-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
The study of plant development is generally carried out in the absence of physical injury. However, damage to plant organs through biotic and abiotic insult is common in nature. Under these conditions the jasmonate pathway that has a low activity in unstressed vegetative tissues imposes its activity on cell division and elongation. Such jasmonate-dependent growth restriction can strongly impact plant productivity. Taking roots as a model, we show that it is possible to manipulate regulatory layers in jasmonate signalling such that cell division and cell elongation can be constrained differently. This approach may lead to future strategies to alter organ growth. Moreover, during this study we identified a novel mutant in a key regulator of the jasmonate pathway. This mutant generated a positive regulator of jasmonate signalling that was so active that we were able to show that hormone synthesis can be completely uncoupled from hormone responses, suggesting ways to modify traits of potential agronomic importance.
Published in the journal: Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth. PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005300
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005300Summary
The study of plant development is generally carried out in the absence of physical injury. However, damage to plant organs through biotic and abiotic insult is common in nature. Under these conditions the jasmonate pathway that has a low activity in unstressed vegetative tissues imposes its activity on cell division and elongation. Such jasmonate-dependent growth restriction can strongly impact plant productivity. Taking roots as a model, we show that it is possible to manipulate regulatory layers in jasmonate signalling such that cell division and cell elongation can be constrained differently. This approach may lead to future strategies to alter organ growth. Moreover, during this study we identified a novel mutant in a key regulator of the jasmonate pathway. This mutant generated a positive regulator of jasmonate signalling that was so active that we were able to show that hormone synthesis can be completely uncoupled from hormone responses, suggesting ways to modify traits of potential agronomic importance.
Introduction
The development, architecture and mass of nascent plant organs are plastic and can be strongly influenced by injury to pre-existing tissues. Wounding reduces plant biomass and damage to young tissues can strongly reduce growth rates, e.g. [1]. In the case of above ground tissues, most of the growth restriction that occurs subsequently to physical damage depends on the activation of the jasmonate (JA) pathway [2–4], which has a pivotal role in controlling herbivore-inducible gene expression and coordinating resource allocation between defence and growth [5, 6]. In contrast to the observation of JA-mediated growth restriction in leaves, root growth responses following damage to aerial organs are so far, not clearly understood. Additionally, there is relatively little knowledge of the cellular organization of JA signalling components in roots. What has emerged to date, however, is that the same basic JA signalling components operate in shoots and roots, although the genetic architecture of the JA pathway appears to be simpler in roots [7].
JA signalling, whether for defence or organ growth restriction, requires the production and perception of low molecular mass lipidic regulators of the JA family, including the biologically active form jasmonoyl-L-isoleucine (JA-Ile) [8, 9]. The transcriptional changes resulting from JA-Ile perception enable plants to modulate the allocation of resources in defense at the expense of growth [3]. In the absence of JA-Ile, JASMONATE ZIM-DOMAIN (JAZ) proteins bind and repress JA-dependent transcription factors (TFs) by recruiting the general co-repressors TOPLESS (TPL) and TPL-Related (TPR) proteins through an interaction with the adaptor protein Novel Interactor of JAZ (NINJA) [10], or directly as in the case of JAZ8 [11]. TPL in turn recruits histone deacetylases and histone methyltransferases to inhibit transcription [12]. Upon stimulation, JA-Ile accumulates and promotes the binding of JAZ proteins to the F-box protein CORONATINE INSENSITIVE 1 (COI1) [13, 14]. This interaction mediates the ubiquitylation and degradation of JAZs [13, 14], liberating TFs to interact with the MED25 subunit of the Mediator complex and recruit RNA polymerase II to JA-responsive genes [15, 16].
Currently, the basic helix-loop-helix (bHLH) MYC2 TF is considered a master regulator of most JA responses and a convergence node with other signalling pathways [17]. It can act as both activator and repressor to regulate diverse aspects of JA-mediated gene expression [18]. Two MYC2-like bHLH proteins (MYC3 and MYC4) also interact with JAZs and act additively with MYC2 in mediating a subset of JA-regulated responses [19, 20]. A simplified scheme for JA signalling is shown in Fig 1. All components shown in the figure can be manipulated to affect the output (defence/growth) of the pathway and, in addition, jasmonate responses can be powerfully activated with exogenous JA. Whatever the upstream manipulation of JA levels or pathway components, much of their activity converges on MYCs. This leads to the theoretical possibility, shown in the dashed box in Fig 1, that strong gain-of-function mutants of MYC2 might be able to recapitulate jasmonate responses in the absence of JA itself. Furthermore, such effects should be facilitated if negative regulatory layers (like that imposed by NINJA) could be removed. Herein, using Arabidopsis seedlings we investigated this possibility in the context of the regulatory organisation of JA signalling components and their contribution to root growth.
Fig. 1. Interventions used in this study to manipulate the jasmonate pathway. 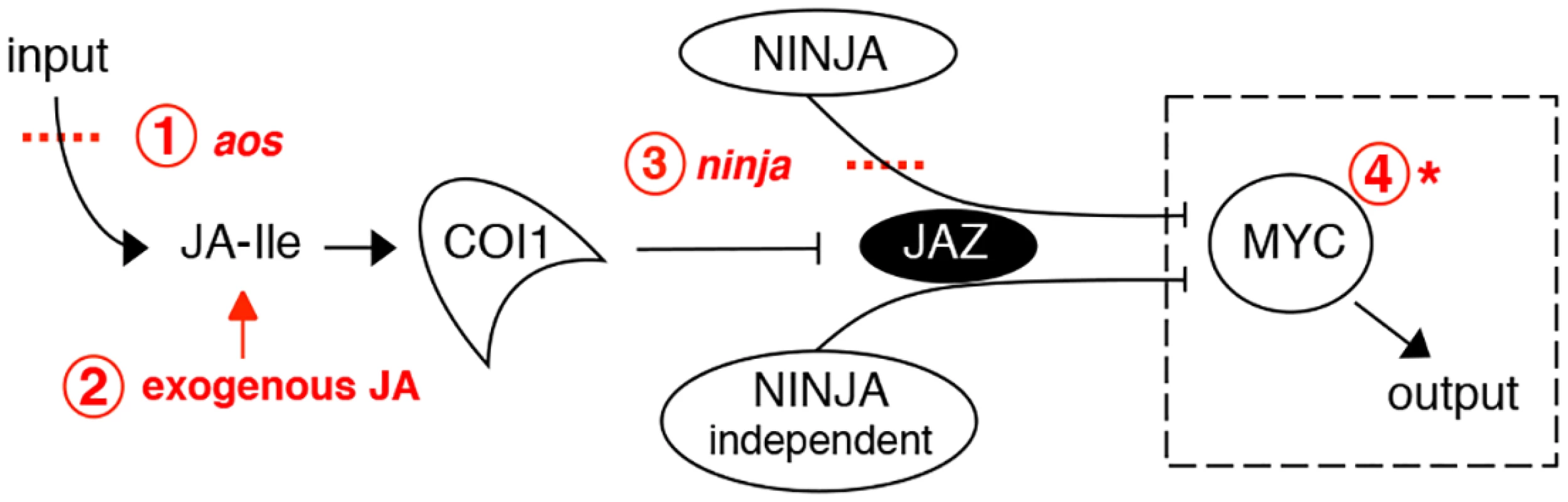
Shown in red are: 1. loss-of-function mutation of the JA synthesis gene allene oxide synthase (aos); 2. treatment with exogenous JA; 3. loss-of-function mutations in the negative regulator NINJA; 4. gain-of-function mutation of MYC transcription factors. The dashed box surrounding MYC indicates that it is conceptually possible to use an overactive MYC to drive JA responses in the absence of JA synthesis (step 1) and of negative regulators like NINJA (steps 1 and 3 combined). This was achieved using a novel myc2 mutant that amplifies JA responses. Although JA-induced root growth inhibition assays have been widely employed to identify JA mutants in Arabidopsis, reviewed in [6, 21], the basal root length of those mutants does not differ from wild-type (WT) [22–24]. To date, mutants in NINJA represent the sole example of a JA signalling component known to affect normal root growth [7]. However, unlike plants grown in medium supplied with exogenous JA where root growth is inhibited as a consequence of reduced meristem cell number and cell elongation [25], ninja mutants display de-repressed JA signalling and shorter roots in the absence of JA due to a defect in cell elongation only [7]. Thus, ninja mutants suggest the existence of additional organ - and cell-specific mechanisms of negative regulation that restrict JA signalling responses in the root. We first characterized the effects of endogenous JA on root growth. NINJA, MYC2, MYC3, and MYC4 distribution maps for the root tip were then established, uncovering their overlapping and unique expression domains. A novel gain-of-function allele of MYC2, either alone or in combination with ninja and JA biosynthesis mutants, revealed the existence of several layers of negative regulation that keep JA responses at bay to allow normal root growth. These results provide a basis for the future engineering of damage - or JA-controlled organ growth, an area of potential importance in agriculture.
Results
A JA-dependent shoot-to-root wound signal reduces primary root growth
We investigated root growth responses at the seedling stage where mechanical wounding of shoots is known to trigger JA signalling in belowground tissues [7, 26]. Repeated wounding of cotyledons in WT seedlings caused the reduction of root length by decreasing meristem cell number and cell elongation in the differentiation zone (Fig 2A–2D and S1 Fig). Furthermore, the treatment significantly decreased root expression of two cell cycle genes CYCB1;1 [27] and PCNA1 [28] (Fig 2E and 2F), confirming the repressive effect of JA signalling on cell division activity in the root meristem. In contrast to the WT, repeated cotyledon wounding did not cause significant morphological or expression changes in roots of the JA-deficient mutant allene oxide synthase (aos) (Fig 2), indicating that the root growth reduction is JA-dependent. Consistent with an increase in bioactive jasmonates, repeated cotyledon wounding also caused a strong reduction in the levels of the JA-biosensor Jas9-VENUS [26] along the root including the meristem (S2 Fig). Thus, a JA-dependent shoot-to-root signal(s) was able to reach the root meristem, activate JA signalling and reduce primary root growth.
