-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
The transcription and export of RNA is a dynamic and highly coordinated process. mRNA species that are selectively mediated by the THO/TRanscription EXport (THO/TREX) complex for their transcription and export remain to be identified. As well, the specific roles of complex components in transcription-coupled export are unclear. We reveal a role for HYPER RECOMBINATION1 (HPR1) [the yeast HYPER RECOMBINATION1 (Hpr1) homolog] in REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1) transcription elongation but not activation or export, which agrees with the role of yeast Hpr1 in transcription elongation. Defects in the THO/TREX component TEX1 but not the RNA-export TREX-2 component SAC3B also reduced the native RTE1 level. Our data suggest a specific role of the THO/TREX component HPR1 in RTE1 expression. Whether TEX1 is involved in RTE1 transcription or stability remains to be determined. The yeast Sub2 protein is an RNA helicase involved in unwinding the inhibitory structure in the nascent RNA, and SUB2 overexpression suppresses yeast Δhpr1 defects; HPR1 could be involved in expression of selected genes with higher-order structure, where RNA polymerase movement could pause. Studies of the gene structure and transcription activity could shed light on roles of these components in gene expression regulation at the transcription-export level.
Published in the journal: HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis. PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004956
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004956Summary
The transcription and export of RNA is a dynamic and highly coordinated process. mRNA species that are selectively mediated by the THO/TRanscription EXport (THO/TREX) complex for their transcription and export remain to be identified. As well, the specific roles of complex components in transcription-coupled export are unclear. We reveal a role for HYPER RECOMBINATION1 (HPR1) [the yeast HYPER RECOMBINATION1 (Hpr1) homolog] in REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1) transcription elongation but not activation or export, which agrees with the role of yeast Hpr1 in transcription elongation. Defects in the THO/TREX component TEX1 but not the RNA-export TREX-2 component SAC3B also reduced the native RTE1 level. Our data suggest a specific role of the THO/TREX component HPR1 in RTE1 expression. Whether TEX1 is involved in RTE1 transcription or stability remains to be determined. The yeast Sub2 protein is an RNA helicase involved in unwinding the inhibitory structure in the nascent RNA, and SUB2 overexpression suppresses yeast Δhpr1 defects; HPR1 could be involved in expression of selected genes with higher-order structure, where RNA polymerase movement could pause. Studies of the gene structure and transcription activity could shed light on roles of these components in gene expression regulation at the transcription-export level.
Introduction
In Arabidopsis, the gaseous plant hormone ethylene is perceived by a family of five ethylene receptor members that structurally resemble prokaryotic “two-component” histidine kinases (HKs). Ethylene receptor signaling is not associated with HK activity, and the biochemical nature of the receptor signal remains to be determined [1]. Without biochemical knowledge of the ethylene receptor signal output, ethylene receptor signaling is largely evaluated by ethylene-induced growth inhibition or altered gene expression.
Through molecular genetic studies, a model for ethylene receptor signaling has been proposed. Without ethylene, the ethylene receptors mediate the receptor signal output to suppress ethylene signaling; ethylene binding to the receptors prevents receptor signaling and suppression of ethylene signaling is relieved [2, 3]. The five members of the ethylene receptor family have common and divergent functions and act cooperatively as complexes [4–7]. The ethylene receptor ETR1 can function largely independently of other family members to suppress the ethylene response to a great extent. In contrast, ETHYLENE RESPONSE SENSOR1 (ERS1) functions differentially depending on other family members [6, 7].
REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1) was isolated from a mutation that suppresses the dominant ethylene-insensitive etr1–2 mutation [8]. RTE1 encodes a membrane protein that is associated with the endoplasmic reticulum (ER) and Golgi apparatus [9, 10]. RTE1 overexpression results in ethylene insensitivity in wild-type plants depending on ETR1 ethylene receptor but not on other family members possibly via the ETR1 N-terminus [8, 10–12]. RTE1 homologs are prevalent in higher eukaryotes, and expression of the rice RTE1 HOMOLOG1 (OsRTH1) complements the rte1–2 loss-of-function mutation and promotes ETR1 receptor signaling in Arabidopsis [13]. Elevated expression of the tomato RTE1 homolog GREEN RIPE (GR), with the dominant Gr mutation or CaMV35S:GR transgene, results in reduced ethylene responsiveness in fruit tissue and thus the non-ripening phenotype [14]. RTE1 functions in ethylene signaling may be highly conserved across higher plant species.
To identify components mediating RTE1 functions, we performed a suppressor screen for RTE1 overexpressor (RTE1ox) and isolated HYPER RECOMBINATION1 (HPR1). HPR1 encodes a protein homologous to yeast (Saccharomyces cerevisiae) HYPER-RECOMBINATION1 (Hpr1) protein, a component of the mRNA transcription export machinery, namely the THO complex. Of note, mutations that affect the mRNA transcription export machinery have been reported in plants and they result in various defects in response to pathogens, stresses, and hormones; however, the involved mRNA species are largely undetermined [15–19]. In yeast, the Δhpr1 mutation results in increased intrachromosomal recombination events that specifically depend on transcription elongation involving RNA polymerase II [20–23]. Proper progression of transcription over specific DNA structures would otherwise pause without Hpr1 [21]. Arabidopsis HPR1 was previously suggested to be involved in trans-acting small interfering RNA (tasi-RNA) production [15, 24], and the mutation also confers pleiotropic phenotypes and reduced ERECTA expression and defects in alternative splicing [15, 25, 26]. Arabidopsis hpr1–4 showed bulk nuclear mRNA accumulation, and the mRNA species that requires HPR1 for nucleocytoplasmic export remains to be determined [26].
Yeast THO comprises Hpr1, Mft1, Tho2, and Thp2, and Hpr1 bridges the Sub2-Yra1 dimer to form the THO/TRanscription EXport (TREX) complex [20, 23, 27, 28]. Other proteins, such as TEX1, were identified in yeast, human and Arabidopsis THO/TREX [24]. Transcription is dynamic and involves coordinated and complicated processes including transcription elongation, docking of various RNA binding proteins to the nascent transcript, 5’ capping, 3’ polyadenylation, and mRNA splicing. During transcription, mRNA binding factors and nascent mRNAs are assembled dynamically into the messenger ribonucleaoprotein particles (mRNPs), which are eventually exported through the nuclear pore complex to the cytoplasm. THO/TREX plays an important role in these coordinated processes [27, 29]. Arabidopsis TEX1 is a component of the TREX complex, and defects in TEX1 result in reduced amounts of tasi-RNA species derived from their TAS1 and TAS2 precursors but not TAS3, which indicates involvement of the THO/TREX complex in the processing or transport of RNA precursors for certain tasi-RNA species [15, 24].
The isolation of HPR1 prompted us to investigate how RTE1 functions were affected in the absence of HPR1. Results from our study provide evidence supporting the involvement of the THO/TREX component HPR1 in RTE1 expression and less likely in its degradation and export, which is consistent with the role of yeast Hpr1 in transcription elongation [20, 23]. We also investigated the roles of other components of the transcription-export machinery in RTE1 expression. Nuclear RTE1 level was reduced in the absence of TEX1, and the role of TEX1 in RTE1 expression remains to be determined.
Results
Loss-of-function Allele of SUPPRESSOR OF RTE1 OVEREXPRESSION1 (SRT1) Suppresses RTE1ox
To isolate components involving RTE1 functions, we performed a suppressor screen for an RTE1 overexpressor [RTE1ox; 35S:RTE1 expressed in the wild type (Col-0)]. Ethylene treatment inhibited the hypocotyl elongation of etiolated wild-type (Col-0) seedlings. RTE1ox was ethylene insensitive, and the seedling produced a long hypocotyl with ethylene treatment [8, 10]. We isolated the mutagenized RTE1ox seedlings, at the M2 generation, showing ethylene growth inhibition. Genes required for ethylene insensitivity conferred by RTE1 overexpression were designated SRT.
Etiolated seedlings of the wild type (Col-0), RTE1ox, and srt1–1 RTE1ox produced a long hypocotyl without ethylene treatment; ethylene treatment inhibited the hypocotyl elongation of wild-type and srt1–1 RTE1ox but not RTE1ox seedlings (Fig. 1A). With ethylene treatment, the hypocotyl was short for wild-type and srt1–1 RTE1ox seedlings and relatively long for RTE1ox seedlings (Fig. 1B).
Fig. 1. Effect of SUPPRESSOR OF RTE1 OVEREXPRESSION1 (SRT1) on the ethylene insensitivity in REVERSION-TO-ETHYLENE SENSITIVITY1 overexpressor (RTE1ox). 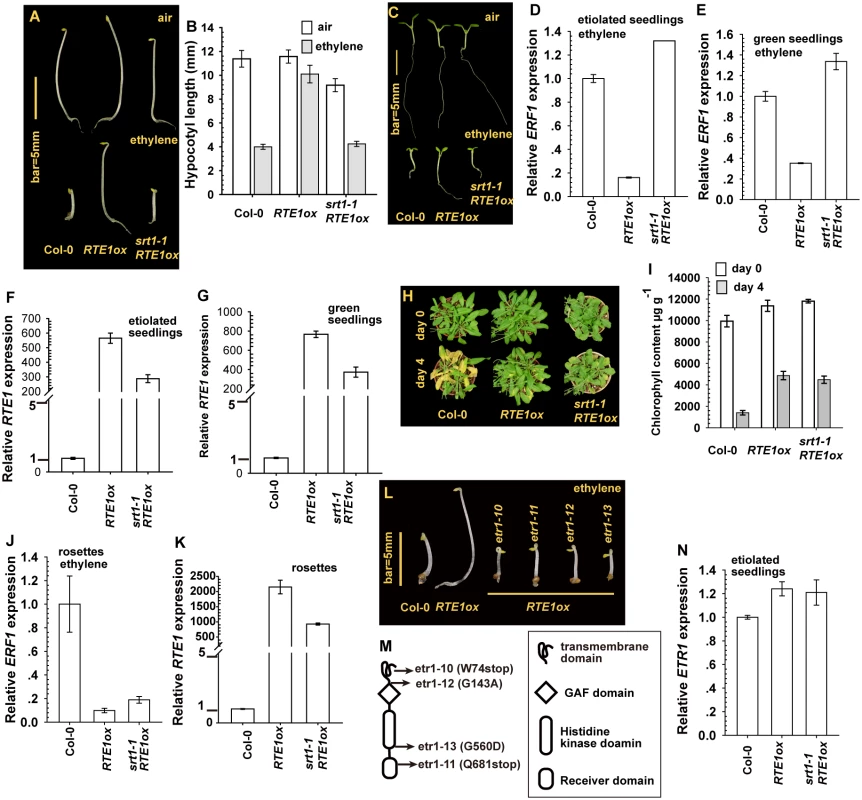
Etiolated seedling phenotype (A), hypocotyl measurement (B), and light-grown seedling phenotype (C) for RTE1ox and srt1–1 RTE1ox seedlings. Expression of ETHYLENE RESPONSE FACTOR1 (ERF1) in RTE1ox with the srt1–1 allele in etiolated (D) and light-grown (E) seedlings on ethylene treatment. RTE1 expression in RTE1ox and srt1–1 RTE1ox seedlings grown in dark (F) and light (G). Leaf senescence phenotype (H) and chlorophyll content (I) in RTE1ox and srt1–1 RTE1ox plants. ERF1 (J) and RTE1 (K) expression in rosette leaves. Phenotype (L) of etiolated RTE1ox seedlings with the indicated etr1 mutations (M). (N) ETR1 expression is not altered by srt1–1. Data are mean±SD for seedling hypocotyls (n>30) and chlorophyll content (n>3), and mean± SE for gene expression (at least 3 independent biological samples with 3 measurement for each). Ethylene concentration is 10 μL L-1 for the seedling growth inhibition test and ERF1 induction, and 100 μL L-1 for the leaf senescence test. Germinated under light, ethylene-treated wild-type and srt1–1 RTE1ox seedlings showed growth inhibition, with small and compact cotyledons and a short root, whereas RTE1ox seedlings produced expanded cotyledons and a long root (Fig. 1C). Without ethylene treatment, all genotypes examined showed a normal growth phenotype. The expression of ETHYLENE RESPONSE FACTOR1 (ERF1) is induced by ethylene treatment [30, 31]; with ethylene, ERF1 level was higher in wild-type and srt1–1 RTE1ox than RTE1ox seedlings (Fig. 1D-1E). The suppression of RTE1ox by srt1–1 may be associated with reduced RTE1 level at the seedling stage (Fig. 1F-1G).
At the adult stage, ethylene did not promote leaf senescence in ethylene-insensitive plants. With 4-day ethylene treatment, wild-type but not RTE1ox and srt1–1 RTE1ox plants showed the leaf senescence phenotype (Fig. 1H). Consistently, after ethylene treatment, the chlorophyll content was more greatly reduced in the wild type (Col-0) than in RTE1ox and srt1–1 RTE1ox plants (Fig. 1I). Ethylene treatment elevated the ERF1 level in rosette leaves of the wild type but not RTE1ox and srt1–1 RTE1ox leaves (Fig. 1J), although RTE1 level was reduced by the srt1–1 allele (Fig. 1K).
