-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
MicroRNAs play pervasive roles in controlling gene expression throughout animal development. Given that individual microRNAs are predicted to regulate hundreds of mRNAs and that most mRNA transcripts are microRNA targets, it is essential that the expression levels of microRNAs be tightly regulated. With the goal of unveiling factors that regulate the expression of microRNAs that control developmental timing, we identified lin-42, the C. elegans homolog of the human and Drosophila period gene implicated in circadian gene regulation, as a negative regulator of microRNA expression. By analyzing the transcriptional expression patterns of representative microRNAs, we found that the transcription of many microRNAs is normally highly dynamic and coupled aspects of post-embryonic growth and behavior. We suggest that lin-42 functions to modulate the transcriptional output of temporally-regulated microRNAs and mRNAs in order to maintain optimal expression of these genes throughout development.
Published in the journal: LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription. PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004486
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004486Summary
MicroRNAs play pervasive roles in controlling gene expression throughout animal development. Given that individual microRNAs are predicted to regulate hundreds of mRNAs and that most mRNA transcripts are microRNA targets, it is essential that the expression levels of microRNAs be tightly regulated. With the goal of unveiling factors that regulate the expression of microRNAs that control developmental timing, we identified lin-42, the C. elegans homolog of the human and Drosophila period gene implicated in circadian gene regulation, as a negative regulator of microRNA expression. By analyzing the transcriptional expression patterns of representative microRNAs, we found that the transcription of many microRNAs is normally highly dynamic and coupled aspects of post-embryonic growth and behavior. We suggest that lin-42 functions to modulate the transcriptional output of temporally-regulated microRNAs and mRNAs in order to maintain optimal expression of these genes throughout development.
Introduction
MicroRNAs (miRNAs) are non-coding RNA molecules that post-transcriptionally regulate gene expression [1]. The maturation of miRNAs is a stepwise process that begins with the RNA polymerase II-dependent transcription of long capped and polyadenylated primary miRNAs (pri-miRNAs) [2], [3]. Most pri-miRNAs are then endonucleolytically cleaved by the nuclear Microprocessor complex, composed of Drosha (an RNase III enzyme) and its binding partner Pasha, to yield a ∼70 nt precursor miRNA hairpin (pre-miRNA) [4]. After export to the cytoplasm, the pre-miRNA is cleaved by Dicer (a second Type III RNase) yielding a ∼22 nt duplex that consists of the mature miRNA and its corresponding passenger RNA [5], [6]. The mature single-stranded ∼22 nt miRNA is then loaded into the Argonaute and GW182 to form the miRNA-induced Silencing Complex (miRISC) [7]–[9]. Through partial complementary base-pairing between the miRNA and target mRNA, the miRISC complex negatively regulates gene expression by either translational repression or mRNA degradation [7], [10]. In vivo, target mRNA down-regulation is directly proportional to the amount of miRNA associated with miRISC [1].
Experimental and computational approaches indicate that an individual miRNA can bind to and regulate hundreds of mRNAs and that the majority of protein-coding genes are miRNA targets [11]–[14]. As such, miRNAs have been implicated in a variety of developmental and cellular processes including cell fate specification, proliferation and apoptosis [15]–[19]. In many of these scenarios, the expression of distinct miRNAs is tightly controlled and/or the individual steps of miRNA biogenesis are actively regulated at either the transcriptional or post-transcriptional level by sequence-specific transcription factors or RNA-binding proteins, respectively. For example, some regulatory proteins control miRNA biogenesis by directly binding structural elements within the pri - or pre-miRNA transcript whereas others broadly impact global miRNA biogenesis by inhibiting enzymes required for general miRNA processing and/or activity [20]. Importantly, many of the proteins that regulate miRNA biogenesis are highly conserved and mutations in these genes result in a variety of developmental disorders and diseases [20].
The C. elegans heterochronic pathway has been instrumental to our understanding of the principles of miRNA-mediated gene regulation and for the identification of components that are required to control miRNA expression, metabolism and activity [21]. Post-embryonic development in C. elegans proceeds through a series of four larval stages, punctuated by molts, in which the temporal and spatial patterns of cell division and differentiation are tightly orchestrated and invariant [22]. Heterochronic genes organize temporal patterns of development by controlling stage-specific gene expression. Defects in heterochronic genes cause animals to display temporal cell fate transformations including either the inappropriate skipping or reiteration of stage-specific patterns of cell divisions [23]. An overarching feature of the heterochronic pathway is that many protein-coding genes that are important for controlling temporal patterning are post-transcriptionally regulated by miRNAs [16], [24]–[28]. In this context, miRNAs are expressed at defined times during post-embryonic development and function as molecular switches to inhibit earlier patterns of development and promote the emergence of later gene expression profiles. Throughout post-embryonic development, the expression of heterochronic miRNAs is regulated at both the transcriptional and post-transcriptional levels [20], [29]–[32]. In addition, mutations that alter heterochronic miRNA expression often display strong temporal patterning and behavioral phenotypes [16], [33]–[36].
While the regulatory strategies that dictate patterns of cell fate specification have rapidly emerged through the identification of conserved heterochronic genes, we still lack a deep understanding of how the temporal expression of heterochronic genes are coordinated with aspects of growth and behavior. This coupling is especially important as many post-embryonic cell division and cell fate specification events are intimately tied to the molting cycle [37], [38]. Surprisingly, most of the known genes required for molting do not dramatically alter temporal cell fates and only a few heterochronic genes disrupt the reiterative process of molting [23], [38]–[45]. The molting phenotypes associated with heterochronic mutants usually result from inappropriate temporal cell fate transformations that lead to either a cessation (for precocious heterochronic mutants) or an inappropriate reiteration (for retarded heterochronic mutants) of molting [16], [23]–[28], [31], [33], [34], [42]–[45]. To date, only a single heterochronic gene, lin-42, is known to alter both temporal patterning of cell fate specification and the precise timing of recurrent developmental events [39]. lin-42 is the C. elegans homolog of human and Drosophila PERIOD and was initially identified as a heterochronic mutant that precociously executes adult-specific patterns of development after the third larval molt [46]–[48]. The lin-42 locus is complex and encodes three protein isoforms (LIN-42A, LIN-42B and LIN-42C) that are expressed from two distinct promoters (Figure 1A) [39], [46]–[48]. During post-embryonic development, lin-42 mRNA levels fluctuate over the molting cycles and peak once during each larval stage [39], [46]–[48]. While its precocious developmental phenotypes are similar to other heterochronic mutants, the periodic expression pattern of LIN-42 distinguishes it from other monotonically expressed heterochronic proteins. Therefore, lin-42 has been proposed to play a more iterative role in developmental timing. However, its relationship to and interplay with other heterochronic genes has been difficult to establish at the molecular level. In addition to altering temporal patterns of development, mutations that disrupt the expression of LIN-42A and LIN-42B isoforms display dramatic defects in behavior and molting [39]. Specifically, lin-42a/b mutants alter the normally synchronous molting patterns displayed by wild-type animals and these defects frequently result in lethality [39]. Given that LIN-42 is a nuclear protein, an attractive hypothesis is that LIN-42 coordinates gene expression programs that control the molting cycles with regulatory pathways that mediate stage-specific cell lineage programs [48]. However, this potential role for LIN-42 remains elusive because 1) the molecular nature of LIN-42 activity is yet to be defined and 2) LIN-42 downstream targets that mediate iterative (molting) and sequential (cell fate patterning) gene regulatory programs are unknown.
Fig. 1. Mutations in lin-42 suppress defects in temporal gene expression and cell lineage phenotypes of heterochronic miRNA mutants. 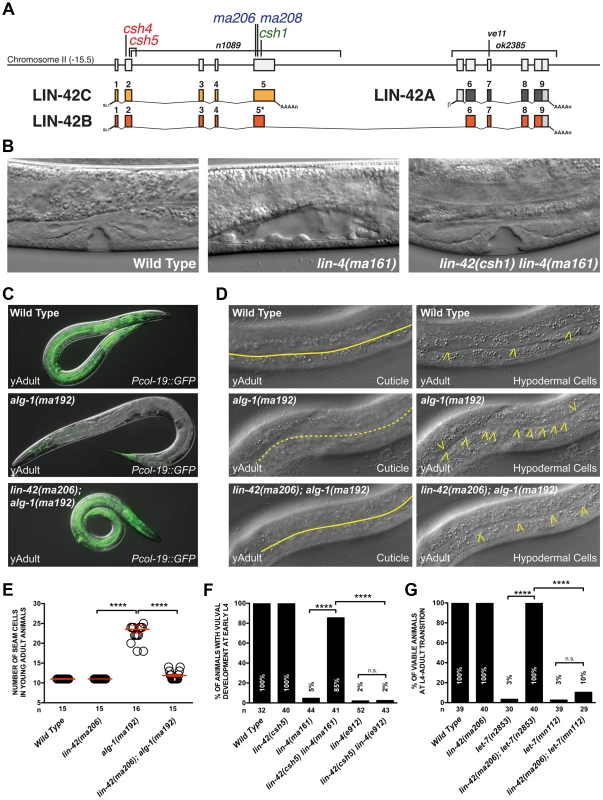
(A) The genomic locus of lin-42 and the corresponding location of the mutations identified in this screen. Red labeled alleles were identified in lin-4(ma161), blue in alg-1(ma192) and green in let-7(n2853) genetic backgrounds. Alleles labeled in black have been previously described [39], [47], [48]. (B) Defects in vulval cell fate specification in lin-4 mutant animals are corrected by lin-42 mutations. (C) Adult-specific Pcol-19::GFP expression patterns in wild-type, alg-1(ma192) and lin-42(ma206); alg-1(ma912) animals. (D) Adult-specific alae phenotypes and seam cell phenotypes of wild-type, alg-1(ma192) and lin-42(ma206); alg-1(ma192) animals. Solid yellow lines indicate complete alae whereas dashed yellow lines indicate an absence of alae structures. Yellow arrowheads indicate lateral seam cells. (E) Quantification of seam cell numbers of young adult wild-type, lin-42(ma206), alg-1(ma192) and lin-42(ma206); alg-1(ma192) animals. Red error bars indicate standard deviation from the mean (SD) (F and G) lin-42 mutations suppress vulval cell lineage and lethality phenotypes of hypomorphic alleles of heterochronic miRNAs but not null mutations of these genes. For E, F and G, four asterisks (****) indicate a highly significant association (the two-tailed P value is less than 0.0001) between groups and/or outcomes as measured by Fisher's exact test and n.s. equals no statistical significance. In this study, we employed multiple forward genetic screens that were collectively geared to identify negative regulators of miRNA expression. As a product of this approach, we identified mutations in lin-42 that suppress multiple stage-specific lineage defects associated with heterochronic miRNAs. Analysis of miRNA expression in lin-42 mutant animals suggests that LIN-42 broadly functions to negatively regulate miRNA expression is therefore is likely to act in a variety of pathways that require miRNAs for proper cell fate specification. Consistent with this hypothesis, we find that lin-42 also plays a role in the miRNA-mediated specification of asymmetric gene expression patterns in gustatory neurons. Analysis of LIN-42 interactions with chromatin suggests that LIN-42 potentially regulates the transcription of both miRNAs and mRNAs. We demonstrate, through the use of transcriptional reporters, that lin-42 mutations alter the transcription of lin-4 and let-7. Surprisingly, mutations that remove LIN-42 isoforms containing the conserved PAS domains (required for circadian gene regulation by human and Drosophila PERIOD) do not uncouple miRNA expression from the molting cycle but, instead, dramatically alter the transcriptional output of miRNA genes. We conclude that a key molecular function of lin-42 is to dynamically inhibit the transcription of post-embryonically expressed miRNAs and mRNAs to ensure the robustness of developmental gene expression.
