-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
Programmed cell death (PCD) is an evolutionarily conserved cellular process that is important for metazoan development and homeostasis. The epidermal growth factor (EGF) promotes cell proliferation, differentiation and survival during animal development. Surprisingly, we found that the EGF-like ligand LIN-3 also promotes the death of specific cells in Caenorhabditis elegans. We found that the LIN-3/EGF signal can be secreted from a cell to facilitate the demise of cells at a distance by activating the transcription of the PCD-promoting gene egl-1 in the doomed cells through the transcription factor LIN-1. LIN-1 binds to the egl-1 promoter in vitro and is positively regulated by the LIN-3/EGF, LET-23/EGF receptor, and the downstream MAPK signaling pathway. To our knowledge, LIN-3/EGF is the first extrinsic signal that has been shown to regulate the intrinsic PCD machinery during C. elegans development. In addition, the transcription factor LIN-31, which binds to LIN-1 and acts downstream of LIN-3/EGF, LET-23/EGF receptor, and the MAPK signaling pathway during vulval development, is dispensable for PCD. Thus, LIN-3/EGF promotes cell proliferation, differentiation, and PCD through common downstream signaling molecules but acts via distinct sets of transcription factors for different target gene expression.
Published in the journal: LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene. PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004513
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004513Summary
Programmed cell death (PCD) is an evolutionarily conserved cellular process that is important for metazoan development and homeostasis. The epidermal growth factor (EGF) promotes cell proliferation, differentiation and survival during animal development. Surprisingly, we found that the EGF-like ligand LIN-3 also promotes the death of specific cells in Caenorhabditis elegans. We found that the LIN-3/EGF signal can be secreted from a cell to facilitate the demise of cells at a distance by activating the transcription of the PCD-promoting gene egl-1 in the doomed cells through the transcription factor LIN-1. LIN-1 binds to the egl-1 promoter in vitro and is positively regulated by the LIN-3/EGF, LET-23/EGF receptor, and the downstream MAPK signaling pathway. To our knowledge, LIN-3/EGF is the first extrinsic signal that has been shown to regulate the intrinsic PCD machinery during C. elegans development. In addition, the transcription factor LIN-31, which binds to LIN-1 and acts downstream of LIN-3/EGF, LET-23/EGF receptor, and the MAPK signaling pathway during vulval development, is dispensable for PCD. Thus, LIN-3/EGF promotes cell proliferation, differentiation, and PCD through common downstream signaling molecules but acts via distinct sets of transcription factors for different target gene expression.
Introduction
PCD is important for proper animal development and tissue homeostasis [1], [2] and its dysregulation can cause aberrant death or survival of cells, which may lead to developmental defects, degenerative diseases, or cancers [1], [2].
Caenorhabditis elegans is an excellent model for studying PCD because of its invariant cell lineage and the conserved cell death pathway [3], [4]. Throughout the development of the C. elegans adult hermaphrodite, 131 somatic cells undergo PCD in an essentially invariant temporal and spatial pattern [5], [6]. Genetic and molecular studies have identified four genes, egl-1 (BH3-only gene), ced-9 (bcl-2), ced-4 (apaf-1), and ced-3 (caspase), that function in the core PCD pathway [7]–[12]. In living cells, CED-9 interacts with, and sequesters, CED-4 at the surface of mitochondria to prevent the cells undergoing PCD [13]. In cells destined to die, EGL-1 binds to CED-9, resulting in a conformational change in CED-9 and the release of bound CED-4 [14]. The released CED-4 translocates from the mitochondrion to the perinuclear membrane and interacts with, and activates, the caspase CED-3, leading to the eventual demise of the cell [15]. A recent study in mid-embryos and the germline suggested the existence of an alternative cell death activation mechanism that does not involve a direct interaction between CED-4 and CED-9 [16]. The transcriptional regulation of egl-1 is a critical step in the induction of most PCD events in the embryo [17]. Several transcription factors controlling egl-1 transcription have been identified and shown to specify the PCD fate of specific cells [4], [18]. For example, two transcription factors HLH-2 and HLH-3 activate egl-1 transcription by direct binding to the egl-1 cis-regulatory region during the specification of the death fate of NSM sister cells [18], [19]. Like HLH-2 and HLH-3, cell death specification genes have been shown to transcriptionally regulate the components of the core PCD machinery in a cell-autonomous manner. It is unclear whether the PCD fate, like many other cell fates, may be regulated by an extrinsic signal.
Extrinsic signals are crucial for a variety of developmental processes and act through receptors to elicit specific biological functions in a cell-nonautonomous manner. One example of such a signal-receptor pair is epidermal growth factor (EGF) and the EGF receptor (EGFR), which are involved in cell proliferation, differentiation, migration, survival, and death [20]–[22]. EGF has long been considered as a growth factor, since it stimulates proliferation in cultured cells, animals, and humans [23]. It also protects cells from apoptosis, as shown in cultured cells and Drosophila [24]–[29]. EGF can signal through the RAS-ERK-mediated and/or PI3K-mediated pathway(s) to activate transcription of various anti-apoptotic proteins, such as Bcl-XL and Mcl-1 [25]–[27], and also regulates post-transcriptional modifications, such as phosphorylation of BAD and caspase-9, to prevent apoptosis [28], [29]. However, in contrast to this cytoprotective function, it has also been shown to promote apoptosis, as exogenous EGF induces apoptosis in several cell lines, such as A431, MDA-MB-468, and MCF-7 [30]–[32]. It is not clear how it exerts different functions in different cells under different conditions or whether the apoptosis-promoting function plays a physiological role during animal development.
In C. elegans, lin-3 and let-23 encode, respectively, the sole EGF-like ligand and the EGFR and control many aspects of development, including ovulation, vulval differentiation, cell specification, and behavioral quiescence [33]–[38]. Activated LET-23 can recruit SEM-5 (orthologous to human Grb2) from the cytosol to the plasma membrane to activate LET-60, a member of the GTP-binding RAS family [39], [40]. LET-60 then triggers a kinase cascade involving the sequential phosphorylation of LIN-45 (a Raf ortholog), MEK-2 (a MAPK kinase kinase), and MPK-1 (an ERK ortholog) [41]–[45]. Once MPK-1 is phosphorylated, it translocates to the nucleus and regulates the transcription of numerous target genes by phosphorylation of specific transcription factors [46]. Besides activating the LET-60-MPK-1 pathway, LET-23 activates the PLC-γ-mediated signaling pathway to regulate ovulation and behavioral quiescence [36], [38]. In addition, the SEM-5-binding protein SOC-1 is involved in both the PLC-γ - and PI3K-mediated signaling pathways [47]. The LET-60-MPK-1 - and PI3K-mediated signaling pathways have been shown to be involved in physiological and genotoxic stress-induced germline cell death [48]–[51]. However, how these pathways are regulated by upstream signals, how they are linked to the intrinsic PCD machinery to control germline cell death, and whether they play a role in somatic PCD remain unknown.
We now report that LIN-3 acts as an extrinsic signal to promote specific somatic PCDs and the identification of components that transmit the LIN-3 signal to the intrinsic PCD machinery. Our data indicate that secreted LIN-3 induces cell death through the receptor LET-23, which acts in a cell-autonomous manner during PCD. Our genetic and biochemical data suggest that the LET-60-MPK-1 pathway transmits the LIN-3 signal from LET-23 to the transcription factor LIN-1 for transcriptional up-regulation of egl-1, which leads to activation of PCD.
Results
lin-3 and let-23 promote specific PCDs in C. elegans
While screening to test the potential involvement of receptor tyrosine kinase genes in PCD (see Materials and Methods), we found that several let-23 mutants have reduced numbers of cell corpses (Figure 1A). The let-23 gene encodes EGFR [34], raising the intriguing possibility that LET-23 might mediate an outside-in signal to affect PCD. lin-3 encodes the only EGF in C. elegans [33] and we found that lin-3 mutants had a similar reduction in numbers of cell corpses as let-23 single mutants or the let-23; lin-3 double mutant at the same embryonic stages (Figure 1A). This shows that lin-3 and let-23 function in the same pathway to affect cell corpse number, probably by acting as a ligand-receptor pair. Inactivation of lin-3 or let-23 by RNA interference (RNAi) reduced the numbers of cell corpses to a similar extent as the lin-3 or let-23 mutations (Figure 1A). Because the lin-3 or let-23 RNAi treatment caused more than 50% rod-like larval lethality, phenocopying the strong loss-of-function lin-3 or let-23 mutants [52], the reduced numbers of cell corpses observed in the lin-3 or let-23 mutants mentioned above likely represented the phenotype caused by nearly complete elimination of lin-3 or let-23, respectively. Interestingly, in these mutants, only a partial reduction of cell corpse numbers from the comma stage to the 2-fold stage was observed, suggesting that lin-3 and let-23 may be essential for some but not all cell deaths.