Of note, in the suppressor screen, 4 alleles of ETR1 ethylene receptor gene were isolated (Fig. 1L-1M), consistent with the notion of ethylene insensitivity conferred by RTE1 overexpression depending on ETR1 [8, 10, 32]. Among these alleles, etr1–10 was previously described with the W74stop early termination [33]. ETR1 expression was not altered by the srt1–1 mutation; srt1–1 did not reduce ETR1 level to suppress RTE1ox (Fig. 1N).
The srt1–1 allele prevented ethylene insensitivity conferred by RTE1 overexpression mainly during the seedling but not adult stage. In RTE1ox, RTE1 level was reduced by the srt1–1 allele throughout development. A lesser amount of RTE1 could be sufficient to confer ethylene insensitivity at the adult but not seedling stage.
Cloning of SRT1 and Isolation of the Loss-of-function Alleles
We performed map-based cloning and mapped SRT1 to a 100-kb region at chromosome 5. This region was sequenced, and we identified only one nucleotide change (the G1665A transition mutation), which resulted in the W220stop early termination of the HPR1 protein (Fig. 2A; locus At5g09860); herein SRT1 is designated HPR1 and the srt1–1 allele hpr1–5. A suppressor screen of the loss-of-function mutant of ENHANCED DISEASE RESISTANCE1 (EDR1) isolated a loss-of-function allele of HPR1 [26] at the ERECTA mRNA UNDER-EXPRESSED (EMU) locus, and the loss-of-function emu mutant produced a reduced amount of ERECTA and several miRNAs [25]. Sequence analysis suggested that HPR1/EMU shared relatively low homology with the yeast (Saccharomyces cerevisiae) Hpr1 protein of the THO/TREX complex involved in mRNA transcription and export [25]. A genomic clone for HPR1 (HPR1p:gHPR1) was introduced into hpr1–5 RTE1ox by transformation, and etiolated seedlings of the resulting lines were ethylene-insensitive and produced a long hypocotyl with ethylene treatment (Fig. 2B-2C). The hpr1–2 mutation [in a Wassilewskija (Ws) ecotype background] resulted from a T-DNA insertion at the 8th intron of HPR1 [15]; etiolated hpr1–2 RTE1ox seedlings also showed growth inhibition with ethylene (Fig. 2A and 2D). Consistently, ethylene treatment inhibited the growth of light-grown hpr1–5 RTE1ox and hpr1–2 RTE1ox seedlings (Fig. 2E).
Fig. 2. The cloning and isolation of HPR1/SRT1 and the mutants. 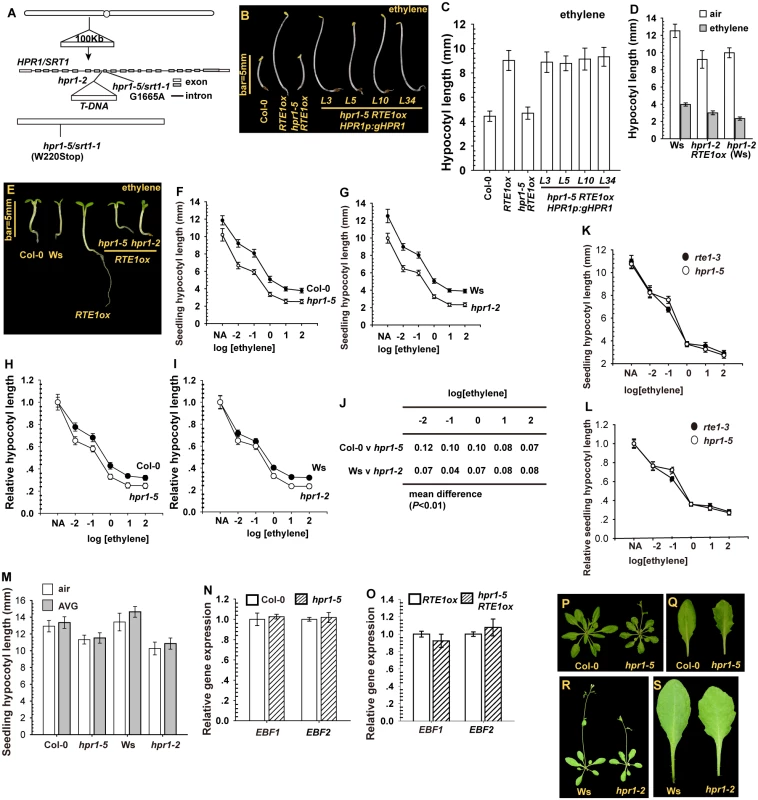
(A) Map-based cloning of HPR1/SRT1, and isolation of the hpr1–5/srt1–1 and hpr1–2 alleles. Etiolated seedling phenotype (B) and hypocotyl measurement (C) for hpr1–5 RTE1ox expressing the HPR1p:gHPR1 transgene. Etiolated seedling hypocotyl measurement (D) and light-grown seedling phenotype (E) for hpr1–2 RTE1ox. Ethylene concentration is 10 μL L-1. Ethylene dose–response assay by etiolated seedling hypocotyl measurement of hpr1–5 (F) and hpr1–2 (G) and by normalized hypocotyl measurement for hpr1–5 (H) and hpr1–2 (I). (J) Mean difference in normalized hypocotyl length between wild-type and hpr1–5 and hpr1–2 seedlings. Ethylene dose-response curves of the absolute (K) and normalized (L) lengths of hypocotyls of etiolated rte1–3 and hpr1–5 seedlings. (M) Hypocotyl length of etiolated seedlings with AVG treatment. Relative gene expression of EIN3-BINDING F-BOX PROTEIN1 (EBF1) and EBF2 in the wild type (Col-0) and hpr1–5 (N) and RTE1ox and hpr1–5 RTE1ox (O). Rosette (P) and leaf (Q) phenotype of the wild type (Col-0) and hpr1–5 plants. Rosette (R) and leaf (S) phenotype of the wild type (Ws) and hpr1–2 plants. Data are mean±SD for seedling hypocotyls (n>30) and mean±SE for gene expression measurement (n = 3 with 3 technical repeats). Student’s t test is for paired comparison for means of 2 measurements. Ethylene concentration presented in logarithm of ethylene (μL L-1) for the dose-response assay. Of note, hpr1–5 and hpr1–2 single mutants produced a slightly shorter seedling hypocotyl than did wild-type seedlings over a wide range of ethylene concentration (Fig. 2F-2G). The seedling hypocotyl length was measured and normalized to that of non-treated seedlings; the relative seedling hypocotyl lengths were slightly shorter for hpr1–5 and hpr1–2 than the wild type (Col-0 and Ws) (Fig. 2H-2J).
RTE1ox was suppressed by hpr1–5, which prompted us to evaluate whether RTE1 function could be suppressed by hpr1–5 such that hpr1 and rte1 seedlings could behave similarly in response to ethylene treatment. Over a wide range of ethylene concentration, hpr1–5 and the loss-of-function rte1–3 seedlings produced a nearly identical dose-response curve for seedling hypocotyl length (Fig. 2K–2L). On treatment with the ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG), hpr1 seedling hypocotyl growth was not rescued to the wild-type level, so the seedling hypocotyl shortening was not due to an elevation in endogenous ethylene biosynthesis (Fig. 2M).
The F-box proteins EIN3-BINDING F-BOX PROTEIN1 (EBF1) and EBF2 have a role in EIN3 degradation via the 26S proteasome, and the mutants are hypersensitive to ethylene, with a weak phenotype of seedling growth inhibition [34, 35]. We investigated whether the hpr1–5 mutation could affect EBF1 and EBF2 expression and therefore seedling growth. qRT-PCR revealed nearly identical expression of EBF1 and EBF2 between wild-type (Col-0) and hpr1–5 seedlings, regardless of the 35S:RTE1 transgene (Fig. 2N-2O). RTE1ox suppression and increased ethylene sensitivity by the hpr1–5 allele was not associated with EBF1/EBF2 levels.
At the adult stage, hpr1–5 and hpr1–2 plants were smaller than their wild-type ecotypes (Col-0 and Ws) in rosette size, and mutant leaves were serrated (Fig. 2P-2S), which is consistent with the emu mutant phenotype [25]. HPR1 could have roles in leaf morphogenesis and other aspects of plant growth and development in addition to its role in RTE1 level.
hpr1–5 Inhibits Ethylene Insensitivity Conferred by etr1–2 but not ein2–50 and ein3–1
The signal output by ETR1 that inhibits ethylene signaling is prevented by ethylene. The etr1–2 mutation, resulting from an A102T substitution, confers dominant ethylene insensitivity [36]. RTE1 is essential to ethylene insensitivity conferred by etr1–2 [8, 10]. Given that hpr1–5 suppressed RTE1ox and that hpr1–5 and rte1–3 seedlings behaved similarly in response to ethylene treatment (Fig. 2K–2L), we wondered whether the hpr1–5 mutation could also suppress etr1–2.
With ethylene treatment, etiolated hpr1–5 etr1–2 but not etr1–2 seedlings showed growth inhibition (Fig. 3A), which agrees with the seedling hypocotyl measurement (Fig. 3B). Consistently, ethylene growth inhibition was stronger for light-grown wild-type (Col-0) and hpr1–5 etr1–2 than etr1–2 seedlings (Fig. 3C). With ethylene treatment, ERF1 level was high for wild-type and hpr1–5 etr1–2 seedlings and low for etr1–2 seedlings (Fig. 3D-3E).
Fig. 3. The hpr1–5 allele suppressed etr1–2 ethylene insensitivity at the seedling stage. 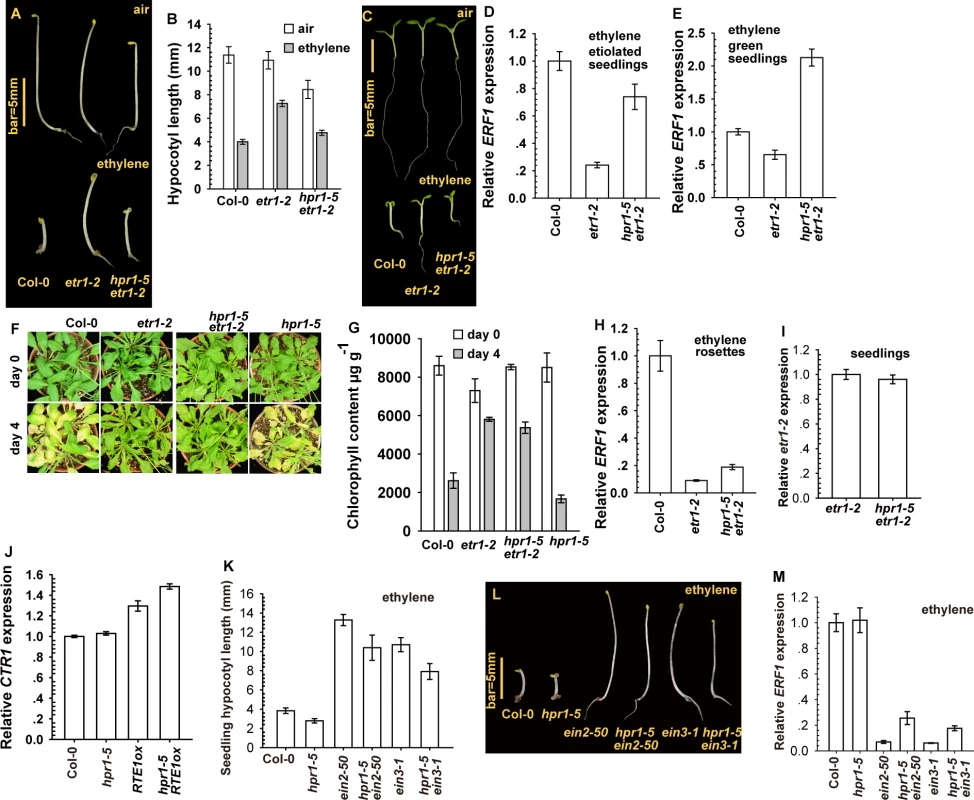
Etiolated seedling phenotype (A), hypocotyl measurement (B), and light-grown seedling phenotype (C) for etr1–2 and hpr1–5 etr1–2. ERF1 expression in etiolated (D) and light-grown (E) seedlings of etr1–2 and hpr1–5 etr1–2. Leaf senescence phenotype (F) and chlorophyll content (G) in etr1–2 and hpr1–5 etr1–2 plants. (H) ERF1 expression in etr1–2 and hpr1–5 etr1–2 rosettes. Expression of etr1–2 (I) and CTR1 (J) is not reduced by the hpr1–5 mutation. Measurement (K) and phenotype (L) of hypocotyls of etiolated seedlings of ein2–50 and ein3–1 mutants with and without the hpr1–5 allele, and ERF1 expression in these genotypes (M). Data are mean±SD seedling hypocotyls (n>30) and chlorophyll content (n>3), and mean±SE for gene expression (at least 3 independent biological samples with 3 measurements for each). Ethylene concentrations are 10 μL L-1 for ERF1 induction and 100 μL L-1 for the leaf senescence test. The hpr1–5 allele had little effect on the ethylene insensitivity conferred by RTE1 overexpression at the adult stage (Fig. 1H-1J). As expected, the hpr1–5 allele had little effect on etr1–2 at the adult stage, and 4-day ethylene treatment did not induce leaf senescence in etr1–2 or hpr1–5 etr1–2 rosette leaves (Fig. 3F). Reduction in chlorophyll content was greater in wild-type (Col-0) and hpr1–5 than etr1–2 and hpr1–5 etr1–2 plants (Fig. 3G). Consistently, ERF1 level that was elevated in the wild type (Col-0) was not elevated with ethylene treatment in etr1–2 or hpr1–5 etr1–2 adult plants (Fig. 3H). qRT-PCR revealed a similar etr1–2 expression in hpr1–5 etr1–2 and etr1–2 seedlings, so hpr1–5 did not affect etr1–2 level to suppress etr1–2 ethylene insensitivity (Fig. 3I). Of note, CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) is a Raf-like protein mediating the ethylene receptor signaling to repress ethylene signaling, and defects in CTR1 result in elevated ethylene responses [37, 38]. CTR1 expression was not reduced by hpr1–5 (Fig. 3J), so the hpr1–5 mutation did not impact CTR1 level to affect the ethylene signaling.