Fig. 2. Root growth reduction after repeated shoot wounding is JA dependent. 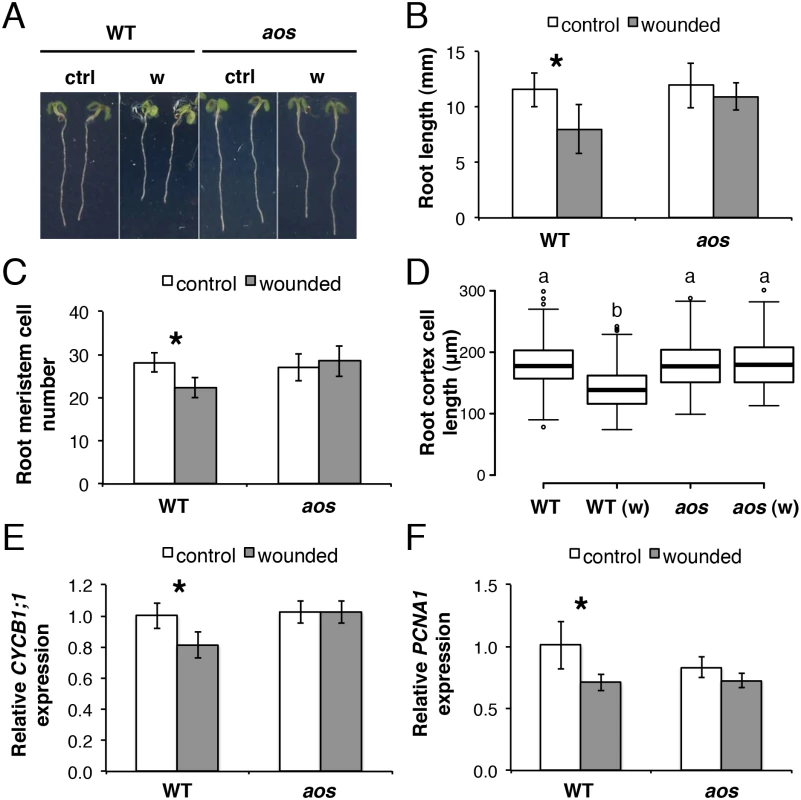
(A-D) Root measurements of 5-day-old (do) WT and aos seedlings grown in control conditions or subjected to repetitive cotyledon wounding: (A) representative control (ctrl) and wounded (w) seedlings; (B) primary root length; (C) cortex cell number in the primary root meristem; (D) box plot of cortex cell length in the differentiation zone of the primary root in control and wounded (w) samples of the indicated genotype. Data shown are means (± SD) from 27–55 (B) or 10 plants (C-D). Asterisks: Student’s t test significance compared with untreated controls (*P < 0.001) and letters in (D) indicate statistically significant differences between pairs as determined by Tukey’s HSD test (P < 0.001). (E-F) qRT-PCR of cell cycle marker genes (E) CYCB1;1 and (F) PCNA1 in 5-do roots of WT and aos seedlings grown in control conditions or subjected to repetitive cotyledon wounding. Root samples were collected 1 h after the 5th cotyledon wound. Transcript levels were normalized to UBC21 and displayed relative to the expression of the WT control. Bars represent means of three biological replicates (±SD), each containing a pool of roots from ~60 individuals. Asterisks: Student’s t test significance compared with untreated controls (*P < 0.01). Organization of JA signalling components in the primary root
As there was no information on the spatial organization of JA signalling in the root meristem, which could potentially mediate root growth responses after cotyledon wounding, we investigated the expression domain of NINJA, the only JA-signalling component affecting root growth in undamaged plants [7], and MYC TFs (MYC2, MYC3 and MYC4) known to have a role in the inhibition of root elongation when seedlings are grown in the presence of exogenous JA [19].
NINJApro-GUSPlus expression was detected in all organs of 5-day-old (do) seedlings: in vascular tissues of cotyledons and hypocotyl, in emerging true leaves, and in the primary root (S3 Fig). The strongest expression was observed in the root particularly in the apical meristem encompassing all cell layers examined. Confirming previous work [25], MYC2pro-GUSPlus was expressed predominantly in the root extending midway into the hypocotyl, and was almost absent in cotyledons. In the division zone of the primary root MYC2pro-GUSPlus activity was found in cells of the epidermis and lateral root cap but not in stele initials, whereas in cells of the elongation and differentiation zones staining was visible in the stele with only a very weak signal from ground tissues (S3 Fig). MYC3pro-GUSPlus and MYC4pro-GUSPlus expression patterns had both distinct and overlapping expression domains to that of MYC2pro-GUSPlus and with respect to each other (S3 Fig). Consistently with [19], MYC3pro-GUSPlus expression was found along the root excluding the division zone with weaker expression in aerial tissues, while the MYC4 promoter was active in the developed vasculature of the root and it extended to other outer cell layers in the hypocotyl and cotyledons. In addition, we observed MYC4pro-GUSPlus expression in outer layers of the columella and lateral root cap, and a strong MYC3pro-GUSPlus expression in the outer margins of cotyledons, where no apparent MYC4pro-GUSPlus activity was detected.
Promoter activities of NINJA and the three TFs were further characterized at the cellular level in the primary root tip with a nuclear localized fluorescent VENUS reporter protein (Fig 3A–3J). NINJApro-NLS3xVENUS was strongly expressed in all cells of the primary root apex (Fig 3A). MYC2pro-NLS3xVENUS was expressed in elongating endodermal and epidermal cells of the elongation zone (Fig 3E) as well as epidermal, lateral root cap and columella cells of the root division zone (Fig 3F). A weaker MYC3pro-NLS3xVENUS signal was present in endodermal, cortex and epidermal cells of the elongation and differentiation zones, while it was not detected in cells of the division zone (Fig 3H). MYC4pro-NLS3xVENUS was absent from the elongation and early differentiation zone of the root (Fig 3I), and its expression was restricted to outer layers of the columella and lateral root cap (Fig 3J). The same expression patterns, although with much weaker fluorescent signals for the three MYC TFs, were observed with functional protein fusion reporters driven by endogenous promoters (S4 and S5 Figs). The localization of NINJApro-NLS3xVENUS, MYC2pro-NLS3xVENUS, MYC3pro-NLS3xVENUS and MYC4pro-NLS3xVENUS florescent reporters did not differ from WT when expressed in the aos background (S6 Fig), indicating that the basal expression of NINJA and the three TFs in the primary root tip is JA-independent.
Fig. 3. Spatial localization of NINJA, MYC2, MYC3 and MYC4 in the primary root meristem and contribution of the three TFs to the ninja mutant phenotype. 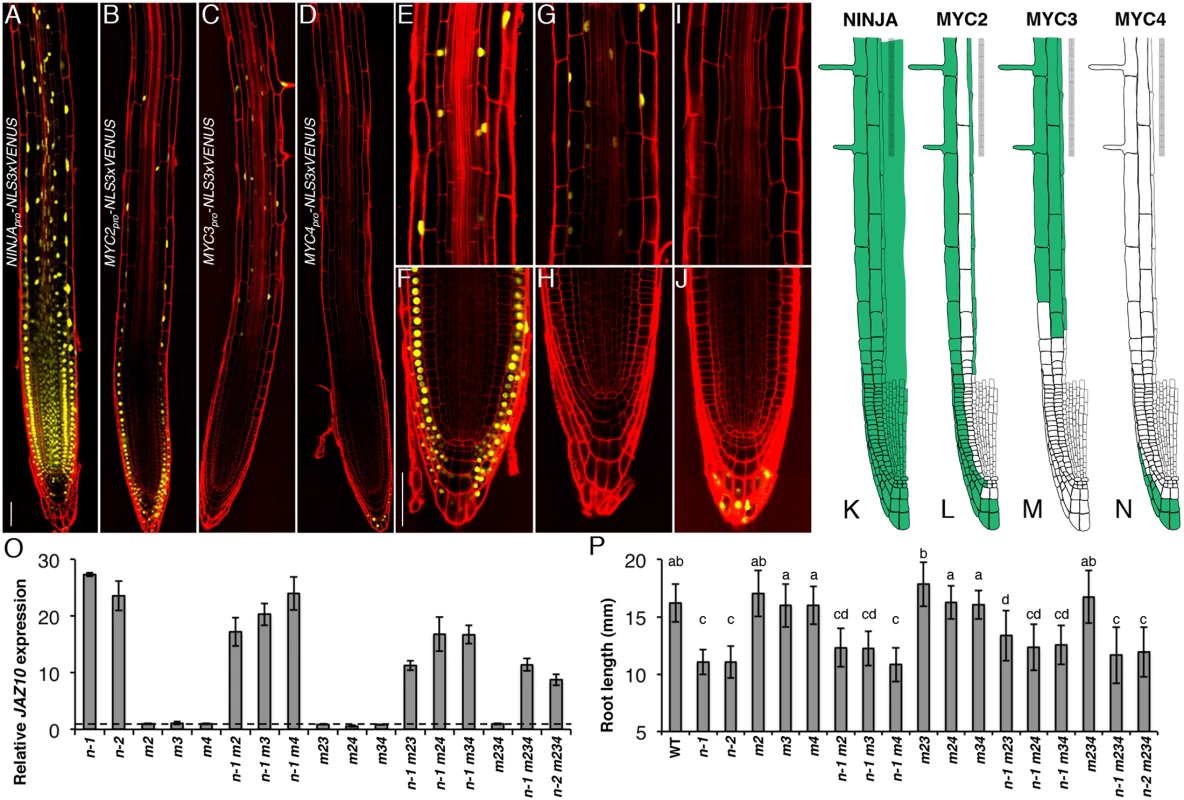
Expression pattern overviews of (A) NINJApro-NLS3xVENUS, (B) MYC2pro-NLS3xVENUS, (C) MYC3pro-NLS3xVENUS and (D) MYC4pro-NLS3xVENUS fluorescent reporters in 5-do WT primary roots. Close-ups of MYC2pro-NLS3xVENUS (E and F), MYC3pro-NLS3xVENUS (G and H) and MYC4pro-NLS3xVENUS (I and J) expression in, respectively, the elongation and division zones of the primary root. Confocal microscopy images (A-J) represent merged overlays of the fluorescent (yellow) and propidium iodide (red) stained roots. Scale bars = 50 μm. (K-N) Distribution maps of NINJA, MYC2, MYC3 and MYC4 expression patterns (green) in the primary root meristem based on promoter and protein fusion reporters (S4 Fig). (O) qRT-PCR of basal JAZ10 expression in 5-do roots of ninja-1 (n-1), ninja-2 (n-2), myc2 (m2), myc3 (m3), myc4 (m4), ninja-1 myc2 (n-1 m2), ninja-1 myc3 (n-1 m3), ninja-1 myc4 (n-1 m4), myc2 myc3 (m23), myc2 myc4 (m24), myc3 myc4 (m34), ninja-1 myc2 myc3 (n-1 m23), ninja-1 myc2 myc4 (n-1 m24), ninja-1 myc3 myc4 (n-1 m34), myc2 myc3 myc4 (m234), ninja-1 myc2 myc3 myc4 (n-1 m234), and ninja-2 myc2 myc3 myc4 (n-2 m234). JAZ10 transcript levels were normalized to those of UBC21 and displayed relative to the expression of WT controls that are set to 1 and indicated with a dashed line. Bars represent the means of three biological replicates (±SD), each containing a pool of ~60 roots. Complete qRT-PCR data are in S1 File. (P) Root length of 7-do seedlings of the same genotype as indicated in (O). Data shown are means (± SD) from 22–48 plants; letters above bars indicate statistically significant differences between samples as determined by Tukey’s HSD test (P < 0.01). A summary of the data is shown in basal distribution maps in the primary root apex (Fig 3K–3N). NINJA was present in all cells of the root tip. MYC2 and MYC3 were coincident in elongating and elongated epidermal and endodermal cells of the elongation and maturation zones of the root meristem. MYC3 was found in cortex cells from the elongation into the maturation zone, while MYC2 was detectable in all epidermal cells in different root zones from the initials on. The expression of MYC2 and MYC4 partially overlapped in the outer layers of the columella and lateral root cap, with MYC2 encompassing also inner cell layers of the two tissues. We did not observe overlapping expression domains for MYC3 and MYC4 in the root tip.