Results
lin-42 functions during post-embryonic development to ensure proper temporal cell fate specification mediated by miRNAs
The inherent dependency of the heterochronic pathway on precisely controlled miRNA activity provides a unique genetic context to identify components that control aspects of miRNA metabolism or expression. To accomplish this, we performed forward genetic screens in either lin-4(ma161), alg-1(ma192) or let-7(n2853) mutant backgrounds to identify novel heterochronic mutations that correct the phenotypes associated with aberrant L1 to L2 (early), L2 to L3 (middle) or L4 to adult (late) cell fate transitions, respectively. These mutants are unique in that they express miRNAs at a much lower level than wild-type animals but do not completely eliminate their expression. lin-4(ma161) and let-7(n2853) mutations alter the conserved seed sequence of the mature miRNA and reduce levels of these miRNAs in vivo [16], [24]. Animals harboring lin-4(ma161) and let-7(n2853) mutations are phenotypically indistinguishable from null mutants and reiterate L1 - and L4-specific cell fates, respectively (Table 1) [16], [24]. alg-1(ma192) mutations alter one of the two miRNA-specific Argonautes and disrupt the ability of processed miRNAs to repress downstream target mRNAs [49]. Animals harboring the alg-1(ma192) mutation inappropriately express hbl-1 (the major miRNA target of miR-48, miR-241, and miR-84) in the L3 stage and reiterate L2-specific seam cell division patterns [49]. Consistent with the defects associated with the misregulation of each of these stage-specific transitions, lin-4(ma161), alg-1(ma192) and let-7(n2853) animals display highly penetrant heterochronic phenotypes and fail to express adult-specific gene regulatory programs, including the expression of the adult-specific Pcol-19::GFP transcriptional reporter (Table 1). Suppressors of the retarded heterochronic phenotypes in each of these genetic backgrounds were identified as F2 progeny of mutagenized animals that were able to restore normal development (Figure 1C). Five mutants (ma206, ma208, csh1, csh4 and csh5) were able to suppress multiple retarded heterochronic phenotypes associated with all three mutant backgrounds. Each mutant mapped to a single locus on chromosome II (Figure 1A and Table 1), and subsequent SNP-SNP mapping and sequencing results demonstrated that all five alleles contain mutations that lie within lin-42 and would be predicted to create a premature truncation of the lin-42b or lin-42c open reading frames (Figure 1A and Table S1) [50]. Consistent with previous analyses of lin-42 mutations, animals harboring the ma206, ma208, csh1, csh4 or csh5 allele display highly-penetrant precocious heterochronic phenotypes (Table 1) which were rescued with a fosmid containing the genomic fragment of the wild-type lin-42 gene [46]–[48]. Mutations in lin-42 have been demonstrated to suppress heterochronic phenotypes associated with multiple heterochronic mutants, including lin-4 and let-7 [16], [46], [47], [51]. In these reports, only terminal cell lineage phenotypes, including a correction of the L4-to-adult vulval bursting phenotypes, restoration of adult-specific expression of Pcol-19::GFP, and formation of adult-specific alae were assayed.
Tab. 1. lin-42 mutations suppress heterochronic phenotypes of multiple heterochronic miRNAs. 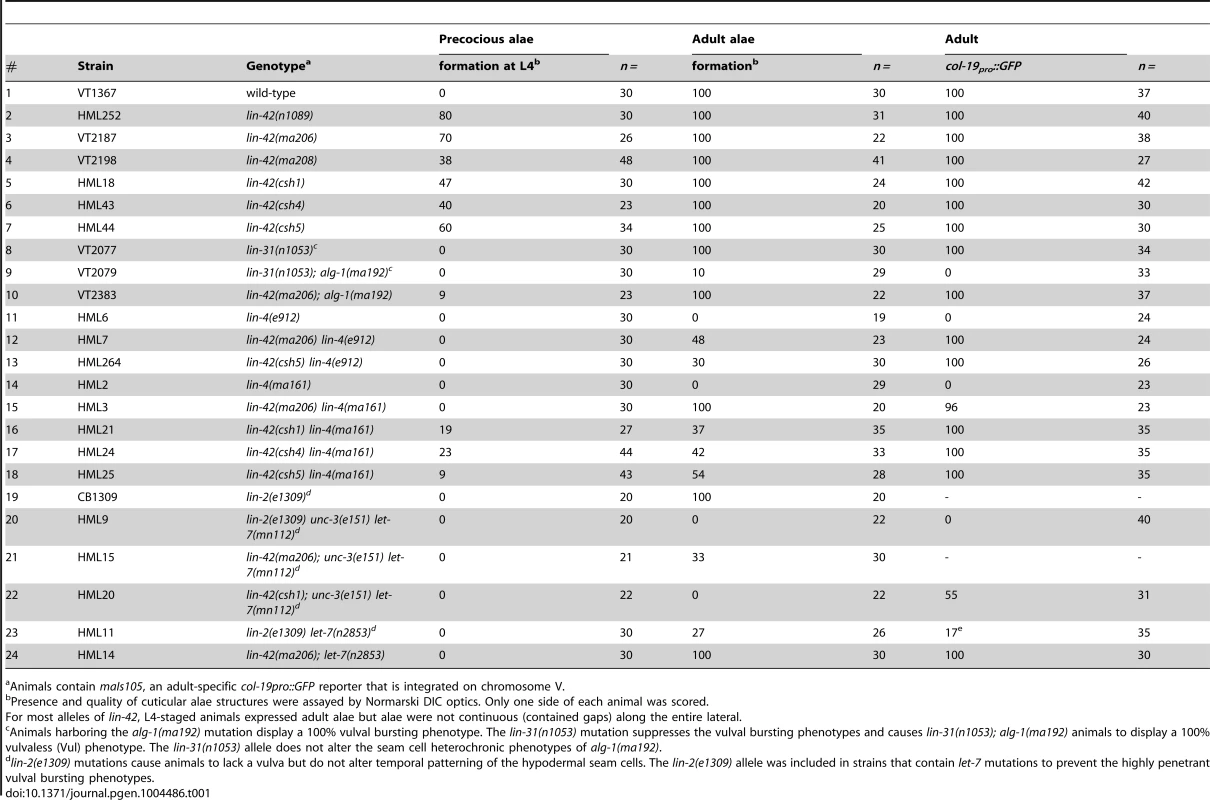
Animals contain maIs105, an adult-specific col-19pro::GFP reporter that is integrated on chromosome V. We next sought to determine whether the lin-42 mutations we isolated suppressed only terminal heterochronic phenotypes or if they corrected additional stage-specific cell lineage defects associated with lin-4(ma161), let-7(n2853) and alg-1(ma192) mutations. To test if our new lin-42 mutants correct retarded cell lineage phenotypes, we compared multiple hypodermal cell lineages in lin-4(ma161), alg-1(ma192), and let-7(n2853) single mutants to double mutants that also harbored the individual lin-42 candidate suppressor mutations. lin-4 animals lack vulval structures as a consequence of reiterating L1-specific developmental programs in the hypodermis and failing to interpret inductive cues from the anchor cell that initiate vulval morphogenesis at the L3 stage [40], [52]. The vulvaless (Vul) phenotypes of lin-4(ma161) animals are highly penetrant (Figure 1B, F) and are almost completely suppressed by lin-42(ma206), lin-42(ma208), lin-42(csh1), lin-42(csh4) and lin-42(csh5) (Figure 1B, F). These results indicate that lin-42 functions to control cell fate specification in at least the mid-L3 stage, when the vulval precursors are spatially patterned.
The ability of several of these suppressors to alleviate hypodermal cell lineage phenotypes in miRNA hypomorphic mutants was not limited to the vulval cell lineage. The lateral seam cells of lin-4(ma161), alg-1(ma192), and let-7(n2853) animals display altered temporal cell fate specification and also fail to terminally differentiate at the L4 molt. As a consequence, lin-4(ma161), alg-1(ma192), and let-7(n2853) animals lack alae structures as young adults (Table 1, Figure 1D). The alae phenotypes in lin-4(ma161), alg-1(ma192), and let-7(n2853) mutants was strongly suppressed by the lin-42(ma206) allele (Table 1). alg-1(ma192) mutants reiterate L2-specific seam cell division programs due to the inappropriate perdurance of hbl-1 expression at the L3 stage [49]. As a consequence, young adult alg-1(m192) animals harbor supernumerary seam cells (23.5+/−3.78; WT = 11) (Figure 1D, E). lin-42(ma206) mutations strongly suppress the L2-to-L3 heterochronic phenotypes of alg-1(ma192) mutants as lin-42(ma206); alg-1(ma192) animals exhibit a significant reduction in the number of supernumerary seam cells (11.9+/−1.3) and display normal adult alae (Figure 1D and E). Therefore, lin-42 has a role in controlling L2-to-L3 temporal cell fate transitions.
lin-42 mutations do not suppress lineage defects and lethal phenotypes associated with lin-4(0) and let-7(0) alleles
We asked whether our new lin-42 alleles could suppress the heterochronic phenotypes associated with lin-4(e912) and let-7(mn112) null mutants to a level similar to that observed with the hypomorphic alleles used in our initial screens. To test this, we compared aspects of vulval cell proliferation and morphogenesis at the early L4 stage in lin-4(ma161), lin-42(csh5) lin-4(ma161), lin-4(e912) and lin-42(csh5) lin-4(e912) mutants to those of similarly staged wild-type and lin-42(ma205) animals. Lowering lin-42 function in the context of the hypomorphic lin-4(ma161) background results in a strong restoration of vulval development with 85% of animals exhibiting induction/proliferation and invagination of P cells from the larval cuticle (Figure 1B and F). Surprisingly, 42% percent of lin-42(csh5) lin-4(ma161) animals exhibited morphologically normal adult vulva and were competent for egg laying (n = 100). In contrast, reducing lin-42 activity in lin-4(e912) animals has little or no effect on P cell proliferation and vulval morphogenesis (Figure 1F). lin-42 exhibits a similar genetic relationship to let-7 mutations. Both hypomorphic (n2853) and null (mn112) alleles of let-7 display highly penetrant vulval bursting phenotypes at the L4-to-adult transition (Figure 1G) [16], [28]. lin-42 mutations almost completely suppress the lethality associated with larval-to-adult transitions in let-7(n2853) animals but do not statistically improve the viability of let-7(mn112) adults (Figure 1G). These results strongly suggest that lin-42 mutations are not bypass suppressors of lin-4 or let-7 mutant phenotypes but likely require a minimum level of lin-4 or let-7 activity for suppression.
lin-42 loss-of-function mutations lead to an overproduction of many C. elegans miRNAs
One mechanism by which lin-42 mutations could suppress multiple hypomorphic miRNA mutants would be that lin-42 normally functions to repress some aspect of miRNA metabolism. To directly test this hypothesis, we measured the abundance of several mature miRNAs when lin-42 function is compromised. Northern blot analysis of total RNA extracted from morphologically-staged, young adult animals demonstrates that the total amount of lin-4 and let-7 miRNAs in alg-1(ma192) mutants is 1–1.5 fold lower than the levels found in wild-type animals (Figure 2A and B). In addition to reducing the levels of mature let-7 miRNA, alg-1(ma192) animals display a slight reduction in pre-miRNA processing and accumulate the pre-let-7 hairpin precursor. This under-accumulation phenotype of mature lin-4 and let-7 miRNAs in alg-1(ma192) mutants is suppressed when lin-42 function is compromised (Figure 2A). Consistent with our hypothesis that lin-42 normally inhibits miRNA biogenesis, similarly-staged lin-42(ma206) mutants over-accumulate both lin-4 and let-7 miRNAs (Figure 2A). While the amount of mature let-7 miRNA increases in lin-42(ma206); alg-1(ma192) double mutants, the ratio of pre-let-7 to mature let-7 miRNA is similar to that detected in alg-1(ma192) single mutants (Figure 2A). Therefore, although mature let-7 miRNA over-accumulates in lin-42(ma206) mutants, there is no change in pre-let-7 to mature let-7 processing efficiency as compared to wild type. These data suggest that lin-42 mutations alter aspects of miRNA expression upstream of pre-miRNA processing.
