-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
Resistance (R) genes play central roles in recognizing pathogens and triggering plant defense responses. CHS3 encodes a TIR-NB-LRR-type R protein harboring a C-terminal LIM domain. A point mutation in CHS3 activates the defense response under chilling stress. Here we identified and characterized ibr5-7, a mutant that suppresses the chilling-induced defense responses of chs3-1. We observed that the enhanced defense responses and cell death in the chs3-1 mutant are synergistically dependent on IBR5 and HSP90. IBR5 physically interacts with CHS3, forming a complex with SGT1b/ HSP90. Moreover, IBR5 is also involved in the R-gene resistance mediated by SNC1, RPS4 and RPM1. Thus, IBR5 plays key roles in regulating defense responses mediated by multiple R proteins.
Published in the journal: IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in. PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005584
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005584Summary
Resistance (R) genes play central roles in recognizing pathogens and triggering plant defense responses. CHS3 encodes a TIR-NB-LRR-type R protein harboring a C-terminal LIM domain. A point mutation in CHS3 activates the defense response under chilling stress. Here we identified and characterized ibr5-7, a mutant that suppresses the chilling-induced defense responses of chs3-1. We observed that the enhanced defense responses and cell death in the chs3-1 mutant are synergistically dependent on IBR5 and HSP90. IBR5 physically interacts with CHS3, forming a complex with SGT1b/ HSP90. Moreover, IBR5 is also involved in the R-gene resistance mediated by SNC1, RPS4 and RPM1. Thus, IBR5 plays key roles in regulating defense responses mediated by multiple R proteins.
Introduction
Plant growth and development are continuously affected by various environmental stresses, including low temperature and pathogen infection. Emerging evidence has shown that the defense responses of plants are regulated by temperatures [1]. Temperature modulates the defense responses induced by certain types of resistance (R)/R-like proteins, including Toll/Interleukin–1 receptor (TIR) - nucleotide-binding (NB) - leucine-rich repeat (LRR) proteins [N (Resistance to tobacco mosaic virus) in tobacco; RPP1 (Recognition of Peronospora parasitica 1)-like, RPP4, RPS4 (Resistance to P. syringae 4) and SNC1 (Suppressor of npr1-1, Constitutive 1) in Arabidopsis], CC-NB-LRR proteins [Rx in tomato; RPM1 (Resistance to Pseudomonas syringae pv. maculicola 1) and RPS2 in Arabidopsis], LRR-TM (transmembrane-domain) proteins (Cf–4 and Cf–9 in tomato), TM-CC proteins [RPW8 (Resistance to Powdery Mildew 8) in Arabidopsis] [2–7], and the TIR-NB protein CHS1 (chilling sensitive 1) in Arabidopsis [8]. A recent study showed that low temperatures (10°C to 23°C) elevate R protein-mediated effector-triggered immunity (ETI), and higher temperatures (23°C to 32°C) lead to a shift in pattern-triggered immunity (PTI) signaling in plants [9]. These studies suggest that temperature largely affects the function of R proteins.
Recent studies have revealed that a number of components regulate the activities of R proteins, which in turn finely tune defense signaling. Chaperone and co-chaperone proteins, such as the HSP90-SGT1b (Suppressor of the G2 allele of skp1)-RAR1 (Required for MLA12 resistance 1) complex, involve in multiple R protein-mediated defense pathways [10]. Earlier reports have indicated that these complexes are required for the correct folding and/or stability of R proteins [11–13]. However, several recent studies have suggested that both SGT1b and HSP90 play positive roles in the degradation of R proteins, including RPM1, RPS2 and SNC1, by the SCF complex [14–16]. The dual functions of R protein regulators ensure that the plant immunity system rapidly and properly responds to pathogen invasion. In addition to these chaperones, many other regulators are also involved in R protein regulation. A series of regulators of SNC1 were identified by screening suppressors and enhancers of snc1-1, have been shown to regulate the chromatin, transcription and protein levels of SNC1. These proteins include E1 and E4 ligases, U-box proteins, acetyltransferases, RNA binding proteins, nuclear pore complex components [13,17–22]. These results suggest that SNC1 and/or other R proteins are regulated by multiple biological processes including nucleo-cytoplasmic trafficking, transcriptional reprogramming, RNA processing and protein modification.
Previous studies have shown that low temperature activates defense responses in plants harboring mutations in R/R-like genes, including CHS1, CHS2/RPP4 and CHS3 [7,8,23]. Arabidopsis CHS3 encodes a TIR-NB-LRR-type R protein harboring a C-terminal LIM domain [23,24]. The chs3-1 mutant exhibits chilling-sensitive phenotypes, including small stature and increased disease resistance. The SGT1b and RAR1 proteins are required for R protein stability [25–27]. The chs3 chilling-sensitive phenotypes are suppressed in sgt1b and rar1 mutants [23]. However, the molecular regulatory mechanism of the temperature-dependent defense responses through CHS3 remains elusive.
In the present study, we identified ibr5-7 as a suppressor of the chilling-sensitive phenotypes of chs3-1. IBR5 (Indole-3-Butyric Acid Response 5) encodes a dual-specificity MAPK phosphatase, which acts as a positive regulator of plant responses to auxin and ABA [28]. IBR5 physically interacts with MPK12, and activated MPK12 is dephosphorylated and inactivated by IBR5, thereby negatively regulating auxin signaling [29]. Here, we observed that ibr5 suppresses the chilling-sensitive phenotypes of chs3-1 independently of MPK12. Biochemical data showed that IBR5 complexes with CHS3 and HSP90-SGT1b to to stabilize CHS3. Moreover, IBR5 is involved in the R-gene mediated resistance specified by SNC1, RPS4 and RPM1. Thus, IBR5 plays an important role in regulating different R protein-mediated defense responses.
Results
Identification of the suppressor of chs3-1, suc5
The chs3-1 plants are dwarfed and have small, curly leaves. The defense responses in chs3-1 are constitutively active at 16°C, but this phenotype is alleviated at higher temperatures (22°C) [23]. To understand the molecular mechanism underlying the temperature-dependent cell death in the chs3-1 mutant, we performed a genetic screen to identify suppressors of chs3-1 (suc). The chs3-1 seeds were mutagenized with ethyl methylsulfonate (EMS), and the M2 population was screened for mutants with wild-type morphology at 16°C. Among the suppressors screened, most of the variations were second-site, loss-of-function mutations in CHS3, thereby rescuing the chs3 chilling-sensitive phenotype. One suppressor harbored a mutation in RAR1, and two suppressors harbored mutations in SGT1b; these genes are important regulators required for CHS3 function [23]. These results indicate that the genetic screen was effective. Here, we characterized suc5 as a new suppressor of chs3-1. The chs3 suc5 mutant plants largely resembled wild-type plants when grown at 16°C, except these plants exhibited a slightly smaller stature compared with the wild type and had serrated true leaves (Fig 1A).
Fig. 1. Identification of a suppressor of chs3-1, suc5. 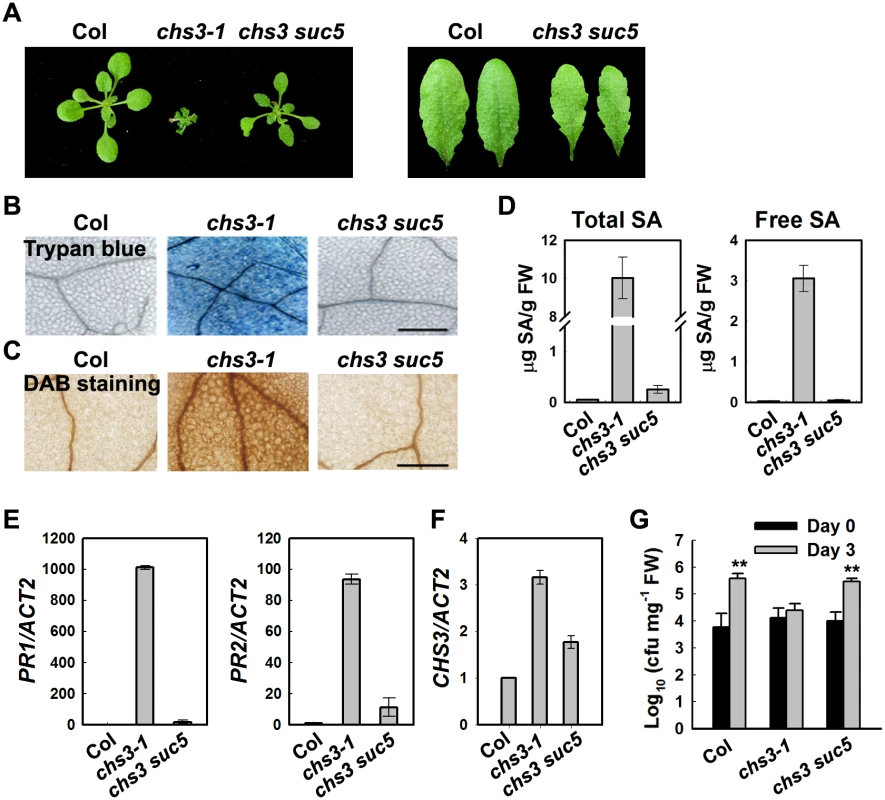
(A) Morphology of wild-type (Col), chs3-1 and chs3 suc5 plants grown at 16°C. The photographs show 4-week-old plants grown in soil (left panel). The rosette leaves of chs3 suc5 have increased leaf serration compared with Col (right panel). (B, C) Trypan blue and DAB staining of leaves from 3-week-old wild type, chs3-1 and chs3 suc5 plants grown at 16°C. Bars: 200 μm. (D) Levels of total and free SA in 4-week-old, wild-type, chs3-1 and chs3 suc5 plants grown at 16°C. (E, F) Expression of PR1, PR2 (E) and CHS3 (F) genes in 3-week-old wild-type, chs3-1 and chs3 suc5 plants grown at 16°C. The values were normalized to the expression of ACTIN2. The error bars represent the SD of three replicates. Similar results were obtained in three independent experiments. (G) Growth of P.s.t. DC3000 on wild-type, chs3-1, and chs3 suc5 plants. Three-week-old plants were dipped with P.s.t. DC3000 (OD600 = 0.05). Bacterial growth was monitored as described in the Materials and Methods. The error bars represent the SD of three replicates, and the asterisks indicate significant differences compared with wild-type Col (**P < 0.01, t-test). suc5 suppresses cell death and defense responses of chs3-1 upon chilling stress
Previous studies have indicated that extensive cell death and strong defense responses occur in chs3-1 mutants grown at 16°C [23]. To determine whether the suc5 mutation affects these cell death-related phenotypes, chs3-1 plants were grown at 16°C, followed by staining with trypan blue and 3, 3’-diaminobenzidine (DAB). The suc5 mutation dramatically reduced the extensive cell death observed in chs3-1 mutants grown at 16°C (Fig 1B). Furthermore, the accumulation of hydrogen peroxide (H2O2) in chs3 suc5 plants grown at 16°C was dramatically reduced compared with chs3-1 (Fig 1C).
