-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
The majority of cancer cells use a special enzyme called telomerase to maintain telomeres. However, some cancer cells do not possess telomerase and use instead the so-called ALT mechanism to maintain telomeres. ALT is a complex pathway that entails the action of many factors, and the telomere DNAs of ALT cancer cells are extremely abnormal (e.g., they are often detached from the rest of the chromosomes and often exist in single-stranded forms). Currently, there are few manipulations that one can use to induce normal cells to engage in the ALT mechanism. The lack of a good “model” system poses a major obstacle to the understanding of this pathway and the development of effective counter-measures against ALT cancer cells. By removing Ku and a checkpoint factor from U. maydis (a yeast-like fungus), we generated clones that exhibit many of the characteristic abnormalities of ALT cancer cells. Moreover, we identified two factors (i.e., Mre11 and Blm) that when deleted, abolished the ALT phenotypes. Further analysis of this model may lead to the development of new strategies for shrinking the telomeres of cancer cells, thereby arresting their proliferation.
Published in the journal: Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient. PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005570
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005570Summary
The majority of cancer cells use a special enzyme called telomerase to maintain telomeres. However, some cancer cells do not possess telomerase and use instead the so-called ALT mechanism to maintain telomeres. ALT is a complex pathway that entails the action of many factors, and the telomere DNAs of ALT cancer cells are extremely abnormal (e.g., they are often detached from the rest of the chromosomes and often exist in single-stranded forms). Currently, there are few manipulations that one can use to induce normal cells to engage in the ALT mechanism. The lack of a good “model” system poses a major obstacle to the understanding of this pathway and the development of effective counter-measures against ALT cancer cells. By removing Ku and a checkpoint factor from U. maydis (a yeast-like fungus), we generated clones that exhibit many of the characteristic abnormalities of ALT cancer cells. Moreover, we identified two factors (i.e., Mre11 and Blm) that when deleted, abolished the ALT phenotypes. Further analysis of this model may lead to the development of new strategies for shrinking the telomeres of cancer cells, thereby arresting their proliferation.
Introduction
Eukaryotic chromosome ends, or telomeres, consist of repetitive DNA sequences and a plethora of protective proteins that are crucial for chromosome stability [1,2]. Aberrations in the DNA repeat tracts or the protective telomere proteins have been shown to induce chromosome re-arrangements. Owing to the “end-replication” problem, chromosome termini experience progressive telomere loss with each cell division [3]. This process eventually generates critically short telomeres that limit further cell proliferation by triggering a DNA-damage response [4]. Progressive telomere loss can be counter-balanced by telomerase reverse transcriptase, which extends telomeres by using an integral RNA component as the template [5,6]. Telomerase expression is repressed in normal somatic cells, but up-regulated in cancer cells, conferring these cells with increased proliferative potential. Telomerase inhibition has been validated as a useful anti-cancer strategy and telomerase inhibitors are in clinical trials for treating a variety of cancers [7].
While the majority of cancer cells utilize telomerase to replenish telomeres, a subset of tumor cells employs an alternative, recombination-based mechanism known as ALT (alternative lengthening of telomeres) [8,9]. The telomere DNAs in ALT cells are characterized by length heterogeneity, accumulation of unpaired telomere C-strand (the strand that is rich in C-residue and that carries the 5’ end), and a substantial increase in extra-chromosomal telomere repeats (ECTR), which comprise both circular and linear repeats. In particular, extra-chromosomal C-circles are believed to be an especially specific and quantifiable marker of ALT activity [10]. In addition, ALT is associated with extensive genome rearrangements, marked micronucleation, defects in the G2/M checkpoint, and altered double-strand break (DSB) repair [11]. While many recombinational repair proteins have been linked to ALT (including e.g., the MRN and the 9-1-1 complex), this pathway is clearly distinguished from the canonical homology-directed DNA repair (HDR) pathway [12,13]. For example, RAD51 is the central catalyst of HDR, yet knockdown of RAD51 either fails to suppress or exacerbates ALT phenotypes [14,15]. In addition, a key feature of the HDR pathway is the generation of long 3’ overhangs, yet ALT is characterized by the accumulation of telomere 5’ overhangs [15]. Despite intensive efforts, including high throughput screening of driver mutations in ALT, the mechanisms that underlie ALT induction and maintenance remain incompletely understood [11]. One potential contributing factor for ALT is a defect in the chromatin remodeling protein ATRX. ATRX mutations are frequently observed in ALT, and exogenous expression of ATRX in some ALT cells can repress ALT phenotypes [16]. However, knockdown of ATRX is not sufficient to trigger the ALT pathway [11], implying the existence of other necessary contributing factors. The current lack of knowledge on the ALT pathway hampers the development of mechanism-based inhibitors against this subset of cancer cells [17].
Recombination-mediated telomere extension pathways have also been analyzed in fungi, including S. cerevisiae and S. pombe [18]. Such pathways are typically activated in telomerase null mutants, and have been classified as Type I or Type II according to the sequence amplified at chromosome ends and the factor requirements [19]. A common weakness of existing fungal models of ALT is that they do not recapitulate the characteristic aberrations of ALT cells. For example, none of the S. cerevisiae or S. pombe telomerase mutant survivor has been reported to accumulate excess C-strand overhangs or C-circles. Alternative and more relevant models are therefore desirable. An attractive model organism is the genetically tractable yeast-like fungus Ustilago maydis, which bears a telomere repeat unit that is identical to the mammalian sequence and an average telomere tract of ~300–400 bp [20,21]. U. maydis also contains a shelterin-like telomere nucleoprotein complex, and a recombination-repair machinery that is remarkably similar to the mammalian machinery [22]. Two recent observations highlight the striking resemblance of the U. maydis telomere regulatory mechanisms to those in humans. First, as in human cells but different from budding or fission yeast, the two central components of HDR in U. maydis (i.e., Rad51 and the BRCA2 ortholog Brh2) are each required for normal telomere maintenance in telomerase-positive cells [21,23]. Second, just like the human Ku complex, the U. maydis Ku complex is essential for cell viability and loss of Ku expression causes massive telomere aberrations [24,25]. In the current study, we further characterized the Ku-deficient U. maydis cells, and found that they initially exhibit telomere abnormalities that are very similar to those observed in ALT cancer cells, including telomere length heterogeneity, the accumulation of C-strands and extrachromosomal telomere repeats (ECTR). The level of C-circles, a highly specific marker of ALT, is dramatically increased in the mutant as well. Moreover, we found that the induction of aberrant telomeres in the Ku-deficient mutant is largely suppressed by deletion of mre11 and fully eliminated by deletion of blm, consistent with previous characterization of ALT cell lines [10,13]. By contrast, deletion of rad51 or other DNA damage response and repair factors had disparate effects on various telomere abnormalities, suggesting that these factors process ALT intermediates or products. Interestingly, in the presence of normal checkpoint function, the ku mutants eventually experience permanent cell cycles arrest, which is accompanied by the disappearance of telomere repeats from chromosome ends and a further increase in short ECTRs. Our findings illustrate the complexity of the ALT-like pathway in U. maydis and begin to suggest factors and mechanisms that may mediate the formation of aberrant telomeres in ALT cells.
Results
Telomere aberrations in the U. maydis ku mutants resemble those in ALT cancer cells
Previous analysis revealed DNA damage signaling from the telomeres of ku70 and ku80-deficient U. maydis cells that leads to permanent cell cycle arrest. (To simplify the discussion, we will refer to the Ustilago maydis uku70 and uku80 genes and mutants as ku70 and ku80.) The investigation made use of U. maydis strains that conditionally express ku70 or ku80 in nitrate-containing medium (named ku70nar1 and ku80nar1). Following 18–20 hr of incubation in YPD (a medium that represses ku expression), the ku mutants cease to proliferate and experience complete G2 arrest as judged by cell morphology (with elongated bud and a single nucleus) and FACS analysis [24]. Drastic telomere restriction fragment (TRF) length heterogeneity and elevated levels of t-circles were also detected [24]. To characterize in greater detail the dynamics of the telomere defects, we further analyzed TRF length distribution at multiple time points following transcriptional repression of the ku70nar1 allele (Fig 1A). Increased total telomere hybridization signal and TRF length heterogeneity became evident ~12 hr post ku70 repression, and these alterations grew more severe over time, reaching a maximum at ~18 hr. This initial TRF length distribution is superseded at ~30 hr by a terminal distribution characterized by the disappearance of extremely long telomere fragments and a further increase (~ 3 fold) in very short telomere fragments (~0.1 to 0.5 kb). To determine if these abnormal TRFs are chromosomal or extra-chromosomal, we compared the Southern hybridization patterns of the DNA samples with and without prior PstI treatment (S1A Fig). Interestingly, omitting PstI digestion reduced but did not eliminate the long TRF signal at 18 hr, indicating that only a portion of this signal is due to ECTR. In contrast, the very short telomere fragments at 30 hr are almost entirely due to ECTRs (and henceforth abbreviated as vsECTRs). Prolonged exonuclease treatment eliminated only the faster migrating portion of the vsECTRs, suggesting that the smallest vsECTRs are linear whereas the remaining ones are circular DNAs (S1B Fig). We also quantified the chromosome-associated telomere signals and total telomere DNA signals using the undigested samples, and found the former were reduced by 50% and 90% after 18 hr and 30 hr of repression, whereas the latter were increased by 50% and 3-fold at the same time points (S1C Fig). Hence, the loss of Ku in U. maydis results in a progressive loss of telomere DNAs from the chromosomes, as well as a drastic accumulation of ECTRs, which are initially long and heterogeneous, but subsequently dominated by very short linear and circular fragments.