Fig. 2. lin-42 mutations lead to the overexpression of several miRNAs. 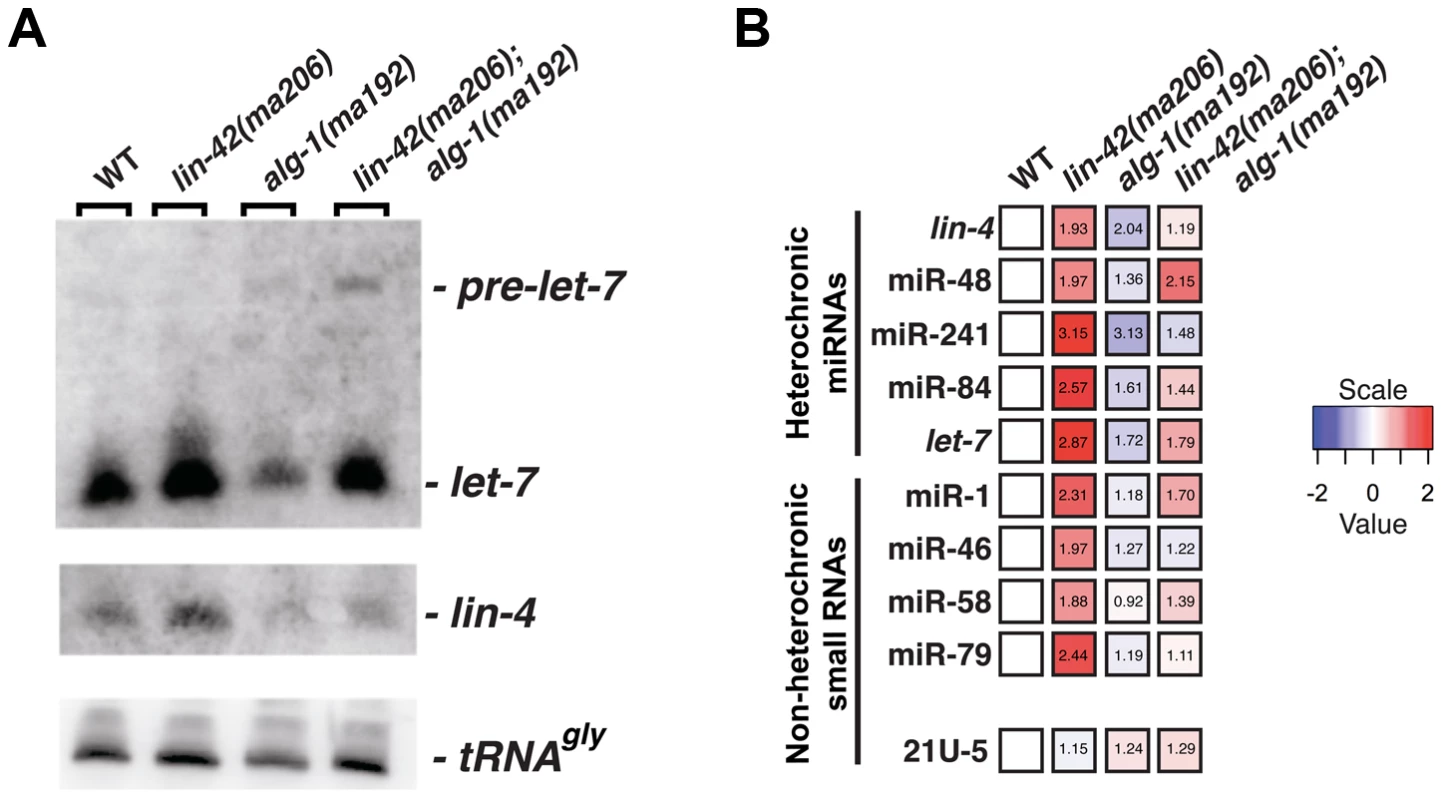
(A) Small RNA northern analysis of 20 µg of total RNA extracted from morphologically staged, young adult wild-type, lin-42(ma206), alg-1(ma192) and lin-42(ma206); alg-1(ma192) animals. Blots were probed sequentially for the indicated miRNAs. tRNAGly serves as a loading control. (B) The results of miR-TaqMan assays to quantify the levels of mature miRNAs in wild-type, lin-42(ma206), alg-1(ma192) and lin-42(ma206); alg-1(ma192) animals. Notice that lin-42(ma206) displays the highest levels of miRNAs relative to the other genotype backgrounds. Data represent 3 biological replicates with 3 technical replicates each. Heat map colors are shown as log 2 scale as indicated and within each individual assay. Red indicates an increase in miRNA expression and blue indicates a reduction in mature miRNA levels. Numbers within each box indicate standard fold change when compared to wild-type samples. To determine if lin-42 plays a more broad role in modulating miRNA expression, we employed real-time quantitative PCR to measure the expression levels of additional miRNAs in morphologically-staged, young adult wild-type, lin-42(ma206), alg-1(ma192) and lin-42(ma206); alg-1(ma192) animals. We measured a variety of miRNAs that display tissue-specific and temporal expression patterns that are distinct from lin-4 and let-7 miRNAs [35], [53]–[58]. For comparison, we also assayed the expression of two additional small nuclear RNAs (U18 and sn2343) as well as two 21U RNAs that associate with PRG-1, a distinct Argonaute involved in the C. elegans piRNA pathway [59]–[61]. Consistent with the observation that alg-1(ma192) mutations broadly affect miRNA expression, the abundance of all miRNAs tested (lin-4, miR-48, miR-241, miR-84, let-7, miR-1, miR-46, miR-58 and miR-79) was decreased in alg-1(ma192) mutants (Figure 2B). The general miRNA under-accumulation phenotype displayed in alg-1(ma192) mutants was suppressed by removing lin-42 function (Figure 2B). Importantly, the expression levels of the 21U-RNA transcripts were not significantly altered in lin-42(ma206) mutants (Figure 2B). Examination of miRNA expression in lin-42(ma206) mutants indicate that all tested miRNAs were overexpressed from ∼1.8 to ∼3.2 fold when compared to similarly-staged wild-type animals (Figure 2B). miRNA stability is dependent on a variety of factors, including the expression levels of the Argonaute components of miRISC [62]. To determine if the increase in miRNA levels in lin-42 mutant backgrounds was due to the overexpression of the C. elegans miRNA-specific Argonautes (ALG-1 and ALG-2), we quantified the levels of functional ALG-1 and ALG-2 fluorescent reporters in animals with reduced lin-42 activity. The results of this analysis, presented in Figure S1, indicate that ALG-1 and ALG-2 expression is not altered in lin-42(RNAi) animals. Collectively, these results indicate that lin-42 functions to negatively regulate the expression of a wide range of miRNAs.
lin-42 suppresses dosage-dependent phenotypes of non-heterochronic miRNA mutants
Because lin-42 regulates the abundance of many miRNAs, we asked if lin-42 functions in other gene regulatory pathways where controlling the expression levels of specific miRNAs is critical for proper cell fate determination. To test this idea, we examined how mutations in lin-42 affected the cell fate specification of two bilaterally symmetric gustatory neurons, ASE left (ASEL) and ASE right (ASER). Normally, a complex gene regulatory network composed of miRNAs and transcription factors form a bi-stable, double-negative feedback loop that ensures mutually exclusive gene expression programs in ASEL and ASER neurons [63], [64]. A major determinant of the exclusive gene expression programs in these two neurons is the ASEL-specific expression of the lsy-6 miRNA and the resulting down-regulation of its target, cog-1. Animals completely lacking lsy-6 fail to down regulate COG-1 in ASEL, and, as a consequence, ASEL neurons in lsy-6(ot71) null mutants adopt an ASER cell fate [63]. These phenotypes can be monitored by a failure to express the Plim-6::GFP transcriptional reporter in ASEL in lsy-6 mutants (Figure 3A). Importantly, lsy-6-mediated repression of cog-1 is dosage-dependent; weak alleles of lsy-6, such as ot150, under-accumulate lsy-6 miRNA as a consequence of reduced lsy-6 transcription and result in a partially penetrant ASEL-to-ASER cell fate transformation phenotype (Fig. 3B) [64]. The ot150 allele of lsy-6 has been used in a variety of contexts as a sensitized genetic background to identify gene products that function in the miRNA pathway [65]–[67]. While 13% of animals harboring only the lsy-6(ot150) allele fail to maintain Plim-6::GFP in ASEL, the penetrance of this phenotype is partially suppressed in lin-42(ma206); lsy-6(ot150) double mutants (Figure 3B), suggesting that lin-42 may play a modulatory role in neuronal cell fate specification. To further explore a potential role for lin-42 in assuring proper neuronal cell fate specification, we developed a more sensitive assay for the lsy-6-mediated repression of cog-1. As previously mentioned, alg-1(ma192) mutants display defects in variety of miRNA-mediated processes, including developmental timing [49]. While the alg-1(ma192) mutation alone does not alter Plim-6::GFP expression in ASEL, combining alg-1(ma192) with lsy-6(ot150) results in a dramatic increase in ASEL to ASER cell fate mis-specification (Figure 3B). As with the suppression of alg-1(ma192) heterochronic phenotypes, reducing lin-42 function significantly restores normal ASEL cell fate specification in lsy-6(ot150); alg-1(ma192) animals (Figure 3B). Because lsy-6-mediated cell fate specification is established during embryonic development, we conclude that lin-42 functions throughout development and is critical for multiple miRNA-mediated developmental processes.