The chs3-1 mutant also accumulates high levels of salicylic acid (SA) [23]. To determine whether suc5 inhibits SA accumulation in chs3-1 at 16°C, the endogenous SA level in chs3 suc5 plants was measured. Both the free and total SA levels were dramatically reduced in chs3 suc5 plants grown at 16°C compared with the chs3 mutant (Fig 1D). Because PR genes were highly expressed in chs3-1, we further examined the expression of the PR genes in chs3 suc5 plants grown at 16°C. Quantitative real-time PCR (qRT-PCR) analysis showed that the expression of PR1, PR2 and CHS3 was significantly reduced in chs3 suc5 plants grown at 16°C (Fig 1E and 1F).
Compared with wild-type plants, chs3-1 plants grown at 16°C exhibit enhanced resistance to a virulent pathogenic strain of Pseudomonas syringae pv tomato (P.s.t.), DC3000 [23]. To investigate the role of SUC5 in the chs3-mediated basal defense response, the response of chs3 suc5 seedlings to P.s.t. DC3000 was analyzed. The suc5 mutation fully suppressed the chs3-conferred constitutive resistance to P.s.t. DC3000, resulting in wild-type-like susceptibility (Fig 1G). Taken together, these results demonstrated that, under chilling stress, the suc5 mutation largely suppresses all known autoimmune phenotypes of chs3.
The SUC5 gene is IBR5
To map the suc5 mutation, chs3 suc5 in Columbia (Col) was crossed with Landsberg erecta (Ler) to generate a mapping population. Among the F2 progeny, 50 plants homozygous or heterozygous at the chs3-1 locus and exhibiting a wild-type morphology at 16°C were used for rough mapping. The suc5 mutation was initially mapped to the top of chromosome II (Fig 2A). Fine mapping using approximately 200 plants refined the mutation to a 500-kb region between markers F3C11 and F5G3 (Fig 2A). Further sequencing analysis of this region in chs3 suc5 revealed a G-to-A substitution in the first exon of At2g04550, resulting in an amino substitution from Arg to Lys (Fig 2A).
Fig. 2. Identification of SUC5. 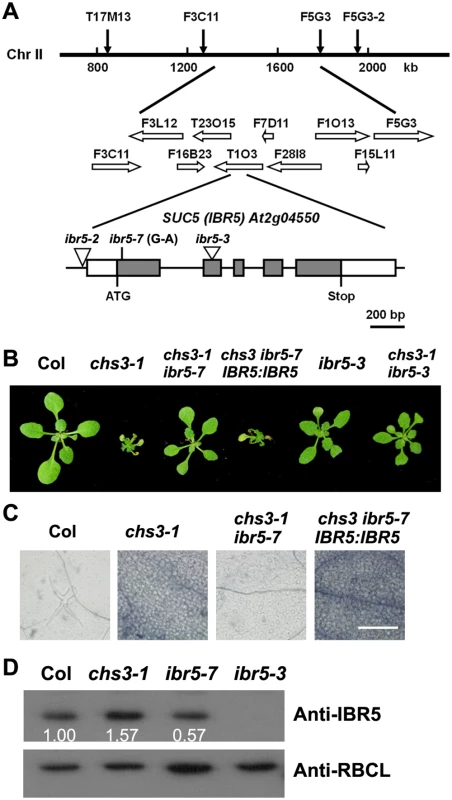
(A) Map position and genomic structure of the SUC5 gene. Exons, introns and UTRs are presented as gray boxes, lines and white boxes, respectively. The point mutation and T-DNA insertions are shown. (B) Morphology of the wild-type, chs3-1, chs3-1 ibr5-7, complemented transgenic plants (IBR5:IBR5 chs3 ibr5-7), T-DNA insertion mutant (ibr5-3) and chs3-1 ibr5-3 double mutant. The photographs show 3-week-old plants grown in soil at 16°C. (C) Trypan blue staining of true leaves from wild-type, chs3-1, chs3-1 ibr5-7 and IBR5:IBR5 chs3-1 ibr5-7 plants. Bars: 200 μm. (D) IBR5 protein levels in wild-type, chs3-1 and ibr5 mutants. Total protein was extracted from 2-week-old seedlings. IBR5 protein was analyzed by immunoblotting using an anti-IBR5 antibody. Anti-RBCL was used as a loading control. At2g04550 was previously identified as IBA RESPONSE5 (IBR5), which encodes a putative dual-specificity protein phosphatase [28]. To determine whether the mutation in IBR5 is responsible for the suppression the phenotypes of chs3, a chs3 ibr5-3 double mutant was generated by crossing chs3-1 with ibr5-3 [29]. The ibr5-3 mutant largely restored the growth defects of chs3-1 mutant plants grown at 16°C (Fig 2B). Furthermore, a wild-type genomic IBR5 fragment containing 5.0 kb of the 5’-promoter region and the 3’ untranslated region was transformed into the chs3 suc5 mutant. All eight T1 transgenic plants displayed chs3-conferred morphological and cell death phenotypes (Fig 2B and 2C). Taken together, these results indicate that SUC5 is indeed IBR5. Therefore, suc5 was renamed ibr5-7.
The ibr5-7 single mutant was isolated by crossing chs3 ibr5-7 with wild-type Col plants. Immunoblot analysis showed that the IBR5 protein level in ibr5-7 was lower than that in the wild-type Col plants, whereas no IBR5 protein was detected in ibr5-3 (Fig 2D). Intriguingly, IBR5 protein accumulated in the chs3-1 mutant (Fig 2D), implying that IBR5 might stabilize CHS3 protein. Consistent with a previous study on ibr5 loss-of-function mutants [28], ibr5-7 displayed serrated leaves and was resistant to high concentrations of exogenous auxins, including IAA, IBA and 2, 4-D (S1 Fig). These results indicate that ibr5-7 is a novel null allele of IBR5.
IBR5 interacts with CHS3
To further investigate whether IBR5 physically interacts with CHS3 in CHS3-mediated signaling, a yeast two-hybrid assay was performed. As a TIR-NB-LRR-LIM-containing protein, the full-length CHS3 protein is approximately 185 kD and is difficult to express in yeast. Therefore, truncated constructs individually carrying the TIR, NB, LRR, unknown and LIM domains of CHS3 were generated and transformed into yeast (Fig 3A). IBR5 directly interacted with the TIR domain of CHS3 in yeast (Fig 3B).