Contribution of MYC2, MYC3, and MYC4 to the ninja root phenotype
Even though MYC2 has a dominant role in root responses to exogenous JA [19, 20, 23], the ninja phenotype was not suppressed in a ninja myc2 double mutant, indicating that other or additional TF(s) are basally repressed by the NINJA-dependent mechanism [7]. Those signalling events are downstream of the COI1 receptor and JA-Ile perception as the ninja short root phenotype was still present in a ninja-1 coi1-1 double mutant background (S7 Fig). We generated multiple ninja myc mutant combinations and evaluated their root phenotypes in terms of the early JA signalling marker JAZ10 [2, 7] and organ length. The 25 times higher-than-WT JAZ10 expression found in ninja roots was only mildly reduced in ninja-1 myc2 and ninja-1 myc3 double mutant combinations, while it remained similar to ninja in a ninja-1 myc4 background (Fig 3O). We have previously shown that the jin1-7 allele of MYC2 did not suppress ectopic JAZ10 expression in ninja [7], which could be attributed to the different allele used (jin1-7 vs jin1-2). On the other hand, JAZ10 levels were almost halved in a ninja-1 myc2 myc3 triple mutant but they were not further reduced in ninja myc2 myc3 myc4 quadruple mutants (Fig 3O). Therefore, the combined activity of MYC2 and MYC3 contributes to approximately 50% of basal JAZ10 expression in ninja roots, and the activity of MYC4 has a minor, if any, role. We also monitored JAZ10 expression in roots 1 h after wounding one cotyledon (S8 Fig). The higher-than-WT JAZ10 accumulation of ninja mutants was abolished in double and triple mutant combinations with myc2 (ninja-1 myc2, ninja-1 myc2 myc3, ninja-1 myc2 myc4), while it was persistently higher in double and triple mutants with a WT copy of MYC2 (ninja-1 myc3, ninja-1 myc4, ninja-1 myc3 myc4).
The roots of ninja mutants are 30% shorter than WT [7] (S7 Fig) and we found that this was maintained in myc2/ myc3/ myc4 double, triple and quadruple mutant combinations with ninja relative to the controls with functional NINJA (Fig 3P). Remaining JA signalling in ninja myc2 myc3 mutants seems sufficient to repress root growth. Alternatively, it is possible that de-repressed JA signalling is not the direct cause of root growth reduction in ninja.
ninja roots display a JA-response transcriptome
To characterize transcriptional changes occurring in the root in the absence of a NINJA-dependent repression mechanism, we conducted a microarray analysis of ninja-1 versus WT roots. Consistent with a major role of NINJA in repressing JA signalling, many of the 113 genes up-regulated in ninja-1 roots encoded typical JA-responsive transcripts and several genes involved in secondary metabolism (notably genes encoding the synthesis of triterpenes such as thalianol) (S1 Table). Only 12 genes were down-regulated (including NINJA; S1 Table). We identified a larger number of differentially expressed genes in roots than found in a previous study that used adult rosettes of a NINJA knock-down line [10], with only 7 genes overlapping between the two datasets (S1 Table). The over-expression of JA response genes in ninja could be the result of de-repressed TF(s) normally inhibited by the NINJA-dependent repression mechanism. In an attempt to identify such TF(s) we generated double mutants between ninja-1 and WRKY38 and bHLH025 T-DNA insertion lines, two TFs found over-expressed in the ninja root transcriptome (S1 Table). However, ninja mutant phenotypes were not suppressed in those double mutant backgrounds (S9 Fig).
myc2-322B: A novel MYC2 allele
MYC2, MYC3 and MYC4 are expressed basally in the root meristem (Fig 3) but insertion alleles in these genes do not show root growth alterations [20]. However, based on the scheme in Fig 1 it is possible that gain-of-function alleles of MYCs constitutively activate JA signalling. One such allele in MYC3 is already known [29]. We extended the genetic screen used by [7] to search for new mutants that, under basal conditions, display ectopic expression of a secretable JAZ10pro-GUSPlussec (JGP) reporter. Unlike the weak JGP activity observed in the WT (Fig 4A) [7], one such mutant displayed basal JGP activity in the early differentiation zone of the primary root tip without reaching mature parts of the upper root (Fig 4B). The mutant segregated recessively from WT MYC2 (S10 Fig) and was mapped by whole-genome sequencing to a G to A transition in position 493 of the MYC2 gene causing a glutamate to lysine (E165K) substitution.
Fig. 4. myc2-322B exhibits enhanced root JA responses in a NINJA-dependent manner. 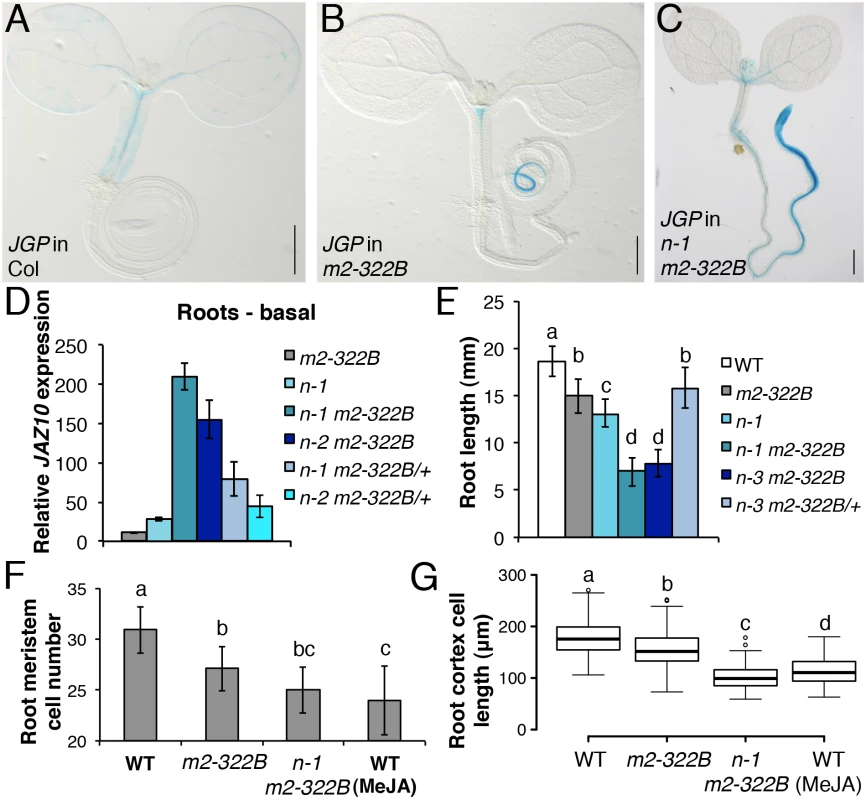
JGP expression in 5-do seedlings of WT (A), myc2-322B (B, m2-322B) and ninja-1 myc2-322B (C, n-1 m2-322B). Note the constitutive reporter activity stained in blue (Scale bars = 0.5 mm). (D) qRT-PCR of basal JAZ10 expression in 5-do roots of myc2-322B, ninja-1 (n-1), ninja-1 myc2-322B (n-1 m2-322B), ninja-2 myc2-322B (n-2 m2-322B), ninja-1 myc2-322B/+ (n-1 m2-322B/+) and ninja-2 myc2-322B/+ (n-2 m2-322B/+). JAZ10 transcript levels were normalized to those of UBC21 and displayed relative to the expression of unwounded WT samples, which are set to 1. Bars represent the means of three biological replicates (±SD), each containing a pool of ~60 roots. Complete qRT-PCR data are in S1 File. (E) Root length quantification of WT and 7-do mutant lines. Data are the means (±SD) from 20–48 plants. Primary root meristem cell number (F) and box plot summary of cortex-cell length (G) in 5-do seedlings of WT and mutant lines grown in control conditions, and of WT grown in presence of 25 μM MeJA (n = 10). Letters above bars and box plots (E-G) indicate statistically significant differences between samples as determined by Tukey’s HSD tests (P < 0.01). The relatively confined JGP expression of myc2-322B (Fig 4B) led us to hypothesize that a transcriptional repression mechanism is still able to inhibit the putative excessive MYC2E165K activity in myc2-322B. Indeed, by removing the NINJA-dependent repression mechanism in ninja-1 myc2-322B double mutants, the effects on JGP activity became more remarkable, extending to a much wider domain in the primary root including the meristem (Fig 4C). To further assess the functionality of this novel myc2 allele we tested its influence on JA-mediated gene expression. myc2-322B showed ~10 fold higher-than-WT JAZ10 expression in 5-do roots (Fig 4D) as well as higher JAZ10 accumulation in roots of cotyledon-wounded seedlings (S11 Fig). By contrast, neither basal JGP nor JAZ10 expression differed from WT in aerial tissues (Fig 4B and S11 Fig). Consistent with our hypothesis that a NINJA-dependent repression mechanism blocks excessive JA responses in myc2-322B, basal JAZ10 expression reached ~200 higher-than-WT levels in roots of ninja myc2-322B double mutants and intermediate JAZ10 and JGP levels in double mutants homozygous for ninja and heterozygous for myc2-322B (ninja myc2-322B/+) (Fig 4D and S12 Fig). Although myc2-322B segregated recessively in the WT background, we found that it is a dosage-dependent, gain-of-function allele once it is released from NINJA-dependent repression mechanisms in specific zones of the primary root.