Fig. 3. lin-42 suppresses neuronal phenotypes associated with lsy-6 miRNA-mediated cell fate specification. 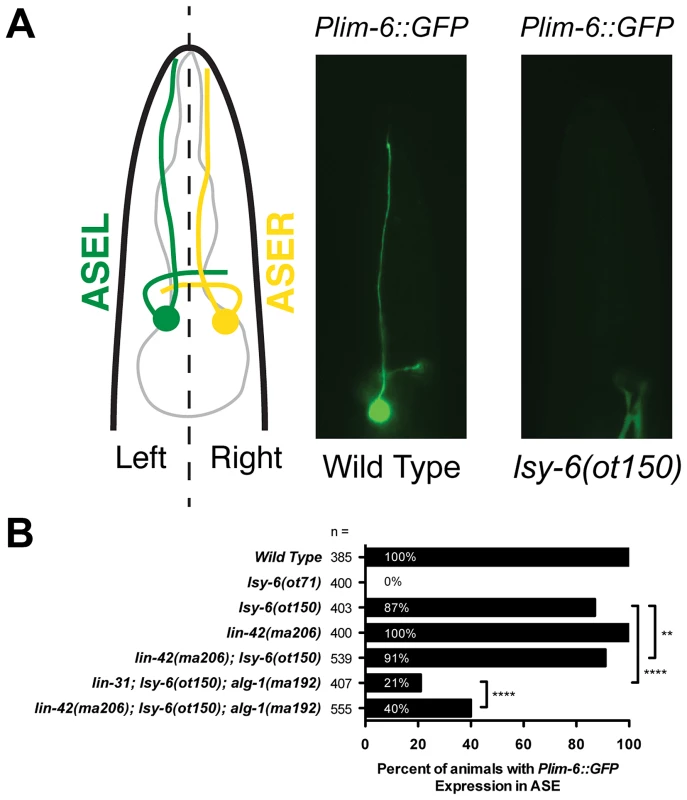
(A) A diagram of a C. elegans larva illustrating the location of the gustatory neurons, ASEL and ASER, whose asymmetric patterns of gene expression are controlled by the ASEL-specific expression of the lsy-6 miRNA. Mutations in lsy-6 result in animals that fail to express the ASEL-specific cell fate reporter Plim-6::GFP. (B) Quantification of Plim-6::GFP expression phenotypes of lsy-6, lin-42, alg-1, and lin-42; alg-1 compound mutants. Four asterisks (****) indicate a highly significant association (the two-tailed P value is less than 0.0001) between groups and/or outcomes as measured by Fisher's exact test. Two asterisks (**) indicate a statistically significant association (P = 0.0242). miRNAs display dynamic expression patterns that are coupled to the molting cycles
To characterize the spatial and temporal expression patterns of lin-42-regulated miRNAs, we generated a series of engineered transcriptional reporters that contain between 2 and 5 kB of genomic upstream regulatory sequence that drives the expression of GFP fused to an optimized proline-glutamate-serine-threonine-rich (PEST) sequence. PEST domains have been demonstrated, in a variety of heterologous systems, to accelerate the degradation of target proteins via the nuclear and cytoplasmic 26S proteasome [41], [68]–[71]. In contrast to transcriptional reporters that drive the expression of stable GFP, analysis of GFP-pest expression in Plin-4::GFP-pest, Plet-7::GFP-pest or PmiR-1::GFP-pest transgenic animals indicates that the expression of each transcriptional reporter is highly dynamic, with peak GFP-pest expression occurring once each larval stage (n>30 animals per time point)(Figure 4A) [29], [53], [55]. The highly dynamic nature of each expression pattern was then monitored in a population of worms that were transiently arrested at the L1 diapause and then developmentally synchronized by restoring bacterial food. For each of the mir::GFP-pest reporters, post-embryonic GFP-pest expression was first detected at approximately 14 hours (Figure 4B, D and F). Once transcriptionally activated, Plin-4::GFP-pest and Plet-7::GFP-pest reporters peak in expression by 18–20 hours and diminish with similar kinetics (Figure 4B and D). For animals expressing the Plet-7::GFP-pest reporter we monitored GFP-pest expression for longer periods after release from L1 arrest. Consistent with the highly pulsatile nature of this expression pattern, GFP-pest expression was reinitiated at 30 hours, which correlates with the later portions of the L2 stage (Figure S3). While transcriptional activation of the Pmir-1::GFP-pest reporter was also initiated at 14 hours post-L1 arrest, the peak of Pmir-1::GFP-pest expression occurred at a later time point, and diminished with slower kinetics, as compared to Plin-4::GFP-pest and Plet-7::GFP-pest expression (Figure 4F).
Fig. 4. The promoters of three different miRNAs display dynamic expression patterns that are coupled to the larval molting cycle. 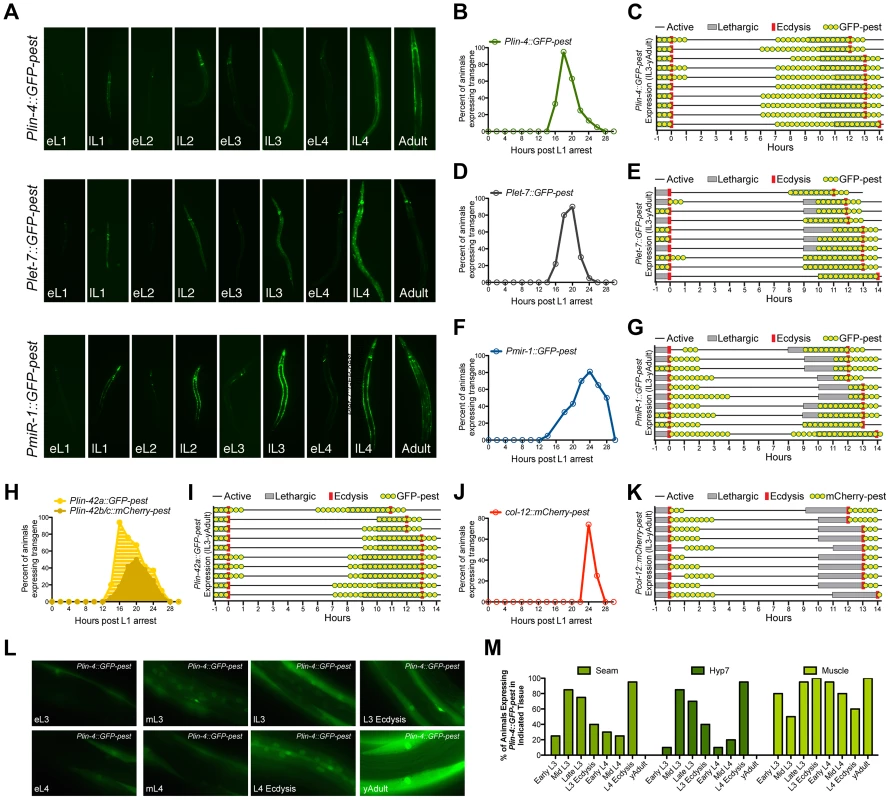
(A) Transcriptional reporters for lin-4, let-7 and miR-1 drive GFP-pest expression in an oscillatory manner during C. elegans post-embryonic development. For each reporter, a single pulse of GFP expression is seen near the end of each inter-molt period of animals grown at 20°C (eL1, early L1 stage; lL1, late L1 stage). (B–K) GFP or mCherry expression profiles of animals expressing the indicated reporters. For panels B, D, F, H and J, larvae were synchronized by starvation-induced L1-diapause, fed and cultivated at 20°C. For panels C, E, G, I and H, a single animal in L3 lethargus (as judged by reduced movement and lack of pharyngeal pumping) was placed on an individual NGM plate seeded with OP50, grown at 20°C, and monitored for GFP or mCherry expression (yellow dots), the lethargus period (grey bars) or ecdysis (red bars). Plin-4::GFP-pest and col-12::mCherry-pest expression were monitored simultaneously in HML168, which co-expresses both reporters. (L) Representative fluorescent images of individual animals expressing the Plin-4::GFP-pest reporter at the indicated stage at 20°C. (M) Graphical representation of the percentage of animals expressing the Plin-4::GFP-pest reporter in any of the hyp7, seam or muscle cells at the indicated stage (n = 20 for each stage). We then asked whether the temporal expression pattern of each Pmir::GFP-pest reporter was synchronized with defined stages of the molting cycle, specifically lethargus and ecdysis. To accomplish this, we isolated late-L3-staged transgenic animals and cultured them on separate nematode growth media (NGM) plates at 20°C. Individual animals were then monitored for GFP-pest expression in relation to the induction and termination of both lethargus and ecdysis (Figure 4C, E, and G). We find that the majority of animals which harbor the Plin-4::GFP-pest transgene cease GFP-pest expression by L3 ecdysis and resume expression by the mid-L4 stage. The pulse of Plin-4::GFP-pest expression at the L4 stage extends through the early portion of young adulthood and completely overlaps with the lethargus period in all animals (Figure 4C). Plet-7::GFP-pest expression followed a similar pattern (Figure 4E). However, GFP-pest expression was more variable at the L3-to-L4 transition and L4-specific induction of this transgene was primarily restricted to the lethargus period (Figure 4E). In contrast to the expression profiles of the lin-4 and let-7 reporters, induction of Pmir-1::GFP-pest expression began during, or immediately after, L3 ecdysis and persisted into the L4 stage. A second pulse of Pmir-1::GFP-pest expression completely overlapped with the L4 lethargus period and continued into early adulthood (Figure 4G). Collectively, these results suggest that the expression patterns of lin-4, let-7 and mir-1 are dynamic throughout development and that the cyclical transcription of these miRNAs is mediated by their cognate promoter sequences. Furthermore, these data show that, while each of the Pmir::GFP-pest reporters display pulsatile expression patterns, the transcriptional dynamics for each gene do not display a complete unity of phase in their expression profiles.
To compare the temporal expression patterns of these three miRNAs with that of lin-42, we constructed transgenic strains that expressed either Plin-42a::GFP-pest or Plin-42b::mCherry-pest and subjected these animals to the same time course analyses. It has been previously demonstrated that two independent promoters drive the expression of LIN-42A, LIN-42B and LIN-42C isoforms [39]. Consistent with these findings, Plin-42a::GFP-pest and Plin-42b::mCherry-pest reporters displayed highly pulsatile expression during the L1 stage with initiation and termination of expression at 12 and 28 hours post L1 arrest, respectively (Figure 4H). In addition, we find that Plin-42a::GFP-pest expression peaks at 16 hrs, immediately preceding the expression of the Pmir::GFP-pest reporters, while the peak of Plin-42b::mCherry-pest expression occurs at 20 hrs (Figure 4H). Detailed analysis of individual L3-to-adult animals indicates that Plin-42a::GFP-pest expression displays a temporal expression pattern that is highly similar to Plin-4::GFP-pest and Plet-7::GFP-pest expression (Figure 4I). Specifically, in all three reporters, GFP-pest expression diminishes prior to L3 ecdysis, resumes prior to the L4 lethargus period, and terminates immediately after L4 ecdysis (Figure 4I). In striking contrast to our mir and lin-42 transcriptional reporters, Pcol-12::mCherry-pest expression does not occur during the molting cycle, but rather is exclusively expressed after each ecdysis (Figures 4J and 4K).