Fig. 3. Interaction of IBR5 with CHS3. 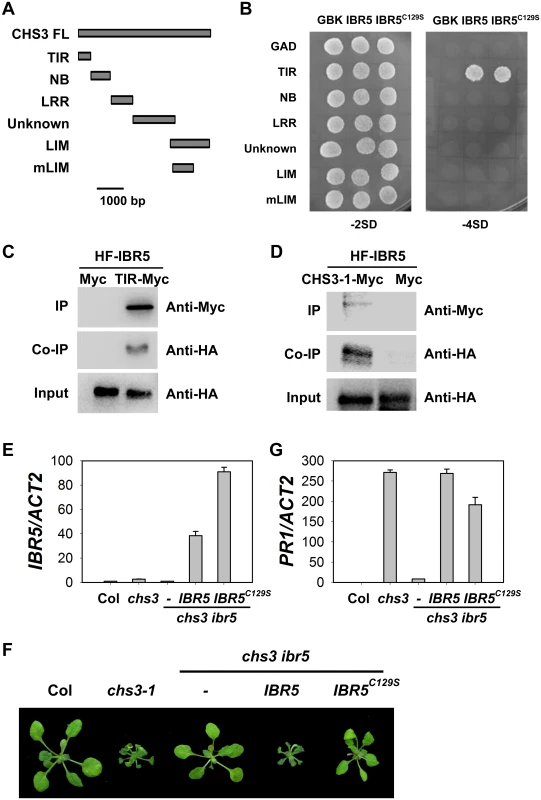
(A) Predicted protein structure of CHS3. TIR, Toll/Interleukin–1 receptor domain (aa 1–138); NB, nucleotide binding domain (aa 139–468); LRR, leucine-rich repeat domain (aa 469–729); Unknown, domain with no known function (aa 730–1240); LIM, LIM (Lin-11, Isl-1 and Mec-3 domains)-containing domain (aa 1135–1614); mLIM, LIM-containing domain with the same mutation in chs3-1, which contains a stop codon at 1422. (B) Interaction between IBR5 and CHS3 in yeast. The different CHS3 domains shown in (A) were fused with the pGADT7 vector. IBR5 and mutated IBR5C129S were fused with the pGBKT7 vector. The constructs were transformed into AH109 yeast cells and spotted onto SD media lacking Trp and Leu (-2SD) or lacking Trp, Leu, His and Ade (-4SD). The experiments were performed four times with similar results. (C) Co-immunoprecipitation of IBR5 and the TIR domain of CHS3. Total proteins were extracted from protoplasts expressing 35S:HA-Flag-IBR5 (35S:HF-IBR5) with Super: TIR domain of CHS3 (Super:TIR-Myc) or Super:Myc, and immunoprecipated with anti-Myc antibody. The proteins were obtained from crude lysates (Input) and the immunoprecipated proteins were detected using an anti-HA antibody. (D) Interaction of IBR5 and full-length CHS3 in vivo. Total proteins were extracted from N. benthamiana leaves transfected with 35S:HF-IBR5 and Super:CHS3-1-Myc, and were immunoprecipated with anti-Myc antibody. The proteins from crude lysates (Input) and the immunoprecipated proteins were detected using an anti-HA antibody. (E) Expression of IBR5 in 3-week-old Super:IBR5-Myc and Super:IBR5C129S-Myc transgenic plants in chs3 ibr5 grown at 16°C. (F) The phenotypes of Super:IBR5-Myc and Super:IBR5C129S-Myc transgenic plants in chs3 ibr5. The photographs show 3-week-old plants grown in soil at 16°C. (G) The expression of PR1 in 3-week-old Super:IBR5-Myc and Super:IBR5C129S-Myc transgenic chs3 ibr5 plants grown at 16°C. Previous studies have shown that CHS3 localizes to the nucleus [24], consistent with the results of the present study (S2B Fig). We also observed that IBR5 localized to both the cytosol and the nucleus (S2A and S2B Fig). To determine whether CHS3 interacts with IBR5 in vivo, 35S:HA-Flag-IBR5 (HF-IBR5) and the Myc-tagged TIR domain of CHS3 driven by the Super promoter (TIR-Myc) were transiently expressed in Arabidopsis mesophyll protoplasts, and subsequently, a co-immunoprecipitation (co-IP) assay was performed. IBR5 precipitated the TIR domain of CHS3 (Fig 3C). Moreover, HF-IBR5 interacted with full-length CHS3-1-Myc (the mutated form of CHS3 containing the same mutation as the chs3-1 mutant) when expressed in N. benthamiana (Fig 3D). These results indicated that CHS3 interacts with IBR5 through the TIR domain of CHS3 in vivo.
IBR5 belongs to a family of dual specificity protein phosphatases (DSPs), and the conserved cysteine in the conserved motif (VxVHCx2GxSRSx5AYLM) of DSPs is necessary for catalytic activity in many DSP proteins, including IBR5 and its homolog DsPTP1 [30]. To examine whether the phosphatase activity of IBR5 is required for interactions with CHS3, we generated the mutated form of IBR5 (IBR5C129S). The yeast two-hybrid assay showed that the mutation did not affect the interaction of IBR5 and CHS3 (Fig 3B), suggesting that the catalytic activity is not necessary for the interaction between IBR5 and CHS3. We next introduced Super:IBR5-Myc and Super:IBR5C129S-Myc into the chs3 ibr5 mutant background (Fig 3E, 3F and 3G). As a control, IBR5-Myc fully recovered the chs3 ibr5 phenotype and PR1 expression. However, the morphological phenotypes and PR1 expression of chs3 ibr5 were partially rescued in the mutated IBR5C129S-Myc plants (Fig 3F and 3G). These results suggest that the catalytic activity of IBR5 is required for full function in the CHS3-mediated defense response.
IBR5 promotes CHS3 accumulation at both transcription and protein levels
Another gain-of-function chs3 mutant, chs3-2D, was dwarfed and exhibited a constitutively active defense phenotype at 22°C [24]. Moreover, chs3:chs3-2D-GFP transgenic plants were generated, exhibiting chs3-2D-like phenotypes [24]. To examine whether IBR5 affects the accumulation of CHS3 in chs3-2D-GFP plants, ibr5-3 chs3-2D-GFP plants were generated by crossing ibr5-3 with chs3-2D-GFP plants (Fig 4A). The dwarf phenotype of chs3-2D-GFP plants was fully restored by ibr5-3 (Fig 4B), and GFP signals were observed in the nuclei of chs3-2D-GFP transgenic plants (Fig 4C). However, no obvious GFP signal was detectable in ibr5-3 chs3-2D-GFP plants (Fig 4C). Consistently, the expression of PR genes in chs3-2D-GFP was also suppressed in ibr5-3 plants (Fig 4D). The expression of CHS3 in ibr5-3 chs3-2D-GFP plants was also examined. As shown in Fig 4E, CHS3 expression in ibr5-3 chs3-2D-GFP plants was dramatically decreased compared with chs3-2D-GFP plants, consistent with the result obtained in chs3 ibr5-7 (Fig 1F). Moreover, the expression of CHS3 was down-regulated in the ibr5-3 mutant (Fig 4F). These results suggest that IBR5 positively regulates the expression of CHS3, which might at least be partially responsible for the decreased CHS3-2D-GFP signals in ibr5-3 chs3-2D-GFP.
Fig. 4. CHS3 is promoted by IBR5 at both mRNA and protein levels. 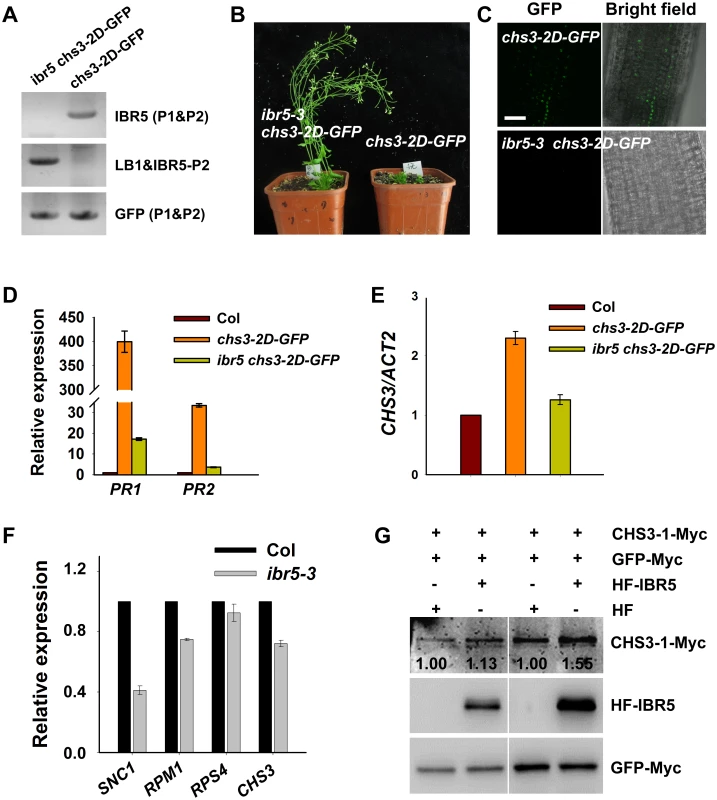
Genotyping of ibr5-3 chs3-2D-GFP plants. The primers used for genotyping the chs3-2D-GFP gene were GFP-P1 and GFP-P2, and the primers used for genotyping ibr5-3 were IBR5-P1, IBR5-P2 and LB1 as listed in S1 Table. The phenotype of chs3-2D-GFP transgenic plants was rescued by ibr5-3. chs3-2D-GFP and ibr5-3 chs3-2D-GFP plants were grown for 5 weeks, and representative plants are shown. Expression of CHS3-2D-GFP protein in 8-d-old chs3-2D-GFP and ibr5-3 chs3-2D-GFP plants. Bar: 100 μm. (D) Expression of CHS3-2D-GFP protein in 8-day-old chs3-2D-GFP and ibr5-3 chs3-2D-GFP plants. Bar: 100 μm. (D, E) Expression of the PR1, PR2 (D) and CHS3 (E) genes in chs3-2D-GFP and ibr5-3 chs3-2D-GFP plants. The values were normalized to the expression of ACTIN2. The error bars represent the SD of three replicates. (F) The expression of several R genes (SNC1, RPM1, RPS4 and CHS3) in 10-day-old ibr5-3 mutant grown at 22°C. The values were normalized to the expression of ACTIN2. The error bars represent the SD of three replicates. Similar results were obtained in three independent experiments. (G) The effect of IBR5 on the CHS3 protein level in Arabidopsis protoplasts. Super:CHS3-1-Myc and 35S:HF-IBR5 or 35S:HF constructs were co-expressed in Arabidopsis protoplasts. IBR5 and CHS3 proteins were detected using anti-HA and anti-Myc antibodies. The GFP-Myc construct was used as a control for transformation efficiency. The numbers show the ratios of CHS3-1-Myc to GFP-Myc protein levels. Two biological repeats are shown with similar results. To dissect the influence of IBR5 on CHS3 at the protein level, we examined the effect of IBR5 on the CHS3 protein level in Arabidopsis protoplasts. The protein level of CHS3-1-Myc in protoplasts co-expressing CHS3-1-Myc and HF-IBR5 was much higher than that in protoplasts expressing CHS3-1-Myc and HF (Fig 4G). This result suggests that IBR5 also promotes the accumulation of CHS3 protein in plant.
