-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
Dimorphic fungal pathogens including Blastomyces are the cause of major fungal diseases in North and South America. The genus Emmonsia includes species infecting small mammals as well as a newly emerging pathogenic species recently reported in HIV-positive patients in South Africa. Here, we synthesize both genome sequencing of four isolates of Blastomyces and two species of Emmonsia as well as deep sequencing of Blastomyces RNA to draw major new insights into the evolution of this group and the pathogen response to infection. We investigate the trajectory of genome evolution of this group, characterizing the phylogenetic relationships of these species, a remarkable genome expansion that formed large isochore-like regions of low GC content in Blastomyces, and variation of gene content, related to host interaction, among the dimorphic fungal pathogens. Using RNA-Seq, we profile the response of Blastomyces to macrophage and mouse pulmonary infection, identifying key pathways and novel virulence factors. The identification of key fungal genes involved in adaptation to the host suggests targets for further study and therapeutic intervention in Blastomyces and related dimorphic fungal pathogens.
Published in the journal: The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative. PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005493
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005493Summary
Dimorphic fungal pathogens including Blastomyces are the cause of major fungal diseases in North and South America. The genus Emmonsia includes species infecting small mammals as well as a newly emerging pathogenic species recently reported in HIV-positive patients in South Africa. Here, we synthesize both genome sequencing of four isolates of Blastomyces and two species of Emmonsia as well as deep sequencing of Blastomyces RNA to draw major new insights into the evolution of this group and the pathogen response to infection. We investigate the trajectory of genome evolution of this group, characterizing the phylogenetic relationships of these species, a remarkable genome expansion that formed large isochore-like regions of low GC content in Blastomyces, and variation of gene content, related to host interaction, among the dimorphic fungal pathogens. Using RNA-Seq, we profile the response of Blastomyces to macrophage and mouse pulmonary infection, identifying key pathways and novel virulence factors. The identification of key fungal genes involved in adaptation to the host suggests targets for further study and therapeutic intervention in Blastomyces and related dimorphic fungal pathogens.
Introduction
Blastomyces is a genus of a thermally dimorphic fungal pathogen, which is the etiological agent of blastomycosis, a lung infection that can become a systemic mycosis. In North America, Blastomyces is endemic in the Ohio and Mississippi river valleys, the Great Lakes region, and the St. Lawrence River [1]. Within Blastomyces, two lineages of B. dermatitidis have been recognized [2], with recent work providing evidence that one lineage is a distinct species, B. gilchristii [3]. Both species can infect humans, and vary in morphology, virulence and immune responses by the host. The primary mode of infection is inhalation of conidia and the subsequent conversion of these conidia into parasitic yeast [4,5]. Clinical manifestations range from asymptomatic infection to symptomatic disease and include pneumonia, acute respiratory distress syndrome, and a rapidly progressive dissemination involving multiple organ systems that is often fatal [5,6]. Diagnosis is often complicated by the similarity of symptoms to those of viral or bacterial respiratory infection and by the aforementioned variety of manifestations [7].
As a thermally dimorphic fungus, Blastomyces has the remarkable ability to switch between two different morphologies in response to external stimuli, predominantly temperature [5]. At 22–25°C, Blastomyces grows as septate hyphae that produce infectious conidia and at 37°C it grows as a budding yeast [8]. Blastomyces is part of a larger group of dimorphic fungal pathogens, including Histoplasma, Paracoccidioides, and Coccidioides, all belonging to the order Onygenales. The dimorphic fungi collectively are the most common cause of invasive fungal disease worldwide and account for several million infections each year [8]. Unlike opportunistic fungi, such as Candida albicans, Cryptococcus neoformans, or Aspergillus fumigatus, the dimorphic fungi can infect immunocompetent and immunocompromised hosts [6,9–11].
Previous work has shown that in Blastomyces, the temperature-dependent switch from hyphae to yeast along with upregulation of yeast-phase specific genes is critical for virulence [12–14]. The dimorphism-regulating kinase-1 (DRK1) promotes the temperature-dependent conversion from mold to yeast, and its deletion renders Blastomyces avirulent during experimental murine pulmonary infection [12]. The upregulation of yeast-phase specific genes, such as the Blastomyces yeast-phase specific gene 1 (BYS1) [15] and the Blastomyces adhesion-1 gene (BAD1) [13,14], is also important for the adaptive response of the yeast cells in the host environment. BAD1 is considered an essential virulence factor in Blastomyces, since it binds yeast cells to host tissue and impairs host immune defenses by inhibiting the production of tumor necrosis factor-α and blocking CD4+ T lymphocyte activation [13].
Within the Onygenales, Blastomyces, Histoplasma and Paracoccidioides belong to the family Ajellomycetaceae. Also within Ajellomycetaceae is the genus Emmonsia, which includes E. crescens and E. parva, the etiological agents of adiaspiromycosis, a pulmonary disease of small mammals and occasionally of humans [16]. Recently, a cluster of systemic infections of HIV-positive patients in South Africa were shown to be caused by Emmonsia isolates [17]. While E. crescens and E. parva also undergo a dimorphic shift at high temperature, they transform into large, thick-walled adiaspores rather than yeast cells [18] (S1 Table). Two phylogenetic studies using 18S ribosomal DNA sequences found that E. parva was the sister species to Blastomyces [19,20]. The positioning of E. crescens was less clear; in one analysis it was a sister group to Paracoccidioides [19] while in the other analysis it was grouped with Blastomyces and E. parva [20]. In neither phylogeny was the alternative positioning of E. crescens strongly supported.
To further investigate the genomic basis of differences observed among the Ajellomycetaceae in terms of pathogenicity, morphology, and the infection process, we sequenced six genomes of Blastomyces and Emmonsia, as well as sequencing the B. dermatitidis transcriptome during macrophage co-cultivation and in vivo pulmonary infection. The newly sequenced genomes included three representative strains of B. dermatitidis (ER-3, ATCC18188, and ATCC26199), and one strain of each of B. gilchristii (SLH14081), E. parva (UAMH139), and E. crescens (UAMH3008). Blastomyces dermatitidis ER-3 was isolated from a woodpile located in a highly endemic region of Wisconsin and is hypovirulent in mice [21,22]. The ATCC18188 strain is the only current example of the 'a' mating type (MAT1-1 locus) available for B. dermatitidis [23]. ATCC26199 is a clinical isolate from South Carolina that is commonly used for in vitro and in vivo laboratory assays [14]. Blastomyces gilchristii SLH14081 is a human clinical isolate that is highly virulent in a murine model of blastomycosis [22,24]. Both Emmonsia strains were isolated from small mammals, E. parva from a weasel in Ravelli County, Montana, and E. crescens from lungs of a rodent (Arvicola terrestris) in Norway.
Utilizing this genomic data, we find that the Blastomyces genomes are much larger than those of their close relatives, and are characterized by large, isochore-like GC-poor regions overrun by repetitive elements. Our whole-genome analyses provide further evidence for the phylogenetic relationships between Blastomyces and Emmonsia and other Onygenales. Finally, we identify novel sets of candidate virulence factors through comparison of the Blastomyces transcription during in vivo pulmonary infection to growth in co-culture with macrophages or in different media or temperature. This combination of genomic and transcriptomic analysis provides a foundation and new candidate genes to further characterize the underlying molecular differences that determine the infectious potency of Blastomyces strains and give rise to the clinical profiles attributable to blastomycosis.
Results
Expanded genomes of Blastomyces species
We sequenced and assembled the genomes of three Blastomyces dermatitidis strains and one B. gilchristii strain, and representatives of two Emmonsia species. The Blastomyces strains were sequenced using either Sanger technology or a hybrid of Sanger and 454 technologies. The Emmonsia strains were sequenced using Illumina technology, and de novo assemblies were generated for each strain (Methods). Comparison of the genomes of four Blastomyces strains, SLH14081, ER-3, ATCC18188 and ATCC26199, revealed they were over twice the size of all other Onygenales. The Blastomyces assemblies range in size from 66.6 Mb for B. dermatitidis strain ER-3 to 75.4 Mb B. gilchristii strain SLH14081 (Table 1). These assemblies were over twice as large as those of other dimorphic pathogens in the order Onygenales including the Emmonsia species (30.4 Mb), although the use of only short reads from a single library for the two Emmonsia may under-represent repetitive sequence (Fig 1). The assemblies of two Blastomyces strains, SLH14081 and ER-3, were sequenced to a higher depth than the other two strains, and as a result contain nearly all of the assembled sequence in a relatively small number of scaffolds, 100 and 25 scaffolds respectively. As an independent assessment of genome size and structure, we generated an optical map of the SLH14081 strain (S1 Fig). Consistent with our assembly of this strain, the map had an estimated size of 79.6 Mb, arranged in eighteen linkage groups. In addition, a total of 65.9 Mb of the 75.4 Mb of the SLH14081 assembly was anchored to the optical map (S2 Table).
Tab. 1. Assembly and annotation statistics for Blastomyces and Emmonsia genomes. 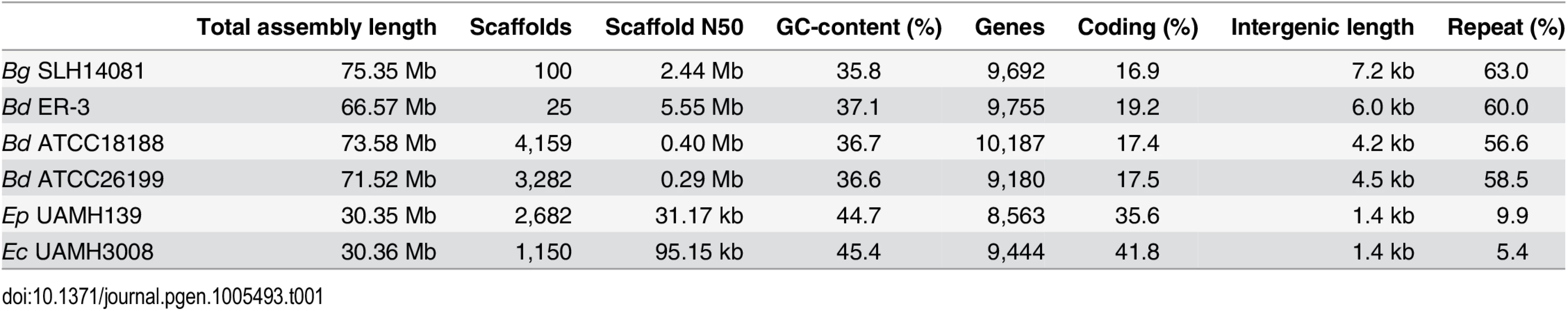
Bd: B. dermatitidis, Bg: B. gilchristii, Ep: E. parva, Ec: E. crescens. Fig. 1. Phylogeny and gene conservation of Blastomyces and Emmonsia spp. 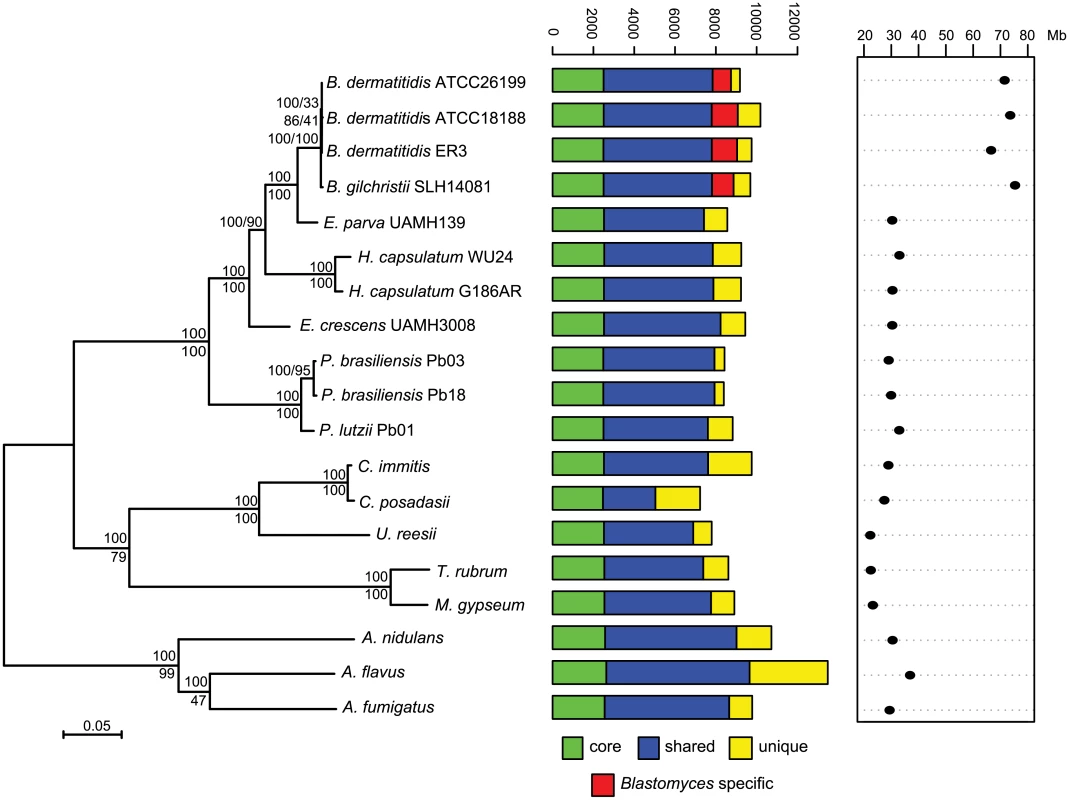
Maximum likelihood tree inferred from concatenated protein alignments of 2,062 core genes based on 1,000 replicates; all bootstrap values (top value for each node) were 100% except for one node within B. dermatitidis, which was 88%. Branch order was also well supported by the consensus of individual gene trees (GSF, lower value for each node). A bar plot of orthology classes is shown to the right, where core genes found in all genomes are shown in green, shared genes present in more than one but not all genomes in blue, genes specific to Blastomyces genomes in red, and genes that were unique to only one of the 19 genomes in yellow. Finally, genome size is plotted for each genome along the x-axis, which ranges from 20 to 80 Mb. The total number of predicted genes in Blastomyces, Emmonsia, and other related fungi was similar despite the large difference in genome size. In Blastomyces, the number of predicted genes varied between 9,180 in ATCC26199 to 10,187 in ATCC18188; for E. parva and E. crescens the counts were similar, 8,563 and 9,444, respectively (Table 1), as were those of other sequenced Onygenales (Fig 1). High representation of core eukaryotic genes in each genome provides evidence that their gene sets are nearly complete; E. parva includes 88% of core eukaryotic genes, while the E. crescens and Blastomyces gene sets include 96–98% (S2 Fig).