We considered whether hpr1–5 could simply suppress ethylene insensitivity without specificity for RTE1. This scenario was evaluated by examining whether ethylene insensitivity by other mutations in the ethylene signaling pathway could be suppressed by hpr1–5. ETHYLENE INSENSITIVE2 (EIN2) acts downstream of CTR1 to mediate ethylene signaling to the transcription factor EIN3 for inducing the ethylene response [39]. The loss-of-function ein2–50 (from T-DNA insertion) [40, 41] and ein3–1 [30] mutants were ethylene-insensitive and each was crossed with hpr1–5 to obtain hpr1–5 ein2–50 and hpr1–5 ein3–1. With ethylene treatment, etiolated ein2–50 and ein3–1 seedlings were ethylene-insensitive and the hypocotyl elongation was not inhibited, whereas wild-type (Col-0) and hpr1–5 seedlings were ethylene-responsive, with hypocotyl growth inhibition (Fig. 3K-3L). Seedlings of hpr1–5 ein2–50 and hpr1–5 ein3–1 were slightly shorter than those of ein2–50 and ein3–1, respectively, possibly because of the intrinsic shortness of hpr1–5 seedlings. Consistently, ERF1 expression was relatively low in genotype with ein2–50 or ein3–1 compared to the wild type (Col-0) and hpr1–5 (Fig. 3M).
The suppression of etr1–2 seedling ethylene insensitivity by hpr1–5 agreed with the suppression of RTE1ox ethylene insensitivity by hpr1–5, which supports HPR1 as essential to RTE1 functions at the seedling stage. With the inability of hpr1–5 to suppress ethylene insensitivity of the ein2–50 and ein3–1 mutations, the suppression of etr1–2 and RTE1 functions by hpr1–5 was not achieved by inhibiting ethylene insensitivity, which indicates specificity of HPR1 for RTE1.
The hpr1–5 Allele Prevents Export of Bulk mRNA but Not RTE1 Transcripts
HPR1 shares homology with yeast Hpr1, so HPR1 may have a role in mRNA processing and export from the nucleus to the cytoplasm. The role of Arabidopsis HPR1 in bulk mRNA export was recently demonstrated in the hpr1–4 allele [26]. We examined whether the suppression of RTE1ox and etr1–2 mutation by the hpr1–5 allele was due to impaired RTE1 transcript export.
With the fluoresceine-labeled oligo(dT)45 as a probe hybridized with polyadenylated mRNA, hpr1–5 and hpr1–5 RTE1ox but not wild-type or RTE1ox cells produced fluorescence within the nucleus (Fig. 4A). The results suggest that the suppression of RTE1ox by hpr1–5 could be associated with impaired bulk mRNA export. The impaired export of mRNA species that involve RTE1 functions could result in the suppression of RTE1ox and etr1–2. Alternatively, HPR1 may directly involve RTE1 transcription or export, and the hpr1–5 mutation suppressed RTE1ox and etr1–2. Therefore, we examined whether RTE1 transcripts could accumulate in the nucleus of the hpr1–5 mutant; none of those genotypes produced fluorescence in the nucleus when hybridized with the fluoresceine-labeled RTE1 probe (Fig. 4B).
Fig. 4. Effect of the hpr1–5 allele on bulk and RTE1 mRNA accumulation in the nucleus. 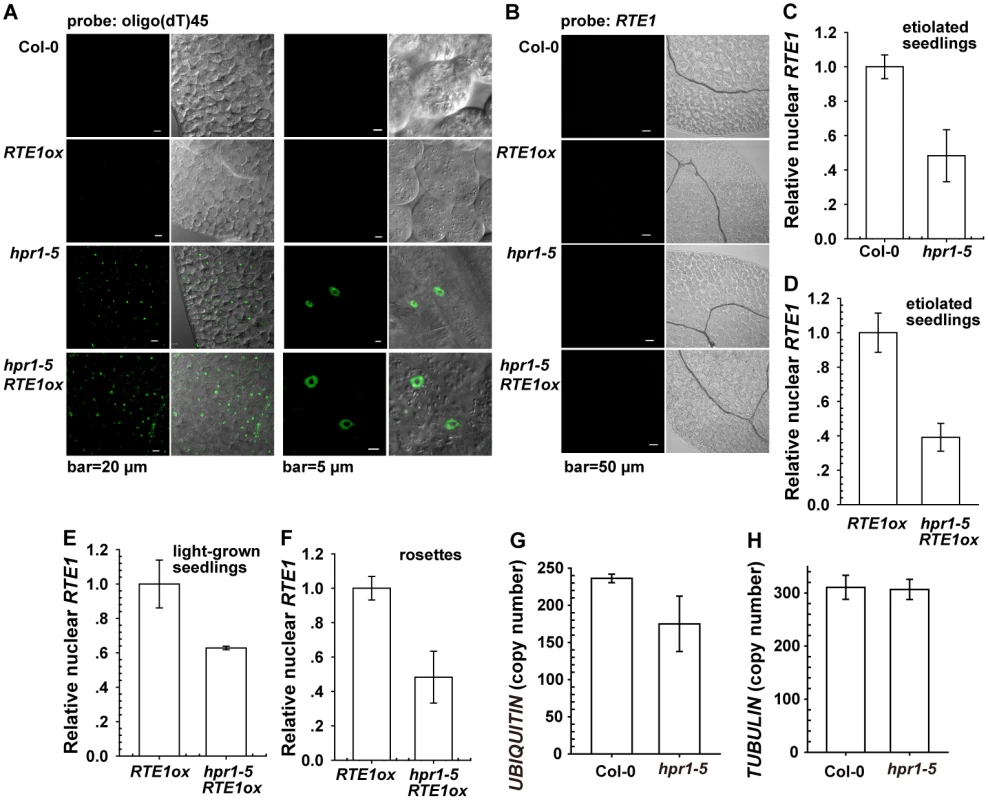
Nuclear fluorescence by fluorescine-labeled oligo(dT)45 (A) and RTE1 (B) probes. Nuclear RTE1 level in etiolated seedlings of the wild-type (Col-0) and hpr1–5 (C); etiolated (D) and light-grown (E) RTE1ox and hpr1–5 RTE1ox seedlings; and RTE1ox and hpr1–5 RTE1ox rosette leaves (F). Measurement of the nuclear UBIQUITIN (G) and TUBULIN (H) transcript copy number in the wild type (Col-0) and hpr1–5. Data are mean±SE, with 3 measurements for each 3 independent biological samples. The lack of evidence for RTE1 transcript accumulation in the nucleus of the hpr1–5 mutant could be due to limited detection sensitivity or hpr1–5 not affecting RTE1 transcript export. Therefore, we measured nuclear RTE1 levels in the wild type (Col-0) and genotypes with the hpr1–5 alleles. Nuclei were isolated from the wild type, RTE1ox, hpr1–5, and hpr1–5 RTE1ox for RNA isolation. The nuclear RTE1 level was reduced to 40% to 60% by the hpr1–5 allele (Fig. 4C - 4F). The total (Fig. 1F, 1G and 1K) and nuclear RTE1 level (Fig. 4C-4F) was reduced to a similar level by the hpr1–5 allele, with no nuclear RTE1 accumulation (Fig. 4B). Therefore, hpr1–5 had little effect on RTE1 transcript export. Of note, considering that defects caused by the HPR1 mutation could affect expression of the calibration gene on qRT-PCR, we examined the expression of 2 widely used normalization genes. By calibrating against the standard curve, with cDNA used as the template, the copy number for the nuclear UBIQUITIN was reduced in hpr1–5 compared to the wild type (Col-0) (Fig. 4G) and unaltered for the nuclear TUBULIN (Fig. 4H). Throughout the work, if not specified, TUBULIN was used as the normalization gene for qRT-PCR.
HPR1/EMU is required for normal expression of ERECTA and several miRNAs [25]. Our results showed that RTE1 level was reduced in the hpr1–5 mutant; these data support HPR1 as required for the normal expression of RTE1.
RTE1 Transcript Degradation Is Not Accelerated by the hpr1–5 Allele
The nuclear RTE1 transcription was reduced by the hpr1–5 allele, which could be caused by reduced transcription activity or accelerated degradation. Cordycepin (or 3'-deoxyadenosine) is structurally analogous to adenosine, and its incorporation into RNA molecules causes premature termination of biosynthesis. Treatment with cordycepin to terminate transcription facilitates kinetics study of the transcript degradation [42–45]. eIF4A has a prolonged, stable half life [46] and was used as an ideal internal reference for measuring RTE1 level with cordycepin treatment.
Light-grown seedlings of Col-0, hpr1–5, RTE1ox, and hpr1–5 RTE1ox were incubated with cordycepin to terminate RNA biosynthesis. As compared with before cordycepin treatment (time 0), after treatment, RTE1 level was reduced over time, with a lower level for hpr1–5 than wild-type seedlings (Fig. 5A). To evaluate the degradation rate, RTE1 levels were normalized to the level at time 0 (before treatment); normalized levels were identical in the wild type and hpr1–5 (Fig. 5B). Consistently, RTE1 level in RTE1ox and hpr1–5 RTE1ox decreased quickly after treatment, with a greater level for RTE1ox than hpr1–5 RTE1ox (Fig. 5C). When normalized to the level at time 0, RTE1 levels were identical in RTE1ox and hpr1–5 RTE1ox (Fig. 5D). Therefore, the hpr1–5 allele did not accelerate RTE1 degradation, so hpr1–5 could possibly affect RTE1 transcription.
Fig. 5. RTE1 degradation with cordycepin treatment. 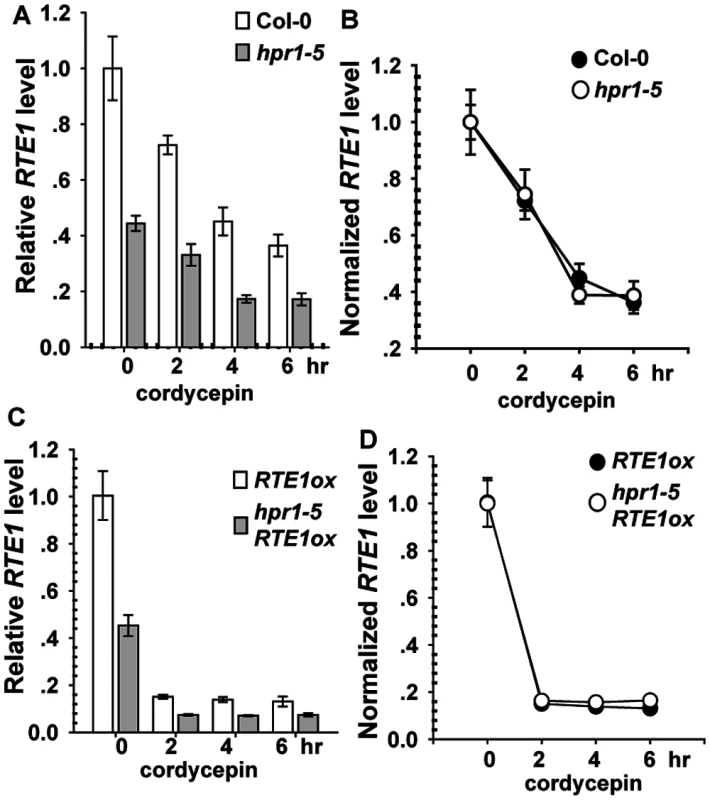
Measured (A) and normalized (B) RTE1 level with cordycepin treatment in wild-type (Col-0) and hpr1–5 seedlings. Measured (C) and normalized (D) RTE1 level with cordycepin treatment in RTE1ox and hpr1–5 RTE1ox seedlings. Data are mean±SE, with 3 measurements for each 3 independent biological samples. HPR1 Is a Nuclear Protein Co-localized with mRNA Splicing Complex
HPR1 was predicted to be a yeast Hpr1 homolog involved in RNA transcription and export. To further our knowledge of HPR1 function, we studied its subcellular localization with the expression of GREEN FLUORESCENT PROTEIN (GFP)-tagged HPR1.