Fig. 1. The induction of ALT telomere features in the Ustilago maydis ku mutants. 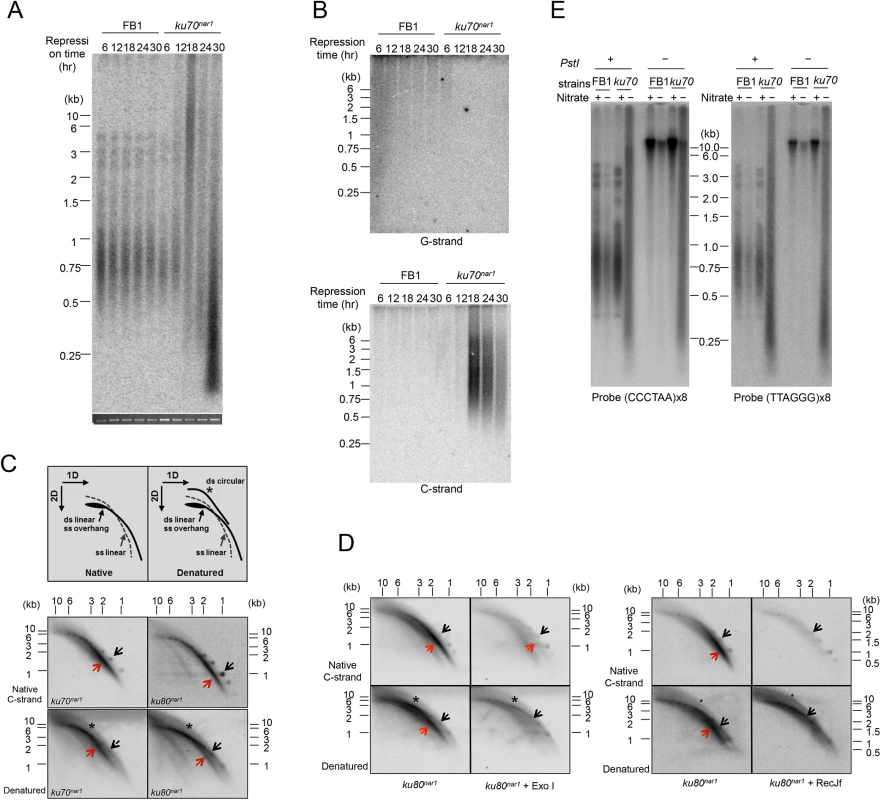
(A) DNA samples were obtained from the indicated strains (FB1 and ku70nar1) grown in restrictive conditions (YPD) for increasing durations (6, 12, 18, 24, and 30 hr), digested with PstI, and subjected to telomere Southern analysis. The total chromosomal DNA levels in the samples were analyzed by gel electrophoresis and EtBr staining prior to PstI digestion to ensure the use of comparable amounts of DNAs (bottom strip). (B) The same PstI-treated samples as in A were subjected to in gel hybridization assays to detect unpaired G-strand (top) or unpaired C-strand (bottom). (C) PstI-digested DNAs from ku70nar1 or ku80nar1 strain were subjected to 2D-gel analysis followed by in-gel hybridization assay. Labeled UmG4 oligonucleotides, corresponding to four copies of the U. maydis telomeric G-strand repeats, were used as the probe to detect unpaired C-strand. Radioactively labeled DNA ladders were used as markers for the double-stranded linear DNA arc (designated by black arrows). The gels were subsequently denatured and reprobed to detect all telomere DNAs. The asterisks in the denatured gels indicate ds circular DNA arcs (t-circles). (D) DNAs were isolated from ku80nar1 after 18 hrs of growth in the restrictive medium, treated with Exo I or RecJf, and then subjected to 2D in-gel hybridization assays to detect unpaired C-strand. The gels were subsequently denatured and reprobed to detect all telomere DNAs. (E) Standard telomere Southern analysis was performed using chromosomal DNA with or without PstI. The DNAs were all isolated from the indicated strains after 18 hrs of growth in permissive or restrictive media. Next, we examined single stranded telomeres in the Ku-deficient mutant at multiple time points using in-gel hybridization assays and observed the disappearance of unpaired G-strand at 6 hr, and the drastic accumulation of unpaired C-strand at 18 hr and thereafter (Fig 1B). We note that while there is a good correlation between the size distribution of TRFs in standard Southern analysis and that of unpaired C-strand, the unpaired G-strand signals are disproportionately concentrated in the high molecular weight region of the gel (Fig 1B). The G-strand signals may therefore arise from a subset of the molecules, perhaps recombination or replication intermediates, which are expected to migrate slower than linear DNAs. More study is required to ascertain the nature of the G-strand signals. Interestingly, despite the dramatic increase in the amount of vsECTRs in the standard telomere Southern analysis, no such increase was detected in either the G-strand or C-strand assays, indicating that vsECTRs are primarily double stranded. The distinct kinetics of the various telomere alterations suggest that the genesis of aberrant telomeres in the ku mutant is a multi-step process that entails the action of many DNA-processing factors.
Preferential accumulation of unpaired C-strand has not been reported for other fungal mutants but is characteristic of ALT cancer cells [15]. To characterize this structure in more detail, we subjected 18 hr DNA samples from the ku70nar1 mutant to 2-D gel analysis followed by in-gel hybridization using a G-strand probe (Fig 1C). We found that a substantial portion of the C-strand signals migrate as an arc below the ds linear arc, and are therefore likely due to ss linear DNA (Fig 1C, marked by red arrows). Following denaturation and re-hybridization, signals corresponding to ds linear and ds circular DNAs are detected as well (Fig 1C, marked by black arrows and asterisks). Treatment of the DNA with E. coli RecJf (a 5’ to 3’ ssDNA exonuclease) prior to 2D-gel fractionation caused a substantial reduction in the C-strand signals, supporting the presence of 5’ ssDNA overhangs (as predicted based on the polarity of the C-strand) (Fig 1D). We also compared the C-strand signals in 1-D native gels before and after subjecting the ku70nar1 DNA to Klenow mediated fill-in synthesis, which is expected to convert 5’ overhangs to duplexes (S2 Fig). Again this treatment eliminated part but not all of the C-strand signals, consistent with the presence of 5’ C-strand overhangs in a fraction of the molecules. Surprisingly, treatment with E. coli Exo I (a 3’ to 5’ ssDNA exonuclease) also reduced the C-strand signal, which is not consistent with unpaired C-strand at the ends of chromosomes. Instead, this results indicates that a portion of the signal may come from ss extra-chromosomal linear DNA; both purely single-stranded DNA and single-stranded DNA bearing an internal duplex should be susceptible to both Exo I and RecJf. This result is also consistent with the finding from 2D gel analysis that a significant fraction of the signals is from ss linear DNA. To confirm the existence of ECTRs, we performed standard telomere Southern analysis on samples that had not been treated with restriction enzyme (Fig 1E). Consistent with our hypothesis, the heterogeneous telomere fragments of the ku70nar1 and ku80nar1 mutants are observed in the absence of PstI cleavage, whereas the typical telomere patterns of control cells are only observed with PstI treatment. Hence we conclude that the U. maydis ku mutants are characterized by the possession of abundant ECTRs that bear a high level of unpaired C-strand, a substantial portion of which exists as ss linear DNA.
As noted before, unpaired C-strand is believed to be a distinguishing feature of ALT [15]. Other features shared by the U. maydis ku mutants and ALT cells include telomere length heterogeneity and ECTR, suggesting substantial mechanistic resemblances between the two systems. Notably, a recent study identified extra-chromosomal C-circles (i.e., circular DNA comprised of continuous C-strand and G-strand nicks/gaps) as an especially specific and quantifiable marker of ALT activity [10]. We therefore examined the levels of C-circles in the U. maydis mutant and found a dramatic increase of such circles in the ku70nar1 strain upon transcriptional repression of ku70 (Fig 2A). The signals are proportional to the amounts of input DNA, and quantification revealed in the mutant at 30 hr post ku repression ~30,000 fold more C-circles than the control cells. Similar to ALT cells, G-circles (i.e., circles comprised of continuous G-strand and C-strand nicks/gaps) are also elevated in the ku mutant, but to a lesser extent (~1,500 fold) (Fig 2A). As expected, the C - and G-circle signals require both input DNA and the Φ29 polymerase, and are unaffected by prior exonuclease treatments (Fig 2B). Thus, the U. maydis ku mutant appears to suffer from very similar telomere defects as ALT cancer cells, and may be engaged in the same set of reactions at telomeres. Moreover, these defects exhibit distinct kinetics (see S3 Fig for a summary of the temporal dynamics of the defects) and may thus represent different intermediates in a complicated pathway.