Phylogenetic position of Blastomyces, Emmonsia parva and E. crescens
To compare gene content and conservation, we identified orthologous gene clusters in the six genomes sequenced here, 10 additional Onygenales genomes, including three other pathogenic species (Histoplasma, Paracoccidioides, and Coccidioides), and three Aspergillus genomes. Using 2,062 single copy core genes present in all strains, we estimated a phylogeny of these organisms using RAxML ([25]; Fig 1). This analysis strongly supports the clustering of Blastomyces with E. parva (100% of bootstrap replicates and 100% Gene Support Frequency (GSF) [26]) as previously reported [19,20]. In contrast to prior work, Histoplasma is strongly supported as sister group to Blastomyces and E. parva (100% of bootstrap replicates and 90% GSF), with E. crescens strongly supported as a sister group to that clade (100% of bootstrap replicates and 100% GSF), and with Paracoccidioides in a basal position (Fig 1). The polyphyletic nature of Emmonsia suggests that the Ajellomycetaceae have undergone multiple evolutionary transitions allowing the infection of humans and other mammals. Within Blastomyces, we found support for strain SLH14081 as an outgroup relative to the other three strains (S3 Fig). This is consistent with the placement of strain SLH14081 within the newly described species B. gilchristii [3]; the other three strains sequenced here are still classified as B. dermatitidis.
Blastomyces genomes show a bimodal GC distribution
A bimodal distribution of GC-content observed in all Blastomyces sequenced, which was less pronounced in E. parva and E. crescens and absent in other Ajellomycetaceae, suggests that these genomes are organized in large isochore-like regions of high and low GC-content. This finding for nuclear DNA explains the GC-poor fraction of the Blastomyces genome initially identified using CsCl gradient analytical ultracentrifugation [27], which the authors hypothesized was due to a large proportion of GC-poor mitochondrial DNA in Blastomyces cells. Examining the genome wide GC content revealed a bimodal distribution for all strains of Blastomyces including ER-3 and SLH14081, the smallest and largest assembly, respectively (Fig 2), and was observed for all window sizes ranging from 2 kb to 256 kb (S4 Fig). The detection of a bimodal signal in larger windows supports the organization of the genomes in large isochore-like regions, with average GC content of 29.6% and 31.0% in GC-poor regions and 45.9% and 46.6% for the rest of the genome in B. gilchristii strain SLH14081 and B. dermatitidis strain ER-3, respectively (Table 2). Analysis of the related pathogens H. capsulatum, P. lutzii, and C. immitis showed no evidence for bimodality of GC content, while both E. parva and E. crescens revealed small peaks of low GC sequence. Read-based analysis and using smaller window sizes (e.g. 128 bp) supported these findings, suggesting they are not due to differences in assembly completeness (S5 Fig).
Tab. 2. Gene and repeat features of <i>Blastomyces</i> GC-rich and GC-poor regions compared to <i>Histoplasma</i>. 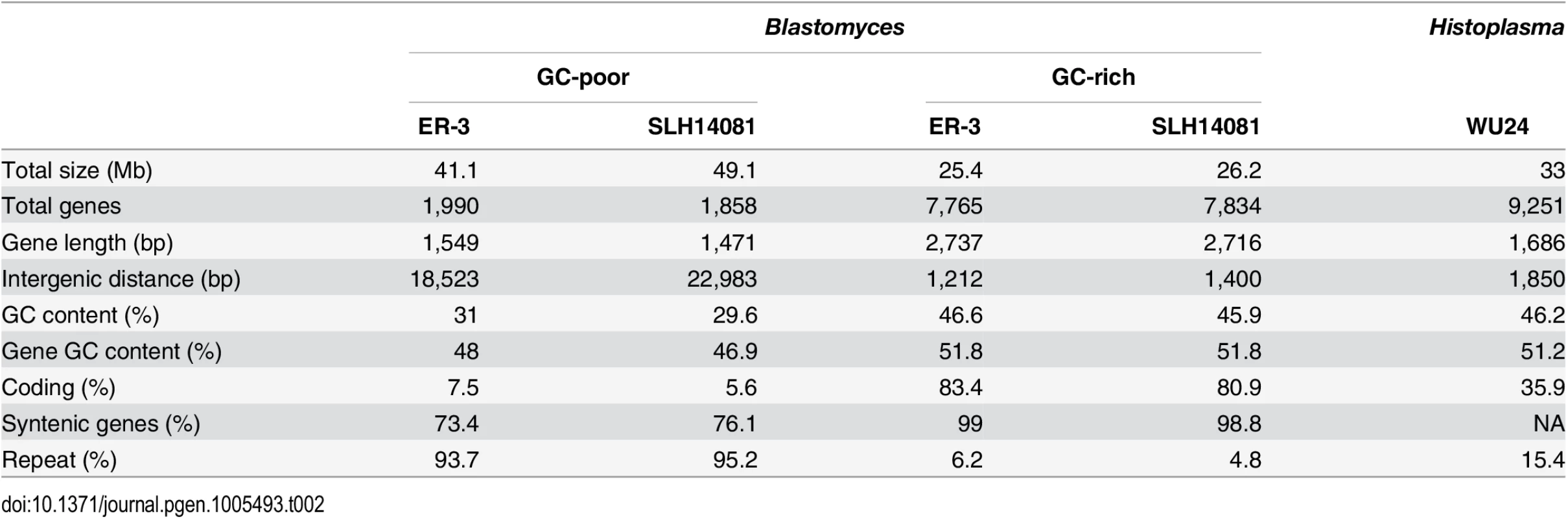
Fig. 2. GC frequency distributions (histograms) of overlapping fragments (windows, of 32 kb) of the genome assemblies of Blastomyces dermatitidis ER-3, B. gilchristii SLH14081, Emmonsia parva (UAMH139), E. crescens (UAMH3008), Histoplasma capsulatum (WU24), Paracoccidioides lutzii (Pb01), Coccidioides immitis (RS3), and Leptosphaeria maculans (v23.1.3). 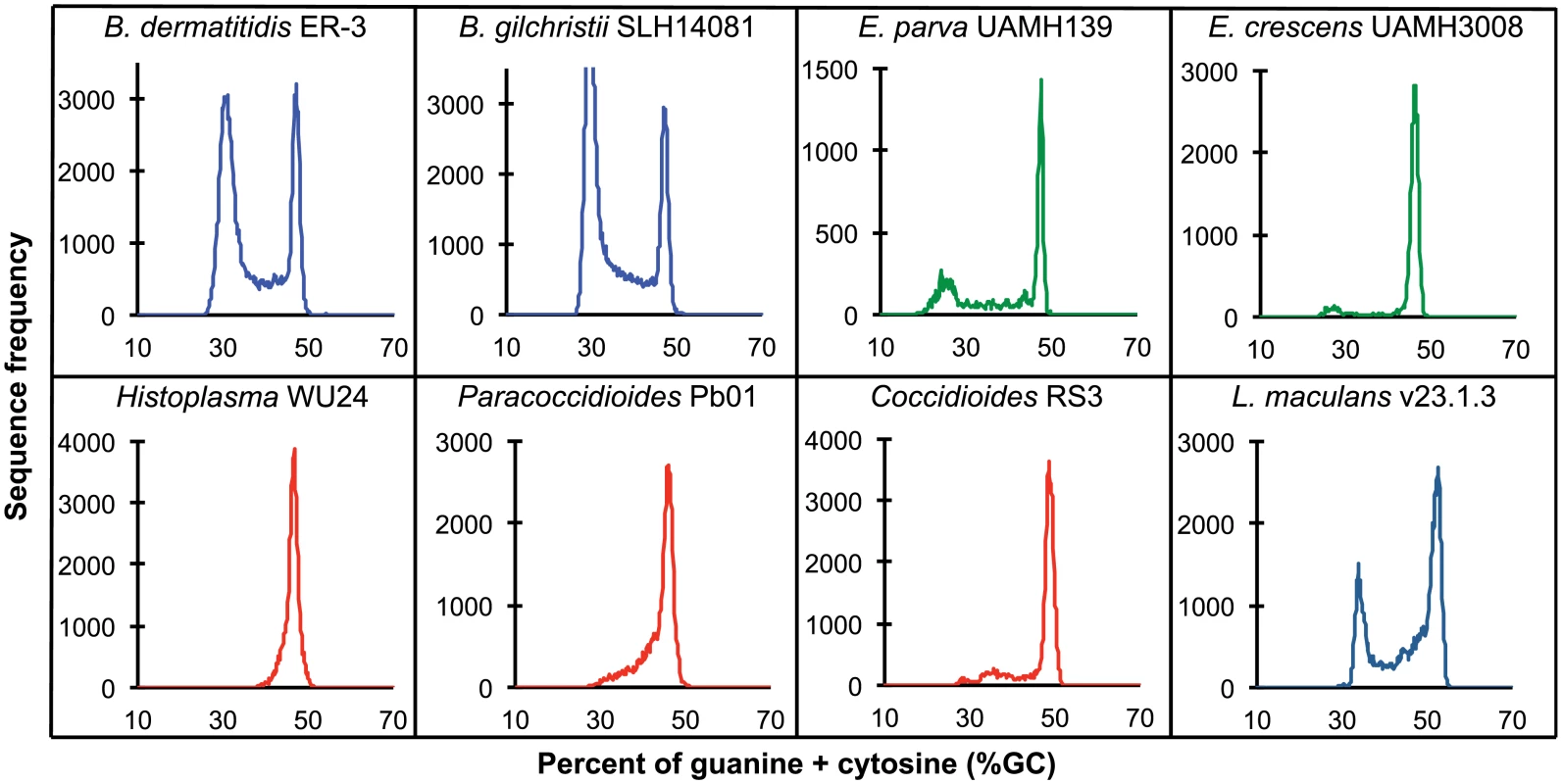
The bin size of the histograms is approximately 0.1% GC. Horizontal axes show GC % and vertical axes show relative frequencies. To further examine the organization of GC-content across the genome, we next defined the boundaries of low GC content regions in Blastomyces. In the smallest assembly, of the ER-3 strain, we identified 221 GC-poor tracts with an average size of 186.0 kb, encompassing a total size of 41.1 Mb (Tables 2 and S3). In the largest assembly, of the SLH14081 strain, we identified 350 GC-poor tracts with an average size of 140.2 kb, encompassing a total size 49.1 Mb (Tables 2 and S3). The 8 Mb difference between the total size of GC-tracts in the genomes of B. dermatitidis ER-3 and B. gilchristii SLH14081 accounts for nearly all of the 8.8 Mb difference in assembly size. Notably, GC-poor tracts in Blastomyces can be quite long, and reach maximal lengths of 1.3 Mb. In the assemblies of E. parva, E. crescens and other Ajellomycetaceae, long GC-poor tracts were rarely observed (e.g., a total of only 4 GC-poor regions larger than 10 kb in E. parva were found adjacent to a long GC-rich region in the same scaffold, and just 1 in E. crescens), corresponding to the less pronounced bimodal GC distribution of the genome assembly. However, more contiguous assemblies would be needed to reveal the overall extent of long GC-poor tracts. The only other fungal genome noted to have an isochore-like structure, Leptosphaeria maculans [28], contains a smaller expansion of GC-poor regions (Fig 2); individual tracts were on average half the size (70.4 kb) of those in Blastomyces, and encompassed a smaller fraction (36%) of the L. maculans genome [28]. This difference is consistent with the lower fraction of long AT blocks we observe by comparing windows of different sizes in Blastomyces and L. maculans (S4 Fig).