To obtain an HPR1 cDNA fragment to be fused with GFP, we cloned HPR1 cDNA and could clone 2 HPR1 cDNA species, HPR1a and HPR1b. Sequence analysis for the 2 cDNA clones suggested that they resulted from alternative splicing, and HPR1a contained an extra codon for lysine. Thus, HPR1a was 1 amino acid residue longer than HPR1b (Fig. 6A). To determine whether either of the HPR1 species could be functional, we fused cDNA clones for HPR1a and HPR1b with GFP and expressed them under regulation of the constitutive 35S promoter in hpr1–5 RTE1ox. With ethylene treatment, etiolated seedlings of hpr1–5 RTE1ox were short and those of RTE1ox were long. Expression of 35S:GFP-HPR1a and - HPR1b rescued the ethylene growth inhibition phenotype to a great extent in hpr1–5 RTE1ox, which agrees with the seedling hypocotyl measurement (Fig. 6B-6C). GFP-HPR1a and-HPR1b both produced fluorescence in the nucleus with a pattern characteristic of nuclear speckle domains in cells of etiolated seedling hypocotyls (Fig. 6D-6E).
Fig. 6. HPR1 produces 2 transcripts and HPR1 subcellular localization. 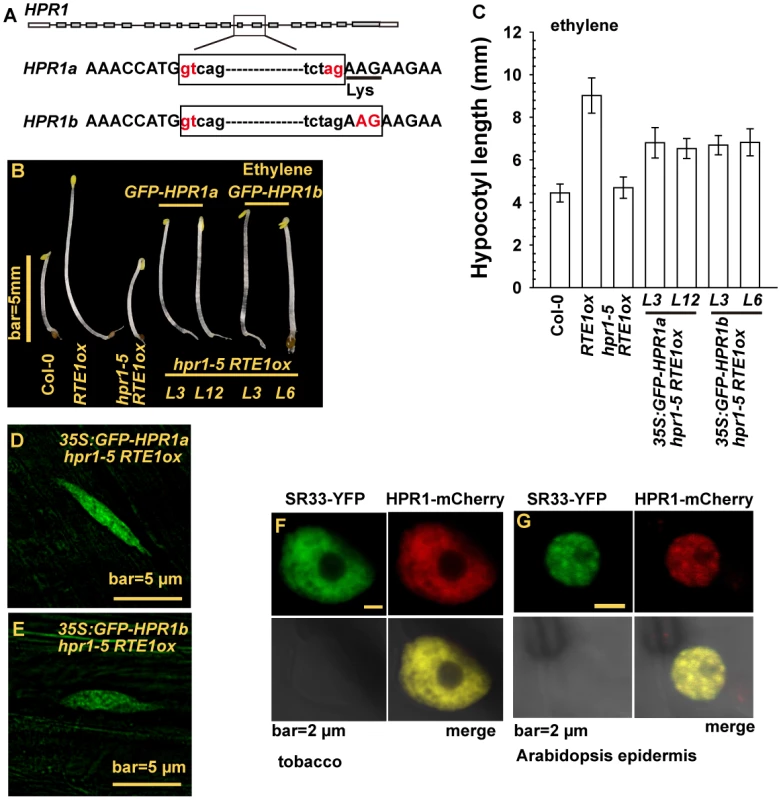
(A) HPR1 produces 2 transcripts by alternative splicing. Phenotype (B) and hypocotyl measurement (C) of etiolated hpr1–5 RTE1ox seedlings expressing GREEN FLUORESCENT PROTEIN (GFP)-fused HPR1a and HPR1b. The fluorescence of GFP-HPR1a (D), GFP-HPR1b (E) in hpr1–5 RTE1ox seedlings. Speckle domain co-localization of the fluorescence by SR33-YFP and HPR1-mCherry in tobacco (F) and Arabidopsis epidermis (G). THO/TREX is involved in mRNA processing, and human THO co-localizes with splicing factors in nuclear speckle domains [47–49]. We examined whether HPR1 could co-localize with mRNA-splicing proteins. SERINE-ARGININE-RICH (SR) proteins have essential roles in mRNA splicing, and SR33 is a member of the SR protein family involved in pre-mRNA splicing [50]. When co-expressed in tobacco and Arabidopsis epidermis by Agrobacterium infiltration, the proteins SR33-YFP and HPR1-mCherry (encoded by a genomic HPR1 clone fused with mCherry) produced fluorescence with an identical speckle pattern in the nucleus but not nucleolus (Fig. 6F-6G). We tried to estimate the relative abundance of HPR1a and HPR1b. On sequencing 56 independent cDNA clones for HPR1, 27 were HPR1a and 29 were HPR1b, for a 1 : 1 ratio (χ2 = 0.62, P = 0.77).
Thus, HPR1 may encode 2 proteins, each involved in RTE1 function. Whether the 2 proteins could act coordinately, independently, or synergistically remains to be investigated. The co-localization of HPR1-mCherry and SR33-YFP provides evidence that in Arabidopsis the THO/TREX complex could co-localize with splicing factors in nuclear speckle domains involving mRNA processing, as observed in human cells [47, 49]. This argument agrees with the HPR1 allele emu producing splicing defects [25].
RTE1 Protein Level Is Reduced in the hpr1–5 Mutant
The hpr1–5 mutation resulted in reduced RTE1 level and, likely, the protein amount required for promoting ETR1 ethylene receptor signaling. We examined RTE1 protein levels in the hpr1–5 mutant.
A GFP-fused RTE1 was previously shown to be localized mainly at the Golgi apparatus [10]. We found that the expression of 35S:GFP-RTE1 conferred ethylene insensitivity in wild-type (Col-0) but not hpr1–5 seedlings, and ethylene-treated etiolated seedlings were longer for 35S:GFP-RTE1 (Col-0) than 35S:GFP-RTE1 hpr1–5 (Fig. 7A). Of note, the 35S:GFP-RTE1 transgene in the wild type was introduced into the hpr1–5 mutant by genetic crossing. The transgene was thus expressed at the same locus in the 2 genotypes, which facilitated study of their expression. We evaluated the association of GFP-RTE1 level and ethylene insensitivity. GFP-RTE1 fluorescence was evident in 35S:GFP-RTE1 (Col-0) seedlings (Fig. 7B) and not detectable in 35S:GFP-RTE1 hpr1–5 seedlings (Fig. 7C). Consistently, relative GFP-RTE1 level was greatly reduced in 35S:GFP-RTE1 hpr1–5 as compared with 35S:GFP-RTE1 (Col-0) (Fig. 7D).
We considered that the GFP-fused RTE1 was an artificially created protein and its accumulation may not necessarily reflect the status of the native RTE1 protein. Immunoassay for RTE1 amount could help address the effect of the hpr1–5 allele on RTE1 level. With a monoclonal antibody against RTE1, we observed a strong immune signal for proteins isolated from RTE1ox. Unfortunately, sensitivity to detect endogenous RTE1 was poor, and no immune signal could be detected for the protein from the wild type (Col-0). As expected, the RTE1 level in hpr1–5 RTE1ox seedlings was reduced throughout development (Fig. 7E). With ACTIN as an internal reference, the amount of RTE1 level was normalized to RTE1ox level; relative RTE1 protein level in RTE1ox was reduced to 25% to 58% by the hpr1–5 allele (Fig. 7F).
Fig. 7. RTE1 level is reduced in the hpr1–5 mutant. 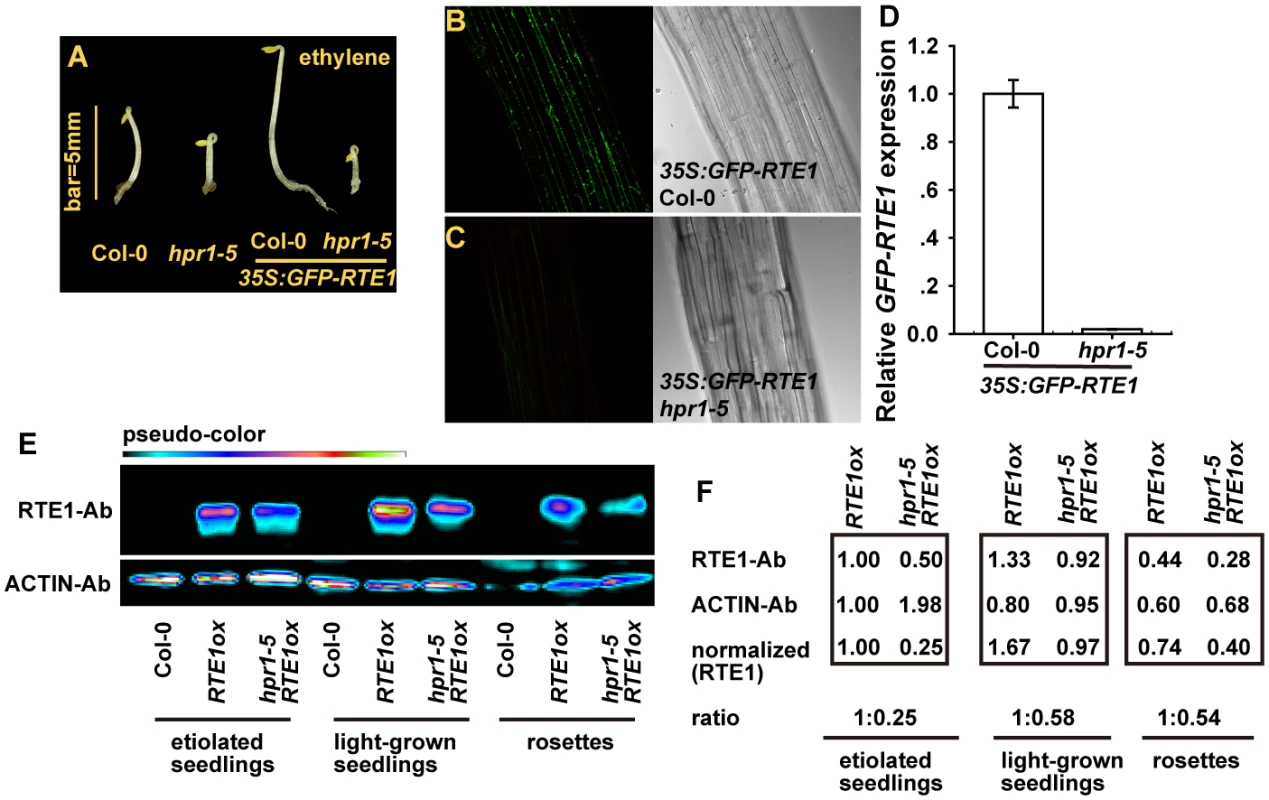
(A) Phenotype of etiolated seedlings of the hpr1–5 mutant with and without 35S:GFP-RTE1. Fluorescence of GFP-RTE1 in wild-type (Col-0) (B) and hpr1–5 (C) seedlings expressing the 35S:GFP-RTE1 transgene and (D) GFP-RTE1 expression measurement. Data are mean±SE, with 3 measurements for each 3 independent biological samples. Immunofluorescence (E) and quantification (F) of RTE1 level in RTE1ox and hpr1–5 RTE1ox at seedling and rosette stages. ACTIN was an internal reference for RTE1 normalization. Pseudo-color indicates immunofluorescence of weak (dark) to strong (bright) signal. RTE1 antibodies (RTE1-Ab) and ACTIN-Ab are monoclonal Abs. Our results showed reduced RTE1 protein level by the hpr1–5 allele, which indicates the basis for why ethylene insensitivity conferred by RTE1ox and possibly etr1–2 was suppressed by the mutation.
Effect of sac3b-2 and tex1–4 Mutations on RTE1ox
In yeast (Saccharomyces cerevisiae), THO (a tetrameric protein complex comprised of Mft1, Tho2, Thp2, and Hpr1) is linked to the Sub2-Yra1 dimer via Hpr1 to form the TREX complex [20, 49, 51]. TEX1 interacts with THO and is a component of the THO/TREX complex; Arabidopsis tex1 mutations result in defects in trans-acting small interfering RNA (tasi-RNA) production [15, 24]. Sac3 is an mRNA export factor in yeast, constituting the TREX-2 complex with Thp1, Sus1 and Cdc31 for RNA export through the nuclear pore complex (NPC). In addition, RNA transcription elongation and export by THO/TREX is coupled to the NPC-bound TREX-2 complex [16, 27]. With impact of the hpr1–5 mutation on RTE1 transcription, we wondered whether other components of the dynamic mRNA transcription export are also involved in RTE1 expression.