Fig. 1. lin-3 and let-23 promote specific PCDs in C. elegans. 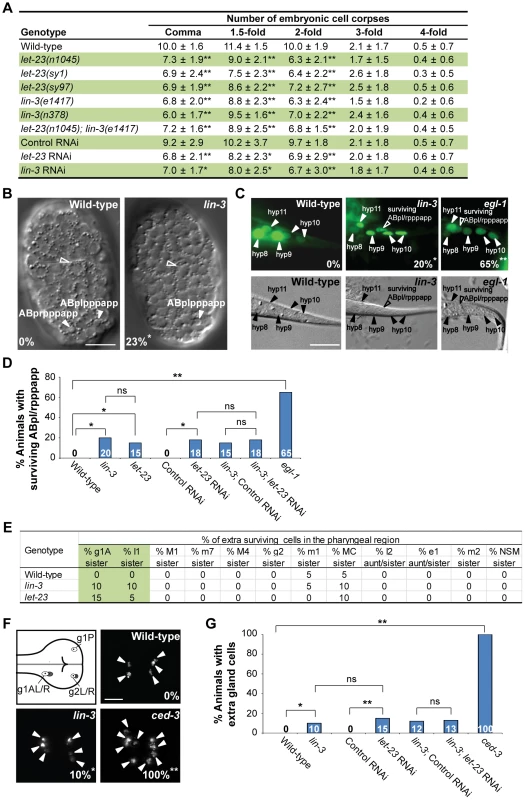
(A) The number of cell corpses was scored at indicated developmental stages and presented as the mean ± SD (n≥30). The rrf-3(pk1426) mutant was used to enhance the RNAi effect in RNAi experiments. Mutants were compared to wild-type, and RNAi treatment of individual genes was compared to control RNAi (*P<0.05 and **P<0.001, two-tailed t test). (B) The DIC images of the wild-type and lin-3(e1417) mutant embryos at approximately 270 min after fertilization. The white arrowheads indicate ABpl/rpppapp cell corpses and the hollow arrowheads indicate excretory cells. The percentage of embryos that did not have ABpl/rpppappcell corpse is shown in the bottom left corner. *P = 0.048 when compared to wild-type (Fisher's exact test). Twenty one and thirteen embryos were scored for wild-type and lin-3 mutants, respectively. Scale bar: 10 µm. (C) The GFP and DIC images of the wild-type, lin-3(e1417), and egl-1(n1084n3082) worms expressing sur-5::gfp at the L1 stage. The arrowheads indicate the nuclei of hyp8, hyp9, hyp10, and hyp11. The hyp10 cell is binuclear. The hollow arrowheads indicate a surviving ABpl/rpppapp. The percentage of worms with surviving ABpl/rpppapp is shown in the bottom right corner. *indicates P<0.05 and **P<0.001 when compared to wild-type (Fisher's exact test). More than thirty worms were scored for each genotype. Scale bar: 10 µm. The GFP protein expressed from the sur-5::gfp transgene is localized to almost all somatic nuclei [86]. (D) The percentage of worms with surviving ABpl/rpppapp of the indicated genotypes was shown. *indicates P<0.05 and **P<0.001 (Fisher's exact test). ns indicates no statistical difference (p>0.05). Alleles used here are lin-3(e1417), let-23(sy1), and egl-1(n1084n3082). More than thirty worms were scored for each genotype. (E) Analysis of surviving cells in the pharyngeal region (n = 20). Alleles used here are lin-3(e1417) and let-23(n1045). (F) The g1 and g2 gland cells in the pharynx are shown in the illustration. The white circles indicate the five gland cells (g1P, g1AL, g1AR, g2L, and g2R) that normally survive and the gray circles the sister cells of g1AL, g1AR, g2L, and g2R that are destined to die. Monomeric red fluorescent protein (mRFP) images of the wild-type, lin-3(e1417), and ced-3(n717) worms expressing PB0280.7::4Xnls::mrfp are shown. The arrowheads indicate cells expressing mRFP. The percentage of animals that have extra gland cell(s) is shown in the bottom right corner. *indicates P<0.05 and **P<0.001 when compared to wild-type (Fisher's exact test). One hundred L4 animals were scored for each genotype. The image stacks were merged by maximum intensity projection using Image J. Scale bar: 10 µm. (G) The percentage of worms with extra gland cells of the indicated genotypes was shown. *indicates P<0.05 and **P<0.001 (Fisher's exact test). ns indicates no statistical difference (p>0.05). Alleles used here are lin-3(e1417) and ced-3(n717). More than forty worms were scored for each genotype. To investigate whether lin-3 may affect specific cell deaths, we examined the first 13 cell deaths that occur in the AB lineage during embryogenesis [6], [53] in the lin-3(e1417) mutant by using four-dimensional DIC microscopy. In three out of thirteen lin-3 embryos we analyzed, only ABplpppapp or its lineal homolog ABprpppapp failed to die, and the other cell deaths occurred normally (Figure 1B and S1). ABplpppapp and ABprpppapp (these cells are collectively termed ABpl/rpppapp hereafter), which are aunt cells of hypodermal cells hyp8/9, are generated at approximately 270 min after fertilization and undergo PCD soon after birth [6]. Consistent with the DIC four-dimensional microscopy data, approximately 20% of surviving ABpl/rpppapp were observed in the tail of the lin-3(e1417) mutant (Figure 1C and D), indicating that the disappearance of ABpl/rpppapp corpses is due to the survival of these cells. A similar result was observed in the let-23(sy1) mutant and in the lin-3(e1417) mutant feeding let-23 RNAi (Figure 1D), showing that lin-3 and let-23 act in the same pathway to promote the death of ABpl/rpppapp.
We also measured the duration time of the first 13 cell corpses derived from the AB lineage in the wild-type and lin-3(e1417) mutant embryos and found that the corpse duration time was statistically undistinguishable in the wild-type and lin-3 embryos (Figure S2, P = 0.24, two-tailed t test). This result indicates that the kinetics of corpse removal is normal in the lin-3(e1417) mutant, and rules out the possibility that the decrease of cell corpse numbers in the lin-3 mutants may be attributed to enhanced cell corpse removal.
We next examined whether lin-3 or let-23 may also be important for the death of other PCDs. We scored the extra surviving cells in the pharynx of the lin-3 or let-23 mutant at the L3 and L4 stages. In wild-type animals, 16 cells in the anterior pharynx and 6 in the posterior pharynx undergo PCD during mid-embryogenesis. Eight out of these 22 cells (NSML/R, I2L/R, g1AL/R and g2L/R) undergo PCD during the comma to 2-fold stage. In virtually all wild-type embryos, no extra surviving cells are observed in these regions at the late larval stages [6], [11], [54]. In lin-3 and let-23 mutants, about 10% of the sister cells of g1A gland cells and I1 interneurons survived, and the death of other 18 cells in the pharynx region was essentially normal (Figure 1E). Furthermore, the percentage of animals with surviving g1A and I1 sisters was enhanced by the weak ced-3(n2427) mutation; however, essentially no enhancement was observed in other 18 cells examined (Table S1). This result shows that lin-3 and let-23 are important for the death of specific cells. Consistently, using an integrated transgene tpIs8, which expresses monomeric red fluorescent protein (mRFP) in the g1A, g1P, and g2 gland cells [55], we found that 10% of the lin-3 animals had extra gland cells, whereas wild-type worms had no extra gland cell and the strong ced-3(n717) mutants all had extra gland cell(s) (Figure 1F and G). This result indicates that, like ced-3, surviving cells in the lin-3 mutant adopt the terminal differentiation fate of their sister cells. A similar result was observed in the wild-type or lin-3(e1417) mutant feeding let-23 RNAi (Figure 1G), showing that lin-3 and let-23 also act in the same pathway to promote the death of gland cells. Our data show that lin-3 and let-23 are important for the death of specific cells, including g1A and I1 sister cells and ABpl/rpppapp and that these genes are dispensable for most cell deaths analyzed in this work.
Overexpression of lin-3 cause ectopic cell deaths
We next examined whether over-activation of lin-3 signaling could cause ectopic cell deaths during embryogenesis. We found that overexpression of lin-3 genomic DNA from the integrated transgene syIs1 [33] increased embryonic cell corpse numbers (Table S2). Using the integrated transgene tpIs8 to label g1 and g2 gland cells, we found that syIs1 caused the loss of one or two gland cells in 19% of transgenic worms (Figure 2A and B). This phenotype of gland cell loss was suppressed by the loss-of-function mutation in egl-1 or ced-3 (Figure 2A and B). This result shows that lin-3 overexpression induces ectopic gland cell death rather than alters gland cell fate, and that the cell death-promoting activity of lin-3 requires the core PCD pathway. In addition, the phenotype of ectopic gland cell death induced by syIs1 was also suppressed by let-23 RNAi (Figure 2B), showing that lin-3 acts through let-23 to promote the ectopic cell death of the gland cells. Consistently, the increased numbers of embryonic cell corpses induced by syIs1 was also suppressed by loss-of-function mutations in let-23 or in components of the core PCD pathway (Table S2).
Fig. 2. Overexpression of lin-3 induces ectopic gland cell death(s) through let-23 and the core PCD pathway. 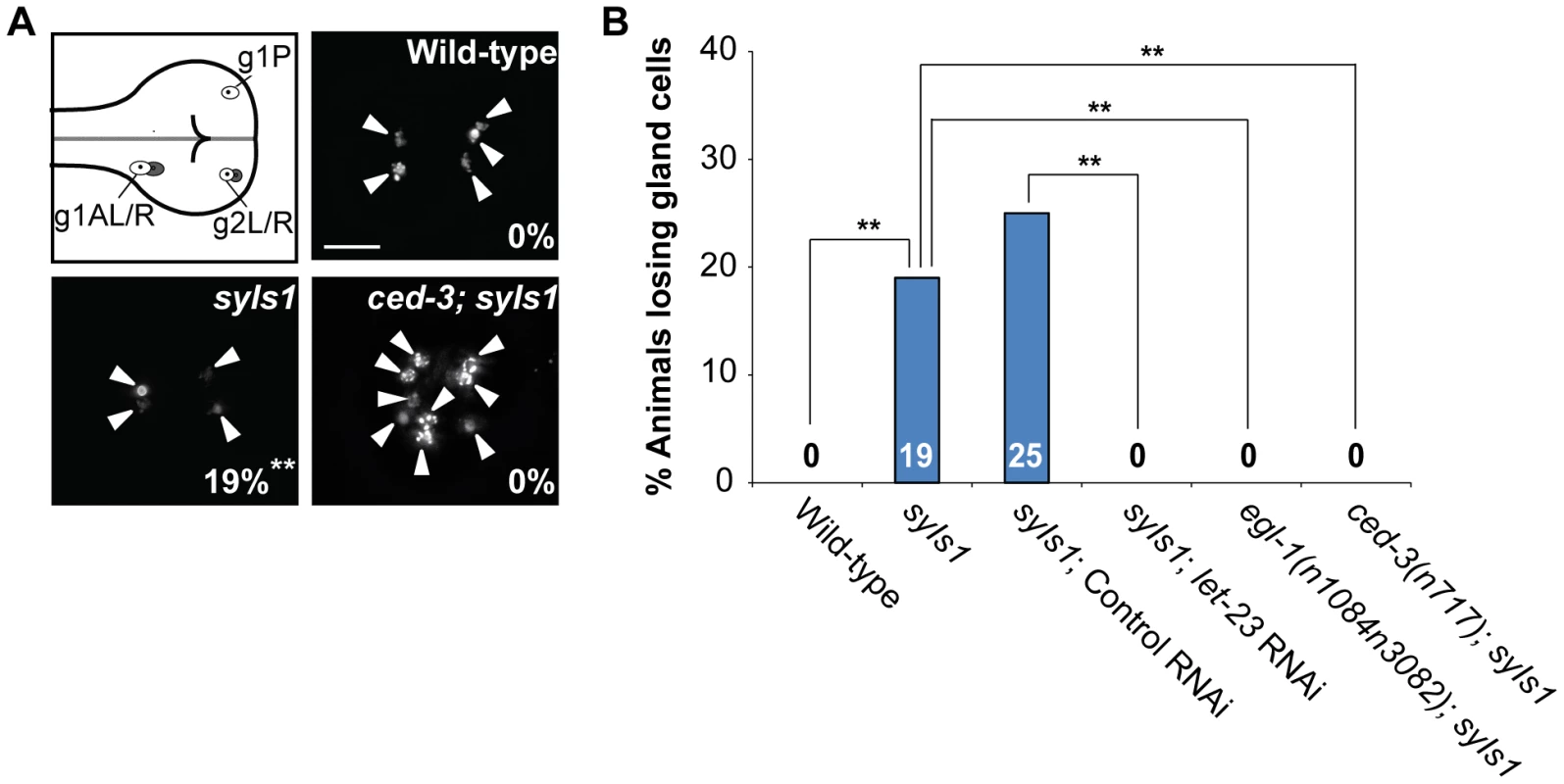
(A) Monomeric red fluorescent protein (mRFP) images of the wild-type, syIs1, and ced-3(n717); syIs1 worms expressing PB0280.7::4Xnls::mrfp are shown. The arrowheads indicate cells expressing mRFP. The percentage of animals losing gland cell(s) is shown in the bottom right corner. **indicates P<0.001 when compared to wild-type (Fisher's exact test). One hundred L4 animals were scored for each genotype. The image stacks were merged by maximum intensity projection using Image J. Scale bar: 10 µm. (B) The percentage of worms losing gland cell(s) of the indicated genotypes was shown. **indicates P<0.001 (Fisher's exact test). More than forty worms were scored for each genotype. let-23 is expressed in dying cells, including ABpl/rpppapp
Next, we examined whether there was a correlation between let-23 expression and PCD using the transgene syEx234[Plet-23::let-23::gfp] [56]. As shown in Figure 3A, the LET-23::GFP fusion protein was detected as a crescent on the surface of some dying cells (indicated by arrowheads). The cause and physiological significance of this crescent-shaped localization pattern is not clear. To be sure that let-23 was expressed in dying cells and not on the membranes of adjacent living cells, we generated the construct Plet-23::4Xnls::gfp and microinjected it into the wild-type to generate transgenic worms. Plet-23::4Xnls::gfp contains the let-23 promoter Plet-23 fused to the GFP cDNA containing four copies of the nuclear localization signal (4XNLS). The GFP signal was detected in the nuclei of the dying cells (Figure 3B), clearly showing that let-23 is expressed in dying cells. Notably, the GFP signal was observed in the ABpl/rpppapp corpses (Figure 3C).