Fig. 2. Elevated levels of C-circles and G-circles in the Ustilago maydis ku mutants. 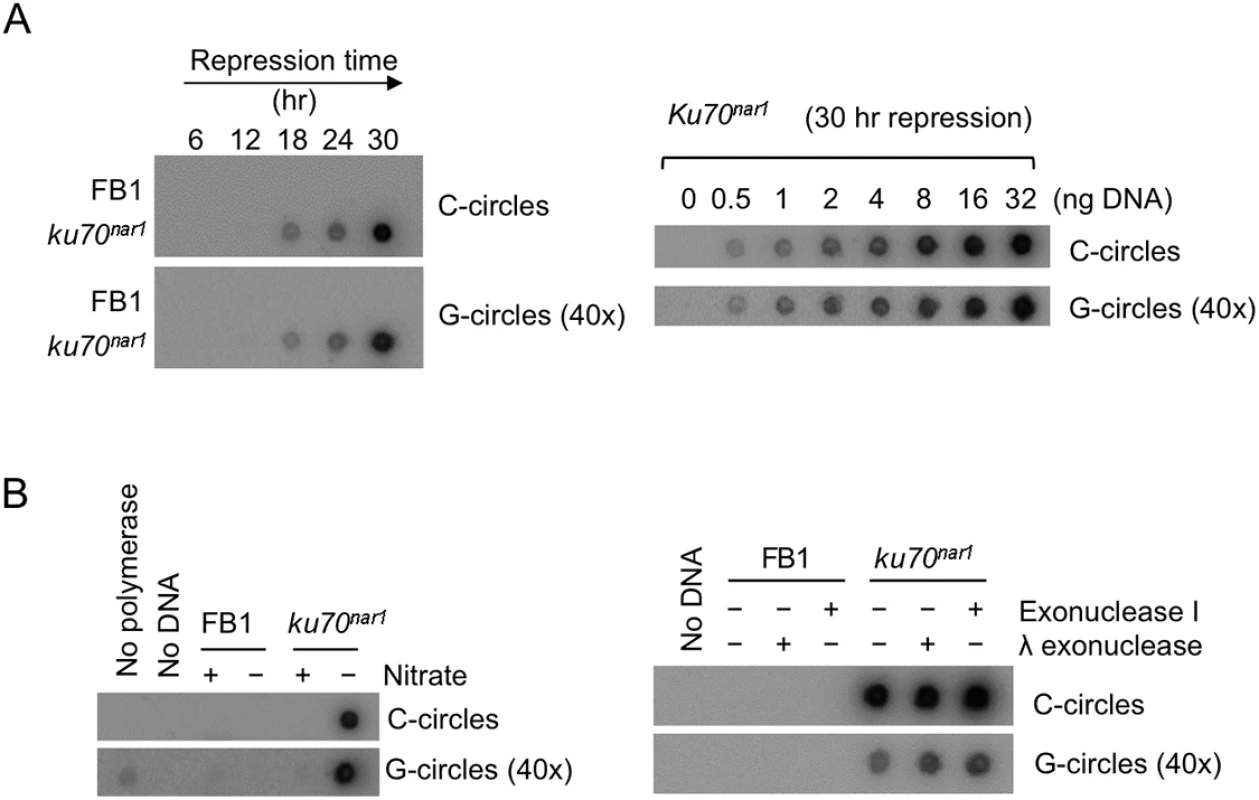
(A) (Left) Genomic DNAs from the indicated strains were isolated after the indicated durations of growth in the restrictive medium, and subjected to C circle or G circle assays followed by Dot blot detection. The intensity of the G circle assay panel was set 40 times higher than that for the C circle panel. (Right) Increasing amounts of the ku70nar1 mutant DNA were assayed for the levels of C- and G-circles. The intensity of the G circle assay panel was set 40 times higher than that for the C circle panel. (B) (Left) Genomic DNAs from indicated cells (FB1 and uku70nar1) were tested for C circles and G circles. Nitrate + and–indicate permissive and restrictive medium, respectively. As negative controls, DNA or polymerase was omitted from some of the assays. (Right) DNAs were isolated from FB1 and uku70nar1 after 18 hrs of growth in YPD (restrictive medium), and subjected to C- or G-circles assays. Some samples were pre-treated with exonuclease I or λ exonuclease to eliminate any signals from linear DNAs. The intensity of the G circle assay panel was set 40 times higher than that for the C circle panel. The telomere abnormalities are reversible
To assess the reversibility of the disparate abnormalities, we first subjected the ku70nar1 and ku80nar1 strains to transcriptional repression for 18 hr to induce the ALT-like defects, and then re-expressed the ku genes by switching the cells to a nitrate-containing medium, followed by telomere analysis at various time-points. Remarkably, the ku mutants rapidly resumed growth when switched from the restrictive to permissive medium. They underwent the first cell division at ~4 hr after the switch, and continue to divide with a generation time of ~210 minutes thereafter. In addition, the telomere aberrations of the mutants are fully reversible, and similar to the induction process described earlier, different aberrations are reversed with distinct kinetics (S4 Fig). The level of extra-long TRFs (5 to >20 kb) becomes substantially reduced at 24 hr following ku re-expression and is similar to control cells at 48 hr, whereas the level of extremely short fragments bearing telomere repeats (<0.5 kb) does not return to normal until 72 hr post re-expression (S4A Fig). A difference in kinetics is also observed between the disappearance of unpaired C-strand and the appearance of unpaired G-strand during phenotypic reversal; the former is complete at 24 hr after ku re-expression, but the latter is only complete at 48 hr (S4B Fig and S4C Fig). These results again support the notion that the generation of the telomere defects is likely to be a multi-step process, and that different telomeric structures may represent distinct intermediates along a complicated pathway.
The formation of the ALT-like telomeres is independent of U. maydis telomerase
In some organisms, Ku has been observed to modulate the activity of telomerase [26,27]. To assess the potential contribution of telomerase activity to the ALT-like telomeres in the U. maydis ku mutants, we analyzed several independent trt1 ku70nar1 double mutants, which were constructed by first deleting the trt1 gene (encoding the telomerase reverse transcriptase in U. maydis [28]) and then introducing the ku70nar1 allele. Similar to the ku70nar1 single mutant, the double mutant experienced growth arrest when switched to the restrictive condition for ku70 expression (Fig 3A and 3B), and exhibited profound telomere aberrations (Fig 3C, 3D and 3E). Interestingly, while the same aberrations are observed in the double mutant, they occur with a slightly delayed kinetics. For example, while high levels of vsECTRs are evident following 18 hr of ku70 repression in the single mutant, they are not observed until 24 hr after ku70 repression in the double mutant (Fig 3C). Similarly, while unpaired C-strand signals peak at 18 hr after ku70 repression and decline thereafter in the single mutant, the C-strand signals remain near the peak level after 30 hr of repression (Fig 3D). A delay or a mild reduction in C - and G-circle formation is also evident in the double mutant relative to the single mutant (Fig 3E). Thus, while none of the ALT telomere aberrations requires trt1, the loss of trt1 (or the shorter initial telomere lengths of the double mutant (Fig 3C)) appears to affect the severity or kinetics of the phenotypes. We conclude that telomerase is not an essential component of the U. maydis ALT pathway, but may act on some of the telomere structures generated during the induction of this pathway.