We used ethylene dose–response assay to evaluate whether the loss-of-function sac3b-2 and tex1–4 mutations [15, 16, 24], resulting from T-DNA insertion, affect the ethylene response. Etiolated sac3b-2 and tex1–4 seedlings showed enhanced ethylene growth inhibition (Fig. 8A). Over a wide range of ethylene concentration, the hypocotyl was slightly shorter for etiolated mutant seedlings than the wild type (Col-0), with a shorter seedling for sac3b-2 than text1–4 (Fig. 8B). When normalized to the untreated seedling hypocotyl length, the relative hypocotyl length was slightly shorter for sac3b-2 than wild-type and tex1–4 seedlings. The sac3b-2 and tex1–4 mutations each conferred ethylene hypersensitivity to various degrees (Fig. 8C).
Fig. 8. Effect of sac3b-2 and tex1–4 alleles on RTE1 expression. 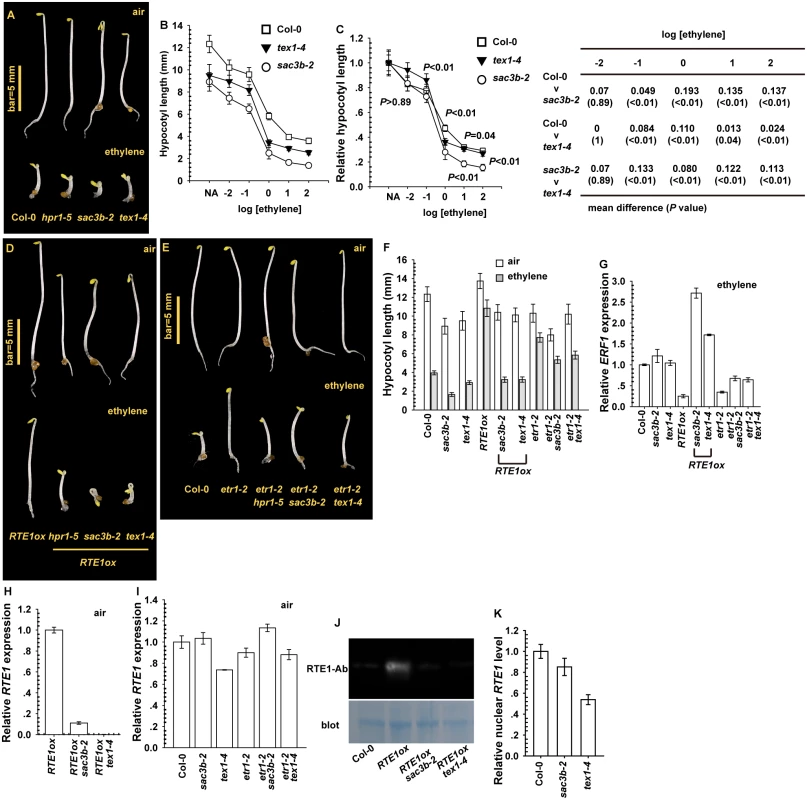
(A) Phenotype of etiolated sac3b-2 and tex1–4 seedlings and wild-type (Col-0) and hpr1–5 seedlings. Ethylene dose–response assay of hypocotyl length (B) and normalized hypocotyl length (C) for wild-type (Col-0), sac3b-2, and tex1–4 seedlings. Mean difference and statistical significance were estimated by Scheffe test (α = 0.01). Ethylene concentration is presented as the logarithm of ethylene (in μL L-1). Phenotype of etiolated seedlings for the indicated genotypes (sac3b-2 and text1–4) in RTE1ox (D) and the etr1–2 allele (E), and hypocotyl length (F) and ERF1 level (G) for the corresponding seedlings. RTE1 level in RTE1ox seedlings with the sac3b-2 and text1–4 alleles (H) and in sac3b-2 and tex1–4 seedlings with and without the etr1–2 allele (I). (J) Immunofluorescence of RTE1 level in RTE1ox and the indicated genotypes with the sac3b-2 and tex1–4 alleles. (K) Nuclear RTE1 level in sac3b-2 and tex1–4. Blot: Commassie Blue staining to indicate the protein amount on the membrane. RTE1-Ab: a monoclonal antibody for RTE1. Data are mean±SD for seedling hypocotyls (n>30), and mean±SE for gene expression (at least 3 independent biological samples with 3 measurements for each). Expression of 35S:RTE1 in the wild type (Col-0) conferred ethylene insensitivity, and ethylene treatment had a very minor effect on seedling hypocotyl elongation. The transgene from RTE1ox (35S:RTE1 expressed in the wild type) was introduced into sac3b-2 and tex1–4 by genetic crossing; homozygous RTE1ox sac3b-2 and RTE1ox tex1–4 were each obtained and confirmed at higher generations by genotyping. The transgene in sac3b-2 and tex1–4 seedlings did not confer ethylene insensitivity, and the hypocotyl was substantially shortened by ethylene treatment (Fig. 8D). The etr1–2 ethylene-insensitive allele conferred ethylene insensitivity depending on RTE1, and the seedlings were moderately shortened by the sac3b-2 and tex1–4 mutations with ethylene treatment (Fig. 8E). The impact of the sac3b-2 and tex1–4 mutations was stronger on RTE1ox than etr1–2, and the seedlings were shortened to a greater degree for RTE1ox than etr1–2 by each of the 2 mutations with ethylene treatment (Fig. 8F).
Consistent with the ethylene growth inhibition phenotype, with ethylene treatment, ERF1 level was increased in wild-type (Col-0), RTE1ox sac3b-2 and RTE1ox tex1–4 but not RTE1ox seedlings (Fig. 8G). Each of the mutations also increased ERF1 level in etr1–2, to a level lower than that for the ethylene-treated wild type (Fig. 8G). Therefore, sac3b-2 and tex1–4 each also affected ethylene insensitivity conferred by the etr1–2 allele. Alternatively, the 2 mutations could confer increased ethylene sensitivity by mechanisms yet to be identified. RTE1 mRNA levels in RTE1ox in both sac3b-2 and tex1–4 were substantially reduced (Fig. 8H). The native RTE1 level was not reduced in etr1–2 by each of the mutations; compared to the wild type (Col-0), RTE1 level was not altered in sac3b-2 but was reduced in tex1–4 (Fig. 8I). As expected, RTE1 protein was detectable in RTE1ox but not RTE1ox with the sac3b-2 or tex1–4 mutation (Fig. 8J).
Whether sac3b-2 and tex1–4 could prevent RTE1 nucleocytoplasmic export to affect RTE1 functions was evaluated by measurement of the nuclear RTE1 level. Nuclear RTE1 was slightly reduced in sac3b-2 compared with the wild type (Col-0); the nuclear RTE1 level in tex1–4 was reduced to a greater extent (Fig. 8K), in line with reduced RTE1 level in the mutant (Fig. 8I). The sac3b-2 mutation does not result in bulk nuclear mRNA accumulation [16]. Our data suggested that SAC3B and TEX1 are not involved in RTE1 export; the reduced nuclear RTE1 level in tex1–4 could be due to reduced gene expression or transcript stability.
Possible Epigenetic Effects on RTE1ox by T-DNA Tag
Of note, on genetic crossing, the T-DNA tag containing the 35S promoter in a mutant could have a dominant epigenetic silencing effect on the 35S promoter-driven transgene in a transgenic line [52]. We evaluated whether the suppression of RTE1ox by sac3b-2 and tex1–4 could result from transgene silencing.
We scored the ethylene response phenotype for the F2 seedlings of genetic crossing of RTE1ox with sac3b-2 and text1–4. Unexpectedly, the ethylene seedling growth-inhibition phenotype was classified into 3 categories: ethylene-insensitive, ethylene-responsive, and intermediate. Individual F2 seedlings that were phenotyped were grown to the adult stage for genotyping, and we determined the association of phenotype and genotype for individuals carrying the 35S:RTE1. The 35S:RTE1-containing F2 generation of the RTE1ox and sac3b-2 cross showed reduced ethylene insensitivity (the intermediate category), with or without the sac3b-2 allele. In total, 2 and 9 of 11 RTE1ox sac3b-2 individuals showed the typical ethylene-response and intermediate phenotype, respectively. For the RTE1ox/ - SAC3B/sac3b-2 heterozygote, 31 and 25 individuals showed the ethylene-insensitive and intermediate phenotypes, respectively; 6 and 2 of 8 RTE1ox/ - sac3b-2 individuals showed the ethylene-responsive and intermediate phenotypes, respectively (Fig. 9A). Ethylene insensitivity conferred by the 35S:RTE1 transgene was reduced after genetic crossing with sac3b-2. The RTE1ox suppression by sac3b-2 observed at higher generations (Fig. 8) was not necessarily caused by the loss of SAC3B.
Fig. 9. Analyses for the ethylene response phenotype and RTE1 expression in 35S:RTE1-containing individuals. 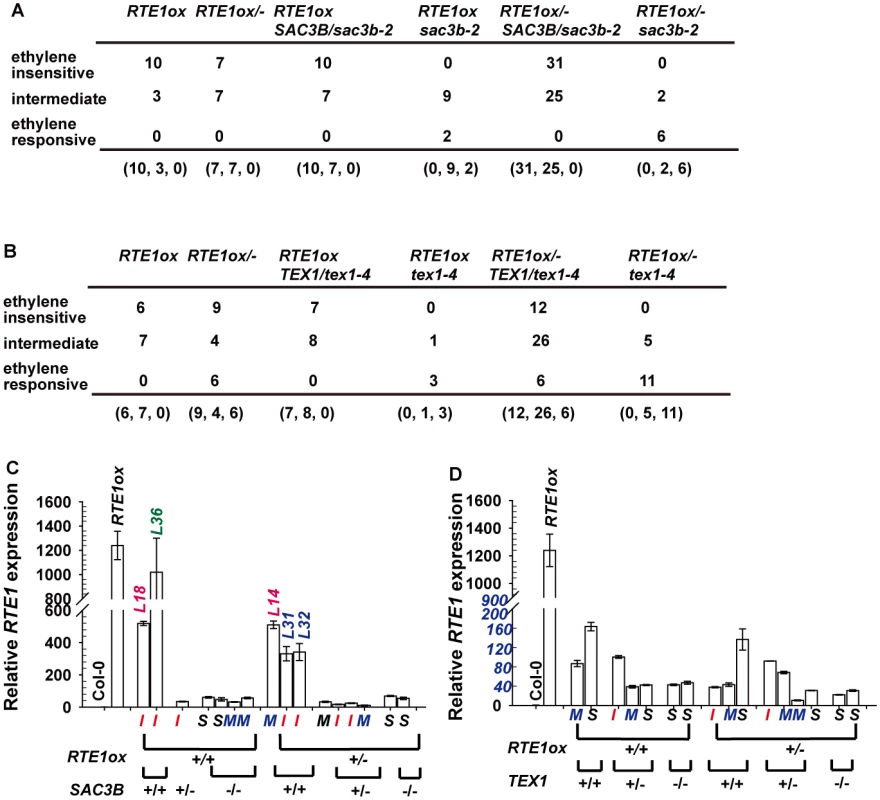
Genotypes and ethylene response phenotypes for 35S:RTE1-containing F2 seedlings from a genetic crossing of RTE1ox and sac3b-2 (A) and RTE1ox and tex1–4 (B). (C) and (D) Rosette RTE1 levels in 35S:RTE1-containing F2 individuals, with their genotypes and seedling ethylene-response phenotypes; I, M, and S indicate ethylene-insensitive, intermediate, and ethylene-responsive seedling phenotypes, respectively. L: line number for individual F2 lines. Data are mean±SD for 3 technical repeats, without biological duplicates. For the F2 generation of the RTE1ox and tex1–4 crossing, reduced ethylene insensitivity (the intermediate phenotype) was not necessarily associated with loss of TEX1: 6 of the 19 F2 RTE1ox/- heterozygotes were ethylene-responsive. Among the 44 F2 RTE1ox/ - TEX1/tex1–4 heterozygotes examined, 12 and 6 had the ethylene-insensitive and-responsive phenotypes, respectively (Fig. 9B). The suppression of RTE1ox by tex1–4 (Fig. 8) observed at higher generations after the genetic crossing was not necessarily caused by the loss of TEX1.