Fig. 3. let-23 is expressed in dying cells, whereas lin-3 acts in a cell-nonautonomous manner to promote PCD. 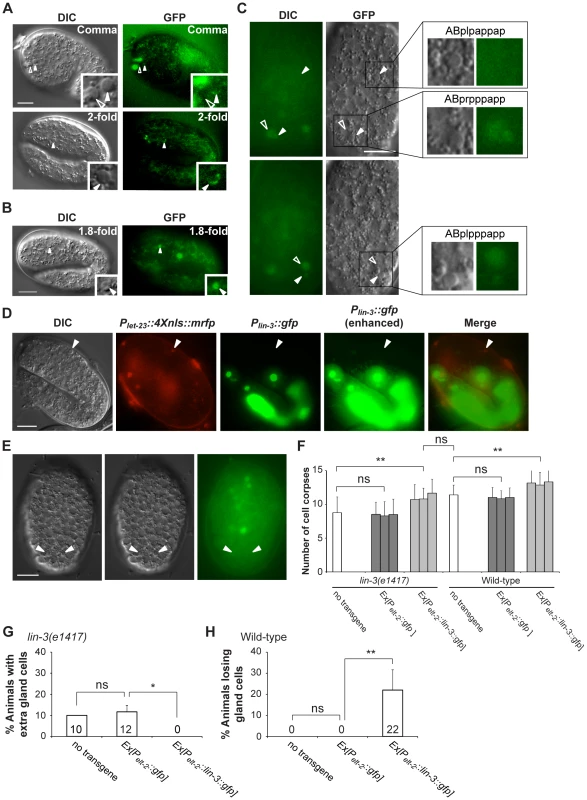
(A and B) Representative DIC and GFP images of embryos carrying (A) the syEx234[Plet-23::let-23::gfp] transgene or (B) the Plet-23::4Xnls::gfp transgene. The white arrowheads indicate cell corpses expressing LET-23::GFP and the hollow arrowheads cell corpses not expressing LET-23::GFP. Scale bar: 10 µm. The insets show a dying cell at a three-fold higher magnification. (C) The DIC and GFP images of an embryo carrying the Plet-23::4Xnls::gfp transgene. Two different focal planes of the same embryo are shown in the upper and lower panels. The white arrowheads indicate ABpl/rpppapp corpses and ABplpappap and the hollow arrowheads sister cells of ABpl/rpppapp. Scale bar: 10 µm. The insets show the indicated cells at a higher magnification. (D) The expression pattern of Plin-3::gfp and Plet-23::4Xnls::mrfp transgenes in a wild-type embryo. The white arrowheads indicate the same cell corpse expressing Plet-23::4Xnls::mrfp. The image stacks of Plin-3::gfp were merged by maximum intensity projection using Image J. The strong GFP expression is from the co-injection marker Pelt-2::gfp. Scale bar: 10 µm. (E) Plin-3::gfp is not expressed in ABpl/rpppapp corpses. The image stacks of Plin-3::gfp were merged by maximum intensity projection using Image J. Scale bar: 10 µm. (F–H) Intestine-specific expression of LIN-3 rescues the cell death defect of the lin-3 mutant and causes ectopic cell deaths in the wild-type. Embryonic cell corpses in the indicated genotypes were scored at the 1.5-fold stage (n≥30), and the numbers of gland cells were scored at the L4 stage (n≥40). Three independent stable transgenic lines were analyzed. *indicates P<0.05 and **P<0.001 in a two-tailed t-test (F) or fisher's exact test (G and H). ns indicates no statistical difference (p>0.05). LIN-3 can act as an extrinsic signal to promote PCD
Like its Drosophila and vertebrate homologs, lin-3 encodes a transmembrane precursor protein, consisting of an extracellular domain containing one EGF motif, a transmembrane domain, and a cytoplasmic domain [33], [57]. Genetic and laser ablation studies have suggested that the LIN-3 precursor protein can be cleaved to form a secreted product [33], [58], [59]. Using the transcriptional reporter Plin-3::gfp [60] and Plet-23::4Xnls::mrfp, no Plin-3::gfp-expressing cell was located adjacent to the Plet-23::4xnls::rfp-expressing cell corpses, as shown in Figure 3D. In addition, no Plin-3::gfp signal was detected near ABpl/rpppapp corpses (Figure 3E). These observations raise a possibility that lin-3 may be secreted and act at a distance to promote PCD.
To test this hypothesis, we generated a DNA construct Pelt-2::lin-3::gfp, in which a lin-3::gfp fusion cDNA was expressed under the control of the elt-2 promoter Pelt-2 and introduced it to the wild-type and lin-3 mutant. Pelt-2 is used here because it is exclusively expressed in intestinal cells and their precursor cells [61] and no PCD occurs in these cells [6]. We found that, in the lin-3 mutant, the Pelt-2::lin-3::gfp transgene not only increased the number of embryonic cell corpses to the wild-type level (Figure 3F) but also rescued the inappropriate survival of gland cells (Figure 3G). Moreover, when introduced to the wild-type, the Pelt-2::lin-3::gfp transgene increased the embryonic cell corpse number (Figure 3F) and caused ectopic gland cell deaths (Figure 3H), as the transgene syIs1 did (Figure 2A, B and Table S2). These results show that LIN-3::GFP secreted from the intestine can act at a distance to induce cell death in other parts of the embryo, e.g. gland cells in the head region. In our study, the adult worms transgenic for Pelt-2::lin-3::gfp had a multi-vulva phenotype, resembling worms carrying syIs1 [33], reinforcing the notion that LIN-3::GFP expressed in the intestine is secreted and active.
lin-3 signaling up-regulates egl-1 transcription
Because the cell death-promoting activity of lin-3 requires the core PCD pathway (Figure 2A, B, and Table S2), we next examined whether lin-3 signaling promotes PCD by regulating the expression of egl-1, ced-9, ced-4, or ced-3. To this end, we measured the transcript levels of egl-1, ced-9, ced-4, or ced-3 in worms with different levels of lin-3 activity by quantitative real-time reverse transcription (RT)-PCR. As shown in Figure 4A, the abundance of egl-1 transcripts correlated well with the level of lin-3 activity, which, in turn, correlated well with the number of cell deaths. egl-1 transcripts were less abundant in lin-3 and let-23 mutants than in wild-type animals (average of 0.53-fold and 0.57-fold, respectively), while lin-3 overexpression using the integrated transgene syIs1 caused an increase in egl-1 transcript levels (average of 2.15-fold). Similarly, levels of the ced-4 transcript, but not those of the ced-9 and ced-3 transcripts, also showed a correlation with the level of lin-3 activity, although to a lesser extent than the egl-1 transcript. These data show that LIN-3 and LET-23 signaling activates transcription of egl-1 and, to a lesser extent, ced-4, in vivo. Interestingly, we found that the lin-3(e1417) mutation reduced the egl-1 transcript level in the ced-3(n717) mutant (average of 0.61-fold) to a similar extent as the lin-3(e1417) mutation did in the wild-type (Figure 4A). This result supports the model that the LIN-3-mediated up-regulation of egl-1 transcription occurs before ced-3 activation during PCD.