Previous analysis of lin-4 and let-7 expression indicates that these miRNAs are expressed in a variety of tissues, including the hypodermis, intestine and muscle [29], [35], [53], [57]. To determine if Plin-4::GFP-pest displays differential temporal expression patterns in a subset of these tissues, we conducted a detailed examination of GFP-pest expression from the early-L3 to the young adult stage. Twenty animals from each of eight morphologically-defined stages were imaged (Figure 4L and Figure S4) and then qualitatively scored for GFP-pest expression in seam cells, hyp7 cells or lateral muscle cells (Figure 4M). Expression of the Plin-4::GFP-pest reporter peaked in hyp7 and seam cells at the mid - and late-L3 stage and then again at the late-L4 stage. In addition, the majority of animals exhibited a cessation of hyp7 and seam cell Plin-4::GFP-pest expression immediately after L4 ecdysis (Figure 4M). In contrast, Plin-4::GFP-pest expression in muscle cells displayed a different transcriptional profile. In the majority of animals, expression of GFP-pest in muscles peaked at L3 ecdysis, gradually diminished throughout the remainder of the L4 stage, and increased again at the young adult stage (Figure 4M). These results suggest that, while lin-4 is dynamically expressed once each larval stage, its promoter activity may be differentially regulated in distinct tissues.
LIN-42 is enriched at promoters of coding and non-coding genes
Analysis of the Plet-7::GFP-pest and Plin-4::GFP-pest reporters in lin-42 loss-of-function (lf) animals demonstrated that mutants that alter either lin-42 b/c (lin-42(n0189)) or lin-42a/b (lin-42(ok2385)) isoforms display elevated Pmir-GFP-pest expression in late larval development (Figure 5A, B and Figure S2). These altered temporal expression patterns suggested that lin-42 may normally function to modulate aspects of miRNA transcription. To investigate the potential interactions between LIN-42 and transcriptional regulatory elements, we performed chromatin immunoprecipitation coupled to high throughput sequencing (ChIP-seq) using extracts prepared from animals harboring a functional, GFP-tagged allele of lin-42 (Figure 5C). From two independent biological ChIP-seq replicates derived from separate L4-staged extracts, we obtained 413 high confidence peaks corresponding to chromosomal regions in which LIN-42 is enriched (see Table S2 and Materials and Methods). In agreement with the hypothesis that LIN-42 regulates let-7 transcriptional activity, we find LIN-42 binding sites at conserved let-7 promoter regions that have been previously demonstrated to control let-7 expression (Figure 5C) [31], [35], [57]. Annotation of additional high confidence peaks revealed that 38% (158/413) of LIN-42 peaks fell within the promoters (defined as 2 kb upstream of each gene) of either coding or non-coding genes, 24% (99/413) fell within the introns of coding genes, 8% (34/413) fell within gene bodies and 29% (121/413) fell within other intergenic regions (Figure 5D). Comparison between LIN-42 peak frequency and their distribution relative to the closest annotated transcription start site (TSS) revealed that LIN-42 has two major regions of enrichment: 1) directly at TSSs and 2) at approximately 750 bp upstream of a TSS (Figure 5E). Of these high confidence peaks, 323 were also detected in LIN-42 ChIP-seq samples obtained using an antibody against endogenous LIN-42, suggesting that this list forms a short, but high confidence, group of LIN-42 target genes. A list describing all high-confidence annotated LIN-42 peaks is provided in Table S2. Using the Generic Gene Ontology Term Mapper, we found that numerous genes with high-confidence LIN-42 peaks can be categorized into groups that function in many diverse biological processes, including development, transport, small molecule metabolism, embryogenesis and growth (Table S3). Collectively, these results strongly suggest that LIN-42 plays a role (either directly or indirectly) in a broad range of biological processes and that it predominately interacts with the promoter regions of coding and non-coding genes to regulate their expression.
Fig. 5. LIN-42 binds to the putative regulatory regions of both miRNAs and mRNAs. 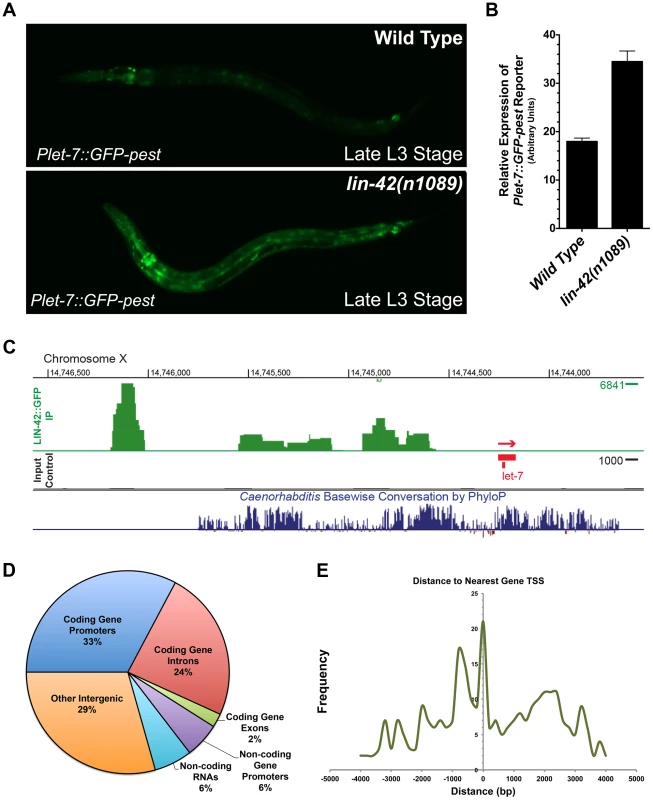
(A) Plet-7::GFP-pest expression is elevated in late L3-staged lin-42(n1089) animals compared to the expression in similarly-staged wild-type animals. (B) Quantification of whole-animal GFP expression in late L3-staged wild-type (n = 20) and lin-42(n1089) animals (n = 20). Error bars indicate the standard deviation from the mean (SD). (C) A representative image of the LIN-42::GFP binding sites within the let-7 genomic region from ChIP-Seq experiments. (D) A pie chart indicating the distribution of the 413 high confidence LIN-42::GFP peaks that were assigned to a RefSeq list of gene models for C. elegans (ce10). (E) LIN-42::GFP binding sites are enriched upstream of the putative transcriptional start sites of both coding and non-coding regions of the genome. lin-42 negatively regulates the transcriptional output of lin-4 and let-7
The genetic and regulatory relationships between lin-42 and lin-4 or let-7, as well as the overlapping temporal expression patterns of these three genes, suggest that lin-42 may play a role in modulating the dynamics of lin-4 and let-7 transcriptional activity. To directly test the idea that lin-42 regulates miRNA levels at the transcriptional level, we quantified the transcriptional profiles of Plin-4::GFP-pest and Plet-7::GFP-pest reporters in wild-type animals and lin-42(n1089) mutants. The n1089 allele of lin-42 deletes genomic sequences that eliminate the coding potential of the lin-42b and c isoforms (Figure 1A) and displays strong heterochronic phenotypes [39], [46], [47]. Importantly, these isoforms contain the domains, PAS-A and PAS-B, that most closely link LIN-42 to PERIOD, a protein involved in controlling the cyclical expression patterns of circadian-regulated genes [39], [47], [48], [72], [73]. We focused on quantifying the GFP intensities of 1) the hypodermal cells in L3-to-adult-staged Plin-4::GFP-pest animals and 2) the seam cells of similarly-staged Plet-7::GFP-pest animals. These tissues and stages were selected for analysis because the majority of well-characterized heterochronic phenotypes are detected in these tissues [16], [24], [39], [42], [46]–[48]. Expression levels for each transcriptional reporter were analyzed throughout eight defined and sequential stages that spanned from early L3 to young adult (Figure 6A–C and Figure S4). In agreement with our previous observations, expression of Plin-4::GFP-pest in hypodermal cells of wild-type animals is dynamic throughout development and displays two main peaks of GFP expression: one at the late-L3 stage and the other at the L4 molt (Figure 6A, B). Similar results are also observed in the seam cells of wild-type animals expressing the Plet-7::GFP-pest reporter (Figure 6B, C). One exception, however, is that that the first peak of Plet-7::GFP-pest expression occurs at the mid-L3 stage (Figure 6B, C). Surprisingly, we find that the cyclical pattern of expression of these reporters is not affected in animals carrying the lin-42(n1089) mutation; both lin-42(n1089) and wild-type animals display nearly identical Plin-4::GFP-pest and Plet-7::GFP-pest temporal expression patterns (Figure 6A, B and C). In contrast, the abundance of GFP-pest expression for each reporter is universally higher in lin-42(n1089) mutants as compared to similarly-staged wild-type animals (Figure 6 A, B and C). In the case of the Plin-4::GFP-pest reporter, higher levels of GFP-pest intensity are observed in hypodermal cells throughout all developmental stages, with the greatest difference occurring between the late-L3 and L3-molt stages (3.1 and 4.3 fold respectively)(Figure 6B). Interestingly, although Plet-7::GFP-pest expression in lin-42(n1089) mutants is also greater in seam cells between the late-L3 and L3-molt stages (2 fold each), Plet-7::GFP-pest expression in lin-42(n1089) and wild-type animals is practically indistinguishable from wild-type during the mid-L4 to the young adult stages (Figure 6C). Taken together, these results suggest that mutations that abolish the expression of PAS domain-containing LIN-42 isoforms do not alter the cyclical expression patterns of miRNA genes during development. Rather, these mutations alter the transcriptional output of miRNAs that display oscillatory expression patterns.
