 minutes and then centrifuged at maximum speed to remove debris. Total protein was quantified by Bradford. 30 µg of total protein were loaded. Samples were run in a 10% SDS gel, followed by wet transfer to nitrocellulose. The HA - or MYC - tag were detected by probing with rat-anti HA horseradish peroxidase HRP coupled HA - antibody (3F10 Roche) or an Anti-MYC mouse antibody (mAb 9E10, Calbiochem) at a 1∶5000 dilution followed by HRP-conjugated anti-mouse antibody at 1∶10,000 dilution. Loading was confirmed by reprobing the membranes with an anti-goat UGP-ase antibody (Agrisera) at 1∶1000 dilution followed by a HRP-conjugated sheep anti-goat antibody (Biorad) at a 1∶5000 dilution. Signal was detected with Amersham ECL kit (GE Health care) according to the manufacturer's protocol. Quantification was performed using Image-J software.
CO2 assimilation measurements and chlorophyll measurements
For adult plant analysis seedlings were sown as indicated in plant material and grown for 1 week under white light 12 h L/12 h D regimes at 22°C. Then plants were transferred to soil and kept under the same photoperiod and temperature for 3 weeks. On week 3, plants were transferred to 12 h L/12 h D regimes at 17°C or 27°C for 7 days before measurements were taken. CO2 assimilation was measured over a 1 minute period for 48 plants using a EGM-4 machine (PP Systems) for 1 minute. Readings were taken in ppm and plants photographed and weighted to calculate area, fresh weigh and total CO2 flux. The same set of plants was used for fresh weight determination and chlorophyll measurements. For chlorophyll extraction 300 mg of fresh tissue from twelve randomly selected plants were used for individual extractions conducted in triplicates. The whole rosette fresh weight was estimated and chlorophyll extracted by the protocol: http://www.nature.com/protocolexchange/protocols/521. Chlorophyll content was measured spectrophotometrically.
Supporting Information
Zdroje
1. LeivarP, QuailPH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16 : 19–28.
2. QuailPH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3 : 85–93.
3. WelschR, BeyerP, HugueneyP, KleinigH, von LintigJ (2000) Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211 : 846–854.
4. TeppermanJM, HudsonME, KhannaR, ZhuT, ChangSH, et al. (2004) Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J 38 : 725–739.
5. FranklinKA, QuailPH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61 : 11–24.
6. ReinbotheS, ReinbotheC, ApelK, LebedevN (1996) Evolution of chlorophyll biosynthesis–the challenge to survive photooxidation. Cell 86 : 703–705.
7. ChenD, XuG, TangW, JingY, JiQ, et al. (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25 : 1657–1673.
8. HuqE, Al-SadyB, HudsonM, KimC, ApelK, et al. (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 : 1937–1941.
9. NiyogiKK (1999) PHOTOPROTECTION REVISITED: Genetic and Molecular Approaches. Annu Rev Plant Physiol Plant Mol Biol 50 : 333–359.
10. HavauxM, NiyogiKK (1999) The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc Natl Acad Sci U S A 96 : 8762–8767.
11. WalterMH, StrackD (2011) Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep 28 : 663–692.
12. CovingtonMF, MaloofJN, StraumeM, KaySA, HarmerSL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130.
13. QuailPH (2010) Phytochromes. Curr Biol 20: R504–507.
14. LeivarP, TeppermanJM, MonteE, CalderonRH, LiuTL, et al. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21 : 3535–3553.
15. ShinJ, KimK, KangH, ZulfugarovIS, BaeG, et al. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A 106 : 7660–7665.
16. LiJ, TerzaghiW, DengXW (2012) Genomic basis for light control of plant development. Protein Cell 3 : 106–116.
17. Toledo-OrtizG, HuqE, Rodriguez-ConcepcionM (2010) Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci U S A 107 : 11626–11631.
18. Martinez-GarciaJF, HuqE, QuailPH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288 : 859–863.
19. ShinJ, ParkE, ChoiG (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49 : 981–994.
20. HornitschekP, KohnenMV, LorrainS, RougemontJ, LjungK, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71 : 699–711.