Fig. 4. lin-3 promotes egl-1 transcription. 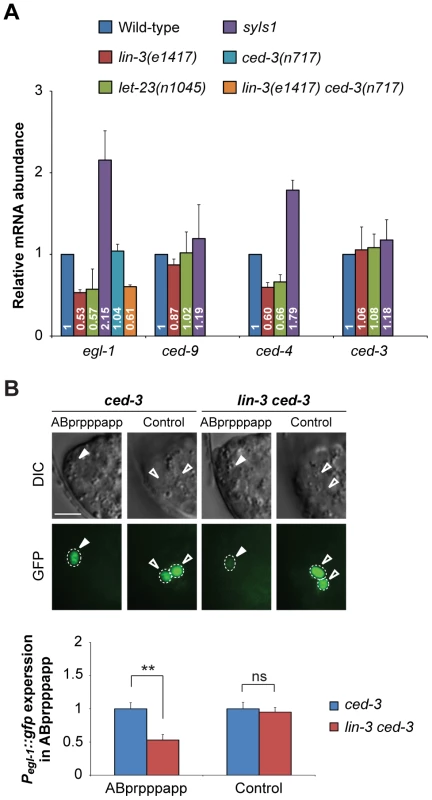
(A) egl-1, ced-9, ced-4, and ced-3 transcripts in embryos of indicated genotypes were measured by quantitative RT-PCR. The relative mRNA abundance is shown as the mean and SD of the results from at least two independent experiments, each performed in triplicates. (B) The DIC and GFP images of ced-3 (left) and lin-3 ced-3 (right) embryos expressing the Pegl-1::gfp transgene. Arrowheads indicate ABprpppapp, and hollow arrowheads indicate two cells that also express egl-1 near ABprpppapp (Control experiment). The bottom figures show the Pegl-1::gfp expression level of the indicated cells in the ced-3 single mutant (blue) and the lin-3 ced-3 double mutant (red). The expression level of Pegl-1::gfp in ABprpppapp of ced-3(n717) (n = 44) and lin-3(e1417) ced-3(n717) (n = 40) mutants was analyzed as described in Materials and Methods. Error bars represent the SEM. **indicates P<0.001 (two-tailed t test). Scale bar: 5 µm. Next, we examined the effect of lin-3 on the egl-1 transcription activity in ABprpppapp using the transcriptional fusion reporter Pegl-1::gfp [62]. The egl-1(n1084n3082) mutation caused frequent survival of ABpl/rpppapp, showing that egl-1 is required for the death of ABprpppapp (Figure 1C and D). We measured and compared the fluorescence intensity of GFP in ABprpppapp of the ced-3 and lin-3 ced-3 mutants. The ced-3(n717) mutation blocks the death of ABprpppapp, hence allowing the observation of the Pegl-1::gfp transgene in the surviving ABprpppapp, and has no effect on the lin-3-mediated egl-1 transcription (Figure 4A). In the ced-3(n717) mutant, ABprpppapp survived and expressed Pegl-1::gfp at the comma stage (Figure 4B). The lin-3(e1417) mutation significantly decreased the intensity of Pegl-1::gfp to about 0.53-fold in ABprpppapp of the ced-3 mutant (Figure 4B, P = 0.0004), consistent with the result from quantitative real-time RT-PCR (Figure 4A). Notably, the lin-3(e1417) mutation did not affect the egl-1 transcription in two adjacent cells that also express egl-1 (Figure 4B, control experiment, P = 0.6839). Therefore, loss of lin-3 reduces the egl-1 transcription level in a cell-specific manner. These data and the aforementioned observation that the egl-1(lf) mutation blocked the syIs1-induced cell death (Figure 2A, B, and Table S2) together support the notion that lin-3 signaling promotes the death of ABprpppapp by transcriptional activation of egl-1.
LET-23 transduces the cell death-promoting signal via the LET-60-MPK-1 pathway
We next investigated how lin-3 and let-23 signaling may up-regulate egl-1 to promote PCD. Activated LET-23 has been shown to act via multiple signaling pathways, including the LET-60-MPK-1 - and PLC-γ-mediated pathways, to control C. elegans development [36], [38]–[45]. SOC-1, a SEM-5-binding protein, has been reported to act not only in the LET-60-MPK-1 pathway, but also in the PLC-γ - and PI3K-mediated signaling pathways [47]. We found that mutants defective in the LET-60-MPK-1 pathway had reduced cell corpse numbers during mid-embryogenesis (Table S3), while mutants defective in either the PI3K-mediated pathway (Table S4A) or PLC genes (Table S4B) had normal cell corpse numbers throughout embryogenesis. We further analyzed whether specific PCDs were affected in the let-60 and mpk-1 mutants using the assays described above. We found that let-60 and mpk-1 are required for the death of ABpl/rpppapp (Figure 5A), glA sisters, and I1 sisters (Figure 5B) but essentially dispensable for most cell deaths examined in this work. These data show that mutants defective in the LET-60-MPK-1 pathway have a PCD-defective phenotype similar to lin-3 and let-23 mutants. Furthermore, the lin-3 RNAi treatment did not enhance the survival of ABpl/rpppapp in the let-60 or mpk-1 mutant (Figure 5A), showing that, at least in ABpl/rpppapp, lin-3 and let-23 transduce the cell death-promoting signal through the LET-60-MPK-1 pathway.
Fig. 5. lin-3, lin-1 and the LET-60-MPK-1 pathway act in the same pathway to promote PCD. 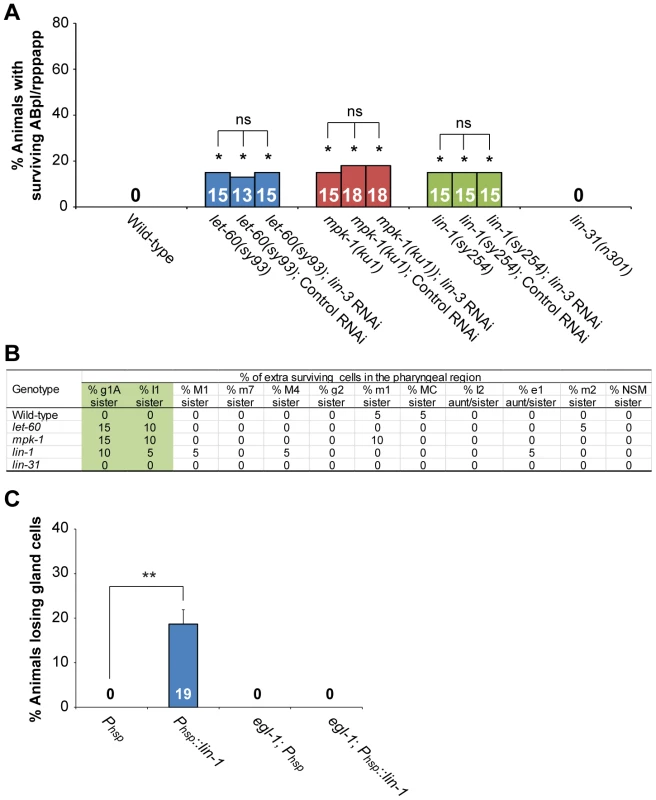
(A) The percentage of worms with surviving ABpl/rpppapp of the indicated genotypes was shown. *indicates P<0.05 when compared to the wild-type (Fisher's exact test). ns indicates no statistical difference (p>0.05). More than thirty worms were scored for each genotype. (B) Analysis of extra surviving cells in the pharyngeal region (n = 20). Alleles used here are let-60(sy93), mpk-1(ku1), lin-1(sy254), and lin-31(n301). (C) The percentage of wild-type or egl-1(n1084n3082) worms carrying the indicated transgene that lost gland cell(s) in the pharynx was shown. **P<0.001 (Fisher's exact test). The gland cells were scored using the tpIs8 transgene, and the worms were treated with heat shock as described in Materials and Methods. Phsp::lin-1 or Phsp indicate the transgenes, in which lin-1 or no cDNA was expressed under the control of the heat-shock promoter, respectively. More than forty worms were scored for each genotype. LET-23 transduces the cell death-promoting signal via the transcription factor LIN-1, but not LIN-31
Two transcription factors, the ETS domain-containing protein LIN-1 and the WH domain-containing protein LIN-31, transduce LET-60-MPK-1 signaling to control vulval differentiation [63], [64]. In the absence of the LET-60-MPK-1 signaling pathway, LIN-1 forms a complex with LIN-31 and inhibits vulval induction [46]. Activation of the LET-60-MPK-1 pathway leads to phosphorylation of LIN-1 and LIN-31 by MPK-1, and phosphorylation of LIN-31 results in dissociation of LIN-1 and LIN-31 from each other and subsequent vulval induction [46]. Interestingly, the lin-1(lf) mutation also caused the survival of ABpl/rpppapp (Figure 5A), g1A sisters, and I1 sisters (Figure 5B), whereas the lin-31(lf) mutation did not (Figure 5A and B). These results show that lin-1, but not lin-31, affects specific PCD, despite both genes being necessary for vulval differentiation. lin-1 mutants also displayed a reduction of embryonic cell corpse numbers to a similar extent as mutants defective in the LET-60-MPK-1 pathway, whereas lin-31 mutants had a normal cell corpse profile during embryogenesis (Table S5). The lin-3 RNAi treatment did not enhance the penetrance of ABpl/rpppapp survival in the lin-1 mutant, showing that lin-3 and lin-1 act in the same pathway to promote the death of ABpl/rpppapp (Figure 5A). Consistently, the cell corpse numbers of the lin-1(sy254) lin-3(e1417) double mutant showed no significant difference when compared to the lin-1(sy254) or lin-3(e1417) single mutant (Table S5). These data and the aforementioned genetic results support the notion that LIN-3 and LET-23 act through the LET-60-MPK-1 pathway and transcription factor LIN-1 to promote PCD.
LIN-1 activates egl-1 transcription by direct binding to the egl-1 promoter
We next studied the mechanism by which LIN-1 promotes PCD. As mentioned above, LIN-3 and LET-23 signaling activates transcription of egl-1 and, to a lesser extent, ced-4, in vivo (Figure 4A). This was confirmed by examining the expression level of the transcriptional fusion reporter Pegl-1::gfp in ABprpppapp (Figure 4B). Moreover, like lin-3 overexpression, heat shock-induced lin-1 overexpression in the wild-type caused the phenotype of gland cell loss, and this phenotype can be suppressed by an egl-1(lf) mutation (Figure 5C). These results support the model that LIN-1 acts upstream of egl-1 to promote the death of gland cells.
LIN-1 contains a highly conserved ETS domain [64], which binds to DNA with the minimal recognition sequence GGAA/T [65]. A potential LIN-1 binding sequence was found in the promoter region of egl-1, but not that of ced-4 (Figure 6A, see Materials and Methods for details), supporting the possibility that LIN-1 binds directly to egl-1. We therefore generated the GST::LIN-1(DBD) fusion protein, in which the DNA binding domain of LIN-1 was fused to glutathione S-transferase (GST), and tested its ability to bind to a 34 bp DNA fragment (named P3) containing the LIN-1 binding sequence in the electrophoretic mobility shift assay (EMSA). The results are shown in Figure 6B. The P3 fragment displayed similar mobility in the presence or absence of GST (lanes 1 and 2), confirming that GST does not bind to the DNA fragment. In contrast, the presence of GST::LIN-1(DBD) resulted in the dose-dependent appearance of a slowly migrating band (lanes 3–5), and this effect was eliminated by addition of an excess of non-labeled wild-type DNA competitor (lanes 6 and 7), but not with an excess of a DNA competitor with a mutation in the core GGAA/T motif (lanes 8 and 9). These results show that the slowly migrating band is a complex of GST::LIN-1(DBD) and P3 and that LIN-1 binds to the P3 region of egl-1 in vitro.