The GC-poor regions include nearly all the repetitive elements in the genome and consequently have a lower density of predicted genes (e.g., see Fig 3). In ER-3, 93.7% of repetitive sequence is found in GC-poor regions (Table 2). The gypsy elements that dominate repetitive sequence in the Blastomyces genomes have low GC-content; on average those in ER-3 and SLH14081 have respective GC-content of 31.0% and 29.9%, matching the overall GC level of the GC-poor regions (Table 2). GC-poor tracts of Blastomyces contain only approximately one fifth of the predicted protein-coding gene set, including some notable genes such as 1,3-beta-glucan synthase component (FKS1), Blastomyces yeast phase-specific gene (BYS1), and one of two BYS1-like proteins we identified (S6 Fig and S4 Table). By contrast, BAD1, which encodes an essential virulence factor involved in host cell interaction and immune evasion [13], is found within a GC-rich region. Intergenic regions are also larger here than for other genes in the genome; the average intergenic region for ER-3 is 18.5 kb in GC-poor regions, a 3-fold expansion compared to the 6.0 kb genome-wide average (Table 2 and Figs 3 and S6).
Fig. 3. Correspondence of GC content and synteny for Blastomyces. 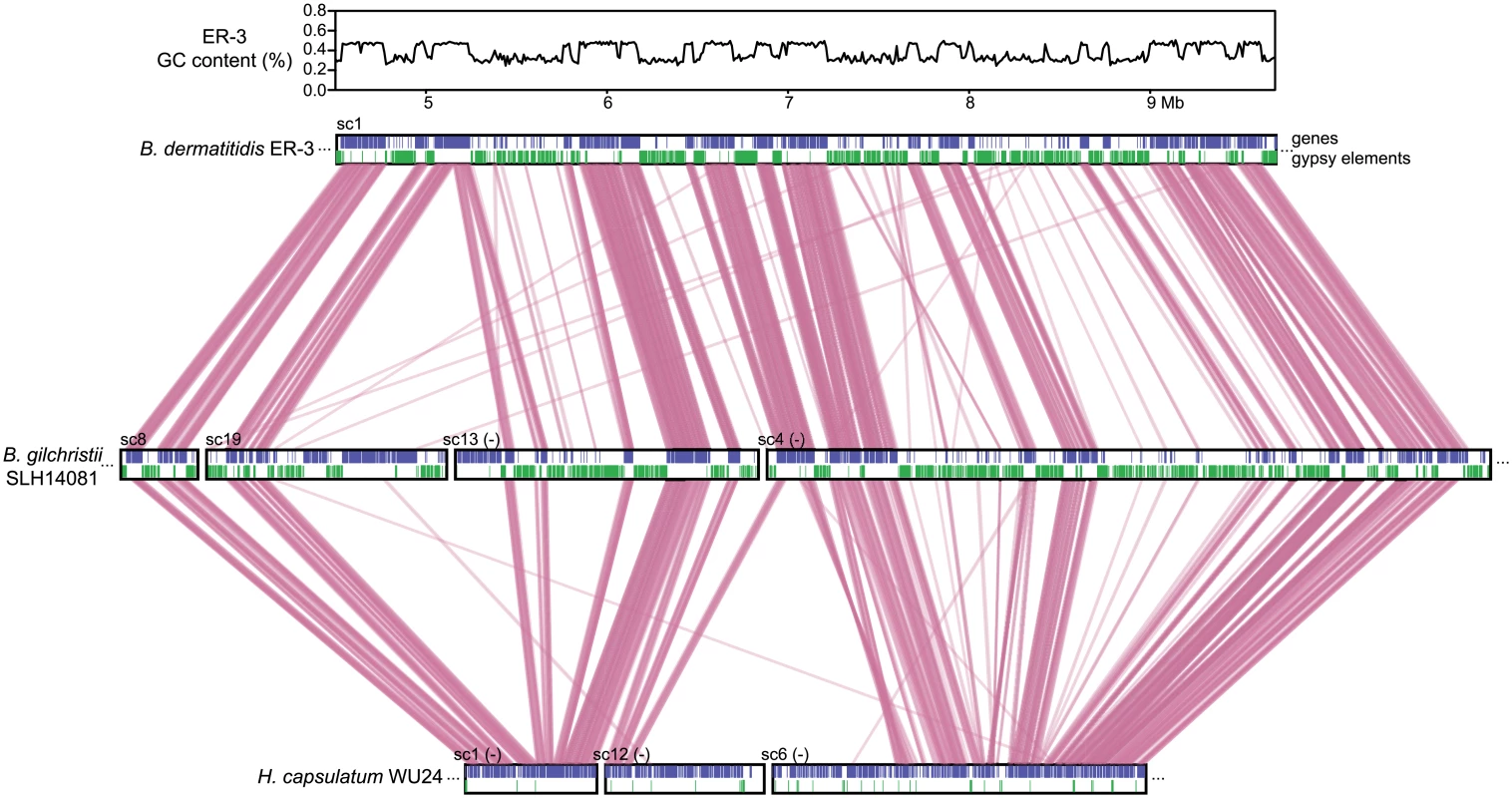
Comparison of GC content (top panel) and genome synteny (lower panel) for a 5.2 Mb region of B. dermatitidis strain ER-3 (scaffold (sc) 1, coordinates from 4.5 to 9.7 Mb) and corresponding syntenic regions of B. gilchristii strain SLH14081 and Histoplasma capsulatum strain WU24. Location of genes (blue boxes) and gypsy elements (green boxes) are depicted across each genomic region. Orthologs between genomes are connected in pink, which are organized into syntenic regions that are disrupted by GC-poor regions in both Blastomyces genomes. The GC-poor regions also show lower synteny between the Blastomyces genomes compared to other regions with more typical GC content (e.g., see Fig 3). Overall, B. dermatitidis strain ER-3 and B. gilchristii strain SLH14081 shared 125 syntenic blocks including 93.8% and 94.5% of genes, encompassing only 69.5% and 69.3% of each assembly. These percentages are much smaller than those observed among strains of related species (such as 95% and 93% synteny between strains of P. brasiliensis [29]). The lower synteny among Blastomyces strains is largely explained by the proportion of genes found in repeat-rich, GC-poor regions (Table 2 and Fig 3). Nearly all (99%) of genes in GC-rich regions are highly syntenic across Blastomyces strains, even between B. dermatitidis strain ER-3 and B. gilchristii strain SLH14081. However, the GC-poor regions have more limited synteny even within strains of Blastomyces encompassing 74 to 76% of genes in those regions (Table 2 and Fig 3).
Overall, the function, expression, and selective pressure of genes in GC-poor regions appear similar to those genes found elsewhere in the genome. Despite the lower synteny, GC-poor regions are not significantly enriched for Blastomyces-specific genes, nor did they show much functional enrichment (S1 Text, S5 Table). Comparing selection pressure on the 7,228 single copy orthologs present in all four Blastomyces genomes also did not find a significant difference in the number of genes with high omega values (omega > 1) (Methods). These analyses suggest that despite the dynamic reorganization due to invading gypsy elements, the GC-poor regions do not appear to be fast evolving by these measures. Furthermore, there is no large-scale difference in the expression levels of genes in GC-poor regions. Comparing transcript levels for genes in GC-poor and GC-rich regions, we found that genes in both GC classes show similar expression levels (S7 Fig), again supporting the general similarity of genes found in these two genomic neighborhoods.
Characterization of repetitive sequence expansion
The 2-fold larger size of the Blastomyces genomes compared to other dimorphic fungi is due largely to an expansion of repetitive sequence. The proportions of the Blastomyces genome assemblies that were covered by repeats ranged from 56.6% (41.6 Mb) for B. dermatitidis ATCC18188 to 63.0% (47.5 Mb) for B. gilchristii SLH14081. SLH14081 had the highest repeat content and the largest assembly size. The E. parva and E. crescens assemblies both had a lower repeat content, 9.9% (3.0 Mb) and 5.4% (1.6 Mb), respectively (Table 1). In all genomes, a small number of transposable element classes as well as AT-rich simple sequence regions were highly represented (Fig 4A).
Fig. 4. Relative contributions from known repeat categories to Blastomyces and Emmonsia genomes. 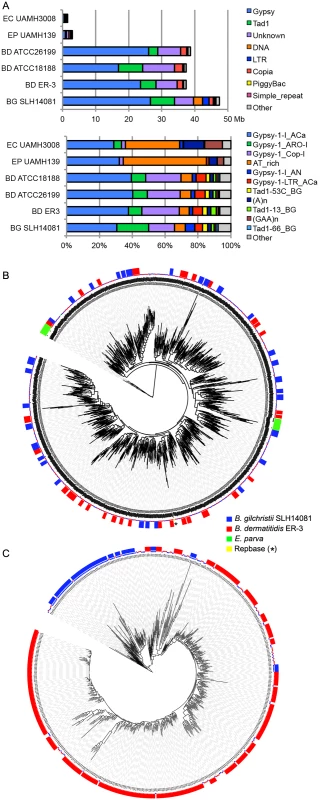
(A) Repetitive elements were identified in each assembly using a combination of de novo classified elements and known elements. The total amount of genome sequence for each element class (top panel) and the relative frequency of known elements (bottom panel) are shown for B. dermatitidis (BD; ATCC26199, ATCC18188, ER-3), B. gilchristii (BG; SLH14081), E. crescens (EC; UAMH3008), and E. parva (EP; UAMH139). (B, C). Phylogenetic relationship of two subgroups of gypsy elements was inferred using FastTreeDP from alignments of reverse transcriptase domains. The largest subgroup of 922 sequences (B) includes domains from the Blastomyces strains ER-3 and SLH14081, E. parva strain UAMH139, and the Repbase ACa Gypsy element, whereas the other subgroup of 544 sequences (C) is specific to the two B. dermatitidis and B. gilchristii. The outer ring indicates strain specific duplications of four or more sequences. More specifically, the genome expansion in Blastomyces strains has resulted from a proliferation of gypsy LTR retrotransposons, including both ancestral and lineage-specific proliferation. In the Blastomyces genomes, Gypsy elements account for almost all repetitive DNA, with a lower frequency of sequences similar to the non-LTR Tad1 and copia LTR retroelements (Figs 4A and S8). In all Blastomyces and Emmonsia genomes the most frequent Gypsy element subtype matches the “ACa” (Ajellomyces or Histoplasma capsulatum) Gypsy element from Repbase [30] (Fig 4A and 4B). Further phylogenetic characterization of 2,331 Gypsy elements identified four subtypes that appear specific to Blastomyces (S1 Text and Figs 4 and S9). Some subtypes had diversities that were primarily the result of ancestral duplication, resulting in large numbers of orthologous elements between strains (e.g., Fig 4B), while other subtypes appeared to predominantly contain strain-specific paralogous expansions, consistent with the cryptic speciation in the Blastomyces genus (e.g., Fig 4C). Gypsy elements were also detected in the Emmonsia and Histoplasma assemblies, but in far fewer copies (Figs 3 and 4), consistent with the recent expansion in Blastomyces. Gypsy elements are frequently observed in fungal genomes [31], including Coccidioides [32] and Paracoccidioides [29] but in far fewer copies.
Gene family evolution of Blastomyces and other Ajellomycetaceae
To identify gene content that could play a role in the evolution of the dimorphism and pathogenesis within the family Ajellomycetaceae, we searched for expansions or contractions in functionally classified genes compared to the other fungi from the order Onygenales (S6 Table). We identified PFAM domains, KEGG pathways, Gene Ontology (GO) terms, or kinase families that were significantly enriched or depleted. Domains associated with polyketide synthases were depleted in the Ajellomycetaceae, and an independent prediction of secondary metabolite enzymes confirmed that Blastomyces and other fungi from the Ajellomycetaceae contain fewer PKS gene clusters than other Onygenales (S7 Table, S1 Text). Other differences between the Ajellomycetaceae and other Onygenales include fewer copies of multiple classes of peptidases (M36, M43, S8) as well as an associated inhibitor (I9, inhibitor of S8 protease), variable copy number of LysM-domain proteins potentially involved in chitin binding, which are most expanded in dermatophytes but at next highest levels among the human pathogens in Blastomyces, and a higher number of protein kinases (S6 Table and S10 Fig), including an expansion of the FunK1 family similar to that previously noted in Paracoccidioides [29].
We next identified 140 gene clusters conserved in Blastomyces, Emmonsia, Histoplasma, and Paracoccidioides, but absent from other Onygenales and Aspergillus (S8 Table). These gene clusters include a predicted heme oxygenase (BDBG_02689), which could produce free iron from host heme. In addition to the 140 gene clusters, we also identified conserved genes in subsets of the Ajellomycetaceae including homologs of two previously typed antigens; a gene similar to the 27 kDa antigen of Paracoccidioides [33] is present in Blastomyces and one Histoplasma genome, and a gene cluster specific to Blastomyces and Paracoccidioides shares similarity with the antigenic Aspergillus cell wall mannoprotein [34].
Genes depleted in or absent from Emmonsia with possible roles in virulence or phase transitions
To identify potential genetic features of the Ajellomycetaceae pathogenic to immunocompetent humans (Blastomyces, Histoplasma, and Paracoccidioides) relative to E. parva and E. crescens, we conducted a second enrichment analysis as described above (S9 Table). The primary pathogens showed enrichment of only two PFAM domains, a phosphorylase and endonuclease (S9 Table). The phosphorylase domain over-represented in Blastomyces is associated with nucleoside phosphorylases; many of these proteins also contain Ankyrin repeats and NACHT domains. Phosphorylases are involved in nucleotide and amino acid salvage, and could allow pathogens greater metabolic versatility when certain building blocks are unavailable. The absence of any larger pattern of gain or loss of functional classes suggests that smaller changes in gene content, independent gain and loss between the species, or expression differences may account for differences in pathogenesis.