21. ZhangY, MaybaO, PfeifferA, ShiH, TeppermanJM, et al. (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244.
22. ShenH, MoonJ, HuqE (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44 : 1023–1035.
23. NozueK, CovingtonMF, DuekPD, LorrainS, FankhauserC, et al. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448 : 358–361.
24. SoyJ, LeivarP, Gonzalez-SchainN, SentandreuM, PratS, et al. (2012) Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J 71 : 390–401.
25. NiwaY, YamashinoT, MizunoT (2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol 50 : 838–854.
26. KunihiroA, YamashinoT, NakamichiN, NiwaY, NakanishiH, et al. (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol 52 : 1315–1329.
27. NomotoY, KubozonoS, YamashinoT, NakamichiN, MizunoT (2012) Circadian clock - and PIF4-controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol 53 : 1950–1964.
28. KoiniMA, AlveyL, AllenT, TilleyCA, HarberdNP, et al. (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19 : 408–413.
29. StavangJA, Gallego-BartolomeJ, GomezMD, YoshidaS, AsamiT, et al. (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60 : 589–601.
30. ForemanJ, JohanssonH, HornitschekP, JosseEM, FankhauserC, et al. (2011) Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J 65 : 441–452.
31. FranklinKA, LeeSH, PatelD, KumarSV, SpartzAK, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A 108 : 20231–20235.
32. KumarSV, LucyshynD, JaegerKE, AlosE, AlveyE, et al. (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484 : 242–245.
33. KamiC, LorrainS, HornitschekP, FankhauserC (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91 : 29–66.
34. LauOS, DengXW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13 : 571–577.
35. LeeJ, HeK, StolcV, LeeH, FigueroaP, et al. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 : 731–749.
36. OyamaT, ShimuraY, OkadaK (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11 : 2983–2995.
37. ZhangH, HeH, WangX, WangX, YangX, et al. (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65 : 346–358.
38. ChattopadhyayS, AngLH, PuenteP, DengXW, WeiN (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10 : 673–683.
39. OsterlundMT, HardtkeCS, WeiN, DengXW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 : 462–466.
40. HardtkeCS, GohdaK, OsterlundMT, OyamaT, OkadaK, et al. (2000) HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19 : 4997–5006.
41. PokhilkoA, RamosJA, HoltanH, MaszleDR, KhannaR, et al. (2011) Ubiquitin ligase switch in plant photomorphogenesis: A hypothesis. J Theor Biol 270 : 31–41.
42. ShinJ, AnwerMU, DavisSJ (2013) Phytochrome-interacting factors (PIFs) as bridges between environmental signals and the circadian clock: diurnal regulation of growth and development. Mol Plant 6 : 592–595.
43. CatalaR, MedinaJ, SalinasJ (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci U S A 108 : 16475–16480.
44. ZhangY, ZhengS, LiuZ, WangL, BiY (2011) Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol 168 : 367–374.
45. ZhangY, LiuZ, LiuR, HaoH, BiY (2011) Gibberellins negatively regulate low temperature-induced anthocyanin accumulation in a HY5/HYH-dependent manner. Plant Signal Behav 6 : 632–634.
46. AlabadiD, BlazquezMA (2008) Integration of light and hormone signals. Plant Signal Behav 3 : 448–449.
47. NomotoY, KubozonoS, MiyachiM, YamashinoT, NakamichiN, et al. (2012) A circadian clock - and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol 53 : 1965–1973.
48. TohS, McCourtP, TsuchiyaY (2012) HY5 is involved in strigolactone-dependent seed germination in Arabidopsis. Plant Signal Behav 7 : 556–558.
49. SunJ, QiL, LiY, ChuJ, LiC (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet 8: e1002594.
50. Nomoto Y, Kubozono S, Miyachi M, Yamashino T, Nakamichi N, et al. (2012) Circadian clock and PIF4-mediated external coincidence mechanism coordinately integrates both of the cues from seasonal changes in photoperiod and temperature to regulate plant growth in Arabidopsis thaliana. Plant Signal Behav 8, 2, e22863.
51. StephensonPG, FankhauserC, TerryMJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci U S A 106 : 7654–7659.
52. RockholmDC, YamamotoHY (1996) Violaxanthin de-epoxidase. Plant Physiol 110 : 697–703.
53. MochizukiN, BrusslanJA, LarkinR, NagataniA, ChoryJ (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci U S A 98 : 2053–2058.