Fig. 3. The growth and telomere phenotypes Ustilago maydis ku mutants do not depend on telomerase. 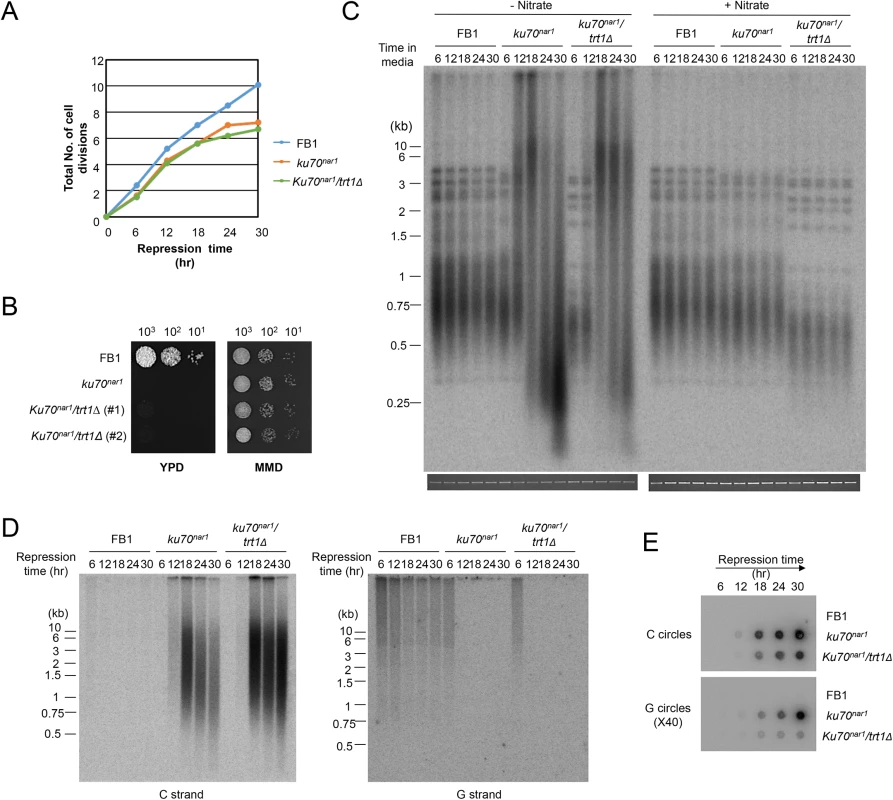
(A) The growth rates of the indicated strains in YPD (ku repression medium) were monitored by successive OD600 measurements and plotted. (B) The long-term growth of the indicated strains in YPD and MMD were monitored by spotting serial ten-fold dilutions of cultures solid media. The YPD plate was photographed after 2 days of incubation and the MMD plate after 3 days of incubation at 30°C. (C) Genomic DNAs from the indicated strains were isolated after the indicated durations of growth in the restrictive or permissive medium (for ku70 expression), and subjected to telomere length analysis. The total chromosomal DNA levels in the samples were analyzed by gel electrophoresis and EtBr staining prior to PstI digestion to ensure the use of comparable amounts of DNAs (bottom strip). (D) Genomic DNAs from the indicated strains were isolated after the indicated durations of growth in YPD, and subjected to in gel hybridization analysis for unpaired C- and G-strand. (E) The same DNAs as in D were assayed for the levels of C- and G-circles. ALT telomere aberrations are slightly ameliorated and stabilized by atr1 and chk1 deletion
ALT cells invariably exhibit checkpoint defects [11], and we have previously shown that atr1 or chk1 deletion enables the U. maydis ku70 and ku80-deficient cells to proliferate without suppressing the telomere length heterogeneity in these cells [29]. To determine in greater detail how the combined loss of checkpoint and ku proteins affects telomeres, we measured the levels of chromosome-associated telomere repeats and other aberrant telomere structures in atr1 ku70nar1 and chk1 ku70nar1 double mutants at multiple time points following ku repression. Interestingly, unlike the ku70nar1 single mutant, the double mutants retained substantial amounts of chromosome-associated telomere repeats and accumulated lower levels of ECTRs after 30 hr of repression (S1C Fig). Even after more than ~225 generations (~ 9 streaks) of passage on YPD plates, the double mutants do not progress to the “terminal telomere phenotype” characterized by the predominance of vsECTRs (Fig 4A), implying that the checkpoint proteins are necessary for this progression. Interestingly, we reproducibly detected higher telomere DNA content in the chk1 ku70nar1 mutant in comparison with the atr1 ku70nar1 mutant (Fig 4A and S1C Fig), suggesting some mechanistic differences between the two checkpoint proteins in the ALT pathway. The nature of this difference remains to be determined.
Fig. 4. Partial suppression of the ALT telomere phenotypes by atr1 and chk1 deletions. 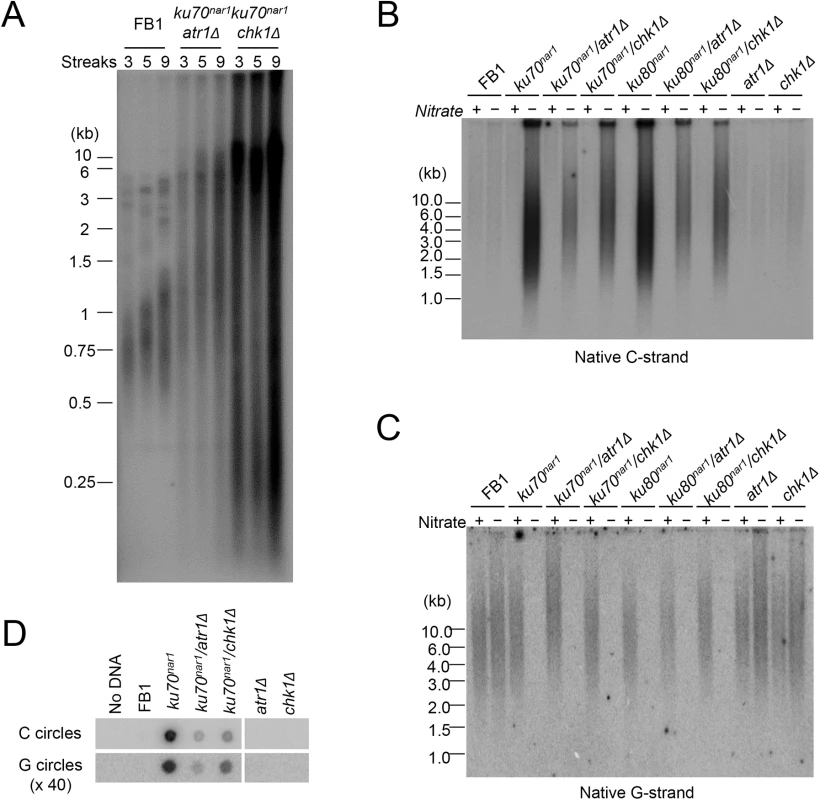
(A) TRF analysis of the uku70nar1 chk1Δ and uku70nar1 atr1Δ double mutants after multiple re-streaks on restrictive medium. The indicated strains were passaged at 30°C on YPD. Each streak represents approximately 25 generations of growth. DNAs from streak 3, 5, and 9 of each strain were isolated, treated with PstI, and analyzed by standard telomere Southern analysis. (B) and (C) Pst1-treated DNA was subjected to in-gel hybridization assay to detect unpaired C-strand (B) and unpaired G-strand telomere DNA (C). (D) DNAs from the indicated strains were subjected to C-circle and G-circle assays. With respect to the accumulation of ss telomere DNAs, deleting either atr1 or chk1 reduced the accumulation of unpaired C-strand by ~ 2 fold in the ku70nar1 strain, but had little effect on the disappearance of unpaired G-strand (Fig 4B and 4C). The C - and G-circle levels of the double mutants were also lower than that of the ku70nar1 single mutant (Fig 4D). Thus the checkpoint proteins appear to play a role in exacerbating a subset of the telomere aberrations in the ku mutant. As expected, deleting atr1 or chk1 alone does not induce any obvious telomere defects (Fig 4B, 4C and 4D), suggesting that these checkpoint factors enhance the telomere aberrations only in the context of ku loss.
Telomere aberrations are altered by 9-1-1 and rad51 mutations, and largely suppressed by deletion of mre11
The foregoing analysis hints at the complexity of ALT-like pathway in the U. maydis ku mutants. To begin to identify factors involved in the pathway, we first surveyed a number of factors involved in DDR and repair. These factors, including 9-1-1 subunits, Rad51, and Mre11, were previously examined in regard to their roles in the non-viability and telomere length heterogeneity of the ku70nar1 mutant, but not their roles in other ALT-like phenotypes. Specifically, the previous study showed that deleting subunits of the rad9-hus1-rec1 complex or rad51 could not suppress the lethality or telomere length heterogeneity of the ku70nar1 mutant [29]. More detailed and comprehensive characterization of the telomeres of these combination mutants revealed in fact exacerbation of selected telomere phenotypes (Fig 5). For example, in each of the 9-1-1 ku70nar1 combination mutants, the levels of extremely long telomeres are further elevated relative to the ku70nar1 single mutant (Fig 5A and 5B), whereas the levels of G - and C-strand are comparable to those of the single mutants (Fig 5C and 5D). The effect of deleting rad51 in the ku70nar1 mutant is even more complex (S5 Fig). Both the levels of very long and short telomeres are increased in the double mutant. The level of unpaired C-strand is also increased, but that of C-circle slightly reduced.
Fig. 5. The telomere phenotypes of the 9-1-1 ku70nar1 double mutants. 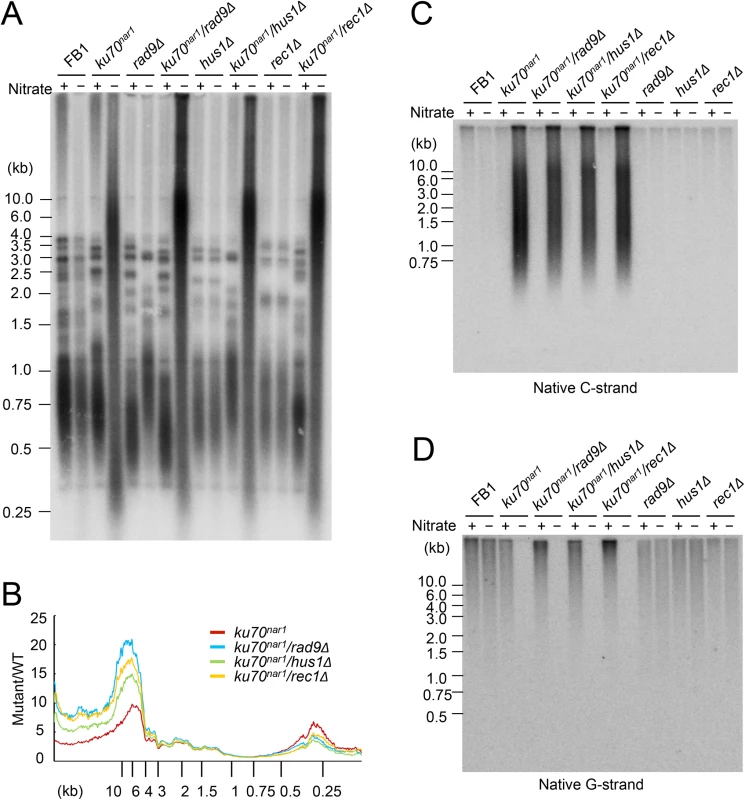
(A) DNAs were isolated from the indicated strains after 18 hrs of growth in MMD (Nitrate +) or YPD (Nitrate -), digested with PstI, and subjected to telomere Southern analysis. (B) The hybridization signals for a subset of the assays shown in A were quantified and plotted against TRF lengths. (C) and (D) The DNA samples from the indicated strains were digested with PstI and subjected to in-gel hybridization to determine the levels of C- and G-strand, respectively. Previous analysis also revealed strong suppression of the lethality and telomere heterogeneity of the ku70nar1 mutant by mre11 deletion, but not the mre11-H228N (nuclease deficient) allele. More detailed characterization of the combination mutants revealed suppression of almost all the telomere aberrations of the ku70nar1 mutant by mre11 deletion; the levels of G-strand, C-strand and C-circles in the combination mutant are all quite similar to the wild type parental strain (Fig 6A, 6B and 6C). In contrast, the mre11-H228N ku70nar1 double mutants have telomeres that closely mimic those of the ku70nar1 single mutant, indicating that a non-nuclease activity of Mre11 confers lethality and is required for the induction of telomere defects upon the loss of ku.