HSP90 and IBR5 synergistically regulate chs3-conferred phenotypes
HSP90 plays an important role in the stability of R proteins [31]. A previous study showed that the hsp90.3–1 mutant was a suppressor of the chs2/rpp4-1d mutant [32]. Therefore, we examined whether hsp90.3–1 rescues the chilling-sensitive phenotype of chs3-1. The chs3-1 hsp90.3–1 double mutant was generated and largely showed wild-type morphology but was smaller at 16°C (Fig 5A), and cell death was dramatically suppressed (Fig 5B). PR1 expression was also significantly inhibited in the double mutant (Fig 5C). Further analysis showed that the F1 progeny of ibr5 chs3 and hsp90.3 chs3 could partially rescue the morphology, cell death and PR1 gene expression of chs3 (Fig 5A, 5B and 5C). Moreover, the chs3 hsp90.3 ibr5 triple mutant more closely resembled wild-type plants than the chs3 hsp90 and chs3 ibr5 mutants in terms of growth, cell death and PR1 gene expression (Fig 5A, 5B and 5C). These data indicate that HSP90 and IBR5 synergistically modulate the chs3-conferred growth and defense responses.
Fig. 5. Genetic analysis of CHS3, IBR5 and HSP90.3. 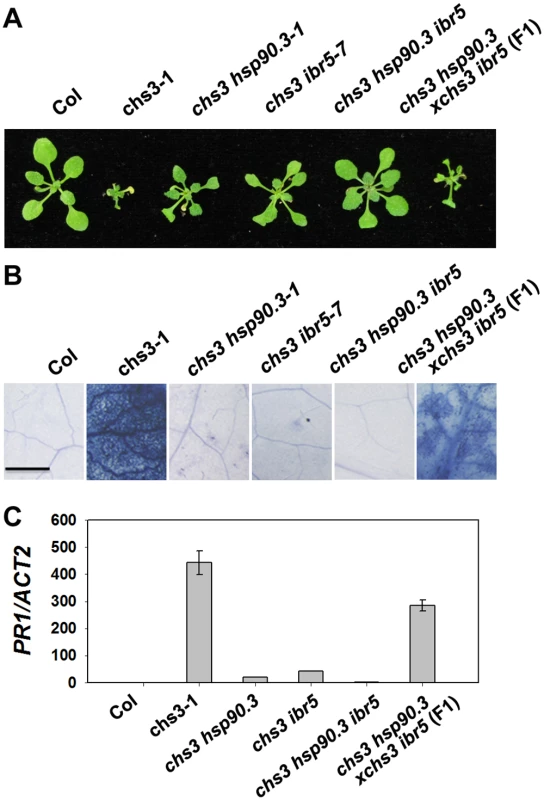
(A) Morphology of 4-week-old wild-type Col, chs3-1, chs3 hsp90.3–1, chs3 ibr5-7 chs3 hsp90.3 ibr5 and F1 progeny of chs3 hsp90.3–1 crossed with chs3 ibr5-7 grown in soil at 16°C. (B) Trypan blue staining of the leaves from the plants described in (A). Bar: 50 μm. (C) PR1 gene expression in the plants described in (A). The values were normalized to the expression of ACTIN2. The error bars represent the SD of three replicates. The experiments were performed three times with similar results. IBR5 interacts with HSP90 and SGT1b
HSP90 functions as a complex with SGT1b and RAR1 [11,33–35] and we previously showed that sgt1b and rar1 suppressed the phenotypes of chs3-1 [23], similar to ibr5 (Fig 1). Next we determined whether IBR5 physically interacts with HSP90 and SGT1b in plants. To this end, a luciferase complementation imaging (LCI) assay was performed in Nicotiana benthamiana. IBR5 interacted with HSP90 and SGT1b in N. benthamiana leaves (Fig 6A and 6B). Furthermore, co-IP assays showed that endogenous HSP90 successfully immunoprecipitated IBR5-Myc in transgenic plants expressing IBR5-Myc, but not in Myc transgenic plants (Fig 6C). Similarly, SGT1b could be co-immunoprecipitated with IBR5 in transgenic plants expressing IBR5-Myc and SGT1b-FLAG, but not in transgenic plants expressing Myc and SGT1b-FLAG (Fig 6D). We also examined the association between SGT1b and CHS3. Co-IP assays showed that SGT1b associated with the TIR domain of CHS3 and full-length CHS3-1 in vivo (S3 Fig). Furthermore, we examined the interaction of CHS3-1 with IBR5 and SGT1b in N. benthamiana leaves. As shown in Fig 6E, CHS3-1 could simultaneously pull down IBR5 and SGT1b in plants. These results indicate that IBR5 forms complex(es) with CHS3, HSP90 and SGT1b in vivo.
Fig. 6. Interaction of IBR5 with HSP90 and SGT1b. 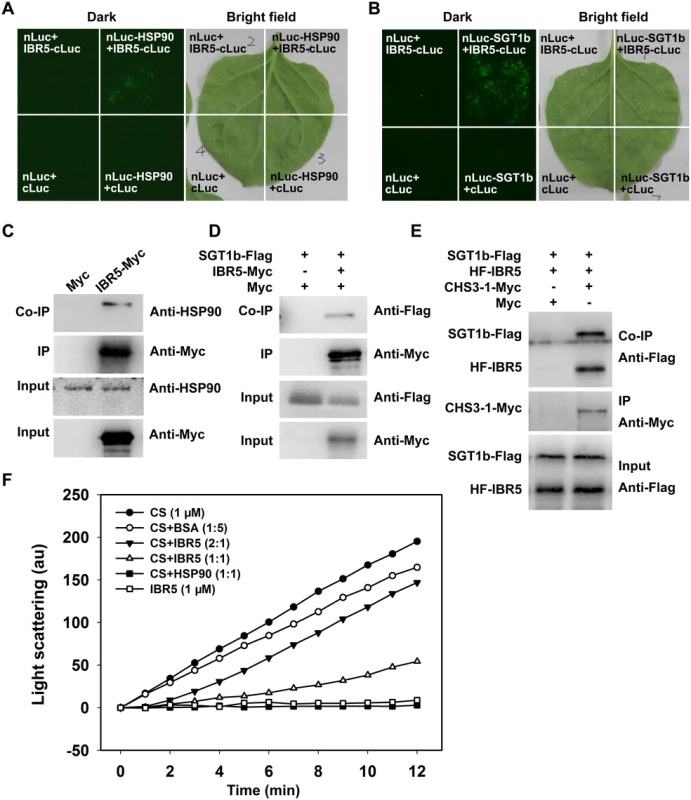
(A) Interaction of IBR5 with HSP90 as revealed via firefly luciferase complementation imaging assay in N. benthamiana leaves. nLuc+cLuc-IBR5, HSP90-nLuc+cLuc, and nLuc+cLuc were used as negative controls. (B) Interaction of IBR5 with SGT1b as revealed by a firefly luciferase complementation imaging assay in N. benthamiana leaves. nLuc+cLuc-IBR5, SGT1b-nLuc+cLuc, and nLuc+cLuc were used as negative controls. (C) Co-immunoprecipitation of IBR5 and HSP90. Total protein was extracted from 2-week-old transgenic seedlings expressing Super:IBR5-Myc or Super:Myc. Anti-Myc antibody was used for immunoprecipitation, and the immunoprecipitated proteins were analyzed by immunoblotting using an anti-HSP90 antibody. (D) Co-immunoprecipitation of IBR5 and SGT1b. Total protein was extracted from transgenic plants expressing Super:Myc/Super:SGT1b-Flag or Super:IBR5-Myc/Super:SGT1b-Flag. An anti-Myc antibody was used for immunoprecipitation, and the immunoprecipitated proteins were analyzed by immunoblotting using an anti-Flag antibody. (E) Interaction of CHS3 with IBR5 and SGT1b in plant. Total proteins were extracted from N. benthamiana leaves transfected with 35S:HF-IBR5, Super:SGT1b-Flag and Super:CHS3-1-Myc, and were immunoprecipated with anti-Myc antibody. The proteins from crude lysates (Input) and the immunoprecipated proteins were detected using an anti-Flag antibody. (F) IBR5 can stabilize CS under thermally denaturing conditions. Citrate synthase (1 μM) was incubated alone or with 5 μM BSA (1:5), 0.5 μM IBR5 (2:1), 1 μM IBR5 (1:1) or 1 μM HSP90 (1:1) at 43°C. The kinetics was determined by light scattering using a fluorescence spectrophotometer. IBR5 exhibits holdase activity in vitro
We subsequently investigated whether IBR5 protects proteins in vitro using a thermal aggregation assay with citrate synthase (CS) as a model substrate [36]. CS aggregation was examined after measuring the absorbance at 500 nm under thermally denaturing conditions (40°C or above). Heat-induced aggregation of CS was inhibited by HSP90 (Fig 6F), consistent with the results of previous study [37]. IBR5 showed a weaker but more significant effect than HSP90 on the inhibition of CS aggregation. When IBR5 was incubated with CS at 43°C, the aggregation of CS was partially suppressed in a dose-dependent manner (Fig 6F). This result indicates that IBR5 protects proteins against aggregation in vitro.
Genetic and physical interactions between IBR5 and SNC1
The TIR domain of CHS3 shares amino acid similarity with TIR-NB-LRR R proteins SNC1 and RPP4 (S4 Fig). Gain-of-function mutants of SNC1 (snc1-1 and bal/snc1-2) activate the defense response in a temperature-dependent manner [6,38]. The loss of BON1 function in Arabidopsis leads to temperature-sensitive growth and autoimmune phenotypes resulting from the activation of SNC1 [6]. Phenotype analyses showed that the bon1-1 ibr5-3 (bon1 ibr5) and bal ibr5-3 (bal ibr5) double mutants were slightly larger than the bon1-1 and bal single mutants (Fig 7A and 7B). Moreover, the cell death in bon1 ibr5 and bal ibr5 was reduced (Fig 7C). Consistently, the expression of PR1 and PR2 genes in bon1 ibr5 and bal ibr5 was also lower than that in the bon1-1 and bal/snc1-2 mutants (Fig 7D). We also examined the basal defense response of bon1 ibr5 and bal ibr5 to P.s.t. DC3000. The pathogen resistance of bon1-1 and bal/snc1-2 mutants to P.s.t. DC3000 was slightly suppressed in the ibr5-3 mutant (Fig 7E and 7F). These genetic data suggest that IBR5 is partially implicated in the SNC1-mediated defense response.