Fig. 6. lin-42 controls the output of lin-4 and let-7 transcription. 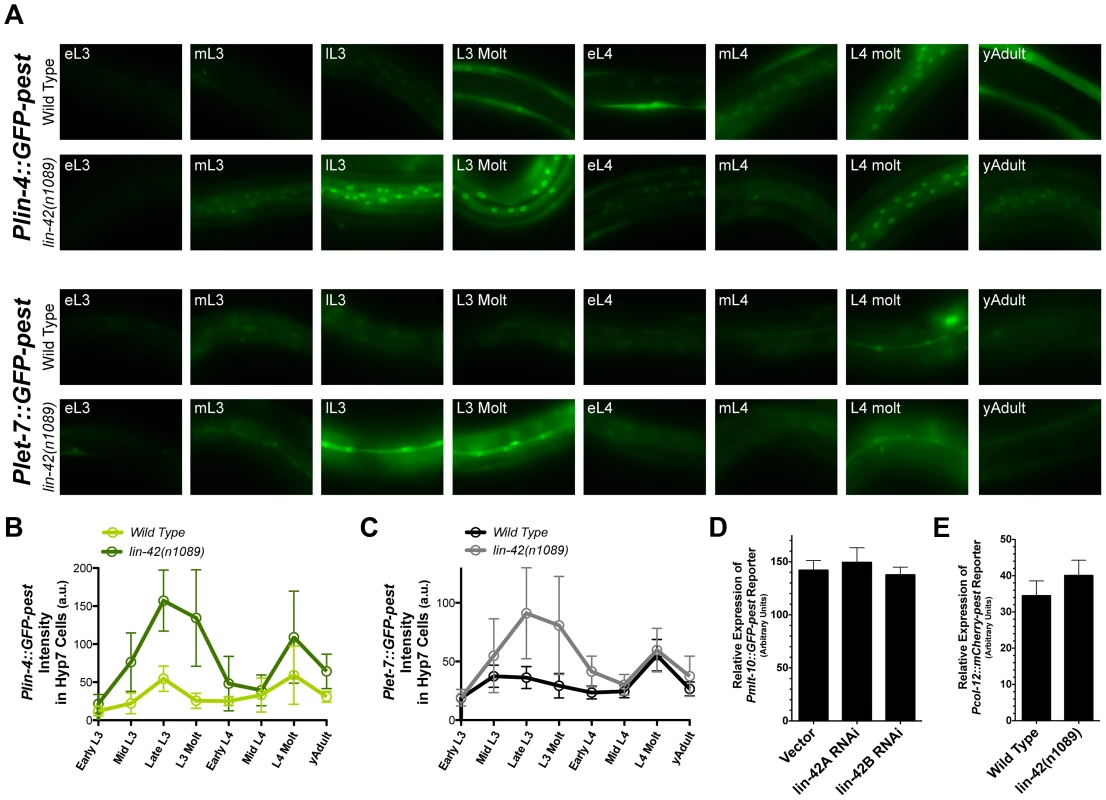
(A) Representative expression patterns of the Plin-4::GFP-pest and Plet-7::GFP-pest transcriptional reporters in morphologically-staged wild-type and lin-42(n1089) animals grown at 20°C (early L3 to young adult). See Figure S4 for details. (B and C) A quantitative representation of gene expression profiles measured at the cellular level for both transcriptional reporters. GFP intensities for Plin-4::GFP-pest and Plet-7::GFP-pest were measured in the nuclei of hypodermal cells and seam cells, respectively. For Plin-4::GFP-pest, each data point in the graph represents the average from 200 nuclei. For Plet-7::GFP-pest, each data point represents the average from 50 seam cells. Error bars indicate the standard deviation from the mean (SD). (D) Quantitation of Pmlt-10::GFP-pest reporter expression in the hyp7 cells of lL4-staged F1 animals that have been exposed to bacteria expressing control RNAi (pDP129.36) or bacteria expressing dsRNAs against two isoforms of the lin-42 gene (n = 20 for each experiment). Error bars indicate the standard error of the mean (SEM). (E) Quantitation of Pcol-12::mCherry-pest reporter expression in seam cells of young adult, wild-type (n = 20) or lin-42(n1089) animals (n = 20). Error bars indicate the standard error of the mean (SEM). As demonstrated in Figure 5D and Table S2, LIN-42 binds the putative regulatory regions of multiple protein coding genes. This observation raises the possibility that LIN-42 may modulate the transcriptional output of other developmentally regulated genes, including those whose expression, like that of lin-4 and let-7, is also linked to the molting cycle. To determine if lin-42 mutants alter the transcriptional output of other cyclically expressed mRNAs, we observed the expression of two transcriptional reporters for genes involved in the molting process, Pmlt-10::GFP-pest and Pcol-12::mCherry-pest. In wild-type animals, Pmlt-10::GFP-pest transcription begins at the end of each larval period when the new cuticle is being synthesized [39], [41]. We monitored the expression of Pmlt-10::GFP-pest in F1 animals that had been exposed to control RNAi or two RNAi constructs that target all major isoforms of lin-42 and induce precocious expression of Pcol-19::GFP and adult alae [74]. As with the expression of Plin-4::GFP-pest and Plet-7::GFP-pest reporters, the Pmlt-10::GFP-pest reporter maintained its normal, oscillatory pattern of expression in lin-42(RNAi) animals. Quantification of Pmlt-10::GFP-pest reporter expression at the late-L4 stage (where Pmlt-10::GFP-pest normally peaks [39], [41]) indicates that lin-42 depletion does not alter the transcriptional output of the mlt-10 promoter (Figure 6D). In addition, quantification of the Pcol-12::mCherry-pest reporter in young adult lin-42(n1089) animals also indicates that mutations in lin-42 do not alter the temporal expression patterns or levels of the col-12 promoter (Figure 6E). Therefore, while lin-42 mutations alter the transcriptional output of the lin-4 and let-7 genes, lin-42 does not play an essential role in controlling the oscillatory expression patterns or transcriptional output of all genes whose expression is tied to the molting cycle.
Discussion
Using an unbiased genetic approach, we sought to identify factors that modulate the expression of miRNAs that are critical for controlling temporal patterns of development throughout post-embryonic development. Our strategy was two-fold: 1) we sought to identify suppressors of heterochronic miRNA mutant phenotypes characterized by stage-specific alterations in temporal patterning and 2) we focused on identifying suppressors that preferentially alleviate phenotypes that result from a reduction in, rather than a complete loss of, miRNA expression. These efforts identified lin-42, the C. elegans homolog of the circadian period gene, as a component that not only modulates heterochronic miRNA expression, but also regulates the expression of a wide range of broadly expressed, and functionally distinct, C. elegans miRNAs. Previous genetic analyses implicated lin-42 as a heterochronic gene that normally inhibits the precocious expression of adult characteristics [39], [46]–[48]. The precise placement of lin-42 in the developmental timing pathway has been difficult to incorporate due to the observation that lin-42 mutations alter cell lineage programs that occur exclusively in late development, namely the transition from the L3 to the L4 stage [46]–[48], [51]. In addition, epistasis experiments with other developmental timing mutants suggest that its interaction with other heterochronic genes is complex [46], [47], [51], [75], [76]. Furthermore, unlike other components that control discrete aspects of temporal patterning and display monotonic expression patterns, lin-42 expression is highly dynamic, suggesting a reiterative role for it in the heterochronic pathway.
Results from our screens have identified five new alleles of lin-42 that suppress the adult-specific gene expression defects of hypomorphic alleles of heterochronic miRNAs. We also find that lin-42 corrects stage-specific cell fate specification defects present throughout larval and adult development in these miRNA mutants. These results indicate that lin-42 functions iteratively to control temporal cell fate specification by controlling the transcription of distinct miRNAs. In addition, we demonstrate that our newly-identified lin-42(lf) mutants precociously express adult-specific programs and that these defects are suppressed by mutations in components of the miRNA machinery. Accordingly, these data suggest that lin-42(lf) heterochronic phenotypes are due to an overexpression of specific miRNAs that control temporal patterning. Also, our results demonstrate that lin-42 mutations are not bypass suppressors of the heterochronic phenotypes displayed by lin-4 and let-7 null mutants, suggesting that lin-42 suppresses retarded heterochronic phenotypes by increasing the expression of heterochronic miRNAs or enhancing their effectiveness in regulating miRNA targets.
Multiple lines of evidence described in this manuscript support the conclusion that LIN-42 regulates the transcription of a wide array of miRNAs. First, we investigated how our lin-42 suppressor alleles affected the overall levels of a subset of miRNAs involved in developmental timing. alg-1(ma192) animals display profound defects in temporal cell fate specification and also under-accumulate both lin-4 and let-7 miRNAs. Genetic and molecular experiments indicate that lin-42 suppresses alg-1(ma192)-dependent phenotypes by increasing the available amount of mature miRNAs. Second, lin-42(ma206) mutants over-accumulate multiple miRNAs, including those with no apparent role in developmental timing. Consistent with the hypothesis that lin-42 functions in additional gene regulatory pathways that require miRNA activity, we demonstrated that lin-42(lf) mutants suppress phenotypes associated with the under-accumulation of a miRNA that is essential for proper neuronal cell fate specification. Because lsy-6-mediated regulation of cog-1 expression is dosage-dependent, we speculate that lin-42(lf) mutations suppress neuronal cell fate specification defects by de-repressing lsy-6 transcription in ASEL neurons.
In order to understand how lin-42 may modulate miRNA expression, we pursued two lines of inquiry. First, we constructed a series of reporters that allowed us to measure, in detail, the transcriptional dynamics of multiple miRNAs in developing animals. Using these reporters, we found that several heterochronic miRNAs, such as lin-4 and let-7, exhibit highly dynamic expression patterns that are synchronized with the expression of genes required for each molting cycle. Importantly, the expression of the Plin-4::GFP-pest and Plet-7::GFP-pest reporters coincided with the transcriptional activation of lin-42. Further analysis of the lin-4 and let-7 reporters in a lin-42 mutant background indicated that one function of lin-42 is to negatively regulate the transcriptional output of miRNA promoters. Therefore, LIN-42 functions in a manner similar to the human and Drosophila PERIOD proteins, which inhibit the transcription of circadian regulated genes [77], [78]. Second, it has previously been shown that LIN-42 is a nuclear protein, which suggests that it may play a role in directly regulating the pulsatile expression patterns of its downstream targets [48]. In order to explore potential roles for LIN-42 in directly controlling aspects of miRNA transcription, we performed ChIP-seq experiments to determine if LIN-42 interacts with the putative regulatory regions thought to control the expression of miRNAs and mRNAs. These experiments demonstrated that LIN-42 interacts with the promoters of non-coding genes (including let-7) as well as protein-coding genes, suggesting that lin-42 may regulate the temporal expression of broad class of genes.
Given the role of human and Drosophila period in regulating circadian gene expression, we were surprised to find that animals harboring the lin-42(n1089) allele, which abolishes the expression of PAS-containing lin-42 isoforms, maintained lin-4 and let-7 periodic expression patterns in later larval development. The PAS domains of human and Drosophila PERIOD are absolutely required to maintain the oscillatory expression patterns of circadian-regulated genes [72], [77], [78]. In our experiments, peak expression of the lin-4 and let-7 transcriptional reporters occurred at roughly the same developmental stages in both wild-type and lin-42(n1089) animals. Interestingly, although the temporal expression patterns were similar, the levels of each reporter were elevated (as high as four fold) in lin-42(n1089) mutants as compared to wild-type animals (Figure 7A and B). Notably, lin-42(n1089) mutations do not alter the expression of the lin-42a isoform, which has been implicated in controlling the periodicity of the molting cycle [39]. While the dissection of lin-42 function will require further study, these findings are consistent with the modular nature of LIN-42 activities and suggest a novel role for the PAS domains of LIN-42 in regulating the transcriptional output of periodically expressed genes.