Fig. 6. Suppression of the ALT telomere phenotypes of ku70nar1 mutant by mre11 deletion. 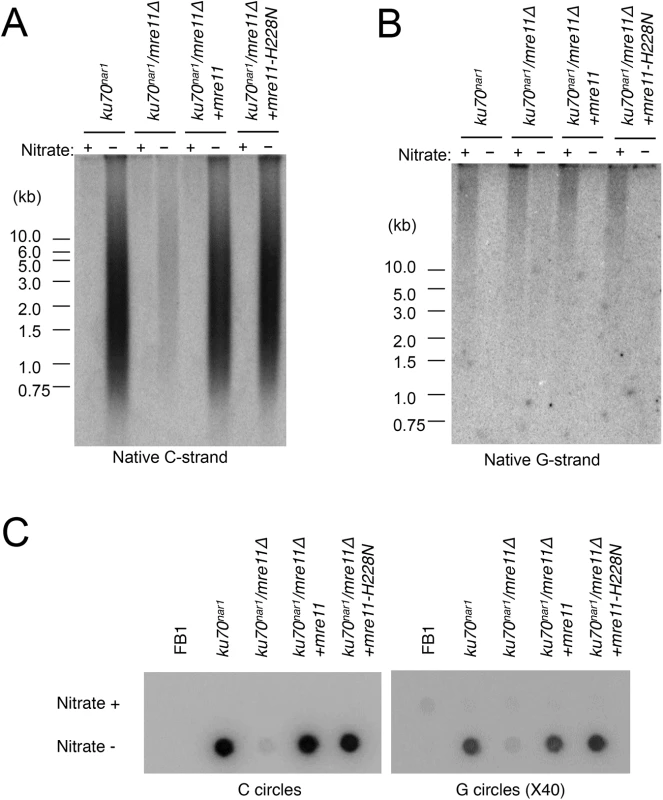
(A) DNAs were isolated from the indicated strains after 18 hr of growth in MMD (Nitrate +) or YPD (Nitrate -), digested with PstI, and subjected to in-gel hybridization assay for unpaired C-strand. During the 18 hr repression, the wild type and mre11 ku70nar1 mutant underwent ~ 7 cell divisions, whereas the ku70nar1 and mre11-H228N ku70nar1 mutant underwent ~ 5.5 cell divisions. (B) The same digested DNAs were analyzed for the levels of unpaired G-strand. (C) The levels of C-circles in the indicated strains after 18 hrs of growth in YPD were analyzed. The contributions of multiple DNA processing factors to the U. maydis ALT pathway
The apparently antagonistic effect of the Ku and MRN complexes at the U. maydis telomeres is reminiscent of their relationships in DSB resection in S. cerevisiae [30]. In particular, the resection defect of the S. cerevisiae MRN mutants can be suppressed by concurrent mutations in yKu subunits. However, a significant distinction between S. cerevisiae Ku and U. maydis Ku is that the former suppresses the formation of 3’ overhangs at DSBs, whereas the latter reduces the production of 5’ overhangs at telomeres by MRN. While the basis for this difference is unclear, MRN is known to interact with many DDR and DNA processing factors, and the distinct nucleoprotein structures of broken DNAs and telomeres could play a role in the distinct outcomes of the two systems.
To address the roles of factors implicated in resection and other processing events downstream of the resection on the ALT pathway in U. maydis, we analyzed additional combination mutants. For this study, the ku70nar1 allele was introduced into a set of largely isogenic strains (initially derived from the UCM350 parental strain), each harboring a single deletion (of blm, ctip, dna2, exo1, mus81, top3). These combination mutants, as well as the single mutants, were subjected to growth analysis and the panel of telomere assays (Fig 7). Remarkably, the deletion of two genes, namely blm and exo1, significantly restored growth to the ku70nar1 mutant, whereas deletion of others did not (Fig 7A). Interestingly, there is only partial correlation between the suppression of growth and telomere defects (Fig 7B, 7C and 7D). In the case of blm deletion, we observed complete suppression of telomere length heterogeneity, as well as C-strand and C-circle accumulation (Fig 7B, 7C and 7D). Consistent with these findings, the blm ku70nar1 double mutant grows as well as wild type cells in YPD (Fig 7A). In the case of exo1 deletion, we observed only partial reduction of the telomere phenotypes, and accordingly only partial suppression of the growth defect (compare colony sizes of the blm ku70nar1 and exo1 ku70nar1 mutant, Fig 7A). In addition, exo1 deletion could not suppress the lethality of ku70 in a different strain background [29]. Exo1 is thus not necessary for the manifestation of all ku70 phenotypes. On the other hand, the dna2 ku70nar1 and ctip ku70nar1 mutants exhibit telomere aberrations that are similar to or milder than those in the exo1 ku70nar1 mutant, yet the former two grow considerably worse than the latter (Fig 7B, 7C and 7D). Finally, none of the deletions except that of blm substantially reduced the high level of vsECTRs that is characteristic of the terminal stage of ku mutant (S6 Fig). Overall, the results suggest that Blm plays a critical role in the ALT pathways, and some blm-dependent telomere aberration is needed to trigger the signals and damages that fully arrest growth. In addition, many factors implicated in DSB resection play significant roles in the U. maydis ALT pathway. However, it seems likely that not all telomere aberrations contribute equally to the loss of cell viability.
Fig. 7. The roles of various DNA processing factors in the induction of ALT telomere features in the ku70nar1 mutant. 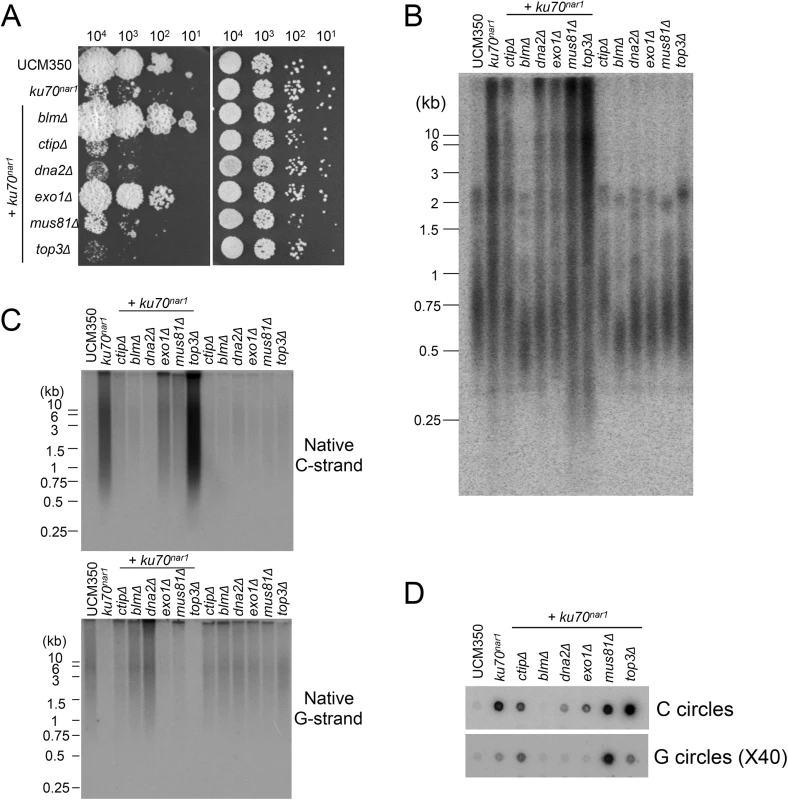
(A) Serial ten-fold dilutions of the indicated strain cultures were spotted on solid rich medium (YPD) and minimal medium with nitrate (MMD+). YPD plates were incubated for 2 days and the MMD+ plates for 3 days at 30°C. (B) and (C) DNAs were isolated from each strain after 20 hrs of growth in YPD, digested with PstI, and subjected to telomere Southern analysis (B) and in-gel hybridization assays for unpaired C-strand and G-strand (C). (D) DNAs from the same set of strains were subjected to C-circle and G-circle assays. A critical function for the Blm helicase activity in the U. maydis ALT pathway
The apparently complete suppression of the U. maydis ALT pathway by blm deletion prompted us to examine the role of this helicase further. First, we validated the suppression in two independent clones of blm ku70nar1 double mutants, and showed that these clones exhibit the same degree of ku repression as the ku70nar1 single mutant when grown in YPD (Fig 8A and 8B). Hence the effects of blm deletion cannot be attributed to unrelated genetic mutations or leaky ku70 expression. Second, we analyzed the effect of abolishing just the helicase activity of Blm on the ALT pathway. Based on alignment with well-characterized homologues, we surmise that the K443R substitution in U. maydis Blm should render the protein without helicase activity [31]. Consistent with this hypothesis, we found previously that introducing this point mutation compromised the DNA-damage sensitivity of U. maydis [31]. Notably, repressing ku70 in strains with this point mutation failed to induce any ALT telomere phenotypes (Fig 8C, 8D, 8E and 8F), implying that the blm helicase activity is essential for the ALT pathway.