Fig. 7. The defense phenotypes of bon1-1 ibr5-3 and bal ibr5-3. 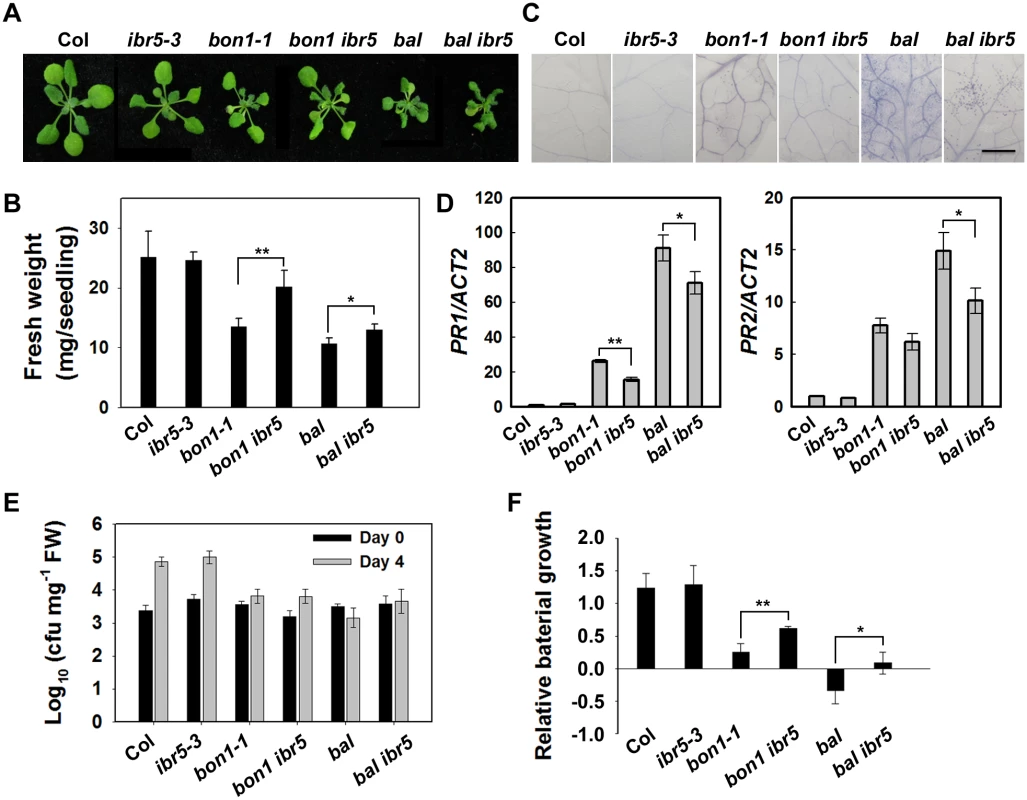
Morphology of 3-week-old Col, ibr5-3, bon1-1, bon1-1 ibr5-3, bal, bal ibr5-3 grown at 22°C. (B) Fresh weight of plants in (A). The error bars represent the SD of three replicates, and the asterisks indicate significant differences (**P < 0.01, *P < 0.05, t-test). (C) Trypan blue staining of leaves from the plants described in (A). Bar: 0.5 mm. The expression of PR genes in plants in (A). The error bars represent the SD of three replicates, and the asterisks indicate significant differences (**P < 0.01, *P < 0.05, t-test). (E) The effect of IBR5 on pathogen resistance of bon1-1 and bal to P.s.t. DC3000. Three-week-old seedlings were dipped with P.s.t. DC3000 (OD600 = 0.05), and leaves were taken immediately (day 0) and at day 4 after infection. The log-transformed values presented are the mean values of three replicates ± SD. The experiments were performed three times with similar results. (F) Relative bacterial growth of plants in (E) after infection with P.s.t. DC3000. The values are the ratios of bacteria number at day 4 to day 0. The error bars represent the SD of three replicates, and the asterisks indicate significant differences (**P < 0.01, *P < 0.05, t-test). To further dissect the function of IBR5 on SNC1, we examined the expression of SNC1 in the ibr5-3 mutant. As shown in Fig 4F, the SNC1 transcript levels in the ibr5-3 mutant were much lower than those in Col, indicating that SNC1 expression is positively regulated by IBR5. We next explored whether IBR5 physically interacts with SNC1. A yeast two-hybrid assay showed that IBR5 interacted with the TIR domain of SNC1 (Fig 8A). The interaction of IBR5 and full-length SNC1-1 (the mutated form of SNC1 containing the same mutation as the snc1-1 mutant) was confirmed by a co-IP assay in N. benthamiana leaves (Fig 8B). Moreover, the protein level of SNC1-1-Myc increased when co-expressed with HF-IBR5 in Arabidopsis protoplasts (Fig 8C). Taken together, these data suggest that IBR5 might also promote SNC1 protein accumulation.
Fig. 8. Interaction of IBR5 with SNC1 and RPP4. 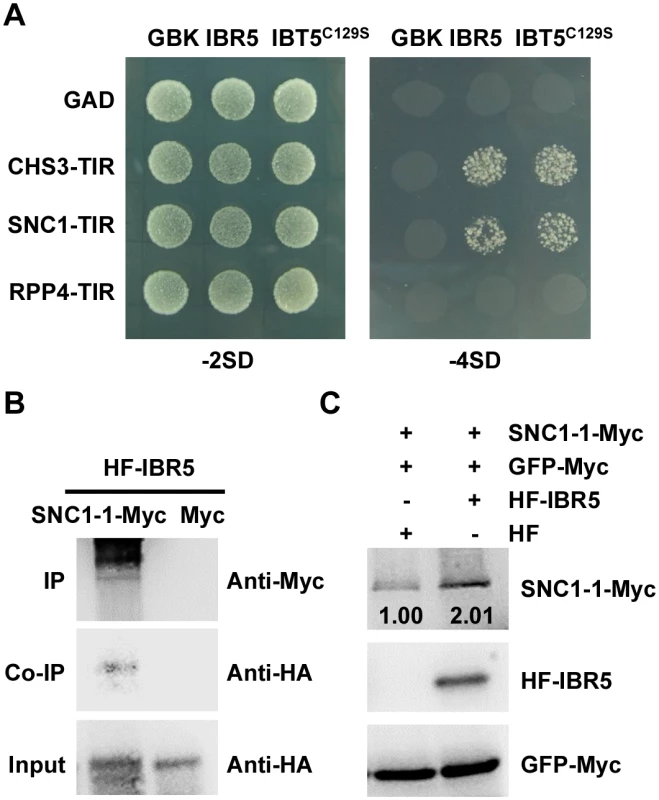
(A) Interaction of IBR5 with TIR domains of CHS3, SNC1 and RPP4 in yeast. The experiments were performed three times with similar results. (B) Interaction of IBR5 and full-length SNC1 in vivo. Total proteins were extracted from N. benthamiana leaves transfected with 35S: HF-IBR5 and Super:SNC1-1-Myc and were immunoprecipated with an anti-Myc antibody. The proteins from crude lysates (Input) and the immunoprecipated proteins were detected using an anti-HA antibody. (C) The effect of IBR5 on SNC1 protein level in Arabidopsis protoplasts. Super:SNC1-1-Myc and 35S:HF-IBR5 or 35S:HF constructs were co-expressed in Arabidopsis protoplasts. IBR5 was detected with anti-HA antibody and SNC1-1 was detected using an anti-Myc antibody. GFP-Myc construct was used as a control for transformation efficiency. Our previous studies showed that the mutations in R protein CHS2/RPP4 or R-like protein CHS1 induce plant sensitivity and activate defense responses under chilling stress [7,8]. To investigate whether ibr5 also suppresses the chilling sensitivity of chs1-2 and chs2-1/rpp4-1d, we generated chs1-2 ibr5-3 (chs1 ibr5) and chs2-1 ibr5-4 (chs2 ibr5) double mutants. In terms of morphology and cell death, chs1 ibr5 and chs2 ibr5 mutants showed chs1-2 and chs2-1 single mutant phenotypes under chilling stress (S5A and S5B Fig). However, PR1 expression in chs1 ibr5 was partially suppressed, whereas the expression of this protein in chs2 ibr5 was not obviously affected. A yeast two-hybrid assay showed that IBR5 did not interact with RPP4 (Fig 8A). We also examined the pathogen resistance of ibr5 mutants to RPP4-specific Hyaloperonospora arabidopsidis (H.a.) Emwa1. As controls, Col was completely resistant to H.a. Emwa1, whereas rpp4-r26, which contains a mutation in RPP4 resulting in the introduction of a stop codon in front of the LRR domain [32], was susceptible to H.a. Emwa1 (S6 Fig). The ibr5-3 and ibr5-7 mutants showed complete resistance to H.a. Emwa1 (S6 Fig), suggesting that IBR5 might not be involved in RPP4-mediated oomycete resistance.
IBR5 is involved in RPM1 - and RPS4-mediated disease resistance
To examine whether basal defense was affected in the ibr5 single mutant, we infected wild-type Col and ibr5 mutants with virulent P.s.t. DC3000. The bacterial growth in ibr5-3 and ibr5-7 was comparable to that in wild-type plants, whereas as the control, pad4-1, supported 100 times more bacterial growth than the wild-type plants (Fig 9A), suggesting that IBR5 does not play an important role in basal defense.