When grown in control conditions, myc2-322B had up to 20% shorter roots than WT (Fig 4E), associated with decreased meristem cell number and cell elongation in the differentiation zone (Fig 4F and 4G). Root growth was more strongly affected in ninja myc2-322B double mutants: primary root length in control conditions reached only 50% of the WT length, and both meristem cell number and cell elongation in the differentiation zone were markedly reduced, and were similar to those of WT treated with JA (Fig 4F and 4G).
Given that both MYC2 and MYC3 contribute to JA signalling in ninja mutants (Fig 3O), we extended our studies to the atr2D allele of MYC3 in which a D94N missense mutation in the JAZ-interacting domain (JID) causes released repression from most JAZ proteins and activation of stress-responsive genes [19, 29, 30]. The atr2D mutant accumulated ~3 times higher-than-WT JAZ10 transcript levels in roots of seedlings (S13 Fig) without altering root length (S14 Fig). Conversely, roots of ninja-1 atr2D double mutants accumulated ~100 fold higher JAZ10 levels than WT but their length was similar to ninja mutants. JAZ10 transcripts were ~30 fold higher-than-WT in roots of myc2-322B atr2D double mutants (S13 Fig) but this did not reduce root length beyond that in myc2-322B alone (S14 Fig).
MYC2E165K results in enhanced transcriptional activity and partial release from JAZ repression
The myc2-322B mutant presumably produces a MYC2 protein affected in the transcriptional activation domain (TAD, residues 149–188) responsible for recruiting the Mediator transcription initiation complex [15, 31]. Embedded in the TAD of MYC2, a stretch of acidic amino acids (MYC2154–165) constitutes the destruction element (DE) required for proteasome-dependent degradation and MYC2 functionality [31]. Deleting the entire DE resulted in a more stable MYC2 protein that could no longer induce the transcription of MYC2 regulated genes, such as LOX2 [31]. The putative MYC2E165K variant found in myc2-322B is altered in the last amino acid of the DE and could result in defective proteolysis and/or transcriptional activity. A functional MYC2E165K-CITRINE transgene driven by the endogenous MYC2pro was expressed in the same root cells as WT MYC2-CITRINE but with a much stronger reporter signal (Fig 5A and S15 Fig). This apparently higher MYC2E165K protein accumulation was not the result of increased transcripts as both basal and wound induced MYC2 levels were the same between WT and myc2-322B mutant roots (S16 Fig). Furthermore, after inhibition of de novo protein synthesis with cycloheximide (CHX), MYC2E165K-CITRINE levels decreased over time, suggesting that the mutant protein is subjected to degradation just as WT MYC2-CITRINE (S17 Fig). Probably due to higher initial levels, MYC2E165K-CITRINE was still visible 60 min after CHX treatment, while MYC2-CITRINE had almost disappeared (S17 Fig). Concomitantly, we could also not detect aberrations in LOX2 accumulation 1 h after wounding in myc2-322B (S18 Fig), implying that MYC2E165K is functional.
Fig. 5. MYC2 E165K is a MYC2 gain-of-function allele. 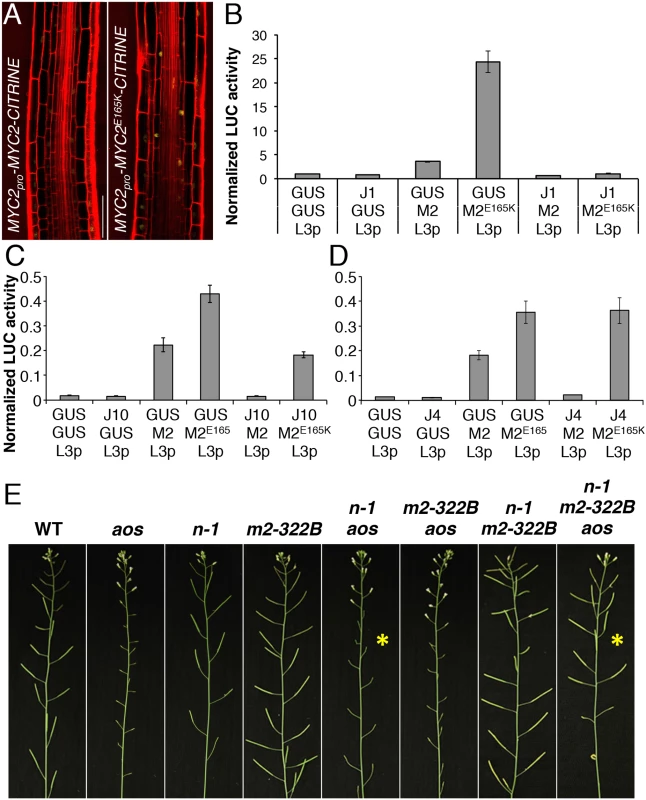
(A) Confocal microscopy images representing the expression pattern of functional MYC2pro-MYC2-CITRINE and MYC2pro-MYC2E165K-CITRINE fluorescent reporters (yellow) in the elongation zone of 5-do jin1-7 roots stained with propidium iodide (red). Scale bar = 50 μm. (B-D) Transactivation of the LOX3 promoter by transient expression of MYC2 or MYC2E165K in the presence or absence of JAZ1, JAZ10 or JAZ4 repressors. Tobacco protoplasts were transfected with a LOX3pro-fLUC (L3p) reporter construct, a 35Spro-MYC2 (M2) or 35Spro-MYC2E165K (M2E165K) effector constructs in the presence or absence of 35Spro-JAZ1 (JAZ1), 35Spro-JAZ10 (J10) or 35Spro-JAZ4 (JAZ4) constructs, and a 35Spro-rLUC normalization construct. The 35Spro-GUS (GUS) was used as control. Bars represent the means of 8 biological replicates (±SE) of normalized fLUC:rLUC activities. (E) Main inflorescences from 5 week-old plants of WT, aos, ninja-1 (n-1), myc2-322B (m2-322B), ninja-1 aos (n-1 aos), myc2-322B aos (m2-322B aos), ninja-1 myc2-322B (n-1 m2-322B) and ninja-1 myc2-322B aos (n-1 m2-322B aos). Note the restored fertility in the ninja-1 myc2-322B aos triple mutant compared to the sterility of ninja-1 aos with a WT MYC2 protein (yellow asterisks). We then assessed the transactivation capacity of MYC2E165K and its regulation by JAZ repressors. In transient expression assays in tobacco protoplasts, MYC2E165K had higher than WT activity in inducing the MYC2 responsive promoter of LOX3 (LOX3pro) driving the expression of a FIREFLY LUCIFERASE (fLUC) reporter (Fig 5B–5D and S19 Fig). The transactivation of LOX3pro by WT MYC2 was counteracted by co-expression with all 7 JAZ proteins tested (Fig 5B–5D and S19 Fig). However, the transactivation of LOX3pro by the MYC2E165K mutant protein was inhibited by co-expression with JAZ1 only, while it was less repressed by JAZ8, JAZ9 and JAZ10, and it failed to be inhibited by JAZ4, JAZ6 and JAZ12 (Fig 5B–5D and S19 Fig). We further compared the ability of WT MYC2 and MYC2E165K to interact with 12 JAZ proteins in Yeast two-hybrid (Y2H) assays. As reported previously [19, 30], WT MYC2 was able to interact with all JAZs, except with JAZ7 (S20 Fig). On the other hand, MYC2E165K was able to strongly interact only with JAZ1 in Y2H (S20 Fig). Thus, MYC2E165K promotes the expression of early JA responsive genes as a consequence of both increased transactivation capacity and reduced inhibition by JAZ proteins. The gain-of-function behaviour of MYC2E165K was then tested using an in planta genetic approach.
The ninja-1 and myc2-322B mutations were introgressed into backgrounds that are fully male-sterile as a consequence of abolished JA production (aos) or signalling (coi1-1). Remarkably, when MYC2E165K was liberated from NINJA-dependent repression, it was able to restore fertility of aos and of coi1-1 mutants in ninja-1 myc2-322B aos and ninja-1 myc2-322B coi1-1 combinations, whereas the WT copy of MYC2 failed to do so (Fig 5E and S21 Fig). Similarly to roots where basal MYC2 expression was aos-independent (S6 Fig), MYC2 transcript levels in stage 12 flowers were similar between WT and aos, whereas they were increased in the ninja-1 myc2-322B aos triple mutant (S22 Fig). Furthermore, we tested whether the restored fertility was a consequence of MYB21 and MYB24 induction, two TFs essential for male fertility whose expression is impaired in aos flowers [32]. In the ninja-1 myc2-322B aos triple mutant the expression of MYB21 rose to WT levels and that of MYB24 was intermediate between that of aos and WT. Conversely, the expression of both TFs was lower-than-WT in ninja-1 aos mutants with WT MYC2 (S22 Fig). The growth effects of MYC2E165K are therefore not seedling specific, but extend into reproductive organs and adult-phase rosettes (S23 Fig). To consolidate this, we used a repetitive leaf-wounding assay that is known to cause a JA-dependent reduction in WT rosette growth [2, 4]. In this assay, the myc2-322B mutant was more sensitive than the WT to wound-induced growth reduction (S24 Fig). Finally, myc2-322B was more susceptible than WT when challenged with the generalist herbivore Spodoptera littoralis (S25 Fig).
MYC2E165K renders roots hypersensitive to exogenous JA
Loss-of-function myc2 alleles are partly insensitive to JA-mediated root growth inhibition while the overexpression of MYC2 causes mild hypersensitivity [23] with 75% reduction in root length compared to the 50% reduction observed for the WT [20]. The myc2-322B mutant responded far more strongly to exogenous JA: its root length was up to 99% shorter in media supplemented with methyl JA (MeJA) compared to control conditions (Fig 6A–6C). The JA-mediated hypersensitivity phenotype was specific to myc2-322B and did not extend to the atr2D allele of MYC3 (S14 Fig) or to plants overexpressing a MYCD105N variant with diminished JAZ binding ability [30]. Moreover, all mutant combinations with myc2-322B displayed a hypersensitive phenotype in JA-mediated root growth inhibition assays (Fig 6C), with almost no measurable root meristem (Fig 6B). Triple mutant combinations of ninja-1 myc2-322B aos and ninja-1 myc2 coi1-1 showed that the extreme root growth reduction constitutively observed in ninja-1 myc2-322B double mutants partly relies on de novo JA synthesis and signalling as triple mutants had intermediate root lengths between the ninja - 1 myc2-322B and myc2-322B mutants (Fig 6C). Consistently, ninja-1 myc2-322B aos was hypersensitive to MeJA treatment, while ninja-1 myc2-322B coi1-1 was completely insensitive (Fig 6C).