Fig. 8. The Blm helicase activity is essential for the induction of ALT-like telomeres in U. maydis. 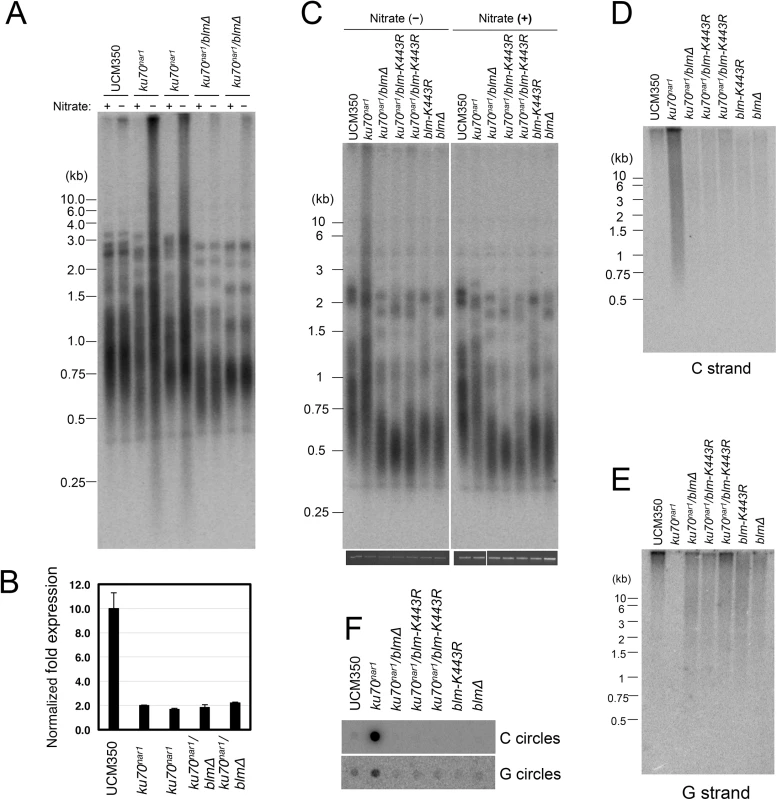
(A) DNAs were isolated from each indicated strain after 20 hrs of growth in YPD, digested with PstI, and subjected to telomere Southern analysis. (B) The levels of ku70 transcripts in the indicated strains after 20 hrs of growth in YPD were measured qRT-PCR. The levels of ku70 was normalized to those of tubulin. n = 3 per genotype. (C) DNAs were isolated from each indicated strain after 20 hrs of growth in MMD or YPD, digested with PstI, and subjected to telomere Southern analysis. The total chromosomal DNA levels in the samples were analyzed by gel electrophoresis and EtBr staining prior to PstI digestion to ensure the use of comparable amounts of DNAs (bottom strip). (D) The same set of PstI-treated DNAs as in (C) was subjected to in-gel hybridization analysis to measure the levels of unpaired C-strand. (E) Same as (D) except that the levels of unpaired G-strand were analyzed. (F) The DNAs from the indicated strains were assayed for the levels of C- and G-circles. It is worth noting that the blm null and blm-K443R mutations alone caused significant telomere shortening (Fig 8A and 8C), suggesting that the helicase helps to maintain telomeres in telomerase-positive U. maydis. While the mechanistic basis for this phenomenon remains to be worked out, we speculate based on studies with mammalian BLM that the U. maydis Blm may play a role in facilitating telomere replication [32]. Failure to fully replicate the telomere tracts, if not adequately compensated by telomerase-mediated extension, may lead to the shortening of average telomere length.
Discussion
The telomere phenotypes of the U. maydis ku mutants described in this report bear remarkable resemblance to those in the human ALT cancer cells, making these mutants valuable tools for understanding the DNA processing reactions that underlie ALT induction and maintenance. Below we highlight the unique features of the current model that are not present in previously characterized yeast survivors. We also discuss the potential mechanisms of ALT in light of the central roles of Ku, MRN, and Blm in our model (see Fig 9 for a model of the U. maydis ALT pathway).
Fig. 9. Model for the ALT-like pathway in the U. maydis ku mutants. 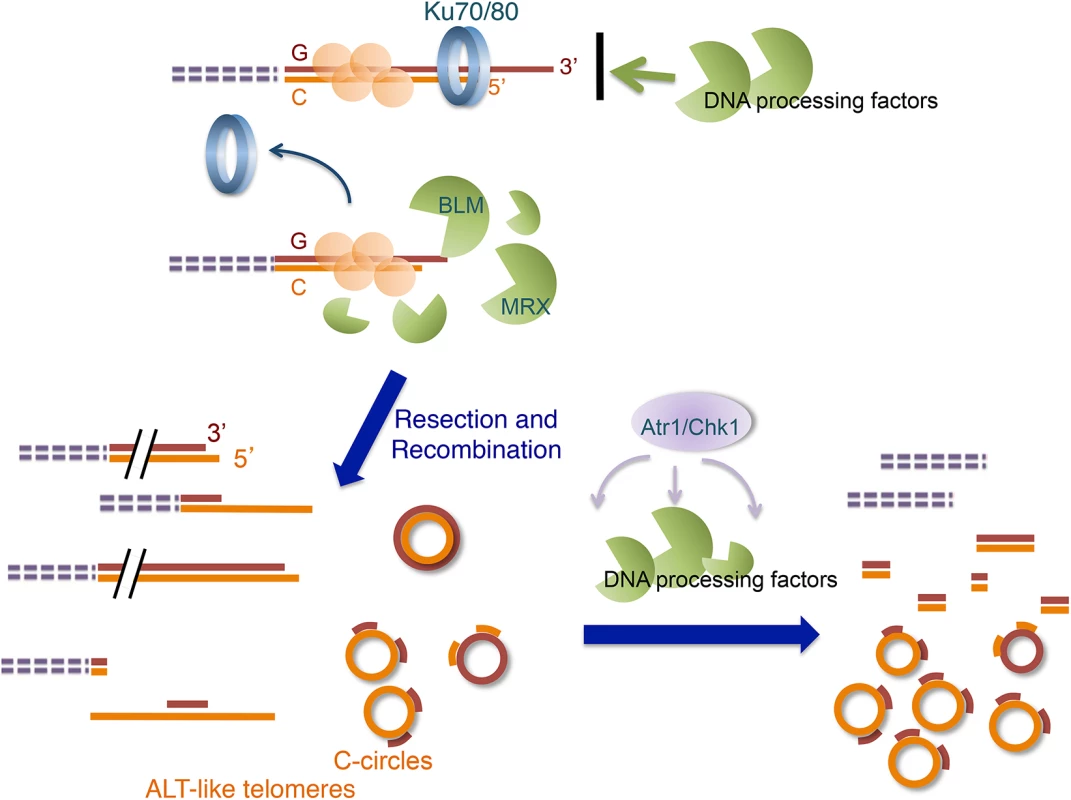
The ALT-like pathway is normally restrained by the Ku70/Ku80 heterodimers at telomeres. When the Ku complex is lost from chromosome ends, the activities of multiple DNA processing factors (especially those of Blm and the MRN complex) become mis-regulated, resulting in aberrant resection and recombination, which in turn produce ALT-like telomeres. Subsequently, the activation of the checkpoint pathway alters the activities of the processing factors and perhaps recruits other proteins to telomeres, leading eventually to the terminal telomere phenotypes. Similarities of the U. maydis ku mutants to ALT cancer cells
The telomeres of ALT cancer cells are characterized by complex aberrations including extreme telomere length heterogeneity, accumulation of unpaired telomere C-strand, and substantial increase in extra-chromosomal telomere repeats (ECTR). Recent studies have accentuated the utility of C-circles as a specific and quantifiable marker of ALT [10]. The abundance of C-circles is reported to be 100 times that of the complementary G-circle, highlighting the specific nature of the former abnormality [10]. Notably, all of the ALT telomere features, including the drastic and specific elevation of C-circles, are recapitulated in the U. maydis ku mutants. The resemblance of the telomere phenotypes of the Ustilago mutants to those of ALT cancer cells clearly distinguishes the current model from all previously characterized fungal “survivors” that employ recombination-based telomere maintenance mechanisms. For example, none of the budding or fission yeast survivors have been reported to accumulate preferentially C-strand overhangs or C-circles, even though many do manifest highly heterogeneous telomere lengths [18].