Fig. 9. Pathogen resistance analysis of ibr5 mutants. 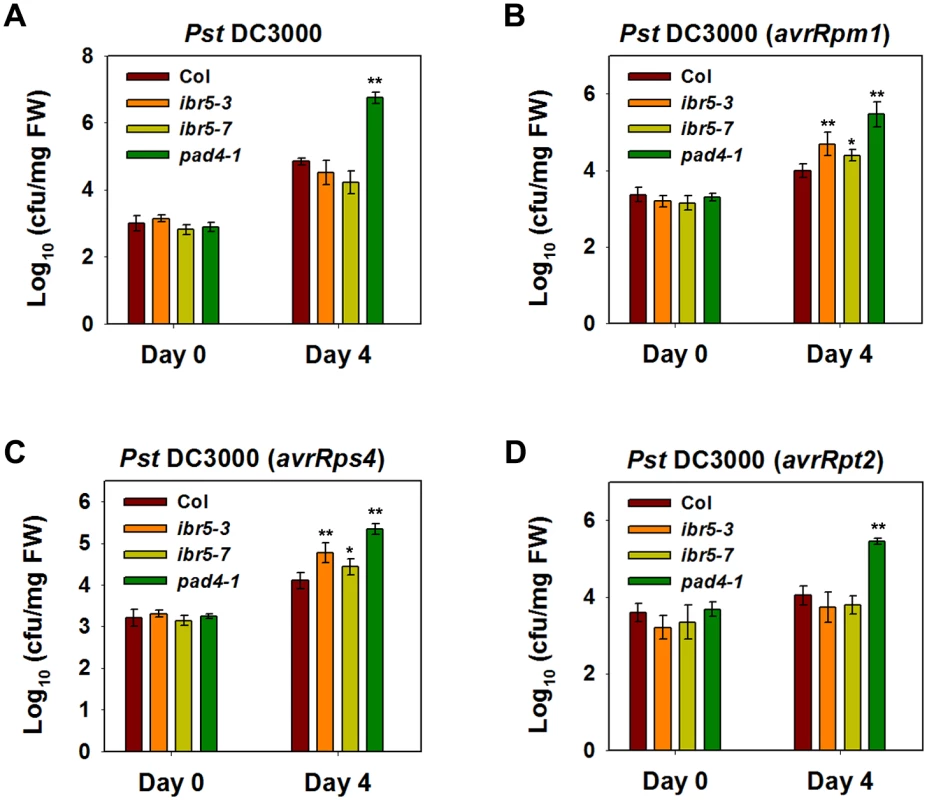
(A) Response of ibr5 to P.s.t. DC3000. Three-week-old plants were dipped with P.s.t. DC 3000 (OD600 = 0.05), and leaves were taken immediately (day 0) and at day 4 after infection. The log-transformed values presented are the mean values of three replicates ± SD. (B-D) Responses of ibr5 to avirulent bacteria. Three-week-old plants were dipped with P.s.t. DC3000 (avrRpt2) (B), P.s.t. DC3000 (avrRpm1) (C) and P.s.t. DC3000 (avrRps4) (D) (OD600 = 0.2). In (A-D), the log-transformed values presented are the mean values of three replicates ± SD, and the asterisks indicate significant differences compared with wild-type Col (**P < 0.01, *P < 0.05, t-test). All experiments were repeated at least three times with similar results. As the role of CHS3 is compromised in the ibr5 mutant, we further investigated whether IBR5 is required for other R-gene-mediated disease resistance. The ibr5 mutants were inoculated with P.s.t. DC3000 carrying either avrRpm1, avrRps4, or avrRpt2. The ibr5 mutants showed enhanced susceptibility to P.s.t. DC3000 (avrRpm1) and P.s.t. DC3000 (avrRps4) (Fig 9B and 9C). In contrast, no remarkable difference in the growth of P.s.t. DC3000 (avrRpt2) was detected between the ibr5 mutants and wild-type Col (Fig 8D). As a control, pad4-1 showed enhanced disease susceptibility to all three avirulent bacterial strains (Fig 9B, 9C and 9D). These results suggest that IBR5 might also contribute to disease resistance mediated by RPM1 and RPS4.
We next examined the expression of RPM1 and RPS4 in the ibr5-3 mutant. The transcription levels of RPM1 and RPS4 were slightly down-regulated in the ibr5-3 mutant compared with wild type plants (Fig 4F). Furthermore, we examined whether IBR5 interacts with TIR-NB-LRR protein RPS4 or CC-NB-LRR protein RPM1. The results of a yeast two-hybrid assay showed that neither wild-type IBR5 nor mutated IBR5C129S interacted with the TIR domain of RPS4 or CC domain of RPM1 (S7A Fig). However, interestingly, we observed the direct interaction of IBR5 and IBR5C129S with RPS4-interacting protein RRS1 [39] in yeast (S7B Fig). The results of additional co-IP assays showed that IBR5 could pull down RPS4 (S7C Fig), suggesting that IBR5 might form a complex with RPS4 and RRS1. In contrast, no interaction between IBR5 and RPM1 was observed in plants (S7D Fig). Furthermore, no obvious change in the RPM1 protein level was detected when IBR5 and RPM1 were co-expressed (S7E Fig).
Discussion
CHS3 is involved in temperature-dependent defense responses [23]. In the present study, we identified ibr5-7 as a suppressor of chs3-1. IBR5 interacts with CHS3 and HSP90/SGT1b chaperones to stabilize the CHS3 protein, thereby modulating temperature-dependent defense responses. In addition, IBR5 is invovled in defense responses mediated by several other R genes, including SNC1, RPS4 and RPM1.
IBR5 is a MAP kinase phosphatase (MKP) that dephosphorylates activated MAPKs [40,41]. Emerging evidence has shown that MKPs play important roles in modulating plant defense responses [41]. For example, the mkp1 null mutation in the Col accession exhibits constitutive stress response phenotypes, causing a dwarf stature dependent on SNC1 [42]. However, it remains unclear whether the substrates of MKP1, MPK3 and MPK6 [42,43] are involved in the regulation of SNC1. Additional studies have revealed that MKP1 functions as a negative regulator of MPK6-mediated pathogen-associated molecular pattern (PAMP) responses and disease resistance [44]. The mkp2 null mutant displays delayed symptoms of disease induced by Ralstonia solanacearum, and shows increased susceptibility to Botrytis cinerea [45]. MKP2 might regulate MPK3/MPK6 activity during different pathogen defense responses. Moreover, MKP2 reverses hypersensitive-like responses triggered by MPK6 upon plant treatment with fungal elicitors [45,46]. It is likely that these MKPs have negative functions in defense signaling pathways through their MPK substrates. The data obtained in the present study showed that the loss of IBR5 function suppresses chs3-conferred, temperature-dependent defense responses and slightly weakens the autoimmunity of bal/snc1-2, a gain-of-function mutant of a TIR-NB-LRR protein SNC1. Moreover, resistance mediated by R proteins, such as RPS4 and RPM1, is compromised in the ibr5 mutants. These results suggest that IBR5 plays a positive role in the ETI pathway mediated by multiple R proteins. Therefore, different MKPs play either positive or negative roles in plant immunity. As IBR5 is a MAP kinase phosphatase, we also examined whether the dephosphorylation activity of IBR5 is required for the regulation of CHS3. The inactive form of IBR5 (IBR5C129S) partially rescues the phenotype of chs3 ibr5, suggesting that the dephosphorylation activity of IBR5 is necessary for the complete function of this protein in the CHS3-mediated defense response.
MPK12 is a substrate of IBR5 [29], and the reduced expression of MPK12 partially complements auxin, but not ABA insensitivity, in the ibr5 mutant, suggesting that there might be other substrate(s) of IBR5 involved in IBR5-mediated ABA responses [29]. In the present study, the loss-of-function mutant mpk12-3 displayed an auxin-sensitive phenotype (S8A, S8B and S8C Fig), similar to that of mpk12 RNAi lines [29]. However, mpk12-3 cannot rescue the phenotype of chs3 ibr5 (S8D Fig), indicating that the CHS3-mediated defense response does not require MPK12. Moreover, the mutant of auxin receptor tir1 enhances the auxin insensitive phenotype of ibr5, implying that IBR5 is involved in the auxin response independently of the TIR1-mediated auxin signaling pathway [30]. Therefore, the role of IBR5 in R-mediated resistance might not occur through the function of IBR5 in the auxin response. ibr5 mutant is shown to be insensitive to ABA, but the underlying mechanism is completely unknown [28]. ABA signaling plays a role in defense responses in a complicated manner. Recent studies have reported that the overexpression of ABA receptors maintains stomatal closure during P.s.t. DC3000 infections, leading to enhanced resistance after dipping, but enhanced susceptibility to the pathogen after infiltration [47]. In contrast, several ABA signaling mutants, such as abi1 and abi4, do not exhibit prominent phenotypes under the snc1-1 background or during pathogen infection [48]. The ABA-insensitive mutant cpr22, containing a deletion on two cyclic nucleotide-gated ion channel genes, shows constitutive PR gene expression, enhanced pathogen resistance and SA accumulation [49]. WRKY40, WRKY18, and WRKY60 play negative roles in ABA signaling [50]. The single and triple mutants show diverse phenotypes to different pathogens, as these mutants are more resistant to P. syringae but more susceptible to B. cinerea compared with the wild type [50]. Whether the role of IBR5 in the ABA response is involved in CHS3-mediated defense pathway remains unknown.