We then identified specific orthologs present in all four strains of Blastomyces but absent from both non-pathogenic Emmonsia species. Comparing the ortholog set of Blastomyces to E. parva and E. crescens, we found a total of 552 ortholog clusters that were present in all Blastomyces strains but absent in both Emmonsia genomes (S10 Table). Most of these (393 clusters) were present only in Blastomyces, and while most of these proteins (92% in B. gilchristii strain SLH14081) had no PFAM domain assignment, the set did include the Blastomyces yeast phase-specific protein 1 (BYS1). This gene is a marker of the phase transition to and from the yeast phase [15], although it has recently been shown not to be required for virulence in studied strains [24].
While both E. parva and E. crescens are not reported to be primary human pathogens, phylogenetic analysis suggests that the transition to this lifestyle may have been independent, resulting in differential gene loss. One of the genes absent only in E. crescens is the siderophore iron transporter mirB (MIRB). Many pathogenic microorganisms have evolved high affinity iron acquisition mechanisms, which include the production and uptake of siderophores. In B. dermatitidis, the expression of genes involved in the biosynthesis of siderophores and uptake of siderophores, including two iron transporters (MIRB and MIRC), are induced under iron-poor conditions [35]. While MIRB appears to be absent in E. crescens, siderophore uptake may be still enabled by the second transporter, MIRC, which is conserved in this species.
Transcriptional profiling of Blastomyces in macrophages
To better understand which Blastomyces genes play roles in pathogenicity and virulence, we carried out RNA-Seq of B. dermatitidis strain ATCC26199 to profile expression under varying temperature, nutrient availability, and infection status. Combining this data allowed us to disambiguate expression variability due solely to differences in temperature and media-specific nutrient availability from those specific to macrophage interactions in vitro or host infection in vivo. Five conditions were sampled: 37°C with macrophages in RPMI media, 37°C in RPMI media, 37°C in HMM media, 22°C in HMM media, and in vivo pulmonary infection with yeast in a mouse model (Fig 5A). For each condition, two biological replicates were performed, and the read counts per transcript were highly correlated between replicates (R> 0.99, S11 Fig). Gene expression levels and mapping statistics are presented in S11 and S12 Tables, respectively.
Fig. 5. Transcriptional response of B. dermatitidis strain ATCC26199 to infection. 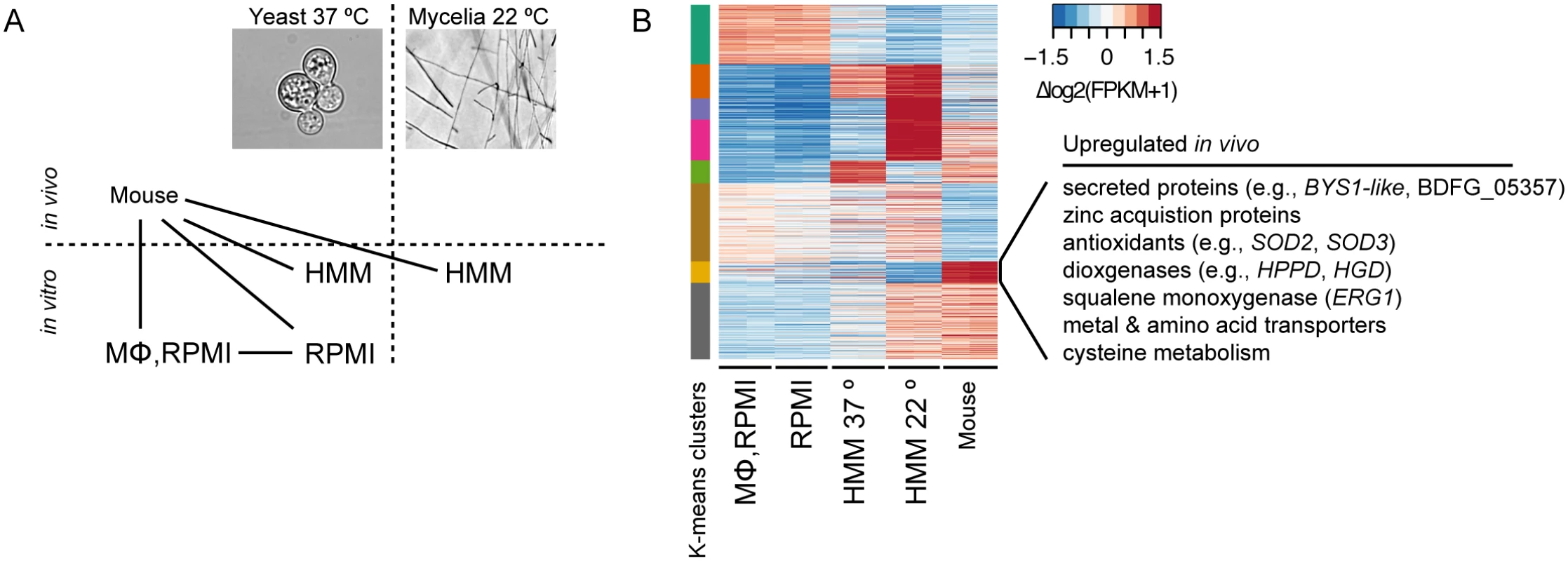
(A) Schematic of samples compared by RNA-Seq analysis and (B) Heatmap of differentially expressed genes, where the cluster of genes specifically induced in vivo during mouse infection is highlighted. When B. dermatitidis yeast cells were co-cultured with human bone marrow derived macrophages, the majority of yeast cells (59%) were internalized by macrophages. Comparison of yeast co-cultured with and without macrophages identified 140 genes differentially expressed between these two conditions, 112 of which were upregulated in the presence of macrophages (S13 Table). This upregulation suggested a small, specific response to macrophages in this experiment. Examination of this set of genes revealed numerous genes that have the potential to facilitate adaptation to the host environment. The 20 most significantly upregulated genes (Table 3) include a predicted secreted endo-1,3(4)-β-glucanase (BDFG_03060) involved in cell separation after cytokinesis in C. albicans [36], transporters, including an ABC transporter (BDFG_05060) homologous to Aspergillus fumigatus MDR1 and a zinc transporter (BDFG_02462) similar to the vacuolar zinc transporter ZRT3 in S. cerevisiae, dehydrogenases involved in amino acid catabolism, and antioxidants peroxisomal catalase (CATP, BDFG_02965) and superoxide dismutases (SOD3, BDFG_01204; SOD2, BDFG_07895), which may protect against reactive oxygen species (ROS). The induction of endo-1,3(4)-β-glucanase and CATP in the presence of macrophages was also confirmed by qRT-PCR (S12 Fig and Methods).
Tab. 3. <i>Blastomyces</i> transcripts most significantly induced during macrophage infection. 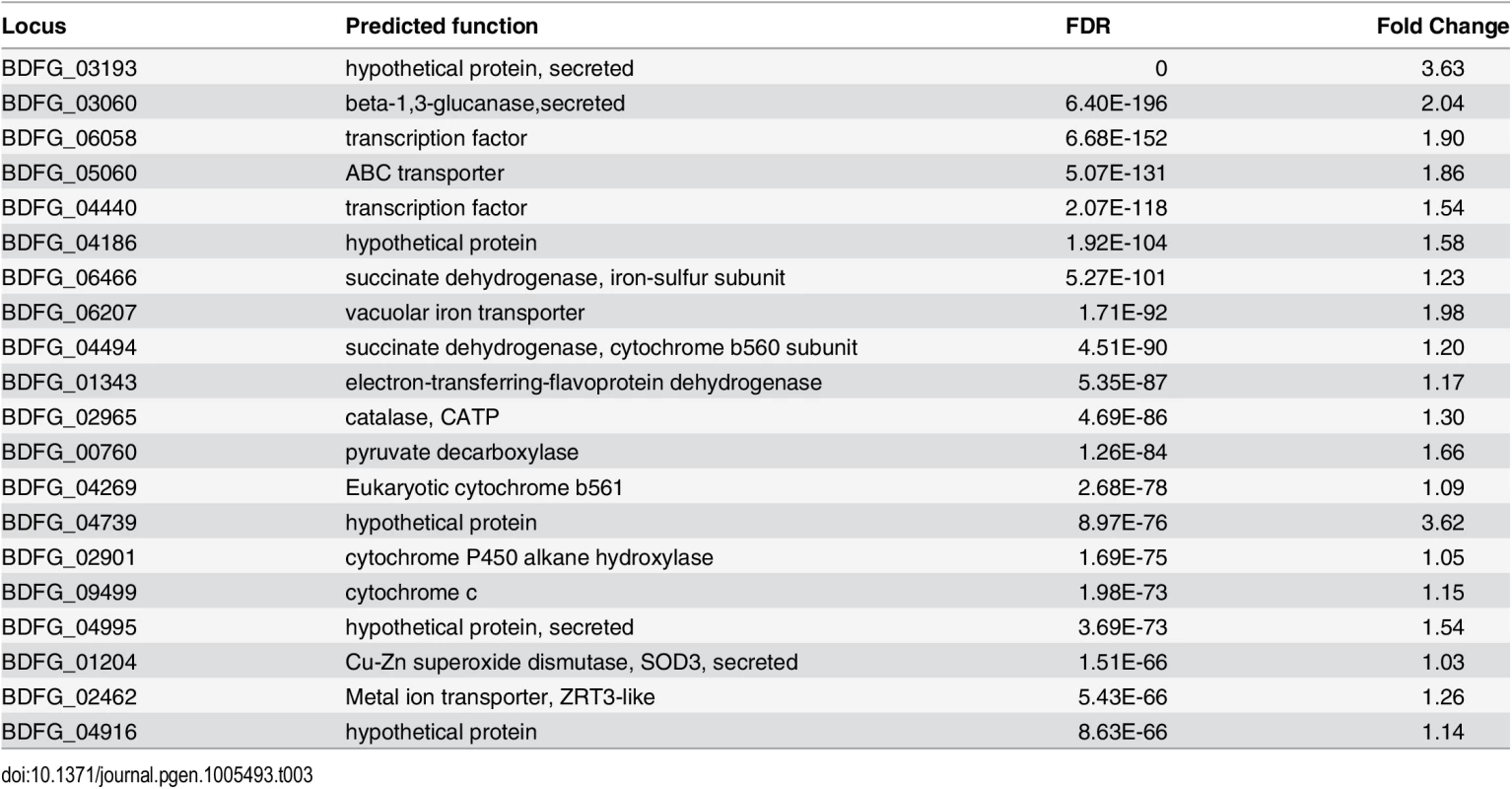
Transcriptional profiling of Blastomyces in a mouse model
We also identified gene expression changes specific to in vivo murine pulmonary infection from our transcriptomic data of B. dermatitidis strain ATCC26199. By k-means clustering of expression values, we identified a set of 72 genes that are upregulated in vivo during mouse infection relative to all other conditions, regardless of temperature, media, and in vitro macrophage interactions (Fig 5B and S14 Table). Using all conditions for this comparison helped eliminate from consideration differences observed, for example, between the yeast samples cultured in different media. Genes in this set with greater than 2-fold upregulation in vivo are highlighted in Table 4, and primarily fell into five functional categories: 1) secreted proteins, 2) zinc acquisition, 3) antioxidants and oxygenases, 4) amino acid metabolism, and 5) transporters.
Tab. 4. Blastomyces genes induced during mouse infection* 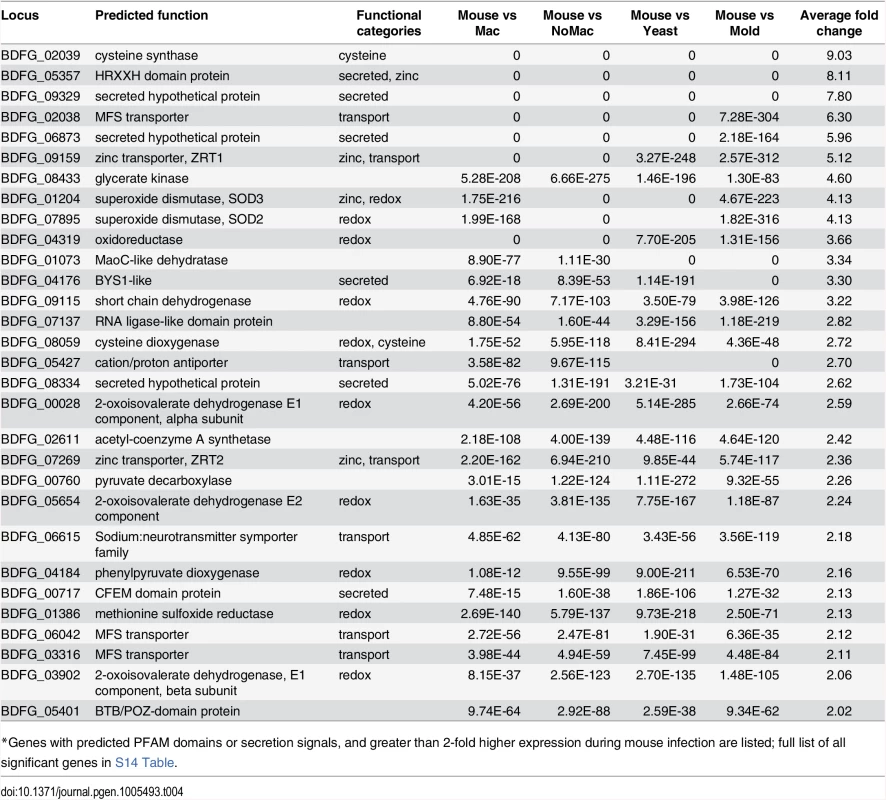
*Genes with predicted PFAM domains or secretion signals, and greater than 2-fold higher expression during mouse infection are listed; full list of all significant genes in S14 Table. The most highly expressed gene in vivo was BAD1 (BDFG_03370; S11 Table), which encodes a yeast-phase specific virulence factor that facilitates adhesion to host cells and evasion of host immune defenses [13]. BAD1 also had the highest transcript abundance for yeast co-cultured with macrophages and yeast without macrophages at 37°C (S13 Table). Thus, BAD1 was not identified among the set of 72 differentially expressed genes because the transcription of BAD1 is influenced by temperature [37]. The effect of temperature during the mold to yeast transition on the transcriptome of dimorphic fungal pathogens has been the topic of previous studies [38–41] and was therefore not further evaluated here.