With respect to the factors involved in the generation and processing of ALT telomeres, the U. maydis model likewise exhibits genetic requirements that are consistent with previous studies of ALT cells. For example, a previous report indicates that RAD17 and the 9-1-1 complex are localized to APB and may be involved in ALT telomere metabolism [33]. In support of this notion, we found that deletion of the U. maydis 9-1-1 subunits exacerbated the telomere length heterogeneity of the ku mutant. In addition, despite the central role of RAD51 in standard homologous recombination, previous studies indicate that this protein is not required for ALT [10,14]. Interestingly, we found that deletion of U. maydis rad51 reduced the formation of C-circles, but increased the levels of extra-long telomeres, suggesting that while Rad51 is not essential for ALT induction, it may act in intermediate steps of the complex ALT pathway to influence the levels of various products. Finally, analysis of ALT cancer cells strongly suggests that MRN and BLM complex are both required for telomere recombination in these cells [10,13,34,35]. Similarly, we found that U. maydis mre11 and blm deletion dramatically reduced all of the telomere abnormalities associated with the ku mutants, implying a key role for these proteins. Importantly, some residual defects can still be detected in the ku mre11 double mutants, implying the existence of an alternative, albeit minor pathway for generating abnormal telomeres.
Similarities between the U. maydis and human Ku mutants and the action of checkpoint proteins in these mutants
While we have shown earlier that the U. maydis and human Ku proteins share the property of being essential for viability and for telomere protection, the telomere phenotypes of the human and U. maydis mutants do not appear to be the same. The human KU mutant was reported to suffer massive deletion through excision of t-circles, resulting in signal-free ends (i.e., chromosome ends that are free of telomere FISH signals) [25]. In contrast, we reported previously that the U. maydis ku70nar1 mutant, while possessing numerous t-circles, also accumulates long and heterogeneous telomeres [29]. However, as reported in the current work, these constitute an initial phenotype that is eventually superseded by the terminal phenotype characterized by the loss of telomere repeats from chromosome ends and appearance of abundant vsECTRs (Fig 9). It is unclear how such high levels of ECTRs can be generated in the mutant. Perhaps ECTRs can be replicated autonomously through an aberrant reaction. Alternatively, long telomere repeat tracts may first be added to chromosome ends (e.g., through rolling circle replication of telomere circles) and then released from the ends through cleavage or recombination. Regardless of the precise mechanisms, our observation indicates that the terminal phenotype of the ku70nar1 mutant exhibits greater resemblance to the human KU mutant than previously realized, further highlighting the relevance of the U. maydis model.
A surprising observation is that the terminal telomere phenotypes of the ku70nar1 mutant do not materialize in the absence of chk1 or atr1. Checkpoint proteins may thus render the ku70nar1 mutant non-viable not only by signaling DNA damage, but also by contributing to the excision of telomere repeats from chromosome ends, and the accumulation of vsECTRs (Fig 7). Flynn et al. recently showed that an ATR inhibitor selectively reduced the proliferation of ALT cancer cells in comparison with telomerase-positive cells [36], suggesting that ATR can confer benefits to ALT cells. While this result may reflect a significant difference between the roles of checkpoint proteins in ALT cancer cells and the U. maydis model, it is worth noting that the U. maydis ku70nar1 atr1 mutant also grows substantially slower than the wild type strain and slower than the ku70nar1 chk1 mutant [24]. Perhaps Atr1 in U. maydis can affect ku70 mutant growth both positively and negatively through multiple pathways. More studies will be necessary to determine the detailed mechanisms of Atr1 in Ku-deficient U. maydis and to assess the extent of similarity between the fungal model and ALT cancer cells.
Similarities between ALT and DSB resection
Interestingly, the production of ALT phenotypes appears to share many features of DSB resection (DSBR). For example, the antagonism of the Ku and MRN complex in the production of ALT telomeres is quite reminiscent of the interplay between the two complexes in DSBR [30]. Specifically, the radiation sensitivity defect of S. cerevisiae mre11∆ can be suppressed by eliminating Ku, suggesting that Ku antagonizes the function of Mre11 in DSB repair. Another parallel between ALT and DSBR concerns the Mre11 nuclease activity, which is not essential for either pathway [37]. In DBSR, the main function of the MRN complex is to recruit other resection factors such as Exo1, Dna2, and Blm. This may also be the function of MRN in ALT [38]. An implication of these similarities is that similar “downstream” factors may be responsible for both DSBR and ALT telomere production. A significant conundrum for this hypothesis is presented by the distinct overhangs produced by ALT and DSBR; the former generates primarily 5’ and the latter 3’ overhangs. One way to rationalize the distinct overhangs is to postulate that the specific nucleoprotein structure of telomeres or telomere factors alters the activity of the resection factors. For example, the intrinsic nuclease activity of Dna2 can act on ssDNA with a free 5’ or 3’ end, but in the presence of RPA, the activity on the free 3’ end is repressed, resulting in the production of 3’ overhangs in DSBR [39,40]. It is conceivable that Dna2 and other potential resection factors may act differently on telomere repeats because of the presence of specific telomere DNA-binding proteins. Such a model would have to be addressed by a combination of biochemical assays in vitro and genetic analysis in vivo.
In summary, we have shown that elimination of ku in U. maydis leads to an attractive model for ALT. The two special strengths of the model are (i) its reversibility and accurate recapitulation of many characteristic features of ALT telomeres, and (ii) its ready amenability to molecular genetic manipulations, including the construction of null mutants to access the role of putative ALT factors. In contrast to previous knockdown studies in ALT cell lines or ALT models, the current use of null mutants reveals complete (or nearly complete) suppression of this pathway with the loss of Mre11 and Blm, implicating their products as prime targets for inhibitor development. Continued analysis of this model should provide mechanistic insights on the complex reactions that mediate the production of ALT telomeres, and suggest useful strategies for therapeutic intervention.
Methods
Ustilago maydis strains and growth conditions
U. maydis strains were derived from the FB1or UCM350 genetic background [41,42] and are listed in S1 Table. Cells were grown in YPD (restrictive medium), which represses the nar1 promoter, or MMD (permissive medium), which induces the nar1 promoter [43]. Additional nutritional supplements (0.2 μg/ml of pantothenic acid and 50 mM of proline) were added in MMD (MMD+) for UCM350-drived cells. Controlled expression of ku genes under the nar1 promoter was performed as described previously [21,44]. Null mutants were constructed by replacing the entire open reading frames with cassettes expressing resistance to hygromycin (HygR), nourseothricin (NatR), or geneticin (G418R) [45,46]. Disruption cassettes for ctip and dna2 null strains were freshly generated for this study using Gibson Assembly (NEB) according to the manufacturer’s instructions. Briefly, three sets of primers (S2 Table) were used to generate three overlapping fragments: (i) 5’ UTR of dna2 (or ctip) linked to 5’ end of the hygromycin ORF, (ii) the complete hygromycin ORF flanked by short segments from the 5’ and 3’UTR of dna2 (or ctip), and (iii) 3’ end of the hygromycin ORF linked to 3’ UTR of dna2 (or ctip). Genomic DNAs and a plasmid for hygromycin resistance cassettes were used as the source of template for PCR. The PCR fragments were used as starting substrates for the Gibson Assembly reaction, and the full-length reaction products were cloned into a vector for further application.
Growth of U. maydis strains for telomere analysis
U. maydis strains were grown for ~ 36 hr in MMD and diluted into fresh YPD (at OD600 of 0.005 ~ 0.01) to repress the expression of Ku proteins, or into fresh MMD (at OD600 of 0.04) as controls. Typically, after growth for 18~20 hrs, the cultures were collected for DNA isolation. For time course experiments, the cultures were grown in YPD for varying durations (6, 12, 18, 24, and 30 hr), and then harvested for DNA isolation. For de-repression studies, cells were first grown for 18 hr in YPD to repress Ku expression, and then transferred into fresh MMD medium. After varying durations (0, 24, 48, 72, and 96 hr), the cultures were harvested, and their DNAs isolated for telomere analysis.
Growth analysis
Serial ten-fold dilutions samples of U. maydis cultures (containing 104, 103, 102, and 10 cells) were applied to agar plates containing YPD, MMD, or MMD supplemented with pantothenic acid and proline (MMD+). The YPD plates were incubated at 30°C for 2 days, and the MMD and MMD+ plates for 3 days, and then photographed.
Telomere length analyses
Standard telomere Southern analysis was performed as described previously with some modifications [21,47]. Briefly, chromosomal DNAs were digested with Pst I (or without Pst I) and fractionated in 1.2% agarose gels. Following transfer to nylon membranes, the telomere restriction fragments were detected using 32P-labeled UmC8 or UmG8 probe that corresponds to eight copies of the Ustilago C-strand or G-strand telomere repeat. Signals obtained by scanning the Phosphor plates were quantified using ImageQuant software (Molecular Dynamics Inc.). For this analysis as well as for other telomere assays, we examined the telomeres of at least two independent clones of the same genotype, and the results are quite reproducible.