Fig. 6. MYC2 E165K confers extreme hypersensitivity to exogenous JA. 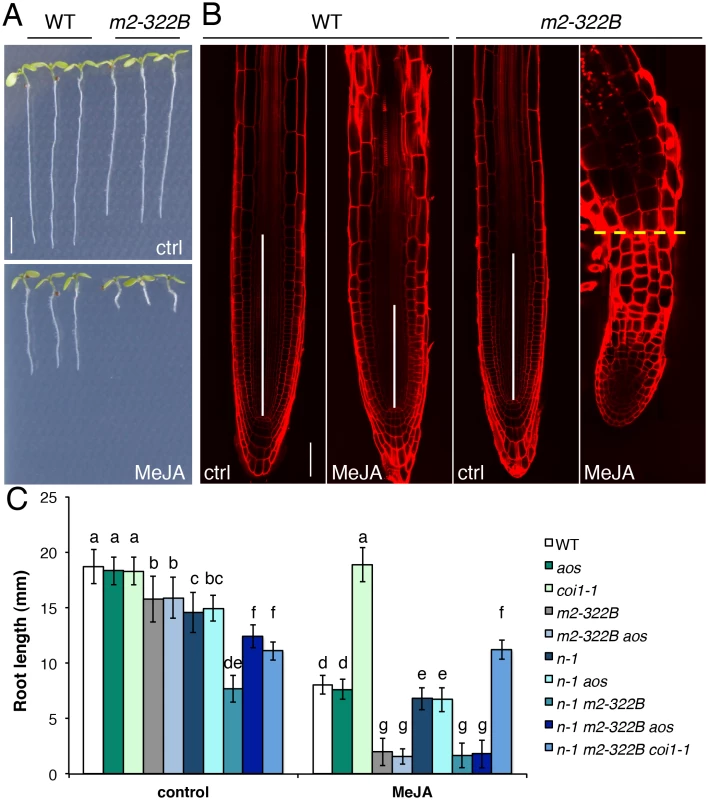
(A) Representative 7-do seedlings of WT and myc2-322B (m2-322B) mutants grown in control conditions (ctrl) or on media supplemented with 25 μM MeJA. Scale bar = 0.5 cm (B) Confocal microscopy images of propidium iodide stained primary root meristems of WT and myc2-322B 5-do seedlings grown in the absence (ctrl) or presence of 25 μM MeJA. Scale bar = 50 μm. Vertical white bars represent the root division zone and the horizontal yellow dashed line marks the root—hypocotyl boundary of myc2-322B grown in the presence of MeJA. (C) Root length of 7-do seedlings of the indicated genotype grown in the absence (control) or presence of 25 μM MeJA. n-1 refers to ninja-1. Data shown are means (± SD) from 20–49 plants. Letters above bars indicate statistically significant differences between samples as determined by Tukey’s HSD test (P < 0.01). Discussion
Single wounding treatments or simulated herbivory in aerial organs transiently reduced root growth in Arabidopsis [33] and Nicotiana attenuata [34] although the effect was reported to occur independently of aos in Arabidopsis. Herein we show that, with a repetitive wounding treatment, a shoot-to-root signal reaches below ground organs and, through JA action, restricts growth by inhibiting both cell proliferation and cell elongation in the primary root tip. These two basic cellular processes influencing root growth are similarly affected when plants are grown in media supplemented with JA [31]. Likewise, JA was shown to control leaf growth by inhibiting cell proliferation and the onset of endoreduplication [35]. Our data reveal a primary function of endogenous JA in coordinating root growth after distal wounding and establish a robust assay with which to investigate this phenomenon.
Distinct expression domains for JA signalling components in the root apex
NINJA is expressed ubiquitously in the primary root tip. This included not only cells in the differentiated parts of the root where a cell elongation defect and de-repressed JA signalling is observed in ninja mutants [7], but also cells of the root division zone where no apparent morphological defect is found in ninja mutants. Therefore, the lack of evident phenotypes in the root division zone of ninja mutants is not the result of differences in spatial distribution of the NINJA repressor, but likely the result of the presence of additional regulatory mechanisms such as direct recruitment of TPL by one or more NINJA-independent JAZs [11, 36]; direct recruitment of HISTONE DEACETYLASE 6 (HDA6) by JAZ1 to inhibit transcription [36]; MYC2 stability, turnover and phosphorylation [31]; and regulation of MYC-MED interactions to promote transcription. The enhanced phenotype of the ninja-1 myc2-322B mutant supports the control of MYC2 levels or activity as a cause of the restricted ninja-1 phenotype.
Mutants in NINJA display shorter roots in the absence of JA due to a defect in cell elongation only [7]. Consistent with a specific role of NINJA in repressing TF(s) mediating JA signalling in the root [7], a whole class of up-regulated transcripts found in roots of ninja was typically JA-responsive, with no major trends from other hormonal pathways. Several studies showed that the hormone ethylene (ET) does not affect root meristem cell division activity but strongly reduces cell elongation [37–39]. Thus, the root growth defect observed in ninja mutants could potentially arise from activated ET signalling. However, transcriptome analysis of ninja roots did not identify ET-responsive transcripts or markers for enhanced ET signalling, such as ACETYL-COA CARBOXYLASE 2 (ACC2), ETHYLENE RESPONSE SENSOR 1 or MYB59 [40]. Instead, the abundance of up-regulated transcripts of genes involved in secondary metabolism is probably the consequence of activated JA signalling in ninja roots. Likewise, the up-regulation of ILL6 that is involved in JA-Ile turnover [41, 42] is indicative of induced JA responses in ninja roots.
TF(s) freed from NINJA-dependent repression in ninja mutants are expected to activate JA-signalling and root growth responses. However, double mutants between ninja and T-DNA alleles of WRKY38 and bHLH025, two TFs that we found were overexpressed in ninja roots, did not suppress ninja root phenotypes. This indicated that, at least alone, these TFs do not strongly regulate JA signalling and root length in ninja. It is likely that several TF(s) act redundantly leading to ectopic JAZ10 accumulation and root length reduction. Nonetheless, ninja myc2 myc3 triple and ninja myc2 myc3 myc4 quadruple mutants revealed that the combined activity of MYC2 and MYC3 (but not, or less so, that of MYC4), contributes to transcribe approximately half of the JAZ10 transcripts overexpressed in ninja roots. MYC3 and MYC4 were shown to have cis-element binding specificity similar to that of MYC2 and to act additively with MYC2 in JA-mediated restriction of root growth [19]. These three TFs can form homo - and heterodimers in vivo to regulate gene expression [19]. It is conceivable that MYC2 and MYC3 form hetero-dimers and are subjected to NINJA-dependent repression specifically in epidermal and endodermal cells of the elongation and maturation zones where they are co-expressed. MYC2 and MYC4 could also form a similar module in the outer layers of the columella and lateral root cap where their expression domains coincide, while it is unlikely that MYC3 and MYC4 operate in such manner because their promoter activities did not overlap in any cell type of the root apex. The TF expression patterns also correlate with the stronger contribution of MYC2 and MYC3 to JAZ10 expression in ninja roots, and the lack of a detectable effect of MYC4. Since loss-of-function of all three TFs did not suppress the root growth phenotype of ninja mutants, it is possible that the remaining JA signalling levels of triple and quadruple mutants are sufficient to repress root growth or that de-repressed JA signalling is not the direct cause of root growth reduction in ninja.
myc2-322B is a gain-of-function MYC2 mutant
In addition to ninja [7], myc2-322B represents a second JA signalling mutant with increased JAZ10 expression in the root that is associated with reduced root growth. However, the phenotypes of ninja and myc2-322B are different: while JGP reporter expression coincides with reduced cell elongation in the root differentiation zone of ninja mutants, JGP expression in myc2-322B correlates only with reduced cell elongation in the root differentiation zone but not with the reduced cell proliferation observed in the root division zone. JA-induced MYC2 inhibits cell proliferation in the root division zone by directly repressing the expression of PLETHORA (PLT) genes that mediate auxin regulation of stem cell niche maintenance in the root division zone [25]. Because the regulatory function of TFs may depend on the cell-type specific network of interactions, the transcriptional outputs of MYC2 might differ in different cell-types and root areas: repression of PLT genes in the root division zone without activating JAZ10 transcription and JAZ10 activation in the root differentiation zone leading to compromised cell elongation. The different growth phenotypes of the mutants indicate that individual elements of the JA signalling pathway affect organ growth in different manners.
Although MYC2E165K is mutated in the transcriptional activation domain (TAD), it lost the ability to interact with JAZ repressors with the exception of JAZ1, as indicated by Y2H assays. This suggests that, in addition to the previously defined JAZ-interacting domain (JID) of MYC2 [13, 14], the destruction element (DE) within the TAD of MYC2 [31] might influence JAZ binding. Accordingly, JAZ1 was able to fully repress MYC2E165K transcriptional activity in protoplast transient expression assays; while JAZ8/-9/-10 were able to only partly repress MYC2E165K transcriptional activity, and JAZ4-6-12 did not show repressor capacity on MYC2E165K. The observed repressor activity of JAZ8/-9/-10 on MYC2E165K in plant protoplasts is likely due to the ability of JAZ proteins to form hetero-dimers among each other [43]. Specifically, JAZ1 was shown to interact with JAZ8, JAZ9 and JAZ10 [44, 45], suggesting that the repressor activity of JAZ8/-9/-10 observed in transient expression assays may rely on hetero-dimerization with a tobacco JAZ1 orthologue and consequent binding to MYC2E165K. The results emphasize the diversity among JAZ proteins in interacting with MYC2 and with one another [43]. Similarly to MYC2E165K, a recently identified MYC2D105N allele mutated in the JID of MYC2 causes impaired protein interactions with most JAZ repressors [30]. However, the transactivation potential of MYC2D105N did not differ from WT MYC2 in inducing pLOX3-fLUC in tobacco protoplasts [30], while that of MYC2E165K from the myc2-322B mutant was much greater than WT MYC2. Since the TAD of MYC2 is also necessary for MED25 binding [15, 16], it also remains possible that the MYC2E165K -MED25 interaction is altered in myc2-322B, favouring a more efficient docking of the Mediator complex to recruit RNA polymerase II and initiate gene transcription.