A total of nine secreted proteins were identified in this set of 72, including five of the ten most highly upregulated genes by fold change, potentially playing roles in host-pathogen interactions. Another highly up-regulated secreted protein (BDFG_00717) contains a predicted CFEM domain as well as a GPI-anchor; these features, as well as small size (236 amino acids), are shared with member of the haemoglobin-receptor gene family in C. albicans [42]. The most highly upregulated gene, BDFG_05357, encodes a HRXXH domain-containing secreted protein that may function as a zinc scavenging protein (Tables 4 and S14). This gene is present in the genomes of Blastomyces and Coccidioides, but absent from those of Emmonsia, Histoplasma and Paracoccidioides. BDFG_05357 is a homolog of C. albicans PRA1 (pH-regulated antigen-1) [43] and S. cerevisiae ZPS1 (zinc-pH-regulated protein). In C. albicans, the transcription of PRA1 and ZPS1 is induced under alkaline pH and zinc-deplete conditions [44,45], and PRA1 is co-regulated with its upstream gene, ZRT1, which encodes a high-affinity zinc transporter that interacts with zinc-bound PRA1 [45]. Similarly, the B. dermatitidis homolog of ZRT1, BDFG_09159, is highly expressed in vivo; the induced expression of both PRA1 and ZRT1 was confirmed by qRT-PCR (S12 Fig). However unlike in C. albicans, ZRT1 is not adjacent to PRA1 in the B. dermatitidis genome. While PRA1 is conserved in all four Blastomyces genomes, there is no copy of this gene in Histoplasma as previously noted [45], nor in Emmonsia or Paracoccidioides, suggesting differences in how zinc is acquired within the Ajellomycetaceae.
In addition to PRA1/ZPS1 and ZRT1, a larger module of genes that regulate zinc acquisition is co-regulated in Blastomyces. The transcript abundance of BDFG_07269, which encodes a low-affinity zinc transporter (ZRT2), is also significantly upregulated in the mouse model. In S. cerevisiae, the zinc-responsive transcription factor ZAP1 regulates expression of ZRT1 and ZRT2, along with ZPS1. We identified the ortholog of ZAP1 in strain ATCC26199 as BDFG_07048, which was also significantly upregulated in vivo relative to all other conditions (S14 Table) despite not being identified by k-means clustering. These results suggest that zinc acquisition and homeostasis may play a critical role for survival of B. dermatitidis in vivo.
Genes that convert reactive oxygen species to dioxygen and dioxygen to metabolites were also highly upregulated in vivo. These include two superoxide dismutases (SOD3: BDFG_01204 and SOD2: BDFG_07895), which were even more upregulated in vivo than in macrophages. Four dioxygenases (BDFG_04184, BDFG_04185, BDFG_08059, BDFG_06504) were also upregulated in vivo, representing almost half of the dioxygenases found in the genome, which utilize dioxygen to drive amino acid catabolism. This set includes 4-hydroxyphenylpyruvate dioxygenase, (4-HPPD; BDFG_04184) and homogentisate 1,2-dioxygenase (BDFG_04185), which are involved with tyrosine catabolism [46]. Other upregulated oxygenases include indoleamine 2,3-dioxygenase (BDFG_06504) and squalene monooxygenase (ERG1—BDFG_07857), which are involved with tryptophan catabolism and ergosterol biosynthesis respectively. ERG1 is a target of current antifungal drugs, including terbinafine. High in vivo expression of this gene may suggest that drugs targeting it may be more effective in vivo than in vitro.
Genes involved in cysteine biosynthesis and catabolism were highly upregulated during infection including cysteine synthase A (BDFG_02039) and cysteine dioxygenase (BDFG_08059). Cysteine synthase A may provide a large pool of synthesized cysteine for B. dermatitidis metabolism; the induced expression during infection was confirmed by qRT-PCR (S12 Fig). Based on orthology analysis, cysteine synthase A is absent from the genome of H. capsulatum, and previous studies have shown that Histoplasma yeast are auxotrophic for cysteine [47,48]. Cysteine dioxygenase catabolizes cysteine to L-cysteinesulfinic acid, an intermediate that can be used for taurine biosynthesis or metabolized to sulfite and pyruvate. A homolog of C. albicans SSU1 (BDFG_06814), which encodes a sulfite efflux pump and is co-regulated with cysteine dioxygenase in C. albicans [49], was also upregulated in B. dermatitidis.
Transporters in fungi have been associated with an enhanced ability to remove harmful products as well as to survive on diverse nutrient sources, both of which could contribute to virulence and pathogenicity. Of the 72 genes upregulated in vivo during mouse infection, 11 are predicted transporters. These included the major facilitator type (MFS; BDFG_06068 –unknown function, BDFG_06042 –glycose transport, BDFG_02038 –unknown function), amino acid transporters (BDFG_02310, BDFG_07447) and metal transporters (zinc/iron transporters discussed above, BDFG_09159, BDFG_07269, and NIC1 nickel transporter, BDFG_02449; S14 Table). This upregulation potentially reflects differences in metabolite and cofactor availability in the host relative to in vitro conditions.
Discussion
Phylogenetic position of Blastomyces spp. and Emmonsia parva and E. crescens
Our whole-genome based phylogenetic analysis recovered a sister-group relationship between Blastomyces spp. and Emmonsia parva, as previously reported from ribosomal DNA sequences [19,20]. However, our study placed Histoplasma as the next most basal species, and uniquely placed E. crescens between Histoplasma and the basal Paracoccidioides with strong bootstrap support. This more external position of Paracoccidioides compared to Histoplasma agrees with an earlier rDNA tree without Emmonsia [50]. Furthermore, gene support frequencies (GSF) were relatively high, and increased when we subsampled only well-supported genes, providing additional support for the topology presented here.
The polyphyletic nature of the non-human pathogen Emmonsia suggests substantial plasticity in regard to human pathogenesis in this group. Ancestral variation in the ability of these species to infect other mammals could then be associated with exaptation to human hosts. Additional diversity of Emmonsia, including the third described species, E. pasteuriana [51,52] and other closely related isolates [17] suggests that the full breadth of the Emmonsia genus may not be captured by the two isolates sequenced here. Interestingly, both E. pasteuriana and related isolates produce yeast cells at high temperature, rather than the adiaspores produced by E. parva and E. crescens. Further sequencing of Emmonsia species and other related strains may reveal additional patterns and trends in the evolution of the dimorphic fungi.
Genome expansion and segmentation: GC-poor isochore-like regions
The mosaicism observed here in the genome of Blastomyces differs substantially from that observed in other fungi and larger eukaryote genomes. While isochore-like GC-poor regions are unprecedented at this scale in fungal genomes described to date, there are parallels to the organization of L. maculans, though GC-poor regions occupy a smaller fraction of that genome [28]. Longer GC-poor isochores of more than 300 kb are commonly found in mammals and other vertebrates [53–55]. GC-poor isochores in vertebrates are often more stable over long evolutionary times [55,56] and have other properties such as lower gene expression [55] that do not appear to be shared by the GC-poor tracts of B. dermatitidis and B. gilchristii (S1 Text).
Characterization of repetitive sequence in GC-poor regions suggests these originated with a dramatic expansion of elements of the LTR/Gypsy category. Phylogenetic analysis suggests these elements swept through a lineage leading to the present-day B. dermatitidis and B. gilchristii, and to a lesser extent Emmonsia parva, and have further expanded during the diversification of Blastomyces. While H. capsulatum does not have such an expanded genome, or a sizable GC-poor component, and so appears less affected by gypsy expansion, Histoplasma may alternatively have more robust defense against repetitive elements or be less able to accommodate large amounts of repeats in its genome.
While GC-poor tracts have been particularly dynamic areas due to Gypsy element insertions during the recent evolution of Blastomyces, these regions appear typical in gene content and expression. Perhaps due to their recent origin, the GC-poor regions do not appear to have sequestered particular classes of genes such as secreted proteins or have other hallmarks of rapidly evolving gene content. The long GC-poor regions also include some well characterized genes involved in phase transitions and pathogenesis, including the Blastomyces yeast-specific gene BYS1, a marker of the phase transition to and from the yeast phase [15,24]. Reduced levels of synteny in the GC poor regions between B. dermatitidis and B. gilchristii could prevent effective meiotic recombination between the two lineages, further supporting their designation as separate species.
Functional diversity of gene content in Blastomyces and the other Ajellomycetaceae
Despite the large increase in genome size in Blastomyces, the total number of protein coding genes is only modestly expanded. Blastomyces and other sequenced species from the Ajellomycetaceae family, including the human primary pathogens Histoplasma and Paracoccidioides, have similar gene content with only a few gene loss or gain events that map to common functional classes. This stability suggests that more modest differences in gene content and sequence, as well as potential divergence of gene regulation, contribute to phenotypic differences between the species. Larger differences exist between the Ajellomycetaceae and other more divergent members of the Onygenales. There is no expansion of degradative proteases as previously noted for Coccidioides [57]; in fact, three peptidase families (M36, M43, and S8) are present at lower copy number in Blastomyces and the other Ajellomycetaceae. While Blastomyces contains a higher number of LysM proteins than the dimorphic Onygenales, the number is small relative to that found in Dermatophytes [58]. This analysis also identified candidate genes involved in host interaction, including proteins homologous to antigens in related fungi and a heme oxygenase that could release iron from host heme.
Features of Blastomyces gene expression in macrophages and in vivo
For yeast co-cultured with macrophages and yeast in vivo, some aspects of the transcriptional response were shared including response to oxidative stress and amino acid catabolism. Yeast co-cultured with macrophages showed upregulation of numerous genes involved in oxidative reduction, which may play a major role in protecting Blastomyces from ROS. The macrophage phagosome is poor in glucose and amino acids, but rich in ROS [59,60]. Blastomyces is relatively resistant to ROS and virulence correlates with the ability to minimize ROS generation in innate immune cells [61,62]. The upregulation of superoxide dismutases (SOD3, SOD2) and catalase P may protect B. dermatitidis yeast against oxidative stress. In H. capsulatum, extracellular SOD3 and intracellular catalase P, contribute to survival within macrophages [63,64]. Moreover, SOD3 promotes H. capsulatum virulence in a murine model of pulmonary infection [63]. The upregulation of 4-HPPD, which is involved with pyomelanin biosynthesis, contributes to antioxidant defense and intracellular survival of Penicillium marneffei [65]. Inhibition of 4-HPPD in P. brasiliensis and P. marneffei blocks the phase transition to yeast at 37°C [65,66]. Furthermore, in vivo numerous dioxygenases were upregulated, suggesting that dioxide produced in response to ROS may be utilized for amino acid metabolism.
Changes in amino acid metabolism were prevalent in both the macrophage co-cultured and in vivo Blastomyces, suggesting the recycling of amino acids as an energy source (Results, S1 Text). In particular, the very specific increase in cysteine catabolism (cysteine dioxygenase) and biosynthesis (cysteine synthase A) during in vivo infection suggests that cysteine may be critical to virulence. In mice, deletion of cysteine dioxygenase (CDG1) in C. albicans results in attenuated virulence [49]. Furthermore, upregulation of sulfite efflux pump (SSU1), which is co-regulated with CDG1 in C. albicans, could play a role in B. dermatitidis virulence during in vivo infection. Exported sulfite can destabilize host proteins by reducing disulfide bonds and facilitates the growth of dermatophytes on keratinized tissue [67]. How breakdown of tryptophan by indoleamine 2,3-dioxygenase (IDO), which can supply de novo nicotinamide adenine dinucleotide (NAD+), alters the fungal-host interaction is unknown. In cancer, tumor cells with increased expression of IDO may facilitate tryptophan depletion in the microenvironment to suppress the host immune response [68]. Infection with H. capsulatum, P. brasiliensis, and C. albicans upregulates host IDO activity, reduces fungal growth, impairs Th17 T-cell differentiation, and blunts excessive tissue inflammation [69–71].