Fig. 7. A model for lin-42 function in regulating post-embryonic miRNA expression. 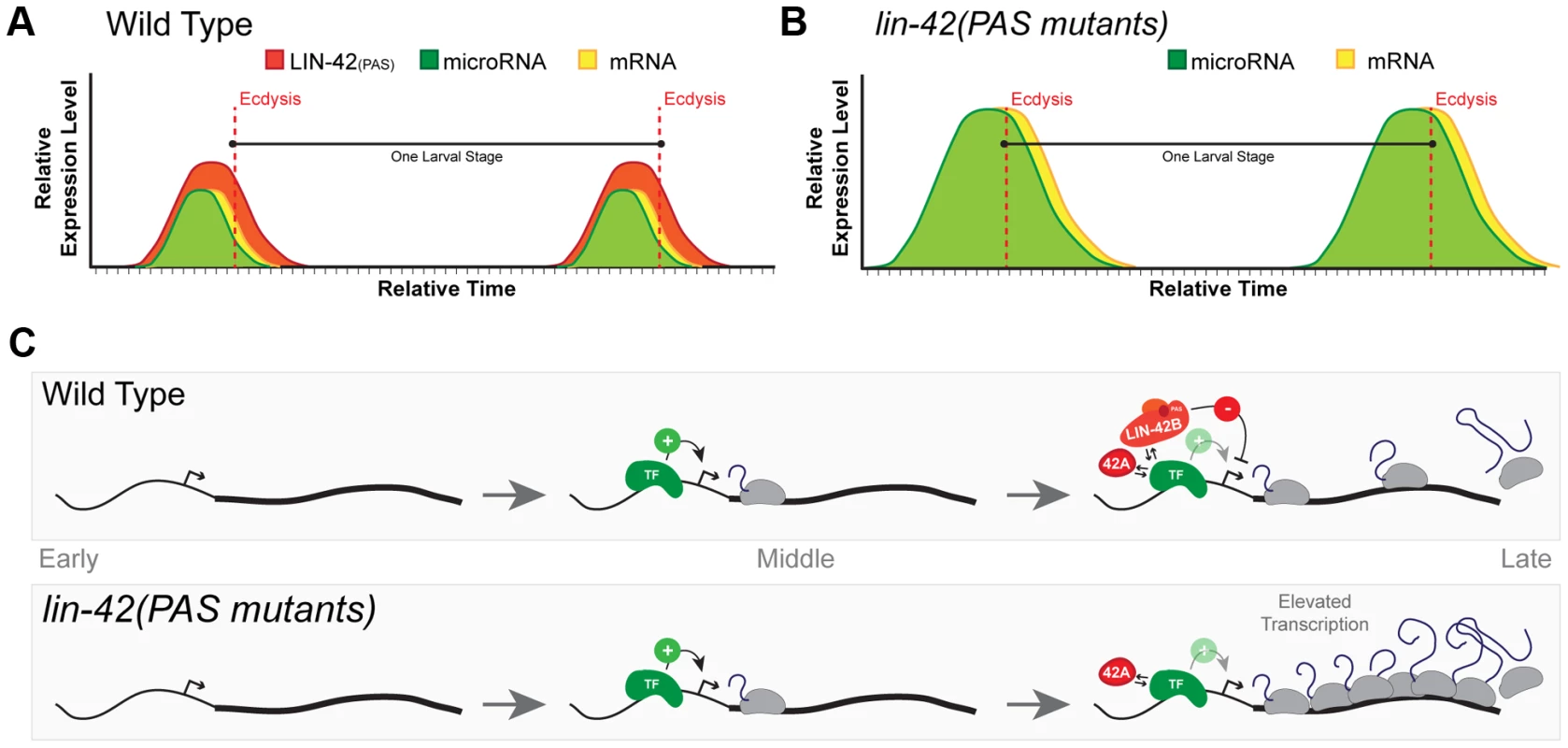
(A) In wild-type animals, transcriptional activation of miRNAs and lin-42-regulated mRNAs is pulsatile and displays a peak of expression once each larval stage. The temporal expression of these miRNAs and mRNAs begins in the later portions of each stage and are coincident with behavioral and morphological events that demarcate the end of each larval stage. Several temporally-regulated miRNAs and mRNAs share similar patterns of expression that are coupled to the molting cycle. By ecdysis and initiation of a new larval stage, periodic expression of miRNAs, mRNAs and lin-42 ceases. (B) The oscillatory expression patterns of miRNAs and mRNAs are maintained in animals that lack expression of LIN-42 isoforms containing the PAS domains. Only the transcriptional output of lin-42-regulated genes is altered, leading to the precocious phenotypes observed in these lin-42 mutants. (C) Analyses of LIN-42 ChIP data and the expression patterns of the lin-4 and let-7 transcriptional reporters in lin-42 mutants indicate that LIN-42 negatively regulates the transcriptional output of miRNA genes. We hypothesize that LIN-42 normally counteracts the transcriptional activity of one or more sequence-specific transcription factors (TF) that normally promote temporal expression. We would also predict that LIN-42 alters multiple aspects of transcription through its direct interaction with the TF or by binding to other cis-regulatory elements or to components of the transcriptional machinery. Mutations that alter only PAS domain-containing isoforms of LIN-42 fail to dampen transcriptional output. Importantly, this class of LIN-42 mutants would leave LIN-42A expression intact. Expression of LIN-42A would ensure normal periodic transcription of target genes and normal molting cycles and behaviors. Based on our current observations, we propose a model in which each of the lin-42 isoform functions to sculpt the dynamic transcription of both miRNAs and mRNAs. In out model, cis-regulatory elements within the promoters of specific miRNAs and mRNAs would be sufficient to drive periodic transcription. Regulatory elements within these sequences would be bound by a sequence-specific transcription factor (TF) that would promote the periodic transcription of these genes near the end of each larval stage (Figure 7C). Based on the role of PERIOD in other organisms and our data demonstrating that LIN-42 binds to the putative cis-regulatory elements of several miRNAs and mRNAs, we propose a model in which distinct isoforms of LIN-42 function to regulate the activity of the TF at multiple, genetically separable levels. Our evidence suggests that mutations that specifically disrupt isoforms containing the PAS domain (LIN-42B and LIN-42), fail to properly limit the transcriptional output of genes regulated by the temporal specific TF (Figure 7C). As a consequence, although these mutants display essentially normal temporal patterns of miRNA transcription, the elevated levels of heterochronic miRNAs lead to precocious developmental phenotypes. Importantly, mutations that only alter PAS-domain containing isoforms of LIN-42 retain the expression of LIN-42A (Figure 7B) [39]. Our model would also predict that mutations that disrupt LIN-42 isoforms that contain the conserved SYQ/LT domains (LIN-42A and LIN-42B) would have complex phenotypes with regard to periodic transcription. Indeed, animals harboring the lin-42(ok2385) allele, which disrupts the expression of the LIN-42A isoform (containing the SYQ/LT domains only) and deletes portions of the LIN-42B isoform (containing both the PAS and SYQ/LT domains), precociously execute stage-specific gene expression, fail to maintain periodic molting cycles and overexpress Pmir::GFP-pest transcriptional reporters [39](Figure S2). We interpret the complex phenotypes of lin-42(ok2385) animals as a reduction of the two modular activities of LIN-42 domains. Specifically, a reduction of LIN-42 PAS domain expression alters transcriptional output and deletion of LIN-42 isoforms which contain the SYQ/LT domains results in defects in periodic transcription. Further studies will be needed to define a specific molecular role for LIN-42 isoforms in maintaining normal periodic transcription.
Recent reports suggest that a significant portion of the C. elegans transcriptome is dynamically expressed [79]–[81]. The combined interpretation of these studies suggests that the post-embryonic expression of 5–20% of mRNAs is synchronized with the molting cycles. The conservation of this process implies that these temporal gene expression patterns confer fitness to an organism and raise a number of interesting questions regarding the nature of developmental gene regulation [81]. Because many genes whose oscillatory expression patterns are coupled to the molting cycle control cell fate decisions and cell metabolism in a dosage-dependent manner, it is interesting to speculate how their temporal expression patterns, and levels, are coordinated with their targets. We suggest that lin-42 plays a fundamental role in this process for a wide range of non-coding and protein-coding genes. Because many of the transcriptional targets of lin-42 include miRNAs, each of which may regulate a vast array of genes, the impact on the dynamic nature of the C. elegans transcriptome during development may be immense.
Materials and Methods
Nematode maintenance and genetics
C. elegans strains were grown under standard conditions and mutagenized as previously described [82]. Positional cloning of each suppressor was performed using standard methods [50]. Transformation of animals and integration of extrachromosomal arrays were performed as previously described [83]. See Text S1 for details of transgenic animals used in this manuscript.
Microscopy
Lineage analysis and scoring of adult alae phenotypes were performed by picking staged animals of the indicated genotypes and monitoring seam cells derived from the V lineage as previously described [22]. All images were taken with an Axio Scope.A1 microscope equipped with a monochrome camera (Diagnostic Instruments Inc) and SPOT imaging software (SPOT Imaging Solutions). GFP images of the hypodermal and seam cells were used for further quantification of Pmir::GFP-pest intensity. The average GFP intensity per area (arbitrary units) was quantified using ImageJ64. For each reporter, 20 individual animals were analyzed per developmental stage. For Plin-4::GFP-pest, 10 hypodermal cell nuclei per animal per stage, or a total of 200 nuclei per time point, were used to calculate the average GFP intensity. For Plet-7::GFP-pest, 5 seam cells per animal per stage, or a total of 100 cells per time point, were used to calculate the average GFP intensity.
Northern blots and TaqMan assays
Total RNA was isolated from staged populations of worms, and northern blots were performed as previously described [27]. Multiplex microRNA TaqMan assays were performed according to the manufacturer's specifications (Life Technologies) and quantified using the ABI 7900HT Fast Real-Time PCR system (Applied Biosystems). For each biological replicate (3 total), the means and standard deviations of the raw Ct values were calculated and the representative heatmap demonstrating the fold change signal was created using R packages (www.r-project.org).
Developmental and behavioral assays
For the characterization of behavioral and GFP/mCherry reporter expression, animals were prepared in one of two ways. For analysis of L1-stage expression, embryos were bleached and staged according to standard protocols and then plated on standard NGM media with OP50 [84]. At indicated times after the release from L1 synchronization, L1-staged animals were imaged with an Axio Scope.A1 microscope. For analysis of the molting cycle and GFP/mCherry-pest reporter expression, individual animals (non-motile, non-pharyngeal pumping) were picked to fresh NGM plates seeded with 20 µL of OP50. Time courses were initiated for each animal after each animal ecdysed. To determine the active and lethargic periods of animals at each stage, the pumping rates of individual animals were observed for 30 s of every hour. GFP/mCherry-pest expression was then monitored using a Zeiss SteREO Discovery V12 microscope with appropriate filters. To prevent photo-bleaching, each animal was exposed to <3 s of UV light.
Chip-Seq methods
See Text S1 for details.
Supporting Information
Zdroje
1. PasquinelliAE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13 : 271–282 doi:10.1038/nrg3162
2. LeeY, KimM, HanJ, YeomK-H, LeeS, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. Embo J 23 : 4051–4060 doi:10.1038/sj.emboj.7600385
3. LeeY, JeonK, LeeJT, KimS, KimVN (2002) MicroRNA maturation: stepwise processing and subcellular localization. Embo J 21 : 4663–70.
4. DenliAM, TopsBBJ, PlasterkRHA, KettingRF, HannonGJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432 : 231–235 doi:10.1038/nature03049
5. KettingRF, FischerSE, BernsteinE, SijenT, HannonGJ, et al. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15 : 2654–9.
6. GrishokA, PasquinelliAE, ConteD, LiN, ParrishS, et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106 : 23–34.
7. KuhnC-D, Joshua-TorL (2013) Eukaryotic Argonautes come into focus. Trends Biochem Sci 38 : 263–271 doi:10.1016/j.tibs.2013.02.008
8. HammellCM (2008) The microRNA-argonaute complex: A platform for mRNA modulation. RNA Biol 5 : 123–7.
9. FabianMR, SonenbergN (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19 : 586–593 doi:10.1038/nsmb.2296
10. BartelDP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136 : 215–233 doi:10.1016/j.cell.2009.01.002
11. BaekD, VillénJ, ShinC, CamargoFD, GygiSP, et al. (2008) The impact of microRNAs on protein output. Nature 455 : 64–71 doi:10.1038/nature07242
12. FarhKK-H, GrimsonA, JanC, LewisBP, JohnstonWK, et al. (2005) The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310 : 1817–1821 doi:10.1126/science.1121158
13. FriedmanRC, FarhKK-H, BurgeCB, BartelDP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19 : 92–105 doi:10.1101/gr.082701.108
14. Lim, LP, LauNC, Garrett-EngeleP, GrimsonA, SchelterJM, et al. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 : 769–773.