With lack of sufficient plant material for gene expression measurement and lack of biological replicates for each of the F2 seedling single lines, measurement of seedling RTE1 expression to determine its association with phenotype and genotype of individual F2 seedlings was technically unlikely. Nevertheless, randomly selected single F2 seedlings that were phenotyped were grown to rosette stage for genotyping and RTE1 expression measurement. Among the 35S:RTE1-containing F2 rosettes of the RTE1ox and sac3b-2 cross, RTE1 levels were largely reduced regardless of the genotype and seedling ethylene-response phenotype, with one exception: the rosette RTE1 level was slighted reduced in line L36 as compared with RTE1ox level, and the seedlings were ethylene-insensitive (Fig. 9C). Rosette RTE1 expression of two individual lines (L18 and L14) was reduced to about 42% and that of another two lines (L31 and L32) to about 26% of the RTE1ox level, the other lines showing RTE1 expression 0.8% to 5% of the RTE1ox level (Fig. 9C). RTE1 expression in the 35S:RTE1-containing F2 rosettes of the RTE1ox and tex1–4 cross was highly reduced, regardless of seedling phenotype and genotype (Fig. 9D).
The genetic analysis and gene expression measurement did not support the 2 mutations suppressing 35S:RTE1 expression. Silencing is inheritable over generations even if the silencing insert is removed [52, 53]. With variation in degrees of the ethylene-response phenotype and in RTE1 expression in the F2 generation, we considered the possibility of epigenetic silencing of 35S:RTE1. Whether the suppression was associated with the T-DNA tag or other mechanisms needs to be verified. A rapid increase in methylation of the 35S promoter during vegetative growth leads to transgene silencing in Nicotiana attenuata lines with multiple T-DNA insertions [54]. The highly reduced RTE1 levels in rosettes from seedlings with the ethylene-insensitive or intermediate phenotype might be due to rapid silencing of the 35S:RTE1 transgene during vegetative development but not the seedling stage, with the silencing mechanism remaining to be determined.
Discussion
Previous studies suggested that ETR1 ethylene receptor signaling could be facilitated by RTE1 via the physical interaction of RTE1 and the ETR1 N-terminus [8, 10, 12, 32, 55]. Contrary to the extensive studies of ETR1, little is known about the regulation of RTE1 functions. We aimed to uncover components involved in RTE1 function for ethylene signaling regulation. We found that reduced RTE1 expression was associated with the loss-of-function mutation of HPR1. Ethylene insensitivity prevented by the hpr1–5 allele in RTE1ox was also associated with reduced RTE1 mRNA and protein level at the seedling stage. The promoters for the native RTE1 and the 35S:RTE1 transgene differ. HPR1 may have a role in normal RTE1 transcription but not transcription initiation to produce the necessary amount of the protein to facilitate ETR1 receptor signaling. The RTE1 amount, although reduced, in rosette leaves could still be sufficient to promote ETR1/etr1–2 receptor signaling to prevent ethylene signaling, so the hpr1–5 allele had little effect on ethylene insensitivity by the 35S:RTE1 transgene and etr1–2 at the adult stage. The lack of suppression at the adult stage was not likely due to redundancy because of the absence of other HPR1 homologs in the genome. Ethylene insensitivity of ein2–50 and ein3–1 was not suppressed by hpr1–5, which suggests specificity of HPR1 for RTE1 functions in ethylene signaling.
HPR1 shares a low degree of homology with yeast Hpr1 and is annotated as HPR1 of the THO tetrameric protein complex that forms the TREX complex together with other interacting proteins, including TEX1 [24, 25]. THO/TREX is involved in transcription/export of RNA species that remain largely to be identified; as expected, pleiotropy would occur on disruption of the THO/TREX complex. The growth inhibition and leaf serration phenotype as well as ethylene hypersensitivity in the hpr1–5 and hpr1–2 mutants could result from pleiotropy caused by aberrant gene expression and transcript export. This argument agrees with the pleiotropic phenotype observed in emu, which is an HPR1 allele [25]. Expression of ETR1, etr1–2, EBF1, EBF2 and CTR1 was unaltered by hpr1–5; hpr1–5 did not likely affect their expression to result in ethylene hypersensitivity. Whether the hpr1 mutation could affect functions of these genes at other levels remains to be investigated.
Transcription/export involves various dynamic activities such as transcription elongation, 5’-capping, 3’-polyadenylation, RNA splicing, the assembly of messenger ribonuclearprotein particles (mRNPs), and mRNP nucleocytoplasmic export through the nuclear pore complex [27, 51]. Although bulk unidentified polyadenylated RNAs accumulated in the nucleus of the hpr1–5 mutant, HPR1 involved in RTE1 export was less likely because the total and nuclear RTE1 amount were both reduced to a similar level in the hpr1–5 mutant, so the allele did not result in nuclear RTE1 accumulation. The reduced RTE1 level could be due to defects in transcription initiation, accelerated transcript degradation, or attenuated transcription elongation. RTE1 level in hpr1–5 and hpr1–5 RTE1ox was reduced to a similar extent but not amount as compared with the wild type and RTE1ox, respectively. Therefore, its effect was not associated with the promoter that drove RTE1 expression. The argument that HPR1 was involved in RTE1 transcription initiation was not favored. The termination of RNA biosynthesis with cordycepin facilitated the study of RNA stability. RTE1 reduction was identical between seedlings of the wild type (Col-0) and hpr1–5 as well as RTE1ox and hpr1–5 RTE1ox with cordycepin treatment, so the hpr1–5 allele did not accelerate RTE1 degradation. HPR1 may be involved in RTE1 transcription elongation.
Of note, the yeast Hpr1 gene is required for efficient transcription elongation for the LacZ reporter driven by different promoters, and the transcription elongation in yeast Δhpr1 cells is not affected for other sequences such as yeast PHO5 [20]. Moreover, EMU, the same gene as HPR1, is required for the normal expression of ERECTA and several miRNAs [25]. These results support our argument that HPR1 is involved in RTE1 transcription elongation but not initiation. Interestingly, the transcription elongation defect in yeast Δhpr1 cells can be rescued by excess Sub2, an RNA helicase that may unwind the inhibitory secondary structure of a nascent RNA molecule; the RNA-DNA hybrid could also obstruct transcription progression [23, 56]. Efficient transcription elongation that requires HPR1 may be associated with the gene sequence or higher-order gene structure.
SAC3B is a component of the TREX-2 complex that couples transcription and mRNP export, and TEX1 is a THO-interacting protein and may have a role in the export of tasi-RNA precursors for TAS1 and TAS2 [15, 16, 24]. Defects in each resulted in growth inhibition and ethylene hypersensitivity in etiolated seedlings. Compared to the wild type, sac3b-2 showed no reduction in RTE1 levels, which were moderately reduced in tex1–4. The increase in ethylene sensitivity in sac3b-2 was not associated with RTE1 levels. Of note, nuclear RTE1 level was reduced in tex1–4 but not sac3b-2; TEX1 could be involved in nuclear RTE1 expression or degradation but not nucleocytoplasmic export.
RTE1 overexpression with the 35S:RTE1 transgene was substantially prevented in sac3b-2 and tex1–4 at higher generations after genetic crossing. Genetic analysis did not support the association of RTE1ox suppression with either mutation. Given that individual F2 RTE1ox sac3b-2 and RTE1ox tex1–4 seedling lines showed reduced ethylene insensitivity (the intermediate phenotype) as well as the typical ethylene-response phenotype and that some of the F2 RTE1ox individuals showed reduced ethylene insensitivity, the suppression more likely resulted from epigenetic effects on 35S:RTE1 expression. We observed strong RTE1ox suppression in sac3b-2 and tex1–4 (Fig. 8) at higher generations after genetic crossing; the epigenetic suppression could be strengthened at higher generations or the specific lines may not be representative for a population. The T-DNA tag in sac3b-2 and tex1–4 could confer the epigenetic silencing effect on 35S:RTE1 expression. Most of the individual F2 lines with ethylene-insensitive or intermediate phenotype at seedling stage showed highly reduced RTE1 levels at rosette stage. The seedling RTE1 levels were not measured because of technical limitations; nevertheless, the lack of association of seedling ethylene-response phenotype and rosette RTE1 levels in these individual lines may be due to rapid epigenetic silencing of the 35S:RTE1 transgene during vegetative development, as observed in N. attenuata lines with multiple T-DNA insertions [54]. Mechanisms involved in the differential 35S:RTE1 transgene silencing in sac3b-2 and tex1–4 remain to be determined.
The transcription/export of RNA species involving the THO/TREX complex remains to be determined in plants and other organisms. In yeast, approximately 20% of the genome and 36% of transcription events are associated with the conserved mRNA export factors Yra1 and Mex67, which supports a combinatorial model for involvement of multiple pathways for mRNA export [57]. In Drosophila melanogaster, less than 20% of the transciptome is regulated by THO, as revealed by gene expression profiling in cells depleted of THO components [58]. Of the 20%, less than 12% may require THO for export. THO may not be involved in the transcription and export of most mRNAs in Drosophila. These studies provide an explanation for why defective HPR1 results in pleiotropy but not lethality and for why our study revealed a role of HPR1 in RTE1 transcription elongation but not export. A small fraction of the transcriptome may depend on the THO/TREX complex in Arabidopsis.
The involvement of HPR1 in normal RTE1 transcription reveals a role of transcriptional regulation in ethylene signaling (Fig. 10). The role for HPR1 in RTE1 transcription elongation agrees with that of yeast Hpr1 in LacZ transcriptional elongation but not activation [20, 23] and that of Arabidopsis EMU/HPR1 in the normal expression of ERECTA, TAS1, and TAS2 [25]. The THO/TREX component TEX1 had a role in nuclear RTE1 level as well as TAS1 and TAS2 expression [15, 24]; how TEX1 is involved in RTE1 expression remains to be investigated. Components of the THO/TREX complex have selective roles in the transcription export process of specific genes; the hpr1–5 mutation affected bulk mRNA but not RTE1 export, and normal RTE1 transcription elongation required HPR1. The assembly of mRNA and RNA-binding proteins into mRNPs is dynamic; the THO/TREX complex could be involved in RTE1 transcription but not its export. Conceivably, the nucleocytoplasmic export of RTE1 could be mediated by other pathways. Defect of the RNA-export TREX-2 complex component SAC3B had little effect on nuclear RTE1 level; RTE1 export may not require SAC3B, in agreement with the lack of bulk mRNA accumulation in sac3b-2 [16].
Fig. 10. A model for the involvement of the THO/TREX complex component HPR1 in RTE1 transcription and ethylene signaling regulation. 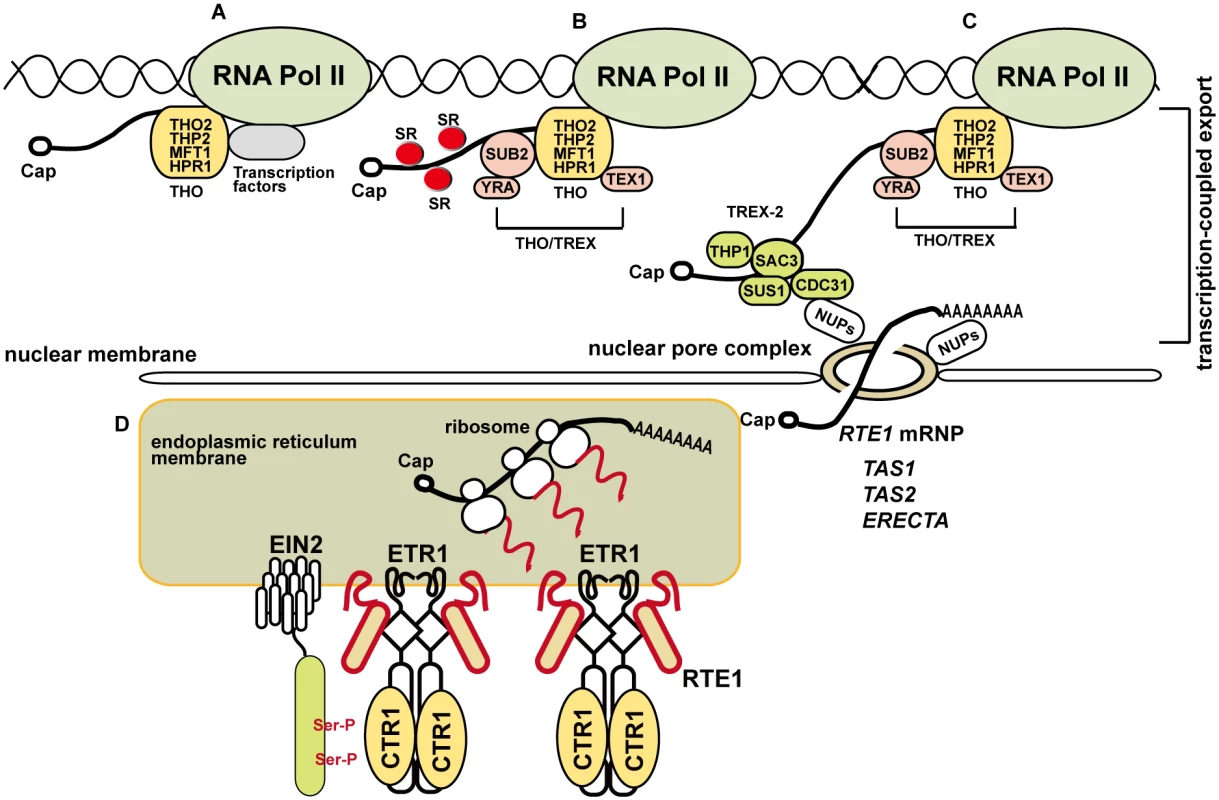
(A) THO is associated with RNA polymerase II, possibly with transcription factors, for transcription. (B) On transcription progression, different components are recruited for mRNA transcription and processing; the SUB2-YRA dimer is recruited to form the THO/TREX complex and TEX1 is a component associated with the complex. SERINE-ARGININE–RICH proteins (SRs) are involved in RNA splicing and co-localize with the THO/TREX complex at nuclear speckle domains. (C) RNA transcription is coupled to TREX-2 (consisting of SAC3, THP1, SUS1, and CDC31) for nucleocytoplamic export through the nuclear pore complex (NPC) via the tethering of TREX-2 with nucleoporins (NUPs). Normal expression of ERECTA [25] and RTE1 requires HPR1, and siRNAs derived from TAS1 and TAS2 involves HPR1 and TEX1 [15, 24]. RTE1 ribonucleoprotein particle (RNP) export could be mediated by other components. (D) Once exported for translation, the produced RTE1 protein facilitates ETR1 receptor signaling to CTR1; EIN2 is retained at the endoplasmic reticulum on phosphorylation by CTR1 and ethylene signaling is prevented. HPR1 could be required for efficient RTE1 transcription elongation through regions with higher-order RNA/DNA structures or a stable RNA–DNA hybrid that obstructs RNA polymerase II movement. Ser-P: phosphorylation of serine residues on EIN2. Except for the possible role of HPR1 in RTE1 transcription, with a bulk mRNA accumulation in the nucleus of hpr1–5, the suppression of RTE1 by hpr1–5 could be due to nuclear accumulation of the affected transcripts that are required for RTE1 functions. Alternatively, normal transcription of genes involved in RTE1 functions could be affected by the hpr1–5 mutation, for an indirect effect on RTE1 functions. One of the possible candidates could be cytochrome b5 (Cb5), which interacts with RTE1 to facilitate ETR1 receptor signaling at the ER membrane [59]; if Cb5 is affected by hpr1–5, RTE1 functions would be weakened.