The specific in vivo upregulation of genes that encode secreted proteins as well as those involved with transmembrane transport (e.g., amino acids, glucose), amino acid metabolism (e.g., cysteine), and metal acquisition (e.g., zinc, nickel) highlights virulence factors potentially being missed by in vitro studies and the importance of understanding nutrient and co-factor availability in any study system. Uptake of zinc and nickel, which serve as enzyme co-factors, contribute to virulence in C. albicans and Cryptococcus neoformans respectively [45,72]. PRA1 encodes a secreted “zincophore” under alkaline and zinc-poor conditions that acts in concert with ZRT1 to promote zinc acquisition and facilitate endothelial cell damage by C. albicans [45]. NIC1-mediated nickel uptake is critical for urease activity, which contributes to C. neoformans invasion of the central nervous system [72]. In C. posadasii, urease induces host tissue damage [73]. While genes involved with the acquisition of zinc (e.g., ZRT1, ZRT2, ZAP1 homologs) and nickel (e.g., NIC1 homolog) are largely conserved with other fungi, the absence of PRA1 in Histoplasma, Paracoccidioides, and Emmonsia highlights recent evolutionary changes in zinc acquisition mechanisms in the family Ajellomycetaceae. This, in conjunction with differences in cysteine metabolism between Blastomyces and Histoplasma, suggest that despite the many common elements of dimorphism and pathogenesis, each genus of dimorphic fungi likely has unique nutritional requirements.
Methods
Selection of isolates for sequencing
Four strains of Blastomyces were sequenced: SLH14081 representing the new species B. gilchristii, and ER-3, ATCC18188 and ATCC26199 representing B. dermatitidis. The SLH14081 strain is a highly virulent, clinical isolate that can cause disease in immunocompetent persons, while ER-3 was isolated from a woodpile and is hypovirulent in mice [21,22]. The remaining two strains are strain ATCC18188, a representative MAT 'alpha' isolate, and ATCC26199, a commonly used laboratory isolate.
Two species that are closely related to Blastomyces, that can cause pulmonary disease in rodents (adiaspiromycosis), were also sequenced: Emmonsia parva UAMH139 and Emmonsia crescens UAMH3008. These isolates were chosen for comparison as these species are not typically human pathogens, yet they are closely related to the three pathogenic dimorphic genera Blastomyces, Histoplasma and Paracoccidioides, with which they form a clade that is nested within the order Onygenales and represents the Ajellomycetaceae family [20].
Sequencing of Blastomyces, E. parva and E. crescens
Genomic DNA for sequencing was prepared from mycelial or yeast culture, using phenol/chloroform extraction. For the Blastomyces SLH14081 and ER-3 strains, whole genome shotgun sequence was obtained using Sanger technology on an ABI 3730 from a Fosmid (epiFOS) and two plasmid (pJAN and pOT) libraries. For B. dermatitidis ATCC18188, whole genome shotgun sequence was obtained from two small insert libraries (fragment and 1.5 kb) using Roche 454 technology and from a Fosmid library using Sanger technology. For B. dermatitidis ATCC26199 20X of sequence was generated using 454 technology from a small insert fragment library. In addition, a plasmid (pOT) and Fosmid (epiFOS) library were constructed and sequenced using Sanger technology by the Washington University Genome Center, generating a total of roughly 3.6X coverage.
For each Emmonsia species, a single library was used to generate 101 bp paired-end reads using Illumina technology on a Genome Analyzer II generating a total of 116X coverage for E. parva UAMH139 and 163X coverage for E. crescens UAMH3009. Libraries of average insert size of 639 bp for E. parva and of 686 bp for E. crescens were chosen based on the electropherograms obtained from Bioanalyzer. Sequencing of both Emmonsia genomes was performed at the Genomic Sequencing Laboratory, University of California, Berkeley.
Genome assemblies
Blastomyces strains SLH14081 and ER-3 were assembled with Arachne [74] (Assemblez Build 20080911). For B. dermatitidis ATCC18188, a hybrid assembly was generated with Newbler version 2.3. For B. dermatitidis ATCC26199, a hybrid assembly of the Sanger and 454 data was generated with Newbler version "MapAsmResearch-03/15/2010" with options-rip and -scaffold.
For the Emmonsia genomes, assemblies were generated using multiple programs, including the SOAPdenovo / GapCloser package [75], ABYSS [76] and Velvet [77]. SOAPdenovo assemblies were selected based on quality metrics. While assemblies with high k values increased the fraction of GC-poor regions represented in the assembly, optimal assembly of gene sequences were achieved using lower k values, based on comparing each assembly to gene sets of Blastomyces and other related dimorphic fungi using TBlastN. The assemblies for the Emmonsia genomes (k = 27 for E. parva and k = 29 for E. crescens) were processed using the program GAEMR (http://www.broadinstitute.org/software/gaemr/), where overall assembly metrics were used to select the best assembly considering both continuity and completeness, though these measures were lower than for genomes assembled from multiple libraries.
Optical mapping of Blastomyces
To validate the assembly of strain SLH14081 and anchor it onto chromosomes, we constructed an optical map, a single-molecule based ordered restriction map. The map of B. gilchristii strain SLH14081 was constructed by OpGen using the restriction enzyme BsiWI (C^GTACG). The optical map consists of 16 linkage groups, with size ranging from 9.728 Mb to 730 kb. The total size of the map was estimated as 79.64 Mb in size, slightly larger than the 75.35 Mb genome assembly, likely due to repetitive element sequence missing from the assembly. A total of 36 assembly scaffolds covering 68.9 Mb were mapped based on shared restriction sites to the optical linkage groups (S2 Table).
RNA-Seq of ATCC26199 from yeast, mold, and infection conditions
To enable more accurate gene prediction and analyze gene expression, RNA was prepared and deeply sequenced from five conditions (yeast with or without macrophages in RPMI media, in vivo during murine pulmonary infection, and in vitro yeast and mold in Histoplasma macrophage media (HMM)) with two biological replicates per condition.
ATCC26199 yeast cells were co-cultured with bone marrow derived murine macrophages from C57BL/6 mice in RPMI media with 10% heat inactivated FBS and supplemented with penicillin (100 U) and streptomycin (100 ug) or incubated in this media alone. Yeast and macrophages were co-cultured using a ratio of one yeast for every two macrophages (MOI 0.5). The use of alveolar macrophages was precluded due to the large numbers of mice that would be needed to harvest these cells. Following inoculation of cell culture flasks with B. dermatitidis yeast, the co-cultures were incubated at 37°C for 24 hrs. The majority of the yeast were either single cells or cells with one bud (average 89%). The extent of macrophage internalization of yeast was measured using Uvitex staining to differentiate between extracellular and intracellular yeast. A total of 1,976 cells were counted across seven individual fields of view, with an average of 59% Uvitex negative (intracellular) and 41% Uvitex positive (extracellular). The majority of B. dermatitidis cells exhibited yeast morphology (> 96%); pseudohyphal growth occurred in 2.4% of co-cultured yeast and 3.7% of yeast grown in RPMI media without macrophages. Harvested yeast cells were flash frozen in liquid nitrogen, ground with a mortar and pestle into a fine powder, and RNA extracted using the phenol-guanidium thiocyanate-1-bromo-3-chloropropane extraction method [78].
For in vivo transcriptional profiling, C57BL/6 mice were infected with 2 x 103 B. dermatitidis ATCC26199 yeast cells intratracheally and monitored for signs and symptoms of infection [79]. Mice with euthanized by carbon dioxide at 17 days post infection and yeast were isolated from murine lung tissue using the technique developed by Marty et al. [80]. Briefly, excised lungs were homogenized in pre-chilled (4°C) double-distilled, sterile water (ddH2O) supplemented with DNase I 10 μg/ml (Roche) using an Omni TH tissue homogenizer (Omni International, Kennesaw, GA), passed through a 40 μm cell strainer (ThermoFisher Scientific, Waltham, MA), and centrifuged at 770g for 5 minutes at 4°C. The supernatant and interface were removed using a serologic pipette and yeast pellet was washed with ice-cold ddH2O and rapidly frozen in liquid nitrogen for RNA extraction. Time ex vivo was less than 30 minutes and samples were near-freezing (4°C) during all isolation steps. Quality control analyses using qRT-PCR demonstrated that the short ex vivo time (< 30 minutes) at 4°C minimized changes in transcript abundance that would have occurred if the samples were processed at higher temperatures or for a longer duration [80]. Total RNA isolated from B. dermatitidis yeast during pulmonary infection was divided into 2 pools of 5 mice each (pool #1 and pool #2).
In vitro yeast were incubated in liquid Histoplasma macrophage media (HMM) at 37°C while shaking [81]. The majority of cells had yeast morphology; less than 3.25% of cells grew as pseudohyphae. To generate mycelia, yeast cells were incubated in liquid HMM for 14 days at 22°C while shaking. Harvested yeast and mycelial cells were flash frozen in liquid nitrogen, ground with a mortar and pestle into a fine powder, and RNA extracted using the phenol-guanidium thiocyanate-1-bromo-3-chloropropane extraction method [78].
Total B. dermatitidis RNA from all samples (in vivo, in vitro, co-cultures) was treated with TurboDNase (Bio-Rad, Hurcules, CA) and cleaned using an RNeasy column (Qiagen). RNA integrity and quality was assessed using Nanodrop spectrophotometry, 0.8% agarose gel electrophoresis, and an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA integrity numbers (RIN) for in vivo samples were > 7.5 (7.6 for pool #1, 7.8 for pool #2). RIN values for in vitro and co-cultures (including yeast only RPMI) were ≥ 8.7.
For RNA-Seq, poly-A mRNA was purified for each total RNA sample and strand-specific libraries prepared as previously described [82,83]; each library was sequenced using Illumina Technology, generating an average of 65,174,908 101 bp reads per sample. RNA-Seq was incorporated into gene prediction and used to detect differentially expressed transcripts as described below.
Genome annotation
For initial gene sets, a total of 38,405 ESTs generated from yeast and mycelial samples of ATCC26199 (Washington University) and from a normalized cDNA library of SLH14081 (Broad Institute) were used for gene prediction. To achieve better transcript coverage, strand-specific RNA-Seq data from 10 samples representing the above yeast, mold, and infection stages was assembled with the Inchworm component of Trinity [84] and processed with PASA [85] to generate a set of transcripts for gene prediction. Gene sets were generated by using EvidenceModeler (EVM) [85] to select the best gene call for a given locus from the gene prediction programs SNAP, Augustus, Geneid, and Genewise and from PASA RNA-Seq transcripts as previously described [85,86].
Project numbers and locus tag prefixes assigned to gene sets are as follows: B. gilchristii SLH14081 (PRJNA41099, locus tag prefix BDBG), B. dermatitidis ER-3 (PRJNA29171, prefix BDCG), ATCC18188 (PRJNA39265, prefix BDDG), and ATCC26199 (PRJNA39263, prefix BDFG); the E. parva UAMH139 (PRJNA178178, prefix EMPG) and E. crescens UAMH3008 (PRJNA178252, EMCG).
Expression profiling
RNA-Seq reads were aligned to the transcript sequences of B. dermatitidis strain ATCC26199 using Bowtie [87]. Transcript abundance was estimated using RSEM [88], TMM-normalized FPKM for each transcript were calculated, and differentially expressed transcripts were identified using edgeR [89], all as implemented in the Trinity package version r2013-2-25 [90]. To identify GO term enrichment of differentially expressed genes, we classified transcripts using Blast2GO [91] and then performed comparisons with Fisher’s exact test. A 2-fold difference in FPKM values and a false discovery rate below 0.05 were used as a criteria for significant differential expression. To identify possible functions of the gene products of significantly differentially expressed parasitic-phase genes, protein homologs were assigned based on BLAST, Gene Ontology (GO) terms and protein family domains (PFAM, TIGRFAM).
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from B. dermatitidis yeast as described above. One microgram of DNase-treated total RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed with SsOFast EvaGreen Supermix (Bio-Rad) using a MyiQ real-time PCR detection system (Bio-Rad). Reactions were performed in triplicate using the following conditions: 1 cycle 95°C x 30 sec, followed by 40 cycles at 95°C for 5 sec, 60°C for 10 sec. Transcript abundance for genes of interest were normalized relative to the transcript abundance of GAPDH. Relative expression (RE) was calculated as RE = 2-ΔCt, ΔCt = Ctgene of interest−CtGAPDH [92].
Primer sequences used were as follows: AATCCTTTGACAGTGAAAC (forward) and CCATAAATCTGCTACAACAG (reverse) for BDFG_03060, ACTGTCGGTGGAGAGAAG (forward) and ACTGGGGTGTTGTTGAAG (reverse) for BDFG 02965, GACTATCCCATCCACAAC (forward) and TACAGAGCGGAATCTTTG (reverse) for BDFG 05357, TTTGGCACTGGAGTTATG (forward) and TGCTTCGTAGTCTAAAGTC (reverse) for BDFG 09159, GTGCTACAACGGAGATAC (forward) and GATAACCACCACGAACAC (reverse) for BDFG 02039, ACCCCCGCTCCTCCATCTTC (forward) and GAGTAGCCCCACTCGTTGTCATACC (reverse) for BDBG_07959 (GAPDH).
Segmentation and identification of genes and repeats located in GC-poor tracts
We used the IsoFinder GC segmentation program (http://bioinfo2.ugr.es/oliver/isofinder; [93]) to segment all ER-3 and SLH14081 scaffolds into long homogeneous genomic regions (LHGRs). The option p2 (parametric/student t-test with different variances), a window size of 5 kb and a p value cutoff of 0.01 (P parameter 0.99) were chosen after evaluating P cutoffs between 0.95 and 0.99, and window sizes ranging between 3 and 5 kb. The final settings were chosen as they accommodated gene synteny between ER-3 and SLH14081 in the GC-poor segments, obviating the need to manually remove narrow GC peaks caused by short genic regions.