15. KasugaH, FukuyamaM, KitazawaA, KontaniK, KatadaT (2013) The microRNA miR-235 couples blast-cell quiescence to the nutritional state. Nature 497 : 503–506 doi:10.1038/nature12117
16. ReinhartBJ, SlackFJ, BassonM, PasquinelliAE, BettingerJC, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 : 901–6.
17. BrenneckeJ, HipfnerDR, StarkA, RussellRB, CohenSM (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113 : 25–36.
18. HeL, HeX, LimLP, de StanchinaE, XuanZ, et al. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447 : 1130–1134 doi:10.1038/nature05939
19. GurtanAM, SharpPA (2013) The Role of miRNAs in Regulating Gene Expression Networks. J Mol Biol 425 : 3582–3600 doi:10.1016/j.jmb.2013.03.007
20. FinneganEF, PasquinelliAE (2013) MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol 48 : 51–68 doi:10.3109/10409238.2012.738643
21. AmbrosV (2011) MicroRNAs and developmental timing. Curr Opin Genet Dev 21 : 511–517 doi:10.1016/j.gde.2011.04.003
22. SulstonJE, HorvitzHR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56 : 110–56.
23. RougvieAE, MossEG (2013) Developmental Transitions in C. elegans Larval Stages. Curr Top Dev Biol 105 : 153–180 doi:10.1016/B978-0-12-396968-2.00006-3
24. LeeRC, FeinbaumRL, AmbrosV (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 : 843–54.
25. WightmanB, HaI, RuvkunG (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 : 855–862.
26. MossEG, LeeRC, AmbrosV (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88 : 637–46.
27. AbbottAL, Alvarez-SaavedraE, MiskaEA, LauNC, BartelDP, et al. (2005) The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 9 : 403–414.
28. SlackFJ, BassonM, LiuZ, AmbrosV, HorvitzHR, et al. (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5 : 659–669.
29. OwMC, MartinezNJ, OlsenPH, SilvermanHS, BarrasaMI, et al. (2008) The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev 22 : 2520–2534.
30. Van WynsberghePM, KaiZS, MassirerKB, BurtonVH, YeoGW, et al. (2011) LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol 18 : 302–308 doi:10.1038/nsmb.1986
31. LinSY, JohnsonSM, AbrahamM, VellaMC, PasquinelliA, et al. (2003) The C. elegans hunchback Homolog, hbl-1, Controls Temporal Patterning and Is a Probable MicroRNA Target. Dev Cell 4 : 639–650.
32. LehrbachNJ, ArmisenJ, LightfootHL, MurfittKJ, BugautA, et al. (2009) LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 16 : 1016–1020 doi:10.1038/nsmb.1675
33. HayesGD, RuvkunG (2006) Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harb Symp Quant Biol 71 : 21–27 doi:10.1101/sqb.2006.71.018
34. FeinbaumR, AmbrosV (1999) The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev Biol 210 : 87–95.
35. RoushSF, SlackFJ (2009) Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Dev Biol 334 : 523–534 doi:10.1016/j.ydbio.2009.07.012
36. JohnsonSM, GrosshansH, ShingaraJ, ByromM, JarvisR, et al. (2005) RAS is regulated by the let-7 microRNA family. Cell 120 : 635–647.
37. KipreosET (2005) C. elegans cell cycles: invariance and stem cell divisions. Nat Rev Mol Cell Biol 6 : 766–776 doi:10.1038/nrm1738
38. MonsalveGC, FrandAR (2012) Toward a unified model of developmental timing: A “molting” approach. Worm 1 : 221–230 doi:10.4161/worm.20874
39. MonsalveGC, Van BuskirkC, FrandAR (2011) LIN-42/PERIOD Controls Cyclical and Developmental Progression of C. elegans Molts. Curr Biol 21 : 2033–2045 doi:10.1016/j.cub.2011.10.054
40. ChalfieM, HorvitzHR, SulstonJE (1981) Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24 : 59–69.
41. FrandAR, RusselS, RuvkunG (2005) Functional genomic analysis of C. elegans molting. PLoS Biol 3: e312 doi:10.1371/journal.pbio.0030312
42. AmbrosV, HorvitzHR (1984) Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226 : 409–416.
43. HayesGD, FrandAR, RuvkunG (2006) The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133 : 4631–4641 doi:10.1242/dev.02655
44. RougvieAE, AmbrosV (1995) The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development 121 : 2491–2500.
45. BettingerJC, LeeK, RougvieAE (1996) Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development 122 : 2517–2527.
46. AbrahanteJE, MillerEA, RougvieAE (1998) Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen::green fluorescent protein fusion gene. Genetics 149 : 1335–1351.
47. JeonM, GardnerHF, MillerEA, DeshlerJ, RougvieAE (1999) Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286 : 1141–1146.
48. TennessenJM, GardnerHF, VolkML, RougvieAE (2006) Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev Biol 289 : 30–43 doi:10.1016/j.ydbio.2005.09.044
49. ZinovyevaAY, BouaskerS, SimardMJ, HammellCM, AmbrosV (2014) Mutations in Conserved Residues of the C. elegans microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression. PLoS Genet 10: e1004286 doi:10.1371/journal.pgen.1004286
50. WicksSR, YehRT, GishWR, WaterstonRH, PlasterkRH (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet 28 : 160–164 doi:10.1038/88878
51. BanerjeeD, KwokA, LinSY, SlackFJ (2005) Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell 8 : 287–295 doi:10.1016/j.devcel.2004.12.006
52. LiJ, GreenwaldI (2010) LIN-14 inhibition of LIN-12 contributes to precision and timing of C. elegans vulval fate patterning. Curr Biol 20 : 1875–1879 doi:10.1016/j.cub.2010.09.055
53. MartinezNJ, OwMC, Reece-HoyesJS, BarrasaMI, AmbrosVR, et al. (2008) Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res 18 : 2005–2015.
54. BrennerJL, KempBJ, AbbottAL (2012) The mir-51 family of microRNAs functions in diverse regulatory pathways in Caenorhabditis elegans. PLoS One 7: e37185 doi:10.1371/journal.pone.0037185
55. SimonDJ, MadisonJM, ConeryAL, Thompson-PeerKL, SoskisM, et al. (2008) The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 133 : 903–915.
56. Esquela-KerscherA, JohnsonSM, BaiL, SaitoK, PartridgeJ, et al. (2005) Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn 234 : 868–877.
57. KaiZS, FinneganEF, HuangS, PasquinelliAE (2013) Multiple cis-elements and trans-acting factors regulate dynamic spatio-temporal transcription of let-7 in Caenorhabditis elegans. Dev Biol 374 : 223–233 doi:10.1016/j.ydbio.2012.11.021
58. Lim, LP, LauNC, WeinsteinEG, AbdelhakimA, YektaS, et al. (2003) The microRNAs of Caenorhabditis elegans. Genes Dev 17 : 991–1008.
59. WangG, ReinkeV (2008) A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol 18 : 861–867 doi:10.1016/j.cub.2008.05.009
60. BatistaPJ, RubyJG, ClaycombJM, ChiangR, FahlgrenN, et al. (2008) PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31 : 67–78 doi:10.1016/j.molcel.2008.06.002
61. GuW, LeeH-C, ChavesD, YoungmanEM, PazourGJ, et al. (2012) CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151 : 1488–1500 doi:10.1016/j.cell.2012.11.023
62. DiederichsS, HaberDA (2007) Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131 : 1097–1108 doi:10.1016/j.cell.2007.10.032
63. JohnstonRJ, HobertO (2003) A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 : 845–849.
64. SarinS, O'MearaMM, FlowersEB, AntonioC, PooleRJ, et al. (2007) Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176 : 2109–2130.
65. HammellCM, LubinI, BoagPR, BlackwellTK, AmbrosV (2009) nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136 : 926–938.
66. Vasquez-RifoA, BosséGD, RondeauEL, JannotG, DallaireA, et al. (2013) A new role for the GARP complex in microRNA-mediated gene regulation. PLoS Genet 9: e1003961 doi:10.1371/journal.pgen.1003961
67. ZhangP, ZhangH (2013) Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep 14 : 568–576 doi:10.1038/embor.2013.53
68. CorishP, Tyler-SmithC (1999) Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 12 : 1035–1040.
69. BarnesJA, GomesAV (1995) PEST sequences in calmodulin-binding proteins. Mol Cell Biochem 149–150 : 17–27.
70. LiX, ZhaoX, FangY, JiangX, DuongT, et al. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 273 : 34970–34975.
71. RechsteinerM, RogersSW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21 : 267–271.
72. HuangZJ, EderyI, RosbashM (1993) PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364 : 259–262 doi:10.1038/364259a0
73. McIntoshBE, HogeneschJB, BradfieldCA (2010) Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol 72 : 625–645 doi:10.1146/annurev-physiol-021909-135922
74. NiwaR, ZhouF, LiC, SlackFJ (2008) The expression of the Alzheimer's amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev Biol 315 : 418–425 doi:10.1016/j.ydbio.2007.12.044
75. FielenbachN, GuardavaccaroD, NeubertK, ChanT, LiD, et al. (2007) DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell 12 : 443–455 doi:10.1016/j.devcel.2007.01.018
76. HornM, GeisenC, CermakL, BeckerB, NakamuraS, et al. (2014) DRE-1/FBXO11-Dependent Degradation of BLMP-1/BLIMP-1 Governs C. elegans Developmental Timing and Maturation. Dev Cell 28 doi:10.1016/j.devcel.2014.01.028
77. ZhengB, LarkinDW, AlbrechtU, SunZS, SageM, et al. (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400 : 169–173 doi:10.1038/22118
78. TeiH, OkamuraH, ShigeyoshiY, FukuharaC, OzawaR, et al. (1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389 : 512–516 doi:10.1038/39086
79. HendriksG-J, GaidatzisD, AeschimannF, GroβhansH (2014) Extensive Oscillatory Gene Expression during C. elegans Larval Development. Mol Cell 53 : 380–392 doi:10.1016/j.molcel.2013.12.013
80. KimDH, GrünD, van OudenaardenA (2013) Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat Genet 45 : 1337–1344 doi:10.1038/ng.2763
81. GrünD, KirchnerM, ThierfelderN, StoeckiusM, SelbachM, et al. (2014) Conservation of mRNA and Protein Expression during Development of C. elegans. Cell Rep 6 : 565–577 doi:10.1016/j.celrep.2014.01.001
82. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
83. MelloCC, KramerJM, StinchcombD, AmbrosV (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J 10 : 3959–3970.
84. Epstein HF, Shakes DC (1995) Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press. 1 pp.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 7- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