Materials and Methods
Plant Materials and Phenotyping
Arabidopsis seeds were stratified at 4°C for 3 days (72 hr) before germination at 22°C for 80 hr in the dark (for phenotyping etiolated seedlings) or 7 days at 25°C with illumination (for phenotyping light-grown seedlings). Adult plants were grown at 25°C with fluorescent light (16-hr light/ 8-hr dark) and phenotyped 4 weeks after germination. The ethylene concentration for treatment was 10 μL L-1, unless otherwise specified. For the ethylene dose–response assay, stratified seeds were germinated in the dark with the necessary amount of ethylene for 80 hr, then seedling hypocotyls were measured as described [33, 55] by use of Video tesT (Moscow). For the senescence test, 4-week-old adult plants were placed in an air-tight container with ethylene (100 μL L-1) for 4 days, and chlorophyll content was measured as described [60]. For cordecypin treatment [46], Arabidopsis seedlings were pre-incubated for 30 min in incubation buffer, and aqueous cordecypin solution was added to a final concentration of 0.6 mM for incubation for the necessary time. hpr1–5 35S:GFP-RTE1 was obtained by crossing hpr1–5 and a wild type (Col-0) carrying the 35S:GFP-RTE1 transgene; thus, the transgene was expressed at the same locus.
Mutagenesis
About 20,000 RTE1ox seeds were treated with ethyl methanesulfonate (EMS; 0.1% w/v) in the dark for 14 hr at room temperature, then washed with continuous flowing water for 2 to 4 hr. The treated seeds were grown, and M2 seeds were harvested for genetic screening.
Nuclei Isolation
Nuclei were enriched and purified as described [61]. In brief, 1 g Arabidopsis tissues was ground into fine powder in liquid nitrogen, homogenized with 3-ml extraction buffer, and filtrated through miracloth (Calbiochem). An amount of 10% Triton X-100 was gradually added into the resulting filtrate to a final concentration of 1%, and the filtrate was incubated on ice for 15 min. An amount of 30% and 80% percoll was prepared with the gradient buffer, with 250 μL 30% percoll at the bottom of a 2-ml centrifuge tube, then 80% percoll was added under the 30% percoll. An amount of 1.4-ml infiltrate was added to the top of the percoll layers. The tube was centrifuged at 1,000×g for 30 min at 4°C, and nuclei were enriched at the interface of the 2 percoll solutions. The nuclei were collected carefully and added with gradient buffer to a volume of 500 μL. An amount of 250 μL 30% percoll was added to the bottom of a tube and the solution was centrifuged at 1,000×g for 10 min at 4°C. After centrifugation, the nuclei were collected as the pellet and resuspended with 200 μL nuclei storage buffer.
Gene Expression Measurement
Gene expression was measured by quantitative RT-PCR (qRT-PCR) with StepOne Plus (Applied Biosystems) and TaKaRa SYBR Premix Ex Taq. To measure ethylene-induced ERF1 expression, plants were treated with ethylene (10 μL L-1) for 4 hr. The primer sequences were for ERF1, F (5’-TTTCTCGATGAGAGGGTC-3’) and R (5’-AAGCTCCTCAAGGTACTG-3’); RTE1, F (5’-GATAGAACCAAGTGTTGC-3’) and R (5’-GCAACAAACGAATGGCAG-3’); GFP-RTE1c-RT, F (5’-ACAGCTGCTGGGATTACACAT-3’) and R (5’-ACACCACCCTGTCTTCAACAT-3’); ETR1, F (5’-GCCATCTCCAAGAGGTTTGTGAA-3’) and R (5’-CCGTTCTCATCCATGACAAGA-3’); CTR1, F (5’-GGCTTATGATGTGGCTAAGG-3’) and R (5’-GCAGCTACAACCTGAGC); EBF1, F (5’-GGAGATTGATGTTCCTTCCAAGA-3’) and R (5’-CAATAGACCGAAGACCAAGATC-3’); EBF2, F (5’ - CTTCAGATTTAGTGGTGATGAAG-3’) and R (5’ - CAAGCACTCCTCTCTTGTCCA-3’), and tubulin (TUB), F (5’-TCAAGAGGTTCTCAGCAGTA-3’) and R (5’-TCACCTTCTTCATCCGCAGTT-3’) (the calibrator). The calibrator for gene expression by cordycepin treatment was eIF4A and the primers were F (5’-TGACCACACAGTCTCTGCAA-3’) and R (5’-ACCAGGGAGACTTGTTGGAC-3’). For measuring UBIQUITIN and TUBULIN transcript copy number, cDNAs of known amount for each gene were serially diluted as the template, and a standard curve (R2>0.99) was drawn for qRT-PCR. UBIQUITIN and TUBULIN cDNAs reverse-transcribed from 500 ng RNA with use of oligo(dT) were estimated for copy number according to the standard curves (R2>0.99). The primer sequences for UBIQUITIN were UBI-F (5’-ATGGAAAATCCCACCTACTAAATT-3’) and UBI-R (5’-TTGAACAACTCGTAGCAACTCATC-3’).
Transformation and Transgenes
Transformation by Agrobacterium was performed as described [62], and the resulting homozygous transformation lines were characterized with T3 and higher generations. For transient expression, tobacco or Arabidopsis leaves were infiltrated with Agrobacterium containing the necessary clones; expression of the fluorescent proteins with the transgenes was observed 2 to 3 days after infiltration. To clone the HPR1 promoter, the primers for PCR were HPR1p-F-EcoRI (5’-CGGAATTCAGTTTTTCTTCAAGTGACGAGAA-3’) and HPR1p-R-HindIII (5’-AGAAGCTTTAATTTAGGGTGGATCCGGAT-3’). The genomic HPR1 fragment (gHPR1) was cloned by PCR with the primers gHPR1-F-HindIII (5’-AGAAGCTTAAATTGTTCTTCCTCCACTCTT-3’) and gHPR1-R-HindIII (5’-AGAAGCTTTCATGAGACGGGCATAGGAGGAT-3’). The HPR1 promoter was cloned to the vector pBJ36, and the genomic HPR1 clone gHPR1 was cloned to the HindIII site of the promoter clone. HPR1p:gHPR1 was released by Not I and cloned to the binary vector pMLBART. HPR1 cDNA was cloned with the primers HPR1-c-F-BglII (5’-ATAGATCTATGGATGCATTTAGAGATGCTAT-3’) and HPR1-c-R-BglII (5’-ATAGATCTTCATGAGACGGGCATAGGAGGAT-3’), and sequence analysis identified the HPR1a and HPR1b cDNAs; each was cloned to the pRTL2 vector containing 35S:mGFP. The resulting clones were each restricted with HindIII to release 35S:mGFP-HPR1a and 35S:mGFP-HPR1b, and the fragments were each cloned to the binary vector pCGN1547.
Laser Scanning Confocal Microscopy (LSCM)
LSCM for the co-localization study of SR33-YFP and HPR1-mCherry involved use of an Olympus FV1000 microscope at our institution’s facility. The fluorescence produced by GFP-HPR1a and GFP-HPR1b was acquired with a DeltaVision Personal DV system (Applied Precision) as described [63]. The image was subjected to constrained iterative deconvolution by use of softWoRx (Applied Precision).
In Vivo mRNA Hybridization [19]
Light-grown Arabidopsis seedlings 7 days old were fixed for 30 min (fixation buffer:heptane = 1 : 1) and washed with methanol twice (5 min for each wash) and with absolute ethanol 3 times (5 min for each wash). After the wash, the seedlings were incubated in ethanol:xylene (1 : 1, v:v) for 30 min, washed with absolute ethanol twice (5 min for each), methanol twice (5 min for each), methanol:fixation solution (1 : 1; without formaldehyde) for 5 min, and incubated in fixation solution for 30 min. Then seedlings were washed with fixation solution (without formaldehyde) twice (5 min each) and with Perfect Hyb Plus hybridization buffer (Sigma, Cat. H7033) for 5 min. With pre-treatment with the Perfect Hyb Plus hybridization buffer for 1 h, fluoresceine-labeled oligo(dT)45 or RTE1 oligos (Invitrogen) was added to the hybridization to a final concentration of 10 nM. Two RTE1 oligonucleotide sequences were used for hybridization, 5’-FITC-RTE1a (5’-CTCGATAGAACCAAGTGTTGCTTACCACCA-3’) and 5’-FITC-RTE1b (5’-AACATAATTACTTGATTTTTTTTTTTGA-3’), at 50°C in the dark overnight. Then seedlings were washed with 2×SSC with 0.1% SDS at room temperature, in the dark for 5 min, then twice with 0.5×SSC with 0.1% SDS at 50°C (20 min for each wash) in the dark. Hybridization was observed by LSCM.
Immunoassay
Immunoassay and enhanced chemiluminescence (ECL) was performed as described [6, 32]. The mouse monoclonal antibody for RTE1 (RTE1-Ab) was custom-made by Abmart (Shanghai). The mouse monoclonal antibody for ACTIN (ACTIN-Ab) was from Abmart. The chemiluminance was captured by use of a cold charge-coupled device (Tanon 5500 Chemiluminescent Imaging System, Tanon, Shanghai, or VersArray System, Princeton Instruments) and analyzed by use of MetaMorph (Molecular Devices) as described [6, 32] for quantification and to normalize the signal strength.
Statistical Analysis
Data are expressed as mean±SD or mean±SE. Student’s t test (α=0.01) was used to compare 2 groups and Scheffe test (α=0.01) for multiple groups. For each seedling hypocotyl measurement, at least 30 seedlings were measured (n≥30).