54. MochizukiN, TanakaR, GrimmB, MasudaT, MoulinM, et al. (2010) The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci 15 : 488–498.
55. Ben-ShemA, FrolowF, NelsonN (2003) Crystal structure of plant photosystem I. Nature. 426 : 630–635.
56. AlboresiA, BallottariM, HienerwadelR, GiacomettiGM, MorosinottoT (2009) Antenna complexes protect Photosystem I from photoinhibition. BMC Plant Biol 9 : 71.
57. FranklinKA, WhitelamGC (2005) Phytochromes and shade-avoidance responses in plants. Ann Bot 96 : 169–175.
58. MoonJ, ZhuL, ShenH, HuqE (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci U S A 105 : 9433–9438.
59. KobayashiK, ObayashiT, MasudaT (2012) Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal Behav 7 : 922–926.
60. ParkE, ParkJ, KimJ, NagataniA, LagariasJC, et al. (2012) Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J 72 : 537–546.
61. LeivarP, MonteE, CohnMM, QuailPH (2012) Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop. Mol Plant 5 : 734–749.
62. Al-SadyB, NiW, KircherS, SchaferE, QuailPH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23 : 439–446.
63. de LucasM, DaviereJM, Rodriguez-FalconM, PontinM, Iglesias-PedrazJM, et al. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451 : 480–484.
64. HenriquesR, JangIC, ChuaNH (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12 : 49–56.
65. AndronisC, BarakS, KnowlesSM, SuganoS, TobinEM (2008) The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol Plant 1 : 58–67.
66. ItoS, MatsushikaA, YamadaH, SatoS, KatoT, et al. (2003) Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44 : 1237–1245.
67. MakinoS, MatsushikaA, KojimaM, YamashinoT, MizunoT (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43 : 58–69.
68. LegnaioliT, CuevasJ, MasP (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28 : 3745–3757.
69. YamashinoT, MatsushikaA, FujimoriT, SatoS, KatoT, et al. (2003) A Link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44 : 619–629.
70. HuangW, Perez-GarciaP, PokhilkoA, MillarAJ, AntoshechkinI, et al. (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336 : 75–79.
71. FukushimaA, KusanoM, NakamichiN, KobayashiM, HayashiN, et al. (2009) Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci U S A 106 : 7251–7256.
72. ShenH, ZhuL, CastillonA, MajeeM, DownieB, et al. (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20 : 1586–1602.
73. PokhilkoA, FernandezAP, EdwardsKD, SouthernMM, HallidayKJ, et al. (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8 : 574.
74. ProveniersMC, van ZantenM (2013) High temperature acclimation through PIF4 signaling. Trends Plant Sci 18 : 59–64.
75. Lopez-JuezE, DillonE, MagyarZ, KhanS, HazeldineS, et al. (2008) Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20 : 947–968.
76. del PozoJC, BoniottiMB, GutierrezC (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14 : 3057–3071.
77. BerckmansB, LammensT, Van Den DaeleH, MagyarZ, BogreL, et al. (2011) Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc. Plant Physiol 157 : 1440–1451.
78. LeivarP, MonteE, OkaY, LiuT, CarleC, et al. (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18 : 1815–1823.
79. HornitschekP, LorrainS, ZoeteV, MichielinO, FankhauserC (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28 : 3893–3902.
80. LorrainS, AllenT, DuekPD, WhitelamGC, FankhauserC (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53 : 312–323.