An alternative scenario is provided by our finding that MYC2E165K seems to accumulate at higher levels than WT MYC2 in basal conditions. It is possible that MYC2E165K is translated more rapidly than WT MYC2, as we did not detect differences in MYC2 transcript levels between WT and myc2-322B. If this was the case, JAZ repression might be relieved if a higher number of MYC2E165K molecules outcompetes the available number of JAZ repressors. This supports the control of MYC2 levels within strict limits as a regulatory layer of JA signalling. Such a mechanism is responsible for the strong effects of shade and light signalling in JA-regulated responses [46].
Finally, at the reproductive stage, myc2-322B in the appropriate background (ninja-1 myc2-322B aos or ninja-1 myc2-322B coi1-1) revealed that it is possible to recapitulate archetype hormone response phenotypes (in this case male fertility) in the absence of the hormone (JA) or of its receptor (COI1). These results also imply a putative role of MYC2 in male fertility as we found that basal MYC2 expression in flowers is aos-independent. Thus, the gain-of-function myc2-322B allele released from NINJA-dependent repression could induce the expression of MYB21 and MYB24, while WT MYC2 was unable to do so.
Myc2-322B represents a novel allele that may find many uses, for example in amplifying JA responses after mild stimulation. Moreover, this mutant will be useful for dissecting JA signalling events in both the adult and reproductive phases. In fact, the mutant rendered rosette leaves hypersensitive to wounding, although it displayed decreased resistance to a chewing herbivore relative to the WT. It is possible that the herbivore susceptibility phenotype of myc2-322B is a consequence of increased MYC2E165K repressor activity on some defense genes (e.g. PDF1.2) [23]. Increasing JA signalling may not always lead to enhanced defence against herbivores.
Layers of root growth regulation by JA signalling
The constitutive JAZ10 expression and root length phenotypes in myc2-322B were relatively mild due to NINJA-dependent and -independent repression mechanisms, particularly the direct recruitment of HDA6 by JAZ1 [36]. Indeed, removing NINJA-dependent repression from MYC2E165K in ninja myc2-322B double mutants led to much stronger phenotypes: extended JGP reporter activity along the whole root, 200 fold higher-than-WT root JAZ10 expression and root length and cellular phenotypes analogous to those of the WT treated with JA. NINJA-dependent repression mechanisms probably inhibit the basal activity of MYC TFs expressed in the root, explaining the lack of morphological phenotypes of myc KO mutants. This was further confirmed in heterozygous myc2-322B/+ mutants that showed no defects in JGP reporter activity or root length phenotypes in the presence of functional NINJA, but that displayed increased JAZ10 levels and reduced growth in a ninja background. A synergistic effect on JAZ10 expression was also observed in a double mutant between ninja-1 and a gain-of-function allele of MYC3, atr2D. However, the ninja-1 atr2D morphological root phenotypes did not differ from those of ninja, indicating that this mutant version of MYC3 induces JA signalling without affecting root growth.
It is likely that the activity of MYC2 is tightly controlled by yet additional NINJA-independent repression mechanisms because, except in coi1-1 backgrounds, all mutant combinations with myc2-322B were hypersensitive to JA in root growth assays. These findings imply that such repression mechanisms are still able to partly repress basal JA responses in the ninja myc2-322B double mutant to allow root growth, and that they rely on JAZ repressors, particularly JAZ1, which are readily degraded in the presence of JA [13, 14]. NINJA-dependent and -independent mechanisms, together with tight controls on MYC2 levels or activity, represent multiple regulatory levels adopted to repress JA responses and guarantee normal root development in the absence of JA. However, MYCs are present in precise cell types and parts of the root, ready to activate JA responses in the event of a JA stimulus. Such a multilayered organization of JA signalling repression mechanisms also explains the lack of a JA-hypersensitive phenotype in ninja mutants. The NINJA-dependent repression mechanism requires docking onto JAZ repressors, thus JAZ stability is epistatic to NINJA-dependent repression and lack of a functional NINJA protein cannot render the roots more sensitive than WT in JA-mediated root growth inhibition assays.
Conclusion
The study of plant development is generally carried out in the absence of physical injury. However, damage to plant tissues through herbivory and environmental insult is common if not omnipresent in nature. Under these conditions, the JA pathway, which has a low activity in unstressed vegetative tissues, is stimulated and imposes its activity on cell division and elongation. The resulting growth restriction can strongly impact plant productivity and is therefore of both fundamental and agronomic importance. Taking roots as a model, and using the simplified scheme for canonical JA signalling shown in Fig 1, we show that it is possible to manipulate regulatory layers in the JA pathway such that cell division and cell elongation can be constrained differentially. This type of approach may lead to future strategies to alter organ growth and, potentially, uncouple it from defense responses that occur when JA signalling is initiated. Our results underline the importance of cell-specific expression of JA signalling components and this will help generate new hypotheses. For example, of the three MYC TFs analysed herein, only MYC2 has been shown to be JA-inducible [19], as well as being expressed in the epidermis in the division zone in the root meristem (Fig 2F). This raises the intriguing possibility of MYC2 decoding very mild and localized JA-derived signals, resulting perhaps from microarthropod attack or during root penetration of soil where MYC2-expressing cells could be mildly squeezed resulting in local and transient JA production [47]. Such scenarios remain to be tested and may be facilitated with the use of myc2-322B, which amplifies JA-mediated growth responses.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana accession Columbia (Col) was the WT line as well as the genetic background of previously described mutants and transgenic lines used in this study: aos [48], coi1-1 [22], JGP, ninja-1, ninja-2 and ninja-3 [7], Jas9-VENUS [26] myc2 (jin1-2) and jin1-7 [23], atr2D [29] and myc3, myc4, myc23, myc24, myc34, myc234 [19]. The myc2-322B mutant allele was identified in a forward genetic screen for ectopic expression of a secretable JAZ10pro-GUSPlussec (JGP) reporter and was identified with whole-genome sequencing of bulk segregants, as described [7]. For experiments with myc2-322B, the WT control was the JGP reporter line shown to have the same root length, basal and wound induced JAZ10 expression as Col [7]. The T-DNA lines of wrky38 (SAIL_749_B02) and bhlh025 (SALK_080900) were obtained from the Nottingham Arabidopsis Stock Centre.
After seed stratification for 2 d at 4°C, plants were grown at 21°C under 100 μE m-2 s-1 of light with photoperiod depending on the application (seedlings: 14 h light, 10 h dark; soil-grown plants for herbivory assays: 10 h light, 14 h dark; soil-grown plants for crosses and phenotyping: 24 h light). For seedling growth, seeds were surfaced-sterilized and grown on half-strength Murashige and Skoog solid medium (0.5X MS, 0.5 g/L MES hydrate, pH 5.7) supplemented with 0.7% agar (for horizontally-grown seedlings sawn on 200 μm pore size nylon mesh) or 0.85% agar (for vertically-grown seedlings), as described previously [7].
Plant treatments
For repetitive wounding, cotyledons of vertically grown seedlings were pierced with a micro-needle (36 gauge beveled needle) in aseptic conditions under a stereomicroscope. Wounding started in the morning (7–8 am) of the third day after transfer to the phytotron (3-do seedlings) and was repeated every 12 h on alternate cotyledons, for a total of 5 wounds per seedling.
Single cotyledon wounding of seedling, MeJA treatments, root phenotypic measurements (total length of primary root and cellular measurements), herbivory assays with S. littoralis were performed as described [7].
Histochemical detection of GUS activity in the primary root
GUS staining and histology of horizontally - grown entire seedlings were performed as in [7]. For GUS staining and sectioning of the primary root, vertically grown 5-do seedlings were carefully transferred to GUS staining solution (50 mM sodium phosphate buffer pH 7.0, 0.1% Triton X-100, 3 mM K4Fe(CN)6, 3 mM K3Fe(CN)6, 0.5 mg/ml X-Gluc) and incubated at 37°C in the dark for 2–4 h. For imaging the primary root tip, the reaction was stopped by replacing the staining solution with 50 mM sodium phosphate buffer pH 7.0. Roots were then immediately mounted in freshly prepared chloral hydrate: glycerol: water solution (8 : 2:1). For cross-sections of GUSPlus reporter lines, the staining solution was replaced with 15% EtOH in water for 30 min, followed by 30 min incubation in 30% EtOH at RT. Seedling were then transferred to a 1% agarose support, bunches of 10–15 roots were closely aligned at the root tip and submerged with 1% warm agarose. Hardened agarose blocks containing the aligned roots were excised in ~0.4 cm3 cubes that were dehydrated through an ethanol series (30%, 50%, 70%, 90% and twice absolute) under agitation for 30 min each at 4°C. Samples were embedded in Technovit 7100 resin (Haslab GmbH, Ostermundigen, Switzerland) according to the manufacturer’s instructions. Briefly, samples were vacuum infiltrated with infiltration solution: absolute EtOH (1 : 1) for 2 h and with infiltration solution for 3 h. Finally, samples were hardened with embedding solution and hardened agarose blocks were aligned at the root tip, arranged in sectioning moulds and covered with additional embedding solution under anaerobic conditions. Samples were sectioned on a Leica RM2255 microtome using disposable Leica TC-65 blades into 6 μm sections and visualized with an upright Leica DM5500 microscope fitted with a DFC420 camera.
Gene expression analyses
For qRT-PCR experiments of JAZ10, MYC2, SUR1, VSP2 and LOX2 5-do seedlings were grown horizontally, separated in shoots and roots or kept intact and collected for basal and 1 h after cotyledon wounding expression analysis. For PCNA1 and CYCB1;1 expression, root samples were collected from 5-do vertically grown seedling subjected to repetitive cotyledon wounding. To determine the expression levels of MYB21 and MYB24, stage 12 flowers (largest closed buds) from plants grown in continuous light were separated from the rest of the inflorescence and frozen in liquid N2. Each biological replica consisted of equivalent flower buds from 3–4 inflorescences of the same plant. For genotypes in which flower maturation was impaired (aos, aos ninja-1) or delayed (aos myc2-322B ninja-1), equivalent stage flowers were identified according to their position in the inflorescence stem. RNA and cDNA were prepared as in [49]. Quantitative RT-PCR was performed as described [50]. Primers for qRT-PCR have been previously reported: JAZ10 (At5g13220) and UBC21 (At5g25760) in [49], SUR1 (At2g20610) in [51], LOX2 (At3g45140) in [52], and VSP2 (At5g24770) in [7]. To quantify other transcripts the following primers were used: MYB21 (At3g27810, ggggaaacaggtggtcgaaa and tgcttgcagcttgatcgttg); MYB24 (At5g40350, tctcgccaaatctgcaggac and ccacctatttccccattttgcat); PCNA1 (At1g07370, tgggttacattcgttactac and atacaaaggaatctcacca), CYCB1;1 (At4g37490, ccggaactgaatctgcttagg and gcgactcattagacttgttca) and MYC2 (At1g32640, gtgcgggattagctggtaaa and atgcatcccaaacactcctc). RT-PCR in S9 Fig was performed with GoTaq DNA polymerase (Promega) on 5-do WT and T-DNA lines as indicated by the manufacturer, with the following primer pairs: for WRKY38 cggtgcaagctatccgttat and ctttcactgccagatgacga, for bHLH025 cgaccaacaatgatccctct and tgaaattcgacaaagcagacc.