To identify the coordinates of the longer GC-poor and GC-rich tracts on the assemblies of Blastomyces strains ER-3 and SLH14081, we set the boundary between GC-poor and GC-rich at 38% GC, a value that is in the deep valley between the two components of these genomes’ bimodal GC distribution. The deep valley is robust and persists over a wide range of window/segment sizes ranging up to > 60 kb (S4 Fig). We then grouped adjacent segments located between successive transitions (regime switches) across the 38% GC border. Islands of N’s in the interior of the GC-poor tracts were retained, but those at the tract fringes (i.e., next to a jump across the 38% GC threshold) were not. This procedure yields a large-scale segmentation of all scaffolds into strictly alternating “GC-poor” and “GC-rich” tracts. The GC-poor tracts and genes in those regions are listed in S3 and S4 Tables, respectively; GC-rich tracts form the remainder of the assemblies. We performed MySQL joins to identify the genes and repeats (GFF files produced by RepeatMasker of elements from RepeatModeler) located entirely or partly in the GC-poor tracts.
Synteny analyses
DAGchainer [94] was used to identify syntenic blocks with a minimum of 6 genes, which were required to be in the same order and orientations in the compared genomes. Synteny plots were generated using a custom perl script, using the GDgraph library; code is available at https://github.com/gustavo11/syntenia. Geneious Pro was used to visualize smaller-scale syntenies within and among genome assemblies.
Recognition and characterization of repeats
De novo repetitive sequence in each assembly was identified using RepeatModeler version open-1.0.7 (www.repeatmasker.org/RepeatModeler.html). Copies of de novo repeats and fungal sequences from RepBase [95] were mapped in each assembly using RepeatMasker version open-3.2.8 (www.repeatmasker.org/). For phylogenetic analysis of gypsy elements, reverse transcriptase domains were identified from each element; matches to the PFAM RVT_1 domain were identified with HMMER (version 3.1b1) [96] for 6-frame translations of each element generated by EMBOSS transeq (version 6.5.7 with parameters-frame 6-clean Y) [97]. The best domain match for each element was selected, requiring 50% alignment coverage and c-Evalue < 1e-5. The domains identified in Blastomyces SLH14081 (991 total) and ER-3 (1,296 total), E. parva (40 total), and similar Repbase gypsy elements (4 total) were aligned with MAFFT (version 6.717) [98], and a phylogeny estimated using FastTreeDP (version 2.1.8) [99]. Four large subgroups were identified and visualized using iTOL [100].
Identification and analysis of orthologs and phylogenetic analysis
A total of 16 genomes from the Onygenales order and three Aspergillus genomes were chosen for comparative analyses (S15 Table). These include the four Blastomyces (SLH14081, ATCC26199, ATCC18188, ER-3) and two Emmonsia species (UAMH139, UAMH3008) as well as the following: Histoplasma capsulatum WU24 (AAJI01000000), H. capsulatum G186AR (ABBS01000000), Paracoccidioides lutzii Pb01 (ABKH02000000), P. brasiliensis Pb03 (ABHV02000000), and P. brasiliensis Pb18 (ABKI02000000), Coccidioides immitis RS (AAEC00000000), C. posadasii C735 delta SOWgp (ACFW00000000), Uncinocarpus reesii 1704 (AAIW00000000), Microsporum gypseum CBS118893 (ABQE00000000), Trichophyton rubrum CBS118892 (ACPH01000000), Aspergillus nidulans FGSC A4 (AACD00000000), A. flavus NRRL3357 (AAIH00000000), A. fumigatus Af293 (AAHF01000000). OrthoMCL was used to cluster the protein-coding genes of the 19 chosen genomes by similarity.
To estimate the species phylogeny, a total of 2,062 orthologs present in a single copy in all of the 19 genomes were identified. Protein sequences of the 2,062 genes were aligned using MUSCLE, and a phylogeny was estimated from the concatenated alignments using RAxML v7.7.8 with model PROTCATWAG. To more closely examine the relationship of the Blastomyces isolates, single copy orthologs were identified in all four strains of Blastomyces and E. parva; the protein sequences of a total of 6,605 single copy orthologs were aligned using MUSCLE, and the resulting sequences replaced with the corresponding codons. A phylogeny was estimated from this nucleotide alignment using RAxML v7.3.3 with model GTRCAT. A total of 1,000 bootstrap replicates were used for each analysis. The level of support for the best RAxML tree was also evaluated using individual gene trees, by calculating the gene support frequency (GSF, [26]). A phylogeny was estimated and bootstrapped using the same parameters as for the concatenated sequence matrix, and gene trees with high bootstrap support at all nodes were then selected. A total of 162 gene trees were supported by at least 70% of bootstrap replicates at all nodes; the percent of gene trees supporting the RAxML best tree was calculated using RAxML and is shown in Fig 1. We also evaluated larger subsets of trees including those with 60% bootstrap support at all nodes, 50% bootstrap support, or all trees regardless of support, and found lower support respectively in each subset for our best tree.
To examine selective pressure on genes in GC-poor regions, we identified 7228 genes that were single copy in the four Blastomyces genomes from the OrthoMCL run. dN/dS values for each gene were computed on codon-based nucleotide alignments with the codeml module of PAML [101], using the one-ratio (M0) model.
Gene family and protein domain analysis
Genes were functionally annotated by assigning PFAM domains, GO terms, and KEGG classification. HMMER3 [96] was used to identify PFAM domains using release 27. GO terms were assigned using Blast2GO [91], with a minimum e-value of 1x10-10. Protein kinases were identified using Kinannote [102] and divergent FunK1 kinases were further identified using HMMER3. Secondary metabolite gene clusters were predicted with antiSMASH version 2.0.2 [103]. Genes were clustered using OrthoMCL [104] with a Markov inflation index of 1.5 and a maximum e-value of 1x10-5.
To identify functional enrichments in Blastomyces and other subsets of the 19 compared genomes, we used four gene classifications: OrthoMCL similarity clusters (see above), PFAM domains, KEGG pathways, and Gene Ontology (GO), including different hierarchy levels. A gene was considered to be a member of a given gene class when, respectively, the gene (a) belonged to the given OrthoMCL cluster, (b) contained at least one instance of the given PFAM domain in the encoded protein, (c) belonged to the given KEGG pathway, or (d) was tagged by the given GO label. Using a matrix of gene class counts for each classification type, we identified enrichment comparing two subsets of queried genomes using Fisher’s exact test. Fisher’s exact test was used to detect enrichment of PFAM, KEGG, or GO terms between groups of interest, and p-values were corrected for multiple comparisons [105]. Significant (corrected p-value < 0.05) PFAM and GO terms expansion or depletion was examined for three comparisons: Ajellomycetaceae compared to other Onygenales (S6 Table), pathogenic compared to non-pathogenic from Ajellomycetaceae (S9 Table), and Blastomyces compared to other Ajellomycetaceae; the only terms found to be expanded only in Blastomyces included nucleosome and zinc ion binding. No significant enrichment in KEGG terms was detected for these comparisons.
Supporting Information
Zdroje
1. Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36 : 1–53. doi: 10.3109/10408410903241444 20088682
2. Meece JK, Anderson JL, Fisher MC, Henk DA, Sloss BL, Reed KD. Population Genetic Structure of Clinical and Environmental Isolates of Blastomyces dermatitidis, Based on 27 Polymorphic Microsatellite Markers. Appl Environ Microbiol. 2011;77 : 5123–5131. doi: 10.1128/AEM.00258-11 21705544
3. Brown EM, McTaggart LR, Zhang SX, Low DE, Stevens DA, Richardson SE. Phylogenetic Analysis Reveals a Cryptic Species Blastomyces gilchristii, sp. nov. within the Human Pathogenic Fungus Blastomyces dermatitidis. PLoS ONE. 2013;8: e59237. doi: 10.1371/journal.pone.0059237 23533607
4. Schwarz J, Baum GL. Blastomycosis. Am J Clin Pathol. 1951;21 : 999–1029. 14885118
5. Gauthier G, Klein BS. Insights into Fungal Morphogenesis and Immune Evasion: Fungal conidia, when situated in mammalian lungs, may switch from mold to pathogenic yeasts or spore-forming spherules. Microbe Wash DC. 2008;3 : 416–423. 20628478
6. Gauthier GM, Safdar N, Klein BS, Andes DR. Blastomycosis in solid organ transplant recipients. Transpl Infect Dis Off J Transplant Soc. 2007;9 : 310–317. doi: 10.1111/j.1399-3062.2007.00227.x
7. Pfister JR, Archer JR, Hersil S, Boers T, Reed KD, Meece JK, et al. Non-rural point source blastomycosis outbreak near a yard waste collection site. Clin Med Res. 2011;9 : 57–65. doi: 10.3121/cmr.2010.958 20974888
8. Klein BS, Tebbets B. Dimorphism and virulence in fungi. Curr Opin Microbiol. 2007;10 : 314–9. doi: 10.1016/j.mib.2007.04.002 17719267
9. Marr KA, Patterson T, Denning D. Aspergillosis: Pathogenesis, clinical manifestations, and therapy. Infect Dis Clin North Am. 2002;16 : 875–894. 12512185
10. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial Bloodstream Infections in US Hospitals: Analysis of 24,179 Cases from a Prospective Nationwide Surveillance Study. Clin Infect Dis. 2004;39 : 309–317. doi: 10.1086/421946 15306996
11. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS Lond Engl. 2009;23 : 525–530. doi: 10.1097/QAD.0b013e328322ffac
12. Nemecek JC, Wuthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312 : 583–8. doi: 10.1126/science.1124105 16645097
13. Brandhorst TT, Roy R, Wüthrich M, Nanjappa S, Filutowicz H, Galles K, et al. Structure and function of a fungal adhesin that binds heparin and mimics thrombospondin-1 by blocking T cell activation and effector function. PLoS Pathog. 2013;9: e1003464. doi: 10.1371/journal.ppat.1003464 23853587
14. Brandhorst TT, Wüthrich M, Warner T, Klein B. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med. 1999;189 : 1207–1216. 10209038
15. Burg EF, Smith LH. Cloning and characterization of bys1, a temperature-dependent cDNA specific to the yeast phase of the pathogenic dimorphic fungus Blastomyces dermatitidis. Infect Immun. 1994;62 : 2521–2528. 8188377
16. England DM, Hochholzer L. Adiaspiromycosis: an unusual fungal infection of the lung. Report of 11 cases. Am J Surg Pathol. 1993;17 : 876–886. 8352373
17. Kenyon C, Bonorchis K, Corcoran C, Meintjes G, Locketz M, Lehloenya R, et al. A Dimorphic Fungus Causing Disseminated Infection in South Africa. N Engl J Med. 2013;369 : 1416–1424. doi: 10.1056/NEJMoa1215460 24106934
18. Anstead GM, Sutton DA, Graybill JR. Adiaspiromycosis causing respiratory failure and a review of human infections due to Emmonsia and Chrysosporium spp. J Clin Microbiol. 2012;50 : 1346–1354. doi: 10.1128/JCM.00226-11 22259200
19. Peterson SW, Sigler L. Molecular genetic variation in Emmonsia crescens and Emmonsia parva, etiologic agents of adiaspiromycosis, and their phylogenetic relationship to Blastomyces dermatitidis (Ajellomyces dermatitidis) and other systemic fungal pathogens. J Clin Microbiol. 1998;36 : 2918–2925. 9738044
20. Untereiner WA, Scott JA, Naveau FA, Sigler L, Bachewich J, Angus A. The Ajellomycetaceae, a new family of vertebrate-associated Onygenales. Mycologia. 2004;96 : 812–821. 21148901
21. Baumgardner DJ, Paretsky DP. The in vitro isolation of Blastomyces dermatitidis from a woodpile in north central Wisconsin, USA. Med Mycol. 1999;37 : 163–168. 10421847
22. Sullivan TD, Rooney PJ, Klein BS. Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot Cell. 2002;1 : 895–905. 12477790
23. Li W, Sullivan TD, Walton E, Averette AF, Sakthikumar S, Cuomo CA, et al. Identification of the mating-type (MAT) locus that controls sexual reproduction of Blastomyces dermatitidis. Eukaryot Cell. 2013;12 : 109–117. doi: 10.1128/EC.00249-12 23143684
24. Krajaejun T, Wüthrich M, Gauthier GM, Warner TF, Sullivan TD, Klein BS. Discordant influence of Blastomyces dermatitidis yeast-phase-specific gene BYS1 on morphogenesis and virulence. Infect Immun. 2010;78 : 2522–2528. doi: 10.1128/IAI.01328-09 20368350
25. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22 : 2688–90. doi: 10.1093/bioinformatics/btl446 16928733
26. Salichos L, Rokas A. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature. 2013;497 : 327–331. doi: 10.1038/nature12130 23657258
27. Bawdon RE, Garrison RG, Fina LR. Deoxyribonucleic acid base composition of the yeastlike and mycelial phases of Histoplasma capsulatum and Blastomyces dermatitidis. J Bacteriol. 1972;111 : 593–596. 5053471
28. Rouxel T, Grandaubert J, Hane JK, Hoede C, van de Wouw AP, Couloux A, et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat Commun. 2011;2 : 202. doi: 10.1038/ncomms1189 21326234