Fig. 6. lin-3 promotes PCD through transcriptional activation of egl-1 by LIN-1. 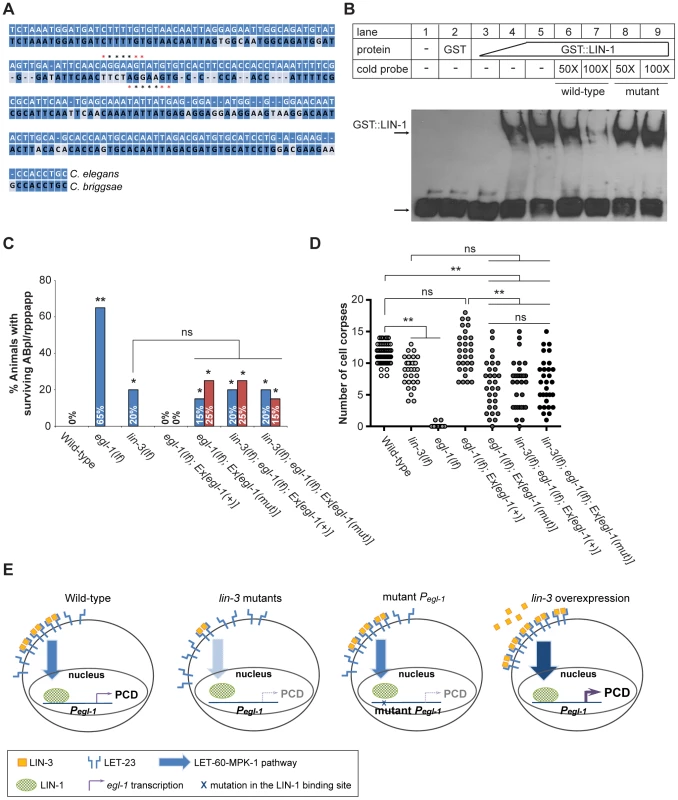
(A) The alignment between the C. elegans and C. briggsae genomic sequences near the P3 fragment. *indicates the LIN-1 minimal recognition sequence GGAA/T. (B) GST::LIN-1(DBD) directly binds to the egl-1 promoter in an EMSA assay. Lane 1, no protein; lane 2, 200 ng of GST; lanes 3–5, 10, 50, or 200 ng of GST::LIN-1(DBD); lane 6: 200 ng of GST::LIN-1(DBD)+50× wild-type cold probe; lane 7, 200 ng of GST::LIN-1(DBD)+100× wild-type cold probe; lane 8, 200 ng of GST::LIN-1(DBD)+50× mutant cold probe; lane 9, 200 ng of GST::LIN-1(DBD)+100× mutant cold probe. The bottom arrow indicates non bound probe and the top arrow DNA bound to GST::LIN-1(DBD) and displaying retarded mobility. (C) The percentage of animals with surviving ABpl/rpppapp of the indicated genotypes was scored (n≥30). Ex[egl-1(+)] and Ex[egl-1(mut)] indicate wild-type and mutant egl-1 transgenes, respectively, as described in the text. The surviving ABpl/rpppapp was scored using the sur-5::gfp reporter as described in Figure 1B. Two independent stably transmitted lines were analyzed for each transgene. Alleles used: lin-3(e1417) and egl-1(n1084n3082). *indicates P<0.05 and **P<0.001 when egl-1(lf) and lin-3(lf) were compared to wild-type or the transgenic strains were compared to egl-1(lf); Ex[egl-1(+)] (Fisher's exact test). ns, no significance. (D) Cell corpses in the embryos of the indicated genotypes carrying the wild-type or mutant egl-1 gene were scored at the 1.5-fold stage (n≥30). Each circle represents a single embryo. At least three independent stably transmitted lines were analyzed for each transgene. Alleles used: lin-3(e1417) and egl-1(n1084n3082). **indicates P<0.001 in a two-tailed t-test. ns indicates no statistical difference (p>0.05). (E) Proposed model of how lin-3 promotes PCD (see text for details). To test the physiological importance of the interaction between LIN-1 and the egl-1 promoter, we mutated the P3 sequence in egl-1 genomic DNA and assessed the ability of the mutant egl-1 gene to rescue the cell death defect of the egl-1 mutant. Specifically, we examined the cell death phenotype by scoring the surviving ABpl/rpppapp in the L1 tail (Figure 6C) and by counting the number of cell corpses at the 1.5-fold stage (Figure 6D). In the control experiment, egl-1 mutants carrying the wild-type egl-1 transgene showed no surviving ABpl/rpppapp (Figure 6C), indicating that the wild-type egl-1 transgene Ex[egl-1(+)] fully rescued the egl-1 cell death defect. When the LIN-1 recognition sequence CAGGAA in the P3 region of the egl-1 promoter was mutated into GCTACT [65], the mutant egl-1 transgene Ex[egl-1(mut)] only partially rescued the egl-1 cell death defect. In the two transgenic lines, the percentages of ABpl/rpppapp survival were 15% and 25%, similar to that of the lin-3 mutant (Figure 6C). The egl-1; Ex[egl-1(mut)] transgenic worms also had a similar number of embryonic cell corpses as the lin-3 mutant at the 1.5-fold stage (Figure 6D). These results show that the binding of LIN-1 to the egl-1 promoter is important for the cell-killing activity of egl-1.
To test whether this LIN-1-mediated cell-killing activity of egl-1 may depend on LIN-3 signaling, we examined whether a lin-3(lf) mutation may reduce the rescuing activity of the mutant egl-1 transgene by scoring surviving ABpl/rpppapp (Figure 6C) and embryonic cell corpses (Figure 6D). As shown in Figure 6C and 6D, the lin-3(lf) mutation reduced the rescuing activity of the wild-type egl-1 transgene, but failed to reduce the rescuing activity of the mutant egl-1 transgene. These results show that binding of LIN-1 to the egl-1 promoter is critical for LIN-3 signaling to promote PCD. These data support the idea that LIN-3 acts through LIN-1 to promote PCD by transcriptional activation of egl-1.
Discussion
LIN-3 signaling promotes PCD through LET-60-MPK-1 signaling and LIN-1 to activate egl-1 transcription
LIN-3-induced LET-60-MPK-1 signaling is important in multiple aspects of development in C. elegans [39]–[45]. In this study, we provide evidence that it is also essential for specific somatic PCD. Mutants defective in genes coding for proteins in this pathway showed reduced PCD in specific cells, ABpl/rpppapp, g1A sisters, and I1 sisters. In contrast, over-activation of this pathway resulted in ectopic deaths of specific cells, such as gland cells. These results show that LIN-3-induced LET-60-MPK-1 signaling is crucial for the death-versus-life fate of specific cells. In addition, the downstream transcription factor LIN-1, but not LIN-31, was shown to act in the LIN-3-induced LET-60-MPK-1 signaling pathway to regulate PCD via the target gene egl-1. Furthermore, we showed that LIN-1 bound directly to the egl-1 promoter in vitro and that the LIN-1 binding site of the egl-1 promoter was important for the cell death-promoting function of LIN-3 in vivo.
Out of 35 embryonic PCDs we analyzed, including the first 13 PCDs in the AB lineage and 22 pharyngeal PCDs, the lin-3-to-lin-1 signaling pathway is important for the death of ABpl/rpppapp, g1A sisters, and I1 sisters. ABpl/rpppapp and I1 sisters undergo PCD before the comma stage, and g1A sisters die during the comma stage [6]. Our embryonic cell corpse data revealed that mutants defective in the lin-3-to-lin-1 signaling pathway had less cell corpses at not only the comma stage, but also 1.5-fold and 2-fold stages. However, the identities of dying cells affected at the latter two stages have not yet been determined, partly because no cell-specific markers are available to test for their survival in the mutants. It is possible that those lineages where PCDs might be more severely affected have not yet been analyzed in our work and, if so, the lin-3 signaling pathway might play a predominant role in these PCDs as compared to the death of ABpl/rpppapp, g1A sisters, and I1 sisters. Since the mutants defective in the lin-3 signaling pathway showed a partially penetrant phenotype in the survival of ABpl/rpppapp, g1A sisters, and I1 sisters, other genes or pathways, in addition to the lin-3 signaling pathway, are required to efficiently promote the death of, at least, ABpl/rpppapp, g1A sisters, and I1 sisters.
Although we have not yet characterized all affected embryonic PCDs, the data obtained using either the specific cell death assays or the embryonic cell corpse counting assays were similar. For example, the lin-3, let-23, let-60, mpk-1 and lin-1 single mutants had the same phenotype as their respective single mutants in the lin-3 or let-23 mutant background or feeding lin-3 or let-23 RNAi (Figure 1A, 1D, 1G, 5A, and Table S5). Overexpression of lin-3 using its endogenous promoter or misexpression of lin-3 in the intestine resulted in ectopic gland cells and increased numbers of embryonic cell corpses during comma to two-fold stages. Both phenotypes could be suppressed by loss of let-23 or components of the core PCD pathway (Figure 2 and Table S2). In addition, the wild-type egl-1 genomic DNA fully rescued the egl-1 cell death defect, but the mutant egl-1 genomic DNA with a mutation in the LIN-1 binding site could only rescue the egl-1 cell death defect to a similar extent as the lin-3 mutant (Figure 6C and 6D). Furthermore, loss of lin-3 did not aggravate the PCD defect of the egl-1; Ex[egl-1(mut)] transgenic animals (Figure 6C and 6D). Therefore, the lin-3-to-lin-1 signaling pathway may promote the death of ABpl/rpppapp and g1A sisters as well as other yet uncharacterized doomed cells by the same molecular mechanism. However, we cannot rule out the possibility that let-23, mpk-1, let-60 or lin-1 may affect other aspects of PCD, rather than promoting the PCD fate, in the yet unidentified doomed cells.
In the genetic interaction experiments, the let-23(n1045) mutation does not completely eliminate the cell death-promoting function of syIs1: the mutation reduced the numbers of ectopic embryonic cell deaths (Table S2) to the wild-type level, but not to the let-23(n1045) mutant level. The let-23(n1045) allele may still have a residual let-23 activity [52] and therefore cannot fully block the lin-3 signaling from syIs1. Alternatively, syIs1 may exert its PCD-promoting function through both LET-23-dependent and LET-23 independent pathways to cause ectopic cell deaths.
Previous studies showed that LIN-1 is negatively regulated by the upstream LIN-3-induced LET-60-MPK-1 signaling pathway during vulval induction [46]. We found that the LIN-1 function is positively regulated by this signaling pathway to promote the PCD fate of specific cells. During vulval development, although lin-1 inhibits most target gene expression downstream of the LET-60-MPK-1 pathway, lin-1 is also required positively for the expression of several downstream genes, such as egl-17 [66]. Our data indicated that lin-1 activates egl-1 expression in promoting PCD of specific cells. Interestingly, both of these lin-1-mediated gene expression processes are positively regulated by the LET-60-MPK-1 pathway and do not require lin-31. It appears that LIN-1 could have either a positive or negative function on gene expression, depending on the cellular context and target genes.