81. YamashinoT, NomotoY, LorrainS, MiyachiM, ItoS, et al. (2013) Verification at the protein level of the PIF4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signal Behav 8: e23390.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Inflammation: Gone with Translation
- Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1
- Caspase Inhibition in Select Olfactory Neurons Restores Innate Attraction Behavior in Aged
- Accurate, Model-Based Tuning of Synthetic Gene Expression Using Introns in
- A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in
- Ancient DNA Analysis of 8000 B.C. Near Eastern Farmers Supports an Early Neolithic Pioneer Maritime Colonization of Mainland Europe through Cyprus and the Aegean Islands
- The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy
- Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
- Netrins and Wnts Function Redundantly to Regulate Antero-Posterior and Dorso-Ventral Guidance in
- Coordination of Wing and Whole-Body Development at Developmental Milestones Ensures Robustness against Environmental and Physiological Perturbations
- Phenotypic Dissection of Bone Mineral Density Reveals Skeletal Site Specificity and Facilitates the Identification of Novel Loci in the Genetic Regulation of Bone Mass Attainment
- Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of -Factors in Two Independent Origins of C Photosynthesis
- Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity
- Translational Regulation of Specific mRNAs Controls Feedback Inhibition and Survival during Macrophage Activation
- Rosa26-GFP Direct Repeat (RaDR-GFP) Mice Reveal Tissue- and Age-Dependence of Homologous Recombination in Mammals
- Abnormal Type I Collagen Post-translational Modification and Crosslinking in a Cyclophilin B KO Mouse Model of Recessive Osteogenesis Imperfecta
- : Clonal Reinforcement Drives Evolution of a Simple Microbial Community
- Reviving the Dead: History and Reactivation of an Extinct L1
- Defective iA37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA
- Early Back-to-Africa Migration into the Horn of Africa
- Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants
- Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia
- Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
- Palaeosymbiosis Revealed by Genomic Fossils of in a Strongyloidean Nematode
- Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- Genetic Background Drives Transcriptional Variation in Human Induced Pluripotent Stem Cells
- Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes
- N-WASP Is Required for Structural Integrity of the Blood-Testis Barrier
- The Transcription Factor TFII-I Promotes DNA Translesion Synthesis and Genomic Stability
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
- Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats
- ATRA-Induced Cellular Differentiation and CD38 Expression Inhibits Acquisition of BCR-ABL Mutations for CML Acquired Resistance
- The EJC Binding and Dissociating Activity of PYM Is Regulated in
- JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling
- Mouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
- Rasa3 Controls Megakaryocyte Rap1 Activation, Integrin Signaling and Differentiation into Proplatelet
- Transcriptional Control of Steroid Biosynthesis Genes in the Prothoracic Gland by Ventral Veins Lacking and Knirps
- Souffle/Spastizin Controls Secretory Vesicle Maturation during Zebrafish Oogenesis
- The POU Factor Ventral Veins Lacking/Drifter Directs the Timing of Metamorphosis through Ecdysteroid and Juvenile Hormone Signaling
- The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses
- Sequence of a Complete Chicken BG Haplotype Shows Dynamic Expansion and Contraction of Two Gene Lineages with Particular Expression Patterns
- Background Selection as Baseline for Nucleotide Variation across the Genome
- CPF-Associated Phosphatase Activity Opposes Condensin-Mediated Chromosome Condensation
- The Effects of Codon Context on Translation Speed
- Glycogen Synthase Kinase (GSK) 3β Phosphorylates and Protects Nuclear Myosin 1c from Proteasome-Mediated Degradation to Activate rDNA Transcription in Early G1 Cells
- Regulation of Gene Expression in Autoimmune Disease Loci and the Genetic Basis of Proliferation in CD4 Effector Memory T Cells
- Muscle Structure Influences Utrophin Expression in Mice
- BLMP-1/Blimp-1 Regulates the Spatiotemporal Cell Migration Pattern in
- Identification of Late Larval Stage Developmental Checkpoints in Regulated by Insulin/IGF and Steroid Hormone Signaling Pathways
- Transport of Magnesium by a Bacterial Nramp-Related Gene
- Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
- The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
- The Rim15-Endosulfine-PP2A Signalling Module Regulates Entry into Gametogenesis and Quiescence Distinct Mechanisms in Budding Yeast
- Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
- Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions
- Positive Feedback of Expression Ensures Irreversible Meiotic Commitment in Budding Yeast
- Hecate/Grip2a Acts to Reorganize the Cytoskeleton in the Symmetry-Breaking Event of Embryonic Axis Induction
- Regulatory Mechanisms That Prevent Re-initiation of DNA Replication Can Be Locally Modulated at Origins by Nearby Sequence Elements
- Speciation and Introgression between and
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Early Back-to-Africa Migration into the Horn of Africa
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