For microarray analysis, seedlings were grown horizontally in aseptic conditions as indicated above. Total RNA was extracted from 5-do roots and purified with RNeasy Plant Mini Kit (Qiagen). Three biological replicates, each consisting of 120 roots, were performed per genotype. RNA quality was analysed with the Agilent 2100 Bioanalyzer (Agilent). RNA amplification and hybridization on Affymetrix ATH1 arrays were performed as described [53]. Microarray data analysis was performed in R. Raw data was normalized using GCRMA algorithm [54] to reduce variability between samples. Comparisons were performed on normalized data using a linear model [55]. Differentially expressed genes were identified using both a p-value cut-off, set to 0.05 after adjustment for false discovery rate [56], and fold change set to 2.
Genotyping
Genotyping of T-DNA insertion lines was performed with the following oligonucleotides: tcttgtccggcaataaaaatg and aattaagtgagccgcgtactg for the WRKY38 WT allele (1.1 kb); tagcatctgaatttcataaccaatctcgatacac and aattaagtgagccgcgtactg for the wrky38 T-DNA allele (600 bp); tagcgagatctttggttggtg and gctgttgcctctgaaaatctg for the bHLH025 WT allele (1.2 kb); attttgccgatttcggaac and gctgttgcctctgaaaatctg for the bhlh025 T-DNA allele (700 bp). For selection of multiple mutant combinations from segregating populations ninja-1 was amplified with ggaggatgagtcacggaaag and gggagctggactggtgagta primers and digested with AciI (WT = 359, 142 bp; mutant = 501 bp); ninja-2 was amplified with tggtggttcttcttccaacc and gcaacaggttgtttgccttc primers and digested with Hpy188I (WT = 284, 209 bp; mutant = 284, 108, 101 bp); ninja-3 was amplified with caacgggagacaacagcaac and tggcttgagagtttgatccg primers and digested with TspRI (WT = 302, 132, 2 bp; mutant = 436 bp); myc2-322B was amplified with gtcatcgaaaccaagaaaaacgatt and gagacggagatcgagttcgc primers and digested with HinfI (WT = 143, 25 bp; mutant = 168 bp); atr2D was amplified with caccacaacaaccacctcag and tgaagcagagaggcagagaag and digested with BccI (WT = 269, 162 bp; mutant = 431 bp).
Transgenic lines
Promoters were amplified from WT genomic DNA with indicated oligonucleotides for NINJA (cggggtaccaatgctcatcctctgctgct and ttcccccccgggagcaaactctgagcaggtcaa, 2.7 kb), MYC2 (cggggtacctcgtgtatttgtgtctgcatgt and ttcccccccgggtccataaaccggtgaccggtaa, 2.1 kb), MYC3 (cggggtaccgcaaagaggatcgcttgaaa and ttcccccccggggtgaacatacgccggttgaaaag, 2 kb) and MYC4 (cggggtaccacagtactaacgtttgatggaaac and ttcccccccgggaacagttctctgacgtagttataaaag, 1.5 kb) and cloned by restriction with XmaI and KpnI into a modified pUC57 [50] to create pEN-L4-promoter-R1 clones. Underlined sequences represent XmaI and KpnI sites. For promoter fusions, pEN-L4-promoter-R1 - plasmids were recombined using the Gateway Cloning Technology with either pEN-L1-GUSPlus-L2 (non-secretable) or pEN-L1-NLS3xVENUS-L2 plasmids into pEDO097pFR7m24GW [50] to obtain pDEST-B4-promoter-B1-GUSPlus-B2 or pDEST-B4-promoter-B1-NLS3xVENUS-B2 clones. Coding DNA sequences (CDS) of NINJA (ggggacaagtttgtacaaaaaagcaggctgcatggacgatgataatgggctc and ggggaccactttgtacaagaaagctgggttggtgtgagctgacgctgcag), MYC2 and MYC2E165K (ggggacaagtttgtacaaaaaagcaggctgcatgactgattaccggctaca and ggggaccactttgtacaagaaagctgggttaccgatttttgaaatcaaacttgc), MYC3 (ggggacaagtttgtacaaaaaagcaggctgcatgaacggcacaacatcatc and ggggaccactttgtacaagaaagctgggttatagttttctccgactttcgtca), MYC4 (ggggacaagtttgtacaaaaaagcaggctgcatgtctccgacgaatgttcaa and ggggaccactttgtacaagaaagctgggtttggacattctccaactttctcc) were amplified with oligonucleotides specified in parenthesis containing the appropriate att sites (underlined). CDSs of NINJA were amplified from WT cDNA, of MYC2E165K from myc2-322B genomic DNA, and of MYC2, MYC3 and MYC4 from WT genomic DNA. Amplification products were recombined into pDONR221 (Invitrogen) to produce pEN-L1-gene-L2 clones. To generate protein fusions under the control of endogenous promoters, pEN-L4-promoter-R1 - plasmids were recombined with pEN-L1-CDS-L2 and pEN-R2-CITRINE-L3 plasmids into pB7m34gw by Multisite Gateway Technology to obtain pDEST-B4-promoter-B1-CDS-B2-CITRINE-B3 clones. All constructs were introduced into Arabidopsis backgrounds by floral dip Agrobacterium-mediated transformation. For promoter fusions, transformed seeds expressing red fluorescence protein in T1, T2 and T3 lines were selected by fluorescence microscopy, whereas for protein fusions, lines were selected on media containing DL-Phosphinothricin 40 μg/ml (Duchefa Biochemie B.V., Haarlem, The Netherlands). A minimum of two independent transgenic lines were used for each construct to perform experiments and verify reproducibility.
Confocal microscopy
Confocal laser scanning microscopy was performed on a Zeiss LSM 700 confocal microscope with vertically grown 5-do seedlings. Roots were mounted in 0.5X MS with or without 30 μg/ml propidium iodide (Sigma). Excitation and detection windows were set as follows: VENUS 488 nm (dye EYFP), 490–555 nm (BP 490–555 filter); CITRINE 488 nm (dye Citr), 490–555 nm (BP 490–555 filter); propidium iodide 555 nm (dye PI), 615–700 nm (LP 615 filter). All images shown within one experiment were taken with identical settings. Image processing was done with FIJI (http://fiji.sc/Fiji).
Transient expression assay in Nicotiana tabacum protoplasts
Transient expression assays were performed as described [57, 58]. Protoplasts were prepared from a Bright Yellow-2 tobacco cell culture and co-transfected with a reporter plasmid containing the firefly luciferase (fLUC) reporter gene driven by the LOX3 promoter [59], a normalization construct expressing Renilla luciferase (rLUC) under control of the 35S promoter [57] and effector constructs. Effector constructs were made by Gateway cloning of MYC2, MYC2E165K, JAZ1, JAZ4, JAZ6, JAZ8, JAZ9, JAZ10 and JAZ12 into the destination vector p2GW7 under control of the 35S promoter. The p2GW7-GUS effector plasmid was used as mock [10]. For each transfection, 2 μg of each plasmid was used. After transfection, protoplasts were incubated overnight and then lysed; fLUC and rLUC activities were determined with the Dual-Luciferase reporter assay system (Promega). Variations in transfection efficiency and technical error were corrected by normalization of fLUC by rLUC activities. All transactivation assays were conducted in an automated experimental set-up with 8 biological replicates for each effector combination.
Yeast two-hybrid analysis
Y2H analysis was performed as described [60]. Bait and prey were fused to the GAL4-AD or GAL4-BD via cloning into pGAL424gate or pGBT9gate (PSB, Ghent), respectively. The Saccharomyces cerevisiae PJ69-4A yeast strain [61] was co-transformed with bait and prey using the polyethylene glycol (PEG)/lithium acetate method. Transformants were selected on Synthetic Defined (SD) media lacking Leu and Trp (-2) (Clontech). Three individual colonies were grown overnight in liquid cultures (-2) at 30°C and 10-fold or 100-fold dilutions were dropped on control media (-2) and selective media lacking Leu, Trp and His (-3) (Clontech).
Data access
Microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE65840.
Supporting Information
Zdroje
1. Poveda K, Steffan-Dewenter I, Scheu S, Tscharntke T. Effects of below - and above-ground herbivores on plant growth, flower visitation and seed set. Oecologia. 2003;135(4):601–5. Epub 2005/10/18. doi: 10.1007/s00442-003-1228-1 16228257.
2. Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19(8):2470–83. Epub 2007/08/07. doi: 10.1105/tpc.107.050708 17675405; PubMed Central PMCID: PMC2002611.
3. Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(19):E1192–200. Epub 2012/04/25. doi: 10.1073/pnas.1201616109 22529386; PubMed Central PMCID: PMC3358897.
4. Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PloS one. 2008;3(11):e3699. Epub 2008/11/13. doi: 10.1371/journal.pone.0003699 19002244; PubMed Central PMCID: PMC2577035.
5. Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60 : 183–205. Epub 2008/11/26. doi: 10.1146/annurev.arplant.043008.092007 19025383.
6. Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111(6):1021–58. Epub 2013/04/06. doi: 10.1093/aob/mct067 23558912; PubMed Central PMCID: PMC3662512.
7. Acosta IF, Gasperini D, Chetelat A, Stolz S, Santuari L, Farmer EE. Role of NINJA in root jasmonate signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(38):15473–8. Epub 2013/09/05. doi: 10.1073/pnas.1307910110 24003128; PubMed Central PMCID: PMC3780868.
8. Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5(5):344–50. Epub 2009/04/08. doi: 10.1038/nchembio.161 19349968.
9. Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16(8):2117–27. Epub 2004/07/20. doi: 10.1105/tpc.104.023549 15258265; PubMed Central PMCID: PMC519202.
10. Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Perez AC, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464(7289):788–91. Epub 2010/04/03. doi: 10.1038/nature08854 20360743; PubMed Central PMCID: PMC2849182.
11. Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, et al. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. 2012;24(2):536–50. Epub 2012/02/14. doi: 10.1105/tpc.111.093005 22327740; PubMed Central PMCID: PMC3315231.
12. Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(2):761–6. Epub 2012/12/26. doi: 10.1073/pnas.1215010110 23267111; PubMed Central PMCID: PMC3545823.
13. Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–71. Epub 2007/07/20. doi: 10.1038/nature06006 17637675.
14. Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448(7154):661–5. Epub 2007/07/20. doi: 10.1038/nature05960 17637677.
15. Cevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012;160(1):541–55. Epub 2012/07/24. doi: 10.1104/pp.112.202697 22822211; PubMed Central PMCID: PMC3440227.
16. Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell. 2012;24(7):2898–916. Epub 2012/07/24. doi: 10.1105/tpc.112.098277 22822206; PubMed Central PMCID: PMC3426122.
17. Kazan K, Manners JM. MYC2: the master in action. Molecular plant. 2013;6(3):686–703. Epub 2012/11/13. doi: 10.1093/mp/sss128 23142764.
18. Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19(7):2225–45. Epub 2007/07/10. doi: 10.1105/tpc.106.048017 17616737; PubMed Central PMCID: PMC1955694.
19. Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–15. Epub 2011/02/22. doi: 10.1105/tpc.110.080788 21335373; PubMed Central PMCID: PMC3077776.
20. Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot. 2011;62(6):2143–54. Epub 2011/02/16. doi: 10.1093/jxb/erq408 21321051; PubMed Central PMCID: PMC3060693.
21. Browse J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry. 2009;70(13–14):1539–46. Epub 2009/09/11. doi: 10.1016/j.phytochem.2009.08.004 19740496.
22. Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. Plant Cell. 1994;6(5):751–9. Epub 1994/05/01. doi: 10.1105/tpc.6.5.751 12244256; PubMed Central PMCID: PMC160473.
23. Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16(7):1938–50. Epub 2004/06/23. doi: 10.1105/tpc.022319 15208388; PubMed Central PMCID: PMC514172.
24. Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(15):6837–40. Epub 1992/08/01. 11607311; PubMed Central PMCID: PMC49599.
25. Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell. 2011;23(9):3335–52. Epub 2011/09/29. doi: 10.1105/tpc.111.089870 21954460; PubMed Central PMCID: PMC3203420.
26. Larrieu A, Champion A, Legrand J, Lavenus J, Mast D, Brunoud G, et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nature communications. 2015;6 : 6043. Epub 2015/01/17. doi: 10.1038/ncomms7043 25592181.
27. Fuerst RA, Soni R, Murray JA, Lindsey K. Modulation of cyclin transcript levels in cultured cells of Arabidopsis thaliana. Plant Physiol. 1996;112(3):1023–33. Epub 1996/11/01. 8938409; PubMed Central PMCID: PMC158029.
28. Egelkrout EM, Mariconti L, Settlage SB, Cella R, Robertson D, Hanley-Bowdoin L. Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell. 2002;14(12):3225–36. Epub 2002/12/07. 12468739; PubMed Central PMCID: PMC151214.
29. Smolen GA, Pawlowski L, Wilensky SE, Bender J. Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics. 2002;161(3):1235–46. Epub 2002/07/24. 12136026; PubMed Central PMCID: PMC1462177.
30. Goossens J, Swinnen G, Vanden Bossche R, Pauwels L, Goossens A. Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 2015. doi: 10.1111/nph.13398
31. Zhai Q, Yan L, Tan D, Chen R, Sun J, Gao L, et al. Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 2013;9(4):e1003422. Epub 2013/04/18. doi: 10.1371/journal.pgen.1003422 23593022; PubMed Central PMCID: PMC3616909.
32. Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, et al. A regulatory network for coordinated flower maturation. PLoS Genet. 2012;8(2):e1002506. Epub 2012/02/22. doi: 10.1371/journal.pgen.1002506 22346763; PubMed Central PMCID: PMC3276552.
33. Schmidt L, Hummel GM, Schottner M, Schurr U, Walter A. Jasmonic acid does not mediate root growth responses to wounding in Arabidopsis thaliana. Plant, cell & environment. 2010;33(1):104–16. Epub 2009/11/10. doi: 10.1111/j.1365-3040.2009.02062.x 19895400.
34. Hummel GM, Naumann M, Schurr U, Walter A. Root growth dynamics of Nicotiana attenuata seedlings are affected by simulated herbivore attack. Plant, cell & environment. 2007;30(10):1326–36. Epub 2007/08/31. doi: 10.1111/j.1365-3040.2007.01718.x 17727422.
35. Noir S, Bomer M, Takahashi N, Ishida T, Tsui TL, Balbi V, et al. Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 2013;161(4):1930–51. Epub 2013/02/27. doi: 10.1104/pp.113.214908 23439917; PubMed Central PMCID: PMC3613466.
36. Zhu Z, An F, Feng Y, Li P, Xue L, A M, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(30):12539–44. Epub 2011/07/09. doi: 10.1073/pnas.1103959108 21737749; PubMed Central PMCID: PMC3145709.
37. Ruzicka K, Ljung K, Vanneste S, Podhorska R, Beeckman T, Friml J, et al. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell. 2007;19(7):2197–212. Epub 2007/07/17. doi: 10.1105/tpc.107.052126 17630274; PubMed Central PMCID: PMC1955700.
38. Stepanova AN, Yun J, Likhacheva AV, Alonso JM. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell. 2007;19(7):2169–85. Epub 2007/07/17. doi: 10.1105/tpc.107.052068 17630276; PubMed Central PMCID: PMC1955696.
39. Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GT, Sandberg G, et al. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell. 2007;19(7):2186–96. Epub 2007/07/17. doi: 10.1105/tpc.107.052100 17630275; PubMed Central PMCID: PMC1955695.
40. Zhong GY, Burns JK. Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol Biol. 2003;53(1–2):117–31. Epub 2004/02/06. 14756311.
41. Bhosale R, Jewell JB, Hollunder J, Koo AJ, Vuylsteke M, Michoel T, et al. Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants. Plant Cell. 2013;25(8):2865–77. Epub 2013/08/15. doi: 10.1105/tpc.113.112268 23943861; PubMed Central PMCID: PMC3784585.
42. Widemann E, Miesch L, Lugan R, Holder E, Heinrich C, Aubert Y, et al. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J Biol Chem. 2013;288(44):31701–14. Epub 2013/09/21. doi: 10.1074/jbc.M113.499228 24052260; PubMed Central PMCID: PMC3814765.
43. Pauwels L, Goossens A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23(9):3089–100. Epub 2011/10/04. doi: 10.1105/tpc.111.089300 21963667; PubMed Central PMCID: PMC3203442.
44. Chini A, Fonseca S, Chico JM, Fernandez-Calvo P, Solano R. The ZIM domain mediates homo - and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009;59(1):77–87. Epub 2009/03/25. doi: 10.1111/j.1365-313X.2009.03852.x 19309455.
45. Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21(1):131–45. Epub 2009/01/20. doi: 10.1105/tpc.108.064097 19151223; PubMed Central PMCID: PMC2648087.
46. Chico JM, Fernandez-Barbero G, Chini A, Fernandez-Calvo P, Diez-Diaz M, Solano R. Repression of Jasmonate-Dependent Defenses by Shade Involves Differential Regulation of Protein Stability of MYC Transcription Factors and Their JAZ Repressors in Arabidopsis. Plant Cell. 2014;26(5):1967–80. Epub 2014/05/16. doi: 10.1105/tpc.114.125047 24824488; PubMed Central PMCID: PMC4079362.
47. Farmer EE, Gasperini D, Acosta IF. The squeeze cell hypothesis for the activation of jasmonate synthesis in response to wounding. New Phytol. 2014;204(2):282–8. Epub 2014/12/03. doi: 10.1111/nph.12897 25453132.
48. Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31(1):1–12. Epub 2002/07/09. 12100478.
49. Gfeller A, Baerenfaller K, Loscos J, Chetelat A, Baginsky S, Farmer EE. Jasmonate controls polypeptide patterning in undamaged tissue in wounded Arabidopsis leaves. Plant Physiol. 2011;156(4):1797–807. Epub 2011/06/23. doi: 10.1104/pp.111.181008 21693672; PubMed Central PMCID: PMC3149931.
50. Chauvin A, Caldelari D, Wolfender JL, Farmer EE. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013;197(2):566–75. Epub 2012/11/23. doi: 10.1111/nph.12029 23171345.
51. Schweizer F, Fernandez-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 2013;25(8):3117–32. Epub 2013/08/15. doi: 10.1105/tpc.113.115139 23943862; PubMed Central PMCID: PMC3784603.
52. Glauser G, Dubugnon L, Mousavi SA, Rudaz S, Wolfender JL, Farmer EE. Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J Biol Chem. 2009;284(50):34506–13. Epub 2009/10/23. doi: 10.1074/jbc.M109.061432 19846562; PubMed Central PMCID: PMC2787311.
53. Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, Farmer EE. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature. 2013;500(7463):422–6. Epub 2013/08/24. doi: 10.1038/nature12478 23969459.
54. Wu Z, Irizarry R, Gentleman R, Murillo FM, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99 : 909–17.
55. Smyth GK. Limma: linear models for microarray data. In Gentleman R, Carey V, Dudoit S, Irizarry R and Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor: Springer, New York; 2005, pp. 397–420.
56. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57 : 289–300.
57. De Sutter V, Vanderhaeghen R, Tilleman S, Lammertyn F, Vanhoutte I, Karimi M, et al. Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J. 2005;44(6):1065–76. Epub 2005/12/20. doi: 10.1111/j.1365-313X.2005.02586.x 16359398.
58. Vanden Bossche R, Demedts B, Vanderhaeghen R, Goossens A. Transient expression assays in tobacco protoplasts. Methods in molecular biology (Clifton, NJ). 2013;1011 : 227–39. Epub 2013/04/26. doi: 10.1007/978-1-62703-414-2_18 23616000.
59. Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, et al. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(4):1380–5. Epub 2008/01/25. doi: 10.1073/pnas.0711203105 18216250; PubMed Central PMCID: PMC2234147.
60. Cuellar Perez A, Nagels Durand A, Vanden Bossche R, De Clercq R, Persiau G, Van Wees SC, et al. The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PloS one. 2014;9(1):e84891. Epub 2014/01/15. doi: 10.1371/journal.pone.0084891 24416306; PubMed Central PMCID: PMC3885651.
61. James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144(4):1425–36. Epub 1996/12/01. 8978031; PubMed Central PMCID: PMC1207695.
Štítky
Genetika Reprodukčná medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of andČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological LifespanČlánek The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