LIN-3 signaling was also found to activate ced-4 transcription, but egl-1 transcription probably accounted for most of the LIN-3 signaling-mediated cell death, as, in the egl-1 mutant, the mutant egl-1 transgene lacking the LIN-1 binding site could only restore the penetrance of ABpl/rpppapp death and the embryonic cell corpse number to the levels seen in the lin-3 mutant.
A previous study showed that dying cells that inappropriately survive generally adopt the fate of the sister or lineal homolog if the latter cells are terminally differentiated [11]. We observed that the g1A sister, when survived in the lin-3 or ced-3 mutant, expressed the gland cell marker PB0280.7::4Xnls::mrfp, indicating that the g1A sister adopted its sister cell fate. To rule out the possibility that the lin-3 mutation may cause cell fate transformation and hence block cell death, we examined surviving ABpl/rpppapp in detail in the lin-3 mutant. ABpl/rpppapp was chosen because the sister of ABpl/rpppapp divides once and generates a hypodermal cell and a phasmid sheath cell [6]. It has been shown that dying cells that inappropriately survive do not adopt their sister cell fates, if the latter cells undergo cell division and the resulting descendent cells then differentiate [11]. The observation that surviving ABpl/rpppapp in the lin-3 or egl-1 mutant did not express the hypodermal cell-specific genes dpy-7 and ajm-1, as assayed by their respective reporters Pdpy-7::yfp and ajm::gfp, suggesting that surviving ABpl/rpppapp did not further divide or differentiate to generate a hypodermal cell. This result argues against the possibility that the lin-3 mutation affects ABpl/rpppapp survival indirectly through cell fate transformation to its sister or niece cell.
An appropriate level of extrinsic LIN-3 signaling is important for the precise fine-tuning of the life-versus-death fate of cells
Our data support the following model explaining how different levels of LIN-3 expression or activity might regulate PCD (Figure 6E). In the wild-type, after binding LIN-3, LET-23 transduces the cell death-promoting signal through the LET-60-MPK-1 pathway, leading to LIN-1 activation, followed by egl-1 transcription, and the demise of specific cells. However, mutations in either the LIN-3-induced LET-60-MPK-1 signaling pathway or LIN-1 or a defect in the LIN-1-binding site of the egl-1 promoter reduces egl-1 transcription and thus the probability that a cell undergoes PCD. In contrast, overexpression of lin-3 enhances egl-1 transcription and leads to ectopic cell deaths. Therefore, LIN-3 signaling may act to specify the death of specific cells and fine-tunes the life-versus-death fate of these cells by modulating transcription of egl-1.
Although EGF has long been known to function in the process of cell proliferation and survival, some reports showed that it also promotes apoptosis [30]–[32]. In some cases, the difference in cellular outcome appears to depend on its concentration, with a low EGF concentration promoting cell proliferation and a high concentration inducing apoptosis [67], [68]. Cells expressing different levels of EGFR also show different responses to the same amount of EGF, as it has been reported that exogenous EGF attenuates cell death in rats, which express low levels of EGFR, but promotes cell death in mice, which express high levels of EGFR [69]. Thus, it is likely that overactivation of EGF signaling triggers apoptosis. This may be a conserved physiological mechanism for preventing unrestricted proliferation when cells respond to persistent or hyperactivated EGF signaling. In C. elegans, cells may use this strategy to fine-tune PCD: cells that are doomed to die may express a high level of LET-23 or its downstream signaling components. This may either avoid inappropriate survival of cells that occasionally escape from the cell death machinery or preset a cell in a sensitized stage to respond quickly when it receives the death signal. This may also explain why some cells are more sensitive to apoptotic stimuli than others.
Our data show that different cells appear to show different susceptibility to lin-3 signaling in PCD at, at least, three different levels. First, ABpl/rpppapp, g1A sisters, and I1 sisters require lin-3 signaling for PCD and are sensitive to the wild-type level of lin-3 signaling. Second, g1 and g2 cells are sensitive to a high level of lin-3 signaling in PCD, since they survive in the wild-type but can be induced to undergo PCD by the syIs1 transgene. Third, cells, such as PLM, survive even in the presence of the transgene syIs1 and are therefore insensitive to lin-3 signaling for their life vs death fate. These results together show a complexity of extrinsic signaling in PCD regulation.
The LET-60-MPK-1 pathway promotes germline and developmental cell deaths
Consistent with our data, mutants defective in ksr-1, a positive regulator of the RAS signaling pathway, have reduced embryonic cell corpse numbers [70]. The LET-60-MPK-1 - and PI3K-mediated signaling pathways are also known to be involved in physiological or genotoxic stress-induced germ cell death in C. elegans [48]–[51], although the ligand and receptor for these pathways have not yet been identified. The LET-60-MPK-1 pathway regulates the apoptotic process of irradiation-induced germ cell death by restricting CEP-1 protein expression to cells in late pachytene. More importantly, MPK-1, the C. elegans ERK, is activated following irradiation and is required for egl-1 transcription in irradiation-induced germ cell death [50]. This result and our present findings show that the LET-60-MPK-1 pathway regulates egl-1 transcription to promote both physiological somatic PCD and genotoxic stress-induced germline cell death. In mammalian cells, activation of the RAS-ERK pathway upregulates Bcl-XL transcription [26]. It has also been shown that EGF regulates the expression of Mcl-1, a member of the anti-apoptotic Bcl-2 family, through activation of MAPK signaling and the direct binding of Elk-1, a member of the ETS gene family, to the Mcl-1 promoter [71]. Thus, the interaction of the RAS-ERK pathway and Bcl-2 family members, such as egl-1, Bcl-XL, and Mcl-1, may be a conserved strategy for regulating cell death or survival in evolution.
Pathological and physiological roles of EGFR in promoting cell death
It has been reported that cisplatin, a widely used chemotherapeutic agent for human cancers, causes nephrotoxicity in patients and induces extensive death of the proximal tubules in mice or cultured proximal tubule cells [72]–[74]. Cisplatin activates EGFR and ERK to trigger apoptosis in cultured proximal tubule cells [74]. In addition, in the autoimmune cutaneous disease pemphigus vulgaris, the body produces autoantibodies that can induce EGFR activation and ERK phosphorylation, leading to activation of caspase-3 [75]. EGFR activation-induced RAS signaling has also been implicated in age-related brain degeneration in Drosophila [76]. Taken together, these results and our data show that EGFR activation-induced cell death is a conversed phenomenon and plays an important role in pathological and physiological conditions.
Mutations that cause constitutive activation of EGFR are commonly observed in human cancers. Specific antibodies or inhibitors against activated EGFR have been developed and used as targeted therapeutics [77]. However, previous studies in human cell lines [30]–[32] and our present results show that overactivation of EGF signaling can promote apoptosis, raising the concern that drugs that antagonize EGFR signaling might have an anti-apoptotic effect and thus a potential tumor-promoting side-effect, depending on the cellular context. Further information on the molecular basis of the distinct divergent cellular responses of cell proliferation, cell survival, and death to EGF signaling is important for our understanding of the physiological roles of EGF and EGFR signaling during development and homeostasis and for providing perspectives for optimizing the strategy of anticancer drug design.
Materials and Methods
General methods and strains
Unless otherwise stated, C. elegans strains were maintained at 20°C as described previously [78]. The N2 Bristol strain was used as the wild-type strain. Alleles used:
Linkage group (LG)I: aap-1(m889), aap-1(ok282), age-2(yw23), daf-16(m26), and mek-2(ku114).
LGII: age-1(hx546), age-1(m333), let-23(n1045), let-23(sy1), let-23(sy97), lin-31(gk569), lin-31(n301), lin-31(n1053), plc-3(tm1340), and rrf-3(pk1426).
LGIII: ced-4(n1162), ced-9(n1950), mpk-1(ku1), mpk-1(n2521), unc-32(e189), unc-79(e1068), and syIs107[Plin-3::pes-10::gfp].
LGIV: ced-3(n717), let-60(n1046), let-60(sy93), lin-1(n1761), lin-1(n303), lin-1(n1047), lin-1(sy254), lin-3(e1417), lin-3(n378), lin-45(n2506), and plc-4(ok1215).
LGV: egl-1(n1084n3082), plc-2(ok1761), sos-1(cs41), and bcIs37[Pegl-1::his-24::gfp].
LGX: plc-1(rx1), sem-5(n1779), and syIs1[lin-3(+)].
All alleles are described in WormBase (http://www.wormbase.org/). tpIs8 is an integrated version of tpEx270[PB0280.7::4Xnls::mrfp; unc-119(+)] (this report). PB0280.7::4Xnls::mrfp is expressed in g1 and g2 gland cells [55].
RNA interference (RNAi)
There are about 40 receptor tyrosin kinase (rtk) genes in the C. elegans genome. To test the involvement of these genes in promoting PCD, RNAi of the individual rtk genes was performed by feeding [79], and the cell corpses of treated embryos were scored. In this screen, only let-23 RNAi significantly decreased cell corpse numbers at the comma and 1.5-fold stages. To characterize the effect of the lin-3 signaling pathway on the number of embryonic cell corpses, the rrf-3(pk1426) mutant, which is sensitive to RNAi [80], was treated by feeding RNAi at 20°C, as previously described [81]. In other RNAi experiments, animals were treated with RNAi without the rrf-3(pk1426) mutation. The lin-3 or let-23 RNAi treatment caused more than 50% rod-like larval lethality both in the wild-type and rrf-3(pk1426) mutant, confirming that lin-3 and let-23 RNAi works well in these experiments. RNAi plasmids were obtained from the J. Ahringer RNAi library. L4 hermaphrodites were put onto the RNAi plates, and F1 were scored for the number of embryonic cell corpses, surviving ABpl/rpppapp, or extra gland cells. The empty vector L4440 was used as negative control.