Zdroje
1. Wang W, Hall AE, O’Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci U S A 100 : 352–357. doi: 10.1073/pnas.0237085100 12509505
2. Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 : 261–271. doi: 10.1016/S0092-8674(00)81425-7 9695954
3. Wang W, Esch JJ, Shiu S-H, Agula H, Binder BM, et al. (2006) Identification of Important Regions for Ethylene Binding and Signaling in the Transmembrane Domain of the ETR1 Ethylene Receptor of Arabidopsis. Plant Cell 18 : 3429–3442. doi: 10.1105/tpc.106.044537 17189345
4. Gao Z, Wen C-K, Binder BM, Chen Y-F, Chang J, et al. (2008) Heteromeric Interactions among Ethylene Receptors Mediate Signaling in Arabidopsis. J Biol Chem 283 : 23801–23810. doi: 10.1074/jbc.M800641200 18577522
5. Chen Y-F, Gao Z, Kerris RJ III, Wang W, Binder BM, et al. (2010) Ethylene Receptors Function as Components of High-Molecular-Mass Protein Complexes in Arabidopsis. PLoS ONE 5: e8640. doi: 10.1371/journal.pone.0008640 20062808
6. Liu Q, Xu C, Wen C-K (2010) Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol 10 : 60. doi: 10.1186/1471-2229-10-60 20374664
7. Liu Q, WenC-K (2012) Arabidopsis ETR1 and ERS1 Differentially Repress the Ethylene Response in Combination with Other Ethylene Receptor Genes. Plant Physiol 158 : 1193–1207. doi: 10.1104/pp.111.187757 22227969
8. Resnick JS, Wen C-K, Shockey JA, Chang C (2006) From The Cover: REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA 103 : 7917–7922. doi: 10.1073/pnas.0602239103 16682642
9. Dong C-H, Rivarola M, Resnick JS, Maggin BD, Chang C (2008) Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. The Plant Journal 53 : 275–286. doi: 10.1111/j.1365-313X.2007.03339.x 17999643
10. Zhou X, Liu Q, Xie F, Wen C-K (2007) RTE1 Is a Golgi-Associated and ETR1-Dependent Negative Regulator of Ethylene Responses. Plant Physiol 145 : 75–86. doi: 10.1104/pp.107.104299 17644624
11. Resnick JS, Rivarola M, Chang C (2008) Involvement of RTE1 in conformational changes promoting ETR1 ethylene receptor signaling in Arabidopsis. The Plant Journal 56 : 423–431. doi: 10.1111/j.1365-313X.2008.03615.x 18643990
12. Dong C-H, Jang M, Scharein B, Malach A, Rivarola M, et al. (2010) Molecular Association of the Arabidopsis ETR1 Ethylene Receptor and a Regulator of Ethylene Signaling, RTE1. J Biol Chem 285 : 40706–40713. doi: 10.1074/jbc.M110.146605 20952388
13. Zhang W, Zhou X, Wen C-K (2012) Modulation of ethylene responses by OsRTH1 overexpression reveals the biological significance of ethylene in rice seedling growth and development. Journal of Experimental Botany 63 : 4151–4164. doi: 10.1093/jxb/ers098 22451723
14. Barry CS, Giovannoni JJ (2006) From The Cover: Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc Natl Acad Sci USA 103 : 7923–7928. doi: 10.1073/pnas.0602319103 16682641
15. Jauvion V, Elmayan T, Vaucheret H (2010) The Conserved RNA Trafficking Proteins HPR1 and TEX1 Are Involved in the Production of Endogenous and Exogenous Small Interfering RNA in Arabidopsis. The Plant Cell 22 : 2697–2709. doi: 10.1105/tpc.110.076638 20798330
16. Lu Q, Tang X, Tian G, Wang F, Liu K, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. The Plant Journal 61 : 259–270. doi: 10.1111/j.1365-313X.2009.04048.x 19843313
17. Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M (2006) The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE Proteins Are Nucleoporins with an Important Role in Hormone Signaling and Development. The Plant Cell 18 : 1590–1603. doi: 10.1105/tpc.106.041566 16751346
18. Germain H, Qu N, Cheng YT, Lee E, Huang Y, et al. (2010) MOS11: A New Component in the mRNA Export Pathway. PLoS Genet 6: e1001250. doi: 10.1371/journal.pgen.1001250 21203492
19. Gong Z, Dong C-H, Lee H, Zhu J, Xiong L, et al. (2005) A DEAD Box RNA Helicase Is Essential for mRNA Export and Important for Development and Stress Responses in Arabidopsis. The Plant Cell 17 : 256–267. doi: 10.1105/tpc.104.027557 15598798
20. Chávez S, Aguilera A (1997) The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes and Development 11 : 3459–3470. doi: 10.1101/gad.11.24.3459 9407037
21. Prado F, Piruat JI, Aguilera A (1997) Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. The EMBO journal 16 : 2826–2835. doi: 10.1093/emboj/16.10.2826 9184227
22. Fan H-Y, Cheng KK, Klein HL (1996) Mutations in the RNA Polymerase II Transcription Machinery Suppress the Hyperrecombination Mutant hpr1 Δ of Saccharomyces cerevisiae. Genetics 142 : 749–759. 8849885
23. Merker RJ, Klein HL (2002) Role of Transcription in Plasmid Maintenance in the hpr1Δ Mutant of Saccharomyces cerevisiae. Molecular and Cellular Biology 22 : 8763–8773. doi: 10.1128/MCB.22.24.8763-8773.2002 12446793
24. Yelina NE, Smith LM, Jones AME, Patel K, Kelly KA, et al. (2010) Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci USA 107 : 13948–13953. doi: 10.1073/pnas.0911341107 20634427
25. Furumizu C, Tsukaya H, Komeda Y (2010) Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA 16 : 1809–1817. doi: 10.1261/rna.2265710 20668032
26. Pan H, Liu S, Tang D (2012) HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. The Plant Journal 69 : 831–843. doi: 10.1111/j.1365-313X.2011.04835.x 22035198
27. Kohler A, Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8 : 761–773. doi: 10.1038/nrm2255 17786152
28. Straszer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, et al. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417 : 304–308. doi: 10.1038/nature746
29. Cheng H, Dufu K, Lee C-S, Hsu JL, Dias A, et al. Human mRNA Export Machinery Recruited to the 5′ End of mRNA. Cell 127 : 1389–1400. doi: 10.1016/j.cell.2006.10.044 17190602
30. Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, et al. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89 : 1133–1144. doi: 10.1016/S0092-8674(00)80300-1 9215635
31. Solano R, Stepanova A, Chao Q, Ecker J (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12 : 3703–3714. doi: 10.1101/gad.12.23.3703 9851977
32. Qiu L, Xie F, Yu J, Wen C-K (2012) Arabidopsis RTE1 Is Essential to Ethylene Receptor ETR1 Amino-Terminal Signaling Independent of CTR1. Plant Physiology 159 : 1263–1276. doi: 10.1104/pp.112.193979 22566492
33. Xie F, Liu Q, Wen C-K (2006) Receptor Signal Output Mediated by the ETR1 N Terminus Is Primarily Subfamily I Receptor Dependent. Plant Physiol 142 : 492–508. doi: 10.1104/pp.106.082628 16891553
34. Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, et al. (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 : 679–689. doi: 10.1016/S0092-8674(03)00968-1 14675533
35. Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 : 667–677. doi: 10.1016/S0092-8674(03)00969-3 14675532
36. Rivarola M, McClellan CA, Resnick JS, Chang C (2009) ETR1-Specific Mutations Distinguish ETR1 from Other Arabidopsis Ethylene Receptors as Revealed by Genetic Interaction with RTE1. Plant Physiol 150 : 547–551. doi: 10.1104/pp.109.138461 19369589
37. Kieber J, Rothenberg M, Roman G, Feldmann K, Ecker J (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72 : 427–441. doi: 10.1016/0092-8674(93)90119-B 8431946
38. Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33 : 221–233. doi: 10.1046/j.1365-313X.2003.01620.x 12535337
39. Ju C, Chang C (2012) Advances in ethylene signalling: protein complexes at the endoplasmic reticulum membrane. AoB Plants 2012.
40. Xu S-L, Rahman A, Baskin TI, Kieber JJ (2008) Two Leucine-Rich Repeat Receptor Kinases Mediate Signaling, Linking Cell Wall Biosynthesis and ACC Synthase in Arabidopsis. Plant Cell 20 : 3065–3079. doi: 10.1105/tpc.108.063354 19017745
41. Xu C, Gao X, Sun X, Wen C-K (2012) The Basal Level Ethylene Response is Important to the Wall and Endomembrane Structure in the Hypocotyl Cells of Etiolated Arabidopsis Seedlings. Journal of Integrative Plant Biology 54 : 434–455. doi: 10.1111/j.1744-7909.2012.01130.x 22591458
42. Siev M, Weinberg R, Penman S (1969) THE SELECTIVE INTERRUPTION OF NUCLEOLAR RNA SYNTHESIS IN HELA CELLS BY CORDYCEPIN. The Journal of Cell Biology 41 : 510–520. doi: 10.1083/jcb.41.2.510 5783871
43. Holtorf H, Kunz HS Christian, Waldvogel R, Meins F (1999) Stochastic and Nonstochastic Post-Transcriptional Silencing of Chitinase and β-1,3-Glucanase Genes Involves Increased RNA Turnover—Possible Role for Ribosome-Independent RNA Degradation. The Plant Cell 11 : 471–483. doi: 10.1105/tpc.11.3.471 10072405
44. Tilghman SM, Hanson RW, Reshef L, Hopgood MF, Ballard FJ (1974) Rapid Loss of Translatable Messenger RNA of Phosphoenolpyruvate Carboxykinase During Glucose Repression in Liver. Proc Natl Acad Sci USA 71 : 1304–1308. doi: 10.1073/pnas.71.4.1304 4364533
45. Souret FF, Kastenmayer JP, Green PJ (2004) AtXRN4 Degrades mRNA in Arabidopsis and Its Substrates Include Selected miRNA Targets. Molecular Cell 15 : 173–183. doi: 10.1016/j.molcel.2004.06.006 15260969
46. Gutiérrez RA, Ewing RM, Cherry JM, and Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch - and specific clock-controlled genes Proc Natl Acad Sci USA 99 : 11513–11518.
47. Masuda S, Das R, Cheng H, Hurt E, Dorman N, et al. (2005) Recruitment of the human TREX complex to mRNA during splicing. Genes & Development 19 : 1512–1517. doi: 10.1101/gad.1302205
48. Reed R (2003) Coupling transcription, splicing and mRNA export. Current Opinion in Cell Biology 15 : 326–331. doi: 10.1016/S0955-0674(03)00048-6 12787775
49. Dias AP, Dufu K, Lei H, Reed R (2010) A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun 1 : 97. doi: 10.1038/ncomms1103 20981025
50. Fang Y, Hearn S, Spector DL (2004) Tissue-specific Expression and Dynamic Organization of SR Splicing Factors in Arabidopsis. Molecular Biology of the Cell 15 : 2664–2673. doi: 10.1091/mbc.E04-02-0100 15034145
51. Grunwald D, Singer RH, Rout M (2011) Nuclear export dynamics of RNA-protein complexes. Nature 475 : 333–341. doi: 10.1038/nature10318 21776079
52. Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, et al. (2008) Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends in Plant Science 13 : 4–6. doi: 10.1016/j.tplants.2007.10.007 18178509
53. Matzke M, Matzke AJM (1993) Genomic Imprinting in Plants: Parental Effects and Trans-Inactivation Phenomena. Annual Review of Plant Physiology and Plant Molecular Biology 44 : 53–76. doi: 10.1146/annurev.pp.44.060193.000413
54. Weinhold A, Kallenbach M, Baldwin IT (2013) Progressive 35S promoter methylation increases rapidly during vegetative development in transgenic Nicotiana attenuata plants. BMC Plant Biology 13 : 99. doi: 10.1186/1471-2229-13-99 23837904
55. Xu A, Zhang W, Wen C-K (2014) ENHANCING CTR1–10 ETHYLENE RESPONSE2 is a novel allele involved in CONSTITUTIVE TRIPLE-RESPONSE1-mediated ethylene receptor signaling in Arabidopsis. BMC Plant Biology 14 : 48. doi: 10.1186/1471-2229-14-48 24529183
56. Fan H-Y, Merker RJ, Klein HL (2001) High-Copy-Number Expression of Sub2p, a Member of the RNA Helicase Superfamily, Suppresses hpr1-Mediated Genomic Instability. Molecular and Cellular Biology 21 : 5459–5470. doi: 10.1128/MCB.21.16.5459-5470.2001 11463828
57. Hieronymus H, Silver PA (2003) Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet 33 : 155–161. doi: 10.1038/ng1080 12524544
58. Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, et al. (2004) Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11 : 558–566. doi: 10.1038/nsmb759 15133499
59. Chang J, Clay JM, Chang C (2014) Association of cytochrome b5 with ETR1 ethylene receptor signaling through RTE1 in Arabidopsis. The Plant Journal 77 : 558–567. doi: 10.1111/tpj.12401 24635651
60. Zhang W, Wen C-K (2010) Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiol Biochem 48 : 45–53. doi: 10.1016/j.plaphy.2009.10.002 19836254
61. Folta KM, Kaufman LS (2006) Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat Protocols 1 : 3094–3100. doi: 10.1038/nprot.2006.471
62. Clough SJ, Bent AF (1998) Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. The Plant Journal 16 : 735–743. doi: 10.1046/j.1365-313x.1998.00343.x 10069079
63. Fang Y, Spector DL (2010) Live Cell Imaging of Plants. Cold Spring Harbor Protocols 2010: pdb.top68.
Štítky
Genetika Reprodukčná medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