In plants, the TIR domain is indispensable for defense signal transduction [51]. For example, the transient expression of the TIR domain of RPS4 in N. benthamiana leaves triggers cell death in a manner dependent on both SGT1 and HSP90 [13,51]. The TIR domain of L6, a TIR-NB-LRR R protein in flax, forms a homodimer required for immune signaling [52]. A recent study showed that TIR domain heterodimerization is necessary to form a functional RRS1/RPS4 effector recognition complex [9], indicating the importance of the TIR domain of R proteins in signal recognition and transduction. Moreover, the TIR domain of R proteins interacts with other proteins [39,53–56]. In the present study, we observed that the TIR domain of CHS3 interacts with IBR5 in vitro and in vivo. The mutation of IBR5C129S does not affect this interaction, suggesting that the dephosphatase activity of IBR5 is not a prerequisite for the interaction between IBR5 and CHS3. We also examined the interaction between IBR5 and four other TIR-NB-LRR R proteins: SNC1, RPP4, RPS4 and its interacting R protein RRS1 [39], and CC-NB-LRR, RPM1. Under these conditions, IBR5 interacts with SNC1 and RRS1/RPS4 in plant, but not with RPP4 and RPM1. Consistently, ibr5 partially suppresses the cell death and PR gene expression in bal/snc1-2 but does not suppress the expression of these proteins in chs2/rpp4-1d. These data suggest that IBR5 might specifically interact with certain TIR-NB-LRR proteins to regulate defense signaling. However, the mechanism underlying the involvement of IBR5 in defense responses mediated by the CC-type R protein remain unclear.
Increasing evidence has shown that HSP90, SGT1 and RAR1 form a protein complex with chaperone activity required for the proper folding and stabilization of R proteins [11,23,33,34,57–60]. In this complex, HSP90 and RAR1 increase R protein levels [57]. The phosphatase PP5 interacts with HSP90 and the tomato R protein I–2, and together with HSP90, PP5 acts as a co-chaperone [61]. In the present study, we found an interaction between the phosphatase IBR5 and the HSP90/SGT1B/RAR1 complex in vivo, suggesting that IBR5 complexes with these chaperone proteins and stabilizes CHS3. Increasing evidence further supports the notion: (1) The in vitro holdase activity of IBR5 indicates that this enzyme markedly inhibits temperature-dependent CS degradation. (2) The F1 progeny of chs3 ibr5 and chs3 hsp90 partially inhibits the chs3 phenotype. Moreover, the chs3 ib5 hsp90 triple mutant is larger than both chs3 hsp90 and chs3 ibr5 and is reminiscent of the wild type. We speculate that different complexes comprising IBR5 and HSP90 might play roles in CHS3-mediated signaling. This involvement might reflect the dosage effect of IBR5 and HSP90. A similar mechanism was observed for the COI1 (CORONATINE INSENSITIVE 1) and HSP90 proteins in regulating RPM1-mediated disease resistance [62]. (3) chs3-dependent phenotypes are suppressed by sgt1b, rar1 [23] and hsp90.3–1, indicating that HSP90, RAR1 and SGT1b are required for CHS3 activation. (4) IBR5 interacts with the HSP90, SGT1b and CHS3 proteins in vivo. Collectively, these data show that IBR5 associates with HSP90/RAR1/SGT1b to stabilize the CHS3 protein. Because SGT1b interacts with a core component of the SKP1-CULLIN1-F-box (SCF) E3 ligase complex, S-phase kinase-associated protein 1 (SKP1), SGT1b has been considered as a subunit of the SCF complex [63,64]. Recent studies have shown that SNC1 is degraded by the SCFCPR1 complex [65,66]. In a protein-protein interactome study, IBR5 was also shown to interact with SKP1 [67]. These results suggest that IBR5, in concert with HSP90/SGT1B proteins, might affect the function of SCF E3 ubiquitin ligase complexes, thereby regulating the ubiquitination and degradation of some R proteins. Further studies will elucidate the molecular mechanisms underlying the roles of IBR5 in R-mediated defense responses.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana Col–0, chs3-1 [23], ibr5-3 [29], pad4-1 [68], hsp90.3–1, rpp4-r26 [32], and mpk12-3 (SAIL_543_F07) were used in this study. Plants were grown in soil or on Murashige and Skoog (MS) medium containing 2% (w/v) Suc and 0.8% (w/v) agar at 16°C or 22°C with a 16-h light/8-h dark cycle under white light. Salicylic acid measurement and were described previously [23].
Suppressor screening and map-based cloning of SUC5
The suppressors of chs3-1 were screened and mapped as described previously [23]. Approximately 200 plants with chs3-1 background with wild-type morphology at 16°C were used for mapping to a 500-kb region between markers F3C11 and F5G3. Full length genomic DNA of candidate genes was amplified by PCR and sequenced to find the mutant site. To confirm the mutated gene, genomic complementation and T-DNA insertion mutant were used.
Trypan blue staining and DAB staining
Trypan blue and DAB staining was performed as previously described [69,70].
Pathogen resistance assay
The pathogen resistance assay on Pseudomonas syringae pv tomato (P.s.t.) strain DC3000 was performed as described previously [32]. Briefly, 3-week-old plants were dipped by the bacterial suspension containing 10 mM MgCl2 and 0.025% Silwet L–77. The concentrations of pathogen for virulent strain (P.s.t. DC3000) and avirulent strains (P.s.t. DC3000 with avrRmp1, avrRpt2 and avrRps4) are OD600 of 0.05 and 0.2, respectively. The dipped leaves were collected at 2 hour and 4 day post-inoculation. Three replicate samples were collected to measure the resistance to pathogen.
qRT-PCR analysis
Total RNA isolated from 2-week-old plants grown at 22°C or 3-week-old plants grown at 16°C using TRIzol reagent (Invitrogen). The quantitative real-time PCR was performed using a SYBR Green PCR Master Mix kit (Takara). The relative expression levels were calculated as described (Huang et al., 2010). Gene-specific primers were listed in S1 Table.
Plasmid construction and plant transformation
To obtain the IBR5::IBR5 construct, the genome fragment of IBR5 (from ~2kb upstream of ATG to stop codon) was amplified and cloned into pCAMBIA1300 vector. To generate Super:IBR5 and Super:SGT1b constructs, cDNAs of IBR5 and SGT1b were amplified and cloned into the pSuper1300-Myc/GFP/Flag vectors containing a Super promoter.
To generate Super:CHS3-1-Myc, Super:SNC1-1-Myc, Super:RPM1-Myc and Super:RPS4-Myc, genomic DNA of chs3-1, snc1-1, RPM1 and RPS4 were amplified and cloned into the pSuper1300-Myc vectors containing a Super promoter. The primers were listed in S1 Table.
Agrobacterium tumefaciens stain of GV3101 carrying different constructs was transformed into wild-type or mutant plants via standard floral dipping method [71].
Yeast two-hybrid assay
To create bait and prey plasmids, fragments were amplified by different primers and cloned into pGBKT7 and pGADT7. All primer sequences are listed in S1 Table. Bait and prey plasmids were co-transformed into yeast strain AH109. Yeast strains containing the bait and prey plasmids were cultured in SD-Trp-Leu liquid medium to OD600 = 0.4–0.6. Interaction was tested on SD-Trp-Leu-His-Ade medium.
LCI assay
cDNAs of HSP90 and SGT1b were amplified and cloned into nLuc vector. IBR5 cDNA was amplified and cloned into cLuc vector. A. tumefaciens strain GV3101 containing different constructs was infiltrated into N. benthamiana leaves according to the protocol described previously [72]. Firefly Luciferase Complementation Imaging (LCI) assay was performed as described [73].
Thermal aggregation assay
To clone the IBR5-His construct, IBR5 cDNA was amplified and cloned into pQE80L vector. HSP90 fragment was amplified and cloned into pMalC2 vector. The thermal aggregation assay was performed as described [74] with some modifications. Citrate synthase from porcine heart (Sigma-Aldrich) was used as substrate. Light scattering was measured by a thermostatically controlled fluorescence spectrophotometer (F–7000). Proteins were diluted by 40 mM HEPES/KOH (pH 7.5) and the reaction temperature was kept at 43°C.
Coimmunoprecipitation
The plasmids were purified and transformed into Arabidopsis mesophyll protoplasts [75] or N. benthamiana leaves [72]. Proteins were extracted with extracting buffer containing 150 mM NaCl, 10 mM Tris-HCl pH 7.6, 2 mM EDTA, 0.5% NP–40 and 1 x protease inhibitor cocktail (Roche). The protein extracts were then incubated with anti-Myc beads (Sigma-Aldrich) at 4°C for 2 hour. After washing five times with extraction buffer, the samples were used for immunoblotting with anti-HA (Sigma-Aldrich) or anti-Flag (Sigma-Aldrich) antibodies.