Transgenic animals
Germline transformation experiments were performed as described previously [82]. To observe the expression pattern of let-23, Plet-23::4Xnls::gfp (50 ng/µl) was injected into unc-119(ed3) worms with the coinjection markers unc-119 [83] (50 ng/µl) and Pced-1::1Xnls::mrfp (50 ng/µl). To examine the expression pattern of lin-3 and let-23, Plet-23::4Xnls::mrfp (200 ng/µl) was injected into an integrated transgene syIs107[Plin-3::pes-10::gfp] with the coinjection markers Pelt-2::gfp (50 ng/µl) [61]. To test whether secreted LIN-3 rescued the cell death defect of the lin-3 mutant and caused ectopic cell deaths, Pelt-2::gfp or Pelt-2::lin-3::gfp (50 ng/µl) was injected into the lin-3(e1417) mutant and wild-type with the coinjection marker sur-5::gfp (50 ng/µl). To test the effect of LIN-1 overexpression on PCD, plasmids pPD49.78 and pPD49.83 (containing Phsp without an insert) or Phsp::lin-1 (50 ng/µl) was co-injected with the coinjection marker sur-5::gfp (50 ng/µl) into animals carrying the integrated transgene tpIs8. To examine the importance of the LIN-1 binding site in the egl-1 promoter, wild-type (pBC08) [7] or mutant egl-1 (2 ng/µl) was injected into either egl-1(n1084n3082) or lin-3(e1417); egl-1(n1084n3082) mutants with the coinjection marker sur-5::gfp (50 ng/µl).
Heat shock treatment
Embryos were incubated at 20°C or heat shocked at 33°C for 10–60 min and then moved to 20°C. The number of g1 and g2 gland cells was scored at the L4 stage using the tpIs8 transgene after three days.
Molecular biology
Standard methods of cloning, sequencing, and PCR amplification were used. To generate Plet-23::4xnls::gfp, the let-23 promoter was amplified by PCR using primers 5′-gtctagagcatctgcacttggg-3′ and 5′-gggatccgcctcccag-3′ and the resulting PCR fragment was inserted into the pPD122.56 vector to generate pYW1242. Full-length lin-3 cDNA was obtained by RT-PCR using worm total RNA and the primers 5′-CCACCGGTATGCGGAAAATGCTAC-3′ and 5′-CCACCGGTTTTGTGTGTCGAATCATTGG-3′. To obtain Pelt-2::lin-3::gfp, the elt-2 promoter, previously amplified by PCR using 5′-TTGGATCCCGGTGAAACTCTCTTGG-3′ and 5′-CCGGATCCCAGTGGCACCTAAAACATC-3′, was cloned into the BamHI site of pPD95.75 and lin-3 cDNA was cloned into the AgeI site to generate pYW1243. To generate Phsp::lin-1, a full-length lin-1 cDNA was amplified from the yk1150h12 plasmid by PCR using 5′-ccggatccatgaatcacattgaccttttg-3′ and 5′-ttccatggctacaaagttggcatttttatg-3′, and the resulting PCR fragment was inserted into the heat-shock vectors pPD49.78 and pPD49.83 to generate pYW1245 and pYW1246, respectively. The DNA fragment corresponding to the LIN-1 DNA binding domain was amplified by PCR using the yk1150h12 plasmid as template and 5′-CCGGATCCATGAATCACATTGACCTTTTG-3′ and 5′-TTGGATCCTTTCGTTGGCGGCTGCGG-3′ as primers [65], then the PCR product was inserted into vector pGEX-4T-1 to generate plasmid pYW1247. The pBC08 plasmid contains the egl-1 genomic fragment, which rescues the cell death defect of egl-1 mutants [7]. To generate mutant egl-1 with a defect in the LIN-1 binding site, two partial fragments of egl-1 from pBC08 were first amplified by PCR using primer pairs 5′-caatttatagtaaagttaacttca-3′ plus 5′-cacatacagtagcttgaattcaactatacatctg-3′ and 5′-aattcaagctactgtatgtgtcacttccaccacc-3′ plus 5′-ggccgctctagaactagtgg-3′. The products were then used as templates for fusion PCR to generate a larger fragment to replace the corresponding region in pBC08 to generate pYW1248. This mutant egl-1 was identical to wild-type egl-1 except that the core LIN-1 binding motif was changed from CAGGAA to GCTACT.
Cell death assays
Cell corpses were counted at the indicated developmental stages and the number of extra cells in the anterior and posterior pharynx of L4 hermaphrodites was scored using Nomarski optics as described previously [54]. To analyze the first 13 cell deaths that occur in the AB lineage during embryogenesis, embryos were mounted onto 4% agar pads and recorded using a four-dimensional DIC microscopy analysis as described previously [84]. The survival of ABpl/rpppapp was examined by its localization in the tail with the aid of sur-5::gfp. To count the number of gland cells in the pharynx, the integrated transgene tpIs8[PB0280.7::4Xnls::mrfp] was used as a marker for these gland cells [55], and the number of the mRFP-positive cells was scored at L4 stage by fluorescence microscopy.
Quantitative real-time reverse transcriptase (RT)-PCR
Total RNA was isolated from embryos developing up to the comma stage and purified by chloroform extraction and isopropanol precipitation, then 500 ng was reverse transcribed into cDNA using RevertAid H Minus First Strand cDNA Synthesis Kits (Fermentas). Equal amounts of the different cDNAs were used as the template for gene-specific PCR with appropriate primers. Water was used as negative control and tbg-1 RT-PCR was performed as an internal control. The CT values of the test genes were normalized to that of tbg-1 and relative expression levels were derived using the comparative CT method [85].
Quantification of Pegl-1::gfp intensity in ABprpppapp
To measure the Pegl-1::gfp expression within ABprpppapp, fluorescence micrographs were captured using a Zeiss AxioImager M2 microscope (Zeiss) equipped with a charge-coupled device camera. GFP intensity was determined using Image J by measuring the fluorescence signals within ABprpppapp (Ia) and its neighboring region (Ib) of ced-3(717) and lin-3(e1417) ced-3(n717) mutants. As a control, the fluorescence signals within two cells near ABprpppapp (Ia) and its neighboring region (Ib) of ced-3(717) and lin-3(e1417) ced-3(n717) mutants was determined. The value of Ia minus the value of Ib was used as the value for relative GFP intensity, and the relative GFP intensity of the ced-3(n717) mutant was used for normalization.
Prediction of potential LIN-1 binding sites
The prediction of potential LIN-1 binding sites was performed at http://www.cbrc.jp/research/db/TFSEARCH.html by submitting the sequence from +174 to +5820 (5′-3′) downstream of the stop codon of the egl-1 gene and the sequence from −1914 to −837 (5′-3′) upstream of the stop codon [62]. Four binding sites for Elk-1, the mammal LIN-1 ortholog, were predicted; these were AGCTTTCCGGCTCA (P1), CGATCCGGAAATCC (P2), TAGTTGAATTCAACAGGAAGTATGTGTCACTTCC (P3), and CAATTTCCGGTAAT (P4). P2 and P3, but not P1 or P4, are conserved in C. briggsae. P2 has been shown to be important for the PCD of the neurosecretory-motor (NSM) neuron [19], but lin-3 and let-23 do not seem to be involved in the death of NSM sister cell (Figure 1D), we therefore focused on P3 in this study.
Electrophoretic mobility shift assay (EMSA)
Expression of GST and GST::LIN-1(DBD) was induced in Escherichia coli strain BL21 carrying the respective plasmids pGEX-4T-1 and pYW1247 by addition of 0.4 mm isopropyl thiogalactoside and incubation for 1 h at 37°C. The cells were then harvested and disrupted by sonication in phosphate-buffered saline containing protease inhibitor cocktail (Calbiochem) and 1% Triton X-100, and the proteins were purified on glutathione Sepharose (Amersham Biosciences) according to the manufacturer's protocol. The amount of protein was measured by the Bradford method (Biorad). The sense and antisense oligonucleotides containing the predicted LIN-1 binding sequences, which were synthesized and biotinylated at the 3′ end (Genomics), were mixed in annealing buffer (5 mM NaCl, 1 mM Tris-HCl, 1 mM MgCl2, and 0.1 mM dithiothreitol, pH 7.9) at a final concentration of 1 µM, denatured at 98°C for 2 min, and allowed to anneal by slowly cooling the mixture to 25°C. Unlabeled probes were used as competitors in competition experiments. The sequences of the probes were: wild-type: 5′-gaattcaacaggaagtatgtgtcacttccaccac-3′ and 5′-gtggtggaagtgacacatacttcctgttgaattc-3′; mutant: 5′ - gaattcaagctactgtatgtgtcacttccaccac-3′ and 5′-gtggtggaagtgacacatacagtagcttgaattc-3′. EMSA were performed using LightShift Chemiluminescent EMSA kits (Pierce), following the manufacturer's instruction. The mixture was separated at 100 V at 4°C on a 5.5% non-denaturing polyacrylamide gel, then the DNA was transferred to a charged nylon membrane (Millipore) at 150 mA for 30 min and cross-linked with UV light using the auto crosslink option on a UV Stratalinker 1800 (Stratagene). The membrane was then blocked, washed, and exposed to X-ray film following the manufacturer's protocol.
Statistical analysis
In histograms, the values are shown as the mean and standard deviation (SD). Data derived from different genetic backgrounds at a specific developmental stage were compared using Student's two-tailed unpaired t-test. Data were considered statistically different at P<0.05. Fisher's exact test was used when comparing the percentages of surviving or missing cells in different genetic backgrounds. Data were considered statistically different at P<0.05.
Supporting Information
Zdroje
1. FuchsY, StellerH (2011) Programmed cell death in animal development and disease. Cell 147 : 742–758.
2. BaehreckeEH (2002) How death shapes life during development. Nat Rev Mol Cell Biol 3 : 779–787.
3. LettreG, HengartnerMO (2006) Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol 7 : 97–108.
4. PottsMB, CameronS (2011) Cell lineage and cell death: Caenorhabditis elegans and cancer research. Nat Rev Cancer 11 : 50–58.
5. SulstonJE, HorvitzHR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56 : 110–156.
6. SulstonJE, SchierenbergE, WhiteJG, ThomsonJN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100 : 64–119.
7. ConradtB, HorvitzHR (1998) The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93 : 519–529.
8. HengartnerMO, HorvitzHR (1994) C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 76 : 665–676.
9. YuanJ, ShahamS, LedouxS, EllisHM, HorvitzHR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75 : 641–652.
10. ZouH, HenzelWJ, LiuX, LutschgA, WangX (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90 : 405–413.
11. EllisHM, HorvitzHR (1986) Genetic control of programmed cell death in the nematode C. elegans. Cell 44 : 817–829.
12. HengartnerMO, EllisRE, HorvitzHR (1992) Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356 : 494–499.
13. ChenF, HershBM, ConradtB, ZhouZ, RiemerD, et al. (2000) Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287 : 1485–1489.