29. Desjardins CA, Champion MD, Holder JW, Muszewska A, Goldberg J, Bailao AM, et al. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genet. 2011;7: e1002345. doi: 10.1371/journal.pgen.1002345 22046142
30. Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9 : 411–2; author reply 414. doi: 10.1038/nrg2165-c1 18421312
31. Daboussi MJ, Capy P. Transposable elements in filamentous fungi. Annu Rev Microbiol. 2003;57 : 275–99. 14527280
32. Muszewska A, Hoffman-Sommer M, Grynberg M. LTR Retrotransposons in Fungi. PLoS ONE. 2011;6: e29425. doi: 10.1371/journal.pone.0029425 22242120
33. McEwen JG, Ortiz BL, García AM, Florez AM, Botero S, Restrepo A. Molecular cloning, nucleotide sequencing, and characterization of a 27-kDa antigenic protein from Paracoccidioides brasiliensis. Fungal Genet Biol FG B. 1996;20 : 125–131. 8810517
34. Yuen KY, Chan CM, Chan KM, Woo PC, Che XY, Leung AS, et al. Characterization of AFMP1: a novel target for serodiagnosis of aspergillosis. J Clin Microbiol. 2001;39 : 3830–3837. 11682494
35. Gauthier GM, Sullivan TD, Gallardo SS, Brandhorst TT, Vanden Wymelenberg AJ, Cuomo CA, et al. SREB, a GATA transcription factor that directs disparate fates in Blastomyces dermatitidis including morphogenesis and siderophore biosynthesis. PLoS Pathog. 2010;6: e1000846. doi: 10.1371/journal.ppat.1000846 20368971
36. Esteban PF, Ríos I, García R, Dueñas E, Plá J, Sánchez M, et al. Characterization of the CaENG1 gene encoding an endo-1,3-beta-glucanase involved in cell separation in Candida albicans. Curr Microbiol. 2005;51 : 385–392. 16328626
37. Rooney PJ, Sullivan TD, Klein BS. Selective expression of the virulence factor BAD1 upon morphogenesis to the pathogenic yeast form of Blastomyces dermatitidis: evidence for transcriptional regulation by a conserved mechanism. Mol Microbiol. 2001;39 : 875–889. 11251809
38. Beyhan S, Gutierrez M, Voorhies M, Sil A. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 2013;11: e1001614. doi: 10.1371/journal.pbio.1001614 23935449
39. Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U A. 2008;105 : 4880–5. doi: 10.1073/pnas.0710448105
40. Webster RH, Sil A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad Sci U A. 2008;105 : 14573–8. doi: 10.1073/pnas.0806221105
41. Boyce KJ, Schreider L, Kirszenblat L, Andrianopoulos A. The two-component histidine kinases DrkA and SlnA are required for in vivo growth in the human pathogen Penicillium marneffei. Mol Microbiol. 2011;82 : 1164–1184. doi: 10.1111/j.1365-2958.2011.07878.x 22059885
42. Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol. 2004;53 : 1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x 15306022
43. Sentandreu M, Elorza MV, Sentandreu R, Fonzi WA. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J Bacteriol. 1998;180 : 282–289. 9440517
44. Lamb TM, Xu W, Diamond A, Mitchell AP. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem. 2001;276 : 1850–1856. 11050096
45. Citiulo F, Jacobsen ID, Miramón P, Schild L, Brunke S, Zipfel P, et al. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog. 2012;8: e1002777. doi: 10.1371/journal.ppat.1002777 22761575
46. Schmaler-Ripcke J, Sugareva V, Gebhardt P, Winkler R, Kniemeyer O, Heinekamp T, et al. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2009;75 : 493–503. doi: 10.1128/AEM.02077-08 19028908
47. Boguslawski G, Akagi JM, Ward LG. Possible role for cysteine biosynthesis in conversion from mycelial to yeast form of Histoplasma capsulatum. Nature. 1976;261 : 336–338. 1272413
48. Howard DH, Dabrowa N, Otto V, Rhodes J. Cysteine transport and sulfite reductase activity in a germination-defective mutant of Histoplasma capsulatum. J Bacteriol. 1980;141 : 417–421. 7354005
49. Hennicke F, Grumbt M, Lermann U, Ueberschaar N, Palige K, Böttcher B, et al. Factors supporting cysteine tolerance and sulfite production in Candida albicans. Eukaryot Cell. 2013;12 : 604–613. doi: 10.1128/EC.00336-12 23417561
50. Herr RA, Tarcha EJ, Taborda PR, Taylor JW, Ajello L, Mendoza L. Phylogenetic Analysis of Lacazia loboi Places This Previously Uncharacterized Pathogen within the Dimorphic Onygenales. J Clin Microbiol. 2001;39 : 309–314. 11136789
51. Gori S, Drouhet E, Gueho E, Huerre M, Lofaro A, Parenti M, et al. Cutaneous disseminated mycosis in a patient with AIDS due to a new dimorphic fungus. J Mycol Med. 1998;8 : 57–63.
52. Drouhet E, Gu Ho E, Gori S, Huerre M, Provost F, Borgers M, et al. Mycological, Ultrastructural and Experimental Aspects of a New Dimorphic Fungus Emmonsia Pasteuriana sp. nov. Isolated From a Cutaneous Disseminated Mycosis in AIDS. J Mycol Médicale. 2008;8 : 64–77.
53. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409 : 860–921. 11237011
54. Costantini M, Clay O, Auletta F, Bernardi G. An isochore map of human chromosomes. Genome Res. 2006;16 : 536–541. doi: 10.1101/gr.4910606 16597586
55. Bernardi G. The neoselectionist theory of genome evolution. Proc Natl Acad Sci. 2007;104 : 8385–8390. doi: 10.1073/pnas.0701652104 17494746
56. Pavlícek A, Paces J, Clay O, Bernardi G. A compact view of isochores in the draft human genome sequence. FEBS Lett. 2002;511 : 165–169. 11821069
57. Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, et al. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 2009;19 : 1722–31. doi: 10.1101/gr.087551.108 19717792
58. Martinez DA, Oliver BG, Gräser Y, Goldberg JM, Li W, Martinez-Rossi NM, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio. 2012;3: e00259–00212. doi: 10.1128/mBio.00259-12 22951933
59. Tavares AHFP, Silva SS, Dantas A, Campos EG, Andrade RV, Maranhão AQ, et al. Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect Inst Pasteur. 2007;9 : 583–590. doi: 10.1016/j.micinf.2007.01.024
60. Isaac DT, Coady A, Van Prooyen N, Sil A. The 3-hydroxy-methylglutaryl coenzyme A lyase HCL1 is required for macrophage colonization by human fungal pathogen Histoplasma capsulatum. Infect Immun. 2013;81 : 411–420. doi: 10.1128/IAI.00833-12 23184522
61. Sugar AM, Chahal RS, Brummer E, Stevens DA. Susceptibility of Blastomyces dermatitidis strains to products of oxidative metabolism. Infect Immun. 1983;41 : 908–912. 6885169
62. Morrison CJ, Stevens DA. Mechanisms of fungal pathogenicity: correlation of virulence in vivo, susceptibility to killing by polymorphonuclear neutrophils in vitro, and neutrophil superoxide anion induction among Blastomyces dermatitidis isolates. Infect Immun. 1991;59 : 2744–2749. 1649799
63. Youseff BH, Holbrook ED, Smolnycki KA, Rappleye CA. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 2012;8: e1002713. doi: 10.1371/journal.ppat.1002713 22615571
64. Holbrook ED, Smolnycki KA, Youseff BH, Rappleye CA. Redundant catalases detoxify phagocyte reactive oxygen and facilitate Histoplasma capsulatum pathogenesis. Infect Immun. 2013;81 : 2334–2346. doi: 10.1128/IAI.00173-13 23589579
65. Boyce KJ, McLauchlan A, Schreider L, Andrianopoulos A. Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen Penicillium marneffei. PLoS Pathog. 2015;11: e1004790. doi: 10.1371/journal.ppat.1004790 25812137
66. Nunes LR, Costa de Oliveira R, Leite DB, da Silva VS, dos Reis Marques E, da Silva Ferreira ME, et al. Transcriptome analysis of Paracoccidioides brasiliensis cells undergoing mycelium-to-yeast transition. Eukaryot Cell. 2005;4 : 2115–28. 16339729
67. Grumbt M, Monod M, Yamada T, Hertweck C, Kunert J, Staib P. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J Invest Dermatol. 2013;133 : 1550–1555. doi: 10.1038/jid.2013.41 23353986
68. Uyttenhove C, Pilotte L, Théate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9 : 1269–1274. 14502282
69. De Luca A, Carvalho A, Cunha C, Iannitti RG, Pitzurra L, Giovannini G, et al. IL-22 and IDO1 affect immunity and tolerance to murine and human vaginal candidiasis. PLoS Pathog. 2013;9: e1003486. doi: 10.1371/journal.ppat.1003486 23853597
70. Hage CA, Horan DJ, Durkin M, Connolly P, Desta Z, Skaar TC, et al. Histoplasma capsulatum preferentially induces IDO in the lung. Med Mycol. 2013;51 : 270–279. doi: 10.3109/13693786.2012.710857 23181600
71. Araújo EF, Loures FV, Bazan SB, Feriotti C, Pina A, Schanoski AS, et al. Indoleamine 2,3-dioxygenase controls fungal loads and immunity in Paracoccidioidomicosis but is more important to susceptible than resistant hosts. PLoS Negl Trop Dis. 2014;8: e3330. doi: 10.1371/journal.pntd.0003330 25411790
72. Singh A, Panting RJ, Varma A, Saijo T, Waldron KJ, Jong A, et al. Factors required for activation of urease as a virulence determinant in Cryptococcus neoformans. mBio. 2013;4: e00220–00213. doi: 10.1128/mBio.00220-13 23653445
73. Mirbod-Donovan F, Schaller R, Hung C-Y, Xue J, Reichard U, Cole GT. Urease produced by Coccidioides posadasii contributes to the virulence of this respiratory pathogen. Infect Immun. 2006;74 : 504–515. 16369007
74. Jaffe DB, Butler J, Gnerre S, Mauceli E, Lindblad-Toh K, Mesirov JP, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13 : 91–6. 12529310
75. Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1 : 18. doi: 10.1186/2047-217X-1-18 23587118
76. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19 : 1117–1123. doi: 10.1101/gr.089532.108 19251739
77. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18 : 821–9. doi: 10.1101/gr.074492.107 18349386
78. Sambrook J, Russell DW. Molecular cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 7.4–7.8.
79. Wüthrich M, Filutowicz HI, Warner T, Klein BS. Requisite Elements in Vaccine Immunity to Blastomyces dermatitidis: Plasticity Uncovers Vaccine Potential in Immune-Deficient Hosts. J Immunol. 2002;169 : 6969–6976. doi: 10.4049/jimmunol.169.12.6969 12471131
80. Marty AJ, Wüthrich M, Carmen JC, Sullivan TD, Klein BS, Cuomo CA, et al. Isolation of Blastomyces dermatitidis yeast from lung tissue during murine infection for in vivo transcriptional profiling. Fungal Genet Biol FG B. 2013; doi: 10.1016/j.fgb.2013.03.001 23499858
81. Worsham PL, Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol Bi-Mon Publ Int Soc Hum Anim Mycol. 1988;26 : 137–143.
82. Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37: e123. doi: 10.1093/nar/gkp596 19620212
83. Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7 : 709–715. doi: 10.1038/nmeth.1491 20711195
84. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29 : 644–652. doi: 10.1038/nbt.1883 21572440
85. Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9: R7. doi: 10.1186/gb-2008-9-1-r7 18190707
86. Haas BJ, Zeng Q, Pearson MD, Cuomo CA, Wortman JR. Approaches to Fungal Genome Annotation. Mycology. 2011;2 : 118–141. 22059117
87. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10: R25. doi: 10.1186/gb-2009-10-3-r25 19261174
88. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12 : 323. doi: 10.1186/1471-2105-12-323 21816040
89. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26 : 139–40. doi: 10.1093/bioinformatics/btp616 19910308
90. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8 : 1494–1512. doi: 10.1038/nprot.2013.084 23845962
91. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21 : 3674–6. doi: 10.1093/bioinformatics/bti610 16081474
92. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001;25 : 402–408.
93. Oliver JL, Carpena P, Hackenberg M, Bernaola-Galván P. IsoFinder: computational prediction of isochores in genome sequences. Nucleic Acids Res. 2004;32: W287–292. doi: 10.1093/nar/gkh399 15215396
94. Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20 : 3643–6. doi: 10.1093/bioinformatics/bth397 15247098
95. Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110 : 462–7. doi: 10.1159/000084979 16093699
96. Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7: e1002195. doi: 10.1371/journal.pcbi.1002195 22039361
97. Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16 : 276–7. 10827456
98. Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9 : 286–298. doi: 10.1093/bib/bbn013 18372315
99. Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PloS One. 2010;5: e9490. doi: 10.1371/journal.pone.0009490 20224823
100. Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39: W475–W478. doi: 10.1093/nar/gkr201 21470960
101. Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13 : 555–6. 9367129
102. Goldberg JM, Griggs AD, Smith JL, Haas BJ, Wortman JR, Zeng Q. Kinannote, a computer program to identify and classify members of the eukaryotic protein kinase superfamily. Bioinforma Oxf Engl. 2013;29 : 2387–2394. doi: 10.1093/bioinformatics/btt419
103. Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, et al. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41: W204–212. doi: 10.1093/nar/gkt449 23737449
104. Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13 : 2178–89. 12952885
105. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U A. 2003;100 : 9440–5.
Štítky
Genetika Reprodukčná medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