Supporting Information
Zdroje
1. Hua J (2013) Modulation of plant immunity by light, circadian rhythm, and temperature. Curr Opin Plant Biol 16 : 406–413. doi: 10.1016/j.pbi.2013.06.017 23856082
2. Alcazar R, Garcia AV, Parker JE, Reymond M (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci U S A 106 : 334–339. doi: 10.1073/pnas.0811734106 19106299
3. de Jong CF, Takken FL, Cai X, de Wit PJ, Joosten MH (2002) Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Mol Plant Microbe Interact 15 : 1040–1049. 12437302
4. Li Y, Pennington BO, Hua J (2009) Multiple R-like genes are negatively regulated by BON1 and BON3 in Aarabidopsis. Mol Plant Microbe Interact 22 : 840–848. doi: 10.1094/MPMI-22-7-0840 19522566
5. Xiao S, Charoenwattana P, Holcombe L, Turner JG (2003) The Arabidopsis genes RPW8.1 and RPW8.2 confer induced resistance to powdery mildew diseases in tobacco. Mol Plant Microbe Interact 16 : 289–294. 12744457
6. Yang S, Hua J (2004) A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16 : 1060–1071. 15031411
7. Huang X, Li J, Bao F, Zhang X, Yang S (2010) A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol 154 : 796–809. doi: 10.1104/pp.110.157610 20699401
8. Wang Y, Zhang Y, Wang Z, Zhang X, Yang S (2013) A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis. Plant J 75 : 553–565. doi: 10.1111/tpj.12232 23651299
9. Cheng C, Gao X, Feng B, Sheen J, Shan L, et al. (2013) Plant immune response to pathogens differs with changing temperatures. Nat Commun 4 : 2530. doi: 10.1038/ncomms3530 24067909
10. Kadota Y, Shirasu K (2012) The HSP90 complex of plants. Biochim Biophys Acta 1823 : 689–697. doi: 10.1016/j.bbamcr.2011.09.016 22001401
11. Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, et al. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22 : 5679–5689. 14592967
12. Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem 279 : 2101–2108. 14583611
13. Zhang Y, Dorey S, Swiderski M, Jones JD (2004) Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J 40 : 213–224. 15447648
14. Holt BF 3rd, Belkhadir Y, Dangl JL (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 : 929–932. 15976272
15. Li Y, Li S, Bi D, Cheng YT, Li X, et al. (2010) SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111. doi: 10.1371/journal.ppat.1001111 20862316
16. Huang S, Monaghan J, Zhong X, Lin L, Sun T, et al. (2014) HSP90s are required for NLR immune receptor accumulation in Arabidopsis. Plant J 79 : 427–439. doi: 10.1111/tpj.12573 24889324
17. Li Y, Tessaro MJ, Li X, Zhang Y (2010) Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol 153 : 1425–1434. doi: 10.1104/pp.110.156240 20439546
18. Zhang Y, Li X (2005) A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17 : 1306–1316. 15772285
19. Cheng YT, Germain H, Wiermer M, Bi D, Xu F, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21 : 2503–2516. doi: 10.1105/tpc.108.064519 19700630
20. Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, et al. (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A 107 : 13960–13965. doi: 10.1073/pnas.1002828107 20647385
21. Huang Y, Minaker S, Roth C, Huang S, Hieter P, et al. (2014) An E4 ligase facilitates polyubiquitination of plant immune receptor resistance proteins in Arabidopsis. Plant Cell 26 : 485–496. doi: 10.1105/tpc.113.119057 24449689
22. Cheng YT, Li Y, Huang S, Huang Y, Dong X, et al. (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci U S A 108 : 14694–14699. doi: 10.1073/pnas.1105685108 21873230
23. Yang H, Shi Y, Liu J, Guo L, Zhang X, et al. (2010) A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J 63 : 283–296. doi: 10.1111/j.1365-313X.2010.04241.x 20444230
24. Bi D, Johnson K, Huang Y, Zhu Z, Li X, et al. (2011) Mutations in an atypical TIR-NB-LRR-LIM resistance protein confers autoimmunity. Front Plant Sci: doi: 10.3389/fpls.2011.00071
25. Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, et al. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295 : 2077–2080. 11847308
26. Shirasu K, Lahaye T, Tan MW, Zhou F, Azevedo C, et al. (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99 : 355–366. 10571178
27. Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, et al. (2002) RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14 : 1005–1015. 12034893
28. Monroe-Augustus M, Zolman BK, Bartel B (2003) IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15 : 2979–2991. 14630970
29. Lee JS, Wang S, Sritubtim S, Chen JG, Ellis BE (2009) Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J 57 : 975–985. doi: 10.1111/j.1365-313X.2008.03741.x 19000167
30. Strader LC, Monroe-Augustus M, Bartel B (2008) The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol 8 : 41. doi: 10.1186/1471-2229-8-41 18423007
31. Pearl LH, Prodromou C (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75 : 271–294. 16756493
32. Bao F, Huang X, Zhu C, Zhang X, Li X, et al. (2014) Arabidopsis HSP90 protein modulates RPP4-mediated temperature-dependent cell death and defense responses. New Phytol 202 : 1320–1334. doi: 10.1111/nph.12760 24611624
33. Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem 279 : 2101–2108. 14583611
34. Takahashi A, Casais C, Ichimura K, Shirasu K (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci U S A 100 : 11777–11782. 14504384
35. Kadota Y, Amigues B, Ducassou L, Madaoui H, Ochsenbein F, et al. (2008) Structural and functional analysis of SGT1-HSP90 core complex required for innate immunity in plants. EMBO Rep 9 : 1209–1215. doi: 10.1038/embor.2008.185 18833289
36. Schumacher RJ, Hansen WJ, Freeman BC, Alnemri E, Litwack G, et al. (1996) Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 35 : 14889–14898. 8942653
37. Shaknovich R, Shue G, Kohtz DS (1992) Conformational activation of a basic helix-loop-helix protein (MyoD1) by the C-terminal region of murine HSP90 (HSP84). Mol Cell Biol 12 : 5059–5068. 1406681
38. Zhang Y, Goritschnig S, Dong X, Li X (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 : 2636–2646. 14576290
39. Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344 : 299–303. doi: 10.1126/science.1247357 24744375
40. Dickinson RJ, Keyse SM (2006) Diverse physiological functions for dual-specificity MAP kinase phosphatases. J Cell Sci 119 : 4607–4615. 17093265
41. Bartels S, Gonzalez Besteiro MA, Lang D, Ulm R (2010) Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci 15 : 322–329. doi: 10.1016/j.tplants.2010.04.003 20452268
42. Bartels S, Anderson JC, Gonzalez Besteiro MA, Carreri A, Hirt H, et al. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21 : 2884–2897. doi: 10.1105/tpc.109.067678 19789277
43. Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, et al. (2002) Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J 21 : 6483–6493. 12456655
44. Anderson JC, Bartels S, Gonzalez Besteiro MA, Shahollari B, Ulm R, et al. (2011) Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J 67 : 258–268. doi: 10.1111/j.1365-313X.2011.04588.x 21447069
45. Lumbreras V, Vilela B, Irar S, Sole M, Capellades M, et al. (2010) MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J 63 : 1017–1030. doi: 10.1111/j.1365-313X.2010.04297.x 20626661
46. Vilela B, Pages M, Lumbreras V (2010) Regulation of MAPK signaling and cell death by MAPK phosphatase MKP2. Plant Signal Behav 5 : 1497–1500. 21057191
47. Lim CW, Luan S, Lee SC (2014) A prominent role for RCAR3-mediated ABA signaling in response to Pseudomonas syringae pv. tomato DC3000 infection in Arabidopsis. Plant Cell Physiol 55 : 1691–1703. doi: 10.1093/pcp/pcu100 25063782
48. Mang HG, Qian W, Zhu Y, Qian J, Kang HG, et al. (2012) Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24 : 1271–1284. doi: 10.1105/tpc.112.096198 22454454
49. Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, et al. (2010) The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol 152 : 1901–1913. doi: 10.1104/pp.109.152603 20164209
50. Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 : 1310–1326. 16603654
51. Swiderski MR, Birker D, Jones JD (2009) The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact 22 : 157–165. doi: 10.1094/MPMI-22-2-0157 19132868
52. Bernoux M, Ve T, Williams S, Warren C, Hatters D, et al. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9 : 200–211. doi: 10.1016/j.chom.2011.02.009 21402359
53. Feys BJ, Parker JE (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16 : 449–455. 11050331
54. Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132 : 449–462. doi: 10.1016/j.cell.2007.12.031 18267075
55. Burch-Smith TM, Dinesh-Kumar SP (2007) The functions of plant TIR domains. Sci STKE 2007: pe46. 17726177
56. Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, et al. (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68. 17298188
57. Shirasu K (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60 : 139–164. doi: 10.1146/annurev.arplant.59.032607.092906 19014346
58. Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, et al. (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25 : 2007–2016. 16619029
59. Bieri S, Mauch S, Shen QH, Peart J, Devoto A, et al. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16 : 3480–3495. 15548741
60. Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, et al. (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17 : 1268–1278. 15749757
61. de la Fuente van Bentem S, Vossen JH, de Vries KJ, van Wees S, Tameling WI, et al. (2005) Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I–2 disease resistance protein. Plant J 43 : 284–298. 15998314
62. He Y, Chung EH, Hubert DA, Tornero P, Dangl JL (2012) Specific missense alleles of the arabidopsis jasmonic acid co-receptor COI1 regulate innate immune receptor accumulation and function. PLoS Genet 8: e1003018. doi: 10.1371/journal.pgen.1003018 23093946
63. Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4 : 21–33. 10445024
64. Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15 : 1310–1319. 12782725
65. Gou M, Shi Z, Zhu Y, Bao Z, Wang G, et al. (2012) The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J 69 : 411–420. doi: 10.1111/j.1365-313X.2011.04799.x 21967323
66. Cheng YT, Li Y, Huang S, Huang Y, Dong X, et al. (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci U S A 108 : 14694–14699. doi: 10.1073/pnas.1105685108 21873230
67. Consortium AIM (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333 : 601–607. doi: 10.1126/science.1203877 21798944
68. Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, et al. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci U S A 96 : 13583–13588. 10557364
69. Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9 : 1573–1584. 9338960
70. Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11 : 1187–1194.
71. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743. 10069079
72. Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 : 428–438. 15469500
73. Chen H, Zou Y, Shang Y, Lin H, Wang Y, et al. (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146 : 368–376. 18065554
74. Buchner J, Grallert H, Jakob U (1998) Analysis of chaperone function using citrate synthase as nonnative substrate protein. Methods Enzymol 290 : 323–338. 9534173
75. Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2 : 1565–1572. 17585298
Štítky
Genetika Reprodukčná medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