14. YanN, GuL, KokelD, ChaiJ, LiW, et al. (2004) Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell 15 : 999–1006.
15. YangX, ChangHY, BaltimoreD (1998) Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281 : 1355–1357.
16. PourkarimiE, GreissS, GartnerA (2012) Evidence that CED-9/Bcl2 and CED-4/Apaf-1 localization is not consistent with the current model for C. elegans apoptosis induction. Cell Death Differ 19 : 406–415.
17. NehmeR, ConradtB (2008) egl-1: a key activator of apoptotic cell death in C. elegans. Oncogene 27 Suppl 1: S30–40.
18. PedenE, KillianDJ, XueD (2008) Cell death specification in C. elegans. Cell Cycle 7 : 2479–2484.
19. ThellmannM, HatzoldJ, ConradtB (2003) The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development 130 : 4057–4071.
20. GrantS, QiaoL, DentP (2002) Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front Biosci 7: d376–389.
21. DanielsenAJ, MaihleNJ (2002) The EGF/ErbB receptor family and apoptosis. Growth Factors 20 : 1–15.
22. WieduwiltMJ, MoasserMM (2008) The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci 65 : 1566–1584.
23. XianCJ, ZhouXF (2004) EGF family of growth factors: essential roles and functional redundancy in the nerve system. Front Biosci 9 : 85–92.
24. BergmannA, TugentmanM, ShiloBZ, StellerH (2002) Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell 2 : 159–170.
25. HensonES, GibsonEM, VillanuevaJ, BristowNA, HaneyN, et al. (2003) Increased expression of Mcl-1 is responsible for the blockage of TRAIL-induced apoptosis mediated by EGF/ErbB1 signaling pathway. J Cell Biochem 89 : 1177–1192.
26. JostM, HuggettTM, KariC, BoiseLH, RodeckU (2001) Epidermal growth factor receptor-dependent control of keratinocyte survival and Bcl-xL expression through a MEK-dependent pathway. J Biol Chem 276 : 6320–6326.
27. LeuCM, ChangC, HuC (2000) Epidermal growth factor (EGF) suppresses staurosporine-induced apoptosis by inducing mcl-1 via the mitogen-activated protein kinase pathway. Oncogene 19 : 1665–1675.
28. AllanLA, MorriceN, BradyS, MageeG, PathakS, et al. (2003) Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol 5 : 647–654.
29. FangX, YuS, EderA, MaoM, BastRCJr, et al. (1999) Regulation of BAD phosphorylation at serine 112 by the Ras-mitogen-activated protein kinase pathway. Oncogene 18 : 6635–6640.
30. GulliLF, PalmerKC, ChenYQ, ReddyKB (1996) Epidermal growth factor-induced apoptosis in A431 cells can be reversed by reducing the tyrosine kinase activity. Cell Growth Differ 7 : 173–178.
31. ArmstrongDK, KaufmannSH, OttavianoYL, FuruyaY, BuckleyJA, et al. (1994) Epidermal growth factor-mediated apoptosis of MDA-MB-468 human breast cancer cells. Cancer Res 54 : 5280–5283.
32. GarciaR, FranklinRA, McCubreyJA (2006) Cell death of MCF-7 human breast cancer cells induced by EGFR activation in the absence of other growth factors. Cell Cycle 5 : 1840–1846.
33. HillRJ, SternbergPW (1992) The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358 : 470–476.
34. AroianRV, KogaM, MendelJE, OhshimaY, SternbergPW (1990) The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348 : 693–699.
35. ChamberlinHM, SternbergPW (1994) The lin-3/let-23 pathway mediates inductive signalling during male spicule development in Caenorhabditis elegans. Development 120 : 2713–2721.
36. ClandininTR, DeModenaJA, SternbergPW (1998) Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell 92 : 523–533.
37. JiangLI, SternbergPW (1998) Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development 125 : 2337–2347.
38. Van BuskirkC, SternbergPW (2007) Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci 10 : 1300–1307.
39. HanM, SternbergPW (1990) let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell 63 : 921–931.
40. ClarkSG, SternMJ, HorvitzHR (1992) C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature 356 : 340–344.
41. HanM, GoldenA, HanY, SternbergPW (1993) C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature 363 : 133–140.
42. WuY, HanM, GuanKL (1995) MEK-2, a Caenorhabditis elegans MAP kinase kinase, functions in Ras-mediated vulval induction and other developmental events. Genes Dev 9 : 742–755.
43. KornfeldK, GuanKL, HorvitzHR (1995) The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes & Development 9 : 756–768.
44. LacknerMR, KornfeldK, MillerLM, HorvitzHR, KimSK (1994) A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes & development 8 : 160–173.
45. WuY, HanM (1994) Suppression of activated Let-60 ras protein defines a role of Caenorhabditis elegans Sur-1 MAP kinase in vulval differentiation. Genes Dev 8 : 147–159.
46. TanPB, LacknerMR, KimSK (1998) MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93 : 569–580.
47. HopperNA (2006) The adaptor protein soc-1/Gab1 modifies growth factor receptor output in Caenorhabditis elegans. Genetics 173 : 163–175.
48. GumiennyTL, LambieE, HartwiegE, HorvitzHR, HengartnerMO (1999) Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126 : 1011–1022.
49. PerrinAJ, GundaM, YuB, YenK, ItoS, et al. (2013) Noncanonical control of C. elegans germline apoptosis by the insulin/IGF-1 and Ras/MAPK signaling pathways. Cell Death Differ 20 : 97–107.
50. RutkowskiR, DickinsonR, StewartG, CraigA, SchimplM, et al. (2011) Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLoS Genet 7: e1002238.
51. QuevedoC, KaplanDR, DerryWB (2007) AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr Biol 17 : 286–292.
52. FergusonEL, HorvitzHR (1985) Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110 : 17–72.
53. HoeppnerDJ, HengartnerMO, SchnabelR (2001) Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412 : 202–206.
54. EllisRE, JacobsonDM, HorvitzHR (1991) Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129 : 79–94.
55. Hunt-NewburyR, ViveirosR, JohnsenR, MahA, AnastasD, et al. (2007) High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5: e237.
56. ChangC, NewmanAP, SternbergPW (1999) Reciprocal EGF signaling back to the uterus from the induced C. elegans vulva coordinates morphogenesis of epithelia. Curr Biol 9 : 237–246.
57. LiuJ, TzouP, HillRJ, SternbergPW (1999) Structural requirements for the tissue-specific and tissue-general functions of the Caenorhabditis elegans epidermal growth factor LIN-3. Genetics 153 : 1257–1269.
58. KatzWS, HillRJ, ClandininTR, SternbergPW (1995) Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell 82 : 297–307.
59. SternbergPW, HorvitzHR (1986) Pattern formation during vulval development in C. elegans. Cell 44 : 761–772.
60. HwangBJ, SternbergPW (2004) A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development 131 : 143–151.
61. FukushigeT, HawkinsMG, McGheeJD (1998) The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol 198 : 286–302.
62. ConradtB, HorvitzHR (1999) The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98 : 317–327.
63. MillerLM, GallegosME, MorisseauBA, KimSK (1993) lin-31, a Caenorhabditis elegans HNF-3/fork head transcription factor homolog, specifies three alternative cell fates in vulval development. Genes Dev 7 : 933–947.
64. BeitelGJ, TuckS, GreenwaldI, HorvitzHR (1995) The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev 9 : 3149–3162.
65. MileyGR, FantzD, GlossipD, LuX, SaitoRM, et al. (2004) Identification of residues of the Caenorhabditis elegans LIN-1 ETS domain that are necessary for DNA binding and regulation of vulval cell fates. Genetics 167 : 1697–1709.
66. TiensuuT, LarsenMK, VernerssonE, TuckS (2005) lin-1 has both positive and negative functions in specifying multiple cell fates induced by Ras/MAP kinase signaling in C. elegans. Dev Biol 286 : 338–351.
67. ChiuB, MirkinB, MadonnaMB (2006) Mitogenic and apoptotic actions of epidermal growth factor on neuroblastoma cells are concentration-dependent. J Surg Res 135 : 209–212.
68. WangH, GuoD, YeF, XiG, WangB, et al. (2006) Effect and mechanism of epidermal growth factor on proliferation of GL15 gliomas cell line. J Huazhong Univ Sci Technolog Med Sci 26 : 604–606.
69. KileySC, ChevalierRL (2007) Species differences in renal Src activity direct EGF receptor regulation in life or death response to EGF. Am J Physiol Renal Physiol 293: F895–903.
70. SugimotoA, KusanoA, HozakRR, DerryWB, ZhuJ, et al. (2001) Many genomic regions are required for normal embryonic programmed cell death in Caenorhabditis elegans. Genetics 158 : 237–252.
71. BooyEP, HensonES, GibsonSB (2011) Epidermal growth factor regulates Mcl-1 expression through the MAPK-Elk-1 signalling pathway contributing to cell survival in breast cancer. Oncogene 30 : 2367–2378.
72. BernsJS, FordPA (1997) Renal toxicities of antineoplastic drugs and bone marrow transplantation. Semin Nephrol 17 : 54–66.
73. PricePM, SafirsteinRL, MegyesiJ (2004) Protection of renal cells from cisplatin toxicity by cell cycle inhibitors. Am J Physiol Renal Physiol 286: F378–384.
74. AranyI, MegyesiJK, KanetoH, PricePM, SafirsteinRL (2004) Cisplatin-induced cell death is EGFR/src/ERK signaling dependent in mouse proximal tubule cells. Am J Physiol Renal Physiol 287: F543–549.
75. Frusic-ZlotkinM, RaichenbergD, WangX, DavidM, MichelB, et al. (2006) Apoptotic mechanism in pemphigus autoimmunoglobulins-induced acantholysis–possible involvement of the EGF receptor. Autoimmunity 39 : 563–575.
76. BotellaJA, KretzschmarD, KiermayerC, FeldmannP, HughesDA, et al. (2003) Deregulation of the Egfr/Ras signaling pathway induces age-related brain degeneration in the Drosophila mutant vap. Mol Biol Cell 14 : 241–250.
77. PaoW, ChmieleckiJ (2010) Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 10 : 760–774.
78. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
79. TimmonsL, CourtDL, FireA (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 : 103–112.
80. SimmerF, TijstermanM, ParrishS, KoushikaSP, NonetML, et al. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12 : 1317–1319.
81. KamathRS, FraserAG, DongY, PoulinG, DurbinR, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 : 231–237.
82. MelloC, FireA (1995) DNA transformation. Methods Cell Biol 48 : 451–482.
83. MaduroM, PilgrimD (1995) Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141 : 977–988.
84. SchnabelR, HutterH, MoermanD, SchnabelH (1997) Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev Biol 184 : 234–265.
85. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 : 402–408.
86. GuT, OritaS, HanM (1998) Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol 18 : 4556–4564.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