G - and C-strand analysis
The standard in-gel hybridization analysis was performed using a combination of established protocols for U. maydis [21] with minor modification. The labeled UmG4 and UmC4 oligonucleotides (S2 Table), corresponding to four copies of the U. maydis telomeric G-strand and C-strand repeats, were used as the probes, and hybridization was performed in the Church Mix at 45°C. For more detailed analysis of single - and double-stranded telomeric DNA in the ku mutants, two-dimensional gel electrophoresis was performed in a combination with in-gel hybridization assay. The samples applied to the 2-D gels include both PstI-digested genomic DNA and labeled double stranded DNA ladders (used as markers for ds DNA arc). In some experiments, chromosomal DNAs were treated either Exonuclease I (2.5 U/μg) or RecJf (4 U/μg) prior to PstI digestion. The first dimension (0.5% agarose) was run at 0.5 V/cm for 16 h in the absence of ethidium bromide (EtBr). The gel was stained with 0.3 μg/ml EtBr to visualize the size standards and bulk chromosomal DNA. Gel strips containing DNA in the 0.5 kb to 15 kb size range are excised and impregnated in a 1.2% agarose gel containing 0.3 μg/ml EtBr. Electrophoresis was then performed in the orthogonal direction at 5 V/cm for 5 h. The gel was further processed according to the standard in-gel hybridization analysis as described above.
C-circle and G-circle assays
The C-circle assay was carried out by the rolling circle amplification method as described previously [10] with some modificatons. Genomic DNAs (10 ng for C-circles and 20 ng for G-circles) were combined with 10 μl 0.2 mg/ml BSA, 0.1% Tween, 1 mM each dATP, dGTP and dTTP, 1× Φ29 Buffer and 5 U Φ29 DNA polymerase (NEB) and incubated at 30°C for 16 h. Φ29 DNA polymerase was then inactivated at 65°C for 20 min. For quantification, the reaction products were dot-blotted onto a 2× SSC-soaked nylon membrane and the DNAs were UV-cross-linked onto the membrane. G-strand products of C-circle assays were hybridized at 45°C with 32P-labeled UmC8 probe (S2 Table). The membranes were further processed according to the standard telomere Southern protocol as described above. Signals obtained by scanning the Phosphor plates were quantified using ImageQuant software (Molecular Dynamics Inc.). The G-circle assay was carried out in the same way except that dATP, dCTP and dTTP were used for polymerization and the reaction products were probed with labeled UmG8 (S2 Table).
In some experiments, chromosomal DNAs were first treated with exonuclease λ (2.5 U/μg) or exonuclease I (2.5 U/μg) to eliminate linear DNAs, and then subjected to rolling circle amplification. The linearity of the C-circle and G-circle assays was also confirmed with serial dilutions of genomic DNA. However, we do not know if the two types of circles are amplified with identical efficiency by the Φ29 DNA polymerase.
Supporting Information
Zdroje
1. de Lange T (2009) How telomeres solve the end-protection problem. Science 326 : 948–952. doi: 10.1126/science.1170633 19965504
2. Jain D, Cooper JP (2011) Telomeric strategies: means to an end. Annu Rev Genet 44 : 243–269.
3. Harley C, Futcher A, Greider C (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345: p458–460.
4. d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, et al. (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426 : 194–198. 14608368
5. Autexier C, Lue NF (2006) The Structure And Function of Telomerase Reverse Transcriptase. Annu Rev Biochem 75 : 493–517. 16756500
6. Blackburn EH, Collins K (2011) Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3.
7. Buseman CM, Wright WE, Shay JW (2012) Is telomerase a viable target in cancer? Mutat Res 730 : 90–97. doi: 10.1016/j.mrfmmm.2011.07.006 21802433
8. Cesare AJ, Reddel RR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11 : 319–330. doi: 10.1038/nrg2763 20351727
9. Shay JW, Reddel RR, Wright WE (2012) Cancer. Cancer and telomeres—an ALTernative to telomerase. Science 336 : 1388–1390. doi: 10.1126/science.1222394 22700908
10. Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, et al. (2009) DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol 27 : 1181–1185. doi: 10.1038/nbt.1587 19935656
11. Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, et al. (2012) Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 8: e1002772. 22829774
12. Nabetani A, Ishikawa F (2011) Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem 149 : 5–14. doi: 10.1093/jb/mvq119 20937668
13. Zhong ZH, Jiang WQ, Cesare AJ, Neumann AA, Wadhwa R, et al. (2007) Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem 282 : 29314–29322. 17693401
14. Potts PR, Yu H (2007) The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol 14 : 581–590. 17589526
15. Oganesian L, Karlseder J (2011) Mammalian 5' C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell 42 : 224–236. doi: 10.1016/j.molcel.2011.03.015 21504833
16. Clynes D, Jelinska C, Xella B, Ayyub H, Scott C, et al. (2015) Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat Commun 6 : 7538. doi: 10.1038/ncomms8538 26143912
17. Hu J, Hwang SS, Liesa M, Gan B, Sahin E, et al. (2012) Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148 : 651–663. doi: 10.1016/j.cell.2011.12.028 22341440
18. McEachern MJ, Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75 : 111–135. 16756487
19. Teng S, Chang J, McCowan B, Zakian V (2000) Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell 6: p947–952.
20. Guzman PA, Sanchez JG (1994) Characterization of telomeric regions from Ustilago maydis. Microbiology 140 (Pt 3): 551–557.
21. Yu EY, Kojic M, Holloman WK, Lue NF (2013) Brh2 and Rad51 promote telomere maintenance in Ustilago maydis, a new model system of DNA repair proteins at telomeres. DNA Repair (Amst) 12 : 472–479.
22. Holloman WK (2011) Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 18 : 748–754. doi: 10.1038/nsmb.2096 21731065
23. Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, et al. (2010) BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17 : 1461–1469. doi: 10.1038/nsmb.1943 21076401
24. de Sena-Tomas C, Yu EY, Calzada A, Holloman WK, Lue NF, et al. (2015) Fungal Ku prevents permanent cell cycle arrest by suppressing DNA damage signaling at telomeres. Nucleic Acids Res 43 : 2138–2151. doi: 10.1093/nar/gkv082 25653166
25. Wang Y, Ghosh G, Hendrickson EA (2009) Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A 106 : 12430–12435. doi: 10.1073/pnas.0903362106 19581589
26. Fisher TS, Zakian VA (2005) Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 4 : 1215–1226.
27. Downs JA, Jackson SP (2004) A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol 5 : 367–378. 15122350
28. Bautista-Espana D, Anastacio-Marcelino E, Horta-Valerdi G, Celestino-Montes A, Kojic M, et al. (2014) The telomerase reverse transcriptase subunit from the dimorphic fungus Ustilago maydis. PLoS One 9: e109981. doi: 10.1371/journal.pone.0109981 25299159
29. de Sena-Tomas C, Yu EY, Calzada A, Holloman WK, Lue NF, et al. (2015) Fungal Ku prevents permanent cell cycle arrest by suppressing DNA damage signaling at telomeres. Nucleic Acids Res.
30. Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45 : 247–271. doi: 10.1146/annurev-genet-110410-132435 21910633
31. Mao N, Kojic M, Holloman WK (2009) Role of Blm and collaborating factors in recombination and survival following replication stress in Ustilago maydis. DNA Repair (Amst) 8 : 752–759.
32. Zimmermann M, Kibe T, Kabir S, de Lange T (2014) TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev 28 : 2477–2491. doi: 10.1101/gad.251611.114 25344324
33. Nabetani A, Yokoyama O, Ishikawa F (2004) Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J Biol Chem 279 : 25849–25857. 15075340
34. Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, et al. (2002) The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet 11 : 3135–3144. 12444098
35. O'Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, et al. (2014) Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol 21 : 167–174. doi: 10.1038/nsmb.2754 24413054
36. Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, et al. (2015) Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 347 : 273–277. doi: 10.1126/science.1257216 25593184
37. Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24 : 9682–9694. 15485933
38. Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, et al. (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29 : 3370–3380. doi: 10.1038/emboj.2010.219 20834227
39. Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, et al. (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25 : 350–362. doi: 10.1101/gad.2003811 21325134
40. Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, et al. (2010) DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467 : 112–116. doi: 10.1038/nature09355 20811461
41. Banuett F, Herskowitz I (1989) Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci U S A 86 : 5878–5882. 16594058
42. Kojic M, Kostrub CF, Buchman AR, Holloman WK (2002) BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell 10 : 683–691. 12408834
43. Holliday R (1974) Ustilago maydis. In: King, editor. Handbook of Genetics. New York: Plenum Press. pp. 575–595.
44. Brachmann A, Weinzierl G, Kamper J, Kahmann R (2001) Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol 42 : 1047–1063. 11737646
45. Brachmann A, Konig J, Julius C, Feldbrugge M (2004) A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Genet Genomics 272 : 216–226. 15316769
46. Kamper J (2004) A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol Genet Genomics 271 : 103–110. 14673645
47. Yu EY, Wang F, Lei M, Lue NF (2008) A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol 15 : 985–989. 19172753
Štítky
Genetika Reprodukčná medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



